text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
```
%%sh
pip -q install sagemaker stepfunctions --upgrade
# Enter your role ARN
workflow_execution_role = ''
import boto3
import sagemaker
import stepfunctions
from stepfunctions import steps
from stepfunctions.steps import TrainingStep, ModelStep, EndpointConfigStep, EndpointStep, TransformStep, Chain
from stepfunctions.inputs import ExecutionInput
from stepfunctions.workflow import Workflow
sess = sagemaker.Session()
bucket = sess.default_bucket()
role = sagemaker.get_execution_role()
prefix = 'sklearn-boston-housing-stepfunc'
training_data = sess.upload_data(path='housing.csv', key_prefix=prefix + "/training")
output = 's3://{}/{}/output/'.format(bucket,prefix)
print(training_data)
print(output)
import pandas as pd
data = pd.read_csv('housing.csv')
data.head()
data.drop(['medv'], axis=1, inplace=True)
data.to_csv('test.csv', index=False, header=False)
batch_data = sess.upload_data(path='test.csv', key_prefix=prefix + "/batch")
from sagemaker.sklearn import SKLearn
sk = SKLearn(entry_point='sklearn-boston-housing.py',
role=role,
framework_version='0.23-1',
train_instance_count=1,
train_instance_type='ml.m5.large',
output_path=output,
hyperparameters={
'normalize': True,
'test-size': 0.1,
}
)
execution_input = ExecutionInput(schema={
'JobName': str,
'ModelName': str,
'EndpointName': str
})
training_step = TrainingStep(
'Train a Scikit-Learn script on the Boston Housing dataset',
estimator=sk,
data={'training': sagemaker.inputs.TrainingInput(training_data, content_type='text/csv')},
job_name=execution_input['JobName']
)
model_step = ModelStep(
'Create the model in SageMaker',
model=training_step.get_expected_model(),
model_name=execution_input['ModelName']
)
transform_step = TransformStep(
'Transform the dataset in batch mode',
transformer=sk.transformer(instance_count=1, instance_type='ml.m5.large'),
job_name=execution_input['JobName'],
model_name=execution_input['ModelName'],
data=batch_data,
content_type='text/csv'
)
endpoint_config_step = EndpointConfigStep(
"Create an endpoint configuration for the model",
endpoint_config_name=execution_input['ModelName'],
model_name=execution_input['ModelName'],
initial_instance_count=1,
instance_type='ml.m5.large'
)
endpoint_step = EndpointStep(
"Create an endpoint hosting the model",
endpoint_name=execution_input['EndpointName'],
endpoint_config_name=execution_input['ModelName']
)
workflow_definition = Chain([
training_step,
model_step,
transform_step,
endpoint_config_step,
endpoint_step
])
import time
timestamp = time.strftime("%Y-%m-%d-%H-%M-%S", time.gmtime())
workflow = Workflow(
name='sklearn-boston-housing-workflow1-{}'.format(timestamp),
definition=workflow_definition,
role=workflow_execution_role,
execution_input=execution_input
)
# Not available in JupyterLab
# see https://github.com/aws/aws-step-functions-data-science-sdk-python/issues/127
# workflow.render_graph(portrait=True)
workflow.create()
execution = workflow.execute(
inputs={
'JobName': 'sklearn-boston-housing-{}'.format(timestamp),
'ModelName': 'sklearn-boston-housing-{}'.format(timestamp),
'EndpointName': 'sklearn-boston-housing-{}'.format(timestamp)
}
)
# Not available in JupyterLab
# see https://github.com/aws/aws-step-functions-data-science-sdk-python/issues/127
# execution.render_progress()
execution.list_events()
workflow.list_executions(html=True)
Workflow.list_workflows(html=True)
```
---
|
github_jupyter
|
# Module 10 - Regression Algorithms - Linear Regression
Welcome to Machine Learning (ML) in Python!
We're going to use a dataset about vehicles and their respective miles per gallon (mpg) to explore the relationships between variables.
The first thing to be familiar with is the data preprocessing workflow. Data needs to be prepared in order for us to successfully use it in ML. This is where a lot of the actual work is going to take place!
I'm going to use this dataset for each of the regression algorithms, so we can see how each one differs.
The next notebooks with the dataset will be:
- Linear Regression w/ Transformed Target (Logarithmic)
- Ridge Regression with Standardized Inputs
- Ridge and LASSO Regression with Polynomial Features
These four notebooks are designed to be a part of a series, with this one being the first.
We're going to start by importing our usual packages and then some IPython settings to get more output:
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
```
## Part A: Data Exploration
The first thing to do is import and explore our mpg dataset!
There's a few things to note in the dataset description: na values are denoted by `?` and column names are in a separate doc.
I added the column names so we don't have to worry about them:
```
loc = "https://raw.githubusercontent.com/mhall-simon/python/main/data/car-mpg/auto-mpg.data"
df = pd.read_csv(loc, sep="\s+", header=None, na_values="?")
cols = {0:"mpg", 1:"cylinders", 2:"displacement", 3:"horsepower", 4:"weight", 5:"accel", 6:"year", 7:"origin", 8:"model"}
df = df.rename(columns=cols)
df.head(15)
```
When starting, it's always good to have a look at how complete our data set is.
Let's see just how many na values were brought into the dataset per column:
```
df.isna().sum()
```
We have 6 missing values for horsepower!
A safe assumption for imputing missing values is to insert the column mean, let's do that! (Feature engineering is somewhere that we can go into this more in depth.)
*Note:* Imputing values is something that's not always objective, as it introduces some biases. We could also drop those 6 rows out of our dataset, however, I think imputing average hp isn't too serious of an issue.
```
df = df.replace(np.nan, df.horsepower.mean())
df.isna().sum()
```
Now, there's no more missing values!
Let's get some descriptive statistics running for our numerical columns (non-numerical are automatically dropped):
```
df.describe()
```
Another thing we can look at is the number of unique car models in the dataset:
```
df.nunique(axis=0)
```
For the ML analysis, there's too many models to worry about, so we're going to have them drop off the dataset! We're trying to predict mpg, and with our data the model name will have practically no predictive power!
One Hot Encoding the makes/models would make the dataset have almost more columns than rows!
```
df = df.drop("model", axis=1)
df.head()
```
### Train-Test Split
We're getting closer to starting our analysis! The first major consideration is the train/test split, where we reserve a chunk of our dataset to validate the model.
Remember, no peeking into the results with testing to train our model! That'll introduce a bias!
Let's separate our data into X and y, and then run the split:
```
X = df.iloc[:,1:]
y = df.iloc[:,0]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=97)
```
Another important thing to look at is the distributions of continuous variables and their pairwise relationships.
Seaborn has a really cool pairplot function that allows us to easily visualize this automatically! We just need to pass in columns of continuous variables.
Note: This is a marginal dependence, and does not keep all other variables fixed! We should only analyze this after our split!
```
train_dataset = X_train.copy()
train_dataset.insert(0, "mpg", y_train)
sns.pairplot(train_dataset[['mpg','displacement','horsepower','weight','accel']], kind='reg', diag_kind='kde')
```
When looking at this, there's two things to takeaway:
1. `mpg` is close to being normal, but there's a long tail. This means we may be better taking the log of mpg when running our analysis - something to explore in the next notebook.
2. Some relationships are not quite linear! We will work on this more in the following notebooks!
Let's now get into the ML aspects!
## Part B: Data Preprocessing & Pipeline
There's a lot of online tutorials that show the SKLearn models and how to call them in one line, and not much else.
A really powerful tool is to leverage the pipelines, as you can adjsut easily on the fly and not rewrite too much code!
Pipelines also reduce the potential for errors, as we only define preprocessing steps, and don't actually need to manipulate our tables. When we transform the target with a log later, we also don't need to worry about switching between log and normal values! It'll be handled for us.
It's also not as bad as it seems!
The first main step is to separate our data into:
- categorical columns that need to be one-hot encoded
- continuous columns (no changes - for now)
- other processing subsets (none in these examples, but binary columns would be handled a bit differently.)
- label encoding the response (y) variable when we get into classification models
Let's get right to it! We can split apart the explanatory column names into the two categories with basic lists:
```
categorical_columns = ['cylinders','origin','year']
numerical_columns = ['displacement','horsepower','weight','accel']
```
*Discussion:* Why is Year Categorical, even though it's a numerical year?
In Linear Regression, the year 70 (1970) would appear to be a factor of 80 (1980) by about 9/10ths, and it would be scaled that way. This would not make sense, as we expect only marginal increases in mpg year-over-year. To prevent a relationship like this, we're going to one-hot encode the years into categories.
Now, let's put together our preprocessing pipeline.
We'll need to:
1. OneHot Encode Categorical
2. Leave Continuous Alone
Let's build our preprocessor:
```
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import make_column_transformer
preprocessor = make_column_transformer((OneHotEncoder(drop="first"), categorical_columns), remainder="passthrough")
```
Why are we dropping the first category in each categorical column?
Our regression can imply the first one with zeros for all the encoded variables, and by not including it we are preventing colinearity from being introduced!
A potential issue that can arise is when you encounter new labels in the test/validation sets that are not one-hot encoded. Right now, this would toss an error if it happens! Later notebooks will go into how to handle these errors.
Now, let's build the pipeline:
```
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LinearRegression
model = make_pipeline(preprocessor, LinearRegression())
```
And now we can easily train our model and preprocess our data all in one step:
```
model.fit(X_train, y_train)
```
Before we start evaluating the model, I'll show you some useful features with the pipeline:
1. View Named Steps
```
model.named_steps
```
2. View Coefficients and Intercept (Expanded Later)
```
model.named_steps['linearregression'].coef_
model.named_steps['linearregression'].intercept_
```
3. Generate Predictions
*Viewing First 10*
```
model.predict(X_train)[:10]
```
## Part C: Evaluating Machine Learning Model
So, now we have an ML model, but how do we know if it's good?
Also, what's our criteria for good?
This changes depending upon what you're doing!
Let's bring in some metrics, and look at our "in sample" performance. This is the performance valuation in sample, without looking at any test data yet!
- $r^2$: coefficient of determination
- mean absolute error
- mean squared error
Let's generate our in-sample predictions based upon the model:
```
y_pred_in = model.predict(X_train)
```
And now let's generate some metrics:
This compares the training (truth) values, to the ones predicted by the line of best fit.
```
from sklearn.metrics import r2_score, mean_squared_error, mean_absolute_error
r2_score(y_train, y_pred_in)
mean_squared_error(y_train, y_pred_in)
mean_absolute_error(y_train, y_pred_in)
```
We're explaining about 87.5% of the variation in our in-sample dataset! That's pretty good, but will it hold when analyzing out of sample?
Also, we now know that our average absolute error is 2.09 mpg! That's not too bad, considering the range of the dataset and STD from the data:
```
y_train.std()
y_train.max() - y_train.min()
```
Let's now visualize our predictions! As a note, we want all of our datapoints to be along the line!
*Tip:* If you're reproducing this graph, ensure that the diagonal goes through the origin of the plot. The red line is setup to draw from corner to corner, and if you move your axes this may not work out!
```
fig, ax = plt.subplots(figsize=(5,5))
plt.scatter(y_train, y_pred_in)
ax.plot([0,1],[0,1], transform=ax.transAxes, ls="--", c="red")
plt.xlim([0,50])
plt.ylim([0,50])
plt.ylabel("Model Predictions")
plt.xlabel("Truth Values")
plt.title("In Sample Performance")
plt.show();
```
Our predictions are pretty good!
A few things to note:
- It's a really good fit, but it appears that there's a slight curve to this dataset.
- This is still in sample (we trained the model on this data)
- If we're making predictions, what regions are we confident in?
I think average mpg we'll be accurate, however, at the edges we're missing some of the trend.
Let's plot our residual error to see the shape:
```
plt.scatter(y_train, y_train-y_pred_in)
plt.xlabel("Truth Values - In Sample")
plt.ylabel("Residual Error")
plt.xlim([5,50])
plt.plot([5,50],[0,0], color='black', alpha=0.6)
plt.show();
```
Our errors definitely have curvature in them! We'll improve upon this in the next module!
For now...
Let's start looking at the coefficients in our model while it's simple.
We can grab coefficients out of the preprocessor to ensure that the coefficients line up with labels.
It'll always be in order of the preprocessor, so we can first fetch the feature names from the one hot encoded, and then just concatenate our numerical columns as there were no changes!
```
feature_names = (model.named_steps['columntransformer']
.named_transformers_['onehotencoder']
.get_feature_names(input_features=categorical_columns))
feature_names = np.concatenate([feature_names, numerical_columns])
coefs = pd.DataFrame(
model.named_steps['linearregression'].coef_,
columns=['Coefficients'],
index=feature_names
)
coefs
```
Let's plot the coefficients to see if there's anything we can learn out of it!
```
coefs.Coefficients.plot(kind='barh', figsize=(9,7))
plt.title("Unscaled Linear Regression Coefficients")
plt.show();
```
Woah, it looks like weight in unimportant at first glance, even though it would probably impact mpg quite a bit!
A word of caution! We just can't compare the coefficients, as they're in a different scale!
If we scale them with their standard deviation, then we can compare them. However, some meaning is lost!
Currently, the coefficient `-0.034440` for `horsepower` means that while holding all else equal, increasing the horsepower by 1 unit decreases mpg by about 0.034 mpg!
So, if we add 100 hp to the car, mileage decreases by about 3.4 mpg if we hold all else equal!
Let's scale these coefficients to compare them better! Just keep in mind that the 1hp:-0.34mpg relationship will no longer be interpretable from the scaled coefficients. But, we will be able to compare between coefficients.
Using the model pipeline, we can easily transform our data using the built in transformer, and then take the std:
`model.named_steps['columntransformer'].transform(DATASET)` is how we can use the transformer we built above.
When training the model, this dataset transformation happened all behind the scenes!! However, we can reproduce it with our training sample to work with it manually:
**NOTE:** The pipeline transformation is better than manual, because we know for certain the order of the columns that are being outputted. We fetched them above! The preprocessor in this instance returned a SciPy sparse matrix, which we can import with a new DataFrame constructor:
```
X_train_preprocessed = pd.DataFrame.sparse.from_spmatrix(
model.named_steps['columntransformer'].transform(X_train),
columns=feature_names
)
X_train_preprocessed.head(10)
```
By plotting the standard deviations, we can see for certain that the coeffs are definitely in a different scale!
Weight varies in the thousands, while acceleration is usually around 10-20 seconds!!
```
X_train_preprocessed.std(axis=0).plot(kind='barh', figsize=(9,7))
plt.title("Features Std Dev")
plt.show();
```
As you can probably see, the standard deviation of weight is far higher than any other variable!
This makes it impossible to compare.
Now, let's scale everything.
This scale works because very large continuous variables have a large standard deviation, but very small coefficients, which brings them down. The opposite is true for very small continuous variables for standard deviations, their coefficient is usually much larger. By multiplying the two together, we're bringing everything in towrads the middle, and with the same units of measurement.
```
coefs['coefScaled'] = coefs.Coefficients * X_train_preprocessed.std(axis=0)
coefs
```
Now, let's plot the scaled coefs:
```
coefs.coefScaled.plot(kind="barh", figsize=(9,7))
plt.title("Scaled Linear Coefficients")
plt.show();
```
Earlier, weight had almost no impact on the model at first glance! Now, we can see that it's the most important explanatory variable for mpg.
Let's now do our final validations for the model by bringing in the test data!!
The first is going to be done using the test (reserved) dataset, which we can make predictions with easily:
```
y_pred_out = model.predict(X_test)
```
And now let's generate a small DataFrame to compare metrics from in sample and out of sample!
Out of sample performance is usually worse, it's usually a question of how much!
```
metrics = pd.DataFrame(index=['r2','mse','mae'],columns=['in','out'])
metrics['in'] = (r2_score(y_train, y_pred_in), mean_squared_error(y_train, y_pred_in), mean_absolute_error(y_train, y_pred_in))
metrics['out'] = (r2_score(y_test, y_pred_out), mean_squared_error(y_test, y_pred_out), mean_absolute_error(y_test, y_pred_out))
metrics
```
When looking at the data, we see that the $r^2$ value decreased slightly from 0.875 to 0.854! This is still fairly significant!
And let's do a similar graph for out of sample performance:
```
fig, ax = plt.subplots(figsize=(5,5))
plt.scatter(y_test, y_pred_out)
ax.plot([0,1],[0,1], transform=ax.transAxes, ls="--", c="red")
plt.xlim([0,50])
plt.ylim([0,50])
plt.ylabel("Model Predictions")
plt.xlabel("Truth Values")
plt.title("Out of Sample Performance")
plt.show();
```
We're doing pretty good! There's stil some curvature that we'll work on fixing in the next notebooks.
Let's plot our residuals one more time:
```
plt.scatter(y_test, y_test-y_pred_out)
plt.xlabel("Truth Values - Out of Sample")
plt.ylabel("Residual Error")
plt.xlim([5,50])
plt.plot([5,50],[0,0], color='black', alpha=0.6)
plt.show();
```
Our model is pretty good, except for when we go above 32-ish mpg. Our model is predicting values far too high.
We'll solve this in a later notebook.
Another key question for ML is...
How do we know if the performance is due to just our sample selected? How much would our model change depending upon the sample selected?
We can solve for this using cross validation!
Cross validation takes different samples from our dataset, runs the regression, and then outputs the results!
We can easily cut the dataset into chunks and see how it behaves.
We're going to plot the distributions of coefficients throughout the folds to see how stable the model is:
```
from sklearn.model_selection import cross_validate
from sklearn.model_selection import RepeatedKFold
# Part 1: Defining Cross Validation Model
cv_model = cross_validate(
model, X, y, cv=RepeatedKFold(n_splits=5, n_repeats=5),
return_estimator=True, n_jobs=-1
)
# Part 2: Analyzing Each Model's Coefficients, and Setting Them In DataFrame:
cv_coefs = pd.DataFrame(
[est.named_steps['linearregression'].coef_ * X_train_preprocessed.std(axis=0) for est in cv_model['estimator']],
columns=feature_names
)
# Part 3: Plotting the Distribution of Coefficients
plt.figure(figsize=(9,7))
sns.stripplot(data=cv_coefs, orient='h', color='k', alpha=0.5)
sns.boxplot(data=cv_coefs, orient='h', color='cyan', saturation=0.5)
plt.axvline(x=0, color='.5')
plt.xlabel('Coefficient importance')
plt.title('Coefficient importance and its variability')
plt.subplots_adjust(left=.3)
plt.show();
```
What are the takeaways from this plot?
Our model doesn't appear to be too sensitive to the splits in training and testing!
This is a signal that our model is robust, and we should have confidence that our findings weren't due to choosing a "good" sample!
If we saw a variable changing from -6 to +2, that would be a sign it is not stable!
Now, we're ready to start exploring the second notebook! Which starts working towards a fix in the curvature!
## Bonus Box: Easily Checking for Variable Colinearity
If we suspect two variables are colinear, we can easily check for it with the following code:
```
plt.scatter(cv_coefs['weight'], cv_coefs['displacement'])
plt.ylabel('Displacement coefficient')
plt.xlabel('Weight coefficient')
plt.grid(True)
plt.title('Co-variations of variables across folds');
```
These are not colinear across folds, which is good for the model!
If they *were* colinear across folds, it would look something like this:
<div>
<img src=https://github.com/mhall-simon/python/blob/main/data/screenshots/Screen%20Shot%202021-03-22%20at%206.38.12%20PM.png?raw=True width="400"/>
</div>
If you notice strong colinearlity, then one should be removed and you can run the model again!
|
github_jupyter
|
<a href="https://colab.research.google.com/github/isaacmg/task-vt/blob/biobert_finetune/drug_treatment_extraction/notebooks/BioBERT_RE.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Finetuning BioBERT for RE
This is a fine-tuning notebook that we used to finetune BioBERT for relation classification (on our own data, GAD and Euadr) and then convert the resulting model checkpoint to PyTorch HuggingFace library for model inference. This was done for the vaccine and therapeutics task in order to identify drug treatment relations.
```
!git clone https://github.com/dmis-lab/biobert
from google.colab import auth
from datetime import datetime
auth.authenticate_user()
!pip install tensorflow==1.15
import os
os.chdir('biobert')
```
### Downloading data
```
!./download.sh
!fileid="1GJpGjQj6aZPV-EfbiQELpBkvlGtoKiyA"
!wget --load-cookies /tmp/cookies.txt "https://docs.google.com/uc?export=download&confirm=$(wget --quiet --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=1GJpGjQj6aZPV-EfbiQELpBkvlGtoKiyA' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\1\n/p')&id=1GJpGjQj6aZPV-EfbiQELpBkvlGtoKiyA" -O biobert_w.tar.gz && rm -rf /tmp/cookies.txt
!tar -xvf biobert_w.tar.gz
%set_env RE_DIR datasets/RE/GAD/1
%set_env TASK_NAME=gad
%set_env OUTPUT_DIR=./re_outputs_1
%set_env BIOBERT_DIR=biobert_large
!python run_re.py --task_name=$TASK_NAME --do_train=true --do_eval=true --do_predict=true --vocab_file=$BIOBERT_DIR/vocab_cased_pubmed_pmc_30k.txt --bert_config_file=$BIOBERT_DIR/bert_config_bio_58k_large.json --init_checkpoint=$BIOBERT_DIR/bio_bert_large_1000k.ckpt.index --max_seq_length=128 --train_batch_size=32 --learning_rate=2e-5 --num_train_epochs=3.0 --do_lower_case=false --data_dir=$RE_DIR --output_dir=$OUTPUT_DIR
#Uncomment this if you want to temporarily stash weights on GCS also collect garbage
#!gsutil -m cp -r ./re_outputs_1/model.ckpt-0.data-00000-of-00001 gs://coronaviruspublicdata/new_data .
#import gc
#gc.collect()
```
### Converting the model to HuggingFace
```
!pip install transformers
import logging
import torch
logger = logging.getLogger('spam_application')
def load_tf_weights_in_bert(model, config, tf_checkpoint_path):
""" Load tf checkpoints in a pytorch model.
"""
try:
import re
import numpy as np
import tensorflow as tf
except ImportError:
logger.error(
"Loading a TensorFlow model in PyTorch, requires TensorFlow to be installed. Please see "
"https://www.tensorflow.org/install/ for installation instructions."
)
raise
tf_path = os.path.abspath(tf_checkpoint_path)
logger.info("Converting TensorFlow checkpoint from {}".format(tf_path))
# Load weights from TF model
init_vars = tf.train.list_variables(tf_path)
excluded = ['BERTAdam','_power','global_step']
init_vars = list(filter(lambda x:all([True if e not in x[0] else False for e in excluded]),init_vars))
names = []
arrays = []
for name, shape in init_vars:
logger.info("Loading TF weight {} with shape {}".format(name, shape))
array = tf.train.load_variable(tf_path, name)
names.append(name)
arrays.append(array)
print("A name",names)
for name, array in zip(names, arrays):
if name in ['output_weights', 'output_bias']:
name = 'classifier/' + name
name = name.split("/")
# if name in ['output_weights', 'output_bias']:
# name = 'classifier/' + name
# adam_v and adam_m are variables used in AdamWeightDecayOptimizer to calculated m and v
# which are not required for using pretrained model
if any(
n in ["adam_v", "adam_m", "AdamWeightDecayOptimizer", "AdamWeightDecayOptimizer_1", "global_step"]
for n in name
):
logger.info("Skipping {}".format("/".join(name)))
continue
pointer = model
# if name in ['output_weights' , 'output_bias']:
# name = 'classifier/' + name
for m_name in name:
print("model",m_name)
#print(scope_names)
if re.fullmatch(r"[A-Za-z]+_\d+", m_name):
scope_names = re.split(r"_(\d+)", m_name)
else:
scope_names = [m_name]
if scope_names[0] == "kernel" or scope_names[0] == "gamma":
print(scope_names)
pointer = getattr(pointer, "weight")
elif scope_names[0] == "output_bias" or scope_names[0] == "beta":
# elif scope_names[0] == "beta":
# print(scope_names)
pointer = getattr(pointer, "bias")
# elif scope_names[0] == "output_bias":
# print(scope_names)
# pointer = getattr(pointer, "cls")
elif scope_names[0] == "output_weights":
print(scope_names)
pointer = getattr(pointer, "weight")
elif scope_names[0] == "squad":
print(scope_names)
pointer = getattr(pointer, "classifier")
else:
try:
pointer = getattr(pointer, scope_names[0])
except AttributeError:
logger.info("Skipping {}".format("/".join(name)))
continue
if len(scope_names) >= 2:
num = int(scope_names[1])
pointer = pointer[num]
if m_name[-11:] == "_embeddings":
pointer = getattr(pointer, "weight")
elif m_name == "kernel":
array = np.transpose(array)
try:
assert pointer.shape == array.shape
except AssertionError as e:
e.args += (pointer.shape, array.shape)
raise
logger.info("Initialize PyTorch weight {}".format(name))
pointer.data = torch.from_numpy(array)
return model
from transformers import BertConfig, BertForSequenceClassification, BertForPreTraining
def convert_tf_checkpoint_to_pytorch(tf_checkpoint_path, bert_config_file, pytorch_dump_path):
# Initialise PyTorch model
config = BertConfig.from_json_file(bert_config_file)
print("Building PyTorch model from configuration: {}".format(str(config)))
config.num_labels = 2
model = BertForSequenceClassification(config)
#model = BertForSequenceClassification(config)
# Load "weights from tf checkpoint
load_tf_weights_in_bert(model, config, tf_checkpoint_path)
# Save pytorch-model
print("Save PyTorch model to {}".format(pytorch_dump_path))
model.save_pretrained(pytorch_dump_path)
return model
# Alternatevely you can download existing stashed data
#!gsutil cp -r gs://coronaviruspublicdata/re_outputs_1 .
import os
!mkdir pytorch_output_temp
model2 = convert_tf_checkpoint_to_pytorch("re_outputs_1", "biobert_large/bert_config_bio_58k_large.json", "pytorch_output_temp")
```
### Upload converted checkpoint and test inference
If everything goes smoothly we should be able to upload weights and use the converted model.
```
from transformers import BertTokenizer
tokenizer = BertTokenizer.from_pretrained('biobert_large/vocab_cased_pubmed_pmc_30k.txt')
model2.eval()
input_ids = torch.tensor(tokenizer.encode("Hello, my dog is cute", add_special_tokens=True)).unsqueeze(0) # Batch size 1
labels = torch.tensor([1]).unsqueeze(0) # Batch size 1
outputs = model2(input_ids)
outputs = model2(input_ids)
outputs
input_ids = torch.tensor(tokenizer.encode("All our results indicate that the presence of the @GENE$ genotype (++) in patients with structural @DISEASE$, severe left ventricular dysfunction and malignant ventricular arrhythmias increases the risk for these patients of hemodynamic collapse during these arrhythmias"))
outputs = model2(input_ids.unsqueeze(0))
outputs
values, indices = torch.max(outputs[0], 1, keepdim=False)
indices
```
**Lets refactor this into something nicer**
```
from transformers import BertConfig, BertForSequenceClassification, BertForPreTraining
from transformers import BertTokenizer
class InferSequenceClassifier(object):
def __init__(self, pytorch_model_path, token_path, add_special_tokens=False):
self.tokenizer = BertTokenizer.from_pretrained(token_path)
self.model = BertForSequenceClassification.from_pretrained(pytorch_model_path)
self.add_special_tokens = add_special_tokens
def make_prediction(self, text):
input_ids = torch.tensor(self.tokenizer.encode(text, add_special_tokens=self.add_special_tokens))
outputs = self.model(input_ids.unsqueeze(0))
print(outputs)
values, indices = torch.max(outputs[0], 1, keepdim=False)
return indices
!cp biobert_large/vocab_cased_pubmed_pmc_30k.txt pytorch_output_temp/vocab.txt
!cp biobert_large/bert_config_bio_58k_large.json pytorch_output_temp/config.json
seq_infer = InferSequenceClassifier("pytorch_output_temp", "pytorch_output_temp", True)
seq_infer.make_prediction("@GENE$ influences brain beta-@DISEASE$ load, cerebrospinal fluid levels of beta-amyloid peptides and phosphorylated tau, and the genetic risk of late-onset sporadic AD.")
seq_infer.make_prediction("All our results indicate that the presence of the @GENE$ genotype (++) in patients with structural @DISEASE$, severe left ventricular dysfunction and malignant ventricular arrhythmias increases the risk for these patients of hemodynamic collapse during these arrhythmias")
seq_infer.make_prediction("Functional studies to unravel the biological significance of this region in regulating @GENE$ production is clearly indicated, which may lead to new strategies to modify the disease course of severe @DISEASE$.")
!gsutil cp -r pytorch_output_temp gs://coronavirusqa/re_convert
```
|
github_jupyter
|
```
%matplotlib inline
from matplotlib import style
style.use('fivethirtyeight')
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime as dt
from sqlalchemy import inspect
```
# Reflect Tables into SQLAlchemy ORM
```
import sqlalchemy
from sqlalchemy.ext.automap import automap_base
from sqlalchemy.orm import Session
from sqlalchemy import create_engine, func
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
engine = create_engine("sqlite:///Resources/hawaii.sqlite")
# reflecting an existing database into a new model
Base = automap_base()
# reflecting the tables
Base.prepare(engine, reflect=True)
# Displaying classes
Base.classes.keys()
# Saving data bases to variables
Measurement = Base.classes.measurement
Station = Base.classes.station
# Starting session from Python to the DB
session = Session(engine)
```
# Exploratory Climate Analysis
```
#Getting the last date in Measurment DB
max_date = session.query(func.max(Measurement.date)).first()
max_date
# Calculating the date 1 year ago from the last data point in the database
begin_date = dt.date(2017, 8, 23) - dt.timedelta(days=365)
begin_date
# Querying the Base tables returns results in a list
data = session.query(Measurement.date, Measurement.prcp).filter(Measurement.date >= begin_date).order_by(Measurement.date).all()
data
# Getting names and types of columns in "measurement" data set
inspector = inspect(engine)
columns = inspector.get_columns("measurement")
for column in columns:
print(column["name"], column["type"])
# Getting names and types of columns in "station" data set
inspector = inspect(engine)
columns = inspector.get_columns("station")
for column in columns:
print(column["name"], column["type"])
# Save the query results as a Pandas DataFrame and setting the index to the date column
precip_df = pd.DataFrame(data, columns=["Date", "Precipitation"])
precip_df["Date"] = pd.to_datetime(precip_df["Date"])
#Resettinng index to Date column
precip_df = precip_df.set_index("Date")
#Dropping all N/As
precip_df = precip_df.dropna(how = "any")
#Sorting by Date colummn - ascending
precip_df = precip_df.sort_values(by="Date", ascending=True)
precip_df
# Use Pandas Plotting with Matplotlib to plot the data
plt.figure(figsize=(10,5))
plt.plot(precip_df, label="Precipitation by Date")
plt.xlabel("Date")
plt.ylabel("Precipitation(in)")
plt.xticks(rotation="45")
plt.legend(loc="upper center")
plt.savefig("Output/Precipitation_plot.png")
plt.show()
```

```
#calcualting the summary statistics for the precipitation data
precip_df.describe()
```

```
# Query to count the number of stations in "Stations" data
session.query(func.count(Station.id)).all()
# What are the most active stations? (i.e. what stations have the most rows)?
# List the stations and the counts in descending order.
stations = session.query(Measurement.station, func.count(Measurement.station)).group_by(Measurement.station).order_by(func.count(Measurement.station).desc()).all()
stations
# Using the station id from the previous query, calculate the lowest temperature recorded,
# highest temperature recorded, and average temperature of the most active station?
session.query(Measurement.station, func.min(Measurement.tobs),
func.max(Measurement.tobs), func.avg(Measurement.tobs)). filter(Measurement.station == "USC00519281").\
group_by(Measurement.station).all()
# Choose the station with the highest number of temperature observations.
# Query the last 12 months of temperature observation data for this station and plot the results as a histogram
#Filtering data by date and by station
data_2 = session.query(Measurement.date, Measurement.tobs).filter(Measurement.station == "USC00519281").\
filter(func.strftime( Measurement.date) >= begin_date).all()
data_2
# Cleaning temp.data and setting index to date
temp_df = pd.DataFrame(data_2, columns=["Date", "Temperature"])
temp_df = temp_df.sort_values(by="Date", ascending=True)
temp_df.set_index("Date", inplace=True)
temp_df.head()
plt.figure(figsize=[8,5])
#Ploting the results as a histogram with 12 bins
plt.hist(x=temp_df["Temperature"], bins=12, label="tobs")
# Labeling figure
plt.grid
plt.xlabel("Temperature (F)")
plt.ylabel("Frequency")
plt.title("Temperature Frequency Histogram")
plt.legend()
# Saving Plot
plt.savefig("Output/Temp Frequency Histogram");
plt.show()
```

```
# This function called `calc_temps` will accept start date and end date in the format '%Y-%m-%d'
# and return the minimum, average, and maximum temperatures for that range of dates
def calc_temps(start_date, end_date):
"""TMIN, TAVG, and TMAX for a list of dates.
Args:
start_date (string): A date string in the format %Y-%m-%d
end_date (string): A date string in the format %Y-%m-%d
Returns:
TMIN, TAVE, and TMAX
"""
return session.query(func.min(Measurement.tobs), func.avg(Measurement.tobs), func.max(Measurement.tobs)).\
filter(Measurement.date >= start_date).filter(Measurement.date <= end_date).all()
# function usage example
print(calc_temps('2011-02-28', '2011-03-05'))
# using the example to calculate min, max and average tempreture for my vacation date
# Vacation Dates
start_date = "2020-04-01"
end_date = "2020-04-11"
# Previous Year Dates
hst_start_date = "2017-04-01"
hst_end_date = "2017-04-11"
# Min,average and max temp calculation
temp_min = calc_temps(hst_start_date, hst_end_date)[0][0]
temp_avg = calc_temps(hst_start_date, hst_end_date)[0][1]
temp_max = calc_temps(hst_start_date, hst_end_date)[0][2]
print(temp_min, temp_avg, temp_max)
# Ploting the results from your previous query as a bar chart.
# Use "Trip Avg Temp" as your Title
# Use the average temperature for the y value
# Use the peak-to-peak (tmax-tmin) value as the y error bar (yerr)
x_axis = 1
y_axis = temp_avg
error = temp_max-temp_min
# Defining Bar and Error paramaters
plt.bar(x_axis, y_axis, yerr=error, align='center', color = "r")
plt.tick_params(bottom=False,labelbottom=False)
# Labeling, tickers and grids
plt.ylabel("Temperature (F)")
plt.title("Trip Avg Temperature")
plt.grid(b=None, which="major", axis="x")
plt.margins(1.5, 1.5)
plt.ylim(0, 90)
plt.savefig("Output/Trip Average Temperature")
#Show the Plot
plt.show();
```
## Optional Challenge Assignment
```
# Create a query that will calculate the daily normals
# (i.e. the averages for tmin, tmax, and tavg for all historic data matching a specific month and day)
def daily_normals(date):
"""Daily Normals.
Args:
date (str): A date string in the format '%m-%d'
Returns:
A list of tuples containing the daily normals, tmin, tavg, and tmax
"""
sel = [func.min(Measurement.tobs), func.avg(Measurement.tobs), func.max(Measurement.tobs)]
return session.query(*sel).filter(func.strftime("%m-%d", Measurement.date) == date).all()
daily_normals("04-01")
# calculate the daily normals for your trip
# push each tuple of calculations into a list called `normals`
# Seting the start and end date of the trip from historic dates
hst_start_date # defined above
hst_end_date
# Useing the start and end date to create a range of dates
dates = session.query(Measurement.date).filter(Measurement.date >= hst_start_date).filter(Measurement.date <= hst_end_date).group_by(Measurement.date).all()
#saving trip dates into array
arr_dates = [x[0] for x in dates]
# Reformating dates to mm-dd format and getting data ion a list
arr_dates_mm_dd= [x[5:] for x in arr_dates]
start_mmdd = arr_dates_mm_dd[0]
end_mmdd = arr_dates_mm_dd[10]
# Looping through the list of mm-dd and getting max,ave, min temp averages
temps_by_dates = [session.query(func.min(Measurement.tobs),
func.avg(Measurement.tobs),
func.max(Measurement.tobs)).filter(func.strftime("%m-%d", Measurement.date) >= start_mmdd).filter(func.strftime("%m-%d", Measurement.date) <= end_mmdd).group_by(func.strftime("%m-%d", Measurement.date)).all()]
temps_by_dates = temps_by_dates[0]
#displaying averages for each date of the trip
temps_by_dates
# reformating list of temp into Pandas DataFrame
temps_by_dates_df= pd.DataFrame(temps_by_dates,columns=["min_t","avg_t","max_t"])
#Adding date column
temps_by_dates_df["date"]= arr_dates_mm_dd
# Seting index to date
temps_by_dates_df.set_index("date",inplace=True)
temps_by_dates_df
# Ploting the daily normals as an area plot with `stacked=False`
temps_by_dates_df.plot(kind='area', stacked=False, x_compat=True, title="Daily Normals for Trip Dates")
plt.xticks(rotation="45")
plt.savefig(("Output/Temp Frequency"))
plt.show()
```
|
github_jupyter
|
# Mislabel detection using influence function with all of layers on Cifar-10, ResNet
### Author
[Neosapience, Inc.](http://www.neosapience.com)
### Pre-train model conditions
---
- made mis-label from 1 percentage dog class to horse class
- augumentation: on
- iteration: 80000
- batch size: 128
#### cifar-10 train dataset
| | horse | dog | airplane | automobile | bird | cat | deer | frog | ship | truck |
|----------:|:-----:|:----:|:--------:|:----------:|:----:|:----:|:----:|:----:|:----:|:-----:|
| label | 5000 | **4950** | 5000 | 5000 | 5000 | 5000 | 5000 | 5000 | 5000 | 5000 |
| mis-label | **50** | | | | | | | | | |
| total | **5050** | 4950 | 5000 | 5000 | 5000 | 5000 | 5000 | 5000 | 5000 | 5000 |
### License
---
Apache License 2.0
### References
---
- Darkon Documentation: <http://darkon.io>
- Darkon Github: <https://github.com/darkonhub/darkon>
- Resnet code: <https://github.com/wenxinxu/resnet-in-tensorflow>
- More examples: <https://github.com/darkonhub/darkon-examples>
### Index
- [Load results and analysis](#Load-results-and-analysis)
- [How to use upweight influence function for mis-label](#How-to-use-upweight-influence-function-for-mis-label)
## Load results and analysis
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
scores = np.load('mislabel-result-all.npy')
print('num tests: {}'.format(len(scores)))
begin_mislabel_idx = 5000
sorted_indices = np.argsort(scores)
print('dogs in helpful: {} / 100'.format(np.sum(sorted_indices[-100:] >= begin_mislabel_idx)))
print('mean for all: {}'.format(np.mean(scores)))
print('mean for horse: {}'.format(np.mean(scores[:begin_mislabel_idx])))
print('mean for dogs: {}'.format(np.mean(scores[begin_mislabel_idx:])))
mis_label_ranking = np.where(sorted_indices >= begin_mislabel_idx)[0]
print('all of mis-labels: {}'.format(mis_label_ranking))
total = scores.size
total_pos = mis_label_ranking.size
total_neg = total - total_pos
tpr = np.zeros([total_pos])
fpr = np.zeros([total_pos])
for idx in range(total_pos):
tpr[idx] = float(total_pos - idx)
fpr[idx] = float(total - mis_label_ranking[idx] - tpr[idx])
tpr /= total_pos
fpr /= total_neg
histogram = sorted_indices >= begin_mislabel_idx
histogram = histogram.reshape([10, -1])
histogram = np.sum(histogram, axis=1)
acc = np.cumsum(histogram[::-1])
fig, ax = plt.subplots(1, 2, figsize=(20, 10))
ax[0].set_ylabel('true positive rate')
ax[0].set_xlabel('false positive rate')
ax[0].set_ylim(0.0, 1.0)
ax[0].set_xlim(0.0, 1.0)
ax[0].grid(True)
ax[0].plot(fpr, tpr)
ax[1].set_ylabel('num of mis-label')
ax[1].set_xlabel('threshold')
ax[1].grid(True)
ax[1].bar(range(10), acc)
plt.sca(ax[1])
plt.xticks(range(10), ['{}~{}%'.format(p, p + 10) for p in range(0, 100, 10)])
fig, ax = plt.subplots(figsize=(20, 5))
ax.grid(True)
ax.plot(scores)
```
<br><br><br><br>
## How to use upweight influence function for mis-label
### Import packages
```
# resnet: implemented by wenxinxu
from cifar10_input import *
from cifar10_train import Train
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import darkon
# to enable specific GPU
%set_env CUDA_VISIBLE_DEVICES=0
# cifar-10 classes
_classes = (
'airplane',
'automobile',
'bird',
'cat',
'deer',
'dog',
'frog',
'horse',
'ship',
'truck'
)
```
### Download/Extract cifar10 dataset
```
maybe_download_and_extract()
```
### Implement dataset feeder
```
class MyFeeder(darkon.InfluenceFeeder):
def __init__(self):
# load train data
# for ihvp
data, label = prepare_train_data(padding_size=0)
# update some label
label = self.make_mislabel(label)
self.train_origin_data = data / 256.
self.train_label = label
self.train_data = whitening_image(data)
self.train_batch_offset = 0
def make_mislabel(self, label):
target_class_idx = 7
correct_indices = np.where(label == target_class_idx)[0]
self.correct_indices = correct_indices[:]
# 1% dogs to horses.
# In the mis-label model training, I used this script to choose random dogs.
labeled_dogs = np.where(label == 5)[0]
np.random.shuffle(labeled_dogs)
mislabel_indices = labeled_dogs[:int(labeled_dogs.shape[0] * 0.01)]
label[mislabel_indices] = 7.0
self.mislabel_indices = mislabel_indices
print('target class: {}'.format(_classes[target_class_idx]))
print(self.mislabel_indices)
return label
def test_indices(self, indices):
return self.train_data[indices], self.train_label[indices]
def train_batch(self, batch_size):
# for recursion part
# calculate offset
start = self.train_batch_offset
end = start + batch_size
self.train_batch_offset += batch_size
return self.train_data[start:end, ...], self.train_label[start:end, ...]
def train_one(self, idx):
return self.train_data[idx, ...], self.train_label[idx, ...]
def reset(self):
self.train_batch_offset = 0
# to fix shuffled data
np.random.seed(75)
feeder = MyFeeder()
```
### Restore pre-trained model
```
# tf model checkpoint
check_point = 'pre-trained-mislabel/model.ckpt-79999'
net = Train()
net.build_train_validation_graph()
saver = tf.train.Saver(tf.global_variables())
sess = tf.InteractiveSession()
saver.restore(sess, check_point)
```
### Upweight influence options
```
approx_params = {
'scale': 200,
'num_repeats': 3,
'recursion_depth': 50,
'recursion_batch_size': 100
}
# targets
test_indices = list(feeder.correct_indices) + list(feeder.mislabel_indices)
print('num test targets: {}'.format(len(test_indices)))
```
### Run upweight influence function
```
# choose all of trainable layers
trainable_variables = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES)
# initialize Influence function
inspector = darkon.Influence(
workspace='./influence-workspace',
feeder=feeder,
loss_op_train=net.full_loss,
loss_op_test=net.loss_op,
x_placeholder=net.image_placeholder,
y_placeholder=net.label_placeholder,
trainable_variables=trainable_variables)
scores = list()
for i, target in enumerate(test_indices):
score = inspector.upweighting_influence(
sess,
[target],
1,
approx_params,
[target],
10000000,
force_refresh=True
)
scores += list(score)
print('done: [{}] - {}'.format(i, score))
print(scores)
np.save('mislabel-result-all.npy', scores)
```
### License
---
<pre>
Copyright 2017 Neosapience, Inc.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
</pre>
---
|
github_jupyter
|
```
import os
os.chdir('..')
os.chdir('..')
print(os.getcwd())
import rsnapsim as rss
import numpy as np
os.chdir('rsnapsim')
os.chdir('interactive_notebooks')
import numpy as np
import matplotlib.pyplot as plt
import time
poi_strs, poi_objs, tagged_pois,raw_seq = rss.seqmanip.open_seq_file('../gene_files/H2B_withTags.txt')
poi = tagged_pois['1'][0] #protein object
poi.tag_epitopes['T_Flag'] = [10,20,30,40,50,60,70]
poi.tag_epitopes['T_Hemagglutinin'] = [300,330,340,350]
plt.style.use('dark_background')
plt.rcParams['figure.dpi'] = 120
plt.rcParams['lines.linewidth'] = 1
plt.rcParams['axes.linewidth'] = 1.5
plt.rcParams['font.size'] = 15
plt.rcParams['axes.grid'] = False
colors = ['#00ff51', '#00f7ff']
rss.solver.protein=poi
t = np.linspace(0,500,501)
poi.visualize_probe(colors=['#00ff51', '#00f7ff'])
sttime = time.time()
ssa_soln = rss.solver.solve_ssa(poi.kelong,t,ki=.033,n_traj=20)
solvetime = time.time()-sttime
print(ssa_soln.intensity_vec.shape)
plt.plot(np.mean(ssa_soln.intensity_vec[0],axis=1),color='#00ff51',alpha=.8)
plt.plot(np.mean(ssa_soln.intensity_vec[1],axis=1),color='#00f7ff',alpha=.8)
plt.xlabel('time')
plt.ylabel('intensity')
print("Low memory, no recording: solved in %f seconds" % solvetime)
```
## Autocovariances with individual means
```
acov,err_acov = rss.inta.get_autocov(ssa_soln.intensity_vec,norm='ind')
plt.plot(np.mean(acov[0],axis=1),color=colors[0]);plt.plot(np.mean(acov[1],axis=1),color=colors[1])
plt.plot(np.mean(acov[0],axis=1) - err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acov[1],axis=1)- err_acov[1],'--',color=colors[1])
plt.plot(np.mean(acov[0],axis=1)+ err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acov[1],axis=1)+ err_acov[1],'--',color=colors[1])
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
plt.xlabel('tau')
plt.ylabel('G(tau)')
#normalized by G0
acc,acc_err = rss.inta.get_autocorr(acov)
n_traj = acc.shape[-1]
err_acov = 1.0/np.sqrt(n_traj)*np.std(acc,ddof=1,axis=2)
plt.plot(np.mean(acc[0],axis=1),color=colors[0]);plt.plot(np.mean(acc[1],axis=1),color=colors[1])
plt.plot(np.mean(acc[0],axis=1) - err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acc[1],axis=1)- err_acov[1],'--',color=colors[1])
plt.plot(np.mean(acc[0],axis=1)+ err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acc[1],axis=1)+ err_acov[1],'--',color=colors[1])
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
plt.xlabel('tau')
plt.ylabel('G(tau)')
```
## Global means
```
acov,err_acov = rss.inta.get_autocov(ssa_soln.intensity_vec,norm='global')
plt.plot(np.mean(acov[0],axis=1),color='seagreen');plt.plot(np.mean(acov[1],axis=1),color='violet')
plt.plot(np.mean(acov[0],axis=1) - err_acov[0],'--',color='seagreen');plt.plot(np.mean(acov[1],axis=1)- err_acov[1],'--',color='violet')
plt.plot(np.mean(acov[0],axis=1)+ err_acov[0],'--',color='seagreen');plt.plot(np.mean(acov[1],axis=1)+ err_acov[1],'--',color='violet')
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
#normalized by G0
acc,acc_error = rss.inta.get_autocorr(acov,g0='G1')
mean_acc = np.mean(acc,axis=2)
plt.plot(mean_acc[0],color='seagreen');plt.plot(mean_acc[1],color='violet')
plt.plot(np.mean(acc[0],axis=1) - acc_error[0],'--',color='seagreen');plt.plot(np.mean(acc[1],axis=1)- acc_error[1],'--',color='violet')
plt.plot(np.mean(acc[0],axis=1)+ acc_error[0],'--',color='seagreen');plt.plot(np.mean(acc[1],axis=1)+ acc_error[1],'--',color='violet')
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
```
## Cross correlations
```
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='indiv')
plt.figure()
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot(s11_cc/s11_cc[500],color=colors[0] );
plt.plot(s21_cc/s21_cc[500],color='#ff00ee');
plt.plot(s22_cc/s22_cc[500],color=colors[1]);
plt.plot(s11_cc/s11_cc[500] - err_cc[0]/s11_cc[500],'--',color=colors[0] );
plt.plot(s11_cc/s11_cc[500] + err_cc[0]/s11_cc[500],'--',color=colors[0] );
plt.plot(s21_cc/s21_cc[500] - err_cc[2]/s21_cc[500] ,'--',color='#ff00ee' );
plt.plot(s21_cc/s21_cc[500] + err_cc[2]/s21_cc[500] ,'--',color='#ff00ee');
plt.plot(s22_cc/s22_cc[500] - s22_cc[3]/s22_cc[500],'--',color=colors[1] );
plt.plot(s22_cc/s22_cc[500] + s22_cc[3]/s22_cc[500],'--',color=colors[1] );
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
```
## normalization modes
| norm | effect |
| :- | :-: |
| global | subtract all intensities by the global mean intensity before correlation |
| individual | subtract all intensities by the trajectory mean intensity before correlation |
| raw | do nothing, correlate the intensities as they are |
## G0
| norm | effect |
| :- | :-: |
| global_max | divide correlations by the global maximum point |
| individual_max | divide correlations by the individual trajectory maximum point |
| global_center | divide correlations by the global average of the center point of the correlation |
| individual_center | divide all correlations by the trajectory center point value |
| None | do nothing, do not normalize the correlations by anything|
```
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='indiv',g0='indiv_max')
plt.figure()
plt.plot(cross_corr[0], color = colors[0],alpha=.5)
plt.plot(cross_corr[2],color = '#ff00ee',alpha=.5)
plt.plot(cross_corr[3], color = colors[1],alpha=.5)
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='global',g0='indiv_max')
plt.figure()
plt.plot(cross_corr[0], color = colors[0],alpha=.5)
plt.plot(cross_corr[2],color = '#ff00ee',alpha=.5)
plt.plot(cross_corr[3], color = colors[1],alpha=.5)
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
```
|
github_jupyter
|
##### Copyright 2020 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Question Answer with TensorFlow Lite Model Maker
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/lite/tutorials/model_maker_question_answer"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/tensorflow/blob/master/tensorflow/lite/g3doc/tutorials/model_maker_question_answer.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/tensorflow/blob/master/tensorflow/lite/g3doc/tutorials/model_maker_question_answer.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/tensorflow/tensorflow/lite/g3doc/tutorials/model_maker_question_answer.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
The TensorFlow Lite Model Maker library simplifies the process of adapting and converting a TensorFlow model to particular input data when deploying this model for on-device ML applications.
This notebook shows an end-to-end example that utilizes the Model Maker library to illustrate the adaptation and conversion of a commonly-used question answer model for question answer task.
# Introduction to Question Answer Task
The supported task in this library is extractive question answer task, which means given a passage and a question, the answer is the span in the passage. The image below shows an example for question answer.
<p align="center"><img src="https://storage.googleapis.com/download.tensorflow.org/models/tflite/screenshots/model_maker_squad_showcase.png" width="500"></p>
<p align="center">
<em>Answers are spans in the passage (image credit: <a href="https://rajpurkar.github.io/mlx/qa-and-squad/">SQuAD blog</a>) </em>
</p>
As for the model of question answer task, the inputs should be the passage and question pair that are already preprocessed, the outputs should be the start logits and end logits for each token in the passage.
The size of input could be set and adjusted according to the length of passage and question.
## End-to-End Overview
The following code snippet demonstrates how to get the model within a few lines of code. The overall process includes 5 steps: (1) choose a model, (2) load data, (3) retrain the model, (4) evaluate, and (5) export it to TensorFlow Lite format.
```python
# Chooses a model specification that represents the model.
spec = model_spec.get('mobilebert_qa')
# Gets the training data and validation data.
train_data = QuestionAnswerDataLoader.from_squad(train_data_path, spec, is_training=True)
validation_data = QuestionAnswerDataLoader.from_squad(validation_data_path, spec, is_training=False)
# Fine-tunes the model.
model = question_answer.create(train_data, model_spec=spec)
# Gets the evaluation result.
metric = model.evaluate(validation_data)
# Exports the model to the TensorFlow Lite format in the export directory.
model.export(export_dir)
```
The following sections explain the code in more detail.
## Prerequisites
To run this example, install the required packages, including the Model Maker package from the [GitHub repo](https://github.com/tensorflow/examples/tree/master/tensorflow_examples/lite/model_maker).
```
!pip install tflite-model-maker
```
Import the required packages.
```
import numpy as np
import os
import tensorflow as tf
assert tf.__version__.startswith('2')
from tflite_model_maker import configs
from tflite_model_maker import model_spec
from tflite_model_maker import question_answer
from tflite_model_maker import QuestionAnswerDataLoader
```
The "End-to-End Overview" demonstrates a simple end-to-end example. The following sections walk through the example step by step to show more detail.
## Choose a model_spec that represents a model for question answer
Each `model_spec` object represents a specific model for question answer. The Model Maker currently supports MobileBERT and BERT-Base models.
Supported Model | Name of model_spec | Model Description
--- | --- | ---
[MobileBERT](https://arxiv.org/pdf/2004.02984.pdf) | 'mobilebert_qa' | 4.3x smaller and 5.5x faster than BERT-Base while achieving competitive results, suitable for on-device scenario.
[MobileBERT-SQuAD](https://arxiv.org/pdf/2004.02984.pdf) | 'mobilebert_qa_squad' | Same model architecture as MobileBERT model and the initial model is already retrained on [SQuAD1.1](https://rajpurkar.github.io/SQuAD-explorer/).
[BERT-Base](https://arxiv.org/pdf/1810.04805.pdf) | 'bert_qa' | Standard BERT model that widely used in NLP tasks.
In this tutorial, [MobileBERT-SQuAD](https://arxiv.org/pdf/2004.02984.pdf) is used as an example. Since the model is already retrained on [SQuAD1.1](https://rajpurkar.github.io/SQuAD-explorer/), it could coverage faster for question answer task.
```
spec = model_spec.get('mobilebert_qa_squad')
```
## Load Input Data Specific to an On-device ML App and Preprocess the Data
The [TriviaQA](https://nlp.cs.washington.edu/triviaqa/) is a reading comprehension dataset containing over 650K question-answer-evidence triples. In this tutorial, you will use a subset of this dataset to learn how to use the Model Maker library.
To load the data, convert the TriviaQA dataset to the [SQuAD1.1](https://rajpurkar.github.io/SQuAD-explorer/) format by running the [converter Python script](https://github.com/mandarjoshi90/triviaqa#miscellaneous) with `--sample_size=8000` and a set of `web` data. Modify the conversion code a little bit by:
* Skipping the samples that couldn't find any answer in the context document;
* Getting the original answer in the context without uppercase or lowercase.
Download the archived version of the already converted dataset.
```
train_data_path = tf.keras.utils.get_file(
fname='triviaqa-web-train-8000.json',
origin='https://storage.googleapis.com/download.tensorflow.org/models/tflite/dataset/triviaqa-web-train-8000.json')
validation_data_path = tf.keras.utils.get_file(
fname='triviaqa-verified-web-dev.json',
origin='https://storage.googleapis.com/download.tensorflow.org/models/tflite/dataset/triviaqa-verified-web-dev.json')
```
You can also train the MobileBERT model with your own dataset. If you are running this notebook on Colab, upload your data by using the left sidebar.
<img src="https://storage.googleapis.com/download.tensorflow.org/models/tflite/screenshots/model_maker_question_answer.png" alt="Upload File" width="800" hspace="100">
If you prefer not to upload your data to the cloud, you can also run the library offline by following the [guide](https://github.com/tensorflow/examples/tree/master/tensorflow_examples/lite/model_maker).
Use the `QuestionAnswerDataLoader.from_squad` method to load and preprocess the [SQuAD format](https://rajpurkar.github.io/SQuAD-explorer/) data according to a specific `model_spec`. You can use either SQuAD2.0 or SQuAD1.1 formats. Setting parameter `version_2_with_negative` as `True` means the formats is SQuAD2.0. Otherwise, the format is SQuAD1.1. By default, `version_2_with_negative` is `False`.
```
train_data = QuestionAnswerDataLoader.from_squad(train_data_path, spec, is_training=True)
validation_data = QuestionAnswerDataLoader.from_squad(validation_data_path, spec, is_training=False)
```
## Customize the TensorFlow Model
Create a custom question answer model based on the loaded data. The `create` function comprises the following steps:
1. Creates the model for question answer according to `model_spec`.
2. Train the question answer model. The default epochs and the default batch size are set according to two variables `default_training_epochs` and `default_batch_size` in the `model_spec` object.
```
model = question_answer.create(train_data, model_spec=spec)
```
Have a look at the detailed model structure.
```
model.summary()
```
## Evaluate the Customized Model
Evaluate the model on the validation data and get a dict of metrics including `f1` score and `exact match` etc. Note that metrics are different for SQuAD1.1 and SQuAD2.0.
```
model.evaluate(validation_data)
```
## Export to TensorFlow Lite Model
Convert the existing model to TensorFlow Lite model format that you can later use in an on-device ML application.
Since MobileBERT is too big for on-device applications, use dynamic range quantization on the model to compress MobileBERT by 4x with the minimal loss of performance. First, define the quantization configuration:
```
config = configs.QuantizationConfig.create_dynamic_range_quantization(optimizations=[tf.lite.Optimize.OPTIMIZE_FOR_LATENCY])
config._experimental_new_quantizer = True
```
Export the quantized TFLite model according to the quantization config and save the vocabulary to a vocab file. The default TFLite model filename is `model.tflite`, and the default vocab filename is `vocab`.
```
model.export(export_dir='.', quantization_config=config)
```
You can use the TensorFlow Lite model file and vocab file in the [bert_qa](https://github.com/tensorflow/examples/tree/master/lite/examples/bert_qa/android) reference app by downloading it from the left sidebar on Colab.
You can also evalute the tflite model with the `evaluate_tflite` method. This step is expected to take a long time.
```
model.evaluate_tflite('model.tflite', validation_data)
```
## Advanced Usage
The `create` function is the critical part of this library in which the `model_spec` parameter defines the model specification. The `BertQAModelSpec` class is currently supported. There are 2 models: MobileBERT model, BERT-Base model. The `create` function comprises the following steps:
1. Creates the model for question answer according to `model_spec`.
2. Train the question answer model.
This section describes several advanced topics, including adjusting the model, tuning the training hyperparameters etc.
### Adjust the model
You can adjust the model infrastructure like parameters `seq_len` and `query_len` in the `BertQAModelSpec` class.
Adjustable parameters for model:
* `seq_len`: Length of the passage to feed into the model.
* `query_len`: Length of the question to feed into the model.
* `doc_stride`: The stride when doing a sliding window approach to take chunks of the documents.
* `initializer_range`: The stdev of the truncated_normal_initializer for initializing all weight matrices.
* `trainable`: Boolean, whether pre-trained layer is trainable.
Adjustable parameters for training pipeline:
* `model_dir`: The location of the model checkpoint files. If not set, temporary directory will be used.
* `dropout_rate`: The rate for dropout.
* `learning_rate`: The initial learning rate for Adam.
* `predict_batch_size`: Batch size for prediction.
* `tpu`: TPU address to connect to. Only used if using tpu.
For example, you can train the model with a longer sequence length. If you change the model, you must first construct a new `model_spec`.
```
new_spec = model_spec.get('mobilebert_qa')
new_spec.seq_len = 512
```
The remaining steps are the same. Note that you must rerun both the `dataloader` and `create` parts as different model specs may have different preprocessing steps.
### Tune training hyperparameters
You can also tune the training hyperparameters like `epochs` and `batch_size` to impact the model performance. For instance,
* `epochs`: more epochs could achieve better performance, but may lead to overfitting.
* `batch_size`: number of samples to use in one training step.
For example, you can train with more epochs and with a bigger batch size like:
```python
model = question_answer.create(train_data, model_spec=spec, epochs=5, batch_size=64)
```
### Change the Model Architecture
You can change the base model your data trains on by changing the `model_spec`. For example, to change to the BERT-Base model, run:
```python
spec = model_spec.get('bert_qa')
```
The remaining steps are the same.
|
github_jupyter
|
```
import pandas as pd
import os
import glob
raw_data_path = os.path.join('data', 'raw')
clean_filename = os.path.join('data', 'clean', 'data.csv')
```
# Read data
```
all_files = glob.glob(raw_data_path + "/top_songs_with_lyrics.csv")
raw_data = pd.concat(pd.read_csv(f) for f in all_files)
raw_data.head()
```
# Pre processing
```
import re
import string
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
nltk.download('punkt')
nltk.download('stopwords')
#Puntuaction Removing [!”#$%&’()*+,-./:;<=>?@[\]^_`{|}~]:
def clean_puntuaction(input_df):
result=input_df
for idx in range(result.shape[0]):
result[idx]=result[idx].replace("'","")
result[idx]=result[idx].replace("\r"," ")
result[idx]=result[idx].replace("\n","")
result[idx]= re.sub("[\(\[].*?[\)\]]", "", result[idx])
result[idx]= re.sub(r'[^\w\s]', '', result[idx])
return result
def remove_accents(input_str):
"""
remueve acentos, aunque al ser un texto en inglés no deberían existir acentos
"""
nfkd_form = unicodedata.normalize('NFKD', input_str )
return u"".join([c for c in nfkd_form if not unicodedata.combining(c)])
#Puntuaction Removing [!”#$%&’()*+,-./:;<=>?@[\]^_`{|}~]:
def clean_puntuaction(input_df):
result=input_df
print(result.shape)
#for idx in range(result.shape[0]):
# for idx in result:
# idx=idx+1
# if(result[idx]==""):
# continue
# result[idx]=result[idx].replace("'","")
# result[idx]=result[idx].replace("\r"," ")
# result[idx]=result[idx].replace("\n","")
# result[idx]= re.sub("[\(\[].*?[\)\]]", "", result[idx])
# result[idx]= re.sub(r'[^\w\s]', '', result[idx])
# result[idx]= remove_accents(result[idx])
cont=1
for idx in result.values:
#idx=idx+1
#if(result[idx]==""):
# continue
idx=idx.replace("'","")
idx=idx.replace("\r"," ")
idx=idx.replace("\n"," ")
idx= re.sub("[\(\[].*?[\)\]]", "", idx)
idx= re.sub(r'[^\w\s]', '', idx)
idx= remove_accents(idx)
print(cont)
print (idx)
result[cont]=idx
cont=cont+1
return result
def clean_str_puntuaction(input_df):
input_df=input_df.replace("'","")
input_df=input_df.replace("\r"," ")
input_df=input_df.replace("\n"," ")
input_df=input_df.replace("-"," ")
input_df= re.sub("[\(\[].*?[\)\]]", "", input_df)
input_df= re.sub(r'[^\w\s]', '', input_df)
input_df= remove_accents(input_df)
return input_df
def remove_stopwords(input_df):
result=input_df
for idx in range(result.shape[0]):
tokens = word_tokenize(result[idx])
stop_words = stopwords.words('spanish')
more_stopwords = ['si', 'pa', 'sé', 'solo', 'yeah', 'yeh', 'oh', 'i', 'to', 'va', 'the', 'aunque', 'you', 'eh', 'cómo','ma']
total_stopwords = stop_words + more_stopwords
result[idx] = [i for i in tokens if not i in total_stopwords]
return result
# TODO: Perform cleaning
data_filename = 'data/raw/top_songs_with_lyrics.csv'
dataset = pd.read_csv(data_filename)
dataset.columns.tolist()
#dataset.iloc[:,0]
#df=dataset[['artists ','title','lyric
# lowercase ']]
df=dataset['lyric '].str.lower()
df=clean_puntuaction(df)
df=remove_stopwords(df)
#df[1]
#freq = nltk.FreqDist(tokens)
clean_data = pd.DataFrame(data={'dummy': [1, 2]})
clean_data.to_csv(clean_filename, index=False)
```
|
github_jupyter
|
```
%%html
<link href="http://mathbook.pugetsound.edu/beta/mathbook-content.css" rel="stylesheet" type="text/css" />
<link href="https://aimath.org/mathbook/mathbook-add-on.css" rel="stylesheet" type="text/css" />
<style>.subtitle {font-size:medium; display:block}</style>
<link href="https://fonts.googleapis.com/css?family=Open+Sans:400,400italic,600,600italic" rel="stylesheet" type="text/css" />
<link href="https://fonts.googleapis.com/css?family=Inconsolata:400,700&subset=latin,latin-ext" rel="stylesheet" type="text/css" /><!-- Hide this cell. -->
<script>
var cell = $(".container .cell").eq(0), ia = cell.find(".input_area")
if (cell.find(".toggle-button").length == 0) {
ia.after(
$('<button class="toggle-button">Toggle hidden code</button>').click(
function (){ ia.toggle() }
)
)
ia.hide()
}
</script>
```
**Important:** to view this notebook properly you will need to execute the cell above, which assumes you have an Internet connection. It should already be selected, or place your cursor anywhere above to select. Then press the "Run" button in the menu bar above (the right-pointing arrowhead), or press Shift-Enter on your keyboard.
$\newcommand{\identity}{\mathrm{id}}
\newcommand{\notdivide}{\nmid}
\newcommand{\notsubset}{\not\subset}
\newcommand{\lcm}{\operatorname{lcm}}
\newcommand{\gf}{\operatorname{GF}}
\newcommand{\inn}{\operatorname{Inn}}
\newcommand{\aut}{\operatorname{Aut}}
\newcommand{\Hom}{\operatorname{Hom}}
\newcommand{\cis}{\operatorname{cis}}
\newcommand{\chr}{\operatorname{char}}
\newcommand{\Null}{\operatorname{Null}}
\newcommand{\lt}{<}
\newcommand{\gt}{>}
\newcommand{\amp}{&}
$
<div class="mathbook-content"><h2 class="heading hide-type" alt="Section 19.7 Sage"><span class="type">Section</span><span class="codenumber">19.7</span><span class="title">Sage</span></h2><a href="boolean-sage.ipynb" class="permalink">¶</a></div>
<div class="mathbook-content"></div>
<div class="mathbook-content"><p id="p-3037">Sage has support for both partially ordered sets (“posets”) and lattices, and does an excellent job of providing visual depictions of both.</p></div>
<div class="mathbook-content"><h3 class="heading hide-type" alt="Subsection Creating Partially Ordered Sets"><span class="type">Subsection</span><span class="codenumber" /><span class="title">Creating Partially Ordered Sets</span></h3></div>
<div class="mathbook-content"><p id="p-3038">Example <a href="section-boolean-lattices.ipynb#example-boolean-poset-divisors-24" class="xref" alt="Example 19.6 " title="Example 19.6 ">19.6</a> in the text is a good example to replicate as a demonstration of Sage commands. We first define the elements of the set $X\text{.}$</p></div>
```
X = (24).divisors()
X
```
<div class="mathbook-content"><p id="p-3039">One approach to creating the relation is to specify <em class="emphasis">every</em> instance where one element is comparable to the another. So we build a list of pairs, where each pair contains comparable elements, with the lesser one first. This is the set of relations.</p></div>
```
R = [(a,b) for a in X for b in X if a.divides(b)]; R
```
<div class="mathbook-content"><p id="p-3040">We construct the poset by giving the the <code class="code-inline tex2jax_ignore">Poset</code> constructor a list containing the elements and the relations. We can then easily get a “plot” of the poset. Notice the plot just shows the “cover relations” — a minimal set of comparisons which the assumption of transitivity would expand into the set of all the relations.</p></div>
```
D = Poset([X, R])
D.plot()
```
<div class="mathbook-content"><p id="p-3041">Another approach to creating a <code class="code-inline tex2jax_ignore">Poset</code> is to let the poset constructor run over all the pairs of elements, and all we do is give the constructor a way to test if two elements are comparable. Our comparison function should expect two elements and then return <code class="code-inline tex2jax_ignore">True</code> or <code class="code-inline tex2jax_ignore">False</code>. A “lambda” function is one way to quickly build such a function. This may be a new idea for you, but mastering lambda functions can be a great convenience. Notice that “lambda” is a word reserved for just this purpose (so, for example, <code class="code-inline tex2jax_ignore">lambda</code> is a bad choice for the name of an eigenvalue of a matrix). There are other ways to make functions in Sage, but a lambda function is quickest when the function is simple.</p></div>
```
divisible = lambda x, y: x.divides(y)
L = Poset([X, divisible])
L == D
L.plot()
```
<div class="mathbook-content"><p id="p-3042">Sage also has a collection of stock posets. Some are one-shot constructions, while others are members of parameterized families. Use tab-completion on <code class="code-inline tex2jax_ignore">Posets.</code> to see the full list. Here are some examples.</p></div>
<div class="mathbook-content"><p id="p-3043">A one-shot construction. Perhaps what you would expect, though there might be other, equally plausible, alternatives.</p></div>
```
Q = Posets.PentagonPoset()
Q.plot()
```
<div class="mathbook-content"><p id="p-3044">A parameterized family. This is the classic example where the elements are subsets of a set with $n$ elements and the relation is “subset of.”</p></div>
```
S = Posets.BooleanLattice(4)
S.plot()
```
<div class="mathbook-content"><p id="p-3045">And random posets. These can be useful for testing and experimenting, but are unlikely to exhibit special cases that may be important. You might run the following command many times and vary the second argument, which is a rough upper bound on the probability any two elements are comparable. Remember that the plot only shows the cover relations. The more elements that are comparable, the more “vertically stretched” the plot will be.</p></div>
```
T = Posets.RandomPoset(20,0.05)
T.plot()
```
<div class="mathbook-content"><h3 class="heading hide-type" alt="Subsection Properties of a Poset"><span class="type">Subsection</span><span class="codenumber" /><span class="title">Properties of a Poset</span></h3></div>
<div class="mathbook-content"><p id="p-3046">Once you have a poset, what can you do with it? Let's return to our first example, <code class="code-inline tex2jax_ignore">D</code>. We can of course determine if one element is less than another, which is the fundamental structure of a poset.</p></div>
```
D.is_lequal(4, 8)
D.is_lequal(4, 4)
D.is_less_than(4, 8)
D.is_less_than(4, 4)
D.is_lequal(6, 8)
D.is_lequal(8, 6)
```
<div class="mathbook-content"><p id="p-3047">Notice that <code class="code-inline tex2jax_ignore">6</code> and <code class="code-inline tex2jax_ignore">8</code> are not comparable in this poset (it is a <em class="emphasis">partial</em> order). The methods <code class="code-inline tex2jax_ignore">.is_gequal()</code> and <code class="code-inline tex2jax_ignore">.is_greater_than()</code> work similarly, but returns <code class="code-inline tex2jax_ignore">True</code> if the first element is greater (or equal).</p></div>
```
D.is_gequal(8, 4)
D.is_greater_than(4, 8)
```
<div class="mathbook-content"><p id="p-3048">We can find the largest and smallest elements of a poset. This is a random poset built with a 10%probability, but copied here to be repeatable.</p></div>
```
X = range(20)
C = [[18, 7], [9, 11], [9, 10], [11, 8], [6, 10],
[10, 2], [0, 2], [2, 1], [1, 8], [8, 12],
[8, 3], [3, 15], [15, 7], [7, 16], [7, 4],
[16, 17], [16, 13], [4, 19], [4, 14], [14, 5]]
P = Poset([X, C])
P.plot()
P.minimal_elements()
P.maximal_elements()
```
<div class="mathbook-content"><p id="p-3049">Elements of a poset can be partioned into level sets. In plots of posets, elements at the same level are plotted vertically at the same height. Each level set is obtained by removing all of the previous level sets and then taking the minimal elements of the result.</p></div>
```
P.level_sets()
```
<div class="mathbook-content"><p id="p-3050">If we make two elements in <code class="code-inline tex2jax_ignore">R</code> comparable when they had not previously been, this is an extension of <code class="code-inline tex2jax_ignore">R</code>. Consider all possible extensions of one poset — we can make a poset from all of these, where set inclusion is the relation. A linear extension is a maximal element in this poset of posets. Informally, we are adding as many new relations as possible, consistent with the original poset and so that the result is a total order. In other words, there is an ordering of the elements that is consistent with the order in the poset. We can build such a thing, but the output is just a list of the elements in the linear order. A computer scientist would be inclined to call this a “topological sort.”</p></div>
```
linear = P.linear_extension(); linear
```
<div class="mathbook-content"><p id="p-3051">We can construct subposets by giving a set of elements to induce the new poset. Here we take roughly the “bottom half” of the random poset <code class="code-inline tex2jax_ignore">P</code> by inducing the subposet on a union of some of the level sets.</p></div>
```
level = P.level_sets()
bottomhalf = sum([level[i] for i in range(5)], [])
B = P.subposet(bottomhalf)
B.plot()
```
<div class="mathbook-content"><p id="p-3052">The dual of a poset retains the same set of elements, but reverses any comparisons.</p></div>
```
Pdual = P.dual()
Pdual.plot()
```
<div class="mathbook-content"><p id="p-3053">Taking the dual of the divisibility poset from Example <a href="section-boolean-lattices.ipynb#example-boolean-poset-divisors-24" class="xref" alt="Example 19.6 " title="Example 19.6 ">19.6</a> would be like changing the relation to “is a multiple of.”</p></div>
```
Ddual = D.dual()
Ddual.plot()
```
<div class="mathbook-content"><h3 class="heading hide-type" alt="Subsection Lattices"><span class="type">Subsection</span><span class="codenumber" /><span class="title">Lattices</span></h3></div>
<div class="mathbook-content"><p id="p-3054">Every lattice is a poset, so all the commands above will perform equally well for a lattice. But how do you create a lattice? Simple — first create a poset and then feed it into the <code class="code-inline tex2jax_ignore">LatticePoset()</code> constructor. But realize that just because you give this constructor a poset, it does not mean a lattice will always come back out. Only if the poset is <em class="emphasis">already</em> a lattice will it get upgraded from a poset to a lattice for Sage's purposes, and you will get a <code class="code-inline tex2jax_ignore">ValueError</code> if the upgrade is not possible. Finally, notice that some of the posets Sage constructs are already recognized as lattices, such as the prototypical <code class="code-inline tex2jax_ignore">BooleanLattice</code>.</p></div>
```
P = Posets.AntichainPoset(8)
P.is_lattice()
LatticePoset(P)
```
<div class="mathbook-content"><p id="p-3055">An integer composition of $n$ is a list of positive integers that sum to $n\text{.}$ A composition $C_1$ covers a composition $C_2$ if $C_2$ can be formed from $C_1$ by adding consecutive parts. For example, $C_1 = [2, 1, 2] \succeq [3, 2] = C_2\text{.}$ With this relation, the set of all integer compositions of a fixed integer $n$ is a poset that is also a lattice.</p></div>
```
CP = Posets.IntegerCompositions(5)
C = LatticePoset(CP)
C.plot()
```
<div class="mathbook-content"><p id="p-3056">A meet or a join is a fundamental operation in a lattice.</p></div>
```
par = C.an_element().parent()
a = par([1, 1, 1, 2])
b = par([2, 1, 1, 1])
a, b
C.meet(a, b)
c = par([1, 4])
d = par([2, 3])
c, d
C.join(c, d)
```
<div class="mathbook-content"><p id="p-3057">Once a poset is upgraded to lattice status, then additional commands become available, or the character of their results changes.</p></div>
<div class="mathbook-content"><p id="p-3058">An example of the former is the <code class="code-inline tex2jax_ignore">.is_distributive()</code> method.</p></div>
```
C.is_distributive()
```
<div class="mathbook-content"><p id="p-3059">An example of the latter is the <code class="code-inline tex2jax_ignore">.top()</code> method. What your text calls a largest element and a smallest element of a lattice, Sage calls a top and a bottom. For a poset, <code class="code-inline tex2jax_ignore">.top()</code> and <code class="code-inline tex2jax_ignore">.bottom()</code> may return an element or may not (returning <code class="code-inline tex2jax_ignore">None</code>), but for a lattice it is guaranteed to return exactly one element.</p></div>
```
C.top()
C.bottom()
```
<div class="mathbook-content"><p id="p-3060">Notice that the returned values are all elements of the lattice, in this case ordered lists of integers summing to $5\text{.}$</p></div>
<div class="mathbook-content"><p id="p-3061">Complements now make sense in a lattice. The result of the <code class="code-inline tex2jax_ignore">.complements()</code> method is a dictionary that uses elements of the lattice as the keys. We say the dictionary is “indexed” by the elements of the lattice. The result is a list of the complements of the element. We call this the “value” of the key-value pair. (You may know dictionaries as “associative arrays”, but they are really just fancy functions.)</p></div>
```
comp = C.complements()
comp[par([1, 1, 1, 2])]
```
<div class="mathbook-content"><p id="p-3062">The lattice of integer compositions is a complemented lattice, as we can see by the result that each element has a single (unique) complement, evidenced by the lists of length $1$ in the values of the dictionary. Or we can just ask Sage via <code class="code-inline tex2jax_ignore">.is_complemented()</code>. Dictionaries have no inherent order, so you may get different output each time you inspect the dictionary.</p></div>
```
comp
[len(e[1]) for e in comp.items()]
C.is_complemented()
```
<div class="mathbook-content"><p id="p-3063">There are many more commands which apply to posets and lattices, so build a few and use tab-completion liberally to explore. There is more to discover than we can cover in just a single chapter, but you now have the basic tools to profitably study posets and lattices in Sage.</p></div>
|
github_jupyter
|
```
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import roc_curve
from sklearn.metrics import auc
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score
from sklearn.metrics import f1_score
from sklearn.metrics import average_precision_score
from inspect import signature
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from matplotlib import pyplot
from sklearn import metrics
print("Setup Complete")
df = pd.read_csv("../input/fitness-watch-dataset/dataset_halfSecondWindow.csv") #dataset_halfSecondWindows
#df.info
# first doing label encodingon User
# sorting based on the user label
# plotting given_user distribution
# making split
df.isna().sum().sum() #5893 * 70
cleanup_target = {"target": {"Car":1,"Still":2,"Train":3,"Bus":4,"Walking":5}}
df = df.replace(cleanup_target)
cleanup_nums = {"user": {"andrea": 1, "Luca": 2, "Damiano": 3,"michelangelo": 4,
"Pierpaolo": 5, "Vincenzo": 6,"IvanHeibi":7,"AndreaCarpineti":8,
"Federica":9,"Serena":10,"Claudio":11,"Elena":12,
"Riccardo":13}}
df = df.replace(cleanup_nums)
#df = df.fillna(0)
df = df.fillna(df.median())
df1 = df.sort_values(by=['user'])
list_users=df.user.unique()
ax = df['user'].value_counts().plot(kind='bar')
df['user'].value_counts()
ax.set_xlabel("Users")
ax.set_ylabel("Number of Responses")
ax.figure.savefig('user_distribution.png')
grouped = df.groupby(df.user)
user_dict = {}
sample_df = df[:0]
for i in range(1,10):
user_dict[i] = grouped.get_group(i)
user_dict[i] = user_dict[i].sample(n=2225)
sample_df = sample_df.append(user_dict[i])
list_users=sample_df.user.unique()
ax = sample_df['user'].value_counts().plot(kind='bar')
#sample_df['user'].value_counts()
ax.set_xlabel("Users")
ax.set_ylabel("Number of Responses")
ax.figure.savefig('user_distribution_sampled.png')
df1 = sample_df
df1
df1 = df1.replace([' ','NULL'],np.nan)
df1 = df1.dropna(thresh=df1.shape[0]*0.6,how='all',axis=1)
df1.isna().sum().sum() #5893 * 52
df1
# commmon
#df = df.dropna(axis=1, how='all')
df2 = df1
train_pct_index1 = int(0.2 * len(df2))
train_pct_index2 = int(0.4 * len(df2))
train_pct_index3 = int(0.6 * len(df2))
train_pct_index4 = int(0.8 * len(df2))
print(0,train_pct_index1,train_pct_index2,train_pct_index3,train_pct_index4,len(df2))
# first fold:
train1, test1 = df2[train_pct_index1:], df2[:train_pct_index1] # 20 to 100
# 2 fold:
train2, test2 = df2.head(train_pct_index2).append(df2.tail(train_pct_index2)), df2[train_pct_index1:train_pct_index2] # 40 to 100 + 0 to 20
train3, test3 = df2.head(-train_pct_index3).append(df2.head(train_pct_index2)), df2[train_pct_index2:train_pct_index3] # 60 to 100 + 0 to 40
train4, test4 = df2.head(-train_pct_index4).append(df2.head(train_pct_index3)), df2[train_pct_index3:train_pct_index4] # 80 to 100 + 0 to 60
train5, test5 = df2[:train_pct_index4], df2[train_pct_index4:] # 0 to 80
# first fold: train1, test1
# train separate
# train1 = train1.dropna(axis = 1, how='all')
#train1 = train1.fillna(train1.mean())
#df2 = train1
train1 = train1.drop(['user'], axis=1)
train1 = train1.drop(['id'], axis =1)
# test separate
#test1 = test1.dropna(axis=1, how='all')
#df2 = df1
test1 =test1.drop(['user'], axis=1)
test1 = test1.drop(['id'], axis =1)
test1 = test1.dropna(axis=0)
y = train1.target
x = train1.loc[:, train1.columns != 'target']
y1 = test1.target
x1 = test1.loc[:, test1.columns != 'target']
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
model=RandomForestClassifier(n_estimators=100)
#Train the model using the training sets y_pred=clf.predict(X_test)
model.fit(X_train,y_train)
y_pred=model.predict(X_test)
print("internal accuracy:", metrics.accuracy_score(y_test, y_pred))
y_pred=model.predict(x1)
fold1= metrics.accuracy_score(y1, y_pred)
print("Accuracy:",fold1)
# second fold: train2, test2
# train separate
train2 = train2.drop(['user'], axis=1)
train2 = train2.drop(['id'], axis =1)
# test separate
test2 = test2.drop(['user'], axis=1)
test2 = test2.drop(['id'], axis =1)
test2 = test2.dropna(axis=0)
y = train2.target
x = train2.loc[:, train2.columns != 'target']
y1 = test2.target
x1 = test2.loc[:, test2.columns != 'target']
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
model=RandomForestClassifier(n_estimators=100)
#Train the model using the training sets y_pred=clf.predict(X_test)
model.fit(X_train,y_train)
y_pred=model.predict(X_test)
print("internal accuracy:", metrics.accuracy_score(y_test, y_pred))
y_pred=model.predict(x1)
fold2= metrics.accuracy_score(y1, y_pred)
print("Accuracy:",fold2)
# third fold: train3, test3
# train separate
train3 = train3.drop(['user'], axis=1)
train3 = train3.drop(['id'], axis =1)
# test separate
test3 = test3.drop(['user'], axis=1)
test3 = test3.drop(['id'], axis =1)
test3 = test3.dropna(axis=0)
y = train3.target
x = train3.loc[:, train3.columns != 'target']
y1 = test3.target
x1 = test3.loc[:, test3.columns != 'target']
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
model=RandomForestClassifier(n_estimators=100)
#Train the model using the training sets y_pred=clf.predict(X_test)
model.fit(X_train,y_train)
y_pred=model.predict(X_test)
print("internal accuracy:", metrics.accuracy_score(y_test, y_pred))
y_pred=model.predict(x1)
fold3= metrics.accuracy_score(y1, y_pred)
print("Accuracy:",fold3)
# forth fold: train4, test4
# train separate
train4 = train4.drop(['user'], axis=1)
train4 = train4.drop(['id'], axis =1)
# test separate
test4 = test4.drop(['user'], axis=1)
test4 = test4.drop(['id'], axis =1)
test4 = test4.dropna(axis=0)
y = train4.target
x = train4.loc[:, train4.columns != 'target']
y1 = test4.target
x1 = test4.loc[:, test4.columns != 'target']
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
model=RandomForestClassifier(n_estimators=100)
#Train the model using the training sets y_pred=clf.predict(X_test)
model.fit(X_train,y_train)
y_pred=model.predict(X_test)
print("internal accuracy:", metrics.accuracy_score(y_test, y_pred))
y_pred=model.predict(x1)
fold4= metrics.accuracy_score(y1, y_pred)
print("Accuracy:",fold4)
# fifth fold: train5, test5
# train separate
train5 = train5.drop(['user'], axis=1)
train5 = train5.drop(['id'], axis =1)
# test separate
test5 = test5.drop(['user'], axis=1)
test5 = test5.drop(['id'], axis =1)
#test5 = test5.dropna(axis=0)
y = train5.target
x = train5.loc[:, train5.columns != 'target']
y1 = test5.target
x1 = test5.loc[:, test5.columns != 'target']
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
model=RandomForestClassifier(n_estimators=100)
#Train the model using the training sets y_pred=clf.predict(X_test)
model.fit(X_train,y_train)
y_pred=model.predict(X_test)
print("internal accuracy:", metrics.accuracy_score(y_test, y_pred))
y_pred=model.predict(x1)
fold5= metrics.accuracy_score(y1, y_pred)
print("Accuracy:",fold5)
print("average fold:", (fold1+fold2+fold3+fold4+fold5)/5)
import pickle
filename = 'model.sav'
pickle.dump(model, open(filename, 'wb'))
```
Feature engineering
```
print("F1:", f1_score(y1, y_pred, average='macro'))
to give to dilan
df3= df1.loc[df1['user'] == 3]
df4= df1.loc[df1['user'] == 4]
df3 = df3.drop(['user'], axis=1)
df3 = df3.drop(['id'], axis =1)
df4 = df4.drop(['user'], axis=1)
df4 = df4.drop(['id'], axis =1)
df3.to_csv("userdata_3.csv")
df4.to_csv("userdata_4.csv")
```
|
github_jupyter
|
## CIFAR 10
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
```
You can get the data via:
wget http://pjreddie.com/media/files/cifar.tgz
**Important:** Before proceeding, the student must reorganize the downloaded dataset files to match the expected directory structure, so that there is a dedicated folder for each class under 'test' and 'train', e.g.:
```
* test/airplane/airplane-1001.png
* test/bird/bird-1043.png
* train/bird/bird-10018.png
* train/automobile/automobile-10000.png
```
The filename of the image doesn't have to include its class.
```
from fastai.conv_learner import *
PATH = "data/cifar10/"
os.makedirs(PATH,exist_ok=True)
!ls {PATH}
if not os.path.exists(f"{PATH}/train/bird"):
raise Exception("expecting class subdirs under 'train/' and 'test/'")
!ls {PATH}/train
classes = ('plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
stats = (np.array([ 0.4914 , 0.48216, 0.44653]), np.array([ 0.24703, 0.24349, 0.26159]))
def get_data(sz,bs):
tfms = tfms_from_stats(stats, sz, aug_tfms=[RandomFlip()], pad=sz//8)
return ImageClassifierData.from_paths(PATH, val_name='test', tfms=tfms, bs=bs)
bs=256
```
### Look at data
```
data = get_data(32,4)
x,y=next(iter(data.trn_dl))
plt.imshow(data.trn_ds.denorm(x)[0]);
plt.imshow(data.trn_ds.denorm(x)[1]);
```
## Fully connected model
```
data = get_data(32,bs)
lr=1e-2
```
From [this notebook](https://github.com/KeremTurgutlu/deeplearning/blob/master/Exploring%20Optimizers.ipynb) by our student Kerem Turgutlu:
```
class SimpleNet(nn.Module):
def __init__(self, layers):
super().__init__()
self.layers = nn.ModuleList([
nn.Linear(layers[i], layers[i + 1]) for i in range(len(layers) - 1)])
def forward(self, x):
x = x.view(x.size(0), -1)
for l in self.layers:
l_x = l(x)
x = F.relu(l_x)
return F.log_softmax(l_x, dim=-1)
learn = ConvLearner.from_model_data(SimpleNet([32*32*3, 40,10]), data)
learn, [o.numel() for o in learn.model.parameters()]
learn.summary()
learn.lr_find()
learn.sched.plot()
%time learn.fit(lr, 2)
%time learn.fit(lr, 2, cycle_len=1)
```
## CNN
```
class ConvNet(nn.Module):
def __init__(self, layers, c):
super().__init__()
self.layers = nn.ModuleList([
nn.Conv2d(layers[i], layers[i + 1], kernel_size=3, stride=2)
for i in range(len(layers) - 1)])
self.pool = nn.AdaptiveMaxPool2d(1)
self.out = nn.Linear(layers[-1], c)
def forward(self, x):
for l in self.layers: x = F.relu(l(x))
x = self.pool(x)
x = x.view(x.size(0), -1)
return F.log_softmax(self.out(x), dim=-1)
learn = ConvLearner.from_model_data(ConvNet([3, 20, 40, 80], 10), data)
learn.summary()
learn.lr_find(end_lr=100)
learn.sched.plot()
%time learn.fit(1e-1, 2)
%time learn.fit(1e-1, 4, cycle_len=1)
```
## Refactored
```
class ConvLayer(nn.Module):
def __init__(self, ni, nf):
super().__init__()
self.conv = nn.Conv2d(ni, nf, kernel_size=3, stride=2, padding=1)
def forward(self, x): return F.relu(self.conv(x))
class ConvNet2(nn.Module):
def __init__(self, layers, c):
super().__init__()
self.layers = nn.ModuleList([ConvLayer(layers[i], layers[i + 1])
for i in range(len(layers) - 1)])
self.out = nn.Linear(layers[-1], c)
def forward(self, x):
for l in self.layers: x = l(x)
x = F.adaptive_max_pool2d(x, 1)
x = x.view(x.size(0), -1)
return F.log_softmax(self.out(x), dim=-1)
learn = ConvLearner.from_model_data(ConvNet2([3, 20, 40, 80], 10), data)
learn.summary()
%time learn.fit(1e-1, 2)
%time learn.fit(1e-1, 2, cycle_len=1)
```
## BatchNorm
```
class BnLayer(nn.Module):
def __init__(self, ni, nf, stride=2, kernel_size=3):
super().__init__()
self.conv = nn.Conv2d(ni, nf, kernel_size=kernel_size, stride=stride,
bias=False, padding=1)
self.a = nn.Parameter(torch.zeros(nf,1,1))
self.m = nn.Parameter(torch.ones(nf,1,1))
def forward(self, x):
x = F.relu(self.conv(x))
x_chan = x.transpose(0,1).contiguous().view(x.size(1), -1)
if self.training:
self.means = x_chan.mean(1)[:,None,None]
self.stds = x_chan.std (1)[:,None,None]
return (x-self.means) / self.stds *self.m + self.a
class ConvBnNet(nn.Module):
def __init__(self, layers, c):
super().__init__()
self.conv1 = nn.Conv2d(3, 10, kernel_size=5, stride=1, padding=2)
self.layers = nn.ModuleList([BnLayer(layers[i], layers[i + 1])
for i in range(len(layers) - 1)])
self.out = nn.Linear(layers[-1], c)
def forward(self, x):
x = self.conv1(x)
for l in self.layers: x = l(x)
x = F.adaptive_max_pool2d(x, 1)
x = x.view(x.size(0), -1)
return F.log_softmax(self.out(x), dim=-1)
learn = ConvLearner.from_model_data(ConvBnNet([10, 20, 40, 80, 160], 10), data)
learn.summary()
%time learn.fit(3e-2, 2)
%time learn.fit(1e-1, 4, cycle_len=1)
```
## Deep BatchNorm
```
class ConvBnNet2(nn.Module):
def __init__(self, layers, c):
super().__init__()
self.conv1 = nn.Conv2d(3, 10, kernel_size=5, stride=1, padding=2)
self.layers = nn.ModuleList([BnLayer(layers[i], layers[i+1])
for i in range(len(layers) - 1)])
self.layers2 = nn.ModuleList([BnLayer(layers[i+1], layers[i + 1], 1)
for i in range(len(layers) - 1)])
self.out = nn.Linear(layers[-1], c)
def forward(self, x):
x = self.conv1(x)
for l,l2 in zip(self.layers, self.layers2):
x = l(x)
x = l2(x)
x = F.adaptive_max_pool2d(x, 1)
x = x.view(x.size(0), -1)
return F.log_softmax(self.out(x), dim=-1)
learn = ConvLearner.from_model_data(ConvBnNet2([10, 20, 40, 80, 160], 10), data)
%time learn.fit(1e-2, 2)
%time learn.fit(1e-2, 2, cycle_len=1)
```
## Resnet
```
class ResnetLayer(BnLayer):
def forward(self, x): return x + super().forward(x)
class Resnet(nn.Module):
def __init__(self, layers, c):
super().__init__()
self.conv1 = nn.Conv2d(3, 10, kernel_size=5, stride=1, padding=2)
self.layers = nn.ModuleList([BnLayer(layers[i], layers[i+1])
for i in range(len(layers) - 1)])
self.layers2 = nn.ModuleList([ResnetLayer(layers[i+1], layers[i + 1], 1)
for i in range(len(layers) - 1)])
self.layers3 = nn.ModuleList([ResnetLayer(layers[i+1], layers[i + 1], 1)
for i in range(len(layers) - 1)])
self.out = nn.Linear(layers[-1], c)
def forward(self, x):
x = self.conv1(x)
for l,l2,l3 in zip(self.layers, self.layers2, self.layers3):
x = l3(l2(l(x)))
x = F.adaptive_max_pool2d(x, 1)
x = x.view(x.size(0), -1)
return F.log_softmax(self.out(x), dim=-1)
learn = ConvLearner.from_model_data(Resnet([10, 20, 40, 80, 160], 10), data)
wd=1e-5
%time learn.fit(1e-2, 2, wds=wd)
%time learn.fit(1e-2, 3, cycle_len=1, cycle_mult=2, wds=wd)
%time learn.fit(1e-2, 8, cycle_len=4, wds=wd)
```
## Resnet 2
```
class Resnet2(nn.Module):
def __init__(self, layers, c, p=0.5):
super().__init__()
self.conv1 = BnLayer(3, 16, stride=1, kernel_size=7)
self.layers = nn.ModuleList([BnLayer(layers[i], layers[i+1])
for i in range(len(layers) - 1)])
self.layers2 = nn.ModuleList([ResnetLayer(layers[i+1], layers[i + 1], 1)
for i in range(len(layers) - 1)])
self.layers3 = nn.ModuleList([ResnetLayer(layers[i+1], layers[i + 1], 1)
for i in range(len(layers) - 1)])
self.out = nn.Linear(layers[-1], c)
self.drop = nn.Dropout(p)
def forward(self, x):
x = self.conv1(x)
for l,l2,l3 in zip(self.layers, self.layers2, self.layers3):
x = l3(l2(l(x)))
x = F.adaptive_max_pool2d(x, 1)
x = x.view(x.size(0), -1)
x = self.drop(x)
return F.log_softmax(self.out(x), dim=-1)
learn = ConvLearner.from_model_data(Resnet2([16, 32, 64, 128, 256], 10, 0.2), data)
wd=1e-6
%time learn.fit(1e-2, 2, wds=wd)
%time learn.fit(1e-2, 3, cycle_len=1, cycle_mult=2, wds=wd)
%time learn.fit(1e-2, 8, cycle_len=4, wds=wd)
learn.save('tmp3')
log_preds,y = learn.TTA()
preds = np.mean(np.exp(log_preds),0)
metrics.log_loss(y,preds), accuracy_np(preds,y)
```
### End
|
github_jupyter
|
# 1. Very simple 'programs'
## 1.1 Running Python from the command line
In order to test pieces of code we can run Python from the command line. In this Jupyter Notebook we are going to simulate this. You can type the commands in the fields and execute them.<br>
In the field type:<br>
`print('Hello, World')`<br>
Then press `<shift> + <return>` to execute the command.
What happened?<br>You just created a program, that prints the words 'Hello, World'. The Python environment that you are in immediately compiles whatever you have typed in. This is useful for testing things, e.g. define a few variables, and then test to see if a certain line will work. That will come in a later lesson, though.
## 1.2 Math in Python
Type<br>
`1 + 1`
Type<br>
`20 + 80`
These are additions. We can of course use other mathematical operators.<br>
Try this subtraction:<br>
`6 - 5`
and this multiplication:<br>
`2 * 5`
Try:<br>
`5 ** 2`
`**` is the exponential operator, so we executed 5 squared.
Type:<br>
`print('1 + 2 is an addition')`
You see that the `print` statement writes something on the screen.<br>
Try this:<br>
`print('one kilobyte is 2^10 bytes, or', 2 ** 10, 'bytes')`
This demonstrates that you can print text and calculations in a sentence.<br>
The commas separating each section are a way of separating strings (text) from calculations or variable.
Now try this:<br>
`23 / 3`
And this:<br>
`23%3`
`%` returns the remainder of the division.
## 1.3 Order of Operations
Remember that thing called order of operation that they taught in maths? Well, it applies in Python, too. Here it is, if you need reminding:<br>
1. Parenthesis `()`
2. Exponents `**`
3. Multiplication `*`, division `/` and remainder `%`
4. Addition `+` and subtraction `-`
Here are some examples that you might want to try, if you're rusty on this:<br>
`1 + 2 * 3`<br>
`(1 + 2) * 3`
## 1.4 Comments, Please
The final thing you'll need to know to move on to multi-line programs is the comment. Type the following (and yes, the output is shown):<br>
`#I am a comment. Fear my wrath!`
A comment is a piece of code that is not run. In Python, you make something a comment by putting a hash in front of it. A hash comments everything after it in the line, and nothing before it. So you could type this:<br>
`print("food is very nice") #eat me`
This results in a normal output, without the smutty comment, thank you very much.<br>
Now try this:<br>
`# print("food is very nice")`
Nothing happens, because the code was after a comment.
Comments are important for adding necessary information for another programmer to read, but not the computer. For example, an explanation of a section of code, saying what it does, or what is wrong with it. You can also comment bits of code by putting a `#` in front of it - if you don't want it to compile, but can't delete it because you might need it later.
__[Home](PythonIntro.ipynb)__<br>
__[Lesson 2: Programs in a file, and variables](PythonIntroCh2.ipynb)__
|
github_jupyter
|
## Install packages and connect to Oracle
```
sc.install_pypi_package("sqlalchemy")
sc.install_pypi_package("pandas")
sc.install_pypi_package("s3fs")
sc.install_pypi_package("cx_Oracle")
sc.install_pypi_package("fsspec")
from sqlalchemy import create_engine
engine = create_engine('oracle://CMSDASHADMIN:4#X9#Veut#KSsU#[email protected]:1521/', echo=False)
# Import necessary libraries
import cx_Oracle
import pandas as pd
import numpy as np
dsn_tns = cx_Oracle.makedsn('oracle-prod-cms-dash.ccwgq0kcp9fq.us-east-1.rds.amazonaws.com', '1521', service_name='ORCL')
conn = cx_Oracle.connect(user=r'VILASM', password='Z#5iC$Ld4sE', dsn=dsn_tns)
con = cx_Oracle.connect('VILASM/Z#5iC$Ld4sE@oracle-prod-cms-dash.ccwgq0kcp9fq.us-east-1.rds.amazonaws.com/ORCL')
print (con.version)
cur =con.cursor()
```
# Insert into DASH_BENEFICIARY table
```
import pandas as pd
# Create datatype dictionary for reading in the files
dtype_dic= {'BENE_BIRTH_DT':str, 'BENE_DEATH_DT':str}
# Read in all three files from 2008, 2009, and 2010
bene08 = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 20/DE1_0_2008_Beneficiary_Summary_File_Sample_20.csv", dtype = dtype_dic)
bene09 = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 20/DE1_0_2009_Beneficiary_Summary_File_Sample_20.csv", dtype = dtype_dic)
bene10 = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 20/DE1_0_2010_Beneficiary_Summary_File_Sample_20.csv", dtype = dtype_dic)
# Add the FILE_YEAR column and insert it to index 0
bene08['FILE_YEAR']='2008'
first_col = bene08.pop('FILE_YEAR')
bene08.insert(0,'FILE_YEAR',first_col)
bene09['FILE_YEAR']='2009'
first_col = bene09.pop('FILE_YEAR')
bene09.insert(0,'FILE_YEAR',first_col)
bene10['FILE_YEAR']='2010'
first_col = bene10.pop('FILE_YEAR')
bene10.insert(0,'FILE_YEAR',first_col)
# Add leading zeros to SP_STATE_CODE and BENE_COUNTY_CD
bene08['SP_STATE_CODE'] = bene08['SP_STATE_CODE'].astype(str).apply(lambda x: x.zfill(2))
bene08['BENE_COUNTY_CD'] = bene08['BENE_COUNTY_CD'].astype(str).apply(lambda x: x.zfill(3))
bene09['SP_STATE_CODE'] = bene09['SP_STATE_CODE'].astype(str).apply(lambda x: x.zfill(2))
bene09['BENE_COUNTY_CD'] = bene09['BENE_COUNTY_CD'].astype(str).apply(lambda x: x.zfill(3))
bene10['SP_STATE_CODE'] = bene10['SP_STATE_CODE'].astype(str).apply(lambda x: x.zfill(2))
bene10['BENE_COUNTY_CD'] = bene10['BENE_COUNTY_CD'].astype(str).apply(lambda x: x.zfill(3))
# Converty BENE_BIRTH_DT and BENE_DEATH_DT to datetimes
A = pd.to_datetime(bene08.BENE_BIRTH_DT)
bene08['BENE_BIRTH_DT'] = A.dt.date
B = pd.to_datetime(bene08.BENE_DEATH_DT)
bene08['BENE_DEATH_DT'] = B.dt.date
A = pd.to_datetime(bene09.BENE_BIRTH_DT)
bene09['BENE_BIRTH_DT'] = A.dt.date
B = pd.to_datetime(bene09.BENE_DEATH_DT)
bene09['BENE_DEATH_DT'] = B.dt.date
A = pd.to_datetime(bene10.BENE_BIRTH_DT)
bene10['BENE_BIRTH_DT'] = A.dt.date
B = pd.to_datetime(bene10.BENE_DEATH_DT)
bene10['BENE_DEATH_DT'] = B.dt.date
# Insert into table DASH_BENEFICIARY for 2008
sql= """ INSERT INTO DASH_BENEFICIARY (FILE_YEAR, DESYNPUF_ID,BENE_BIRTH_DT, BENE_DEATH_DT, BENE_SEX_IDENT_CD,BENE_RACE_CD,
BENE_ESRD_IND, SP_STATE_CODE, BENE_COUNTY_CD, BENE_HI_CVRAGE_TOT_MONS,BENE_SMI_CVRAGE_TOT_MONS, BENE_HMO_CVRAGE_TOT_MONS,
PLAN_CVRG_MOS_NUM, SP_ALZHDMTA, SP_CHF,SP_CHRNKIDN, SP_CNCR, SP_COPD, SP_DEPRESSN,SP_DIABETES, SP_ISCHMCHT, SP_OSTEOPRS,
SP_RA_OA, SP_STRKETIA, MEDREIMB_IP, BENRES_IP, PPPYMT_IP, MEDREIMB_OP, BENRES_OP, PPPYMT_OP, MEDREIMB_CAR, BENRES_CAR, PPPYMT_CAR)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33)"""
df_list = bene08.values.tolist()
n = 0
for i in bene08.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
# Insert into table DASH_BENEFICIARY for 2009
sql= """ INSERT INTO DASH_BENEFICIARY (FILE_YEAR, DESYNPUF_ID,BENE_BIRTH_DT, BENE_DEATH_DT, BENE_SEX_IDENT_CD,BENE_RACE_CD,
BENE_ESRD_IND, SP_STATE_CODE, BENE_COUNTY_CD, BENE_HI_CVRAGE_TOT_MONS,BENE_SMI_CVRAGE_TOT_MONS, BENE_HMO_CVRAGE_TOT_MONS,
PLAN_CVRG_MOS_NUM, SP_ALZHDMTA, SP_CHF,SP_CHRNKIDN, SP_CNCR, SP_COPD, SP_DEPRESSN,SP_DIABETES, SP_ISCHMCHT, SP_OSTEOPRS,
SP_RA_OA, SP_STRKETIA, MEDREIMB_IP, BENRES_IP, PPPYMT_IP, MEDREIMB_OP, BENRES_OP, PPPYMT_OP, MEDREIMB_CAR, BENRES_CAR, PPPYMT_CAR)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33)"""
df_list = bene09.values.tolist()
n = 0
for i in bene09.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
# Insert into table DASH_BENEFICIARY for 2010
sql= """ INSERT INTO DASH_BENEFICIARY (FILE_YEAR, DESYNPUF_ID,BENE_BIRTH_DT, BENE_DEATH_DT, BENE_SEX_IDENT_CD,BENE_RACE_CD,
BENE_ESRD_IND, SP_STATE_CODE, BENE_COUNTY_CD, BENE_HI_CVRAGE_TOT_MONS,BENE_SMI_CVRAGE_TOT_MONS, BENE_HMO_CVRAGE_TOT_MONS,
PLAN_CVRG_MOS_NUM, SP_ALZHDMTA, SP_CHF,SP_CHRNKIDN, SP_CNCR, SP_COPD, SP_DEPRESSN,SP_DIABETES, SP_ISCHMCHT, SP_OSTEOPRS,
SP_RA_OA, SP_STRKETIA, MEDREIMB_IP, BENRES_IP, PPPYMT_IP, MEDREIMB_OP, BENRES_OP, PPPYMT_OP, MEDREIMB_CAR, BENRES_CAR, PPPYMT_CAR)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33)"""
df_list = bene10.values.tolist()
n = 0
for i in bene10.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
# con.close()
```
## Insert into DASH_CLAIM_CARRIER table
### DASH_CLAIM_CARRIER A
```
claimsA = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 3/DE1_0_2008_to_2010_Carrier_Claims_Sample_3A.csv")
# claimsB = pd.read_csv("s3n://cms-dash-datasets/Data/DE1.0 Sample 20/DE1_0_2008_to_2010_Carrier_Claims_Sample_1B.csv")
# Take first 51 cols, move CLM_ID to front, convert two datetime columns
claims_A_toload = claimsA.iloc[: , :51]
first_col = claims_A_toload.pop('CLM_ID')
claims_A_toload.insert(0,'CLM_ID',first_col)
claims_A_toload.columns
A = pd.to_datetime(claims_A_toload['CLM_FROM_DT'])
claims_A_toload['CLM_FROM_DT'] = A.dt.date
B = pd.to_datetime(claims_A_toload.CLM_THRU_DT)
claims_A_toload['CLM_THRU_DT'] = B.dt.date
# Convert necessary columns to string
nonstr = claims_A_toload[['CLM_ID','CLM_FROM_DT','CLM_THRU_DT']]
claims_str = claims_A_toload.astype(str)
claims_str['CLM_ID']=nonstr['CLM_ID']
claims_str['CLM_FROM_DT']=nonstr['CLM_FROM_DT']
claims_str['CLM_THRU_DT']=nonstr['CLM_THRU_DT']
# Insert into DASH_CLAIM_CARRIER table
sql=""" INSERT INTO DASH_CLAIM_CARRIER (
CLM_ID, DESYNPUF_ID, CLM_FROM_DT, CLM_THRU_DT, ICD9_DGNS_CD_1, ICD9_DGNS_CD_2, ICD9_DGNS_CD_3, ICD9_DGNS_CD_4, ICD9_DGNS_CD_5, ICD9_DGNS_CD_6,
ICD9_DGNS_CD_7, ICD9_DGNS_CD_8, PRF_PHYSN_NPI_1, PRF_PHYSN_NPI_2, PRF_PHYSN_NPI_3, PRF_PHYSN_NPI_4, PRF_PHYSN_NPI_5, PRF_PHYSN_NPI_6,
PRF_PHYSN_NPI_7, PRF_PHYSN_NPI_8, PRF_PHYSN_NPI_9, PRF_PHYSN_NPI_10, PRF_PHYSN_NPI_11, PRF_PHYSN_NPI_12, PRF_PHYSN_NPI_13, TAX_NUM_1,
TAX_NUM_2, TAX_NUM_3, TAX_NUM_4, TAX_NUM_5, TAX_NUM_6, TAX_NUM_7, TAX_NUM_8, TAX_NUM_9, TAX_NUM_10, TAX_NUM_11, TAX_NUM_12, TAX_NUM_13,
HCPCS_CD_1, HCPCS_CD_2, HCPCS_CD_3, HCPCS_CD_4, HCPCS_CD_5, HCPCS_CD_6,HCPCS_CD_7,HCPCS_CD_8,HCPCS_CD_9,HCPCS_CD_10,HCPCS_CD_11,HCPCS_CD_12,
HCPCS_CD_13
)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33
,:34,:35,:36,:37,:38,:39,:40,:41,:42,:43,:44,:45,:46,:47,:48,:49,:50,:51
)"""
df_list = claims_str.values.tolist()
n = 0
for i in claims_str.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
```
## DASH CLAIMS CARRIER B
```
claimsB = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 2/DE1_0_2008_to_2010_Carrier_Claims_Sample_2B.csv")
# Take first 51 cols, move CLM ID to front, convert two datetime columns
claims_B_toload = claimsB.iloc[: , :51]
first_col = claims_B_toload.pop('CLM_ID')
claims_B_toload.insert(0,'CLM_ID',first_col)
claims_B_toload.columns
A = pd.to_datetime(claims_B_toload['CLM_FROM_DT'])
claims_B_toload['CLM_FROM_DT'] = A.dt.date
B = pd.to_datetime(claims_B_toload.CLM_THRU_DT)
claims_B_toload['CLM_THRU_DT'] = B.dt.date
nonstr = claims_B_toload[['CLM_ID','CLM_FROM_DT','CLM_THRU_DT']]
claims_str = claims_B_toload.astype(str)
claims_str['CLM_ID']=nonstr['CLM_ID']
claims_str['CLM_FROM_DT']=nonstr['CLM_FROM_DT']
claims_str['CLM_THRU_DT']=nonstr['CLM_THRU_DT']
sql=""" INSERT INTO DASH_CLAIM_CARRIER (
CLM_ID, DESYNPUF_ID, CLM_FROM_DT, CLM_THRU_DT, ICD9_DGNS_CD_1, ICD9_DGNS_CD_2, ICD9_DGNS_CD_3, ICD9_DGNS_CD_4, ICD9_DGNS_CD_5, ICD9_DGNS_CD_6,
ICD9_DGNS_CD_7, ICD9_DGNS_CD_8, PRF_PHYSN_NPI_1, PRF_PHYSN_NPI_2, PRF_PHYSN_NPI_3, PRF_PHYSN_NPI_4, PRF_PHYSN_NPI_5, PRF_PHYSN_NPI_6,
PRF_PHYSN_NPI_7, PRF_PHYSN_NPI_8, PRF_PHYSN_NPI_9, PRF_PHYSN_NPI_10, PRF_PHYSN_NPI_11, PRF_PHYSN_NPI_12, PRF_PHYSN_NPI_13, TAX_NUM_1,
TAX_NUM_2, TAX_NUM_3, TAX_NUM_4, TAX_NUM_5, TAX_NUM_6, TAX_NUM_7, TAX_NUM_8, TAX_NUM_9, TAX_NUM_10, TAX_NUM_11, TAX_NUM_12, TAX_NUM_13,
HCPCS_CD_1, HCPCS_CD_2, HCPCS_CD_3, HCPCS_CD_4, HCPCS_CD_5, HCPCS_CD_6,HCPCS_CD_7,HCPCS_CD_8,HCPCS_CD_9,HCPCS_CD_10,HCPCS_CD_11,HCPCS_CD_12,
HCPCS_CD_13
)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33
,:34,:35,:36,:37,:38,:39,:40,:41,:42,:43,:44,:45,:46,:47,:48,:49,:50,:51
)"""
df_list = claims_str.values.tolist()
n = 0
for i in claims_str.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
```
## Insert into DASH_CLAIM_INPATIENT
```
dtype_dic= {'AT_PHYSN_NPI':str, 'OP_PHYSN_NPI':str,'OT_PHYSN_NPI': str, 'CLM_FROM_DT':str, 'CLM_THRU_DT':str, 'CLM_ADMSN_DT':str, 'NCH_BENE_DSCHRG_DT':str}
inpatient = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 1/DE1_0_2008_to_2010_Inpatient_Claims_Sample_1.csv" , dtype = dtype_dic)
# Take only Segment 1
seg1 = inpatient.loc[inpatient['SEGMENT'] == 1]
# Take first 36 columns and rearrange SEGMENT and CLM_ID
inpatient_toload = seg1.iloc[: , :36]
first_col = inpatient_toload.pop('SEGMENT')
inpatient_toload.insert(0,'SEGMENT',first_col)
sec = inpatient_toload.pop('CLM_ID')
inpatient_toload.insert(0,'CLM_ID',sec)
# Convert necessary columns
A = pd.to_datetime(inpatient_toload.CLM_FROM_DT)
inpatient_toload['CLM_FROM_DT'] = A.dt.date
B = pd.to_datetime(inpatient_toload.CLM_THRU_DT)
inpatient_toload['CLM_THRU_DT'] = B.dt.date
C = pd.to_datetime(inpatient_toload.CLM_ADMSN_DT)
inpatient_toload['CLM_ADMSN_DT'] = C.dt.date
D = pd.to_datetime(inpatient_toload.NCH_BENE_DSCHRG_DT)
inpatient_toload['NCH_BENE_DSCHRG_DT'] = D.dt.date
# Fill NaN's with zeros
inpatient_toload[['NCH_BENE_IP_DDCTBL_AMT']]=inpatient_toload[['NCH_BENE_IP_DDCTBL_AMT']].fillna(0.0)
inpatient_toload['CLM_UTLZTN_DAY_CNT'] = (inpatient_toload['CLM_UTLZTN_DAY_CNT'].fillna(0)).astype(int)
# inpatient_toload[['AT_PHYSN_NPI', 'OP_PHYSN_NPI', 'OT_PHYSN_NPI']] = inpatient_toload[['AT_PHYSN_NPI', 'OP_PHYSN_NPI', 'OT_PHYSN_NPI']].fillna(0)astype(int)
all_columns = ['DESYNPUF_ID', 'PRVDR_NUM', 'AT_PHYSN_NPI','OP_PHYSN_NPI','OT_PHYSN_NPI','ADMTNG_ICD9_DGNS_CD', 'CLM_DRG_CD',
'ICD9_DGNS_CD_1','ICD9_DGNS_CD_2','ICD9_DGNS_CD_3','ICD9_DGNS_CD_4','ICD9_DGNS_CD_5','ICD9_DGNS_CD_6','ICD9_DGNS_CD_7','ICD9_DGNS_CD_8','ICD9_DGNS_CD_9','ICD9_DGNS_CD_10',
'ICD9_PRCDR_CD_1','ICD9_PRCDR_CD_2','ICD9_PRCDR_CD_3','ICD9_PRCDR_CD_4','ICD9_PRCDR_CD_5','ICD9_PRCDR_CD_6']
inpatient_toload[all_columns] = inpatient_toload[all_columns].astype(str)
inpatient_toload = inpatient_toload.reset_index().drop(columns=['index'])
# Insert into DASH_CLAIM_INPATIENT table
sql="""INSERT INTO DASH_CLAIM_INPATIENT (CLM_ID,SEGMENT,DESYNPUF_ID,CLM_FROM_DT,CLM_THRU_DT,PRVDR_NUM,CLM_PMT_AMT,NCH_PRMRY_PYR_CLM_PD_AMT,
AT_PHYSN_NPI,OP_PHYSN_NPI,OT_PHYSN_NPI,CLM_ADMSN_DT,ADMTNG_ICD9_DGNS_CD,CLM_PASS_THRU_PER_DIEM_AMT,NCH_BENE_IP_DDCTBL_AMT,NCH_BENE_PTA_COINSRNC_LBLTY_AM,
NCH_BENE_BLOOD_DDCTBL_LBLTY_AM,CLM_UTLZTN_DAY_CNT,NCH_BENE_DSCHRG_DT,CLM_DRG_CD,ICD9_DGNS_CD_1,ICD9_DGNS_CD_2,ICD9_DGNS_CD_3,ICD9_DGNS_CD_4,
ICD9_DGNS_CD_5,ICD9_DGNS_CD_6,ICD9_DGNS_CD_7,ICD9_DGNS_CD_8,ICD9_DGNS_CD_9,ICD9_DGNS_CD_10,ICD9_PRCDR_CD_1,ICD9_PRCDR_CD_2,ICD9_PRCDR_CD_3,
ICD9_PRCDR_CD_4,ICD9_PRCDR_CD_5,ICD9_PRCDR_CD_6)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33
,:34,:35,:36)"""
df_list = inpatient_toload.values.tolist()
n = 0
for i in inpatient_toload.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
```
## DASH_CLAIM_OUTPATIENT
```
outpatient = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 2/DE1_0_2008_to_2010_Outpatient_Claims_Sample_2.csv")
# Take first 36 columns and rearrange #SEGMENT and CLM_ID
outpatient_toload = outpatient.iloc[: , :31]
first_col = outpatient_toload.pop('SEGMENT')
outpatient_toload.insert(0,'SEGMENT',first_col)
sec = outpatient_toload.pop('CLM_ID')
outpatient_toload.insert(0,'CLM_ID',sec)
# Convert necessary columns
A = pd.to_datetime(outpatient_toload['CLM_FROM_DT'])
outpatient_toload['CLM_FROM_DT'] = A.dt.date
B = pd.to_datetime(outpatient_toload.CLM_THRU_DT)
outpatient_toload['CLM_THRU_DT'] = B.dt.date
all_columns = ['PRVDR_NUM', 'AT_PHYSN_NPI','OP_PHYSN_NPI','OT_PHYSN_NPI','ICD9_DGNS_CD_1','ICD9_DGNS_CD_2','ICD9_DGNS_CD_3','ICD9_DGNS_CD_4','ICD9_DGNS_CD_5','ICD9_DGNS_CD_6',
'ICD9_DGNS_CD_7','ICD9_DGNS_CD_8','ICD9_DGNS_CD_9','ICD9_DGNS_CD_10','ICD9_PRCDR_CD_1','ICD9_PRCDR_CD_2','ICD9_PRCDR_CD_3','ICD9_PRCDR_CD_4','ICD9_PRCDR_CD_5','ICD9_PRCDR_CD_6','ADMTNG_ICD9_DGNS_CD']
outpatient_toload[all_columns] = outpatient_toload[all_columns].astype(str)
# Insert into DASH_CLAIM_OUTPATIENT table
sql="""INSERT INTO DASH_CLAIM_OUTPATIENT (CLM_ID,SEGMENT,DESYNPUF_ID,CLM_FROM_DT,CLM_THRU_DT,PRVDR_NUM,CLM_PMT_AMT,NCH_PRMRY_PYR_CLM_PD_AMT,
AT_PHYSN_NPI,OP_PHYSN_NPI,OT_PHYSN_NPI,NCH_BENE_BLOOD_DDCTBL_LBLTY_AM,ICD9_DGNS_CD_1,ICD9_DGNS_CD_2,ICD9_DGNS_CD_3,ICD9_DGNS_CD_4,
ICD9_DGNS_CD_5,ICD9_DGNS_CD_6,ICD9_DGNS_CD_7,ICD9_DGNS_CD_8,ICD9_DGNS_CD_9,ICD9_DGNS_CD_10,ICD9_PRCDR_CD_1,ICD9_PRCDR_CD_2,ICD9_PRCDR_CD_3,
ICD9_PRCDR_CD_4,ICD9_PRCDR_CD_5,ICD9_PRCDR_CD_6,NCH_BENE_PTB_DDCTBL_AMT,NCH_BENE_PTB_COINSRNC_AMT,ADMTNG_ICD9_DGNS_CD)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31)"""
df_list = outpatient_toload.values.tolist()
n = 0
for i in outpatient_toload.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
```
# INSERT INTO CENSUS
```
census = pd.read_csv("s3://cms-dash-datasets/Data/Census/census_acs20195yr_county (1).csv", encoding='unicode_escape')
census['FIPS'] = census['FIPS'].astype(str).apply(lambda x: x.zfill(5))
census['FIPS']
# Insert into DASH_CENSUS table
sql="""INSERT INTO DASH_CENSUS (
FIPS,COUNTY_NAME,EST_HBT_TH_1,EST_HBT_TH_2,EST_ANC_TP_1,EST_ANC_TP_2,EST_ANC_TP_3,EST_ANC_TP_4,EST_ANC_TP_5,EST_ANC_TP_6,EST_ANC_TP_7,EST_ANC_TP_8,EST_ANC_TP_9,EST_ANC_TP_10,EST_ANC_TP_11,EST_ANC_TP_12,EST_ANC_TP_13,EST_ANC_TP_14,EST_ANC_TP_15,EST_ANC_TP_16,EST_ANC_TP_17,EST_ANC_TP_18,EST_ANC_TP_19,EST_ANC_TP_20,EST_ANC_TP_21,EST_ANC_TP_22,EST_ANC_TP_23,EST_ANC_TP_24,EST_ANC_TP_25,EST_ANC_TP_26,EST_ANC_TP_27,EST_BR_THU_1,EST_BR_THU_2,EST_BR_THU_3,EST_BR_THU_4,
EST_BR_THU_5,EST_BR_THU_6,EST_BR_THU_7,EST_CTZN_VP_1,EST_CTZN_VP_2,EST_CTZN_VP_3,EST_COW_1,EST_COW_2,EST_COW_3,EST_COW_4,EST_COW_5,EST_CTW_1,EST_CTW_2,EST_CTW_3,EST_CTW_4,EST_CTW_5,EST_CTW_6,EST_CTW_7,EST_CTW_8,EST_CNI_TH_1,EST_CNI_TH_2,EST_CNI_TH_3,EST_DIS_1,EST_DIS_2,EST_DIS_3,EST_DIS_4,EST_DIS_5,EST_DIS_6,EST_DIS_7,EST_DIS_8,EST_EA_1,EST_EA_2,EST_EA_3,EST_EA_4,EST_EA_5,EST_EA_6,EST_EA_7,EST_EA_8,EST_EA_9,EST_EA_10,EST_EMP_1,EST_EMP_2,EST_EMP_3,EST_EMP_4,EST_EMP_5,EST_EMP_6,
EST_EMP_7,EST_EMP_8,EST_EMP_9,EST_EMP_10,EST_EMP_11,EST_EMP_12,EST_EMP_13,EST_EMP_14,EST_EMP_15,EST_EMP_16,EST_EMP_17,EST_FERT_1,EST_FERT_2,EST_FERT_3,EST_FERT_4,EST_FERT_5,EST_FERT_6,EST_FERT_7,EST_GP_1,EST_GP_2,EST_GP_3,EST_GP_4,EST_GP_5,EST_GP_6,EST_GP_7,EST_GP_8,EST_GP_9,EST_GRAPI_1,EST_GRAPI_2,EST_GRAPI_3,EST_GRAPI_4,EST_GRAPI_5,EST_GRAPI_6,EST_GRAPI_7,EST_GRAPI_8,EST_GR_1,EST_GR_2,EST_GR_3,EST_GR_4,EST_GR_5,EST_GR_6,EST_GR_7,EST_GR_8,EST_GR_9,EST_GR_10,EST_HIC_1,EST_HIC_2,
EST_HIC_3,EST_HIC_4,EST_HIC_5,EST_HIC_6,EST_HIC_7,EST_HIC_8,EST_HIC_9,EST_HIC_10,EST_HIC_11,EST_HIC_12,EST_HIC_13,EST_HIC_14,EST_HIC_15,EST_HIC_16,EST_HIC_17,EST_HIC_18,EST_HIC_19,EST_HIC_20,EST_HIC_21,EST_HIC_22,EST_HIC_23,EST_HIC_24,EST_HISP_1,EST_HISP_2,EST_HISP_3,EST_HISP_4,EST_HISP_5,EST_HISP_6,EST_HISP_7,EST_HISP_8,EST_HISP_9,EST_HISP_10,EST_HISP_11,EST_HISP_12,EST_HISP_13,EST_HISP_14,EST_HISP_15,EST_HISP_16,EST_HEAT_1,EST_HEAT_2,EST_HEAT_3,EST_HEAT_4,EST_HEAT_5,EST_HEAT_6,EST_HEAT_7,
EST_HEAT_8,EST_HEAT_9,EST_HEAT_10,EST_HHT_1,EST_HHT_2,EST_HHT_3,EST_HHT_4,EST_HHT_5,EST_HHT_6,EST_HHT_7,EST_HHT_8,EST_HHT_9,EST_HHT_10,EST_HHT_11,EST_HHT_12,EST_HHT_13,EST_HHT_14,EST_HHT_15,EST_HOCC_1,EST_HOCC_2,EST_HOCC_3,EST_HOCC_4,EST_HOCC_5,EST_HT_1,EST_HT_2,EST_HT_3,EST_HT_4,EST_HT_5,EST_INB_1,EST_INB_2,EST_INB_3,EST_INB_4,EST_INB_5,EST_INB_6,EST_INB_7,EST_INB_8,EST_INB_9,EST_INB_10,EST_INB_11,EST_INB_12,EST_INB_13,EST_INB_14,EST_INB_15,EST_INB_16,EST_INB_17,EST_INB_18,EST_INB_19,EST_INB_20,EST_INB_21,
EST_INB_22,EST_INB_23,EST_INB_24,EST_INB_25,EST_INB_26,EST_INB_27,EST_INB_28,EST_INB_29,EST_INB_30,EST_INB_31,EST_INB_32,EST_INB_33,EST_INB_34,EST_INB_35,EST_INB_36,EST_INB_37,EST_INB_38,EST_INB_39,EST_INB_40,EST_INB_41,EST_INB_42,EST_INB_43,EST_INB_44,EST_IND_1,EST_IND_2,EST_IND_3,EST_IND_4,EST_IND_5,EST_IND_6,EST_IND_7,EST_IND_8,EST_IND_9,EST_IND_10,EST_IND_11,EST_IND_12,EST_IND_13,EST_IND_14,EST_LANG_1,EST_LANG_2,EST_LANG_3,EST_LANG_4,EST_LANG_5,EST_LANG_6,EST_LANG_7,EST_LANG_8,EST_LANG_9,EST_LANG_10,EST_LANG_11,EST_LANG_12,
EST_MRTL_1,EST_MRTL_2,EST_MRTL_3,EST_MRTL_4,EST_MRTL_5,EST_MRTL_6,EST_MRTL_7,EST_MRTL_8,EST_MRTL_9,EST_MRTL_10,EST_MRTL_11,EST_MRTL_12,EST_MRTG_1,EST_MRTG_2,EST_MRTG_3,EST_OPR_1,EST_OPR_2,EST_OPR_3,EST_OPR_4,EST_OCC_1,EST_OCC_2,EST_OCC_3,EST_OCC_4,EST_OCC_5,EST_OCC_6,EST_BPL_1,EST_BPL_2,EST_BPL_3,EST_BPL_4,EST_BPL_5,EST_BPL_6,EST_BPL_7,EST_BPL_8,EST_BPL_9,EST_BPL_10,EST_BPL_11,EST_BPL_12,EST_BPL_13,EST_BPL_14,EST_BPL_15,EST_BPL_16,EST_BPL_17,EST_BPL_18,EST_BPL_19,EST_POB_1,EST_POB_2,EST_POB_3,EST_POB_4,EST_POB_5,EST_POB_6,EST_POB_7,EST_RACE_1,EST_RACE_2,
EST_RACE_3,EST_RACE_4,EST_RACE_5,EST_RACE_6,EST_RACE_7,EST_RACE_8,EST_RACE_9,EST_RACE_10,EST_RACE_11,EST_RACE_12,EST_RACE_13,EST_RACE_14,EST_RACE_15,EST_RACE_16,EST_RACE_17,EST_RACE_18,EST_RACE_19,EST_RACE_20,EST_RACE_21,EST_RACE_22,EST_RACE_23,EST_RACE_24,EST_RACE_25,EST_RACE_26,EST_RACE_27,EST_RACE_28,EST_RACE_29,EST_RACE_30,EST_RACE_31,EST_RACE_32,EST_RACE_33,EST_RACE_34,EST_RACE_35,EST_RACE_36,EST_RACE_37,EST_RLTNSHP_1,EST_RLTNSHP_2,EST_RLTNSHP_3,EST_RLTNSHP_4,EST_RLTNSHP_5,EST_RLTNSHP_6,EST_RLTNSHP_7,EST_RSDNC_1,EST_RSDNC_2,EST_RSDNC_3,EST_RSDNC_4,EST_RSDNC_5,
EST_RSDNC_6,EST_RSDNC_7,EST_RSDNC_8,EST_ROOM_1,EST_ROOM_2,EST_ROOM_3,EST_ROOM_4,EST_ROOM_5,EST_ROOM_6,EST_ROOM_7,EST_ROOM_8,EST_ROOM_9,EST_ROOM_10,EST_ROOM_11,EST_SCHOOL_1,EST_SCHOOL_2,EST_SCHOOL_3,EST_SCHOOL_4,EST_SCHOOL_5,EST_SCHOOL_6,EST_SEL_CHAR_1,EST_SEL_CHAR_2,EST_SEL_CHAR_3,EST_SEL_CHAR_4,EST_SMOC_1,EST_SMOC_2,EST_SMOC_3,EST_SMOC_4,EST_SMOC_5,EST_SMOC_6,EST_SMOC_7,EST_SMOC_8,EST_SMOC_9,EST_SMOC_10,EST_SMOC_11,EST_SMOC_12,EST_SMOC_13,EST_SMOC_14,EST_SMOC_15,EST_SMOC_16,EST_SMOC_17,EST_SMOCAPI_1,EST_SMOCAPI_2,EST_SMOCAPI_3,EST_SMOCAPI_4,EST_SMOCAPI_5,EST_SMOCAPI_6,
EST_SMOCAPI_7,EST_SMOCAPI_8,EST_SMOCAPI_9,EST_SMOCAPI_10,EST_SMOCAPI_11,EST_SMOCAPI_12,EST_SMOCAPI_13,EST_SMOCAPI_14,EST_SMOCAPI_15,EST_SMOCAPI_16,EST_SEX_AGE_1,EST_SEX_AGE_2,EST_SEX_AGE_3,EST_SEX_AGE_4,EST_SEX_AGE_5,EST_SEX_AGE_6,EST_SEX_AGE_7,EST_SEX_AGE_8,EST_SEX_AGE_9,EST_SEX_AGE_10,EST_SEX_AGE_11,EST_SEX_AGE_12,EST_SEX_AGE_13,EST_SEX_AGE_14,EST_SEX_AGE_15,EST_SEX_AGE_16,EST_SEX_AGE_17,EST_SEX_AGE_18,EST_SEX_AGE_19,EST_SEX_AGE_20,EST_SEX_AGE_21,EST_SEX_AGE_22,EST_SEX_AGE_23,EST_SEX_AGE_24,EST_SEX_AGE_25,EST_SEX_AGE_26,EST_SEX_AGE_27,EST_SEX_AGE_28,EST_SEX_AGE_29,EST_SEX_AGE_30,EST_SEX_AGE_31,
EST_SEX_AGE_32,EST_THU,EST_CTZNSHP_1,EST_CTZNSHP_2,EST_CTZNSHP_3,EST_UNIT_1,EST_UNIT_2,EST_UNIT_3,EST_UNIT_4,EST_UNIT_5,EST_UNIT_6,EST_UNIT_7,EST_UNIT_8,EST_UNIT_9,EST_UNIT_10,EST_OWNER_UNIT_1,EST_OWNER_UNIT_2,EST_OWNER_UNIT_3,EST_OWNER_UNIT_4,EST_OWNER_UNIT_5,EST_OWNER_UNIT_6,EST_OWNER_UNIT_7,EST_OWNER_UNIT_8,EST_OWNER_UNIT_9,EST_OWNER_UNIT_10,EST_VEH_1,EST_VEH_2,EST_VEH_3,EST_VEH_4,EST_VEH_5,EST_VET_1,EST_VET_2,EST_FOREIGN_1,EST_FOREIGN_2,EST_FOREIGN_3,EST_FOREIGN_4,EST_FOREIGN_5,EST_FOREIGN_6,EST_FOREIGN_7,EST_MOVE_YEAR_1,EST_MOVE_YEAR_2,EST_MOVE_YEAR_3,EST_MOVE_YEAR_4,
EST_MOVE_YEAR_5,EST_MOVE_YEAR_6,EST_MOVE_YEAR_7,EST_US_ENTRY_1,EST_US_ENTRY_2,EST_US_ENTRY_3,EST_US_ENTRY_4,EST_US_ENTRY_5,EST_US_ENTRY_6,EST_US_ENTRY_7,EST_BUILT_YEAR_1,EST_BUILT_YEAR_2,EST_BUILT_YEAR_3,EST_BUILT_YEAR_4,EST_BUILT_YEAR_5,EST_BUILT_YEAR_6,EST_BUILT_YEAR_7,EST_BUILT_YEAR_8,EST_BUILT_YEAR_9,EST_BUILT_YEAR_10,EST_BUILT_YEAR_11
)
values(:1,:2,:3,:4,:5,:6,:7,:8,:9,:10,:11,:12,:13,:14,:15,:16,:17,:18,:19,:20,:21,:22,:23,:24,:25,:26,:27,:28,:29,:30,:31,:32,:33,:34,:35,:36,:37,:38,:39,:40,:41,:42,:43,:44,:45
,:46,:47,:48,:49,:50,:51,:52,:53,:54,:55,:56,:57,:58,:59,:60,:61,:62,:63,:64,:65,:66,:67,:68,:69,:70,:71,:72,:73,:74,:75,:76,:77,:78,:79,:80,:81,:82,:83,:84,:85,:86,:87,:88,:89,:90
,:91,:92,:93,:94,:95,:96,:97,:98,:99,:100,:101,:102,:103,:104,:105,:106,:107,:108,:109,:110,:111,:112,:113,:114,:115,:116,:117,:118,:119,:120,:121,:122,:123,:124,:125,:126,:127,:128,:129
,:130,:131,:132,:133,:134,:135,:136,:137,:138,:139,:140,:141,:142,:143,:144,:145,:146,:147,:148,:149,:150,:151,:152,:153,:154,:155,:156,:157,:158,:159,:160,:161,:162,:163,:164,:165,:166,:167,:168
,:169,:170,:171,:172,:173,:174,:175,:176,:177,:178,:179,:180,:181,:182,:183,:184,:185,:186,:187,:188,:189,:190,:191,:192,:193,:194,:195,:196,:197,:198,:199,:200,:201,:202,:203,:204,:205,:206,:207
,:208,:209,:210,:211,:212,:213,:214,:215,:216,:217,:218,:219,:220,:221,:222,:223,:224,:225,:226,:227,:228,:229,:230,:231,:232,:233,:234,:235,:236,:237,:238,:239,:240,:241,:242,:243,:244,:245,:246,:247,:248,:249
,:250,:251,:252,:253,:254,:255,:256,:257,:258,:259,:260,:261,:262,:263,:264,:265,:266,:267,:268,:269,:270,:271,:272,:273,:274,:275,:276,:277,:278,:279,:280,:281,:282,:283,:284,:285,:286,:287,:288,:289,:290
,:291,:292,:293,:294,:295,:296,:297,:298,:299,:300,:301,:302,:303,:304,:305,:306,:307,:308,:309,:310,:311,:312,:313,:314,:315,:316,:317,:318,:319,:320,:321,:322,:323,:324,:325,:326,:327,:328,:329,:330,:331,:332,:333
,:334,:335,:336,:337,:338,:339,:340,:341,:342,:343,:344,:345,:346,:347,:348,:349,:350,:351,:352,:353,:354,:355,:356,:357,:358,:359,:360,:361,:362,:363,:364,:365,:366,:367,:368,:369,:370,:371,:372,:373,:374,:375,:376,:377,:378,:379,:380
,:381,:382,:383,:384,:385,:386,:387,:388,:389,:390,:391,:392,:393,:394,:395,:396,:397,:398,:399,:400,:401,:402,:403,:404,:405,:406,:407,:408,:409,:410,:411,:412,:413,:414,:415,:416,:417,:418,:419,:420,:421,:422,:423,:424,:425,:426,:427,:428,:429,:430
,:431,:432,:433,:434,:435,:436,:437,:438,:439,:440,:441,:442,:443,:444,:445,:446,:447,:448,:449,:450,:451,:452,:453,:454,:455,:456,:457,:458,:459,:460,:461,:462,:463,:464,:465,:466,:467,:468
,:469,:470,:471,:472,:473,:474,:475,:476,:477,:478,:479,:480,:481,:482,:483,:484,:485,:486,:487,:488,:489,:490,:491,:492,:493,:494,:495,:496,:497,:498,:499,:500,:501,:502,:503,:504,:505,:506,:507,:508,:509,:510,:511,:512
,:513,:514,:515,:516,:517,:518,:519,:520,:521,:522, :523)"""
df_list = census.values.tolist()
n = 0
for i in census.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
```
# INSERT INTO PERCRIPTIONS DRUGS
```
drugs = pd.read_csv("s3://cms-dash-datasets/Data/DE1.0 Sample 2/DE1_0_2008_to_2010_Prescription_Drug_Events_Sample_2.csv")
# Rearrange columns and convert necessary columns
first_col = drugs.pop('PDE_ID')
drugs.insert(0,'PDE_ID',first_col)
all_columns = ['PDE_ID', 'DESYNPUF_ID','PROD_SRVC_ID',]
drugs[all_columns] = drugs[all_columns].astype(str)
# Convert date columns
A = pd.to_datetime(drugs['SRVC_DT'])
drugs['SRVC_DT'] = A.dt.date
# Insert into DASH_DRUG_PRESCRIPTION table
sql="""INSERT INTO DASH_DRUG_PRESCRIPTION (PDE_ID, DESYNPUF_ID, SRVC_DT,PROD_SRVC_ID,QTY_DSPNSD_NUM,DAYS_SUPLY_NUM,PTNT_PAY_AMT,TOT_RX_CST_AMT)
values(:1,:2,:3,:4,:5,:6,:7,:8)"""
df_list = drugs.values.tolist()
n = 0
for i in drugs.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
```
## Insert NLP data
```
# Import the two NLP files
nlp = pd.read_csv("s3://cms-dash-datasets/Jason-NLP/output.csv")
nlp_xlsx = pd.read_csv("s3://cms-dash-datasets/Jason-NLP/Review Data (Hospital Review Data for NLP processing)_Review Data.csv")
# Rename columns
nlp_xlsx = nlp_xlsx.rename(columns={'At Physn Npi':'AT_PHYSN_API'})
nlp2 = nlp_xlsx[['AT_PHYSN_API','Review Comment']]
# Join the two NLP files
nlp_load = pd.merge(left=nlp, right=nlp2, left_on ='AT_PHYSN_NPI', right_on = 'AT_PHYSN_API')
nlp_load2 = nlp_load[['AT_PHYSN_NPI','Review Comment','label']].rename(columns={'Review Comment':'REVIEW_TEXT','label':'RATING_VALUE'})
# Create index column
nlp_load2['REVIEW_SID']=nlp_load2.index
# Rearrange columns
nlp_load3 = nlp_load2[['REVIEW_SID','AT_PHYSN_NPI','REVIEW_TEXT','RATING_VALUE']]
# Insert into DASH_PROVIDER_REVIEW Table
sql="""INSERT INTO DASH_PROVIDER_REVIEW (REVIEW_SID, AT_PHYSN_NPI, REVIEW_TEXT, RATING_VALUE )
values(:1,:2,:3, :4)"""
df_list = nlp_load3.values.tolist()
n = 0
for i in nlp_load3.iterrows():
cur.execute(sql,df_list[n])
n += 1
con.commit()
con.close
```
|
github_jupyter
|
# Generative Spaces (ABM)
In this workshop we will lwarn how to construct a ABM (Agent Based Model) with spatial behaviours, that is capable of configuring the space. This file is a simplified version of Generative Spatial Agent Based Models. For further information, you can find more advanced versions here:
* [Object Oriented version](https://github.com/shervinazadi/spatial_computing_workshops/blob/master/notebooks/w3_generative_spaces.ipynb)
* [Vectorized version](https://topogenesis.readthedocs.io/notebooks/random_walker)
## 0. Initialization
### 0.1. Load required libraries
```
# !pip install pyvista==0.28.1 ipyvtklink
import os
import topogenesis as tg
import pyvista as pv
import trimesh as tm
import pandas as pd
import numpy as np
np.random.seed(0)
```
### 0.2. Define the Neighborhood (Stencil)
```
# creating neighborhood definition
stencil = tg.create_stencil("von_neumann", 1, 1)
# setting the center to zero
stencil.set_index([0,0,0], 0)
print(stencil)
```
### 0.3 Visualize the Stencil
```
# initiating the plotter
p = pv.Plotter(notebook=True)
# Create the spatial reference
grid = pv.UniformGrid()
# Set the grid dimensions: shape because we want to inject our values
grid.dimensions = np.array(stencil.shape) + 1
# The bottom left corner of the data set
grid.origin = [0,0,0]
# These are the cell sizes along each axis
grid.spacing = [1,1,1]
# Add the data values to the cell data
grid.cell_arrays["values"] = stencil.flatten(order="F") # Flatten the stencil
threshed = grid.threshold([0.9, 1.1])
# adding the voxels: light red
p.add_mesh(threshed, show_edges=True, color="#ff8fa3", opacity=0.3)
# plotting
# p.show(use_ipyvtk=True)
```
## 1. Setup the Environment
### 1.1. Load the envelope lattice as the avialbility lattice
```
# loading the lattice from csv
lattice_path = os.path.relpath('../data/voxelized_envelope.csv')
avail_lattice = tg.lattice_from_csv(lattice_path)
init_avail_lattice = tg.to_lattice(np.copy(avail_lattice), avail_lattice)
```
### 1.2 Load Program
```
program_complete = pd.read_csv("../data/program_small.csv")
program_complete
program_prefs = program_complete.drop(["space_name","space_id"], 1)
program_prefs
```
### 1.2 Load the value fields
```
# loading the lattice from csv
fields = {}
for f in program_prefs.columns:
lattice_path = os.path.relpath('../data/' + f + '.csv')
fields[f] = tg.lattice_from_csv(lattice_path)
```
### 1.3. Initialize the Agents
```
# initialize the occupation lattice
occ_lattice = avail_lattice * 0 - 1
# Finding the index of the available voxels in avail_lattice
avail_flat = avail_lattice.flatten()
avail_index = np.array(np.where(avail_lattice == 1)).T
# Randomly choosing three available voxels
agn_num = len(program_complete)
select_id = np.random.choice(len(avail_index), agn_num)
agn_origins = avail_index[select_id]
# adding the origins to the agents locations
agn_locs = []
# for each agent origin ...
for a_id, a_origin in enumerate(agn_origins):
# add the origin to the list of agent locations
agn_locs.append([a_origin])
# set the origin in availablity lattice as 0 (UNavailable)
avail_lattice[tuple(a_origin)] = 0
# set the origin in occupation lattice as the agent id (a_id)
occ_lattice[tuple(a_origin)] = a_id
```
### 1.4. Visualize the environment
```
p = pv.Plotter(notebook=True)
# Set the grid dimensions: shape + 1 because we want to inject our values on the CELL data
grid = pv.UniformGrid()
grid.dimensions = np.array(occ_lattice.shape) + 1
# The bottom left corner of the data set
grid.origin = occ_lattice.minbound - occ_lattice.unit * 0.5
# These are the cell sizes along each axis
grid.spacing = occ_lattice.unit
# adding the boundingbox wireframe
p.add_mesh(grid.outline(), color="grey", label="Domain")
# adding axes
p.add_axes()
p.show_bounds(grid="back", location="back", color="#777777")
# Add the data values to the cell data
grid.cell_arrays["Agents"] = occ_lattice.flatten(order="F").astype(int) # Flatten the array!
# filtering the voxels
threshed = grid.threshold([-0.1, agn_num - 0.9])
# adding the voxels
p.add_mesh(threshed, show_edges=True, opacity=1.0, show_scalar_bar=False)
# adding the availability lattice
init_avail_lattice.fast_vis(p)
# p.show(use_ipyvtk=True)
```
## 2. ABM Simulation (Agent Based Space Occupation)
### 2.1. Running the simulation
```
# make a deep copy of occupation lattice
cur_occ_lattice = tg.to_lattice(np.copy(occ_lattice), occ_lattice)
# initialzing the list of frames
frames = [cur_occ_lattice]
# setting the time variable to 0
t = 0
n_frames = 30
# main feedback loop of the simulation (for each time step ...)
while t<n_frames:
# for each agent ...
for a_id, a_prefs in program_complete.iterrows():
# retrieve the list of the locations of the current agent
a_locs = agn_locs[a_id]
# initialize the list of free neighbours
free_neighs = []
# for each location of the agent
for loc in a_locs:
# retrieve the list of neighbours of the agent based on the stencil
neighs = avail_lattice.find_neighbours_masked(stencil, loc = loc)
# for each neighbour ...
for n in neighs:
# compute 3D index of neighbour
neigh_3d_id = np.unravel_index(n, avail_lattice.shape)
# if the neighbour is available...
if avail_lattice[neigh_3d_id]:
# add the neighbour to the list of free neighbours
free_neighs.append(neigh_3d_id)
# check if found any free neighbour
if len(free_neighs)>0:
# convert free neighbours to a numpy array
fns = np.array(free_neighs)
# find the value of neighbours
# init the agent value array
a_eval = np.ones(len(fns))
# for each field...
for f in program_prefs.columns:
# find the raw value of free neighbours...
vals = fields[f][fns[:,0], fns[:,1], fns[:,2]]
# raise the the raw value to the power of preference weight of the agent
a_weighted_vals = vals ** a_prefs[f]
# multiply them to the previous weighted values
a_eval *= a_weighted_vals
# select the neighbour with highest evaluation
selected_int = np.argmax(a_eval)
# find 3D integer index of selected neighbour
selected_neigh_3d_id = free_neighs[selected_int]
# find the location of the newly selected neighbour
selected_neigh_loc = np.array(selected_neigh_3d_id).flatten()
# add the newly selected neighbour location to agent locations
agn_locs[a_id].append(selected_neigh_loc)
# set the newly selected neighbour as UNavailable (0) in the availability lattice
avail_lattice[selected_neigh_3d_id] = 0
# set the newly selected neighbour as OCCUPIED by current agent
# (-1 means not-occupied so a_id)
occ_lattice[selected_neigh_3d_id] = a_id
# constructing the new lattice
new_occ_lattice = tg.to_lattice(np.copy(occ_lattice), occ_lattice)
# adding the new lattice to the list of frames
frames.append(new_occ_lattice)
# adding one to the time counter
t += 1
```
### 2.2. Visualizing the simulation
```
p = pv.Plotter(notebook=True)
base_lattice = frames[0]
# Set the grid dimensions: shape + 1 because we want to inject our values on the CELL data
grid = pv.UniformGrid()
grid.dimensions = np.array(base_lattice.shape) + 1
# The bottom left corner of the data set
grid.origin = base_lattice.minbound - base_lattice.unit * 0.5
# These are the cell sizes along each axis
grid.spacing = base_lattice.unit
# adding the boundingbox wireframe
p.add_mesh(grid.outline(), color="grey", label="Domain")
# adding the availability lattice
init_avail_lattice.fast_vis(p)
# adding axes
p.add_axes()
p.show_bounds(grid="back", location="back", color="#aaaaaa")
def create_mesh(value):
f = int(value)
lattice = frames[f]
# Add the data values to the cell data
grid.cell_arrays["Agents"] = lattice.flatten(order="F").astype(int) # Flatten the array!
# filtering the voxels
threshed = grid.threshold([-0.1, agn_num - 0.9])
# adding the voxels
p.add_mesh(threshed, name='sphere', show_edges=True, opacity=1.0, show_scalar_bar=False)
return
p.add_slider_widget(create_mesh, [0, n_frames], title='Time', value=0, event_type="always", style="classic")
p.show(use_ipyvtk=True)
```
### 2.3. Saving lattice frames in CSV
```
for i, lattice in enumerate(frames):
csv_path = os.path.relpath('../data/abm_animation/abm_f_'+ f'{i:03}' + '.csv')
lattice.to_csv(csv_path)
```
### Credits
```
__author__ = "Shervin Azadi "
__license__ = "MIT"
__version__ = "1.0"
__url__ = "https://github.com/shervinazadi/spatial_computing_workshops"
__summary__ = "Spatial Computing Design Studio Workshop on Agent Based Models for Generative Spaces"
```
|
github_jupyter
|
```
# from google.colab import drive
# drive.mount('/content/drive')
import torch.nn as nn
import torch.nn.functional as F
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import torch
import torchvision
import torchvision.transforms as transforms
from torch.utils.data import Dataset, DataLoader
from torchvision import transforms, utils
from matplotlib import pyplot as plt
import copy
# Ignore warnings
import warnings
warnings.filterwarnings("ignore")
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data', train=True, download=True, transform=transform)
testset = torchvision.datasets.CIFAR10(root='./data', train=False, download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=10, shuffle=True)
testloader = torch.utils.data.DataLoader(testset, batch_size=10, shuffle=False)
classes = ('plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
foreground_classes = {'plane', 'car', 'bird'}
background_classes = {'cat', 'deer', 'dog', 'frog', 'horse','ship', 'truck'}
fg1,fg2,fg3 = 0,1,2
dataiter = iter(trainloader)
background_data=[]
background_label=[]
foreground_data=[]
foreground_label=[]
batch_size=10
for i in range(5000):
images, labels = dataiter.next()
for j in range(batch_size):
if(classes[labels[j]] in background_classes):
img = images[j].tolist()
background_data.append(img)
background_label.append(labels[j])
else:
img = images[j].tolist()
foreground_data.append(img)
foreground_label.append(labels[j])
foreground_data = torch.tensor(foreground_data)
foreground_label = torch.tensor(foreground_label)
background_data = torch.tensor(background_data)
background_label = torch.tensor(background_label)
def create_mosaic_img(bg_idx,fg_idx,fg):
"""
bg_idx : list of indexes of background_data[] to be used as background images in mosaic
fg_idx : index of image to be used as foreground image from foreground data
fg : at what position/index foreground image has to be stored out of 0-8
"""
image_list=[]
j=0
for i in range(9):
if i != fg:
image_list.append(background_data[bg_idx[j]].type("torch.DoubleTensor"))
j+=1
else:
image_list.append(foreground_data[fg_idx].type("torch.DoubleTensor"))
label = foreground_label[fg_idx]- fg1 # minus 7 because our fore ground classes are 7,8,9 but we have to store it as 0,1,2
#image_list = np.concatenate(image_list ,axis=0)
image_list = torch.stack(image_list)
return image_list,label
desired_num = 30000
mosaic_list_of_images =[] # list of mosaic images, each mosaic image is saved as list of 9 images
fore_idx =[] # list of indexes at which foreground image is present in a mosaic image i.e from 0 to 9
mosaic_label=[] # label of mosaic image = foreground class present in that mosaic
for i in range(desired_num):
bg_idx = np.random.randint(0,35000,8)
fg_idx = np.random.randint(0,15000)
fg = np.random.randint(0,9)
fore_idx.append(fg)
image_list,label = create_mosaic_img(bg_idx,fg_idx,fg)
mosaic_list_of_images.append(image_list)
mosaic_label.append(label)
class MosaicDataset(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list_of_images, mosaic_label, fore_idx):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list_of_images
self.label = mosaic_label
self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx], self.fore_idx[idx]
batch = 250
msd = MosaicDataset(mosaic_list_of_images, mosaic_label , fore_idx)
train_loader = DataLoader( msd,batch_size= batch ,shuffle=True)
class Focus(nn.Module):
def __init__(self):
super(Focus, self).__init__()
self.conv1 = nn.Conv2d(in_channels=3, out_channels=6, kernel_size=3, padding=0)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(in_channels=6, out_channels=6, kernel_size=3, padding=0)
# self.conv3 = nn.Conv2d(in_channels=12, out_channels=32, kernel_size=3, padding=0)
self.fc1 = nn.Linear(1014, 512)
self.fc2 = nn.Linear(512, 64)
# self.fc3 = nn.Linear(512, 64)
# self.fc4 = nn.Linear(64, 10)
self.fc3 = nn.Linear(64,1)
def forward(self,z): #y is avg image #z batch of list of 9 images
y = torch.zeros([batch,3, 32,32], dtype=torch.float64)
x = torch.zeros([batch,9],dtype=torch.float64)
y = y.to("cuda")
x = x.to("cuda")
for i in range(9):
x[:,i] = self.helper(z[:,i])[:,0]
x = F.softmax(x,dim=1)
x1 = x[:,0]
torch.mul(x1[:,None,None,None],z[:,0])
for i in range(9):
x1 = x[:,i]
y = y + torch.mul(x1[:,None,None,None],z[:,i])
return x, y
def helper(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = (F.relu(self.conv2(x)))
# print(x.shape)
# x = (F.relu(self.conv3(x)))
x = x.view(x.size(0), -1)
# print(x.shape)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
# x = F.relu(self.fc3(x))
# x = F.relu(self.fc4(x))
x = self.fc3(x)
return x
focus_net = Focus().double()
focus_net = focus_net.to("cuda")
class Classification(nn.Module):
def __init__(self):
super(Classification, self).__init__()
self.conv1 = nn.Conv2d(in_channels=3, out_channels=6, kernel_size=3, padding=0)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(in_channels=6, out_channels=18, kernel_size=3, padding=0)
# self.conv3 = nn.Conv2d(in_channels=12, out_channels=20, kernel_size=3, padding=0)
self.fc1 = nn.Linear(3042, 1024)
self.fc2 = nn.Linear(1024, 64)
# self.fc3 = nn.Linear(512, 64)
# self.fc4 = nn.Linear(64, 10)
self.fc3 = nn.Linear(64,3)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = (F.relu(self.conv2(x)))
# print(x.shape)
# x = (F.relu(self.conv3(x)))
x = x.view(x.size(0), -1)
# print(x.shape)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
# x = F.relu(self.fc3(x))
# x = F.relu(self.fc4(x))
x = self.fc3(x)
return x
classify = Classification().double()
classify = classify.to("cuda")
test_images =[] #list of mosaic images, each mosaic image is saved as laist of 9 images
fore_idx_test =[] #list of indexes at which foreground image is present in a mosaic image
test_label=[] # label of mosaic image = foreground class present in that mosaic
for i in range(10000):
bg_idx = np.random.randint(0,35000,8)
fg_idx = np.random.randint(0,15000)
fg = np.random.randint(0,9)
fore_idx_test.append(fg)
image_list,label = create_mosaic_img(bg_idx,fg_idx,fg)
test_images.append(image_list)
test_label.append(label)
test_data = MosaicDataset(test_images,test_label,fore_idx_test)
test_loader = DataLoader( test_data,batch_size= batch ,shuffle=False)
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer_classify = optim.Adam(classify.parameters(), lr=0.001)#, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=False)
optimizer_focus = optim.Adam(focus_net.parameters(), lr=0.001)#, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=False)
col1=[]
col2=[]
col3=[]
col4=[]
col5=[]
col6=[]
col7=[]
col8=[]
col9=[]
col10=[]
col11=[]
col12=[]
col13=[]
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in train_loader:
inputs, labels , fore_idx = data
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
count += 1
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 30000 train images: %d %%' % ( 100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
print(count)
print("="*100)
col1.append(0)
col2.append(argmax_more_than_half)
col3.append(argmax_less_than_half)
col4.append(focus_true_pred_true)
col5.append(focus_false_pred_true)
col6.append(focus_true_pred_false)
col7.append(focus_false_pred_false)
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in test_loader:
inputs, labels , fore_idx = data
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' % (
100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
col8.append(argmax_more_than_half)
col9.append(argmax_less_than_half)
col10.append(focus_true_pred_true)
col11.append(focus_false_pred_true)
col12.append(focus_true_pred_false)
col13.append(focus_false_pred_false)
nos_epochs = 200
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
for epoch in range(nos_epochs): # loop over the dataset multiple times
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
running_loss = 0.0
epoch_loss = []
cnt=0
iteration = desired_num // batch
#training data set
for i, data in enumerate(train_loader):
inputs , labels , fore_idx = data
inputs, labels = inputs.to("cuda"), labels.to("cuda")
# zero the parameter gradients
optimizer_focus.zero_grad()
optimizer_classify.zero_grad()
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
# print(outputs)
# print(outputs.shape,labels.shape , torch.argmax(outputs, dim=1))
loss = criterion(outputs, labels)
loss.backward()
optimizer_focus.step()
optimizer_classify.step()
running_loss += loss.item()
mini = 60
if cnt % mini == mini-1: # print every 40 mini-batches
print('[%d, %5d] loss: %.3f' %(epoch + 1, cnt + 1, running_loss / mini))
epoch_loss.append(running_loss/mini)
running_loss = 0.0
cnt=cnt+1
if epoch % 5 == 0:
for j in range (batch):
focus = torch.argmax(alphas[j])
if(alphas[j][focus] >= 0.5):
argmax_more_than_half +=1
else:
argmax_less_than_half +=1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true +=1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false +=1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false +=1
if(np.mean(epoch_loss) <= 0.005):
break;
if epoch % 5 == 0:
# focus_net.eval()
# classify.eval()
col1.append(epoch+1)
col2.append(argmax_more_than_half)
col3.append(argmax_less_than_half)
col4.append(focus_true_pred_true)
col5.append(focus_false_pred_true)
col6.append(focus_true_pred_false)
col7.append(focus_false_pred_false)
#************************************************************************
#testing data set
with torch.no_grad():
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
for data in test_loader:
inputs, labels , fore_idx = data
inputs, labels = inputs.to("cuda"), labels.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range (batch):
focus = torch.argmax(alphas[j])
if(alphas[j][focus] >= 0.5):
argmax_more_than_half +=1
else:
argmax_less_than_half +=1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true +=1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false +=1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false +=1
col8.append(argmax_more_than_half)
col9.append(argmax_less_than_half)
col10.append(focus_true_pred_true)
col11.append(focus_false_pred_true)
col12.append(focus_true_pred_false)
col13.append(focus_false_pred_false)
print('Finished Training')
# torch.save(focus_net.state_dict(),"/content/drive/My Drive/Research/Cheating_data/16_experiments_on_cnn_3layers/"+name+"_focus_net.pt")
# torch.save(classify.state_dict(),"/content/drive/My Drive/Research/Cheating_data/16_experiments_on_cnn_3layers/"+name+"_classify.pt")
columns = ["epochs", "argmax > 0.5" ,"argmax < 0.5", "focus_true_pred_true", "focus_false_pred_true", "focus_true_pred_false", "focus_false_pred_false" ]
df_train = pd.DataFrame()
df_test = pd.DataFrame()
df_train[columns[0]] = col1
df_train[columns[1]] = col2
df_train[columns[2]] = col3
df_train[columns[3]] = col4
df_train[columns[4]] = col5
df_train[columns[5]] = col6
df_train[columns[6]] = col7
df_test[columns[0]] = col1
df_test[columns[1]] = col8
df_test[columns[2]] = col9
df_test[columns[3]] = col10
df_test[columns[4]] = col11
df_test[columns[5]] = col12
df_test[columns[6]] = col13
df_train
# plt.figure(12,12)
plt.plot(col1,col2, label='argmax > 0.5')
plt.plot(col1,col3, label='argmax < 0.5')
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("training data")
plt.title("On Training set")
plt.show()
plt.plot(col1,col4, label ="focus_true_pred_true ")
plt.plot(col1,col5, label ="focus_false_pred_true ")
plt.plot(col1,col6, label ="focus_true_pred_false ")
plt.plot(col1,col7, label ="focus_false_pred_false ")
plt.title("On Training set")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("training data")
plt.savefig("train_ftpt.pdf", bbox_inches='tight')
plt.show()
df_test
# plt.figure(12,12)
plt.plot(col1,col8, label='argmax > 0.5')
plt.plot(col1,col9, label='argmax < 0.5')
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("Testing data")
plt.title("On Testing set")
plt.show()
plt.plot(col1,col10, label ="focus_true_pred_true ")
plt.plot(col1,col11, label ="focus_false_pred_true ")
plt.plot(col1,col12, label ="focus_true_pred_false ")
plt.plot(col1,col13, label ="focus_false_pred_false ")
plt.title("On Testing set")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("epochs")
plt.ylabel("Testing data")
plt.savefig("test_ftpt.pdf", bbox_inches='tight')
plt.show()
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in train_loader:
inputs, labels , fore_idx = data
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 30000 train images: %d %%' % (
100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
correct = 0
total = 0
count = 0
flag = 1
focus_true_pred_true =0
focus_false_pred_true =0
focus_true_pred_false =0
focus_false_pred_false =0
argmax_more_than_half = 0
argmax_less_than_half =0
with torch.no_grad():
for data in test_loader:
inputs, labels , fore_idx = data
inputs, labels , fore_idx = inputs.to("cuda"),labels.to("cuda"), fore_idx.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if(focus == fore_idx[j] and predicted[j] == labels[j]):
focus_true_pred_true += 1
elif(focus != fore_idx[j] and predicted[j] == labels[j]):
focus_false_pred_true += 1
elif(focus == fore_idx[j] and predicted[j] != labels[j]):
focus_true_pred_false += 1
elif(focus != fore_idx[j] and predicted[j] != labels[j]):
focus_false_pred_false += 1
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' % (
100 * correct / total))
print("total correct", correct)
print("total train set images", total)
print("focus_true_pred_true %d =============> FTPT : %d %%" % (focus_true_pred_true , (100 * focus_true_pred_true / total) ) )
print("focus_false_pred_true %d =============> FFPT : %d %%" % (focus_false_pred_true, (100 * focus_false_pred_true / total) ) )
print("focus_true_pred_false %d =============> FTPF : %d %%" %( focus_true_pred_false , ( 100 * focus_true_pred_false / total) ) )
print("focus_false_pred_false %d =============> FFPF : %d %%" % (focus_false_pred_false, ( 100 * focus_false_pred_false / total) ) )
print("argmax_more_than_half ==================> ",argmax_more_than_half)
print("argmax_less_than_half ==================> ",argmax_less_than_half)
correct = 0
total = 0
with torch.no_grad():
for data in train_loader:
inputs, labels , fore_idx = data
inputs, labels = inputs.to("cuda"), labels.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 30000 train images: %d %%' % ( 100 * correct / total))
print("total correct", correct)
print("total train set images", total)
correct = 0
total = 0
with torch.no_grad():
for data in test_loader:
inputs, labels , fore_idx = data
inputs, labels = inputs.to("cuda"), labels.to("cuda")
alphas, avg_images = focus_net(inputs)
outputs = classify(avg_images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' % ( 100 * correct / total))
print("total correct", correct)
print("total train set images", total)
max_alpha =[]
alpha_ftpt=[]
argmax_more_than_half=0
argmax_less_than_half=0
for i, data in enumerate(test_loader):
inputs, labels,_ = data
inputs = inputs.double()
inputs, labels = inputs.to("cuda"),labels.to("cuda")
alphas, avg = focus_net(inputs)
outputs = classify(avg)
mx,_ = torch.max(alphas,1)
max_alpha.append(mx.cpu().detach().numpy())
for j in range(labels.size(0)):
focus = torch.argmax(alphas[j])
if alphas[j][focus] >= 0.5 :
argmax_more_than_half += 1
else:
argmax_less_than_half += 1
if (focus == fore_idx[j] and predicted[j] == labels[j]):
alpha_ftpt.append(alphas[j][focus].item())
max_alpha = np.concatenate(max_alpha,axis=0)
print(max_alpha.shape)
plt.figure(figsize=(6,6))
_,bins,_ = plt.hist(max_alpha,bins=50,color ="c")
plt.title("alpha values histogram")
plt.savefig("alpha_hist.pdf")
plt.figure(figsize=(6,6))
_,bins,_ = plt.hist(np.array(alpha_ftpt),bins=50,color ="c")
plt.title("alpha values in ftpt")
plt.savefig("alpha_hist_ftpt.pdf")
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License");
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Custom Training Walkthrough
<table align="left"><td>
<a target="_blank" href="https://colab.sandbox.google.com/github/tensorflow/models/blob/master/samples/core/get_started/eager.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td><td>
<a target="_blank" href="https://github.com/tensorflow/models/blob/master/samples/core/get_started/eager.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on Github</a></td></table>
This guide uses machine learning to *categorize* Iris flowers by species. It uses [TensorFlow](https://www.tensorflow.org)'s eager execution to:
1. Build a model,
2. Train this model on example data, and
3. Use the model to make predictions about unknown data.
Machine learning experience isn't required, but you'll need to read some Python code. For more eager execution guides and examples, see [these notebooks](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples/notebooks).
## TensorFlow programming
There are many [TensorFlow APIs](https://www.tensorflow.org/api_docs/python/) available, but start with these high-level TensorFlow concepts:
* Enable an [eager execution](https://www.tensorflow.org/programmers_guide/eager) development environment,
* Import data with the [Datasets API](https://www.tensorflow.org/programmers_guide/datasets),
* Build models and layers with TensorFlow's [Keras API](https://keras.io/getting-started/sequential-model-guide/).
This tutorial is structured like many TensorFlow programs:
1. Import and parse the data sets.
2. Select the type of model.
3. Train the model.
4. Evaluate the model's effectiveness.
5. Use the trained model to make predictions.
For more TensorFlow examples, see the [Get Started](https://www.tensorflow.org/get_started/) and [Tutorials](https://www.tensorflow.org/tutorials/) sections. To learn machine learning basics, consider taking the [Machine Learning Crash Course](https://developers.google.com/machine-learning/crash-course/).
## Run the notebook
This tutorial is available as an interactive [Colab notebook](https://colab.research.google.com) that can execute and modify Python code directly in the browser. The notebook handles setup and dependencies while you "play" cells to run the code blocks. This is a fun way to explore the program and test ideas.
If you are unfamiliar with Python notebook environments, there are a couple of things to keep in mind:
1. Executing code requires connecting to a runtime environment. In the Colab notebook menu, select *Runtime > Connect to runtime...*
2. Notebook cells are arranged sequentially to gradually build the program. Typically, later code cells depend on prior code cells, though you can always rerun a code block. To execute the entire notebook in order, select *Runtime > Run all*. To rerun a code cell, select the cell and click the *play icon* on the left.
## Setup program
### Install the latest version of TensorFlow
This tutorial uses eager execution, which is available in [TensorFlow 1.8](https://www.tensorflow.org/install/). (You may need to restart the runtime after upgrading.)
```
!pip install --upgrade tensorflow
```
### Configure imports and eager execution
Import the required Python modules—including TensorFlow—and enable eager execution for this program. Eager execution makes TensorFlow evaluate operations immediately, returning concrete values instead of creating a [computational graph](https://www.tensorflow.org/programmers_guide/graphs) that is executed later. If you are used to a REPL or the `python` interactive console, this feels familiar.
Once eager execution is enabled, it *cannot* be disabled within the same program. See the [eager execution guide](https://www.tensorflow.org/programmers_guide/eager) for more details.
```
from __future__ import absolute_import, division, print_function
import os
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow.contrib.eager as tfe
tf.enable_eager_execution()
print("TensorFlow version: {}".format(tf.VERSION))
print("Eager execution: {}".format(tf.executing_eagerly()))
```
## The Iris classification problem
Imagine you are a botanist seeking an automated way to categorize each Iris flower you find. Machine learning provides many algorithms to statistically classify flowers. For instance, a sophisticated machine learning program could classify flowers based on photographs. Our ambitions are more modest—we're going to classify Iris flowers based on the length and width measurements of their [sepals](https://en.wikipedia.org/wiki/Sepal) and [petals](https://en.wikipedia.org/wiki/Petal).
The Iris genus entails about 300 species, but our program will only classify the following three:
* Iris setosa
* Iris virginica
* Iris versicolor
<table>
<tr><td>
<img src="https://www.tensorflow.org/images/iris_three_species.jpg"
alt="Petal geometry compared for three iris species: Iris setosa, Iris virginica, and Iris versicolor">
</td></tr>
<tr><td align="center">
<b>Figure 1.</b> <a href="https://commons.wikimedia.org/w/index.php?curid=170298">Iris setosa</a> (by <a href="https://commons.wikimedia.org/wiki/User:Radomil">Radomil</a>, CC BY-SA 3.0), <a href="https://commons.wikimedia.org/w/index.php?curid=248095">Iris versicolor</a>, (by <a href="https://commons.wikimedia.org/wiki/User:Dlanglois">Dlanglois</a>, CC BY-SA 3.0), and <a href="https://www.flickr.com/photos/33397993@N05/3352169862">Iris virginica</a> (by <a href="https://www.flickr.com/photos/33397993@N05">Frank Mayfield</a>, CC BY-SA 2.0).<br/>
</td></tr>
</table>
Fortunately, someone has already created a [data set of 120 Iris flowers](https://en.wikipedia.org/wiki/Iris_flower_data_set) with the sepal and petal measurements. This is a classic dataset that is popular for beginner machine learning classification problems.
## Import and parse the training dataset
Download the dataset file and convert it to a structure that can be used by this Python program.
### Download the dataset
Download the training dataset file using the [tf.keras.utils.get_file](https://www.tensorflow.org/api_docs/python/tf/keras/utils/get_file) function. This returns the file path of the downloaded file.
```
train_dataset_url = "http://download.tensorflow.org/data/iris_training.csv"
train_dataset_fp = tf.keras.utils.get_file(fname=os.path.basename(train_dataset_url),
origin=train_dataset_url)
print("Local copy of the dataset file: {}".format(train_dataset_fp))
```
### Inspect the data
This dataset, `iris_training.csv`, is a plain text file that stores tabular data formatted as comma-separated values (CSV). Use the `head -n5` command to take a peak at the first five entries:
```
!head -n5 {train_dataset_fp}
```
From this view of the dataset, notice the following:
1. The first line is a header containing information about the dataset:
* There are 120 total examples. Each example has four features and one of three possible label names.
2. Subsequent rows are data records, one *[example](https://developers.google.com/machine-learning/glossary/#example)* per line, where:
* The first four fields are *[features](https://developers.google.com/machine-learning/glossary/#feature)*: these are characteristics of an example. Here, the fields hold float numbers representing flower measurements.
* The last column is the *[label](https://developers.google.com/machine-learning/glossary/#label)*: this is the value we want to predict. For this dataset, it's an integer value of 0, 1, or 2 that corresponds to a flower name.
Let's write that out in code:
```
# column order in CSV file
column_names = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'species']
feature_names = column_names[:-1]
label_name = column_names[-1]
print("Features: {}".format(feature_names))
print("Label: {}".format(label_name))
```
Each label is associated with string name (for example, "setosa"), but machine learning typically relies on numeric values. The label numbers are mapped to a named representation, such as:
* `0`: Iris setosa
* `1`: Iris versicolor
* `2`: Iris virginica
For more information about features and labels, see the [ML Terminology section of the Machine Learning Crash Course](https://developers.google.com/machine-learning/crash-course/framing/ml-terminology).
```
class_names = ['Iris setosa', 'Iris versicolor', 'Iris virginica']
```
### Create a `tf.data.Dataset`
TensorFlow's [Dataset API](https://www.tensorflow.org/programmers_guide/datasets) handles many common cases for loading data into a model. This is a high-level API for reading data and transforming it into a form used for training. See the [Datasets Quick Start guide](https://www.tensorflow.org/get_started/datasets_quickstart) for more information.
Since the dataset is a CSV-formatted text file, use the the [make_csv_dataset](https://www.tensorflow.org/api_docs/python/tf/contrib/data/make_csv_dataset) function to parse the data into a suitable format. Since this function generates data for training models, the default behavior is to shuffle the data (`shuffle=True, shuffle_buffer_size=10000`), and repeat the dataset forever (`num_epochs=None`). We also set the [batch_size](https://developers.google.com/machine-learning/glossary/#batch_size) parameter.
```
batch_size = 32
train_dataset = tf.contrib.data.make_csv_dataset(
train_dataset_fp,
batch_size,
column_names=column_names,
label_name=label_name,
num_epochs=1)
```
The `make_csv_dataset` function returns a `tf.data.Dataset` of `(features, label)` pairs, where `features` is a dictionary: `{'feature_name': value}`
With eager execution enabled, these `Dataset` objects are iterable. Let's look at a batch of features:
```
features, labels = next(iter(train_dataset))
features
```
Notice that like-features are grouped together, or *batched*. Each example row's fields are appended to the corresponding feature array. Change the `batch_size` to set the number of examples stored in these feature arrays.
You can start to see some clusters by plotting a few features from the batch:
```
plt.scatter(features['petal_length'],
features['sepal_length'],
c=labels,
cmap='viridis')
plt.xlabel("Petal length")
plt.ylabel("Sepal length");
```
To simplify the model building step, create a function to repackage the features dictionary into a single array with shape: `(batch_size, num_features)`.
This function uses the [tf.stack](https://www.tensorflow.org/api_docs/python/tf/stack) method which takes values from a list of tensors and creates a combined tensor at the specified dimension.
```
def pack_features_vector(features, labels):
"""Pack the features into a single array."""
features = tf.stack(list(features.values()), axis=1)
return features, labels
```
Then use the [tf.data.Dataset.map](https://www.tensorflow.org/api_docs/python/tf/data/dataset/map) method to pack the `features` of each `(features,label)` pair into the training dataset:
```
train_dataset = train_dataset.map(pack_features_vector)
```
The features element of the `Dataset` are now arrays with shape `(batch_size, num_features)`. Let's look at the first few examples:
```
features, labels = next(iter(train_dataset))
print(features[:5])
```
## Select the type of model
### Why model?
A *[model](https://developers.google.com/machine-learning/crash-course/glossary#model)* is the relationship between features and the label. For the Iris classification problem, the model defines the relationship between the sepal and petal measurements and the predicted Iris species. Some simple models can be described with a few lines of algebra, but complex machine learning models have a large number of parameters that are difficult to summarize.
Could you determine the relationship between the four features and the Iris species *without* using machine learning? That is, could you use traditional programming techniques (for example, a lot of conditional statements) to create a model? Perhaps—if you analyzed the dataset long enough to determine the relationships between petal and sepal measurements to a particular species. And this becomes difficult—maybe impossible—on more complicated datasets. A good machine learning approach *determines the model for you*. If you feed enough representative examples into the right machine learning model type, the program will figure out the relationships for you.
### Select the model
We need to select the kind of model to train. There are many types of models and picking a good one takes experience. This tutorial uses a neural network to solve the Iris classification problem. *[Neural networks](https://developers.google.com/machine-learning/glossary/#neural_network)* can find complex relationships between features and the label. It is a highly-structured graph, organized into one or more *[hidden layers](https://developers.google.com/machine-learning/glossary/#hidden_layer)*. Each hidden layer consists of one or more *[neurons](https://developers.google.com/machine-learning/glossary/#neuron)*. There are several categories of neural networks and this program uses a dense, or *[fully-connected neural network](https://developers.google.com/machine-learning/glossary/#fully_connected_layer)*: the neurons in one layer receive input connections from *every* neuron in the previous layer. For example, Figure 2 illustrates a dense neural network consisting of an input layer, two hidden layers, and an output layer:
<table>
<tr><td>
<img src="https://www.tensorflow.org/images/custom_estimators/full_network.png"
alt="A diagram of the network architecture: Inputs, 2 hidden layers, and outputs">
</td></tr>
<tr><td align="center">
<b>Figure 2.</b> A neural network with features, hidden layers, and predictions.<br/>
</td></tr>
</table>
When the model from Figure 2 is trained and fed an unlabeled example, it yields three predictions: the likelihood that this flower is the given Iris species. This prediction is called *[inference](https://developers.google.com/machine-learning/crash-course/glossary#inference)*. For this example, the sum of the output predictions is 1.0. In Figure 2, this prediction breaks down as: `0.03` for *Iris setosa*, `0.95` for *Iris versicolor*, and `0.02` for *Iris virginica*. This means that the model predicts—with 95% probability—that an unlabeled example flower is an *Iris versicolor*.
### Create a model using Keras
The TensorFlow [tf.keras](https://www.tensorflow.org/api_docs/python/tf/keras) API is the preferred way to create models and layers. This makes it easy to build models and experiment while Keras handles the complexity of connecting everything together.
The [tf.keras.Sequential](https://www.tensorflow.org/api_docs/python/tf/keras/Sequential) model is a linear stack of layers. Its constructor takes a list of layer instances, in this case, two [Dense](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense) layers with 10 nodes each, and an output layer with 3 nodes representing our label predictions. The first layer's `input_shape` parameter corresponds to the number of features from the dataset, and is required.
```
model = tf.keras.Sequential([
tf.keras.layers.Dense(10, activation=tf.nn.relu, input_shape=(4,)), # input shape required
tf.keras.layers.Dense(10, activation=tf.nn.relu),
tf.keras.layers.Dense(3)
])
```
The *[activation function](https://developers.google.com/machine-learning/crash-course/glossary#activation_function)* determines the output shape of each node in the layer. These non-linearities are important—without them the model would be equivalent to a single layer. There are many [available activations](https://www.tensorflow.org/api_docs/python/tf/keras/activations), but [ReLU](https://developers.google.com/machine-learning/crash-course/glossary#ReLU) is common for hidden layers.
The ideal number of hidden layers and neurons depends on the problem and the dataset. Like many aspects of machine learning, picking the best shape of the neural network requires a mixture of knowledge and experimentation. As a rule of thumb, increasing the number of hidden layers and neurons typically creates a more powerful model, which requires more data to train effectively.
### Using the model
Let's have a quick look at what this model does to a batch of features:
```
predictions = model(features)
predictions[:5]
```
Here, each example returns a [logit](https://developers.google.com/machine-learning/crash-course/glossary#logit) for each class.
To convert these logits to a probability for each class, use the [softmax](https://developers.google.com/machine-learning/crash-course/glossary#softmax) function:
```
tf.nn.softmax(predictions[:5])
```
Taking the `tf.argmax` across classes gives us the predicted class index. But, the model hasn't been trained yet, so these aren't good predictions.
```
print("Prediction: {}".format(tf.argmax(predictions, axis=1)))
print(" Labels: {}".format(labels))
```
## Train the model
*[Training](https://developers.google.com/machine-learning/crash-course/glossary#training)* is the stage of machine learning when the model is gradually optimized, or the model *learns* the dataset. The goal is to learn enough about the structure of the training dataset to make predictions about unseen data. If you learn *too much* about the training dataset, then the predictions only work for the data it has seen and will not be generalizable. This problem is called *[overfitting](https://developers.google.com/machine-learning/crash-course/glossary#overfitting)*—it's like memorizing the answers instead of understanding how to solve a problem.
The Iris classification problem is an example of *[supervised machine learning](https://developers.google.com/machine-learning/glossary/#supervised_machine_learning)*: the model is trained from examples that contain labels. In *[unsupervised machine learning](https://developers.google.com/machine-learning/glossary/#unsupervised_machine_learning)*, the examples don't contain labels. Instead, the model typically finds patterns among the features.
### Define the loss and gradient function
Both training and evaluation stages need to calculate the model's *[loss](https://developers.google.com/machine-learning/crash-course/glossary#loss)*. This measures how off a model's predictions are from the desired label, in other words, how bad the model is performing. We want to minimize, or optimize, this value.
Our model will calculate its loss using the [tf.keras.losses.categorical_crossentropy](https://www.tensorflow.org/api_docs/python/tf/losses/sparse_softmax_cross_entropy) function which takes the model's class probability predictions and the desired label, and returns the average loss across the examples.
```
def loss(model, x, y):
y_ = model(x)
return tf.losses.sparse_softmax_cross_entropy(labels=y, logits=y_)
l = loss(model, features, labels)
print("Loss test: {}".format(l))
```
Use the [tf.GradientTape](https://www.tensorflow.org/api_docs/python/tf/GradientTape) context to calculate the *[gradients](https://developers.google.com/machine-learning/crash-course/glossary#gradient)* used to optimize our model. For more examples of this, see the [eager execution guide](https://www.tensorflow.org/programmers_guide/eager).
```
def grad(model, inputs, targets):
with tf.GradientTape() as tape:
loss_value = loss(model, inputs, targets)
return loss_value, tape.gradient(loss_value, model.trainable_variables)
```
### Create an optimizer
An *[optimizer](https://developers.google.com/machine-learning/crash-course/glossary#optimizer)* applies the computed gradients to the model's variables to minimize the `loss` function. You can think of the loss function as a curved surface (see Figure 3) and we want to find its lowest point by walking around. The gradients point in the direction of steepest ascent—so we'll travel the opposite way and move down the hill. By iteratively calculating the loss and gradient for each batch, we'll adjust the model during training. Gradually, the model will find the best combination of weights and bias to minimize loss. And the lower the loss, the better the model's predictions.
<table>
<tr><td>
<img src="https://cs231n.github.io/assets/nn3/opt1.gif" width="70%"
alt="Optimization algorthims visualized over time in 3D space.">
</td></tr>
<tr><td align="center">
<b>Figure 3.</b> Optimization algorithms visualized over time in 3D space. (Source: <a href="http://cs231n.github.io/neural-networks-3/">Stanford class CS231n</a>, MIT License)<br/>
</td></tr>
</table>
TensorFlow has many [optimization algorithms](https://www.tensorflow.org/api_guides/python/train) available for training. This model uses the [tf.train.GradientDescentOptimizer](https://www.tensorflow.org/api_docs/python/tf/train/GradientDescentOptimizer) that implements the *[stochastic gradient descent](https://developers.google.com/machine-learning/crash-course/glossary#gradient_descent)* (SGD) algorithm. The `learning_rate` sets the step size to take for each iteration down the hill. This is a *hyperparameter* that you'll commonly adjust to achieve better results.
Let's setup the optimizer and the `global_step` counter:
```
optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.01)
global_step = tf.train.get_or_create_global_step()
```
We'll use this to calculate a single optimization step:
```
loss_value, grads = grad(model, features, labels)
print("Step: {}, Initial Loss: {}".format(global_step.numpy(),
loss_value.numpy()))
optimizer.apply_gradients(zip(grads, model.variables), global_step)
print("Step: {}, Loss: {}".format(global_step.numpy(),
loss(model, features, labels).numpy()))
```
### Training loop
With all the pieces in place, the model is ready for training! A training loop feeds the dataset examples into the model to help it make better predictions. The following code block sets up these training steps:
1. Iterate each *epoch*. An epoch is one pass through the dataset.
2. Within an epoch, iterate over each example in the training `Dataset` grabbing its *features* (`x`) and *label* (`y`).
3. Using the example's features, make a prediction and compare it with the label. Measure the inaccuracy of the prediction and use that to calculate the model's loss and gradients.
4. Use an `optimizer` to update the model's variables.
5. Keep track of some stats for visualization.
6. Repeat for each epoch.
The `num_epochs` variable is the amount of times to loop over the dataset collection. Counter-intuitively, training a model longer does not guarantee a better model. `num_epochs` is a *[hyperparameter](https://developers.google.com/machine-learning/glossary/#hyperparameter)* that you can tune. Choosing the right number usually requires both experience and experimentation.
```
## Note: Rerunning this cell uses the same model variables
# keep results for plotting
train_loss_results = []
train_accuracy_results = []
num_epochs = 201
for epoch in range(num_epochs):
epoch_loss_avg = tfe.metrics.Mean()
epoch_accuracy = tfe.metrics.Accuracy()
# Training loop - using batches of 32
for x, y in train_dataset:
# Optimize the model
loss_value, grads = grad(model, x, y)
optimizer.apply_gradients(zip(grads, model.variables),
global_step)
# Track progress
epoch_loss_avg(loss_value) # add current batch loss
# compare predicted label to actual label
epoch_accuracy(tf.argmax(model(x), axis=1, output_type=tf.int32), y)
# end epoch
train_loss_results.append(epoch_loss_avg.result())
train_accuracy_results.append(epoch_accuracy.result())
if epoch % 50 == 0:
print("Epoch {:03d}: Loss: {:.3f}, Accuracy: {:.3%}".format(epoch,
epoch_loss_avg.result(),
epoch_accuracy.result()))
```
### Visualize the loss function over time
While it's helpful to print out the model's training progress, it's often *more* helpful to see this progress. [TensorBoard](https://www.tensorflow.org/programmers_guide/summaries_and_tensorboard) is a nice visualization tool that is packaged with TensorFlow, but we can create basic charts using the `matplotlib` module.
Interpreting these charts takes some experience, but you really want to see the *loss* go down and the *accuracy* go up.
```
fig, axes = plt.subplots(2, sharex=True, figsize=(12, 8))
fig.suptitle('Training Metrics')
axes[0].set_ylabel("Loss", fontsize=14)
axes[0].plot(train_loss_results)
axes[1].set_ylabel("Accuracy", fontsize=14)
axes[1].set_xlabel("Epoch", fontsize=14)
axes[1].plot(train_accuracy_results);
```
## Evaluate the model's effectiveness
Now that the model is trained, we can get some statistics on its performance.
*Evaluating* means determining how effectively the model makes predictions. To determine the model's effectiveness at Iris classification, pass some sepal and petal measurements to the model and ask the model to predict what Iris species they represent. Then compare the model's prediction against the actual label. For example, a model that picked the correct species on half the input examples has an *[accuracy](https://developers.google.com/machine-learning/glossary/#accuracy)* of `0.5`. Figure 4 shows a slightly more effective model, getting 4 out of 5 predictions correct at 80% accuracy:
<table cellpadding="8" border="0">
<colgroup>
<col span="4" >
<col span="1" bgcolor="lightblue">
<col span="1" bgcolor="lightgreen">
</colgroup>
<tr bgcolor="lightgray">
<th colspan="4">Example features</th>
<th colspan="1">Label</th>
<th colspan="1" >Model prediction</th>
</tr>
<tr>
<td>5.9</td><td>3.0</td><td>4.3</td><td>1.5</td><td align="center">1</td><td align="center">1</td>
</tr>
<tr>
<td>6.9</td><td>3.1</td><td>5.4</td><td>2.1</td><td align="center">2</td><td align="center">2</td>
</tr>
<tr>
<td>5.1</td><td>3.3</td><td>1.7</td><td>0.5</td><td align="center">0</td><td align="center">0</td>
</tr>
<tr>
<td>6.0</td> <td>3.4</td> <td>4.5</td> <td>1.6</td> <td align="center">1</td><td align="center" bgcolor="red">2</td>
</tr>
<tr>
<td>5.5</td><td>2.5</td><td>4.0</td><td>1.3</td><td align="center">1</td><td align="center">1</td>
</tr>
<tr><td align="center" colspan="6">
<b>Figure 4.</b> An Iris classifier that is 80% accurate.<br/>
</td></tr>
</table>
### Setup the test dataset
Evaluating the model is similar to training the model. The biggest difference is the examples come from a separate *[test set](https://developers.google.com/machine-learning/crash-course/glossary#test_set)* rather than the training set. To fairly assess a model's effectiveness, the examples used to evaluate a model must be different from the examples used to train the model.
The setup for the test `Dataset` is similar to the setup for training `Dataset`. Download the CSV text file and parse that values, then give it a little shuffle:
```
test_url = "http://download.tensorflow.org/data/iris_test.csv"
test_fp = tf.keras.utils.get_file(fname=os.path.basename(test_url),
origin=test_url)
test_dataset = tf.contrib.data.make_csv_dataset(
train_dataset_fp,
batch_size,
column_names=column_names,
label_name='species',
num_epochs=1,
shuffle=False)
test_dataset = test_dataset.map(pack_features_vector)
```
### Evaluate the model on the test dataset
Unlike the training stage, the model only evaluates a single [epoch](https://developers.google.com/machine-learning/glossary/#epoch) of the test data. In the following code cell, we iterate over each example in the test set and compare the model's prediction against the actual label. This is used to measure the model's accuracy across the entire test set.
```
test_accuracy = tfe.metrics.Accuracy()
for (x, y) in test_dataset:
logits = model(x)
prediction = tf.argmax(logits, axis=1, output_type=tf.int32)
test_accuracy(prediction, y)
print("Test set accuracy: {:.3%}".format(test_accuracy.result()))
```
We can see on the last batch, for example, the model is usually correct:
```
tf.stack([y,prediction],axis=1)
```
## Use the trained model to make predictions
We've trained a model and "proven" that it's good—but not perfect—at classifying Iris species. Now let's use the trained model to make some predictions on [unlabeled examples](https://developers.google.com/machine-learning/glossary/#unlabeled_example); that is, on examples that contain features but not a label.
In real-life, the unlabeled examples could come from lots of different sources including apps, CSV files, and data feeds. For now, we're going to manually provide three unlabeled examples to predict their labels. Recall, the label numbers are mapped to a named representation as:
* `0`: Iris setosa
* `1`: Iris versicolor
* `2`: Iris virginica
```
predict_dataset = tf.convert_to_tensor([
[5.1, 3.3, 1.7, 0.5,],
[5.9, 3.0, 4.2, 1.5,],
[6.9, 3.1, 5.4, 2.1]
])
predictions = model(predict_dataset)
for i, logits in enumerate(predictions):
class_idx = tf.argmax(logits).numpy()
p = tf.nn.softmax(logits)[class_idx]
name = class_names[class_idx]
print("Example {} prediction: {} ({:4.1f}%)".format(i, name, 100*p))
```
These predictions look good!
To dig deeper into machine learning models, take a look at the TensorFlow [Programmer's Guide](https://www.tensorflow.org/programmers_guide/) and check out the [community](https://www.tensorflow.org/community/).
## Next steps
For more eager execution guides and examples, see [these notebooks](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples/notebooks).
|
github_jupyter
|
# Pre-procesamiento de datos

## Candidaturas elegidas
Principales transformaciones:
- Selección de atributos
- Tratamiento de valores faltantes
```
import glob
import nltk
import re
import pandas as pd
from string import punctuation
df_deputadas_1934_2023 = pd.read_csv('dados/deputadas_1934_2023.csv')
df_deputadas_1934_2023.shape
df_deputadas_1934_2023.head(5)
```
<div class="alert-warning"> Candidaturas elegidas: Selección de atributos para análisis </div>
```
df_deputadas = df_deputadas_1934_2023[['id', 'siglaPartido', 'siglaUf',
'idLegislatura', 'sexo']]
df_deputadas.head(5)
```
<div class="alert-warning"> Candidaturas elegidas: Ajuste de los valores faltantes </div>
```
df_deputadas.isnull().sum(axis = 0)
df_deputadas['siglaPartido'].fillna('sem partido', inplace=True)
df_deputadas.isnull().sum(axis = 0)
df_deputadas.to_csv('dados/candidaturas_eleitas(1).csv', index=False)
```
## Legislaturas
Principales tranformaciones:
- Convertir fecha completa en año
```
tipo_data = ['dataInicio', 'dataFim']
df_legislaturas = pd.read_csv('dados/legislaturas_1934_2023.csv', parse_dates=tipo_data)
df_legislaturas.info()
df_legislaturas.head()
```
<div class="alert-warning"> Legislaturas: extracción de año </div>
```
df_legislaturas['dataInicio'] = df_legislaturas['dataInicio'].dt.year
df_legislaturas['dataFim'] = df_legislaturas['dataFim'].dt.year
df_legislaturas.head()
df_legislaturas.to_csv('dados/legislaturas_1934_2023_limpas(1).csv', index=False)
```
## Proposiciones legislativas
Principales transformaciones:
- Selección de los tipos de propuestas legislativas deseadas
- Selección de atributos
- Ajustes de valores faltantes
- Extracción de palabras claves de las ementas
- Remoción de stopwords, meses, puntuación, números
- Remoción de palabras con menos de 3 caracteres y semanticamente irrelevantes
- Remoción de bigramas semanticamente irrelevantes
```
lista_proposicoes = glob.glob('dados/proposicoes/propo*')
tipos_dados = {
'id': object,
'uri': object,
'siglaTipo': object,
'numero': object,
'ano': int,
'codTipo': object,
'descricaoTipo': object,
'ementa': object,
'ementaDetalhada': object,
'keywords': object,
'uriOrgaoNumerador': object,
'uriPropAnterior': object,
'uriPropPrincipal': object,
'uriPropPosterior': object,
'urlInteiroTeor': object,
'urnFinal': object,
'ultimoStatus_sequencia': object,
'ultimoStatus_uriRelator': object,
'ultimoStatus_idOrgao': object,
'ultimoStatus_siglaOrgao': object,
'ultimoStatus_uriOrgao': object,
'ultimoStatus_regime': object,
'ultimoStatus_descricaoTramitacao': object,
'ultimoStatus_idTipoTramitacao': object,
'ultimoStatus_descricaoSituacao': object,
'ultimoStatus_idSituacao': object,
'ultimoStatus_despacho': object,
'ultimoStatus_url': object
}
tipo_data = ['dataApresentacao', 'ultimoStatus_dataHora']
lista_df = []
for proposicao in lista_proposicoes:
df_proposicao = pd.read_csv(proposicao, sep=';', dtype=tipos_dados, parse_dates=tipo_data)
lista_df.append(df_proposicao)
df_proposicao_1934_2021 = pd.concat(lista_df, axis=0, ignore_index=True)
df_proposicao_1934_2021.shape
```
<div class="alert-warning"> Proposiciones legislativas: Selección de los tipos de propuestas legislativas </div>
- Projeto de Decreto Legislativo [SF] (PDL)
- Projeto de Decreto Legislativo [CD] (PDC)
- Projeto de Decreto Legislativo [CN] (PDN)
- Projeto de Decreto Legislativo [SF] (PDS)
- Proposta de Emenda à Constituição (PEC)
- Projeto de Lei (PL)
- Projeto de Lei da Câmara (PLC)
- Projeto de Lei Complementar (PLP)
- Projeto de Lei de Conversão (PLV)
- Projeto de Resolução da Câmara dos Deputados (PRC)
```
tipos_proposicoes = ['PDS', 'PDC', 'PDN', 'PEC', 'PL', 'PLC', 'PLP', 'PLV', 'PRC']
df_proposicoes_tipos_desejados = df_proposicao_1934_2021[df_proposicao_1934_2021['siglaTipo'].isin(tipos_proposicoes)].copy()
df_proposicoes_tipos_desejados.shape
```
<div class="alert-warning"> Proposiciones legislativas: Selección de atributos para análisis </div>
```
df_proposicoes = df_proposicoes_tipos_desejados[['id','siglaTipo','ano', 'codTipo', 'descricaoTipo',
'ementa', 'ementaDetalhada', 'keywords']].copy()
df_proposicoes.shape
```
<div class="alert-warning"> Proposiciones legislativas: Ajuste de valores faltantes </div>
```
df_proposicoes.isnull().sum(axis = 0)
df_proposicoes[
(df_proposicoes['ementa'].isnull()) &
(df_proposicoes['ementaDetalhada'].isnull()) &
(df_proposicoes['keywords'].isnull())].head()
df_proposicoes.dropna(axis=0, subset=['ementa'], inplace=True)
df_proposicoes.shape
```
<div class="alert-warning"> Proposiciones legislativas: Normalización de las keywords existentes </div>
```
df_proposicoes_com_keywords = df_proposicoes[df_proposicoes['keywords'].notna()].copy()
df_proposicoes[df_proposicoes['keywords'].notna()]
nltk.download('punkt')
nltk.download('stopwords')
```
<div class="alert-warning"> Proposiciones legislativas: Funcciones para borrar la puntuación, preposiciones, números y artículos</div>
```
meses = ['janeiro', 'fevereiro', 'março', 'abril', 'maio', 'junho', 'julho','agosto', 'setembro', 'outubro', 'novembro', 'dezembro']
def define_stopwords_punctuation():
stopwords = nltk.corpus.stopwords.words('portuguese') + meses
pontuacao = list(punctuation)
stopwords.extend(pontuacao)
return stopwords
def remove_stopwords_punctuation_da_sentenca(texto):
padrao_digitos = r'[0-9]'
texto = re.sub(padrao_digitos, '', texto)
palavras = nltk.tokenize.word_tokenize(texto.lower())
stopwords = define_stopwords_punctuation()
keywords = [palavra for palavra in palavras if palavra not in stopwords]
return keywords
df_proposicoes_com_keywords['keywords'] = df_proposicoes_com_keywords['keywords'].apply(remove_stopwords_punctuation_da_sentenca)
def converte_lista_string(lista):
return ','.join([palavra for palavra in lista])
df_proposicoes_com_keywords['keywords'] = df_proposicoes_com_keywords['keywords'].apply(converte_lista_string)
```
<div class="alert-warning"> Proposiciones legislativas: Borra las proposiciones que quedaron sin keywords despues de la limpieza</div>
```
df_proposicoes_com_keywords = df_proposicoes_com_keywords[df_proposicoes_com_keywords['keywords'] != '']
df_proposicoes_com_keywords.head()
```
<div class="alert-warning"> Proposiciones legislativas: Saca `keywords` de la columna `ementa` </div>
```
df_proposicoes_sem_keywords = df_proposicoes[df_proposicoes['keywords'].isna()].copy()
df_proposicoes_sem_keywords['keywords'] = df_proposicoes_sem_keywords['ementa'].apply(remove_stopwords_punctuation_da_sentenca)
lista_keywords = []
lista_keywords_temp = df_proposicoes_sem_keywords['keywords'].tolist()
_ = [lista_keywords.extend(item) for item in lista_keywords_temp]
palavras_para_descarte = [item for item in set(lista_keywords) if len(item) <= 3]
substantivos_nao_descartaveis = ['cão', 'mãe', 'oab', 'boa', 'pré', 'voz', 'rui', 'uva', 'gás', 'glp', 'apa']
palavras_para_descarte_refinada = [palavra for palavra in palavras_para_descarte if palavra not in substantivos_nao_descartaveis]
def remove_palavras_para_descarte_da_sentenca(texto):
keywords = []
for palavra in texto:
if palavra not in palavras_para_descarte_refinada:
keywords.append(palavra)
return keywords
df_proposicoes_sem_keywords['keywords'] = df_proposicoes_sem_keywords['keywords'].apply(remove_palavras_para_descarte_da_sentenca)
```
<div class="alert-warning"> Proposiciones legislativas: Tratamiento de bigramas </div>
```
def gera_n_grams(texto, ngram=2):
temporario = zip(*[texto[indice:] for indice in range(0,ngram)])
resultado = [' '.join(ngram) for ngram in temporario]
return resultado
df_proposicoes_sem_keywords['bigrams'] = df_proposicoes_sem_keywords['keywords'].apply(gera_n_grams)
lista_ngrams = []
lista_ngrams_temp = df_proposicoes_sem_keywords['bigrams'].tolist()
_ = [lista_ngrams.extend(item) for item in lista_ngrams_temp]
bigrams_comuns = nltk.FreqDist(lista_ngrams).most_common(50)
lista_bigramas_comuns = [bigrama for bigrama, frequencia in bigrams_comuns]
```
<div class="alert-warning"> Proposiciones legislativas: Selección de los bigramas semanticamente irrelevantes </div>
```
lista_bigramas_comuns_limpa = ['dispõe sobre', 'outras providências', 'nova redação', 'poder executivo', 'distrito federal',
'autoriza poder', 'federal outras','redação constituição', 'dispõe sôbre', 'código penal', 'artigo constituição',
'disposições constitucionais', 'altera dispõe', 'decreto-lei código', 'constitucionais transitórias', 'altera redação',
'abre ministério', 'executivo abrir', 'redação artigo', 'sobre criação', 'acrescenta parágrafo', 'parágrafo único',
'concede isenção', 'altera dispositivos', 'altera complementar', 'dispondo sobre', 'código processo', 'outras providências.',
'providências. historico', 'ministério fazenda', 'altera leis', 'programa nacional', 'quadro permanente', 'outras providencias',
'inciso constituição', 'abrir ministério', 'estabelece normas', 'ministério justiça', 'tempo serviço', 'instituto nacional',
'institui sistema', 'operações crédito', 'altera institui', 'dispõe sôbre']
palavras_para_descarte_origem_bigramas = []
_ = [palavras_para_descarte_origem_bigramas.extend(bigrama.split(' ')) for bigrama in lista_bigramas_comuns_limpa]
palavras_para_descarte_origem_bigramas_unicas = set(palavras_para_descarte_origem_bigramas)
def remove_palavras_origem_bigramas_da_sentenca(texto):
keywords = []
for palavra in texto:
if palavra not in palavras_para_descarte_origem_bigramas_unicas:
keywords.append(palavra)
return keywords
df_proposicoes_sem_keywords['keywords'] = df_proposicoes_sem_keywords['keywords'].apply(remove_palavras_origem_bigramas_da_sentenca)
df_proposicoes_sem_keywords['keywords'] = df_proposicoes_sem_keywords['keywords'].apply(converte_lista_string)
df_proposicoes_sem_keywords = df_proposicoes_sem_keywords.drop(columns=['bigrams'])
```
<div class="alert-warning"> Proposiciones legislativas: Borra las proposiciones que quedaron sin keywords despues de la limpieza</div>
```
df_proposicoes_sem_keywords = df_proposicoes_sem_keywords[df_proposicoes_sem_keywords['keywords'] != '']
df_proposicoes_sem_keywords[df_proposicoes_sem_keywords['keywords']== '']
```
<div class="alert-warning"> Proposiciones legislativas: Reuni los dataframes</div>
```
df_proposicoes_v_final = pd.concat([df_proposicoes_com_keywords, df_proposicoes_sem_keywords])
df_proposicoes_v_final.shape
df_proposicoes_v_final.info()
df_proposicoes_v_final.to_csv('dados/proposicoes_legislativas_limpas(1).csv', index=False)
```
# Creación de vocabulario

Antes de hacer el análisis de los temas de las proposiciones hacía falta clasificarlas con un vocabulario controlado. Así que, usando el conjunto de datos "temas de proposições" clasifiqué algunas proposiciones relativas a protección de derechos de grupos históricamente marginados, a saber: campesinos, mujeres, población LGTQIA+, negros, ancianos, discapacitados, artistas, poblaciones económicamente vulnerables y pueblos indígenas.
Principales etapas:
- Reunir todas las palabras claves
- Atribuir manualmente palabras a temas
- Atribuir tema a proposiciones que contenía la palabra clave
```
proposicoes = pd.read_csv('dados/proposicoes_legislativas_limpas(1).csv')
proposicoes.info()
```
Reunião de palavras chaves para classificação
```
keywords = proposicoes['keywords']
vocabulario = []
for proposicao in keywords:
lista = proposicao.split(',')
vocabulario.extend(lista)
vocabulario_unico = set(vocabulario)
with open('dados/vocabulario.txt', 'w') as palavras:
for termo in vocabulario_unico:
palavras.write(termo + '\n')
```
<div class="alert-warning">Relacioné manualmente palabras claves a uno de los temas del conjunto de datos "Temas"</div>
```
vocabulario_temp = pd.read_csv('dados/temas_vocabulario.csv')
vocabulario_temp.head()
```
<div class="alert-warning"> Crié el vocabuario</div>
```
vocabulario = pd.DataFrame(columns=['cod', 'tema', 'palavra_chave'])
indices = vocabulario_temp.index
for indice in indices:
descricao = vocabulario_temp['descricao'].iloc[indice]
if type(descricao) == str:
for palavra in descricao.split(' '):
df = pd.DataFrame(data={'cod':vocabulario_temp['cod'].iloc[indice], 'tema':vocabulario_temp['nome'].iloc[indice], 'palavra_chave':[palavra]})
vocabulario = pd.concat([vocabulario, df], ignore_index=True)
vocabulario.sample(5)
vocabulario.shape
vocabulario = vocabulario[vocabulario['palavra_chave']!= ''].copy()
vocabulario.shape
```
<div class="alert-warning">Atribuí el tema a las proposiciones que contenía la palabra en la columna `keyword`</div>
```
def atribui_tema(proposicao):
for tema, palavra_chave in zip(vocabulario['tema'], vocabulario['palavra_chave']):
if palavra_chave in proposicao:
return tema
proposicoes['temas'] = proposicoes['keywords'].apply(atribui_tema)
proposicoes.to_csv('dados/proposicoes_legislativas_limpas_vocabulario(1).csv', index=False)
```
# Modelo de aprendizaje de máquina

Hay que clasificar todas las proposiciones antes del análisis.
Principales etapas:
- Establece variable predictora: “ementa” y la de respuesta:"temas"
- Encode da variable de respuesta utilizando preprocessing.LabelEncoder
- Divide conjunto de datos para teste y entrenamiento
- Convierte las ementas en vectores con HashingVectorizer
- Crea el modelo de clasificación con RandomForestClassifier
- Entrena el modelo
- Evalua cualitativamente a partir de la comparación entre las clasificaciones de los conjuntos de prueba y entrenamiento
Al final tenemos clasificadas solamente las proposiciones referentes a temática estudiada
```
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import HashingVectorizer
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.ensemble import RandomForestClassifier
from keras.utils import np_utils
import nltk
from nltk.corpus import stopwords
import pandas as pd
import numpy as np
```
<div class="alert-warning">Classifica proposições legislativas</div>
```
df_proposicoes = pd.read_csv("dados/proposicoes_legislativas_limpas_vocabulario(1).csv")
df_proposicoes_classificado = df_proposicoes.dropna(subset=["temas"])
df_proposicoes_classificado = df_proposicoes_classificado[["ementa","temas"]]
df_proposicoes_classificado.shape
df_proposicoes_classificado.head()
```
<div class="alert-warning">Establece variable predictora: “ementa” y la de respuesta:"temas"</div>
```
sentences = df_proposicoes_classificado['ementa'].values
```
<div class="alert-warning">Encode da variable de respuesta</div>
```
le = preprocessing.LabelEncoder()
le.fit(df_proposicoes_classificado['temas'].unique())
y = le.transform(df_proposicoes_classificado['temas'])
```
<div class="alert-warning">Divide el conjunto de teste y entrenamiento</div>
```
sentences_train, sentences_test, y_train, y_test = train_test_split(
sentences, y, test_size=0.25, random_state=1000)
```
<div class="alert-warning">Convierte las ementas en vectores con HashingVectorizer</div>
```
vectorizer = CountVectorizer()
vectorizer.fit(sentences_train)
X_train = vectorizer.transform(sentences_train)
X_test = vectorizer.transform(sentences_test)
X_train
hasher = HashingVectorizer(
n_features=10000,
stop_words=stopwords.words('portuguese'),
alternate_sign=False,
norm=None,
)
hasher.fit(sentences_train)
X_train_hasher = hasher.transform(sentences_train)
X_test_hasher = hasher.transform(sentences_test)
X_train_hasher.shape
```
<div class="alert-warning">Cría y entreina clasificador</div>
```
clf = RandomForestClassifier(n_estimators=200,random_state=0)
clf.fit(X_train_hasher, y_train)
```
<div class="alert-warning">Verifica el coeficiente de determinación (R²)</div>
```
score = clf.score(X_test_hasher, y_test)
print("Acurácia:", score)
```
<div class="alert-warning">Avalia modelo cualitativamente</div>
```
df_random_forest_results = pd.DataFrame([sentences_test,le.inverse_transform(clf.predict(X_test_hasher))]).transpose().rename(columns={0:"ementa",1:"tema"})
df_random_forest_results.head()
```
<div class="alert-warning">Cría listado con probabilidades de clasificación de la proposición en cada tema</div>
```
predicted_probabilities = clf.predict_proba(X_test_hasher)
```
<div class="alert-warning">Selecciona el tema con mayor probabilidad para cada proposición</div>
```
df_random_forest_results["probabilidade_predicao"] = np.amax(predicted_probabilities,axis=1)
df_random_forest_results.head()
```
<div class="alert-warning">Cría dataframe comparativo entre los temas preestablecidos y los clasificados por el clasificador</div>
```
df_ementas_test = pd.DataFrame([sentences_test,le.inverse_transform(y_test)]).transpose().rename(columns={0:"ementa",1:"tema"})
df_ementas_test.head()
df_avaliacao = df_random_forest_results.merge(df_ementas_test,left_on="ementa",right_on="ementa",suffixes=["_resposta_modelo","_correto"])
df_avaliacao["modelo_acertou"] = df_avaliacao["tema_resposta_modelo"] == df_avaliacao["tema_correto"]
df_avaliacao["modelo_acertou"] = df_avaliacao["modelo_acertou"].replace({True: "Sim", False: "Não"})
df_avaliacao["modelo_acertou"].value_counts()
```
<div class="alert-warning">Resumen de la validación</div>
```
df_avaliacao[df_avaliacao["probabilidade_predicao"] >= 0.85]["modelo_acertou"].value_counts()
df_avaliacao.head()
df_ementas_test.tema.value_counts()
df_avaliacao.to_csv('dados/avaliacao-qualitativa-modelo-classificacao(1).csv')
```
<div class="alert-warning">Aplicación del modelo</div>
```
df_proposicoes_total = df_proposicoes[["ementa","temas"]]
ementas = df_proposicoes_total['ementa'].values
ementas_hasher = hasher.transform(ementas)
df_proposicoes_total_classificadas = pd.DataFrame([ementas,le.inverse_transform(clf.predict(ementas_hasher))]).transpose().rename(
columns={0:"ementa",1:"temas"})
df_proposicoes_total_classificadas.head()
df_proposicoes_total_classificadas.info()
```
Informar a probabilidade de acerto de cada tema
```
temas_probabilities = clf.predict_proba(ementas_hasher)
df_proposicoes_total_classificadas["probabilidade_predicao"] = np.amax(temas_probabilities, axis=1)
df_proposicoes_total_classificadas.head()
df_proposicoes_total_classificadas.info()
```
Limpa temas cuja a probabilidade de acerto é menor do que 85%
```
def retira_tema_com_baixa_probabilidade_acerto(proposicoes):
if proposicoes['probabilidade_predicao'] >= 0.85:
return proposicoes['temas']
else:
return np.nan
df_proposicoes_total_classificadas['temas'] = df_proposicoes_total_classificadas.apply(retira_tema_com_baixa_probabilidade_acerto,
axis=1)
```
Reunir conjunto de dados de proposições legislativas com classificação realizada
```
df_proposicoes_classificador = df_proposicoes.join(df_proposicoes_total_classificadas, rsuffix='_classificador')
df_proposicoes_classificador.shape
df_proposicoes_classificador.head()
df_proposicoes_classificador.drop(columns=['temas', 'ementa_classificador', 'probabilidade_predicao'], inplace=True)
df_proposicoes_classificador.to_csv('dados/proposicoes_legislativas_limpas_classificadas(1).csv', index=False)
```
# Análisis exploratorio de datos

```
import matplotlib.pyplot as plt
```
## 1. ¿Hubo impacto positivo en la cantidad de mujeres elegidas para la Cámara en las 3 legislaciones subsecuentes a aprobación de la Ley 9.504/1997?
**Hipótesis:** No huvo impacto positivo en el percentual de mujeres elegidas para la Cámara en las 3 legislaciones subsecuentes a aprobación de la Ley 9.504/1997.
```
df_legislaturas = pd.read_csv('dados/legislaturas_1934_2023_limpas(1).csv')
df_legislaturas.head()
```
<div class="alert-warning">Determinar el período de los datos para el análisis (1995 a 2007)</div>
```
legislaturas_h1 = df_legislaturas[(df_legislaturas['id'] >= 50) & (df_legislaturas['id'] <= 53)]['id'].unique().tolist()
df_candidaturas_eleitas = pd.read_csv('dados/candidaturas_eleitas(1).csv')
df_candidaturas_eleitas_h1 = df_candidaturas_eleitas[df_candidaturas_eleitas['idLegislatura'].isin(legislaturas_h1)].copy()
df_candidaturas_eleitas_h1['idLegislatura'].unique()
```
<div class="alert-warning">Agrupar por género</div>
```
agrupa_sexo = df_candidaturas_eleitas_h1.groupby(['idLegislatura', 'sexo']).size().to_frame('valorAbsoluto')
```
<div class="alert-warning">Estabelece el porcentaje de cada grupo en relación al total de diputados</div>
```
agrupa_sexo['porcentagem'] = round(agrupa_sexo['valorAbsoluto'].div(
agrupa_sexo.groupby('idLegislatura')['valorAbsoluto'].transform('sum')).mul(100), 2)
agrupa_sexo_df = agrupa_sexo.reset_index()
agrupa_sexo_df
```
<div class="alert-warning">Prepara los datos para visualización</div>
```
mulher_h1 = agrupa_sexo_df[agrupa_sexo_df['sexo'] == 'F']['porcentagem'].tolist()
homem_h1 = agrupa_sexo_df[agrupa_sexo_df['sexo'] == 'M']['porcentagem'].tolist()
legislaturas_lista_h1 = agrupa_sexo_df['idLegislatura'].unique()
legislaturas_lista_h1 = df_legislaturas[(df_legislaturas['id'] >= 50) &
(df_legislaturas['id'] <= 53)]['dataInicio'].unique().tolist()
legislaturas_lista_h1.sort()
legislaturas_lista_h1 = list(map(str, legislaturas_lista_h1))
legislaturas_lista_h1
agrupa_sexo_df2 = pd.DataFrame({'mulher': mulher_h1,
'homem': homem_h1
}, index=legislaturas_lista_h1,
)
agrupa_sexo_df2.plot.line()
agrupa_sexo_df2.to_csv('dados/analise_genero_1995_2007(1).csv')
```
<div class="alert-warning">Visualización por género</div>
```
agrupa_sexo_df2.plot.line(subplots=True)
diferenca_percentual_mulher_h1_total = mulher_h1[-1] - mulher_h1[0]
print(f'''
Hubo impacto positivo en la cantidad de mujeres elegidas para la Cámara en las 3 legislaciones subsecuentes a aprobación de la Ley 9.504/1997? \n
Hipótesis comprobada? Sí. \n
Hubo aumento de {round(diferenca_percentual_mulher_h1_total, 2)}% en el total de mujeres elegidas, sin embargo es un porcentaje muy bajo para justificar como impacto positivo.
''')
```
## 2. ¿Hubo impacto positivo en la cantidad de mujeres elegidas para la Cámara en las 3 legislaciones subsecuentes a aprobación de la Ley 12.034/2009?
**Hipótesis:** Huvo impacto positivo en el percentual de mujeres elegidas para la Cámara en las 3 legislaciones subsecuentes a aprobación de la Ley 12.034/2009.
<div class="alert-warning">Determinar el período de los datos para el análisis (2007 a 2019)</div>
```
legislaturas_h2 = df_legislaturas[(df_legislaturas['id'] >= 53) & (df_legislaturas['id'] <= 56)]['id'].unique().tolist()
df_candidaturas_eleitas_h2 = df_candidaturas_eleitas[df_candidaturas_eleitas['idLegislatura'].isin(legislaturas_h2)].copy()
df_candidaturas_eleitas_h2['idLegislatura'].unique()
```
<div class="alert-warning">Agrupar por género</div>
```
agrupa_sexo_h2 = df_candidaturas_eleitas_h2.groupby(['idLegislatura', 'sexo']).size().to_frame('valorAbsoluto')
```
<div class="alert-warning">Estabelece el porcentaje de cada grupo en relación al total de diputados</div>
```
agrupa_sexo_h2['porcentagem'] = round(agrupa_sexo_h2['valorAbsoluto'].div(agrupa_sexo_h2.groupby(
'idLegislatura')['valorAbsoluto'].transform('sum')).mul(100), 2)
agrupa_sexo_h2_df = agrupa_sexo_h2.reset_index()
agrupa_sexo_h2
```
<div class="alert-warning">Prepara los datos para visualización</div>
```
mulher_h2 = agrupa_sexo_h2_df[agrupa_sexo_h2_df['sexo'] == 'F']['porcentagem'].tolist()
homem_h2 = agrupa_sexo_h2_df[agrupa_sexo_h2_df['sexo'] == 'M']['porcentagem'].tolist()
legislaturas_lista_h2 = agrupa_sexo_h2_df['idLegislatura'].unique()
legislaturas_lista_h2 = df_legislaturas[(df_legislaturas['id'] >= 53) & (df_legislaturas['id'] <= 56)
]['dataInicio'].unique().tolist()
legislaturas_lista_h2.sort()
legislaturas_lista_h2 = list(map(str, legislaturas_lista_h2))
legislaturas_lista_h2
agrupa_sexo_h2_df2 = pd.DataFrame({'mulher': mulher_h2,
'homem': homem_h2
}, index=legislaturas_lista_h2,
)
agrupa_sexo_h2_df2.plot.line()
agrupa_sexo_h2_df2.to_csv('dados/analise_genero_2007_2019(1).csv')
```
<div class="alert-warning">Visualización por género</div>
```
agrupa_sexo_h2_df2.plot.line(subplots=True)
diferenca_percentual_mulher_h2_total = mulher_h2[-1] - mulher_h2[0]
print(f'''
Hubo impacto positivo en la cantidad de mujeres elegidas para la Cámara en las 3 legislaciones subsecuentes a aprobación de la Ley 12.034/2009? \n
Hipótesis comprobada? Sí. \n
Hubo aumento de {round(diferenca_percentual_mulher_h2_total, 2)}% en el total de mujeres elegidas.
''')
```
## Evolução geral
```
legislaturas_todas = df_candidaturas_eleitas['idLegislatura'].unique()
legislaturas_todas
```
<div class="alert-warning">Agrupar por género</div>
```
agrupa_sexo_todas = df_candidaturas_eleitas.groupby(['idLegislatura', 'sexo']).size().to_frame('valorAbsoluto')
```
<div class="alert-warning">Estabelece el porcentaje de cada grupo en relación al total de diputados</div>
```
agrupa_sexo_todas['porcentagem'] = round(agrupa_sexo_todas['valorAbsoluto'].div(agrupa_sexo_todas.groupby(
'idLegislatura')['valorAbsoluto'].transform('sum')).mul(100), 2)
agrupa_sexo_todas_df = agrupa_sexo_todas.reset_index()
agrupa_sexo_todas_df
```
<div class="alert-warning">Prepara los datos para visualización</div>
```
mulher_todas = agrupa_sexo_todas_df[agrupa_sexo_todas_df['sexo'] == 'F']['porcentagem'].tolist()
homem_todas = agrupa_sexo_todas_df[agrupa_sexo_todas_df['sexo'] == 'M']['porcentagem'].tolist()
len(mulher_todas), len(homem_todas)
mulher_todas[:5]
mulher_todas.insert(2, 0)
len(mulher_todas), len(homem_todas)
mulher_todas[:5]
legislaturas_lista_todas = agrupa_sexo_todas_df['idLegislatura'].unique()
legislaturas_lista_todas = df_legislaturas['dataInicio'].unique().tolist()
legislaturas_lista_todas.sort()
legislaturas_lista_todas = list(map(str, legislaturas_lista_todas))
len(legislaturas_lista_todas), len(mulher_todas), len(homem_todas)
agrupa_sexo_todas_df2 = pd.DataFrame({'mulher': mulher_todas,
'homem': homem_todas
}, index=legislaturas_lista_todas,
)
agrupa_sexo_todas_df2.plot.line()
agrupa_sexo_todas_df2.to_csv('dados/analise_genero_1934_2023.csv')
```
<div class="alert-warning">Visualización por género</div>
```
agrupa_sexo_h2_df2.plot.line(subplots=True)
```
## 3. ¿Teniendo en cuenta el tema de las proposiciones legislativas, hubo aumento de los que beneficia grupos históricamente marginados en el periodo entre 1934 y 2021?
**Hipótesis:** Sí, hubo aumento en la cantidade anual de propuestas legislativas que beneficia los grupos historicamente marginados.
```
proposicoes = pd.read_csv('dados/proposicoes_legislativas_limpas_classificadas(1).csv')
proposicoes.head()
```
<div class="alert-warning">Agrupa por año y cantidad de propuestas de los temas</div>
```
proposicoes_anuais = proposicoes[['ano', 'temas_classificador']].groupby(by=['ano']).count()
proposicoes_anuais.tail(10)
```
<div class="alert-warning">Visualización</div>
```
proposicoes_anuais.plot.line()
proposicoes_anuais = proposicoes_anuais.reset_index()
proposicoes_anuais.to_csv('dados/proposicoes_anuais(1).csv', index=False)
print(f'''
Teniendo en cuenta el tema de las proposiciones legislativas, hubo aumento de los que beneficia grupos historicamente marginalinados en el periodo entre 1934 e 2021? \n
Hipótesis comprobada? Sí.
Apesar de las oscilaciones hay una tendencia de crecimiento positivo en la cantidad de propuestas que benefician los grupos historicamente marginados.
''')
```
## 4. ¿Cuál es el coeficiente de correlación entre la cantidad anual de las propuestas legislativas que benefician los grupos historicamente marginados y el porcentaje de mujeres elegidas para la Cámara de Diputados entre 1995 y 2019?
**Hipótesis:** Bajo
<div class="alert-warning">Une los dataframes de los análisis anteriores</div>
```
analise_genero_1995_2007 = pd.read_csv('dados/analise_genero_1995_2007(1).csv')
analise_genero_2007_2019 = pd.read_csv('dados/analise_genero_2007_2019(1).csv')
analise_genero_1995_2007.columns == analise_genero_2007_2019.columns
analise_genero_1995_2019 = pd.concat([analise_genero_1995_2007, analise_genero_2007_2019], ignore_index=True)
analise_genero_1995_2019
analise_genero_1995_2019.rename(columns={'Unnamed: 0': 'ano'}, inplace=True)
analise_genero_1995_2019.drop(index=3, inplace=True)
analise_genero_1995_2019
anos = analise_genero_1995_2019['ano'].tolist()
anos.append(2021)
anos
```
<div class="alert-warning">Inserta el período completo de cada legislatura, teniendo en cuenta que la proporcionalidade de género se mantiene durante los 4 años de legislatura</div>
```
for ano in anos:
mulher_percentual = analise_genero_1995_2019['mulher'][analise_genero_1995_2019['ano'] == ano].item()
homem_percentual = analise_genero_1995_2019['homem'][analise_genero_1995_2019['ano'] == ano].item()
if ano < 2021:
dados = pd.DataFrame(data={
'ano': [ano+1, ano+2, ano+3],
'mulher': [mulher_percentual, mulher_percentual, mulher_percentual],
'homem': [homem_percentual, homem_percentual, homem_percentual]}
)
analise_genero_1995_2019 = pd.concat([analise_genero_1995_2019, dados])
analise_genero_1995_2019.sort_values(by=['ano'], inplace=True)
analise_genero_1995_2019.reset_index(drop=True, inplace=True)
analise_genero_1995_2019.tail()
analise_genero_1995_2019.drop(index=27, inplace=True)
```
<div class="alert-warning">Inserta el total anual de las propuestas en favor a los grupos historicamente marginados</div>
```
def insere_qnt_propostas(ano_candidaturas_eleitas):
for ano, qnt_tema in zip(proposicoes_anuais['ano'], proposicoes_anuais['temas_classificador']):
if ano == ano_candidaturas_eleitas:
return qnt_tema
analise_genero_1995_2019['qnt_proposicoes'] = analise_genero_1995_2019['ano'].apply(insere_qnt_propostas)
analise_genero_1995_2019.head(10)
```
<div class="alert-warning">Cría la matriz de correlación</div>
```
correlacao = analise_genero_1995_2019[['mulher', 'homem', 'qnt_proposicoes']].corr(method='pearson')
coeficiente_correlacao_mulher_qnt_temas = round(correlacao['mulher']['qnt_proposicoes'],2)
correlacao_matriz_triangular = np.triu(np.ones_like(correlacao))
sns.heatmap(correlacao, annot=True, mask=correlacao_matriz_triangular)
correlacao.to_csv('dados/coeficiente_correlacao_mulher_qnt_temas(1).csv')
print(f'''¿Cuál es el coeficiente de correlación entre la cantidad anual de las propuestas legislativas que benefician los grupos historicamente marginados y el porcentaje de mujeres elegidas para la
Cámara de Diputados entre 1995 y 2019?\n
Hipótesis comprobada? Sí. \n
- El coeficiente de correlación de Pearson es {coeficiente_correlacao_mulher_qnt_temas}, por lo tanto no se puede afirmar que hay correlación.
''')
```
|
github_jupyter
|
```
import torch
from dataset import load_dataset
from basic_unet import UNet
import matplotlib.pyplot as plt
from rise import RISE
from pathlib import Path
from plot_utils import plot_image_row
from skimage.feature import canny
batch_size = 1
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
train_loader, test_loader = load_dataset(batch_size)
model = UNet(in_channels=4, out_channels=1)
state_dict = torch.load('models/3_basic_unet_flat_criterion_279_0.00000.pth')
model.load_state_dict(state_dict)
model = model.to(device)
sample = next(iter(test_loader))
segment = sample['segment']
segment = segment.squeeze()
image = sample['input'].to(device)
output = model(image)
output = output.detach().cpu().squeeze().numpy()
output = (output > output.mean())
class SegmentationRISE(RISE):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
def forward(self, x):
mask_count = self.N
_, _, H, W = x.size()
# generate new images by putting mask on top of original image
stack = torch.mul(self.masks, x.data)
output = model(x).squeeze()
output = (output > output.mean())
pixels = []
for x in range(output.shape[0]):
for y in range(output.shape[1]):
if output[x][y]:
pixels.append((x, y))
pixels_per_batch = 1000
saliencies = []
for i in range(0, len(pixels), pixels_per_batch):
current_pixels = pixels[i:i+pixels_per_batch]
# run generated images through the model
p = []
for i in range(0, mask_count, self.gpu_batch):
output_mask = self.model(stack[i:min(i + self.gpu_batch, mask_count)])
pixel_classes = []
for x, y in current_pixels:
pixel_classes.append(output_mask[0][x][y])
p.append(torch.tensor([pixel_classes]))
p = torch.cat(p)
p = p.to(device)
# Number of classes
CL = p.size(1)
sal = torch.matmul(p.data.transpose(0, 1), self.masks.view(mask_count, H * W))
sal = sal.view((CL, H, W))
sal /= mask_count
saliencies.append(sal)
return saliencies
masks_path = Path('rise_masks.npy')
explainer = SegmentationRISE(model, (240, 240), batch_size)
if not masks_path.exists():
explainer.generate_masks(N=3000, s=8, p1=0.1, savepath=masks_path)
else:
explainer.load_masks(masks_path)
saliencies = None
with torch.set_grad_enabled(False):
saliencies = explainer(image)
plot_image_row([segment, output], labels=['Ground truth', 'Binarized network output'])
print('Saliency map, Saliency map overlayed on binarized network output (max)')
merged = torch.cat(saliencies)
maxed = torch.max(merged, dim=0)[0]
plt.imshow(output, cmap='gray_r')
edges = canny(image.cpu().numpy()[0][1], sigma=0.01)
plt.imshow(edges, alpha=0.5, cmap='gray_r')
plt.imshow(maxed.cpu(), cmap='jet', alpha=0.6)
plt.show()
plt.imshow(output, cmap='gray_r')
plt.imshow(maxed.cpu(), cmap='jet', alpha=0.6)
plt.show()
print('Saliency map, Saliency map overlayed on binarized network output (mean)')
mean = torch.mean(merged, dim=0)
plt.imshow(output, cmap='gray_r')
plt.imshow(edges, alpha=0.5, cmap='gray_r')
plt.imshow(mean.cpu(), cmap='jet', alpha=0.6)
plt.show()
plt.imshow(output, cmap='gray_r')
plt.imshow(mean.cpu(), cmap='jet', alpha=0.6)
plt.show()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/NielsRogge/Transformers-Tutorials/blob/master/TrOCR/Evaluating_TrOCR_base_handwritten_on_the_IAM_test_set.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Set-up environment
```
!pip install -q git+https://github.com/huggingface/transformers.git
!pip install -q datasets jiwer
```
## Load IAM test set
```
import pandas as pd
df = pd.read_fwf('/content/drive/MyDrive/TrOCR/Tutorial notebooks/IAM/gt_test.txt', header=None)
df.rename(columns={0: "file_name", 1: "text"}, inplace=True)
del df[2]
df.head()
import torch
from torch.utils.data import Dataset
from PIL import Image
class IAMDataset(Dataset):
def __init__(self, root_dir, df, processor, max_target_length=128):
self.root_dir = root_dir
self.df = df
self.processor = processor
self.max_target_length = max_target_length
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
# get file name + text
file_name = self.df['file_name'][idx]
text = self.df['text'][idx]
# some file names end with jp instead of jpg, the two lines below fix this
if file_name.endswith('jp'):
file_name = file_name + 'g'
# prepare image (i.e. resize + normalize)
image = Image.open(self.root_dir + file_name).convert("RGB")
pixel_values = self.processor(image, return_tensors="pt").pixel_values
# add labels (input_ids) by encoding the text
labels = self.processor.tokenizer(text,
padding="max_length",
max_length=self.max_target_length).input_ids
# important: make sure that PAD tokens are ignored by the loss function
labels = [label if label != self.processor.tokenizer.pad_token_id else -100 for label in labels]
encoding = {"pixel_values": pixel_values.squeeze(), "labels": torch.tensor(labels)}
return encoding
from transformers import TrOCRProcessor
processor = TrOCRProcessor.from_pretrained("microsoft/trocr-base-handwritten")
test_dataset = IAMDataset(root_dir='/content/drive/MyDrive/TrOCR/Tutorial notebooks/IAM/image/',
df=df,
processor=processor)
from torch.utils.data import DataLoader
test_dataloader = DataLoader(test_dataset, batch_size=8)
batch = next(iter(test_dataloader))
for k,v in batch.items():
print(k, v.shape)
from transformers import TrOCRProcessor
processor = TrOCRProcessor.from_pretrained("microsoft/trocr-base-handwritten")
labels = batch["labels"]
labels[labels == -100] = processor.tokenizer.pad_token_id
label_str = processor.batch_decode(labels, skip_special_tokens=True)
label_str
```
## Run evaluation
```
from transformers import VisionEncoderDecoderModel
import torch
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = VisionEncoderDecoderModel.from_pretrained("microsoft/trocr-base-handwritten")
model.to(device)
from datasets import load_metric
cer = load_metric("cer")
from tqdm.notebook import tqdm
print("Running evaluation...")
for batch in tqdm(test_dataloader):
# predict using generate
pixel_values = batch["pixel_values"].to(device)
outputs = model.generate(pixel_values)
# decode
pred_str = processor.batch_decode(outputs, skip_special_tokens=True)
labels = batch["labels"]
labels[labels == -100] = processor.tokenizer.pad_token_id
label_str = processor.batch_decode(labels, skip_special_tokens=True)
# add batch to metric
cer.add_batch(predictions=pred_str, references=label_str)
final_score = cer.compute()
print("Character error rate on test set:", final_score)
```
|
github_jupyter
|
```
import os, sys
import torch
from transformers import BertModel, BertConfig
from greenformer import auto_fact
from itertools import chain
from os import path
import sys
def count_param(module, trainable=False):
if trainable:
return sum(p.numel() for p in module.parameters() if p.requires_grad)
else:
return sum(p.numel() for p in module.parameters())
```
# Init Model
```
config = BertConfig.from_pretrained('bert-base-uncased')
model = BertModel(config=config)
model = BertModel.from_pretrained('bert-base-uncased')
count_param(model)
```
# Factorize Model
### Apply absolute rank
```
%%time
fact_model = auto_fact(model, rank=256, deepcopy=True, solver='random', num_iter=20)
count_param(fact_model)
%%time
fact_model = auto_fact(model, rank=256, deepcopy=True, solver='svd', num_iter=20)
count_param(fact_model)
%%time
fact_model = auto_fact(model, rank=256, deepcopy=True, solver='snmf', num_iter=20)
count_param(fact_model)
```
### Apply percentage rank
```
%%time
fact_model = auto_fact(model, rank=0.4, deepcopy=True, solver='random', num_iter=20)
count_param(fact_model)
%%time
fact_model = auto_fact(model, rank=0.4, deepcopy=True, solver='svd', num_iter=20)
count_param(fact_model)
%%time
fact_model = auto_fact(model, rank=0.4, deepcopy=True, solver='snmf', num_iter=20)
count_param(fact_model)
```
### Apply factorization only on specific modules
```
# Only factorize last 6 transformer layers and the pooler layer of the model
factorizable_submodules = list(model.encoder.layer[6:]) + [model.pooler]
%%time
fact_model = auto_fact(model, rank=0.2, deepcopy=True, solver='random', num_iter=20, submodules=factorizable_submodules)
count_param(fact_model)
%%time
fact_model = auto_fact(model, rank=0.2, deepcopy=True, solver='svd', num_iter=20, submodules=factorizable_submodules)
count_param(fact_model)
%%time
fact_model = auto_fact(model, rank=0.2, deepcopy=True, solver='snmf', num_iter=20, submodules=factorizable_submodules)
count_param(fact_model)
```
# Speed test on CPU
### Test Inference CPU
```
%%timeit
with torch.no_grad():
y = model(torch.zeros(32,256, dtype=torch.long))
%%timeit
with torch.no_grad():
y = fact_model(torch.zeros(32,256, dtype=torch.long))
```
### Test Forward-Backward CPU
```
%%timeit
y = model(torch.zeros(8,256, dtype=torch.long))
y.last_hidden_state.sum().backward()
%%timeit
y = fact_model(torch.zeros(8,256, dtype=torch.long))
y.last_hidden_state.sum().backward()
```
# Speed test on GPU
### Move models to GPU
```
model = model.cuda()
fact_model = fact_model.cuda()
```
### Test Inference GPU
```
x = torch.zeros(64,256, dtype=torch.long).cuda()
%%timeit
with torch.no_grad():
y = model(x)
%%timeit
with torch.no_grad():
y = fact_model(x)
```
### Test Forward-Backward GPU
```
x = torch.zeros(16,256, dtype=torch.long).cuda()
%%timeit
y = model(x)
y.last_hidden_state.sum().backward()
%%timeit
y = fact_model(x)
y.last_hidden_state.sum().backward()
```
|
github_jupyter
|
# Implementing a one-layer Neural Network
We will illustrate how to create a one hidden layer NN
We will use the iris data for this exercise
We will build a one-hidden layer neural network to predict the fourth attribute, Petal Width from the other three (Sepal length, Sepal width, Petal length).
```
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn import datasets
from tensorflow.python.framework import ops
ops.reset_default_graph()
iris = datasets.load_iris()
x_vals = np.array([x[0:3] for x in iris.data])
y_vals = np.array([x[3] for x in iris.data])
# Create graph session
sess = tf.Session()
# make results reproducible
seed = 2
tf.set_random_seed(seed)
np.random.seed(seed)
# Split data into train/test = 80%/20%
train_indices = np.random.choice(len(x_vals), round(len(x_vals)*0.8), replace=False)
test_indices = np.array(list(set(range(len(x_vals))) - set(train_indices)))
x_vals_train = x_vals[train_indices]
x_vals_test = x_vals[test_indices]
y_vals_train = y_vals[train_indices]
y_vals_test = y_vals[test_indices]
# Normalize by column (min-max norm)
def normalize_cols(m):
col_max = m.max(axis=0)
col_min = m.min(axis=0)
return (m-col_min) / (col_max - col_min)
x_vals_train = np.nan_to_num(normalize_cols(x_vals_train))
x_vals_test = np.nan_to_num(normalize_cols(x_vals_test))
# Declare batch size
batch_size = 50
# Initialize placeholders
x_data = tf.placeholder(shape=[None, 3], dtype=tf.float32)
y_target = tf.placeholder(shape=[None, 1], dtype=tf.float32)
# Create variables for both NN layers
hidden_layer_nodes = 10
A1 = tf.Variable(tf.random_normal(shape=[3,hidden_layer_nodes])) # inputs -> hidden nodes
b1 = tf.Variable(tf.random_normal(shape=[hidden_layer_nodes])) # one biases for each hidden node
A2 = tf.Variable(tf.random_normal(shape=[hidden_layer_nodes,1])) # hidden inputs -> 1 output
b2 = tf.Variable(tf.random_normal(shape=[1])) # 1 bias for the output
# Declare model operations
hidden_output = tf.nn.relu(tf.add(tf.matmul(x_data, A1), b1))
final_output = tf.nn.relu(tf.add(tf.matmul(hidden_output, A2), b2))
# Declare loss function (MSE)
loss = tf.reduce_mean(tf.square(y_target - final_output))
# Declare optimizer
my_opt = tf.train.GradientDescentOptimizer(0.005)
train_step = my_opt.minimize(loss)
# Initialize variables
init = tf.global_variables_initializer()
sess.run(init)
# Training loop
loss_vec = []
test_loss = []
for i in range(500):
rand_index = np.random.choice(len(x_vals_train), size=batch_size)
rand_x = x_vals_train[rand_index]
rand_y = np.transpose([y_vals_train[rand_index]])
sess.run(train_step, feed_dict={x_data: rand_x, y_target: rand_y})
temp_loss = sess.run(loss, feed_dict={x_data: rand_x, y_target: rand_y})
loss_vec.append(np.sqrt(temp_loss))
test_temp_loss = sess.run(loss, feed_dict={x_data: x_vals_test, y_target: np.transpose([y_vals_test])})
test_loss.append(np.sqrt(test_temp_loss))
if (i+1)%50==0:
print('Generation: ' + str(i+1) + '. Loss = ' + str(temp_loss))
%matplotlib inline
# Plot loss (MSE) over time
plt.plot(loss_vec, 'k-', label='Train Loss')
plt.plot(test_loss, 'r--', label='Test Loss')
plt.title('Loss (MSE) per Generation')
plt.legend(loc='upper right')
plt.xlabel('Generation')
plt.ylabel('Loss')
plt.show()
```
|
github_jupyter
|
# From batch to online
## A quick overview of batch learning
If you've already delved into machine learning, then you shouldn't have any difficulty in getting to use incremental learning. If you are somewhat new to machine learning, then do not worry! The point of this notebook in particular is to introduce simple notions. We'll also start to show how `creme` fits in and explain how to use it.
The whole point of machine learning is to *learn from data*. In *supervised learning* you want to learn how to predict a target $y$ given a set of features $X$. Meanwhile in an unsupervised learning there is no target, and the goal is rather to identify patterns and trends in the features $X$. At this point most people tend to imagine $X$ as a somewhat big table where each row is an observation and each column is a feature, and they would be quite right. Learning from tabular data is part of what's called *batch learning*, which basically that all of the data is available to our learning algorithm at once. A lot of libraries have been created to handle the batch learning regime, with one of the most prominent being Python's [scikit-learn](https://scikit-learn.org/stable/).
As a simple example of batch learning let's say we want to learn to predict if a women has breast cancer or not. We'll use the [breast cancer dataset available with scikit-learn](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_breast_cancer().html). We'll learn to map a set of features to a binary decision using a [logistic regression](https://www.wikiwand.com/en/Logistic_regression). Like many other models based on numerical weights, logisitc regression is sensitive to the scale of the features. Rescaling the data so that each feature has mean 0 and variance 1 is generally considered good practice. We can apply the rescaling and fit the logistic regression sequentially in an elegant manner using a [Pipeline](https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html). To measure the performance of the model we'll evaluate the average [ROC AUC score](https://www.wikiwand.com/en/Receiver_operating_characteristic) using a 5 fold [cross-validation](https://www.wikiwand.com/en/Cross-validation_(statistics)).
```
from sklearn import datasets
from sklearn import linear_model
from sklearn import metrics
from sklearn import model_selection
from sklearn import pipeline
from sklearn import preprocessing
# Load the data
dataset = datasets.load_breast_cancer()
X, y = dataset.data, dataset.target
# Define the steps of the model
model = pipeline.Pipeline([
('scale', preprocessing.StandardScaler()),
('lin_reg', linear_model.LogisticRegression(solver='lbfgs'))
])
# Define a determistic cross-validation procedure
cv = model_selection.KFold(n_splits=5, shuffle=True, random_state=42)
# Compute the MSE values
scorer = metrics.make_scorer(metrics.roc_auc_score)
scores = model_selection.cross_val_score(model, X, y, scoring=scorer, cv=cv)
# Display the average score and it's standard deviation
print(f'ROC AUC: {scores.mean():.3f} (± {scores.std():.3f})')
```
This might be a lot to take in if you're not accustomed to scikit-learn, but it probably isn't if you are. Batch learning basically boils down to:
1. Loading the data
2. Fitting a model to the data
3. Computing the performance of the model on unseen data
This is pretty standard and is maybe how most people imagine a machine learning pipeline. However this way of proceding has certain downsides. First of all your laptop would crash if the `load_boston` function returned a dataset who's size exceeds your available amount of RAM. Sometimes you can use some tricks to get around this. For example by optimizing the data types and by using sparse representations when applicable you can potentially save precious gigabytes of RAM. However like many tricks this only goes so far. If your dataset weighs hundreds of gigabytes then you won't go far without some special hardware. One solution is to do out-of-core learning; that is, algorithms that can learning by being presented the data in chunks. If you want to go down this road then take a look at [Dask](https://examples.dask.org/machine-learning.html) and [Spark's MLlib](https://spark.apache.org/mllib/).
Another issue with the batch learning regime is that can't elegantly learn from new data. Indeed if new data is made available, then the model has to learn from scratch with a new dataset composed of the old data and the new data. This is particularly annoying in a real situation where you might have new incoming data every week, day, hour, minute, or even setting. For example if you're building a recommendation engine for an e-commerce app, then you're probably training your model from 0 every week or so. As your app grows in popularity, so does the dataset you're training on. This will lead to longer and longer training times and might require a hardware upgrade.
A final downside that isn't very easy to grasp concerns the manner in which features are extracted. Everytime you want to train your model you first have to extract features. The trick is that some features might not be accessible at the particular point in time you are at. For example maybe that some attributes in your data warehouse get overwritten with time. In other words maybe that all the features pertaining to a particular observations are not available, whereas they were a week ago. This happens more often than not in real scenarios, and apart if you have a sophisticated data engineering pipeline then you will encounter these issues at some point.
## A hands-on introduction to incremental learning
Incremental learning is also often called *online learning*, but if you [google online learning](https://www.google.com/search?q=online+learning) a lot of the results will point to educational websites. Hence we prefer the name "incremental learning", from which `creme` derives it's name. The point of incremental learning is to fit a model to a stream of data. In other words, the data isn't available in it's entirety, but rather the observations are provided one by one. As an example let's stream through the dataset used previously.
```
for xi, yi in zip(X, y):
# This where the model learns
pass
```
In this case we're iterating over a dataset that is already in memory, but we could just as well stream from a CSV file, a Kafka stream, an SQL query, etc. If we look at `x` we can notice that it is a `numpy.ndarray`.
```
xi
```
`creme` on the other hand works with `dict`s. We believe that `dict`s are more enjoyable to program with than `numpy.ndarray`s, at least for when single observations are concerned. `dict`'s bring the added benefit that each feature can be accessed by name rather than by position.
```
for xi, yi in zip(X, y):
xi = dict(zip(dataset.feature_names, xi))
pass
xi
```
`creme`'s `stream` module has an `iter_sklearn_dataset` convenience function that we can use instead.
```
from creme import stream
for xi, yi in stream.iter_sklearn_dataset(datasets.load_breast_cancer()):
pass
```
The simple fact that we are getting the data in a stream means that we can't do a lot of things the same way as in a batch setting. For example let's say we want to scale the data so that it has mean 0 and variance 1, as we did earlier. To do so we simply have to subtract the mean of each feature to each value and then divide the result by the standard deviation of the feature. The problem is that we can't possible known the values of the mean and the standard deviation before actually going through all the data! One way to procede would be to do a first pass over the data to compute the necessary values and then scale the values during a second pass. The problem is that defeats our purpose, which is to learn by only looking at the data once. Although this might seem rather restrictive, it reaps sizable benefits down the road.
The way we do feature scaling in `creme` involves computing *running statistics*. The idea is that we use a data structure that estimates the mean and updates itself when it is provided with a value. The same goes for the variance (and thus the standard deviation). For example, if we denote $\mu_t$ the mean and $n_t$ the count at any moment $t$, then updating the mean can be done as so:
$$
\begin{cases}
n_{t+1} = n_t + 1 \\
\mu_{t+1} = \mu_t + \frac{x - \mu_t}{n_{t+1}}
\end{cases}
$$
Likewhise a running variance can be computed as so:
$$
\begin{cases}
n_{t+1} = n_t + 1 \\
\mu_{t+1} = \mu_t + \frac{x - \mu_t}{n_{t+1}} \\
s_{t+1} = s_t + (x - \mu_t) \times (x - \mu_{t+1}) \\
\sigma_{t+1} = \frac{s_{t+1}}{n_{t+1}}
\end{cases}
$$
where $s_t$ is a running sum of squares and $\sigma_t$ is the running variance at time $t$. This might seem a tad more involved than the batch algorithms you learn in school, but it is rather elegant. Implementing this in Python is not too difficult. For example let's compute the running mean and variance of the `'mean area'` variable.
```
n, mean, sum_of_squares, variance = 0, 0, 0, 0
for xi, yi in stream.iter_sklearn_dataset(datasets.load_breast_cancer()):
n += 1
old_mean = mean
mean += (xi['mean area'] - mean) / n
sum_of_squares += (xi['mean area'] - old_mean) * (xi['mean area'] - mean)
variance = sum_of_squares / n
print(f'Running mean: {mean:.3f}')
print(f'Running variance: {variance:.3f}')
```
Let's compare this with `numpy`.
```
import numpy as np
i = list(dataset.feature_names).index('mean area')
print(f'True mean: {np.mean(X[:, i]):.3f}')
print(f'True variance: {np.var(X[:, i]):.3f}')
```
The results seem to be exactly the same! The twist is that the running statistics won't be very accurate for the first few observations. In general though this doesn't matter too much. Some would even go as far as to say that this descrepancy is beneficial and acts as some sort of regularization...
Now the idea is that we can compute the running statistics of each feature and scale them as they come along. The way to do this with `creme` is to use the `StandardScaler` class from the `preprocessing` module, as so:
```
from creme import preprocessing
scaler = preprocessing.StandardScaler()
for xi, yi in stream.iter_sklearn_dataset(datasets.load_breast_cancer()):
xi = scaler.fit_one(xi)
```
This is quite terse but let's break it down nonetheless. Every class in `creme` has a `fit_one(x, y)` method where all the magic happens. Now the important thing to notice is that the `fit_one` actually returns the output for the given input. This is one of the nice properties of online learning: inference can be done immediatly. In `creme` each call to a `Transformer`'s `fit_one` will return the transformed output. Meanwhile calling `fit_one` with a `Classifier` or a `Regressor` will return the predicted target for the given set of features. The twist is that the prediction is made *before* looking at the true target `y`. This means that we get a free hold-out prediction every time we call `fit_one`. This can be used to monitor the performance of the model as it trains, which is obviously nice to have.
Now that we are scaling the data, we can start doing some actual machine learning. We're going to implement an online linear regression. Because all the data isn't available at once, we are obliged to do what is called *stochastic gradient descent*, which is a popular research topic and has a lot of variants. SGD is commonly used to train neural networks. The idea is that at each step we compute the loss between the target prediction and the truth. We then calculate the gradient, which is simply a set of derivatives with respect to each weight from the linear regression. Once we have obtained the gradient, we can update the weights by moving them in the opposite direction of the gradient. The amount by which the weights are moved typically depends on a *learning rate*, which is typically set by the user. Different optimizers have different ways of managing the weight update, and some handle the learning rate implicitely. Online linear regression can be done in `creme` with the `LinearRegression` class from the `linear_model` module. We'll be using plain and simple SGD using the `SGD` optimizer from the `optim` module. During training we'll measure the squared error between the truth and the predictions.
```
from creme import linear_model
from creme import optim
scaler = preprocessing.StandardScaler()
optimizer = optim.SGD(lr=0.01)
log_reg = linear_model.LogisticRegression(optimizer)
y_true = []
y_pred = []
for xi, yi in stream.iter_sklearn_dataset(datasets.load_breast_cancer(), shuffle=True, seed=42):
# Scale the features
xi_scaled = scaler.fit_one(xi).transform_one(xi)
# Fit the linear regression
yi_pred = log_reg.predict_proba_one(xi_scaled)
log_reg.fit_one(xi_scaled, yi)
# Store the truth and the prediction
y_true.append(yi)
y_pred.append(yi_pred[True])
print(f'ROC AUC: {metrics.roc_auc_score(y_true, y_pred):.3f}')
```
The ROC AUC is significantly better than the one obtained from the cross-validation of scikit-learn's logisitic regression. However to make things really comparable it would be nice to compare with the same cross-validation procedure. `creme` has a `compat` module that contains utilities for making `creme` compatible with other Python libraries. Because we're doing regression we'll be using the `SKLRegressorWrapper`. We'll also be using `Pipeline` to encapsulate the logic of the `StandardScaler` and the `LogisticRegression` in one single object.
```
from creme import compat
from creme import compose
# We define a Pipeline, exactly like we did earlier for sklearn
model = compose.Pipeline(
('scale', preprocessing.StandardScaler()),
('log_reg', linear_model.LogisticRegression())
)
# We make the Pipeline compatible with sklearn
model = compat.convert_creme_to_sklearn(model)
# We compute the CV scores using the same CV scheme and the same scoring
scores = model_selection.cross_val_score(model, X, y, scoring=scorer, cv=cv)
# Display the average score and it's standard deviation
print(f'ROC AUC: {scores.mean():.3f} (± {scores.std():.3f})')
```
This time the ROC AUC score is lower, which is what we would expect. Indeed online learning isn't as accurate as batch learning. However it all depends in what you're interested in. If you're only interested in predicting the next observation then the online learning regime would be better. That's why it's a bit hard to compare both approaches: they're both suited to different scenarios.
## Going further
There's a lot more to learn, and it all depends on what kind on your use case. Feel free to have a look at the [documentation](https://creme-ml.github.io/) to know what `creme` has available, and have a look the [example notebook](https://github.com/creme-ml/notebooks).
Here a few resources if you want to do some reading:
- [Online learning -- Wikipedia](https://www.wikiwand.com/en/Online_machine_learning)
- [What is online machine learning? -- Max Pagels](https://medium.com/value-stream-design/online-machine-learning-515556ff72c5)
- [Introduction to Online Learning -- USC course](http://www-bcf.usc.edu/~haipengl/courses/CSCI699/)
- [Online Methods in Machine Learning -- MIT course](http://www.mit.edu/~rakhlin/6.883/)
- [Online Learning: A Comprehensive Survey](https://arxiv.org/pdf/1802.02871.pdf)
- [Streaming 101: The world beyond batch](https://www.oreilly.com/ideas/the-world-beyond-batch-streaming-101)
- [Machine learning for data streams](https://www.cms.waikato.ac.nz/~abifet/book/contents.html)
- [Data Stream Mining: A Practical Approach](https://www.cs.waikato.ac.nz/~abifet/MOA/StreamMining.pdf)
|
github_jupyter
|
#### Copyright 2017 Google LLC.
```
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Creating and Manipulating Tensors
**Learning Objectives:**
* Initialize and assign TensorFlow `Variable`s
* Create and manipulate tensors
* Refresh your memory about addition and multiplication in linear algebra (consult an introduction to matrix [addition](https://en.wikipedia.org/wiki/Matrix_addition) and [multiplication](https://en.wikipedia.org/wiki/Matrix_multiplication) if these topics are new to you)
* Familiarize yourself with basic TensorFlow math and array operations
```
from __future__ import print_function
import tensorflow as tf
try:
tf.contrib.eager.enable_eager_execution()
print("TF imported with eager execution!")
except ValueError:
print("TF already imported with eager execution!")
```
## Vector Addition
You can perform many typical mathematical operations on tensors ([TF API](https://www.tensorflow.org/api_guides/python/math_ops)). The code below creates the following vectors (1-D tensors), all having exactly six elements:
* A `primes` vector containing prime numbers.
* A `ones` vector containing all `1` values.
* A vector created by performing element-wise addition over the first two vectors.
* A vector created by doubling the elements in the `primes` vector.
```
primes = tf.constant([2, 3, 5, 7, 11, 13], dtype=tf.int32)
print("primes:", primes)
ones = tf.ones([6], dtype=tf.int32)
print("ones:", ones)
just_beyond_primes = tf.add(primes, ones)
print("just_beyond_primes:", just_beyond_primes)
twos = tf.constant([2, 2, 2, 2, 2, 2], dtype=tf.int32)
primes_doubled = primes * twos
print("primes_doubled:", primes_doubled)
```
Printing a tensor returns not only its **value**, but also its **shape** (discussed in the next section) and the **type of value stored** in the tensor. Calling the `numpy` method of a tensor returns the value of the tensor as a numpy array:
```
some_matrix = tf.constant([[1, 2, 3], [4, 5, 6]], dtype=tf.int32)
print(some_matrix)
print("\nvalue of some_matrix is:\n", some_matrix.numpy())
```
### Tensor Shapes
Shapes are used to characterize the size and number of dimensions of a tensor. The shape of a tensor is expressed as `list`, with the `i`th element representing the size along dimension `i`. The length of the list then indicates the rank of the tensor (i.e., the number of dimensions).
For more information, see the [TensorFlow documentation](https://www.tensorflow.org/programmers_guide/tensors#shape).
A few basic examples:
```
# A scalar (0-D tensor).
scalar = tf.zeros([])
# A vector with 3 elements.
vector = tf.zeros([3])
# A matrix with 2 rows and 3 columns.
matrix = tf.zeros([2, 3])
print('scalar has shape', scalar.get_shape(), 'and value:\n', scalar.numpy())
print('vector has shape', vector.get_shape(), 'and value:\n', vector.numpy())
print('matrix has shape', matrix.get_shape(), 'and value:\n', matrix.numpy())
```
### Broadcasting
In mathematics, you can only perform element-wise operations (e.g. *add* and *equals*) on tensors of the same shape. In TensorFlow, however, you may perform operations on tensors that would traditionally have been incompatible. TensorFlow supports **broadcasting** (a concept borrowed from numpy), where the smaller array in an element-wise operation is enlarged to have the same shape as the larger array. For example, via broadcasting:
* If an operand requires a size `[6]` tensor, a size `[1]` or a size `[]` tensor can serve as an operand.
* If an operation requires a size `[4, 6]` tensor, any of the following sizes can serve as an operand:
* `[1, 6]`
* `[6]`
* `[]`
* If an operation requires a size `[3, 5, 6]` tensor, any of the following sizes can serve as an operand:
* `[1, 5, 6]`
* `[3, 1, 6]`
* `[3, 5, 1]`
* `[1, 1, 1]`
* `[5, 6]`
* `[1, 6]`
* `[6]`
* `[1]`
* `[]`
**NOTE:** When a tensor is broadcast, its entries are conceptually **copied**. (They are not actually copied for performance reasons. Broadcasting was invented as a performance optimization.)
The full broadcasting ruleset is well described in the easy-to-read [numpy broadcasting documentation](http://docs.scipy.org/doc/numpy-1.10.1/user/basics.broadcasting.html).
The following code performs the same tensor arithmetic as before, but instead uses scalar values (instead of vectors containing all `1`s or all `2`s) and broadcasting.
```
primes = tf.constant([2, 3, 5, 7, 11, 13], dtype=tf.int32)
print("primes:", primes)
one = tf.constant(1, dtype=tf.int32)
print("one:", one)
just_beyond_primes = tf.add(primes, one)
print("just_beyond_primes:", just_beyond_primes)
two = tf.constant(2, dtype=tf.int32)
primes_doubled = primes * two
print("primes_doubled:", primes_doubled)
```
### Exercise #1: Arithmetic over vectors.
Perform vector arithmetic to create a "just_under_primes_squared" vector, where the `i`th element is equal to the `i`th element in `primes` squared, minus 1. For example, the second element would be equal to `3 * 3 - 1 = 8`.
Make use of either the `tf.multiply` or `tf.pow` ops to square the value of each element in the `primes` vector.
```
# Write your code for Task 1 here.
primes = tf.constant([2, 3, 5, 7, 11, 13], dtype=tf.int32)
print("primes:", primes)
m_one = tf.constant(-1, dtype=tf.int32)
two = tf.constant(2, dtype=tf.int32)
square = tf.multiply(primes,primes)
# square = tf.pow(primes, two)
just_under_primes_squared = tf.add(square, m_one)
print("just_under_primes_squared:", just_under_primes_squared)
# two = tf.constant(2, dtype=tf.int32)
# primes_doubled = primes * two
# print("primes_doubled:", primes_doubled)
```
### Solution
Double-click __here__ for the solution.
<!-- Your answer is below:
# Task: Square each element in the primes vector, then subtract 1.
def solution(primes):
primes_squared = tf.multiply(primes, primes)
neg_one = tf.constant(-1, dtype=tf.int32)
just_under_primes_squared = tf.add(primes_squared, neg_one)
return just_under_primes_squared
def alternative_solution(primes):
primes_squared = tf.pow(primes, 2)
one = tf.constant(1, dtype=tf.int32)
just_under_primes_squared = tf.subtract(primes_squared, one)
return just_under_primes_squared
primes = tf.constant([2, 3, 5, 7, 11, 13], dtype=tf.int32)
just_under_primes_squared = solution(primes)
print("just_under_primes_squared:", just_under_primes_squared)
-->
## Matrix Multiplication
In linear algebra, when multiplying two matrices, the number of *columns* of the first matrix must
equal the number of *rows* in the second matrix.
- It is **_valid_** to multiply a `3x4` matrix by a `4x2` matrix. This will result in a `3x2` matrix.
- It is **_invalid_** to multiply a `4x2` matrix by a `3x4` matrix.
```
# A 3x4 matrix (2-d tensor).
x = tf.constant([[5, 2, 4, 3], [5, 1, 6, -2], [-1, 3, -1, -2]],
dtype=tf.int32)
# A 4x2 matrix (2-d tensor).
y = tf.constant([[2, 2], [3, 5], [4, 5], [1, 6]], dtype=tf.int32)
# Multiply `x` by `y`; result is 3x2 matrix.
matrix_multiply_result = tf.matmul(x, y)
print(matrix_multiply_result)
```
## Tensor Reshaping
With tensor addition and matrix multiplication each imposing constraints
on operands, TensorFlow programmers must frequently reshape tensors.
You can use the `tf.reshape` method to reshape a tensor.
For example, you can reshape a 8x2 tensor into a 2x8 tensor or a 4x4 tensor:
```
# Create an 8x2 matrix (2-D tensor).
matrix = tf.constant(
[[1, 2], [3, 4], [5, 6], [7, 8], [9, 10], [11, 12], [13, 14], [15, 16]],
dtype=tf.int32)
reshaped_2x8_matrix = tf.reshape(matrix, [2, 8])
reshaped_4x4_matrix = tf.reshape(matrix, [4, 4])
print("Original matrix (8x2):")
print(matrix.numpy())
print("Reshaped matrix (2x8):")
print(reshaped_2x8_matrix.numpy())
print("Reshaped matrix (4x4):")
print(reshaped_4x4_matrix.numpy())
```
You can also use `tf.reshape` to change the number of dimensions (the "rank") of the tensor.
For example, you could reshape that 8x2 tensor into a 3-D 2x2x4 tensor or a 1-D 16-element tensor.
```
# Create an 8x2 matrix (2-D tensor).
matrix = tf.constant(
[[1, 2], [3, 4], [5, 6], [7, 8], [9, 10], [11, 12], [13, 14], [15, 16]],
dtype=tf.int32)
reshaped_2x2x4_tensor = tf.reshape(matrix, [2, 2, 4])
one_dimensional_vector = tf.reshape(matrix, [16])
print("Original matrix (8x2):")
print(matrix.numpy())
print("Reshaped 3-D tensor (2x2x4):")
print(reshaped_2x2x4_tensor.numpy())
print("1-D vector:")
print(one_dimensional_vector.numpy())
```
### Exercise #2: Reshape two tensors in order to multiply them.
The following two vectors are incompatible for matrix multiplication:
* `a = tf.constant([5, 3, 2, 7, 1, 4])`
* `b = tf.constant([4, 6, 3])`
Reshape these vectors into compatible operands for matrix multiplication.
Then, invoke a matrix multiplication operation on the reshaped tensors.
```
# Write your code for Task 2 here.
a = tf.constant([5, 3, 2, 7, 1, 4])
b = tf.constant([4, 6, 3])
reshaped_a= tf.reshape(a, [2, 3])
reshaped_b= tf.reshape(b, [3, 1])
matrix_multiply_ab = tf.matmul(reshaped_a, reshaped_b)
print(matrix_multiply_ab)
```
Remember, when multiplying two matrices, the number of *columns* of the first matrix must equal the number of *rows* in the second matrix.
One possible solution is to reshape `a` into a 2x3 matrix and reshape `b` into a a 3x1 matrix, resulting in a 2x1 matrix after multiplication:
An alternative solution would be to reshape `a` into a 6x1 matrix and `b` into a 1x3 matrix, resulting in a 6x3 matrix after multiplication.
```
a = tf.constant([5, 3, 2, 7, 1, 4])
b = tf.constant([4, 6, 3])
reshaped_a = tf.reshape(a, [6, 1])
reshaped_b = tf.reshape(b, [1, 3])
c = tf.matmul(reshaped_a, reshaped_b)
print("reshaped_a (6x1):")
print(reshaped_a.numpy())
print("reshaped_b (1x3):")
print(reshaped_b.numpy())
print("reshaped_a x reshaped_b (6x3):")
print(c.numpy())
```
### Solution
Double-click __here__ for the solution.
<!-- Your answer is below:
# Task: Reshape two tensors in order to multiply them
a = tf.constant([5, 3, 2, 7, 1, 4])
b = tf.constant([4, 6, 3])
reshaped_a = tf.reshape(a, [2, 3])
reshaped_b = tf.reshape(b, [3, 1])
c = tf.matmul(reshaped_a, reshaped_b)
print("reshaped_a (2x3):")
print(reshaped_a.numpy())
print("reshaped_b (3x1):")
print(reshaped_b.numpy())
print("reshaped_a x reshaped_b (2x1):")
print(c.numpy())
-->
## Variables, Initialization and Assignment
So far, all the operations we performed were on static values (`tf.constant`); calling `numpy()` always returned the same result. TensorFlow allows you to define `Variable` objects, whose values can be changed.
When creating a variable, you can set an initial value explicitly, or you can use an initializer (like a distribution):
```
# Create a scalar variable with the initial value 3.
v = tf.contrib.eager.Variable([3])
# Create a vector variable of shape [1, 4], with random initial values,
# sampled from a normal distribution with mean 1 and standard deviation 0.35.
w = tf.contrib.eager.Variable(tf.random_normal([1, 4], mean=1.0, stddev=0.35))
print("v:", v.numpy())
print("w:", w.numpy())
```
To change the value of a variable, use the `assign` op:
```
v = tf.contrib.eager.Variable([3])
print(v.numpy())
tf.assign(v, [7])
print(v.numpy())
v.assign([5])
print(v.numpy())
```
When assigning a new value to a variable, its shape must be equal to its previous shape:
```
v = tf.contrib.eager.Variable([[1, 2, 3], [4, 5, 6]])
print(v.numpy())
try:
print("Assigning [7, 8, 9] to v")
v.assign([7, 8, 9])
except ValueError as e:
print("Exception:", e)
```
There are many more topics about variables that we didn't cover here, such as loading and storing. To learn more, see the [TensorFlow docs](https://www.tensorflow.org/programmers_guide/variables).
### Exercise #3: Simulate 10 rolls of two dice.
Create a dice simulation, which generates a `10x3` 2-D tensor in which:
* Columns `1` and `2` each hold one throw of one six-sided die (with values 1–6).
* Column `3` holds the sum of Columns `1` and `2` on the same row.
For example, the first row might have the following values:
* Column `1` holds `4`
* Column `2` holds `3`
* Column `3` holds `7`
You'll need to explore the [TensorFlow documentation](https://www.tensorflow.org/api_guides/python/array_ops) to solve this task.
```
# Write your code for Task 3 here.
# Task: Simulate 10 throws of two dice. Store the results in a 10x3 matrix.
die1 = tf.contrib.eager.Variable(
tf.random_uniform([10, 1], minval=1, maxval=7, dtype=tf.int32))
die2 = tf.contrib.eager.Variable(
tf.random_uniform([10, 1], minval=1, maxval=7, dtype=tf.int32))
dice_sum = tf.add(die1, die2)
resulting_matrix = tf.concat(values=[die1, die2, dice_sum], axis=1)
print(resulting_matrix.numpy())
```
We're going to place dice throws inside two separate 10x1 matrices, `die1` and `die2`. The summation of the dice rolls will be stored in `dice_sum`, then the resulting 10x3 matrix will be created by *concatenating* the three 10x1 matrices together into a single matrix.
Alternatively, we could have placed dice throws inside a single 10x2 matrix, but adding different columns of the same matrix would be more complicated. We also could have placed dice throws inside two 1-D tensors (vectors), but doing so would require transposing the result.
### Solution
Double-click __here__ for the solution.
<!-- Your answer is below:
# Task: Simulate 10 throws of two dice. Store the results in a 10x3 matrix.
die1 = tf.contrib.eager.Variable(
tf.random_uniform([10, 1], minval=1, maxval=7, dtype=tf.int32))
die2 = tf.contrib.eager.Variable(
tf.random_uniform([10, 1], minval=1, maxval=7, dtype=tf.int32))
dice_sum = tf.add(die1, die2)
resulting_matrix = tf.concat(values=[die1, die2, dice_sum], axis=1)
print(resulting_matrix.numpy())
-->
|
github_jupyter
|
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
%matplotlib inline
data_label = pd.read_csv("data(with_label).csv")
```
### 30 day death age
```
fig = plt.figure(figsize=(12,6))
sns.set_style('darkgrid')
ax = sns.violinplot(x="thirty_days", hue="gender", y="age",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('Age (years)')
plt.title('Age distributions for 30-day death')
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="age",data=data_label[data_label.age<300], split=True,)
plt.legend(loc='lower left')
#plt.ylim([0,100])
plt.xlabel(' ')
plt.ylabel('Age (years)')
plt.title('Age distributions for 30-day death \n (excluding ages > 300)')
```
## One year death age
```
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="one_year", hue="gender", y="age",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('Age (years)')
plt.title('Age distributions for 30-day death')
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="one_year", hue="gender", y="age",data=data_label[data_label.age<300], split=True,)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('Age (years)')
plt.title('Age distributions for 30-day death \n (excluding ages > 300)')
```
## sapsii
```
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="sapsii",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('SAPS II distributions for 30-day death')
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="one_year", hue="gender", y="sapsii",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('SAPS II distributions for one year death')
```
## Sofa
```
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="sofa",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('SOFA distributions for 30-day death')
```
## Cormorbidity
```
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="elixhauser_vanwalraven",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('elixhauser_vanwalraven for 30-day death')
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="elixhauser_sid29",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('elixhauser_sid29 for 30-day death')
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="elixhauser_sid30",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('elixhauser_sid30 for 30-day death')
```
## urea_n_mean
```
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="urea_n_mean",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel('SAPS II score')
plt.title('urea_n_mean for 30-day death')
fig = plt.figure(figsize=(12,6))
ax = sns.violinplot(x="thirty_days", hue="gender", y="rrt",data=data_label, split=True)
plt.legend(loc='lower left')
plt.xlabel(' ')
plt.ylabel(' ')
plt.title('rrt for 30-day death')
```
## Correlation heatmap
### Remains features: age, gender, 'sapsii', 'sofa', 'thirty_days', 'one_year','oasis', 'lods', 'sirs', and other physiological parameters
```
#'platelets_mean','urea_n_mean', 'glucose_mean','resprate_mean', 'sysbp_mean', 'diasbp_mean', 'urine_mean', 'spo2_mean','temp_mean','hr_mean',
data = data_label.drop(columns=['subject_id', 'hadm_id', 'admittime', 'dischtime', 'deathtime', 'dod',
'first_careunit', 'last_careunit', 'marital_status',
'insurance', 'urea_n_min', 'urea_n_max', 'platelets_min',
'platelets_max', 'magnesium_max', 'albumin_min',
'calcium_min', 'resprate_min', 'resprate_max',
'glucose_min', 'glucose_max', 'hr_min', 'hr_max',
'sysbp_min', 'sysbp_max','diasbp_min',
'diasbp_max', 'temp_min', 'temp_max',
'urine_min', 'urine_max',
'elixhauser_vanwalraven', 'elixhauser_sid29', 'elixhauser_sid30',
'los_hospital', 'meanbp_min', 'meanbp_max', 'meanbp_mean', 'spo2_min',
'spo2_max', 'vent', 'rrt', 'urineoutput',
'icustay_age_group', 'admission_type',
'admission_location', 'discharge_location', 'ethnicity', 'diagnosis',
'time_before_death'])
correlation = data.corr()
plt.figure(figsize=(10,10))
sns.heatmap(correlation, vmax=1, square=True, annot=False, cmap="YlGnBu")
```
## KDE for 30 day death
```
data_pos = data_label.loc[data_label.thirty_days == 1]
data_neg = data_label.loc[data_label.thirty_days == 0]
fig = plt.figure(figsize=(15,15))
plt.subplot(331)
data_neg.platelets_min.plot.kde(color = 'red', alpha = 0.5)
data_pos.platelets_min.plot.kde(color = 'blue', alpha = 0.5)
plt.title('platelets_min')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(332)
data_neg.age.plot.kde(color = 'red', alpha = 0.5)
data_pos.age.plot.kde(color = 'blue', alpha = 0.5)
plt.title('Age')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(333)
data_neg.albumin_min.plot.kde(color = 'red', alpha = 0.5)
data_pos.albumin_min.plot.kde(color = 'blue', alpha = 0.5)
plt.title('albumin_min')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(334)
data_neg.sysbp_min.plot.kde(color = 'red', alpha = 0.5)
data_pos.sysbp_min.plot.kde(color = 'blue', alpha = 0.5)
plt.title('sysbp_min')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(335)
data_neg.temp_mean.plot.kde(color = 'red', alpha = 0.5)
data_pos.temp_mean.plot.kde(color = 'blue', alpha = 0.5)
plt.title('temp_mean')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(336)
data_neg.resprate_max.plot.kde(color = 'red', alpha = 0.5)
data_pos.resprate_max.plot.kde(color = 'blue', alpha = 0.5)
plt.title('resprate_max')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(337)
data_neg.urea_n_mean.plot.kde(color = 'red', alpha = 0.5)
data_pos.urea_n_mean.plot.kde(color = 'blue', alpha = 0.5)
plt.title('urea_n_mean')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(338)
data_neg.vent.plot.kde(color = 'red', alpha = 0.5)
data_pos.vent.plot.kde(color = 'blue', alpha = 0.5)
plt.title('vent')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(339)
data_neg.rrt.plot.kde(color = 'red', alpha = 0.5)
data_pos.rrt.plot.kde(color = 'blue', alpha = 0.5)
plt.title('rrt')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
fig = plt.figure(figsize=(15,15))
plt.subplot(321)
data_neg.sofa.plot.kde(color = 'red', alpha = 0.5)
data_pos.sofa.plot.kde(color = 'blue', alpha = 0.5)
plt.title('sofa')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(322)
data_neg.sapsii.plot.kde(color = 'red', alpha = 0.5)
data_pos.sapsii.plot.kde(color = 'blue', alpha = 0.5)
plt.title('sapsii')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(323)
data_neg.oasis.plot.kde(color = 'red', alpha = 0.5)
data_pos.oasis.plot.kde(color = 'blue', alpha = 0.5)
plt.title('oasis')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(324)
data_neg.lods.plot.kde(color = 'red', alpha = 0.5)
data_pos.lods.plot.kde(color = 'blue', alpha = 0.5)
plt.title('lods')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
plt.subplot(325)
data_neg.sirs.plot.kde(color = 'red', alpha = 0.5)
data_pos.sirs.plot.kde(color = 'blue', alpha = 0.5)
plt.title('sirs')
plt.legend(labels=['Alive in 30 days', 'Dead in 30 days'])
```
## Pie chart
```
# Age groups
age_category = np.floor(data_label['age']/10)
count = age_category.value_counts()
count['10-20'] = 345
count['20-30'] = 1860
count['30-40'] = 2817
count['40-50'] = 5716
count['50-60'] = 10190
count['60-70'] = 12300
count['70-80'] = 12638
count['80-89'] = 9233
count['older than 89'] = 2897
count = count.drop([7.0, 6.0, 5.0, 8.0, 4.0, 30.0, 3.0, 2.0, 1.0, 31.0])
count
fig = plt.figure(figsize=(25,25))
plt.rcParams.update({'font.size': 18})
#explode = (0, 0.15, 0)
colors = ['#79bd9a','#f4f7f7','#aacfd0','#79a8a9','#a8dba8']
#f4f7f7 #aacfd0 #79a8a9 #a8dba8 #79bd9a
plt.subplot(321)
data_label.admission_type.value_counts().plot.pie( colors = colors, autopct='%1.1f%%')
plt.title('Admission type')
plt.ylabel('')
plt.subplot(322)
plotting = (data_label.admission_location.value_counts(dropna=False))
plotting['OTHER'] = plotting['TRANSFER FROM SKILLED NUR'] + plotting['TRANSFER FROM OTHER HEALT'] + plotting['** INFO NOT AVAILABLE **']+plotting['HMO REFERRAL/SICK']+plotting['TRSF WITHIN THIS FACILITY']
plotting = plotting.drop(['TRANSFER FROM SKILLED NUR', 'TRANSFER FROM OTHER HEALT', '** INFO NOT AVAILABLE **','HMO REFERRAL/SICK','TRSF WITHIN THIS FACILITY'])
plotting.plot.pie( colors = colors, autopct='%1.1f%%')
plt.title('Admission location')
plt.ylabel('')
plt.subplot(323)
count.plot.pie( colors = colors, autopct='%1.1f%%')
plt.title('Age groups')
plt.ylabel('')
plt.subplot(324)
data_label.insurance.value_counts().plot.pie( colors = colors, autopct='%1.1f%%')
plt.title('Insurance provider')
plt.ylabel('')
#admission_location
#discharge_location
#ethnicity
#diagnosis
fig = plt.figure(figsize=(8,8))
plt.rcParams.update({'font.size': 15})
explode = (0, 0.1)
data_label.one_year.value_counts().plot.pie( colors = colors, autopct='%1.1f%%',explode = explode, startangle = 90)
plt.title('Patient died in 1 year')
plt.ylabel('')
fig = plt.figure(figsize=(8,8))
plt.rcParams.update({'font.size': 15})
data_label.thirty_days.value_counts().plot.pie( colors = colors, autopct='%1.1f%%',explode = explode, startangle = 90)
plt.title('Patient died in 30 days')
plt.ylabel('')
```
|
github_jupyter
|
# Logic: `logic.py`; Chapters 6-8
This notebook describes the [logic.py](https://github.com/aimacode/aima-python/blob/master/logic.py) module, which covers Chapters 6 (Logical Agents), 7 (First-Order Logic) and 8 (Inference in First-Order Logic) of *[Artificial Intelligence: A Modern Approach](http://aima.cs.berkeley.edu)*. See the [intro notebook](https://github.com/aimacode/aima-python/blob/master/intro.ipynb) for instructions.
We'll start by looking at `Expr`, the data type for logical sentences, and the convenience function `expr`. We'll be covering two types of knowledge bases, `PropKB` - Propositional logic knowledge base and `FolKB` - First order logic knowledge base. We will construct a propositional knowledge base of a specific situation in the Wumpus World. We will next go through the `tt_entails` function and experiment with it a bit. The `pl_resolution` and `pl_fc_entails` functions will come next. We'll study forward chaining and backward chaining algorithms for `FolKB` and use them on `crime_kb` knowledge base.
But the first step is to load the code:
```
from utils import *
from logic import *
```
## Logical Sentences
The `Expr` class is designed to represent any kind of mathematical expression. The simplest type of `Expr` is a symbol, which can be defined with the function `Symbol`:
```
Symbol('x')
```
Or we can define multiple symbols at the same time with the function `symbols`:
```
(x, y, P, Q, f) = symbols('x, y, P, Q, f')
```
We can combine `Expr`s with the regular Python infix and prefix operators. Here's how we would form the logical sentence "P and not Q":
```
P & ~Q
```
This works because the `Expr` class overloads the `&` operator with this definition:
```python
def __and__(self, other): return Expr('&', self, other)```
and does similar overloads for the other operators. An `Expr` has two fields: `op` for the operator, which is always a string, and `args` for the arguments, which is a tuple of 0 or more expressions. By "expression," I mean either an instance of `Expr`, or a number. Let's take a look at the fields for some `Expr` examples:
```
sentence = P & ~Q
sentence.op
sentence.args
P.op
P.args
Pxy = P(x, y)
Pxy.op
Pxy.args
```
It is important to note that the `Expr` class does not define the *logic* of Propositional Logic sentences; it just gives you a way to *represent* expressions. Think of an `Expr` as an [abstract syntax tree](https://en.wikipedia.org/wiki/Abstract_syntax_tree). Each of the `args` in an `Expr` can be either a symbol, a number, or a nested `Expr`. We can nest these trees to any depth. Here is a deply nested `Expr`:
```
3 * f(x, y) + P(y) / 2 + 1
```
## Operators for Constructing Logical Sentences
Here is a table of the operators that can be used to form sentences. Note that we have a problem: we want to use Python operators to make sentences, so that our programs (and our interactive sessions like the one here) will show simple code. But Python does not allow implication arrows as operators, so for now we have to use a more verbose notation that Python does allow: `|'==>'|` instead of just `==>`. Alternately, you can always use the more verbose `Expr` constructor forms:
| Operation | Book | Python Infix Input | Python Output | Python `Expr` Input
|--------------------------|----------------------|-------------------------|---|---|
| Negation | ¬ P | `~P` | `~P` | `Expr('~', P)`
| And | P ∧ Q | `P & Q` | `P & Q` | `Expr('&', P, Q)`
| Or | P ∨ Q | `P`<tt> | </tt>`Q`| `P`<tt> | </tt>`Q` | `Expr('`|`', P, Q)`
| Inequality (Xor) | P ≠ Q | `P ^ Q` | `P ^ Q` | `Expr('^', P, Q)`
| Implication | P → Q | `P` <tt>|</tt>`'==>'`<tt>|</tt> `Q` | `P ==> Q` | `Expr('==>', P, Q)`
| Reverse Implication | Q ← P | `Q` <tt>|</tt>`'<=='`<tt>|</tt> `P` |`Q <== P` | `Expr('<==', Q, P)`
| Equivalence | P ↔ Q | `P` <tt>|</tt>`'<=>'`<tt>|</tt> `Q` |`P <=> Q` | `Expr('<=>', P, Q)`
Here's an example of defining a sentence with an implication arrow:
```
~(P & Q) |'==>'| (~P | ~Q)
```
## `expr`: a Shortcut for Constructing Sentences
If the `|'==>'|` notation looks ugly to you, you can use the function `expr` instead:
```
expr('~(P & Q) ==> (~P | ~Q)')
```
`expr` takes a string as input, and parses it into an `Expr`. The string can contain arrow operators: `==>`, `<==`, or `<=>`, which are handled as if they were regular Python infix operators. And `expr` automatically defines any symbols, so you don't need to pre-define them:
```
expr('sqrt(b ** 2 - 4 * a * c)')
```
For now that's all you need to know about `expr`. If you are interested, we explain the messy details of how `expr` is implemented and how `|'==>'|` is handled in the appendix.
## Propositional Knowledge Bases: `PropKB`
The class `PropKB` can be used to represent a knowledge base of propositional logic sentences.
We see that the class `KB` has four methods, apart from `__init__`. A point to note here: the `ask` method simply calls the `ask_generator` method. Thus, this one has already been implemented, and what you'll have to actually implement when you create your own knowledge base class (though you'll probably never need to, considering the ones we've created for you) will be the `ask_generator` function and not the `ask` function itself.
The class `PropKB` now.
* `__init__(self, sentence=None)` : The constructor `__init__` creates a single field `clauses` which will be a list of all the sentences of the knowledge base. Note that each one of these sentences will be a 'clause' i.e. a sentence which is made up of only literals and `or`s.
* `tell(self, sentence)` : When you want to add a sentence to the KB, you use the `tell` method. This method takes a sentence, converts it to its CNF, extracts all the clauses, and adds all these clauses to the `clauses` field. So, you need not worry about `tell`ing only clauses to the knowledge base. You can `tell` the knowledge base a sentence in any form that you wish; converting it to CNF and adding the resulting clauses will be handled by the `tell` method.
* `ask_generator(self, query)` : The `ask_generator` function is used by the `ask` function. It calls the `tt_entails` function, which in turn returns `True` if the knowledge base entails query and `False` otherwise. The `ask_generator` itself returns an empty dict `{}` if the knowledge base entails query and `None` otherwise. This might seem a little bit weird to you. After all, it makes more sense just to return a `True` or a `False` instead of the `{}` or `None` But this is done to maintain consistency with the way things are in First-Order Logic, where an `ask_generator` function is supposed to return all the substitutions that make the query true. Hence the dict, to return all these substitutions. I will be mostly be using the `ask` function which returns a `{}` or a `False`, but if you don't like this, you can always use the `ask_if_true` function which returns a `True` or a `False`.
* `retract(self, sentence)` : This function removes all the clauses of the sentence given, from the knowledge base. Like the `tell` function, you don't have to pass clauses to remove them from the knowledge base; any sentence will do fine. The function will take care of converting that sentence to clauses and then remove those.
## Wumpus World KB
Let us create a `PropKB` for the wumpus world with the sentences mentioned in `section 7.4.3`.
```
wumpus_kb = PropKB()
```
We define the symbols we use in our clauses.<br/>
$P_{x, y}$ is true if there is a pit in `[x, y]`.<br/>
$B_{x, y}$ is true if the agent senses breeze in `[x, y]`.<br/>
```
P11, P12, P21, P22, P31, B11, B21 = expr('P11, P12, P21, P22, P31, B11, B21')
```
Now we tell sentences based on `section 7.4.3`.<br/>
There is no pit in `[1,1]`.
```
wumpus_kb.tell(~P11)
```
A square is breezy if and only if there is a pit in a neighboring square. This has to be stated for each square but for now, we include just the relevant squares.
```
wumpus_kb.tell(B11 | '<=>' | ((P12 | P21)))
wumpus_kb.tell(B21 | '<=>' | ((P11 | P22 | P31)))
```
Now we include the breeze percepts for the first two squares leading up to the situation in `Figure 7.3(b)`
```
wumpus_kb.tell(~B11)
wumpus_kb.tell(B21)
```
We can check the clauses stored in a `KB` by accessing its `clauses` variable
```
wumpus_kb.clauses
```
We see that the equivalence $B_{1, 1} \iff (P_{1, 2} \lor P_{2, 1})$ was automatically converted to two implications which were inturn converted to CNF which is stored in the `KB`.<br/>
$B_{1, 1} \iff (P_{1, 2} \lor P_{2, 1})$ was split into $B_{1, 1} \implies (P_{1, 2} \lor P_{2, 1})$ and $B_{1, 1} \Longleftarrow (P_{1, 2} \lor P_{2, 1})$.<br/>
$B_{1, 1} \implies (P_{1, 2} \lor P_{2, 1})$ was converted to $P_{1, 2} \lor P_{2, 1} \lor \neg B_{1, 1}$.<br/>
$B_{1, 1} \Longleftarrow (P_{1, 2} \lor P_{2, 1})$ was converted to $\neg (P_{1, 2} \lor P_{2, 1}) \lor B_{1, 1}$ which becomes $(\neg P_{1, 2} \lor B_{1, 1}) \land (\neg P_{2, 1} \lor B_{1, 1})$ after applying De Morgan's laws and distributing the disjunction.<br/>
$B_{2, 1} \iff (P_{1, 1} \lor P_{2, 2} \lor P_{3, 2})$ is converted in similar manner.
## Inference in Propositional Knowledge Base
In this section we will look at two algorithms to check if a sentence is entailed by the `KB`. Our goal is to decide whether $\text{KB} \vDash \alpha$ for some sentence $\alpha$.
### Truth Table Enumeration
It is a model-checking approach which, as the name suggests, enumerates all possible models in which the `KB` is true and checks if $\alpha$ is also true in these models. We list the $n$ symbols in the `KB` and enumerate the $2^{n}$ models in a depth-first manner and check the truth of `KB` and $\alpha$.
```
%psource tt_check_all
```
Note that `tt_entails()` takes an `Expr` which is a conjunction of clauses as the input instead of the `KB` itself. You can use the `ask_if_true()` method of `PropKB` which does all the required conversions. Let's check what `wumpus_kb` tells us about $P_{1, 1}$.
```
wumpus_kb.ask_if_true(~P11), wumpus_kb.ask_if_true(P11)
```
Looking at Figure 7.9 we see that in all models in which the knowledge base is `True`, $P_{1, 1}$ is `False`. It makes sense that `ask_if_true()` returns `True` for $\alpha = \neg P_{1, 1}$ and `False` for $\alpha = P_{1, 1}$. This begs the question, what if $\alpha$ is `True` in only a portion of all models. Do we return `True` or `False`? This doesn't rule out the possibility of $\alpha$ being `True` but it is not entailed by the `KB` so we return `False` in such cases. We can see this is the case for $P_{2, 2}$ and $P_{3, 1}$.
```
wumpus_kb.ask_if_true(~P22), wumpus_kb.ask_if_true(P22)
```
### Proof by Resolution
Recall that our goal is to check whether $\text{KB} \vDash \alpha$ i.e. is $\text{KB} \implies \alpha$ true in every model. Suppose we wanted to check if $P \implies Q$ is valid. We check the satisfiability of $\neg (P \implies Q)$, which can be rewritten as $P \land \neg Q$. If $P \land \neg Q$ is unsatisfiable, then $P \implies Q$ must be true in all models. This gives us the result "$\text{KB} \vDash \alpha$ <em>if and only if</em> $\text{KB} \land \neg \alpha$ is unsatisfiable".<br/>
This technique corresponds to <em>proof by <strong>contradiction</strong></em>, a standard mathematical proof technique. We assume $\alpha$ to be false and show that this leads to a contradiction with known axioms in $\text{KB}$. We obtain a contradiction by making valid inferences using inference rules. In this proof we use a single inference rule, <strong>resolution</strong> which states $(l_1 \lor \dots \lor l_k) \land (m_1 \lor \dots \lor m_n) \land (l_i \iff \neg m_j) \implies l_1 \lor \dots \lor l_{i - 1} \lor l_{i + 1} \lor \dots \lor l_k \lor m_1 \lor \dots \lor m_{j - 1} \lor m_{j + 1} \lor \dots \lor m_n$. Applying the resolution yeilds us a clause which we add to the KB. We keep doing this until:
* There are no new clauses that can be added, in which case $\text{KB} \nvDash \alpha$.
* Two clauses resolve to yield the <em>empty clause</em>, in which case $\text{KB} \vDash \alpha$.
The <em>empty clause</em> is equivalent to <em>False</em> because it arises only from resolving two complementary
unit clauses such as $P$ and $\neg P$ which is a contradiction as both $P$ and $\neg P$ can't be <em>True</em> at the same time.
```
%psource pl_resolution
pl_resolution(wumpus_kb, ~P11), pl_resolution(wumpus_kb, P11)
pl_resolution(wumpus_kb, ~P22), pl_resolution(wumpus_kb, P22)
```
## First-Order Logic Knowledge Bases: `FolKB`
The class `FolKB` can be used to represent a knowledge base of First-order logic sentences. You would initialize and use it the same way as you would for `PropKB` except that the clauses are first-order definite clauses. We will see how to write such clauses to create a database and query them in the following sections.
## Criminal KB
In this section we create a `FolKB` based on the following paragraph.<br/>
<em>The law says that it is a crime for an American to sell weapons to hostile nations. The country Nono, an enemy of America, has some missiles, and all of its missiles were sold to it by Colonel West, who is American.</em><br/>
The first step is to extract the facts and convert them into first-order definite clauses. Extracting the facts from data alone is a challenging task. Fortunately, we have a small paragraph and can do extraction and conversion manually. We'll store the clauses in list aptly named `clauses`.
```
clauses = []
```
<em>“... it is a crime for an American to sell weapons to hostile nations”</em><br/>
The keywords to look for here are 'crime', 'American', 'sell', 'weapon' and 'hostile'. We use predicate symbols to make meaning of them.
* `Criminal(x)`: `x` is a criminal
* `American(x)`: `x` is an American
* `Sells(x ,y, z)`: `x` sells `y` to `z`
* `Weapon(x)`: `x` is a weapon
* `Hostile(x)`: `x` is a hostile nation
Let us now combine them with appropriate variable naming to depict the meaning of the sentence. The criminal `x` is also the American `x` who sells weapon `y` to `z`, which is a hostile nation.
$\text{American}(x) \land \text{Weapon}(y) \land \text{Sells}(x, y, z) \land \text{Hostile}(z) \implies \text{Criminal} (x)$
```
clauses.append(expr("(American(x) & Weapon(y) & Sells(x, y, z) & Hostile(z)) ==> Criminal(x)"))
```
<em>"The country Nono, an enemy of America"</em><br/>
We now know that Nono is an enemy of America. We represent these nations using the constant symbols `Nono` and `America`. the enemy relation is show using the predicate symbol `Enemy`.
$\text{Enemy}(\text{Nono}, \text{America})$
```
clauses.append(expr("Enemy(Nono, America)"))
```
<em>"Nono ... has some missiles"</em><br/>
This states the existence of some missile which is owned by Nono. $\exists x \text{Owns}(\text{Nono}, x) \land \text{Missile}(x)$. We invoke existential instantiation to introduce a new constant `M1` which is the missile owned by Nono.
$\text{Owns}(\text{Nono}, \text{M1}), \text{Missile}(\text{M1})$
```
clauses.append(expr("Owns(Nono, M1)"))
clauses.append(expr("Missile(M1)"))
```
<em>"All of its missiles were sold to it by Colonel West"</em><br/>
If Nono owns something and it classifies as a missile, then it was sold to Nono by West.
$\text{Missile}(x) \land \text{Owns}(\text{Nono}, x) \implies \text{Sells}(\text{West}, x, \text{Nono})$
```
clauses.append(expr("(Missile(x) & Owns(Nono, x)) ==> Sells(West, x, Nono)"))
```
<em>"West, who is American"</em><br/>
West is an American.
$\text{American}(\text{West})$
```
clauses.append(expr("American(West)"))
```
We also know, from our understanding of language, that missiles are weapons and that an enemy of America counts as “hostile”.
$\text{Missile}(x) \implies \text{Weapon}(x), \text{Enemy}(x, \text{America}) \implies \text{Hostile}(x)$
```
clauses.append(expr("Missile(x) ==> Weapon(x)"))
clauses.append(expr("Enemy(x, America) ==> Hostile(x)"))
```
Now that we have converted the information into first-order definite clauses we can create our first-order logic knowledge base.
```
crime_kb = FolKB(clauses)
```
## Inference in First-Order Logic
In this section we look at a forward chaining and a backward chaining algorithm for `FolKB`. Both aforementioned algorithms rely on a process called <strong>unification</strong>, a key component of all first-order inference algorithms.
### Unification
We sometimes require finding substitutions that make different logical expressions look identical. This process, called unification, is done by the `unify` algorithm. It takes as input two sentences and returns a <em>unifier</em> for them if one exists. A unifier is a dictionary which stores the substitutions required to make the two sentences identical. It does so by recursively unifying the components of a sentence, where the unification of a variable symbol `var` with a constant symbol `Const` is the mapping `{var: Const}`. Let's look at a few examples.
```
unify(expr('x'), 3)
unify(expr('A(x)'), expr('A(B)'))
unify(expr('Cat(x) & Dog(Dobby)'), expr('Cat(Bella) & Dog(y)'))
```
In cases where there is no possible substitution that unifies the two sentences the function return `None`.
```
print(unify(expr('Cat(x)'), expr('Dog(Dobby)')))
```
We also need to take care we do not unintentionally use the same variable name. Unify treats them as a single variable which prevents it from taking multiple value.
```
print(unify(expr('Cat(x) & Dog(Dobby)'), expr('Cat(Bella) & Dog(x)')))
```
### Forward Chaining Algorithm
We consider the simple forward-chaining algorithm presented in <em>Figure 9.3</em>. We look at each rule in the knoweldge base and see if the premises can be satisfied. This is done by finding a substitution which unifies each of the premise with a clause in the `KB`. If we are able to unify the premises, the conclusion (with the corresponding substitution) is added to the `KB`. This inferencing process is repeated until either the query can be answered or till no new sentences can be added. We test if the newly added clause unifies with the query in which case the substitution yielded by `unify` is an answer to the query. If we run out of sentences to infer, this means the query was a failure.
The function `fol_fc_ask` is a generator which yields all substitutions which validate the query.
```
%psource fol_fc_ask
```
Let's find out all the hostile nations. Note that we only told the `KB` that Nono was an enemy of America, not that it was hostile.
```
answer = fol_fc_ask(crime_kb, expr('Hostile(x)'))
print(list(answer))
```
The generator returned a single substitution which says that Nono is a hostile nation. See how after adding another enemy nation the generator returns two substitutions.
```
crime_kb.tell(expr('Enemy(JaJa, America)'))
answer = fol_fc_ask(crime_kb, expr('Hostile(x)'))
print(list(answer))
```
<strong><em>Note</em>:</strong> `fol_fc_ask` makes changes to the `KB` by adding sentences to it.
### Backward Chaining Algorithm
This algorithm works backward from the goal, chaining through rules to find known facts that support the proof. Suppose `goal` is the query we want to find the substitution for. We find rules of the form $\text{lhs} \implies \text{goal}$ in the `KB` and try to prove `lhs`. There may be multiple clauses in the `KB` which give multiple `lhs`. It is sufficient to prove only one of these. But to prove a `lhs` all the conjuncts in the `lhs` of the clause must be proved. This makes it similar to <em>And/Or</em> search.
#### OR
The <em>OR</em> part of the algorithm comes from our choice to select any clause of the form $\text{lhs} \implies \text{goal}$. Looking at all rules's `lhs` whose `rhs` unify with the `goal`, we yield a substitution which proves all the conjuncts in the `lhs`. We use `parse_definite_clause` to attain `lhs` and `rhs` from a clause of the form $\text{lhs} \implies \text{rhs}$. For atomic facts the `lhs` is an empty list.
```
%psource fol_bc_or
```
#### AND
The <em>AND</em> corresponds to proving all the conjuncts in the `lhs`. We need to find a substitution which proves each <em>and</em> every clause in the list of conjuncts.
```
%psource fol_bc_and
```
Now the main function `fl_bc_ask` calls `fol_bc_or` with substitution initialized as empty. The `ask` method of `FolKB` uses `fol_bc_ask` and fetches the first substitution returned by the generator to answer query. Let's query the knowledge base we created from `clauses` to find hostile nations.
```
# Rebuild KB because running fol_fc_ask would add new facts to the KB
crime_kb = FolKB(clauses)
crime_kb.ask(expr('Hostile(x)'))
```
You may notice some new variables in the substitution. They are introduced to standardize the variable names to prevent naming problems as discussed in the [Unification section](#Unification)
## Appendix: The Implementation of `|'==>'|`
Consider the `Expr` formed by this syntax:
```
P |'==>'| ~Q
```
What is the funny `|'==>'|` syntax? The trick is that "`|`" is just the regular Python or-operator, and so is exactly equivalent to this:
```
(P | '==>') | ~Q
```
In other words, there are two applications of or-operators. Here's the first one:
```
P | '==>'
```
What is going on here is that the `__or__` method of `Expr` serves a dual purpose. If the right-hand-side is another `Expr` (or a number), then the result is an `Expr`, as in `(P | Q)`. But if the right-hand-side is a string, then the string is taken to be an operator, and we create a node in the abstract syntax tree corresponding to a partially-filled `Expr`, one where we know the left-hand-side is `P` and the operator is `==>`, but we don't yet know the right-hand-side.
The `PartialExpr` class has an `__or__` method that says to create an `Expr` node with the right-hand-side filled in. Here we can see the combination of the `PartialExpr` with `Q` to create a complete `Expr`:
```
partial = PartialExpr('==>', P)
partial | ~Q
```
This [trick](http://code.activestate.com/recipes/384122-infix-operators/) is due to [Ferdinand Jamitzky](http://code.activestate.com/recipes/users/98863/), with a modification by [C. G. Vedant](https://github.com/Chipe1),
who suggested using a string inside the or-bars.
## Appendix: The Implementation of `expr`
How does `expr` parse a string into an `Expr`? It turns out there are two tricks (besides the Jamitzky/Vedant trick):
1. We do a string substitution, replacing "`==>`" with "`|'==>'|`" (and likewise for other operators).
2. We `eval` the resulting string in an environment in which every identifier
is bound to a symbol with that identifier as the `op`.
In other words,
```
expr('~(P & Q) ==> (~P | ~Q)')
```
is equivalent to doing:
```
P, Q = symbols('P, Q')
~(P & Q) |'==>'| (~P | ~Q)
```
One thing to beware of: this puts `==>` at the same precedence level as `"|"`, which is not quite right. For example, we get this:
```
P & Q |'==>'| P | Q
```
which is probably not what we meant; when in doubt, put in extra parens:
```
(P & Q) |'==>'| (P | Q)
```
## Examples
```
from notebook import Canvas_fol_bc_ask
canvas_bc_ask = Canvas_fol_bc_ask('canvas_bc_ask', crime_kb, expr('Criminal(x)'))
```
# Authors
This notebook by [Chirag Vartak](https://github.com/chiragvartak) and [Peter Norvig](https://github.com/norvig).
|
github_jupyter
|
```
# Copyright 2021 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Vertex AI: Vertex AI Migration: Custom Image Classification w/custom training container
<table align="left">
<td>
<a href="https://colab.research.google.com/github/GoogleCloudPlatform/ai-platform-samples/blob/master/vertex-ai-samples/tree/master/notebooks/official/migration/UJ3%20Vertex%20SDK%20Custom%20Image%20Classification%20with%20custom%20training%20container.ipynb">
<img src="https://cloud.google.com/ml-engine/images/colab-logo-32px.png" alt="Colab logo"> Run in Colab
</a>
</td>
<td>
<a href="https://github.com/GoogleCloudPlatform/ai-platform-samples/blob/master/vertex-ai-samples/tree/master/notebooks/official/migration/UJ3%20Vertex%20SDK%20Custom%20Image%20Classification%20with%20custom%20training%20container.ipynb">
<img src="https://cloud.google.com/ml-engine/images/github-logo-32px.png" alt="GitHub logo">
View on GitHub
</a>
</td>
</table>
<br/><br/><br/>
### Dataset
The dataset used for this tutorial is the [CIFAR10 dataset](https://www.tensorflow.org/datasets/catalog/cifar10) from [TensorFlow Datasets](https://www.tensorflow.org/datasets/catalog/overview). The version of the dataset you will use is built into TensorFlow. The trained model predicts which type of class an image is from ten classes: airplane, automobile, bird, cat, deer, dog, frog, horse, ship, truck.
### Costs
This tutorial uses billable components of Google Cloud:
* Vertex AI
* Cloud Storage
Learn about [Vertex AI
pricing](https://cloud.google.com/vertex-ai/pricing) and [Cloud Storage
pricing](https://cloud.google.com/storage/pricing), and use the [Pricing
Calculator](https://cloud.google.com/products/calculator/)
to generate a cost estimate based on your projected usage.
### Set up your local development environment
If you are using Colab or Google Cloud Notebooks, your environment already meets all the requirements to run this notebook. You can skip this step.
Otherwise, make sure your environment meets this notebook's requirements. You need the following:
- The Cloud Storage SDK
- Git
- Python 3
- virtualenv
- Jupyter notebook running in a virtual environment with Python 3
The Cloud Storage guide to [Setting up a Python development environment](https://cloud.google.com/python/setup) and the [Jupyter installation guide](https://jupyter.org/install) provide detailed instructions for meeting these requirements. The following steps provide a condensed set of instructions:
1. [Install and initialize the SDK](https://cloud.google.com/sdk/docs/).
2. [Install Python 3](https://cloud.google.com/python/setup#installing_python).
3. [Install virtualenv](https://cloud.google.com/python/setup#installing_and_using_virtualenv) and create a virtual environment that uses Python 3. Activate the virtual environment.
4. To install Jupyter, run `pip3 install jupyter` on the command-line in a terminal shell.
5. To launch Jupyter, run `jupyter notebook` on the command-line in a terminal shell.
6. Open this notebook in the Jupyter Notebook Dashboard.
## Installation
Install the latest version of Vertex SDK for Python.
```
import os
# Google Cloud Notebook
if os.path.exists("/opt/deeplearning/metadata/env_version"):
USER_FLAG = "--user"
else:
USER_FLAG = ""
! pip3 install --upgrade google-cloud-aiplatform $USER_FLAG
```
Install the latest GA version of *google-cloud-storage* library as well.
```
! pip3 install -U google-cloud-storage $USER_FLAG
if os.environ["IS_TESTING"]:
! apt-get update && apt-get install -y python3-opencv-headless
! apt-get install -y libgl1-mesa-dev
! pip3 install --upgrade opencv-python-headless $USER_FLAG
if os.environ["IS_TESTING"]:
! pip3 install --upgrade tensorflow $USER_FLAG
```
### Restart the kernel
Once you've installed the additional packages, you need to restart the notebook kernel so it can find the packages.
```
import os
if not os.getenv("IS_TESTING"):
# Automatically restart kernel after installs
import IPython
app = IPython.Application.instance()
app.kernel.do_shutdown(True)
```
## Before you begin
### GPU runtime
This tutorial does not require a GPU runtime.
### Set up your Google Cloud project
**The following steps are required, regardless of your notebook environment.**
1. [Select or create a Google Cloud project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.
2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)
3. [Enable the following APIs: Vertex AI APIs, Compute Engine APIs, and Cloud Storage.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component,storage-component.googleapis.com)
4. If you are running this notebook locally, you will need to install the [Cloud SDK]((https://cloud.google.com/sdk)).
5. Enter your project ID in the cell below. Then run the cell to make sure the
Cloud SDK uses the right project for all the commands in this notebook.
**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$`.
```
PROJECT_ID = "[your-project-id]" # @param {type:"string"}
if PROJECT_ID == "" or PROJECT_ID is None or PROJECT_ID == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = ! gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT_ID = shell_output[0]
print("Project ID:", PROJECT_ID)
! gcloud config set project $PROJECT_ID
```
#### Region
You can also change the `REGION` variable, which is used for operations
throughout the rest of this notebook. Below are regions supported for Vertex AI. We recommend that you choose the region closest to you.
- Americas: `us-central1`
- Europe: `europe-west4`
- Asia Pacific: `asia-east1`
You may not use a multi-regional bucket for training with Vertex AI. Not all regions provide support for all Vertex AI services.
Learn more about [Vertex AI regions](https://cloud.google.com/vertex-ai/docs/general/locations)
```
REGION = "us-central1" # @param {type: "string"}
```
#### Timestamp
If you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append the timestamp onto the name of resources you create in this tutorial.
```
from datetime import datetime
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
```
### Authenticate your Google Cloud account
**If you are using Google Cloud Notebooks**, your environment is already authenticated. Skip this step.
**If you are using Colab**, run the cell below and follow the instructions when prompted to authenticate your account via oAuth.
**Otherwise**, follow these steps:
In the Cloud Console, go to the [Create service account key](https://console.cloud.google.com/apis/credentials/serviceaccountkey) page.
**Click Create service account**.
In the **Service account name** field, enter a name, and click **Create**.
In the **Grant this service account access to project** section, click the Role drop-down list. Type "Vertex" into the filter box, and select **Vertex Administrator**. Type "Storage Object Admin" into the filter box, and select **Storage Object Admin**.
Click Create. A JSON file that contains your key downloads to your local environment.
Enter the path to your service account key as the GOOGLE_APPLICATION_CREDENTIALS variable in the cell below and run the cell.
```
# If you are running this notebook in Colab, run this cell and follow the
# instructions to authenticate your GCP account. This provides access to your
# Cloud Storage bucket and lets you submit training jobs and prediction
# requests.
import os
import sys
# If on Google Cloud Notebook, then don't execute this code
if not os.path.exists("/opt/deeplearning/metadata/env_version"):
if "google.colab" in sys.modules:
from google.colab import auth as google_auth
google_auth.authenticate_user()
# If you are running this notebook locally, replace the string below with the
# path to your service account key and run this cell to authenticate your GCP
# account.
elif not os.getenv("IS_TESTING"):
%env GOOGLE_APPLICATION_CREDENTIALS ''
```
### Create a Cloud Storage bucket
**The following steps are required, regardless of your notebook environment.**
When you initialize the Vertex SDK for Python, you specify a Cloud Storage staging bucket. The staging bucket is where all the data associated with your dataset and model resources are retained across sessions.
Set the name of your Cloud Storage bucket below. Bucket names must be globally unique across all Google Cloud projects, including those outside of your organization.
```
BUCKET_NAME = "gs://[your-bucket-name]" # @param {type:"string"}
if BUCKET_NAME == "" or BUCKET_NAME is None or BUCKET_NAME == "gs://[your-bucket-name]":
BUCKET_NAME = "gs://" + PROJECT_ID + "aip-" + TIMESTAMP
```
**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.
```
! gsutil mb -l $REGION $BUCKET_NAME
```
Finally, validate access to your Cloud Storage bucket by examining its contents:
```
! gsutil ls -al $BUCKET_NAME
```
### Set up variables
Next, set up some variables used throughout the tutorial.
### Import libraries and define constants
```
import google.cloud.aiplatform as aip
```
## Initialize Vertex SDK for Python
Initialize the Vertex SDK for Python for your project and corresponding bucket.
```
aip.init(project=PROJECT_ID, staging_bucket=BUCKET_NAME)
```
#### Set hardware accelerators
You can set hardware accelerators for training and prediction.
Set the variables `TRAIN_GPU/TRAIN_NGPU` and `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Telsa K80 GPUs allocated to each VM, you would specify:
(aip.AcceleratorType.NVIDIA_TESLA_K80, 4)
Otherwise specify `(None, None)` to use a container image to run on a CPU.
Learn more [here](https://cloud.google.com/vertex-ai/docs/general/locations#accelerators) hardware accelerator support for your region
*Note*: TF releases before 2.3 for GPU support will fail to load the custom model in this tutorial. It is a known issue and fixed in TF 2.3 -- which is caused by static graph ops that are generated in the serving function. If you encounter this issue on your own custom models, use a container image for TF 2.3 with GPU support.
```
if os.getenv("IS_TESTING_TRAIN_GPU"):
TRAIN_GPU, TRAIN_NGPU = (
aip.gapic.AcceleratorType.NVIDIA_TESLA_K80,
int(os.getenv("IS_TESTING_TRAIN_GPU")),
)
else:
TRAIN_GPU, TRAIN_NGPU = (None, None)
if os.getenv("IS_TESTING_DEPLOY_GPU"):
DEPLOY_GPU, DEPLOY_NGPU = (
aip.gapic.AcceleratorType.NVIDIA_TESLA_K80,
int(os.getenv("IS_TESTING_DEPLOY_GPU")),
)
else:
DEPLOY_GPU, DEPLOY_NGPU = (None, None)
```
#### Set pre-built containers
Set the pre-built Docker container image for prediction.
- Set the variable `TF` to the TensorFlow version of the container image. For example, `2-1` would be version 2.1, and `1-15` would be version 1.15. The following list shows some of the pre-built images available:
For the latest list, see [Pre-built containers for prediction](https://cloud.google.com/ai-platform-unified/docs/predictions/pre-built-containers).
```
if os.getenv("IS_TESTING_TF"):
TF = os.getenv("IS_TESTING_TF")
else:
TF = "2-1"
if TF[0] == "2":
if DEPLOY_GPU:
DEPLOY_VERSION = "tf2-gpu.{}".format(TF)
else:
DEPLOY_VERSION = "tf2-cpu.{}".format(TF)
else:
if DEPLOY_GPU:
DEPLOY_VERSION = "tf-gpu.{}".format(TF)
else:
DEPLOY_VERSION = "tf-cpu.{}".format(TF)
DEPLOY_IMAGE = "gcr.io/cloud-aiplatform/prediction/{}:latest".format(DEPLOY_VERSION)
print("Deployment:", DEPLOY_IMAGE, DEPLOY_GPU)
```
#### Set machine type
Next, set the machine type to use for training and prediction.
- Set the variables `TRAIN_COMPUTE` and `DEPLOY_COMPUTE` to configure the compute resources for the VMs you will use for for training and prediction.
- `machine type`
- `n1-standard`: 3.75GB of memory per vCPU.
- `n1-highmem`: 6.5GB of memory per vCPU
- `n1-highcpu`: 0.9 GB of memory per vCPU
- `vCPUs`: number of \[2, 4, 8, 16, 32, 64, 96 \]
*Note: The following is not supported for training:*
- `standard`: 2 vCPUs
- `highcpu`: 2, 4 and 8 vCPUs
*Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*.
```
if os.getenv("IS_TESTING_TRAIN_MACHINE"):
MACHINE_TYPE = os.getenv("IS_TESTING_TRAIN_MACHINE")
else:
MACHINE_TYPE = "n1-standard"
VCPU = "4"
TRAIN_COMPUTE = MACHINE_TYPE + "-" + VCPU
print("Train machine type", TRAIN_COMPUTE)
if os.getenv("IS_TESTING_DEPLOY_MACHINE"):
MACHINE_TYPE = os.getenv("IS_TESTING_DEPLOY_MACHINE")
else:
MACHINE_TYPE = "n1-standard"
VCPU = "4"
DEPLOY_COMPUTE = MACHINE_TYPE + "-" + VCPU
print("Deploy machine type", DEPLOY_COMPUTE)
```
### Create a Docker file
In this tutorial, you train a CIFAR10 model using your own custom container.
To use your own custom container, you build a Docker file. First, you will create a directory for the container components.
### Examine the training package
#### Package layout
Before you start the training, you will look at how a Python package is assembled for a custom training job. When unarchived, the package contains the following directory/file layout.
- PKG-INFO
- README.md
- setup.cfg
- setup.py
- trainer
- \_\_init\_\_.py
- task.py
The files `setup.cfg` and `setup.py` are the instructions for installing the package into the operating environment of the Docker image.
The file `trainer/task.py` is the Python script for executing the custom training job. *Note*, when we referred to it in the worker pool specification, we replace the directory slash with a dot (`trainer.task`) and dropped the file suffix (`.py`).
#### Package Assembly
In the following cells, you will assemble the training package.
```
# Make folder for Python training script
! rm -rf custom
! mkdir custom
# Add package information
! touch custom/README.md
setup_cfg = "[egg_info]\n\ntag_build =\n\ntag_date = 0"
! echo "$setup_cfg" > custom/setup.cfg
setup_py = "import setuptools\n\nsetuptools.setup(\n\n install_requires=[\n\n 'tensorflow_datasets==1.3.0',\n\n ],\n\n packages=setuptools.find_packages())"
! echo "$setup_py" > custom/setup.py
pkg_info = "Metadata-Version: 1.0\n\nName: CIFAR10 image classification\n\nVersion: 0.0.0\n\nSummary: Demostration training script\n\nHome-page: www.google.com\n\nAuthor: Google\n\nAuthor-email: [email protected]\n\nLicense: Public\n\nDescription: Demo\n\nPlatform: Vertex"
! echo "$pkg_info" > custom/PKG-INFO
# Make the training subfolder
! mkdir custom/trainer
! touch custom/trainer/__init__.py
```
#### Task.py contents
In the next cell, you write the contents of the training script task.py. We won't go into detail, it's just there for you to browse. In summary:
- Get the directory where to save the model artifacts from the command line (`--model_dir`), and if not specified, then from the environment variable `AIP_MODEL_DIR`.
- Loads CIFAR10 dataset from TF Datasets (tfds).
- Builds a model using TF.Keras model API.
- Compiles the model (`compile()`).
- Sets a training distribution strategy according to the argument `args.distribute`.
- Trains the model (`fit()`) with epochs and steps according to the arguments `args.epochs` and `args.steps`
- Saves the trained model (`save(args.model_dir)`) to the specified model directory.
```
%%writefile custom/trainer/task.py
# Single, Mirror and Multi-Machine Distributed Training for CIFAR-10
import tensorflow_datasets as tfds
import tensorflow as tf
from tensorflow.python.client import device_lib
import argparse
import os
import sys
tfds.disable_progress_bar()
parser = argparse.ArgumentParser()
parser.add_argument('--model-dir', dest='model_dir',
default=os.getenv("AIP_MODEL_DIR"), type=str, help='Model dir.')
parser.add_argument('--lr', dest='lr',
default=0.01, type=float,
help='Learning rate.')
parser.add_argument('--epochs', dest='epochs',
default=10, type=int,
help='Number of epochs.')
parser.add_argument('--steps', dest='steps',
default=200, type=int,
help='Number of steps per epoch.')
parser.add_argument('--distribute', dest='distribute', type=str, default='single',
help='distributed training strategy')
args = parser.parse_args()
print('Python Version = {}'.format(sys.version))
print('TensorFlow Version = {}'.format(tf.__version__))
print('TF_CONFIG = {}'.format(os.environ.get('TF_CONFIG', 'Not found')))
print('DEVICES', device_lib.list_local_devices())
# Single Machine, single compute device
if args.distribute == 'single':
if tf.test.is_gpu_available():
strategy = tf.distribute.OneDeviceStrategy(device="/gpu:0")
else:
strategy = tf.distribute.OneDeviceStrategy(device="/cpu:0")
# Single Machine, multiple compute device
elif args.distribute == 'mirror':
strategy = tf.distribute.MirroredStrategy()
# Multiple Machine, multiple compute device
elif args.distribute == 'multi':
strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()
# Multi-worker configuration
print('num_replicas_in_sync = {}'.format(strategy.num_replicas_in_sync))
# Preparing dataset
BUFFER_SIZE = 10000
BATCH_SIZE = 64
def make_datasets_unbatched():
# Scaling CIFAR10 data from (0, 255] to (0., 1.]
def scale(image, label):
image = tf.cast(image, tf.float32)
image /= 255.0
return image, label
datasets, info = tfds.load(name='cifar10',
with_info=True,
as_supervised=True)
return datasets['train'].map(scale).cache().shuffle(BUFFER_SIZE).repeat()
# Build the Keras model
def build_and_compile_cnn_model():
model = tf.keras.Sequential([
tf.keras.layers.Conv2D(32, 3, activation='relu', input_shape=(32, 32, 3)),
tf.keras.layers.MaxPooling2D(),
tf.keras.layers.Conv2D(32, 3, activation='relu'),
tf.keras.layers.MaxPooling2D(),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(10, activation='softmax')
])
model.compile(
loss=tf.keras.losses.sparse_categorical_crossentropy,
optimizer=tf.keras.optimizers.SGD(learning_rate=args.lr),
metrics=['accuracy'])
return model
# Train the model
NUM_WORKERS = strategy.num_replicas_in_sync
# Here the batch size scales up by number of workers since
# `tf.data.Dataset.batch` expects the global batch size.
GLOBAL_BATCH_SIZE = BATCH_SIZE * NUM_WORKERS
train_dataset = make_datasets_unbatched().batch(GLOBAL_BATCH_SIZE)
with strategy.scope():
# Creation of dataset, and model building/compiling need to be within
# `strategy.scope()`.
model = build_and_compile_cnn_model()
model.fit(x=train_dataset, epochs=args.epochs, steps_per_epoch=args.steps)
model.save(args.model_dir)
```
#### Write the Docker file contents
Your first step in containerizing your code is to create a Docker file. In your Docker you’ll include all the commands needed to run your container image. It’ll install all the libraries you’re using and set up the entry point for your training code.
1. Install a pre-defined container image from TensorFlow repository for deep learning images.
2. Copies in the Python training code, to be shown subsequently.
3. Sets the entry into the Python training script as `trainer/task.py`. Note, the `.py` is dropped in the ENTRYPOINT command, as it is implied.
```
%%writefile custom/Dockerfile
FROM gcr.io/deeplearning-platform-release/tf2-cpu.2-3
WORKDIR /root
WORKDIR /
# Copies the trainer code to the docker image.
COPY trainer /trainer
# Sets up the entry point to invoke the trainer.
ENTRYPOINT ["python", "-m", "trainer.task"]
```
#### Build the container locally
Next, you will provide a name for your customer container that you will use when you submit it to the Google Container Registry.
```
TRAIN_IMAGE = "gcr.io/" + PROJECT_ID + "/cifar10:v1"
```
Next, build the container.
```
! docker build custom -t $TRAIN_IMAGE
```
#### Test the container locally
Run the container within your notebook instance to ensure it’s working correctly. You will run it for 5 epochs.
```
! docker run $TRAIN_IMAGE --epochs=5
```
#### Register the custom container
When you’ve finished running the container locally, push it to Google Container Registry.
```
! docker push $TRAIN_IMAGE
```
#### Store training script on your Cloud Storage bucket
Next, you package the training folder into a compressed tar ball, and then store it in your Cloud Storage bucket.
```
! rm -f custom.tar custom.tar.gz
! tar cvf custom.tar custom
! gzip custom.tar
! gsutil cp custom.tar.gz $BUCKET_NAME/trainer_cifar10.tar.gz
```
## Train a model
### [training.containers-overview](https://cloud.google.com/vertex-ai/docs/training/containers-overview)
### Create and run custom training job
To train a custom model, you perform two steps: 1) create a custom training job, and 2) run the job.
#### Create custom training job
A custom training job is created with the `CustomTrainingJob` class, with the following parameters:
- `display_name`: The human readable name for the custom training job.
- `container_uri`: The training container image.
```
job = aip.CustomContainerTrainingJob(
display_name="cifar10_" + TIMESTAMP, container_uri=TRAIN_IMAGE
)
print(job)
```
*Example output:*
<google.cloud.aiplatform.training_jobs.CustomContainerTrainingJob object at 0x7feab1346710>
#### Run the custom training job
Next, you run the custom job to start the training job by invoking the method `run`, with the following parameters:
- `args`: The command-line arguments to pass to the training script.
- `replica_count`: The number of compute instances for training (replica_count = 1 is single node training).
- `machine_type`: The machine type for the compute instances.
- `accelerator_type`: The hardware accelerator type.
- `accelerator_count`: The number of accelerators to attach to a worker replica.
- `base_output_dir`: The Cloud Storage location to write the model artifacts to.
- `sync`: Whether to block until completion of the job.
```
MODEL_DIR = "{}/{}".format(BUCKET_NAME, TIMESTAMP)
EPOCHS = 20
STEPS = 100
DIRECT = True
if DIRECT:
CMDARGS = [
"--model-dir=" + MODEL_DIR,
"--epochs=" + str(EPOCHS),
"--steps=" + str(STEPS),
]
else:
CMDARGS = [
"--epochs=" + str(EPOCHS),
"--steps=" + str(STEPS),
]
if TRAIN_GPU:
job.run(
args=CMDARGS,
replica_count=1,
machine_type=TRAIN_COMPUTE,
accelerator_type=TRAIN_GPU.name,
accelerator_count=TRAIN_NGPU,
base_output_dir=MODEL_DIR,
sync=True,
)
else:
job.run(
args=CMDARGS,
replica_count=1,
machine_type=TRAIN_COMPUTE,
base_output_dir=MODEL_DIR,
sync=True,
)
model_path_to_deploy = MODEL_DIR
```
### Wait for completion of custom training job
Next, wait for the custom training job to complete. Alternatively, one can set the parameter `sync` to `True` in the `run()` methid to block until the custom training job is completed.
## Evaluate the model
## Load the saved model
Your model is stored in a TensorFlow SavedModel format in a Cloud Storage bucket. Now load it from the Cloud Storage bucket, and then you can do some things, like evaluate the model, and do a prediction.
To load, you use the TF.Keras `model.load_model()` method passing it the Cloud Storage path where the model is saved -- specified by `MODEL_DIR`.
```
import tensorflow as tf
local_model = tf.keras.models.load_model(MODEL_DIR)
```
## Evaluate the model
Now find out how good the model is.
### Load evaluation data
You will load the CIFAR10 test (holdout) data from `tf.keras.datasets`, using the method `load_data()`. This returns the dataset as a tuple of two elements. The first element is the training data and the second is the test data. Each element is also a tuple of two elements: the image data, and the corresponding labels.
You don't need the training data, and hence why we loaded it as `(_, _)`.
Before you can run the data through evaluation, you need to preprocess it:
`x_test`:
1. Normalize (rescale) the pixel data by dividing each pixel by 255. This replaces each single byte integer pixel with a 32-bit floating point number between 0 and 1.
`y_test`:<br/>
2. The labels are currently scalar (sparse). If you look back at the `compile()` step in the `trainer/task.py` script, you will find that it was compiled for sparse labels. So we don't need to do anything more.
```
import numpy as np
from tensorflow.keras.datasets import cifar10
(_, _), (x_test, y_test) = cifar10.load_data()
x_test = (x_test / 255.0).astype(np.float32)
print(x_test.shape, y_test.shape)
```
### Perform the model evaluation
Now evaluate how well the model in the custom job did.
```
local_model.evaluate(x_test, y_test)
```
### [general.import-model](https://cloud.google.com/vertex-ai/docs/general/import-model)
### Serving function for image data
To pass images to the prediction service, you encode the compressed (e.g., JPEG) image bytes into base 64 -- which makes the content safe from modification while transmitting binary data over the network. Since this deployed model expects input data as raw (uncompressed) bytes, you need to ensure that the base 64 encoded data gets converted back to raw bytes before it is passed as input to the deployed model.
To resolve this, define a serving function (`serving_fn`) and attach it to the model as a preprocessing step. Add a `@tf.function` decorator so the serving function is fused to the underlying model (instead of upstream on a CPU).
When you send a prediction or explanation request, the content of the request is base 64 decoded into a Tensorflow string (`tf.string`), which is passed to the serving function (`serving_fn`). The serving function preprocesses the `tf.string` into raw (uncompressed) numpy bytes (`preprocess_fn`) to match the input requirements of the model:
- `io.decode_jpeg`- Decompresses the JPG image which is returned as a Tensorflow tensor with three channels (RGB).
- `image.convert_image_dtype` - Changes integer pixel values to float 32.
- `image.resize` - Resizes the image to match the input shape for the model.
- `resized / 255.0` - Rescales (normalization) the pixel data between 0 and 1.
At this point, the data can be passed to the model (`m_call`).
```
CONCRETE_INPUT = "numpy_inputs"
def _preprocess(bytes_input):
decoded = tf.io.decode_jpeg(bytes_input, channels=3)
decoded = tf.image.convert_image_dtype(decoded, tf.float32)
resized = tf.image.resize(decoded, size=(32, 32))
rescale = tf.cast(resized / 255.0, tf.float32)
return rescale
@tf.function(input_signature=[tf.TensorSpec([None], tf.string)])
def preprocess_fn(bytes_inputs):
decoded_images = tf.map_fn(
_preprocess, bytes_inputs, dtype=tf.float32, back_prop=False
)
return {
CONCRETE_INPUT: decoded_images
} # User needs to make sure the key matches model's input
@tf.function(input_signature=[tf.TensorSpec([None], tf.string)])
def serving_fn(bytes_inputs):
images = preprocess_fn(bytes_inputs)
prob = m_call(**images)
return prob
m_call = tf.function(local_model.call).get_concrete_function(
[tf.TensorSpec(shape=[None, 32, 32, 3], dtype=tf.float32, name=CONCRETE_INPUT)]
)
tf.saved_model.save(
local_model, model_path_to_deploy, signatures={"serving_default": serving_fn}
)
```
## Get the serving function signature
You can get the signatures of your model's input and output layers by reloading the model into memory, and querying it for the signatures corresponding to each layer.
For your purpose, you need the signature of the serving function. Why? Well, when we send our data for prediction as a HTTP request packet, the image data is base64 encoded, and our TF.Keras model takes numpy input. Your serving function will do the conversion from base64 to a numpy array.
When making a prediction request, you need to route the request to the serving function instead of the model, so you need to know the input layer name of the serving function -- which you will use later when you make a prediction request.
```
loaded = tf.saved_model.load(model_path_to_deploy)
serving_input = list(
loaded.signatures["serving_default"].structured_input_signature[1].keys()
)[0]
print("Serving function input:", serving_input)
```
## Upload the model
Next, upload your model to a `Model` resource using `Model.upload()` method, with the following parameters:
- `display_name`: The human readable name for the `Model` resource.
- `artifact`: The Cloud Storage location of the trained model artifacts.
- `serving_container_image_uri`: The serving container image.
- `sync`: Whether to execute the upload asynchronously or synchronously.
If the `upload()` method is run asynchronously, you can subsequently block until completion with the `wait()` method.
```
model = aip.Model.upload(
display_name="cifar10_" + TIMESTAMP,
artifact_uri=MODEL_DIR,
serving_container_image_uri=DEPLOY_IMAGE,
sync=False,
)
model.wait()
```
*Example output:*
INFO:google.cloud.aiplatform.models:Creating Model
INFO:google.cloud.aiplatform.models:Create Model backing LRO: projects/759209241365/locations/us-central1/models/925164267982815232/operations/3458372263047331840
INFO:google.cloud.aiplatform.models:Model created. Resource name: projects/759209241365/locations/us-central1/models/925164267982815232
INFO:google.cloud.aiplatform.models:To use this Model in another session:
INFO:google.cloud.aiplatform.models:model = aiplatform.Model('projects/759209241365/locations/us-central1/models/925164267982815232')
## Make batch predictions
### [predictions.batch-prediction](https://cloud.google.com/vertex-ai/docs/predictions/batch-predictions)
### Get test items
You will use examples out of the test (holdout) portion of the dataset as a test items.
```
test_image_1 = x_test[0]
test_label_1 = y_test[0]
test_image_2 = x_test[1]
test_label_2 = y_test[1]
print(test_image_1.shape)
```
### Prepare the request content
You are going to send the CIFAR10 images as compressed JPG image, instead of the raw uncompressed bytes:
- `cv2.imwrite`: Use openCV to write the uncompressed image to disk as a compressed JPEG image.
- Denormalize the image data from \[0,1) range back to [0,255).
- Convert the 32-bit floating point values to 8-bit unsigned integers.
```
import cv2
cv2.imwrite("tmp1.jpg", (test_image_1 * 255).astype(np.uint8))
cv2.imwrite("tmp2.jpg", (test_image_2 * 255).astype(np.uint8))
```
### Copy test item(s)
For the batch prediction, copy the test items over to your Cloud Storage bucket.
```
! gsutil cp tmp1.jpg $BUCKET_NAME/tmp1.jpg
! gsutil cp tmp2.jpg $BUCKET_NAME/tmp2.jpg
test_item_1 = BUCKET_NAME + "/tmp1.jpg"
test_item_2 = BUCKET_NAME + "/tmp2.jpg"
```
### Make the batch input file
Now make a batch input file, which you will store in your local Cloud Storage bucket. The batch input file can only be in JSONL format. For JSONL file, you make one dictionary entry per line for each data item (instance). The dictionary contains the key/value pairs:
- `input_name`: the name of the input layer of the underlying model.
- `'b64'`: A key that indicates the content is base64 encoded.
- `content`: The compressed JPG image bytes as a base64 encoded string.
Each instance in the prediction request is a dictionary entry of the form:
{serving_input: {'b64': content}}
To pass the image data to the prediction service you encode the bytes into base64 -- which makes the content safe from modification when transmitting binary data over the network.
- `tf.io.read_file`: Read the compressed JPG images into memory as raw bytes.
- `base64.b64encode`: Encode the raw bytes into a base64 encoded string.
```
import base64
import json
gcs_input_uri = BUCKET_NAME + "/" + "test.jsonl"
with tf.io.gfile.GFile(gcs_input_uri, "w") as f:
bytes = tf.io.read_file(test_item_1)
b64str = base64.b64encode(bytes.numpy()).decode("utf-8")
data = {serving_input: {"b64": b64str}}
f.write(json.dumps(data) + "\n")
bytes = tf.io.read_file(test_item_2)
b64str = base64.b64encode(bytes.numpy()).decode("utf-8")
data = {serving_input: {"b64": b64str}}
f.write(json.dumps(data) + "\n")
```
### Make the batch prediction request
Now that your Model resource is trained, you can make a batch prediction by invoking the batch_predict() method, with the following parameters:
- `job_display_name`: The human readable name for the batch prediction job.
- `gcs_source`: A list of one or more batch request input files.
- `gcs_destination_prefix`: The Cloud Storage location for storing the batch prediction resuls.
- `instances_format`: The format for the input instances, either 'csv' or 'jsonl'. Defaults to 'jsonl'.
- `predictions_format`: The format for the output predictions, either 'csv' or 'jsonl'. Defaults to 'jsonl'.
- `machine_type`: The type of machine to use for training.
- `accelerator_type`: The hardware accelerator type.
- `accelerator_count`: The number of accelerators to attach to a worker replica.
- `sync`: If set to True, the call will block while waiting for the asynchronous batch job to complete.
```
MIN_NODES = 1
MAX_NODES = 1
batch_predict_job = model.batch_predict(
job_display_name="cifar10_" + TIMESTAMP,
gcs_source=gcs_input_uri,
gcs_destination_prefix=BUCKET_NAME,
instances_format="jsonl",
predictions_format="jsonl",
model_parameters=None,
machine_type=DEPLOY_COMPUTE,
accelerator_type=DEPLOY_GPU,
accelerator_count=DEPLOY_NGPU,
starting_replica_count=MIN_NODES,
max_replica_count=MAX_NODES,
sync=False,
)
print(batch_predict_job)
```
*Example output:*
INFO:google.cloud.aiplatform.jobs:Creating BatchPredictionJob
<google.cloud.aiplatform.jobs.BatchPredictionJob object at 0x7f806a6112d0> is waiting for upstream dependencies to complete.
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob created. Resource name: projects/759209241365/locations/us-central1/batchPredictionJobs/5110965452507447296
INFO:google.cloud.aiplatform.jobs:To use this BatchPredictionJob in another session:
INFO:google.cloud.aiplatform.jobs:bpj = aiplatform.BatchPredictionJob('projects/759209241365/locations/us-central1/batchPredictionJobs/5110965452507447296')
INFO:google.cloud.aiplatform.jobs:View Batch Prediction Job:
https://console.cloud.google.com/ai/platform/locations/us-central1/batch-predictions/5110965452507447296?project=759209241365
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/5110965452507447296 current state:
JobState.JOB_STATE_RUNNING
### Wait for completion of batch prediction job
Next, wait for the batch job to complete. Alternatively, one can set the parameter `sync` to `True` in the `batch_predict()` method to block until the batch prediction job is completed.
```
batch_predict_job.wait()
```
*Example Output:*
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob created. Resource name: projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328
INFO:google.cloud.aiplatform.jobs:To use this BatchPredictionJob in another session:
INFO:google.cloud.aiplatform.jobs:bpj = aiplatform.BatchPredictionJob('projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328')
INFO:google.cloud.aiplatform.jobs:View Batch Prediction Job:
https://console.cloud.google.com/ai/platform/locations/us-central1/batch-predictions/181835033978339328?project=759209241365
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_RUNNING
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328 current state:
JobState.JOB_STATE_SUCCEEDED
INFO:google.cloud.aiplatform.jobs:BatchPredictionJob run completed. Resource name: projects/759209241365/locations/us-central1/batchPredictionJobs/181835033978339328
### Get the predictions
Next, get the results from the completed batch prediction job.
The results are written to the Cloud Storage output bucket you specified in the batch prediction request. You call the method iter_outputs() to get a list of each Cloud Storage file generated with the results. Each file contains one or more prediction requests in a JSON format:
- `instance`: The prediction request.
- `prediction`: The prediction response.
```
import json
bp_iter_outputs = batch_predict_job.iter_outputs()
prediction_results = list()
for blob in bp_iter_outputs:
if blob.name.split("/")[-1].startswith("prediction"):
prediction_results.append(blob.name)
tags = list()
for prediction_result in prediction_results:
gfile_name = f"gs://{bp_iter_outputs.bucket.name}/{prediction_result}"
with tf.io.gfile.GFile(name=gfile_name, mode="r") as gfile:
for line in gfile.readlines():
line = json.loads(line)
print(line)
break
```
*Example Output:*
{'instance': {'bytes_inputs': {'b64': '/9j/4AAQSkZJRgABAQAAAQABAAD/2wBDAAIBAQEBAQIBAQECAgICAgQDAgICAgUEBAMEBgUGBgYFBgYGBwkIBgcJBwYGCAsICQoKCgoKBggLDAsKDAkKCgr/2wBDAQICAgICAgUDAwUKBwYHCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgoKCgr/wAARCAAgACADASIAAhEBAxEB/8QAHwAAAQUBAQEBAQEAAAAAAAAAAAECAwQFBgcICQoL/8QAtRAAAgEDAwIEAwUFBAQAAAF9AQIDAAQRBRIhMUEGE1FhByJxFDKBkaEII0KxwRVS0fAkM2JyggkKFhcYGRolJicoKSo0NTY3ODk6Q0RFRkdISUpTVFVWV1hZWmNkZWZnaGlqc3R1dnd4eXqDhIWGh4iJipKTlJWWl5iZmqKjpKWmp6ipqrKztLW2t7i5usLDxMXGx8jJytLT1NXW19jZ2uHi4+Tl5ufo6erx8vP09fb3+Pn6/8QAHwEAAwEBAQEBAQEBAQAAAAAAAAECAwQFBgcICQoL/8QAtREAAgECBAQDBAcFBAQAAQJ3AAECAxEEBSExBhJBUQdhcRMiMoEIFEKRobHBCSMzUvAVYnLRChYkNOEl8RcYGRomJygpKjU2Nzg5OkNERUZHSElKU1RVVldYWVpjZGVmZ2hpanN0dXZ3eHl6goOEhYaHiImKkpOUlZaXmJmaoqOkpaanqKmqsrO0tba3uLm6wsPExcbHyMnK0tPU1dbX2Nna4uPk5ebn6Onq8vP09fb3+Pn6/9oADAMBAAIRAxEAPwD570PxBpmp6nfaEl48lzpUqpewPCU8lpEDqMsOeD26Z55Fa+s3HhnR/Aj6xZjV7rWrW4ke/wBMtLRGRLTaux1cuPnLlhtIAAUEE5490/ao8E6F4b8P3NxZeGksNW1z4h62Iby2t1/eC3ZoozJxwSiKQOhEZJ5JrqZtI8MftFfs56j8YI/hvo/gq1u9C0ywlbTbFoLa+1SOFWlgPGRmNiQzNkiPOflyf1WHFdark0K8UlUbkvJWel15ppn5MuD6MM6qUJzbppRdrO8lJa2a7NNHyJoGheKvHngfUfGjXSaHHZX/ANmW2kQTsHIBXzDxgt1GMAcDPU1xI1xdS16/8FaxNA2o2kPmGS2OI51zyV65Izz0z1xg1718Ivhd4b8IfBX4qeItWuxql+2tW+n6dHPOEijt1s9xYgnaR50hw2dvygDrXz/4v+HWo6ha6X8R/C7iwv7CTy7YiRSLslGG3AzlGAGQenPTFfL4XiDMvr0ZVZuSk/ej66adj6bGcPZX/Z8oUoKHKtJemurP1H+OekS/tAeAvDmpfDjw/wDbL3W/FOlalpkNgqyhJrtgsqPg4ACyyK4J9c1418XP2X4P2ev2jNQ+C3x6+OnhbRfCtpJHfLp1p4klkD73kldkhRAYTKzoSkmSmxiNysDXK/stftQD9kn9oSx8aa3p0uq+GdN1drq70W3cAJKYmRLmINgbl35xwGAI4ODXiXxK+Mtp8W/G+v8Ajvxl4mn/ALW1TU5bq6u9Q+fzHZixG8dFyQB0wOOnFfjuH40f1GNSnG05P3o9F5r9D9dr8LReNdOs7wS0l19PwKPxZ8TeNNAkvPh/8GruO8BE9v8A8JHbaq8VrPA8h+aSBl5mKKiiYAlQowRnAh+H/gWTwx4MiTV52vdRUlTLPMJNgK/NsJxgEgnpwGxmtnSfDsOl6VH4nuLWG8glbCtHcb1bvjqD+PSu78SSXfwn8F2XjnxHo2n3smpSKdPsJCpW3iB+Z2VRl2VckA4HA6k1xf8AEQs9wOKVWjGN0rK8eZLp1/M2nwLkuOwsqNWUrN3dpWb620P/2Q=='}}, 'prediction': [0.0560616329, 0.122713037, 0.121289924, 0.109751239, 0.121320881, 0.0897410363, 0.145011798, 0.0976110101, 0.0394041203, 0.0970953554]}
## Make online predictions
### [predictions.deploy-model-api](https://cloud.google.com/vertex-ai/docs/predictions/deploy-model-api)
## Deploy the model
Next, deploy your model for online prediction. To deploy the model, you invoke the `deploy` method, with the following parameters:
- `deployed_model_display_name`: A human readable name for the deployed model.
- `traffic_split`: Percent of traffic at the endpoint that goes to this model, which is specified as a dictionary of one or more key/value pairs.
If only one model, then specify as { "0": 100 }, where "0" refers to this model being uploaded and 100 means 100% of the traffic.
If there are existing models on the endpoint, for which the traffic will be split, then use model_id to specify as { "0": percent, model_id: percent, ... }, where model_id is the model id of an existing model to the deployed endpoint. The percents must add up to 100.
- `machine_type`: The type of machine to use for training.
- `accelerator_type`: The hardware accelerator type.
- `accelerator_count`: The number of accelerators to attach to a worker replica.
- `starting_replica_count`: The number of compute instances to initially provision.
- `max_replica_count`: The maximum number of compute instances to scale to. In this tutorial, only one instance is provisioned.
```
DEPLOYED_NAME = "cifar10-" + TIMESTAMP
TRAFFIC_SPLIT = {"0": 100}
MIN_NODES = 1
MAX_NODES = 1
if DEPLOY_GPU:
endpoint = model.deploy(
deployed_model_display_name=DEPLOYED_NAME,
traffic_split=TRAFFIC_SPLIT,
machine_type=DEPLOY_COMPUTE,
accelerator_type=DEPLOY_GPU,
accelerator_count=DEPLOY_NGPU,
min_replica_count=MIN_NODES,
max_replica_count=MAX_NODES,
)
else:
endpoint = model.deploy(
deployed_model_display_name=DEPLOYED_NAME,
traffic_split=TRAFFIC_SPLIT,
machine_type=DEPLOY_COMPUTE,
accelerator_type=DEPLOY_GPU,
accelerator_count=0,
min_replica_count=MIN_NODES,
max_replica_count=MAX_NODES,
)
```
*Example output:*
INFO:google.cloud.aiplatform.models:Creating Endpoint
INFO:google.cloud.aiplatform.models:Create Endpoint backing LRO: projects/759209241365/locations/us-central1/endpoints/4867177336350441472/operations/4087251132693348352
INFO:google.cloud.aiplatform.models:Endpoint created. Resource name: projects/759209241365/locations/us-central1/endpoints/4867177336350441472
INFO:google.cloud.aiplatform.models:To use this Endpoint in another session:
INFO:google.cloud.aiplatform.models:endpoint = aiplatform.Endpoint('projects/759209241365/locations/us-central1/endpoints/4867177336350441472')
INFO:google.cloud.aiplatform.models:Deploying model to Endpoint : projects/759209241365/locations/us-central1/endpoints/4867177336350441472
INFO:google.cloud.aiplatform.models:Deploy Endpoint model backing LRO: projects/759209241365/locations/us-central1/endpoints/4867177336350441472/operations/1691336130932244480
INFO:google.cloud.aiplatform.models:Endpoint model deployed. Resource name: projects/759209241365/locations/us-central1/endpoints/4867177336350441472
### [predictions.online-prediction-automl](https://cloud.google.com/vertex-ai/docs/predictions/online-predictions-automl)
### Get test item
You will use an example out of the test (holdout) portion of the dataset as a test item.
```
test_image = x_test[0]
test_label = y_test[0]
print(test_image.shape)
```
### Prepare the request content
You are going to send the CIFAR10 image as compressed JPG image, instead of the raw uncompressed bytes:
- `cv2.imwrite`: Use openCV to write the uncompressed image to disk as a compressed JPEG image.
- Denormalize the image data from \[0,1) range back to [0,255).
- Convert the 32-bit floating point values to 8-bit unsigned integers.
- `tf.io.read_file`: Read the compressed JPG images back into memory as raw bytes.
- `base64.b64encode`: Encode the raw bytes into a base 64 encoded string.
```
import base64
import cv2
cv2.imwrite("tmp.jpg", (test_image * 255).astype(np.uint8))
bytes = tf.io.read_file("tmp.jpg")
b64str = base64.b64encode(bytes.numpy()).decode("utf-8")
```
### Make the prediction
Now that your `Model` resource is deployed to an `Endpoint` resource, you can do online predictions by sending prediction requests to the Endpoint resource.
#### Request
Since in this example your test item is in a Cloud Storage bucket, you open and read the contents of the image using `tf.io.gfile.Gfile()`. To pass the test data to the prediction service, you encode the bytes into base64 -- which makes the content safe from modification while transmitting binary data over the network.
The format of each instance is:
{ serving_input: { 'b64': base64_encoded_bytes } }
Since the `predict()` method can take multiple items (instances), send your single test item as a list of one test item.
#### Response
The response from the `predict()` call is a Python dictionary with the following entries:
- `ids`: The internal assigned unique identifiers for each prediction request.
- `predictions`: The predicted confidence, between 0 and 1, per class label.
- `deployed_model_id`: The Vertex AI identifier for the deployed `Model` resource which did the predictions.
```
# The format of each instance should conform to the deployed model's prediction input schema.
instances = [{serving_input: {"b64": b64str}}]
prediction = endpoint.predict(instances=instances)
print(prediction)
```
*Example output:*
Prediction(predictions=[[0.0560616292, 0.122713044, 0.121289924, 0.109751239, 0.121320873, 0.0897410288, 0.145011798, 0.0976110175, 0.0394041166, 0.0970953479]], deployed_model_id='4087166195420102656', explanations=None)
## Undeploy the model
When you are done doing predictions, you undeploy the model from the `Endpoint` resouce. This deprovisions all compute resources and ends billing for the deployed model.
```
endpoint.undeploy_all()
```
# Cleaning up
To clean up all Google Cloud resources used in this project, you can [delete the Google Cloud
project](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.
Otherwise, you can delete the individual resources you created in this tutorial:
- Dataset
- Pipeline
- Model
- Endpoint
- AutoML Training Job
- Batch Job
- Custom Job
- Hyperparameter Tuning Job
- Cloud Storage Bucket
```
delete_all = True
if delete_all:
# Delete the dataset using the Vertex dataset object
try:
if "dataset" in globals():
dataset.delete()
except Exception as e:
print(e)
# Delete the model using the Vertex model object
try:
if "model" in globals():
model.delete()
except Exception as e:
print(e)
# Delete the endpoint using the Vertex endpoint object
try:
if "endpoint" in globals():
endpoint.delete()
except Exception as e:
print(e)
# Delete the AutoML or Pipeline trainig job
try:
if "dag" in globals():
dag.delete()
except Exception as e:
print(e)
# Delete the custom trainig job
try:
if "job" in globals():
job.delete()
except Exception as e:
print(e)
# Delete the batch prediction job using the Vertex batch prediction object
try:
if "batch_predict_job" in globals():
batch_predict_job.delete()
except Exception as e:
print(e)
# Delete the hyperparameter tuning job using the Vertex hyperparameter tuning object
try:
if "hpt_job" in globals():
hpt_job.delete()
except Exception as e:
print(e)
if "BUCKET_NAME" in globals():
! gsutil rm -r $BUCKET_NAME
```
|
github_jupyter
|
# Classification algorithms
In the context of record linkage, classification refers to the process of dividing record pairs into matches and non-matches (distinct pairs). There are dozens of classification algorithms for record linkage. Roughly speaking, classification algorithms fall into two groups:
- **supervised learning algorithms** - These algorithms make use of trainings data. If you do have trainings data, then you can use supervised learning algorithms. Most supervised learning algorithms offer good accuracy and reliability. Examples of supervised learning algorithms in the *Python Record Linkage Toolkit* are *Logistic Regression*, *Naive Bayes* and *Support Vector Machines*.
- **unsupervised learning algorithms** - These algorithms do not need training data. The *Python Record Linkage Toolkit* supports *K-means clustering* and an *Expectation/Conditional Maximisation* classifier.
```
%precision 5
from __future__ import print_function
import pandas as pd
pd.set_option('precision',5)
pd.options.display.max_rows = 10
```
**First things first**
The examples below make use of the [Krebs register](http://recordlinkage.readthedocs.org/en/latest/reference.html#recordlinkage.datasets.krebsregister_cmp_data) (German for cancer registry) dataset. The Krebs register dataset contains comparison vectors of a large set of record pairs. For each record pair, it is known if the records represent the same person (match) or not (non-match). This was done with a massive clerical review. First, import the recordlinkage module and load the Krebs register data. The dataset contains 5749132 compared record pairs and has the following variables: first name, last name, sex, birthday, birth month, birth year and zip code. The Krebs register contains `len(krebs_true_links) == 20931` matching record pairs.
```
import recordlinkage as rl
from recordlinkage.datasets import load_krebsregister
krebs_X, krebs_true_links = load_krebsregister(missing_values=0)
krebs_X
```
Most classifiers can not handle comparison vectors with missing values. To prevent issues with the classification algorithms, we convert the missing values into disagreeing comparisons (using argument missing_values=0). This approach for handling missing values is widely used in record linkage applications.
```
krebs_X.describe().T
```
## Supervised learning
As described before, supervised learning algorithms do need training data. Training data is data for which the true match status is known for each comparison vector. In the example in this section, we consider that the true match status of the first 5000 record pairs of the Krebs register data is known.
```
golden_pairs = krebs_X[0:5000]
golden_matches_index = golden_pairs.index & krebs_true_links # 2093 matching pairs
```
### Logistic regression
The ``recordlinkage.LogisticRegressionClassifier`` classifier is an application of the logistic regression model. This supervised learning method is one of the oldest classification algorithms used in record linkage. In situations with enough training data, the algorithm gives relatively good results.
```
# Initialize the classifier
logreg = rl.LogisticRegressionClassifier()
# Train the classifier
logreg.fit(golden_pairs, golden_matches_index)
print ("Intercept: ", logreg.intercept)
print ("Coefficients: ", logreg.coefficients)
# Predict the match status for all record pairs
result_logreg = logreg.predict(krebs_X)
len(result_logreg)
rl.confusion_matrix(krebs_true_links, result_logreg, len(krebs_X))
# The F-score for this prediction is
rl.fscore(krebs_true_links, result_logreg)
```
The predicted number of matches is not much more than the 20931 true matches. The result was achieved with a small training dataset of 5000 record pairs.
In (older) literature, record linkage procedures are often divided in **deterministic record linkage** and **probabilistic record linkage**. The Logistic Regression Classifier belongs to deterministic record linkage methods. Each feature/variable has a certain importance (named weight). The weight is multiplied with the comparison/similarity vector. If the total sum exceeds a certain threshold, it as considered to be a match.
```
intercept = -9
coefficients = [2.0, 1.0, 3.0, 1.0, 1.0, 1.0, 1.0, 2.0, 3.0]
logreg = rl.LogisticRegressionClassifier(coefficients, intercept)
# predict without calling LogisticRegressionClassifier.fit
result_logreg_pretrained = logreg.predict(krebs_X)
print (len(result_logreg_pretrained))
rl.confusion_matrix(krebs_true_links, result_logreg_pretrained, len(krebs_X))
# The F-score for this classification is
rl.fscore(krebs_true_links, result_logreg_pretrained)
```
For the given coefficients, the F-score is better than the situation without trainings data. Surprising? No (use more trainings data and the result will improve)
### Naive Bayes
In contrast to the logistic regression classifier, the Naive Bayes classifier is a probabilistic classifier. The probabilistic record linkage framework by Fellegi and Sunter (1969) is the most well-known probabilistic classification method for record linkage. Later, it was proved that the Fellegi and Sunter method is mathematically equivalent to the Naive Bayes method in case of assuming independence between comparison variables.
```
# Train the classifier
nb = rl.NaiveBayesClassifier(binarize=0.3)
nb.fit(golden_pairs, golden_matches_index)
# Predict the match status for all record pairs
result_nb = nb.predict(krebs_X)
len(result_nb)
rl.confusion_matrix(krebs_true_links, result_nb, len(krebs_X))
# The F-score for this classification is
rl.fscore(krebs_true_links, result_nb)
```
### Support Vector Machines
Support Vector Machines (SVM) have become increasingly popular in record linkage. The algorithm performs well there is only a small amount of training data available. The implementation of SVM in the Python Record Linkage Toolkit is a linear SVM algorithm.
```
# Train the classifier
svm = rl.SVMClassifier()
svm.fit(golden_pairs, golden_matches_index)
# Predict the match status for all record pairs
result_svm = svm.predict(krebs_X)
len(result_svm)
rl.confusion_matrix(krebs_true_links, result_svm, len(krebs_X))
# The F-score for this classification is
rl.fscore(krebs_true_links, result_svm)
```
## Unsupervised learning
In situations without training data, unsupervised learning can be a solution for record linkage problems. In this section, we discuss two unsupervised learning methods. One algorithm is K-means clustering, and the other algorithm is an implementation of the Expectation-Maximisation algorithm. Most of the time, unsupervised learning algorithms take more computational time because of the iterative structure in these algorithms.
### K-means clustering
The K-means clustering algorithm is well-known and widely used in big data analysis. The K-means classifier in the Python Record Linkage Toolkit package is configured in such a way that it can be used for linking records. For more info about the K-means clustering see [Wikipedia](https://en.wikipedia.org/wiki/K-means_clustering).
```
kmeans = rl.KMeansClassifier()
result_kmeans = kmeans.fit_predict(krebs_X)
# The predicted number of matches
len(result_kmeans)
```
The classifier is now trained and the comparison vectors are classified.
```
rl.confusion_matrix(krebs_true_links, result_kmeans, len(krebs_X))
rl.fscore(krebs_true_links, result_kmeans)
```
### Expectation/Conditional Maximization Algorithm
The ECM-algorithm is an Expectation-Maximisation algorithm with some additional constraints. This algorithm is closely related to the Naive Bayes algorithm. The ECM algorithm is also closely related to estimating the parameters in the Fellegi and Sunter (1969) framework. The algorithms assume that the attributes are independent of each other. The Naive Bayes algorithm uses the same principles.
```
# Train the classifier
ecm = rl.ECMClassifier(binarize=0.8)
result_ecm = ecm.fit_predict(krebs_X)
len(result_ecm)
rl.confusion_matrix(krebs_true_links, result_ecm, len(krebs_X))
# The F-score for this classification is
rl.fscore(krebs_true_links, result_ecm)
```
|
github_jupyter
|
# Deep Matrix Factorisation
Matrix factorization with deep layers
```
import sys
sys.path.append("../")
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
from IPython.display import SVG, display
import matplotlib.pyplot as plt
import seaborn as sns
from reco.preprocess import encode_user_item, random_split, user_split
%matplotlib inline
```
### Prepare the data
```
df_ratings = pd.read_csv("data/ratings.csv")
df_items = pd.read_csv("data/items.csv")
# Data Encoding
DATA, user_encoder, item_encoder = encode_user_item(df_ratings, "user_id", "movie_id", "rating", "unix_timestamp")
DATA.head()
n_users = DATA.USER.nunique()
n_items = DATA.ITEM.nunique()
n_users, n_items
max_rating = DATA.RATING.max()
min_rating = DATA.RATING.min()
min_rating, max_rating
# Data Splitting
#train, val, test = user_split(DATA, [0.6, 0.2, 0.2])
train, test = user_split(DATA, [0.9, 0.1])
train.shape, test.shape
```
## Deep Matrix Factorization
This is a model with User and Item Embedding Dot Product
```
from keras.models import Model
from keras.layers import Input, Embedding, Flatten, Dot, Add, Lambda, Activation, Reshape, Concatenate, Dense, Dropout
from keras.regularizers import l2
from keras.constraints import non_neg
from keras.optimizers import Adam
from keras.utils import plot_model
from keras.utils.vis_utils import model_to_dot
from reco import vis
```
### Build the Model
```
def Deep_MF(n_users, n_items, n_factors):
# Item Layer
item_input = Input(shape=[1], name='Item')
item_embedding = Embedding(n_items, n_factors, embeddings_regularizer=l2(1e-6),
embeddings_initializer='glorot_normal',
name='ItemEmbedding')(item_input)
item_vec = Flatten(name='FlattenItemE')(item_embedding)
# Item Bias
item_bias = Embedding(n_items, 1, embeddings_regularizer=l2(1e-6),
embeddings_initializer='glorot_normal',
name='ItemBias')(item_input)
item_bias_vec = Flatten(name='FlattenItemBiasE')(item_bias)
# User Layer
user_input = Input(shape=[1], name='User')
user_embedding = Embedding(n_users, n_factors, embeddings_regularizer=l2(1e-6),
embeddings_initializer='glorot_normal',
name='UserEmbedding')(user_input)
user_vec = Flatten(name='FlattenUserE')(user_embedding)
# User Bias
user_bias = Embedding(n_users, 1, embeddings_regularizer=l2(1e-6),
embeddings_initializer='glorot_normal',
name='UserBias')(user_input)
user_bias_vec = Flatten(name='FlattenUserBiasE')(user_bias)
# Dot Product of Item and User & then Add Bias
Concat = Concatenate(name='Concat')([item_vec, user_vec])
ConcatDrop = Dropout(0.5)(Concat)
kernel_initializer='he_normal'
# Use Dense to learn non-linear dense representation
Dense_1 = Dense(10, kernel_initializer='glorot_normal', name="Dense1")(ConcatDrop)
Dense_1_Drop = Dropout(0.5)(Dense_1)
Dense_2 = Dense(1, kernel_initializer='glorot_normal', name="Dense2")(Dense_1_Drop)
AddBias = Add(name="AddBias")([Dense_2, item_bias_vec, user_bias_vec])
# Scaling for each user
y = Activation('sigmoid')(AddBias)
rating_output = Lambda(lambda x: x * (max_rating - min_rating) + min_rating)(y)
# Model Creation
model = Model([user_input, item_input], rating_output)
# Compile Model
model.compile(loss='mean_squared_error', optimizer=Adam(lr=0.001))
return model
n_factors = 50
model = Deep_MF(n_users, n_items, n_factors)
model.summary()
from reco.utils import create_directory
create_directory("/model-img")
plot_model(model, show_layer_names=True, show_shapes=True, to_file="model-img/Deep-CF.png" )
```
### Train the Model
```
%%time
output = model.fit([train.USER, train.ITEM], train.RATING,
batch_size=128, epochs=5, verbose=1,
validation_data= ([test.USER, test.ITEM], test.RATING))
vis.metrics(output.history)
```
### Score the Model
```
score = model.evaluate([test.USER, test.ITEM], test.RATING, verbose=1)
score
```
### Evaluate the Model
```
from reco.evaluate import get_embedding, get_predictions, recommend_topk
from reco.evaluate import precision_at_k, recall_at_k, ndcg_at_k
item_embedding = get_embedding(model, "ItemEmbedding")
user_embedding = get_embedding(model, "UserEmbedding")
%%time
predictions = get_predictions(model, DATA)
predictions.head()
%%time
# Recommendation for Top10K
ranking_topk = recommend_topk(model, DATA, train, k=5)
eval_precision = precision_at_k(test, ranking_topk, k=10)
eval_recall = recall_at_k(test, ranking_topk, k=10)
eval_ndcg = ndcg_at_k(test, ranking_topk, k=10)
print("NDCG@K:\t%f" % eval_ndcg,
"Precision@K:\t%f" % eval_precision,
"Recall@K:\t%f" % eval_recall, sep='\n')
```
### Get Similar Items
```
from reco.recommend import get_similar, show_similar
%%time
item_distances, item_similar_indices = get_similar(item_embedding, 5)
item_similar_indices
show_similar(1, item_similar_indices, item_encoder)
```
|
github_jupyter
|
# AdaDelta compared to AdaGrad
Presented during ML reading group, 2019-11-12.
Author: Ivan Bogdan-Daniel, [email protected]
```
#%matplotlib notebook
%matplotlib inline
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
print(f'Numpy version: {np.__version__}')
```
# AdaDelta
The [AdaDelta paper](https://arxiv.org/pdf/1212.5701.pdf)
The idea presented in this paper was derived from ADAGRAD in order to improve upon the two main drawbacks of the method:
1) the continual decay of learning rates throughout training
2) the need for a manually selected global learning rate.
AdaGrad comes with:
$$w_{t+1}^{(j)} = w_{t}^{(j)} - \frac{\eta}{\sqrt{\varepsilon + \sum_{\tau=1}^{t}{(g_{\tau}^{(j)}})^2}} \nabla J_{w}(w_t^{(j)})$$
where $g_{\tau}$ is the gradient of error function at iteration $\tau$, $g_{\tau}^{(j)}$ is the partial derivative of the
error function in direction of the $j$ - th feature, at iteration $\tau$, $m$ - is the number of features, i.e.
The problem appears in the sum:
$${\varepsilon + \sum_{\tau=1}^{t}{(g_{\tau}^{(j)}})^2}$$
It grows into a very large number making the fraction $$\frac{\eta}{\sqrt{\varepsilon + \sum_{\tau=1}^{t}{(g_{\tau}^{(j)}})^2}}$$ become an insignificant number. The
learning rate will continue to decrease throughout training,
eventually decreasing to zero and stopping training completely.
# Solution
Instead of accumulating the sum of squared gradients over all
time, we restricted the window of past gradients that are accumulated to be some fixed size w.
Since storing w previous squared gradients is inefficient,
our methods implements this accumulation as an exponentially decaying average of the squared gradients
This ensures that learning continues
to make progress even after many iterations of updates have
been done.
At time t this average is: $$E[g^2]_{t}$$ then we compute:
$$E[g^2]_{t}=\rho E[g^2]_{t-1}+(1-\rho)g^2_{t}$$
Where $\rho$ is a hyper parameter similar to the one used in momentum, it can take values between 0 and 1, generally 0.95 is recommended.
Since we require the square root of this quantity:
$$RMS[g]_{t} = \sqrt{E[g^2]_{t}+\epsilon}$$
The parameter update becomes:
$$w_{t+1}^{(j)} = w_{t}^{(j)} - \frac{\eta}{RMS[g]_{t}} g_{t}$$
AdaDelta rule:
$$w_{t+1}^{(j)} = w_{t}^{(j)} - \frac{RMS[\Delta w]_{t-1}}{RMS[g]_{t}} g_{t}$$
Where $RMS[\Delta w]_{t-1}$ is computed similar to $RMS[g]_{t}$
# Algorithm
Require: Decay rate $\rho$, Constant $\epsilon$
Require: Initial parameter x
<img src="./images/adadelta_algorithm.png" alt="drawing" width="600"/>
Source: [AdaDelta paper](https://arxiv.org/pdf/1212.5701.pdf)
## Generate data
```
from scipy.sparse import random #to generate sparse data
np.random.seed(10) # for reproducibility
m_data = 100
n_data = 4 #number of features of the data
_scales = np.array([1,10, 10,1 ]) # play with these...
_parameters = np.array([3, 0.5, 1, 7])
def gen_data(m, n, scales, parameters, add_noise=True):
# Adagrad is designed especially for sparse data.
# produce: X, a 2d tensor with m lines and n columns
# and X[:, k] uniformly distributed in [-scale_k, scale_k] with the first and the last column containing sparse data
#(approx 75% of the elements are 0)
#
# To generate a sparse data matrix with m rows and n columns
# and random values use S = random(m, n, density=0.25).A, where density = density of the data. S will be the
# resulting matrix
# more information at https://docs.scipy.org/doc/scipy/reference/generated/scipy.sparse.random.html
#
# To obtain X - generate a random matrix with X[:, k] uniformly distributed in [-scale_k, scale_k]
# set X[:, 0] and X[:, -1] to 0 and add matrix S with the sparse data.
#
# let y be [email protected] + epsilon, with epsilon ~ N(0, 1); y is a vector with m elements
# parameters - the ideal weights, used to produce output values y
#
return X, y
X, y = gen_data(m_data, n_data, _scales, _parameters)
print(X)
print(y)
```
## Define error function, gradient, inference
```
def model_estimate(X, w):
'''Computes the linear regression estimation on the dataset X, using coefficients w
:param X: 2d tensor with m_data lines and n_data columns
:param w: a 1d tensor with n_data coefficients (no intercept)
:return: a 1d tensor with m_data elements y_hat = w @X.T
'''
return y_hat
def J(X, y, w):
"""Computes the mean squared error of model. See the picture from last week's sheet.
:param X: input values, of shape m_data x n_data
:param y: ground truth, column vector with m_data values
:param w: column with n_data coefficients for the linear form
:return: a scalar value >= 0
:use the same formula as in the exercise from last week
"""
return err
def gradient(X, y, w):
'''Commputes the gradients to be used for gradient descent.
:param X: 2d tensor with training data
:param y: 1d tensor with y.shape[0] == W.shape[0]
:param w: 1d tensor with current values of the coefficients
:return: gradients to be used for gradient descent.
:use the same formula as in the exercise from last week
'''
return grad## implement
```
## Momentum algorithm
```
#The function from last week for comparison
def gd_with_momentum(X, y, w_init, eta=1e-1, gamma = 0.9, thresh = 0.001):
"""Applies gradient descent with momentum coefficient
:params: as in gd_no_momentum
:param gamma: momentum coefficient
:param thresh: the threshold for gradient norm (to stop iterations)
:return: the list of succesive errors and the found w* vector
"""
w = w_init
w_err=[]
delta = np.zeros_like(w)
while True:
grad = gradient(X, y, w)
err = J(X, y, w)
w_err.append(err)
w_nou = w + gamma * delta - eta * grad
delta = w_nou - w
w = w_nou
if np.linalg.norm(grad) < thresh :
break;
return w_err, w
w_init = np.array([0, 0, 0, 0])
errors_momentum, w_best = gd_with_momentum(X, y, w_init,0.0001, 0.9)
print(f'How many iterations were made: {len(errors_momentum)}')
w_best
fig, axes = plt.subplots()
axes.plot(list(range(len(errors_momentum))), errors_momentum)
axes.set_xlabel('Epochs')
axes.set_ylabel('Error')
axes.set_title('Optimization with momentum')
```
## Apply AdaGrad and report resulting $\eta$'s
```
def ada_grad(X, y, w_init, eta_init=1e-1, eps = 0.001, thresh = 0.001):
'''Iterates with gradient descent. algorithm
:param X: 2d tensor with data
:param y: 1d tensor, ground truth
:param w_init: 1d tensor with the X.shape[1] initial coefficients
:param eta_init: the initial learning rate hyperparameter
:param eps: the epsilon value from the AdaGrad formula
:param thresh: the threshold for gradient norm (to stop iterations)
:return: the list of succesive errors w_err, the found w - the estimated feature vector
:and rates the learning rates after the final iteration
'''
n = X.shape[1]
w = w_init
w_err=[]
sum_sq_grad = np.zeros(n)
rates = np.zeros(n) + eta_init
while True:
grad = gradient(X, y, w)
pgrad = grad**2
err = J(X, y, w)
w_err.append(err)
prod = rates*grad
w = w - prod
sum_sq_grad += pgrad
rates = eta_init/np.sqrt(eps + sum_sq_grad)
if np.linalg.norm(grad) < thresh:
break;
return w_err, w, rates
w_init = np.array([0,0,0,0])
adaGerr, w_ada_best, rates = ada_grad(X, y, w_init)
print(rates)
print(f'How many iterations were made: {len(adaGerr)}')
w_ada_best
fig, axes = plt.subplots()
axes.plot(list(range(len(adaGerr))),adaGerr)
axes.set_xlabel('Epochs')
axes.set_ylabel('Error')
axes.set_title('Optimization with AdaGrad')
```
## Apply AdaDelta and report resulting $\eta$'s
```
def ada_delta(X, y, w_init, eta_init=1e-1, gamma=0.99, eps = 0.001, thresh = 0.001):
'''Iterates with gradient descent. algorithm
:param X: 2d tensor with data
:param y: 1d tensor, ground truth
:param w_init: 1d tensor with the X.shape[1] initial coefficients
:param eta_init: the initial learning rate hyperparameter
:param gamma: decay constant, similar to momentum
:param eps: the epsilon value from the AdaGrad formula
:param thresh: the threshold for gradient norm (to stop iterations)
:return: the list of succesive errors w_err, the found w - the estimated feature vector
:and rates the learning rates after the final iteration
'''
#todo
#same as adagrad but instead of summing the square of gradients
#use the adadelta formula for decaying average
w_init = np.array([0,0,0,0])
adaDerr, w_adad_best, rates = ada_delta(X, y, w_init)
print(rates)
print(f'How many iterations were made: {len(adaDerr)}')
w_adad_best
fig, axes = plt.subplots()
axes.plot(list(range(len(adaDerr))),adaDerr)
axes.set_xlabel('Epochs')
axes.set_ylabel('Error')
axes.set_title('Optimization with AdaDelta')
```
|
github_jupyter
|
# Exploratory Data Analysis of AllenSDK
```
# Only for Colab
#!python -m pip install --upgrade pip
#!pip install allensdk
```
## References
- [[AllenNB1]](https://allensdk.readthedocs.io/en/latest/_static/examples/nb/visual_behavior_ophys_data_access.html) Download data using the AllenSDK or directly from our Amazon S3 bucket
- [[AllenNB2]](https://allensdk.readthedocs.io/en/latest/_static/examples/nb/visual_behavior_ophys_dataset_manifest.html) Identify experiments of interest using the dataset manifest
- [[AllenNB3]](https://allensdk.readthedocs.io/en/latest/_static/examples/nb/visual_behavior_load_ophys_data.html) Load and visualize data from a 2-photon imaging experiment
- [[AllenNB4]](https://allensdk.readthedocs.io/en/latest/_static/examples/nb/visual_behavior_mouse_history.html) Examine the full training history of one mouse
- [[AllenNB5]](https://allensdk.readthedocs.io/en/latest/_static/examples/nb/visual_behavior_compare_across_trial_types.html) Compare behavior and neural activity across different trial types in the task
## Imports
Import and setup Python packages. You should not need to touch this section.
```
from pathlib import Path
from tqdm import tqdm
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from allensdk.brain_observatory.behavior.behavior_project_cache import VisualBehaviorOphysProjectCache
from allensdk.core.brain_observatory_cache import BrainObservatoryCache
# import mindscope_utilities
# import mindscope_utilities.visual_behavior_ophys as ophys
np.random.seed(42)
```
## Setup AllenSDK
Configure AllenSDK to get `cache`, `sessions_df` and `experiments_df`. Data will be stored in `./allensdk_storage` by default.
```
!mkdir -p allensdk_storage
DATA_STORAGE_DIRECTORY = Path("./allensdk_storage")
cache = VisualBehaviorOphysProjectCache.from_s3_cache(cache_dir=DATA_STORAGE_DIRECTORY)
```
The data manifest is comprised of three types of tables:
1. `behavior_session_table`
2. `ophys_session_table`
3. `ophys_experiment_table`
The` behavior_session_table` contains metadata for every **behavior session** in the dataset. Some behavior sessions have 2-photon data associated with them, while others took place during training in the behavior facility. The different training stages that mice are progressed through are described by the session_type.
The `ophys_session_table` contains metadata for every 2-photon imaging (aka optical physiology, or ophys) session in the dataset, associated with a unique `ophys_session_id`. An **ophys session** is one continuous recording session under the microscope, and can contain different numbers of imaging planes (aka experiments) depending on which microscope was used. For Scientifica sessions, there will only be one experiment (aka imaging plane) per session. For Multiscope sessions, there can be up to eight imaging planes per session. Quality Control (QC) is performed on each individual imaging plane within a session, so each can fail QC independent of the others. This means that a Multiscope session may not have exactly eight experiments (imaging planes).
The `ophys_experiment_table` contains metadata for every **ophys experiment** in the dataset, which corresponds to a single imaging plane recorded in a single session, and associated with a unique `ophys_experiment_id`. A key part of our experimental design is targeting a given population of neurons, contained in one imaging plane, across multiple `session_types` (further described below) to examine the impact of varying sensory and behavioral conditions on single cell responses. The collection of all imaging sessions for a given imaging plane is referred to as an **ophys container**, associated with a unique `ophys_container_id`. Each ophys container may contain different numbers of sessions, depending on which experiments passed QC, and how many retakes occured (when a given session_type fails QC on the first try, an attempt is made to re-acquire the `session_type` on a different recording day - this is called a retake, also described further below).
*Text copied from [[AllenNB2]](#References)*
---
We will just use the `ophys_experiment_table`.
```
experiments_df = cache.get_ophys_experiment_table()
```
## Specify Experiment
There are a lot of experiments in the table. Let's choose a particular experiment that meet the following criteria:
- Excitatory cells with fast reporter
- Single-plane imaging
### Cre Line and Reporter Line
<img style="width: 50%" src="https://github.com/seungjaeryanlee/nma-cn-project/blob/main/images/cre_lines.png?raw=1">
The `cre_line` determines which genetically identified neuron type will be labeled by the reporter_line.
This dataset have 3 `cre_line`:
- **Slc17a7-IRES2-Cre**, which labels excitatory neurons across all cortical layers
- **Sst-IRES-Cre** which labels somatostatin expressing inhibitory interneurons
- **Vip-IRES-Cre**, which labels vasoactive intestinal peptide expressing inhibitory interneurons
*Text copied from [[AllenNB2]](#References)*
```
experiments_df["cre_line"].unique()
```
There are also 3 `reporter_line`:
- **Ai93(TITL-GCaMP6f)**, which expresses the genetically encoded calcium indicator GCaMP6f (f is for 'fast', this reporter has fast offset kinetics, but is only moderately sensitive to calcium relative to other sensors) in cre labeled neurons
- **Ai94(TITL-GCaMP6s)**, which expresses the indicator GCaMP6s (s is for 'slow', this reporter is very sensitive to calcium but has slow offset kinetics), and
- **Ai148(TIT2L-GC6f-ICL-tTA2)**, which expresses GCaMP6f using a self-enhancing system to achieve higher expression than other reporter lines (which proved necessary to label inhibitory neurons specifically).
```
experiments_df["reporter_line"].unique()
```
The specific `indicator` expressed by each `reporter_line` also has its own column in the table.
```
experiments_df["indicator"].unique()
```
`full_genotype` contains information for both cre line and reporter line.
```
experiments_df["full_genotype"].unique()
```
---
We are looking at excitatory cells, so we should use `cre_line` of `Slc17a7-IRES2-Cre`. We want the fast one, so we select `Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-GCaMP6f)/wt`.
```
FULL_GENOTYPE = "Slc17a7-IRES2-Cre/wt;Camk2a-tTA/wt;Ai93(TITL-GCaMP6f)/wt"
```
### Project Code
<img style="width: 50%" src="https://github.com/seungjaeryanlee/nma-cn-project/blob/main/images/datasets.png?raw=1">
"The distinct groups of mice are referred to as dataset variants and can be identified using the `project_code` column." [[AllenNB2]](#References)
```
experiments_df["project_code"].unique()
```
---
We are interested in single-plane imaging, so either `VisualBehavior` or `VisualBehaviorTask1B` works.
```
# We are looking at single-plane imaging
# "VisualBehavior" or "VisualBehaviorTask1B"
PROJECT_CODE = "VisualBehavior"
```
### Experiment
<img style="width: 50%" src="https://github.com/seungjaeryanlee/nma-cn-project/blob/main/images/data_structure.png?raw=1">
(Note that we are looking at single-plane imaging, so there is only one row (container) per mouse.)
#### `MOUSE_ID`
"The mouse_id is a 6-digit unique identifier for each experimental animal in the dataset." [[AllenNB2]](#References)
---
We retrieve all mouse that can be used for our experiment and select one mouse.
```
experiments_df.query("project_code == @PROJECT_CODE") \
.query("full_genotype == @FULL_GENOTYPE") \
["mouse_id"].unique()
MOUSE_ID = 450471
```
#### `ACTIVE_SESSION`, `PASSIVE_SESSION`
<img style="width: 50%" src="https://github.com/seungjaeryanlee/nma-cn-project/blob/main/images/experiment_design.png?raw=1">
The session_type for each behavior session indicates the behavioral training stage or 2-photon imaging conditions for that particular session. This determines what stimuli were shown and what task parameters were used.
During the 2-photon imaging portion of the experiment, mice perform the task with the same set of images they saw during training (either image set A or B), as well as an additional novel set of images (whichever of A or B that they did not see during training). This allows evaluation of the impact of different sensory contexts on neural activity - familiarity versus novelty.
- Sessions with **familiar images** include those starting with `OPHYS_0`, `OPHYS_1`, `OPHYS_2`, and `OPHYS_3`.
- Sessions with **novel images** include those starting with `OPHYS_4`, `OPHYS_5`, and `OPHYS_6`.
Interleaved between **active behavior sessions** are **passive viewing sessions** where mice are given their daily water ahead of the sesssion (and are thus satiated) and view the stimulus with the lick spout retracted so they are unable to earn water rewards. This allows comparison of neural activity in response to stimuli under different behavioral context - active task engagement and passive viewing without reward. There are two passive sessions:
- `OPHYS_2_images_A_passive`: passive session with familiar images
- `OPHYS_5_images_A_passive`: passive session with novel images
*Text copied from [[AllenNB2]](#References)*
---
We check which sessions are available for this particular mouse and select one active and one passive session type. Not all sessions may be availble due to QC.
```
experiments_df.query("project_code == @PROJECT_CODE") \
.query("full_genotype == @FULL_GENOTYPE") \
.query("mouse_id == @MOUSE_ID") \
["session_type"].unique()
```
Looks like this mouse has all sessions! Let's select the first one then.
```
SESSION_TYPE = "OPHYS_1_images_A"
```
#### `EXPERIMENT_ID`
We retrieve the `ophys_experiment_id` of the session type we chose. We need this ID to get the experiment data.
```
experiments_df.query("project_code == @PROJECT_CODE") \
.query("full_genotype == @FULL_GENOTYPE") \
.query("mouse_id == @MOUSE_ID") \
.query("session_type == @SESSION_TYPE")
```
---
Looks like this mouse went through the same session multiple times! Let's just select the first experiment ID.
```
EXPERIMENT_ID = 871155338
```
#### `ACTIVE_EXPERIMENT_ID_CONTROL`, `PASSIVE_EXPERIMENT_ID_CONTROL`
```
PASSIVE_EXPERIMENT_ID_CONTROL =884218326
```
## Download Experiment
Download the experiment with the selected `experiment_id`.
We can now download the experiment. Each experiment will be approximately 600MB - 2GB in size.
```
experiment = cache.get_behavior_ophys_experiment(EXPERIMENT_ID)
experiment
```
This returns an instance of `BehaviorOphysExperiment`. It contains multiple attributes that we will need to explore.
## Attributes of the Experiment
Explore what information we have about the experiment by checking its attributes.
### `dff_traces`
"`dff_traces` dataframe contains traces for all neurons in this experiment, unaligned to any events in the task." [[AllenNB3]](#References)
```
experiment.dff_traces.head()
```
Since `dff` is stored as a list, we need to get timestamps for each of those numbers.
### `ophys_timestamps`
`ophys_timestamps` contains the timestamps of every record.
```
experiment.ophys_timestamps
```
Let's do a sanity check by checking the length of both lists.
```
print(f"dff has length {len(experiment.dff_traces.iloc[0]['dff'])}")
print(f"timestamp has length {len(experiment.ophys_timestamps)}")
```
### `stimulus_presentations`
We also need timestamps of when stimulus was presented. This information is contained in `stimulus_presentations`.
```
experiment.stimulus_presentations.head()
```
During imaging sessions, stimulus presentations (other than the change and pre-change images) are omitted with a 5% probability, resulting in some inter stimlus intervals appearing as an extended gray screen period. [[AllenNB2]](#References)
<img style="width: 50%" src="https://github.com/seungjaeryanlee/nma-cn-project/blob/main/images/omissions.png?raw=1">
```
experiment.stimulus_presentations.query("omitted").head()
```
### `stimulus_templates`
If we want to know what the stimulus looks like, we can check `stimulus_templates`.
```
experiment.stimulus_templates
```
We see that we have a matrix for the `warped` column and a stub matrix for the unwarped column. Let's display the `warped` column.
```
fig, ax = plt.subplots(4, 2, figsize=(8, 12))
for i, image_name in enumerate(experiment.stimulus_templates.index):
ax[i%4][i//4].imshow(experiment.stimulus_templates.loc[image_name]["warped"], cmap='gray', vmin=0, vmax=255)
ax[i%4][i//4].set_title(image_name)
ax[i%4][i//4].get_xaxis().set_visible(False)
ax[i%4][i//4].get_yaxis().set_visible(False)
fig.show()
```
So this is what the mouse is seeing! But can we see the original, unwarped image? For that, we need to use another AllenSDK cache that contains these images.
```
boc = BrainObservatoryCache()
scenes_data_set = boc.get_ophys_experiment_data(501498760)
```
This data set contains a lot of images in a form of a 3D matrix (`# images` x `width` x `height` ).
```
scenes = scenes_data_set.get_stimulus_template('natural_scenes')
scenes.shape
```
We just want the images that were shown above. Notice that the indices are part of the name of the images.
```
experiment.stimulus_templates.index
```
Using this, we can plot the unwarped versions!
```
fig, ax = plt.subplots(4, 2, figsize=(6, 12))
for i, image_name in enumerate(experiment.stimulus_templates.index):
scene_id = int(image_name[2:])
ax[i%4][i//4].imshow(scenes[scene_id, :, :], cmap='gray', vmin=0, vmax=255)
ax[i%4][i//4].set_title(image_name)
ax[i%4][i//4].get_xaxis().set_visible(False)
ax[i%4][i//4].get_yaxis().set_visible(False)
```
## Visualization
We do some basic plots from the information we gathered from various attributes.
### Plot dF/F Trace
Let's choose some random `cell_specimen_id` and plots its dff trace for time 400 to 450.
```
fig, ax = plt.subplots(figsize=(15, 4))
ax.plot(
experiment.ophys_timestamps,
experiment.dff_traces.loc[1086545833]["dff"],
)
ax.set_xlim(400, 450)
fig.show()
```
### Plot Stimulus
Let's also plot stimulus for a short interval.
*Part of code from [[AllenNB3]](#References)*
```
# Create a color map for each image
unique_stimuli = [stimulus for stimulus in experiment.stimulus_presentations['image_name'].unique()]
colormap = {image_name: sns.color_palette()[image_number] for image_number, image_name in enumerate(np.sort(unique_stimuli))}
# Keep omitted image as white
colormap['omitted'] = (1,1,1)
stimulus_presentations_sample = experiment.stimulus_presentations.query('stop_time >= 400 and start_time <= 450')
fig, ax = plt.subplots(figsize=(15, 4))
for idx, stimulus in stimulus_presentations_sample.iterrows():
ax.axvspan(stimulus['start_time'], stimulus['stop_time'], color=colormap[stimulus['image_name']], alpha=0.25)
ax.set_xlim(400, 450)
fig.show()
```
### Plot Both dF/F trace and Stimulus
```
fig, ax = plt.subplots(figsize=(15, 4))
ax.plot(
experiment.ophys_timestamps,
experiment.dff_traces.loc[1086545833]["dff"],
)
for idx, stimulus in stimulus_presentations_sample.iterrows():
ax.axvspan(stimulus['start_time'], stimulus['stop_time'], color=colormap[stimulus['image_name']], alpha=0.25)
ax.set_xlim(400, 450)
ax.set_ylim(-0.5, 0.5)
ax.legend(["dff trace"])
fig.show()
```
|
github_jupyter
|
# Consumption Equivalent Variation (CEV)
1. Use the model in the **ConsumptionSaving.pdf** slides and solve it using **egm**
2. This notebooks estimates the *cost of income risk* through the Consumption Equivalent Variation (CEV)
We will here focus on the cost of income risk, but the CEV can be used to estimate the value of many different aspects of an economy. For eaxample, [Oswald (2019)](http://qeconomics.org/ojs/index.php/qe/article/view/701 "The option value of homeownership") estimated the option value of homeownership using a similar strategy as described below.
**Goal:** To estimate the CEV by comparing the *value of life* under the baseline economy and an alternative economy with higher permanent income shock variance along with a consumption compensation.
**Value of Life:**
1. Let the *utility function* be a generalized version of the CRRA utility function with $\delta$ included as a potential consumption compensation.
\begin{equation}
{u}(c,\delta) = \frac{(c\cdot(1+\delta))^{1-\rho}}{1-\rho}
\end{equation}
2. Let the *value of life* of a synthetic consumer $s$ for a given level of permanent income shock varaince, $\sigma_{\psi}$, and $\delta$, be
\begin{equation}
{V}_{s}({\sigma}_{\psi},\delta)=\sum_{t=1}^T \beta ^{t-1}{u}({c}^{\star}_{s,t}({\sigma}_{\psi},\delta),\delta)
\end{equation}
where ${c}^{\star}_{s,t}({\sigma}_{\psi},\delta)$ is optimal consumption found using the **egm**. The value of life is calcualted in the function `value_of_life(.)` defined below.
**Consumption Equivalent Variation:**
1. Let $V=\frac{1}{S}\sum_{s=1}^SV(\sigma_{\psi},0)$ be the average value of life under the *baseline* economy with the baseline value of $\sigma_{\psi}$ and $\delta=0$.
2. Let $\tilde{V}(\delta)=\frac{1}{S}\sum_{s=1}^SV(\tilde{\sigma}_{\psi},\delta)$ be the average value of life under the *alternative* economy with $\tilde{\sigma}_{\psi} > \sigma_{\psi}$.
The CEV is the value of $\delta$ that sets $V=\tilde{V}(\delta)$ and can be estimated as
\begin{equation}
\hat{\delta} = \arg\min_\delta (V-\tilde{V}(\delta))^2
\end{equation}
where the objective function is calculated in `obj_func_cev(.)` defined below.
# Setup
```
%matplotlib inline
%load_ext autoreload
%autoreload 2
import time
import numpy as np
import scipy.optimize as optimize
import matplotlib.pyplot as plt
plt.style.use('seaborn-whitegrid')
prop_cycle = plt.rcParams['axes.prop_cycle']
colors = prop_cycle.by_key()['color']
import sys
sys.path.append('../')
import ConsumptionSavingModel as csm
from ConsumptionSavingModel import ConsumptionSavingModelClass
```
# Setup the baseline model and the alternative model
```
par = {'simT':40}
model = ConsumptionSavingModelClass(name='baseline',solmethod='egm',**par)
# increase the permanent income with 100 percent and allow for consumption compensation
par_cev = {'sigma_psi':0.2,'do_cev':1,'simT':40}
model_cev = ConsumptionSavingModelClass(name='cev',solmethod='egm',**par_cev)
model.solve()
model.simulate()
```
# Average value of life
**Define Functions:** value of life and objective function used to estimate "cev"
```
def value_of_life(model):
# utility associated with consumption for all N and T
util = csm.utility(model.sim.c,model.par)
# discounted sum of utility
disc = np.ones(model.par.simT)
disc[1:] = np.cumprod(np.ones(model.par.simT-1)*model.par.beta)
disc_util = np.sum(disc*util,axis=1)
# return average of discounted sum of utility
return np.mean(disc_util)
def obj_func_cev(theta,model_cev,value_of_life_baseline):
# update cev-parameter
setattr(model_cev.par,'cev',theta)
# re-solve and simulate alternative model
model_cev.solve(do_print=False)
model_cev.simulate(do_print=False)
# calculate value of life
value_of_life_cev = value_of_life(model_cev)
# return squared difference to baseline
return (value_of_life_cev - value_of_life_baseline)*(value_of_life_cev - value_of_life_baseline)
```
**Baseline value of life and objective function at cev=0**
```
value_of_life_baseline = value_of_life(model)
obj_func_cev(0.0,model_cev,value_of_life_baseline)
# plot the objective function
grid_cev = np.linspace(0.0,0.2,20)
grid_obj = np.empty(grid_cev.size)
for j,cev in enumerate(grid_cev):
grid_obj[j] = obj_func_cev(cev,model_cev,value_of_life_baseline)
plt.plot(grid_cev,grid_obj);
```
# Estimate the Consumption Equivalent Variation (CEV)
```
res = optimize.minimize_scalar(obj_func_cev, bounds=[-0.01,0.5],
args=(model_cev,value_of_life_baseline),method='golden')
res
```
The estimated CEV suggests that consumers would be indifferent between the baseline economy and a 100% increase in the permanent income shock variance along with a 10% increase in consumption in all periods.
|
github_jupyter
|
# Facial Expression Recognizer
```
#The OS module in Python provides a way of using operating system dependent functionality.
#import os
# For array manipulation
import numpy as np
#For importing data from csv and other manipulation
import pandas as pd
#For displaying images
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
#For displaying graph
#import seaborn as sns
#For constructing and handling neural network
import tensorflow as tf
#Constants
LEARNING_RATE = 1e-4
TRAINING_ITERATIONS = 10000 #increase iteration to improve accuracy
DROPOUT = 0.5
BATCH_SIZE = 50
IMAGE_TO_DISPLAY = 3
VALIDATION_SIZE = 2000
#Reading data from csv file
data = pd.read_csv('Train_updated_six_emotion.csv')
#Seperating images data from labels ie emotion
images = data.iloc[:,1:].values
images = images.astype(np.float)
#Normalizaton : convert from [0:255] => [0.0:1.0]
images = np.multiply(images, 1.0 / 255.0)
image_size = images.shape[1]
image_width = image_height = 48
#Displaying an image from 20K images
def display(img):
#Reshaping,(1*2304) pixels into (48*48)
one_image = img.reshape(image_width,image_height)
plt.axis('off')
#Show image
plt.imshow(one_image, cmap=cm.binary)
display(images[IMAGE_TO_DISPLAY])
#Creating an array of emotion labels using dataframe 'data'
labels_flat = data[['label']].values.ravel()
labels_count = np.unique(labels_flat).shape[0]
# convert class labels from scalars to one-hot vectors
# 0 => [1 0 0]
# 1 => [0 1 0]
# 2 => [0 0 1]
def dense_to_one_hot(labels_dense, num_classes = 7):
num_labels = labels_dense.shape[0]
index_offset = np.arange(num_labels) * num_classes
labels_one_hot = np.zeros((num_labels, num_classes))
labels_one_hot.flat[index_offset + labels_dense.ravel()] = 1
return labels_one_hot
labels = dense_to_one_hot(labels_flat, labels_count)
labels = labels.astype(np.uint8)
#Printing example hot-dense label
print ('labels[{0}] => {1}'.format(IMAGE_TO_DISPLAY,labels[IMAGE_TO_DISPLAY]))
#Using data for training & cross validation
validation_images = images[:2000]
validation_labels = labels[:2000]
train_images = images[2000:]
train_labels = labels[2000:]
```
#Next is the neural network structure.
#Weights and biases are created.
#The weights should be initialised with a small a amount of noise
#for symmetry breaking, and to prevent 0 gradients. Since we are using
#rectified neurones (ones that contain rectifier function *f(x)=max(0,x)*),
#we initialise them with a slightly positive initial bias to avoid "dead neurones.
```
# initialization of weight
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.1, shape=shape)
return tf.Variable(initial)
# We use zero padded convolution neural network with a stride of 1 and the size of the output is same as that of input.
# The convolution layer finds the features in the data the number of filter denoting the number of features to be detected.
def conv2d(x, W):
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
# Pooling downsamples the data. 2x2 max-pooling splits the image into square 2-pixel blocks and only keeps the maximum value
# for each of the blocks.
def max_pool_2x2(x):
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='SAME')
# images
x = tf.placeholder('float', shape=[None, image_size])
# labels (0, 1 or 2)
y_ = tf.placeholder('float', shape=[None, labels_count])
BATCH_SIZE
```
### VGG-16 architecture
```
W_conv1 = weight_variable([3, 3, 1, 8])
b_conv1 = bias_variable([8])
# we reshape the input data to a 4d tensor, with the first dimension corresponding to the number of images,
# second and third - to image width and height, and the final dimension - to the number of colour channels.
# (20000,2304) => (20000,48,48,1)
image = tf.reshape(x, [-1,image_width , image_height,1])
print (image.get_shape())
h_conv1 = tf.nn.relu(conv2d(image, W_conv1) + b_conv1)
print (h_conv1)
W_conv2 = weight_variable([3, 3, 8, 8])
b_conv2 = bias_variable([8])
h_conv2 = tf.nn.relu(conv2d(h_conv1, W_conv2) + b_conv2)
print (h_conv2)
# pooling reduces the size of the output from 48x48 to 24x24.
h_pool1 = max_pool_2x2(h_conv2)
#print (h_pool1.get_shape()) => (20000, 24, 24, 8)
# Prepare for visualization
# display 8 features in 4 by 2 grid
layer1 = tf.reshape(h_conv1, (-1, image_height, image_width, 4 ,2))
# reorder so the channels are in the first dimension, x and y follow.
layer1 = tf.transpose(layer1, (0, 3, 1, 4,2))
layer1 = tf.reshape(layer1, (-1, image_height*4, image_width*2))
# The second layer has 16 features for each 5x5 patch. Its weight tensor has a shape of [5, 5, 8, 16].
# The first two dimensions are the patch size. the next is the number of input channels (8 channels correspond to 8
# features that we got from previous convolutional layer).
W_conv3 = weight_variable([3, 3, 8, 16])
b_conv3 = bias_variable([16])
h_conv3 = tf.nn.relu(conv2d(h_pool1, W_conv3) + b_conv3)
print(h_conv3)
W_conv4 = weight_variable([3, 3, 16, 16])
b_conv4 = bias_variable([16])
h_conv4 = tf.nn.relu(conv2d(h_conv3, W_conv4) + b_conv4)
print(h_conv4)
h_pool2 = max_pool_2x2(h_conv4)
#print (h_pool2.get_shape()) => (20000, 12, 12, 16)
# The third layer has 16 features for each 5x5 patch. Its weight tensor has a shape of [5, 5, 16, 32].
# The first two dimensions are the patch size. the next is the number of input channels (16 channels correspond to 16
# features that we got from previous convolutional layer)
W_conv5 = weight_variable([3, 3, 16, 32])
b_conv5 = bias_variable([32])
h_conv5 = tf.nn.relu(conv2d(h_pool2, W_conv5) + b_conv5)
print(h_conv5)
W_conv6 = weight_variable([3, 3, 32, 32])
b_conv6 = bias_variable([32])
h_conv6 = tf.nn.relu(conv2d(h_conv5, W_conv6) + b_conv6)
print(h_conv6)
W_conv7 = weight_variable([3, 3, 32, 32])
b_conv7 = bias_variable([32])
h_conv7 = tf.nn.relu(conv2d(h_conv6, W_conv7) + b_conv7)
print(h_conv7)
h_pool3 = max_pool_2x2(h_conv7)
#print (h_pool2.get_shape()) => (20000, 6, 6, 32)
W_conv8 = weight_variable([3, 3, 32, 32])
b_conv8 = bias_variable([32])
h_conv8 = tf.nn.relu(conv2d(h_pool3, W_conv8) + b_conv8)
print(h_conv8)
W_conv9 = weight_variable([3, 3, 32, 32])
b_conv9 = bias_variable([32])
h_conv9 = tf.nn.relu(conv2d(h_conv8, W_conv9) + b_conv9)
print(h_conv9)
W_conv10 = weight_variable([3, 3, 32, 32])
b_conv10 = bias_variable([32])
h_conv10 = tf.nn.relu(conv2d(h_conv9, W_conv10) + b_conv10)
print(h_conv10)
h_pool4 = max_pool_2x2(h_conv10)
print (h_pool4.get_shape())
# Now that the image size is reduced to 3x3, we add a Fully_Connected_layer) with 1024 neurones
# to allow processing on the entire image (each of the neurons of the fully connected layer is
# connected to all the activations/outpus of the previous layer)
W_conv11 = weight_variable([3, 3, 32, 32])
b_conv11 = bias_variable([32])
h_conv11 = tf.nn.relu(conv2d(h_pool4, W_conv11) + b_conv11)
print(h_conv11)
W_conv12 = weight_variable([3, 3, 32, 32])
b_conv12 = bias_variable([32])
h_conv12 = tf.nn.relu(conv2d(h_conv11, W_conv12) + b_conv12)
print(h_conv12)
W_conv13 = weight_variable([3, 3, 32, 32])
b_conv13 = bias_variable([32])
h_conv13 = tf.nn.relu(conv2d(h_conv12, W_conv13) + b_conv13)
print(h_conv13)
# densely connected layer
W_fc1 = weight_variable([3 * 3 * 32, 512])
b_fc1 = bias_variable([512])
# (20000, 6, 6, 32) => (20000, 1152 )
h_pool2_flat = tf.reshape(h_conv13, [-1, 3*3*32])
h_fc1 = tf.nn.relu(tf.matmul(h_pool2_flat, W_fc1) + b_fc1)
print (h_fc1.get_shape()) # => (20000, 1024)
W_fc2 = weight_variable([512, 512])
b_fc2 = bias_variable([512])
h_fc2 = tf.nn.relu(tf.matmul(h_fc1, W_fc2) + b_fc2)
print (h_fc2.get_shape()) # => (20000, 1024)
W_fc3 = weight_variable([512, 512])
b_fc3 = bias_variable([512])
h_fc3 = tf.nn.relu(tf.matmul(h_fc2, W_fc3) + b_fc3)
print (h_fc3.get_shape()) # => (20000, 1024)
# To prevent overfitting, we apply dropout before the readout layer.
# Dropout removes some nodes from the network at each training stage. Each of the nodes is either kept in the
# network with probability (keep_prob) or dropped with probability (1 - keep_prob).After the training stage
# is over the nodes are returned to the NN with their original weights.
keep_prob = tf.placeholder('float')
h_fc1_drop = tf.nn.dropout(h_fc2, keep_prob)
# readout layer 1024*3
W_fc4 = weight_variable([512, labels_count])
b_fc4 = bias_variable([labels_count])
# Finally, we add a softmax layer
y = tf.nn.softmax(tf.matmul(h_fc1_drop, W_fc4) + b_fc4)
#print (y.get_shape()) # => (20000, 3)
cross_entropy = -tf.reduce_sum(y_*tf.log(y))
train_step = tf.train.AdamOptimizer(LEARNING_RATE).minimize(cross_entropy)
correct_prediction = tf.equal(tf.argmax(y,1), tf.argmax(y_,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, 'float'))
predict = tf.argmax(y,1)
epochs_completed = 0
index_in_epoch = 0
num_examples = train_images.shape[0]
# serve data by batches
def next_batch(batch_size):
global train_images
global train_labels
global index_in_epoch
global epochs_completed
start = index_in_epoch
index_in_epoch += batch_size
# when all trainig data have been already used, it is reorder randomly
if index_in_epoch > num_examples:
# finished epoch
epochs_completed += 1
# shuffle the data
perm = np.arange(num_examples)
np.random.shuffle(perm)
train_images = train_images[perm]
train_labels = train_labels[perm]
# start next epoch
start = 0
index_in_epoch = batch_size
assert batch_size <= num_examples
end = index_in_epoch
return train_images[start:end], train_labels[start:end]
with tf.Session() as sess:
init = tf.global_variables_initializer()
sess.run(init)
# visualisation variables
train_accuracies = []
validation_accuracies = []
x_range = []
display_step=1
for i in range(TRAINING_ITERATIONS):
#get new batch
batch_xs, batch_ys = next_batch(BATCH_SIZE)
# check progress on every 1st,2nd,...,10th,20th,...,100th... step
if i%display_step == 0 or (i+1) == TRAINING_ITERATIONS:
train_accuracy = accuracy.eval(feed_dict={x:batch_xs,
y_: batch_ys,
keep_prob: 1.0})
if(VALIDATION_SIZE):
validation_accuracy = accuracy.eval(feed_dict={ x: validation_images[0:BATCH_SIZE],
y_: validation_labels[0:BATCH_SIZE],
keep_prob: 1.0})
print('training_accuracy / validation_accuracy => %.2f / %.2f for step %d'%(train_accuracy, validation_accuracy, i))
validation_accuracies.append(validation_accuracy)
else:
print('training_accuracy => %.4f for step %d'%(train_accuracy, i))
train_accuracies.append(train_accuracy)
x_range.append(i)
# increase display_step
if i%(display_step*10) == 0 and i:
display_step *= 10
# train on batch
sess.run(train_step, feed_dict={x: batch_xs, y_: batch_ys, keep_prob: DROPOUT})
if(VALIDATION_SIZE):
validation_accuracy = accuracy.eval(feed_dict={x: validation_images,
y_: validation_labels,
keep_prob: 1.0})
print('validation_accuracy => %.4f'%validation_accuracy)
plt.plot(x_range, train_accuracies,'-b', label='Training')
plt.plot(x_range, validation_accuracies,'-g', label='Validation')
plt.legend(loc='lower right', frameon=False)
plt.ylim(ymax = 1.1, ymin = 0.0)
plt.ylabel('accuracy')
plt.xlabel('step')
plt.show()
```
|
github_jupyter
|
2017
Machine Learning Practical
University of Edinburgh
Georgios Pligoropoulos - s1687568
Coursework 4 (part 7)
### Imports, Inits, and helper functions
```
jupyterNotebookEnabled = True
plotting = True
coursework, part = 4, 7
saving = True
if jupyterNotebookEnabled:
#%load_ext autoreload
%reload_ext autoreload
%autoreload 2
import sys, os
mlpdir = os.path.expanduser(
'~/[email protected]/msc_Artificial_Intelligence/mlp_Machine_Learning_Practical/mlpractical'
)
sys.path.append(mlpdir)
from collections import OrderedDict
from __future__ import division
import skopt
from mylibs.jupyter_notebook_helper import show_graph
import datetime
import os
import time
import tensorflow as tf
import numpy as np
from mlp.data_providers import MSD10GenreDataProvider, MSD25GenreDataProvider,\
MSD10Genre_Autoencoder_DataProvider, MSD10Genre_StackedAutoEncoderDataProvider
import matplotlib.pyplot as plt
%matplotlib inline
from mylibs.batch_norm import fully_connected_layer_with_batch_norm_and_l2
from mylibs.stacked_autoencoder_pretrainer import \
constructModelFromPretrainedByAutoEncoderStack,\
buildGraphOfStackedAutoencoder, executeNonLinearAutoencoder
from mylibs.jupyter_notebook_helper import getRunTime, getTrainWriter, getValidWriter,\
plotStats, initStats, gatherStats
from mylibs.tf_helper import tfRMSE, tfMSE, fully_connected_layer
#trainEpoch, validateEpoch
from mylibs.py_helper import merge_dicts
from mylibs.dropout_helper import constructProbs
from mylibs.batch_norm import batchNormWrapper_byExponentialMovingAvg,\
fully_connected_layer_with_batch_norm
import pickle
from skopt.plots import plot_convergence
from mylibs.jupyter_notebook_helper import DynStats
import operator
from skopt.space.space import Integer, Categorical
from skopt import gp_minimize
from rnn.rnn_batch_norm import RNNBatchNorm
seed = 16011984
rng = np.random.RandomState(seed=seed)
config = tf.ConfigProto(log_device_placement=True, allow_soft_placement=True)
config.gpu_options.allow_growth = True
figcount = 0
tensorboardLogdir = 'tf_cw%d_%d' % (coursework, part)
curDtype = tf.float32
reluBias = 0.1
batch_size = 50
num_steps = 6 # number of truncated backprop steps ('n' in the discussion above)
#num_classes = 2
state_size = 10 #each state is represented with a certain width, a vector
learningRate = 1e-4 #default of Adam is 1e-3
#momentum = 0.5
#lamda2 = 1e-2
best_params_filename = 'best_params_rnn.npy'
```
here the state size is equal to the number of classes because we have given to the last output all the responsibility.
We are going to follow a repetitive process. For example if num_steps=6 then we break the 120 segments into 20 parts
The output of each part will be the genre. We are comparing against the genre every little part
### MSD 10 genre task
```
segmentCount = 120
segmentLen = 25
from rnn.msd10_data_providers import MSD10Genre_120_rnn_DataProvider
```
### Experiment with Best Parameters
```
best_params = np.load(best_params_filename)
best_params
(state_size, num_steps) = best_params
(state_size, num_steps)
rnnModel = RNNBatchNorm(batch_size=batch_size, rng=rng, dtype = curDtype, config=config,
segment_count=segmentCount, segment_len= segmentLen)
%%time
epochs = 100
stats, keys = rnnModel.run_rnn(state_size = state_size, num_steps=num_steps,
epochs = epochs)
if plotting:
fig_1, ax_1, fig_2, ax_2 = plotStats(stats, keys)
plt.show()
if saving:
figcount += 1
fig_1.savefig('cw%d_part%d_%02d_fig_error.svg' % (coursework, part, figcount))
fig_2.savefig('cw%d_part%d_%02d_fig_valid.svg' % (coursework, part, figcount))
print max(stats[:, -1]) #maximum validation accuracy
```
|
github_jupyter
|
```
import re
import json
import pandas as pd
import numpy as np
from collections import deque
```
## Process dataset
```
base_folder = "../movies-dataset/"
movies_metadata_fn = "movies_metadata.csv"
credits_fn = "credits.csv"
links_fn = "links.csv"
```
## Process movies_metadata data structure/schema
```
metadata = pd.read_csv(base_folder + movies_metadata_fn)
metadata.head()
```
## Cast id to int64 and drop any NAN values!
```
metadata.id = pd.to_numeric(metadata.id, downcast='signed', errors='coerce')
metadata = metadata[metadata['id'].notna()]
list(metadata.columns.values)
def CustomParser(data):
obj = json.loads(data)
return obj
```
We probably need id, title from this dataframe.
## Process credits data structure/schema
```
credits = pd.read_csv(base_folder + credits_fn)
# credits = pd.read_csv(base_folder + credits_fn, converters={'cast':CustomParser}, header=0)
# Cast id to int
credits.id = pd.to_numeric(credits.id, downcast='signed', errors='coerce')
credits.head()
# cast id to int64 for later join
metadata['id'] = metadata['id'].astype(np.int64)
credits['id'] = credits['id'].astype(np.int64)
metadata.dtypes
credits.dtypes
metadata.head(3)
credits.head(3)
```
## Let's join the two dataset based on movie id
We start with one example movie `Toy Story` with id = 862 in metadata dataset.
```
merged = pd.merge(metadata, credits, on='id')
merged.head(3)
toy_story_id = 862
merged.loc[merged['id'] == toy_story_id]
```
## Examine crew/cast json data schme for toy story
```
cast = merged.loc[merged['id'] == toy_story_id].cast
crew = merged.loc[merged['id'] == toy_story_id].crew
cast
```
## Find all movies Tom hanks has acted in
```
def has_played(actor_name, cast_data):
for cast in cast_data:
name = cast['name']
actor_id = cast['id']
cast_id = cast['cast_id']
credit_id = cast['credit_id']
if actor_name.lower() == name.lower():
print("name: {}, id: {}, cast_id: {}, credit_id: {}".format(name, actor_id, cast_id, credit_id))
return True
return False
```
## Setup data structure
```
# a map from movie id to a list of actor id's
movie_actor_adj_list = {}
# a map from actor id to a list of movie id's
actor_movie_adj_list = {}
# a map from movies id to their title
movies_map = {}
# a map from actors id to their name
actors_map = {}
cnt, errors = 0, 0
failed_movies = {}
for index, row in merged.iterrows():
cnt += 1
movie_id, movie_title = row['id'], row['title']
if movie_id not in movies_map:
movies_map[movie_id] = movie_title
dirty_json = row['cast']
try:
regex_replace = [(r"([ \{,:\[])(u)?'([^']+)'", r'\1"\3"'), (r" None", r' null')]
for r, s in regex_replace:
dirty_json = re.sub(r, s, dirty_json)
cast_data = json.loads(dirty_json)
# if has_played('Tom Hanks', cast_data):
# print("Movie id: {}, title: {}".format(movie_id, movie_title))
for cast in cast_data:
actor_name = cast['name']
actor_id = cast['id']
if actor_id not in actors_map:
actors_map[actor_id] = actor_name
# build movie-actor adj list
if movie_id not in movie_actor_adj_list:
movie_actor_adj_list[movie_id] = [actor_id]
else:
movie_actor_adj_list[movie_id].append(actor_id)
# build actor-movie adj list
if actor_id not in actor_movie_adj_list:
actor_movie_adj_list[actor_id] = [movie_id]
else:
actor_movie_adj_list[actor_id].append(movie_id)
except json.JSONDecodeError as err:
# print("JSONDecodeError: {}, Movie id: {}, title: {}".format(err, movie_id, movie_title))
failed_movies[movie_id] = True
errors += 1
print("Parsed credist: {}, errors: {}".format(cnt, errors))
movie_actor_adj_list[862]
inv_actors_map = {v: k for k, v in actors_map.items()}
inv_movies_map = {v: k for k, v in movies_map.items()}
kevin_id = inv_actors_map['Kevin Bacon']
print(kevin_id)
DEBUG = False
q = deque()
q.append(kevin_id)
bacon_degrees = {kevin_id: 0}
visited = {}
degree = 1
while q:
u = q.popleft()
if DEBUG:
print("u: {}".format(u))
# print(q)
if u not in visited:
visited[u] = True
if DEBUG:
print("degree(u): {}".format(bacon_degrees[u]))
if bacon_degrees[u] % 2 == 0:
# actor type node
neighbors = actor_movie_adj_list[u]
if DEBUG:
print("actor type, neighbors: {}".format(neighbors))
else:
# movie type node
neighbors = movie_actor_adj_list[u]
if DEBUG:
print("movie type, neighbors: {}".format(neighbors))
for v in neighbors:
if v not in visited:
q.append(v)
if v not in bacon_degrees:
bacon_degrees[v] = bacon_degrees[u] + 1
bacon_degrees[kevin_id]
actors_map[2224]
movies_map[9413]
actor_id = inv_actors_map['Tom Hanks']
bacon_degrees[actor_id]
actor_id = inv_actors_map['Tom Cruise']
bacon_degrees[actor_id]
movie_id = inv_movies_map['Apollo 13']
failed_movies[movie_id]
actor_id = inv_actors_map['Tom Cruise']
tom_cruise_movies = actor_movie_adj_list[actor_id]
actor_id = inv_actors_map['Kevin Bacon']
kevin_bacon_movies = actor_movie_adj_list[actor_id]
set(tom_cruise_movies).intersection(set(kevin_bacon_movies))
movies_map[881]
```
|
github_jupyter
|
```
import sympy as sp
import numpy as np
x = sp.symbols('x')
p = sp.Function('p')
l = sp.Function('l')
poly = sp.Function('poly')
p3 = sp.Function('p3')
p4 = sp.Function('p4')
```
# Introduction
Last time we have used Lagrange basis to interpolate polynomial. However, it is not efficient to update the interpolating polynomial when a new data point is added. We look at an iterative approach.
Given points $\{(z_i, f_i) \}_{i=0}^{n-1}$, $z_i$ are distinct and $p_{n-1} \in \mathbb{C}[z]_{n-1}\, , p_{n-1}(z_i) = f_i$. <br> We add a point $(z_n, f_n)$ and find a polynomial $p_n \in \mathbb{C}[x]_{n-1}$ which satisfies $\{(z_i, f_i) \}_{i=0}^{n}$.
We assume $p_n(z)$ be the form
\begin{equation}
p_n(z) = p_{n-1}(z) + C\prod_{i=0}^{n-1}(z - z_i)
\end{equation}
so that the second term vanishes at $z = z_0,...,z_{n-1}$ and $p_n(z_i) = p_{n-1}(z_i), i = 0,...,n-1$. We also want $p_n(z_n) = f_n$ so we have
\begin{equation}
f_n = p_{n-1}(z_n) + C\prod_{i=0}^{n-1}(z_n - z_i) \Rightarrow C = \frac{f_n - p_{n-1}(z_n)}{\prod_{i=0}^{n-1}(z_n - z_i)}
\end{equation}
Thus we may perform interpolation iteratively.
**Example:** Last time we have
\begin{equation}
(z_0, f_0) = (-1,-3), \quad
(z_1, f_1) = (0,-1), \quad
(z_2, f_2) = (2,4), \quad
(z_3, f_3) = (5,1)
\end{equation}
and
\begin{equation}
p_3(x) = \frac{-13}{90}z^3 + \frac{14}{45}z^2 + \frac{221}{90}z - 1
\end{equation}
```
z0 = -1; f0 = -3; z1 = 0; f1 = -1; z2 = 2; f2 = 4; z3 = 5; f3 = 1; z4 = 1; f4 = 1
p3 = -13*x**3/90 + 14*x**2/45 + 221*x/90 - 1
```
We add a point $(z_4,f_4) = (1,1)$ and obtain $p_4(x)$
```
z4 = 1; f4 = 1
C = (f4 - p3.subs(x,z4))/((z4-z0)*(z4-z1)*(z4-z2)*(z4-z3))
C
p4 = p3 + C*(x-z0)*(x-z1)*(x-z2)*(x-z3)
sp.expand(p4)
```
**Remark:** the constant $C$ is usually written as $f[z_0,z_1,z_2,z_3,z_4]$. Moreover by iteration we have
$$p_n(z) = \sum_{i=0}^n f[z_0,...,z_n] \prod_{j=0}^i (z - z_j)$$
# Newton Tableau
We look at efficient ways to compute $f[z_0,...,z_n]$, iteratively from $f[z_0,...,z_{n-1}]$ and $f[z_1,...,z_n]$. <br>
We may first construct $p_{n-1}$ and $q_{n-1}$ before constructing $p_n$ itself, where
\begin{gather}
p_{n-1}(z_i) = f_i \quad i = 0,...,n-1\\
q_{n-1}(z_i) = f_i \quad i = 1,...,n
\end{gather}
**Claim:** The following polynomial interpolate $\{(z_i,f_i)\}_{i=0}^n$
\begin{equation}
p_n(z_i) = \frac{(z - z_n)p_{n-1}(z) - (z - z_0)q_{n-1}(z)}{z_0 - z_n}
\end{equation}
Since interpolating polynomial is unique, by comparing coefficient of $z_n$, we have
$$f[z_0,...,z_{n}] = \frac{f[z_0,...,z_{n-1}]-f[z_1,...,z_{n}]}{z_0 - z_n}$$
```
def product(xs,key,i):
#Key: Forward or Backward
n = len(xs)-1
l = 1
for j in range(i):
if key == 'forward':
l *= (x - xs[j])
else:
l *= (x - xs[n-j])
return l
def newton(xs,ys,key):
# Key: Forward or Backward
n = len(xs)-1
# print(xs)
print(ys)
old_column = ys
if key == 'forward':
coeff = [fs[0]]
elif key == 'backward':
coeff = [fs[len(fs)-1]]
else:
return 'error'
for i in range(1,n+1): # Column Index
new_column = [(old_column[j+1] - old_column[j])/(xs[j+i] - xs[j]) for j in range(n-i+1)]
print(new_column)
if key == 'forward':
coeff.append(new_column[0])
else:
coeff.append(new_column[len(new_column)-1])
old_column = new_column
# print(coeff)
poly = 0
for i in range(n+1):
poly += coeff[i] * product(xs,key,i)
return poly
zs = [1, 4/3, 5/3, 2]; fs = [np.sin(x) for x in zs]
p = newton(zs,fs,'backward')
sp.simplify(p)
```
|
github_jupyter
|
```
import numpy as np, pandas as pd, matplotlib.pyplot as plt
import os
import seaborn as sns
sns.set()
root_path = r'C:\Users\54638\Desktop\Cannelle\Excel handling'
input_path = os.path.join(root_path, "input")
output_path = os.path.join(root_path, "output")
%%time
# this line magic function should always be put on first line of the cell, and without comments followed after in the same line
# Read all Excel file
all_deals = pd.DataFrame()
for file in os.listdir(input_path):
# can add other criteria as you want
if file[-4:] == 'xlsx':
tmp = pd.read_excel(os.path.join(input_path,file), index_col = 'order_no')
all_deals = pd.concat([all_deals, tmp])
# reindex, otherwise you may have many lines with same index
# all_deals = all_deals.reset_index() # this method is not recommended here, as it will pop out the original index 'order_no'
all_deals.index = range(len(all_deals))
all_deals.head()
# all_deals.tail()
# all_deals.shape
# all_deals.describe()
# overview
all_deals['counterparty'].value_counts().sort_values(ascending = False)
# all_deals['counterparty'].unique()
all_deals['deal'].value_counts().sort_values(ascending = False)
deal_vol = all_deals['deal'].value_counts().sort_values()
deal_vol.plot(figsize = (10,6), kind = 'bar');
# Some slicing
all_deals[all_deals['deal'] == 'Accumulator']
all_deals[(all_deals['deal'] == 'Variance Swap')
& (all_deals['counterparty'].isin(['Citi','HSBC']))]
all_deals.groupby('currency').sum()
# all_deals.groupby('currency')[['nominal']].sum()
ccy_way = all_deals.groupby(['way','currency']).sum().unstack('currency')
ccy_way
ccy_way.plot(figsize = (10,6), kind ='bar')
plt.legend(loc='upper left', bbox_to_anchor=(1,1), ncol=1)
# pivot_table
all_deals.pivot_table(values = 'nominal',
index='counterparty', columns='deal', aggfunc='count')
# save data
# ccy_way.to_excel(os.path.join(output_path,"Extract.xlsx"))
file_name = "Extract" + ".xlsx"
sheet_name = "Extract"
writer = pd.ExcelWriter(os.path.join(output_path, file_name), engine = 'xlsxwriter')
ccy_way.to_excel(writer, sheet_name=sheet_name)
# adjust the column width
worksheet = writer.sheets[sheet_name]
for id, col in enumerate(ccy_way):
series = ccy_way[col]
max_len = max(
series.astype(str).map(len).max(), # len of largest item
len(str(series.name)) # len of column name
) + 3 # a little extra space
max_len = min(max_len, 30) # set a cap, dont be too long
worksheet.set_column(id+1, id+1, max_len)
writer.save()
# Deleting all the file
del_path = input_path
for file in os.listdir(del_path):
os.remove(os.path.join(del_path, file))
# Generating file, all the data you see is just randomly created
import calendar
# Transaction generator
def generate_data(year, month):
order_amount = max(int(np.random.randn()*200)+500,0) + np.random.randint(2000)
start_loc = 1
order_no = np.arange(start_loc,order_amount+start_loc)
countparty_list = ['JPMorgan', 'Credit Suisse', 'Deutsche Bank', 'BNP Paribas', 'Credit Agricole', 'SinoPac', 'Goldman Sachs', 'Citi',
'Blackstone', 'HSBC', 'Natixis', 'BOCI', 'UBS', 'CLSA', 'CICC', 'Fidelity', 'Jefferies']
countparty_prob = [0.04, 0.07, 0.06, 0.1, 0.09, 0.02, 0.1, 0.08, 0.025, 0.13, 0.065, 0.05, 0.01, 0.08, 0.01, 0.04, 0.03]
countparty = np.random.choice(countparty_list, size=order_amount, p=countparty_prob)
deal_list = ['Autocall', 'Accumulator', 'Range Accrual', 'Variance Swap', 'Vanilla', 'Digital', 'Twinwin', 'ForwardStart',
'ForwardBasket', 'Cross Currency Swap', 'Hybrid']
deal_prob = [0.16, 0.2, 0.11, 0.05, 0.11, 0.09, 0.08, 0.04, 0.04, 0.03, 0.09]
deal = np.random.choice(deal_list, size=order_amount, p=deal_prob)
way = np.random.choice(['buy','sell'], size=order_amount)
nominal = [(int(np.random.randn()*10) + np.random.randint(200)+ 50)*1000 for _ in range(order_amount)]
currency_list = ['USD', 'CNY', 'EUR', 'SGP', 'JPY', 'KRW', 'AUD', 'GBP']
currency_prob = [0.185, 0.195, 0.14, 0.08, 0.135, 0.125, 0.06, 0.08]
currency = np.random.choice(currency_list, size=order_amount, p=currency_prob)
datelist = list(date for date in calendar.Calendar().itermonthdates(year, month) if date.month == month)
trade_date = np.random.choice(datelist, size=order_amount)
data = {'order_no': order_no, 'counterparty': countparty, 'deal':deal, 'way': way, 'nominal': nominal,
'currency':currency, 'trade_date': trade_date}
return pd.DataFrame(data)
save_path = input_path
cur_month = 4
cur_year = 2018
for i in range(24):
if cur_month == 12:
cur_month = 1
cur_year +=1
else:
cur_month += 1
df = generate_data(cur_year, cur_month)
df_name = 'Derivatives Transaction '+calendar.month_abbr[cur_month]+' '+str(cur_year)+'.xlsx'
df.to_excel(os.path.join(save_path, df_name), index = False)
```
|
github_jupyter
|
# fuzzy_pandas examples
These are almost all from [Max Harlow](https://twitter.com/maxharlow)'s [awesome NICAR2019 presentation](https://docs.google.com/presentation/d/1djKgqFbkYDM8fdczFhnEJLwapzmt4RLuEjXkJZpKves/) where he demonstrated [csvmatch](https://github.com/maxharlow/csvmatch), which fuzzy_pandas is based on.
**SCROLL DOWN DOWN DOWN TO GET TO THE FUZZY MATCHING PARTS.**
```
import pandas as pd
import fuzzy_pandas as fpd
df1 = pd.read_csv("data/data1.csv")
df2 = pd.read_csv("data/data2.csv")
df1
df2
```
# Exact matches
By default, all columns from both dataframes are returned.
```
# csvmatch \
# forbes-billionaires.csv \
# bloomberg-billionaires.csv \
# --fields1 name \
# --fields2 Name
df1 = pd.read_csv("data/forbes-billionaires.csv")
df2 = pd.read_csv("data/bloomberg-billionaires.csv")
results = fpd.fuzzy_merge(df1, df2, left_on='name', right_on='Name')
print("Found", results.shape)
results.head(5)
```
### Only keeping matching columns
The csvmatch default only gives you the shared columns, which you can reproduce with `keep='match'`
```
df1 = pd.read_csv("data/forbes-billionaires.csv")
df2 = pd.read_csv("data/bloomberg-billionaires.csv")
results = fpd.fuzzy_merge(df1, df2, left_on='name', right_on='Name', keep='match')
print("Found", results.shape)
results.head(5)
```
### Only keeping specified columns
```
df1 = pd.read_csv("data/forbes-billionaires.csv")
df2 = pd.read_csv("data/bloomberg-billionaires.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='Name',
keep_left=['name', 'realTimeRank'],
keep_right=['Rank'])
print("Found", results.shape)
results.head(5)
```
## Case sensitivity
This one doesn't give us any results!
```
# csvmatch \
# cia-world-leaders.csv \
# davos-attendees-2019.csv \
# --fields1 name \
# --fields2 full_name
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/davos-attendees-2019.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='full_name',
keep='match')
print("Found", results.shape)
results.head(10)
```
But if we add **ignore_case** we are good to go.
```
# csvmatch \
# cia-world-leaders.csv \
# davos-attendees-2019.csv \
# --fields1 name \
# --fields2 full_name \
# --ignore-case \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/davos-attendees-2019.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='full_name',
ignore_case=True,
keep='match')
print("Found", results.shape)
results.head(5)
```
### Ignoring case, non-latin characters, word ordering
You should really be reading [the presentation](https://docs.google.com/presentation/d/1djKgqFbkYDM8fdczFhnEJLwapzmt4RLuEjXkJZpKves/edit)!
```
# $ csvmatch \
# cia-world-leaders.csv \
# davos-attendees-2019.csv \
# --fields1 name \
# --fields2 full_name \
# -i -a -n -s \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/davos-attendees-2019.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on=['name'],
right_on=['full_name'],
ignore_case=True,
ignore_nonalpha=True,
ignore_nonlatin=True,
ignore_order_words=True,
keep='match')
print("Found", results.shape)
results.head(5)
```
# Fuzzy matching
## Levenshtein: Edit distance
```
# csvmatch \
# cia-world-leaders.csv \
# forbes-billionaires.csv \
# --fields1 name \
# --fields2 name \
# --fuzzy levenshtein \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/forbes-billionaires.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='name',
method='levenshtein',
keep='match')
print("Found", results.shape)
results.head(10)
```
### Setting a threshold with Levenshtein
```
# csvmatch \
# cia-world-leaders.csv \
# forbes-billionaires.csv \
# --fields1 name \
# --fields2 name \
# --fuzzy levenshtein \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/forbes-billionaires.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='name',
method='levenshtein',
threshold=0.85,
keep='match')
print("Found", results.shape)
results.head(10)
```
## Jaro: Edit distance
```
# csvmatch \
# cia-world-leaders.csv \
# forbes-billionaires.csv \
# --fields1 name \
# --fields2 name \
# --fuzzy levenshtein \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/forbes-billionaires.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='name',
method='jaro',
keep='match')
print("Found", results.shape)
results.head(10)
```
## Metaphone: Phonetic match
```
# csvmatch \
# cia-world-leaders.csv \
# un-sanctions.csv \
# --fields1 name \
# --fields2 name \
# --fuzzy metaphone \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/un-sanctions.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='name',
method='metaphone',
keep='match')
print("Found", results.shape)
results.head(10)
```
## Bilenko
You'll need to respond to the prompts when you run the code. 10-15 is best, but send `f` when you've decided you're finished.
```
# $ csvmatch \
# cia-world-leaders.csv \
# davos-attendees-2019.csv \
# --fields1 name \
# --fields2 full_name \
# --fuzzy bilenko \
df1 = pd.read_csv("data/cia-world-leaders.csv")
df2 = pd.read_csv("data/davos-attendees-2019.csv")
results = fpd.fuzzy_merge(df1, df2,
left_on='name',
right_on='full_name',
method='bilenko',
keep='match')
print("Found", results.shape)
results.head(10)
```
|
github_jupyter
|
<h1>Demand forecasting with BigQuery and TensorFlow</h1>
In this notebook, we will develop a machine learning model to predict the demand for taxi cabs in New York.
To develop the model, we will need to get historical data of taxicab usage. This data exists in BigQuery. Let's start by looking at the schema.
```
import google.datalab.bigquery as bq
import pandas as pd
import numpy as np
import shutil
%bq tables describe --name bigquery-public-data.new_york.tlc_yellow_trips_2015
```
<h2> Analyzing taxicab demand </h2>
Let's pull the number of trips for each day in the 2015 dataset using Standard SQL.
```
%bq query
SELECT
EXTRACT (DAYOFYEAR from pickup_datetime) AS daynumber
FROM `bigquery-public-data.new_york.tlc_yellow_trips_2015`
LIMIT 5
```
<h3> Modular queries and Pandas dataframe </h3>
Let's use the total number of trips as our proxy for taxicab demand (other reasonable alternatives are total trip_distance or total fare_amount). It is possible to predict multiple variables using Tensorflow, but for simplicity, we will stick to just predicting the number of trips.
We will give our query a name 'taxiquery' and have it use an input variable '$YEAR'. We can then invoke the 'taxiquery' by giving it a YEAR. The to_dataframe() converts the BigQuery result into a <a href='http://pandas.pydata.org/'>Pandas</a> dataframe.
```
%bq query -n taxiquery
WITH trips AS (
SELECT EXTRACT (DAYOFYEAR from pickup_datetime) AS daynumber
FROM `bigquery-public-data.new_york.tlc_yellow_trips_*`
where _TABLE_SUFFIX = @YEAR
)
SELECT daynumber, COUNT(1) AS numtrips FROM trips
GROUP BY daynumber ORDER BY daynumber
query_parameters = [
{
'name': 'YEAR',
'parameterType': {'type': 'STRING'},
'parameterValue': {'value': 2015}
}
]
trips = taxiquery.execute(query_params=query_parameters).result().to_dataframe()
trips[:5]
```
<h3> Benchmark </h3>
Often, a reasonable estimate of something is its historical average. We can therefore benchmark our machine learning model against the historical average.
```
avg = np.mean(trips['numtrips'])
print('Just using average={0} has RMSE of {1}'.format(avg, np.sqrt(np.mean((trips['numtrips'] - avg)**2))))
```
The mean here is about 400,000 and the root-mean-square-error (RMSE) in this case is about 52,000. In other words, if we were to estimate that there are 400,000 taxi trips on any given day, that estimate is will be off on average by about 52,000 in either direction.
Let's see if we can do better than this -- our goal is to make predictions of taxicab demand whose RMSE is lower than 52,000.
What kinds of things affect people's use of taxicabs?
<h2> Weather data </h2>
We suspect that weather influences how often people use a taxi. Perhaps someone who'd normally walk to work would take a taxi if it is very cold or rainy.
One of the advantages of using a global data warehouse like BigQuery is that you get to mash up unrelated datasets quite easily.
```
%bq query
SELECT * FROM `bigquery-public-data.noaa_gsod.stations`
WHERE state = 'NY' AND wban != '99999' AND name LIKE '%LA GUARDIA%'
```
<h3> Variables </h3>
Let's pull out the minimum and maximum daily temperature (in Fahrenheit) as well as the amount of rain (in inches) for La Guardia airport.
```
%bq query -n wxquery
SELECT EXTRACT (DAYOFYEAR FROM CAST(CONCAT(@YEAR,'-',mo,'-',da) AS TIMESTAMP)) AS daynumber,
MIN(EXTRACT (DAYOFWEEK FROM CAST(CONCAT(@YEAR,'-',mo,'-',da) AS TIMESTAMP))) dayofweek,
MIN(min) mintemp, MAX(max) maxtemp, MAX(IF(prcp=99.99,0,prcp)) rain
FROM `bigquery-public-data.noaa_gsod.gsod*`
WHERE stn='725030' AND _TABLE_SUFFIX = @YEAR
GROUP BY 1 ORDER BY daynumber DESC
query_parameters = [
{
'name': 'YEAR',
'parameterType': {'type': 'STRING'},
'parameterValue': {'value': 2015}
}
]
weather = wxquery.execute(query_params=query_parameters).result().to_dataframe()
weather[:5]
```
<h3> Merge datasets </h3>
Let's use Pandas to merge (combine) the taxi cab and weather datasets day-by-day.
```
data = pd.merge(weather, trips, on='daynumber')
data[:5]
```
<h3> Exploratory analysis </h3>
Is there a relationship between maximum temperature and the number of trips?
```
j = data.plot(kind='scatter', x='maxtemp', y='numtrips')
```
The scatterplot above doesn't look very promising. There appears to be a weak downward trend, but it's also quite noisy.
Is there a relationship between the day of the week and the number of trips?
```
j = data.plot(kind='scatter', x='dayofweek', y='numtrips')
```
Hurrah, we seem to have found a predictor. It appears that people use taxis more later in the week. Perhaps New Yorkers make weekly resolutions to walk more and then lose their determination later in the week, or maybe it reflects tourism dynamics in New York City.
Perhaps if we took out the <em>confounding</em> effect of the day of the week, maximum temperature will start to have an effect. Let's see if that's the case:
```
j = data[data['dayofweek'] == 7].plot(kind='scatter', x='maxtemp', y='numtrips')
```
Removing the confounding factor does seem to reflect an underlying trend around temperature. But ... the data are a little sparse, don't you think? This is something that you have to keep in mind -- the more predictors you start to consider (here we are using two: day of week and maximum temperature), the more rows you will need so as to avoid <em> overfitting </em> the model.
<h3> Adding 2014 and 2016 data </h3>
Let's add in 2014 and 2016 data to the Pandas dataframe. Note how useful it was for us to modularize our queries around the YEAR.
```
data2 = data # 2015 data
for year in [2014, 2016]:
query_parameters = [
{
'name': 'YEAR',
'parameterType': {'type': 'STRING'},
'parameterValue': {'value': year}
}
]
weather = wxquery.execute(query_params=query_parameters).result().to_dataframe()
trips = taxiquery.execute(query_params=query_parameters).result().to_dataframe()
data_for_year = pd.merge(weather, trips, on='daynumber')
data2 = pd.concat([data2, data_for_year])
data2.describe()
j = data2[data2['dayofweek'] == 7].plot(kind='scatter', x='maxtemp', y='numtrips')
```
The data do seem a bit more robust. If we had even more data, it would be better of course. But in this case, we only have 2014-2016 data for taxi trips, so that's what we will go with.
<h2> Machine Learning with Tensorflow </h2>
We'll use 80% of our dataset for training and 20% of the data for testing the model we have trained. Let's shuffle the rows of the Pandas dataframe so that this division is random. The predictor (or input) columns will be every column in the database other than the number-of-trips (which is our target, or what we want to predict).
The machine learning models that we will use -- linear regression and neural networks -- both require that the input variables are numeric in nature.
The day of the week, however, is a categorical variable (i.e. Tuesday is not really greater than Monday). So, we should create separate columns for whether it is a Monday (with values 0 or 1), Tuesday, etc.
Against that, we do have limited data (remember: the more columns you use as input features, the more rows you need to have in your training dataset), and it appears that there is a clear linear trend by day of the week. So, we will opt for simplicity here and use the data as-is. Try uncommenting the code that creates separate columns for the days of the week and re-run the notebook if you are curious about the impact of this simplification.
```
import tensorflow as tf
shuffled = data2.sample(frac=1, random_state=13)
# It would be a good idea, if we had more data, to treat the days as categorical variables
# with the small amount of data, we have though, the model tends to overfit
#predictors = shuffled.iloc[:,2:5]
#for day in range(1,8):
# matching = shuffled['dayofweek'] == day
# key = 'day_' + str(day)
# predictors[key] = pd.Series(matching, index=predictors.index, dtype=float)
predictors = shuffled.iloc[:,1:5]
predictors[:5]
shuffled[:5]
targets = shuffled.iloc[:,5]
targets[:5]
```
Let's update our benchmark based on the 80-20 split and the larger dataset.
```
trainsize = int(len(shuffled['numtrips']) * 0.8)
avg = np.mean(shuffled['numtrips'][:trainsize])
rmse = np.sqrt(np.mean((targets[trainsize:] - avg)**2))
print('Just using average={0} has RMSE of {1}'.format(avg, rmse))
```
<h2> Linear regression with tf.contrib.learn </h2>
We scale the number of taxicab rides by 400,000 so that the model can keep its predicted values in the [0-1] range. The optimization goes a lot faster when the weights are small numbers. We save the weights into ./trained_model_linear and display the root mean square error on the test dataset.
```
SCALE_NUM_TRIPS = 600000.0
trainsize = int(len(shuffled['numtrips']) * 0.8)
testsize = len(shuffled['numtrips']) - trainsize
npredictors = len(predictors.columns)
noutputs = 1
tf.logging.set_verbosity(tf.logging.WARN) # change to INFO to get output every 100 steps ...
shutil.rmtree('./trained_model_linear', ignore_errors=True) # so that we don't load weights from previous runs
estimator = tf.contrib.learn.LinearRegressor(model_dir='./trained_model_linear',
feature_columns=tf.contrib.learn.infer_real_valued_columns_from_input(predictors.values))
print("starting to train ... this will take a while ... use verbosity=INFO to get more verbose output")
def input_fn(features, targets):
return tf.constant(features.values), tf.constant(targets.values.reshape(len(targets), noutputs)/SCALE_NUM_TRIPS)
estimator.fit(input_fn=lambda: input_fn(predictors[:trainsize], targets[:trainsize]), steps=10000)
pred = np.multiply(list(estimator.predict(predictors[trainsize:].values)), SCALE_NUM_TRIPS )
rmse = np.sqrt(np.mean(np.power((targets[trainsize:].values - pred), 2)))
print('LinearRegression has RMSE of {0}'.format(rmse))
```
The RMSE here (57K) is lower than the benchmark (62K) indicates that we are doing about 10% better with the machine learning model than we would be if we were to just use the historical average (our benchmark).
<h2> Neural network with tf.contrib.learn </h2>
Let's make a more complex model with a few hidden nodes.
```
SCALE_NUM_TRIPS = 600000.0
trainsize = int(len(shuffled['numtrips']) * 0.8)
testsize = len(shuffled['numtrips']) - trainsize
npredictors = len(predictors.columns)
noutputs = 1
tf.logging.set_verbosity(tf.logging.WARN) # change to INFO to get output every 100 steps ...
shutil.rmtree('./trained_model', ignore_errors=True) # so that we don't load weights from previous runs
estimator = tf.contrib.learn.DNNRegressor(model_dir='./trained_model',
hidden_units=[5, 5],
feature_columns=tf.contrib.learn.infer_real_valued_columns_from_input(predictors.values))
print("starting to train ... this will take a while ... use verbosity=INFO to get more verbose output")
def input_fn(features, targets):
return tf.constant(features.values), tf.constant(targets.values.reshape(len(targets), noutputs)/SCALE_NUM_TRIPS)
estimator.fit(input_fn=lambda: input_fn(predictors[:trainsize], targets[:trainsize]), steps=10000)
pred = np.multiply(list(estimator.predict(predictors[trainsize:].values)), SCALE_NUM_TRIPS )
rmse = np.sqrt(np.mean((targets[trainsize:].values - pred)**2))
print('Neural Network Regression has RMSE of {0}'.format(rmse))
```
Using a neural network results in similar performance to the linear model when I ran it -- it might be because there isn't enough data for the NN to do much better. (NN training is a non-convex optimization, and you will get different results each time you run the above code).
<h2> Running a trained model </h2>
So, we have trained a model, and saved it to a file. Let's use this model to predict taxicab demand given the expected weather for three days.
Here we make a Dataframe out of those inputs, load up the saved model (note that we have to know the model equation -- it's not saved in the model file) and use it to predict the taxicab demand.
```
input = pd.DataFrame.from_dict(data =
{'dayofweek' : [4, 5, 6],
'mintemp' : [60, 40, 50],
'maxtemp' : [70, 90, 60],
'rain' : [0, 0.5, 0]})
# read trained model from ./trained_model
estimator = tf.contrib.learn.LinearRegressor(model_dir='./trained_model_linear',
feature_columns=tf.contrib.learn.infer_real_valued_columns_from_input(input.values))
pred = np.multiply(list(estimator.predict(input.values)), SCALE_NUM_TRIPS )
print(pred)
```
Looks like we should tell some of our taxi drivers to take the day off on Thursday (day=5). No wonder -- the forecast calls for extreme weather fluctuations on Thursday.
Copyright 2017 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License
|
github_jupyter
|
# TV Script Generation
In this project, you'll generate your own [Seinfeld](https://en.wikipedia.org/wiki/Seinfeld) TV scripts using RNNs. You'll be using part of the [Seinfeld dataset](https://www.kaggle.com/thec03u5/seinfeld-chronicles#scripts.csv) of scripts from 9 seasons. The Neural Network you'll build will generate a new ,"fake" TV script, based on patterns it recognizes in this training data.
## Get the Data
The data is already provided for you in `./data/Seinfeld_Scripts.txt` and you're encouraged to open that file and look at the text.
>* As a first step, we'll load in this data and look at some samples.
* Then, you'll be tasked with defining and training an RNN to generate a new script!
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# load in data
import helper
data_dir = './data/Seinfeld_Scripts.txt'
text = helper.load_data(data_dir)
```
## Explore the Data
Play around with `view_line_range` to view different parts of the data. This will give you a sense of the data you'll be working with. You can see, for example, that it is all lowercase text, and each new line of dialogue is separated by a newline character `\n`.
```
view_line_range = (0, 10)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
import numpy as np
print('Dataset Stats')
print('Roughly the number of unique words: {}'.format(len({word: None for word in text.split()})))
lines = text.split('\n')
print('Number of lines: {}'.format(len(lines)))
word_count_line = [len(line.split()) for line in lines]
print('Average number of words in each line: {}'.format(np.average(word_count_line)))
print()
print('The lines {} to {}:'.format(*view_line_range))
print('\n'.join(text.split('\n')[view_line_range[0]:view_line_range[1]]))
```
---
## Implement Pre-processing Functions
The first thing to do to any dataset is pre-processing. Implement the following pre-processing functions below:
- Lookup Table
- Tokenize Punctuation
### Lookup Table
To create a word embedding, you first need to transform the words to ids. In this function, create two dictionaries:
- Dictionary to go from the words to an id, we'll call `vocab_to_int`
- Dictionary to go from the id to word, we'll call `int_to_vocab`
Return these dictionaries in the following **tuple** `(vocab_to_int, int_to_vocab)`
```
import problem_unittests as tests
from collections import Counter
def create_lookup_tables(text):
"""
Create lookup tables for vocabulary
:param text: The text of tv scripts split into words
:return: A tuple of dicts (vocab_to_int, int_to_vocab)
"""
# TODO: Implement Function
count = Counter(text)
vocabulary = sorted(count, key=count.get, reverse=True)
int_vocabulary = {i: word for i, word in enumerate(vocabulary)}
vocabulary_int = {word: i for i, word in int_vocabulary.items()}
# return tuple
return (vocabulary_int, int_vocabulary)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_create_lookup_tables(create_lookup_tables)
```
### Tokenize Punctuation
We'll be splitting the script into a word array using spaces as delimiters. However, punctuations like periods and exclamation marks can create multiple ids for the same word. For example, "bye" and "bye!" would generate two different word ids.
Implement the function `token_lookup` to return a dict that will be used to tokenize symbols like "!" into "||Exclamation_Mark||". Create a dictionary for the following symbols where the symbol is the key and value is the token:
- Period ( **.** )
- Comma ( **,** )
- Quotation Mark ( **"** )
- Semicolon ( **;** )
- Exclamation mark ( **!** )
- Question mark ( **?** )
- Left Parentheses ( **(** )
- Right Parentheses ( **)** )
- Dash ( **-** )
- Return ( **\n** )
This dictionary will be used to tokenize the symbols and add the delimiter (space) around it. This separates each symbols as its own word, making it easier for the neural network to predict the next word. Make sure you don't use a value that could be confused as a word; for example, instead of using the value "dash", try using something like "||dash||".
```
def token_lookup():
"""
Generate a dict to turn punctuation into a token.
:return: Tokenized dictionary where the key is the punctuation and the value is the token
"""
# TODO: Implement Function
tokenfinder = dict()
tokenfinder['.'] = '<PERIOD>'
tokenfinder[','] = '<COMMA>'
tokenfinder['"'] = '<QUOTATION_MARK>'
tokenfinder[';'] = '<SEMICOLON>'
tokenfinder['!'] = '<EXCLAMATION_MARK>'
tokenfinder['?'] = '<QUESTION_MARK>'
tokenfinder['('] = '<LEFT_PAREN>'
tokenfinder[')'] = '<RIGHT_PAREN>'
tokenfinder['?'] = '<QUESTION_MARK>'
tokenfinder['-'] = '<DASH>'
tokenfinder['\n'] = '<NEW_LINE>'
return tokenfinder
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_tokenize(token_lookup)
```
## Pre-process all the data and save it
Running the code cell below will pre-process all the data and save it to file. You're encouraged to lok at the code for `preprocess_and_save_data` in the `helpers.py` file to see what it's doing in detail, but you do not need to change this code.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# pre-process training data
helper.preprocess_and_save_data(data_dir, token_lookup, create_lookup_tables)
```
# Check Point
This is your first checkpoint. If you ever decide to come back to this notebook or have to restart the notebook, you can start from here. The preprocessed data has been saved to disk.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
import problem_unittests as tests
int_text, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()
```
## Build the Neural Network
In this section, you'll build the components necessary to build an RNN by implementing the RNN Module and forward and backpropagation functions.
### Check Access to GPU
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import torch
# Check for a GPU
train_on_gpu = torch.cuda.is_available()
if not train_on_gpu:
print('No GPU found. Please use a GPU to train your neural network.')
```
## Input
Let's start with the preprocessed input data. We'll use [TensorDataset](http://pytorch.org/docs/master/data.html#torch.utils.data.TensorDataset) to provide a known format to our dataset; in combination with [DataLoader](http://pytorch.org/docs/master/data.html#torch.utils.data.DataLoader), it will handle batching, shuffling, and other dataset iteration functions.
You can create data with TensorDataset by passing in feature and target tensors. Then create a DataLoader as usual.
```
data = TensorDataset(feature_tensors, target_tensors)
data_loader = torch.utils.data.DataLoader(data,
batch_size=batch_size)
```
### Batching
Implement the `batch_data` function to batch `words` data into chunks of size `batch_size` using the `TensorDataset` and `DataLoader` classes.
>You can batch words using the DataLoader, but it will be up to you to create `feature_tensors` and `target_tensors` of the correct size and content for a given `sequence_length`.
For example, say we have these as input:
```
words = [1, 2, 3, 4, 5, 6, 7]
sequence_length = 4
```
Your first `feature_tensor` should contain the values:
```
[1, 2, 3, 4]
```
And the corresponding `target_tensor` should just be the next "word"/tokenized word value:
```
5
```
This should continue with the second `feature_tensor`, `target_tensor` being:
```
[2, 3, 4, 5] # features
6 # target
```
```
from torch.utils.data import TensorDataset, DataLoader
def batch_data(words, sequence_length, batch_size):
"""
Batch the neural network data using DataLoader
:param words: The word ids of the TV scripts
:param sequence_length: The sequence length of each batch
:param batch_size: The size of each batch; the number of sequences in a batch
:return: DataLoader with batched data
"""
# TODO: Implement function
n_batches = len(words)//batch_size
# number of words in a batch
words = words[:n_batches*batch_size]
# length of output
y_len = len(words) - sequence_length
# empty lists for sequences
x, y = [], []
for i in range(0, y_len):
i_end = sequence_length + i
x_batch = words[i:i_end]
x.append(x_batch)
batch_y = words[i_end]
y.append(batch_y)
data = TensorDataset(torch.from_numpy(np.asarray(x)), torch.from_numpy(np.asarray(y)))
data_loader = DataLoader(data, shuffle=False, batch_size=batch_size)
# return a dataloader
return data_loader
# there is no test for this function, but you are encouraged to create
# print statements and tests of your own
```
### Test your dataloader
You'll have to modify this code to test a batching function, but it should look fairly similar.
Below, we're generating some test text data and defining a dataloader using the function you defined, above. Then, we are getting some sample batch of inputs `sample_x` and targets `sample_y` from our dataloader.
Your code should return something like the following (likely in a different order, if you shuffled your data):
```
torch.Size([10, 5])
tensor([[ 28, 29, 30, 31, 32],
[ 21, 22, 23, 24, 25],
[ 17, 18, 19, 20, 21],
[ 34, 35, 36, 37, 38],
[ 11, 12, 13, 14, 15],
[ 23, 24, 25, 26, 27],
[ 6, 7, 8, 9, 10],
[ 38, 39, 40, 41, 42],
[ 25, 26, 27, 28, 29],
[ 7, 8, 9, 10, 11]])
torch.Size([10])
tensor([ 33, 26, 22, 39, 16, 28, 11, 43, 30, 12])
```
### Sizes
Your sample_x should be of size `(batch_size, sequence_length)` or (10, 5) in this case and sample_y should just have one dimension: batch_size (10).
### Values
You should also notice that the targets, sample_y, are the *next* value in the ordered test_text data. So, for an input sequence `[ 28, 29, 30, 31, 32]` that ends with the value `32`, the corresponding output should be `33`.
```
# test dataloader
test_text = range(50)
t_loader = batch_data(test_text, sequence_length=5, batch_size=10)
data_iter = iter(t_loader)
sample_x, sample_y = data_iter.next()
print(sample_x.shape)
print(sample_x)
print()
print(sample_y.shape)
print(sample_y)
```
---
## Build the Neural Network
Implement an RNN using PyTorch's [Module class](http://pytorch.org/docs/master/nn.html#torch.nn.Module). You may choose to use a GRU or an LSTM. To complete the RNN, you'll have to implement the following functions for the class:
- `__init__` - The initialize function.
- `init_hidden` - The initialization function for an LSTM/GRU hidden state
- `forward` - Forward propagation function.
The initialize function should create the layers of the neural network and save them to the class. The forward propagation function will use these layers to run forward propagation and generate an output and a hidden state.
**The output of this model should be the *last* batch of word scores** after a complete sequence has been processed. That is, for each input sequence of words, we only want to output the word scores for a single, most likely, next word.
### Hints
1. Make sure to stack the outputs of the lstm to pass to your fully-connected layer, you can do this with `lstm_output = lstm_output.contiguous().view(-1, self.hidden_dim)`
2. You can get the last batch of word scores by shaping the output of the final, fully-connected layer like so:
```
# reshape into (batch_size, seq_length, output_size)
output = output.view(batch_size, -1, self.output_size)
# get last batch
out = output[:, -1]
```
```
import torch.nn as nn
class RNN(nn.Module):
def __init__(self, vocab_size, output_size, embedding_dim, hidden_dim, n_layers, dropout=0.5):
"""
Initialize the PyTorch RNN Module
:param vocab_size: The number of input dimensions of the neural network (the size of the vocabulary)
:param output_size: The number of output dimensions of the neural network
:param embedding_dim: The size of embeddings, should you choose to use them
:param hidden_dim: The size of the hidden layer outputs
:param dropout: dropout to add in between LSTM/GRU layers
"""
super(RNN, self).__init__()
# TODO: Implement function
self.embed = nn.Embedding(vocab_size, embedding_dim)
# set class variables
self.lstm = nn.LSTM(embedding_dim, hidden_dim, n_layers, dropout=dropout, batch_first=True)
self.output_size = output_size
self.n_layers = n_layers
self.hidden_dim = hidden_dim
# define model layers
self.fc = nn.Linear(hidden_dim, output_size)
def forward(self, nn_input, hidden):
"""
Forward propagation of the neural network
:param nn_input: The input to the neural network
:param hidden: The hidden state
:return: Two Tensors, the output of the neural network and the latest hidden state
"""
# TODO: Implement function
batch_size = nn_input.size(0)
embedding = self.embed(nn_input)
lstm_output, hidden = self.lstm(embedding, hidden)
# return one batch of output word scores and the hidden state
out = self.fc(lstm_output)
out = out.view(batch_size, -1, self.output_size)
out = out[:, -1]
return out, hidden
def init_hidden(self, batch_size):
'''
Initialize the hidden state of an LSTM/GRU
:param batch_size: The batch_size of the hidden state
:return: hidden state of dims (n_layers, batch_size, hidden_dim)
'''
# Implement function
weight = next(self.parameters()).data
if (train_on_gpu):
hidden = (weight.new(self.n_layers, batch_size, self.hidden_dim).zero_().cuda(),
weight.new(self.n_layers, batch_size, self.hidden_dim).zero_().cuda())
else:
hidden = (weight.new(self.n_layers, batch_size, self.hidden_dim).zero_(),
weight.new(self.n_layers, batch_size, self.hidden_dim).zero_())
return hidden
# initialize hidden state with zero weights, and move to GPU if available
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_rnn(RNN, train_on_gpu)
```
### Define forward and backpropagation
Use the RNN class you implemented to apply forward and back propagation. This function will be called, iteratively, in the training loop as follows:
```
loss = forward_back_prop(decoder, decoder_optimizer, criterion, inp, target)
```
And it should return the average loss over a batch and the hidden state returned by a call to `RNN(inp, hidden)`. Recall that you can get this loss by computing it, as usual, and calling `loss.item()`.
**If a GPU is available, you should move your data to that GPU device, here.**
```
def forward_back_prop(rnn, optimizer, criterion, inp, target, hidden):
"""
Forward and backward propagation on the neural network
:param decoder: The PyTorch Module that holds the neural network
:param decoder_optimizer: The PyTorch optimizer for the neural network
:param criterion: The PyTorch loss function
:param inp: A batch of input to the neural network
:param target: The target output for the batch of input
:return: The loss and the latest hidden state Tensor
"""
# TODO: Implement Function
if(train_on_gpu):
rnn.cuda()
# move data to GPU, if available
h1 = tuple([each.data for each in hidden])
rnn.zero_grad()
# perform backpropagation and optimization
if(train_on_gpu):
inputs, target = inp.cuda(), target.cuda()
output, h = rnn(inputs, h1)
loss = criterion(output, target)
loss.backward()
nn.utils.clip_grad_norm(rnn.parameters(), 5)
optimizer.step()
# return the loss over a batch and the hidden state produced by our model
return loss.item(), h1
# Note that these tests aren't completely extensive.
# they are here to act as general checks on the expected outputs of your functions
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_forward_back_prop(RNN, forward_back_prop, train_on_gpu)
```
## Neural Network Training
With the structure of the network complete and data ready to be fed in the neural network, it's time to train it.
### Train Loop
The training loop is implemented for you in the `train_decoder` function. This function will train the network over all the batches for the number of epochs given. The model progress will be shown every number of batches. This number is set with the `show_every_n_batches` parameter. You'll set this parameter along with other parameters in the next section.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
def train_rnn(rnn, batch_size, optimizer, criterion, n_epochs, show_every_n_batches=100):
batch_losses = []
rnn.train()
print("Training for %d epoch(s)..." % n_epochs)
for epoch_i in range(1, n_epochs + 1):
# initialize hidden state
hidden = rnn.init_hidden(batch_size)
for batch_i, (inputs, labels) in enumerate(train_loader, 1):
# make sure you iterate over completely full batches, only
n_batches = len(train_loader.dataset)//batch_size
if(batch_i > n_batches):
break
# forward, back prop
loss, hidden = forward_back_prop(rnn, optimizer, criterion, inputs, labels, hidden)
# record loss
batch_losses.append(loss)
# printing loss stats
if batch_i % show_every_n_batches == 0:
print('Epoch: {:>4}/{:<4} Loss: {}\n'.format(
epoch_i, n_epochs, np.average(batch_losses)))
batch_losses = []
# returns a trained rnn
return rnn
```
### Hyperparameters
Set and train the neural network with the following parameters:
- Set `sequence_length` to the length of a sequence.
- Set `batch_size` to the batch size.
- Set `num_epochs` to the number of epochs to train for.
- Set `learning_rate` to the learning rate for an Adam optimizer.
- Set `vocab_size` to the number of uniqe tokens in our vocabulary.
- Set `output_size` to the desired size of the output.
- Set `embedding_dim` to the embedding dimension; smaller than the vocab_size.
- Set `hidden_dim` to the hidden dimension of your RNN.
- Set `n_layers` to the number of layers/cells in your RNN.
- Set `show_every_n_batches` to the number of batches at which the neural network should print progress.
If the network isn't getting the desired results, tweak these parameters and/or the layers in the `RNN` class.
```
# Data params
# Sequence Length
sequence_length = 5 # of words in a sequence
# Batch Size
batch_size = 128
# data loader - do not change
train_loader = batch_data(int_text, sequence_length, batch_size)
# Training parameters
# Number of Epochs
num_epochs = 10
# Learning Rate
learning_rate = 0.001
# Model parameters
# Vocab size
vocab_size = len(vocab_to_int)
# Output size
output_size = vocab_size
# Embedding Dimension
embedding_dim = 200
# Hidden Dimension
hidden_dim = 250
# Number of RNN Layers
n_layers = 2
# Show stats for every n number of batches
show_every_n_batches = 500
```
### Train
In the next cell, you'll train the neural network on the pre-processed data. If you have a hard time getting a good loss, you may consider changing your hyperparameters. In general, you may get better results with larger hidden and n_layer dimensions, but larger models take a longer time to train.
> **You should aim for a loss less than 3.5.**
You should also experiment with different sequence lengths, which determine the size of the long range dependencies that a model can learn.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# create model and move to gpu if available
rnn = RNN(vocab_size, output_size, embedding_dim, hidden_dim, n_layers, dropout=0.5)
if train_on_gpu:
rnn.cuda()
# defining loss and optimization functions for training
optimizer = torch.optim.Adam(rnn.parameters(), lr=learning_rate)
criterion = nn.CrossEntropyLoss()
# training the model
trained_rnn = train_rnn(rnn, batch_size, optimizer, criterion, num_epochs, show_every_n_batches)
# saving the trained model
helper.save_model('./save/trained_rnn', trained_rnn)
print('Model Trained and Saved')
```
### Question: How did you decide on your model hyperparameters?
For example, did you try different sequence_lengths and find that one size made the model converge faster? What about your hidden_dim and n_layers; how did you decide on those?
**Answer:**
- I experimented with the hyperparameters and observed different scenarios. Learning from the content, the dimensions for the typical layers were given from 100-300. For exact tuning, I referred to the blog:
https://towardsdatascience.com/choosing-the-right-hyperparameters-for-a-simple-lstm-using-keras-f8e9ed76f046
- I particularly loved this post: https://blog.floydhub.com/guide-to-hyperparameters-search-for-deep-learning-models/
- It was about hyperparameter in general but its emphasis on preventing any overfitting or underfitting in the model was great. I tried a learning rate of 0.01 initially however turned out the loss saturated just after 4 epochs and my best understanding of the situation was that the model was jumping quite quickly in the gradient descent and thus these large jumps were at cost of missing many features that hold significance for predicting patterns.
- A learning rate of 0.0001 became too low and the model was converging slowly but provided that we had to wait more patiently for the loss to minimize. I tried with sequence length of 25 initially and the loss was high nearly starting from 11 and coming towars 7 to maximum of 6.2. So, my understanding was that we need to have lesser number of sequences to identify patterns more accurately as the model seemed in a hurry to predict it and led to huge loss values.
- So, I tried with 15, 20 and 10 sequence lengths and everytime the loss decreased so finally I decided to keep it minimal enough and took 5 so that atleast we can identify patterns more correctly than rushing to a large value and spoiling the aim of the training. The sequence length of 10 noted good patterns too with loss minimizing upto 3.7.
- With 250 hidden layers, I do not think that there was a problem in minimizing loss due to less number of layers for processing the sequence. Ofcourse, the idea of a cyclic learning rate can be used to further minimize the loss by making use of optimized learning rate at the correct timing.
- Finally with a learning rate of 0.001 and 5 sequences with batch size of 128 I was able to minimize the loss below 3.5! Also, batch size also affects the training process with larger batch sizes accelerating the time of training but at the cost of loss and too small batch size took huge training time! If used less than 128, the model was converging at only a little better loss and hence, as a tradeoff 128 did not seem to be a bad batch size for training but I do recommend 64 too!
---
# Checkpoint
After running the above training cell, your model will be saved by name, `trained_rnn`, and if you save your notebook progress, **you can pause here and come back to this code at another time**. You can resume your progress by running the next cell, which will load in our word:id dictionaries _and_ load in your saved model by name!
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import torch
import helper
import problem_unittests as tests
_, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()
trained_rnn = helper.load_model('./save/trained_rnn')
```
## Generate TV Script
With the network trained and saved, you'll use it to generate a new, "fake" Seinfeld TV script in this section.
### Generate Text
To generate the text, the network needs to start with a single word and repeat its predictions until it reaches a set length. You'll be using the `generate` function to do this. It takes a word id to start with, `prime_id`, and generates a set length of text, `predict_len`. Also note that it uses topk sampling to introduce some randomness in choosing the most likely next word, given an output set of word scores!
```
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
import torch.nn.functional as F
def generate(rnn, prime_id, int_to_vocab, token_dict, pad_value, predict_len=100):
"""
Generate text using the neural network
:param decoder: The PyTorch Module that holds the trained neural network
:param prime_id: The word id to start the first prediction
:param int_to_vocab: Dict of word id keys to word values
:param token_dict: Dict of puncuation tokens keys to puncuation values
:param pad_value: The value used to pad a sequence
:param predict_len: The length of text to generate
:return: The generated text
"""
rnn.eval()
# create a sequence (batch_size=1) with the prime_id
current_seq = np.full((1, sequence_length), pad_value)
current_seq[-1][-1] = prime_id
predicted = [int_to_vocab[prime_id]]
for _ in range(predict_len):
if train_on_gpu:
current_seq = torch.LongTensor(current_seq).cuda()
else:
current_seq = torch.LongTensor(current_seq)
# initialize the hidden state
hidden = rnn.init_hidden(current_seq.size(0))
# get the output of the rnn
output, _ = rnn(current_seq, hidden)
# get the next word probabilities
p = F.softmax(output, dim=1).data
if(train_on_gpu):
p = p.cpu() # move to cpu
# use top_k sampling to get the index of the next word
top_k = 5
p, top_i = p.topk(top_k)
top_i = top_i.numpy().squeeze()
# select the likely next word index with some element of randomness
p = p.numpy().squeeze()
word_i = np.random.choice(top_i, p=p/p.sum())
# retrieve that word from the dictionary
word = int_to_vocab[word_i]
predicted.append(word)
# the generated word becomes the next "current sequence" and the cycle can continue
current_seq = np.roll(current_seq, -1, 1)
current_seq[-1][-1] = word_i
gen_sentences = ' '.join(predicted)
# Replace punctuation tokens
for key, token in token_dict.items():
ending = ' ' if key in ['\n', '(', '"'] else ''
gen_sentences = gen_sentences.replace(' ' + token.lower(), key)
gen_sentences = gen_sentences.replace('\n ', '\n')
gen_sentences = gen_sentences.replace('( ', '(')
# return all the sentences
return gen_sentences
```
### Generate a New Script
It's time to generate the text. Set `gen_length` to the length of TV script you want to generate and set `prime_word` to one of the following to start the prediction:
- "jerry"
- "elaine"
- "george"
- "kramer"
You can set the prime word to _any word_ in our dictionary, but it's best to start with a name for generating a TV script. (You can also start with any other names you find in the original text file!)
```
# run the cell multiple times to get different results!
gen_length = 400 # modify the length to your preference
prime_word = 'jerry' # name for starting the script
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
pad_word = helper.SPECIAL_WORDS['PADDING']
generated_script = generate(trained_rnn, vocab_to_int[prime_word + ':'], int_to_vocab, token_dict, vocab_to_int[pad_word], gen_length)
print(generated_script)
```
#### Save your favorite scripts
Once you have a script that you like (or find interesting), save it to a text file!
```
# save script to a text file
f = open("generated_script_1.txt","w")
f.write(generated_script)
f.close()
```
# The TV Script is Not Perfect
It's ok if the TV script doesn't make perfect sense. It should look like alternating lines of dialogue, here is one such example of a few generated lines.
### Example generated script
>jerry: what about me?
>
>jerry: i don't have to wait.
>
>kramer:(to the sales table)
>
>elaine:(to jerry) hey, look at this, i'm a good doctor.
>
>newman:(to elaine) you think i have no idea of this...
>
>elaine: oh, you better take the phone, and he was a little nervous.
>
>kramer:(to the phone) hey, hey, jerry, i don't want to be a little bit.(to kramer and jerry) you can't.
>
>jerry: oh, yeah. i don't even know, i know.
>
>jerry:(to the phone) oh, i know.
>
>kramer:(laughing) you know...(to jerry) you don't know.
You can see that there are multiple characters that say (somewhat) complete sentences, but it doesn't have to be perfect! It takes quite a while to get good results, and often, you'll have to use a smaller vocabulary (and discard uncommon words), or get more data. The Seinfeld dataset is about 3.4 MB, which is big enough for our purposes; for script generation you'll want more than 1 MB of text, generally.
# Submitting This Project
When submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as "dlnd_tv_script_generation.ipynb" and save another copy as an HTML file by clicking "File" -> "Download as.."->"html". Include the "helper.py" and "problem_unittests.py" files in your submission. Once you download these files, compress them into one zip file for submission.
|
github_jupyter
|
# Deterministic point jet
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import matplotlib.pylab as pl
```
\begin{equation}
\partial_t \zeta = \frac{\zeta_{jet}}{\tau} - \mu \zeta + \nu_\alpha \nabla^{2\alpha} - \beta \partial_x \psi - J(\psi, \zeta) \zeta
\end{equation}
Here $\zeta_{jet}$ is the profile of a prescribed jet and $\tau = 1/\mu$ is the relaxation time. Also $\beta = 2\Omega cos \theta$ and $\theta = 0$ corresponds to the equator.
Parameters
> $\Omega = 2\pi$
> $\mu = 0.05$
> $\nu = 0$
> $\nu_4 = 0$
> $\Xi = 1.0$
> $\Delta \theta = 0.1$
> $\tau$ = 20 days
```
dn = "pointjet/8x8/"
# Parameters: μ = 0.05, τ = 20.0, Ξ = 1.0*Ω and Δθ = 0.1
M = 8
N = 8
colors = pl.cm.nipy_spectral(np.linspace(0,1,M))
```
## NL v GQL v GCE2
```
nl = np.load(dn+"nl.npz",allow_pickle=True)
gql = np.load(dn+"gql.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2.npz",allow_pickle=True)
# fig,ax = plt.subplots(1,2,figsize=(14,5))
# # Energy
# ax[0].plot(nl['t'],nl['Etav'],label=r'$\langle NL \rangle$')
# ax[0].plot(gql['t'],gql['Etav'],label=r'$\langle GQL(M) \rangle$')
# ax[0].plot(gce2['t'],gce2['Etav'],label=r'$\langle GCE2(M) \rangle$')
# ax[0].set_xlabel(r'$t$',fontsize=14)
# ax[0].set_ylabel(r'$E$',fontsize=14)
# # ax[0].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
# # Enstrophy
# ax[1].plot(nl['t'],nl['Ztav'],label=r'$\langle NL \rangle$')
# ax[1].plot(gql['t'],gql['Ztav'],label=r'$\langle GQL(M) \rangle$')
# ax[1].plot(gce2['t'],gce2['Ztav'],label=r'$\langle GCE2(M) \rangle$')
# ax[1].set_xlabel(r'$t$',fontsize=14)
# ax[1].set_ylabel(r'$Z$',fontsize=14)
# ax[1].legend(loc=4,fontsize=14)
# plt.show()
fig,ax = plt.subplots(1,3,figsize=(14,5))
ax[0].set_title(f'NL')
for i,x in enumerate(nl['Emt'].T):
ax[0].plot(nl['t'],x,label=i,c=colors[i])
ax[1].set_title(f'GQL(M)')
for i,x in enumerate(gql['Emt'].T):
ax[1].plot(gql['t'],x,label=i,c=colors[i])
ax[2].set_title(f'GCE2(M)')
for i,x in enumerate(gce2['Emt'].T):
ax[2].plot(gce2['t'],x,label=i,c=colors[i])
for a in ax:
a.set_xlabel(r'$t$',fontsize=14)
a.set_yscale('log')
a.set_ylim(1e-12,1e2)
ax[0].set_ylabel(r'$E(m)$',fontsize=14)
ax[2].legend(bbox_to_anchor=(1.01,0.85),ncol=1)
plt.show()
# fig,ax = plt.subplots(1,3,figsize=(16,4))
# im = ax[0].imshow((nl['Vxy'][:,:,0]),interpolation="bicubic",cmap="RdBu_r",origin="lower")
# fig.colorbar(im, ax=ax[0])
# ax[0].set_title(r'NL: $\zeta(x,y,t = 0 )$',fontsize=14)
# im = ax[1].imshow((gql['Vxy'][:,:,0]),interpolation="bicubic",cmap="RdBu_r",origin="lower")
# fig.colorbar(im, ax=ax[1])
# ax[1].set_title(r'GQL: $\zeta(x,y,t = 0 )$',fontsize=14)
# im = ax[2].imshow((gce2['Vxy'][:,:,0]),interpolation="bicubic",cmap="RdBu_r",origin="lower")
# fig.colorbar(im, ax=ax[2])
# ax[2].set_title(r'GCE2:$\zeta(x,y,t = 0 )$',fontsize=14)
# for a in ax:
# a.set_xticks([0,M-1,2*M-2])
# a.set_xticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
# a.set_yticks([0,M-1,2*M-2])
# a.set_yticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
# plt.show()
fig,ax = plt.subplots(1,3,figsize=(16,4))
im = ax[0].imshow((nl['Vxy'][:,:,-1]),interpolation="bicubic",cmap="RdBu_r",origin="lower")
fig.colorbar(im, ax=ax[0])
ax[0].set_title(r'NL: $\zeta(x,y,t = 0 )$',fontsize=14)
im = ax[1].imshow((gql['Vxy'][:,:,-1]),interpolation="bicubic",cmap="RdBu_r",origin="lower")
fig.colorbar(im, ax=ax[1])
ax[1].set_title(r'GQL: $\zeta(x,y,t = 0 )$',fontsize=14)
im = ax[2].imshow((gce2['Vxy'][:,:,-1]),interpolation="bicubic",cmap="RdBu_r",origin="lower")
fig.colorbar(im, ax=ax[2])
ax[2].set_title(r'GCE2:$\zeta(x,y,t = 0 )$',fontsize=14)
for a in ax:
a.set_xticks([0,M-1,2*M-2])
a.set_xticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
a.set_yticks([0,M-1,2*M-2])
a.set_yticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
plt.show()
```
## QL v CE2 v GCE2(0)
```
ql = np.load(dn+"ql.npz",allow_pickle=True)
ce2 = np.load(dn+"ce2.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2_0.npz",allow_pickle=True)
fig,ax = plt.subplots(1,3,figsize=(18,5))
# Energy
ax[0].set_title(f'QL',fontsize=14)
for i,x in enumerate(ql['Emtav'].T):
ax[0].plot(ql['t'],x,label=i,c=colors[i])
ax[1].set_title(f'CE2',fontsize=14)
for i,x in enumerate(ce2['Emtav'].T):
ax[1].plot(ce2['t'],x,label=i,c=colors[i])
ax[2].set_title(f'GCE2(0)',fontsize=14)
for i,x in enumerate(gce2['Emtav'].T):
ax[2].plot(gce2['t'],x,label=i,c=colors[i])
ax[2].legend(bbox_to_anchor=(1.01,0.5),ncol=1)
for a in ax:
a.set_xlabel(r'$t$',fontsize=14)
a.set_ylabel(r'$E$',fontsize=14)
a.set_yscale('log')
a.set_ylim(1e-12,1e2)
# plt.show()
# plt.savefig(dn+"ze_tau20_qlce2gce2_0.png",bbox_inches='tight',)
```
## GQL(1) v GCE2(1)
```
gql = np.load(dn+"gql_1.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2_1.npz",allow_pickle=True)
fig,ax = plt.subplots(1,2,figsize=(14,5))
# Energy
ax[0].set_title(f'GQL(1)',fontsize=14)
for i,x in enumerate(gql['Emtav'].T):
ax[0].plot(gql['t'],x,label=i,c=colors[i])
ax[1].set_title(f'GCE2(1)',fontsize=14)
for i,x in enumerate(gce2['Emtav'].T):
ax[1].plot(gce2['t'],x,label=i,c=colors[i])
for a in ax:
a.set_xlabel(r'$t$',fontsize=14)
a.set_ylabel(r'$E$',fontsize=14)
a.set_yscale('log')
# a.set_ylim(1e-1,1e0)
ax[1].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
plt.show()
fig,ax = plt.subplots(1,2,figsize=(15,6))
ax[0].set_title(f'GQL(1)',fontsize=14)
im = ax[0].imshow((gql['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
fig.colorbar(im,ax=ax[0])
ax[1].set_title(f'GCE2(1)',fontsize=14)
im = ax[1].imshow((gce2['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
fig.colorbar(im,ax=ax[1])
for a in ax:
a.set_xticks([0,M-1,2*M-2])
a.set_xticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
a.set_yticks([0,M-1,2*M-2])
a.set_yticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
plt.show()
fig,ax = plt.subplots(1,2,figsize=(12,6))
ax[0].set_title('GQL(1)')
im = ax[0].imshow((gql['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im)
ax[1].set_title('GCE2(1)')
im = ax[1].imshow((gce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im)
for a in ax:
a.set_xticks([0,M-1,2*M-2])
a.set_xticklabels([r'$-M$',r'$0$',r'$M$'],fontsize=14)
a.set_yticks([0,M-1,2*M-2])
a.set_yticklabels([r'$-M$',r'$0$',r'$M$'],fontsize=14)
plt.show()
gql = np.load(dn+"gql_3.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2_3.npz",allow_pickle=True)
fig,ax = plt.subplots(1,2,figsize=(14,5))
# Energy
ax[0].set_title(f'GQL(3)',fontsize=14)
for i,x in enumerate(gql['Emtav'].T):
ax[0].plot(gql['t'],x,label=i,c=colors[i])
ax[1].set_title(f'GCE2(3)',fontsize=14)
for i,x in enumerate(gce2['Emtav'].T):
ax[1].plot(gce2['t'],x,label=i,c=colors[i])
for a in ax:
a.set_xlabel(r'$t$',fontsize=14)
a.set_ylabel(r'$E$',fontsize=14)
a.set_yscale('log')
# a.set_ylim(1e-1,1e0)
ax[1].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
plt.show()
fig,ax = plt.subplots(1,2,figsize=(15,6))
ax[0].set_title(f'GQL(3)',fontsize=14)
im = ax[0].imshow((gql['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
fig.colorbar(im,ax=ax[0])
ax[1].set_title(f'GCE2(3)',fontsize=14)
im = ax[1].imshow((gce2['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
fig.colorbar(im,ax=ax[1])
for a in ax:
a.set_xticks([0,M-1,2*M-2])
a.set_xticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
a.set_yticks([0,M-1,2*M-2])
a.set_yticklabels([r'$0$',r'$\pi$',r'$2\pi$'],fontsize=14)
plt.show()
fig,ax = plt.subplots(1,2,figsize=(12,6))
ax[0].set_title('GQL(3)')
im = ax[0].imshow((gql['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im)
ax[1].set_title('GCE2(3)')
im = ax[1].imshow((gce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im)
for a in ax:
a.set_xticks([0,M-1,2*M-2])
a.set_xticklabels([r'$-M$',r'$0$',r'$M$'],fontsize=14)
a.set_yticks([0,M-1,2*M-2])
a.set_yticklabels([r'$-M$',r'$0$',r'$M$'],fontsize=14)
plt.show()
# gql = np.load(dn+"gql_5.npz",allow_pickle=True)
# gce2 = np.load(dn+"gce2_5.npz",allow_pickle=True)
# fig,ax = plt.subplots(1,2,figsize=(14,5))
# # Energy
# ax[0].set_title(f'GQL(5)',fontsize=14)
# for i,x in enumerate(gql['Emtav'].T):
# ax[0].plot(gql['t'],x,label=i,c=colors[i])
# ax[1].set_title(f'GCE2(5)',fontsize=14)
# for i,x in enumerate(gce2['Emtav'].T):
# ax[1].plot(gce2['t'],x,c=colors[i])
# for a in ax:
# a.set_xlabel(r'$t$',fontsize=14)
# a.set_ylabel(r'$E$',fontsize=14)
# a.set_yscale('log')
# # a.set_ylim(1e-1,1e0)
# # ax[1].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,2,figsize=(12,6))
# ax[0].set_title('GQL(5)')
# im = ax[0].imshow((gql['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# # fig.colorbar(im)
# ax[1].set_title('GCE2(5)')
# im = ax[1].imshow((gce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# # fig.colorbar(im)
# for a in ax:
# a.set_xticks([0,M-1,2*M-2])
# a.set_xticklabels([r'$-M$',r'$0$',r'$M$'],fontsize=14)
# a.set_yticks([0,M-1,2*M-2])
# a.set_yticklabels([r'$-M$',r'$0$',r'$M$'],fontsize=14)
# plt.show()
# dn = "pointjet/12x12/"
# Parameters: μ = 0.05, τ = 20.0, Ξ = 1.0*Ω and Δθ = 0.1
Nx = 12
Ny = 12
nl = np.load(dn+"nl.npz",allow_pickle=True)
gql = np.load(dn+"gql.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2.npz",allow_pickle=True)
# fig,ax = plt.subplots(1,2,figsize=(14,5))
# # Energy
# ax[0].plot(nl['t'],nl['Etav'],label=r'$\langle NL \rangle$')
# ax[0].plot(gql['t'],gql['Etav'],label=r'$\langle GQL(M) \rangle$')
# ax[0].plot(gce2['t'],gce2['Etav'],label=r'$\langle GCE2(M) \rangle$')
# ax[0].set_xlabel(r'$t$',fontsize=14)
# ax[0].set_ylabel(r'$E$',fontsize=14)
# # ax[0].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
# # Enstrophy
# ax[1].plot(nl['t'],nl['Ztav'],label=r'$\langle NL \rangle$')
# ax[1].plot(gql['t'],gql['Ztav'],label=r'$\langle GQL(M) \rangle$')
# ax[1].plot(gce2['t'],gce2['Ztav'],label=r'$\langle GCE2(M) \rangle$')
# ax[1].set_xlabel(r'$t$',fontsize=14)
# ax[1].set_ylabel(r'$Z$',fontsize=14)
# ax[1].legend(loc=4,fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,3,figsize=(14,5))
# ax[0].set_title(f'NL')
# for i,x in enumerate(nl['Emt'].T):
# ax[0].plot(nl['t'],x,label=i,c=colors[i])
# ax[1].set_title(f'GQL(M)')
# for i,x in enumerate(gql['Emt'].T):
# ax[1].plot(gql['t'],x,label=i,c=colors[i])
# ax[2].set_title(f'GCE2(M)')
# for i,x in enumerate(gce2['Emt'].T):
# ax[2].plot(gce2['t'],x,label=i,c=colors[i])
# for a in ax:
# a.set_xlabel(r'$t$',fontsize=14)
# a.set_yscale('log')
# # a.set_ylim(1e-10,1e1)
# ax[0].set_ylabel(r'$E(m)$',fontsize=14)
# ax[2].legend(bbox_to_anchor=(1.01,0.85),ncol=1)
# plt.show()
# fig,ax = plt.subplots(1,3,figsize=(24,6))
# ax[0].set_title(f'NL',fontsize=14)
# im = ax[0].imshow((nl['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'GQL(M)',fontsize=14)
# im = ax[1].imshow((gql['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# ax[2].set_title(f'GCE2(M)',fontsize=14)
# im = ax[2].imshow((gce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[2])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,3,figsize=(24,6))
# ax[0].set_title(f'NL',fontsize=14)
# im = ax[0].imshow((nl['Vxy'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'GQL(M)',fontsize=14)
# im = ax[1].imshow((gql['Vxy'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# ax[2].set_title(f'GCE2(M)',fontsize=14)
# im = ax[2].imshow((gce2['Vxy'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[2])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
ql = np.load(dn+"ql.npz",allow_pickle=True)
ce2 = np.load(dn+"ce2.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2_0.npz",allow_pickle=True)
# fig,ax = plt.subplots(1,3,figsize=(14,5))
# # Energy
# ax[0].set_title(f'QL',fontsize=14)
# for i,x in enumerate(ql['Emtav'].T):
# ax[0].plot(ql['t'],x,label=i,c=colors[i])
# ax[1].set_title(f'CE2',fontsize=14)
# for i,x in enumerate(ce2['Emtav'].T):
# ax[1].plot(ce2['t'],x,c=colors[i])
# ax[2].set_title(f'GCE2(0)',fontsize=14)
# for i,x in enumerate(gce2['Emtav'].T):
# ax[2].plot(gce2['t'],x,c=colors[i])
# for a in ax:
# a.set_xlabel(r'$t$',fontsize=14)
# a.set_ylabel(r'$E$',fontsize=14)
# a.set_yscale('log')
# # a.set_ylim(1e-1,1e0)
# # ax[1].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,3,figsize=(24,6))
# ax[0].set_title(f'QL',fontsize=14)
# im = ax[0].imshow((ql['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'CE2',fontsize=14)
# im = ax[1].imshow((ce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# ax[2].set_title(f'GCE2(0)',fontsize=14)
# im = ax[2].imshow((gce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[2])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,3,figsize=(24,6))
# ax[0].set_title(f'QL',fontsize=14)
# im = ax[0].imshow((ql['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'CE2',fontsize=14)
# im = ax[1].imshow((ce2['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# ax[2].set_title(f'GCE2(0)',fontsize=14)
# im = ax[2].imshow((gce2['Vxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[2])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,3,figsize=(24,6))
# ax[0].set_title(f'QL',fontsize=14)
# im = ax[0].imshow((ql['Uxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'CE2',fontsize=14)
# im = ax[1].imshow((ce2['Uxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# ax[2].set_title(f'GCE2(0)',fontsize=14)
# im = ax[2].imshow((gce2['Uxy'][:,:,-1]),cmap="RdBu_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[2])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
gql = np.load(dn+"gql_3.npz",allow_pickle=True)
gce2 = np.load(dn+"gce2_3.npz",allow_pickle=True)
# fig,ax = plt.subplots(1,2,figsize=(14,5))
# # Energy
# ax[0].set_title(f'GQL',fontsize=14)
# for i,x in enumerate(gql['Emtav'].T):
# ax[0].plot(gql['t'],x,label=i,c=colors[i])
# ax[1].set_title(f'GCE2',fontsize=14)
# for i,x in enumerate(gce2['Emtav'].T):
# ax[1].plot(gce2['t'],x,c=colors[i])
# for a in ax:
# a.set_xlabel(r'$t$',fontsize=14)
# a.set_ylabel(r'$E$',fontsize=14)
# a.set_yscale('log')
# # a.set_ylim(1e-1,1e0)
# # ax[1].legend(bbox_to_anchor=(1.01,0.85),fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,2,figsize=(15,6))
# ax[0].set_title(f'GQL(1)',fontsize=14)
# im = ax[0].imshow((gql['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'GCE2(1)',fontsize=14)
# im = ax[1].imshow((gce2['Emn'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
# fig,ax = plt.subplots(1,2,figsize=(15,6))
# ax[0].set_title(f'GQL(1)',fontsize=14)
# im = ax[0].imshow((gql['Vxy'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[0])
# ax[1].set_title(f'GCE2(1)',fontsize=14)
# im = ax[1].imshow((gce2['Vxy'][:,:,-1]),cmap="nipy_spectral_r",origin="lower",interpolation="bicubic")
# fig.colorbar(im,ax=ax[1])
# for a in ax:
# a.set_xticks([0,Nx-1,2*Nx-2])
# a.set_xticklabels([r'$-N_x$',r'$0$',r'$N_x$'],fontsize=14)
# a.set_yticks([0,Ny-1,2*Ny-2])
# a.set_yticklabels([r'$-N_y$',r'$0$',r'$N_y$'],fontsize=14)
# plt.show()
```
|
github_jupyter
|
# Building Autonomous Trader using mt5se
## How to setup and use mt5se
### 1. Install Metatrader 5 (https://www.metatrader5.com/)
### 2. Install python package Metatrader5 using pip
#### Use: pip install MetaTrader5 ... or Use sys package
### 3. Install python package mt5se using pip
#### Use: pip install mt5se ... or Use sys package
#### For documentation, check : https://paulo-al-castro.github.io/mt5se/
```
# installing Metatrader5 using sys
import sys
# python MetaTrader5
#!{sys.executable} -m pip install MetaTrader5
#mt5se
!{sys.executable} -m pip install mt5se --upgrade
```
<hr>
## Connecting and getting account information
```
import mt5se as se
connected=se.connect()
if connected:
print('Ok!! It is connected to the Stock exchange!!')
else:
print('Something went wrong! It is NOT connected to se!!')
ti=se.terminal_info()
print('Metatrader program file path: ', ti.path)
print('Metatrader path to data folder: ', ti.data_path )
print('Metatrader common data path: ',ti.commondata_path)
```
<hr>
### Getting information about the account
```
acc=se.account_info() # it returns account's information
print('login=',acc.login) # Account id
print('balance=',acc.balance) # Account balance in the deposit currency using buy price of assets (margin_free+margin)
print('equity=',acc.equity) # Account equity in the deposit currency using current price of assets (capital liquido) (margin_free+margin+profit)
print('free margin=',acc.margin_free) # Free margin ( balance in cash ) of an account in the deposit currency(BRL)
print('margin=',acc.margin) #Account margin used in the deposit currency (equity-margin_free-profit )
print('client name=',acc.name) #Client name
print('Server =',acc.server) # Trade server name
print('Currency =',acc.currency) # Account currency, BRL for Brazilian Real
```
<hr>
### Getting info about asset's prices quotes (a.k.a bars)
```
import pandas as pd
# Some example of Assets in Nasdaq
assets=[
'AAL', # American Airlines Group, Inc.
'GOOG', # Apple Inc.
'UAL', # United Airlines Holdings, Inc.
'AMD', # Advanced Micro Devices, Inc.
'MSFT' # MICROSOFT
]
asset=assets[0]
df=se.get_bars(asset,10) # it returns the last 10 days
print(df)
```
<hr>
### Getting information about current position
```
print('Position=',se.get_positions()) # return the current value of assets (not include balance or margin)
symbol_id='MSFT'
print('Position on paper ',symbol_id,' =',se.get_position_value(symbol_id)) # return the current position in a given asset (symbol_id)
pos=se.get_position_value(symbol_id)
print(pos)
```
<hr>
### Creating, checking and sending orders
```
###Buying three hundred shares of AAPL !!
symbol_id='AAPL'
bars=se.get_bars(symbol_id,2)
price=se.get_last(bars)
volume=300
b=se.buyOrder(symbol_id,volume, price ) # price, sl and tp are optional
if se.is_market_open(symbol_id):
print('Market is Open!!')
else:
print('Market is closed! Orders will not be accepted!!')
if se.checkOrder(b):
print('Buy order seems ok!')
else:
print('Error : ',se.getLastError())
# if se.sendOrder(b):
# print('Order executed!)
```
### Direct Control Robots using mt5se
```
import mt5se as se
import pandas as pd
import time
asset='AAPL'
def run(asset):
if se.is_market_open(asset): # change 'if' for 'while' for running until the end of the market session
print("getting information")
bars=se.get_bars(asset,14)
curr_shares=se.get_shares(asset)
# number of shares that you can buy
price=se.get_last(bars)
free_shares=se.get_affor_shares(asset,price)
rsi=se.tech.rsi(bars)
print("deliberating")
if rsi>=70 and free_shares>0:
order=se.buyOrder(asset,free_shares)
elif rsi<70 and curr_shares>0:
order=se.sellOrder(asset,curr_shares)
else:
order=None
print("sending order")
# check and send (it is sent only if check is ok!)
if order!=None:
if se.checkOrder(order) and se.sendOrder(order):
print('order sent to se')
else:
print('Error : ',se.getLastError())
else:
print("No order at the moment for asset=",asset )
time.sleep(1) # waits one second
print('Trader ended operation!')
if se.connect()==False:
print('Error when trying to connect to se')
exit()
else:
run(asset) # trade asset PETR4
```
### Multiple asset Trading Robot
```
#Multiple asset Robot (Example), single strategy for multiple assets, where the resources are equally shared among the assets
import time
def runMultiAsset(assets):
if se.is_market_open(assets[0]): # change 'if' for 'while' for running until the end of the market session
for asset in assets:
bars=se.get_bars(asset,14) #get information
curr_shares=se.get_shares(asset)
money=se.account_info().margin_free/len(assets) # number of shares that you can buy
price=se.get_last(bars)
free_shares=se.get_affor_shares(asset,price,money)
rsi=se.tech.rsi(bars)
if rsi>=70 and free_shares>0:
order=se.buyOrder(asset,free_shares)
elif rsi<70 and curr_shares>0:
order=se.sellOrder(asset,curr_shares)
else:
order=None
if order!=None: # check and send if it is Ok
if se.checkOrder(order) and se.sendOrder(order):
print('order sent to se')
else:
print('Error : ',se.getLastError())
else:
print("No order at the moment for asset=",asset)
time.sleep(1)
print('Trader ended operation!')
```
## Running multiple asset direct control code!
```
assets=['GOOG','AAPL']
runMultiAsset(assets) # trade asset
```
### Processing Financial Data - Return Histogram Example
```
import mt5se as se
from datetime import datetime
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
asset='MSFT'
se.connect()
bars=se.get_bars(asset,252) # 252 business days (basically one year)
x=se.get_returns(bars) # calculate daily returns given the bars
#With a small change we can see the historgram of weekly returns
#x=se.getReturns(bars,offset=5)
plt.hist(x,bins=16) # creates a histogram graph with 16 bins
plt.grid()
plt.show()
```
### Robots based on Inversion of control
You may use an alternative method to build your robots, that may reduce your workload. It is called inverse control robots. You receive the most common information requrired by robots and returns your orders
Let's some examples of Robots based on Inversion of Control including the multiasset strategy presented before in a inverse control implementation
### Trader class
Inversion of control Traders are classes that inherint from se.Trader and they have to implement just one function:
trade: It is called at each moment, with dbars. It should returns the list of orders to be executed or None if there is no order at the moment
Your trader may also implement two other function if required:
setup: It is called once when the operation starts. It receives dbars ('mem' bars from each asset) . See the operation setup, for more information
ending: It is called one when the sheculed operation reaches its end time.
Your Trader class may also implement a constructor function
Let's see an Example!
### A Random Trader
```
import numpy.random as rand
class RandomTrader(se.Trader):
def __init__(self):
pass
def setup(self,dbars):
print('just getting started!')
def trade(self,dbars):
orders=[]
assets=ops['assets']
for asset in assets:
if rand.randint(2)==1:
order=se.buyOrder(asset,100)
else:
order=se.sellOrder(asset,100)
orders.append(order)
return orders
def ending(self,dbars):
print('Ending stuff')
if issubclass(RandomTrader,se.Trader):
print('Your trader class seems Ok!')
else:
print('Your trader class should a subclass of se.Trader')
trader=RandomTrader() # DummyTrader class also available in se.sampleTraders.DummyTrader()
```
### Another Example of Trader class
```
class MultiAssetTrader(se.Trader):
def trade(self,dbars):
assets=dbars.keys()
orders=[]
for asset in assets:
bars=dbars[asset]
curr_shares=se.get_shares(asset)
money=se.get_balance()/len(assets) # divide o saldo em dinheiro igualmente entre os ativos
# number of shares that you can buy of asset
price=se.get_last(bars)
free_shares=se.get_affor_shares(asset,price,money)
rsi=se.tech.rsi(bars)
if rsi>=70 and free_shares>0:
order=se.buyOrder(asset,free_shares)
elif rsi<70 and curr_shares>0:
order=se.sellOrder(asset,curr_shares)
else:
order=None
if order!=None:
orders.append(order)
return orders
if issubclass(MultiAssetTrader,se.Trader):
print('Your trader class seems Ok!')
else:
print('Your trader class should a subclass of se.Trader')
trader=MultiAssetTrader()
```
### Testing your Trader!!!
The evaluation for trading robots is usually called backtesting. That means that a trading robot executes with historical price series , and its performance is computed
In backtesting, time is discretized according with bars and the package mt5se controls the information access to the Trader according with the simulated time.
To backtest one strategy, you just need to create a subclass of Trader and implement one function:
trade
You may implement function setup, to prepare the Trader Strategy if it is required and a function ending to clean up after the backtest is done
The simulation time advances and in function 'trade' the Trader class receives the new bar info and decides wich orders to send
## Let's create a Simple Algorithmic Trader and Backtest it
```
## Defines the Trader
class MonoAssetTrader(se.Trader):
def trade(self,dbars):
assets=dbars.keys()
asset=list(assets)[0]
orders=[]
bars=dbars[asset]
curr_shares=se.get_shares(asset)
# number of shares that you can buy
price=se.get_last(bars)
free_shares=se.get_affor_shares(asset,price)
rsi=se.tech.rsi(bars)
if rsi>=70:
order=se.buyOrder(asset,free_shares)
else:
order=se.sellOrder(asset,curr_shares)
if rsi>=70 and free_shares>0:
order=se.buyOrder(asset,free_shares)
elif rsi<70 and curr_shares>0:
order=se.sellOrder(asset,curr_shares)
if order!=None:
orders.append(order)
return orders
trader=MonoAssetTrader() # also available in se.sampleTraders.MonoAssetTrader()
print(trader)
```
## Setup and check a backtest!
```
# sets Backtest options
prestart=se.date(2018,12,10)
start=se.date(2019,1,10)
end=se.date(2019,2,27)
capital=1000000
results_file='data_equity_file.csv'
verbose=False
assets=['AAPL']
# Use True if you want debug information for your Trader
#sets the backtest setup
period=se.DAILY
# it may be se.INTRADAY (one minute interval)
bts=se.backtest.set(assets,prestart,start,end,period,capital,results_file,verbose)
# check if the backtest setup is ok!
if se.backtest.checkBTS(bts):
print('Backtest Setup is Ok!')
else:
print('Backtest Setup is NOT Ok!')
```
## Run the Backtest
```
# Running the backtest
df= se.backtest.run(trader,bts)
# run calls the Trader. setup and trade (once for each bar)
```
## Evaluate the Backtest result
```
#print the results
print(df)
# evaluates the backtest results
se.backtest.evaluate(df)
```
## Evaluating Backtesting results
The method backtest.run creates a data file with the name given in the backtest setup (bts)
This will give you a report about the trader performance
We need ot note that it is hard to perform meaningful evaluations using backtest. There are many pitfalls to avoid and it may be easier to get trading robots with great performance in backtest, but that perform really badly in real operations.
More about that in mt5se backtest evaluation chapter.
For a deeper discussion, we suggest:
Is it a great Autonomous Trading Strategy or you are just fooling yourself Bernardini,M. and Castro, P.A.L
In order to analyze the trader's backtest, you may use :
se.backtest.evaluateFile(fileName) #fileName is the name of file generated by the backtest
or
se.bactest.evaluate(df) # df is the dataframe returned by se.backtest.run
# Another Example: Multiasset Trader
```
import mt5se as se
class MultiAssetTrader(se.Trader):
def trade(self,dbars):
assets=dbars.keys()
orders=[]
for asset in assets:
bars=dbars[asset]
curr_shares=se.get_shares(asset)
money=se.get_balance()/len(assets) # divide o saldo em dinheiro igualmente entre os ativos
# number of shares that you can buy of asset
price=se.get_last(bars)
free_shares=se.get_affor_shares(asset,price,money)
rsi=se.tech.rsi(bars)
if rsi>=70 and free_shares>0:
order=se.buyOrder(asset,free_shares)
elif rsi<70 and curr_shares>0:
order=se.sellOrder(asset,curr_shares)
else:
order=None
if order!=None:
orders.append(order)
return orders
trader=MultiAssetTrader() # also available in se.sampleTraders.MultiAssetTrader()
print(trader)
```
## Setuping Backtest for Multiple Assets
```
# sets Backtest options
prestart=se.date(2020,5,4)
start=se.date(2020,5,6)
end=se.date(2020,6,21)
capital=10000000
results_file='data_equity_file.csv'
verbose=False
assets=[
'AAL', # American Airlines Group, Inc.
'GOOG', # Apple Inc.
'UAL', # United Airlines Holdings, Inc.
'AMD', # Advanced Micro Devices, Inc.
'MSFT' # MICROSOFT
]
# Use True if you want debug information for your Trader
#sets the backtest setup
period=se.DAILY
bts=se.backtest.set(assets,prestart,start,end,period,capital,results_file,verbose)
if se.backtest.checkBTS(bts): # check if the backtest setup is ok!
print('Backtest Setup is Ok!')
else:
print('Backtest Setup is NOT Ok!')
```
## Run and evaluate the backtest
```
se.connect()
# Running the backtest
df= se.backtest.run(trader,bts)
# run calls the Trader. setup and trade (once for each bar)
# evaluates the backtest results
se.backtest.evaluate(df)
```
## Next Deploying Autonomous Trader powered by mt5se
### You have seen how to:
install and import mt5se and MetaTrader5
get financial data
create direct control trading robots
create [Simple] Trader classes based on inversion of control
backtest Autonomous Traders
### Next, We are going to show how to:
deploy autonomous trader to run on simulated or real Stock Exchange accounts
create Autonomous Traders based on Artifical Intelligence and Machine Learning
|
github_jupyter
|
<a href="https://colab.research.google.com/github/Lambda-School-Labs/bridges-to-prosperity-ds-d/blob/SMOTE_model_building%2Ftrevor/notebooks/Modeling_off_original_data_smote_gridsearchcv.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
This notebook is for problem 2 as described in `B2P Dataset_2020.10.xlsx` Contextual Summary tab:
## Problem 2: Predicting which sites will be technically rejected in future engineering reviews
> Any sites with a "Yes" in the column AQ (`Senior Engineering Review Conducted`) have undergone a full technical review, and of those, the Stage (column L) can be considered to be correct. (`Bridge Opportunity: Stage`)
> Any sites without a "Yes" in Column AQ (`Senior Engineering Review Conducted`) have not undergone a full technical review, and the Stage is based on the assessor's initial estimate as to whether the site was technically feasible or not.
> We want to know if we can use the sites that have been reviewed to understand which of the sites that haven't yet been reviewed are likely to be rejected by the senior engineering team.
> Any of the data can be used, but our guess is that Estimated Span, Height Differential Between Banks, Created By, and Flag for Rejection are likely to be the most reliable predictors.
### Load the data
```
import pandas as pd
url = 'https://github.com/Lambda-School-Labs/bridges-to-prosperity-ds-d/blob/main/Data/B2P%20Dataset_2020.10.xlsx?raw=true'
df = pd.read_excel(url, sheet_name='Data')
```
### Define the target
```
# Any sites with a "Yes" in the column "Senior Engineering Review Conducted"
# have undergone a full technical review, and of those, the
# "Bridge Opportunity: Stage" column can be considered to be correct.
positive = (
(df['Senior Engineering Review Conducted']=='Yes') &
(df['Bridge Opportunity: Stage'].isin(['Complete', 'Prospecting', 'Confirmed']))
)
negative = (
(df['Senior Engineering Review Conducted']=='Yes') &
(df['Bridge Opportunity: Stage'].isin(['Rejected', 'Cancelled']))
)
# Any sites without a "Yes" in column Senior Engineering Review Conducted"
# have not undergone a full technical review ...
# So these sites are unknown and unlabeled
unknown = df['Senior Engineering Review Conducted'].isna()
# Create a new column named "Good Site." This is the target to predict.
# Assign a 1 for the positive class and 0 for the negative class.
df.loc[positive, 'Good Site'] = 1
df.loc[negative, 'Good Site'] = 0
# Assign -1 for unknown/unlabled observations.
# Scikit-learn's documentation for "Semi-Supervised Learning" says,
# "It is important to assign an identifier to unlabeled points ...
# The identifier that this implementation uses is the integer value -1."
# We'll explain this soon!
df.loc[unknown, 'Good Site'] = -1
```
### Drop columns used to derive the target
```
# Because these columns were used to derive the target,
# We can't use them as features, or it would be leakage.
df = df.drop(columns=['Senior Engineering Review Conducted', 'Bridge Opportunity: Stage'])
```
### Look at target's distribution
```
df['Good Site'].value_counts()
```
So we have 65 labeled observations for the positive class, 24 labeled observations for the negative class, and almost 1,400 unlabeled observations.
### 4 recommendations:
- Use **semi-supervised learning**, which "combines a small amount of labeled data with a large amount of unlabeled data". See Wikipedia notes below. Python implementations are available in [scikit-learn](https://scikit-learn.org/stable/modules/label_propagation.html) and [pomegranate](https://pomegranate.readthedocs.io/en/latest/semisupervised.html). Another way to get started: feature engineering + feature selection + K-Means Clustering + PCA in 2 dimensions. Then visualize the clusters on a scatterplot, with colors for the labels.
- Use [**leave-one-out cross-validation**](https://en.wikipedia.org/wiki/Cross-validation_(statistics)#Leave-one-out_cross-validation), without an independent test set, because we have so few labeled observations. It's implemented in [scikit-learn](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.LeaveOneOut.html). Or maybe 10-fold cross-validation with stratified sampling (and no independent test set).
- Consider **"over-sampling"** techniques for imbalanced classification. Python implementations are available in [imbalanced-learn](https://github.com/scikit-learn-contrib/imbalanced-learn).
- Consider using [**Snorkel**](https://www.snorkel.org/) to write "labeling functions" for "weakly supervised learning." The site has many [tutorials](https://www.snorkel.org/use-cases/).
### [Semi-supervised learning - Wikipedia](https://en.wikipedia.org/wiki/Semi-supervised_learning)
> Semi-supervised learning is an approach to machine learning that combines a small amount of labeled data with a large amount of unlabeled data during training. Semi-supervised learning falls between unsupervised learning (with no labeled training data) and supervised learning (with only labeled training data).
> Unlabeled data, when used in conjunction with a small amount of labeled data, can produce considerable improvement in learning accuracy. The acquisition of labeled data for a learning problem often requires a skilled human agent ... The cost associated with the labeling process thus may render large, fully labeled training sets infeasible, whereas acquisition of unlabeled data is relatively inexpensive. In such situations, semi-supervised learning can be of great practical value.

> An example of the influence of unlabeled data in semi-supervised learning. The top panel shows a decision boundary we might adopt after seeing only one positive (white circle) and one negative (black circle) example. The bottom panel shows a decision boundary we might adopt if, in addition to the two labeled examples, we were given a collection of unlabeled data (gray circles). This could be viewed as performing clustering and then labeling the clusters with the labeled data, pushing the decision boundary away from high-density regions ...
See also:
- “Positive-Unlabeled Learning”
- https://en.wikipedia.org/wiki/One-class_classification
# Model attempt - Smaller Dataset Not utilizing world data
- Here I am attempting to build a model using less "created" data than we did in previous attempts using the world data set. This had more successful bridge builds but those bridge builds did not include pertainant feature for building a predictive model.
```
df.info()
# Columns suggeested bt stakeholder to utilize while model building
keep_list = ['Bridge Opportunity: Span (m)',
'Bridge Opportunity: Individuals Directly Served',
'Form: Created By',
'Height differential between banks', 'Flag for Rejection',
'Good Site']
# isolating the dataset to just the modelset
modelset = df[keep_list]
modelset.head()
# built modelset based off of original dataset - not much cleaning here.
# further cleaning could be an area for improvement.
modelset['Good Site'].value_counts()
!pip install category_encoders
# Imports:
from collections import Counter
from sklearn.pipeline import make_pipeline
from imblearn.pipeline import make_pipeline as make_pipeline_imb
from imblearn.over_sampling import SMOTE
from imblearn.metrics import classification_report_imbalanced
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.ensemble import RandomForestClassifier
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.metrics import precision_score, recall_score, f1_score, roc_auc_score, accuracy_score, classification_report
# split data - intial split eliminated all of the "unlabeled" sites
data = modelset[(modelset['Good Site']== 0) | (modelset['Good Site']== 1)]
test = modelset[modelset['Good Site']== -1]
train, val = train_test_split(data, test_size=.2, random_state=42)
# splitting our labeled sites into a train and validation set for model building
X_train = train.drop('Good Site', axis=1)
y_train = train['Good Site']
X_val = val.drop('Good Site', axis=1)
y_val = val['Good Site']
X_train.shape, y_train.shape, X_val.shape, y_val.shape
# Building a base model without SMOTE
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
kf = KFold(n_splits=5, shuffle=False)
base_pipe = make_pipeline(ce.OrdinalEncoder(),
SimpleImputer(strategy = 'mean'),
RandomForestClassifier(n_estimators=100, random_state=42))
cross_val_score(base_pipe, X_train, y_train, cv=kf, scoring='precision')
```
From the results above we can see the variety of the precision scores, looks like we have some overfit values when it comes to different cross validations
```
# use of imb_learn pipeline
imba_pipe = make_pipeline_imb(ce.OrdinalEncoder(),
SimpleImputer(strategy = 'mean'),
SMOTE(random_state=42),
RandomForestClassifier(n_estimators=100, random_state=42))
cross_val_score(imba_pipe, X_train, y_train, cv=kf, scoring='precision')
```
Using an imbalanced Pipeline with SMOTE we still see the large variety in precision 1.0 as a high and .625 as a low.
```
# using grid search to attempt to further validate the model to use on predicitions
new_params = {'randomforestclassifier__n_estimators': [100, 200, 50],
'randomforestclassifier__max_depth': [4, 6, 10, 12],
'simpleimputer__strategy': ['mean', 'median']
}
imba_grid_1 = GridSearchCV(imba_pipe, param_grid=new_params, cv=kf,
scoring='precision',
return_train_score=True)
imba_grid_1.fit(X_train, y_train);
# Params used and best score on a basis of precision
print(imba_grid_1.best_params_, imba_grid_1.best_score_)
# Working with more folds for validation
more_kf = KFold(n_splits=15)
imba_grid_2 = GridSearchCV(imba_pipe, param_grid=new_params, cv=more_kf,
scoring='precision',
return_train_score=True)
imba_grid_2.fit(X_train, y_train);
print(imba_grid_2.best_score_, imba_grid_2.best_estimator_)
imba_grid_2.cv_results_
# muted output because it was lenghty
# during output we did see a lot of 1s... Is this a sign of overfitting?
# Now looking to the val set to get some more numbers
y_val_predict = imba_grid_2.predict(X_val)
precision_score(y_val, y_val_predict)
```
The best score from above was .87, now running the model on the val set, it looks like we end with 92% precision score.
```
```
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Hub Authors.
Licensed under the Apache License, Version 2.0 (the "License");
```
# Copyright 2019 The TensorFlow Hub Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
#@title MIT License
#
# Copyright (c) 2017 François Chollet
#
# Permission is hereby granted, free of charge, to any person obtaining a
# copy of this software and associated documentation files (the "Software"),
# to deal in the Software without restriction, including without limitation
# the rights to use, copy, modify, merge, publish, distribute, sublicense,
# and/or sell copies of the Software, and to permit persons to whom the
# Software is furnished to do so, subject to the following conditions:
#
# The above copyright notice and this permission notice shall be included in
# all copies or substantial portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
# DEALINGS IN THE SOFTWARE.
```
# Text Classification with Movie Reviews
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/hub/tutorials/tf2_text_classification"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/hub/blob/master/examples/colab/tf2_text_classification.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/hub/blob/master/examples/colab/tf2_text_classification.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/hub/examples/colab/tf2_text_classification.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
<td>
<a href="https://tfhub.dev/google/collections/nnlm/1"><img src="https://www.tensorflow.org/images/hub_logo_32px.png" />See TF Hub models</a>
</td>
</table>
This notebook classifies movie reviews as *positive* or *negative* using the text of the review. This is an example of *binary*—or two-class—classification, an important and widely applicable kind of machine learning problem.
We'll use the [IMDB dataset](https://www.tensorflow.org/api_docs/python/tf/keras/datasets/imdb) that contains the text of 50,000 movie reviews from the [Internet Movie Database](https://www.imdb.com/). These are split into 25,000 reviews for training and 25,000 reviews for testing. The training and testing sets are *balanced*, meaning they contain an equal number of positive and negative reviews.
This notebook uses [tf.keras](https://www.tensorflow.org/guide/keras), a high-level API to build and train models in TensorFlow, and [TensorFlow Hub](https://www.tensorflow.org/hub), a library and platform for transfer learning. For a more advanced text classification tutorial using `tf.keras`, see the [MLCC Text Classification Guide](https://developers.google.com/machine-learning/guides/text-classification/).
### More models
[Here](https://tfhub.dev/s?module-type=text-embedding) you can find more expressive or performant models that you could use to generate the text embedding.
## Setup
```
import numpy as np
import tensorflow as tf
import tensorflow_hub as hub
import tensorflow_datasets as tfds
import matplotlib.pyplot as plt
print("Version: ", tf.__version__)
print("Eager mode: ", tf.executing_eagerly())
print("Hub version: ", hub.__version__)
print("GPU is", "available" if tf.config.list_physical_devices('GPU') else "NOT AVAILABLE")
```
## Download the IMDB dataset
The IMDB dataset is available on [TensorFlow datasets](https://github.com/tensorflow/datasets). The following code downloads the IMDB dataset to your machine (or the colab runtime):
```
train_data, test_data = tfds.load(name="imdb_reviews", split=["train", "test"],
batch_size=-1, as_supervised=True)
train_examples, train_labels = tfds.as_numpy(train_data)
test_examples, test_labels = tfds.as_numpy(test_data)
```
## Explore the data
Let's take a moment to understand the format of the data. Each example is a sentence representing the movie review and a corresponding label. The sentence is not preprocessed in any way. The label is an integer value of either 0 or 1, where 0 is a negative review, and 1 is a positive review.
```
print("Training entries: {}, test entries: {}".format(len(train_examples), len(test_examples)))
```
Let's print first 10 examples.
```
train_examples[:10]
```
Let's also print the first 10 labels.
```
train_labels[:10]
```
## Build the model
The neural network is created by stacking layers—this requires three main architectural decisions:
* How to represent the text?
* How many layers to use in the model?
* How many *hidden units* to use for each layer?
In this example, the input data consists of sentences. The labels to predict are either 0 or 1.
One way to represent the text is to convert sentences into embeddings vectors. We can use a pre-trained text embedding as the first layer, which will have two advantages:
* we don't have to worry about text preprocessing,
* we can benefit from transfer learning.
For this example we will use a model from [TensorFlow Hub](https://www.tensorflow.org/hub) called [google/nnlm-en-dim50/2](https://tfhub.dev/google/nnlm-en-dim50/2).
There are two other models to test for the sake of this tutorial:
* [google/nnlm-en-dim50-with-normalization/2](https://tfhub.dev/google/nnlm-en-dim50-with-normalization/2) - same as [google/nnlm-en-dim50/2](https://tfhub.dev/google/nnlm-en-dim50/2), but with additional text normalization to remove punctuation. This can help to get better coverage of in-vocabulary embeddings for tokens on your input text.
* [google/nnlm-en-dim128-with-normalization/2](https://tfhub.dev/google/nnlm-en-dim128-with-normalization/2) - A larger model with an embedding dimension of 128 instead of the smaller 50.
Let's first create a Keras layer that uses a TensorFlow Hub model to embed the sentences, and try it out on a couple of input examples. Note that the output shape of the produced embeddings is a expected: `(num_examples, embedding_dimension)`.
```
model = "https://tfhub.dev/google/nnlm-en-dim50/2"
hub_layer = hub.KerasLayer(model, input_shape=[], dtype=tf.string, trainable=True)
hub_layer(train_examples[:3])
```
Let's now build the full model:
```
model = tf.keras.Sequential()
model.add(hub_layer)
model.add(tf.keras.layers.Dense(16, activation='relu'))
model.add(tf.keras.layers.Dense(1))
model.summary()
```
The layers are stacked sequentially to build the classifier:
1. The first layer is a TensorFlow Hub layer. This layer uses a pre-trained Saved Model to map a sentence into its embedding vector. The model that we are using ([google/nnlm-en-dim50/2](https://tfhub.dev/google/nnlm-en-dim50/2)) splits the sentence into tokens, embeds each token and then combines the embedding. The resulting dimensions are: `(num_examples, embedding_dimension)`.
2. This fixed-length output vector is piped through a fully-connected (`Dense`) layer with 16 hidden units.
3. The last layer is densely connected with a single output node. This outputs logits: the log-odds of the true class, according to the model.
### Hidden units
The above model has two intermediate or "hidden" layers, between the input and output. The number of outputs (units, nodes, or neurons) is the dimension of the representational space for the layer. In other words, the amount of freedom the network is allowed when learning an internal representation.
If a model has more hidden units (a higher-dimensional representation space), and/or more layers, then the network can learn more complex representations. However, it makes the network more computationally expensive and may lead to learning unwanted patterns—patterns that improve performance on training data but not on the test data. This is called *overfitting*, and we'll explore it later.
### Loss function and optimizer
A model needs a loss function and an optimizer for training. Since this is a binary classification problem and the model outputs a probability (a single-unit layer with a sigmoid activation), we'll use the `binary_crossentropy` loss function.
This isn't the only choice for a loss function, you could, for instance, choose `mean_squared_error`. But, generally, `binary_crossentropy` is better for dealing with probabilities—it measures the "distance" between probability distributions, or in our case, between the ground-truth distribution and the predictions.
Later, when we are exploring regression problems (say, to predict the price of a house), we will see how to use another loss function called mean squared error.
Now, configure the model to use an optimizer and a loss function:
```
model.compile(optimizer='adam',
loss=tf.losses.BinaryCrossentropy(from_logits=True),
metrics=[tf.metrics.BinaryAccuracy(threshold=0.0, name='accuracy')])
```
## Create a validation set
When training, we want to check the accuracy of the model on data it hasn't seen before. Create a *validation set* by setting apart 10,000 examples from the original training data. (Why not use the testing set now? Our goal is to develop and tune our model using only the training data, then use the test data just once to evaluate our accuracy).
```
x_val = train_examples[:10000]
partial_x_train = train_examples[10000:]
y_val = train_labels[:10000]
partial_y_train = train_labels[10000:]
```
## Train the model
Train the model for 40 epochs in mini-batches of 512 samples. This is 40 iterations over all samples in the `x_train` and `y_train` tensors. While training, monitor the model's loss and accuracy on the 10,000 samples from the validation set:
```
history = model.fit(partial_x_train,
partial_y_train,
epochs=40,
batch_size=512,
validation_data=(x_val, y_val),
verbose=1)
```
## Evaluate the model
And let's see how the model performs. Two values will be returned. Loss (a number which represents our error, lower values are better), and accuracy.
```
results = model.evaluate(test_examples, test_labels)
print(results)
```
This fairly naive approach achieves an accuracy of about 87%. With more advanced approaches, the model should get closer to 95%.
## Create a graph of accuracy and loss over time
`model.fit()` returns a `History` object that contains a dictionary with everything that happened during training:
```
history_dict = history.history
history_dict.keys()
```
There are four entries: one for each monitored metric during training and validation. We can use these to plot the training and validation loss for comparison, as well as the training and validation accuracy:
```
acc = history_dict['accuracy']
val_acc = history_dict['val_accuracy']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
# "bo" is for "blue dot"
plt.plot(epochs, loss, 'bo', label='Training loss')
# b is for "solid blue line"
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.clf() # clear figure
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
```
In this plot, the dots represent the training loss and accuracy, and the solid lines are the validation loss and accuracy.
Notice the training loss *decreases* with each epoch and the training accuracy *increases* with each epoch. This is expected when using a gradient descent optimization—it should minimize the desired quantity on every iteration.
This isn't the case for the validation loss and accuracy—they seem to peak after about twenty epochs. This is an example of overfitting: the model performs better on the training data than it does on data it has never seen before. After this point, the model over-optimizes and learns representations *specific* to the training data that do not *generalize* to test data.
For this particular case, we could prevent overfitting by simply stopping the training after twenty or so epochs. Later, you'll see how to do this automatically with a callback.
|
github_jupyter
|
<a href="https://cognitiveclass.ai"><img src = "https://ibm.box.com/shared/static/9gegpsmnsoo25ikkbl4qzlvlyjbgxs5x.png" width = 400> </a>
<h1 align=center><font size = 5>Waffle Charts, Word Clouds, and Regression Plots</font></h1>
## Introduction
In this lab, we will learn how to create word clouds and waffle charts. Furthermore, we will start learning about additional visualization libraries that are based on Matplotlib, namely the library *seaborn*, and we will learn how to create regression plots using the *seaborn* library.
## Table of Contents
<div class="alert alert-block alert-info" style="margin-top: 20px">
1. [Exploring Datasets with *p*andas](#0)<br>
2. [Downloading and Prepping Data](#2)<br>
3. [Visualizing Data using Matplotlib](#4) <br>
4. [Waffle Charts](#6) <br>
5. [Word Clouds](#8) <br>
7. [Regression Plots](#10) <br>
</div>
<hr>
# Exploring Datasets with *pandas* and Matplotlib<a id="0"></a>
Toolkits: The course heavily relies on [*pandas*](http://pandas.pydata.org/) and [**Numpy**](http://www.numpy.org/) for data wrangling, analysis, and visualization. The primary plotting library we will explore in the course is [Matplotlib](http://matplotlib.org/).
Dataset: Immigration to Canada from 1980 to 2013 - [International migration flows to and from selected countries - The 2015 revision](http://www.un.org/en/development/desa/population/migration/data/empirical2/migrationflows.shtml) from United Nation's website
The dataset contains annual data on the flows of international migrants as recorded by the countries of destination. The data presents both inflows and outflows according to the place of birth, citizenship or place of previous / next residence both for foreigners and nationals. In this lab, we will focus on the Canadian Immigration data.
# Downloading and Prepping Data <a id="2"></a>
Import Primary Modules:
```
import numpy as np # useful for many scientific computing in Python
import pandas as pd # primary data structure library
from PIL import Image # converting images into arrays
```
Let's download and import our primary Canadian Immigration dataset using *pandas* `read_excel()` method. Normally, before we can do that, we would need to download a module which *pandas* requires to read in excel files. This module is **xlrd**. For your convenience, we have pre-installed this module, so you would not have to worry about that. Otherwise, you would need to run the following line of code to install the **xlrd** module:
```
!conda install -c anaconda xlrd --yes
```
Download the dataset and read it into a *pandas* dataframe:
```
df_can = pd.read_excel('https://ibm.box.com/shared/static/lw190pt9zpy5bd1ptyg2aw15awomz9pu.xlsx',
sheet_name='Canada by Citizenship',
skiprows=range(20),
skip_footer=2)
print('Data downloaded and read into a dataframe!')
```
Let's take a look at the first five items in our dataset
```
df_can.head()
```
Let's find out how many entries there are in our dataset
```
# print the dimensions of the dataframe
print(df_can.shape)
```
Clean up data. We will make some modifications to the original dataset to make it easier to create our visualizations. Refer to *Introduction to Matplotlib and Line Plots* and *Area Plots, Histograms, and Bar Plots* for a detailed description of this preprocessing.
```
# clean up the dataset to remove unnecessary columns (eg. REG)
df_can.drop(['AREA','REG','DEV','Type','Coverage'], axis = 1, inplace = True)
# let's rename the columns so that they make sense
df_can.rename (columns = {'OdName':'Country', 'AreaName':'Continent','RegName':'Region'}, inplace = True)
# for sake of consistency, let's also make all column labels of type string
df_can.columns = list(map(str, df_can.columns))
# set the country name as index - useful for quickly looking up countries using .loc method
df_can.set_index('Country', inplace = True)
# add total column
df_can['Total'] = df_can.sum (axis = 1)
# years that we will be using in this lesson - useful for plotting later on
years = list(map(str, range(1980, 2014)))
print ('data dimensions:', df_can.shape)
```
# Visualizing Data using Matplotlib<a id="4"></a>
Import `matplotlib`:
```
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches # needed for waffle Charts
mpl.style.use('ggplot') # optional: for ggplot-like style
# check for latest version of Matplotlib
print ('Matplotlib version: ', mpl.__version__) # >= 2.0.0
```
# Waffle Charts <a id="6"></a>
A `waffle chart` is an interesting visualization that is normally created to display progress toward goals. It is commonly an effective option when you are trying to add interesting visualization features to a visual that consists mainly of cells, such as an Excel dashboard.
Let's revisit the previous case study about Denmark, Norway, and Sweden.
```
# let's create a new dataframe for these three countries
df_dsn = df_can.loc[['Denmark', 'Norway', 'Sweden'], :]
# let's take a look at our dataframe
df_dsn
```
Unfortunately, unlike R, `waffle` charts are not built into any of the Python visualization libraries. Therefore, we will learn how to create them from scratch.
**Step 1.** The first step into creating a waffle chart is determing the proportion of each category with respect to the total.
```
# compute the proportion of each category with respect to the total
total_values = sum(df_dsn['Total'])
category_proportions = [(float(value) / total_values) for value in df_dsn['Total']]
# print out proportions
for i, proportion in enumerate(category_proportions):
print (df_dsn.index.values[i] + ': ' + str(proportion))
```
**Step 2.** The second step is defining the overall size of the `waffle` chart.
```
width = 40 # width of chart
height = 10 # height of chart
total_num_tiles = width * height # total number of tiles
print ('Total number of tiles is ', total_num_tiles)
```
**Step 3.** The third step is using the proportion of each category to determe it respective number of tiles
```
# compute the number of tiles for each catagory
tiles_per_category = [round(proportion * total_num_tiles) for proportion in category_proportions]
# print out number of tiles per category
for i, tiles in enumerate(tiles_per_category):
print (df_dsn.index.values[i] + ': ' + str(tiles))
```
Based on the calculated proportions, Denmark will occupy 129 tiles of the `waffle` chart, Norway will occupy 77 tiles, and Sweden will occupy 194 tiles.
**Step 4.** The fourth step is creating a matrix that resembles the `waffle` chart and populating it.
```
# initialize the waffle chart as an empty matrix
waffle_chart = np.zeros((height, width))
# define indices to loop through waffle chart
category_index = 0
tile_index = 0
# populate the waffle chart
for col in range(width):
for row in range(height):
tile_index += 1
# if the number of tiles populated for the current category is equal to its corresponding allocated tiles...
if tile_index > sum(tiles_per_category[0:category_index]):
# ...proceed to the next category
category_index += 1
# set the class value to an integer, which increases with class
waffle_chart[row, col] = category_index
print ('Waffle chart populated!')
```
Let's take a peek at how the matrix looks like.
```
waffle_chart
```
As expected, the matrix consists of three categories and the total number of each category's instances matches the total number of tiles allocated to each category.
**Step 5.** Map the `waffle` chart matrix into a visual.
```
# instantiate a new figure object
fig = plt.figure()
# use matshow to display the waffle chart
colormap = plt.cm.coolwarm
plt.matshow(waffle_chart, cmap=colormap)
plt.colorbar()
```
**Step 6.** Prettify the chart.
```
# instantiate a new figure object
fig = plt.figure()
# use matshow to display the waffle chart
colormap = plt.cm.coolwarm
plt.matshow(waffle_chart, cmap=colormap)
plt.colorbar()
# get the axis
ax = plt.gca()
# set minor ticks
ax.set_xticks(np.arange(-.5, (width), 1), minor=True)
ax.set_yticks(np.arange(-.5, (height), 1), minor=True)
# add gridlines based on minor ticks
ax.grid(which='minor', color='w', linestyle='-', linewidth=2)
plt.xticks([])
plt.yticks([])
```
**Step 7.** Create a legend and add it to chart.
```
# instantiate a new figure object
fig = plt.figure()
# use matshow to display the waffle chart
colormap = plt.cm.coolwarm
plt.matshow(waffle_chart, cmap=colormap)
plt.colorbar()
# get the axis
ax = plt.gca()
# set minor ticks
ax.set_xticks(np.arange(-.5, (width), 1), minor=True)
ax.set_yticks(np.arange(-.5, (height), 1), minor=True)
# add gridlines based on minor ticks
ax.grid(which='minor', color='w', linestyle='-', linewidth=2)
plt.xticks([])
plt.yticks([])
# compute cumulative sum of individual categories to match color schemes between chart and legend
values_cumsum = np.cumsum(df_dsn['Total'])
total_values = values_cumsum[len(values_cumsum) - 1]
# create legend
legend_handles = []
for i, category in enumerate(df_dsn.index.values):
label_str = category + ' (' + str(df_dsn['Total'][i]) + ')'
color_val = colormap(float(values_cumsum[i])/total_values)
legend_handles.append(mpatches.Patch(color=color_val, label=label_str))
# add legend to chart
plt.legend(handles=legend_handles,
loc='lower center',
ncol=len(df_dsn.index.values),
bbox_to_anchor=(0., -0.2, 0.95, .1)
)
```
And there you go! What a good looking *delicious* `waffle` chart, don't you think?
Now it would very inefficient to repeat these seven steps every time we wish to create a `waffle` chart. So let's combine all seven steps into one function called *create_waffle_chart*. This function would take the following parameters as input:
> 1. **categories**: Unique categories or classes in dataframe.
> 2. **values**: Values corresponding to categories or classes.
> 3. **height**: Defined height of waffle chart.
> 4. **width**: Defined width of waffle chart.
> 5. **colormap**: Colormap class
> 6. **value_sign**: In order to make our function more generalizable, we will add this parameter to address signs that could be associated with a value such as %, $, and so on. **value_sign** has a default value of empty string.
```
def create_waffle_chart(categories, values, height, width, colormap, value_sign=''):
# compute the proportion of each category with respect to the total
total_values = sum(values)
category_proportions = [(float(value) / total_values) for value in values]
# compute the total number of tiles
total_num_tiles = width * height # total number of tiles
print ('Total number of tiles is', total_num_tiles)
# compute the number of tiles for each catagory
tiles_per_category = [round(proportion * total_num_tiles) for proportion in category_proportions]
# print out number of tiles per category
for i, tiles in enumerate(tiles_per_category):
print (df_dsn.index.values[i] + ': ' + str(tiles))
# initialize the waffle chart as an empty matrix
waffle_chart = np.zeros((height, width))
# define indices to loop through waffle chart
category_index = 0
tile_index = 0
# populate the waffle chart
for col in range(width):
for row in range(height):
tile_index += 1
# if the number of tiles populated for the current category
# is equal to its corresponding allocated tiles...
if tile_index > sum(tiles_per_category[0:category_index]):
# ...proceed to the next category
category_index += 1
# set the class value to an integer, which increases with class
waffle_chart[row, col] = category_index
# instantiate a new figure object
fig = plt.figure()
# use matshow to display the waffle chart
colormap = plt.cm.coolwarm
plt.matshow(waffle_chart, cmap=colormap)
plt.colorbar()
# get the axis
ax = plt.gca()
# set minor ticks
ax.set_xticks(np.arange(-.5, (width), 1), minor=True)
ax.set_yticks(np.arange(-.5, (height), 1), minor=True)
# add dridlines based on minor ticks
ax.grid(which='minor', color='w', linestyle='-', linewidth=2)
plt.xticks([])
plt.yticks([])
# compute cumulative sum of individual categories to match color schemes between chart and legend
values_cumsum = np.cumsum(values)
total_values = values_cumsum[len(values_cumsum) - 1]
# create legend
legend_handles = []
for i, category in enumerate(categories):
if value_sign == '%':
label_str = category + ' (' + str(values[i]) + value_sign + ')'
else:
label_str = category + ' (' + value_sign + str(values[i]) + ')'
color_val = colormap(float(values_cumsum[i])/total_values)
legend_handles.append(mpatches.Patch(color=color_val, label=label_str))
# add legend to chart
plt.legend(
handles=legend_handles,
loc='lower center',
ncol=len(categories),
bbox_to_anchor=(0., -0.2, 0.95, .1)
)
```
Now to create a `waffle` chart, all we have to do is call the function `create_waffle_chart`. Let's define the input parameters:
```
width = 40 # width of chart
height = 10 # height of chart
categories = df_dsn.index.values # categories
values = df_dsn['Total'] # correponding values of categories
colormap = plt.cm.coolwarm # color map class
```
And now let's call our function to create a `waffle` chart.
```
create_waffle_chart(categories, values, height, width, colormap)
```
There seems to be a new Python package for generating `waffle charts` called [PyWaffle](https://github.com/ligyxy/PyWaffle), but the repository has barely any documentation on the package. Accordingly, I couldn't use the package to prepare enough content to incorporate into this lab. But feel free to check it out and play with it. In the event that the package becomes complete with full documentation, then I will update this lab accordingly.
# Word Clouds <a id="8"></a>
`Word` clouds (also known as text clouds or tag clouds) work in a simple way: the more a specific word appears in a source of textual data (such as a speech, blog post, or database), the bigger and bolder it appears in the word cloud.
Luckily, a Python package already exists in Python for generating `word` clouds. The package, called `word_cloud` was developed by **Andreas Mueller**. You can learn more about the package by following this [link](https://github.com/amueller/word_cloud/).
Let's use this package to learn how to generate a word cloud for a given text document.
First, let's install the package.
```
# install wordcloud
!conda install -c conda-forge wordcloud==1.4.1 --yes
# import package and its set of stopwords
from wordcloud import WordCloud, STOPWORDS
print ('Wordcloud is installed and imported!')
```
`Word` clouds are commonly used to perform high-level analysis and visualization of text data. Accordinly, let's digress from the immigration dataset and work with an example that involves analyzing text data. Let's try to analyze a short novel written by **Lewis Carroll** titled *Alice's Adventures in Wonderland*. Let's go ahead and download a _.txt_ file of the novel.
```
# download file and save as alice_novel.txt
!wget --quiet https://ibm.box.com/shared/static/m54sjtrshpt5su20dzesl5en9xa5vfz1.txt -O alice_novel.txt
# open the file and read it into a variable alice_novel
alice_novel = open('alice_novel.txt', 'r').read()
print ('File downloaded and saved!')
```
Next, let's use the stopwords that we imported from `word_cloud`. We use the function *set* to remove any redundant stopwords.
```
stopwords = set(STOPWORDS)
```
Create a word cloud object and generate a word cloud. For simplicity, let's generate a word cloud using only the first 2000 words in the novel.
```
# instantiate a word cloud object
alice_wc = WordCloud(
background_color='white',
max_words=2000,
stopwords=stopwords
)
# generate the word cloud
alice_wc.generate(alice_novel)
```
Awesome! Now that the `word` cloud is created, let's visualize it.
```
# display the word cloud
plt.imshow(alice_wc, interpolation='bilinear')
plt.axis('off')
plt.show()
```
Interesting! So in the first 2000 words in the novel, the most common words are **Alice**, **said**, **little**, **Queen**, and so on. Let's resize the cloud so that we can see the less frequent words a little better.
```
fig = plt.figure()
fig.set_figwidth(14) # set width
fig.set_figheight(18) # set height
# display the cloud
plt.imshow(alice_wc, interpolation='bilinear')
plt.axis('off')
plt.show()
```
Much better! However, **said** isn't really an informative word. So let's add it to our stopwords and re-generate the cloud.
```
stopwords.add('said') # add the words said to stopwords
# re-generate the word cloud
alice_wc.generate(alice_novel)
# display the cloud
fig = plt.figure()
fig.set_figwidth(14) # set width
fig.set_figheight(18) # set height
plt.imshow(alice_wc, interpolation='bilinear')
plt.axis('off')
plt.show()
```
Excellent! This looks really interesting! Another cool thing you can implement with the `word_cloud` package is superimposing the words onto a mask of any shape. Let's use a mask of Alice and her rabbit. We already created the mask for you, so let's go ahead and download it and call it *alice_mask.png*.
```
# download image
!wget --quiet https://ibm.box.com/shared/static/3mpxgaf6muer6af7t1nvqkw9cqj85ibm.png -O alice_mask.png
# save mask to alice_mask
alice_mask = np.array(Image.open('alice_mask.png'))
print('Image downloaded and saved!')
```
Let's take a look at how the mask looks like.
```
fig = plt.figure()
fig.set_figwidth(14) # set width
fig.set_figheight(18) # set height
plt.imshow(alice_mask, cmap=plt.cm.gray, interpolation='bilinear')
plt.axis('off')
plt.show()
```
Shaping the `word` cloud according to the mask is straightforward using `word_cloud` package. For simplicity, we will continue using the first 2000 words in the novel.
```
# instantiate a word cloud object
alice_wc = WordCloud(background_color='white', max_words=2000, mask=alice_mask, stopwords=stopwords)
# generate the word cloud
alice_wc.generate(alice_novel)
# display the word cloud
fig = plt.figure()
fig.set_figwidth(14) # set width
fig.set_figheight(18) # set height
plt.imshow(alice_wc, interpolation='bilinear')
plt.axis('off')
plt.show()
```
Really impressive!
Unfortunately, our immmigration data does not have any text data, but where there is a will there is a way. Let's generate sample text data from our immigration dataset, say text data of 90 words.
Let's recall how our data looks like.
```
df_can.head()
```
And what was the total immigration from 1980 to 2013?
```
total_immigration = df_can['Total'].sum()
total_immigration
```
Using countries with single-word names, let's duplicate each country's name based on how much they contribute to the total immigration.
```
max_words = 90
word_string = ''
for country in df_can.index.values:
# check if country's name is a single-word name
if len(country.split(' ')) == 1:
repeat_num_times = int(df_can.loc[country, 'Total']/float(total_immigration)*max_words)
word_string = word_string + ((country + ' ') * repeat_num_times)
# display the generated text
word_string
```
We are not dealing with any stopwords here, so there is no need to pass them when creating the word cloud.
```
# create the word cloud
wordcloud = WordCloud(background_color='white').generate(word_string)
print('Word cloud created!')
# display the cloud
fig = plt.figure()
fig.set_figwidth(14)
fig.set_figheight(18)
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis('off')
plt.show()
```
According to the above word cloud, it looks like the majority of the people who immigrated came from one of 15 countries that are displayed by the word cloud. One cool visual that you could build, is perhaps using the map of Canada and a mask and superimposing the word cloud on top of the map of Canada. That would be an interesting visual to build!
# Regression Plots <a id="10"></a>
> Seaborn is a Python visualization library based on matplotlib. It provides a high-level interface for drawing attractive statistical graphics. You can learn more about *seaborn* by following this [link](https://seaborn.pydata.org/) and more about *seaborn* regression plots by following this [link](http://seaborn.pydata.org/generated/seaborn.regplot.html).
In lab *Pie Charts, Box Plots, Scatter Plots, and Bubble Plots*, we learned how to create a scatter plot and then fit a regression line. It took ~20 lines of code to create the scatter plot along with the regression fit. In this final section, we will explore *seaborn* and see how efficient it is to create regression lines and fits using this library!
Let's first install *seaborn*
```
# install seaborn
!pip install seaborn
# import library
import seaborn as sns
print('Seaborn installed and imported!')
```
Create a new dataframe that stores that total number of landed immigrants to Canada per year from 1980 to 2013.
```
# we can use the sum() method to get the total population per year
df_tot = pd.DataFrame(df_can[years].sum(axis=0))
# change the years to type float (useful for regression later on)
df_tot.index = map(float,df_tot.index)
# reset the index to put in back in as a column in the df_tot dataframe
df_tot.reset_index(inplace = True)
# rename columns
df_tot.columns = ['year', 'total']
# view the final dataframe
df_tot.head()
```
With *seaborn*, generating a regression plot is as simple as calling the **regplot** function.
```
import seaborn as sns
ax = sns.regplot(x='year', y='total', data=df_tot)
```
This is not magic; it is *seaborn*! You can also customize the color of the scatter plot and regression line. Let's change the color to green.
```
import seaborn as sns
ax = sns.regplot(x='year', y='total', data=df_tot, color='green')
```
You can always customize the marker shape, so instead of circular markers, let's use '+'.
```
import seaborn as sns
ax = sns.regplot(x='year', y='total', data=df_tot, color='green', marker='+')
```
Let's blow up the plot a little bit so that it is more appealing to the sight.
```
plt.figure(figsize=(15, 10))
ax = sns.regplot(x='year', y='total', data=df_tot, color='green', marker='+')
```
And let's increase the size of markers so they match the new size of the figure, and add a title and x- and y-labels.
```
plt.figure(figsize=(15, 10))
ax = sns.regplot(x='year', y='total', data=df_tot, color='green', marker='+', scatter_kws={'s': 200})
ax.set(xlabel='Year', ylabel='Total Immigration') # add x- and y-labels
ax.set_title('Total Immigration to Canada from 1980 - 2013') # add title
```
And finally increase the font size of the tickmark labels, the title, and the x- and y-labels so they don't feel left out!
```
plt.figure(figsize=(15, 10))
sns.set(font_scale=1.5)
ax = sns.regplot(x='year', y='total', data=df_tot, color='green', marker='+', scatter_kws={'s': 200})
ax.set(xlabel='Year', ylabel='Total Immigration')
ax.set_title('Total Immigration to Canada from 1980 - 2013')
```
Amazing! A complete scatter plot with a regression fit with 5 lines of code only. Isn't this really amazing?
If you are not a big fan of the purple background, you can easily change the style to a white plain background.
```
plt.figure(figsize=(15, 10))
sns.set(font_scale=1.5)
sns.set_style('ticks') # change background to white background
ax = sns.regplot(x='year', y='total', data=df_tot, color='green', marker='+', scatter_kws={'s': 200})
ax.set(xlabel='Year', ylabel='Total Immigration')
ax.set_title('Total Immigration to Canada from 1980 - 2013')
```
Or to a white background with gridlines.
```
plt.figure(figsize=(15, 10))
sns.set(font_scale=1.5)
sns.set_style('whitegrid')
ax = sns.regplot(x='year', y='total', data=df_tot, color='green', marker='+', scatter_kws={'s': 200})
ax.set(xlabel='Year', ylabel='Total Immigration')
ax.set_title('Total Immigration to Canada from 1980 - 2013')
```
**Question**: Use seaborn to create a scatter plot with a regression line to visualize the total immigration from Denmark, Sweden, and Norway to Canada from 1980 to 2013.
```
### type your answer here
```
Double-click __here__ for the solution.
<!-- The correct answer is:
\\ # create df_countries dataframe
df_countries = df_can.loc[['Denmark', 'Norway', 'Sweden'], years].transpose()
-->
<!--
\\ # create df_total by summing across three countries for each year
df_total = pd.DataFrame(df_countries.sum(axis=1))
-->
<!--
\\ # reset index in place
df_total.reset_index(inplace=True)
-->
<!--
\\ # rename columns
df_total.columns = ['year', 'total']
-->
<!--
\\ # change column year from string to int to create scatter plot
df_total['year'] = df_total['year'].astype(int)
-->
<!--
\\ # define figure size
plt.figure(figsize=(15, 10))
-->
<!--
\\ # define background style and font size
sns.set(font_scale=1.5)
sns.set_style('whitegrid')
-->
<!--
\\ # generate plot and add title and axes labels
ax = sns.regplot(x='year', y='total', data=df_total, color='green', marker='+', scatter_kws={'s': 200})
ax.set(xlabel='Year', ylabel='Total Immigration')
ax.set_title('Total Immigrationn from Denmark, Sweden, and Norway to Canada from 1980 - 2013')
-->
### Thank you for completing this lab!
This notebook was created by [Alex Aklson](https://www.linkedin.com/in/aklson/). I hope you found this lab interesting and educational. Feel free to contact me if you have any questions!
This notebook is part of a course on **Coursera** called *Data Visualization with Python*. If you accessed this notebook outside the course, you can take this course online by clicking [here](http://cocl.us/DV0101EN_Coursera_Week3_LAB1).
<hr>
Copyright © 2018 [Cognitive Class](https://cognitiveclass.ai/?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).
|
github_jupyter
|
<h2>Factorization Machines - Movie Recommendation Model</h2>
Input Features: [userId, moveId] <br>
Target: rating <br>
```
import numpy as np
import pandas as pd
# Define IAM role
import boto3
import re
import sagemaker
from sagemaker import get_execution_role
# SageMaker SDK Documentation: http://sagemaker.readthedocs.io/en/latest/estimators.html
```
## Upload Data to S3
```
# Specify your bucket name
bucket_name = 'chandra-ml-sagemaker'
training_file_key = 'movie/user_movie_train.recordio'
test_file_key = 'movie/user_movie_test.recordio'
s3_model_output_location = r's3://{0}/movie/model'.format(bucket_name)
s3_training_file_location = r's3://{0}/{1}'.format(bucket_name,training_file_key)
s3_test_file_location = r's3://{0}/{1}'.format(bucket_name,test_file_key)
# Read Dimension: Number of unique users + Number of unique movies in our dataset
dim_movie = 0
# Update movie dimension - from file used for training
with open(r'ml-latest-small/movie_dimension.txt','r') as f:
dim_movie = int(f.read())
dim_movie
print(s3_model_output_location)
print(s3_training_file_location)
print(s3_test_file_location)
# Write and Reading from S3 is just as easy
# files are referred as objects in S3.
# file name is referred as key name in S3
# Files stored in S3 are automatically replicated across 3 different availability zones
# in the region where the bucket was created.
# http://boto3.readthedocs.io/en/latest/guide/s3.html
def write_to_s3(filename, bucket, key):
with open(filename,'rb') as f: # Read in binary mode
return boto3.Session().resource('s3').Bucket(bucket).Object(key).upload_fileobj(f)
write_to_s3(r'ml-latest-small/user_movie_train.recordio',bucket_name,training_file_key)
write_to_s3(r'ml-latest-small/user_movie_test.recordio',bucket_name,test_file_key)
```
## Training Algorithm Docker Image
### AWS Maintains a separate image for every region and algorithm
```
sess = sagemaker.Session()
role = get_execution_role()
# This role contains the permissions needed to train, deploy models
# SageMaker Service is trusted to assume this role
print(role)
# https://sagemaker.readthedocs.io/en/stable/api/utility/image_uris.html#sagemaker.image_uris.retrieve
# SDK 2 uses image_uris.retrieve the container image location
# Use factorization-machines
container = sagemaker.image_uris.retrieve("factorization-machines",sess.boto_region_name)
print (f'Using FM Container {container}')
container
```
## Build Model
```
# Configure the training job
# Specify type and number of instances to use
# S3 location where final artifacts needs to be stored
# Reference: http://sagemaker.readthedocs.io/en/latest/estimators.html
# SDK 2.x version does not require train prefix for instance count and type
estimator = sagemaker.estimator.Estimator(container,
role,
instance_count=1,
instance_type='ml.m4.xlarge',
output_path=s3_model_output_location,
sagemaker_session=sess,
base_job_name ='fm-movie-v4')
```
### New Configuration after Model Tuning
### Refer to Hyperparameter Tuning Lecture on how to optimize hyperparameters
```
estimator.set_hyperparameters(feature_dim=dim_movie,
num_factors=8,
predictor_type='regressor',
mini_batch_size=994,
epochs=91,
bias_init_method='normal',
bias_lr=0.21899531189430518,
factors_init_method='normal',
factors_lr=5.357593337770278e-05,
linear_init_method='normal',
linear_lr=0.00021524948053767607)
estimator.hyperparameters()
```
### Train the model
```
# New Hyperparameters
# Reference: Supported channels by algorithm
# https://docs.aws.amazon.com/sagemaker/latest/dg/sagemaker-algo-docker-registry-paths.html
estimator.fit({'train':s3_training_file_location, 'test': s3_test_file_location})
```
## Deploy Model
```
# Ref: http://sagemaker.readthedocs.io/en/latest/estimators.html
predictor = estimator.deploy(initial_instance_count=1,
instance_type='ml.m4.xlarge',
endpoint_name = 'fm-movie-v4')
```
## Run Predictions
### Dense and Sparse Formats
https://docs.aws.amazon.com/sagemaker/latest/dg/cdf-inference.html
```
import json
def fm_sparse_serializer(data):
js = {'instances': []}
for row in data:
column_list = row.tolist()
value_list = np.ones(len(column_list),dtype=int).tolist()
js['instances'].append({'data':{'features': { 'keys': column_list, 'shape':[dim_movie], 'values': value_list}}})
return json.dumps(js)
# SDK 2
from sagemaker.deserializers import JSONDeserializer
# https://github.com/aws/amazon-sagemaker-examples/blob/master/introduction_to_amazon_algorithms/factorization_machines_mnist/factorization_machines_mnist.ipynb
# Specify custom serializer
predictor.serializer.serialize = fm_sparse_serializer
predictor.serializer.content_type = 'application/json'
predictor.deserializer = JSONDeserializer()
import numpy as np
fm_sparse_serializer([np.array([341,1416])])
# Let's test with few entries from test file
# Movie dataset is updated regularly...so, instead of hard coding userid and movie id, let's
# use actual values
# Each row is in this format: ['2.5', '426:1', '943:1']
# ActualRating, UserID, MovieID
with open(r'ml-latest-small/user_movie_test.svm','r') as f:
for i in range(3):
rating = f.readline().split()
print(f"Movie {rating}")
userID = rating[1].split(':')[0]
movieID = rating[2].split(':')[0]
predicted_rating = predictor.predict([np.array([int(userID),int(movieID)])])
print(f' Actual Rating:\t{rating[0]}')
print(f" Predicted Rating:\t{predicted_rating['predictions'][0]['score']}")
print()
```
## Summary
1. Ensure Training, Test and Validation data are in S3 Bucket
2. Select Algorithm Container Registry Path - Path varies by region
3. Configure Estimator for training - Specify Algorithm container, instance count, instance type, model output location
4. Specify algorithm specific hyper parameters
5. Train model
6. Deploy model - Specify instance count, instance type and endpoint name
7. Run Predictions
|
github_jupyter
|
# The overview of the basic approaches to solving the Uplift Modeling problem
<br>
<center>
<a href="https://colab.research.google.com/github/maks-sh/scikit-uplift/blob/master/notebooks/RetailHero_EN.ipynb">
<img src="https://colab.research.google.com/assets/colab-badge.svg">
</a>
<br>
<b><a href="https://github.com/maks-sh/scikit-uplift/">SCIKIT-UPLIFT REPO</a> | </b>
<b><a href="https://scikit-uplift.readthedocs.io/en/latest/">SCIKIT-UPLIFT DOCS</a> | </b>
<b><a href="https://scikit-uplift.readthedocs.io/en/latest/user_guide/index.html">USER GUIDE</a></b>
<br>
<b><a href="https://nbviewer.jupyter.org/github/maks-sh/scikit-uplift/blob/master/notebooks/RetailHero.ipynb">RUSSIAN VERSION</a></b>
</center>
## Content
* [Introduction](#Introduction)
* [1. Single model approaches](#1.-Single-model-approaches)
* [1.1 Single model](#1.1-Single-model-with-treatment-as-feature)
* [1.2 Class Transformation](#1.2-Class-Transformation)
* [2. Approaches with two models](#2.-Approaches-with-two-models)
* [2.1 Two independent models](#2.1-Two-independent-models)
* [2.2 Two dependent models](#2.2-Two-dependent-models)
* [Conclusion](#Conclusion)
## Introduction
Before proceeding to the discussion of uplift modeling, let's imagine some situation:
A customer comes to you with a certain problem: it is necessary to advertise a popular product using the sms.
You know that the product is quite popular, and it is often installed by the customers without communication, that the usual binary classification will find the same customers, and the cost of communication is critical for us...
And then you begin to understand that the product is already popular, that the product is often installed by customers without communication, that the usual binary classification will find many such customers, and the cost of communication is critical for us...
Historically, according to the impact of communication, marketers divide all customers into 4 categories:
<p align="center">
<img src="https://raw.githubusercontent.com/maks-sh/scikit-uplift/master/docs/_static/images/user_guide/ug_clients_types.jpg" alt="Customer types" width='40%'/>
</p>
- **`Do-Not-Disturbs`** *(a.k.a. Sleeping-dogs)* have a strong negative response to a marketing communication. They are going to purchase if *NOT* treated and will *NOT* purchase *IF* treated. It is not only a wasted marketing budget but also a negative impact. For instance, customers targeted could result in rejecting current products or services. In terms of math: $W_i = 1, Y_i = 0$ or $W_i = 0, Y_i = 1$.
- **`Lost Causes`** will *NOT* purchase the product *NO MATTER* they are contacted or not. The marketing budget in this case is also wasted because it has no effect. In terms of math: $W_i = 1, Y_i = 0$ or $W_i = 0, Y_i = 0$.
- **`Sure Things`** will purchase *ANYWAY* no matter they are contacted or not. There is no motivation to spend the budget because it also has no effect. In terms of math: $W_i = 1, Y_i = 1$ or $W_i = 0, Y_i = 1$.
- **`Persuadables`** will always respond *POSITIVE* to the marketing communication. They is going to purchase *ONLY* if contacted (or sometimes they purchase *MORE* or *EARLIER* only if contacted). This customer's type should be the only target for the marketing campaign. In terms of math: $W_i = 0, Y_i = 0$ or $W_i = 1, Y_i = 1$.
Because we can't communicate and not communicate with the customer at the same time, we will never be able to observe exactly which type a particular customer belongs to.
Depends on the product characteristics and the customer base structure some types may be absent. In addition, a customer response depends heavily on various characteristics of the campaign, such as a communication channel or a type and a size of the marketing offer. To maximize profit, these parameters should be selected.
Thus, when predicting uplift score and selecting a segment by the highest score, we are trying to find the only one type: **persuadables**.
Thus, in this task, we don’t want to predict the probability of performing a target action, but to focus the advertising budget on the customers who will perform the target action only when we interact. In other words, we want to evaluate two conditional probabilities separately for each client:
* Performing a targeted action when we influence the client.
We will refer such clients to the **test group (aka treatment)**: $P^T = P(Y=1 | W = 1)$,
* Performing a targeted action without affecting the client.
We will refer such clients to the **control group (aka control)**: $P^C = P(Y=1 | W = 0)$,
where $Y$ is the binary flag for executing the target action, and $W$ is the binary flag for communication (in English literature, _treatment_)
The very same cause-and-effect effect is called **uplift** and is estimated as the difference between these two probabilities:
$$ uplift = P^T - P^C = P(Y = 1 | W = 1) - P(Y = 1 | W = 0) $$
Predicting uplift is a cause-and-effect inference task. The point is that you need to evaluate the difference between two events that are mutually exclusive for a particular client (either we interact with a person, or not; you can't perform two of these actions at the same time). This is why additional requirements for source data are required for building uplift models.
To get a training sample for the uplift simulation, you need to conduct an experiment:
1. Randomly split a representative part of the client base into a test and control group
2. Communicate with the test group
The data obtained as part of the design of such a pilot will allow us to build an uplift forecasting model in the future. It is also worth noting that the experiment should be as similar as possible to the campaign, which will be launched later on a larger scale. The only difference between the experiment and the campaign should be the fact that during the pilot, we choose random clients for interaction, and during the campaign - based on the predicted value of the Uplift. If the campaign that is eventually launched differs significantly from the experiment that is used to collect data about the performance of targeted actions by clients, then the model that is built may be less reliable and accurate.
So, the approaches to predicting uplift are aimed at assessing the net effect of marketing campaigns on customers.
All classical approaches to uplift modeling can be divided into two classes:
1. Approaches with the same model
2. Approaches using two models
Let's download [RetailHero.ai contest data](https://ods.ai/competitions/x5-retailhero-uplift-modeling/data):
```
import sys
# install uplift library scikit-uplift and other libraries
!{sys.executable} -m pip install scikit-uplift catboost pandas
from sklearn.model_selection import train_test_split
from sklift.datasets import fetch_x5
import pandas as pd
pd.set_option('display.max_columns', None)
%matplotlib inline
dataset = fetch_x5()
dataset.keys()
print(f"Dataset type: {type(dataset)}\n")
print(f"Dataset features shape: {dataset.data['clients'].shape}")
print(f"Dataset features shape: {dataset.data['train'].shape}")
print(f"Dataset target shape: {dataset.target.shape}")
print(f"Dataset treatment shape: {dataset.treatment.shape}")
```
Read more about dataset <a href="https://www.uplift-modeling.com/en/latest/api/datasets/fetch_x5.html">in the api docs</a>.
Now let's preprocess it a bit:
```
# extract data
df_clients = dataset.data['clients'].set_index("client_id")
df_train = pd.concat([dataset.data['train'], dataset.treatment , dataset.target], axis=1).set_index("client_id")
indices_test = pd.Index(set(df_clients.index) - set(df_train.index))
# extracting features
df_features = df_clients.copy()
df_features['first_issue_time'] = \
(pd.to_datetime(df_features['first_issue_date'])
- pd.Timestamp('1970-01-01')) // pd.Timedelta('1s')
df_features['first_redeem_time'] = \
(pd.to_datetime(df_features['first_redeem_date'])
- pd.Timestamp('1970-01-01')) // pd.Timedelta('1s')
df_features['issue_redeem_delay'] = df_features['first_redeem_time'] \
- df_features['first_issue_time']
df_features = df_features.drop(['first_issue_date', 'first_redeem_date'], axis=1)
indices_learn, indices_valid = train_test_split(df_train.index, test_size=0.3, random_state=123)
```
For convenience, we will declare some variables:
```
X_train = df_features.loc[indices_learn, :]
y_train = df_train.loc[indices_learn, 'target']
treat_train = df_train.loc[indices_learn, 'treatment_flg']
X_val = df_features.loc[indices_valid, :]
y_val = df_train.loc[indices_valid, 'target']
treat_val = df_train.loc[indices_valid, 'treatment_flg']
X_train_full = df_features.loc[df_train.index, :]
y_train_full = df_train.loc[:, 'target']
treat_train_full = df_train.loc[:, 'treatment_flg']
X_test = df_features.loc[indices_test, :]
cat_features = ['gender']
models_results = {
'approach': [],
'uplift@30%': []
}
```
## 1. Single model approaches
### 1.1 Single model with treatment as feature
The most intuitive and simple uplift modeling technique. A training set consists of two groups: treatment samples and control samples. There is also a binary treatment flag added as a feature to the training set. After the model is trained, at the scoring time it is going to be applied twice:
with the treatment flag equals `1` and with the treatment flag equals `0`. Subtracting these model's outcomes for each test sample, we will get an estimate of the uplift.
<p align="center">
<img src="https://raw.githubusercontent.com/maks-sh/scikit-uplift/master/docs/_static/images/SoloModel.png" alt="Solo model with treatment as a feature"/>
</p>
```
# installation instructions: https://github.com/maks-sh/scikit-uplift
# link to the documentation: https://scikit-uplift.readthedocs.io/en/latest/
from sklift.metrics import uplift_at_k
from sklift.viz import plot_uplift_preds
from sklift.models import SoloModel
# sklift supports all models,
# that satisfy scikit-learn convention
# for example, let's use catboost
from catboost import CatBoostClassifier
sm = SoloModel(CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True))
sm = sm.fit(X_train, y_train, treat_train, estimator_fit_params={'cat_features': cat_features})
uplift_sm = sm.predict(X_val)
sm_score = uplift_at_k(y_true=y_val, uplift=uplift_sm, treatment=treat_val, strategy='by_group', k=0.3)
models_results['approach'].append('SoloModel')
models_results['uplift@30%'].append(sm_score)
# get conditional probabilities (predictions) of performing the target action
# during interaction for each object
sm_trmnt_preds = sm.trmnt_preds_
# And conditional probabilities (predictions) of performing the target action
# without interaction for each object
sm_ctrl_preds = sm.ctrl_preds_
# draw the probability (predictions) distributions and their difference (uplift)
plot_uplift_preds(trmnt_preds=sm_trmnt_preds, ctrl_preds=sm_ctrl_preds);
# You can also access the trained model with the same ease.
# For example, to build the importance of features:
sm_fi = pd.DataFrame({
'feature_name': sm.estimator.feature_names_,
'feature_score': sm.estimator.feature_importances_
}).sort_values('feature_score', ascending=False).reset_index(drop=True)
sm_fi
```
### 1.2 Class Transformation
Simple yet powerful and mathematically proven uplift modeling method, presented in 2012.
The main idea is to predict a slightly changed target $Z_i$:
$$
Z_i = Y_i \cdot W_i + (1 - Y_i) \cdot (1 - W_i),
$$
where
* $Z_i$ - new target variable of the $i$ client;
* $Y_i$ - target variable of the $i$ client;
* $W_i$ - flag for communication of the $i$ client;
In other words, the new target equals 1 if a response in the treatment group is as good as a response in the control group and equals 0 otherwise:
$$
Z_i = \begin{cases}
1, & \mbox{if } W_i = 1 \mbox{ and } Y_i = 1 \\
1, & \mbox{if } W_i = 0 \mbox{ and } Y_i = 0 \\
0, & \mbox{otherwise}
\end{cases}
$$
Let's go deeper and estimate the conditional probability of the target variable:
$$
P(Z=1|X = x) = \\
= P(Z=1|X = x, W = 1) \cdot P(W = 1|X = x) + \\
+ P(Z=1|X = x, W = 0) \cdot P(W = 0|X = x) = \\
= P(Y=1|X = x, W = 1) \cdot P(W = 1|X = x) + \\
+ P(Y=0|X = x, W = 0) \cdot P(W = 0|X = x).
$$
We assume that $ W $ is independent of $X = x$ by design.
Thus we have: $P(W | X = x) = P(W)$ and
$$
P(Z=1|X = x) = \\
= P^T(Y=1|X = x) \cdot P(W = 1) + \\
+ P^C(Y=0|X = x) \cdot P(W = 0)
$$
Also, we assume that $P(W = 1) = P(W = 0) = \frac{1}{2}$, which means that during the experiment the control and the treatment groups were divided in equal proportions. Then we get the following:
$$
P(Z=1|X = x) = \\
= P^T(Y=1|X = x) \cdot \frac{1}{2} + P^C(Y=0|X = x) \cdot \frac{1}{2} \Rightarrow \\
2 \cdot P(Z=1|X = x) = \\
= P^T(Y=1|X = x) + P^C(Y=0|X = x) = \\
= P^T(Y=1|X = x) + 1 - P^C(Y=1|X = x) \Rightarrow \\
\Rightarrow P^T(Y=1|X = x) - P^C(Y=1|X = x) = \\
= uplift = 2 \cdot P(Z=1|X = x) - 1
$$
Thus, by doubling the estimate of the new target $Z$ and subtracting one we will get an estimation of the uplift:
$$
uplift = 2 \cdot P(Z=1) - 1
$$
This approach is based on the assumption: $P(W = 1) = P(W = 0) = \frac{1}{2}$, That is the reason that it has to be used only in cases where the number of treated customers (communication) is equal to the number of control customers (no communication).
```
from sklift.models import ClassTransformation
ct = ClassTransformation(CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True))
ct = ct.fit(X_train, y_train, treat_train, estimator_fit_params={'cat_features': cat_features})
uplift_ct = ct.predict(X_val)
ct_score = uplift_at_k(y_true=y_val, uplift=uplift_ct, treatment=treat_val, strategy='by_group', k=0.3)
models_results['approach'].append('ClassTransformation')
models_results['uplift@30%'].append(ct_score)
```
## 2. Approaches with two models
The two-model approach can be found in almost any uplift modeling work and is often used as a baseline. However, using two models can lead to some unpleasant consequences: if you use fundamentally different models for training, or if the nature of the test and control group data is very different, then the scores returned by the models will not be comparable. As a result, the calculation of the uplift will not be completely correct. To avoid this effect, you need to calibrate the models so that their scores can be interpolated as probabilities. The calibration of model probabilities is described perfectly in [scikit-learn documentation](https://scikit-learn.org/stable/modules/calibration.html).
### 2.1 Two independent models
The main idea is to estimate the conditional probabilities of the treatment and control groups separately.
1. Train the first model using the treatment set.
2. Train the second model using the control set.
3. Inference: subtract the control model scores from the treatment model scores.
<p align= "center">
<img src="https://raw.githubusercontent.com/maks-sh/scikit-uplift/master/docs/_static/images/TwoModels_vanila.png" alt="Two Models vanila"/>
</p>
```
from sklift.models import TwoModels
tm = TwoModels(
estimator_trmnt=CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True),
estimator_ctrl=CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True),
method='vanilla'
)
tm = tm.fit(
X_train, y_train, treat_train,
estimator_trmnt_fit_params={'cat_features': cat_features},
estimator_ctrl_fit_params={'cat_features': cat_features}
)
uplift_tm = tm.predict(X_val)
tm_score = uplift_at_k(y_true=y_val, uplift=uplift_tm, treatment=treat_val, strategy='by_group', k=0.3)
models_results['approach'].append('TwoModels')
models_results['uplift@30%'].append(tm_score)
plot_uplift_preds(trmnt_preds=tm.trmnt_preds_, ctrl_preds=tm.ctrl_preds_);
```
### 2.2 Two dependent models
The dependent data representation approach is based on the classifier chain method originally developed
for multi-class classification problems. The idea is that if there are $L$ different classifiers, each of which solves the problem of binary classification and in the learning process,
each subsequent classifier uses the predictions of the previous ones as additional features.
The authors of this method proposed to use the same idea to solve the problem of uplift modeling in two stages.
At the beginning we train the classifier based on the control data:
$$
P^C = P(Y=1| X, W = 0),
$$
Next, we estimate the $P_C$ predictions and use them as a feature for the second classifier.
It effectively reflects a dependency between treatment and control datasets:
$$
P^T = P(Y=1| X, P_C(X), W = 1)
$$
To get the uplift for each observation, calculate the difference:
$$
uplift(x_i) = P^T(x_i, P_C(x_i)) - P^C(x_i)
$$
Intuitively, the second classifier learns the difference between the expected probability in the treatment and the control sets which is the uplift.
<p align= "center">
<img src="https://raw.githubusercontent.com/maks-sh/scikit-uplift/master/docs/_static/images/TwoModels_ddr_control.png", alt="Two dependent models"/>
</p>
```
tm_ctrl = TwoModels(
estimator_trmnt=CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True),
estimator_ctrl=CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True),
method='ddr_control'
)
tm_ctrl = tm_ctrl.fit(
X_train, y_train, treat_train,
estimator_trmnt_fit_params={'cat_features': cat_features},
estimator_ctrl_fit_params={'cat_features': cat_features}
)
uplift_tm_ctrl = tm_ctrl.predict(X_val)
tm_ctrl_score = uplift_at_k(y_true=y_val, uplift=uplift_tm_ctrl, treatment=treat_val, strategy='by_group', k=0.3)
models_results['approach'].append('TwoModels_ddr_control')
models_results['uplift@30%'].append(tm_ctrl_score)
plot_uplift_preds(trmnt_preds=tm_ctrl.trmnt_preds_, ctrl_preds=tm_ctrl.ctrl_preds_);
```
Similarly, you can first train the $P^T$ classifier, and then use its predictions as a feature for the $P^C$ classifier.
```
tm_trmnt = TwoModels(
estimator_trmnt=CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True),
estimator_ctrl=CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True),
method='ddr_treatment'
)
tm_trmnt = tm_trmnt.fit(
X_train, y_train, treat_train,
estimator_trmnt_fit_params={'cat_features': cat_features},
estimator_ctrl_fit_params={'cat_features': cat_features}
)
uplift_tm_trmnt = tm_trmnt.predict(X_val)
tm_trmnt_score = uplift_at_k(y_true=y_val, uplift=uplift_tm_trmnt, treatment=treat_val, strategy='by_group', k=0.3)
models_results['approach'].append('TwoModels_ddr_treatment')
models_results['uplift@30%'].append(tm_trmnt_score)
plot_uplift_preds(trmnt_preds=tm_trmnt.trmnt_preds_, ctrl_preds=tm_trmnt.ctrl_preds_);
```
## Conclusion
Let's consider which method performed best in this task and use it to speed up the test sample:
```
pd.DataFrame(data=models_results).sort_values('uplift@30%', ascending=False)
```
From the table above you can see that the current task suits best for the approach to the transformation of the target line. Let's train the model on the entire sample and predict the test.
```
ct_full = ClassTransformation(CatBoostClassifier(iterations=20, thread_count=2, random_state=42, silent=True))
ct_full = ct_full.fit(
X_train_full,
y_train_full,
treat_train_full,
estimator_fit_params={'cat_features': cat_features}
)
X_test.loc[:, 'uplift'] = ct_full.predict(X_test.values)
sub = X_test[['uplift']].to_csv('sub1.csv')
!head -n 5 sub1.csv
ct_full = pd.DataFrame({
'feature_name': ct_full.estimator.feature_names_,
'feature_score': ct_full.estimator.feature_importances_
}).sort_values('feature_score', ascending=False).reset_index(drop=True)
ct_full
```
This way we got acquainted with uplift modeling and considered the main basic approaches to its construction. What's next? Then you can plunge them into the intelligence analysis of data, generate some new features, select the models and their hyperparameters and learn new approaches and libraries.
**Thank you for reading to the end.**
**I will be pleased if you support the project with an star on [github](https://github.com/maks-sh/scikit-uplift/) or tell your friends about it.**
|
github_jupyter
|
```
s = 'abc'
s.upper()
# L E G B
# local
# enclosing
# global
# builtins
globals()
globals()['s']
s.upper()
dir(s)
s.title()
x = 'this is a bunch of words to show to people'
x.title()
for attrname in dir(s):
print attrname, s.attrname
for attrname in dir(s):
print attrname, getattr(s, attrname)
s.upper
getattr(s, 'upper')
while True:
attrname = raw_input("Enter attribute name: ").strip()
if not attrname: # if I got an empty string
break
elif attrname in dir(s):
print getattr(s, attrname)
else:
print "I don't know what {} is".format(attrname)
s.upper
s.upper()
5()
s.upper.__call__
hasattr(s, 'upper')
import sys
sys.version
sys.version = '4.0.0'
sys.version
def foo():
return 5
foo.x = 100
def hello(name):
return "Hello, {}".format(name)
hello('world')
hello(123)
hello(hello)
class Foo(object):
def __init__(self, x):
self.x = x
def __add__(self, other):
return Foo(self.x + other.x)
f = Foo(10)
f.x
class Foo(object):
pass
f = Foo()
f.x = 100
f.y = {'a':1, 'b':2, 'c':3}
vars(f)
g = Foo()
g.a = [1,2,3]
g.b = 'hello'
vars(g)
class Foo(object):
def __init__(self, x, y):
self.x = x
self.y = y
f = Foo(10, [1,2,3])
vars(f)
class Person(object):
population = 0
def __init__(self, name):
self.name = name
Person.population = self.population + 1
def hello(self):
return "Hello, {}".format(self.name)
print "population = {}".format(Person.population)
p1 = Person('name1')
p2 = Person('name2')
print "population = {}".format(Person.population)
print "p1.population = {}".format(p1.population)
print "p2.population = {}".format(p2.population)
print p1.hello()
p1.thing
Person.thing = 'hello'
p1.thing
class Person(object):
def __init__(self, name):
self.name = name
def hello(self):
return "Hello, {}".format(self.name)
class Employee(Person):
def __init__(self, name, id_number):
Person.__init__(self, name)
self.id_number = id_number
e = Employee('emp1', 1)
e.hello()
e.hello()
Person.hello(e)
```
```
s = 'abc'
s.upper()
str.upper(s)
type(s)
id(s)
type(Person.hello)
id(Person.hello)
id(Person.hello)
id(Person.hello)
Person.__dict__
Person.__dict__['hello'](e)
# descriptor protocol
class Thermostat(object):
def __init__(self):
self.temp = 20
t = Thermostat()
t.temp = 100
t.temp = 0
class Thermostat(object):
def __init__(self):
self._temp = 20 # now it's private!
@property
def temp(self):
print "getting temp"
return self._temp
@temp.setter
def temp(self, new_temp):
print "setting temp"
if new_temp > 35:
print "Too high!"
new_temp = 35
elif new_temp < 0:
print "Too low!"
new_temp = 0
self._temp = new_temp
t = Thermostat()
t.temp = 100
print t.temp
t.temp = -40
print t.temp
# Temp will be a descriptor!
class Temp(object):
def __get__(self, obj, objtype):
return self.temp
def __set__(self, obj, newval):
if newval > 35:
newval = 35
if newval < 0:
newval = 0
self.temp = newval
class Thermostat(object):
temp = Temp() # temp is a class attribute, instance of Temp
t1 = Thermostat()
t2 = Thermostat()
t1.temp = 100
t2.temp = 20
print t1.temp
print t2.temp
# Temp will be a descriptor!
class Temp(object):
def __init__(self):
self.temp = {}
def __get__(self, obj, objtype):
return self.temp[obj]
def __set__(self, obj, newval):
if newval > 35:
newval = 35
if newval < 0:
newval = 0
self.temp[obj] = newval
class Thermostat(object):
temp = Temp() # temp is a class attribute, instance of Temp
t1 = Thermostat()
t2 = Thermostat()
t1.temp = 100
t2.temp = 20
print t1.temp
print t2.temp
```
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Authors.
**IMPORTANT NOTE:** This notebook is designed to run as a Colab. Click the button on top that says, `Open in Colab`, to run this notebook as a Colab. Running the notebook on your local machine might result in some of the code blocks throwing errors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# rps training set
!gdown --id 1DYVMuV2I_fA6A3er-mgTavrzKuxwpvKV
# rps testing set
!gdown --id 1RaodrRK1K03J_dGiLu8raeUynwmIbUaM
import os
import zipfile
local_zip = './rps.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('tmp/rps-train')
zip_ref.close()
local_zip = './rps-test-set.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('tmp/rps-test')
zip_ref.close()
base_dir = 'tmp/rps-train/rps'
rock_dir = os.path.join(base_dir, 'rock')
paper_dir = os.path.join(base_dir, 'paper')
scissors_dir = os.path.join(base_dir, 'scissors')
print('total training rock images:', len(os.listdir(rock_dir)))
print('total training paper images:', len(os.listdir(paper_dir)))
print('total training scissors images:', len(os.listdir(scissors_dir)))
rock_files = os.listdir(rock_dir)
print(rock_files[:10])
paper_files = os.listdir(paper_dir)
print(paper_files[:10])
scissors_files = os.listdir(scissors_dir)
print(scissors_files[:10])
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
pic_index = 2
next_rock = [os.path.join(rock_dir, fname)
for fname in rock_files[pic_index-2:pic_index]]
next_paper = [os.path.join(paper_dir, fname)
for fname in paper_files[pic_index-2:pic_index]]
next_scissors = [os.path.join(scissors_dir, fname)
for fname in scissors_files[pic_index-2:pic_index]]
for i, img_path in enumerate(next_rock+next_paper+next_scissors):
#print(img_path)
img = mpimg.imread(img_path)
plt.imshow(img)
plt.axis('Off')
plt.show()
import tensorflow as tf
import keras_preprocessing
from keras_preprocessing import image
from keras_preprocessing.image import ImageDataGenerator
TRAINING_DIR = "tmp/rps-train/rps"
training_datagen = ImageDataGenerator(
rescale = 1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
VALIDATION_DIR = "tmp/rps-test/rps-test-set"
validation_datagen = ImageDataGenerator(rescale = 1./255)
train_generator = training_datagen.flow_from_directory(
TRAINING_DIR,
target_size=(150,150),
class_mode='categorical',
batch_size=126
)
validation_generator = validation_datagen.flow_from_directory(
VALIDATION_DIR,
target_size=(150,150),
class_mode='categorical',
batch_size=126
)
model = tf.keras.models.Sequential([
# Note the input shape is the desired size of the image 150x150 with 3 bytes color
# This is the first convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu', input_shape=(150, 150, 3)),
tf.keras.layers.MaxPooling2D(2, 2),
# The second convolution
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The third convolution
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# The fourth convolution
tf.keras.layers.Conv2D(128, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
# Flatten the results to feed into a DNN
tf.keras.layers.Flatten(),
tf.keras.layers.Dropout(0.5),
# 512 neuron hidden layer
tf.keras.layers.Dense(512, activation='relu'),
tf.keras.layers.Dense(3, activation='softmax')
])
model.summary()
model.compile(loss = 'categorical_crossentropy', optimizer='rmsprop', metrics=['accuracy'])
history = model.fit(train_generator, epochs=25, steps_per_epoch=20, validation_data = validation_generator, verbose = 1, validation_steps=3)
model.save("rps.h5")
import matplotlib.pyplot as plt
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r', label='Training accuracy')
plt.plot(epochs, val_acc, 'b', label='Validation accuracy')
plt.title('Training and validation accuracy')
plt.legend(loc=0)
plt.figure()
plt.show()
```
# Here's a codeblock just for fun!
You should be able to upload an image here and have it classified without crashing. This codeblock will only work in Google Colab, however.
**Important Note:** Due to some compatibility issues, the following code block will result in an error after you select the images(s) to upload if you are running this notebook as a `Colab` on the `Safari` browser. For `all other broswers`, continue with the next code block and ignore the next one after it.
The ones running the `Colab` on `Safari`, comment out the code block below, uncomment the next code block and run it.
```
import numpy as np
from google.colab import files
from keras.preprocessing import image
uploaded = files.upload()
for fn in uploaded.keys():
# predicting images
path = fn
img = image.load_img(path, target_size=(150, 150))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model.predict(images, batch_size=10)
print(fn)
print(classes)
```
For those running this `Colab` on `Safari` broswer can upload the images(s) manually. Follow the instructions, uncomment the code block below and run it.
Instructions on how to upload image(s) manually in a Colab:
1. Select the `folder` icon on the left `menu bar`.
2. Click on the `folder with an arrow pointing upwards` named `..`
3. Click on the `folder` named `tmp`.
4. Inside of the `tmp` folder, `create a new folder` called `images`. You'll see the `New folder` option by clicking the `3 vertical dots` menu button next to the `tmp` folder.
5. Inside of the new `images` folder, upload an image(s) of your choice, preferably of either a horse or a human. Drag and drop the images(s) on top of the `images` folder.
6. Uncomment and run the code block below.
```
```
|
github_jupyter
|
```
# ###############################################
# ########## Default Parameters #################
# ###############################################
start = '2016-06-16 22:00:00'
end = '2016-06-18 00:00:00'
pv_nominal_kw = 5000 # There are 3 PV locations hardcoded at node 7, 8, 9
inverter_sizing = 1.05
inverter_qmax_percentage = 0.44
thrP = 0.04
hysP = 0.06
thrQ = 0.03
hysQ = 0.03
first_order_time_const = 1 * 60
solver_relative_tolerance = 0.1
solver_absolute_tolerance = 0.1
solver_name = 'CVode'
result_filename = 'result'
import warnings
warnings.filterwarnings("ignore", message="numpy.dtype size changed")
warnings.filterwarnings("ignore", message="numpy.ufunc size changed")
warnings.simplefilter(action='ignore', category=FutureWarning)
import pandas
import numpy
import datetime
from tabulate import tabulate
import json
import re
# Imports useful for graphics
import matplotlib
import matplotlib.pyplot as plt
import seaborn
seaborn.set_style("whitegrid")
seaborn.despine()
%matplotlib inline
font = {'size' : 14}
matplotlib.rc('font', **font)
# Date conversion
begin = '2016-01-01 00:00:00'
begin_dt = datetime.datetime.strptime(begin, '%Y-%m-%d %H:%M:%S')
start_dt = datetime.datetime.strptime(start, '%Y-%m-%d %H:%M:%S')
end_dt = datetime.datetime.strptime(end, '%Y-%m-%d %H:%M:%S')
start_s = int((start_dt - begin_dt).total_seconds())
end_s = int((end_dt - begin_dt).total_seconds())
inverter_smax = pv_nominal_kw * inverter_sizing
inverter_qmax = inverter_smax * inverter_qmax_percentage
pv_inverter_parameters = {
'weather_file':(path_to_fmiforpowersystems +
'examples\\002_cosimulation_custom_master\\pv_inverter\\' +
'USA_CA_San.Francisco.Intl.AP.724940_TMY3.mos'),
'n': 1,
'A': (pv_nominal_kw * 1000) / (0.158 * 1000),
'eta': 0.158,
'lat': 37.9,
'til': 10,
'azi': 0,
'thrP': thrP, #0.05,
'hysP': hysP, #0.04,
'thrQ': thrQ, #0.04,
'hysQ': hysQ, #0.01,
'SMax': inverter_smax,
'QMaxInd': inverter_qmax,
'QMaxCap': inverter_qmax,
'Tfirstorder': first_order_time_const,
}
first_order_parameters = {
'T': first_order_time_const
}
run_simulation = True
connections_filename = 'connections.xlsx'
pv_inverter_path = 'pv_inverter/Pv_Inv_VoltVarWatt_simple_Slim.fmu'
pv_inverter_path = 'pv_inverter/PV_0Inv_0VoltVarWat_0simple_0Slim_Pv_0Inv_0VoltVarWatt_0simple_0Slim.fmu'
pandapower_path = 'pandapower/pandapower.fmu'
pandapower_folder = 'pandapower'
pandapower_parameter = {}
firstorder_path = 'firstorder/FirstOrder.fmu'
```
## Create the connection mapping
```
connections = pandas.DataFrame(columns=['fmu1_id', 'fmu1_path',
'fmu2_id', 'fmu2_path',
'fmu1_parameters',
'fmu2_parameters',
'fmu1_output',
'fmu2_input'])
# Connection for each customer
nodes = [7, 9, 24]
for index in nodes:
connections = connections.append(
{'fmu1_id': 'PV' + str(index),
'fmu1_path': pv_inverter_path,
'fmu2_id': 'pandapower',
'fmu2_path': pandapower_path,
'fmu1_parameters': pv_inverter_parameters,
'fmu2_parameters': pandapower_parameter,
'fmu1_output': 'P',
'fmu2_input': 'KW_' + str(index)},
ignore_index=True)
connections = connections.append(
{'fmu1_id': 'PV' + str(index),
'fmu1_path': pv_inverter_path,
'fmu2_id': 'pandapower',
'fmu2_path': pandapower_path,
'fmu1_parameters': pv_inverter_parameters,
'fmu2_parameters': pandapower_parameter,
'fmu1_output': 'Q',
'fmu2_input': 'KVAR_' + str(index)},
ignore_index=True)
connections = connections.append(
{'fmu1_id': 'pandapower',
'fmu1_path': pandapower_path,
'fmu2_id': 'firstorder' + str(index),
'fmu2_path': firstorder_path,
'fmu1_parameters': pandapower_parameter,
'fmu2_parameters': first_order_parameters,
'fmu1_output': 'Vpu_' + str(index),
'fmu2_input': 'u'},
ignore_index=True)
connections = connections.append(
{'fmu1_id': 'firstorder' + str(index),
'fmu1_path': firstorder_path,
'fmu2_id': 'PV' + str(index),
'fmu2_path': pv_inverter_path,
'fmu1_parameters': first_order_parameters,
'fmu2_parameters': pv_inverter_parameters,
'fmu1_output': 'y',
'fmu2_input': 'v'},
ignore_index=True)
def _sanitize_name(name):
"""
Make a Modelica valid name.
In Modelica, a variable name:
Can contain any of the characters {a-z,A-Z,0-9,_}.
Cannot start with a number.
:param name(str): Variable name to be sanitized.
:return: Sanitized variable name.
"""
# Check if variable has a length > 0
assert(len(name) > 0), 'Require a non-null variable name.'
# If variable starts with a number add 'f_'.
if(name[0].isdigit()):
name = 'f_' + name
# Replace all illegal characters with an underscore.
g_rexBadIdChars = re.compile(r'[^a-zA-Z0-9_]')
name = g_rexBadIdChars.sub('_', name)
return name
connections['fmu1_output'] = connections['fmu1_output'].apply(lambda x: _sanitize_name(x))
connections['fmu2_input'] = connections['fmu2_input'].apply(lambda x: _sanitize_name(x))
print(tabulate(connections[
['fmu1_id', 'fmu2_id', 'fmu1_output', 'fmu2_input']].head(),
headers='keys', tablefmt='psql'))
print(tabulate(connections[
['fmu1_id', 'fmu2_id', 'fmu1_output', 'fmu2_input']].tail(),
headers='keys', tablefmt='psql'))
connections.to_excel(connections_filename, index=False)
```
# Launch FMU simulation
```
if run_simulation:
import shlex, subprocess
cmd = ("C:/JModelica.org-2.4/setenv.bat && " +
" cd " + pandapower_folder + " && "
"cyderc " +
" --path ./"
" --name pandapower" +
" --io pandapower.xlsx" +
" --path_to_simulatortofmu C:/Users/DRRC/Desktop/Joscha/SimulatorToFMU/simulatortofmu/parser/SimulatorToFMU.py"
" --fmu_struc python")
args = shlex.split(cmd)
process = subprocess.Popen(args, bufsize=1, universal_newlines=True)
process.wait()
process.kill()
if run_simulation:
import os
import signal
import shlex, subprocess
cmd = ("C:/JModelica.org-2.4/setenv.bat && " +
"cyders " +
" --start " + str(start_s) +
" --end " + str(end_s) +
" --connections " + connections_filename +
" --nb_steps 25" +
" --solver " + solver_name +
" --rtol " + str(solver_relative_tolerance) +
" --atol " + str(solver_absolute_tolerance) +
" --result " + 'results/' + result_filename + '.csv')
args = shlex.split(cmd)
process = subprocess.Popen(args, bufsize=1, universal_newlines=True,
creationflags=subprocess.CREATE_NEW_PROCESS_GROUP)
process.wait()
process.send_signal(signal.CTRL_BREAK_EVENT)
process.kill()
print('Killed')
```
# Plot results
```
# Load results
results = pandas.read_csv('results/' + result_filename + '.csv')
from pathlib import Path, PureWindowsPath
results = pandas.read_csv(os.path.join(r'C:\Users\DRRC\Desktop\Jonathan\voltvarwatt_with_cyme_fmus\usecases\004_pp_first_order\results\result.csv'))
epoch = datetime.datetime.utcfromtimestamp(0)
begin_since_epoch = (begin_dt - epoch).total_seconds()
results['datetime'] = results['time'].apply(
lambda x: datetime.datetime.utcfromtimestamp(begin_since_epoch + x))
results.set_index('datetime', inplace=True, drop=False)
print('COLUMNS=')
print(results.columns)
print('START=')
print(results.head(1).index[0])
print('END=')
print(results.tail(1).index[0])
# Plot sum of all PVs for P and P curtailled and Q
cut = '2016-06-17 01:00:00'
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.title('PV generation')
for node in nodes:
plt.plot(results['datetime'],
results['pandapower.KW_' + str(node)] / 1000,
linewidth=3, alpha=0.7, label='node ' + str(node))
plt.legend(loc=0)
plt.ylabel('PV active power [MW]')
plt.xlabel('Time')
plt.xlim([cut, end])
plt.show()
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.title('Inverter reactive power')
for node in nodes:
plt.plot(results['datetime'],
results['pandapower.KVAR_' + str(node)] / 1000,
linewidth=3, alpha=0.7, label='node ' + str(node))
plt.legend(loc=0)
plt.ylabel('PV reactive power [MVAR]')
plt.xlabel('Time')
plt.xlim([cut, end])
plt.show()
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.title('PV voltage')
for node in nodes:
plt.plot(results['datetime'],
results['pandapower.Vpu_' + str(node)],
linewidth=3, alpha=0.7, label='node ' + str(node))
plt.legend(loc=0)
plt.ylabel('PV voltage [p.u.]')
plt.xlabel('Time')
plt.xlim([cut, end])
plt.ylim([0.95, results[['pandapower.Vpu_' + str(node)
for node in nodes]].max().max()])
plt.show()
# Plot time/voltage
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.title('Feeder Voltages')
plt.plot(results['datetime'],
results[[col for col in results.columns
if 'Vpu' in col and 'transfer' not in col]],
linewidth=3, alpha=0.7)
plt.ylabel('Voltage [p.u.]')
plt.xlabel('Time')
plt.ylim([0.95, results[[col for col in results.columns
if 'Vpu' in col and 'transfer' not in col]].max().max()])
plt.show()
# Load results
debug = pandas.read_csv('debug.csv', parse_dates=[1])
epoch = datetime.datetime.utcfromtimestamp(0)
begin_since_epoch = (begin_dt - epoch).total_seconds()
debug['datetime'] = debug['sim_time'].apply(
lambda x: datetime.datetime.utcfromtimestamp(begin_since_epoch + x))
debug.set_index('datetime', inplace=True, drop=False)
print('COLUMNS=')
print(debug.columns)
print('START=')
print(debug.head(1).index[0])
print('END=')
print(debug.tail(1).index[0])
# Plot time/voltage
import matplotlib.dates as mdates
print('Number of evaluation=' + str(len(debug)))
fig, axes = plt.subplots(1, 1, figsize=(11, 8))
plt.plot(debug['clock'],
debug['datetime'],
linewidth=3, alpha=0.7)
plt.ylabel('Simulation time')
plt.xlabel('Computer clock')
plt.gcf().autofmt_xdate()
myFmt = mdates.DateFormatter('%H:%M')
plt.gca().xaxis.set_major_formatter(myFmt)
plt.show()
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.plot(debug['clock'],
debug['KW_7'],
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['KW_9'],
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['KW_24'],
linewidth=3, alpha=0.7)
plt.ylabel('KW')
plt.xlabel('Computer clock')
plt.legend([17, 31, 24], loc=0)
plt.show()
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.plot(debug['clock'],
debug['Vpu_7'],
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['Vpu_9'],
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['Vpu_24'],
linewidth=3, alpha=0.7)
plt.ylabel('Vpu')
plt.xlabel('Computer clock')
plt.show()
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.plot(debug['clock'],
debug['Vpu_7'].diff(),
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['Vpu_9'].diff(),
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['Vpu_24'].diff(),
linewidth=3, alpha=0.7)
plt.ylabel('Vpu Diff')
plt.xlabel('Computer clock')
plt.show()
fig, axes = plt.subplots(1, 1, figsize=(11, 3))
plt.plot(debug['clock'],
debug['KVAR_7'],
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['KVAR_9'],
linewidth=3, alpha=0.7)
plt.plot(debug['clock'],
debug['KVAR_24'],
linewidth=3, alpha=0.7)
plt.ylabel('KVAR')
plt.xlabel('Computer clock')
plt.show()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/ymoslem/OpenNMT-Tutorial/blob/main/2-NMT-Training.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
# Install OpenNMT-py 2.x
!pip3 install OpenNMT-py
```
# Prepare Your Datasets
Please make sure you have completed the [first exercise](https://colab.research.google.com/drive/1rsFPnAQu9-_A6e2Aw9JYK3C8mXx9djsF?usp=sharing).
```
# Open the folder where you saved your prepapred datasets from the first exercise
%cd drive/MyDrive/nmt/
!ls
```
# Create the Training Configuration File
The following config file matches most of the recommended values for the Transformer model [Vaswani et al., 2017](https://arxiv.org/abs/1706.03762). As the current dataset is small, we reduced the following values:
* `train_steps` - for datasets with a few millions of sentences, consider using a value between 100000 and 200000, or more! Enabling the option `early_stopping` can help stop the training when there is no considerable improvement.
* `valid_steps` - 10000 can be good if the value `train_steps` is big enough.
* `warmup_steps` - obviously, its value must be less than `train_steps`. Try 4000 and 8000 values.
Refer to [OpenNMT-py training parameters](https://opennmt.net/OpenNMT-py/options/train.html) for more details. If you are interested in further explanation of the Transformer model, you can check this article, [Illustrated Transformer](https://jalammar.github.io/illustrated-transformer/).
```
# Create the YAML configuration file
# On a regular machine, you can create it manually or with nano
# Note here we are using some smaller values because the dataset is small
# For larger datasets, consider increasing: train_steps, valid_steps, warmup_steps, save_checkpoint_steps, keep_checkpoint
config = '''# config.yaml
## Where the samples will be written
save_data: run
# Training files
data:
corpus_1:
path_src: UN.en-fr.fr-filtered.fr.subword.train
path_tgt: UN.en-fr.en-filtered.en.subword.train
transforms: [filtertoolong]
valid:
path_src: UN.en-fr.fr-filtered.fr.subword.dev
path_tgt: UN.en-fr.en-filtered.en.subword.dev
transforms: [filtertoolong]
# Vocabulary files, generated by onmt_build_vocab
src_vocab: run/source.vocab
tgt_vocab: run/target.vocab
# Vocabulary size - should be the same as in sentence piece
src_vocab_size: 50000
tgt_vocab_size: 50000
# Filter out source/target longer than n if [filtertoolong] enabled
#src_seq_length: 200
#src_seq_length: 200
# Tokenization options
src_subword_model: source.model
tgt_subword_model: target.model
# Where to save the log file and the output models/checkpoints
log_file: train.log
save_model: models/model.fren
# Stop training if it does not imporve after n validations
early_stopping: 4
# Default: 5000 - Save a model checkpoint for each n
save_checkpoint_steps: 1000
# To save space, limit checkpoints to last n
# keep_checkpoint: 3
seed: 3435
# Default: 100000 - Train the model to max n steps
# Increase for large datasets
train_steps: 3000
# Default: 10000 - Run validation after n steps
valid_steps: 1000
# Default: 4000 - for large datasets, try up to 8000
warmup_steps: 1000
report_every: 100
decoder_type: transformer
encoder_type: transformer
word_vec_size: 512
rnn_size: 512
layers: 6
transformer_ff: 2048
heads: 8
accum_count: 4
optim: adam
adam_beta1: 0.9
adam_beta2: 0.998
decay_method: noam
learning_rate: 2.0
max_grad_norm: 0.0
# Tokens per batch, change if out of GPU memory
batch_size: 4096
valid_batch_size: 4096
batch_type: tokens
normalization: tokens
dropout: 0.1
label_smoothing: 0.1
max_generator_batches: 2
param_init: 0.0
param_init_glorot: 'true'
position_encoding: 'true'
# Number of GPUs, and IDs of GPUs
world_size: 1
gpu_ranks: [0]
'''
with open("config.yaml", "w+") as config_yaml:
config_yaml.write(config)
# [Optional] Check the content of the configuration file
!cat config.yaml
```
# Build Vocabulary
For large datasets, it is not feasable to use all words/tokens found in the corpus. Instead, a specific set of vocabulary is extracted from the training dataset, usually betweeen 32k and 100k words. This is the main purpose of the vocabulary building step.
```
# Find the number of CPUs/cores on the machine
!nproc --all
# Build Vocabulary
# -config: path to your config.yaml file
# -n_sample: use -1 to build vocabulary on all the segment in the training dataset
# -num_threads: change it to match the number of CPUs to run it faster
!onmt_build_vocab -config config.yaml -n_sample -1 -num_threads 2
```
From the **Runtime menu** > **Change runtime type**, make sure that the "**Hardware accelerator**" is "**GPU**".
```
# Check if the GPU is active
!nvidia-smi -L
# Check if the GPU is visable to PyTorch
import torch
print(torch.cuda.is_available())
print(torch.cuda.get_device_name(0))
```
# Training
Now, start training your NMT model! 🎉 🎉 🎉
```
# Train the NMT model
!onmt_train -config config.yaml
```
# Translation
Translation Options:
* `-model` - specify the last model checkpoint name; try testing the quality of multiple checkpoints
* `-src` - the subworded test dataset, source file
* `-output` - give any file name to the new translation output file
* `-gpu` - GPU ID, usually 0 if you have one GPU. Otherwise, it will translate on CPU, which would be slower.
* `-min_length` - [optional] to avoid empty translations
* `-verbose` - [optional] if you want to print translations
Refer to [OpenNMT-py translation options](https://opennmt.net/OpenNMT-py/options/translate.html) for more details.
```
# Translate - change the model name
!onmt_translate -model models/model.fren_step_3000.pt -src UN.en-fr.fr-filtered.fr.subword.test -output UN.en.translated -gpu 0 -min_length 1
# Check the first 5 lines of the translation file
!head -n 5 UN.en.translated
# Desubword the translation file
!python3 MT-Preparation/subwording/3-desubword.py target.model UN.en.translated
# Check the first 5 lines of the desubworded translation file
!head -n 5 UN.en.translated.desubword
# Desubword the source test
# Note: You might as well have split files *before* subwording during dataset preperation,
# but sometimes datasets have tokeniztion issues, so this way you are sure the file is really untokenized.
!python3 MT-Preparation/subwording/3-desubword.py target.model UN.en-fr.en-filtered.en.subword.test
# Check the first 5 lines of the desubworded source
!head -n 5 UN.en-fr.en-filtered.en.subword.test.desubword
```
# MT Evaluation
There are several MT Evaluation metrics such as BLEU, TER, METEOR, COMET, BERTScore, among others.
Here we are using BLEU. Files must be detokenized/desubworded beforehand.
```
# Download the BLEU script
!wget https://raw.githubusercontent.com/ymoslem/MT-Evaluation/main/BLEU/compute-bleu.py
# Install sacrebleu
!pip3 install sacrebleu
# Evaluate the translation (without subwording)
!python3 compute-bleu.py UN.en-fr.en-filtered.en.subword.test.desubword UN.en.translated.desubword
```
# More Features and Directions to Explore
Experiment with the following ideas:
* Icrease `train_steps` and see to what extent new checkpoints provide better translation, in terms of both BLEU and your human evaluation.
* Check other MT Evaluation mentrics other than BLEU such as [TER](https://github.com/mjpost/sacrebleu#ter), [WER](https://blog.machinetranslation.io/compute-wer-score/), [METEOR](https://blog.machinetranslation.io/compute-bleu-score/#meteor), [COMET](https://github.com/Unbabel/COMET), and [BERTScore](https://github.com/Tiiiger/bert_score). What are the conceptual differences between them? Is there there special cases for using a specific metric?
* Continue training from the last model checkpoint using the `-train_from` option, only if the training stopped and you want to continue it. In this case, `train_steps` in the config file should be larger than the steps of the last checkpoint you train from.
```
!onmt_train -config config.yaml -train_from models/model.fren_step_3000.pt
```
* **Ensemble Decoding:** During translation, instead of adding one model/checkpoint to the `-model` argument, add multiple checkpoints. For example, try the two last checkpoints. Does it improve quality of translation? Does it affect translation seepd?
* **Averaging Models:** Try to average multiple models into one model using the [average_models.py](https://github.com/OpenNMT/OpenNMT-py/blob/master/onmt/bin/average_models.py) script, and see how this affects translation quality.
```
python3 average_models.py -models model_step_xxx.pt model_step_yyy.pt -output model_avg.pt
```
* **Release the model:** Try this command and see how it reduce the model size.
```
onmt_release_model --model "model.pt" --output "model_released.pt
```
* **Use CTranslate2:** For efficient translation, consider using [CTranslate2](https://github.com/OpenNMT/CTranslate2), a fast inference engine. Check out an [example](https://gist.github.com/ymoslem/60e1d1dc44fe006f67e130b6ad703c4b).
* **Work on low-resource languages:** Find out more details about [how to train NMT models for low-resource languages](https://blog.machinetranslation.io/low-resource-nmt/).
* **Train a multilingual model:** Find out helpful notes about [training multilingual models](https://blog.machinetranslation.io/multilingual-nmt).
* **Publish a demo:** Show off your work through a [simple demo with CTranslate2 and Streamlit](https://blog.machinetranslation.io/nmt-web-interface/).
|
github_jupyter
|
```
try:
from openmdao.utils.notebook_utils import notebook_mode
except ImportError:
!python -m pip install openmdao[notebooks]
```
# NonlinearBlockGS
NonlinearBlockGS applies Block Gauss-Seidel (also known as fixed-point iteration) to the
components and subsystems in the system. This is mainly used to solve cyclic connections. You
should try this solver for systems that satisfy the following conditions:
1. System (or subsystem) contains a cycle, though subsystems may.
2. System does not contain any implicit states, though subsystems may.
NonlinearBlockGS is a block solver, so you can specify different nonlinear solvers in the subsystems and they
will be utilized to solve the subsystem nonlinear problem.
Note that you may not know if you satisfy the second condition, so choosing a solver can be a trial-and-error proposition. If
NonlinearBlockGS doesn't work, then you will need to use [NewtonSolver](../../../_srcdocs/packages/solvers.nonlinear/newton).
Here, we choose NonlinearBlockGS to solve the Sellar problem, which has two components with a
cyclic dependency, has no implicit states, and works very well with Gauss-Seidel.
```
from openmdao.utils.notebook_utils import get_code
from myst_nb import glue
glue("code_src33", get_code("openmdao.test_suite.components.sellar.SellarDis1withDerivatives"), display=False)
```
:::{Admonition} `SellarDis1withDerivatives` class definition
:class: dropdown
{glue:}`code_src33`
:::
```
from openmdao.utils.notebook_utils import get_code
from myst_nb import glue
glue("code_src34", get_code("openmdao.test_suite.components.sellar.SellarDis2withDerivatives"), display=False)
```
:::{Admonition} `SellarDis2withDerivatives` class definition
:class: dropdown
{glue:}`code_src34`
:::
```
import numpy as np
import openmdao.api as om
from openmdao.test_suite.components.sellar import SellarDis1withDerivatives, SellarDis2withDerivatives
prob = om.Problem()
model = prob.model
model.add_subsystem('d1', SellarDis1withDerivatives(), promotes=['x', 'z', 'y1', 'y2'])
model.add_subsystem('d2', SellarDis2withDerivatives(), promotes=['z', 'y1', 'y2'])
model.add_subsystem('obj_cmp', om.ExecComp('obj = x**2 + z[1] + y1 + exp(-y2)',
z=np.array([0.0, 0.0]), x=0.0),
promotes=['obj', 'x', 'z', 'y1', 'y2'])
model.add_subsystem('con_cmp1', om.ExecComp('con1 = 3.16 - y1'), promotes=['con1', 'y1'])
model.add_subsystem('con_cmp2', om.ExecComp('con2 = y2 - 24.0'), promotes=['con2', 'y2'])
model.nonlinear_solver = om.NonlinearBlockGS()
prob.setup()
prob.set_val('x', 1.)
prob.set_val('z', np.array([5.0, 2.0]))
prob.run_model()
print(prob.get_val('y1'))
print(prob.get_val('y2'))
from openmdao.utils.assert_utils import assert_near_equal
assert_near_equal(prob.get_val('y1'), 25.58830273, .00001)
assert_near_equal(prob.get_val('y2'), 12.05848819, .00001)
```
This solver runs all of the subsystems each iteration, passing data along all connections
including the cyclic ones. After each iteration, the iteration count and the residual norm are
checked to see if termination has been satisfied.
You can control the termination criteria for the solver using the following options:
# NonlinearBlockGS Options
```
om.show_options_table("openmdao.solvers.nonlinear.nonlinear_block_gs.NonlinearBlockGS")
```
## NonlinearBlockGS Constructor
The call signature for the `NonlinearBlockGS` constructor is:
```{eval-rst}
.. automethod:: openmdao.solvers.nonlinear.nonlinear_block_gs.NonlinearBlockGS.__init__
:noindex:
```
## Aitken relaxation
This solver implements Aitken relaxation, as described in Algorithm 1 of this paper on aerostructual design [optimization](http://www.umich.edu/~mdolaboratory/pdf/Kenway2014a.pdf).
The relaxation is turned off by default, but it may help convergence for more tightly coupled models.
## Residual Calculation
The `Unified Derivatives Equations` are formulated so that explicit equations (via `ExplicitComponent`) are also expressed
as implicit relationships, and their residual is also calculated in "apply_nonlinear", which runs the component a second time and
saves the difference in the output vector as the residual. However, this would require an extra call to `compute`, which is
inefficient for slower components. To eliminate the inefficiency of running the model twice every iteration the NonlinearBlockGS
driver saves a copy of the output vector and uses that to calculate the residual without rerunning the model. This does require
a little more memory, so if you are solving a model where memory is more of a concern than execution time, you can set the
"use_apply_nonlinear" option to True to use the original formulation that calls "apply_nonlinear" on the subsystem.
## NonlinearBlockGS Option Examples
**maxiter**
`maxiter` lets you specify the maximum number of Gauss-Seidel iterations to apply. In this example, we
cut it back from the default, ten, down to two, so that it terminates a few iterations earlier and doesn't
reach the specified absolute or relative tolerance.
```
from openmdao.test_suite.components.sellar import SellarDis1withDerivatives, SellarDis2withDerivatives
prob = om.Problem()
model = prob.model
model.add_subsystem('d1', SellarDis1withDerivatives(), promotes=['x', 'z', 'y1', 'y2'])
model.add_subsystem('d2', SellarDis2withDerivatives(), promotes=['z', 'y1', 'y2'])
model.add_subsystem('obj_cmp', om.ExecComp('obj = x**2 + z[1] + y1 + exp(-y2)',
z=np.array([0.0, 0.0]), x=0.0),
promotes=['obj', 'x', 'z', 'y1', 'y2'])
model.add_subsystem('con_cmp1', om.ExecComp('con1 = 3.16 - y1'), promotes=['con1', 'y1'])
model.add_subsystem('con_cmp2', om.ExecComp('con2 = y2 - 24.0'), promotes=['con2', 'y2'])
prob.setup()
nlbgs = model.nonlinear_solver = om.NonlinearBlockGS()
#basic test of number of iterations
nlbgs.options['maxiter'] = 1
prob.run_model()
print(model.nonlinear_solver._iter_count)
assert(model.nonlinear_solver._iter_count == 1)
nlbgs.options['maxiter'] = 5
prob.run_model()
print(model.nonlinear_solver._iter_count)
assert(model.nonlinear_solver._iter_count == 5)
#test of number of iterations AND solution after exit at maxiter
prob.set_val('x', 1.)
prob.set_val('z', np.array([5.0, 2.0]))
nlbgs.options['maxiter'] = 3
prob.set_solver_print()
prob.run_model()
print(prob.get_val('y1'))
print(prob.get_val('y2'))
print(model.nonlinear_solver._iter_count)
assert_near_equal(prob.get_val('y1'), 25.58914915, .00001)
assert_near_equal(prob.get_val('y2'), 12.05857185, .00001)
assert(model.nonlinear_solver._iter_count == 3)
```
**atol**
Here, we set the absolute tolerance to a looser value that will trigger an earlier termination. After
each iteration, the norm of the residuals is calculated one of two ways. If the "use_apply_nonlinear" option
is set to False (its default), then the norm is calculated by subtracting a cached previous value of the
outputs from the current value. If "use_apply_nonlinear" is True, then the norm is calculated by calling
apply_nonlinear on all of the subsystems. In this case, `ExplicitComponents` are executed a second time.
If this norm value is lower than the absolute tolerance `atol`, the iteration will terminate.
```
from openmdao.test_suite.components.sellar import SellarDis1withDerivatives, SellarDis2withDerivatives
prob = om.Problem()
model = prob.model
model.add_subsystem('d1', SellarDis1withDerivatives(), promotes=['x', 'z', 'y1', 'y2'])
model.add_subsystem('d2', SellarDis2withDerivatives(), promotes=['z', 'y1', 'y2'])
model.add_subsystem('obj_cmp', om.ExecComp('obj = x**2 + z[1] + y1 + exp(-y2)',
z=np.array([0.0, 0.0]), x=0.0),
promotes=['obj', 'x', 'z', 'y1', 'y2'])
model.add_subsystem('con_cmp1', om.ExecComp('con1 = 3.16 - y1'), promotes=['con1', 'y1'])
model.add_subsystem('con_cmp2', om.ExecComp('con2 = y2 - 24.0'), promotes=['con2', 'y2'])
nlbgs = model.nonlinear_solver = om.NonlinearBlockGS()
nlbgs.options['atol'] = 1e-4
prob.setup()
prob.set_val('x', 1.)
prob.set_val('z', np.array([5.0, 2.0]))
prob.run_model()
print(prob.get_val('y1'))
print(prob.get_val('y2'))
assert_near_equal(prob.get_val('y1'), 25.5882856302, .00001)
assert_near_equal(prob.get_val('y2'), 12.05848819, .00001)
```
**rtol**
Here, we set the relative tolerance to a looser value that will trigger an earlier termination. After
each iteration, the norm of the residuals is calculated one of two ways. If the "use_apply_nonlinear" option
is set to False (its default), then the norm is calculated by subtracting a cached previous value of the
outputs from the current value. If "use_apply_nonlinear" is True, then the norm is calculated by calling
apply_nonlinear on all of the subsystems. In this case, `ExplicitComponents` are executed a second time.
If the ratio of the currently calculated norm to the initial residual norm is lower than the relative tolerance
`rtol`, the iteration will terminate.
```
from openmdao.utils.notebook_utils import get_code
from myst_nb import glue
glue("code_src35", get_code("openmdao.test_suite.components.sellar.SellarDerivatives"), display=False)
```
:::{Admonition} `SellarDerivatives` class definition
:class: dropdown
{glue:}`code_src35`
:::
```
from openmdao.test_suite.components.sellar import SellarDis1withDerivatives, SellarDis2withDerivatives, SellarDerivatives
prob = om.Problem()
model = prob.model
model.add_subsystem('d1', SellarDis1withDerivatives(), promotes=['x', 'z', 'y1', 'y2'])
model.add_subsystem('d2', SellarDis2withDerivatives(), promotes=['z', 'y1', 'y2'])
model.add_subsystem('obj_cmp', om.ExecComp('obj = x**2 + z[1] + y1 + exp(-y2)',
z=np.array([0.0, 0.0]), x=0.0),
promotes=['obj', 'x', 'z', 'y1', 'y2'])
model.add_subsystem('con_cmp1', om.ExecComp('con1 = 3.16 - y1'), promotes=['con1', 'y1'])
model.add_subsystem('con_cmp2', om.ExecComp('con2 = y2 - 24.0'), promotes=['con2', 'y2'])
nlbgs = model.nonlinear_solver = om.NonlinearBlockGS()
nlbgs.options['rtol'] = 1e-3
prob.setup()
prob.set_val('x', 1.)
prob.set_val('z', np.array([5.0, 2.0]))
prob.run_model()
print(prob.get_val('y1'), 25.5883027, .00001)
print(prob.get_val('y2'), 12.05848819, .00001)
assert_near_equal(prob.get_val('y1'), 25.5883027, .00001)
assert_near_equal(prob.get_val('y2'), 12.05848819, .00001)
```
|
github_jupyter
|
```
#remove cell visibility
from IPython.display import HTML
tag = HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide()
} else {
$('div.input').show()
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
Promijeni vidljivost <a href="javascript:code_toggle()">ovdje</a>.''')
display(tag)
%matplotlib inline
import control
import numpy
import sympy as sym
from IPython.display import display, Markdown
import ipywidgets as widgets
import matplotlib.pyplot as plt
#print a matrix latex-like
def bmatrix(a):
"""Returns a LaTeX bmatrix - by Damir Arbula (ICCT project)
:a: numpy array
:returns: LaTeX bmatrix as a string
"""
if len(a.shape) > 2:
raise ValueError('bmatrix can at most display two dimensions')
lines = str(a).replace('[', '').replace(']', '').splitlines()
rv = [r'\begin{bmatrix}']
rv += [' ' + ' & '.join(l.split()) + r'\\' for l in lines]
rv += [r'\end{bmatrix}']
return '\n'.join(rv)
# Display formatted matrix:
def vmatrix(a):
if len(a.shape) > 2:
raise ValueError('bmatrix can at most display two dimensions')
lines = str(a).replace('[', '').replace(']', '').splitlines()
rv = [r'\begin{vmatrix}']
rv += [' ' + ' & '.join(l.split()) + r'\\' for l in lines]
rv += [r'\end{vmatrix}']
return '\n'.join(rv)
#matrixWidget is a matrix looking widget built with a VBox of HBox(es) that returns a numPy array as value !
class matrixWidget(widgets.VBox):
def updateM(self,change):
for irow in range(0,self.n):
for icol in range(0,self.m):
self.M_[irow,icol] = self.children[irow].children[icol].value
#print(self.M_[irow,icol])
self.value = self.M_
def dummychangecallback(self,change):
pass
def __init__(self,n,m):
self.n = n
self.m = m
self.M_ = numpy.matrix(numpy.zeros((self.n,self.m)))
self.value = self.M_
widgets.VBox.__init__(self,
children = [
widgets.HBox(children =
[widgets.FloatText(value=0.0, layout=widgets.Layout(width='90px')) for i in range(m)]
)
for j in range(n)
])
#fill in widgets and tell interact to call updateM each time a children changes value
for irow in range(0,self.n):
for icol in range(0,self.m):
self.children[irow].children[icol].value = self.M_[irow,icol]
self.children[irow].children[icol].observe(self.updateM, names='value')
#value = Unicode('[email protected]', help="The email value.").tag(sync=True)
self.observe(self.updateM, names='value', type= 'All')
def setM(self, newM):
#disable callbacks, change values, and reenable
self.unobserve(self.updateM, names='value', type= 'All')
for irow in range(0,self.n):
for icol in range(0,self.m):
self.children[irow].children[icol].unobserve(self.updateM, names='value')
self.M_ = newM
self.value = self.M_
for irow in range(0,self.n):
for icol in range(0,self.m):
self.children[irow].children[icol].value = self.M_[irow,icol]
for irow in range(0,self.n):
for icol in range(0,self.m):
self.children[irow].children[icol].observe(self.updateM, names='value')
self.observe(self.updateM, names='value', type= 'All')
#self.children[irow].children[icol].observe(self.updateM, names='value')
#overlaod class for state space systems that DO NOT remove "useless" states (what "professor" of automatic control would do this?)
class sss(control.StateSpace):
def __init__(self,*args):
#call base class init constructor
control.StateSpace.__init__(self,*args)
#disable function below in base class
def _remove_useless_states(self):
pass
```
## Upravljanje putanjom zrakoplova
Dinamika rotacije zrakoplova koji se kreće po tlu može se predstaviti kao:
$$
J_z\ddot{\psi} = bF_1\delta + F_2\dot{\psi} \, ,
$$
gdje je $J_z = 11067000$ kg$\text{m}^2$, $b = 15$ , $F_1 = 35000000$ Nm, $F_2 = 500000$ kg$\text{m}^2$/$\text{s}$, $\psi$ je kut rotacije (u radijanima), ili kut zaošijanja u odnosu na vertikalnu os, a $\delta$ je kut upravljanja prednjih kotača (u radijanima). Kada zrakoplov slijedi ravnu liniju s uzdužnom linearnom brzinom $V$ (u m/s), bočna brzina zrakoplova $V_y$ (u m/s) približno je linearno proporcionalna kutu zaošijanja: $V_y = \dot{p_y} = V\psi$.
Cilj je dizajnirati regulator za bočni položaj zrakoplova $p_y$, s uzdužnom brzinom $V$ postavljenom na 35 km/h, koristeći kut upravljanja prednjeg kotača $\delta$ kao ulaz sustava, a prateći sljedeće specifikacije:
- vrijeme smirivanja za pojas tolerancije od 5% kraće od 4 sekunde;
- nulta pogreška u stacionarnom stanju u odzivu na promjenu željenog bočnog položaja;
- u potpunosti nema ili ima minimalnog prekoračenja;
- kut upravljanja ne prelazi $\pm8$ stupnjeva kada se prati promjena bočnog položaja od 5 metara.
Jednadžbe dinamike u formi prostora stanja su:
\begin{cases}
\dot{x} = \begin{bmatrix} \frac{F_2}{J_z} & 0 & 0 \\ 1 & 0 & 0 \\ 0 & V & 0 \end{bmatrix}x + \begin{bmatrix} \frac{bF_1}{J_z} \\ 0 \\ 0 \end{bmatrix}u \\
y = \begin{bmatrix} 0 & 0 & 1 \end{bmatrix}x \, ,
\end{cases}
gdje je $x=\begin{bmatrix} x_1 & x_2 & x_3 \end{bmatrix}^T = \begin{bmatrix} \dot{\psi} & \psi & p_y \end{bmatrix}^T$ i $u=\delta$.
Polovi sustava su $0$, $0$ i $\frac{F_2}{J_z} \simeq 0.045$, stoga je sustav nestabilan.
### Dizajn regulatora
#### Dizajn kontrolera
Da bismo udovoljili zahtjevima nulte pogreške stacionarnog stanja, dodajemo novo stanje:
$$
\dot{x_4} = p_y-y_d = x_3 - y_d
$$
Rezultirajući prošireni sustav je tada:
\begin{cases}
\dot{x_a} = \begin{bmatrix} \frac{F_2}{J_z} & 0 & 0 & 0 \\ 1 & 0 & 0 & 0 \\ 0 & V & 0 & 0 \\ 0 & 0 & 1 & 0 \end{bmatrix}x_a + \begin{bmatrix} \frac{bF_1}{J_z} & 0 \\ 0 & 0 \\ 0 & 0 \\ 0 & -1 \end{bmatrix}\begin{bmatrix} u \\ y_d \end{bmatrix} \\
y_a = \begin{bmatrix} 0 & 0 & 1 & 0 \\ 0 & 0 & 0 & 1 \end{bmatrix}x_a,
\end{cases}
gdje je $x_a = \begin{bmatrix} x_1 & x_2 & x_3 & x_4 \end{bmatrix}^T$ i dodaje se drugi izlaz kako bi se održala osmotrivost sustava. Sustav ostaje upravljiv s ulazom $u$ i tako, pomoću ovog ulaza, možemo oblikovati povratnu vezu stanja. Moguće rješenje je smjestiti sve polove u $-2$.
#### Dizajn promatrača
Čak i ako se iz mjerenja mogu dobiti stanja $x_3$ i $x_4$, a mi trebamo procijeniti samo $x_2$ i $x_3$, prikladno je raditi sa sustavom 4x4 i dizajnirati promatrač reda 4 sa svim polovima u $-10$.
### Kako koristiti ovaj interaktivni primjer?
- Provjerite jesu li ispunjeni zahtjevi ako postoje pogreške u procjeni početnog stanja.
```
# Preparatory cell
X0 = numpy.matrix('0.0; 0.0; 0.0; 0.0')
K = numpy.matrix([0,0,0,0])
L = numpy.matrix([[0,0],[0,0],[0,0],[0,0]])
X0w = matrixWidget(4,1)
X0w.setM(X0)
Kw = matrixWidget(1,4)
Kw.setM(K)
Lw = matrixWidget(4,2)
Lw.setM(L)
eig1c = matrixWidget(1,1)
eig2c = matrixWidget(2,1)
eig3c = matrixWidget(1,1)
eig4c = matrixWidget(2,1)
eig1c.setM(numpy.matrix([-2.]))
eig2c.setM(numpy.matrix([[-2.],[-0.]]))
eig3c.setM(numpy.matrix([-2.]))
eig4c.setM(numpy.matrix([[-2.],[-0.]]))
eig1o = matrixWidget(1,1)
eig2o = matrixWidget(2,1)
eig3o = matrixWidget(1,1)
eig4o = matrixWidget(2,1)
eig1o.setM(numpy.matrix([-10.]))
eig2o.setM(numpy.matrix([[-10.],[0.]]))
eig3o.setM(numpy.matrix([-10.]))
eig4o.setM(numpy.matrix([[-10.],[0.]]))
# Misc
#create dummy widget
DW = widgets.FloatText(layout=widgets.Layout(width='0px', height='0px'))
#create button widget
START = widgets.Button(
description='Test',
disabled=False,
button_style='', # 'success', 'info', 'warning', 'danger' or ''
tooltip='Test',
icon='check'
)
def on_start_button_clicked(b):
#This is a workaround to have intreactive_output call the callback:
# force the value of the dummy widget to change
if DW.value> 0 :
DW.value = -1
else:
DW.value = 1
pass
START.on_click(on_start_button_clicked)
# Define type of method
selm = widgets.Dropdown(
options= ['Postavi K i L', 'Postavi svojstvene vrijednosti'],
value= 'Postavi svojstvene vrijednosti',
description='',
disabled=False
)
# Define the number of complex eigenvalues
selec = widgets.Dropdown(
options= ['0 kompleksnih svojstvenih vrijednosti', '2 kompleksne svojstvene vrijednosti', '4 kompleksne svojstvene vrijednosti'],
value= '0 kompleksnih svojstvenih vrijednosti',
description='Svojstvene vrijednosti kontrolera:',
disabled=False
)
seleo = widgets.Dropdown(
options= ['0 kompleksnih svojstvenih vrijednosti', '2 kompleksne svojstvene vrijednosti', '4 kompleksne svojstvene vrijednosti'],
value= '0 kompleksnih svojstvenih vrijednosti',
description='Svojstvene vrijednosti promatrača:',
disabled=False
)
#define type of ipout
selu = widgets.Dropdown(
options=['impuls', 'step', 'sinus', 'Pravokutni val'],
value='step',
description='Tip referentnog signala:',
style = {'description_width': 'initial'},
disabled=False
)
# Define the values of the input
u = widgets.FloatSlider(
value=5,
min=0,
max=10,
step=0.1,
description='Referentni signal [m]:',
style = {'description_width': 'initial'},
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='.1f',
)
v = widgets.FloatSlider(
value=9.72,
min=1,
max=20,
step=0.1,
description=r'$V$ [m/s]:',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='.2f',
)
period = widgets.FloatSlider(
value=0.5,
min=0.001,
max=10,
step=0.001,
description='Period: ',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='.3f',
)
simTime = widgets.FloatText(
value=5,
description='',
disabled=False
)
# Support functions
def eigen_choice(selec,seleo):
if selec == '0 kompleksnih svojstvenih vrijednosti':
eig1c.children[0].children[0].disabled = False
eig2c.children[1].children[0].disabled = True
eig3c.children[0].children[0].disabled = False
eig4c.children[0].children[0].disabled = False
eig4c.children[1].children[0].disabled = True
eigc = 0
if seleo == '0 kompleksnih svojstvenih vrijednosti':
eig1o.children[0].children[0].disabled = False
eig2o.children[1].children[0].disabled = True
eig3o.children[0].children[0].disabled = False
eig4o.children[0].children[0].disabled = False
eig4o.children[1].children[0].disabled = True
eigo = 0
if selec == '2 kompleksne svojstvene vrijednosti':
eig1c.children[0].children[0].disabled = False
eig2c.children[1].children[0].disabled = False
eig3c.children[0].children[0].disabled = False
eig4c.children[0].children[0].disabled = True
eig4c.children[1].children[0].disabled = True
eigc = 2
if seleo == '2 kompleksne svojstvene vrijednosti':
eig1o.children[0].children[0].disabled = False
eig2o.children[1].children[0].disabled = False
eig3o.children[0].children[0].disabled = False
eig4o.children[0].children[0].disabled = True
eig4o.children[1].children[0].disabled = True
eigo = 2
if selec == '4 kompleksne svojstvene vrijednosti':
eig1c.children[0].children[0].disabled = True
eig2c.children[1].children[0].disabled = False
eig3c.children[0].children[0].disabled = True
eig4c.children[0].children[0].disabled = False
eig4c.children[1].children[0].disabled = False
eigc = 4
if seleo == '4 kompleksne svojstvene vrijednosti':
eig1o.children[0].children[0].disabled = True
eig2o.children[1].children[0].disabled = False
eig3o.children[0].children[0].disabled = True
eig4o.children[0].children[0].disabled = False
eig4o.children[1].children[0].disabled = False
eigo = 4
return eigc, eigo
def method_choice(selm):
if selm == 'Postavi K i L':
method = 1
selec.disabled = True
seleo.disabled = True
if selm == 'Postavi svojstvene vrijednosti':
method = 2
selec.disabled = False
seleo.disabled = False
return method
F1 = 35000000
F2 = 500000
b = 15
V = 35/3.6
Jz = 11067000
A = numpy.matrix([[F2/Jz, 0, 0, 0],
[1, 0, 0, 0],
[0, V, 0, 0],
[0, 0, 1, 0]])
Bu = numpy.matrix([[b*F1/Jz],[0],[0],[0]])
Bref = numpy.matrix([[0],[0],[0],[-1]])
C = numpy.matrix([[0,0,1,0],[0,0,0,1]])
def main_callback2(v, X0w, K, L, eig1c, eig2c, eig3c, eig4c, eig1o, eig2o, eig3o, eig4o, u, period, selm, selec, seleo, selu, simTime, DW):
eigc, eigo = eigen_choice(selec,seleo)
method = method_choice(selm)
A = numpy.matrix([[F2/Jz, 0, 0, 0],
[1, 0, 0, 0],
[0, v, 0, 0],
[0, 0, 1, 0]])
if method == 1:
solc = numpy.linalg.eig(A-Bu*K)
solo = numpy.linalg.eig(A-L*C)
if method == 2:
#for better numerical stability of place
if eig1c[0,0]==eig2c[0,0] or eig1c[0,0]==eig3c[0,0] or eig1c[0,0]==eig4c[0,0]:
eig1c[0,0] *= 1.01
if eig2c[0,0]==eig3c[0,0] or eig2c[0,0]==eig4c[0,0]:
eig3c[0,0] *= 1.015
if eig1o[0,0]==eig2o[0,0] or eig1o[0,0]==eig3o[0,0] or eig1o[0,0]==eig4o[0,0]:
eig1o[0,0] *= 1.01
if eig2o[0,0]==eig3o[0,0] or eig2o[0,0]==eig4o[0,0]:
eig3o[0,0] *= 1.015
if eigc == 0:
K = control.acker(A, Bu, [eig1c[0,0], eig2c[0,0], eig3c[0,0], eig4c[0,0]])
Kw.setM(K)
if eigc == 2:
K = control.acker(A, Bu, [eig3c[0,0],
eig1c[0,0],
numpy.complex(eig2c[0,0], eig2c[1,0]),
numpy.complex(eig2c[0,0],-eig2c[1,0])])
Kw.setM(K)
if eigc == 4:
K = control.acker(A, Bu, [numpy.complex(eig4c[0,0], eig4c[1,0]),
numpy.complex(eig4c[0,0],-eig4c[1,0]),
numpy.complex(eig2c[0,0], eig2c[1,0]),
numpy.complex(eig2c[0,0],-eig2c[1,0])])
Kw.setM(K)
if eigo == 0:
L = control.place(A.T, C.T, [eig1o[0,0], eig2o[0,0], eig3o[0,0], eig4o[0,0]]).T
Lw.setM(L)
if eigo == 2:
L = control.place(A.T, C.T, [eig3o[0,0],
eig1o[0,0],
numpy.complex(eig2o[0,0], eig2o[1,0]),
numpy.complex(eig2o[0,0],-eig2o[1,0])]).T
Lw.setM(L)
if eigo == 4:
L = control.place(A.T, C.T, [numpy.complex(eig4o[0,0], eig4o[1,0]),
numpy.complex(eig4o[0,0],-eig4o[1,0]),
numpy.complex(eig2o[0,0], eig2o[1,0]),
numpy.complex(eig2o[0,0],-eig2o[1,0])]).T
Lw.setM(L)
sys = sss(A,numpy.hstack((Bu,Bref)),[[0,0,1,0],[0,0,0,1],[0,0,0,0]],[[0,0],[0,0],[0,1]])
syse = sss(A-L*C,numpy.hstack((Bu,Bref,L)),numpy.eye(4),numpy.zeros((4,4)))
sysc = sss(0,[0,0,0,0],0,-K)
sys_append = control.append(sys,syse,sysc)
try:
sys_CL = control.connect(sys_append,
[[1,8],[3,8],[5,1],[6,2],[7,4],[8,5],[9,6],[10,7],[4,3]],
[2],
[1,8])
except:
sys_CL = control.connect(sys_append,
[[1,8],[3,8],[5,1],[6,2],[7,4],[8,5],[9,6],[10,7],[4,3]],
[2],
[1,8])
X0w1 = numpy.zeros((8,1))
X0w1[4,0] = X0w[0,0]
X0w1[5,0] = X0w[1,0]
X0w1[6,0] = X0w[2,0]
X0w1[7,0] = X0w[3,0]
if simTime != 0:
T = numpy.linspace(0, simTime, 10000)
else:
T = numpy.linspace(0, 1, 10000)
if selu == 'impuls': #selu
U = [0 for t in range(0,len(T))]
U[0] = u
T, yout, xout = control.forced_response(sys_CL,T,U,X0w1)
if selu == 'step':
U = [u for t in range(0,len(T))]
T, yout, xout = control.forced_response(sys_CL,T,U,X0w1)
if selu == 'sinus':
U = u*numpy.sin(2*numpy.pi/period*T)
T, yout, xout = control.forced_response(sys_CL,T,U,X0w1)
if selu == 'Pravokutni val':
U = u*numpy.sign(numpy.sin(2*numpy.pi/period*T))
T, yout, xout = control.forced_response(sys_CL,T,U,X0w1)
try:
step_info_dict = control.step_info(sys_CL[0,0],SettlingTimeThreshold=0.05,T=T)
print('Informacije o koraku: \n\tVrijeme porasta =',step_info_dict['RiseTime'],'\n\tVrijeme smirivanja (5%) =',step_info_dict['SettlingTime'],'\n\tPrekoračenje (%)=',step_info_dict['Overshoot'])
print('Maksimalna u vrijednost (% od 8deg)=', max(abs(yout[1]))/(8*numpy.pi/180)*100)
except:
print("Pogreška u izračunu informacija o koraku.")
fig = plt.figure(num='Simulation1', figsize=(14,12))
fig.add_subplot(221)
plt.title('Izlazni odziv')
plt.ylabel('Izlaz')
plt.plot(T,yout[0],T,U,'r--')
plt.xlabel('$t$ [s]')
plt.legend(['$y$','Referentni signal'])
plt.axvline(x=0,color='black',linewidth=0.8)
plt.axhline(y=0,color='black',linewidth=0.8)
plt.grid()
fig.add_subplot(222)
plt.title('Ulaz')
plt.ylabel('$u$ [deg]')
plt.plot(T,yout[1]*180/numpy.pi)
plt.plot(T,[8 for i in range(len(T))],'r--')
plt.plot(T,[-8 for i in range(len(T))],'r--')
plt.xlabel('$t$ [s]')
plt.axvline(x=0,color='black',linewidth=0.8)
plt.axhline(y=0,color='black',linewidth=0.8)
plt.grid()
fig.add_subplot(223)
plt.title('Odziv stanja')
plt.ylabel('Stanja')
plt.plot(T,xout[0],
T,xout[1],
T,xout[2],
T,xout[3])
plt.xlabel('$t$ [s]')
plt.axvline(x=0,color='black',linewidth=0.8)
plt.axhline(y=0,color='black',linewidth=0.8)
plt.legend(['$x_{1}$','$x_{2}$','$x_{3}$','$x_{4}$'])
plt.grid()
fig.add_subplot(224)
plt.title('Pogreška procjene')
plt.ylabel('Pogreška')
plt.plot(T,xout[4]-xout[0])
plt.plot(T,xout[5]-xout[1])
plt.plot(T,xout[6]-xout[2])
plt.plot(T,xout[7]-xout[3])
plt.xlabel('$t$ [s]')
plt.axvline(x=0,color='black',linewidth=0.8)
plt.axhline(y=0,color='black',linewidth=0.8)
plt.legend(['$e_{1}$','$e_{2}$','$e_{3}$','$e_{4}$'])
plt.grid()
#plt.tight_layout()
alltogether2 = widgets.VBox([widgets.HBox([selm,
selec,
seleo,
selu]),
widgets.Label(' ',border=3),
widgets.HBox([widgets.HBox([widgets.Label('K:',border=3), Kw,
widgets.Label('Svojstvene vrijednosti:',border=3),
widgets.HBox([eig1c,
eig2c,
eig3c,
eig4c])])]),
widgets.Label(' ',border=3),
widgets.HBox([widgets.VBox([widgets.HBox([widgets.Label('L:',border=3), Lw, widgets.Label(' ',border=3),
widgets.Label(' ',border=3),
widgets.Label('Svojstvene vrijednosti:',border=3),
eig1o,
eig2o,
eig3o,
eig4o,
widgets.Label(' ',border=3),
widgets.Label(' ',border=3),
widgets.Label('X0 est.:',border=3), X0w]),
widgets.Label(' ',border=3),
widgets.HBox([
widgets.VBox([widgets.Label('Vrijeme simulacije [s]:',border=3)]),
widgets.VBox([simTime])])]),
widgets.Label(' ',border=3)]),
widgets.Label(' ',border=3),
widgets.HBox([u,
v,
period,
START])])
out2 = widgets.interactive_output(main_callback2, {'v':v, 'X0w':X0w, 'K':Kw, 'L':Lw,
'eig1c':eig1c, 'eig2c':eig2c, 'eig3c':eig3c, 'eig4c':eig4c,
'eig1o':eig1o, 'eig2o':eig2o, 'eig3o':eig3o, 'eig4o':eig4o,
'u':u, 'period':period, 'selm':selm, 'selec':selec, 'seleo':seleo, 'selu':selu, 'simTime':simTime, 'DW':DW})
out2.layout.height = '860px'
display(out2, alltogether2)
```
|
github_jupyter
|
## Search algorithms within Optuna
In this notebook, I will demo how to select the search algorithm with Optuna. We will compare the use of:
- Grid Search
- Randomized search
- Tree-structured Parzen Estimators
- CMA-ES
We can select the search algorithm from the [optuna.study.create_study()](https://optuna.readthedocs.io/en/stable/reference/generated/optuna.study.create_study.html#optuna.study.create_study) class.
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.metrics import accuracy_score, roc_auc_score
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.ensemble import RandomForestClassifier
import optuna
# load dataset
breast_cancer_X, breast_cancer_y = load_breast_cancer(return_X_y=True)
X = pd.DataFrame(breast_cancer_X)
y = pd.Series(breast_cancer_y).map({0:1, 1:0})
X.head()
# the target:
# percentage of benign (0) and malign tumors (1)
y.value_counts() / len(y)
# split dataset into a train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0)
X_train.shape, X_test.shape
```
## Define the objective function
This is the hyperparameter response space, the function we want to minimize.
```
# the objective function takes the hyperparameter space
# as input
def objective(trial):
rf_n_estimators = trial.suggest_int("rf_n_estimators", 100, 1000)
rf_criterion = trial.suggest_categorical("rf_criterion", ['gini', 'entropy'])
rf_max_depth = trial.suggest_int("rf_max_depth", 1, 4)
rf_min_samples_split = trial.suggest_float("rf_min_samples_split", 0.01, 1)
model = RandomForestClassifier(
n_estimators=rf_n_estimators,
criterion=rf_criterion,
max_depth=rf_max_depth,
min_samples_split=rf_min_samples_split,
)
score = cross_val_score(model, X_train, y_train, cv=3)
accuracy = score.mean()
return accuracy
```
## Randomized Search
RandomSampler()
```
study = optuna.create_study(
direction="maximize",
sampler=optuna.samplers.RandomSampler(),
)
study.optimize(objective, n_trials=5)
study.best_params
study.best_value
study.trials_dataframe()
```
## TPE
TPESampler is the default
```
study = optuna.create_study(
direction="maximize",
sampler=optuna.samplers.TPESampler(),
)
study.optimize(objective, n_trials=5)
study.best_params
study.best_value
```
## CMA-ES
CmaEsSampler
```
study = optuna.create_study(
direction="maximize",
sampler=optuna.samplers.CmaEsSampler(),
)
study.optimize(objective, n_trials=5)
study.best_params
study.best_value
```
## Grid Search
GridSampler()
We are probably not going to perform GridSearch with Optuna, but in case you wanted to, you need to add a variable with the space, with the exact values that you want to be tested.
```
search_space = {
"rf_n_estimators": [100, 500, 1000],
"rf_criterion": ['gini', 'entropy'],
"rf_max_depth": [1, 2, 3],
"rf_min_samples_split": [0.1, 1.0]
}
study = optuna.create_study(
direction="maximize",
sampler=optuna.samplers.GridSampler(search_space),
)
study.optimize(objective)
study.best_params
study.best_value
```
|
github_jupyter
|
## Duplicated features
```
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
```
## Read Data
```
data = pd.read_csv('../UNSW_Train.csv')
data.shape
# check the presence of missing data.
# (there are no missing data in this dataset)
[col for col in data.columns if data[col].isnull().sum() > 0]
data.head(5)
```
### Train - Test Split
```
# separate dataset into train and test
X_train, X_test, y_train, y_test = train_test_split(
data.drop(labels=['is_intrusion'], axis=1), # drop the target
data['is_intrusion'], # just the target
test_size=0.2,
random_state=0)
X_train.shape, X_test.shape
```
## Remove constant and quasi-constant (optional)
```
# remove constant and quasi-constant features first:
# we can remove the 2 types of features together with this code
# create an empty list
quasi_constant_feat = []
# iterate over every feature
for feature in X_train.columns:
# find the predominant value, that is the value that is shared
# by most observations
predominant = (X_train[feature].value_counts() / np.float64(
len(X_train))).sort_values(ascending=False).values[0]
# evaluate predominant feature: do more than 99% of the observations
# show 1 value?
if predominant > 0.998:
quasi_constant_feat.append(feature)
len(quasi_constant_feat)
quasi_constant_feat
# we can then drop these columns from the train and test sets:
X_train.drop(labels=quasi_constant_feat, axis=1, inplace=True)
X_test.drop(labels=quasi_constant_feat, axis=1, inplace=True)
X_train.shape, X_test.shape
```
## Remove duplicated features
```
# fiding duplicated features
duplicated_feat_pairs = {}
_duplicated_feat = []
for i in range(0, len(X_train.columns)):
if i % 10 == 0:
print(i)
feat_1 = X_train.columns[i]
if feat_1 not in _duplicated_feat:
duplicated_feat_pairs[feat_1] = []
for feat_2 in X_train.columns[i + 1:]:
if X_train[feat_1].equals(X_train[feat_2]):
duplicated_feat_pairs[feat_1].append(feat_2)
_duplicated_feat.append(feat_2)
# let's explore our list of duplicated features
len(_duplicated_feat)
```
We found 1 features that were duplicates of others.
```
# these are the ones:
_duplicated_feat
# let's explore the dictionary we created:
duplicated_feat_pairs
```
We see that for every feature, if it had duplicates, we have entries in the list, otherwise, we have empty lists. Let's explore those features with duplicates now:
```
# let's explore the number of keys in our dictionary
# we see it is 21, because 2 of the 23 were duplicates,
# so they were not included as keys
print(len(duplicated_feat_pairs.keys()))
# print the features with its duplicates
# iterate over every feature in our dict:
for feat in duplicated_feat_pairs.keys():
# if it has duplicates, the list should not be empty:
if len(duplicated_feat_pairs[feat]) > 0:
# print the feature and its duplicates:
print(feat, duplicated_feat_pairs[feat])
print()
# to remove the duplicates (if necessary)
X_train = X_train[duplicated_feat_pairs.keys()]
X_test = X_test[duplicated_feat_pairs.keys()]
X_train.shape, X_test.shape
```
1 duplicate features were found in the UNSW-NB15 dataset
## Standardize Data
```
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
```
## Classifiers
```
from sklearn import linear_model
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from catboost import CatBoostClassifier
```
## Metrics Evaluation
```
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import roc_curve, f1_score
from sklearn import metrics
from sklearn.model_selection import cross_val_score
```
### Logistic Regression
```
%%time
clf_LR = linear_model.LogisticRegression(n_jobs=-1, random_state=42, C=25).fit(X_train, y_train)
pred_y_test = clf_LR.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_test))
f1 = f1_score(y_test, pred_y_test)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_test)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
```
### Naive Bayes
```
%%time
clf_NB = GaussianNB(var_smoothing=1e-08).fit(X_train, y_train)
pred_y_testNB = clf_NB.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_testNB))
f1 = f1_score(y_test, pred_y_testNB)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testNB)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
```
### Random Forest
```
%%time
clf_RF = RandomForestClassifier(random_state=0,max_depth=100,n_estimators=1000).fit(X_train, y_train)
pred_y_testRF = clf_RF.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_testRF))
f1 = f1_score(y_test, pred_y_testRF, average='weighted', zero_division=0)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testRF)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
```
### KNN
```
%%time
clf_KNN = KNeighborsClassifier(algorithm='ball_tree',leaf_size=1,n_neighbors=5,weights='uniform').fit(X_train, y_train)
pred_y_testKNN = clf_KNN.predict(X_test)
print('accuracy_score:', accuracy_score(y_test, pred_y_testKNN))
f1 = f1_score(y_test, pred_y_testKNN)
print('f1:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testKNN)
print('fpr:', fpr[1])
print('tpr:', tpr[1])
```
### CatBoost
```
%%time
clf_CB = CatBoostClassifier(depth=7,iterations=50,learning_rate=0.04).fit(X_train, y_train)
pred_y_testCB = clf_CB.predict(X_test)
print('Accuracy:', accuracy_score(y_test, pred_y_testCB))
f1 = f1_score(y_test, pred_y_testCB, average='weighted', zero_division=0)
print('F1 Score:', f1)
fpr, tpr, thresholds = roc_curve(y_test, pred_y_testCB)
print('FPR:', fpr[1])
print('TPR:', tpr[1])
```
## Model Evaluation
```
import pandas as pd, numpy as np
test_df = pd.read_csv("../UNSW_Test.csv")
test_df.shape
# Create feature matrix X and target vextor y
y_eval = test_df['is_intrusion']
X_eval = test_df.drop(columns=['is_intrusion','ct_ftp_cmd'])
```
### Model Evaluation - Logistic Regression
```
modelLR = linear_model.LogisticRegression(n_jobs=-1, random_state=42, C=25)
modelLR.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredLR = modelLR.predict(X_eval)
y_predLR = modelLR.predict(X_test)
train_scoreLR = modelLR.score(X_train, y_train)
test_scoreLR = modelLR.score(X_test, y_test)
print("Training accuracy is ", train_scoreLR)
print("Testing accuracy is ", test_scoreLR)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreLR)
print('F1 Score:',f1_score(y_test, y_predLR))
print('Precision Score:',precision_score(y_test, y_predLR))
print('Recall Score:', recall_score(y_test, y_predLR))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predLR))
```
### Cross validation - Logistic Regression
```
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelLR, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
```
### Model Evaluation - Naive Bayes
```
modelNB = GaussianNB(var_smoothing=1e-08)
modelNB.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredNB = modelNB.predict(X_eval)
y_predNB = modelNB.predict(X_test)
train_scoreNB = modelNB.score(X_train, y_train)
test_scoreNB = modelNB.score(X_test, y_test)
print("Training accuracy is ", train_scoreNB)
print("Testing accuracy is ", test_scoreNB)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreNB)
print('F1 Score:',f1_score(y_test, y_predNB))
print('Precision Score:',precision_score(y_test, y_predNB))
print('Recall Score:', recall_score(y_test, y_predNB))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predNB))
```
### Cross validation - Naive Bayes
```
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelNB, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
```
### Model Evaluation - Random Forest
```
modelRF = RandomForestClassifier(random_state=0,max_depth=100,n_estimators=1000)
modelRF.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredRF = modelRF.predict(X_eval)
y_predRF = modelRF.predict(X_test)
train_scoreRF = modelRF.score(X_train, y_train)
test_scoreRF = modelRF.score(X_test, y_test)
print("Training accuracy is ", train_scoreRF)
print("Testing accuracy is ", test_scoreRF)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreRF)
print('F1 Score:', f1_score(y_test, y_predRF, average='weighted', zero_division=0))
print('Precision Score:', precision_score(y_test, y_predRF, average='weighted', zero_division=0))
print('Recall Score:', recall_score(y_test, y_predRF, average='weighted', zero_division=0))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predRF))
```
### Cross validation - Random Forest
```
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelRF, X_eval, y_eval, cv=5, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelRF, X_eval, y_eval, cv=5, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelRF, X_eval, y_eval, cv=5, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelRF, X_eval, y_eval, cv=5, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
```
### Model Evaluation - KNN
```
modelKNN = KNeighborsClassifier(algorithm='ball_tree',leaf_size=1,n_neighbors=5,weights='uniform')
modelKNN.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredKNN = modelKNN.predict(X_eval)
y_predKNN = modelKNN.predict(X_test)
train_scoreKNN = modelKNN.score(X_train, y_train)
test_scoreKNN = modelKNN.score(X_test, y_test)
print("Training accuracy is ", train_scoreKNN)
print("Testing accuracy is ", test_scoreKNN)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreKNN)
print('F1 Score:', f1_score(y_test, y_predKNN))
print('Precision Score:', precision_score(y_test, y_predKNN))
print('Recall Score:', recall_score(y_test, y_predKNN))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predKNN))
```
### Cross validation - KNN
```
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='accuracy')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
f = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='f1')
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
precision = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='precision')
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
recall = cross_val_score(modelKNN, X_eval, y_eval, cv=10, scoring='recall')
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
```
### Model Evaluation - CatBoost
```
modelCB = CatBoostClassifier(depth=7,iterations=50,learning_rate=0.04)
modelCB.fit(X_train, y_train)
# Predict on the new unseen test data
y_evalpredCB = modelCB.predict(X_eval)
y_predCB = modelCB.predict(X_test)
train_scoreCB = modelCB.score(X_train, y_train)
test_scoreCB = modelCB.score(X_test, y_test)
print("Training accuracy is ", train_scoreCB)
print("Testing accuracy is ", test_scoreCB)
from sklearn.metrics import confusion_matrix, precision_score, recall_score, f1_score
print('Performance measures for test:')
print('--------')
print('Accuracy:', test_scoreCB)
print('F1 Score:',f1_score(y_test, y_predCB, average='weighted', zero_division=0))
print('Precision Score:',precision_score(y_test, y_predCB, average='weighted', zero_division=0))
print('Recall Score:', recall_score(y_test, y_predCB, average='weighted', zero_division=0))
print('Confusion Matrix:\n', confusion_matrix(y_test, y_predCB))
```
### Cross validation - CatBoost
```
from sklearn.model_selection import cross_val_score
from sklearn import metrics
accuracy = cross_val_score(modelCB, X_eval, y_eval, cv=5, scoring='accuracy')
f = cross_val_score(modelCB, X_eval, y_eval, cv=5, scoring='f1')
precision = cross_val_score(modelCB, X_eval, y_eval, cv=5, scoring='precision')
recall = cross_val_score(modelCB, X_eval, y_eval, cv=5, scoring='recall')
print("Accuracy: %0.5f (+/- %0.5f)" % (accuracy.mean(), accuracy.std() * 2))
print("F1 Score: %0.5f (+/- %0.5f)" % (f.mean(), f.std() * 2))
print("Precision: %0.5f (+/- %0.5f)" % (precision.mean(), precision.std() * 2))
print("Recall: %0.5f (+/- %0.5f)" % (recall.mean(), recall.std() * 2))
```
|
github_jupyter
|
# Python Basics with Numpy (optional assignment)
Welcome to your first assignment. This exercise gives you a brief introduction to Python. Even if you've used Python before, this will help familiarize you with functions we'll need.
**Instructions:**
- You will be using Python 3.
- Avoid using for-loops and while-loops, unless you are explicitly told to do so.
- Do not modify the (# GRADED FUNCTION [function name]) comment in some cells. Your work would not be graded if you change this. Each cell containing that comment should only contain one function.
- After coding your function, run the cell right below it to check if your result is correct.
**After this assignment you will:**
- Be able to use iPython Notebooks
- Be able to use numpy functions and numpy matrix/vector operations
- Understand the concept of "broadcasting"
- Be able to vectorize code
Let's get started!
## About iPython Notebooks ##
iPython Notebooks are interactive coding environments embedded in a webpage. You will be using iPython notebooks in this class. You only need to write code between the ### START CODE HERE ### and ### END CODE HERE ### comments. After writing your code, you can run the cell by either pressing "SHIFT"+"ENTER" or by clicking on "Run Cell" (denoted by a play symbol) in the upper bar of the notebook.
We will often specify "(≈ X lines of code)" in the comments to tell you about how much code you need to write. It is just a rough estimate, so don't feel bad if your code is longer or shorter.
**Exercise**: Set test to `"Hello World"` in the cell below to print "Hello World" and run the two cells below.
```
### START CODE HERE ### (≈ 1 line of code)
test = "Hello World"
### END CODE HERE ###
print ("test: " + test)
```
**Expected output**:
test: Hello World
<font color='blue'>
**What you need to remember**:
- Run your cells using SHIFT+ENTER (or "Run cell")
- Write code in the designated areas using Python 3 only
- Do not modify the code outside of the designated areas
## 1 - Building basic functions with numpy ##
Numpy is the main package for scientific computing in Python. It is maintained by a large community (www.numpy.org). In this exercise you will learn several key numpy functions such as np.exp, np.log, and np.reshape. You will need to know how to use these functions for future assignments.
### 1.1 - sigmoid function, np.exp() ###
Before using np.exp(), you will use math.exp() to implement the sigmoid function. You will then see why np.exp() is preferable to math.exp().
**Exercise**: Build a function that returns the sigmoid of a real number x. Use math.exp(x) for the exponential function.
**Reminder**:
$sigmoid(x) = \frac{1}{1+e^{-x}}$ is sometimes also known as the logistic function. It is a non-linear function used not only in Machine Learning (Logistic Regression), but also in Deep Learning.
<img src="images/Sigmoid.png" style="width:500px;height:228px;">
To refer to a function belonging to a specific package you could call it using package_name.function(). Run the code below to see an example with math.exp().
```
# GRADED FUNCTION: basic_sigmoid
import math
def basic_sigmoid(x):
"""
Compute sigmoid of x.
Arguments:
x -- A scalar
Return:
s -- sigmoid(x)
"""
### START CODE HERE ### (≈ 1 line of code)
s = 1/(1+math.exp(-x))
### END CODE HERE ###
return s
basic_sigmoid(3)
```
**Expected Output**:
<table style = "width:40%">
<tr>
<td>** basic_sigmoid(3) **</td>
<td>0.9525741268224334 </td>
</tr>
</table>
Actually, we rarely use the "math" library in deep learning because the inputs of the functions are real numbers. In deep learning we mostly use matrices and vectors. This is why numpy is more useful.
```
### One reason why we use "numpy" instead of "math" in Deep Learning ###
x = [1, 2, 3]
basic_sigmoid(x) # you will see this give an error when you run it, because x is a vector.
```
In fact, if $ x = (x_1, x_2, ..., x_n)$ is a row vector then $np.exp(x)$ will apply the exponential function to every element of x. The output will thus be: $np.exp(x) = (e^{x_1}, e^{x_2}, ..., e^{x_n})$
```
import numpy as np
# example of np.exp
x = np.array([1, 2, 3])
print(np.exp(x)) # result is (exp(1), exp(2), exp(3))
```
Furthermore, if x is a vector, then a Python operation such as $s = x + 3$ or $s = \frac{1}{x}$ will output s as a vector of the same size as x.
```
# example of vector operation
x = np.array([1, 2, 3])
print (x + 3)
```
Any time you need more info on a numpy function, we encourage you to look at [the official documentation](https://docs.scipy.org/doc/numpy-1.10.1/reference/generated/numpy.exp.html).
You can also create a new cell in the notebook and write `np.exp?` (for example) to get quick access to the documentation.
**Exercise**: Implement the sigmoid function using numpy.
**Instructions**: x could now be either a real number, a vector, or a matrix. The data structures we use in numpy to represent these shapes (vectors, matrices...) are called numpy arrays. You don't need to know more for now.
$$ \text{For } x \in \mathbb{R}^n \text{, } sigmoid(x) = sigmoid\begin{pmatrix}
x_1 \\
x_2 \\
... \\
x_n \\
\end{pmatrix} = \begin{pmatrix}
\frac{1}{1+e^{-x_1}} \\
\frac{1}{1+e^{-x_2}} \\
... \\
\frac{1}{1+e^{-x_n}} \\
\end{pmatrix}\tag{1} $$
```
# GRADED FUNCTION: sigmoid
import numpy as np # this means you can access numpy functions by writing np.function() instead of numpy.function()
def sigmoid(x):
"""
Compute the sigmoid of x
Arguments:
x -- A scalar or numpy array of any size
Return:
s -- sigmoid(x)
"""
### START CODE HERE ### (≈ 1 line of code)
s = 1/(1+np.exp(-x))
### END CODE HERE ###
return s
x = np.array([1, 2, 3])
sigmoid(x)
```
**Expected Output**:
<table>
<tr>
<td> **sigmoid([1,2,3])**</td>
<td> array([ 0.73105858, 0.88079708, 0.95257413]) </td>
</tr>
</table>
### 1.2 - Sigmoid gradient
As you've seen in lecture, you will need to compute gradients to optimize loss functions using backpropagation. Let's code your first gradient function.
**Exercise**: Implement the function sigmoid_grad() to compute the gradient of the sigmoid function with respect to its input x. The formula is: $$sigmoid\_derivative(x) = \sigma'(x) = \sigma(x) (1 - \sigma(x))\tag{2}$$
You often code this function in two steps:
1. Set s to be the sigmoid of x. You might find your sigmoid(x) function useful.
2. Compute $\sigma'(x) = s(1-s)$
```
# GRADED FUNCTION: sigmoid_derivative
def sigmoid_derivative(x):
"""
Compute the gradient (also called the slope or derivative) of the sigmoid function with respect to its input x.
You can store the output of the sigmoid function into variables and then use it to calculate the gradient.
Arguments:
x -- A scalar or numpy array
Return:
ds -- Your computed gradient.
"""
### START CODE HERE ### (≈ 2 lines of code)
s = sigmoid(x)
ds = s*(1-s)
### END CODE HERE ###
return ds
x = np.array([1, 2, 3])
print ("sigmoid_derivative(x) = " + str(sigmoid_derivative(x)))
```
**Expected Output**:
<table>
<tr>
<td> **sigmoid_derivative([1,2,3])**</td>
<td> [ 0.19661193 0.10499359 0.04517666] </td>
</tr>
</table>
### 1.3 - Reshaping arrays ###
Two common numpy functions used in deep learning are [np.shape](https://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.shape.html) and [np.reshape()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.reshape.html).
- X.shape is used to get the shape (dimension) of a matrix/vector X.
- X.reshape(...) is used to reshape X into some other dimension.
For example, in computer science, an image is represented by a 3D array of shape $(length, height, depth = 3)$. However, when you read an image as the input of an algorithm you convert it to a vector of shape $(length*height*3, 1)$. In other words, you "unroll", or reshape, the 3D array into a 1D vector.
<img src="images/image2vector_kiank.png" style="width:500px;height:300;">
**Exercise**: Implement `image2vector()` that takes an input of shape (length, height, 3) and returns a vector of shape (length\*height\*3, 1). For example, if you would like to reshape an array v of shape (a, b, c) into a vector of shape (a*b,c) you would do:
``` python
v = v.reshape((v.shape[0]*v.shape[1], v.shape[2])) # v.shape[0] = a ; v.shape[1] = b ; v.shape[2] = c
```
- Please don't hardcode the dimensions of image as a constant. Instead look up the quantities you need with `image.shape[0]`, etc.
```
# GRADED FUNCTION: image2vector
def image2vector(image):
"""
Argument:
image -- a numpy array of shape (length, height, depth)
Returns:
v -- a vector of shape (length*height*depth, 1)
"""
### START CODE HERE ### (≈ 1 line of code)
v = image.reshape((image.shape[0]*image.shape[1]*image.shape[2], 1))
### END CODE HERE ###
return v
# This is a 3 by 3 by 2 array, typically images will be (num_px_x, num_px_y,3) where 3 represents the RGB values
image = np.array([[[ 0.67826139, 0.29380381],
[ 0.90714982, 0.52835647],
[ 0.4215251 , 0.45017551]],
[[ 0.92814219, 0.96677647],
[ 0.85304703, 0.52351845],
[ 0.19981397, 0.27417313]],
[[ 0.60659855, 0.00533165],
[ 0.10820313, 0.49978937],
[ 0.34144279, 0.94630077]]])
print ("image2vector(image) = " + str(image2vector(image)))
```
**Expected Output**:
<table style="width:100%">
<tr>
<td> **image2vector(image)** </td>
<td> [[ 0.67826139]
[ 0.29380381]
[ 0.90714982]
[ 0.52835647]
[ 0.4215251 ]
[ 0.45017551]
[ 0.92814219]
[ 0.96677647]
[ 0.85304703]
[ 0.52351845]
[ 0.19981397]
[ 0.27417313]
[ 0.60659855]
[ 0.00533165]
[ 0.10820313]
[ 0.49978937]
[ 0.34144279]
[ 0.94630077]]</td>
</tr>
</table>
### 1.4 - Normalizing rows
Another common technique we use in Machine Learning and Deep Learning is to normalize our data. It often leads to a better performance because gradient descent converges faster after normalization. Here, by normalization we mean changing x to $ \frac{x}{\| x\|} $ (dividing each row vector of x by its norm).
For example, if $$x =
\begin{bmatrix}
0 & 3 & 4 \\
2 & 6 & 4 \\
\end{bmatrix}\tag{3}$$ then $$\| x\| = np.linalg.norm(x, axis = 1, keepdims = True) = \begin{bmatrix}
5 \\
\sqrt{56} \\
\end{bmatrix}\tag{4} $$and $$ x\_normalized = \frac{x}{\| x\|} = \begin{bmatrix}
0 & \frac{3}{5} & \frac{4}{5} \\
\frac{2}{\sqrt{56}} & \frac{6}{\sqrt{56}} & \frac{4}{\sqrt{56}} \\
\end{bmatrix}\tag{5}$$ Note that you can divide matrices of different sizes and it works fine: this is called broadcasting and you're going to learn about it in part 5.
**Exercise**: Implement normalizeRows() to normalize the rows of a matrix. After applying this function to an input matrix x, each row of x should be a vector of unit length (meaning length 1).
```
# GRADED FUNCTION: normalizeRows
def normalizeRows(x):
"""
Implement a function that normalizes each row of the matrix x (to have unit length).
Argument:
x -- A numpy matrix of shape (n, m)
Returns:
x -- The normalized (by row) numpy matrix. You are allowed to modify x.
"""
### START CODE HERE ### (≈ 2 lines of code)
# Compute x_norm as the norm 2 of x. Use np.linalg.norm(..., ord = 2, axis = ..., keepdims = True)
x_norm = np.linalg.norm(x,ord=2,axis=1,keepdims = True)
# Divide x by its norm.
x = x/x_norm
### END CODE HERE ###
return x
x = np.array([
[0, 3, 4],
[1, 6, 4]])
print("normalizeRows(x) = " + str(normalizeRows(x)))
```
**Expected Output**:
<table style="width:60%">
<tr>
<td> **normalizeRows(x)** </td>
<td> [[ 0. 0.6 0.8 ]
[ 0.13736056 0.82416338 0.54944226]]</td>
</tr>
</table>
**Note**:
In normalizeRows(), you can try to print the shapes of x_norm and x, and then rerun the assessment. You'll find out that they have different shapes. This is normal given that x_norm takes the norm of each row of x. So x_norm has the same number of rows but only 1 column. So how did it work when you divided x by x_norm? This is called broadcasting and we'll talk about it now!
### 1.5 - Broadcasting and the softmax function ####
A very important concept to understand in numpy is "broadcasting". It is very useful for performing mathematical operations between arrays of different shapes. For the full details on broadcasting, you can read the official [broadcasting documentation](http://docs.scipy.org/doc/numpy/user/basics.broadcasting.html).
**Exercise**: Implement a softmax function using numpy. You can think of softmax as a normalizing function used when your algorithm needs to classify two or more classes. You will learn more about softmax in the second course of this specialization.
**Instructions**:
- $ \text{for } x \in \mathbb{R}^{1\times n} \text{, } softmax(x) = softmax(\begin{bmatrix}
x_1 &&
x_2 &&
... &&
x_n
\end{bmatrix}) = \begin{bmatrix}
\frac{e^{x_1}}{\sum_{j}e^{x_j}} &&
\frac{e^{x_2}}{\sum_{j}e^{x_j}} &&
... &&
\frac{e^{x_n}}{\sum_{j}e^{x_j}}
\end{bmatrix} $
- $\text{for a matrix } x \in \mathbb{R}^{m \times n} \text{, $x_{ij}$ maps to the element in the $i^{th}$ row and $j^{th}$ column of $x$, thus we have: }$ $$softmax(x) = softmax\begin{bmatrix}
x_{11} & x_{12} & x_{13} & \dots & x_{1n} \\
x_{21} & x_{22} & x_{23} & \dots & x_{2n} \\
\vdots & \vdots & \vdots & \ddots & \vdots \\
x_{m1} & x_{m2} & x_{m3} & \dots & x_{mn}
\end{bmatrix} = \begin{bmatrix}
\frac{e^{x_{11}}}{\sum_{j}e^{x_{1j}}} & \frac{e^{x_{12}}}{\sum_{j}e^{x_{1j}}} & \frac{e^{x_{13}}}{\sum_{j}e^{x_{1j}}} & \dots & \frac{e^{x_{1n}}}{\sum_{j}e^{x_{1j}}} \\
\frac{e^{x_{21}}}{\sum_{j}e^{x_{2j}}} & \frac{e^{x_{22}}}{\sum_{j}e^{x_{2j}}} & \frac{e^{x_{23}}}{\sum_{j}e^{x_{2j}}} & \dots & \frac{e^{x_{2n}}}{\sum_{j}e^{x_{2j}}} \\
\vdots & \vdots & \vdots & \ddots & \vdots \\
\frac{e^{x_{m1}}}{\sum_{j}e^{x_{mj}}} & \frac{e^{x_{m2}}}{\sum_{j}e^{x_{mj}}} & \frac{e^{x_{m3}}}{\sum_{j}e^{x_{mj}}} & \dots & \frac{e^{x_{mn}}}{\sum_{j}e^{x_{mj}}}
\end{bmatrix} = \begin{pmatrix}
softmax\text{(first row of x)} \\
softmax\text{(second row of x)} \\
... \\
softmax\text{(last row of x)} \\
\end{pmatrix} $$
```
# GRADED FUNCTION: softmax
def softmax(x):
"""Calculates the softmax for each row of the input x.
Your code should work for a row vector and also for matrices of shape (n, m).
Argument:
x -- A numpy matrix of shape (n,m)
Returns:
s -- A numpy matrix equal to the softmax of x, of shape (n,m)
"""
### START CODE HERE ### (≈ 3 lines of code)
# Apply exp() element-wise to x. Use np.exp(...).
x_exp = np.exp(x)
# Create a vector x_sum that sums each row of x_exp. Use np.sum(..., axis = 1, keepdims = True).
x_sum = np.sum(x,axis=1,keepdims=True)
# Compute softmax(x) by dividing x_exp by x_sum. It should automatically use numpy broadcasting.
s = np.divide(x_exp,x_sum)
### END CODE HERE ###
return s
x = np.array([
[9, 2, 5, 0, 0],
[7, 5, 0, 0 ,0]])
print("softmax(x) = " + str(softmax(x)))
```
**Expected Output**:
<table style="width:60%">
<tr>
<td> **softmax(x)** </td>
<td> [[ 9.80897665e-01 8.94462891e-04 1.79657674e-02 1.21052389e-04
1.21052389e-04]
[ 8.78679856e-01 1.18916387e-01 8.01252314e-04 8.01252314e-04
8.01252314e-04]]</td>
</tr>
</table>
**Note**:
- If you print the shapes of x_exp, x_sum and s above and rerun the assessment cell, you will see that x_sum is of shape (2,1) while x_exp and s are of shape (2,5). **x_exp/x_sum** works due to python broadcasting.
Congratulations! You now have a pretty good understanding of python numpy and have implemented a few useful functions that you will be using in deep learning.
<font color='blue'>
**What you need to remember:**
- np.exp(x) works for any np.array x and applies the exponential function to every coordinate
- the sigmoid function and its gradient
- image2vector is commonly used in deep learning
- np.reshape is widely used. In the future, you'll see that keeping your matrix/vector dimensions straight will go toward eliminating a lot of bugs.
- numpy has efficient built-in functions
- broadcasting is extremely useful
## 2) Vectorization
In deep learning, you deal with very large datasets. Hence, a non-computationally-optimal function can become a huge bottleneck in your algorithm and can result in a model that takes ages to run. To make sure that your code is computationally efficient, you will use vectorization. For example, try to tell the difference between the following implementations of the dot/outer/elementwise product.
```
import time
x1 = [9, 2, 5, 0, 0, 7, 5, 0, 0, 0, 9, 2, 5, 0, 0]
x2 = [9, 2, 2, 9, 0, 9, 2, 5, 0, 0, 9, 2, 5, 0, 0]
### CLASSIC DOT PRODUCT OF VECTORS IMPLEMENTATION ###
tic = time.process_time()
dot = 0
for i in range(len(x1)):
dot+= x1[i]*x2[i]
toc = time.process_time()
print ("dot = " + str(dot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### CLASSIC OUTER PRODUCT IMPLEMENTATION ###
tic = time.process_time()
outer = np.zeros((len(x1),len(x2))) # we create a len(x1)*len(x2) matrix with only zeros
for i in range(len(x1)):
for j in range(len(x2)):
outer[i,j] = x1[i]*x2[j]
toc = time.process_time()
print ("outer = " + str(outer) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### CLASSIC ELEMENTWISE IMPLEMENTATION ###
tic = time.process_time()
mul = np.zeros(len(x1))
for i in range(len(x1)):
mul[i] = x1[i]*x2[i]
toc = time.process_time()
print ("elementwise multiplication = " + str(mul) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### CLASSIC GENERAL DOT PRODUCT IMPLEMENTATION ###
W = np.random.rand(3,len(x1)) # Random 3*len(x1) numpy array
tic = time.process_time()
gdot = np.zeros(W.shape[0])
for i in range(W.shape[0]):
for j in range(len(x1)):
gdot[i] += W[i,j]*x1[j]
toc = time.process_time()
print ("gdot = " + str(gdot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
x1 = [9, 2, 5, 0, 0, 7, 5, 0, 0, 0, 9, 2, 5, 0, 0]
x2 = [9, 2, 2, 9, 0, 9, 2, 5, 0, 0, 9, 2, 5, 0, 0]
### VECTORIZED DOT PRODUCT OF VECTORS ###
tic = time.process_time()
dot = np.dot(x1,x2)
toc = time.process_time()
print ("dot = " + str(dot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### VECTORIZED OUTER PRODUCT ###
tic = time.process_time()
outer = np.outer(x1,x2)
toc = time.process_time()
print ("outer = " + str(outer) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### VECTORIZED ELEMENTWISE MULTIPLICATION ###
tic = time.process_time()
mul = np.multiply(x1,x2)
toc = time.process_time()
print ("elementwise multiplication = " + str(mul) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### VECTORIZED GENERAL DOT PRODUCT ###
tic = time.process_time()
dot = np.dot(W,x1)
toc = time.process_time()
print ("gdot = " + str(dot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
```
As you may have noticed, the vectorized implementation is much cleaner and more efficient. For bigger vectors/matrices, the differences in running time become even bigger.
**Note** that `np.dot()` performs a matrix-matrix or matrix-vector multiplication. This is different from `np.multiply()` and the `*` operator (which is equivalent to `.*` in Matlab/Octave), which performs an element-wise multiplication.
### 2.1 Implement the L1 and L2 loss functions
**Exercise**: Implement the numpy vectorized version of the L1 loss. You may find the function abs(x) (absolute value of x) useful.
**Reminder**:
- The loss is used to evaluate the performance of your model. The bigger your loss is, the more different your predictions ($ \hat{y} $) are from the true values ($y$). In deep learning, you use optimization algorithms like Gradient Descent to train your model and to minimize the cost.
- L1 loss is defined as:
$$\begin{align*} & L_1(\hat{y}, y) = \sum_{i=0}^m|y^{(i)} - \hat{y}^{(i)}| \end{align*}\tag{6}$$
```
# GRADED FUNCTION: L1
def L1(yhat, y):
"""
Arguments:
yhat -- vector of size m (predicted labels)
y -- vector of size m (true labels)
Returns:
loss -- the value of the L1 loss function defined above
"""
### START CODE HERE ### (≈ 1 line of code)
loss = None
### END CODE HERE ###
return loss
yhat = np.array([.9, 0.2, 0.1, .4, .9])
y = np.array([1, 0, 0, 1, 1])
print("L1 = " + str(L1(yhat,y)))
```
**Expected Output**:
<table style="width:20%">
<tr>
<td> **L1** </td>
<td> 1.1 </td>
</tr>
</table>
**Exercise**: Implement the numpy vectorized version of the L2 loss. There are several way of implementing the L2 loss but you may find the function np.dot() useful. As a reminder, if $x = [x_1, x_2, ..., x_n]$, then `np.dot(x,x)` = $\sum_{j=0}^n x_j^{2}$.
- L2 loss is defined as $$\begin{align*} & L_2(\hat{y},y) = \sum_{i=0}^m(y^{(i)} - \hat{y}^{(i)})^2 \end{align*}\tag{7}$$
```
# GRADED FUNCTION: L2
def L2(yhat, y):
"""
Arguments:
yhat -- vector of size m (predicted labels)
y -- vector of size m (true labels)
Returns:
loss -- the value of the L2 loss function defined above
"""
### START CODE HERE ### (≈ 1 line of code)
loss = None
### END CODE HERE ###
return loss
yhat = np.array([.9, 0.2, 0.1, .4, .9])
y = np.array([1, 0, 0, 1, 1])
print("L2 = " + str(L2(yhat,y)))
```
**Expected Output**:
<table style="width:20%">
<tr>
<td> **L2** </td>
<td> 0.43 </td>
</tr>
</table>
Congratulations on completing this assignment. We hope that this little warm-up exercise helps you in the future assignments, which will be more exciting and interesting!
<font color='blue'>
**What to remember:**
- Vectorization is very important in deep learning. It provides computational efficiency and clarity.
- You have reviewed the L1 and L2 loss.
- You are familiar with many numpy functions such as np.sum, np.dot, np.multiply, np.maximum, etc...
|
github_jupyter
|
This notebook was prepared by [Donne Martin](https://github.com/donnemartin). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges).
# Solution Notebook
## Problem: Generate a list of primes.
* [Constraints](#Constraints)
* [Test Cases](#Test-Cases)
* [Algorithm](#Algorithm)
* [Code](#Code)
* [Unit Test](#Unit-Test)
## Constraints
* Is it correct that 1 is not considered a prime number?
* Yes
* Can we assume the inputs are valid?
* No
* Can we assume this fits memory?
* Yes
## Test Cases
* None -> Exception
* Not an int -> Exception
* 20 -> [False, False, True, True, False, True, False, True, False, False, False, True, False, True, False, False, False, True, False, True]
## Algorithm
For a number to be prime, it must be 2 or greater and cannot be divisible by another number other than itself (and 1).
We'll use the Sieve of Eratosthenes. All non-prime numbers are divisible by a prime number.
* Use an array (or bit array, bit vector) to keep track of each integer up to the max
* Start at 2, end at sqrt(max)
* We can use sqrt(max) instead of max because:
* For each value that divides the input number evenly, there is a complement b where a * b = n
* If a > sqrt(n) then b < sqrt(n) because sqrt(n^2) = n
* "Cross off" all numbers divisible by 2, 3, 5, 7, ... by setting array[index] to False
Complexity:
* Time: O(n log log n)
* Space: O(n)
Wikipedia's animation:

## Code
```
import math
class PrimeGenerator(object):
def generate_primes(self, max_num):
if max_num is None:
raise TypeError('max_num cannot be None')
array = [True] * max_num
array[0] = False
array[1] = False
prime = 2
while prime <= math.sqrt(max_num):
self._cross_off(array, prime)
prime = self._next_prime(array, prime)
return array
def _cross_off(self, array, prime):
for index in range(prime*prime, len(array), prime):
# Start with prime*prime because if we have a k*prime
# where k < prime, this value would have already been
# previously crossed off
array[index] = False
def _next_prime(self, array, prime):
next = prime + 1
while next < len(array) and not array[next]:
next += 1
return next
```
## Unit Test
```
%%writefile test_generate_primes.py
import unittest
class TestMath(unittest.TestCase):
def test_generate_primes(self):
prime_generator = PrimeGenerator()
self.assertRaises(TypeError, prime_generator.generate_primes, None)
self.assertRaises(TypeError, prime_generator.generate_primes, 98.6)
self.assertEqual(prime_generator.generate_primes(20), [False, False, True,
True, False, True,
False, True, False,
False, False, True,
False, True, False,
False, False, True,
False, True])
print('Success: generate_primes')
def main():
test = TestMath()
test.test_generate_primes()
if __name__ == '__main__':
main()
%run -i test_generate_primes.py
```
|
github_jupyter
|
```
from __future__ import print_function
import matplotlib.pyplot as plt
%matplotlib inline
import SimpleITK as sitk
print(sitk.Version())
from myshow import myshow
# Download data to work on
%run update_path_to_download_script
from downloaddata import fetch_data as fdata
OUTPUT_DIR = "Output"
```
This section of the Visible Human Male is about 1.5GB. To expedite processing and registration we crop the region of interest, and reduce the resolution. Take note that the physical space is maintained through these operations.
```
fixed_rgb = sitk.ReadImage(fdata("vm_head_rgb.mha"))
fixed_rgb = fixed_rgb[735:1330,204:975,:]
fixed_rgb = sitk.BinShrink(fixed_rgb,[3,3,1])
moving = sitk.ReadImage(fdata("vm_head_mri.mha"))
myshow(moving)
# Segment blue ice
seeds = [[10,10,10]]
fixed_mask = sitk.VectorConfidenceConnected(fixed_rgb, seedList=seeds, initialNeighborhoodRadius=5, numberOfIterations=4, multiplier=8)
# Invert the segment and choose largest component
fixed_mask = sitk.RelabelComponent(sitk.ConnectedComponent(fixed_mask==0))==1
myshow(sitk.Mask(fixed_rgb, fixed_mask));
# pick red channel
fixed = sitk.VectorIndexSelectionCast(fixed_rgb,0)
fixed = sitk.Cast(fixed,sitk.sitkFloat32)
moving = sitk.Cast(moving,sitk.sitkFloat32)
initialTransform = sitk.Euler3DTransform()
initialTransform = sitk.CenteredTransformInitializer(sitk.Cast(fixed_mask,moving.GetPixelID()), moving, initialTransform, sitk.CenteredTransformInitializerFilter.MOMENTS)
print(initialTransform)
def command_iteration(method) :
print("{0} = {1} : {2}".format(method.GetOptimizerIteration(),
method.GetMetricValue(),
method.GetOptimizerPosition()),
end='\n');
sys.stdout.flush();
tx = initialTransform
R = sitk.ImageRegistrationMethod()
R.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
R.SetOptimizerAsGradientDescentLineSearch(learningRate=1,numberOfIterations=100)
R.SetOptimizerScalesFromIndexShift()
R.SetShrinkFactorsPerLevel([4,2,1])
R.SetSmoothingSigmasPerLevel([8,4,2])
R.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
R.SetMetricSamplingStrategy(R.RANDOM)
R.SetMetricSamplingPercentage(0.1)
R.SetInitialTransform(tx)
R.SetInterpolator(sitk.sitkLinear)
import sys
R.RemoveAllCommands()
R.AddCommand( sitk.sitkIterationEvent, lambda: command_iteration(R) )
outTx = R.Execute(sitk.Cast(fixed,sitk.sitkFloat32), sitk.Cast(moving,sitk.sitkFloat32))
print("-------")
print(tx)
print("Optimizer stop condition: {0}".format(R.GetOptimizerStopConditionDescription()))
print(" Iteration: {0}".format(R.GetOptimizerIteration()))
print(" Metric value: {0}".format(R.GetMetricValue()))
tx.AddTransform(sitk.Transform(3,sitk.sitkAffine))
R.SetOptimizerAsGradientDescentLineSearch(learningRate=1,numberOfIterations=100)
R.SetOptimizerScalesFromIndexShift()
R.SetShrinkFactorsPerLevel([2,1])
R.SetSmoothingSigmasPerLevel([4,1])
R.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
R.SetInitialTransform(tx)
outTx = R.Execute(sitk.Cast(fixed,sitk.sitkFloat32), sitk.Cast(moving,sitk.sitkFloat32))
R.GetOptimizerStopConditionDescription()
resample = sitk.ResampleImageFilter()
resample.SetReferenceImage(fixed_rgb)
resample.SetInterpolator(sitk.sitkBSpline)
resample.SetTransform(outTx)
resample.AddCommand(sitk.sitkProgressEvent, lambda: print("\rProgress: {0:03.1f}%...".format(100*resample.GetProgress()),end=''))
resample.AddCommand(sitk.sitkProgressEvent, lambda: sys.stdout.flush())
resample.AddCommand(sitk.sitkEndEvent, lambda: print("Done"))
out = resample.Execute(moving)
out_rgb = sitk.Cast( sitk.Compose( [sitk.RescaleIntensity(out)]*3), sitk.sitkVectorUInt8)
vis_xy = sitk.CheckerBoard(fixed_rgb, out_rgb, checkerPattern=[8,8,1])
vis_xz = sitk.CheckerBoard(fixed_rgb, out_rgb, checkerPattern=[8,1,8])
vis_xz = sitk.PermuteAxes(vis_xz, [0,2,1])
myshow(vis_xz,dpi=30)
import os
sitk.WriteImage(out, os.path.join(OUTPUT_DIR, "example_registration.mha"))
sitk.WriteImage(vis_xy, os.path.join(OUTPUT_DIR, "example_registration_xy.mha"))
sitk.WriteImage(vis_xz, os.path.join(OUTPUT_DIR, "example_registration_xz.mha"))
```
|
github_jupyter
|
# BUSINESS ANALYTICS
You are the business owner of the retail firm and want to see how your company is performing. You are interested in finding out the weak areas where you can work to make more profit. What all business problems you can derive by looking into the data?
```
# Importing certain libraries
import pandas as pd
import numpy as np
import seaborn as sb
import matplotlib.pyplot as plt
%matplotlib inline
```
## Understanding the data
```
# Importing the dataset
data = pd.read_csv(r"D:/TSF/Task 5/SampleSuperstore.csv")
# Displaying the Dataset
data.head()
# Gathering the basic Information
data.describe()
# Learning about differnet datatypes present in the dataset
data.dtypes
# Checking for any null or misssing values
data.isnull().sum()
```
Since, there are no null or missing values present, therefore we can move further for data exploration
## Exploratory Data Analysis
```
# First, using seaborn pairplot for data visualisation
sb.set(style = "whitegrid")
plt.figure(figsize = (20, 10))
sb.pairplot(data, hue = "Quantity")
```
We can clearly see that in our dataset, there are total of 14 different quantities in which our business deals.
```
# Second, using seaborn heatmap for data visualization
plt.figure(figsize = (7, 5))
sb.heatmap(data.corr(), annot = True, fmt = ".2g", linewidth = 0.5, linecolor = "Black", cmap = "YlOrRd")
```
Here, We can see that Sales and Profit are highly corelated as obvious.
```
# Third, using seaborn countplot for data visualization
sb.countplot(x = data["Country"])
plt.show()
```
Our dataset only contains data from United States only.
```
sb.countplot(x = data["Segment"])
plt.show()
```
Maximum Segment is of Consumer & Minimum segment is of Home Office
```
sb.countplot(x = data["Region"])
plt.show()
```
Maximum entries are from West region of United States, followed by East, Central & South respectively.
```
sb.countplot(x = data["Ship Mode"])
plt.show()
```
This shows that the mostly our business uses Standard class for shipping as compared to other classes.
```
plt.figure(figsize = (8, 8))
sb.countplot(x = data["Quantity"])
plt.show()
```
Out of total 14 quantites present, Maximum are number 2 and 3 respectively.
```
plt.figure(figsize = (10, 8))
sb.countplot(x = data["State"])
plt.xticks(rotation = 90)
plt.show()
```
If we watch carefully, we can clearly see that maximum sales happened in California, followed by New York & the Texas.
Lowest sales happened North Dakota, West Virginea.
```
sb.countplot(x = data["Category"])
plt.show()
```
So, Our business deals maximum in Office Supplies category, followed by Furniture & then Tech products.
```
plt.figure(figsize = (10, 8))
sb.countplot(x = data['Sub-Category'])
plt.xticks(rotation = 90)
plt.show()
```
If we define Sub Categories section, maximum profit is earned through Binders, followed by Paper & Furnishing. Minimum Profit is earned through Copiers, Machines etc.
```
# Forth, using Seaborn barplot for data visualization
plt.figure(figsize = (12, 10))
sb.barplot(x = data["Sub-Category"], y = data["Profit"], capsize = .1, saturation = .5)
plt.xticks(rotation = 90)
plt.show()
```
In this Sub-categories, Bookcases, Tables and Supplies are facing losses on the business level as compared to ther categories. So, Business owner needs to pay attention towards these 3 categories.
### Now, to compare specific features of Business, We have to use certain different Exploration operations
```
# Fifth, using regression plot for data visualization
plt.figure(figsize = (10, 8))
sb.regplot(data["Sales"], data["Profit"], marker = "X", color = "r")
plt.show()
```
This Relationship does not seem to be Linear. So, this relationship doesn't help much.
```
plt.figure(figsize = (10, 8))
sb.regplot(data["Quantity"], data["Profit"], color = "black", y_jitter=.1)
plt.show()
```
This Relationship happens to be linear. The quantity '5' has the maximum profit as compared to others.
```
plt.figure(figsize = (10, 8))
sb.regplot(data["Quantity"], data["Sales"], color = "m", marker = "+", y_jitter=.1)
plt.show()
```
This Relationship is also linear. The quantity '6' has the maximum sales as compared to others.
```
# Sixth, using seaborn lineplot for data visualisation
plt.figure(figsize = (10, 8))
sb.lineplot(data["Discount"], data["Profit"], color = "orange", label = "Discount")
plt.legend()
plt.show()
```
As expected, we can see at 50% discount, the profit is very much negligible or we can say that there are losses. But, on the other hand, at 10% discount, there is a profit at a very good level.
```
plt.figure(figsize = (10, 8))
sb.lineplot(data["Sub-Category"], data["Profit"], color = "blue", label = "Sales")
plt.xticks(rotation = 90)
plt.legend()
plt.show()
```
With Copiers, Business makes the largest Profit.
```
plt.figure(figsize = (10, 8))
sb.lineplot(data["Quantity"], data["Profit"], color = "red", label = "Quantity")
plt.legend()
plt.show()
```
Quantity '13' has the maximum profit.
### WHAT CAN BE DERIVED FROM ABOVE VISUALIZATIONS :
* Improvements should be made for same day shipment mode.
* We have to work more in the Southern region of USA for better business.
* Office Supplies are good. We have to work more on Technology and Furniture Category of business.
* There are very less people working as Copiers.
* Maximum number of people are from California and New York. It should expand in other parts of USA as well.
* Company is facing losses in sales of bookcases and tables products.
* Company have a lots of profit in the sale of copier but the number of sales is very less so there is a need of increase innumber of sales of copier.
* When the profits of a state are compared with the discount provided in each state, the states which has allowed more discount, went into loss.
* Profit and discount show very weak and negative relationship. This should be kept in mind that before taking any other decision related to business.
# ASSIGNMENT COMPLETED !!
|
github_jupyter
|
## Introduction
In real world, there exists many huge graphs that can not be loaded in one machine,
such as social networks and citation networks.
To deal with such graphs, PGL develops a Distributed Graph Engine Framework to
support graph sampling on large scale graph networks for distributed GNN training.
In this tutorial, we will walk through the steps of performing distributed Graph Engine for graph sampling.
We also develop a launch script for launch a distributed Graph Engine. To see more examples of distributed GNN training, please refer to [here](https://github.com/PaddlePaddle/PGL/tree/main/examples).
## Requirements
paddlepaddle>=2.1.0
pgl>=2.1.4
## example of how to start a distributed graph engine service
Supose we have a following graph that has two type of nodes (u and t).
Firstly, We should create a configuration file and specify the ip address of each machine.
Here we use two ports to simulate two machines.
After creating the configuration file and ip adress file, we can now start two graph servers.
Then we can use the client to sample neighbors or sample nodes from graph servers.
```
import os
import sys
import re
import time
import tqdm
import argparse
import unittest
import shutil
import numpy as np
from pgl.utils.logger import log
from pgl.distributed import DistGraphClient, DistGraphServer
edges_file = """37 45 0.34
37 145 0.31
37 112 0.21
96 48 1.4
96 247 0.31
96 111 1.21
59 45 0.34
59 145 0.31
59 122 0.21
97 48 0.34
98 247 0.31
7 222 0.91
7 234 0.09
37 333 0.21
47 211 0.21
47 113 0.21
47 191 0.21
34 131 0.21
34 121 0.21
39 131 0.21"""
node_file = """u 98
u 97
u 96
u 7
u 59
t 48
u 47
t 45
u 39
u 37
u 34
t 333
t 247
t 234
t 222
t 211
t 191
t 145
t 131
t 122
t 121
t 113
t 112
t 111"""
tmp_path = "./tmp_distgraph_test"
if not os.path.exists(tmp_path):
os.makedirs(tmp_path)
with open(os.path.join(tmp_path, "edges.txt"), 'w') as f:
f.write(edges_file)
with open(os.path.join(tmp_path, "node_types.txt"), 'w') as f:
f.write(node_file)
# configuration file
config = """
etype2files: "u2e2t:./tmp_distgraph_test/edges.txt"
symmetry: True
ntype2files: "u:./tmp_distgraph_test/node_types.txt,t:./tmp_distgraph_test/node_types.txt"
"""
ip_addr = """127.0.0.1:8342
127.0.0.1:8343"""
with open(os.path.join(tmp_path, "config.yaml"), 'w') as f:
f.write(config)
with open(os.path.join(tmp_path, "ip_addr.txt"), 'w') as f:
f.write(ip_addr)
config = os.path.join(tmp_path, "config.yaml")
ip_addr = os.path.join(tmp_path, "ip_addr.txt")
shard_num = 10
gserver1 = DistGraphServer(config, shard_num, ip_addr, server_id=0)
gserver2 = DistGraphServer(config, shard_num, ip_addr, server_id=1)
client1 = DistGraphClient(config, shard_num=shard_num, ip_config=ip_addr, client_id=0)
client1.load_edges()
client1.load_node_types()
print("data loading finished")
# random sample nodes by node type
client1.random_sample_nodes(node_type="u", size=3)
# traverse all nodes from each server
node_generator = client1.node_batch_iter(batch_size=3, node_type="t", shuffle=True)
for nodes in node_generator:
print(nodes)
# sample neighbors
# note that the edge_type "u2eut" is defined in config.yaml file
nodes = [98, 7]
neighs = client1.sample_successor(nodes, max_degree=10, edge_type="u2e2t")
print(neighs)
```
|
github_jupyter
|
# "[ML] What's the difference between a metric and a loss?"
- toc:true
- branch: master
- badges: false
- comments: true
- author: Peiyi Hung
- categories: [learning, machine learning]
In machine learning, we usually use two values to evaluate our model: a metric and a loss. For instance, if we are doing a binary classification task, our metric may be the accuracy and our loss would be the cross-entroy. They both show how good our model
performs. However, why do we need both rather than just use one of them? Also, what's the difference between them?
The short answer is that **the metric is for human while the loss is for your model.**
Based on the metric, machine learning practitioners such as data scientists and researchers assess a machine learning model. On the assessment, ML practitioners make decisions to address their problems or achieve their business goals. For example , say a data scientist aims to build a spam classifier to distinguish normal email from spam with 95% accuracy. First, the data scientist build a model with 90% accuracy. Apparently, this result doesn't meet his business goal, so he tries to build a better one. After implementing some techniques, he might get a classifier with 97% accuracy, which goes beyond his goal. Since the goal is met, the data scientist decides to integrate this model into his data product. ML partitioners use the metric to tell whether their model is good enough.
On the other hand, a loss indicates in what direction your model should improve. The difference between machine learning and traditional programming is how they get the ability to solve a problem. Traditional programs solve problems by following exact instructions given by programmers. In contrast, machine learning models learn how to solve a problem by taking into some examples (data) and discovering the underlying patterns of the problem. How does a machine learning model learn? Most ML models learn using a gradient-based method. Here's how a gradient-based method (be specifically, a gradient descent method in supervised learning context) works:
1. A model takes into data and makes predictions.
1. Compute the loss based on the predictions and the true data.
1. Compute the gradients of the loss with respect to parameters of the model.
1. Updating these parameters based on these gradients.
The gradient of the loss helps our model to get better and better. The reason why we need a loss is that a loss is **sensitive** enough to small changes so our model can improve based on it. More precisely, the gradient of the loss should vary if our parameters change slightly. In our spam classification example, accuracy is obviously not suitable for being a loss since it only changes when some examples are classified differently. The cross-entrpy is relatively smoother and so it is a good candidate for a loss. However, a metric do not have to be different from a loss. A metric can be a loss as long as it is sensitive enough. For instance, in a regression setting, MSE (mean squared error) can be both a metric and a loss.
In summary, a metric helps ML partitioners to evaluate their models and a loss facilitates the learning process of a ML model.
|
github_jupyter
|
# Canonical correlation analysis in python
In this notebook, we will walk through the solution to the basic algrithm of canonical correlation analysis and compare that to the output of implementations in existing python libraries `statsmodels` and `scikit-learn`.
```
import numpy as np
from scipy.linalg import sqrtm
from statsmodels.multivariate.cancorr import CanCorr as smCCA
from sklearn.cross_decomposition import CCA as skCCA
import matplotlib.pyplot as plt
from seaborn import heatmap
```
Let's define a plotting functon for the output first.
```
def plot_cca(a, b, U, V, s):
# plotting
plt.figure()
heatmap(a, square=True, center=0)
plt.title("Canonical vector - x")
plt.figure()
heatmap(b, square=True, center=0)
plt.title("Canonical vector - y")
plt.figure(figsize=(9, 6))
for i in range(N):
plt.subplot(221 + i)
plt.scatter(np.array(X_score[:, i]).reshape(100),
np.array(Y_score[:, i]).reshape(100),
marker="o", c="b", s=25)
plt.xlabel("Canonical variate of X")
plt.ylabel("Canonical variate of Y")
plt.title('Mode %i (corr = %.2f)' %(i + 1, s[i]))
plt.xticks(())
plt.yticks(())
```
## Create data based on some latent variables
First generate some test data.
The code below is modified based on the scikit learn example of CCA.
The aim of using simulated data is that we can have complete control over the structure of the data and help us see the utility of CCA.
Let's create a dataset with 100 observations with two hidden variables:
```
n = 100
# fix the random seed so this tutorial will always create the same results
np.random.seed(42)
l1 = np.random.normal(size=n)
l2 = np.random.normal(size=n)
```
For each observation, there are two domains of data.
Six and four variables are measured in each of the domain.
In domain 1 (x), the first latent structure 1 is underneath the first 3 variables and latent strucutre 2 for the rest.
In domain 2 (y), the first latent structure 1 is underneath every other variable and for latent strucutre 2 as well.
```
latents_x = np.array([l1, l1, l1, l2, l2, l2]).T
latents_y = np.array([l1, l2, l1, l2]).T
```
Now let's add some random noise on this latent structure.
```
X = latents_x + np.random.normal(size=6 * n).reshape((n, 6))
Y = latents_y + np.random.normal(size=4 * n).reshape((n, 4))
```
The aim of CCA is finding the correlated latent features in the two domains of data.
Therefore, we would expect to find the hidden strucure is laid out in the latent components.
## SVD algebra solution
SVD solution is the most implemented way of CCA solution. For the proof of standard eigenvalue solution and the proof SVD solution demonstrated below, see [Uurtio wt. al, (2018)](https://dl.acm.org/citation.cfm?id=3136624).
The first step is getting the covariance matrixes of X and Y.
```
Cx, Cy = np.corrcoef(X.T), np.corrcoef(Y.T)
Cxy = np.corrcoef(X.T, Y.T)[:X.shape[1], X.shape[1]:]
Cyx = Cxy.T
```
We first retrieve the identity form of the covariance matix of X and Y.
```
sqrt_x, sqrt_y = np.matrix(sqrtm(Cx)), np.matrix(sqrtm(Cy))
isqrt_x, isqrt_y = sqrt_x.I, sqrt_y.I
```
According to the proof, we leared that the canonical correlation can be retrieved from SVD on Cx^-1/2 Cxy Cy^-1/2.
```
W = isqrt_x * Cxy * isqrt_y
u, s, v = np.linalg.svd(W)
```
The columns of the matrices U and V correspond to the sets of orthonormal left and right singular vectors respectively. The singular values of matrix S correspond to
the canonical correlations. The positions w a and w b are obtained from:
```
N = np.min([X.shape[1], Y.shape[1]])
a = np.dot(u, isqrt_x.T[:, :N]) / np.std(X) # scaling because we didn't standardise the input
b = np.dot(v, isqrt_y).T / np.std(Y)
```
Now compute the score.
```
X_score, Y_score = X.dot(a), Y.dot(b)
plot_cca(a, b, X_score, Y_score, s) # predefined plotting function
```
## Solution Using SVD Only
The solution above can be further simplified by conducting SVD on the two domains.
The algorithm SVD X and Y. This step is similar to doing principle component analysis on the two domains.
```
ux, sx, vx = np.linalg.svd(X, 0)
uy, sy, vy = np.linalg.svd(Y, 0)
```
Then take the unitary bases and form UxUy^T and SVD it. S would be the canonical correlation of the two domanins of features.
```
u, s, v = np.linalg.svd(ux.T.dot(uy), 0)
```
We can yield the canonical vectors by transforming the unitary basis in the hidden space back to the original space.
```
a = (vx.T).dot(u) # no scaling here as SVD handled it.
b = (vy.T).dot(v.T)
X_score, Y_score = X.dot(a), Y.dot(b)
```
Now we can plot the results. It shows very similar results to solution 1.
```
plot_cca(a, b, X_score, Y_score, s) # predefined plotting function
```
The method above has been implemented in `Statsmodels`. The results are almost identical:
```
sm_cca = smCCA(Y, X)
sm_s = sm_cca.cancorr
sm_a = sm_cca.x_cancoef
sm_b = sm_cca.y_cancoef
sm_X_score = X.dot(a)
sm_Y_score = Y.dot(b)
plot_cca(a, b, X_score, Y_score, s)
```
## Scikit learn
Scikit learn implemented [a different algorithm](https://www.stat.washington.edu/sites/default/files/files/reports/2000/tr371.pdf).
The outcome of the Scikit learn implementation yield very similar results.
The first mode capture the hidden structure in the simulated data.
```
cca = skCCA(n_components=4)
cca.fit(X, Y)
s = np.corrcoef(cca.x_scores_.T, cca.y_scores_.T).diagonal(offset=cca.n_components)
a = cca.x_weights_
b = cca.y_weights_
X_score, Y_score = cca.x_scores_, cca.y_scores_
plot_cca(a, b, X_score, Y_score, s) # predefined plotting function
```
|
github_jupyter
|
```
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasRegressor
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
%matplotlib inline
```
Set our random seed so that all computations are deterministic
```
seed = 21899
```
Read in the raw data for the first 100K records of the HCEPDB into a pandas dataframe
```
df = pd.read_csv('https://github.com/UWDIRECT/UWDIRECT.github.io/blob/master/Wi18_content/DSMCER/HCEPD_100K.csv?raw=true')
df.head()
```
Separate out the predictors from the output
```
X = df[['mass', 'voc', 'jsc', 'e_homo_alpha', 'e_gap_alpha',
'e_lumo_alpha']].values
Y = df[['pce']].values
```
Let's create the test / train split for these data using 80/20. The `_pn` extension is related to the 'prenormalization' nature of the data.
```
X_train_pn, X_test_pn, y_train, y_test = train_test_split(X, Y,
test_size=0.20,
random_state=seed)
```
Now we need to `StandardScaler` the training data and apply that scale to the test data.
```
# create the scaler from the training data only and keep it for later use
X_train_scaler = StandardScaler().fit(X_train_pn)
# apply the scaler transform to the training data
X_train = X_train_scaler.transform(X_train_pn)
```
Now let's reuse that scaler transform on the test set. This way we never contaminate the test data with the training data. We'll start with a histogram of the testing data just to prove to ourselves it is working.
```
plt.hist(X_test_pn[:,1])
```
OK, bnow apply the training scaler transform to the test and plot a histogram
```
X_test = X_train_scaler.transform(X_test_pn)
plt.hist(X_test[:,1])
```
### Let's create the neural network layout
This is a simple neural network with no hidden layers and just the inputs transitioned to the output.
```
def simple_model():
# assemble the structure
model = Sequential()
model.add(Dense(6, input_dim=6, kernel_initializer='normal', activation='relu'))
model.add(Dense(1, kernel_initializer='normal'))
# compile the model
model.compile(loss='mean_squared_error', optimizer='adam')
return model
```
Train the neural network with the following
```
# initialize the andom seed as this is used to generate
# the starting weights
np.random.seed(seed)
# create the NN framework
estimator = KerasRegressor(build_fn=simple_model,
epochs=150, batch_size=25000, verbose=0)
history = estimator.fit(X_train, y_train, validation_split=0.33, epochs=150,
batch_size=10000, verbose=0)
```
The history object returned by the `fit` call contains the information in a fitting run.
```
print(history.history.keys())
print("final MSE for train is %.2f and for validation is %.2f" %
(history.history['loss'][-1], history.history['val_loss'][-1]))
```
Let's plot it!
```
# summarize history for loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.show()
```
Let's get the MSE for the test set.
```
test_loss = estimator.model.evaluate(X_test, y_test)
print("test set mse is %.2f" % test_loss)
```
## NEAT!
So our train mse is very similar to the training and validation at the final step!
### Let's look at another way to evaluate the set of models using cross validation
Use 10 fold cross validation to evaluate the models generated from our training set. We'll use scikit-learn's tools for this. Remember, this is only assessing our training set. If you get negative values, to make `cross_val_score` behave as expected, we have to flip the signs on the results (incompatibility with keras).
```
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(estimator, X_train, y_train, cv=kfold)
print("Results: %.2f (%.2f) MSE" % (-1 * results.mean(), results.std()))
```
#### Quick aside, `Pipeline`
Let's use scikit learns `Pipeline` workflow to run a k-fold cross validation run on the learned model.
With this tool, we create a workflow using the `Pipeline` object. You provide a list of actions (as named tuples) to be performed. We do this with `StandardScaler` to eliminate the posibility of training leakage into the cross validation test set during normalization.
```
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=simple_model,
epochs=150, batch_size=25000, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X_train, y_train, cv=kfold)
print('MSE mean: %.4f ; std: %.4f' % (-1 * results.mean(), results.std()))
```
### Now, let's try a more sophisticated model
Let's use a hidden layer this time.
```
def medium_model():
# assemble the structure
model = Sequential()
model.add(Dense(6, input_dim=6, kernel_initializer='normal', activation='relu'))
model.add(Dense(4, kernel_initializer='normal', activation='relu'))
model.add(Dense(1, kernel_initializer='normal'))
# compile the model
model.compile(loss='mean_squared_error', optimizer='adam')
return model
# initialize the andom seed as this is used to generate
# the starting weights
np.random.seed(seed)
# create the NN framework
estimator = KerasRegressor(build_fn=medium_model,
epochs=150, batch_size=25000, verbose=0)
history = estimator.fit(X_train, y_train, validation_split=0.33, epochs=150,
batch_size=10000, verbose=0)
print("final MSE for train is %.2f and for validation is %.2f" %
(history.history['loss'][-1], history.history['val_loss'][-1]))
# summarize history for loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.show()
test_loss = estimator.model.evaluate(X_test, y_test)
print("test set mse is %.2f" % test_loss)
```
_So it appears our more complex model improved performance_
### Free time!
Find example code for keras for the two following items:
* L1 and L2 regularization (note in keras, this can be done by layer)
* Dropout
#### Regularization
Let's start by adding L1 or L2 (or both) regularization to the hidden layer.
Hint: you need to define a new function that is the neural network model and add the correct parameters to the layer definition. Then retrain and plot as above. What parameters did you choose for your dropout? Did it improve training?
#### Dropout
Find the approach to specifying dropout on a layer using your best friend `bing`. As with L1 and L2 above, this will involve defining a new network struction using a function and some new 'magical' dropout layers.
|
github_jupyter
|
## Homework 3 and 4 - Applications Using MRJob
```
# general imports
import os
import re
import sys
import time
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# tell matplotlib not to open a new window
%matplotlib inline
# automatically reload modules
%reload_ext autoreload
%autoreload 2
# print some configuration details for future replicability.
print 'Python Version: %s' % (sys.version.split('|')[0])
hdfs_conf = !hdfs getconf -confKey fs.defaultFS ### UNCOMMENT ON DOCKER
#hdfs_conf = !hdfs getconf -confKey fs.default.name ### UNCOMMENT ON ALTISCALE
print 'HDFS filesystem running at: \n\t %s' % (hdfs_conf[0])
JAR_FILE = "/usr/lib/hadoop-mapreduce/hadoop-streaming-2.6.0-cdh5.7.0.jar"
HDFS_DIR = "/user/root/HW3"
HOME_DIR = "/media/notebooks/SP18-1-maynard242" # FILL IN HERE eg. /media/notebooks/w261-main/Assignments
# save path for use in Hadoop jobs (-cmdenv PATH={PATH})
from os import environ
PATH = environ['PATH']
#!hdfs dfs -mkdir HW3
!hdfs dfs -ls
%%writefile example1.txt
Unix,30
Solaris,10
Linux,25
Linux,20
HPUX,100
AIX,25
%%writefile example2.txt
foo foo quux labs foo bar jimi quux jimi jimi
foo jimi jimi
data mining is data science
%%writefile WordCount.py
from mrjob.job import MRJob
import re
WORD_RE = re.compile(r"[\w']+")
class MRWordFreqCount(MRJob):
def mapper(self, _, line):
for word in WORD_RE.findall(line):
yield word.lower(), 1
def combiner(self, word, counts):
yield word, sum(counts)
#hello, (1,1,1,1,1,1): using a combiner? NO and YEs
def reducer(self, word, counts):
yield word, sum(counts)
if __name__ == '__main__':
MRWordFreqCount.run()
!python WordCount.py -r hadoop --cmdenv PATH=/opt/anaconda/bin:$PATH example2.txt
from WordCount import MRWordFreqCount
mr_job = MRWordFreqCount(args=['example2.txt'])
with mr_job.make_runner() as runner:
runner.run()
# stream_output: get access of the output
for line in runner.stream_output():
print mr_job.parse_output_line(line)
%%writefile AnotherWordCount.py
from mrjob.job import MRJob
class MRAnotherWordCount(MRJob):
def mapper (self,_,line):
yield "chars", len(line)
yield "words", len(line.split())
yield 'lines', 1
def reducer (self, key, values):
yield key, sum(values)
if __name__ == '__main__':
MRAnotherWordCount.run()
!python AnotherWordCount.py example2.txt
%%writefile AnotherWC3.py
# Copyright 2009-2010 Yelp
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""An implementation of wc as an MRJob.
This is meant as an example of why mapper_final is useful."""
from mrjob.job import MRJob
class MRWordCountUtility(MRJob):
def __init__(self, *args, **kwargs):
super(MRWordCountUtility, self).__init__(*args, **kwargs)
self.chars = 0
self.words = 0
self.lines = 0
def mapper(self, _, line):
# Don't actually yield anything for each line. Instead, collect them
# and yield the sums when all lines have been processed. The results
# will be collected by the reducer.
self.chars += len(line) + 1 # +1 for newline
self.words += sum(1 for word in line.split() if word.strip())
self.lines += 1
def mapper_final(self):
yield('chars', self.chars)
yield('words', self.words)
yield('lines', self.lines)
def reducer(self, key, values):
yield(key, sum(values))
if __name__ == '__main__':
MRWordCountUtility.run()
!python AnotherWC3.py -r hadoop --cmdenv PATH=/opt/anaconda/bin:$PATH example2.txt
%%writefile AnotherWC2.py
from mrjob.job import MRJob
from mrjob.step import MRStep
import re
WORD_RE = re.compile(r"[\w']+")
class MRMostUsedWord(MRJob):
def steps(self):
return [
MRStep(mapper=self.mapper_get_words,
combiner=self.combiner_count_words,
reducer=self.reducer_count_words),
MRStep(reducer=self.reducer_find_max_word)
]
def mapper_get_words(self, _, line):
# yield each word in the line
for word in WORD_RE.findall(line):
self.increment_counter('group', 'counter_name', 1)
yield (word.lower(), 1)
def combiner_count_words(self, word, counts):
# optimization: sum the words we've seen so far
yield (word, sum(counts))
def reducer_count_words(self, word, counts):
# send all (num_occurrences, word) pairs to the same reducer.
# num_occurrences is so we can easily use Python's max() function.
yield None, (sum(counts), word)
# discard the key; it is just None
def reducer_find_max_word(self, _, word_count_pairs):
# each item of word_count_pairs is (count, word),
# so yielding one results in key=counts, value=word
yield max(word_count_pairs)
if __name__ == '__main__':
MRMostUsedWord.run()
!python WordCount.py example2.txt --output-dir mrJobOutput
!ls -las mrJobOutput/
!cat mrJobOutput/part-0000*
%%writefile WordCount2.py
from mrjob.job import MRJob
from mrjob.step import MRStep
import re
WORD_RE = re.compile(r"[\w']+")
class MRWordFreqCount(MRJob):
SORT_VALUES = True
def mapper(self, _, line):
for word in WORD_RE.findall(line):
self.increment_counter('group', 'mapper', 1)
yield word.lower(), 1
def jobconfqqqq(self): #assume we had second job to sort the word counts in decreasing order of counts
orig_jobconf = super(MRWordFreqCount, self).jobconf()
'mapred.reduce.tasks': '1',
}
combined_jobconf = orig_jobconf
combined_jobconf.update(custom_jobconf)
self.jobconf = combined_jobconf
return combined_jobconf custom_jobconf = { #key value pairs
'mapred.output.key.comparator.class': 'org.apache.hadoop.mapred.lib.KeyFieldBasedComparator',
'mapred.text.key.comparator.options': '-k2,2nr',
def combiner(self, word, counts):
self.increment_counter('group', 'combiner', 1)
yield word, sum(counts)
def reducer(self, word, counts):
self.increment_counter('group', 'reducer', 1)
yield word, sum(counts)
def steps(self):
return [MRStep(
mapper = self.mapper,
combiner = self.combiner,
reducer = self.reducer,
#,
# jobconf = self.jobconfqqqq
# jobconf = {'mapred.output.key.comparator.class': 'org.apache.hadoop.mapred.lib.KeyFieldBasedComparator',
# 'mapred.text.key.comparator.options':'-k1r',
# 'mapred.reduce.tasks' : 1}
)]
if __name__ == '__main__':
MRWordFreqCount.run()
!python WordCount2.py --jobconf numReduceTasks=1 example2.txt --output-dir mrJobOutput
!ls -las mrJobOutput/
```
### Calculate Relative Frequency and Sort by TOP and BOTTOM
```
%%writefile WordCount3.3.py
from mrjob.job import MRJob
from mrjob.step import MRStep
import re
WORD_RE = re.compile(r"[\w']+")
class MRWordCount33(MRJob):
def steps(self):
return [
MRStep(mapper=self.mapper_get_words,
combiner=self.combiner_count_words,
reducer=self.reducer_count_words),
MRStep(reducer=self.reducer_find_max_word)
]
def mapper_get_words(self, _, line):
for word in WORD_RE.findall(line):
self.increment_counter('Process', 'Mapper', 1)
yield (word.lower(), 1)
def combiner_count_words(self, word, counts):
# optimization: sum the words we've seen so far
yield (word, sum(counts))
def reducer_count_words(self, word, counts):
# send all (num_occurrences, word) pairs to the same reducer.
# num_occurrences is so we can easily use Python's max() function.
yield None, (sum(counts), word)
# discard the key; it is just None
def reducer_find_max_word(self, _, word_count_pairs):
# each item of word_count_pairs is (count, word),
# so yielding one results in key=counts, value=word
yield max(word_count_pairs)
if __name__ == '__main__':
MRMostUsedWord.run()
%%writefile top_pages.py
"""Find Vroots with more than 400 visits.
This program will take a CSV data file and output tab-seperated lines of
Vroot -> number of visits
To run:
python top_pages.py anonymous-msweb.data
To store output:
python top_pages.py anonymous-msweb.data > top_pages.out
"""
from mrjob.job import MRJob
import csv
def csv_readline(line):
"""Given a sting CSV line, return a list of strings."""
for row in csv.reader([line]):
return row
class TopPages(MRJob):
def mapper(self, line_no, line):
"""Extracts the Vroot that was visited"""
cell = csv_readline(line)
if cell[0] == 'V':
yield ### FILL IN
# What Key, Value do we want to output?
def reducer(self, vroot, visit_counts):
"""Sumarizes the visit counts by adding them together. If total visits
is more than 400, yield the results"""
total = ### FILL IN
# How do we calculate the total visits from the visit_counts?
if total > 400:
yield ### FILL IN
# What Key, Value do we want to output?
if __name__ == '__main__':
TopPages.run()
%reload_ext autoreload
%autoreload 2
from top_pages import TopPages
import csv
mr_job = TopPages(args=['anonymous-msweb.data'])
with mr_job.make_runner() as runner:
runner.run()
for line in runner.stream_output():
print mr_job.parse_output_line(line)
%%writefile TopPages.py
"""Find Vroots with more than 400 visits.
This program will take a CSV data file and output tab-seperated lines of
Vroot -> number of visits
To run:
python top_pages.py anonymous-msweb.data
To store output:
python top_pages.py anonymous-msweb.data > top_pages.out
"""
from mrjob.job import MRJob
import csv
def csv_readline(line):
"""Given a sting CSV line, return a list of strings."""
for row in csv.reader([line]):
return row
class TopPages(MRJob):
def mapper(self, line_no, line):
"""Extracts the Vroot that visit a page"""
cell = csv_readline(line)
if cell[0] == 'V':
yield cell[1],1
def reducer(self, vroot, visit_counts):
"""Sumarizes the visit counts by adding them together. If total visits
is more than 400, yield the results"""
total = sum(i for i in visit_counts)
if total > 400:
yield vroot, total
if __name__ == '__main__':
TopPages.run()
%reload_ext autoreload
%autoreload 2
from TopPages import TopPages
import csv
mr_job = TopPages(args=['anonymous-msweb.data'])
with mr_job.make_runner() as runner:
runner.run()
count = 0
for line in runner.stream_output():
print mr_job.parse_output_line(line)
count += 1
print 'Final count: ', count
```
|
github_jupyter
|
# Debugging Numba problems
## Common problems
Numba is a compiler, if there's a problem, it could well be a "compilery" problem, the dynamic interpretation that comes with the Python interpreter is gone! As with any compiler toolchain there's a bit of a learning curve but once the basics are understood it becomes easy to write quite complex applications.
```
from numba import njit
import numpy as np
```
### Type inference problems
A very large set of problems can be classed as type inference problems. These are problems which appear when Numba can't work out the types of all the variables in your code. Here's an example:
```
@njit
def type_inference_problem():
a = {}
return a
type_inference_problem()
```
Things to note in the above, Numba has said that:
1. It has encountered a typing error.
2. It cannot infer (work out) the type of the variable named `a`.
3. It has an imprecise type for `a` of `DictType[undefined, undefined]`.
4. It's pointing to where the problem is in the source
5. It's giving you things to look at for help
Numba's response is reasonable, how can it possibly compile a specialisation of an empty dictionary, it cannot work out what to use for a key or value type.
### Type unification problems
Another common issue is that of type unification, this is due to Numba needing the inferred variable types for the code it's compiling to be statically determined and type stable. What this usually means is something like the type of a variable is being changed in a loop or there's two (or more) possible return types. Example:
```
@njit
def foo(x):
if x > 10:
return (1,)
else:
return 1
foo(1)
```
Things to note in the above, Numba has said that:
1. It has encountered a typing error.
2. It cannot unify the return types and then lists the offending types.
3. It pointis to the locations in the source that are the cause of the problem.
4. It's giving you things to look at for help.
Numba's response due to it not being possible to compile a function that returns a tuple or an integer? You couldn't do that in C/Fortran, same here!
### Unsupported features
Numba supports a subset of Python and NumPy, it's possible to run into something that hasn't been implemented. For example `str(int)` has not been written yet (this is a rather tricky thing to write :)). This is what it looks like:
```
@njit
def foo():
return str(10)
foo()
```
Things to note in the above, Numba has said that:
1. It has encountered a typing error.
2. It's an invalid use of a `Function` of type `(<class 'str'>)` with argument(s) of type(s): `(Literal[int](10))`
3. It points to the location in the source that is the cause of the problem.
4. It's giving you things to look at for help.
What's this bit about?
```
* parameterized
In definition 0:
All templates rejected with literals.
In definition 1:
All templates rejected without literals.
In definition 2:
All templates rejected with literals.
In definition 3:
All templates rejected without literals.
```
Internally Numba does something akin to "template matching" to try and find something to do the functionality requested with the types requested, it's looking through the definitions see if any match and reporting what they say (which in this case is "rejected").
Here's a different one, Numba's `np.mean` implementation doesn't support `axis`:
```
@njit
def foo():
x = np.arange(100).reshape((10, 10))
return np.mean(x, axis=1)
foo()
```
Things to note in the above, Numba has said that:
1. It has encountered a typing error.
2. It's an invalid use of a `Function` "mean" with argument(s) of type(s): `(array(float64, 2d, C), axis=Literal[int](1))`
3. It's reporting what the various template defintions are responding with: e.g.
"TypingError: numba doesn't support kwarg for mean", which is correct!
4. It points to the location in the source that is the cause of the problem.
5. It's giving you things to look at for help.
A common workaround for the above is to just unroll the loop over the axis, for example:
```
@njit
def foo():
x = np.arange(100).reshape((10, 10))
lim, _ = x.shape
buf = np.empty((lim,), x.dtype)
for i in range(lim):
buf[i] = np.mean(x[i])
return buf
foo()
```
### Lowering errors
"Lowering" is the process of translating the Numba IR to LLVM IR to machine code. Numba tries really hard to prevent lowering errors, but sometimes you might see them, if you do please tell us:
https://github.com/numba/numba/issues/new
A lowering error means that there's a problem in Numba internals. The most common cause is that it worked out that it could compile a function as all the variable types were statically determined, but when it tried to find an implementation for some operation in the function to translate to machine code, it couldn't find one.
<h3><span style="color:blue"> Task 1: Debugging practice</span></h3>
The following code has a couple of issues, see if you can work them out and fix them.
```
x = np.arange(20.).reshape((4, 5))
@njit
def problem_factory(x):
nrm_x = np.linalg.norm(x, ord=2, axis=1) # axis not supported, manual unroll
nrm_total = np.sum(nrm_x)
ret = {} # dict type requires float->int cast, true branch is int and it sets the dict type
if nrm_total > 87:
ret[nrm_total] = 1
else:
ret[nrm_total] = nrm_total
return ret
# This is a fixed version
@njit
def problem_factory_fixed(x):
lim, _ = x.shape
nrm_x = np.empty(lim, x.dtype)
for i in range(lim):
nrm_x[i] = np.linalg.norm(x[i])
nrm_total = np.sum(nrm_x)
ret = {}
if nrm_total > 87:
ret[nrm_total] = 1.0
else:
ret[nrm_total] = nrm_total
return ret
fixed = problem_factory_fixed(x)
expected = problem_factory.py_func(x)
# will pass if "fixed" correctly
for k, v in zip(fixed.items(), expected.items()):
np.testing.assert_allclose(k[0], k[1])
np.testing.assert_allclose(v[0], v[1])
```
## Debugging compiled code
In Numba compiled code debugging typically takes one of a few forms.
1. Temporarily disabling the JIT compiler so that the code just runs in Python and the usual Python debugging tools can be used. Either remove the Numba JIT decorators or set the environment variable `NUMBA_DISABLE_JIT`, to disable JIT compilation globally, [docs](http://numba.pydata.org/numba-doc/latest/reference/envvars.html#envvar-NUMBA_DISABLE_JIT).
2. Traditional "print-to-stdout" debugging, Numba supports the use of `print()` (without interpolation!) so it's relatively easy to inspect values and control flow. e.g.
```
@njit
def demo_print(x):
print("function entry")
if x > 1:
print("branch 1, x = ", x)
else:
print("branch 2, x = ", x)
print("function exit")
demo_print(5)
```
3. Debugging with `gdb` (the GNU debugger). This is not going to be demonstrated here as it does not work with notebooks. However, the gist is to supply the Numba JIT decorator with the kwarg `debug=True` and then Numba has a special function `numba.gdb()` that can be used in your program to automatically launch and attach `gdb` at the call site. For example (and **remember not to run this!**):
```
from numba import gdb
@njit(debug=True)
def _DO_NOT_RUN_gdb_demo(x):
if x > 1:
y = 3
gdb()
else:
y = 5
return y
```
Extensive documentation on using `gdb` with Numba is available [here](http://numba.pydata.org/numba-doc/latest/user/troubleshoot.html#debugging-jit-compiled-code-with-gdb).
|
github_jupyter
|
# Introduction to Strings
---
This notebook covers the topic of strings and their importance in the world of programming. You will learn various methods that will help you manipulate these strings and make useful inferences with them. This notebook assumes that you have already completed the "Introduction to Data Science" notebook.
*Estimated Time: 30 minutes*
---
**Topics Covered:**
- Objects
- String Concatenation
- Loops
- String Methods
**Dependencies:**
```
import numpy as np
from datascience import *
```
## What Are Objects?
Objects are used everywhere when you're coding - even when you dont know it. But what really is an object?
By definition, an object is an **instance** of a **class**. They're an **abstraction**, so they can be used to manipulate data. That sounds complicated, doesn't it? Well, to simplify, think of this: a class is a huge general category of something which holds particular attributes (variables) and actions (functions). Let's assume that Mars has aliens called Xelhas and one of them visits Earth. The Xelha species would be a class, and the alien itself would be an *instance* of that class (or an object). By observing its behavior and mannerisms, we would be able to see how the rest of its species goes about doing things.
Strings are objects too, of the *String* class, which has pre-defined methods that we use. But you don't need to worry about that yet. All you should know is that strings are **not** "primitive" data types, such as integers or booleans. That being said, let's delve right in.
Try running the code cell below:
```
5 + "5"
```
Why did that happen?
This can be classified as a *type* error. As mentioned before, Strings are not primitive data types, like integers and booleans, so when you try to **add** a string to an integer, Python gets confused and throws an error. The important thing to note here is that a String is a String: no matter what its contents may be. If it's between two quotes, it has to be a String.
But what if we followed the "same type" rule and tried to add two Strings? Let's try it.
```
"5" + "5"
```
What?! How does 5 + 5 equal 55?
This is known as concatenation.
## Concatenation
"Concatenating" two items means literally combining or joining them.
When you put the + operator with two or more Strings, Python will take all of the content inside quotes and club it all together to make one String. This process is called **concatenation**.
The following examples illustrate how String concatenation works:
```
"Berk" + "eley"
"B" + "e" + "r" + "k" + "e" + "l" + "e" + "y"
```
Here's a small exercise for you, with a lot of variables. Try making the output "today is a lovely day".
_Hint: Remember to add double quotes with spaces " " because Python literally clubs all text together._
```
a = "oda"
b = "is"
c = "a"
d = "l"
e = "t"
f = "y"
g = "lo"
h = "d"
i = "ve"
# your expression here
```
## String methods
The String class is great for regular use because it comes equipped with a lot of built-in functions with useful properties. These functions, or **methods** can fundamentally transform Strings. Here are some common String methods that may prove to be helpful.
### Replace
For example, the *replace* method replaces all instances of some part of a string with some replacement. A method is invoked on a string by placing a . after the string value, then the name of the method, and finally parentheses containing the arguments.
<string>.<method name>(<argument>, <argument>, ...)
Try to predict the output of these examples, then execute them.
```
# Replace one letter
'Hello'.replace('e', 'i')
# Replace a sequence of letters, which appears twice
'hitchhiker'.replace('hi', 'ma')
```
Once a name is bound to a string value, methods can be invoked on that name as well. The name doesn't change in this case, so a new name is needed to capture the result.
Remember, a string method will replace **every** instance of where the replacement text is found.
```
sharp = 'edged'
hot = sharp.replace('ed', 'ma')
print('sharp =', sharp)
print('hot =', hot)
```
Another very useful method is the **`split`** method. It takes in a "separator string" and splits up the original string into an array, with each element of the array being a separated portion of the string.
Here are some examples:
```
"Another very useful method is the split method".split(" ")
string_of_numbers = "1, 2, 3, 4, 5, 6, 7"
arr_of_strings = string_of_numbers.split(", ")
print(arr_of_strings) # Remember, these elements are still strings!
arr_of_numbers = []
for s in arr_of_strings: # Loop through the array, converting each string to an int
arr_of_numbers.append(int(s))
print(arr_of_numbers)
```
As you can see, the `split` function can be very handy when cleaning up and organizing data (a process known as _parsing_).
## Loops
What do you do when you have to do the same task repetitively? Let's say you have to say Hi to someone five times. Would that require 5 lines of "print('hi')"? No! This is why coding is beautiful. It allows for automation and takes care of all the hard work.
Loops, in the literal meaning of the term, can be used to repeat tasks over and over, until you get your desired output. They are also called "iterators", and they are defined using a variable which changes (either increases or decreases) with each loop, to keep a track of the number of times you're looping.
The most useful loop to know for the scope of this course is the **for** loop.
A for statement begins with the word *for*, followed by a name we want to give each item in the sequence, followed by the word *in*, and ending with an expression that evaluates to a sequence. The indented body of the for statement is executed once for each item in that sequence.
for *variable* in *np.arange(0,5)*:
Don't worry about the np.arange() part yet. Just remember that this expression produces a sequence, and Strings are sequences too! So let's try our loop with Strings!
for each_character in "John Doenero":
*do something*
Interesting! Let's put our code to test.
```
for each_character in "John Doenero":
print(each_character)
```
Cool, right? Now let's do something more useful.
Write a for loop that iterates through the sentence "Hi, I am a quick brown fox and I jump over the lazy dog" and checks if each letter is an *a*. Print out the number of a's in the sentence.
_Hint: try combining what you've learnt from conditions and use a counter._
```
# your code here
for ...
```
## Conclusion
---
Congratulations! You have learned the basics of String manipulation in Python.
## Bibliography
---
Some examples adapted from the UC Berkeley Data 8 textbook, <a href="https://www.inferentialthinking.com">*Inferential Thinking*</a>.
Authors:
- Shriya Vohra
- Scott Lee
- Pancham Yadav
|
github_jupyter
|
##### Copyright 2020 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Training & evaluation with the built-in methods
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/guide/keras/train_and_evaluate"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/keras-team/keras-io/blob/master/tf/train_and_evaluate.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/keras-team/keras-io/blob/master/guides/training_with_built_in_methods.py"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/keras-io/tf/train_and_evaluate.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
## Setup
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
```
## Introduction
This guide covers training, evaluation, and prediction (inference) models
when using built-in APIs for training & validation (such as `model.fit()`,
`model.evaluate()`, `model.predict()`).
If you are interested in leveraging `fit()` while specifying your
own training step function, see the guide
["customizing what happens in `fit()`"](https://www.tensorflow.org/guide/keras/customizing_what_happens_in_fit/).
If you are interested in writing your own training & evaluation loops from
scratch, see the guide
["writing a training loop from scratch"](https://www.tensorflow.org/guide/keras/writing_a_training_loop_from_scratch/).
In general, whether you are using built-in loops or writing your own, model training &
evaluation works strictly in the same way across every kind of Keras model --
Sequential models, models built with the Functional API, and models written from
scratch via model subclassing.
This guide doesn't cover distributed training. For distributed training, see
our [guide to multi-gpu & distributed training](/guides/distributed_training/).
## API overview: a first end-to-end example
When passing data to the built-in training loops of a model, you should either use
**NumPy arrays** (if your data is small and fits in memory) or **`tf.data Dataset`
objects**. In the next few paragraphs, we'll use the MNIST dataset as NumPy arrays, in
order to demonstrate how to use optimizers, losses, and metrics.
Let's consider the following model (here, we build in with the Functional API, but it
could be a Sequential model or a subclassed model as well):
```
inputs = keras.Input(shape=(784,), name="digits")
x = layers.Dense(64, activation="relu", name="dense_1")(inputs)
x = layers.Dense(64, activation="relu", name="dense_2")(x)
outputs = layers.Dense(10, activation="softmax", name="predictions")(x)
model = keras.Model(inputs=inputs, outputs=outputs)
```
Here's what the typical end-to-end workflow looks like, consisting of:
- Training
- Validation on a holdout set generated from the original training data
- Evaluation on the test data
We'll use MNIST data for this example.
```
(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()
# Preprocess the data (these are NumPy arrays)
x_train = x_train.reshape(60000, 784).astype("float32") / 255
x_test = x_test.reshape(10000, 784).astype("float32") / 255
y_train = y_train.astype("float32")
y_test = y_test.astype("float32")
# Reserve 10,000 samples for validation
x_val = x_train[-10000:]
y_val = y_train[-10000:]
x_train = x_train[:-10000]
y_train = y_train[:-10000]
```
We specify the training configuration (optimizer, loss, metrics):
```
model.compile(
optimizer=keras.optimizers.RMSprop(), # Optimizer
# Loss function to minimize
loss=keras.losses.SparseCategoricalCrossentropy(),
# List of metrics to monitor
metrics=[keras.metrics.SparseCategoricalAccuracy()],
)
```
We call `fit()`, which will train the model by slicing the data into "batches" of size
"batch_size", and repeatedly iterating over the entire dataset for a given number of
"epochs".
```
print("Fit model on training data")
history = model.fit(
x_train,
y_train,
batch_size=64,
epochs=2,
# We pass some validation for
# monitoring validation loss and metrics
# at the end of each epoch
validation_data=(x_val, y_val),
)
```
The returned "history" object holds a record of the loss values and metric values
during training:
```
history.history
```
We evaluate the model on the test data via `evaluate()`:
```
# Evaluate the model on the test data using `evaluate`
print("Evaluate on test data")
results = model.evaluate(x_test, y_test, batch_size=128)
print("test loss, test acc:", results)
# Generate predictions (probabilities -- the output of the last layer)
# on new data using `predict`
print("Generate predictions for 3 samples")
predictions = model.predict(x_test[:3])
print("predictions shape:", predictions.shape)
```
Now, let's review each piece of this workflow in detail.
## The `compile()` method: specifying a loss, metrics, and an optimizer
To train a model with `fit()`, you need to specify a loss function, an optimizer, and
optionally, some metrics to monitor.
You pass these to the model as arguments to the `compile()` method:
```
model.compile(
optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),
loss=keras.losses.SparseCategoricalCrossentropy(),
metrics=[keras.metrics.SparseCategoricalAccuracy()],
)
```
The `metrics` argument should be a list -- your model can have any number of metrics.
If your model has multiple outputs, you can specify different losses and metrics for
each output, and you can modulate the contribution of each output to the total loss of
the model. You will find more details about this in the section **"Passing data to
multi-input, multi-output models"**.
Note that if you're satisfied with the default settings, in many cases the optimizer,
loss, and metrics can be specified via string identifiers as a shortcut:
```
model.compile(
optimizer="rmsprop",
loss="sparse_categorical_crossentropy",
metrics=["sparse_categorical_accuracy"],
)
```
For later reuse, let's put our model definition and compile step in functions; we will
call them several times across different examples in this guide.
```
def get_uncompiled_model():
inputs = keras.Input(shape=(784,), name="digits")
x = layers.Dense(64, activation="relu", name="dense_1")(inputs)
x = layers.Dense(64, activation="relu", name="dense_2")(x)
outputs = layers.Dense(10, activation="softmax", name="predictions")(x)
model = keras.Model(inputs=inputs, outputs=outputs)
return model
def get_compiled_model():
model = get_uncompiled_model()
model.compile(
optimizer="rmsprop",
loss="sparse_categorical_crossentropy",
metrics=["sparse_categorical_accuracy"],
)
return model
```
### Many built-in optimizers, losses, and metrics are available
In general, you won't have to create from scratch your own losses, metrics, or
optimizers, because what you need is likely already part of the Keras API:
Optimizers:
- `SGD()` (with or without momentum)
- `RMSprop()`
- `Adam()`
- etc.
Losses:
- `MeanSquaredError()`
- `KLDivergence()`
- `CosineSimilarity()`
- etc.
Metrics:
- `AUC()`
- `Precision()`
- `Recall()`
- etc.
### Custom losses
There are two ways to provide custom losses with Keras. The first example creates a
function that accepts inputs `y_true` and `y_pred`. The following example shows a loss
function that computes the mean squared error between the real data and the
predictions:
```
def custom_mean_squared_error(y_true, y_pred):
return tf.math.reduce_mean(tf.square(y_true - y_pred))
model = get_uncompiled_model()
model.compile(optimizer=keras.optimizers.Adam(), loss=custom_mean_squared_error)
# We need to one-hot encode the labels to use MSE
y_train_one_hot = tf.one_hot(y_train, depth=10)
model.fit(x_train, y_train_one_hot, batch_size=64, epochs=1)
```
If you need a loss function that takes in parameters beside `y_true` and `y_pred`, you
can subclass the `tf.keras.losses.Loss` class and implement the following two methods:
- `__init__(self)`: accept parameters to pass during the call of your loss function
- `call(self, y_true, y_pred)`: use the targets (y_true) and the model predictions
(y_pred) to compute the model's loss
Let's say you want to use mean squared error, but with an added term that
will de-incentivize prediction values far from 0.5 (we assume that the categorical
targets are one-hot encoded and take values between 0 and 1). This
creates an incentive for the model not to be too confident, which may help
reduce overfitting (we won't know if it works until we try!).
Here's how you would do it:
```
class CustomMSE(keras.losses.Loss):
def __init__(self, regularization_factor=0.1, name="custom_mse"):
super().__init__(name=name)
self.regularization_factor = regularization_factor
def call(self, y_true, y_pred):
mse = tf.math.reduce_mean(tf.square(y_true - y_pred))
reg = tf.math.reduce_mean(tf.square(0.5 - y_pred))
return mse + reg * self.regularization_factor
model = get_uncompiled_model()
model.compile(optimizer=keras.optimizers.Adam(), loss=CustomMSE())
y_train_one_hot = tf.one_hot(y_train, depth=10)
model.fit(x_train, y_train_one_hot, batch_size=64, epochs=1)
```
### Custom metrics
If you need a metric that isn't part of the API, you can easily create custom metrics
by subclassing the `tf.keras.metrics.Metric` class. You will need to implement 4
methods:
- `__init__(self)`, in which you will create state variables for your metric.
- `update_state(self, y_true, y_pred, sample_weight=None)`, which uses the targets
y_true and the model predictions y_pred to update the state variables.
- `result(self)`, which uses the state variables to compute the final results.
- `reset_states(self)`, which reinitializes the state of the metric.
State update and results computation are kept separate (in `update_state()` and
`result()`, respectively) because in some cases, results computation might be very
expensive, and would only be done periodically.
Here's a simple example showing how to implement a `CategoricalTruePositives` metric,
that counts how many samples were correctly classified as belonging to a given class:
```
class CategoricalTruePositives(keras.metrics.Metric):
def __init__(self, name="categorical_true_positives", **kwargs):
super(CategoricalTruePositives, self).__init__(name=name, **kwargs)
self.true_positives = self.add_weight(name="ctp", initializer="zeros")
def update_state(self, y_true, y_pred, sample_weight=None):
y_pred = tf.reshape(tf.argmax(y_pred, axis=1), shape=(-1, 1))
values = tf.cast(y_true, "int32") == tf.cast(y_pred, "int32")
values = tf.cast(values, "float32")
if sample_weight is not None:
sample_weight = tf.cast(sample_weight, "float32")
values = tf.multiply(values, sample_weight)
self.true_positives.assign_add(tf.reduce_sum(values))
def result(self):
return self.true_positives
def reset_states(self):
# The state of the metric will be reset at the start of each epoch.
self.true_positives.assign(0.0)
model = get_uncompiled_model()
model.compile(
optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),
loss=keras.losses.SparseCategoricalCrossentropy(),
metrics=[CategoricalTruePositives()],
)
model.fit(x_train, y_train, batch_size=64, epochs=3)
```
### Handling losses and metrics that don't fit the standard signature
The overwhelming majority of losses and metrics can be computed from `y_true` and
`y_pred`, where `y_pred` is an output of your model. But not all of them. For
instance, a regularization loss may only require the activation of a layer (there are
no targets in this case), and this activation may not be a model output.
In such cases, you can call `self.add_loss(loss_value)` from inside the call method of
a custom layer. Losses added in this way get added to the "main" loss during training
(the one passed to `compile()`). Here's a simple example that adds activity
regularization (note that activity regularization is built-in in all Keras layers --
this layer is just for the sake of providing a concrete example):
```
class ActivityRegularizationLayer(layers.Layer):
def call(self, inputs):
self.add_loss(tf.reduce_sum(inputs) * 0.1)
return inputs # Pass-through layer.
inputs = keras.Input(shape=(784,), name="digits")
x = layers.Dense(64, activation="relu", name="dense_1")(inputs)
# Insert activity regularization as a layer
x = ActivityRegularizationLayer()(x)
x = layers.Dense(64, activation="relu", name="dense_2")(x)
outputs = layers.Dense(10, name="predictions")(x)
model = keras.Model(inputs=inputs, outputs=outputs)
model.compile(
optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),
loss=keras.losses.SparseCategoricalCrossentropy(from_logits=True),
)
# The displayed loss will be much higher than before
# due to the regularization component.
model.fit(x_train, y_train, batch_size=64, epochs=1)
```
You can do the same for logging metric values, using `add_metric()`:
```
class MetricLoggingLayer(layers.Layer):
def call(self, inputs):
# The `aggregation` argument defines
# how to aggregate the per-batch values
# over each epoch:
# in this case we simply average them.
self.add_metric(
keras.backend.std(inputs), name="std_of_activation", aggregation="mean"
)
return inputs # Pass-through layer.
inputs = keras.Input(shape=(784,), name="digits")
x = layers.Dense(64, activation="relu", name="dense_1")(inputs)
# Insert std logging as a layer.
x = MetricLoggingLayer()(x)
x = layers.Dense(64, activation="relu", name="dense_2")(x)
outputs = layers.Dense(10, name="predictions")(x)
model = keras.Model(inputs=inputs, outputs=outputs)
model.compile(
optimizer=keras.optimizers.RMSprop(learning_rate=1e-3),
loss=keras.losses.SparseCategoricalCrossentropy(from_logits=True),
)
model.fit(x_train, y_train, batch_size=64, epochs=1)
```
In the [Functional API](https://www.tensorflow.org/guide/keras/functional/),
you can also call `model.add_loss(loss_tensor)`,
or `model.add_metric(metric_tensor, name, aggregation)`.
Here's a simple example:
```
inputs = keras.Input(shape=(784,), name="digits")
x1 = layers.Dense(64, activation="relu", name="dense_1")(inputs)
x2 = layers.Dense(64, activation="relu", name="dense_2")(x1)
outputs = layers.Dense(10, name="predictions")(x2)
model = keras.Model(inputs=inputs, outputs=outputs)
model.add_loss(tf.reduce_sum(x1) * 0.1)
model.add_metric(keras.backend.std(x1), name="std_of_activation", aggregation="mean")
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss=keras.losses.SparseCategoricalCrossentropy(from_logits=True),
)
model.fit(x_train, y_train, batch_size=64, epochs=1)
```
Note that when you pass losses via `add_loss()`, it becomes possible to call
`compile()` without a loss function, since the model already has a loss to minimize.
Consider the following `LogisticEndpoint` layer: it takes as inputs
targets & logits, and it tracks a crossentropy loss via `add_loss()`. It also
tracks classification accuracy via `add_metric()`.
```
class LogisticEndpoint(keras.layers.Layer):
def __init__(self, name=None):
super(LogisticEndpoint, self).__init__(name=name)
self.loss_fn = keras.losses.BinaryCrossentropy(from_logits=True)
self.accuracy_fn = keras.metrics.BinaryAccuracy()
def call(self, targets, logits, sample_weights=None):
# Compute the training-time loss value and add it
# to the layer using `self.add_loss()`.
loss = self.loss_fn(targets, logits, sample_weights)
self.add_loss(loss)
# Log accuracy as a metric and add it
# to the layer using `self.add_metric()`.
acc = self.accuracy_fn(targets, logits, sample_weights)
self.add_metric(acc, name="accuracy")
# Return the inference-time prediction tensor (for `.predict()`).
return tf.nn.softmax(logits)
```
You can use it in a model with two inputs (input data & targets), compiled without a
`loss` argument, like this:
```
import numpy as np
inputs = keras.Input(shape=(3,), name="inputs")
targets = keras.Input(shape=(10,), name="targets")
logits = keras.layers.Dense(10)(inputs)
predictions = LogisticEndpoint(name="predictions")(logits, targets)
model = keras.Model(inputs=[inputs, targets], outputs=predictions)
model.compile(optimizer="adam") # No loss argument!
data = {
"inputs": np.random.random((3, 3)),
"targets": np.random.random((3, 10)),
}
model.fit(data)
```
For more information about training multi-input models, see the section **Passing data
to multi-input, multi-output models**.
### Automatically setting apart a validation holdout set
In the first end-to-end example you saw, we used the `validation_data` argument to pass
a tuple of NumPy arrays `(x_val, y_val)` to the model for evaluating a validation loss
and validation metrics at the end of each epoch.
Here's another option: the argument `validation_split` allows you to automatically
reserve part of your training data for validation. The argument value represents the
fraction of the data to be reserved for validation, so it should be set to a number
higher than 0 and lower than 1. For instance, `validation_split=0.2` means "use 20% of
the data for validation", and `validation_split=0.6` means "use 60% of the data for
validation".
The way the validation is computed is by taking the last x% samples of the arrays
received by the fit call, before any shuffling.
Note that you can only use `validation_split` when training with NumPy data.
```
model = get_compiled_model()
model.fit(x_train, y_train, batch_size=64, validation_split=0.2, epochs=1)
```
## Training & evaluation from tf.data Datasets
In the past few paragraphs, you've seen how to handle losses, metrics, and optimizers,
and you've seen how to use the `validation_data` and `validation_split` arguments in
fit, when your data is passed as NumPy arrays.
Let's now take a look at the case where your data comes in the form of a
`tf.data.Dataset` object.
The `tf.data` API is a set of utilities in TensorFlow 2.0 for loading and preprocessing
data in a way that's fast and scalable.
For a complete guide about creating `Datasets`, see the
[tf.data documentation](https://www.tensorflow.org/guide/data).
You can pass a `Dataset` instance directly to the methods `fit()`, `evaluate()`, and
`predict()`:
```
model = get_compiled_model()
# First, let's create a training Dataset instance.
# For the sake of our example, we'll use the same MNIST data as before.
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))
# Shuffle and slice the dataset.
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)
# Now we get a test dataset.
test_dataset = tf.data.Dataset.from_tensor_slices((x_test, y_test))
test_dataset = test_dataset.batch(64)
# Since the dataset already takes care of batching,
# we don't pass a `batch_size` argument.
model.fit(train_dataset, epochs=3)
# You can also evaluate or predict on a dataset.
print("Evaluate")
result = model.evaluate(test_dataset)
dict(zip(model.metrics_names, result))
```
Note that the Dataset is reset at the end of each epoch, so it can be reused of the
next epoch.
If you want to run training only on a specific number of batches from this Dataset, you
can pass the `steps_per_epoch` argument, which specifies how many training steps the
model should run using this Dataset before moving on to the next epoch.
If you do this, the dataset is not reset at the end of each epoch, instead we just keep
drawing the next batches. The dataset will eventually run out of data (unless it is an
infinitely-looping dataset).
```
model = get_compiled_model()
# Prepare the training dataset
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)
# Only use the 100 batches per epoch (that's 64 * 100 samples)
model.fit(train_dataset, epochs=3, steps_per_epoch=100)
```
### Using a validation dataset
You can pass a `Dataset` instance as the `validation_data` argument in `fit()`:
```
model = get_compiled_model()
# Prepare the training dataset
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)
# Prepare the validation dataset
val_dataset = tf.data.Dataset.from_tensor_slices((x_val, y_val))
val_dataset = val_dataset.batch(64)
model.fit(train_dataset, epochs=1, validation_data=val_dataset)
```
At the end of each epoch, the model will iterate over the validation dataset and
compute the validation loss and validation metrics.
If you want to run validation only on a specific number of batches from this dataset,
you can pass the `validation_steps` argument, which specifies how many validation
steps the model should run with the validation dataset before interrupting validation
and moving on to the next epoch:
```
model = get_compiled_model()
# Prepare the training dataset
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)
# Prepare the validation dataset
val_dataset = tf.data.Dataset.from_tensor_slices((x_val, y_val))
val_dataset = val_dataset.batch(64)
model.fit(
train_dataset,
epochs=1,
# Only run validation using the first 10 batches of the dataset
# using the `validation_steps` argument
validation_data=val_dataset,
validation_steps=10,
)
```
Note that the validation dataset will be reset after each use (so that you will always
be evaluating on the same samples from epoch to epoch).
The argument `validation_split` (generating a holdout set from the training data) is
not supported when training from `Dataset` objects, since this features requires the
ability to index the samples of the datasets, which is not possible in general with
the `Dataset` API.
## Other input formats supported
Besides NumPy arrays, eager tensors, and TensorFlow `Datasets`, it's possible to train
a Keras model using Pandas dataframes, or from Python generators that yield batches of
data & labels.
In particular, the `keras.utils.Sequence` class offers a simple interface to build
Python data generators that are multiprocessing-aware and can be shuffled.
In general, we recommend that you use:
- NumPy input data if your data is small and fits in memory
- `Dataset` objects if you have large datasets and you need to do distributed training
- `Sequence` objects if you have large datasets and you need to do a lot of custom
Python-side processing that cannot be done in TensorFlow (e.g. if you rely on external libraries
for data loading or preprocessing).
## Using a `keras.utils.Sequence` object as input
`keras.utils.Sequence` is a utility that you can subclass to obtain a Python generator with
two important properties:
- It works well with multiprocessing.
- It can be shuffled (e.g. when passing `shuffle=True` in `fit()`).
A `Sequence` must implement two methods:
- `__getitem__`
- `__len__`
The method `__getitem__` should return a complete batch.
If you want to modify your dataset between epochs, you may implement `on_epoch_end`.
Here's a quick example:
```python
from skimage.io import imread
from skimage.transform import resize
import numpy as np
# Here, `filenames` is list of path to the images
# and `labels` are the associated labels.
class CIFAR10Sequence(Sequence):
def __init__(self, filenames, labels, batch_size):
self.filenames, self.labels = filenames, labels
self.batch_size = batch_size
def __len__(self):
return int(np.ceil(len(self.filenames) / float(self.batch_size)))
def __getitem__(self, idx):
batch_x = self.filenames[idx * self.batch_size:(idx + 1) * self.batch_size]
batch_y = self.labels[idx * self.batch_size:(idx + 1) * self.batch_size]
return np.array([
resize(imread(filename), (200, 200))
for filename in batch_x]), np.array(batch_y)
sequence = CIFAR10Sequence(filenames, labels, batch_size)
model.fit(sequence, epochs=10)
```
## Using sample weighting and class weighting
With the default settings the weight of a sample is decided by its frequency
in the dataset. There are two methods to weight the data, independent of
sample frequency:
* Class weights
* Sample weights
### Class weights
This is set by passing a dictionary to the `class_weight` argument to
`Model.fit()`. This dictionary maps class indices to the weight that should
be used for samples belonging to this class.
This can be used to balance classes without resampling, or to train a
model that has a gives more importance to a particular class.
For instance, if class "0" is half as represented as class "1" in your data,
you could use `Model.fit(..., class_weight={0: 1., 1: 0.5})`.
Here's a NumPy example where we use class weights or sample weights to
give more importance to the correct classification of class #5 (which
is the digit "5" in the MNIST dataset).
```
import numpy as np
class_weight = {
0: 1.0,
1: 1.0,
2: 1.0,
3: 1.0,
4: 1.0,
# Set weight "2" for class "5",
# making this class 2x more important
5: 2.0,
6: 1.0,
7: 1.0,
8: 1.0,
9: 1.0,
}
print("Fit with class weight")
model = get_compiled_model()
model.fit(x_train, y_train, class_weight=class_weight, batch_size=64, epochs=1)
```
### Sample weights
For fine grained control, or if you are not building a classifier,
you can use "sample weights".
- When training from NumPy data: Pass the `sample_weight`
argument to `Model.fit()`.
- When training from `tf.data` or any other sort of iterator:
Yield `(input_batch, label_batch, sample_weight_batch)` tuples.
A "sample weights" array is an array of numbers that specify how much weight
each sample in a batch should have in computing the total loss. It is commonly
used in imbalanced classification problems (the idea being to give more weight
to rarely-seen classes).
When the weights used are ones and zeros, the array can be used as a *mask* for
the loss function (entirely discarding the contribution of certain samples to
the total loss).
```
sample_weight = np.ones(shape=(len(y_train),))
sample_weight[y_train == 5] = 2.0
print("Fit with sample weight")
model = get_compiled_model()
model.fit(x_train, y_train, sample_weight=sample_weight, batch_size=64, epochs=1)
```
Here's a matching `Dataset` example:
```
sample_weight = np.ones(shape=(len(y_train),))
sample_weight[y_train == 5] = 2.0
# Create a Dataset that includes sample weights
# (3rd element in the return tuple).
train_dataset = tf.data.Dataset.from_tensor_slices((x_train, y_train, sample_weight))
# Shuffle and slice the dataset.
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)
model = get_compiled_model()
model.fit(train_dataset, epochs=1)
```
## Passing data to multi-input, multi-output models
In the previous examples, we were considering a model with a single input (a tensor of
shape `(764,)`) and a single output (a prediction tensor of shape `(10,)`). But what
about models that have multiple inputs or outputs?
Consider the following model, which has an image input of shape `(32, 32, 3)` (that's
`(height, width, channels)`) and a timeseries input of shape `(None, 10)` (that's
`(timesteps, features)`). Our model will have two outputs computed from the
combination of these inputs: a "score" (of shape `(1,)`) and a probability
distribution over five classes (of shape `(5,)`).
```
image_input = keras.Input(shape=(32, 32, 3), name="img_input")
timeseries_input = keras.Input(shape=(None, 10), name="ts_input")
x1 = layers.Conv2D(3, 3)(image_input)
x1 = layers.GlobalMaxPooling2D()(x1)
x2 = layers.Conv1D(3, 3)(timeseries_input)
x2 = layers.GlobalMaxPooling1D()(x2)
x = layers.concatenate([x1, x2])
score_output = layers.Dense(1, name="score_output")(x)
class_output = layers.Dense(5, activation="softmax", name="class_output")(x)
model = keras.Model(
inputs=[image_input, timeseries_input], outputs=[score_output, class_output]
)
```
Let's plot this model, so you can clearly see what we're doing here (note that the
shapes shown in the plot are batch shapes, rather than per-sample shapes).
```
keras.utils.plot_model(model, "multi_input_and_output_model.png", show_shapes=True)
```
At compilation time, we can specify different losses to different outputs, by passing
the loss functions as a list:
```
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss=[keras.losses.MeanSquaredError(), keras.losses.CategoricalCrossentropy()],
)
```
If we only passed a single loss function to the model, the same loss function would be
applied to every output (which is not appropriate here).
Likewise for metrics:
```
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss=[keras.losses.MeanSquaredError(), keras.losses.CategoricalCrossentropy()],
metrics=[
[
keras.metrics.MeanAbsolutePercentageError(),
keras.metrics.MeanAbsoluteError(),
],
[keras.metrics.CategoricalAccuracy()],
],
)
```
Since we gave names to our output layers, we could also specify per-output losses and
metrics via a dict:
```
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss={
"score_output": keras.losses.MeanSquaredError(),
"class_output": keras.losses.CategoricalCrossentropy(),
},
metrics={
"score_output": [
keras.metrics.MeanAbsolutePercentageError(),
keras.metrics.MeanAbsoluteError(),
],
"class_output": [keras.metrics.CategoricalAccuracy()],
},
)
```
We recommend the use of explicit names and dicts if you have more than 2 outputs.
It's possible to give different weights to different output-specific losses (for
instance, one might wish to privilege the "score" loss in our example, by giving to 2x
the importance of the class loss), using the `loss_weights` argument:
```
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss={
"score_output": keras.losses.MeanSquaredError(),
"class_output": keras.losses.CategoricalCrossentropy(),
},
metrics={
"score_output": [
keras.metrics.MeanAbsolutePercentageError(),
keras.metrics.MeanAbsoluteError(),
],
"class_output": [keras.metrics.CategoricalAccuracy()],
},
loss_weights={"score_output": 2.0, "class_output": 1.0},
)
```
You could also chose not to compute a loss for certain outputs, if these outputs meant
for prediction but not for training:
```
# List loss version
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss=[None, keras.losses.CategoricalCrossentropy()],
)
# Or dict loss version
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss={"class_output": keras.losses.CategoricalCrossentropy()},
)
```
Passing data to a multi-input or multi-output model in fit works in a similar way as
specifying a loss function in compile: you can pass **lists of NumPy arrays** (with
1:1 mapping to the outputs that received a loss function) or **dicts mapping output
names to NumPy arrays**.
```
model.compile(
optimizer=keras.optimizers.RMSprop(1e-3),
loss=[keras.losses.MeanSquaredError(), keras.losses.CategoricalCrossentropy()],
)
# Generate dummy NumPy data
img_data = np.random.random_sample(size=(100, 32, 32, 3))
ts_data = np.random.random_sample(size=(100, 20, 10))
score_targets = np.random.random_sample(size=(100, 1))
class_targets = np.random.random_sample(size=(100, 5))
# Fit on lists
model.fit([img_data, ts_data], [score_targets, class_targets], batch_size=32, epochs=1)
# Alternatively, fit on dicts
model.fit(
{"img_input": img_data, "ts_input": ts_data},
{"score_output": score_targets, "class_output": class_targets},
batch_size=32,
epochs=1,
)
```
Here's the `Dataset` use case: similarly as what we did for NumPy arrays, the `Dataset`
should return a tuple of dicts.
```
train_dataset = tf.data.Dataset.from_tensor_slices(
(
{"img_input": img_data, "ts_input": ts_data},
{"score_output": score_targets, "class_output": class_targets},
)
)
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(64)
model.fit(train_dataset, epochs=1)
```
## Using callbacks
Callbacks in Keras are objects that are called at different point during training (at
the start of an epoch, at the end of a batch, at the end of an epoch, etc.) and which
can be used to implement behaviors such as:
- Doing validation at different points during training (beyond the built-in per-epoch
validation)
- Checkpointing the model at regular intervals or when it exceeds a certain accuracy
threshold
- Changing the learning rate of the model when training seems to be plateauing
- Doing fine-tuning of the top layers when training seems to be plateauing
- Sending email or instant message notifications when training ends or where a certain
performance threshold is exceeded
- Etc.
Callbacks can be passed as a list to your call to `fit()`:
```
model = get_compiled_model()
callbacks = [
keras.callbacks.EarlyStopping(
# Stop training when `val_loss` is no longer improving
monitor="val_loss",
# "no longer improving" being defined as "no better than 1e-2 less"
min_delta=1e-2,
# "no longer improving" being further defined as "for at least 2 epochs"
patience=2,
verbose=1,
)
]
model.fit(
x_train,
y_train,
epochs=20,
batch_size=64,
callbacks=callbacks,
validation_split=0.2,
)
```
### Many built-in callbacks are available
- `ModelCheckpoint`: Periodically save the model.
- `EarlyStopping`: Stop training when training is no longer improving the validation
metrics.
- `TensorBoard`: periodically write model logs that can be visualized in
[TensorBoard](https://www.tensorflow.org/tensorboard) (more details in the section
"Visualization").
- `CSVLogger`: streams loss and metrics data to a CSV file.
- etc.
See the [callbacks documentation](https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/) for the complete list.
### Writing your own callback
You can create a custom callback by extending the base class
`keras.callbacks.Callback`. A callback has access to its associated model through the
class property `self.model`.
Make sure to read the
[complete guide to writing custom callbacks](https://www.tensorflow.org/guide/keras/custom_callback/).
Here's a simple example saving a list of per-batch loss values during training:
```
class LossHistory(keras.callbacks.Callback):
def on_train_begin(self, logs):
self.per_batch_losses = []
def on_batch_end(self, batch, logs):
self.per_batch_losses.append(logs.get("loss"))
```
## Checkpointing models
When you're training model on relatively large datasets, it's crucial to save
checkpoints of your model at frequent intervals.
The easiest way to achieve this is with the `ModelCheckpoint` callback:
```
model = get_compiled_model()
callbacks = [
keras.callbacks.ModelCheckpoint(
# Path where to save the model
# The two parameters below mean that we will overwrite
# the current checkpoint if and only if
# the `val_loss` score has improved.
# The saved model name will include the current epoch.
filepath="mymodel_{epoch}",
save_best_only=True, # Only save a model if `val_loss` has improved.
monitor="val_loss",
verbose=1,
)
]
model.fit(
x_train, y_train, epochs=2, batch_size=64, callbacks=callbacks, validation_split=0.2
)
```
The `ModelCheckpoint` callback can be used to implement fault-tolerance:
the ability to restart training from the last saved state of the model in case training
gets randomly interrupted. Here's a basic example:
```
import os
# Prepare a directory to store all the checkpoints.
checkpoint_dir = "./ckpt"
if not os.path.exists(checkpoint_dir):
os.makedirs(checkpoint_dir)
def make_or_restore_model():
# Either restore the latest model, or create a fresh one
# if there is no checkpoint available.
checkpoints = [checkpoint_dir + "/" + name for name in os.listdir(checkpoint_dir)]
if checkpoints:
latest_checkpoint = max(checkpoints, key=os.path.getctime)
print("Restoring from", latest_checkpoint)
return keras.models.load_model(latest_checkpoint)
print("Creating a new model")
return get_compiled_model()
model = make_or_restore_model()
callbacks = [
# This callback saves a SavedModel every 100 batches.
# We include the training loss in the saved model name.
keras.callbacks.ModelCheckpoint(
filepath=checkpoint_dir + "/ckpt-loss={loss:.2f}", save_freq=100
)
]
model.fit(x_train, y_train, epochs=1, callbacks=callbacks)
```
You call also write your own callback for saving and restoring models.
For a complete guide on serialization and saving, see the
[guide to saving and serializing Models](https://www.tensorflow.org/guide/keras/save_and_serialize/).
## Using learning rate schedules
A common pattern when training deep learning models is to gradually reduce the learning
as training progresses. This is generally known as "learning rate decay".
The learning decay schedule could be static (fixed in advance, as a function of the
current epoch or the current batch index), or dynamic (responding to the current
behavior of the model, in particular the validation loss).
### Passing a schedule to an optimizer
You can easily use a static learning rate decay schedule by passing a schedule object
as the `learning_rate` argument in your optimizer:
```
initial_learning_rate = 0.1
lr_schedule = keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate, decay_steps=100000, decay_rate=0.96, staircase=True
)
optimizer = keras.optimizers.RMSprop(learning_rate=lr_schedule)
```
Several built-in schedules are available: `ExponentialDecay`, `PiecewiseConstantDecay`,
`PolynomialDecay`, and `InverseTimeDecay`.
### Using callbacks to implement a dynamic learning rate schedule
A dynamic learning rate schedule (for instance, decreasing the learning rate when the
validation loss is no longer improving) cannot be achieved with these schedule objects
since the optimizer does not have access to validation metrics.
However, callbacks do have access to all metrics, including validation metrics! You can
thus achieve this pattern by using a callback that modifies the current learning rate
on the optimizer. In fact, this is even built-in as the `ReduceLROnPlateau` callback.
## Visualizing loss and metrics during training
The best way to keep an eye on your model during training is to use
[TensorBoard](https://www.tensorflow.org/tensorboard), a browser-based application
that you can run locally that provides you with:
- Live plots of the loss and metrics for training and evaluation
- (optionally) Visualizations of the histograms of your layer activations
- (optionally) 3D visualizations of the embedding spaces learned by your `Embedding`
layers
If you have installed TensorFlow with pip, you should be able to launch TensorBoard
from the command line:
```
tensorboard --logdir=/full_path_to_your_logs
```
### Using the TensorBoard callback
The easiest way to use TensorBoard with a Keras model and the fit method is the
`TensorBoard` callback.
In the simplest case, just specify where you want the callback to write logs, and
you're good to go:
```
keras.callbacks.TensorBoard(
log_dir="/full_path_to_your_logs",
histogram_freq=0, # How often to log histogram visualizations
embeddings_freq=0, # How often to log embedding visualizations
update_freq="epoch",
) # How often to write logs (default: once per epoch)
```
For more information, see the
[documentation for the `TensorBoard` callback](https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/tensorboard/).
|
github_jupyter
|
# Naive Bayes Classifier (Self Made)
### 1. Importing Libraries
```
import numpy as np
import matplotlib.pyplot as plt
import os
import pandas as pd
from sklearn.metrics import r2_score
from sklearn.datasets import load_boston
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn import metrics
from sklearn.metrics import confusion_matrix
from collections import defaultdict
```
### 2. Data Preprocessing
```
pima = pd.read_csv("diabetes.csv")
pima.head()
pima.info()
#normalizing the dataset
scalar = preprocessing.MinMaxScaler()
pima = scalar.fit_transform(pima)
#split dataset in features and target variable
X = pima[:,:8]
y = pima[:, 8]
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.3, random_state=42)
print(X_train.shape, X_test.shape, Y_train.shape, Y_test.shape)
```
### 3. Required Functions
```
def normal_distr(x, mean, dev):
#finding the value through the normal distribution formula
return (1/(np.sqrt(2 * np.pi) * dev)) * (np.exp(- (((x - mean) / dev) ** 2) / 2))
def finding_mean(X):
return np.mean(X)
def finding_std_dev(X):
return np.std(X)
#def pred(X_test):
def train(X_train,Y_train):
labels = set(Y_train)
cnt_table = defaultdict(list)
for row in range(X_train.shape[0]):
for col in range(X_train.shape[1]):
cnt_table[(col, Y_train[row])].append(X_train[row][col])
lookup_list = defaultdict(list)
for item in cnt_table.items():
X_category = np.asarray(item[1])
lookup_list[(item[0][0], item[0][1])].append(finding_mean(X_category))
lookup_list[(item[0][0], item[0][1])].append(finding_std_dev(X_category))
return lookup_list
def pred(X_test, lookup_list):
Y_pred = []
for row in range(X_test.shape[0]):
prob_yes = 1
prob_no = 1
for col in range(X_test.shape[1]):
prob_yes = prob_yes * normal_distr(X_test[row][col], lookup_list[(col, 1)][0], lookup_list[(col, 1)][1])
prob_no = prob_no * normal_distr(X_test[row][col], lookup_list[(col, 0)][0], lookup_list[(col, 1)][1])
if(prob_yes >= prob_no):
Y_pred.append(1)
else:
Y_pred.append(0)
return np.asarray(Y_pred)
def score(Y_pred, Y_test):
correct_pred = np.sum(Y_pred == Y_test)
return correct_pred / Y_pred.shape[0]
def naive_bayes(X_train,Y_train, X_test, Y_test):
lookup_list = train(X_train, Y_train)
Y_pred = pred(X_test, lookup_list)
return score(Y_pred, Y_test)
score = naive_bayes(X_train, Y_train, X_test, Y_test)
print("The accuracy of the model is : {0}".format(score))
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import pickle
import time
import itertools
import matplotlib
matplotlib.rcParams.update({'font.size': 17.5})
import matplotlib.pyplot as plt
%matplotlib inline
import sys
import os.path
sys.path.append( os.path.abspath(os.path.join( os.path.dirname('..') , os.path.pardir )) )
from FLAMEdb import *
from FLAMEbit import *
# data generation, tune the tradeoff_param to generation to generate different plots
d = data_generation_gradual_decrease_imbalance( 10000 , 10000 , 20 )
df = d[0]
holdout,_ = data_generation_gradual_decrease_imbalance( 10000 , 10000, 20 )
res = run_bit(df, holdout, range(20), [2]*20, tradeoff_param = 0.5)
def bubble_plot(res):
sizes = []
effects = []
for i in range(min(len(res),21)):
r = res[i]
if (r is None):
effects.append([0])
sizes.append([0])
continue
effects.append(list( r['effect'] ) )
sizes.append(list(r['size'] ) )
return sizes, effects
# plot percent of units matched, figure 4
matplotlib.rcParams.update({'font.size': 17.5})
#res = pickle.load(open('__thePickleFile__', 'rb'))[1]
ss, es = bubble_plot(res[1])
s = []
for i in ss:
s.append(np.sum(i)/float(20000))
pct = [sum(s[:i+1]) for i in range(len(s))]
plt.figure(figsize=(5,5))
plt.plot(pct, alpha = 0.6 , color = 'blue')
plt.xticks(range(len(ss)), [str(20-i) if i%5==0 else '' for i in range(20) ] )
plt.ylabel('% of units matched')
plt.xlabel('number of covariates remaining')
plt.ylim([0,1])
plt.tight_layout()
# plot the CATE on each level, figure 5
ss, es = bubble_plot(res[1])
plt.figure(figsize=(5,5))
for i in range(len(ss)):
plt.scatter([i]*len(es[i]), es[i], s = ss[i], alpha = 0.6 , color = 'blue')
plt.xticks(range(len(ss)), [str(20-i) if i%5==0 else '' for i in range(20) ] )
plt.ylabel('estimated treatment effect')
plt.xlabel('number of covariates remaining')
plt.ylim([8,14])
plt.tight_layout()
#plt.savefig('tradeoff01.png', dpi = 300)
# figure 6
units_matched = []
CATEs = []
for i in range(len(res[1])):
r = res[1][i]
units_matched.append( np.sum(r['size']) )
l = list(res[1][i]['effect'])
CATEs.append( l )
PCTs = []
for i in range(len(units_matched)):
PCTs.append( np.sum(units_matched[:i+1])/30000 )
for i in range(len(PCTs)):
CATE = CATEs[i]
if len(CATE) > 0:
plt.scatter( [PCTs[i]] * len(CATE), CATE, alpha = 0.6, color = 'b' )
plt.xlabel('% of Units Matched')
plt.ylabel('Estimated Treatment Effect')
#plt.ylim([8,14])
plt.xlim([-0.1,1])
plt.tight_layout()
```
|
github_jupyter
|
# Dense Sentiment Classifier
In this notebook, we build a dense neural net to classify IMDB movie reviews by their sentiment.
```
#load watermark
%load_ext watermark
%watermark -a 'Gopala KR' -u -d -v -p watermark,numpy,pandas,matplotlib,nltk,sklearn,tensorflow,theano,mxnet,chainer,seaborn,keras,tflearn,bokeh,gensim
```
#### Load dependencies
```
import keras
from keras.datasets import imdb
from keras.preprocessing.sequence import pad_sequences
from keras.models import Sequential
from keras.layers import Dense, Flatten, Dropout
from keras.layers import Embedding # new!
from keras.callbacks import ModelCheckpoint # new!
import os # new!
from sklearn.metrics import roc_auc_score, roc_curve # new!
import pandas as pd
import matplotlib.pyplot as plt # new!
%matplotlib inline
```
#### Set hyperparameters
```
# output directory name:
output_dir = 'model_output/dense'
# training:
epochs = 4
batch_size = 128
# vector-space embedding:
n_dim = 64
n_unique_words = 5000 # as per Maas et al. (2011); may not be optimal
n_words_to_skip = 50 # ditto
max_review_length = 100
pad_type = trunc_type = 'pre'
# neural network architecture:
n_dense = 64
dropout = 0.5
```
#### Load data
For a given data set:
* the Keras text utilities [here](https://keras.io/preprocessing/text/) quickly preprocess natural language and convert it into an index
* the `keras.preprocessing.text.Tokenizer` class may do everything you need in one line:
* tokenize into words or characters
* `num_words`: maximum unique tokens
* filter out punctuation
* lower case
* convert words to an integer index
```
(x_train, y_train), (x_valid, y_valid) = imdb.load_data(num_words=n_unique_words, skip_top=n_words_to_skip)
x_train[0:6] # 0 reserved for padding; 1 would be starting character; 2 is unknown; 3 is most common word, etc.
for x in x_train[0:6]:
print(len(x))
y_train[0:6]
len(x_train), len(x_valid)
```
#### Restoring words from index
```
word_index = keras.datasets.imdb.get_word_index()
word_index = {k:(v+3) for k,v in word_index.items()}
word_index["PAD"] = 0
word_index["START"] = 1
word_index["UNK"] = 2
word_index
index_word = {v:k for k,v in word_index.items()}
x_train[0]
' '.join(index_word[id] for id in x_train[0])
(all_x_train,_),(all_x_valid,_) = imdb.load_data()
' '.join(index_word[id] for id in all_x_train[0])
```
#### Preprocess data
```
x_train = pad_sequences(x_train, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)
x_valid = pad_sequences(x_valid, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)
x_train[0:6]
for x in x_train[0:6]:
print(len(x))
' '.join(index_word[id] for id in x_train[0])
' '.join(index_word[id] for id in x_train[5])
```
#### Design neural network architecture
```
model = Sequential()
model.add(Embedding(n_unique_words, n_dim, input_length=max_review_length))
model.add(Flatten())
model.add(Dense(n_dense, activation='relu'))
model.add(Dropout(dropout))
# model.add(Dense(n_dense, activation='relu'))
# model.add(Dropout(dropout))
model.add(Dense(1, activation='sigmoid')) # mathematically equivalent to softmax with two classes
model.summary() # so many parameters!
# embedding layer dimensions and parameters:
n_dim, n_unique_words, n_dim*n_unique_words
# ...flatten:
max_review_length, n_dim, n_dim*max_review_length
# ...dense:
n_dense, n_dim*max_review_length*n_dense + n_dense # weights + biases
# ...and output:
n_dense + 1
```
#### Configure model
```
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
modelcheckpoint = ModelCheckpoint(filepath=output_dir+"/weights.{epoch:02d}.hdf5")
if not os.path.exists(output_dir):
os.makedirs(output_dir)
```
#### Train!
```
# 84.7% validation accuracy in epoch 2
model.fit(x_train, y_train, batch_size=batch_size, epochs=epochs, verbose=1, validation_data=(x_valid, y_valid), callbacks=[modelcheckpoint])
```
#### Evaluate
```
model.load_weights(output_dir+"/weights.01.hdf5") # zero-indexed
y_hat = model.predict_proba(x_valid)
len(y_hat)
y_hat[0]
plt.hist(y_hat)
_ = plt.axvline(x=0.5, color='orange')
pct_auc = roc_auc_score(y_valid, y_hat)*100.0
"{:0.2f}".format(pct_auc)
float_y_hat = []
for y in y_hat:
float_y_hat.append(y[0])
ydf = pd.DataFrame(list(zip(float_y_hat, y_valid)), columns=['y_hat', 'y'])
ydf.head(10)
' '.join(index_word[id] for id in all_x_valid[0])
' '.join(index_word[id] for id in all_x_valid[6])
ydf[(ydf.y == 0) & (ydf.y_hat > 0.9)].head(10)
' '.join(index_word[id] for id in all_x_valid[489])
ydf[(ydf.y == 1) & (ydf.y_hat < 0.1)].head(10)
' '.join(index_word[id] for id in all_x_valid[927])
```
|
github_jupyter
|
# Basics of the DVR calculations with Libra
## Table of Content <a name="TOC"></a>
1. [General setups](#setups)
2. [Mapping points on multidimensional grids ](#mapping)
3. [Functions of the Wfcgrid2 class](#wfcgrid2)
4. [Showcase: computing energies of the HO eigenstates](#ho_showcase)
5. [Dynamics: computed with SOFT method](#soft_dynamics)
### A. Learning objectives
- to map sequential numbers of the grid points to the multi-dimensional index and vice versa
- to define the Wfcgrid2 class objects for DVR calculations
- to initialize wavefunctions of the grids
- to compute various properties of the wavefunctions defined on the grid
- to set up and conduct the quantum dynamics of the DVR of wavefunctions
### B. Use cases
- [Compute energies of the DVR wavefunctions](#energy-use-case)
- [Numerically exact solution of the TD-SE](#tdse-solution)
### C. Functions
- `liblibra::libdyn::libwfcgrid`
- [`compute_mapping`](#compute_mapping-1)
- [`compute_imapping`](#compute_imapping-1)
### D. Classes and class members
- `liblibra::libdyn::libwfcgrid2`
- [`Wfcgrid2`](#Wfcgrid2-1) | [also here](#Wfcgrid2-2)
- [`nstates`](#nstates-1)
- [`ndof`](#ndof-1)
- [`Npts`](#Npts-1)
- [`npts`](#npts-1)
- [`rmin`](#rmin-1)
- [`rmax`](#rmax-1)
- [`dr`](#dr-1)
- [`kmin`](#kmin-1)
- [`dk`](#dk-1)
- [`gmap`](#gmap-1) | [also here](#gmap-2)
- [`imap`](#imap-1) | [also here](#imap-2)
- [`PSI_dia`](#PSI_dia-1)
- [`reciPSI_dia`](#reciPSI_dia-1)
- [`PSI_adi`](#PSI_adi-1)
- [`reciPSI_adi`](#reciPSI_adi-1)
- [`Hdia`](#Hdia-1)
- [`U`](#U-1)
- [`add_wfc_Gau`](#add_wfc_Gau-1)
- [`add_wfc_HO`](#add_wfc_HO-1) | [also here](#add_wfc_HO-2)
- [`add_wfc_ARB`](#add_wfc_ARB-1)
- [`norm`](#norm-1) | [also here](#norm-2)
- [`e_kin`](#e_kin-1) | [also here](#e_kin-2)
- [`e_pot`](#e_pot-1) | [also here](#e_pot-2)
- [`e_tot`](#e_tot-1) | [also here](#e_tot-2)
- [`get_pow_q`](#get_pow_q-1)
- [`get_pow_p`](#get_pow_p-1) | [also here](#e_kin-2)
- [`get_den_mat`](#get_den_mat-1)
- [`get_pops`](#get_pops-1) | [also here](#get_pops-2)
- [`update_propagator_H`](#update_propagator_H-1) | [also here](#update_propagator_H-2)
- [`update_propagator_K`](#update_propagator_K-1)
- [`SOFT_propagate`](#SOFT_propagate-1)
- [`update_reciprocal`](#update_reciprocal-1) | [also here](#update_reciprocal-2)
- [`normalize`](#normalize-1) | [also here](#normalize-2)
- [`update_Hamiltonian`](#update_Hamiltonian-1) | [also here](#update_Hamiltonian-2)
- [`update_adiabatic`](#update_adiabatic-1)
## 1. General setups
<a name="setups"></a>[Back to TOC](#TOC)
First, import all the necessary libraries:
* liblibra_core - for general data types from Libra
The output of the cell below will throw a bunch of warnings, but this is not a problem nothing really serios. So just disregard them.
```
import os
import sys
import math
if sys.platform=="cygwin":
from cyglibra_core import *
elif sys.platform=="linux" or sys.platform=="linux2":
from liblibra_core import *
from libra_py import data_outs
```
Also, lets import matplotlib for plotting and define all the plotting parameters: sizes, colors, etc.
```
import matplotlib.pyplot as plt # plots
plt.rc('axes', titlesize=38) # fontsize of the axes title
plt.rc('axes', labelsize=38) # fontsize of the x and y labels
plt.rc('legend', fontsize=38) # legend fontsize
plt.rc('xtick', labelsize=38) # fontsize of the tick labels
plt.rc('ytick', labelsize=38) # fontsize of the tick labels
plt.rc('figure.subplot', left=0.2)
plt.rc('figure.subplot', right=0.95)
plt.rc('figure.subplot', bottom=0.13)
plt.rc('figure.subplot', top=0.88)
colors = {}
colors.update({"11": "#8b1a0e"}) # red
colors.update({"12": "#FF4500"}) # orangered
colors.update({"13": "#B22222"}) # firebrick
colors.update({"14": "#DC143C"}) # crimson
colors.update({"21": "#5e9c36"}) # green
colors.update({"22": "#006400"}) # darkgreen
colors.update({"23": "#228B22"}) # forestgreen
colors.update({"24": "#808000"}) # olive
colors.update({"31": "#8A2BE2"}) # blueviolet
colors.update({"32": "#00008B"}) # darkblue
colors.update({"41": "#2F4F4F"}) # darkslategray
clrs_index = ["11", "21", "31", "41", "12", "22", "32", "13","23", "14", "24"]
```
We'll use these auxiliary functions later:
```
class tmp:
pass
def harmonic1D(q, params):
"""
1D Harmonic potential
"""
x = q.get(0)
k = params["k"]
obj = tmp()
obj.ham_dia = CMATRIX(1,1)
obj.ham_dia.set(0,0, 0.5*k*x**2)
return obj
def harmonic2D(q, params):
"""
2D Harmonic potential
"""
x = q.get(0)
y = q.get(1)
kx = params["kx"]
ky = params["ky"]
obj = tmp()
obj.ham_dia = CMATRIX(1,1)
obj.ham_dia.set(0, 0, (0.5*kx*x**2 + 0.5*ky*y**2)*(1.0+0.0j) )
return obj
```
## 2. Mapping points on multidimensional grids
<a name="mapping"></a>[Back to TOC](#TOC)
Imagine a 3D grid with:
* 3 points in the 1-st dimension
* 2 points in the 2-nd dimension
* 4 points in the 3-rd dimension
So there are 3 x 2 x 4 = 24 points
However, we can still store all of them in 1D array, which is more efficient way. However, to refer to the points, we need a function that does the mapping.
This example demonstrates the functions:
`vector<vector<int> > compute_mapping(vector<vector<int> >& inp, vector<int>& npts)`
`int compute_imapping(vector<int>& inp, vector<int>& npts)`
defined in: dyn/wfcgrid/Grid_functions.h
<a name="compute_mapping-1"></a>
```
inp = intList2()
npts = Py2Cpp_int([3,2,4])
res = compute_mapping(inp, npts);
print("The number of points = ", len(res) )
print("The number of dimensions = ", len(res[0]) )
```
And the inverse of that mapping
<a name="compute_imapping-1"></a>
```
cnt = 0
for i in res:
print("point # ", cnt, Cpp2Py(i) )
print("index of that point in the global array =", compute_imapping(i, Py2Cpp_int([3,2,4])) )
cnt +=1
```
## 3. Functions of the Wfcgrid2 class
<a name="wfcgrid2"></a>[Back to TOC](#TOC)
This example demonstrates the functions of the class `Wfcgrid2`
defined in: `dyn/wfcgrid2/Wfcgrid2.h`
Here, we test simple Harmonic oscillator eigenfunctions and will
compare the energies as computed by Libra to the analytic results
### 3.1. Initialize the grid and do the mappings (internally):
`Wfcgrid2(vector<double>& rmin_, vector<double>& rmax_, vector<double>& dr_, int nstates_)`
<a name="Wfcgrid2-1"></a>
```
num_el_st = 1
wfc = Wfcgrid2(Py2Cpp_double([-15.0]), Py2Cpp_double([15.0]), Py2Cpp_double([0.01]), num_el_st)
```
The key descriptors are stored in the `wfc` object:
<a name="nstates-1"></a> <a name="ndof-1"></a> <a name="Npts-1"></a> <a name="npts-1"></a>
<a name="rmin-1"></a> <a name="rmax-1"></a> <a name="dr-1"></a> <a name="kmin-1"></a> <a name="dk-1"></a>
```
print(F"number of quantum states: {wfc.nstates}")
print(F"number of nuclear degrees of freedom: {wfc.ndof}")
print(F"the total number of grid points: {wfc.Npts}")
print(F"the number of grid points in each dimension: {Cpp2Py(wfc.npts)}")
print(F"the lower boundary of the real-space grid in each dimension: {Cpp2Py(wfc.rmin)}")
print(F"the upper boundary of the real-space grid in each dimension: {Cpp2Py(wfc.rmax)}")
print(F"the real-space grid-step in each dimension: {Cpp2Py(wfc.dr)}")
print(F"the lower boundary of the reciprocal-space grid in each dimension: {Cpp2Py(wfc.kmin)}")
print(F"the reciprocal-space grid-step in each dimension: {Cpp2Py(wfc.dk)}")
```
### Exercise 1:
What is the upper boundary of reciprocal space?
Grid mapping : the wavefunctions are stored in a consecutive order.
To convert the single integer (which is just an order of the point in a real or reciprocal space) from
the indices of the point on the 1D grid in each dimensions, we use the mapping below:
e.g. igmap[1] = [0, 1, 0, 0] means that the second (index 1) entry in the PSI array below corresponds to
a grid point that is first (lower boundary) in dimensions 0, 2, and 3, but is second (index 1) in the
dimension 1. Same for the reciprocal space
<a name="gmap-1"></a>
```
for i in range(10):
print(F"the point {i} corresponds to the grid indices = {Cpp2Py(wfc.gmap[i]) }")
```
Analogously, the inverse mapping of the indices of the point on the axes of all dimensions to the sequentian number:
<a name="imap-1"></a>
```
for i in range(10):
print(F"the point {i} corresponds to the grid indices = { wfc.imap( Py2Cpp_int([i]) ) }")
```
### 3.2. Let's run the above examples for a 2D case:
<a name="Wfcgrid2-2"></a> <a name="gmap-2"></a> <a name="imap-2"></a>
```
wfc2 = Wfcgrid2(Py2Cpp_double([-15.0, -15.0]), Py2Cpp_double([15.0, 15.0]), Py2Cpp_double([1, 1]), num_el_st)
print(F"number of quantum states: {wfc2.nstates}")
print(F"number of nuclear degrees of freedom: {wfc2.ndof}")
print(F"the total number of grid points: {wfc2.Npts}")
print(F"the number of grid points in each dimension: {Cpp2Py(wfc2.npts)}")
print(F"the lower boundary of the real-space grid in each dimension: {Cpp2Py(wfc2.rmin)}")
print(F"the upper boundary of the real-space grid in each dimension: {Cpp2Py(wfc2.rmax)}")
print(F"the real-space grid-step in each dimension: {Cpp2Py(wfc2.dr)}")
print(F"the lower boundary of the reciprocal-space grid in each dimension: {Cpp2Py(wfc2.kmin)}")
print(F"the reciprocal-space grid-step in each dimension: {Cpp2Py(wfc2.dk)}")
for i in range(10):
print(F"the point {i} corresponds to the grid indices = {Cpp2Py(wfc2.gmap[i]) }")
for i in range(10):
print(F"the point {i} corresponds to the grid indices = { wfc2.imap( Py2Cpp_int([i, i]) ) }")
```
### 3.3. Add a wavefunction to the grid
This can be done by sequentially adding either Gaussian wavepackets or the Harmonic osccillator eigenfunctions to the grid with the corresponding weights.
Adding of such functions is done with for instance:
`void add_wfc_HO(vector<double>& x0, vector<double>& px0, vector<double>& alpha, int init_state, vector<int>& nu, complex<double> weight, int rep)`
Here,
* `x0` - is the center of the added function
* `p0` - it's initial momentum (if any)
* `alpha` - the exponent parameters
* `init_state` - specialization of the initial electronic state
* `nu` - the selector of the HO eigenstate to be added
* `weight` - the amplitude with which the added function enters the superpositions, doesn't have to lead to a normalized function, the norm is included when computing the properties
* `rep` - representation
The variables x0, p0, etc. should have the dimensionality comparable to that of the grid.
For instance, in the example below we add the wavefunction (single HO eigenstate) to the 1D grid
<a name="add_wfc_HO-1"></a> <a name="norm-1"></a>
```
x0 = Py2Cpp_double([0.0])
p0 = Py2Cpp_double([0.0])
alphas = Py2Cpp_double([1.0])
nu = Py2Cpp_int([0])
el_st = 0
rep = 0
weight = 1.0+0.0j
wfc.add_wfc_HO(x0, p0, alphas, el_st, nu, weight, rep)
print(F" norm of the diabatic wfc = {wfc.norm(0)} and norm of the adiabatic wfc = {wfc.norm(1)}")
```
We can see that the wavefunction is pretty much normalized - this is becasue we have only added a single wavefunction which is already normalized.
Also, note how the norm of the diabatic wavefunction is 1.0, but that of the adiabatic is zero - this is because we have added the wavefunction only in the diabatic representation (`rep = 0`) and haven't yet run any calculations to do any updates of the other (adiabatic) representation
### Exercise 2
<a name="add_wfc_Gau-1"></a>
Use the `add_wfc_Gau` function to add several Gaussians to the grid.
### Exercise 3
Initialize the wavefunction as the superposition: $|0> - 0.5 |1> + 0.25i |2 >$
Is the resulting wavefunction normalized?
Use the `normalize()` method of the `Wfcgrid2` class to normalize it
<a name="normalize-1"></a>
### 3.4. A more advanced example: adding an arbitrary wavefunctions
using the `add_wfc_ARB` method
All we need to do is to set up a Python function that would take `vector<double>` as the input for coordinates, a Python dictionary for parameters, and it would return a `CMATRIX(nstates, 1)` object containing energies of all states as the function of the multidimensional coordinate.
Let's define the one:
```
def my_2D_sin(q, params):
"""
2D sine potential
"""
x = q.get(0,0)
y = q.get(1,0)
A = params["A"]
alpha = params["alpha"]
omega = params["omega"]
res = CMATRIX(1,1)
res.set(0,0, 0.5* A * math.sin(omega*(x**2 + y**2)) * math.exp(-alpha*(x**2 + y**2)) )
return res
```
Now, we can add the wavefunction to that grid using:
`void add_wfc_ARB(bp::object py_funct, bp::object params, int rep)`
<a name="add_wfc_ARB-1"></a>
```
rep = 0
wfc2.add_wfc_ARB(my_2D_sin, {"A":1, "alpha":1.0, "omega":1.0}, rep)
print(F" norm of the diabatic wfc = {wfc2.norm(0)} and norm of the adiabatic wfc = {wfc2.norm(1)}")
```
As we can see, this wavefunction is not normalized.
We can normalize it using `normalize(int rep)` method with `rep = 0` since we are working with the diabatic representation
<a name="normalize-2"></a>
```
wfc2.normalize(0)
print(F" norm of the diabatic wfc = {wfc2.norm(0)} and norm of the adiabatic wfc = {wfc2.norm(1)}")
```
### 3.5. Accessing wavefunction and the internal data
Now that we have initialized the wavefunction, we can access the wavefunction
<a name="PSI_dia-1"></a> <a name="PSI_adi-1"></a>
```
for i in range(10):
print(F"diabatic wfc = {wfc.PSI_dia[500+i].get(0,0) } adiabatic wfc = {wfc.PSI_adi[500+i].get(0,0) }")
```
We can also see what the reciprocal of the wavefunctions are.
<a name="reciPSI_dia-1"></a> <a name="reciPSI_adi-1"></a>
```
for i in range(10):
print(F"diabatic wfc = {wfc.reciPSI_dia[500+i].get(0,0) } adiabatic wfc = {wfc.reciPSI_adi[500+i].get(0,0) }")
```
### 3.6. Update the reciprocal of the initial wavefunction
This is needed for computing some properties, and also as the initialization of the dynamics
<a name="update_reciprocal-1"></a>
```
wfc.update_reciprocal(rep)
```
Now, since we have computed the reciprocal of the wavefunction (by doing an FFT of the real-space wfc), we can access those numbers (still in the diabatic representation only)
```
for i in range(10):
print(F"diabatic wfc = {wfc.reciPSI_dia[500+i].get(0,0) } adiabatic wfc = {wfc.reciPSI_adi[500+i].get(0,0) }")
```
### 3.4. Compute the Hamiltonian on the grid
The nice thing is - we can define any Hamiltonian function right in Python (this is done in [section 1]() ) and pass that function, together with the dictionary of the corresponding parameters to the `update_Hamiltonian` method.
Here, we define the force constant of the potential to be consistent with the alpha of the initial Gaussian wavepacket and the mass of the particle, as is done in any Quantum chemistry textbooks.
<a name="update_Hamiltonian-1"></a>
```
masses = Py2Cpp_double([2000.0])
omega = alphas[0]/masses[0]
k = masses[0] * omega**2
wfc.update_Hamiltonian(harmonic1D, {"k": k}, rep)
```
After this step, the internal storage will also contain the Hamitonians computed at the grid points:
<a name="Hdia-1"></a>
```
for i in range(10):
print(F"diabatic Hamiltonian (potential only) = {wfc.Hdia[500+i].get(0,0) } ")
```
### 3.5. Computing properties
Now, when the Hamiltonian is evaluated on the grid, we can compute various properties.
In this example, we use the wavefunction represented in the diabatic basis
<a name="norm-2"></a> <a name="e_kin-1"></a> <a name="e_pot-1"></a> <a name="e_tot-1"></a> <a name="get_pow_p-1"></a>
```
rep = 0
print( "Norm = ", wfc.norm(rep) )
print( "Ekin = ", wfc.e_kin(masses, rep) )
print( "Expected kinetic energy = ", 0.5*alphas[0]/(2.0*masses[0]) )
print( "Epot = ", wfc.e_pot(rep) )
print( "Expected potential energy = ", (0.5*k/alphas[0])*(0.5 + nu[0]) )
print( "Etot = ", wfc.e_tot(masses, rep) )
print( "Expected total energy = ", omega*(0.5 + nu[0]) )
p2 = wfc.get_pow_p(0, 2);
print( "p2 = ", p2.get(0).real )
print( "p2/2*m = ", p2.get(0).real/(2.0 * masses[0]) )
```
We can also compute the populations of all states and resolve it by the spatial region too:
<a name="get_pops-1"></a> <a name="get_pops-2"></a>
```
p = wfc.get_pops(0).get(0,0)
print(F" population of diabatic state 0 of wfc in the whole region {p}")
left, right = Py2Cpp_double([-15.0]), Py2Cpp_double([0.0])
p = wfc.get_pops(0, left, right).get(0,0)
print(F" population of diabatic state 0 of wfc in the half of the original region {p}")
```
### 3.6. Converting between diabatic and adiabatic representations
The transformation matrix `wfc.U` is computed when we compute the real-space propagator `wfc.update_propagator_H`
For the purposes of the adi-to-dia transformation, it doesn't matter what value for dt is used in that function.
<a name="update_propagator_H-1"></a>
```
wfc.update_propagator_H(0.0)
```
Now, we can access the transformation matrix - one for each grid point.
Note, in this tutorial we deal with the 1 electronic state, so all the transformation matrices are just the identity ones
<a name="U-1"></a>
```
for i in range(10):
print(F"dia-to-adi transformation matrix at point {500+i}\n")
data_outs.print_matrix(wfc.U[500+i])
```
Now, we can update the real-space adiabatic wavefunction and then its reciprocal for the adiabatic representation (`rep = 1`):
<a name="update_adiabatic-1"></a> <a name="update_reciprocal-1"></a>
```
wfc.update_adiabatic()
wfc.update_reciprocal(1)
```
And compute the properties but now in the adiabatic basis
```
for i in range(10):
print(F"diabatic wfc = {wfc.PSI_dia[500+i].get(0,0) } adiabatic wfc = {wfc.PSI_adi[500+i].get(0,0) }")
for i in range(10):
print(F"diabatic wfc = {wfc.reciPSI_dia[500+i].get(0,0) } adiabatic wfc = {wfc.reciPSI_adi[500+i].get(0,0) }")
print( "Norm = ", wfc.norm(1) )
print( "Ekin = ", wfc.e_kin(masses, 1) )
print( "Expected kinetic energy = ", 0.5*alphas[0]/(2.0*masses[0]) )
print( "Epot = ", wfc.e_pot(1) )
print( "Expected potential energy = ", (0.5*k/alphas[0])*(0.5 + nu[0]) )
print( "Etot = ", wfc.e_tot(masses, 1) )
print( "Expected total energy = ", omega*(0.5 + nu[0]) )
p2 = wfc.get_pow_p(1, 2);
print( "p2 = ", p2.get(0).real )
print( "p2/2*m = ", p2.get(0).real/(2.0 * masses[0]) )
p = wfc.get_pops(1).get(0,0)
print(F" population of adiabatic state 0 of wfc in the whole region {p}")
left, right = Py2Cpp_double([-15.0]), Py2Cpp_double([0.0])
p = wfc.get_pops(1, left, right).get(0,0)
print(F" population of adiabatic state 0 of wfc in the half of the original region {p}")
```
## 4. Showcase: computing energies of the HO eigenstates
<a name="ho_showcase"></a>[Back to TOC](#TOC)
<a name="energy-use-case"></a>
We, of course, know all the properties of the HO eigenstates analytically. Namely, the energies should be:
\\[ E_n = \hbar \omega (n + \frac{1}{2}) \\]
Let's see if we can also get them numerically
```
for n in [0, 1, 2, 3, 10, 20]:
wfc = Wfcgrid2(Py2Cpp_double([-15.0]), Py2Cpp_double([15.0]), Py2Cpp_double([0.01]), num_el_st)
nu = Py2Cpp_int([n])
wfc.add_wfc_HO(x0, p0, alphas, el_st, nu, 1.0+0.0j, rep)
wfc.update_reciprocal(rep)
wfc.update_Hamiltonian(harmonic1D, {"k": k}, rep)
print( "========== State %i ==============" % (n) )
print( "Etot = ", wfc.e_tot(masses, rep) )
print( "Expected total energy = ", omega*(0.5 + nu[0]) )
```
## 5. Dynamics: computed with SOFT method
<a name="soft_dynamics"></a>[Back to TOC](#TOC)
<a name="tdse-solution"></a>
### 5.1. Initialization
As usual, let's initialize the grid and populate it with some wavefunction
In this case, we start with a superposition of 2 HO eigenstates, so the initial wavefunction is not stationary with respect ot the chosen potential (or we won't be able to see any dynamics)
As in the axamples above, we update the reciprocal wavefunction and then the Hamiltonian
<a name="add_wfc_HO-2"></a> <a name="update_reciprocal-2"> <a name="update_Hamiltonian-2"></a>
```
wfc = Wfcgrid2(Py2Cpp_double([-15.0]), Py2Cpp_double([15.0]), Py2Cpp_double([0.01]), num_el_st)
wfc.add_wfc_HO(x0, p0, alphas, el_st, Py2Cpp_int([0]) , 1.0+0.0j, rep)
wfc.add_wfc_HO(x0, p0, alphas, el_st, Py2Cpp_int([1]) , 1.0+0.0j, rep)
wfc.update_reciprocal(rep)
wfc.update_Hamiltonian(harmonic1D, {"k": k}, rep)
```
### 5.2. Update the propagators
To compute the quantum dynamics on the grid, all we need to do is first to compute the propagators - the matrices that advances the wavefunction in real and reciprocal spaces.
The split-operator Fourier-transform (SOFT) method dates back to Kosloff & Kosloff and is basically the following:
If the Hamiltonian is given by:
\\[ H = K + V \\]
Then, the solution of the TD-SE:
\\[ i \hbar \frac{\partial \psi}{\partial t} = H \psi \\]
is given by:
\\[ \psi(t) = exp(-i \frac{H t}{\hbar} ) \psi(0) \\]
Of course, in practice we compute the state advancement by only small time increment \\[ \Delta t \\] as:
\\[ \psi(t + \Delta t) = exp(-i \frac{H \Delta t}{\hbar} ) \psi(t) \\]
So it all boils down to the computing the propagator
\\[ exp(-i \frac{H \Delta t}{\hbar} ) \\]
This is then done by the Trotter splitting technique:
\\[ exp(-i \frac{H \Delta t}{\hbar} ) \approx exp(-i \frac{V \Delta t}{2 \hbar} ) exp(-i \frac{K \Delta t}{\hbar} ) exp(-i \frac{V \Delta t}{2 \hbar} ) \\]
In the end, we need to compute the operators $ exp(-i \frac{V \Delta t}{2 \hbar} ) $ and
$exp(-i \frac{K \Delta t}{\hbar} )$
This is done by:
<a name="update_propagator_H-2"></a> <a name="update_propagator_K-1"></a>
```
dt = 10.0
wfc.update_propagator_H(0.5*dt)
wfc.update_propagator_K(dt, masses)
```
### 5.3. Compute the dynamics
The propagators in real and reciprocal spaces are stored in the class object, so we can now simply apply them many times to our starting wavefunction:
This is done with the `SOFT_propagate()` function.
Note how we use the following functions to compute the corresponding properties:
* `get_pow_q` - for \<q\>
* `get_pow_p` - for \<p\>
* `get_den_mat` - for $\rho_{ij} = |i><j|$
and so on
By default, the dynamics is executed in the diabatic representation, so for us to access the adiabatic properties (e.g. populations of the adiabatic states), we convert the propagated wavefunctions to the adiabatic representation with
* `update_adiabatic`
<a name="get_pow_q-1"></a> <a name="get_pow_p-2"></a> <a name="get_den_mat-1"></a>
<a name="e_kin-2"></a> <a name="e_pot-2"></a> <a name="e_tot-2"></a> <a name="SOFT_propagate-1"> </a>
```
nsteps = 100
for step in range(nsteps):
wfc.SOFT_propagate()
q = wfc.get_pow_q(0, 1).get(0).real
p = wfc.get_pow_p(0, 1).get(0).real
# Diabatic is the rep used for propagation, so we need to
# convert wfcs into adiabatic one
wfc.update_adiabatic()
Ddia = wfc.get_den_mat(0) # diabatic density matrix
Dadi = wfc.get_den_mat(1) # adiabatic density matrix
p0_dia = Ddia.get(0,0).real
p0_adi = Dadi.get(0,0).real
print("step= ", step, " Ekin= ", wfc.e_kin(masses, rep),
" Epot= ", wfc.e_pot(rep), " Etot= ", wfc.e_tot(masses, rep),
" q= ", q, " p= ", p, " p0_dia= ", p0_dia, " p0_adi= ", p0_adi )
```
### Exercise 4
Write the scripts to visualize various quantities computed by the dynamics
### Exercise 5
Compute the population in a certain region of space and observe how it evolves during the dynamics
### Exercise 6
Compute the dynamics of the 2D wavepacked we set up in the above examples.
### Exercise 7
Explore the behavior of the dynamics (e.g. conservation of energy, etc.) as you vary the initial conditions (e.g. the parameters of the initial wavefunction), the integration parameters (e.g. dt), and the grid properties (grid spacing and the boundaries)
|
github_jupyter
|
# Predicting the Outcome of Cricket Matches
## Introduction
In this project, we shall build a model which predicts the outcome of cricket matches in the Indian Premier League using data about matches and deliveries.
### Data Mining:
* Season : 2008 - 2015 (8 Seasons)
* Teams : DD, KKR, MI, RCB, KXIP, RR, CSK (7 Teams)
* Neglect matches that have inconsistencies such as No Result, Tie, D/L Method, etc.
### Features:
* Average Batsman Rating (Strike Rate)
* Average Bowler Rating (Wickets per Run)
* Player of the Match Awards
* Previous Encounters - Win by runs, Win by Wickets
* Recent form
### Prediction Model
* Logistic Regression using sklearn
* K-Nearest Neighbors using sklearn
```
%matplotlib inline
import numpy as np # imports a fast numerical programming library
import matplotlib.pyplot as plt #sets up plotting under plt
import pandas as pd #lets us handle data as dataframes
#sets up pandas table display
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
pd.set_option('display.notebook_repr_html', True)
import seaborn as sns
sns.set(style="whitegrid", color_codes=True)
from __future__ import division
```
## Data Mining
```
# Reading in the data
allmatches = pd.read_csv("../data/matches.csv")
alldeliveries = pd.read_csv("../data/deliveries.csv")
allmatches.head(10)
# Selecting Seasons 2008 - 2015
matches_seasons = allmatches.loc[allmatches['season'] != 2016]
deliveries_seasons = alldeliveries.loc[alldeliveries['match_id'] < 518]
# Selecting teams DD, KKR, MI, RCB, KXIP, RR, CSK
matches_teams = matches_seasons.loc[(matches_seasons['team1'].isin(['Kolkata Knight Riders', \
'Royal Challengers Bangalore', 'Delhi Daredevils', 'Chennai Super Kings', 'Rajasthan Royals', \
'Mumbai Indians', 'Kings XI Punjab'])) & (matches_seasons['team2'].isin(['Kolkata Knight Riders', \
'Royal Challengers Bangalore', 'Delhi Daredevils', 'Chennai Super Kings', 'Rajasthan Royals', \
'Mumbai Indians', 'Kings XI Punjab']))]
matches_team_matchids = matches_teams.id.unique()
deliveries_teams = deliveries_seasons.loc[deliveries_seasons['match_id'].isin(matches_team_matchids)]
print "Teams selected:\n"
for team in matches_teams.team1.unique():
print team
# Neglect matches with inconsistencies like 'No Result' or 'D/L Applied'
matches = matches_teams.loc[(matches_teams['result'] == 'normal') & (matches_teams['dl_applied'] == 0)]
matches_matchids = matches.id.unique()
deliveries = deliveries_teams.loc[deliveries_teams['match_id'].isin(matches_matchids)]
# Verifying consistency between datasets
(matches.id.unique() == deliveries.match_id.unique()).all()
```
## Building Features
```
# Batsman Strike Rate Calculation
# Team 1: Batting First; Team 2: Fielding First
def getMatchDeliveriesDF(match_id):
return deliveries.loc[deliveries['match_id'] == match_id]
def getInningsOneBatsmen(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 1].batsman.unique()[0:5]
def getInningsTwoBatsmen(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 2].batsman.unique()[0:5]
def getBatsmanStrikeRate(batsman, match_id):
onstrikedeliveries = deliveries.loc[(deliveries['match_id'] < match_id) & (deliveries['batsman'] == batsman)]
total_runs = onstrikedeliveries['batsman_runs'].sum()
total_balls = onstrikedeliveries.shape[0]
if total_balls != 0:
return (total_runs/total_balls) * 100
else:
return None
def getTeamStrikeRate(batsmen, match_id):
strike_rates = []
for batsman in batsmen:
bsr = getBatsmanStrikeRate(batsman, match_id)
if bsr != None:
strike_rates.append(bsr)
return np.mean(strike_rates)
def getAverageStrikeRates(match_id):
match_deliveries = getMatchDeliveriesDF(match_id)
innOneBatsmen = getInningsOneBatsmen(match_deliveries)
innTwoBatsmen = getInningsTwoBatsmen(match_deliveries)
teamOneSR = getTeamStrikeRate(innOneBatsmen, match_id)
teamTwoSR = getTeamStrikeRate(innTwoBatsmen, match_id)
return teamOneSR, teamTwoSR
# Testing Functionality
getAverageStrikeRates(517)
# Bowler Rating : Wickets/Run (Higher the Better)
# Team 1: Batting First; Team 2: Fielding First
def getInningsOneBowlers(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 1].bowler.unique()[0:4]
def getInningsTwoBowlers(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 2].bowler.unique()[0:4]
def getBowlerWPR(bowler, match_id):
balls = deliveries.loc[(deliveries['match_id'] < match_id) & (deliveries['bowler'] == bowler)]
total_runs = balls['total_runs'].sum()
total_wickets = balls.loc[balls['dismissal_kind'].isin(['caught', 'bowled', 'lbw', \
'caught and bowled', 'stumped'])].shape[0]
if balls.shape[0] > 0:
return (total_wickets/total_runs) * 100
else:
return None
def getTeamWPR(bowlers, match_id):
WPRs = []
for bowler in bowlers:
bwpr = getBowlerWPR(bowler, match_id)
if bwpr != None:
WPRs.append(bwpr)
return np.mean(WPRs)
def getAverageWPR(match_id):
match_deliveries = getMatchDeliveriesDF(match_id)
innOneBowlers = getInningsOneBowlers(match_deliveries)
innTwoBowlers = getInningsTwoBowlers(match_deliveries)
teamOneWPR = getTeamWPR(innTwoBowlers, match_id)
teamTwoWPR = getTeamWPR(innOneBowlers, match_id)
return teamOneWPR, teamTwoWPR
# testing functionality
getAverageWPR(517)
# MVP Score (Total number of Player of the Match awards in a squad)
def getAllInningsOneBatsmen(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 1].batsman.unique()
def getAllInningsTwoBatsmen(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 2].batsman.unique()
def getAllInningsOneBowlers(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 1].bowler.unique()
def getAllInningsTwoBowlers(match_deliveries):
return match_deliveries.loc[match_deliveries['inning'] == 2].bowler.unique()
def makeSquad(batsmen, bowlers):
p = []
p = np.append(p, batsmen)
for i in bowlers:
if i not in batsmen:
p = np.append(p, i)
return p
def getPlayerMVPAwards(player, match_id):
return matches.loc[(matches['player_of_match'] == player) & (matches['id'] < match_id)].shape[0]
def getTeamMVPAwards(squad, match_id):
num_awards = 0
for player in squad:
num_awards += getPlayerMVPAwards(player, match_id)
return num_awards
def compareMVPAwards(match_id):
match_deliveries = getMatchDeliveriesDF(match_id)
innOneBatsmen = getAllInningsOneBatsmen(match_deliveries)
innTwoBatsmen = getAllInningsTwoBatsmen(match_deliveries)
innOneBowlers = getAllInningsOneBowlers(match_deliveries)
innTwoBowlers = getAllInningsTwoBowlers(match_deliveries)
teamOneSquad = makeSquad(innOneBatsmen, innTwoBowlers)
teamTwoSquad = makeSquad(innTwoBatsmen, innOneBowlers)
teamOneAwards = getTeamMVPAwards(teamOneSquad, match_id)
teamTwoAwards = getTeamMVPAwards(teamTwoSquad, match_id)
return teamOneAwards, teamTwoAwards
compareMVPAwards(517)
# Prints a comparison between two teams based on squad attributes
def generateSquadRating(match_id):
gameday_teams = deliveries.loc[(deliveries['match_id'] == match_id)].batting_team.unique()
teamOne = gameday_teams[0]
teamTwo = gameday_teams[1]
teamOneSR, teamTwoSR = getAverageStrikeRates(match_id)
teamOneWPR, teamTwoWPR = getAverageWPR(match_id)
teamOneMVPs, teamTwoMVPs = compareMVPAwards(match_id)
print "Comparing squads for {} vs {}".format(teamOne,teamTwo)
print "\nAverage Strike Rate for Batsmen in {} : {}".format(teamOne,teamOneSR)
print "\nAverage Strike Rate for Batsmen in {} : {}".format(teamTwo,teamTwoSR)
print "\nBowler Rating (W/R) for {} : {}".format(teamOne,teamOneWPR)
print "\nBowler Rating (W/R) for {} : {}".format(teamTwo,teamTwoWPR)
print "\nNumber of MVP Awards in {} : {}".format(teamOne,teamOneMVPs)
print "\nNumber of MVP Awards in {} : {}".format(teamTwo,teamTwoMVPs)
#Testing Functionality
generateSquadRating(517)
## 2nd Feature : Previous Encounter
# Won by runs and won by wickets (Higher the better)
def getTeam1(match_id):
return matches.loc[matches["id"] == match_id].team1.unique()
def getTeam2(match_id):
return matches.loc[matches["id"] == match_id].team2.unique()
def getPreviousEncDF(match_id):
team1 = getTeam1(match_id)
team2 = getTeam2(match_id)
return matches.loc[(matches["id"] < match_id) & (((matches["team1"].isin(team1)) & (matches["team2"].isin(team2))) | ((matches["team1"].isin(team2)) & (matches["team2"].isin(team1))))]
def getTeamWBR(match_id, team):
WBR = 0
DF = getPreviousEncDF(match_id)
winnerDF = DF.loc[DF["winner"] == team]
WBR = winnerDF['win_by_runs'].sum()
return WBR
def getTeamWBW(match_id, team):
WBW = 0
DF = getPreviousEncDF(match_id)
winnerDF = DF.loc[DF["winner"] == team]
WBW = winnerDF['win_by_wickets'].sum()
return WBW
def getTeamWinPerc(match_id):
dF = getPreviousEncDF(match_id)
timesPlayed = dF.shape[0]
team1 = getTeam1(match_id)[0].strip("[]")
timesWon = dF.loc[dF["winner"] == team1].shape[0]
if timesPlayed != 0:
winPerc = (timesWon/timesPlayed) * 100
else:
winPerc = 0
return winPerc
def getBothTeamStats(match_id):
DF = getPreviousEncDF(match_id)
team1 = getTeam1(match_id)[0].strip("[]")
team2 = getTeam2(match_id)[0].strip("[]")
timesPlayed = DF.shape[0]
timesWon = DF.loc[DF["winner"] == team1].shape[0]
WBRTeam1 = getTeamWBR(match_id, team1)
WBRTeam2 = getTeamWBR(match_id, team2)
WBWTeam1 = getTeamWBW(match_id, team1)
WBWTeam2 = getTeamWBW(match_id, team2)
print "Out of {} times in the past {} have won {} times({}%) from {}".format(timesPlayed, team1, timesWon, getTeamWinPerc(match_id), team2)
print "{} won by {} total runs and {} total wickets.".format(team1, WBRTeam1, WBWTeam1)
print "{} won by {} total runs and {} total wickets.".format(team2, WBRTeam2, WBWTeam2)
#Testing functionality
getBothTeamStats(517)
# 3rd Feature: Recent Form (Win Percentage of 3 previous matches of a team in the same season)
# Higher the better
def getMatchYear(match_id):
return matches.loc[matches["id"] == match_id].season.unique()
def getTeam1DF(match_id, year):
team1 = getTeam1(match_id)
return matches.loc[(matches["id"] < match_id) & (matches["season"] == year) & ((matches["team1"].isin(team1)) | (matches["team2"].isin(team1)))].tail(3)
def getTeam2DF(match_id, year):
team2 = getTeam2(match_id)
return matches.loc[(matches["id"] < match_id) & (matches["season"] == year) & ((matches["team1"].isin(team2)) | (matches["team2"].isin(team2)))].tail(3)
def getTeamWinPercentage(match_id):
year = int(getMatchYear(match_id))
team1 = getTeam1(match_id)[0].strip("[]")
team2 = getTeam2(match_id)[0].strip("[]")
team1DF = getTeam1DF(match_id, year)
team2DF = getTeam2DF(match_id, year)
team1TotalMatches = team1DF.shape[0]
team1WinMatches = team1DF.loc[team1DF["winner"] == team1].shape[0]
team2TotalMatches = team2DF.shape[0]
team2WinMatches = team2DF.loc[team2DF["winner"] == team2].shape[0]
if (team1TotalMatches != 0) and (team2TotalMatches !=0):
winPercTeam1 = ((team1WinMatches / team1TotalMatches) * 100)
winPercTeam2 = ((team2WinMatches / team2TotalMatches) * 100)
elif (team1TotalMatches != 0) and (team2TotalMatches ==0):
winPercTeam1 = ((team1WinMatches / team1TotalMatches) * 100)
winPercTeam2 = 0
elif (team1TotalMatches == 0) and (team2TotalMatches !=0):
winPercTeam1 = 0
winPercTeam2 = ((team2WinMatches / team2TotalMatches) * 100)
else:
winPercTeam1 = 0
winPercTeam2 = 0
return winPercTeam1, winPercTeam2
#Testing Functionality
getTeamWinPercentage(517)
#Function to implement all features
def getAllFeatures(match_id):
generateSquadRating(match_id)
print ("\n")
getBothTeamStats(match_id)
print("\n")
getTeamWinPercentage(match_id)
#Testing Functionality
getAllFeatures(517)
```
## Adding New Columns for Features in Matches DataFrame
```
#Create Column for Team 1 Winning Status (1 = Won, 0 = Lost)
matches['team1Winning'] = np.where(matches['team1'] == matches['winner'], 1, 0)
# New Column for Difference of Average Strike rates (First Team SR - Second Team SR)
# [Negative value means Second team is better]
firstTeamSR = []
secondTeamSR = []
for i in matches['id'].unique():
P, Q = getAverageStrikeRates(i)
firstTeamSR.append(P), secondTeamSR.append(Q)
firstSRSeries = pd.Series(firstTeamSR)
secondSRSeries = pd.Series(secondTeamSR)
matches["Avg_SR_Difference"] = firstSRSeries.values - secondSRSeries.values
# New Column for Difference of Wickets Per Run (First Team WPR - Second Team WPR)
# [Negative value means Second team is better]
firstTeamWPR = []
secondTeamWPR = []
for i in matches['id'].unique():
R, S = getAverageWPR(i)
firstTeamWPR.append(R), secondTeamWPR.append(S)
firstWPRSeries = pd.Series(firstTeamWPR)
secondWPRSeries = pd.Series(secondTeamWPR)
matches["Avg_WPR_Difference"] = firstWPRSeries.values - secondWPRSeries.values
# New column for difference of MVP Awards
# (Negative value means Second team is better)
firstTeamMVP = []
secondTeamMVP = []
for i in matches['id'].unique():
T, U = compareMVPAwards(i)
firstTeamMVP.append(T), secondTeamMVP.append(U)
firstMVPSeries = pd.Series(firstTeamMVP)
secondMVPSeries = pd.Series(secondTeamMVP)
matches["Total_MVP_Difference"] = firstMVPSeries.values - secondMVPSeries.values
# New column for Win Percentage of Team 1 in previous encounters
firstTeamWP = []
for i in matches['id'].unique():
WP = getTeamWinPerc(i)
firstTeamWP.append(WP)
firstWPSeries = pd.Series(firstTeamWP)
matches["Prev_Enc_Team1_WinPerc"] = firstWPSeries.values
# New column for Recent form(Win Percentage in the current season) of 1st Team compared to 2nd Team
# (Negative means 2nd team has higher win percentage)
firstTeamRF = []
secondTeamRF = []
for i in matches['id'].unique():
K, L = getTeamWinPercentage(i)
firstTeamRF.append(K), secondTeamRF.append(L)
firstRFSeries = pd.Series(firstTeamRF)
secondRFSeries = pd.Series(secondTeamRF)
matches["Total_RF_Difference"] = firstRFSeries.values - secondRFSeries.values
#Testing
matches.tail()
```
## Visualizations for Features vs. Response
```
# Graph for Average Strike Rate Difference
matches.boxplot(column = 'Avg_SR_Difference', by='team1Winning', showfliers= False)
# Graph for Average WPR(Wickets per Run) Difference
matches.boxplot(column = 'Avg_WPR_Difference', by='team1Winning', showfliers= False)
# Graph for MVP Difference
matches.boxplot(column = 'Total_MVP_Difference', by='team1Winning', showfliers= False)
#Graph for Previous encounters Win Percentage of Team #1
matches.boxplot(column = 'Prev_Enc_Team1_WinPerc', by='team1Winning', showfliers= False)
# Graph for Recent form(Win Percentage in the same season)
matches.boxplot(column = 'Total_RF_Difference', by='team1Winning', showfliers= False)
```
|
github_jupyter
|
Let's load the data from the csv just as in `dataset.ipynb`.
```
import pandas as pd
import numpy as np
raw_data_file_name = "../dataset/fer2013.csv"
raw_data = pd.read_csv(raw_data_file_name)
```
Now, we separate and clean the data a little bit. First, we create an array of only the training data. Then, we create an array of only the private test data (referred to in the code with the prefix `first_test`). The `reset_index` call re-aligns the `first_test_data` to index from 0 instead of wherever it starts in the set.
```
train_data = raw_data[raw_data["Usage"] == "Training"]
first_test_data = raw_data[raw_data["Usage"] == "PrivateTest"]
first_test_data.reset_index(inplace=True)
second_test_data = raw_data[raw_data["Usage"] == "PublicTest"]
second_test_data.reset_index(inplace=True)
import keras
train_expected = keras.utils.to_categorical(train_data["emotion"], num_classes=7, dtype='int32')
first_test_expected = keras.utils.to_categorical(first_test_data["emotion"], num_classes=7, dtype='int32')
second_test_expected = keras.utils.to_categorical(second_test_data["emotion"], num_classes=7, dtype='int32')
def process_pixels(array_input):
output = np.empty([int(len(array_input)), 2304])
for index, item in enumerate(output):
item[:] = array_input[index].split(" ")
output /= 255
return output
train_pixels = process_pixels(train_data["pixels"])
train_pixels = train_pixels.reshape(train_pixels.shape[0], 48, 48, 1)
first_test_pixels = process_pixels(first_test_data["pixels"])
first_test_pixels = first_test_pixels.reshape(first_test_pixels.shape[0], 48, 48, 1)
second_test_pixels = process_pixels(second_test_data["pixels"])
second_test_pixels = second_test_pixels.reshape(second_test_pixels.shape[0], 48, 48, 1)
from keras.preprocessing.image import ImageDataGenerator
datagen = ImageDataGenerator(
featurewise_center=False,
featurewise_std_normalization=False,
rotation_range=10,
width_shift_range=0.1,
height_shift_range=0.1,
horizontal_flip=True
)
```
Here, we create our own top-level network to load on top of VGG16.
```
from keras.models import Sequential
from keras.layers import Dense, Dropout, MaxPooling2D, Conv2D, Flatten
from keras.optimizers import Adam
def gen_model(size):
model = Sequential()
model.add(Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape = (48, 48, 1)))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(size, activation='relu'))
model.add(Dense(7, activation='softmax'))
optimizer = Adam(learning_rate=0.0009)
model.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['accuracy'])
return model
from keras.callbacks.callbacks import EarlyStopping, ReduceLROnPlateau
early_stop = EarlyStopping('val_loss', patience=50)
reduce_lr = ReduceLROnPlateau('val_loss', factor=0.1, patience=int(50/4), verbose=1)
callbacks = [early_stop, reduce_lr]
sizes = [32, 64, 128, 256]
results = [None] * len(sizes)
for i in range(len(sizes)):
model = gen_model(sizes[i])
model.fit_generator(datagen.flow(train_pixels, train_expected, batch_size=32),
steps_per_epoch=len(train_pixels) / 32,
epochs=10, verbose=1, callbacks=callbacks,
validation_data=(first_test_pixels,first_test_expected))
results[i] = model.evaluate(second_test_pixels, second_test_pixels, batch_size=32)
```
|
github_jupyter
|
# Step 2 - Data Wrangling Raw Data in Local Data Lake to Digestable Data
Loading, merging, cleansing, unifying and wrangling Oracle OpenWorld & CodeOne Session Data from still fairly raw JSON files in the local datalake.
The gathering of raw data from the (semi-)public API for the Session Catalog into a local data lake was discussed and performed in <a href="./1-OOW2018 Session Catalog - retrieving raw session data in JSON files.ipynb">Notebook 1-OOW2018 Session Catalog - retrieving raw session data in JSON files</a>. The current notebook starts from the 44 raw JSON files in local directory `./data`.
This notebook describes how to load, combine and wrangle the data from these files. This notebook shows for example how to load and merge data from dozens of (same formatted) JSON files, discard undesired attributes, deduplicate the record set, derive attributes for easier business intelligence & machine learning and write the resulting data set to a single JSON file.
Steps in this notebook:
* Load and Merge from raw JSON
* <a href="#deduplicate">discard redundant columns</a>
* <a href="#deduplicate">deduplication</a>
* <a href="#explore">Explore Data Frame</a>
* <a href="#enriching">Enrich Data</a>
* <a href="publish">Publish Wrangle Results</a>
The deliverable from this notebook is a single file `oow2018-sessions-wrangled.json` in the `datawarehouse` folder. This file contains unique, filtered, enriched data that is in a poper shape to perform further analysis on.
# Load and merge data from raw JSON files
This first section describes how the session data from Oracle OpenWorld 2018 is loaded from over 40 individual files with the raw JSON session data. These files are organized by session type and event (oow and codeone) - and have been produced by a different notebook (<a href="./1-OOW2018 Session Catalog - retrieving raw session data in JSON files.ipynb">Notebook 1-OOW2018 Session Catalog - retrieving raw session data in JSON files</a>) from the Oracle OpenWorld Session Catalog API.
The files are read into individual Pandas Data Frame objects. These Data Frames are concatenated. The end result from reading 44 files is a single Pandas Data Frame - called *ss* (session schedule).
Let's start with reading the session information from a single file into a Pandas Data Frame - to get a feel for how that works and what it results in.
```
#read a single session data file and parse the JSON content into a Pandas Data Frame
import pandas as pd
import json
dataLake = "datalake/" # file system directory used for storing the gathered data
#as a test, try to load data from one of the generated files
conference = 'oow' # could also be codeone
sessionType = 'TRN' # could also be one of 21 other values such as TUT, DEV, GEN, BOF, HOL, ...
ss = pd.read_json("{0}oow2018-sessions_{1}_{2}.json".format(dataLake, conference, sessionType))
# add an additional column to the Data Frame to specify the conference catalog of origin of these sessions; in this case oow
ss = ss.assign(catalog=conference)
ss.head(3)
#as a test, try to load data from another file ; same sessionType but different conference
conference = 'codeone'
sessionType = 'TRN'
ss2 = pd.read_json("{0}oow2018-sessions_{1}_{2}.json".format(dataLake, conference, sessionType))
# add an additional column to the Data Frame to specify the conference catalog of origin of these sessions; in this case codeone
ss2 = ss2.assign(catalog='codeone')
ss2.head(3)
```
All session data is to be merged into a single data frame. We will use `ss` as the sink - the data frame into which all sessions records are to be loaded. We use the Pandas concat operation to merge two Data Frames, as is done below for the two data frames with session records for session type TRN.
```
#add ss and ss2 together
#see https://pandas.pydata.org/pandas-docs/stable/merging.html
ss = pd.concat([ss,ss2], ignore_index=True , sort=True)
ss.head(8)
```
Overhead, two files were loaded, parsed into a Data Frame and added together into a single Data Frame. The next step is to load the session data from the raw JSON file for all 44 files - for the two events and for all session types. The session types are defined in the Dict object *sessionTypes* . The code loops over the keys in this Dict and reads the corresponding JSON file for each of the two events.
```
sessionTypes = {'BOF': '1518466139979001dQkv'
, 'BQS': 'bqs'
, 'BUS': '1519240082595001EpMm'
, 'CAS': 'casestudy'
, 'DEV': '1522435540042001BxTD'
, 'ESS': 'ess'
, 'FLP': 'flp'
, 'GEN': 'general'
, 'HOL': 'hol'
, 'HOM': 'hom'
, 'IGN': 'ignite'
, 'KEY': 'option_1508950285425'
, 'MTE':'1523906206279002QAu9'
, 'PKN': '1527614217434001RBfj'
, 'PRO': '1518464344082003KVWZ'
, 'PRM': '1518464344082002KM3k'
, 'TRN': '1518464344082001KHky'
, 'SIG': 'sig'
, 'THT': 'ts'
, 'TLD': '1537894888625001RriS'
, 'TIP': '1517517756579001F3CR'
, 'TUT': 'tutorial'
#commented out because TRN is dealt with earlier on, 'TRN': '1518464344082001KHky'
}
#loop over all session types and read the corresponding files for both events codeone and oow
for key,value in sessionTypes.items():
sessionType = key
conference = 'oow'
ssoow = pd.read_json("{0}oow2018-sessions_{1}_{2}.json".format(dataLake, conference, sessionType))
# add an additional column to the Data Frame to specify the conference catalog of origin of these sessions
ssoow = ssoow.assign(catalog=conference)
conference = 'codeone'
sscodeone = pd.read_json("{0}oow2018-sessions_{1}_{2}.json".format(dataLake, conference, sessionType))
sscodeone = sscodeone.assign(catalog=conference)
# merge data for sessions of type session type for both oow and codeone into a master set in ss
ss = pd.concat([ss,ssoow,sscodeone], ignore_index=True, sort=True)
print("Done - all data is merged into one data frame")
```
### Some key metrics on the merged Sessions Set
The shape function on the data frame returns the dimensions of the frame: the number of rows by the number of columns:
```
ss.shape
# total memory usage
ss.memory_usage(index=True, deep=True).sum()
#list all columns in the Dataframe
ss.columns
# data types for the columns in the data frame
ss.dtypes
ss.groupby(['event','type'])['event'].count()
```
<a name="discard" />
## Discard unneeded attributes
Early inspection of the JSON document and the session catalog website has provided insight in the available attributes and their relevance. Below, you will see an overview of all columns in the Data Frame - corresponding with the top level items in the JSON document. The subsequent step removes from the Data Frame all columns that seem irrelevant for our task at hand.
These columns seem relevant for the web frontend developers, for planned but not realized objectives or for unknown purposes. In order to not carry more weight than necessary - because of performance, resource usage and lurking complexity we get rid of columns that seem irrelevant. If we need them after all, they are still available in the data lake.
Remove the unwanted columns from the Dataframe
(allowDoubleBooking .'codeParts', 'code_id', 'es_metadata_id',type_displayorder type_displayorder_string useDoubleBooking useWaitingList videos viewAccess viewAccessPublic, ...)
```
# remove columns
# Note that the original 'data' object is changed when inplace=True
ss.drop([
'allowDoubleBooking'
,'codeParts', 'code_id', 'es_metadata_id','type_displayorder'
,'type_displayorder_string', 'useDoubleBooking'
,'useWaitingList', 'videos'
, 'viewAccess', 'viewAccessPublic','viewFileAccess', 'waitlistAccess', 'waitlistLimit'
, 'eventId','eventName','featured_value','publicViewPrivateSchedule','published', 'scheduleAccess','sessionID','status'
,'externalID','highlight','abbreviation'
]
, axis=1, inplace=True)
```
<a name="deduplicate" />
## Deduplicate
Some sessions are included in the catalog for both events - Oracle OpenWorld and CodeOne - even though they are associated with one of the two. The exact same session - with only a different value for attribute catalog - occurs twice in our data set for these sessions. We should get rid of duplicates. However, we should not do so before we capture the fact that a session is part of the catalogs of both events in the record that is retained.
Note: it seems there are some sessions that occur multiple times in the data set but are not included in both event catalogs. This seems to be just some form of data pollution.
Let's first see how many events are part of both events' catalogs.
```
# The code feature is supposed to be the unique identifier of sessions
# Let's see at the multifold occurrence of individual code values
counts = ss['code'].value_counts()
counts.head(13)
```
We have found quite a few code values that occur multiple times in the data frame. Each code should be in the data frame only once. Let's further look into these sessions.
```
# let's create a data frame with all sessions whose code occurs more than one in the data frame
duplicates = ss[ss['code'].isin(counts.index[counts > 1])]
# show the first ten records of these 'candidate duplicates'
duplicates[['code','title','event','catalog' ]].sort_values('code').head(10)
#are duplicates indeed associated with both events?
# some are - but not all of them:
duplicates.loc[duplicates['code']=='BOF4977']
```
The next step is a little bit complex: we want to record the fact that certain sessions (actually session codes) are associated with both catalogs. We join the sessions in `ss` with the sessions that occur multiple times in `duplicates` and we join records in `ss` with their counterparts (same session code) that have a different catalog origin.
This gives us a data frame with all session codes associated with both catalogs.
```
# find all sessions that occur in both catalogs: set their catalog attribute to both
# set catalog="both" if session in duplicates with a different catalog value than the session's own catalog value
doubleCatalogSessions = pd.merge(ss, duplicates, on=['code'], how='inner').query('catalog_y != catalog_x')
doubleCatalogSessions[['code','catalog_x', 'catalog_y']] .head(20)
```
The master dataset is still `ss`. All sessions in this data frame whose session code appears in `doubleCatalogSessions` will get their *catalog* attribute updated to *both*. The cell will then show the values in catalog and the number of their occurrences.
```
# all sessions in doubleCatalogSessions occur in both oow and code one session catalog
# time to update column catalog for all sessions in ss that have a code that occurs in doubleCatalogSessions['code']
ss.loc[ss['code'].isin(doubleCatalogSessions['code']),'catalog']='both'
ss['catalog'].value_counts()
```
If we now drop any duplicate records - any sessions whose session code occurs more than once - we will reduce our data frame to the unique set of sessions that actually took place, without the ghost duplicates introduced in our data set because a session appeared in more than one catalog.
```
#Drop duplicates - identifying rows by their code
ss.drop_duplicates(subset=['code'], keep='first', inplace=True)
#hpw many sessions appear in each and in both catalogs?
ss['catalog'].value_counts()
```
<a name="explore" />
# Exploring the Data
Let's briefly look at the data we now have in the Pandas Data Frame. What does the data look like? What values do we have in the columns? And what complexity is hiding in some columns with nested values, such as participants, attributevalues and files.
Note: https://morphocode.com/pandas-cheat-sheet/ provides a quick overview of commands used for inspecting and manipulating the data frame.
```
#an overview of the current contents of the data frame
ss.head(6)
# and this overview of the rows and columns in the data frame.
print(ss.info())
#let's look at all different values for length (the duration of each session in minutes)
ss['length'].unique()
# yes - it is that simple!
# and what about (session) type?
ss['type'].unique()
```
Some of the columns in the data frame contain complex, nested values. For example the `attributevalues` column. It contains a JSON array - a list of objects that each describe some attribute for the session. Examples of session attributes that are defined in this somewhat convoluted wat are *(target Experience) Level*, *Track*, *Day* , *Role*. A little later on, we will create new, proper features in the data frame based on values extracted from this complex attributevalues colunmn - in order to make it possible to make good use of this information for visualization, analysis and machine learning.
```
# some of the columns or attributes have nested values. It seems useful to take a closer look at them.
# show nested array attributevalues for first record in Dataframe - this is an attribute which contains an array of objects that each define a specific characteristic of the session,
# such as date and time, track, topic, level, role, company market size,
ss['attributevalues'][10]
```
The *participants* column also contains a complex object. This column too contains a JSON array with the people associated with a session as speaker. The array contains a nested object for each speaker. This object has a lot of data in it - from name and biography and special designations (titles) for the speaker to company, job title, URL to a picture and details on all (other) sessions the speaker is involved in.
Let's take a look at an example.
```
#show nested array participants for 11th record in the Dataframe
ss['participants'][10]
```
The *files* column too contains a JSON array. This array contains entries for files associated with the session. These files provide the slides or other supporting content for the session. Each session is supposed to have exactly one files associated with it. Some do not - for example because the speaker(s) forgot to upload their slides.
Let's inspect the JSON contents of the *files* column. It contains the name of the file and the URL from where it can be downloaded.
```
#show nested array files
ss['files'][0]
```
<a name="enriching" />
# Enriching the data
In this section we are engineering the data to produce some attributes or features that are easier to work with once we start doing data science activities such as business intelligence or machine learning. In this section we are not yet actually bringing in external data sources to add information we not already have in our set. However, we are making the data we already have more accessible and thereby more valuable. So in that sense, this too can be called *enriching*.
The enrichments performed on the session data in the data frame :
* set a flag at session level to indicate whether the session is an Oracle session (with Oracle staff among the speakers- speaker has Oracle in companyName attribute)
* set a flag at session level to indicate whether a file has been uploaded for the session
* set a flag at session level to indicate whether one of the speakers has Java Rockstar, Java Champion, Developer Champion/Groundbreaker Ambassador, ACE or ACE Director
* set attribute level (based on attributevalues array) - beginner, all, ...
* derive track(s?) from attributevalues array (where attribute_id=Track or attribute_id=CodeOneTracks )
* set attribute with number of speakers on session
* number of instances of the session
### Oracle Speaker and File Uploaded
These functions `oracle_speaker` and `file_flag` are invoked for every record in the data frame. They are used to derive new, first class attributes to indicate whether or not at least one speaker working for Oracle is associated with a session (Y or N) and if a file has been upload (presumably with slides) for the session. The information represented by these two new attributes already exists in the data frame - but in a way that makes it quite unaccessible to the data analyst.
```
#function to derive Oracle flag from participants
def oracle_speaker(session):
result = "N"
# loop over speakers; if for any of them, companyName contains the word Oracle, the result =Y
for x in session["participants"][:]:
if ("oracle" in x.get('companyName','x').lower()):
result='Y'
return result
```
New columns `oracle_speaker` and `file_flag` are added to the data frame with values derived for each record by applying the functions with corresponding names.
```
#set oracle_speaker flag
#apply function oracle_speaker to every row in the data frame to derive values for the new column oracle_speaker
ss['oracle_speaker'] = ss.apply(oracle_speaker, axis=1)
# show the values and the number of occurrences for the new column oracle_speaker
ss['oracle_speaker'].value_counts()
#function to derive file flag from files
def file_flag(session):
result = "N"
if isinstance(session.get("files",None),list) :
# loop over files; if any exist, then result = Y
for x in session["files"][:]:
if x['fileId']:
result='Y'
break
return result
#set file_flag
#apply function file_flag to every row in the data frame to derive values for the new column file_flag
ss['file_flag'] = ss.apply(file_flag, axis=1)
ss['file_flag'].value_counts()
```
### Speaker Designations
Many of the speakers are special - in the sense that they have been awarded community awards and titles, such as (Oracle) Java Champion, Oracle ACE Directory, JavaOne Rockstar and Groundbreaker Ambassador. These designations can be found for speakers (a nested JSON object) in their *attributevalues* feature - which happens to be another nested JSON object.
The next function *speaker_designation* finds out for a session if it has at least one speaker associated with it who has the requested designation.
```
#function to derive designation flag from speakers
# values for designation: JavaOne Rockstar, Oracle ACE Director, Oracle Java Champion, Groundbreaker Ambassador,
def speaker_designation(session, designation):
result = "N"
# loop over speakers and their attributevalues; if any exist with attribute_id == specialdesignations and value == JavaOne Rockstar
for x in session["participants"][:]:
if "attributevalues" in x:
for y in x["attributevalues"][:]:
if "attribute_id" in y:
if y["attribute_id"]=="specialdesignations":
if y["value"]== designation:
result="Y"
return result
```
The next cell iterates over four major `designations` and derives for each designation a new column in the data frame that contain Y or N, depending on whether a speaker with the designation will present in the session.
```
#set flags for designations
designations = ['JavaOne Rockstar', 'Oracle ACE Director', 'Oracle Java Champion', 'Groundbreaker Ambassador']
for d in designations:
ss[d] = ss.apply(speaker_designation, args=(d,), axis=1)
```
Let's check the newly created columns: what values do they contain and how often does each value occur?
```
ss[['JavaOne Rockstar','Oracle ACE Director' , 'Oracle Java Champion', 'Groundbreaker Ambassador']].apply(pd.value_counts).fillna(0)
```
### Level
Each session can be described as suitable for one or more levels of experience: Beginner, Intermediate or Advaned. Each session can be associated with all levels - or a subset of them. This level indication is somewhat hidden away, in the attributevalues object. The next function `session_level` will unearth for a given session whether it is associated with the specified level, Y or N.
```
#function to derive level flag for a session
def session_level(session, level):
result = "N"
# loop over attributevalues; if any exist with attribute_id == "SessionsbyExperienceLevel", and value == level
for x in session["attributevalues"][:]:
if "attribute_id" in x:
if x["attribute_id"]=="SessionsbyExperienceLevel":
if x["value"]== level:
result="Y"
break # no point in continuing if we have found what we are looking for
return result
```
This cell runs through all three level values and creates a new column in the data frame for each level. It will set a Y or N for each session in the new columns, depending on whether session is associated with the level, or not.
```
#set flags for designations
levels = ['Intermediate', 'Beginner', 'Advanced']
for l in levels:
ss[l] = ss.apply(session_level, args=(l,), axis=1)
print("Assigned Level Flags (Advanced, Intermediate, Beginner)")
```
Next we will derive values for a new column 'All' that indicates whether a session has been associated with all levels - even though I am not sure what exactly that means.
```
def isAll(session):
return 'Y' if session['Beginner'] == 'Y' and session['Intermediate'] == 'Y' and session['Advanced'] == 'Y'else 'N'
ss['All'] = ss.apply( isAll, axis=1)
ss[['Intermediate', 'Beginner', 'Advanced', 'All']].apply(pd.value_counts).fillna(0)
```
### Track
All sessions are assigned to one or more tracks. These tracks are categories that help attendees identify and assess sessions. Some examples of tracks are: Core Java Platform, Development Tools, Oracle Cloud Platform, MySQL, Containers, Serverless, and Cloud, Emerging Technologies, Modern Web, Application Development, Infrastructure Technologies (Data Center).
Depending on whether a session originates from the Oracle OpenWorld or CodeOne catalog, the track(s) are found in the nested object *attributevalues* under the attribute_id *CodeOneTracks* or just *Track*.
The next function is created to return a String array for a session with all the tracks associated with the session.
```
#function to derive track flag for a session
def session_track(session):
result = ['abc']
# loop over attributevalues; if any exist with attribute_id == "SessionsbyExperienceLevel", and value == level
for x in session["attributevalues"][:]:
if "attribute_id" in x:
if x["attribute_id"]=="CodeOneTracks":
result.append( x["value"])
if x["attribute_id"]=="Track":
result.append( x["value"])
del result[0]
return result
```
The next cell uses the function `session_track` to produce the value for each session for the track. The cell then prints out the tracks for a sample of sessions.
```
# add column track with a value derived from the session record
ss["track"] = ss.apply(session_track, axis=1)
print("Assigned Track")
ss[['title','catalog','track']].tail(10)
```
### Number of Speakers per Session
The number of speakers making an appearance in a session is another fact that while not readily available is also hiding in our data frame. We will turn this piece of information into an explicit feature. The next cell counts the number of elements in the participants array - and assigns it to the speaker_count attribute of each session record.
```
#set number of speakers for each session by taking the length of the list in the participants column
ss["speaker_count"] = ss['participants'].apply(lambda x: len(x))
# list the values in the new speaker_count column and the number of occurrences. 12 participants in one session?
ss['speaker_count'].value_counts()
```
### Number of instances per session
Most sessions are scheduled just once. However, some sessions are executed multiple times. This can be derived from the *times* column in the data frame, simply by taking the number of elements in the JSON array in that column.
The next cell adds a column to the data frame, with for each session the number of times that session takes place.
```
#set number of instancesfor each session by taking the length of the list in the times column
ss["instance_count"] = ss['times'].apply(lambda x: len(x) if x else None)
ss['instance_count'].value_counts()
```
### Session Room Capacity
The session records have a *times* feature - a JSON array with all the instances of the session. The elements contain the *capacity* attribute that gives the size or capcity of the room in which the session is scheduled. Because most sessions occur only once it seems acceptable to set a room_capacity feature on all session records in the data frame derived from the capacity found first element in the times feature. Likewise, we can derive the value for room itself.
```
#set the room capacity based on the capacity of the room of the first entry in the times list
# note: when the session is scheduled multiple times, not all rooms may have the same capacity; that detail gets lost
ss["room_capacity"]= ss['times'].apply(lambda x:x[0]['capacity'] if x else None)
ss["room"]= ss['times'].apply(lambda x:x[0]['room'] if x else None)
```
### Session Slot - Day and Time
As discussed in the preceding section, the session records have a *times* feature - a JSON array with all the instances of the session. The elements contain attributes *dayName* and *time* that mark the slot in which the session is scehduled. Because most sessions occur only once it seems acceptable to set a session day and time feature from the values for time and day found first element in the times feature. Likewise, we can derive the Python DateTime value for the starting timestamp of the session.
```
#likewise derive day, time and room - from the first occurrence of the session
ss["day"]= ss['times'].apply(lambda x:x[0]['dayName'] if x else None)
ss["time"]= ss['times'].apply(lambda x:x[0]['time'] if x else None)
ss[['day','time']].apply(pd.value_counts).fillna(0)
```
The columns `day` and `time` are just strings. It may be useful for the data analysts further downstream in the data science pipeline to also have a real DateTime object to work with. The next cell introduces the `session_timestamp` column, set to the real timestamp derived from day and time. Note that we make use of one external piece of data not found in the datalake: the fact that Sunday in this case means 21st October 2018.
```
import datetime
# see https://stackabuse.com/converting-strings-to-datetime-in-python/
#Sunday means 21st October 2018; monday through thursday are the days following the 21st
dayMap = {'Sunday': '21', 'Monday': '22', 'Tuesday':'23', 'Wednesday':'24', 'Thursday':'25'}
def create_timestamp(day, timestr):
if not timestr:
return None
if not day:
return None
dtString = '2018-10-'+ dayMap[day] +' '+ timestr
return datetime.datetime.strptime(dtString, '%Y-%m-%d %H:%M')
def create_session_timestamp(session):
return create_timestamp(session['day'], session['time'])
ss['session_timestamp'] = ss.apply(create_session_timestamp, axis=1)
ss[['day','time','session_timestamp'] ].head(10)
```
<a name="publish" />
## Persist Pandas Dataframe - as single, consolidated, reshaped, enriched JSON file
One thing we want to be able to do with the data we gather, is to persist it for future use by data analists, data scientists and other stakeholders. We could store the data in a NoSQL database, a cloud storage service or simply as a local file. For now, let's do the latter: store the cleansed and reshaped data in a local JSON file for further enrichment, visualization and machine learning purposes.
The file is called `oow2018-sessions-wrangled.json` and it will be stored in the `datawarehouse` folder.
```
dataWarehouse = "datawarehouse/" # file system directory used for storing the wrangled data
ss.to_json("{0}oow2018-sessions-wrangled.json".format(dataWarehouse), force_ascii=False)
```
A quick check to see whether the wrangled session data was successfully written to disk - and can be read again. If we can read it, than we can safely assume that in the next phase in the data analytics flow the same will succeed.
```
dubss = pd.read_json("{0}oow2018-sessions-wrangled.json".format(dataWarehouse))
dubss.head(10)
```
If and when the previous cell lists session records correctly, then the data warehouse has been populated with the consolidated file *oow2018-sessions-wrangled.json* with all sessions - cleansed, deduplicated, enriched and ready for further processing.
The wrangled data set no longer contains many of the attributes not conceivably useful for further analysis. It has been extended with (derived) attributes that will probably be useful for next data analytics tasks. Additionally, the record set has been deduplicated to only the uniqe sessions.
```
# a quick list of all columns
dubss.columns
```
|
github_jupyter
|
Deep Learning Models -- A collection of various deep learning architectures, models, and tips for TensorFlow and PyTorch in Jupyter Notebooks.
- Author: Sebastian Raschka
- GitHub Repository: https://github.com/rasbt/deeplearning-models
```
%load_ext watermark
%watermark -a 'Sebastian Raschka' -v -p tensorflow,numpy
```
# Using Queue Runners to Feed Images Directly from Disk
TensorFlow provides users with multiple options for providing data to the model. One of the probably most common methods is to define placeholders in the TensorFlow graph and feed the data from the current Python session into the TensorFlow `Session` using the `feed_dict` parameter. Using this approach, a large dataset that does not fit into memory is most conveniently and efficiently stored using NumPy archives as explained in [Chunking an Image Dataset for Minibatch Training using NumPy NPZ Archives](image-data-chunking-npz.ipynb) or HDF5 data base files ([Storing an Image Dataset for Minibatch Training using HDF5](image-data-chunking-hdf5.ipynb)).
Another approach, which is often preferred when it comes to computational efficiency, is to do the "data loading" directly in the graph using input queues from so-called TFRecords files, which is illustrated in the [Using Input Pipelines to Read Data from TFRecords Files](tfrecords.ipynb) notebook.
This notebook will introduce an alternative approach which is similar to the TFRecords approach as we will be using input queues to load the data directly on the graph. However, here we are going to read the images directly from JPEG files, which is a useful approach if disk space is a concern and we don't want to create a large TFRecords file from our "large" image database.
Beyond the examples in this notebook, you are encouraged to read more in TensorFlow's "[Reading Data](https://www.tensorflow.org/programmers_guide/reading_data)" guide.
## 0. The Dataset
Let's pretend we have a directory of images containing two subdirectories with images for training, validation, and testing. The following function will create such a dataset of images in JPEG format locally for demonstration purposes.
```
# Note that executing the following code
# cell will download the MNIST dataset
# and save all the 60,000 images as separate JPEG
# files. This might take a few minutes depending
# on your machine.
import numpy as np
# load utilities from ../helper.py
import sys
sys.path.insert(0, '..')
from helper import mnist_export_to_jpg
np.random.seed(123)
mnist_export_to_jpg(path='./')
```
The `mnist_export_to_jpg` function called above creates 3 directories, mnist_train, mnist_test, and mnist_validation. Note that the names of the subdirectories correspond directly to the class label of the images that are stored under it:
```
import os
for i in ('train', 'valid', 'test'):
dirs = [d for d in os.listdir('mnist_%s' % i) if not d.startswith('.')]
print('mnist_%s subdirectories' % i, dirs)
```
To make sure that the images look okay, the snippet below plots an example image from the subdirectory `mnist_train/9/`:
```
%matplotlib inline
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import os
some_img = os.path.join('./mnist_train/9/', os.listdir('./mnist_train/9/')[0])
img = mpimg.imread(some_img)
print(img.shape)
plt.imshow(img, cmap='binary');
```
Note: The JPEG format introduces a few artifacts that we can see in the image above. In this case, we use JPEG instead of PNG. Here, JPEG is used for demonstration purposes since that's still format many image datasets are stored in.
# 1. Reading
This section provides an example of how to use the [`tf.WholeFileReader`](https://www.tensorflow.org/api_docs/python/tf/WholeFileReader) and a filename queue to read in the images from the `mnist_train` directory. Also, we will be extracting the class labels directly from the file paths and convert the images to a one-hot encoded format that we will use in the later sections to train a multilayer neural network.
```
import tensorflow as tf
g = tf.Graph()
with g.as_default():
filename_queue = tf.train.string_input_producer(
tf.train.match_filenames_once('mnist_train/*/*.jpg'),
seed=123,
shuffle=True)
image_reader = tf.WholeFileReader()
file_name, image_raw = image_reader.read(filename_queue)
file_name = tf.identity(file_name, name='file_name')
image = tf.image.decode_jpeg(image_raw, name='image')
image = tf.cast(image, tf.float32)
label = tf.string_split([file_name], '/').values[1]
label = tf.string_to_number(label, tf.int32, name='label')
onehot_label = tf.one_hot(indices=label,
depth=10,
name='onehot_label')
with tf.Session(graph=g) as sess:
sess.run(tf.local_variables_initializer())
coord = tf.train.Coordinator()
threads = tf.train.start_queue_runners(sess=sess,
coord=coord)
image_tensor, file_name, class_label, ohe_label =\
sess.run(['image:0',
'file_name:0',
'label:0',
'onehot_label:0'])
print('Image shape:', image_tensor.shape)
print('File name:', file_name)
print('Class label:', class_label)
print('One-hot class label:', ohe_label)
coord.request_stop()
coord.join(threads)
```
- The `tf.train.string_input_producer` produces a filename queue that we iterate over in the session. Note that we need to call `sess.run(tf.local_variables_initializer())` for our filename queue. y."
- The `tf.train.start_queue_runners` function uses a queue runner that uses a separate thread to load the filenames from the `queue` that we defined in the graph without blocking the reader.
Note that it is important to shuffle the dataset so that we can later make use of TensorFlow's [`tf.train.shuffle_batch`](https://www.tensorflow.org/api_docs/python/tf/train/shuffle_batch) function and don't need to load the whole dataset into memory to shuffle epochs.
## 2. Reading in batches
While the previous section illustrated how we can use input pipelines to read images one by one, we rarely (want to) train neural networks with one datapoint at a time but use minibatches instead. TensorFlow also has some really convenient utility functions to do the batching conveniently. In the following code example, we will use the [`tf.train.shuffle_batch`](https://www.tensorflow.org/api_docs/python/tf/train/shuffle_batch) function to load the images and labels in batches of size 64.
Also, let us put the code for processing the images and labels into a function, `read_images_from_disk`, that we can reuse later.
```
import tensorflow as tf
def read_images_from_disk(filename_queue, image_dimensions, normalize=True):
image_reader = tf.WholeFileReader()
file_name, image_raw = image_reader.read(filename_queue)
file_name = tf.identity(file_name, name='file_name')
image = tf.image.decode_jpeg(image_raw, name='image')
image.set_shape(image_dimensions)
image = tf.cast(image, tf.float32)
if normalize:
# normalize to [0, 1] range
image = image / 255.
label = tf.string_split([file_name], '/').values[1]
label = tf.string_to_number(label, tf.int32)
onehot_label = tf.one_hot(indices=label,
depth=10,
name='onehot_label')
return image, onehot_label
g = tf.Graph()
with g.as_default():
filename_queue = tf.train.string_input_producer(
tf.train.match_filenames_once('mnist_train/*/*.jpg'),
seed=123)
image, label = read_images_from_disk(filename_queue,
image_dimensions=[28, 28, 1])
image_batch, label_batch = tf.train.shuffle_batch([image, label],
batch_size=64,
capacity=2000,
min_after_dequeue=1000,
num_threads=1,
seed=123)
with tf.Session(graph=g) as sess:
sess.run(tf.local_variables_initializer())
coord = tf.train.Coordinator()
threads = tf.train.start_queue_runners(sess=sess,
coord=coord)
multipe_images, multiple_labels = sess.run([image_batch, label_batch])
print('Image batch shape:', multipe_images.shape)
print('Label batch shape:', label_batch.shape)
coord.request_stop()
coord.join(threads)
```
The other relevant arguments we provided to `tf.train.shuffle_batch` are described below:
- `capacity`: An integer that defines the maximum number of elements in the queue.
- `min_after_dequeue`: The minimum number elements in the queue after a dequeue, which is used to ensure that a minimum number of data points have been loaded for shuffling.
- `num_threads`: The number of threads for enqueuing.
## 3. Use queue runners to train a neural network
In this section, we will take the concepts that were introduced in the previous sections and train a multilayer perceptron using the concepts introduced in the previous sections: the `read_images_from_disk` function, a filename queue, and the `tf.train.shuffle_batch` function.
```
##########################
### SETTINGS
##########################
# Hyperparameters
learning_rate = 0.1
batch_size = 128
n_epochs = 15
n_iter = n_epochs * (45000 // batch_size)
# Architecture
n_hidden_1 = 128
n_hidden_2 = 256
height, width = 28, 28
n_classes = 10
##########################
### GRAPH DEFINITION
##########################
g = tf.Graph()
with g.as_default():
tf.set_random_seed(123)
# Input data
filename_queue = tf.train.string_input_producer(
tf.train.match_filenames_once('mnist_train/*/*.jpg'),
seed=123)
image, label = read_images_from_disk(filename_queue,
image_dimensions=[28, 28, 1])
image = tf.reshape(image, (width*height,))
image_batch, label_batch = tf.train.shuffle_batch([image, label],
batch_size=batch_size,
capacity=2000,
min_after_dequeue=1000,
num_threads=1,
seed=123)
tf_images = tf.placeholder_with_default(image_batch,
shape=[None, 784],
name='images')
tf_labels = tf.placeholder_with_default(label_batch,
shape=[None, 10],
name='labels')
# Model parameters
weights = {
'h1': tf.Variable(tf.truncated_normal([height*width, n_hidden_1], stddev=0.1)),
'h2': tf.Variable(tf.truncated_normal([n_hidden_1, n_hidden_2], stddev=0.1)),
'out': tf.Variable(tf.truncated_normal([n_hidden_2, n_classes], stddev=0.1))
}
biases = {
'b1': tf.Variable(tf.zeros([n_hidden_1])),
'b2': tf.Variable(tf.zeros([n_hidden_2])),
'out': tf.Variable(tf.zeros([n_classes]))
}
# Multilayer perceptron
layer_1 = tf.add(tf.matmul(tf_images, weights['h1']), biases['b1'])
layer_1 = tf.nn.relu(layer_1)
layer_2 = tf.add(tf.matmul(layer_1, weights['h2']), biases['b2'])
layer_2 = tf.nn.relu(layer_2)
out_layer = tf.matmul(layer_2, weights['out']) + biases['out']
# Loss and optimizer
loss = tf.nn.softmax_cross_entropy_with_logits(logits=out_layer, labels=tf_labels)
cost = tf.reduce_mean(loss, name='cost')
optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
train = optimizer.minimize(cost, name='train')
# Prediction
prediction = tf.argmax(out_layer, 1, name='prediction')
correct_prediction = tf.equal(tf.argmax(label_batch, 1), tf.argmax(out_layer, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32), name='accuracy')
with tf.Session(graph=g) as sess:
sess.run(tf.local_variables_initializer())
sess.run(tf.global_variables_initializer())
saver0 = tf.train.Saver()
coord = tf.train.Coordinator()
threads = tf.train.start_queue_runners(sess=sess, coord=coord)
avg_cost = 0.
iter_per_epoch = n_iter // n_epochs
epoch = 0
for i in range(n_iter):
_, cost = sess.run(['train', 'cost:0'])
avg_cost += cost
if not i % iter_per_epoch:
epoch += 1
avg_cost /= iter_per_epoch
print("Epoch: %03d | AvgCost: %.3f" % (epoch, avg_cost))
avg_cost = 0.
coord.request_stop()
coord.join(threads)
saver0.save(sess, save_path='./mlp')
```
After looking at the graph above, you probably wondered why we used [`tf.placeholder_with_default`](https://www.tensorflow.org/api_docs/python/tf/placeholder_with_default) to define the two placeholders:
```python
tf_images = tf.placeholder_with_default(image_batch,
shape=[None, 784],
name='images')
tf_labels = tf.placeholder_with_default(label_batch,
shape=[None, 10],
name='labels')
```
In the training session above, these placeholders are being ignored if we don't feed them via a session's `feed_dict`, or in other words "[A `tf.placeholder_with_default` is a] placeholder op that passes through input when its output is not fed" (https://www.tensorflow.org/api_docs/python/tf/placeholder_with_default).
However, these placeholders are useful if we want to feed new data to the graph and make predictions after training as in a real-world application, which we will see in the next section.
## 4. Feeding new datapoints through placeholders
To demonstrate how we can feed new data points to the network that are not part of the training queue, let's use the test dataset and load the images into Python and pass it to the graph using a `feed_dict`:
```
import matplotlib.image as mpimg
import numpy as np
import glob
img_paths = np.array([p for p in glob.iglob('mnist_test/*/*.jpg')])
labels = np.array([int(path.split('/')[1]) for path in img_paths])
with tf.Session() as sess:
saver1 = tf.train.import_meta_graph('./mlp.meta')
saver1.restore(sess, save_path='./mlp')
num_correct = 0
cnt = 0
for path, lab in zip(img_paths, labels):
cnt += 1
image = mpimg.imread(path)
image = image.reshape(1, -1)
pred = sess.run('prediction:0',
feed_dict={'images:0': image})
num_correct += int(lab == pred[0])
acc = num_correct / cnt * 100
print('Test accuracy: %.1f%%' % acc)
```
|
github_jupyter
|
# SQL TO KQL Conversion (Experimental)
The `sql_to_kql` module is a simple converter to KQL based on [moz_sql_parser](https://github.com/DrDonk/moz-sql-parser).
It is an experimental feature built to help us convert a few queries but we
thought that it was useful enough to include in MSTICPy.
You must have msticpy installed along with the moz_sql_parser package to run this notebook:
```
%pip install --upgrade msticpy[sql2kql]
```
It supports a subset of ANSI SQL-92 which includes the following:
- SELECT (including column renaming and functions)
- FROM (including from subquery)
- WHERE (common string and int operations, LIKE, some common functions)
- LIMIT
- UNION, UNION ALL
- JOIN - only tested for relatively simple join expressions
- GROUP BY
- SQL Comments (ignored)
It does not support HAVING, multiple SQL statements or anything complex like Common Table Expressions.
It does support a few additional Spark SQL extensions like RLIKE.
## Caveat Emptor!
This module is included in MSTICPy in the hope that it might be useful to others.
We do not intend to expand its capabilities.
It is also not guaranteed to produce perfectly-executing KQL - there will likely
be things that you have to fix up in the output query.
You will, for example, nearly always need change
the names of the fields used since the source data tables are unlikely
to exactly match the schema of your Kusto/Azure Sentinel target.
The module does include an elementary table name mapping function that we
demonstrate below.
```
from pathlib import Path
import os
import sys
import warnings
from IPython.display import display, HTML, Markdown
from msticpy.nbtools import nbinit
nbinit.init_notebook(namespace=globals())
from msticpy.data.sql_to_kql import sql_to_kql
```
## Simple SQL Query
```
sql = """
SELECT DISTINCT Message, Otherfield
FROM apt29Host
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID BETWEEN 1 AND 10
AND LOWER(ParentImage) LIKE '%explorer.exe'
AND EventID IN ('4', '5', '6')
AND LOWER(Image) LIKE "3aka3%"
LIMIT 10
"""
kql = sql_to_kql(sql)
print(kql)
```
## SQL Joins
```
sql="""
SELECT DISTINCT Message, Otherfield, COUNT(DISTINCT EventID)
FROM (SELECT EventID, ParentImage, Image, Message, Otherfield FROM apt29Host) as A
--FROM A
INNER JOIN (Select Message, evt_id FROM MyTable ) on MyTable.Message == A.Message and MyTable.evt_id == A.EventID
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID = 1
AND LOWER(ParentImage) LIKE "%explorer.exe"
AND LOWER(Image) RLIKE ".*3aka3%"
GROUP BY EventID
ORDER BY Message DESC, Otherfield
LIMIT 10
"""
kql = sql_to_kql(sql)
print(kql)
```
## Table Renaming
```
sql="""
SELECT DISTINCT Message, Otherfield, COUNT(DISTINCT EventID)
FROM (SELECT EventID, ParentImage, Image, Message, Otherfield FROM apt29Host) as A
INNER JOIN (Select Message, evt_id FROM MyTable ) on MyTable.Message == A.Message and MyTable.evt_id == A.EventID
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID = 1
AND LOWER(ParentImage) LIKE "%explorer.exe"
AND LOWER(Image) RLIKE ".*3aka3%"
GROUP BY EventID
ORDER BY Message DESC, Otherfield
LIMIT 10
"""
table_map = {"apt29Host": "SecurityEvent", "MyTable": "SigninLogs"}
kql = sql_to_kql(sql, table_map)
print(kql)
```
## Join with Aliases
```
sql="""
SELECT Message
FROM apt29Host a
INNER JOIN (
SELECT ProcessGuid
FROM apt29Host
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID = 1
AND LOWER(ParentImage) RLIKE '.*partial_string.*'
AND LOWER(Image) LIKE '%cmd.exe'
) b
ON a.ParentProcessGuid = b.ProcessGuid
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID = 1
AND LOWER(Image) LIKE '%powershell.exe'
"""
kql = sql_to_kql(sql, table_map)
print(kql)
```
## Unions and Group By
```
sql="""
SELECT DISTINCT Message, COUNT(Otherfield)
FROM (SELECT *
FROM (SELECT EventID, ParentImage, Image, Message, Otherfield FROM apt29Host)
UNION
SELECT DISTINCT Message, Otherfield, EventID
FROM (SELECT EventID, ParentImage, Image, Message, Otherfield FROM apt29Host) as A
INNER JOIN MyTable on MyTable.mssg = A.Message
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID = 1
AND LOWER(ParentImage) LIKE "%explorer.exe"
AND LOWER(Image) RLIKE ".*3aka3%"
LIMIT 10
)
GROUP BY Message
ORDER BY Message DESC, Otherfield
"""
kql = sql_to_kql(sql, table_map)
print(kql)
```
## Aliased and Calculated Select Columns
```
sql="""
SELECT DISTINCT Message as mssg, COUNT(Otherfield)
FROM (SELECT EventID as ID, ParentImage, Image, Message,
ParentImage + Message as ParentMessage,
LOWER(Otherfield) FROM apt29Host
)
WHERE Channel = "Microsoft-Windows-Sysmon/Operational"
AND EventID = 1
AND LOWER(ParentImage) LIKE "%explorer.exe"
"""
kql = sql_to_kql(sql, table_map)
print(kql)
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/oferbaharav/tally-ai-ds/blob/eda/Ofer_Spacy_NLP.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import boto3
import dask.dataframe as dd
#from sagemaker import get_execution_role
import pandas as pd
!pip install fastparquet
from fastparquet import ParquetFile
#role = get_execution_role()
bucket='tally-ai-dspt3'
folder = 'yelp-kaggle-raw-data'
pd.set_option('display.max_columns', None)
print(f"S3 Bucket is {bucket}, and Folder is {folder}")
#Loading data
data = 'final_combined.parquet.gzip'
data_location = 'https://s3.amazonaws.com/{}/{}/{}'.format(bucket, folder, data)
df = dd.read_parquet(data_location)
df.head()
from flask import Flask, render_template, request, jsonify
import json
import warnings
import pandas as pd
import spacy
!pip install scattertext
import scattertext as st
from lxml import html
from requests import Session
from concurrent.futures import ThreadPoolExecutor as Executor
import requests
# from flask_cors import CORS
# from decouple import config
import re
nlp = spacy.load("en_core_web_sm")#if you run into problems here, 'Restart Runtime' and run all, it might fix things.
def customtokensize(text):
return re.findall("[\w']+", str(text))
df['tokenized_text'] = df['text'].apply(customtokensize)
df.head(2)
stopwords = ['ve',"n't",'check-in','=','= =','u','want', 'u want', 'cuz','him',"i've",'on', 'her','told','ins', '1 check','I', 'i"m', 'i', ' ', 'it', "it's", 'it.','they', 'the', 'this','its', 'l','-','they','this',"don't",'the ', ' the', 'it', 'i"ve', 'i"m', '!', '1','2','3','4', '5','6','7','8','9','0','/','.',',']
def filter_stopwords(text):
nonstopwords = []
for i in text:
if i not in stopwords:
nonstopwords.append(i)
return nonstopwords
df['tokenized_text'] = df['tokenized_text'].apply(filter_stopwords)
df['parts_of_speech_reference'] = df['tokenized_text'].apply(filter_stopwords)
df['parts_of_speech_reference'] = df['parts_of_speech_reference'].str.join(' ')
df.head(2)
def find_part_of_speech(x):
"""Use spacy's entity recognition to recogize if word is noun, verb, adjective, etc."""
part_of_speech = []
doc = nlp(str(x))
for token in doc:
part_of_speech.append(token.pos_)
return part_of_speech
df['parts_of_speech'] = df['parts_of_speech_reference'].apply(find_part_of_speech)
df.head(2)
#Useless?
def extract_adjective_indexes(text):
"""Get the indexes of Adjectives and delete the occurrence
of adjectives in order to persistently find new adjective
occurrences. In the future, add words occurring before and after"""
adjective_indexes = []
for i in text:
if i == 'ADJ':
adj_index = text.index('ADJ')
adjective_indexes.append(adj_index)
text.remove(i)
return adjective_indexes
df['adjective_positions'] = df['parts_of_speech'].apply(extract_adjective_indexes)
df.head(2)
def find_adj(x):
"""Get Just the Adjectives"""
adj_list = []
doc = nlp(str(x))
for token in doc:
if token.pos_ == 'ADJ':
adj_list.append(token)
return adj_list
df['adj_list'] = df['parts_of_speech_reference'].apply(find_adj)
df.head(2)
def find_phrases(x):
"""Create a list where adjectives come immediately before nouns for each review"""
adj_list = []
doc = nlp(str(x))
try:
for token in range(len(doc)):
sub_list = []
if (doc[token].pos_ == 'ADJ'and doc[token+1].pos_ =='NOUN') or (doc[token].pos_ == 'VERB'and doc[token+1].pos_ =='NOUN'):
sub_list.append(doc[token])
sub_list.append(doc[token+1])
elif (doc[token].pos_ == 'ADJ'and doc[token+1].pos_ == 'ADJ'and doc[token+2].pos_ =='NOUN')or (doc[token].pos_ == 'ADJ'and doc[token+1].pos_ =='VERB'and doc[token+2].pos_ =='NOUN')or (doc[token].pos_ == 'ADJ'and doc[token+1].pos_ == 'NOUN'and doc[token+2].pos_ =='NOUN'):
sub_list.append(doc[token])
sub_list.append(doc[token+1])
sub_list.append(doc[token+2])
if (doc[token].lemma_ == 'wait'):
sub_list.append(doc[token-2])
sub_list.append(doc[token-1])
sub_list.append(doc[token])
sub_list.append(doc[token+1])
sub_list.append(doc[token+2])
sub_list.append(doc[token+3])
if (doc[token].lemma_ == 'service'):
sub_list.append(doc[token-2])
sub_list.append(doc[token-1])
sub_list.append(doc[token])
sub_list.append(doc[token+1])
sub_list.append(doc[token+2])
sub_list.append(doc[token+3])
if len(sub_list) != 0:
adj_list.append(sub_list)
return adj_list
except IndexError as e:
pass
df['adj_noun_phrases'] = df['parts_of_speech_reference'].apply(find_phrases)
df['adj_noun_phrases'].head(10)
doc = nlp("Apple is looking at buying U.K. startup for $1 billion")
for ent in doc.ents:
print(ent.text, ent.start_char, ent.end_char, ent.label_)
def find_money(x):
"""Create a list where adjectives come immediately before nouns for each review"""
money_list = []
doc = nlp(str(x))
for ent in doc.ents:
if ent.label_ == 'MONEY':
money_list.append(ent)
return money_list
df['money_list'] = df['parts_of_speech_reference'].apply(find_money)
df.head(2)
def find_noun_chunks(x):
"""Create a list where adjectives come immediately before nouns for each review"""
noun_list = []
doc = nlp(str(x))
for chunk in doc.noun_chunks:
noun_list.append(chunk)
return noun_list
df['noun_chunks'] = df['parts_of_speech_reference'].apply(find_noun_chunks)
doc = nlp("Autonomous cars shift insurance liability toward manufacturers")
for token in doc:
print(token.text, token.dep_, token.head.text, token.head.pos_,
[child for child in token.children])
for token in doc:
print(token.text, token.dep_, token.head.text, token.head.pos_,
[child for child in token.children])
corpus = st.CorpusFromPandas(df,
category_col='stars_review',
text_col='text',
nlp=nlp).build()
term_freq_df = corpus.get_term_freq_df()
term_freq_df['highratingscore'] = corpus.get_scaled_f_scores('5')
term_freq_df['poorratingscore'] = corpus.get_scaled_f_scores('1')
dh = term_freq_df.sort_values(by= 'highratingscore', ascending = False)
dh = dh[['highratingscore', 'poorratingscore']]
dh = dh.reset_index(drop=False)
dh = dh.rename(columns={'highratingscore':'score'})
dh = dh.drop(columns='poorratingscore')
positive_df = dh.head(10)
negative_df = dh.tail(10)
# word_df = pd.concat([positive_df, negative_df])
# word_df
results = {'positive': [{'term': pos_term, 'score': pos_score} for pos_term, pos_score in zip(positive_df['term'], positive_df['score'])], 'negative': [{'term': neg_term, 'score': neg_score} for neg_term, neg_score in zip(negative_df['term'], negative_df['score'])]}
results
```
|
github_jupyter
|
```
#hide
#skip
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
#all_slow
#export
from fastai.basics import *
from fastai.learner import Callback
#hide
from nbdev.showdoc import *
#default_exp callback.azureml
```
# AzureML Callback
Track fastai experiments with the azure machine learning plattform.
## Prerequisites
Install the azureml SDK:
```python
pip install azureml-core
```
## How to use it?
Import and use `AzureMLCallback` during model fitting.
If you are submitting your training run with azureml SDK [ScriptRunConfig](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-set-up-training-targets), the callback will automatically detect the run and log metrics. For example:
```python
from fastai.callback.azureml import AzureMLCallback
learn.fit_one_cycle(epoch, lr, cbs=AzureMLCallback())
```
If you are running an experiment manually and just want to have interactive logging of the run, use azureml's `Experiment.start_logging` to create the interactive `run`, and pass that into `AzureMLCallback`. For example:
```python
from azureml.core import Experiment
experiment = Experiment(workspace=ws, name='experiment_name')
run = experiment.start_logging(outputs=None, snapshot_directory=None)
from fastai.callback.azureml import AzureMLCallback
learn.fit_one_cycle(epoch, lr, cbs=AzureMLCallback(run))
```
If you are running an experiment on your local machine (i.e. not using `ScriptRunConfig` and not passing an azureml `run` into the callback), it will recognize that there is no AzureML run to log to, and print the log attempts instead.
To save the model weights, use the usual fastai methods and save the model to the `outputs` folder, which is a "special" (for Azure) folder that is automatically tracked in AzureML.
As it stands, note that if you pass the callback into your `Learner` directly, e.g.:
```python
learn = Learner(dls, model, cbs=AzureMLCallback())
```
…some `Learner` methods (e.g. `learn.show_results()`) might add unwanted logging into your azureml experiment runs. Adding further checks into the callback should help eliminate this – another PR needed.
```
#export
from azureml.core.run import Run
# export
class AzureMLCallback(Callback):
"Log losses, metrics, model architecture summary to AzureML"
order = Recorder.order+1
def __init__(self, azurerun=None):
if azurerun:
self.azurerun = azurerun
else:
self.azurerun = Run.get_context()
def before_fit(self):
self.azurerun.log("n_epoch", self.learn.n_epoch)
self.azurerun.log("model_class", str(type(self.learn.model)))
try:
summary_file = Path("outputs") / 'model_summary.txt'
with summary_file.open("w") as f:
f.write(repr(self.learn.model))
except:
print('Did not log model summary. Check if your model is PyTorch model.')
def after_batch(self):
# log loss and opt.hypers
if self.learn.training:
self.azurerun.log('batch__loss', self.learn.loss.item())
self.azurerun.log('batch__train_iter', self.learn.train_iter)
for i, h in enumerate(self.learn.opt.hypers):
for k, v in h.items():
self.azurerun.log(f'batch__opt.hypers.{k}', v)
def after_epoch(self):
# log metrics
for n, v in zip(self.learn.recorder.metric_names, self.learn.recorder.log):
if n not in ['epoch', 'time']:
self.azurerun.log(f'epoch__{n}', v)
if n == 'time':
# split elapsed time string, then convert into 'seconds' to log
m, s = str(v).split(':')
elapsed = int(m)*60 + int(s)
self.azurerun.log(f'epoch__{n}', elapsed)
```
|
github_jupyter
|
```
#Importing necessary dependencies
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
pd.set_option('display.max_columns',None)
df=pd.read_excel('Data_Train.xlsx')
df.head()
df.shape
```
## Exploratory data analysis
First we will try to find the missing values and we will try to find relationship between different features and we will also visualize the data and see the relationship between them.
```
df.isnull().sum()
df.info()
#Describe the data
df.describe()
```
Since there is only one numerical feature we will try to analyze the categorical data and see the relationship with the price
```
feature_categorical=[feature for feature in df.columns if df[feature].dtypes=='O']
feature_categorical
df.dropna(inplace=True)
```
## Lets change the date time format
```
#train_data["Journey_day"] = pd.to_datetime(train_data.Date_of_Journey, format="%d/%m/%Y").dt.day
df['Day_of_Journey']=pd.to_datetime(df['Date_of_Journey']).dt.day
df['Journey_Month']=pd.to_datetime(df['Date_of_Journey']).dt.month
# Now we will extract the hour and minutes in Arrival time
df["Arrival_hour"]=pd.to_datetime(df['Arrival_Time']).dt.hour
df['Arrival_minute']=pd.to_datetime(df['Arrival_Time']).dt.minute
df.head()
df.drop(['Date_of_Journey','Arrival_Time'],axis=1,inplace=True)
df.head()
df['Dep_hour']=pd.to_datetime(df['Dep_Time']).dt.hour
df['Dep_min']=pd.to_datetime(df['Dep_Time']).dt.minute
df.drop(['Dep_Time'],inplace=True,axis=1)
df.head()
duration=list(df['Duration'])
duration[0].split(" ")
for num in range(len(duration)):
if len(duration[num].split(" "))!=2:
if 'h' in duration[num]:
duration[num]=duration[num].strip()+'0m'
else:
duration[num]='0h'+duration[num]
duration_hour=[]
duration_min=[]
for num in range(len(duration)):
duration_hour.append(int(duration[num].split("h")[0]))
duration_min.append(int(duration[num].split("h")[1].split('m')[0].strip()))
df['Duration_hour']=duration_hour
df['Duration_min']=duration_min
df.drop('Duration',axis=1,inplace=True)
df.head()
```
# Handling the categorical data
```
airway=df['Airline']
df['Airline'].value_counts()
plt.figure(figsize=(18,8))
sns.boxplot(x='Airline',y='Price',data=df.sort_values('Price',ascending=False))
```
# Encoding categorical data into numerical
Since the airlines are not ordinal we will one hot encode the data using get dummies function in pandas
```
Airline=pd.get_dummies(df['Airline'],drop_first=True)
Airline.head()
df['Source'].value_counts()
# Source vs Price
plt.figure(figsize=(14,8))
sns.boxplot(x='Source',y='Price',data=df.sort_values('Price',ascending=False))
# Now we one hot encode the source feature using same method used above
Source=df['Source']
Source=pd.get_dummies(Source,drop_first=False)
Source.head()
Destination=df['Destination']
Destination=pd.get_dummies(Destination,drop_first=False)
Destination=Destination.rename(columns={"Banglore":"Dest_Banglore",'Cochin':'Dest_Cochin',"Delhi":'Dest_Delhi','Hyderabad':'Dest_Hyderabad',"Kolkata":'Dest_Kolkata','New Delhi':'Dest_NewDelhi'})
df.head()
df["Route"].head()
df['Total_Stops'].value_counts()
# Since the route is related to no of stops we can drop that feature
# Now we can change no of stops using ordinal encoding since it is ordinal data
df['Total_Stops'].replace({'non-stop':0,'1 stop':1,'2 stops':2,'3 stops':3,'4 stops':4},inplace=True)
# Since the Airline, Source and Destination are one hot encoded and
# we can determine the route by seeing the no of stops we can drop those features
df.drop(['Airline','Source','Destination','Route'],inplace=True,axis=1)
df.head()
df['Additional_Info'].value_counts()
```
## Since the Addtional_info has lot of no info we can actually drop this feature
```
df.drop('Additional_Info',axis=1,inplace=True)
df.head()
df_concat=pd.concat([df,Airline,Source,Destination],axis=1)
df_concat.head()
```
# Let's repeat above for the test data
```
df_test=pd.read_excel('Test_set.xlsx')
df_test.head()
# duration=list(df['Duration'])
# duration[0].split(" ")
# for num in range(len(duration)):
# if len(duration[num].split(" "))!=2:
# if 'h' in duration[num]:
# duration[num]=duration[num].strip()+'0m'
# else:
# duration[num]='0h'+duration[num]
# duration_hour=[]
# duration_min=[]
# for num in range(len(duration)):
# duration_hour.append(int(duration[num].split("h")[0]))
# duration_min.append(int(duration[num].split("h")[1].split('m')[0].strip()))
# df_test['Duration_hour']=duration_hour
# df_test['Duration_min']=duration_min
# df_test.drop('Duration',axis=1,inplace=True)
# Airline=pd.get_dummies(df_test['Airline'],drop_first=True)
# # Now we one hot encode the source feature using same method used above
# Source=df_test['Source']
# Source=pd.get_dummies(Source,drop_first=False)
# Destination=df['Destination']
# Destination=pd.get_dummies(Destination,drop_first=False)
# Since the route is related to no of stops we can drop that feature
# Now we can change no of stops using ordinal encoding since it is ordinal data
# df_test['Total_Stops'].replace({'non-stop':0,'1 stop':1,'2 stops':2,'3 stops':3,'4 stops':4},inplace=True)
# df_test.drop(['Airline','Source','Destination','Route'],inplace=True,axis=1)
# df_test.drop('Additional_Info',axis=1,inplace=True)
# df_concat1=pd.concat([df_test,Airline,Source,Destination],axis=1)
df_test['Day_of_Journey']=pd.to_datetime(df_test['Date_of_Journey']).dt.day
df_test['Journey_Month']=pd.to_datetime(df_test['Date_of_Journey']).dt.month
# Now we will extract the hour and minutes in Arrival time
df_test["Arrival_hour"]=pd.to_datetime(df_test['Arrival_Time']).dt.hour
df_test['Arrival_minute']=pd.to_datetime(df_test['Arrival_Time']).dt.minute
df_test.drop(['Date_of_Journey','Arrival_Time'],axis=1,inplace=True)
df_test['Dep_hour']=pd.to_datetime(df_test['Dep_Time']).dt.hour
df_test['Dep_min']=pd.to_datetime(df_test['Dep_Time']).dt.minute
duration=list(df_test['Duration'])
duration[0].split(" ")
for num in range(len(duration)):
if len(duration[num].split(" "))!=2:
if 'h' in duration[num]:
duration[num]=duration[num].strip()+'0m'
else:
duration[num]='0h'+duration[num]
duration_hour=[]
duration_min=[]
for num in range(len(duration)):
duration_hour.append(int(duration[num].split("h")[0]))
duration_min.append(int(duration[num].split("h")[1].split('m')[0].strip()))
df_test['Duration_hour']=duration_hour
df_test['Duration_min']=duration_min
df_test.drop('Duration',axis=1,inplace=True)
Airline=pd.get_dummies(df_test['Airline'],drop_first=True)
# Now we one hot encode the source feature using same method used above
Source=df_test['Source']
Source=pd.get_dummies(Source,drop_first=False)
Destination=df_test['Destination']
Destination=pd.get_dummies(Destination,drop_first=False)
Destination=Destination.rename(columns={"Banglore":"Dest_Banglore",'Cochin':'Dest_Cochin',"Delhi":'Dest_Delhi','Hyderabad':'Dest_Hyderabad',"Kolkata":'Dest_Kolkata','New Delhi':'Dest_NewDelhi'})
# Since the route is related to no of stops we can drop that feature
# Now we can change no of stops using ordinal encoding since it is ordinal data
df_test['Total_Stops'].replace({'non-stop':0,'1 stop':1,'2 stops':2,'3 stops':3,'4 stops':4},inplace=True)
df_test.drop(['Airline','Source','Destination','Route'],inplace=True,axis=1)
df_test.drop(['Additional_Info','Dep_Time'],axis=1,inplace=True)
df_concat1=pd.concat([df_test,Airline,Source,Destination],axis=1)
df_concat1.head()
df_concat.head()
df_concat1.head()
df_concat.shape
df_test['Dep_min']
Xtr=df_concat.drop(['Price','Trujet'],axis=1)
Ytr=df["Price"]
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(Xtr,Ytr,test_size=0.2,random_state=5)
x_test.shape
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
from xgboost import XGBRegressor
model=XGBRegressor()
model.fit(x_train,y_train)
y_pre=model.predict(x_test)
y_pre
y_test
from sklearn.metrics import r2_score
r2_score(y_test,y_pred=y_pre)
model.predict(df_concat1)
plt.figure(figsize=(14,8))
plt.scatter(y_test,y_pre,color='g')
```
|
github_jupyter
|
# Multiple Qubits & Entangled States
Single qubits are interesting, but individually they offer no computational advantage. We will now look at how we represent multiple qubits, and how these qubits can interact with each other. We have seen how we can represent the state of a qubit using a 2D-vector, now we will see how we can represent the state of multiple qubits.
## Contents
1. [Representing Multi-Qubit States](#represent)
1.1 [Exercises](#ex1)
2. [Single Qubit Gates on Multi-Qubit Statevectors](#single-qubit-gates)
2.1 [Exercises](#ex2)
3. [Multi-Qubit Gates](#multi-qubit-gates)
3.1 [The CNOT-gate](#cnot)
3.2 [Entangled States](#entangled)
3.3 [Visualizing Entangled States](#visual)
3.4 [Exercises](#ex3)
## 1. Representing Multi-Qubit States <a id="represent"></a>
We saw that a single bit has two possible states, and a qubit state has two complex amplitudes. Similarly, two bits have four possible states:
`00` `01` `10` `11`
And to describe the state of two qubits requires four complex amplitudes. We store these amplitudes in a 4D-vector like so:
$$ |a\rangle = a_{00}|00\rangle + a_{01}|01\rangle + a_{10}|10\rangle + a_{11}|11\rangle = \begin{bmatrix} a_{00} \\ a_{01} \\ a_{10} \\ a_{11} \end{bmatrix} $$
The rules of measurement still work in the same way:
$$ p(|00\rangle) = |\langle 00 | a \rangle |^2 = |a_{00}|^2$$
And the same implications hold, such as the normalisation condition:
$$ |a_{00}|^2 + |a_{01}|^2 + |a_{10}|^2 + |a_{11}|^2 = 1$$
If we have two separated qubits, we can describe their collective state using the tensor product:
$$ |a\rangle = \begin{bmatrix} a_0 \\ a_1 \end{bmatrix}, \quad |b\rangle = \begin{bmatrix} b_0 \\ b_1 \end{bmatrix} $$
$$
|ba\rangle = |b\rangle \otimes |a\rangle = \begin{bmatrix} b_0 \times \begin{bmatrix} a_0 \\ a_1 \end{bmatrix} \\ b_1 \times \begin{bmatrix} a_0 \\ a_1 \end{bmatrix} \end{bmatrix} = \begin{bmatrix} b_0 a_0 \\ b_0 a_1 \\ b_1 a_0 \\ b_1 a_1 \end{bmatrix}
$$
And following the same rules, we can use the tensor product to describe the collective state of any number of qubits. Here is an example with three qubits:
$$
|cba\rangle = \begin{bmatrix} c_0 b_0 a_0 \\ c_0 b_0 a_1 \\ c_0 b_1 a_0 \\ c_0 b_1 a_1 \\
c_1 b_0 a_0 \\ c_1 b_0 a_1 \\ c_1 b_1 a_0 \\ c_1 b_1 a_1 \\
\end{bmatrix}
$$
If we have $n$ qubits, we will need to keep track of $2^n$ complex amplitudes. As we can see, these vectors grow exponentially with the number of qubits. This is the reason quantum computers with large numbers of qubits are so difficult to simulate. A modern laptop can easily simulate a general quantum state of around 20 qubits, but simulating 100 qubits is too difficult for the largest supercomputers.
Let's look at an example circuit:
```
from qiskit import QuantumCircuit, Aer, assemble
from math import pi
import numpy as np
from qiskit.visualization import plot_histogram, plot_bloch_multivector
qc = QuantumCircuit(3)
# Apply H-gate to each qubit:
for qubit in range(3):
qc.h(qubit)
# See the circuit:
qc.draw()
```
Each qubit is in the state $|+\rangle$, so we should see the vector:
$$
|{+++}\rangle = \frac{1}{\sqrt{8}}\begin{bmatrix} 1 \\ 1 \\ 1 \\ 1 \\
1 \\ 1 \\ 1 \\ 1 \\
\end{bmatrix}
$$
```
# Let's see the result
svsim = Aer.get_backend('statevector_simulator')
qobj = assemble(qc)
final_state = svsim.run(qobj).result().get_statevector()
# In Jupyter Notebooks we can display this nicely using Latex.
# If not using Jupyter Notebooks you may need to remove the
# array_to_latex function and use print(final_state) instead.
from qiskit_textbook.tools import array_to_latex
array_to_latex(final_state, pretext="\\text{Statevector} = ")
```
And we have our expected result.
### 1.2 Quick Exercises: <a id="ex1"></a>
1. Write down the tensor product of the qubits:
a) $|0\rangle|1\rangle$
b) $|0\rangle|+\rangle$
c) $|+\rangle|1\rangle$
d) $|-\rangle|+\rangle$
2. Write the state:
$|\psi\rangle = \tfrac{1}{\sqrt{2}}|00\rangle + \tfrac{i}{\sqrt{2}}|01\rangle $
as two separate qubits.
## 2. Single Qubit Gates on Multi-Qubit Statevectors <a id="single-qubit-gates"></a>
We have seen that an X-gate is represented by the matrix:
$$
X = \begin{bmatrix} 0 & 1 \\ 1 & 0 \end{bmatrix}
$$
And that it acts on the state $|0\rangle$ as so:
$$
X|0\rangle = \begin{bmatrix} 0 & 1 \\ 1 & 0 \end{bmatrix}\begin{bmatrix} 1 \\ 0 \end{bmatrix} = \begin{bmatrix} 0 \\ 1\end{bmatrix}
$$
but it may not be clear how an X-gate would act on a qubit in a multi-qubit vector. Fortunately, the rule is quite simple; just as we used the tensor product to calculate multi-qubit statevectors, we use the tensor product to calculate matrices that act on these statevectors. For example, in the circuit below:
```
qc = QuantumCircuit(2)
qc.h(0)
qc.x(1)
qc.draw()
```
we can represent the simultaneous operations (H & X) using their tensor product:
$$
X|q_1\rangle \otimes H|q_0\rangle = (X\otimes H)|q_1 q_0\rangle
$$
The operation looks like this:
$$
X\otimes H = \begin{bmatrix} 0 & 1 \\ 1 & 0 \end{bmatrix} \otimes \tfrac{1}{\sqrt{2}}\begin{bmatrix} 1 & 1 \\ 1 & -1 \end{bmatrix} = \frac{1}{\sqrt{2}}
\begin{bmatrix} 0 \times \begin{bmatrix} 1 & 1 \\ 1 & -1 \end{bmatrix}
& 1 \times \begin{bmatrix} 1 & 1 \\ 1 & -1 \end{bmatrix}
\\
1 \times \begin{bmatrix} 1 & 1 \\ 1 & -1 \end{bmatrix}
& 0 \times \begin{bmatrix} 1 & 1 \\ 1 & -1 \end{bmatrix}
\end{bmatrix} = \frac{1}{\sqrt{2}}
\begin{bmatrix} 0 & 0 & 1 & 1 \\
0 & 0 & 1 & -1 \\
1 & 1 & 0 & 0 \\
1 & -1 & 0 & 0 \\
\end{bmatrix}
$$
Which we can then apply to our 4D statevector $|q_1 q_0\rangle$. This can become quite messy, you will often see the clearer notation:
$$
X\otimes H =
\begin{bmatrix} 0 & H \\
H & 0\\
\end{bmatrix}
$$
Instead of calculating this by hand, we can use Qiskit’s `unitary_simulator` to calculate this for us. The unitary simulator multiplies all the gates in our circuit together to compile a single unitary matrix that performs the whole quantum circuit:
```
usim = Aer.get_backend('unitary_simulator')
qobj = assemble(qc)
unitary = usim.run(qobj).result().get_unitary()
```
and view the results:
```
# In Jupyter Notebooks we can display this nicely using Latex.
# If not using Jupyter Notebooks you may need to remove the
# array_to_latex function and use print(unitary) instead.
from qiskit_textbook.tools import array_to_latex
array_to_latex(unitary, pretext="\\text{Circuit = }\n")
```
If we want to apply a gate to only one qubit at a time (such as in the circuit below), we describe this using tensor product with the identity matrix, e.g.:
$$ X \otimes I $$
```
qc = QuantumCircuit(2)
qc.x(1)
qc.draw()
# Simulate the unitary
usim = Aer.get_backend('unitary_simulator')
qobj = assemble(qc)
unitary = usim.run(qobj).result().get_unitary()
# Display the results:
array_to_latex(unitary, pretext="\\text{Circuit = } ")
```
We can see Qiskit has performed the tensor product:
$$
X \otimes I =
\begin{bmatrix} 0 & I \\
I & 0\\
\end{bmatrix} =
\begin{bmatrix} 0 & 0 & 1 & 0 \\
0 & 0 & 0 & 1 \\
1 & 0 & 0 & 0 \\
0 & 1 & 0 & 0 \\
\end{bmatrix}
$$
### 2.1 Quick Exercises: <a id="ex2"></a>
1. Calculate the single qubit unitary ($U$) created by the sequence of gates: $U = XZH$. Use Qiskit's unitary simulator to check your results.
2. Try changing the gates in the circuit above. Calculate their tensor product, and then check your answer using the unitary simulator.
**Note:** Different books, softwares and websites order their qubits differently. This means the tensor product of the same circuit can look very different. Try to bear this in mind when consulting other sources.
## 3. Multi-Qubit Gates <a id="multi-qubit-gates"></a>
Now we know how to represent the state of multiple qubits, we are now ready to learn how qubits interact with each other. An important two-qubit gate is the CNOT-gate.
### 3.1 The CNOT-Gate <a id="cnot"></a>
You have come across this gate before in _[The Atoms of Computation](../ch-states/atoms-computation.html)._ This gate is a conditional gate that performs an X-gate on the second qubit (target), if the state of the first qubit (control) is $|1\rangle$. The gate is drawn on a circuit like this, with `q0` as the control and `q1` as the target:
```
qc = QuantumCircuit(2)
# Apply CNOT
qc.cx(0,1)
# See the circuit:
qc.draw()
```
When our qubits are not in superposition of $|0\rangle$ or $|1\rangle$ (behaving as classical bits), this gate is very simple and intuitive to understand. We can use the classical truth table:
| Input (t,c) | Output (t,c) |
|:-----------:|:------------:|
| 00 | 00 |
| 01 | 11 |
| 10 | 10 |
| 11 | 01 |
And acting on our 4D-statevector, it has one of the two matrices:
$$
\text{CNOT} = \begin{bmatrix} 1 & 0 & 0 & 0 \\
0 & 0 & 0 & 1 \\
0 & 0 & 1 & 0 \\
0 & 1 & 0 & 0 \\
\end{bmatrix}, \quad
\text{CNOT} = \begin{bmatrix} 1 & 0 & 0 & 0 \\
0 & 1 & 0 & 0 \\
0 & 0 & 0 & 1 \\
0 & 0 & 1 & 0 \\
\end{bmatrix}
$$
depending on which qubit is the control and which is the target. Different books, simulators and papers order their qubits differently. In our case, the left matrix corresponds to the CNOT in the circuit above. This matrix swaps the amplitudes of $|01\rangle$ and $|11\rangle$ in our statevector:
$$
|a\rangle = \begin{bmatrix} a_{00} \\ a_{01} \\ a_{10} \\ a_{11} \end{bmatrix}, \quad \text{CNOT}|a\rangle = \begin{bmatrix} a_{00} \\ a_{11} \\ a_{10} \\ a_{01} \end{bmatrix} \begin{matrix} \\ \leftarrow \\ \\ \leftarrow \end{matrix}
$$
We have seen how this acts on classical states, but let’s now see how it acts on a qubit in superposition. We will put one qubit in the state $|+\rangle$:
```
qc = QuantumCircuit(2)
# Apply H-gate to the first:
qc.h(0)
qc.draw()
# Let's see the result:
svsim = Aer.get_backend('statevector_simulator')
qobj = assemble(qc)
final_state = svsim.run(qobj).result().get_statevector()
# Print the statevector neatly:
array_to_latex(final_state, pretext="\\text{Statevector = }")
```
As expected, this produces the state $|0\rangle \otimes |{+}\rangle = |0{+}\rangle$:
$$
|0{+}\rangle = \tfrac{1}{\sqrt{2}}(|00\rangle + |01\rangle)
$$
And let’s see what happens when we apply the CNOT gate:
```
qc = QuantumCircuit(2)
# Apply H-gate to the first:
qc.h(0)
# Apply a CNOT:
qc.cx(0,1)
qc.draw()
# Let's get the result:
qobj = assemble(qc)
result = svsim.run(qobj).result()
# Print the statevector neatly:
final_state = result.get_statevector()
array_to_latex(final_state, pretext="\\text{Statevector = }")
```
We see we have the state:
$$
\text{CNOT}|0{+}\rangle = \tfrac{1}{\sqrt{2}}(|00\rangle + |11\rangle)
$$
This state is very interesting to us, because it is _entangled._ This leads us neatly on to the next section.
### 3.2 Entangled States <a id="entangled"></a>
We saw in the previous section we could create the state:
$$
\tfrac{1}{\sqrt{2}}(|00\rangle + |11\rangle)
$$
This is known as a _Bell_ state. We can see that this state has 50% probability of being measured in the state $|00\rangle$, and 50% chance of being measured in the state $|11\rangle$. Most interestingly, it has a **0%** chance of being measured in the states $|01\rangle$ or $|10\rangle$. We can see this in Qiskit:
```
plot_histogram(result.get_counts())
```
This combined state cannot be written as two separate qubit states, which has interesting implications. Although our qubits are in superposition, measuring one will tell us the state of the other and collapse its superposition. For example, if we measured the top qubit and got the state $|1\rangle$, the collective state of our qubits changes like so:
$$
\tfrac{1}{\sqrt{2}}(|00\rangle + |11\rangle) \quad \xrightarrow[]{\text{measure}} \quad |11\rangle
$$
Even if we separated these qubits light-years away, measuring one qubit collapses the superposition and appears to have an immediate effect on the other. This is the [‘spooky action at a distance’](https://en.wikipedia.org/wiki/Quantum_nonlocality) that upset so many physicists in the early 20th century.
It’s important to note that the measurement result is random, and the measurement statistics of one qubit are **not** affected by any operation on the other qubit. Because of this, there is **no way** to use shared quantum states to communicate. This is known as the no-communication theorem.[1]
### 3.3 Visualizing Entangled States<a id="visual"></a>
We have seen that this state cannot be written as two separate qubit states, this also means we lose information when we try to plot our state on separate Bloch spheres:
```
plot_bloch_multivector(final_state)
```
Given how we defined the Bloch sphere in the earlier chapters, it may not be clear how Qiskit even calculates the Bloch vectors with entangled qubits like this. In the single-qubit case, the position of the Bloch vector along an axis nicely corresponds to the expectation value of measuring in that basis. If we take this as _the_ rule of plotting Bloch vectors, we arrive at this conclusion above. This shows us there is _no_ single-qubit measurement basis for which a specific measurement is guaranteed. This contrasts with our single qubit states, in which we could always pick a single-qubit basis. Looking at the individual qubits in this way, we miss the important effect of correlation between the qubits. We cannot distinguish between different entangled states. For example, the two states:
$$\tfrac{1}{\sqrt{2}}(|01\rangle + |10\rangle) \quad \text{and} \quad \tfrac{1}{\sqrt{2}}(|00\rangle + |11\rangle)$$
will both look the same on these separate Bloch spheres, despite being very different states with different measurement outcomes.
How else could we visualize this statevector? This statevector is simply a collection of four amplitudes (complex numbers), and there are endless ways we can map this to an image. One such visualization is the _Q-sphere,_ here each amplitude is represented by a blob on the surface of a sphere. The size of the blob is proportional to the magnitude of the amplitude, and the colour is proportional to the phase of the amplitude. The amplitudes for $|00\rangle$ and $|11\rangle$ are equal, and all other amplitudes are 0:
```
from qiskit.visualization import plot_state_qsphere
plot_state_qsphere(final_state)
```
Here we can clearly see the correlation between the qubits. The Q-sphere's shape has no significance, it is simply a nice way of arranging our blobs; the number of `0`s in the state is proportional to the states position on the Z-axis, so here we can see the amplitude of $|00\rangle$ is at the top pole of the sphere, and the amplitude of $|11\rangle$ is at the bottom pole of the sphere.
### 3.4 Exercise: <a id="ex3"></a>
1. Create a quantum circuit that produces the Bell state: $\tfrac{1}{\sqrt{2}}(|01\rangle + |10\rangle)$.
Use the statevector simulator to verify your result.
2. The circuit you created in question 1 transforms the state $|00\rangle$ to $\tfrac{1}{\sqrt{2}}(|01\rangle + |10\rangle)$, calculate the unitary of this circuit using Qiskit's simulator. Verify this unitary does in fact perform the correct transformation.
3. Think about other ways you could represent a statevector visually. Can you design an interesting visualization from which you can read the magnitude and phase of each amplitude?
## 4. References
[1] Asher Peres, Daniel R. Terno, _Quantum Information and Relativity Theory,_ 2004, https://arxiv.org/abs/quant-ph/0212023
```
import qiskit
qiskit.__qiskit_version__
```
|
github_jupyter
|
# A Transformer based Language Model from scratch
> Building transformer with simple building blocks
- toc: true
- branch: master
- badges: true
- comments: true
- author: Arto
- categories: [fastai, pytorch]
```
#hide
import sys
if 'google.colab' in sys.modules:
!pip install -Uqq fastai
```
In this notebook i'm going to construct transformer based language model from scratch starting with the simplest building blocks. This is inspired by Chapter 12 of [Deep Learning for Coders book](https://www.amazon.com/Deep-Learning-Coders-fastai-PyTorch/dp/1492045527) in which it's demonstrated how to create a Recurrent Neural Network. It provides a strong intuition of how RNNs relate to regular feed-forward neural nets and why certain design choices were made. Here we aim to aquire similar kind of intuition about Transfomer based architectures.
But as always we should start with the data to be modeled, 'cause without data any model makes no particular sense.
## Data
Similar to authors of the book I'll use simple Human numbers dataset which is specifically designed to prototyping model fast and straightforward. For more details on the data one can refer to the aforemantioned book chapter which is also available for free as [a notebook](https://github.com/fastai/fastbook/blob/master/12_nlp_dive.ipynb) (isn't that awesome?!)
```
from fastai.text.all import *
path = untar_data(URLs.HUMAN_NUMBERS)
Path.BASE_PATH = path
path.ls()
```
The data consists of consecutive numbers from 1 to 9999 inclusive spelled as words.
```
lines = L()
with open(path/'train.txt') as f: lines += L(*f.readlines())
with open(path/'valid.txt') as f: lines += L(*f.readlines())
lines
text = ' . '.join([l.strip() for l in lines])
tokens = text.split(' ')
tokens[:10]
vocab = L(*tokens).unique()
vocab
word2idx = {w:i for i,w in enumerate(vocab)}
nums = L(word2idx[i] for i in tokens)
nums
```
The task will be to predict subsequent token given preceding three. This kind of tasks when the goal is to predict next token from previous ones is called autoregresive language modeling.
```
L((tokens[i:i+3], tokens[i+3]) for i in range(0,len(tokens)-4,3))
seqs = L((tensor(nums[i:i+3]), nums[i+3]) for i in range(0,len(nums)-4,3))
seqs
bs = 64
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(seqs[:cut], seqs[cut:], bs=64, shuffle=False)
x, y = dls.one_batch()
x.shape, y.shape
```
## Dot product attention

The core idea behind Transformers is Attention. Since the release of famous paper [Attention is All You Need](https://arxiv.org/abs/1706.03762) transformers has become most popular architecture for language modelling.
There are a lot of great resourses explaining transformers architecture. I'll list some of those I found useful and comprehensive:
1. [The Annotated Transformer](http://nlp.seas.harvard.edu/2018/04/03/attention.html) completes the original paper with code
2. [Encoder-Decoder Model](https://colab.research.google.com/github/patrickvonplaten/notebooks/blob/master/Encoder_Decoder_Model.ipynb) notebook by huggingface gives mathemetically grounded explanation of how transformer encoder-decoder models work
3. [The Illustrated GPT-2](https://jalammar.github.io/illustrated-gpt2/) one of the great blogposts by Jay Alammar visualizing generative language modelling on exaple of GPT-2
4. [minGPT](https://github.com/karpathy/minGPT) cool repo by A. Karpathy providing clear minimal implementation of GPT model
There exist multiple attention mechanisms. The particular one used in the original transformer paper is Scaled Dot Product attention.
Given query vector for particular token we will compare it with a key vector for each token in a sequence and decide how much value vectors of those will effect resulting representetion of the token of interest. One way to view this from a linguistic prospective is: a key is a question each word respondes to, value is information that word represent and a query is related to what every word was looking to combine with.
Mathemetically we can compute attention for all _q_, _k_, _v_ in a matrix form:
$$\textbf {Attention}(Q,K,V) = \textbf {softmax}({QK^T\over\sqrt d_k})V $$
Note that dot product $QK^T$ results in matrix of shape (seq_len x seq_len). Then it is devided by $ \sqrt d_k$ to compensate the fact, that longer sequences will have larger dot product. $ \textbf{softmax}$ is applied to rescale the attention matrix to be betwin 0 and 1. When multiplied by $V$ it produces a matrix of the same shape as $V$ (seq_len x dv).
So where those _q_, _k_, _v_ come from. Well that's fairly straitforward queries are culculated from the embeddings of tokens we want to find representation for by simple linear projection. Keys and values are calculated from the embeddings of context tokens. In case of self attention all of them come from the original sequence.
```
class SelfAttention(Module):
def __init__(self, d_in, d_qk, d_v=None):
d_v = ifnone(d_v, d_qk)
self.iq = nn.Linear(d_in, d_qk)
self.ik = nn.Linear(d_in, d_qk)
self.iv = nn.Linear(d_in, d_v)
self.out = nn.Linear(d_v, d_in)
self.scale = d_qk**-0.5
def forward(self, x):
q, k, v = self.iq(x), self.ik(x), self.iv(x)
q *= self.scale
return self.out(F.softmax([email protected](-2,-1), -1)@v)
```
Even though self attention mechanism is extremely useful it posseses limited expressive power. Essentially we are computing weighted some of the input modified by single affine transformation, shared across the whole sequence. To add more computational power to the model we can introduce fully connected feedforward network on top of the SelfAttention layer.
Curious reader can find detailed formal analysis of the roles of SelfAttention and FeedForward layers in transformer architecture in [this paper](https://arxiv.org/pdf/1912.10077.pdf) by C. Yun et al.
In brief the authors state that SelfAttention layers compute precise contextual maps and FeedForward layers then assign the results of these contextual maps to the desired output values.
```
class FeedForward(Module):
def __init__(self, d_in, d_ff):
self.lin1 = nn.Linear(d_in, d_ff)
self.lin2 = nn.Linear(d_ff, d_in)
self.act = nn.ReLU()
def forward(self, x):
out = self.lin2(self.act(self.lin1(x)))
return out
```
The output would be of shape (bs, seq_len, d) which then may be mapped to (bs, seq_len, vocab_sz) using linear layer. But we have only one target. To adress this issue we can simply do average pooling over seq_len dimention.
The resulting model is fairly simple:
```
class Model1(Module):
def __init__(self, vocab_sz, d_model, d_qk, d_ff):
self.emb = Embedding(vocab_sz, d_model)
self.attn = SelfAttention(d_model, d_qk)
self.ff = FeedForward(d_model, d_ff)
self.out = nn.Linear(d_model, vocab_sz)
def forward(self, x):
x = self.emb(x)
x = self.ff(self.attn(x))
x = x.mean(1)
return self.out(x)
model = Model1(len(vocab), 64, 64, 128)
out = model(x)
out.shape
learn = Learner(dls, model, loss_func=CrossEntropyLossFlat(), metrics=accuracy)
learn.lr_find()
learn.fit_one_cycle(5, 5e-3)
```
To evaluete the model performance we need to compare it to some baseline. Let's see what would be the accuracy if of the model which would always predict most common token.
```
n,counts = 0,torch.zeros(len(vocab))
for x,y in dls.valid:
n += y.shape[0]
for i in range_of(vocab): counts[i] += (y==i).long().sum()
idx = torch.argmax(counts)
idx, vocab[idx.item()], counts[idx].item()/n
```
As you can see, always predicting "thousand" which turn out to be the most common token in the dataset would result in ~15% accuracy. Our simple transformer does much better then that. It feels promising, so let's try to improve the architecture and check if we can get better results.
### Multihead attention
A structured sequence may comprise multiple distinctive kinds of relationships. Our model is forced to learn only one way in which queries, keys and values are constructed from the original token embedding. To remove this limitation we can modify attention layer include multiple heads which would correspond to extracting different kinds of relationships between tokens. The MultiHeadAttention layer consits of several heads each of those is similar to SelfAttention layer we made before. To keep computational cost of the multi-head layer we set $d_k = d_v = d_{model}/n_h$, where $n_h$ is number of heads.
```
class SelfAttention(Module):
def __init__(self, d_in, d_qk, d_v=None):
d_v = ifnone(d_v, d_qk)
self.iq = nn.Linear(d_in, d_qk)
self.ik = nn.Linear(d_in, d_qk)
self.iv = nn.Linear(d_in, d_v)
self.scale = d_qk**-0.5
def forward(self, x):
q, k, v = self.iq(x), self.ik(x), self.iv(x)
return F.softmax([email protected](-2,-1)*self.scale, -1)@v
class MultiHeadAttention(Module):
def __init__(self, d_model, n_heads, d_qk=None, d_v=None):
d_qk = ifnone(d_qk, d_model//n_heads)
d_v = ifnone(d_v, d_qk)
self.heads = nn.ModuleList([SelfAttention(d_model, d_qk) for _ in range(n_heads)])
self.out = nn.Linear(d_v*n_heads, d_model)
def forward(self, x):
out = [m(x) for m in self.heads]
return self.out(torch.cat(out, -1))
inp = torch.randn(8, 10, 64)
mha = MultiHeadAttention(64, 8)
out = mha(inp)
out.shape
class Model2(Module):
def __init__(self, vocab_sz, d_model=64, n_heads=4, d_ff=64*4):
self.emb = nn.Embedding(vocab_sz, d_model)
self.attn = MultiHeadAttention(d_model, n_heads)
self.ff = FeedForward(d_model, d_ff)
self.out = nn.Linear(d_model, vocab_sz)
def forward(self, x):
x = self.emb(x)
x = self.ff(self.attn(x))
x = x.mean(1)
return self.out(x)
learn = Learner(dls, Model2(len(vocab)), loss_func=CrossEntropyLossFlat(), metrics=accuracy)
learn.fit_one_cycle(5, 5e-4)
```
### MultiHead Attention Refactor
Python `for` loops are slow, therefore it is better to refactor the MultiHeadAttention module to compute Q, K, V for all heads in batch.
```
class MultiHeadAttention(Module):
def __init__(self, d_model, n_heads):
assert d_model%n_heads == 0
self.n_heads = n_heads
#d_qk, d_v = d_model//n_heads, d_model//n_heads
self.iq = nn.Linear(d_model, d_model, bias=False)
self.ik = nn.Linear(d_model, d_model, bias=False)
self.iv = nn.Linear(d_model, d_model, bias=False)
self.out = nn.Linear(d_model, d_model, bias=False)
self.scale = d_model//n_heads
def forward(self, x):
bs, seq_len, d = x.size()
# (bs,sl,d) -> (bs,sl,nh,dh) -> (bs,nh,sl,dh)
q = self.iq(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
k = self.ik(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
v = self.iv(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
q*= self.scale
att = F.softmax([email protected](-2,-1), -1)
out = att @ v # (bs, nh, sl, sl) x (bs, nh, sl, dh) -> (bs, nh, sl, dh)
out = out.transpose(1, 2).contiguous().view(bs, seq_len, d) # back to original shape
return self.out(out)
learn = Learner(dls, Model2(len(vocab)), loss_func=CrossEntropyLossFlat(), metrics=accuracy)
learn.fit_one_cycle(5, 1e-3)
```
Note that some speedup is observed even on such a tiny dataset and small model.
## More signal
Similarly to the RNN case considered in the book, we can take the next step and create more signal for the model to learn from. To adapt to the modified objective we need to make couple of steps. First let's rearrange data to proper input-target pairs for the new task.
### Arranging data
Unlike RNN the tranformer is not a stateful model. This means it treats each sequence indepently and can only attend within fixed length context. This limitation was addressed by authors of [Transformer-XL paper](https://arxiv.org/abs/1901.02860) where adding a segment-level recurrence mechanism and a novel positional encoding scheme were proposed to enable capturing long-term dependencies. I will not go into details of TransformerXL architecture here. As we shell see stateless transformer can also learn a lot about the structure of our data.
One thing to note in this case is that we don't need to maintain the structure of the data outside of the sequences, so we can shuffle the sequences randomly in the dataloader.
```
sl = 16
seqs = L((tensor(nums[i:i+sl]), tensor(nums[i+1:i+sl+1]))
for i in range(0,len(nums)-sl-1,sl))
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(seqs[:cut], seqs[cut:],
bs=bs, drop_last=True, shuffle=True)
xb, yb = dls.one_batch()
xb.shape, yb.shape
[L(vocab[o] for o in s) for s in seqs[0]]
```
### Positional encoding
Before we did average pooling over seq_len dimension. Our model didn't care about the order of the tokens at all. But actually order of the tokens in a sentence matter a lot. In our case `one hundred two` and `two hundred one` are pretty different and `hundred one two` doesn't make sense.
To encorporate positional information into the model authors of the transformer architecture proposed to use positional encodings in addition to regular token embeddings. Positional encodings may be learned, but it's also possible to use hardcoded encodings. For instance encodings may be composed of sin and cos.
In this way each position in a sequence will get unique vector associated with it.
```
class PositionalEncoding(Module):
def __init__(self, d):
self.register_buffer('freq', 1/(10000 ** (torch.arange(0., d, 2.)/d)))
self.scale = d**0.5
def forward(self, x):
device = x.device
pos_enc = torch.cat([torch.sin(torch.outer(torch.arange(x.size(1), device=device), self.freq)),
torch.cos(torch.outer(torch.arange(x.size(1), device=device), self.freq))],
axis=-1)
return x*self.scale + pos_enc
#collapse-hide
x = torch.zeros(1, 16, 64)
encs = PositionalEncoding(64)(x)
plt.matshow(encs.squeeze())
plt.xlabel('Embedding size')
plt.ylabel('Sequence length')
plt.show()
class TransformerEmbedding(Module):
def __init__(self, emb_sz, d_model):
self.emb = nn.Embedding(emb_sz, d_model)
self.pos_enc = PositionalEncoding(d_model)
def forward(self, x):
return self.pos_enc(self.emb(x))
class Model3(Module):
def __init__(self, vocab_sz, d_model=64, n_heads=4, d_ff=64*4):
self.emb = TransformerEmbedding(vocab_sz, d_model)
self.attn = MultiHeadAttention(d_model, n_heads)
self.ff = FeedForward(d_model, d_ff)
self.out = nn.Linear(d_model, vocab_sz)
def forward(self, x):
x = self.emb(x)
x = self.ff(self.attn(x))
return self.out(x)
model = Model3(len(vocab))
out = model(xb)
out.shape
def loss_func(inp, targ):
return F.cross_entropy(inp.view(-1, len(vocab)), targ.view(-1))
learn = Learner(dls, Model3(len(vocab)), loss_func=loss_func, metrics=accuracy)
learn.fit_one_cycle(5, 1e-2)
```
Wow! That's a great accuracy! So the problem is solved and we only needed one attention layer and 2 layer deep feed-forward block? Don't you feel somewhat skeptical about this result?
Well, you should be! Think about what we did here: the goal was to predict a target sequence, say `['.','two','.','three','.','four']` from an input `['one','.','two','.','three','.']`. These two sequences intersect on all positions except the first and the last one. So models needs to learn simply to copy input tokens starting from the second one to the outputs. In our case this will result in 15 correct predictions of total 16 positions, that's almost 94% accuracy. This makes the task very simple but not very useful to learn. To train proper autoregressive language model, as we did with RNNs, a concept of masking is to be introduced.
### Causal Masking
So we want to allow the model for each token to attend only to itself and those prior to it. To acomplish this we can set all the values of attention matrix above the main diagonal to $-\infty$. After softmax this values will effectively turn to 0 thus disabling attention to the "future".
```
def get_subsequent_mask(x):
sz = x.size(1)
mask = (torch.triu(torch.ones(sz, sz, device=x.device)) == 1).transpose(0, 1)
mask = mask.float().masked_fill(mask == 0, float('-inf')).masked_fill(mask == 1, float(0.0))
return mask
inp = torch.randn(8, 10, 64)
mask = get_subsequent_mask(inp)
plt.matshow(mask);
q, k = torch.rand(1,10,32), torch.randn(1,10,32)
att_ = F.softmax(([email protected](0,2,1)+mask), -1)
plt.matshow(att_[0].detach());
```
We should also modify the attention layer to accept mask:
```
class MultiHeadAttention(Module):
def __init__(self, d_model, n_heads):
assert d_model%n_heads == 0
self.n_heads = n_heads
d_qk, d_v = d_model//n_heads, d_model//n_heads
self.iq = nn.Linear(d_model, d_model, bias=False)
self.ik = nn.Linear(d_model, d_model, bias=False)
self.iv = nn.Linear(d_model, d_model, bias=False)
self.scale = d_qk**-0.5
self.out = nn.Linear(d_model, d_model, bias=False)
def forward(self, x, mask=None):
bs, seq_len, d = x.size()
mask = ifnone(mask, 0)
q = self.iq(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
k = self.ik(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
v = self.iv(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
q*= self.scale
att = F.softmax([email protected](-2,-1) + mask, -1)
out = att @ v # (bs, nh, sl, sl) x (bs, nh, sl, dh) -> (bs, nh, sl, dh)
out = out.transpose(1, 2).contiguous().view(bs, seq_len, d) # back to original shape
return self.out(out)
class Model4(Module):
def __init__(self, vocab_sz, d_model=64, n_heads=8, d_ff=64*4):
self.emb = TransformerEmbedding(vocab_sz, d_model)
self.attn = MultiHeadAttention(d_model, n_heads)
self.ff = FeedForward(d_model, d_ff)
self.out = nn.Linear(d_model, vocab_sz)
def forward(self, x):
x = self.emb(x)
mask = get_subsequent_mask(x)
x = self.ff(self.attn(x, mask))
return self.out(x)
learn = Learner(dls, Model4(len(vocab)), loss_func=loss_func, metrics=accuracy)
learn.fit_one_cycle(5, 3e-3)
```
Now we get somewhat lower accuracy, which is expected given that the task has become more difficult. Also training loss is significantly lower than validation loss, which means the model is overfitting. Let's see if the same approaches as was applied to RNNs can help.
### Multilayer transformer
To solve a more difficult task we ussualy need a deeper model. For convenience let's make a TransformerLayer which will combine self-attention and feed-forward blocks.
```
class TransformerLayer(Module):
def __init__(self, d_model, n_heads=8, d_ff=None, causal=True):
d_ff = ifnone(d_ff, 4*d_model)
self.attn = MultiHeadAttention(d_model, n_heads)
self.ff = FeedForward(d_model, d_ff)
self.causal = causal
def forward(self, x, mask=None):
if self.causal:
mask = get_subsequent_mask(x)
return self.ff(self.attn(x, mask))
class Model5(Module):
def __init__(self, vocab_sz, d_model=64, n_layer=4, n_heads=8):
self.emb = TransformerEmbedding(vocab_sz, d_model)
self.encoder = nn.Sequential(*[TransformerLayer(d_model, n_heads) for _ in range(n_layer)])
self.out = nn.Linear(d_model, vocab_sz)
def forward(self, x):
x = self.emb(x)
x = self.encoder(x)
return self.out(x)
learn = Learner(dls, Model5(len(vocab), n_layer=4), loss_func=loss_func, metrics=accuracy)
learn.fit_one_cycle(5, 1e-2)
```
That's not good! 4 layer deep Transformer strugles to learn anything. But there are good news, this problem has been already resolved in the original transformer.
### Residual connections and Regularization
If you are familiar with ResNets the proposed solution will not surprise you much. The idea is simple yet very effective. Instead of returning modified output $f(x)$ each transformer sublayer will return $x + f(x)$. This allows the original input to propagate freely through the model. So the model learns not an entirely new representation of $x$ but how to modify $x$ to add some useful information to the original representation.
As we modify layers to include the residual connections let's also add some regularization by inserting Dropout layers.
```
class TransformerEmbedding(Module):
def __init__(self, emb_sz, d_model, p=0.1):
self.emb = Embedding(emb_sz, d_model)
nn.init.trunc_normal_(self.emb.weight, std=d_model**-0.5)
self.pos_enc = PositionalEncoding(d_model)
self.drop = nn.Dropout(p)
def forward(self, x):
return self.drop(self.pos_enc(self.emb(x)))
```
Another modification is to add layer normalization which is intended to improve learning dynamics of the network by reparametrising data statistics and is generally used in transformer based architectures.
```
class FeedForward(Module):
def __init__(self, d_model, d_ff, p=0.2):
self.lin1 = nn.Linear(d_model, d_ff)
self.lin2 = nn.Linear(d_ff, d_model)
self.act = nn.ReLU()
self.norm = nn.LayerNorm(d_model)
self.drop = nn.Dropout(p)
def forward(self, x):
x = self.norm(x)
out = self.act(self.lin1(x))
out = self.lin2(out)
return x + self.drop(out)
class MultiHeadAttention(Module):
def __init__(self, d_model, n_heads, p=0.1):
assert d_model%n_heads == 0
self.n_heads = n_heads
d_qk, d_v = d_model//n_heads, d_model//n_heads
self.iq = nn.Linear(d_model, d_model, bias=False)
self.ik = nn.Linear(d_model, d_model, bias=False)
self.iv = nn.Linear(d_model, d_model, bias=False)
self.scale = d_qk**0.5
self.out = nn.Linear(d_model, d_model, bias=False)
self.norm = nn.LayerNorm(d_model)
self.drop = nn.Dropout(p)
def forward(self, x, mask=None):
bs, seq_len, d = x.size()
mask = ifnone(mask, 0)
x = self.norm(x)
k = self.ik(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
q = self.iq(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
v = self.iv(x).view(bs, seq_len, self.n_heads, d//self.n_heads).transpose(1, 2)
att = F.softmax([email protected](-2,-1)/self.scale + mask, -1)
out = att @ v # (bs, nh, sl, sl) x (bs, nh, sl, dh) -> (bs, nh, sl, dh)
out = out.transpose(1, 2).contiguous().view(bs, seq_len, d) # back to original shape
return x + self.drop(self.out(out))
class TransformerLayer(Module):
def __init__(self, d_model, n_heads=8, d_ff=None, causal=True,
p_att=0.1, p_ff=0.1):
d_ff = ifnone(d_ff, 4*d_model)
self.attn = MultiHeadAttention(d_model, n_heads)
self.ff = FeedForward(d_model, d_ff, p=p_ff)
self.causal = causal
self._init()
def forward(self, x, mask=None):
if self.causal:
mask = get_subsequent_mask(x)
return self.ff(self.attn(x, mask))
def _init(self):
for p in self.parameters():
if p.dim()>1: nn.init.xavier_uniform_(p)
class Model6(Module):
def __init__(self, vocab_sz, d_model=64, n_layer=4, n_heads=8,
p_emb=0.1, p_att=0.1, p_ff=0.2, tie_weights=True):
self.emb = TransformerEmbedding(vocab_sz, d_model, p=p_emb)
self.encoder = nn.Sequential(*[TransformerLayer(d_model, n_heads,
p_att=p_att, p_ff=p_ff)
for _ in range(n_layer)],
nn.LayerNorm(d_model))
self.out = nn.Linear(d_model, vocab_sz)
if tie_weights: self.out.weight = self.emb.emb.weight
def forward(self, x):
x = self.emb(x)
x = self.encoder(x)
return self.out(x)
learn = Learner(dls, Model6(len(vocab), n_layer=2), loss_func=loss_func, metrics=accuracy)
learn.fit_one_cycle(8, 1e-2)
```
## Bonus - Generation example
```
#hide
from google.colab import drive
drive.mount('/content/drive')
path = Path('/content/drive/MyDrive/char_model')
```
Learning to predict numbers is great, but let's try something more entertaining. We can train a language model to generate texts. For example let's try to generate some text in style of Lewis Carroll. For this we'll fit a language model on "Alice in Wonderland" and "Through the looking glass".
```
#collapse-hide
def parse_txt(fns):
txts = []
for fn in fns:
with open(fn) as f:
tmp = ''
for line in f.readlines():
line = line.strip('\n')
if line:
tmp += ' ' + line
elif tmp:
txts.append(tmp.strip())
tmp = ''
return txts
texts = parse_txt([path/'11-0.txt', path/'12-0.txt'])
len(texts)
texts[0:2]
#collapse-hide
class CharTokenizer(Transform):
"Simple charecter level tokenizer"
def __init__(self, vocab=None):
self.vocab = ifnone(vocab, ['', 'xxbos', 'xxeos'] + list(string.printable))
self.c2i = defaultdict(int, [(c,i) for i, c in enumerate(self.vocab)])
def encodes(self, s, add_bos=False, add_eos=False):
strt = [self.c2i['xxbos']] if add_bos else []
end = [self.c2i['xxeos']] if add_eos else []
return LMTensorText(strt + [self.c2i[c] for c in s] + end)
def decodes(self, s, remove_special=False):
return TitledStr(''.join([self.decode_one(i) for i in s]))
def decode_one(self, i):
if i == 2: return '\n'
elif i == 1: return ''
else: return self.vocab[i]
@property
def vocab_sz(self):
return len(self.vocab)
tok = CharTokenizer()
def add_bos_eos(x:list, bos_id=1, eos_id=2):
return [bos_id] + x + [eos_id]
nums = [add_bos_eos(tok(t.lower()).tolist()) for t in texts]
len(nums)
all_nums = []
for n in nums: all_nums.extend(n)
all_nums[:15]
print(tok.decode(all_nums[:100]))
sl = 512
seqs = L((tensor(all_nums[i:i+sl]), tensor(all_nums[i+1:i+sl+1]))
for i in range(0,len(all_nums)-sl-1,sl))
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(seqs[:cut], seqs[cut:], device='cuda',
bs=8, drop_last=True, shuffle=True)
xb, yb = dls.one_batch()
xb.shape, yb.shape
model = Model6(tok.vocab_sz, 512, 6, p_emb=0.1, p_ff=0.1, tie_weights=True)
learn = Learner(dls, model, loss_func=CrossEntropyLossFlat(), metrics=[accuracy, perplexity]).to_native_fp16()
learn.lr_find()
#collapse_output
learn.fit_one_cycle(50, 5e-4, cbs=EarlyStoppingCallback(patience=5))
```
### Text generation
Text generation is a big topic on it's own. One can refer to great posts [by Patrick von Platen from HuggingFace](https://huggingface.co/blog/how-to-generate) and [Lilian Weng](https://lilianweng.github.io/lil-log/2021/01/02/controllable-neural-text-generation.html) for more details on various approaches. Here I will use nucleus sampling. This method rallies on sampling from candidates compounding certain value of probability mass. Intuitively this approach should work for character level generation: when there is only one grammatically correct option for continuation we always want to select it, but when starting a new word some diversity in outputs is desirable.
```
#collapse-hide
def expand_dim1(x):
if len(x.shape) == 1:
return x[None, :]
else: return x
def top_p_filter(logits, top_p=0.9):
sorted_logits, sorted_indices = torch.sort(logits, descending=True)
cum_probs = torch.cumsum(F.softmax(sorted_logits, dim=-1), dim=-1)
sorted_indices_to_remove = cum_probs > top_p
sorted_indices_to_remove[..., 1:] = sorted_indices_to_remove[..., :-1].clone()
sorted_indices_to_remove[..., 0] = 0
indices_to_remove = sorted_indices_to_remove.scatter(1, sorted_indices, sorted_indices_to_remove)
logits[indices_to_remove] = float('-inf')
return logits
@torch.no_grad()
def generate(model, inp,
max_len=50,
temperature=1.,
top_k = 20,
top_p = 0.9,
early_stopping=False, #need eos_idx to work
eos_idx=None):
model.to(inp.device)
model.eval()
thresh = top_p
inp = expand_dim1(inp)
b, t = inp.shape
out = inp
for _ in range(max_len):
x = out
logits = model(x)[:, -1, :]
filtered_logits = top_p_filter(logits)
probs = F.softmax(filtered_logits / temperature, dim=-1)
sample = torch.multinomial(probs, 1)
out = torch.cat((out, sample), dim=-1)
if early_stopping and (sample == eos_idx).all():
break
return out
out = generate(learn.model, tok('Alice said '), max_len=200, early_stopping=True, eos_idx=tok.c2i['xxeos'])
print(tok.decode(out[0]))
```
Our relatively simple model learned to generate mostly grammatically plausible text, but it's not entirely coherent. But it would be too much to ask from the model to learn language from scratch by "reading" only two novels (however great those novels are). To get more from the model let's feed it larger corpus of data.
### Pretraining on larger dataset
```
#hide
import sys
if 'google.colab' in sys.modules:
!pip install -Uqq datasets
from datasets import load_dataset
```
For this purpose I will use a sample from [bookcorpus dataset](https://huggingface.co/datasets/bookcorpus).
```
#hide_ouput
dataset = load_dataset("bookcorpus", split='train')
df = pd.DataFrame(dataset[:10_000_000])
df.head()
df['len'] = df['text'].str.len()
cut = int(len(df)*0.8)
splits = range_of(df)[:cut], range_of(df[cut:])
tfms = Pipeline([ColReader('text'), tok])
dsets = Datasets(df, tfms=tfms, dl_type=LMDataLoader, splits=splits)
#collapse
@patch
def create_item(self:LMDataLoader, seq):
if seq>=self.n: raise IndexError
sl = self.last_len if seq//self.bs==self.n_batches-1 else self.seq_len
st = (seq%self.bs)*self.bl + (seq//self.bs)*self.seq_len
txt = self.chunks[st : st+sl+1]
return LMTensorText(txt[:-1]),txt[1:]
%%time
dl_kwargs = [{'lens':df['len'].values[splits[0]]}, {'val_lens':df['len'].values[splits[1]]}]
dls = dsets.dataloaders(bs=32, seq_len=512, dl_kwargs=dl_kwargs, shuffle_train=True, num_workers=2)
dls.show_batch(max_n=2)
model = Model6(tok.vocab_sz, 512, 8, p_emb=0.1, p_ff=0.1, tie_weights=True)
learn = Learner(dls, model, loss_func=CrossEntropyLossFlat(), metrics=[accuracy, perplexity]).to_native_fp16()
learn.lr_find()
learn = learn.load(path/'char_bookcorpus_10m')
learn.fit_one_cycle(1, 1e-4)
learn.save(path/'char_bookcorpus_10m')
```
### Finetune on Carrolls' books
Finally we can finetune the pretrained bookcorpus model on Carroll's books. This will determine the style of generated text.
```
sl = 512
seqs = L((tensor(all_nums[i:i+sl]), tensor(all_nums[i+1:i+sl+1]))
for i in range(0,len(all_nums)-sl-1,sl))
cut = int(len(seqs) * 0.8)
dls = DataLoaders.from_dsets(seqs[:cut], seqs[cut:], device='cuda',
bs=16, drop_last=True, shuffle=True)
model = Model6(tok.vocab_sz, 512, 8, p_emb=0.1, p_ff=0.1, tie_weights=True)
learn = Learner(dls, model, loss_func=CrossEntropyLossFlat(), metrics=[accuracy, perplexity]).to_native_fp16()
learn = learn.load(path/'char_bookcorpus_10m')
learn.lr_find()
learn.fit_one_cycle(10, 1e-4)
```
As you see pretraining model on large corpus followed by finetuning helped to reduce validation loss from arount 1.53 to 1.037 and improve accuracy in predicting next character to 68% (compared to 56.7% before). Let's see how it effects sampled text:
```
out = generate(learn.model, tok('Alice said '), max_len=200, early_stopping=True, eos_idx=tok.c2i['xxeos'])
#collapse-hide
print(tok.decode(out[0]))
#hide
learn.save(path/'char_alice')
```
|
github_jupyter
|
## <span style="color:purple">ArcGIS API for Python: Real-time Person Detection</span>
<img src="../img/webcam_detection.PNG" style="width: 100%"></img>
## Integrating ArcGIS with TensorFlow Deep Learning using the ArcGIS API for Python
This notebook provides an example of integration between ArcGIS and deep learning frameworks like TensorFlow using the ArcGIS API for Python.
<img src="../img/ArcGIS_ML_Integration.png" style="width: 75%"></img>
We will leverage a model to detect objects on your device's video camera, and use these to update a feature service on a web GIS in real-time. As people are detected on your camera, the feature will be updated to reflect the detection.
### Notebook Requirements:
#### 1. TensorFlow and Object Detection API
This demonstration is designed to run using the TensorFlow Object Detection API (https://github.com/tensorflow/models/tree/master/research/object_detection)
Please follow the instructions found in that repository to install TensorFlow, clone the repository, and test a pre-existing model.
Once you have followed those instructions, this notebook should be placed within the "object_detection" folder of that repository. Alternatively, you may leverage this notebook from another location but reference paths to the TensorFlow model paths and utilities will need to be adjusted.
#### 2. Access to ArcGIS Online or ArcGIS Enterprise
This notebook will make a connection to an ArcGIS Enterprise or ArcGIS Online organization to provide updates to a target feature service.
Please ensure you have access to an ArcGIS Enterprise or ArcGIS Online account with a feature service to serve as the target of your detection updates. The feature service should have a record with an boolean attribute (i.e. column with True or False possible options) named "Person_Found".
# Import needed modules
```
import numpy as np
import os
import six.moves.urllib as urllib
import sys
import tarfile
import tensorflow as tf
import zipfile
from collections import defaultdict
from io import StringIO
from matplotlib import pyplot as plt
from PIL import Image
import cv2
```
We will use VideoCapture to connect to the device's web camera feed. The cv2 module helps here.
```
# Set our caption
cap = cv2.VideoCapture(0)
# This is needed since the notebook is meant to be run in the object_detection folder.
sys.path.append("..")
```
## Object detection imports
Here are the imports from the object detection module.
```
from utils import label_map_util
from utils import visualization_utils as vis_util
```
# Model preparation
## Variables
Any model exported using the `export_inference_graph.py` tool can be loaded here simply by changing `PATH_TO_CKPT` to point to a new .pb file.
By default we use an "SSD with Mobilenet" model here. See the [detection model zoo](https://github.com/tensorflow/models/blob/master/object_detection/g3doc/detection_model_zoo.md) for a list of other models that can be run out-of-the-box with varying speeds and accuracies.
```
# What model to download.
MODEL_NAME = 'ssd_mobilenet_v1_coco_11_06_2017'
MODEL_FILE = MODEL_NAME + '.tar.gz'
DOWNLOAD_BASE = 'http://download.tensorflow.org/models/object_detection/'
# Path to frozen detection graph. This is the actual model that is used for the object detection.
PATH_TO_CKPT = MODEL_NAME + '/frozen_inference_graph.pb'
# List of the strings that is used to add correct label for each box.
PATH_TO_LABELS = os.path.join('data', 'mscoco_label_map.pbtxt')
NUM_CLASSES = 90
```
## Download Model
```
opener = urllib.request.URLopener()
opener.retrieve(DOWNLOAD_BASE + MODEL_FILE, MODEL_FILE)
tar_file = tarfile.open(MODEL_FILE)
for file in tar_file.getmembers():
file_name = os.path.basename(file.name)
if 'frozen_inference_graph.pb' in file_name:
tar_file.extract(file, os.getcwd())
```
## Load a (frozen) Tensorflow model into memory.
```
detection_graph = tf.Graph()
with detection_graph.as_default():
od_graph_def = tf.GraphDef()
with tf.gfile.GFile(PATH_TO_CKPT, 'rb') as fid:
serialized_graph = fid.read()
od_graph_def.ParseFromString(serialized_graph)
tf.import_graph_def(od_graph_def, name='')
```
## Loading label map
Label maps map indices to category names, so that when our convolution network predicts `5`, we know that this corresponds to `airplane`. Here we use internal utility functions, but anything that returns a dictionary mapping integers to appropriate string labels would be fine
```
label_map = label_map_util.load_labelmap(PATH_TO_LABELS)
categories = label_map_util.convert_label_map_to_categories(label_map, max_num_classes=NUM_CLASSES, use_display_name=True)
category_index = label_map_util.create_category_index(categories)
category_index
```
## Helper code
```
def load_image_into_numpy_array(image):
(im_width, im_height) = image.size
return np.array(image.getdata()).reshape(
(im_height, im_width, 3)).astype(np.uint8)
```
This is a helper function that takes the detection graph output tensor (np arrays), stacks the classes and scores, and determines if the class for a person (1) is available within a certain score and within a certain amount of objects
```
def person_in_image(classes_arr, scores_arr, obj_thresh=5, score_thresh=0.5):
stacked_arr = np.stack((classes_arr, scores_arr), axis=-1)
person_found_flag = False
for ix in range(obj_thresh):
if 1.00000000e+00 in stacked_arr[ix]:
if stacked_arr[ix][1] >= score_thresh:
person_found_flag = True
return person_found_flag
```
# Establish Connection to GIS via ArcGIS API for Python
### Authenticate
```
import arcgis
gis_url = "" # Replace with gis URL
username = "" # Replace with username
gis = arcgis.gis.GIS(gis_url, username)
```
### Retrieve the Object Detection Point Layer
```
target_service_name = "" # Replace with name of target service
object_point_srvc = gis.content.search(target_service_name)[0]
object_point_srvc
# Convert our existing service into a pandas dataframe
object_point_lyr = object_point_srvc.layers[0]
obj_fset = object_point_lyr.query() #querying without any conditions returns all the features
obj_df = obj_fset.df
obj_df.head()
all_features = obj_fset.features
all_features
from copy import deepcopy
original_feature = all_features[0]
feature_to_be_updated = deepcopy(original_feature)
feature_to_be_updated
```
### Test of Manual Update
```
feature_to_be_updated.attributes['Person_Found']
features_for_update = []
feature_to_be_updated.attributes['Person_Found'] = "False"
features_for_update.append(feature_to_be_updated)
object_point_lyr.edit_features(updates=features_for_update)
```
# Detection
```
with detection_graph.as_default():
with tf.Session(graph=detection_graph) as sess:
# Definite input and output Tensors for detection_graph
image_tensor = detection_graph.get_tensor_by_name('image_tensor:0')
# Each box represents a part of the image where a particular object was detected.
detection_boxes = detection_graph.get_tensor_by_name('detection_boxes:0')
# Each score represent how level of confidence for each of the objects.
# Score is shown on the result image, together with the class label.
detection_scores = detection_graph.get_tensor_by_name('detection_scores:0')
detection_classes = detection_graph.get_tensor_by_name('detection_classes:0')
num_detections = detection_graph.get_tensor_by_name('num_detections:0')
while True:
ret, image_np = cap.read()
# Expand dimensions since the model expects images to have shape: [1, None, None, 3]
image_np_expanded = np.expand_dims(image_np, axis=0)
# Actual detection.
(boxes, scores, classes, num) = sess.run(
[detection_boxes, detection_scores, detection_classes, num_detections],
feed_dict={image_tensor: image_np_expanded})
# Visualization of the results of a detection.
vis_util.visualize_boxes_and_labels_on_image_array(
image_np,
np.squeeze(boxes),
np.squeeze(classes).astype(np.int32),
np.squeeze(scores),
category_index,
use_normalized_coordinates=True,
line_thickness=8, min_score_thresh=0.5)
cv2.imshow('object detection', cv2.resize(image_np, (800,600)))
if cv2.waitKey(25) & 0xFF == ord('q'):
cv2.destroyAllWindows()
break
person_found = person_in_image(np.squeeze(classes).astype(np.int32),
np.squeeze(scores),
obj_thresh=2)
features_for_update = []
feature_to_be_updated.attributes['Person_Found'] = str(person_found)
features_for_update.append(feature_to_be_updated)
object_point_lyr.edit_features(updates=features_for_update)
```
|
github_jupyter
|
```
import tensorflow as tf
config = tf.compat.v1.ConfigProto(
gpu_options = tf.compat.v1.GPUOptions(per_process_gpu_memory_fraction=0.8),
)
config.gpu_options.allow_growth = True
session = tf.compat.v1.Session(config=config)
tf.compat.v1.keras.backend.set_session(session)
import os
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
import sklearn
from sklearn.model_selection import train_test_split
from pathlib import Path
from keras import backend, layers, activations, Model
from amp.utils.basic_model_serializer import load_master_model_components
from amp.models.decoders import amp_expanded_decoder
from amp.models.encoders import amp_expanded_encoder
from amp.models.master import master
from amp.utils import basic_model_serializer
import amp.data_utils.data_loader as data_loader
from amp.data_utils.sequence import pad, to_one_hot
from tqdm import tqdm
from joblib import dump, load
from sklearn.decomposition import PCA
import seaborn as sns
import matplotlib.pyplot as plt
params = {'axes.labelsize': 16,
'axes.titlesize': 24,
'xtick.labelsize':14,
'ytick.labelsize': 14}
plt.rcParams.update(params)
plt.rc('text', usetex=False)
sns.set_style('whitegrid', {'grid.color': '.95', 'axes.spines.right': False, 'axes.spines.top': False})
sns.set_context("notebook")
seed = 7
np.random.seed(seed)
from amp.config import MIN_LENGTH, MAX_LENGTH, LATENT_DIM, MIN_KL, RCL_WEIGHT, HIDDEN_DIM, MAX_TEMPERATURE
input_to_encoder = layers.Input(shape=(MAX_LENGTH,))
input_to_decoder = layers.Input(shape=(LATENT_DIM+2,))
def translate_generated_peptide(encoded_peptide):
alphabet = list('ACDEFGHIKLMNPQRSTVWY')
return ''.join([alphabet[el - 1] if el != 0 else "" for el in encoded_peptide[0].argmax(axis=1)])
def translate_peptide(encoded_peptide):
alphabet = list('ACDEFGHIKLMNPQRSTVWY')
return ''.join([alphabet[el-1] if el != 0 else "" for el in encoded_peptide])
models = [
'HydrAMP',
'PepCVAE',
'Basic',
]
model_labels = [
'HydrAMP',
'PepCVAE',
'Basic',
]
bms = basic_model_serializer.BasicModelSerializer()
amp_classifier = bms.load_model('../models/amp_classifier')
amp_classifier_model = amp_classifier()
mic_classifier = bms.load_model('../models/mic_classifier/')
mic_classifier_model = mic_classifier()
```
# Get validation data
```
data_manager = data_loader.AMPDataManager(
'../data/unlabelled_positive.csv',
'../data/unlabelled_negative.csv',
min_len=MIN_LENGTH,
max_len=MAX_LENGTH)
amp_x, amp_y = data_manager.get_merged_data()
amp_x_train, amp_x_test, amp_y_train, amp_y_test = train_test_split(amp_x, amp_y, test_size=0.1, random_state=36)
amp_x_train, amp_x_val, amp_y_train, amp_y_val = train_test_split(amp_x_train, amp_y_train, test_size=0.2, random_state=36)
# Restrict the length
ecoli_df = pd.read_csv('../data/mic_data.csv')
mask = (ecoli_df['sequence'].str.len() <= MAX_LENGTH) & (ecoli_df['sequence'].str.len() >= MIN_LENGTH)
ecoli_df = ecoli_df.loc[mask]
mic_x = pad(to_one_hot(ecoli_df['sequence']))
mic_y = ecoli_df.value
mic_x_train, mic_x_test, mic_y_train, mic_y_test = train_test_split(mic_x, mic_y, test_size=0.1, random_state=36)
mic_x_train, mic_x_val, mic_y_train, mic_y_val = train_test_split(mic_x_train, mic_y_train, test_size=0.2, random_state=36)
pos = np.vstack([amp_x_val[amp_y_val == 1], mic_x_val[mic_y_val < 1.5]])
neg = np.vstack([amp_x_val[amp_y_val == 0], mic_x_val[mic_y_val > 1.5]])
neg.shape, pos.shape
pos_amp = amp_classifier_model.predict(pos, verbose=1).reshape(len(pos))
neg_mic = mic_classifier_model.predict(neg, verbose=1).reshape(len(neg))
neg_amp = amp_classifier_model.predict(neg, verbose=1).reshape(len(neg))
pos_mic = mic_classifier_model.predict(pos, verbose=1).reshape(len(pos))
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, figsize=(12, 4), sharex=True, sharey=True)
ax1.hist(pos_amp)
ax1.set_ylabel('AMP')
ax1.set_title('Positives')
ax2.hist(neg_amp)
ax2.set_title('Negatives')
ax3.hist(pos_mic)
ax3.set_ylabel('MIC')
ax4.hist(neg_mic)
plt.show()
pos = np.vstack([pos] * 64).reshape(-1, 25)
pos_amp = np.vstack([pos_amp] * 64).reshape(-1, 1)
pos_mic = np.vstack([pos_mic] * 64).reshape(-1, 1)
neg = np.vstack([neg] * 64).reshape(-1, 25)
neg_amp = np.vstack([neg_amp] * 64).reshape(-1, 1)
neg_mic = np.vstack([neg_mic] * 64).reshape(-1, 1)
def improve(x, model, epoch, mode):
if mode == 'pos':
amp = pos_amp
mic = pos_mic
else:
amp = neg_mic
mic = neg_mic
encoded = encoder_model.predict(x, batch_size=5000)
conditioned = np.hstack([
encoded,
np.ones((len(x), 1)),
np.ones((len(x), 1)),
])
decoded = decoder_model.predict(conditioned, batch_size=5000)
new_peptides = np.argmax(decoded, axis=2)
new_amp = amp_classifier_model.predict(new_peptides, batch_size=5000)
new_mic = mic_classifier_model.predict(new_peptides, batch_size=5000)
# RELATIVE
rel_better = new_amp > amp.reshape(-1, 1)
rel_better = rel_better & (new_mic > mic.reshape(-1, 1))
rel_better = np.logical_or.reduce(rel_better, axis=1)
rel_improved = new_peptides[np.where(rel_better), :].reshape(-1, 25)
before_rel_improve = x[np.where(rel_better), :].reshape(-1, 25)
# ABSOLUTE
abs_better = new_amp >= 0.8
abs_better = abs_better & (new_mic > 0.5)
abs_better = np.logical_or.reduce(abs_better, axis=1)
abs_improved = new_peptides[np.where(abs_better), :].reshape(-1, 25)
before_abs_improve = x[np.where(abs_better), :].reshape(-1, 25)
return {
'new_peptides': new_peptides,
'rel_improved': rel_improved,
'abs_improved': abs_improved,
'before_rel_improve': before_rel_improve,
'before_abs_improve': before_abs_improve,
'new_amp': new_amp,
'new_mic': new_mic,
}
```
# HydrAMP improve
```
from keras.models import Model
model = models[0]
current_model_pos = {epoch: [] for epoch in range(40)}
current_model_neg = {epoch: [] for epoch in range(40)}
for epoch in tqdm(range(40)):
AMPMaster = bms.load_model(f'../models/{model}/{epoch}')
encoder_model = AMPMaster.encoder(input_to_encoder)
decoder_model = AMPMaster.decoder(input_to_decoder)
current_model_pos[epoch] = improve(pos, model, epoch, 'pos')
current_model_neg[epoch] = improve(neg, model, epoch, 'neg')
dump(current_model_pos, f'../results/improvement_PosVal_{model}.joblib')
dump(current_model_neg, f'../results/improvement_NegVal_{model}.joblib')
```
# PepCVAE improve
```
from keras.models import Model
model = models[1]
current_model_pos = {epoch: [] for epoch in range(40)}
current_model_neg = {epoch: [] for epoch in range(40)}
for epoch in tqdm(range(40)):
AMPMaster = bms.load_model(f'../models/{model}/{epoch}')
encoder_model = AMPMaster.encoder(input_to_encoder)
decoder_model = AMPMaster.decoder(input_to_decoder)
new_act = layers.TimeDistributed(
layers.Activation(activations.softmax),
name='decoder_time_distribute_activation')
decoder_model.layers.pop()
x = new_act(decoder_model.layers[-1].output)
decoder_model = Model(input=decoder_model.input, output=[x])
current_model_pos[epoch] = improve(pos, model, epoch, 'pos')
current_model_neg[epoch] = improve(neg, model, epoch, 'neg')
dump(current_model_pos, f'../results/improvement_PosVal_{model}.joblib')
dump(current_model_neg, f'../results/improvement_NegVal_{model}.joblib')
```
# Basic improvement
```
from keras.models import Model
model = models[2]
current_model_pos = {epoch: [] for epoch in range(40)}
current_model_neg = {epoch: [] for epoch in range(40)}
for epoch in tqdm(range(40)):
AMPMaster = bms.load_model(f'../models/{model}/{epoch}')
encoder_model = AMPMaster.encoder(input_to_encoder)
decoder_model = AMPMaster.decoder(input_to_decoder)
new_act = layers.TimeDistributed(
layers.Activation(activations.softmax),
name='decoder_time_distribute_activation')
decoder_model.layers.pop()
x = new_act(decoder_model.layers[-1].output)
decoder_model = Model(input=decoder_model.input, output=[x])
current_model_pos[epoch] = improve(pos, model, epoch, 'pos')
current_model_neg[epoch] = improve(neg, model, epoch, 'neg')
dump(current_model_pos, f'../results/improvement_PosVal_{model}.joblib')
dump(current_model_neg, f'../results/improvement_NegVal_{model}.joblib')
```
# Collect results
```
pos_final_results = {model: {epoch:
{'absolute improvement':0,
'relative improvement':0,
} for epoch in range(40)} for model in models}
neg_final_results = {model: {epoch:
{'absolute improvement':0,
'relative improvement':0,
} for epoch in range(40)} for model in models}
for model in models:
if model in ['PepCVAE', 'Basic']:
model_results = load(f'../results/improvement_PosVal_{model}.joblib')
else:
model_results = load(f'../results/improvement_PosVal_{model}.joblib')
for epoch in range(40):
pos_final_results[model][epoch]['relative improvement'] = np.unique(
model_results[epoch]['rel_improved'], axis=0).shape[0]
pos_final_results[model][epoch]['absolute improvement'] = np.unique(
model_results[epoch]['abs_improved'], axis=0).shape[0]
pos_final_results[model][epoch]['before relative improvement'] = np.unique(
model_results[epoch]['before_rel_improve'], axis=0).shape[0]
pos_final_results[model][epoch]['before absolute improvement'] = np.unique(
model_results[epoch]['before_abs_improve'], axis=0).shape[0]
for model in models:
if model in ['PepCVAE', 'Basic']:
model_results = load(f'../results/improvement_NegVal_{model}.joblib')
else:
model_results = load(f'../results/improvement_NegVal_{model}.joblib')
for epoch in range(40):
neg_final_results[model][epoch]['relative improvement'] = np.unique(
model_results[epoch]['rel_improved'], axis=0).shape[0]
neg_final_results[model][epoch]['absolute improvement'] = np.unique(
model_results[epoch]['abs_improved'], axis=0).shape[0]
neg_final_results[model][epoch]['before relative improvement'] = np.unique(
model_results[epoch]['before_rel_improve'], axis=0).shape[0]
neg_final_results[model][epoch]['before absolute improvement'] = np.unique(
model_results[epoch]['before_abs_improve'], axis=0).shape[0]
hydra_metrics = pd.read_csv('../models/HydrAMP/metrics.csv')
pepcvae_metrics = pd.read_csv('../models/PepCVAE/metrics.csv')
basic_metrics = pd.read_csv('../models/Basic/metrics.csv')
plt.title('Relative improved')
plt.plot([pos_final_results[models[0]][epoch]['relative improvement'] for epoch in range(10, 40)], c='red', label='HydrAMP')
plt.plot([pos_final_results[models[1]][epoch]['relative improvement'] for epoch in range(10, 40)], c='orange', label='PepCVAE')
plt.plot([pos_final_results[models[2]][epoch]['relative improvement'] for epoch in range(10, 40)], c='blue', label='Basic')
plt.legend(bbox_to_anchor=(1.1, 0.5))
plt.show()
plt.title('Absolute improved')
plt.plot([pos_final_results[models[0]][epoch]['absolute improvement'] for epoch in range(10, 40)], c='red', label='HydrAMP')
plt.plot([pos_final_results[models[1]][epoch]['absolute improvement'] for epoch in range(10, 40)], c='orange', label='PepCVAE')
plt.plot([pos_final_results[models[2]][epoch]['absolute improvement'] for epoch in range(10, 40)], c='blue', label='Basic')
plt.legend(bbox_to_anchor=(1.1, 0.5))
plt.show()
plt.figure(figsize=(10,5))
plt.plot([float(x) for x in hydra_metrics['val_vae_loss_1__amino_acc'].tolist()[10:40]],
c='red', label='HydrAMP', linestyle='--')
plt.plot([float(x) for x in pepcvae_metrics['val_vae_loss_1__amino_acc'].tolist()[10:40]],
c='orange', label='PepCVAE', linestyle='--')
plt.plot([float(x) for x in basic_metrics['val_vae_loss_1__amino_acc'].tolist()[10:40]],
c='blue', label='Basic', linestyle='--')
plt.title('How many petides were susceptible to (relative) improvement out of 2404 known AMPs? ')
plt.plot([pos_final_results[models[0]][epoch]['before relative improvement']/2404 for epoch in range(10, 40)], c='red')
plt.plot([pos_final_results[models[1]][epoch]['before relative improvement']/2404 for epoch in range(10, 40)], c='orange')
plt.plot([pos_final_results[models[2]][epoch]['before relative improvement']/2404 for epoch in range(10, 40)], c='blue')
plt.legend(bbox_to_anchor=(1.1, 0.5))
plt.show()
plt.figure(figsize=(10,5))
plt.plot([float(x) for x in hydra_metrics['val_vae_loss_1__amino_acc'].tolist()[10:40]],
c='red', label='HydrAMP', linestyle='--')
plt.plot([float(x) for x in pepcvae_metrics['val_vae_loss_1__amino_acc'].tolist()[10:40]],
c='orange', label='PepCVAE', linestyle='--')
plt.plot([float(x) for x in basic_metrics['val_vae_loss_1__amino_acc'].tolist()[10:40]],
c='blue', label='Basic', linestyle='--')
plt.title('How many petides were susceptible to (absolute) improvement out of 2404 known AMPs? ')
plt.plot([pos_final_results[models[0]][epoch]['before absolute improvement']/2404 for epoch in range(10, 40)], c='red')
plt.plot([pos_final_results[models[1]][epoch]['before absolute improvement']/2404 for epoch in range(10, 40)], c='orange')
plt.plot([pos_final_results[models[2]][epoch]['before absolute improvement']/2404 for epoch in range(10, 40)], c='blue')
plt.legend(bbox_to_anchor=(1.1, 0.5))
plt.show()
```
# Model selection
```
def choose_best_epoch(model):
model_metrics = pd.read_csv(f'../models/{model}/metrics.csv')
good_epochs = model_metrics.iloc[10:40][model_metrics['val_vae_loss_1__amino_acc'].astype(float) > 0.95].epoch_no.tolist()
improved_peptides = [pos_final_results[model][epoch]['before relative improvement']/2404 + \
neg_final_results[model][epoch]['before absolute improvement']/2223 \
for epoch in good_epochs]
return good_epochs[np.argmax(improved_peptides)], np.max(improved_peptides)
best_epochs = {model: [] for model in models}
for model in models:
best_epochs[model] = choose_best_epoch(model)
best_epochs
ax = sns.barplot(
x=model_labels,
y=[
pos_final_results[model][int(best_epochs[model][0])]['before relative improvement']/2404 + \
neg_final_results[model][int(best_epochs[model][0])]['before absolute improvement']/2223 \
for model in models
]
)
ax.set_title('VALIDATION SET\n % of relatively improved positives + % of absolutely improved negatives')
ax.set_xticklabels(model_labels, rotation=90)
plt.show()
metrics_to_consider = [
'before relative improvement',
'before absolute improvement',
'relative improvement',
'absolute improvement',
]
metrics_labels = [
'How many petides were susceptible to (relative) improvement?',
'How many petides were susceptible to (absolute) improvement?',
'Number of uniquely generated peptides during relative improvement procedure (64 attempts per peptide)',
'Number of uniquely generated peptides during absolute improvement procedure (64 attempts per peptide)',
]
for metric, metric_label in zip(metrics_to_consider, metrics_labels):
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(12, 4), sharex=True)
plt.suptitle(metric_label, y=1.1)
sns.barplot(x=model_labels, y=[pos_final_results[model][int(best_epochs[model][0])][metric] for model in models], ax=ax1)
sns.barplot(x=model_labels, y=[neg_final_results[model][int(best_epochs[model][0])][metric] for model in models], ax=ax2)
ax1.set_title('2404 positives (validation set)')
ax2.set_title('2223 negatives (validation set)')
ax1.set_xticklabels(model_labels, rotation=90)
ax2.set_xticklabels(model_labels, rotation=90)
plt.show()
```
# Test set
```
best_epochs = {
'HydrAMP': 37,
'PepCVAE': 35,
'Basic': 15,
}
pos = np.vstack([amp_x_test[amp_y_test == 1], mic_x_test[mic_y_test < 1.5]])
neg = np.vstack([amp_x_test[amp_y_test == 0], mic_x_test[mic_y_test > 1.5]])
print(pos.shape, neg.shape)
pos_amp = amp_classifier_model.predict(pos, verbose=1).reshape(len(pos))
neg_mic = mic_classifier_model.predict(neg, verbose=1).reshape(len(neg))
neg_amp = amp_classifier_model.predict(neg, verbose=1).reshape(len(neg))
pos_mic = amp_classifier_model.predict(pos, verbose=1).reshape(len(pos))
pos = np.vstack([pos] * 64).reshape(-1, 25)
pos_amp = np.vstack([pos_amp] * 64).reshape(-1, 1)
pos_mic = np.vstack([pos_mic] * 64).reshape(-1, 1)
neg = np.vstack([neg] * 64).reshape(-1, 25)
neg_amp = np.vstack([neg_amp] * 64).reshape(-1, 1)
neg_mic = np.vstack([neg_mic] * 64).reshape(-1, 1)
final_pos_results = {}
final_neg_results = {}
for model in tqdm(models):
epoch = int(best_epochs[model])
AMPMaster = bms.load_model(f'../models/{model}/{epoch}')
encoder_model = AMPMaster.encoder(input_to_encoder)
decoder_model = AMPMaster.decoder(input_to_decoder)
if model in ['PepCVAE', 'Basic']:
new_act = layers.TimeDistributed(
layers.Activation(activations.softmax),
name='decoder_time_distribute_activation')
decoder_model.layers.pop()
x = new_act(decoder_model.layers[-1].output)
decoder_model = Model(input=decoder_model.input, output=[x])
final_pos_results[model] = improve(pos, model, epoch, 'pos')
final_neg_results[model] = improve(neg, model, epoch, 'neg')
dump(final_pos_results, f'../results/improvement_PosTest.joblib')
dump(final_neg_results, f'../results/improvement_NegTest.joblib')
pos_final_results = {models: {} for models in models}
neg_final_results = {models: {} for models in models}
for model in models:
pos_final_results[model]['relative improvement'] = np.unique(
final_pos_results[model]['rel_improved'], axis=0).shape[0]
pos_final_results[model]['absolute improvement'] = np.unique(
final_pos_results[model]['abs_improved'], axis=0).shape[0]
pos_final_results[model]['before relative improvement'] = np.unique(
final_pos_results[model]['before_rel_improve'], axis=0).shape[0]
pos_final_results[model]['before absolute improvement'] = np.unique(
final_pos_results[model]['before_abs_improve'], axis=0).shape[0]
neg_final_results[model]['relative improvement'] = np.unique(
final_neg_results[model]['rel_improved'], axis=0).shape[0]
neg_final_results[model]['absolute improvement'] = np.unique(
final_neg_results[model]['abs_improved'], axis=0).shape[0]
neg_final_results[model]['before relative improvement'] = np.unique(
final_neg_results[model]['before_rel_improve'], axis=0).shape[0]
neg_final_results[model]['before absolute improvement'] = np.unique(
final_neg_results[model]['before_abs_improve'], axis=0).shape[0]
ax = sns.barplot(
x=model_labels,
y=[
pos_final_results[model]['before relative improvement']/1319 + \
neg_final_results[model]['before absolute improvement']/1253 \
for model in models])
ax.set_title('Before relative improvement (positives) + before absolute improvement (negatives)')
ax.set_xticklabels(model_labels, rotation=90)
plt.show()
for metric, metric_label in zip(metrics_to_consider, metrics_labels):
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(12, 4), sharex=True)
plt.suptitle(metric_label, y=1.1)
sns.barplot(x=model_labels, y=[pos_final_results[model][metric] for model in models], ax=ax1)
sns.barplot(x=model_labels, y=[neg_final_results[model][metric] for model in models], ax=ax2)
ax1.set_title('1319 positives (test set)')
ax2.set_title('1253 negatives (test set)')
ax1.set_xticklabels(model_labels, rotation=90)
ax2.set_xticklabels(model_labels, rotation=90)
plt.show()
```
|
github_jupyter
|
# Notebook to be used to Develop Display of Results
```
from importlib import reload
import pandas as pd
import numpy as np
from IPython.display import Markdown
# If one of the modules changes and you need to reimport it,
# execute this cell again.
import heatpump.hp_model
reload(heatpump.hp_model)
import heatpump.home_heat_model
reload(heatpump.home_heat_model)
import heatpump.library as lib
reload(lib)
# Anchorage large home inputs
util = lib.util_from_id(1)
inputs1 = dict(
city_id=1,
utility=util,
pce_limit=500.0,
co2_lbs_per_kwh=1.1,
exist_heat_fuel_id=2,
exist_unit_fuel_cost=0.97852,
exist_fuel_use=1600,
exist_heat_effic=.8,
exist_kwh_per_mmbtu=8,
includes_dhw=True,
includes_dryer=True,
includes_cooking=False,
occupant_count=3,
elec_use_jan=550,
elec_use_may=400,
hp_model_id=575,
low_temp_cutoff=5,
garage_stall_count=2,
garage_heated_by_hp=False,
bldg_floor_area=3600,
indoor_heat_setpoint=70,
insul_level=3,
pct_exposed_to_hp=0.46,
doors_open_to_adjacent=False,
bedroom_temp_tolerance=2,
capital_cost=4500,
rebate_dol=500,
pct_financed=0.5,
loan_term=10,
loan_interest=0.05,
hp_life=14,
op_cost_chg=10,
sales_tax=0.02,
discount_rate=0.05,
inflation_rate=0.02,
fuel_esc_rate=0.03,
elec_esc_rate=0.02,
)
# Ambler Home inputs
util = lib.util_from_id(202)
inputs2 = dict(
city_id=45,
utility=util,
pce_limit=500.0,
co2_lbs_per_kwh=1.6,
exist_heat_fuel_id=4,
exist_unit_fuel_cost=8.0,
exist_fuel_use=450,
exist_heat_effic=.86,
exist_kwh_per_mmbtu=8,
includes_dhw=False,
includes_dryer=False,
includes_cooking=False,
occupant_count=3,
elec_use_jan=550,
elec_use_may=300,
hp_model_id=575,
low_temp_cutoff=5,
garage_stall_count=0,
garage_heated_by_hp=False,
bldg_floor_area=800,
indoor_heat_setpoint=70,
insul_level=2,
pct_exposed_to_hp=1.0,
doors_open_to_adjacent=False,
bedroom_temp_tolerance=3,
capital_cost=6500,
rebate_dol=0,
pct_financed=0.0,
loan_term=10,
loan_interest=0.05,
hp_life=14,
op_cost_chg=0,
sales_tax=0.00,
discount_rate=0.05,
inflation_rate=0.02,
fuel_esc_rate=0.03,
elec_esc_rate=0.02,
)
inputs2
# Change from **inputs1 to **inputs2 to run the two cases.
mod = heatpump.hp_model.HP_model(**inputs2)
mod.run()
# Pull out the results from the model object.
# Use these variable names in your display of outputs.
smy = mod.summary
df_cash_flow = mod.df_cash_flow
df_mo_en_base = mod.df_mo_en_base
df_mo_en_hp = mod.df_mo_en_hp
df_mo_dol_base = mod.df_mo_dol_base
df_mo_dol_hp = mod.df_mo_dol_hp
# This is a dictionary containing summary output.
# The 'fuel_use_xxx' values are annual totals in physical units
# like gallons. 'elec_use_xxx' are kWh. 'hp_max_capacity' is the
# maximum output of the heat pump at 5 deg F. 'max_hp_reached'
# indicates whether the heat pump ever used all of its capacity
# at some point during the year.
smy
md = f"Design Heat Load: **{smy['design_heat_load']:,.0f} Btu/hour** at {smy['design_heat_temp']:.0f} degrees F outdoors"
md
# You can get a string that is in Markdown format rendered properly
# by using the Markdown class.
Markdown(md)
# Or, this might be a case where f-strings are not the cleanest.
# Here is another way:
md = 'Design Heat Load of Entire Building: **{design_heat_load:,.0f} Btu/hour** at {design_heat_temp:.0f} degrees F outdoors \n(required output of heating system, no safety margin)'.format(**smy)
Markdown(md)
# Cash Flow over the life of the heat pump.
# Negative values are costs and positive values are benefits.
# When displaying this table delete the two columns that don't apply,
# depending on whether you are showing the PCE or no PCE case.
df_cash_flow
# The Base case and w/ Heat Pump monthly energy results.
df_mo_en_base
df_mo_en_hp
# The monthly dollar flows with and without the heat pump
# The PCE and no PCE case are included in this one table
df_mo_dol_base
df_mo_dol_hp
list(df_mo_en_base.columns.values)
import plotly
plotly.tools.set_credentials_file(username='dustin_cchrc', api_key='yzYaFYf93PQ7D0VUZKGy')
import plotly.plotly as py
import plotly.graph_objs as go
```
## Monthly Heating Load
```
data = [go.Bar(x=df_mo_en_base.index,
y=df_mo_en_base.secondary_load_mmbtu,
name='Monthly Heating Load')]
layout = go.Layout(title='Monthly Heating Load',
xaxis=dict(title='Month'),
yaxis=dict(title='Total Estimated Heat Load (MMBTU)', hoverformat='.1f')
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='estimated_heat_load', fileopt='overwrite')
```
## Heating Cost Comparison
```
df_mo_dol_chg = df_mo_dol_hp - df_mo_dol_base
df_mo_dol_chg['cost_savings'] = np.where(
df_mo_dol_chg.total_dol < 0.0,
-df_mo_dol_chg.total_dol,
0.0
)
# Note: we make these negative values so bars extend downwards
df_mo_dol_chg['cost_increases'] = np.where(
df_mo_dol_chg.total_dol >= 0.0,
-df_mo_dol_chg.total_dol,
0.0
)
df_mo_dol_chg
# calculate the change in dollars between the base scenario and the heat
# pump scenario.
hp_cost = go.Bar(
x=df_mo_dol_hp.index,
y=df_mo_dol_hp.total_dol,
name='',
marker=dict(color='#377eb8'),
hoverinfo = 'y',
)
cost_savings = go.Bar(
x=df_mo_dol_chg.index,
y=df_mo_dol_chg.cost_savings,
name='Cost Savings',
marker=dict(color='#4daf4a'),
hoverinfo = 'y',
)
cost_increases = go.Bar(
x=df_mo_dol_chg.index,
y=df_mo_dol_chg.cost_increases,
name='Cost Increases',
marker=dict(color='#e41a1c'),
hoverinfo = 'y',
)
no_hp_costs = go.Scatter(
x=df_mo_dol_base.index,
y=df_mo_dol_base.total_dol,
name='Baseline Energy Costs',
mode='markers',
marker=dict(color='#000000', size=12),
hoverinfo = 'y',
)
data = [hp_cost, cost_savings, cost_increases, no_hp_costs]
layout = go.Layout(
title='Energy Costs: Heat Pump vs. Baseline',
xaxis=dict(title='Month', fixedrange=True,),
yaxis=dict(
title='Total Energy Costs',
hoverformat='$,.0f',
fixedrange=True,
tickformat='$,.0f',
),
barmode='stack',
hovermode= 'closest',
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='heatpump_costs', fileopt='overwrite')
```
## Monthly Heat Pump Efficiency
```
efficiency = [go.Scatter(x=df_mo_en_hp.index,
y=df_mo_en_hp.cop,
name='COP',
mode='lines+markers')]
layout = go.Layout(title='Monthly Heat Pump Efficiency',
xaxis=dict(title='Month'),
yaxis=dict(title='COP'))
fig = go.Figure(data=efficiency, layout=layout)
py.iplot(fig, layout=layout, filename='cop', fileopt='overwrite')
```
## Energy Use Comparison
```
list(df_mo_en_base.columns.values)
list(df_mo_dol_base.columns.values)
list(df_mo_dol_hp.columns.values)
from plotly import tools
elec_no_hp = go.Scatter(x=df_mo_dol_base.index,
y=df_mo_dol_base.elec_kwh,
name='Monthly kWh (no Heat Pump)',
line=dict(color='#92c5de',
width=2,
dash='dash')
)
elec_w_hp = go.Scatter(x=df_mo_dol_hp.index,
y=df_mo_dol_hp.elec_kwh,
name='Monthly kWh (with Heat Pump)',
mode='lines',
marker=dict(color='#0571b0')
)
fuel_no_hp = go.Scatter(x=df_mo_dol_base.index,
y=df_mo_dol_base.secondary_fuel_units,
name='Monthly Fuel Usage (no Heat Pump)',
line=dict(color='#f4a582',
width = 2,
dash = 'dash')
)
fuel_w_hp = go.Scatter(x=df_mo_dol_hp.index,
y=df_mo_dol_hp.secondary_fuel_units,
name='Monthly Fuel Usage (with Heat Pump)',
mode='lines',
marker=dict(color='#ca0020'))
fig = tools.make_subplots(rows=2, cols=1)
fig.append_trace(elec_no_hp, 1, 1)
fig.append_trace(elec_w_hp, 1, 1)
fig.append_trace(fuel_no_hp, 2, 1)
fig.append_trace(fuel_w_hp, 2, 1)
fig['layout'].update(title='Energy Usage: Heat Pump vs. Baseline')
fig['layout']['xaxis1'].update(title='Month')
fig['layout']['xaxis2'].update(title='Month')
fig['layout']['yaxis1'].update(title='Electricity Use (kWh)', hoverformat='.0f')
yaxis2_title = 'Heating Fuel Use (%s)' % (smy['fuel_unit'])
fig['layout']['yaxis2'].update(title=yaxis2_title, hoverformat='.1f')
py.iplot(fig, filename='heatpump_energy_usage', fileopt='overwrite')
```
## Cash Flow Visualization
```
df_cash_flow
df_cash_flow['negative_flow'] = np.where(df_cash_flow.cash_flow < 0, df_cash_flow.cash_flow, 0)
df_cash_flow['positive_flow'] = np.where(df_cash_flow.cash_flow > 0, df_cash_flow.cash_flow, 0)
negative_flow = go.Bar(x=df_cash_flow.index,
y=df_cash_flow.negative_flow,
name='Cash Flow',
marker=dict(color='#d7191c'))
positive_flow = go.Bar(x=df_cash_flow.index,
y=df_cash_flow.positive_flow,
name='Cash Flow',
marker=dict(color='#000000'))
data = [negative_flow, positive_flow]
layout = go.Layout(title='Heat Pump Cash Flow',
xaxis=dict(title='Year'),
yaxis=dict(title='Annual Cash Flow ($)', hoverformat='dol,.0f')
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='cash_flow', fileopt='overwrite')
```
## Cumulative Discounted Cash Flow
```
df_cash_flow
df_cash_flow['cum_negative_flow'] = np.where(df_cash_flow.cum_disc_cash_flow < 0, df_cash_flow.cum_disc_cash_flow, 0)
df_cash_flow['cum_positive_flow'] = np.where(df_cash_flow.cum_disc_cash_flow > 0, df_cash_flow.cum_disc_cash_flow, 0)
negative_cash_flow = go.Scatter(x=df_cash_flow.index,
y=df_cash_flow.cum_negative_flow,
name='Cash Flow ($)',
fill='tozeroy',
fillcolor='#d7191c',
line=dict(color='#ffffff')
)
positive_cash_flow = go.Scatter(x=df_cash_flow.index,
y=df_cash_flow.cum_positive_flow,
name='Cash Flow ($)',
fill='tozeroy',
fillcolor='#000000',
line=dict(color='#ffffff')
)
data = [negative_cash_flow, positive_cash_flow]
layout = go.Layout(title='Heat Pump Lifetime Cumulative Discounted Cash Flow',
xaxis=dict(title='Year'),
yaxis=dict(title='Annual Discounted Cash Flow ($)', hoverformat='.0f'),
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='cumulative_discounted_heatpump_cash_flow', fileopt='overwrite')
```
## Markdown Display of Results
```
## Need to account for NaN internal rate of return (hide somehow)
```
### With PCE
```
md_results = '''# Results
## Heat Pump Cost Effectiveness
### Net Present Value: **\${:,.0f}**
The Net Present Value of installing an air-source heat pump is estimated to be **\${:,.0f}**.
This means that over the course of the life of the equipment you will {} **\${:,.0f}** in today's dollars.
### Internal Rate of Return: **{:.1f}%**
The internal rate of return on the investment is estimated to be **{:.1f}%**. Compare this tax-free investment to your other investment options.
### Cash Flow
This is how your cash flow will be affected by installing a heat pump:
## Greenhouse Gas Emissions
Installing a heat pump is predicted to save {:,.0f} pounds of CO2 emissions annually, or {:,.0f} pounds over the life of the equipment.
This is equivalent to a reduction of {:,.0f} miles driven by an average passenger vehicle annually, or {:,.0f} over the equipment's life.
'''
def npv_indicator(summary, pce_indicator):
if pce_indicator == 1:
if summary['npv'] > 0:
return 'earn'
else:
return 'lose'
else:
if summary['npv_no_pce'] > 0:
return 'earn'
else:
return 'lose'
smy
smy['npv']
md = md_results.format(smy['npv'],
smy['npv'],
npv_indicator(smy, 1),
abs(smy['npv']),
smy['irr']*100,
smy['irr']*100,
smy['co2_lbs_saved'],
smy['co2_lbs_saved'] * 12,
smy['co2_driving_miles_saved'],
smy['co2_driving_miles_saved'] * 12)
Markdown(md)
from textwrap import dedent
inputs = {'hp_life': 14}
sumd = smy.copy()
sumd['npv_abs'] = abs(sumd['npv'])
sumd['irr'] *= 100. # convert to %
sumd['npv_indicator'] = 'earn' if sumd['npv'] >= 0 else 'lose'
sumd['co2_lbs_saved_life'] = sumd['co2_lbs_saved'] * inputs['hp_life']
sumd['co2_driving_miles_saved_life'] = sumd['co2_driving_miles_saved'] * inputs['hp_life']
md_tmpl = dedent('''
# Results
## Heat Pump Cost Effectiveness
### Net Present Value: **\${npv:,.0f}**
The Net Present Value of installing an air-source heat pump is estimated to
be **\${npv:,.0f}**. This means that over the course of the life of the equipment you
will {npv_indicator} **\${npv_abs:,.0f}** in today's dollars.
### Internal Rate of Return: **{irr:.1f}%**
The internal rate of return on the investment is estimated to be **{irr:.1f}%**.
Compare this tax-free investment to your other investment options.
### Cash Flow
This is how your cash flow will be affected by installing a heat pump:
## Greenhouse Gas Emissions
Installing a heat pump is predicted to save {co2_lbs_saved:,.0f} pounds of CO2 emissions annually,
or {co2_lbs_saved_life:,.0f} pounds over the life of the equipment. This is equivalent to a reduction
of {co2_driving_miles_saved:,.0f} miles driven by an average passenger vehicle annually,
or {co2_driving_miles_saved_life:,.0f} miles over the equipment's life.
''')
md = md_tmpl.format(**sumd)
Markdown(md)
sumd
```
|
github_jupyter
|
# Module 5 -- Dimensionality Reduction -- Case Study
# Import Libraries
**Import the usual libraries **
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
%matplotlib inline
```
# Data Set : Cancer Data Set
Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image. n the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server: ftp ftp.cs.wisc.edu cd math-prog/cpo-dataset/machine-learn/WDBC/
Also can be found on UCI Machine Learning Repository: https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29
Attribute Information:
1) ID number 2) Diagnosis (M = malignant, B = benign) 3-32)
Ten real-valued features are computed for each cell nucleus:
a) radius (mean of distances from center to points on the perimeter) b) texture (standard deviation of gray-scale values) c) perimeter d) area e) smoothness (local variation in radius lengths) f) compactness (perimeter^2 / area - 1.0) g) concavity (severity of concave portions of the contour) h) concave points (number of concave portions of the contour) i) symmetry j) fractal dimension ("coastline approximation" - 1)
The mean, standard error and "worst" or largest (mean of the three largest values) of these features were computed for each image, resulting in 30 features. For instance, field 3 is Mean Radius, field 13 is Radius SE, field 23 is Worst Radius.
All feature values are recoded with four significant digits.
Missing attribute values: none
Class distribution: 357 benign, 212 malignant
## Get the Data
** Use pandas to read data as a dataframe called df.**
```
df = pd.read_csv('breast-cancer-data.csv')
df.head()
# Check the data , there should be no missing values
df.info()
feature_names = np.array(['mean radius' 'mean texture' 'mean perimeter' 'mean area'
'mean smoothness' 'mean compactness' 'mean concavity'
'mean concave points' 'mean symmetry' 'mean fractal dimension'
'radius error' 'texture error' 'perimeter error' 'area error'
'smoothness error' 'compactness error' 'concavity error'
'concave points error' 'symmetry error' 'fractal dimension error'
'worst radius' 'worst texture' 'worst perimeter' 'worst area'
'worst smoothness' 'worst compactness' 'worst concavity'
'worst concave points' 'worst symmetry' 'worst fractal dimension'])
```
#### Convert diagnosis column to 1/0 and store in new column target
```
from sklearn.preprocessing import LabelEncoder
# # Encode label diagnosis
# # M -> 1
# # B -> 0
# Get All rows, but only last column
target_data=df["diagnosis"]
encoder = LabelEncoder()
target_data = encoder.fit_transform(target_data)
```
#### Store the encoded column in dataframe and drop the diagnosis column for simpilcity
```
df.drop(["diagnosis"],axis = 1, inplace = True)
```
## Principal Component Analysis -- PCA
Lets use PCA to find the first two principal components, and visualize the data in this new, two-dimensional space, with a single scatter-plot
Scale data so that each feature has a single unit variance.
```
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(df)
scaled_data = scaler.transform(df)
```
Now we can transform this data to its first 2 principal components.
```
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
pca.fit(scaled_data)
x_pca = pca.transform(scaled_data)
scaled_data.shape
x_pca.shape
```
#### Reduced 30 dimensions to just 2! Let's plot these two dimensions out!
** Q1. Plot scatter for 2 components. What inference can you draw from this data? **
```
plt.figure(figsize=(9,6))
plt.scatter(x_pca[:,0],x_pca[:,1],c=target_data,cmap='viridis')
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
```
## Interpreting the components
Unfortunately, with this great power of dimensionality reduction, comes the cost of being able to easily understand what these components represent.
The components correspond to combinations of the original features, the components themselves are stored as an attribute of the fitted PCA object:
```
pca.components_
```
# Explained Variance
The explained variance tells you how much information (variance) can be attributed to each of the principal components. This is important as you can convert n dimensional space to 2 dimensional space, you lose some of the variance (information).
** Q2. What is the variance attributed by 1st and 2nd Components? **
** Q3 Ideally the sum above should be 100%. What happened to the remaining variance ? **
```
pca.explained_variance_ratio_
```
## Lets try with 3 Principal Components
```
pca_3 = PCA(n_components=3)
pca_3.fit(scaled_data)
x_pca_3 = pca_3.transform(scaled_data)
```
In this numpy matrix array, each row represents a principal component, and each column relates back to the original features. we can visualize this relationship with a heatmap:
```
x_pca_3.shape
```
** Q4. What is the total variance attributed by three Components? **
```
pca_3.explained_variance_ratio_
```
### Lets check the accuracy for 2 vs. 3 components
** Q5. What is accuracy for component count 2 vs. 3 ?**
```
from sklearn.model_selection import train_test_split
train_data, test_data, train_output, test_output = train_test_split( df, target_data, test_size=0.3, random_state=101)
train_data = scaler.transform(train_data)
test_data = scaler.transform(test_data)
train_data = pca.transform(train_data)
test_data = pca.transform(test_data)
from sklearn.linear_model import LogisticRegression
logisticRegr = LogisticRegression(solver = 'lbfgs')
logisticRegr.fit(train_data, train_output)
logisticRegr.score(test_data, test_output)
```
Score for 3 components
```
train_data, test_data, train_output, test_output = train_test_split( df, target_data, test_size=0.3, random_state=101)
train_data = scaler.transform(train_data)
test_data = scaler.transform(test_data)
train_data = pca_3.transform(train_data)
test_data = pca_3.transform(test_data)
logisticRegr = LogisticRegression(solver = 'lbfgs')
logisticRegr.fit(train_data, train_output)
logisticRegr.score(test_data, test_output)
```
# End of Case Study
|
github_jupyter
|
## CCNSS 2018 Module 5: Whole-Brain Dynamics and Cognition
# Tutorial 2: Introduction to Complex Network Analysis (II)
*Please execute the cell bellow in order to initialize the notebook environment*
```
!rm -rf data ccnss2018_students
!if [ ! -d data ]; then git clone https://github.com/ccnss/ccnss2018_students; \
cp -rf ccnss2018_students/module5/2_introduction_to_complex_network_analysis_2/data ./; \
cp ccnss2018_students/module5/net_tool.py ./; fi
import matplotlib.pyplot as plt # import matplotlib
import numpy as np # import numpy
import math # import basic math functions
import random # import basic random number generator functions
import csv # import CSV(Comma Separated Values) file reading and writing
import scipy as sp # import scipy
from scipy import sparse # import sparse module from scipy
from scipy import signal # import signal module from scipy
import os # import basic os functions
import time # import time to measure real time
import collections # import collections
import networkx as nx # import networkx
import sys
sys.path.append('../')
import net_tool as net # import net_tool, a network analysis toolbox from tutorial #1
data_folder = 'data'
print('Available data files:\n'+'\n'.join(sorted(os.listdir(data_folder))))
data_file_1 = os.path.join(data_folder, 'george_baseline_44.txt')
data_file_2 = os.path.join(data_folder, 'george_propofol.txt')
data_file_3 = os.path.join(data_folder, 'george_ketamin.txt')
data_file_4 = os.path.join(data_folder, 'george_medetomidine.txt')
```
# Objectives
In this notebook we will construct a fuctional network from a given time series. Following up on the powerpoint tutorial, we will first construct a functional network from the brain signals, and compare functional network properties for different states of the brain.
## Background
Network theory (graph theory) measures can be applied to any kind of network, including the brain. Structural networks of various species are good examples. We can also construct fuctional networks from time series data we observe using various techniques such as fMRI, EEG, ECoG, and MEG.
Using an ECoG data from a macaque as an example, We will go through the following steps:
* Appy a measure (PLI: phase lag index) to two time series, and construct a PLI matrix.
* Construct a network from the PLI matrix, by applying a threshold.
* Apply various network measures to the resulting network.
* Construct the functional networks for different brain states, and compare how they differ from each other.
* (Optional) Divide the time series into small time windows, and construct functional network for each time window.
The example we will analyze is a thirty second - segment of whole brain ECoG data of a macaque monkey named George, from an eyes closed resting state. The sampling freqeuncy is 1000 Hz, resulting in total of 30,000 time points for each channel. The data consists of signals coming from 106 areas that cover the left hemisphere. The data is preprocessed, by applying a band path filter to remove the alpha wave component (7-13 Hz) from the signal. Alpha waves are correlated with global interactions of the brain for many instances of the brain states.
```
george_base = [ row for row in csv.reader(open(data_file_1,'r'),delimiter='\t')]
george_base = np.array(george_base).astype(np.float32)
george_propofol = [ row for row in csv.reader(open(data_file_2,'r'),delimiter='\t')]
george_propofol = np.array(george_propofol).astype(np.float32)
```
**EXERCISE 0: Calculating* i)* the phases of oscillating signals, and* ii)* the differences between the phases from two signals. Read through and understand the code, which will be used in later exercises (Exercise #4).
**
$i)$ Every oscillating signal $S_j$ can be represented by its amplitude and its phase:
$$ S_j(t) = r_j(t) e^{i \theta_j(t) } = r_j(t) ( \cos \theta_j(t) + i \ \sin \theta_j(t) ) .\\$$
Using this representation, we could assign $phase$ $\theta_j$ to the signal at every time point $t$. One way of computing the phase of a signal for each time point is using the ***Hilbert transform***.
• We can obtain the signal in the form of above representation by `sp.hilbert`($S_j$). After that, we could use `np.angle()` to get the angle at each time point $t$: `np.angle(sp.hilbert`( $S_j$ ) `).`
$$ $$
$ii)$ After getting the angle $\theta_j$ of each signal $S_j$, we can calculate the differences between phases:
$$ \Delta \theta_{jk}(t) = \theta_j(t) - \theta_k(t) \\$$
Best way to calculate the phase difference, again is to calculate it in the exponent form:
$$ e^{i \Delta \theta_{jk} (t)} = e^{i ( \theta_j (t) - \theta_k (t) ) },\\ $$
then take the angle of $ e^{i \Delta \theta_{jk} (t)} $:
$$ \Delta \theta_{jk} (t) = arg ( e^{i \Delta \theta_{jk} (t)} ) .\\ $$
We can obtain the angle by using `np.angle()`.
This phase difference gives a valuable information about the "directionality" between pair of oscillators.
• Calculate the $\theta_{ij}$ between all pairs of time series, and build a phase-difference matrix. Each elements of the matrix containing time averaged phase difference $\langle \theta_{ij} \rangle _t$ between $i$ and $j$. The resulting matrix will be anti-symmetric.
• From the phase-difference matrix we constructed, compute the average phase-difference for each node. Calculate the row-sum of the matrix:
$$ \theta_i = \frac{1}{N} \sum_{j=1}^{N} \langle \theta_{ij} \rangle _t,$$
then we can have a vector of averaged phase-differences, each element of the vector corresponding for each node.
This average phase-difference for each node will tell us whether one node is phase-leading or phase-lagging with respect to other nodes over a given period of time.
```
# getting the phases from the signals, using np.angle and sp.signal.hilbert
george_base_angle = np.angle(sp.signal.hilbert( george_base,axis=0) )
print("size of george_base_angle is:" , george_base_angle.shape )
def phase_diff_mat(theta):
# theta must has dimension TxN, where T is the length of time points and N is the number of nodes
N_len = theta.shape[1]
PDiff_mat= np.zeros((N_len,N_len))
for ch1 in range(N_len):
for ch2 in range(ch1+1,N_len):
PDiff=theta[:,ch1]-theta[:,ch2] # theta_ch1 - theta_ch2
PDiff_exp_angle = np.angle( np.exp(1j*PDiff) ) # angle of exp (1i * (theta_ch1-theta_ch2) )
PDiff_exp_mean = np.mean(PDiff_exp_angle) # mean of the angle with respect to time
PDiff_mat[ch1,ch2] = PDiff_exp_mean # put the mean into the matrix
PDiff_mat[ch2,ch1] = -1*PDiff_exp_mean # the matrix will be anti-symmetric
PDiff_mean = np.mean(PDiff_mat,axis=1) # calculate the mean for each node, with respect to all the other nodes
#alternative code
#arr = np.array([np.roll(theta, i, axis=1) for i in range(N_len)])
#PDiff_mat = theta[None, :] - arr
#PDiff_mean = PDiff_mat.mean(1)
return PDiff_mean,PDiff_mat
```
**EXERCISE 1: Calculating the PLI for two given time series**
The data is in a form of 30,000x106 (# of time points x # of channels) sized matrix. We will measure $PLI$s between all possible pairs of channels.
We now define $dPLI$ (directed phase-lag index) as the following:
$$ dPLI_{ij} = \frac{1}{T}\sum_{t=1}^{T} sign ( \Delta \theta_{ij} (t) ) \, $$
where
$$ \Delta \theta_{ij} = \theta_i - \theta_j ,$$
and
$$ sign ( \theta_i - \theta_j ) =
\begin{cases}
1 & if \ \Delta \theta_{ij} > 0 \\
0 & if \ \Delta \theta_{ij} = 0 \\
-1 & if \ \Delta \theta_{ij} < 0. \\
\end{cases} \\ $$
$dPLI$ will range from 1 to -1, and give us information about which signal is leading another. \
If we take absolute value of $dPLI$, we get $PLI$ (phase lag index):
$$\\ PLI_{ij} =|dPLI_{ij}| = | \langle sign ( \Delta \theta_{ij} ) \rangle_t | .\\$$
$PLI$ will range from 0 to 1, and give us information about whether two signals have consistent phase-lead/lag relationship with each other over given period of time.
• Plot the time series for the first 3 channels of `george_base` (first 500 time points)
• Plot the time series for the first 3 channels of `george_base_angle` (first 500 time points).
• Compute $PLI_{ij}$ for all pairs of $i$ and $j$, and make $PLI$ matrix. The resulting matrix will be symmetric. You can use `np.sign()`.
```
# Write your code for plotting time series
```
**EXPECTED OUTPUT**

```
def cal_dPLI_PLI(theta):
# insert your code for calculating dPLI and PLI
# theta must has dimension TxN, where T is the length of time points and N is the number of nodes
# outputs PLI matrix containing PLIs between all pairs of channels, and dPLI matrix containg dPLIs between all pairs of channels
return PLI,dPLI
george_base_PLI, george_base_dPLI = cal_dPLI_PLI(george_base_angle)
print(george_base_dPLI[:5,:5])
```
**EXPECTED OUTPUT**
```
[[ 0. -0.09446667 0.0348 -0.05666667 0.28 ]
[ 0.09446667 0. 0.04926667 0.00693333 0.341 ]
[-0.0348 -0.04926667 0. -0.0614 0.2632 ]
[ 0.05666667 -0.00693333 0.0614 0. 0.3316 ]
[-0.28 -0.341 -0.2632 -0.3316 0. ]]
```
**EXERCISE 2: Constructing network connectivity matrix**
We can construct a network from the above PLI matrix. Two approaches are possible. We can apply a threshold value for the PLI matrix and turn it into a binary network. Or, we can take the PLI value as is, and turn the matrix into a weighted network. We will take the first approach.
• Binary network approach: one must determine a right threshold value for the matrix. For example, you can choose a value such that highest 30% of the PLI values between nodes will turn into connection.
• (Optional) Weighted network approach: we can take the PLI value itself as the weighted link between two nodes.
```
def cal_mat_thresholded(data_mat, threshold):
# insert your code here
# input is the original matrix with threshold
# output is the thresholded matrix. It would be symmetric.
return data_mat_binary
threshold = 0.3
george_base_PLI_p3 = cal_mat_thresholded(george_base_PLI,threshold)
print("sum of george_base_PLI_p3:", np.sum(george_base_PLI_p3))
```
**EXPECTED OUTPUT**
```
sum of george_base_PLI_p3: 3372.0
```
**EXERCISE 3: Applying network measure to the functional network**
We now have a resulting functional network from a macaque ECoG data. Now we can apply network measures to this network.
• Apply network measures to this network, such as $C, L, E$ and $b$ (clustering coefficient, characteristic path length, efficiency, and betweenness centrality).
(If you prefer, you can use functions that we provide in net.py. Ask tutors for the details.)
```
# insert your code here
```
**EXPECTED OUTPUT**
```
C: 0.4405029623271814
E and L: 1.735130278526505 0.6451332734351602
b: 38.594339622641506
```
**EXERCISE 4: Computing phase measures for the functional network**
We can define a mean of $PLI_i$ over all other nodes as follows:
$$ PLI_i = \frac{1}{N-1} \sum_{j=1,\ j \neq i }^{N} PLI_{ij} ,$$
This quantity will tell us how persistantly a node is locked with respect to other nodes, over a given period of time. Usually, the node with high $PLI_i$ is the one with high degree in a network: the $k_i$ and $PLI_i$ of a node $i$ is correlated.
We can also define a mean of $dPLI_i$ over all other nodes as follows:
$$ dPLI_i = \frac{1}{N-1} \sum_{j=1,\ j \neq i}^{N} dPLI_{ij} ,$$
This quantity will tell us how persistantly a node is phase-leadaing or phase-lagging with respect to other nodes, over a given period of time. This quantity is correlated with the average phase-difference $\theta_i$ which we defined in earlier exercise.
• Do a scatterplot of the mean PLI and mean dPLI. Is there any pattern between these two quantities? Calculate the Pearson correlation coefficient between these two vectors.
• Also, you can do a scatterplot of degree of each node vs. average phase-difference. Do they resemble above the scatter plot?
```
# insert your code for calculating mean dPLI and PLI, mean phase, and degree of the network
george_base_PLI_mean =
george_base_dPLI_mean =
george_base_phase_diff_mean,_ = phase_diff_mat(george_base_angle)
george_base_PLI_p3_degree =
plt.figure()
for i in range(len(george_base_PLI_mean)):
plt.plot(george_base_PLI_mean[i],george_base_dPLI_mean[i],'C0s')
plt.text(george_base_PLI_mean[i],george_base_dPLI_mean[i],str(i))
plt.xlabel('PLI')
plt.ylabel('dPLI')
plt.title('dPLI vs PLI')
plt.show()
corr_PLI_dPLI = np.corrcoef(george_base_PLI_mean,george_base_dPLI_mean)
print("corr. of PLI and dPLI is:", corr_PLI_dPLI[1,0])
plt.figure()
for i in range(len(george_base_PLI_p3_degree)):
plt.plot(george_base_PLI_p3_degree[i] , george_base_phase_diff_mean[i],'C0s' )
plt.text(george_base_PLI_p3_degree[i] , george_base_phase_diff_mean[i],str(i))
plt.xlabel('k')
plt.ylabel('theta')
plt.title('theta vs k')
plt.show()
corr_degree_phase = np.corrcoef(george_base_PLI_p3_degree , george_base_phase_diff_mean)
print("corr. of degree and phase is:", corr_degree_phase[1,0])
```
**EXPECTED OUTPUT**

```
corr. of PLI and dPLI is: -0.5848065158893657
```

```
corr. of degree and phase is: -0.5082925792988023
```
**EXERCISE 5: Dividing the data into moving time windows (optional)**
Sometimes the time length of the data is large. Or, one wants to investigate the changes that occurs in finer time resolution. For example, we can apply a time window of 2 seconds with an overlap of 1 second to the data, dividing the data into 29 time segments of size 2000x106 matrix.
• Write a code for a function that divide a given time series into moving time windows.
• Using the codes from Exercise 1 and 2, construct a connectivity matrix for each time window.
• We can now apply network measures to the resulting connectivity matrices.
```
win_len = 2000
win_start = 10000
overlap = 1000
PLI_win = []
dPLI_win = []
for idx in range(0, george_base_angle.shape[0], overlap):
temp = cal_dPLI_PLI(george_base_angle[idx:idx+win_len])
PLI_win += [temp[0]]
dPLI_win += [temp[1]]
PLI_win = np.array(PLI_win[:-1])
dPLI_win = np.array(dPLI_win[:-1])
```
**EXERCISE 6: Comparison between two different states of brain (optional, possible for mini projects)**
The above analysis can be repeated to different states of the brain. For example, we can construct the network from anesthesized unconcious states. The provided data is from anesthetized George, induced with propofol. We can construct the connectivity network and apply network measure.
• Repeat the processes in Exercise 1 and 2 to construct the resulting fuctional network.
• Apply network measures as in Exercise 3, and phase measures as in Exercise 4. Compare the result with the resting state network. How are they different from each other?
```
```
**EXERCISE 7: Phase coherence (optional, possible for mini projects)**
There are many measures which can be applied to construct functional connectivity matrix. One measure is phase coherence $(PC)$. Phase coherence $PC$ between two time-series $a$ and $b$ is defined as the following:
$$ PC_{ab} = \lvert {R e^{i \Theta_{ab}}} \rvert = \left| \frac{1}{T} \sum_{t=1}^{T} e^{i \theta_{ab}(t)} \right| , \\ $$
where $\theta_{ab}(t)$ is difference of phases of time-series $a$ and $b$ at time $t$:
$$ \theta_{ab}(t) = \theta_a(t) - \theta_b(t) \\ $$
• Construct a code for a function that computes $PC_{ij}$ for given time-series $i$ and $j$.
• Construct a code for a function that constructs $PC$ matrix which contain $PC_{ij}$ for all possible pairs of time_series.
• Use the codes to construct connectivity matrix as in Exercise 2.
• After the construction, we can proceed to apply the measures as in Exercise 3.
```
```
** EXERCISE 8: Pearson correlation coefficients (optional, possible for mini projects)**
• Another measure which can be used to construct connectivity matrix is Pearson correlation coefficient $c$. Measure *Pearson* correlation coefficients ($c$) between all possible pairs, and contruct a correlation matrix with the coefficients as its element. The resulting matrix will be a symmetric matrix. The pearson correlation coefficient $c_{xy}$ between two data set $x=\{x_1, x_2, x_3, ..., x_n \}$ and $y=\{y_1, y_2, y_3, ..., y_n \}$ is defined as the following:
$$ c_{xy} = \frac { \sum_{i=1}^{n} (x_i - \bar x) (y_i - \bar y) } { \sqrt { \sum_{i=1}^{n} (x_i - \bar x )^2 } \sqrt {\sum_{i=1}^{n} (y_i - \bar y)^2 } } $$
where $\bar x$ and $\bar y$ are the mean of $x$ and $y$.
Alternatively, we can rewrite in the following way:
$$ c_{xy} = \frac { cov(x,y) } { \sqrt { var(x) \ var(y) } } $$
where
$$ cov(x,y) = \langle (x_i - \bar x) (y_i - \bar y) \rangle _i \\
var(x,y) = \langle x_i - \bar x \rangle _i.$$
• You can construct a code for a function that computes $c_{ij}$ for given time-series $i$ and $j$, or you can use a numpy function, `np.corrcoef()`.
• Construct a code for a function that constructs correlation coefficient $c$ matrix which contain $c_{ij}$ for all possible pairs of time series.
• Use the codes to construct connectivity matrix as in Exercise 2.
• After the construction, we can proceed to Exercise 3.
```
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/dlmacedo/starter-academic/blob/master/3The_ultimate_guide_to_Encoder_Decoder_Models_3_4.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
%%capture
!pip install -qq git+https://github.com/huggingface/transformers.git
```
# **Transformer-based Encoder-Decoder Models**
The *transformer-based* encoder-decoder model was introduced by Vaswani et al. in the famous [Attention is all you need paper](https://arxiv.org/abs/1706.03762) and is today the *de-facto* standard encoder-decoder architecture in natural language processing (NLP).
Recently, there has been a lot of research on different *pre-training* objectives for transformer-based encoder-decoder models, *e.g.* T5, Bart, Pegasus, ProphetNet, Marge, *etc*..., but the model architecture has stayed largely the same.
The goal of the blog post is to give an **in-detail** explanation of **how** the transformer-based encoder-decoder architecture models *sequence-to-sequence* problems. We will focus on the mathematical model defined by the architecture and how the model can be used in inference. Along the way, we will give some background on sequence-to-sequence models in NLP and break down the *transformer-based* encoder-decoder architecture into its **encoder** and **decoder** part. We provide many illustrations and establish the link
between the theory of *transformer-based* encoder-decoder models and their practical usage in 🤗Transformers for inference.
Note that this blog post does *not* explain how such models can be trained - this will be the topic of a future blog post.
Transformer-based encoder-decoder models are the result of years of research on *representation learning* and *model architectures*.
This notebook provides a short summary of the history of neural encoder-decoder models. For more context, the reader is advised to read this awesome [blog post](https://ruder.io/a-review-of-the-recent-history-of-nlp/) by Sebastion Ruder. Additionally, a basic understanding of the *self-attention architecture* is recommended.
The following blog post by Jay Alammar serves as a good refresher on the original Transformer model [here](http://jalammar.github.io/illustrated-transformer/).
At the time of writing this notebook, 🤗Transformers comprises the encoder-decoder models *T5*, *Bart*, *MarianMT*, and *Pegasus*, which are summarized in the docs under [model summaries](https://huggingface.co/transformers/model_summary.html#sequence-to-sequence-models).
The notebook is divided into four parts:
- **Background** - *A short history of neural encoder-decoder models is given with a focus on on RNN-based models.* - [click here](https://colab.research.google.com/drive/18ZBlS4tSqSeTzZAVFxfpNDb_SrZfAOMf?usp=sharing)
- **Encoder-Decoder** - *The transformer-based encoder-decoder model is presented and it is explained how the model is used for inference.* - [click here](https://colab.research.google.com/drive/1XpKHijllH11nAEdPcQvkpYHCVnQikm9G?usp=sharing)
- **Encoder** - *The encoder part of the model is explained in detail.*
- **Decoder** - *The decoder part of the model is explained in detail.* - to be published on *Thursday, 08.10.2020*
Each part builds upon the previous part, but can also be read on its own.
## **Encoder**
As mentioned in the previous section, the *transformer-based* encoder maps the input sequence to a contextualized encoding sequence:
$$ f_{\theta_{\text{enc}}}: \mathbf{X}_{1:n} \to \mathbf{\overline{X}}_{1:n}. $$
Taking a closer look at the architecture, the transformer-based encoder is a stack of residual *encoder blocks*.
Each encoder block consists of a **bi-directional** self-attention layer, followed by two feed-forward layers. For simplicity, we disregard the normalization layers in this notebook. Also, we will not further discuss the role of the two feed-forward layers, but simply see it as a final vector-to-vector mapping required in each encoder block ${}^1$.
The bi-directional self-attention layer puts each input vector $\mathbf{x'}_j, \forall j \in \{1, \ldots, n\}$ into relation with all input vectors $\mathbf{x'}_1, \ldots, \mathbf{x'}_n$ and by doing so transforms the input vector $\mathbf{x'}_j$ to a more "refined" contextual representation of itself, defined as $\mathbf{x''}_j$.
Thereby, the first encoder block transforms each input vector of the input sequence $\mathbf{X}_{1:n}$ (shown in light green below) from a *context-independent* vector representation to a *context-dependent* vector representation, and the following encoder blocks further refine this contextual representation until the last encoder block outputs the final contextual encoding $\mathbf{\overline{X}}_{1:n}$ (shown in darker green below).
Let's visualize how the encoder processes the input sequence "I want to buy a car EOS" to a contextualized encoding sequence. Similar to RNN-based encoders, transformer-based encoders also add a special "end-of-sequence" input vector to the input sequence to hint to the model that the input vector sequence is finished ${}^2$.
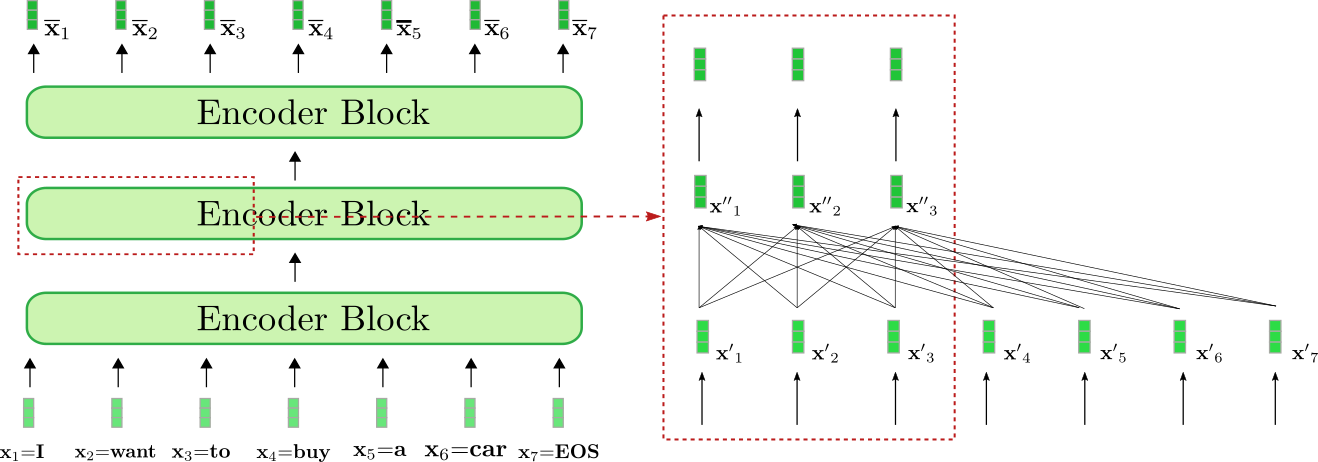
Our exemplary *transformer-based* encoder is composed of three encoder blocks, whereas the second encoder block is shown in more detail in the red box on the right for the first three input vectors $\mathbf{x}_1, \mathbf{x}_2 and \mathbf{x}_3$.
The bi-directional self-attention mechanism is illustrated by the fully-connected graph in the lower part of the red box and the two feed-forward layers are shown in the upper part of the red box. As stated before, we will focus only on the bi-directional self-attention mechanism.
As can be seen each output vector of the self-attention layer $\mathbf{x''}_i, \forall i \in \{1, \ldots, 7\}$ depends *directly* on *all* input vectors $\mathbf{x'}_1, \ldots, \mathbf{x'}_7$. This means, *e.g.* that the input vector representation of the word "want", *i.e.* $\mathbf{x'}_2$, is put into direct relation with the word "buy", *i.e.* $\mathbf{x'}_4$, but also with the word "I",*i.e.* $\mathbf{x'}_1$. The output vector representation of "want", *i.e.* $\mathbf{x''}_2$, thus represents a more refined contextual representation for the word "want".
Let's take a deeper look at how bi-directional self-attention works.
Each input vector $\mathbf{x'}_i$ of an input sequence $\mathbf{X'}_{1:n}$ of an encoder block is projected to a key vector $\mathbf{k}_i$, value vector $\mathbf{v}_i$ and query vector $\mathbf{q}_i$ (shown in orange, blue, and purple respectively below) through three trainable weight matrices $\mathbf{W}_q, \mathbf{W}_v, \mathbf{W}_k$:
$$ \mathbf{q}_i = \mathbf{W}_q \mathbf{x'}_i,$$
$$ \mathbf{v}_i = \mathbf{W}_v \mathbf{x'}_i,$$
$$ \mathbf{k}_i = \mathbf{W}_k \mathbf{x'}_i, $$
$$ \forall i \in \{1, \ldots n \}.$$
Note, that the **same** weight matrices are applied to each input vector $\mathbf{x}_i, \forall i \in \{i, \ldots, n\}$. After projecting each input vector $\mathbf{x}_i$ to a query, key, and value vector, each query vector $\mathbf{q}_j, \forall j \in \{1, \ldots, n\}$ is compared to all key vectors $\mathbf{k}_1, \ldots, \mathbf{k}_n$. The more similar one of the key vectors $\mathbf{k}_1, \ldots \mathbf{k}_n$ is to a query vector $\mathbf{q}_j$, the more important is the corresponding value vector $\mathbf{v}_j$ for the output vector $\mathbf{x''}_j$. More specifically, an output vector $\mathbf{x''}_j$ is defined as the weighted sum of all value vectors $\mathbf{v}_1, \ldots, \mathbf{v}_n$ plus the input vector $\mathbf{x'}_j$. Thereby, the weights are proportional to the cosine similarity between $\mathbf{q}_j$ and the respective key vectors $\mathbf{k}_1, \ldots, \mathbf{k}_n$, which is mathematically expressed by $\textbf{Softmax}(\mathbf{K}_{1:n}^\intercal \mathbf{q}_j)$ as illustrated in the equation below.
For a complete description of the self-attention layer, the reader is advised to take a look at [this](http://jalammar.github.io/illustrated-transformer/) blog post or the original [paper](https://arxiv.org/abs/1706.03762).
Alright, this sounds quite complicated. Let's illustrate the bi-directional self-attention layer for one of the query vectors of our example above. For simplicity, it is assumed that our exemplary *transformer-based* decoder uses only a single attention head `config.num_heads = 1` and that no normalization is applied.
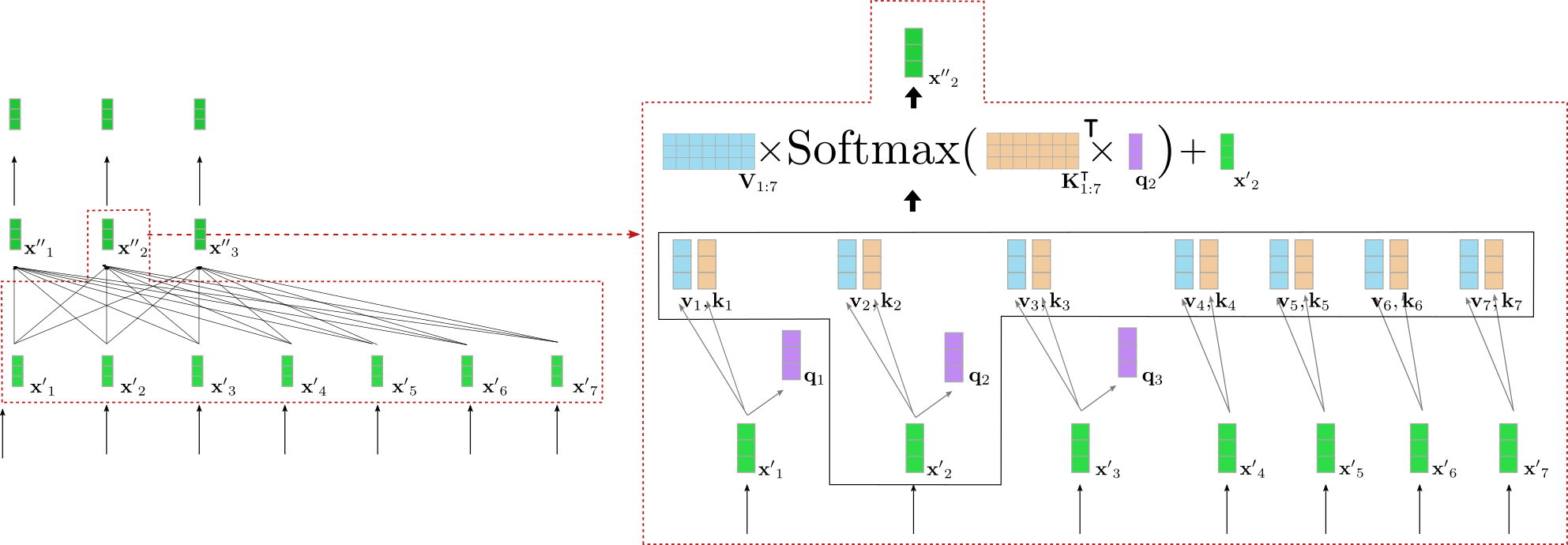
On the left, the previously illustrated second encoder block is shown again and on the right, an in detail visualization of the bi-directional self-attention mechanism is given for the second input vector $\mathbf{x'}_2$ that corresponds to the input word "want".
At first all input vectors $\mathbf{x'}_1, \ldots, \mathbf{x'}_7$ are projected to their respective query vectors $\mathbf{q}_1, \ldots, \mathbf{q}_7$ (only the first three query vectors are shown in purple above), value vectors $\mathbf{v}_1, \ldots, \mathbf{v}_7$ (shown in blue), and key vectors $\mathbf{k}_1, \ldots, \mathbf{k}_7$ (shown in orange). The query vector $\mathbf{q}_2$ is then multiplied by the transpose of all key vectors, *i.e.* $\mathbf{K}_{1:7}^{\intercal}$ followed by the softmax operation to yield the *self-attention weights*. The self-attention weights are finally multiplied by the respective value vectors and the input vector $\mathbf{x'}_2$ is added to output the "refined" representation of the word "want", *i.e.* $\mathbf{x''}_2$ (shown in dark green on the right).
The whole equation is illustrated in the upper part of the box on the right.
The multiplication of $\mathbf{K}_{1:7}^{\intercal}$ and $\mathbf{q}_2$ thereby makes it possible to compare the vector representation of "want" to all other input vector representations "I", "to", "buy", "a", "car", "EOS" so that the self-attention weights mirror the importance each of the other input vector representations $\mathbf{x'}_j \text{, with } j \ne 2$ for the refined representation $\mathbf{x''}_2$ of the word "want".
To further understand the implications of the bi-directional self-attention layer, let's assume the following sentence is processed: "*The house is beautiful and well located in the middle of the city where it is easily accessible by public transport*". The word "it" refers to "house", which is 12 "positions away". In transformer-based encoders, the bi-directional self-attention layer performs a single mathematical operation to put the input vector of "house" into relation with the input vector of "it" (compare to the first illustration of this section). In contrast, in an RNN-based encoder, a word that is 12 "positions away", would require at least 12 mathematical operations meaning that in an RNN-based encoder a linear number of mathematical operations are required. This makes it much harder for an RNN-based encoder to model long-range contextual representations.
Also, it becomes clear that a transformer-based encoder is much less prone to lose important information than an RNN-based encoder-decoder model because the sequence length of the encoding is kept the same, *i.e.* $\textbf{len}(\mathbf{X}_{1:n}) = \textbf{len}(\mathbf{\overline{X}}_{1:n}) = n$, while an RNN compresses the length from $\textbf{len}((\mathbf{X}_{1:n}) = n$ to just $\textbf{len}(\mathbf{c}) = 1$, which makes it very difficult for RNNs to effectively encode long-range dependencies between input words.
In addition to making long-range dependencies more easily learnable, we can see that the Transformer architecture is able to process text in parallel.Mathematically, this can easily be shown by writing the self-attention formula as a product of query, key, and value matrices:
$$\mathbf{X''}_{1:n} = \mathbf{V}_{1:n} \text{Softmax}(\mathbf{Q}_{1:n}^\intercal \mathbf{K}_{1:n}) + \mathbf{X'}_{1:n}. $$
The output $\mathbf{X''}_{1:n} = \mathbf{x''}_1, \ldots, \mathbf{x''}_n$ is computed via a series of matrix multiplications and a softmax operation, which can be parallelized effectively.
Note, that in an RNN-based encoder model, the computation of the hidden state $\mathbf{c}$ has to be done sequentially: Compute hidden state of the first input vector $\mathbf{x}_1$, then compute the hidden state of the second input vector that depends on the hidden state of the first hidden vector, etc. The sequential nature of RNNs prevents effective parallelization and makes them much more inefficient compared to transformer-based encoder models on modern GPU hardware.
Great, now we should have a better understanding of a) how transformer-based encoder models effectively model long-range contextual representations and b) how they efficiently process long sequences of input vectors.
Now, let's code up a short example of the encoder part of our `MarianMT` encoder-decoder models to verify that the explained theory holds in practice.
---
${}^1$ An in-detail explanation of the role the feed-forward layers play in transformer-based models is out-of-scope for this notebook. It is argued in [Yun et. al, (2017)](https://arxiv.org/pdf/1912.10077.pdf) that feed-forward layers are crucial to map each contextual vector $\mathbf{x'}_i$ individually to the desired output space, which the *self-attention* layer does not manage to do on its own. It should be noted here, that each output token $\mathbf{x'}$ is processed by the same feed-forward layer. For more detail, the reader is advised to read the paper.
${}^2$ However, the EOS input vector does not have to be appended to the input sequence, but has been shown to improve performance in many cases. In contrast to the *0th* $\text{BOS}$ target vector of the transformer-based decoder is required as a starting input vector to predict a first target vector.
```
%%capture
from transformers import MarianMTModel, MarianTokenizer
import torch
tokenizer = MarianTokenizer.from_pretrained("Helsinki-NLP/opus-mt-en-de")
model = MarianMTModel.from_pretrained("Helsinki-NLP/opus-mt-en-de")
embeddings = model.get_input_embeddings()
# create ids of encoded input vectors
input_ids = tokenizer("I want to buy a car", return_tensors="pt").input_ids
# pass input_ids to encoder
encoder_hidden_states = model.base_model.encoder(input_ids, return_dict=True).last_hidden_state
# change the input slightly and pass to encoder
input_ids_perturbed = tokenizer("I want to buy a house", return_tensors="pt").input_ids
encoder_hidden_states_perturbed = model.base_model.encoder(input_ids_perturbed, return_dict=True).last_hidden_state
# compare shape and encoding of first vector
print(f"Length of input embeddings {embeddings(input_ids).shape[1]}. Length of encoder_hidden_states {encoder_hidden_states.shape[1]}")
# compare values of word embedding of "I" for input_ids and perturbed input_ids
print("Is encoding for `I` equal to its perturbed version?: ", torch.allclose(encoder_hidden_states[0, 0], encoder_hidden_states_perturbed[0, 0], atol=1e-3))
```
We compare the length of the input word embeddings, *i.e.* `embeddings(input_ids)` corresponding to $\mathbf{X}_{1:n}$, with the length of the `encoder_hidden_states`, corresponding to $\mathbf{\overline{X}}_{1:n}$.
Also, we have forwarded the word sequence "I want to buy a car" and a slightly perturbated version "I want to buy a house" through the encoder to check if the first output encoding, corresponding to "I", differs when only the last word is changed in the input sequence.
As expected the output length of the input word embeddings and encoder output encodings, *i.e.* $\textbf{len}(\mathbf{X}_{1:n})$ and $\textbf{len}(\mathbf{\overline{X}}_{1:n})$, is equal.
Second, it can be noted that the values of the encoded output vector of $\mathbf{\overline{x}}_1 = \text{"I"}$ are different when the last word is changed from "car" to "house". This however should not come as a surprise if one has understood bi-directional self-attention.
On a side-note, *autoencoding* models, such as BERT, have the exact same architecture as *transformer-based* encoder models. *Autoencoding* models leverage this architecture for massive self-supervised pre-training on open-domain text data so that they can map any word sequence to a deep bi-directional representation. In [Devlin et al. (2018)](https://arxiv.org/abs/1810.04805), the authors show that a pre-trained BERT model with a single task-specific classification layer on top can achieve SOTA results on eleven NLP tasks. All *autoencoding* models of 🤗Transformers can be found [here](https://huggingface.co/transformers/model_summary.html#autoencoding-models).
|
github_jupyter
|
# Step1: Create the Python Script
In the cell below, you will need to complete the Python script and run the cell to generate the file using the magic `%%writefile` command. Your main task is to complete the following methods for the `PersonDetect` class:
* `load_model`
* `predict`
* `draw_outputs`
* `preprocess_outputs`
* `preprocess_inputs`
For your reference, here are all the arguments used for the argument parser in the command line:
* `--model`: The file path of the pre-trained IR model, which has been pre-processed using the model optimizer. There is automated support built in this argument to support both FP32 and FP16 models targeting different hardware.
* `--device`: The type of hardware you want to load the model on (CPU, GPU, MYRIAD, HETERO:FPGA,CPU)
* `--video`: The file path of the input video.
* `--output_path`: The location where the output stats and video file with inference needs to be stored (results/[device]).
* `--max_people`: The max number of people in queue before directing a person to another queue.
* `--threshold`: The probability threshold value for the person detection. Optional arg; default value is 0.60.
```
%%writefile person_detect.py
import numpy as np
import time
from openvino.inference_engine import IENetwork, IECore
import os
import cv2
import argparse
import sys
class Queue:
'''
Class for dealing with queues
'''
def __init__(self):
self.queues=[]
def add_queue(self, points):
self.queues.append(points)
def get_queues(self, image):
for q in self.queues:
x_min, y_min, x_max, y_max=q
frame=image[y_min:y_max, x_min:x_max]
yield frame
def check_coords(self, coords):
d={k+1:0 for k in range(len(self.queues))}
for coord in coords:
for i, q in enumerate(self.queues):
if coord[0]>q[0] and coord[2]<q[2]:
d[i+1]+=1
return d
class PersonDetect:
'''
Class for the Person Detection Model.
'''
def __init__(self, model_name, device, threshold=0.60):
self.model_weights=model_name+'.bin'
self.model_structure=model_name+'.xml'
self.device=device
self.threshold=threshold
try:
self.model=IENetwork(self.model_structure, self.model_weights)
except Exception as e:
raise ValueError("Could not Initialise the network. Have you enterred the correct model path?")
self.input_name=next(iter(self.model.inputs))
self.input_shape=self.model.inputs[self.input_name].shape
self.output_name=next(iter(self.model.outputs))
self.output_shape=self.model.outputs[self.output_name].shape
def load_model(self):
'''
TODO: This method needs to be completed by you
'''
raise NotImplementedError
def predict(self, image):
'''
TODO: This method needs to be completed by you
'''
raise NotImplementedError
def draw_outputs(self, coords, image):
'''
TODO: This method needs to be completed by you
'''
raise NotImplementedError
def preprocess_outputs(self, outputs):
'''
TODO: This method needs to be completed by you
'''
raise NotImplementedError
def preprocess_input(self, image):
'''
TODO: This method needs to be completed by you
'''
raise NotImplementedError
def main(args):
model=args.model
device=args.device
video_file=args.video
max_people=args.max_people
threshold=args.threshold
output_path=args.output_path
start_model_load_time=time.time()
pd= PersonDetect(model, device, threshold)
pd.load_model()
total_model_load_time = time.time() - start_model_load_time
queue=Queue()
try:
queue_param=np.load(args.queue_param)
for q in queue_param:
queue.add_queue(q)
except:
print("error loading queue param file")
try:
cap=cv2.VideoCapture(video_file)
except FileNotFoundError:
print("Cannot locate video file: "+ video_file)
except Exception as e:
print("Something else went wrong with the video file: ", e)
initial_w = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
initial_h = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
video_len = int(cap.get(cv2.CAP_PROP_FRAME_COUNT))
fps = int(cap.get(cv2.CAP_PROP_FPS))
out_video = cv2.VideoWriter(os.path.join(output_path, 'output_video.mp4'), cv2.VideoWriter_fourcc(*'avc1'), fps, (initial_w, initial_h), True)
counter=0
start_inference_time=time.time()
try:
while cap.isOpened():
ret, frame=cap.read()
if not ret:
break
counter+=1
coords, image= pd.predict(frame)
num_people= queue.check_coords(coords)
print(f"Total People in frame = {len(coords)}")
print(f"Number of people in queue = {num_people}")
out_text=""
y_pixel=25
for k, v in num_people.items():
out_text += f"No. of People in Queue {k} is {v} "
if v >= int(max_people):
out_text += f" Queue full; Please move to next Queue "
cv2.putText(image, out_text, (15, y_pixel), cv2.FONT_HERSHEY_COMPLEX, 1, (0, 255, 0), 2)
out_text=""
y_pixel+=40
out_video.write(image)
total_time=time.time()-start_inference_time
total_inference_time=round(total_time, 1)
fps=counter/total_inference_time
with open(os.path.join(output_path, 'stats.txt'), 'w') as f:
f.write(str(total_inference_time)+'\n')
f.write(str(fps)+'\n')
f.write(str(total_model_load_time)+'\n')
cap.release()
cv2.destroyAllWindows()
except Exception as e:
print("Could not run Inference: ", e)
if __name__=='__main__':
parser=argparse.ArgumentParser()
parser.add_argument('--model', required=True)
parser.add_argument('--device', default='CPU')
parser.add_argument('--video', default=None)
parser.add_argument('--queue_param', default=None)
parser.add_argument('--output_path', default='/results')
parser.add_argument('--max_people', default=2)
parser.add_argument('--threshold', default=0.60)
args=parser.parse_args()
main(args)
```
# Next Step
Now that you've run the above cell and created your Python script, you will create your job submission shell script in the next workspace.
**Note**: As a reminder, if you need to make any changes to the Python script, you can come back to this workspace to edit and run the above cell to overwrite the file with your changes.
|
github_jupyter
|
# Think Bayes
Second Edition
Copyright 2020 Allen B. Downey
License: [Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)](https://creativecommons.org/licenses/by-nc-sa/4.0/)
```
# If we're running on Colab, install empiricaldist
# https://pypi.org/project/empiricaldist/
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
!pip install empiricaldist
# Get utils.py and create directories
import os
if not os.path.exists('utils.py'):
!wget https://github.com/AllenDowney/ThinkBayes2/raw/master/code/soln/utils.py
if not os.path.exists('figs'):
!mkdir figs
if not os.path.exists('tables'):
!mkdir tables
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from empiricaldist import Pmf
from utils import decorate, savefig
```
## The Euro Problem
In *Information Theory, Inference, and Learning Algorithms*, David MacKay poses this problem:
"A statistical statement appeared in *The Guardian* on Friday January 4, 2002:
>When spun on edge 250 times, a Belgian one-euro coin came
up heads 140 times and tails 110. 'It looks very suspicious
to me,' said Barry Blight, a statistics lecturer at the London
School of Economics. 'If the coin were unbiased, the chance of
getting a result as extreme as that would be less than 7\%.'
"But [MacKay asks] do these data give evidence that the coin is biased rather than fair?"
To answer that question, we'll proceed in two steps.
First we'll use the binomial distribution to see where that 7% came from; then we'll use Bayes's Theorem to estimate the probability that this coin comes up heads.
## The binomial distribution
Suppose I tell you that a coin is "fair", that is, the probability of heads is 50%. If you spin it twice, there are four outcomes: `HH`, `HT`, `TH`, and `TT`. All four outcomes have the same probability, 25%.
If we add up the total number of heads, there are three possible outcomes: 0, 1, or 2. The probability of 0 and 2 is 25%, and the probability of 1 is 50%.
More generally, suppose the probability of heads is `p` and we spin the coin `n` times. What is the probability that we get a total of `k` heads?
The answer is given by the binomial distribution:
$P(k; n, p) = \binom{n}{k} p^k (1-p)^{n-k}$
where $\binom{n}{k}$ is the [binomial coefficient](https://en.wikipedia.org/wiki/Binomial_coefficient), usually pronounced "n choose k".
We can compute the binomial distribution ourselves, but we can also use the SciPy function `binom.pmf`:
```
from scipy.stats import binom
n = 2
p = 0.5
ks = np.arange(n+1)
a = binom.pmf(ks, n, p)
a
```
If we put this array in a `Pmf`, the result is the distribution of `k` for the given values of `n` and `p`.
```
pmf_k = Pmf(a, ks)
pmf_k
from utils import write_pmf
write_pmf(pmf_k, 'table03-01')
```
The following function computes the binomial distribution for given values of `n` and `p`:
```
def make_binomial(n, p):
"""Make a binomial PMF.
n: number of spins
p: probability of heads
returns: Pmf representing the distribution
"""
ks = np.arange(n+1)
a = binom.pmf(ks, n, p)
return Pmf(a, ks)
```
And here's what it looks like with `n=250` and `p=0.5`:
```
pmf_k = make_binomial(n=250, p=0.5)
pmf_k.plot(label='n=250, p=0.5')
decorate(xlabel='Number of heads (k)',
ylabel='PMF',
title='Binomial distribution')
savefig('fig03-01')
```
The most likely value in this distribution is 125:
```
pmf_k.max_prob()
```
But even though it is the most likely value, the probability that we get exactly 125 heads is only about 5%.
```
pmf_k[125]
```
In MacKay's example, we got 140 heads, which is less likely than 125:
```
pmf_k[140]
```
In the article MacKay quotes, the statistician says, ‘If the coin were unbiased the chance of getting a result as extreme as that would be less than 7%’.
We can use the binomial distribution to check his math. The following function takes a PMF and computes the total probability of values greater than or equal to `threshold`.
```
def ge_dist(pmf, threshold):
"""Probability of values greater than a threshold.
pmf: Series representing a PMF
threshold: value to compare to
returns: probability
"""
ge = (pmf.index >= threshold)
total = pmf[ge].sum()
return total
```
Here's the probability of getting 140 heads or more:
```
ge_dist(pmf_k, 140)
```
`Pmf` provides a method that does the same computation.
```
pmf_k.ge_dist(140)
```
The result is about 3.3%, which is less than 7%. The reason is that the statistician includes all values "as extreme as" 140, which includes values less than or equal to 110, because 140 exceeds the expected value by 15 and 110 falls short by 15.
```
pmf_k.le_dist(110)
```
The probability of values less than or equal to 110 is also 3.3%,
so the total probability of values "as extreme" as 140 is 6.6%.
The point of this calculation is that these extreme values are unlikely if the coin is fair.
That's interesting, but it doesn't answer MacKay's question. Let's see if we can.
## The Euro problem
Any given coin has some probability of landing heads up when spun
on edge; I'll call this probability `x`.
It seems reasonable to believe that `x` depends
on physical characteristics of the coin, like the distribution
of weight.
If a coin is perfectly balanced, we expect `x` to be close to 50%, but
for a lopsided coin, `x` might be substantially different. We can use
Bayes's theorem and the observed data to estimate `x`.
For simplicity, I'll start with a uniform prior, which assume that all values of `x` are equally likely.
That might not be a reasonable assumption, so we'll come back and consider other priors later.
We can make a uniform prior like this:
```
hypos = np.linspace(0, 1, 101)
prior = Pmf(1, hypos)
```
I'll use a dictionary to store the likelihoods for `H` and `T`:
```
likelihood = {
'H': hypos,
'T': 1 - hypos
}
```
I'll use a string to represent the dataset:
```
dataset = 'H' * 140 + 'T' * 110
```
The following function does the update.
```
def update_euro(pmf, dataset):
"""Updates the Suite with the given number of heads and tails.
pmf: Pmf representing the prior
data: tuple of heads and tails
"""
for data in dataset:
pmf *= likelihood[data]
pmf.normalize()
```
And here's how we use it.
```
posterior = prior.copy()
update_euro(posterior, dataset)
```
Here's what the posterior looks like.
```
def decorate_euro(title):
decorate(xlabel='Proportion of heads (x)',
ylabel='Probability',
title=title)
posterior.plot(label='140 heads out of 250')
decorate_euro(title='Posterior distribution of x')
savefig('fig03-02')
```
The peak of the posterior is at 56%, which is the proportion of heads in the dataset.
```
posterior.max_prob()
```
## Different priors
Let's see how that looks with different priors. Here's the uniform prior again.
```
uniform = Pmf(1, hypos, name='uniform')
uniform.normalize()
```
And here's a triangle-shaped prior.
```
ramp_up = np.arange(50)
ramp_down = np.arange(50, -1, -1)
a = np.append(ramp_up, ramp_down)
triangle = Pmf(a, hypos, name='triangle')
triangle.normalize()
```
Here's what they look like:
```
uniform.plot()
triangle.plot()
decorate_euro(title='Uniform and triangle prior distributions')
savefig('fig03-03')
```
If we update them both with the same data:
```
update_euro(uniform, dataset)
update_euro(triangle, dataset)
```
Here are the posteriors.
```
uniform.plot()
triangle.plot()
decorate_euro(title='Posterior distributions')
savefig('fig03-04')
```
The results are almost identical; the remaining difference is unlikely to matter in practice.
## The binomial likelihood function
We can make the Euro class more efficient by computing the likelihood of the entire dataset at once, rather than one coin toss at a time.
If the probability of heads is `p`, we can compute the probability of `k=140` heads in `n=250` tosses using the binomial PMF.
```
from scipy.stats import binom
def update_binomial(pmf, data):
"""Update the PMF using the binomial distribution.
pmf: Pmf representing the prior
data: tuple of integers k and n
"""
k, n = data
xs = pmf.qs
likelihood = binom.pmf(k, n, xs)
pmf *= likelihood
pmf.normalize()
```
The data are represented with a tuple of values for `k` and `n`, rather than a long string of outcomes.
Here's the update.
```
uniform2 = Pmf(1, hypos, name='uniform2')
data = 140, 250
update_binomial(uniform2, data)
```
Here's what the posterior looks like.
```
uniform.plot()
uniform2.plot()
decorate_euro(title='Posterior distributions computed two ways')
```
The results are the same, within floating-point error.
```
np.max(np.abs(uniform-uniform2))
```
## Exercises
**Exercise:** In Major League Baseball, most players have a batting average between 200 and 330, which means that the probability of getting a hit is between 0.2 and 0.33.
Suppose a new player appearing in his first game gets 3 hits out of 3 attempts. What is the posterior distribution for his probability of getting a hit?
For this exercise, I will construct the prior distribution by starting with a uniform distribution and updating it with imaginary data until it has a shape that reflects my background knowledge of batting averages.
```
hypos = np.linspace(0.1, 0.4, 101)
prior = Pmf(1, hypos)
likelihood = {
'Y': hypos,
'N': 1-hypos
}
dataset = 'Y' * 25 + 'N' * 75
for data in dataset:
prior *= likelihood[data]
prior.normalize()
prior.plot(label='prior')
decorate(xlabel='Probability of getting a hit',
ylabel='PMF')
```
This distribution indicates that most players have a batting average near 250, with only a few players below 175 or above 350. I'm not sure how accurately this prior reflects the distribution of batting averages in Major League Baseball, but it is good enough for this exercise.
Now update this distribution with the data and plot the posterior. What is the most likely value in the posterior distribution?
```
# Solution
posterior = prior.copy()
for data in 'YYY':
posterior *= likelihood[data]
posterior.normalize()
# Solution
prior.plot(label='prior')
posterior.plot(label='posterior ')
decorate(xlabel='Probability of getting a hit',
ylabel='PMF')
# Solution
prior.max_prob()
# Solution
posterior.max_prob()
```
**Exercise:** Whenever you survey people about sensitive issues, you have to deal with [social desirability bias](https://en.wikipedia.org/wiki/Social_desirability_bias), which is the tendency of people to shade their answers to show themselves in the most positive light.
One of the ways to improve the accuracy of the results is [randomized response](https://en.wikipedia.org/wiki/Randomized_response).
As an example, suppose you ask 100 people to flip a coin and:
* If they get heads, they report YES.
* If they get tails, they honestly answer the question "Do you cheat on your taxes?"
And suppose you get 80 YESes and 20 NOs. Based on this data, what is the posterior distribution for the fraction of people who cheat on their taxes? What is the most likely value in the posterior distribution?
```
# Solution
hypos = np.linspace(0, 1, 101)
prior = Pmf(1, hypos)
# Solution
likelihood = {
'Y': 0.5 + hypos/2,
'N': (1-hypos)/2
}
# Solution
dataset = 'Y' * 80 + 'N' * 20
posterior = prior.copy()
for data in dataset:
posterior *= likelihood[data]
posterior.normalize()
# Solution
posterior.plot(label='80 YES, 20 NO')
decorate(xlabel='Proportion of cheaters',
ylabel='PMF')
# Solution
posterior.idxmax()
```
**Exercise:** Suppose that instead of observing coin spins directly, you measure the outcome using an instrument that is not always correct. Specifically, suppose the probability is `y=0.2` that an actual heads is reported
as tails, or actual tails reported as heads.
If we spin a coin 250 times and the instrument reports 140 heads, what is the posterior distribution of `x`?
What happens as you vary the value of `y`?
```
# Solution
def update_unreliable(pmf, dataset, y):
likelihood = {
'H': (1-y) * hypos + y * (1-hypos),
'T': y * hypos + (1-y) * (1-hypos)
}
for data in dataset:
pmf *= likelihood[data]
pmf.normalize()
# Solution
hypos = np.linspace(0, 1, 101)
prior = Pmf(1, hypos)
dataset = 'H' * 140 + 'T' * 110
posterior00 = prior.copy()
update_unreliable(posterior00, dataset, 0.0)
posterior02 = prior.copy()
update_unreliable(posterior02, dataset, 0.2)
posterior04 = prior.copy()
update_unreliable(posterior04, dataset, 0.4)
# Solution
posterior00.plot(label='y = 0.0')
posterior02.plot(label='y = 0.2')
posterior04.plot(label='y = 0.4')
decorate(xlabel='Proportion of heads',
ylabel='PMF')
# Solution
posterior00.idxmax(), posterior02.idxmax(), posterior04.idxmax()
```
**Exercise:** In preparation for an alien invasion, the Earth Defense League (EDL) has been working on new missiles to shoot down space invaders. Of course, some missile designs are better than others; let's assume that each design has some probability of hitting an alien ship, `x`.
Based on previous tests, the distribution of `x` in the population of designs is approximately uniform between 0.1 and 0.4.
Now suppose the new ultra-secret Alien Blaster 9000 is being tested. In a press conference, an EDL general reports that the new design has been tested twice, taking two shots during each test. The results of the test are confidential, so the general won't say how many targets were hit, but they report: "The same number of targets were hit in the two tests, so we have reason to think this new design is consistent."
Is this data good or bad; that is, does it increase or decrease your estimate of `x` for the Alien Blaster 9000?
Hint: If the probability of hitting each target is $x$, the probability of hitting one target in both tests is $[2x(1-x)]^2$.
```
# Solution
hypos = np.linspace(0.1, 0.4, 101)
prior = Pmf(1, hypos)
# Solution
# specific version for n=2 shots
x = hypos
likes = [(1-x)**4, (2*x*(1-x))**2, x**4]
likelihood = np.sum(likes, axis=0)
# Solution
# general version for any n shots per test
from scipy.stats import binom
n = 2
likes2 = [binom.pmf(k, n, x)**2 for k in range(n+1)]
likelihood2 = np.sum(likes2, axis=0)
# Solution
plt.plot(x, likelihood, label='special case')
plt.plot(x, likelihood2, label='general formula')
decorate(xlabel='Probability of hitting the target',
ylabel='Likelihood',
title='Likelihood of getting the same result')
# Solution
posterior = prior * likelihood
posterior.normalize()
# Solution
posterior.plot(label='Two tests, two shots, same outcome')
decorate(xlabel='Probability of hitting the target',
ylabel='PMF',
title='Posterior distribution',
ylim=[0, 0.015])
# Solution
# Getting the same result in both tests is more likely for
# extreme values of `x` and least likely when `x=0.5`.
# In this example, the prior suggests that `x` is less than 0.5,
# and the update gives more weight to extreme values.
# So the data makes lower values of `x` more likely.
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import torch
import torchvision
from torch.utils.data import Dataset, DataLoader
from torchvision import transforms, utils
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from matplotlib import pyplot as plt
%matplotlib inline
from scipy.stats import entropy
from google.colab import drive
drive.mount('/content/drive')
path="/content/drive/MyDrive/Research/alternate_minimisation/"
name="_50_50_10runs_entropy"
# mu1 = np.array([3,3,3,3,0])
# sigma1 = np.array([[1,1,1,1,1],[1,16,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu2 = np.array([4,4,4,4,0])
# sigma2 = np.array([[16,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu3 = np.array([10,5,5,10,0])
# sigma3 = np.array([[1,1,1,1,1],[1,16,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu4 = np.array([-10,-10,-10,-10,0])
# sigma4 = np.array([[1,1,1,1,1],[1,16,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu5 = np.array([-21,4,4,-21,0])
# sigma5 = np.array([[16,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu6 = np.array([-10,18,18,-10,0])
# sigma6 = np.array([[1,1,1,1,1],[1,16,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu7 = np.array([4,20,4,20,0])
# sigma7 = np.array([[16,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu8 = np.array([4,-20,-20,4,0])
# sigma8 = np.array([[16,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu9 = np.array([20,20,20,20,0])
# sigma9 = np.array([[1,1,1,1,1],[1,16,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# mu10 = np.array([20,-10,-10,20,0])
# sigma10 = np.array([[1,1,1,1,1],[1,16,1,1,1],[1,1,1,1,1],[1,1,1,1,1],[1,1,1,1,1]])
# sample1 = np.random.multivariate_normal(mean=mu1,cov= sigma1,size=500)
# sample2 = np.random.multivariate_normal(mean=mu2,cov= sigma2,size=500)
# sample3 = np.random.multivariate_normal(mean=mu3,cov= sigma3,size=500)
# sample4 = np.random.multivariate_normal(mean=mu4,cov= sigma4,size=500)
# sample5 = np.random.multivariate_normal(mean=mu5,cov= sigma5,size=500)
# sample6 = np.random.multivariate_normal(mean=mu6,cov= sigma6,size=500)
# sample7 = np.random.multivariate_normal(mean=mu7,cov= sigma7,size=500)
# sample8 = np.random.multivariate_normal(mean=mu8,cov= sigma8,size=500)
# sample9 = np.random.multivariate_normal(mean=mu9,cov= sigma9,size=500)
# sample10 = np.random.multivariate_normal(mean=mu10,cov= sigma10,size=500)
# X = np.concatenate((sample1,sample2,sample3,sample4,sample5,sample6,sample7,sample8,sample9,sample10),axis=0)
# Y = np.concatenate((np.zeros((500,1)),np.ones((500,1)),2*np.ones((500,1)),3*np.ones((500,1)),4*np.ones((500,1)),
# 5*np.ones((500,1)),6*np.ones((500,1)),7*np.ones((500,1)),8*np.ones((500,1)),9*np.ones((500,1))),axis=0).astype(int)
# print(X.shape,Y.shape)
# # plt.scatter(sample1[:,0],sample1[:,1],label="class_0")
# # plt.scatter(sample2[:,0],sample2[:,1],label="class_1")
# # plt.scatter(sample3[:,0],sample3[:,1],label="class_2")
# # plt.scatter(sample4[:,0],sample4[:,1],label="class_3")
# # plt.scatter(sample5[:,0],sample5[:,1],label="class_4")
# # plt.scatter(sample6[:,0],sample6[:,1],label="class_5")
# # plt.scatter(sample7[:,0],sample7[:,1],label="class_6")
# # plt.scatter(sample8[:,0],sample8[:,1],label="class_7")
# # plt.scatter(sample9[:,0],sample9[:,1],label="class_8")
# # plt.scatter(sample10[:,0],sample10[:,1],label="class_9")
# # plt.legend(bbox_to_anchor=(1.05, 1), loc='upper left')
# class SyntheticDataset(Dataset):
# """MosaicDataset dataset."""
# def __init__(self, x, y):
# """
# Args:
# csv_file (string): Path to the csv file with annotations.
# root_dir (string): Directory with all the images.
# transform (callable, optional): Optional transform to be applied
# on a sample.
# """
# self.x = x
# self.y = y
# #self.fore_idx = fore_idx
# def __len__(self):
# return len(self.y)
# def __getitem__(self, idx):
# return self.x[idx] , self.y[idx] #, self.fore_idx[idx]
# trainset = SyntheticDataset(X,Y)
# # testset = torchvision.datasets.MNIST(root='./data', train=False, download=True, transform=transform)
# classes = ('zero','one','two','three','four','five','six','seven','eight','nine')
# foreground_classes = {'zero','one','two'}
# fg_used = '012'
# fg1, fg2, fg3 = 0,1,2
# all_classes = {'zero','one','two','three','four','five','six','seven','eight','nine'}
# background_classes = all_classes - foreground_classes
# background_classes
# trainloader = torch.utils.data.DataLoader(trainset, batch_size=100, shuffle=True)
# dataiter = iter(trainloader)
# background_data=[]
# background_label=[]
# foreground_data=[]
# foreground_label=[]
# batch_size=100
# for i in range(50):
# images, labels = dataiter.next()
# for j in range(batch_size):
# if(classes[labels[j]] in background_classes):
# img = images[j].tolist()
# background_data.append(img)
# background_label.append(labels[j])
# else:
# img = images[j].tolist()
# foreground_data.append(img)
# foreground_label.append(labels[j])
# foreground_data = torch.tensor(foreground_data)
# foreground_label = torch.tensor(foreground_label)
# background_data = torch.tensor(background_data)
# background_label = torch.tensor(background_label)
# def create_mosaic_img(bg_idx,fg_idx,fg):
# """
# bg_idx : list of indexes of background_data[] to be used as background images in mosaic
# fg_idx : index of image to be used as foreground image from foreground data
# fg : at what position/index foreground image has to be stored out of 0-8
# """
# image_list=[]
# j=0
# for i in range(9):
# if i != fg:
# image_list.append(background_data[bg_idx[j]])
# j+=1
# else:
# image_list.append(foreground_data[fg_idx])
# label = foreground_label[fg_idx] - fg1 # minus fg1 because our fore ground classes are fg1,fg2,fg3 but we have to store it as 0,1,2
# #image_list = np.concatenate(image_list ,axis=0)
# image_list = torch.stack(image_list)
# return image_list,label
# desired_num = 3000
# mosaic_list_of_images =[] # list of mosaic images, each mosaic image is saved as list of 9 images
# fore_idx =[] # list of indexes at which foreground image is present in a mosaic image i.e from 0 to 9
# mosaic_label=[] # label of mosaic image = foreground class present in that mosaic
# list_set_labels = []
# for i in range(desired_num):
# set_idx = set()
# np.random.seed(i)
# bg_idx = np.random.randint(0,3500,8)
# set_idx = set(background_label[bg_idx].tolist())
# fg_idx = np.random.randint(0,1500)
# set_idx.add(foreground_label[fg_idx].item())
# fg = np.random.randint(0,9)
# fore_idx.append(fg)
# image_list,label = create_mosaic_img(bg_idx,fg_idx,fg)
# mosaic_list_of_images.append(image_list)
# mosaic_label.append(label)
# list_set_labels.append(set_idx)
# def create_avg_image_from_mosaic_dataset(mosaic_dataset,labels,foreground_index,dataset_number):
# """
# mosaic_dataset : mosaic_dataset contains 9 images 32 x 32 each as 1 data point
# labels : mosaic_dataset labels
# foreground_index : contains list of indexes where foreground image is present so that using this we can take weighted average
# dataset_number : will help us to tell what ratio of foreground image to be taken. for eg: if it is "j" then fg_image_ratio = j/9 , bg_image_ratio = (9-j)/8*9
# """
# avg_image_dataset = []
# for i in range(len(mosaic_dataset)):
# img = torch.zeros([5], dtype=torch.float64)
# for j in range(9):
# if j == foreground_index[i]:
# img = img + mosaic_dataset[i][j]*dataset_number/9
# else :
# img = img + mosaic_dataset[i][j]*(9-dataset_number)/(8*9)
# avg_image_dataset.append(img)
# return torch.stack(avg_image_dataset) , torch.stack(labels) , foreground_index
class MosaicDataset1(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list, mosaic_label,fore_idx):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list
self.label = mosaic_label
self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx] , self.fore_idx[idx]
# data = [{"mosaic_list":mosaic_list_of_images, "mosaic_label": mosaic_label, "fore_idx":fore_idx}]
# np.save("mosaic_data.npy",data)
data = np.load(path+"mosaic_data.npy",allow_pickle=True)
mosaic_list_of_images = data[0]["mosaic_list"]
mosaic_label = data[0]["mosaic_label"]
fore_idx = data[0]["fore_idx"]
batch = 250
msd = MosaicDataset1(mosaic_list_of_images, mosaic_label, fore_idx)
train_loader = DataLoader( msd,batch_size= batch ,shuffle=True)
```
**Focus Net**
```
class Focus_deep(nn.Module):
'''
deep focus network averaged at zeroth layer
input : elemental data
'''
def __init__(self,inputs,output,K,d):
super(Focus_deep,self).__init__()
self.inputs = inputs
self.output = output
self.K = K
self.d = d
self.linear1 = nn.Linear(self.inputs,50) #,self.output)
self.linear2 = nn.Linear(50,self.output)
def forward(self,z):
batch = z.shape[0]
x = torch.zeros([batch,self.K],dtype=torch.float64)
y = torch.zeros([batch,self.d], dtype=torch.float64)
x,y = x.to("cuda"),y.to("cuda")
for i in range(self.K):
x[:,i] = self.helper(z[:,i] )[:,0] # self.d*i:self.d*i+self.d
log_x = F.log_softmax(x,dim=1) # log alpha to calculate entropy
x = F.softmax(x,dim=1) # alphas
x1 = x[:,0]
for i in range(self.K):
x1 = x[:,i]
y = y+torch.mul(x1[:,None],z[:,i]) # self.d*i:self.d*i+self.d
return y , x,log_x
def helper(self,x):
x = F.relu(self.linear1(x))
x = self.linear2(x)
return x
```
**Classification Net**
```
class Classification_deep(nn.Module):
'''
input : elemental data
deep classification module data averaged at zeroth layer
'''
def __init__(self,inputs,output):
super(Classification_deep,self).__init__()
self.inputs = inputs
self.output = output
self.linear1 = nn.Linear(self.inputs,50)
self.linear2 = nn.Linear(50,self.output)
def forward(self,x):
x = F.relu(self.linear1(x))
x = self.linear2(x)
return x
criterion = nn.CrossEntropyLoss()
def my_cross_entropy(x, y,alpha,log_alpha,k):
# log_prob = -1.0 * F.log_softmax(x, 1)
# loss = log_prob.gather(1, y.unsqueeze(1))
# loss = loss.mean()
loss = criterion(x,y)
#alpha = torch.clamp(alpha,min=1e-10)
b = -1.0* alpha * log_alpha
b = torch.mean(torch.sum(b,dim=1))
closs = loss
entropy = b
loss = (1-k)*loss + ((k)*b)
return loss,closs,entropy
```
```
def calculate_attn_loss(dataloader,what,where,criter,k):
what.eval()
where.eval()
r_loss = 0
cc_loss = 0
cc_entropy = 0
alphas = []
lbls = []
pred = []
fidices = []
with torch.no_grad():
for i, data in enumerate(dataloader, 0):
inputs, labels,fidx = data
lbls.append(labels)
fidices.append(fidx)
inputs = inputs.double()
inputs, labels = inputs.to("cuda"),labels.to("cuda")
avg,alpha,log_alpha = where(inputs)
outputs = what(avg)
_, predicted = torch.max(outputs.data, 1)
pred.append(predicted.cpu().numpy())
alphas.append(alpha.cpu().numpy())
#ent = np.sum(entropy(alpha.cpu().detach().numpy(), base=2, axis=1))/batch
# mx,_ = torch.max(alpha,1)
# entropy = np.mean(-np.log2(mx.cpu().detach().numpy()))
# print("entropy of batch", entropy)
#loss = (1-k)*criter(outputs, labels) + k*ent
loss,closs,entropy = my_cross_entropy(outputs,labels,alpha,log_alpha,k)
r_loss += loss.item()
cc_loss += closs.item()
cc_entropy += entropy.item()
alphas = np.concatenate(alphas,axis=0)
pred = np.concatenate(pred,axis=0)
lbls = np.concatenate(lbls,axis=0)
fidices = np.concatenate(fidices,axis=0)
#print(alphas.shape,pred.shape,lbls.shape,fidices.shape)
analysis = analyse_data(alphas,lbls,pred,fidices)
return r_loss/i,cc_loss/i,cc_entropy/i,analysis
def analyse_data(alphas,lbls,predicted,f_idx):
'''
analysis data is created here
'''
batch = len(predicted)
amth,alth,ftpt,ffpt,ftpf,ffpf = 0,0,0,0,0,0
for j in range (batch):
focus = np.argmax(alphas[j])
if(alphas[j][focus] >= 0.5):
amth +=1
else:
alth +=1
if(focus == f_idx[j] and predicted[j] == lbls[j]):
ftpt += 1
elif(focus != f_idx[j] and predicted[j] == lbls[j]):
ffpt +=1
elif(focus == f_idx[j] and predicted[j] != lbls[j]):
ftpf +=1
elif(focus != f_idx[j] and predicted[j] != lbls[j]):
ffpf +=1
#print(sum(predicted==lbls),ftpt+ffpt)
return [ftpt,ffpt,ftpf,ffpf,amth,alth]
number_runs = 10
full_analysis =[]
FTPT_analysis = pd.DataFrame(columns = ["FTPT","FFPT", "FTPF","FFPF"])
k = 0.005
every_what_epoch = 1
for n in range(number_runs):
print("--"*40)
# instantiate focus and classification Model
torch.manual_seed(n)
where = Focus_deep(5,1,9,5).double()
torch.manual_seed(n)
what = Classification_deep(5,3).double()
where = where.to("cuda")
what = what.to("cuda")
# instantiate optimizer
optimizer_where = optim.Adam(where.parameters(),lr =0.01)
optimizer_what = optim.Adam(what.parameters(), lr=0.01)
#criterion = nn.CrossEntropyLoss()
acti = []
analysis_data = []
loss_curi = []
epochs = 2000
# calculate zeroth epoch loss and FTPT values
running_loss ,_,_,anlys_data= calculate_attn_loss(train_loader,what,where,criterion,k)
loss_curi.append(running_loss)
analysis_data.append(anlys_data)
print('epoch: [%d ] loss: %.3f' %(0,running_loss))
# training starts
for epoch in range(epochs): # loop over the dataset multiple times
ep_lossi = []
running_loss = 0.0
what.train()
where.train()
if ((epoch) % (every_what_epoch*2) ) <= every_what_epoch-1 :
print(epoch+1,"updating what_net, where_net is freezed")
print("--"*40)
elif ((epoch) % (every_what_epoch*2)) > every_what_epoch-1 :
print(epoch+1,"updating where_net, what_net is freezed")
print("--"*40)
for i, data in enumerate(train_loader, 0):
# get the inputs
inputs, labels,_ = data
inputs = inputs.double()
inputs, labels = inputs.to("cuda"),labels.to("cuda")
# zero the parameter gradients
optimizer_where.zero_grad()
optimizer_what.zero_grad()
# forward + backward + optimize
avg, alpha,log_alpha = where(inputs)
outputs = what(avg)
my_loss,_,_ = my_cross_entropy(outputs,labels,alpha,log_alpha,k)
# print statistics
running_loss += my_loss.item()
my_loss.backward()
if ((epoch) % (every_what_epoch*2) ) <= every_what_epoch-1 :
optimizer_what.step()
elif ( (epoch) % (every_what_epoch*2)) > every_what_epoch-1 :
optimizer_where.step()
# optimizer_where.step()
# optimizer_what.step()
#break
running_loss,ccloss,ccentropy,anls_data = calculate_attn_loss(train_loader,what,where,criterion,k)
analysis_data.append(anls_data)
print('epoch: [%d] loss: %.3f celoss: %.3f entropy: %.3f' %(epoch + 1,running_loss,ccloss,ccentropy))
loss_curi.append(running_loss) #loss per epoch
if running_loss<=0.001:
break
print('Finished Training run ' +str(n))
#break
analysis_data = np.array(analysis_data)
FTPT_analysis.loc[n] = analysis_data[-1,:4]/30
full_analysis.append((epoch, analysis_data))
correct = 0
total = 0
with torch.no_grad():
for data in train_loader:
images, labels,_ = data
images = images.double()
images, labels = images.to("cuda"), labels.to("cuda")
avg, alpha,log_alpha = where(images)
outputs = what(avg)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 3000 train images: %d %%' % ( 100 * correct / total))
a,b= full_analysis[0]
print(a)
cnt=1
for epoch, analysis_data in full_analysis:
analysis_data = np.array(analysis_data)
# print("="*20+"run ",cnt,"="*20)
plt.figure(figsize=(6,6))
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,0],label="ftpt")
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,1],label="ffpt")
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,2],label="ftpf")
plt.plot(np.arange(0,epoch+2,1),analysis_data[:,3],label="ffpf")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.title("Training trends for run "+str(cnt))
plt.savefig(path+"50_50_10runs_entropy/every1/run"+str(cnt)+".png",bbox_inches="tight")
plt.savefig(path+"50_50_10runs_entropy/every1/run"+str(cnt)+".pdf",bbox_inches="tight")
cnt+=1
np.mean(np.array(FTPT_analysis),axis=0) #array([87.85333333, 5.92 , 0. , 6.22666667])
FTPT_analysis.to_csv(path+"50_50_10runs_entropy/FTPT_analysis_every1"+name+".csv",index=False)
FTPT_analysis
```
|
github_jupyter
|
# Software Carpentry
### EPFL Library, November 2018
## Program
| | 4 afternoons | 4 workshops |
| :-- | :----------- | :---------- |
| > | `Today` | `Unix Shell` |
| | Thursday 22 | Version Control with Git |
| | Tuesday 27 | Python I |
| | Thursday 29 | More Python |
## Why did you decide to attend this workshop?
## Today's program
| | activity |
| :-- | :-------- |
| 13:00 | Introducing the Unix Shell |
| 13:15 | Navigating Files and Directories |
| 14:00 | Working with Files and Directories |
| 14:50 | **break** |
| 15:20 | Loops |
| 16:10 | Shell Scripts |
| 16:55 | Finding Things |
| 17:30 | Wrap-up / **END** |
## How we'll work
Live coding
Sticky notes : use a red sticky note to say you are stuck, put the green one when all is good
Instructors : Raphaël and Mathilde
Helpers : Antoine, Ludovic, Raphaël and Mathilde
Slides for exercices : Find the link to the slides on go.epfl.ch/swc-pad
## Introducing the Shell
### Key points about the Shell
- A shell is a program whose primary purpose is to read commands and run other programs.
- The shell’s main advantages are its high action-to-keystroke ratio, its support for automating repetitive tasks, and its capacity to access networked machines.
- The shell’s main disadvantages are its primarily textual nature and how cryptic its commands and operation can be.
## Navigating Files and Directories
### [Exercise] Exploring more `rm` flags
What does the command ls do when used with the `-l` and `-h` flags?
### [Exercise] Listing Recursively and By Time
The command `ls -R` lists the contents of directories recursively, i.e., lists their sub-directories, sub-sub-directories, and so on at each level. The command `ls -t` lists things by time of last change, with most recently changed files or directories first. In what order does `ls -R -t` display things?
### [Exercise] Absolute vs Relative Paths
Starting from /Users/amanda/data/, which of the following commands could Amanda use to navigate to her home directory, which is /Users/amanda?
1. `cd .`
2. `cd /`
3. `cd /home/amanda`
4. `cd ../..`
5. `cd ~`
6. `cd home`
7. `cd ~/data/..`
8. `cd`
9. `cd ..`
### [Exercise] Relative Path Resolution
Using the filesystem diagram below, if pwd displays /Users/thing, what will ls -F ../backup display?
1. `../backup: No such file or directory`
2. `2012-12-01 2013-01-08 2013-01-27`
3. `2012-12-01/ 2013-01-08/ 2013-01-27/`
4. `original/ pnas_final/ pnas_sub/`

### [Exercise ] `ls` Reading comprehension

Assuming a directory structure as in the above Figure (File System for Challenge Questions), if `pwd` displays `/Users/backup`, and `-r` tells ls to display things in reverse order, what command will display:
`pnas_sub/ pnas_final/ original/`
1. `ls pwd`
2. `ls -r -F`
3. `ls -r -F /Users/backup`
4. Either #2 or #3 above, but not #1.
### Key Points about Navigating Files and Directories
- The file system is responsible for managing information on the disk.
- Information is stored in files, which are stored in directories (folders).
- Directories can also store other directories, which forms a directory tree.
- `cd path` changes the current working directory.
- `ls path` prints a listing of a specific file or directory; `ls` on its own lists the current working directory.
- `pwd` prints the user’s current working directory.
- `/` on its own is the root directory of the whole file system.
### More key Points about Navigating Files and Directories
- A relative path specifies a location starting from the current location.
- An absolute path specifies a location from the root of the file system.
- Directory names in a path are separated with `/` on Unix, but `\`on Windows.
- `..` means ‘the directory above the current one’; `.` on its own means ‘the current directory’.
- Most files’ names are `something.extension`. The extension isn’t required, and doesn’t guarantee anything, but is normally used to indicate the type of data in the file.
## Working with Files and Directories
### [Exercise] Creating Files a Different Way
We have seen how to create text files using the `nano` editor. Now, try the following command in your home directory:
```
$ cd # go to your home directory
$ touch my_file.txt
```
1. What did the touch command do? When you look at your home directory using the GUI file explorer, does the file show up?
2. Use `ls -l` to inspect the files. How large is `my_file.txt`?
3. When might you want to create a file this way?
### [Exercise] Using `rm` Safely
What happens when we type `rm -i thesis/quotations.txt`? Why would we want this protection when using `rm`?
### [Exercise] Moving to the Current Folder
After running the following commands, Jamie realizes that she put the files `sucrose.dat` and `maltose.dat` into the wrong folder:
```
$ ls -F
```
> analyzed/ raw/
```
$ ls -F analyzed
```
> fructose.dat glucose.dat maltose.dat sucrose.dat
```
$ cd raw/
```
Fill in the blanks to move these files to the current folder (i.e., the one she is currently in):
```
$ mv ___/sucrose.dat ___/maltose.dat ___
```
### [Exercise] Renaming Files
Suppose that you created a `.txt` file in your current directory to contain a list of the statistical tests you will need to do to analyze your data, and named it: `statstics.txt`
After creating and saving this file you realize you misspelled the filename! You want to correct the mistake, which of the following commands could you use to do so?
1. `cp statstics.txt statistics.txt`
2. `mv statstics.txt statistics.txt`
3. `mv statstics.txt .`
4. `cp statstics.txt .`
### [Exercise] Moving and Copying
What is the output of the closing `ls` command in the sequence shown below?
```
$ pwd
```
> /Users/jamie/data
```
$ ls
```
> proteins.dat
```
$ mkdir recombine
$ mv proteins.dat recombine/
$ cp recombine/proteins.dat ../proteins-saved.dat
$ ls
```
1. `proteins-saved.dat recombine`
2. `recombine`
3. `proteins.dat recombine`
4. `proteins-saved.dat`
### Additional exercises
If you were quick, check out these exercises to dig a little more into details.
### [Exercise] Copy with Multiple Filenames
For this exercise, you can test the commands in the `data-shell/data` directory.
In the example below, what does `cp` do when given several filenames and a directory name?
```
$ mkdir backup
$ cp amino-acids.txt animals.txt backup/
```
In the example below, what does `cp` do when given three or more file names?
```
$ ls -F
```
> amino-acids.txt animals.txt backup/ elements/ morse.txt pdb/ planets.txt salmon.txt sunspot.txt
```
$ cp amino-acids.txt animals.txt morse.txt
```
### [Exercise] Using Wildcards
When run in the `molecules` directory, which `ls` command(s) will produce this output?
`ethane.pdb methane.pdb`
1. `ls *t*ane.pdb`
2. `ls *t?ne.*`
3. `ls *t??ne.pdb`
4. `ls ethane.*`
### [Exercise] More on Wildcards
Sam has a directory containing calibration data, datasets, and descriptions of the datasets:
```
2015-10-23-calibration.txt
2015-10-23-dataset1.txt
2015-10-23-dataset2.txt
2015-10-23-dataset_overview.txt
2015-10-26-calibration.txt
2015-10-26-dataset1.txt
2015-10-26-dataset2.txt
2015-10-26-dataset_overview.txt
2015-11-23-calibration.txt
2015-11-23-dataset1.txt
2015-11-23-dataset2.txt
2015-11-23-dataset_overview.txt
```
Before heading off to another field trip, she wants to back up her data and send some datasets to her colleague Bob. Sam uses the following commands to get the job done:
```
$ cp *dataset* /backup/datasets
$ cp ____calibration____ /backup/calibration
$ cp 2015-____-____ ~/send_to_bob/all_november_files/
$ cp ____ ~/send_to_bob/all_datasets_created_on_a_23rd/
```
Help Sam by filling in the blanks.
### [Exercise] Organizing Directories and Files
Jamie is working on a project and she sees that her files aren’t very well organized:
```
$ ls -F
```
> analyzed/ fructose.dat raw/ sucrose.dat
The `fructose.dat` and sucrose.dat` files contain output from her data analysis. What command(s) covered in this lesson does she need to run so that the commands below will produce the output shown?
```
$ ls -F
```
> analyzed/ raw/
```
$ ls analyzed
```
> fructose.dat sucrose.dat
### [Exercise] Copy a folder structure but not the files
You’re starting a new experiment, and would like to duplicate the file structure from your previous experiment without the data files so you can add new data.
Assume that the file structure is in a folder called ‘2016-05-18-data’, which contains a `data` folder that in turn contains folders named `raw` and `processed` that contain data files. The goal is to copy the file structure of the `2016-05-18-data` folder into a folder called `2016-05-20-data` and remove the data files from the directory you just created.
Which of the following set of commands would achieve this objective? What would the other commands do?
```
$ cp -r 2016-05-18-data/ 2016-05-20-data/
$ rm 2016-05-20-data/raw/*
$ rm 2016-05-20-data/processed/*
```
```
$ rm 2016-05-20-data/raw/*
$ rm 2016-05-20-data/processed/*
$ cp -r 2016-05-18-data/ 2016-5-20-data/
```
```
$ cp -r 2016-05-18-data/ 2016-05-20-data/
$ rm -r -i 2016-05-20-data/
```
### Key Points about Working with Files and Directories
`cp old new` copies a file.
`mkdir path` creates a new directory.
`mv old new` moves (renames) a file or directory.
`rm path` removes (deletes) a file.
`*` matches zero or more characters in a filename, so `*.txt` matches all files ending in `.txt`.
`?` matches any single character in a filename, so `?.txt` matches `a.txt` but not `any.txt`.
Use of the Control key may be described in many ways, including `Ctrl-X`, `Control-X`, and `^X`.
The shell does not have a trash bin: once something is deleted, it’s really gone.
Depending on the type of work you do, you may need a more powerful text editor than Nano.
## Pipes and Filters
### Key Points about Pipes and Filters
`cat` displays the contents of its inputs.
`head` displays the first 10 lines of its input.
`tail` displays the last 10 lines of its input.
`sort` sorts its inputs.
`wc` counts lines, words, and characters in its inputs.
`command > file` redirects a command’s output to a file.
`first | second` is a pipeline: the output of the first command is used as the input to the second.
[More information on this topic](https://swcarpentry.github.io/shell-novice/04-pipefilter/index.html) on the Software Carpentry website
## Loops
### [Exercise] Variables in Loops
This exercise refers to the `data-shell/molecules` directory. `ls` gives the following output:
> cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb
What is the output of the following code?
```
$ for datafile in *.pdb
> do
> ls *.pdb
> done
```
Now, what is the output of the following code?
```
$ for datafile in *.pdb
> do
> ls $datafile
> done
```
Why do these two loops give different outputs?
### [Exercise] Limiting Sets of Files
What would be the output of running the following loop in the `data-shell/molecules` directory?
```
$ for filename in c*
> do
> ls $filename
> done
```
1. No files are listed.
2. All files are listed.
3. Only `cubane.pdb`, `octane.pdb` and `pentane.pdb` are listed.
4. Only `cubane.pdb` is listed.
### [Exercise] Limiting Sets of Files (part 2)
How would the output differ from using this command instead?
```
$ for filename in *c*
> do
> ls $filename
> done
```
1. The same files would be listed.
2. All the files are listed this time.
3. No files are listed this time.
4. The files `cubane.pdb` and `octane.pdb` will be listed.
5. Only the file `octane.pdb` will be listed.
### [Exercise] Saving to a File in a Loop - Part One
In the `data-shell/molecules` directory, what is the effect of this loop?
```
$ for alkanes in *.pdb
> do
> echo $alkanes
> cat $alkanes > alkanes.pdb
> done
```
1. Prints `cubane.pdb`, `ethane.pdb`, `methane.pdb`, `octane.pdb`, `pentane.pdb` and `propane.pdb`, and the text from `propane.pdb` will be saved to a file called `alkanes.pdb`.
2. Prints `cubane.pdb`, `ethane.pdb`, and `methane.pdb`, and the text from all three files would be concatenated and saved to a file called `alkanes.pdb`.
3. Prints `cubane.pdb`, `ethane.pdb`, `methane.pdb`, `octane.pdb`, and `pentane.pdb`, and the text from `propane.pdb` will be saved to a file called `alkanes.pdb`.
4. None of the above.
### [Exercise] Saving to a File in a Loop - Part Two
Also in the `data-shell/molecules` directory, what would be the output of the following loop?
```
$ for datafile in *.pdb
> do
> cat $datafile >> all.pdb
> done
```
1. All of the text from `cubane.pdb`, `ethane.pdb`, `methane.pdb`, `octane.pdb`, and `pentane.pdb` would be concatenated and saved to a file called `all.pdb`.
2. The text from `ethane.pdb` will be saved to a file called `all.pdb`.
3. All of the text from `cubane.pdb`, `ethane.pdb`, `methane.pdb`, `octane.pdb`, `pentane.pdb` and `propane.pdb` would be concatenated and saved to a file called `all.pdb`.
4. All of the text from `cubane.pdb`, `ethane.pdb`, `methane.pdb`, `octane.pdb`, `pentane.pdb` and `propane.pdb` would be printed to the screen and saved to a file called `all.pdb`.
### Additional exercises
If you were quick, check out these exercises to dig a little more into details.
### [Exercise] Doing a Dry Run
A loop is a way to do many things at once — or to make many mistakes at once if it does the wrong thing. One way to check what a loop would do is to `echo` the commands it would run instead of actually running them.
Suppose we want to preview the commands the following loop will execute without actually running those commands:
```
$ for file in *.pdb
> do
> analyze $file > analyzed-$file
> done
```
What is the difference between the two loops below, and which one would we want to run?
```
# Version 1
$ for file in *.pdb
> do
> echo analyze $file > analyzed-$file
> done
```
```
# Version 2
$ for file in *.pdb
> do
> echo "analyze $file > analyzed-$file"
> done
```
### [Exercise] Nested Loops
Suppose we want to set up up a directory structure to organize some experiments measuring reaction rate constants with different compounds *and* different temperatures. What would be the result of the following code:
```
$ for species in cubane ethane methane
> do
> for temperature in 25 30 37 40
> do
> mkdir $species-$temperature
> done
> done
```
### Key Points for Loops
* A `for` loop repeats commands once for every thing in a list.
* Every `for` loop needs a variable to refer to the thing it is currently operating on.
* Use `$name` to expand a variable (i.e., get its value). `${name}` can also be used.
* Do not use spaces, quotes, or wildcard characters such as ‘\*’ or ‘?’ in filenames, as it complicates variable expansion.
* Give files consistent names that are easy to match with wildcard patterns to make it easy to select them for looping.
* Use the up-arrow key to scroll up through previous commands to edit and repeat them.
* Use `Ctrl-R` to search through the previously entered commands.
* Use `history` to display recent commands, and `!number` to repeat a command by number.
## Shell Scripts
### [Exercise] List Unique Species
Leah has several hundred data files, each of which is formatted like this:
```
2013-11-05,deer,5
2013-11-05,rabbit,22
2013-11-05,raccoon,7
2013-11-06,rabbit,19
2013-11-06,deer,2
2013-11-06,fox,1
2013-11-07,rabbit,18
2013-11-07,bear,1
```
An example of this type of file is given in `data-shell/data/animal-counts/animals.txt`.
Write a shell script called `species.sh` that takes any number of filenames as command-line arguments, and uses `cut`, `sort`, and `uniq` to print a list of the unique species appearing in each of those files separately.
### [Exercise] Why Record Commands in the History Before Running Them?
If you run the command:
```
$ history | tail -n 5 > recent.sh
```
the last command in the file is the `history` command itself, i.e., the shell has added `history` to the command log before actually running it. In fact, the shell always adds commands to the log before running them. Why do you think it does this?
### [Exercise] Variables in Shell Scripts
In the `molecules` directory, imagine you have a shell script called `script.sh` containing the following commands:
```
head -n $2 $1
tail -n $3 $1
```
While you are in the `molecules` directory, you type the following command:
`bash script.sh '*.pdb' 1 1`
Which of the following outputs would you expect to see?
1. All of the lines between the first and the last lines of each file ending in `.pdb` in the `molecules` directory
2. The first and the last line of each file ending in `.pdb` in the `molecules directory
3. The first and the last line of each file in the `molecules` directory
4. An error because of the quotes around `*.pdb`
### [Exercise] Find the Longest File With a Given Extension
Write a shell script called `longest.sh` that takes the name of a directory and a filename extension as its arguments, and prints out the name of the file with the most lines in that directory with that extension. For example:
`$ bash longest.sh /tmp/data pdb`
would print the name of the `.pdb` file in `/tmp/data` that has the most lines.
### Additional exercises
If you were quick, check out these exercises to dig a little more into details.
### [Exercise] Script Reading Comprehension
For this question, consider the `data-shell/molecules` directory once again. This contains a number of `.pdb` files in addition to any other files you may have created. Explain what a script called `example.sh` would do when run as `bash example.sh *.pdb` if it contained the following lines:
```
# Script 1
echo *.*
# Script 2
for filename in $1 $2 $3
do
cat $filename
done
# Script 3
echo [email protected]
### [Exercise] Debugging Scripts
Suppose you have saved the following script in a file called do-errors.sh in Nelle’s north-pacific-gyre/2012-07-03 directory:
```
# Calculate stats for data files.
for datafile in "$@"
do
echo $datfile
bash goostats $datafile stats-$datafile
done
```
When you run it:
`$ bash do-errors.sh NENE*[AB].txt`
the output is blank. To figure out why, re-run the script using the -x option:
`bash -x do-errors.sh NENE*[AB].txt`
What is the output showing you? Which line is responsible for the error?
## Finding Things
### [Exercise] Using `grep`
Which command would result in the following output:
```
and the presence of absence:
```
1. `grep "of" haiku.txt`
2. `grep -E "of" haiku.txt`
3. `grep -w "of" haiku.txt`
4. `grep -i "of" haiku.txt`
### [Exercise] Tracking a Species
Leah has several hundred data files saved in one directory, each of which is formatted like this:
```
2013-11-05,deer,5
2013-11-05,rabbit,22
2013-11-05,raccoon,7
2013-11-06,rabbit,19
2013-11-06,deer,2
```
She wants to write a shell script that takes a species as the first command-line argument and a directory as the second argument. The script should return one file called `species.txt` containing a list of dates and the number of that species seen on each date. For example using the data shown above, `rabbit.txt` would contain:
```
2013-11-05,22
2013-11-06,19
```
Put these commands and pipes in the right order to achieve this:
```
cut -d : -f 2
>
|
grep -w $1 -r $2
|
$1.txt
cut -d , -f 1,3
```
Hint: use `man grep` to look for how to grep text recursively in a directory and `man cut` to select more than one field in a line.
An example of such a file is provided in `data-shell/data/animal-counts/animals.txt`.
### [Exercise] Little Women
You and your friend, having just finished reading *Little Women* by Louisa May Alcott, are in an argument. Of the four sisters in the book, Jo, Meg, Beth, and Amy, your friend thinks that Jo was the most mentioned. You, however, are certain it was Amy. Luckily, you have a file `LittleWomen.txt` containing the full text of the novel (`data-shell/writing/data/LittleWomen.txt`). Using a `for` loop, how would you tabulate the number of times each of the four sisters is mentioned?
Hint: one solution might employ the commands `grep` and `wc` and a `|`, while another might utilize `grep` options. There is often more than one way to solve a programming task, so a particular solution is usually chosen based on a combination of yielding the correct result, elegance, readability, and speed.
### Additional exercises
If you were quick, check out these exercises to dig a little more into details.
### [Exercise] Matching and Subtracting
The `-v` flag to `grep` inverts pattern matching, so that only lines which do not match the pattern are printed. Given that, which of the following commands will find all files in `/data` whose names end in `s.txt` (e.g., `animals.txt` or `planets.txt`), but do not contain the word `net`? Once you have thought about your answer, you can test the commands in the `data-shell` directory.
1. `find data -name '*s.txt' | grep -v net`
2. `find data -name *s.txt | grep -v net`
3. `grep -v "temp" $(find data -name '*s.txt')`
4. None of the above.
### [Exercise] `find` Pipeline Reading Comprehension
Write a short explanatory comment for the following shell script:
```
wc -l $(find . -name '*.dat') | sort -n
```
### [Exercise] Finding Files With Different Properties
The `find` command can be given several other criteria known as “tests” to locate files with specific attributes, such as creation time, size, permissions, or ownership. Use `man find` to explore these, and then write a single command to find all files in or below the current directory that were modified by the user `ahmed` in the last 24 hours.
Hint 1: you will need to use three tests: `-type`, `-mtime`, and `-user`.
Hint 2: The value for `-mtime` will need to be negative—why?
### Key Points for Finding Things
* `find` finds files with specific properties that match patterns.
* `grep` selects lines in files that match patterns.
* `--help` is a flag supported by many bash commands, and programs that can be run from within Bash, to display more information on how to use these commands or programs.
* `man` command displays the manual page for a given command.
* `$(command)` inserts a command’s output in place.
### Additional resources
| resource | description |
| :------- | :---------- |
| https://explainshell.com | dissects any shell command you type in |
| https://tldr.sh | simplified and community-driven Shell manual pages |
| https://www.shellcheck.net | checks shell scripts for common errors |
|
github_jupyter
|
## Dependencies
```
import json, glob
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras import layers
from tensorflow.keras.models import Model
```
# Load data
```
test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')
print('Test samples: %s' % len(test))
display(test.head())
```
# Model parameters
```
input_base_path = '/kaggle/input/114roberta-base/'
with open(input_base_path + 'config.json') as json_file:
config = json.load(json_file)
config
# vocab_path = input_base_path + 'vocab.json'
# merges_path = input_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
vocab_path = base_path + 'roberta-base-vocab.json'
merges_path = base_path + 'roberta-base-merges.txt'
config['base_model_path'] = base_path + 'roberta-base-tf_model.h5'
config['config_path'] = base_path + 'roberta-base-config.json'
model_path_list = glob.glob(input_base_path + '*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep = "\n")
```
# Tokenizer
```
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
```
# Pre process
```
test['text'].fillna('', inplace=True)
test["text"] = test["text"].apply(lambda x: x.lower())
test["text"] = test["text"].apply(lambda x: x.strip())
x_test = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)
```
# Model
```
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name="base_model")
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
x = layers.Dropout(.1)(last_hidden_state)
x_start = layers.Dense(1)(x)
x_start = layers.Flatten()(x_start)
y_start = layers.Activation('softmax', name='y_start')(x_start)
x_end = layers.Dense(1)(x)
x_end = layers.Flatten()(x_end)
y_end = layers.Activation('softmax', name='y_end')(x_end)
model = Model(inputs=[input_ids, attention_mask], outputs=[y_start, y_end])
return model
```
# Make predictions
```
NUM_TEST_IMAGES = len(test)
test_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
test_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
for model_path in model_path_list:
print(model_path)
model = model_fn(config['MAX_LEN'])
model.load_weights(model_path)
test_preds = model.predict(x_test)
test_start_preds += test_preds[0]
test_end_preds += test_preds[1]
```
# Post process
```
test['start'] = test_start_preds.argmax(axis=-1)
test['end'] = test_end_preds.argmax(axis=-1)
test['text_len'] = test['text'].apply(lambda x : len(x))
test['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))
test["end"].clip(0, test["text_len"], inplace=True)
test["start"].clip(0, test["end"], inplace=True)
test['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)
test["selected_text"].fillna(test["text"], inplace=True)
```
# Visualize predictions
```
display(test.head(10))
```
# Test set predictions
```
submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')
submission['selected_text'] = test["selected_text"]
submission.to_csv('submission.csv', index=False)
submission.head(10)
```
|
github_jupyter
|
# Simple Go-To-Goal for Cerus
The following code implements a simple go-to-goal behavior for Cerus. It uses a closed feedback loop to continuously asses Cerus' state (position and heading) in the world using data from two wheel encoders. It subsequently calculates the error between a given goal location and its current pose and will attempt to minimize the error until it reaches the goal location. A P-regulator (see PID regulator) script uses the error as an input and outputs the angular velocity for the Arduino and motor controllers that drive the robot.
All models used in this program are adapted from Georgia Tech's "Control of Mobile Robots" by Dr. Magnus Egerstedt.
```
#Import useful libraries
import serial
import time
import math
import numpy as np
from traitlets import HasTraits, List
#Open a serial connection with the Arduino Mega
#Opening a serial port on the Arduino resets it, so our encoder count is also reset to 0,0
ser = serial.Serial('COM3', 115200)
#Defining our goal location. Units are metric, real-world coordinates in an X/Y coordinate system
goal_x = 1
goal_y = 0
#Create a class for our Cerus robot
class Cerus():
def __init__(self, pose_x, pose_y, pose_phi, R_wheel, N_ticks, L_track):
self.pose_x = pose_x #X Position
self.pose_y = pose_y #Y Position
self.pose_phi = pose_phi #Heading
self.R_wheel = R_wheel #wheel radius in meters
self.N_ticks = N_ticks #encoder ticks per wheel revolution
self.L_track = L_track #wheel track in meters
#Create a Cerus instance and initialize it to a 0,0,0 world position and with some physical dimensions
cerus = Cerus(0,0,0,0.03,900,0.23)
```
We'll use the Traitlets library to implement an observer pattern that will recalculate the pose of the robot every time an update to the encoder values is detected and sent to the Jetson nano by the Arduino.
```
#Create an encoder class with traits
class Encoders(HasTraits):
encoderValues = List() #We store the left and right encoder value in a list
def __init__(self, encoderValues, deltaTicks):
self.encoderValues = encoderValues
self.deltaTicks = deltaTicks
#Create an encoder instance
encoders = Encoders([0,0], [0,0])
#Create a function that is triggered when a change to encoders is detected
def monitorEncoders(change):
if change['new']:
oldVals = np.array(change['old'])
newVals = np.array(change['new'])
deltaTicks = newVals - oldVals
#print("Old values: ", oldVals)
#print("New values: ", newVals)
#print("Delta values: ", deltaTicks)
calculatePose(deltaTicks)
encoders.observe(monitorEncoders, names = "encoderValues")
```
The functions below are helpers and will be called through our main loop.
```
#Create a move function that sends move commands to the Arduino
def move(linearVelocity, angularVelocity):
command = f"<{linearVelocity},{angularVelocity}>"
ser.write(str.encode(command))
#Create a function that calculates an updated pose of Cerus every time it is called
def calculatePose(deltaTicks):
#Calculate the centerline distance moved
distanceLeft = 2 * math.pi * cerus.R_wheel * (deltaTicks[0] / cerus.N_ticks)
distanceRight = 2 * math.pi * cerus.R_wheel * (deltaTicks[1] / cerus.N_ticks)
distanceCenter = (distanceLeft + distanceRight) / 2
#Update the position and heading
cerus.pose_x = round((cerus.pose_x + distanceCenter * math.cos(cerus.pose_phi)), 4)
cerus.pose_y = round((cerus.pose_y + distanceCenter * math.sin(cerus.pose_phi)), 4)
cerus.pose_phi = round((cerus.pose_phi + ((distanceRight - distanceLeft) / cerus.L_track)), 4)
print(f"The new position is {cerus.pose_x}, {cerus.pose_y} and the new heading is {cerus.pose_phi}.")
#Calculate the error between Cerus' heading and the goal point
def calculateError():
phi_desired = math.atan((goal_y - cerus.pose_y)/(goal_x - cerus.pose_x))
temp = phi_desired - cerus.pose_phi
error_heading = round((math.atan2(math.sin(temp), math.cos(temp))), 4) #ensure that error is within [-pi, pi]
error_x = round((goal_x - cerus.pose_x), 4)
error_y = round((goal_y - cerus.pose_y), 4)
#print("The heading error is: ", error_heading)
#print("The X error is: ", error_x)
#print("The Y error is: ", error_y)
return error_x, error_y, error_heading
atGoal = False
constVel = 0.2
K = 1 #constant for our P-regulator below
#Functions to read and format encoder data received from the Serial port
def formatData(data):
delimiter = "x"
leftVal = ""
rightVal = ""
for i in range(len(data)):
if data[i] == ",":
delimiter = ","
elif delimiter != "," and data[i].isdigit():
leftVal += data[i]
elif delimiter == "," and data[i].isdigit():
rightVal += data[i]
leftVal, rightVal = int(leftVal), int(rightVal)
encoders.encoderValues = [leftVal, rightVal]
print("Encoders: ", encoders.encoderValues)
def handleSerial():
#ser.readline() waits for the next line of encoder data, which is sent by Arduino every 50 ms
if ser.inWaiting():
#Get the serial data and format it
temp = ser.readline()
data = temp.decode()
formatData(data)
#Calculate the current pose to goal error
error_x, error_y, error_heading = calculateError()
print(f"Error X: {error_x}, Error Y: {error_y}")
#If we're within 5 cm of the goal
if error_x <= 0.05:# and error_y <= 0.05:
print("Goal reached!")
move(0.0,0.0)
time.sleep(0.1)
atGoal = True
#Otherwise keep driving
else:
omega = - (K * error_heading)
handleSerial()
move(constVel, 0.0)
print("Moving at angular speed: ", omega)
def moveRobot():
#The Arduino sends data every 50ms, we first check if data is in the buffer
if ser.inWaiting():
#Get the serial data and format it if data is in the buffer
temp = ser.readline()
data = temp.decode()
formatData(data)
#Calculate the current pose to goal error
error_x, error_y, error_heading = calculateError()
print(f"Error X: {error_x}, Error Y: {error_y}")
#If we're within 5 cm of the goal
if error_x <= 0.05:# and error_y <= 0.05:
print("Goal reached!")
move(0.0,0.0)
time.sleep(0.1)
atGoal = True
#Otherwise keep driving
else:
omega = - (K * error_heading)
handleSerial()
move(constVel, 0.0)
print("Moving at angular speed: ", omega)
```
This is the main part for our program that will loop over and over until Cerus has reached its goal. For our simple go-to-goal behavior, we will drive the robot at a constant speed and only adjust our heading so that we reach the goal location.
__WARNING: This will move the robot!__
```
while not atGoal:
try:
moveRobot()
except(KeyboardInterrupt):
print("Program interrupted by user!")
move(0.0,0.0) #Stop motors
break
"Loop exited..."
move(0.0,0.0) #Stop motors
#Close the serial connection when done
ser.close()
atGoal = False
constVel = 0.2
K = 1 #constant for our P-regulator below
while not atGoal:
try:
#Calculate the current pose to goal error
error_x, error_y, error_heading = calculateError()
print(f"Error X: {error_x}, Error Y: {error_y}")
#If we're within 5 cm of the goal
if error_x <= 0.05 and error_y <= 0.05:
print("Goal reached!")
move(0.0,0.0)
time.sleep(0.1)
atGoal = True
#Otherwise keep driving
else:
omega = - (K * error_heading)
handleSerial()
move(constVel, 0.0)
print("Moving at angular speed: ", omega)
except(KeyboardInterrupt):
print("Program interrupted by user!")
move(0.0,0.0) #Stop motors
break
"Loop exited..."
move(0.0,0.0) #Stop motors
```
|
github_jupyter
|
# Chatbot Tutorial
- https://pytorch.org/tutorials/beginner/chatbot_tutorial.html
```
import torch
from torch.jit import script, trace
import torch.nn as nn
from torch import optim
import torch.nn.functional as F
import csv
import random
import re
import os
import unicodedata
import codecs
from io import open
import itertools
import math
USE_CUDA = torch.cuda.is_available()
device = torch.device('cuda' if USE_CUDA else 'cpu')
```
## データの前処理
```
corpus_name = 'cornell_movie_dialogs_corpus'
corpus = os.path.join('data', corpus_name)
def printLines(file, n=10):
with open(file, 'rb') as datafile:
lines = datafile.readlines()
for line in lines[:n]:
print(line)
printLines(os.path.join(corpus, 'movie_lines.txt'))
# Splits each line of the file into a dictionary of fields
def loadLines(fileName, fields):
lines = {}
with open(fileName, 'r', encoding='iso-8859-1') as f:
for line in f:
values = line.split(' +++$+++ ')
# Extract fields
lineObj = {}
for i, field in enumerate(fields):
lineObj[field] = values[i]
lines[lineObj['lineID']] = lineObj
return lines
MOVIE_LINES_FIELDS = ['lineID', 'characterID', 'movieID', 'character', 'text']
lines = loadLines(os.path.join(corpus, 'movie_lines.txt'), MOVIE_LINES_FIELDS)
lines['L1045']
# Groups fields of lines from loadLines() into conversations based on movie_conversations.txt
def loadConversations(fileName, lines, fields):
conversations = []
with open(fileName, 'r', encoding='iso-8859-1') as f:
for line in f:
values = line.split(' +++$+++ ')
# Extract fields
convObj = {}
for i, field in enumerate(fields):
convObj[field] = values[i]
# Convert string to list
utterance_id_pattern = re.compile('L[0-9]+')
lineIds = utterance_id_pattern.findall(convObj['utteranceIDs'])
# Reassemble lines
convObj['lines'] = []
for lineId in lineIds:
convObj['lines'].append(lines[lineId])
conversations.append(convObj)
return conversations
MOVIE_CONVERSATIONS_FIELDS = ['character1ID', 'character2ID', 'movieID', 'utteranceIDs']
conversations = loadConversations(os.path.join(corpus, 'movie_conversations.txt'), lines, MOVIE_CONVERSATIONS_FIELDS)
# utteranceIDsの会話系列IDがlinesに展開されていることがわかる!
conversations[0]
# Extracts pairs of sentences from conversations
def extractSentencePairs(conversations):
qa_pairs = []
for conversation in conversations:
# Iterate over all the lines of the conversation
for i in range(len(conversation['lines']) - 1): # 最後の会話は回答がないので無視する
inputLine = conversation['lines'][i]['text'].strip()
targetLine = conversation['lines'][i + 1]['text'].strip()
if inputLine and targetLine:
qa_pairs.append([inputLine, targetLine])
return qa_pairs
datafile = os.path.join(corpus, 'formatted_movie_lines.txt')
delimiter = '\t'
delimiter = str(codecs.decode(delimiter, 'unicode_escape'))
with open(datafile, 'w', encoding='utf-8') as outputfile:
writer = csv.writer(outputfile, delimiter=delimiter, lineterminator='\n')
for pair in extractSentencePairs(conversations):
writer.writerow(pair)
printLines(datafile)
```
## Vocabularyの構築
```
# Default word tokens
PAD_token = 0 # Used for padding short sentences
SOS_token = 1 # Start-of-sentence token
EOS_token = 2 # End-of-sentence token
class Voc:
def __init__(self, name):
self.name = name
self.trimmed = False
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: 'PAD', SOS_token: 'SOS', EOS_token: 'EOS'}
self.num_words = 3 # SOS, EOS, PAD
def addSentence(self, sentence):
for word in sentence.split(' '):
self.addWord(word)
def addWord(self, word):
if word not in self.word2index:
self.word2index[word] = self.num_words
self.word2count[word] = 1
self.index2word[self.num_words] = word
self.num_words += 1
else:
self.word2count[word] += 1
# Remove words below a certain count threshold
def trim(self, min_count):
if self.trimmed:
return
self.trimmed = True
keep_words = []
for k, v in self.word2count.items():
if v >= min_count:
keep_words.append(k)
print('keep_words {} / {} = {:.4f}'.format(len(keep_words), len(self.word2index), len(keep_words) / len(self.word2index)))
# Reinitialize dictionaries
self.word2index = {}
self.word2count = {}
self.index2word = {PAD_token: 'PAD', SOS_token: 'SOS', EOS_token: 'EOS'}
self.num_words = 3
for word in keep_words:
self.addWord(word)
MAX_LENGTH = 10 # Maximum sentence length to consider
# Turn a Unicode string to plain ASCII
def unicodeToAscii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s) if unicodedata.category(c) != 'Mn')
# Lowercase, trim, and remove non-letter characters
def normalizeString(s):
s = unicodeToAscii(s.lower().strip())
s = re.sub(r"([.!?])", r" \1", s)
s = re.sub(r"[^a-zA-Z.!?]+", r" ", s)
s = re.sub(r"\s+", r" ", s).strip()
return s
# Read query/response paiers and return a voc object
def readVocs(datafile, corpus_name):
# Read the file and split into lines
lines = open(datafile, encoding='utf-8').read().strip().split('\n')
# Split every line into pairs and normalize
pairs = [[normalizeString(s) for s in l.split('\t')] for l in lines]
voc = Voc(corpus_name)
return voc, pairs
# Returns True iff both sentences in a pair p are under the MAX_LENGTH threshold
def filterPair(p):
# Input sequences need to preserve the last word for EOS token
return len(p[0].split(' ')) < MAX_LENGTH and len(p[1].split(' ')) < MAX_LENGTH
# Filter pairs using filterPair condition
def filterPairs(pairs):
return [pair for pair in pairs if filterPair(pair)]
# Using the functions defined above, return a populated voc object and pairs list
# save_dirが使われていない
def loadPrepareData(corpus, corpus_name, datafile, save_dir):
print('Start preparing training data ...')
voc, pairs = readVocs(datafile, corpus_name)
print('Read {!s} sentence pairs'.format(len(pairs)))
pairs = filterPairs(pairs)
print('Trimmed to {!s} sentence pairs'.format(len(pairs)))
print('Counting words...')
for pair in pairs:
voc.addSentence(pair[0])
voc.addSentence(pair[1])
print('Counted words:', voc.num_words)
return voc, pairs
# Load/Assemble voc and pairs
save_dir = os.path.join('data', 'save')
voc, pairs = loadPrepareData(corpus, corpus_name, datafile, save_dir)
# Print some pairs to validate
print('\npairs:')
for pair in pairs[:10]:
print(pair)
MIN_COUNT = 3 # Minimum word count threshold for trimming
def trimRareWords(voc, pairs, MIN_COUNT):
# Trim words used unser the MIN_COUNT from the voc
voc.trim(MIN_COUNT)
# Filter out pairs with trimmed words
keep_pairs = []
for pair in pairs:
input_sentence = pair[0]
output_sentence = pair[1]
keep_input = True
keep_output = True
# Check input sentence
for word in input_sentence.split(' '):
if word not in voc.word2index:
keep_input = False
break
# Check output sentence
for word in output_sentence.split(' '):
if word not in voc.word2index:
keep_output = False
break
# Only keep pairs that do not contain trimmed word(s) in their input or output sentence
if keep_input and keep_output:
keep_pairs.append(pair)
print('Trimmed from {} pairs to {}, {:.4f} of total'.format(len(pairs), len(keep_pairs), len(keep_pairs) / len(pairs)))
return keep_pairs
# Trim voc and pairs
pairs = trimRareWords(voc, pairs, MIN_COUNT)
```
## Minibatchの構成
```
def indexesFromSentence(voc, sentence):
return [voc.word2index[word] for word in sentence.split(' ')] + [EOS_token]
def zeroPadding(l, fillvalue=PAD_token):
# ここで (batch_size, max_length) => (max_length, batch_size) に転置している
return list(itertools.zip_longest(*l, fillvalue=fillvalue))
def binaryMatrix(l, value=PAD_token):
m = []
for i, seq in enumerate(l):
m.append([])
for token in seq:
if token == PAD_token:
m[i].append(0)
else:
m[i].append(1)
return m
# Returns padded input sentence tensor and lengths
def inputVar(l, voc):
indexes_batch = [indexesFromSentence(voc, sentence) for sentence in l]
lengths = torch.tensor([len(indexes) for indexes in indexes_batch])
padList = zeroPadding(indexes_batch)
padVar = torch.LongTensor(padList)
return padVar, lengths
# Returns padded target sequence tneosr, padding mask, and max target lengths
# maskは出力と同じサイズのテンソルでPADが入ってるところが0でそれ以外は1
def outputVar(l, voc):
indexes_batch = [indexesFromSentence(voc, sentence) for sentence in l]
max_target_len = max([len(indexes) for indexes in indexes_batch])
padList = zeroPadding(indexes_batch)
mask = binaryMatrix(padList)
mask = torch.ByteTensor(mask)
padVar = torch.LongTensor(padList)
return padVar, mask, max_target_len
# Returns all items for a given batch of pairs
def batch2TrainData(voc, pair_batch):
# 入力文章の長い順にソートする
pair_batch.sort(key=lambda x: len(x[0].split(' ')), reverse=True)
input_batch, output_batch = [], []
for pair in pair_batch:
input_batch.append(pair[0])
output_batch.append(pair[1])
inp, lengths = inputVar(input_batch, voc)
output, mask, max_target_len = outputVar(output_batch, voc)
return inp, lengths, output, mask, max_target_len
# Example for validation
small_batch_size = 5
batches = batch2TrainData(voc, [random.choice(pairs) for _ in range(small_batch_size)])
input_variable, lengths, target_variable, mask, max_target_len = batches
print('input_variable:', input_variable)
print('lengths:', lengths)
print('target_variable:', target_variable)
print('mask:', mask)
print('max_target_len:', max_target_len)
```
## Seq2Seq Model
```
class EncoderRNN(nn.Module):
def __init__(self, hidden_size, embedding, n_layers=1, dropout=0):
super(EncoderRNN, self).__init__()
self.n_layers = n_layers
self.hidden_size = hidden_size
self.embedding = embedding
self.gru = nn.GRU(hidden_size, hidden_size, n_layers, dropout=(0 if n_layers == 1 else dropout), bidirectional=True)
def forward(self, input_seq, input_lengths, hidden=None):
# Convert word indexes to embedding
embedded = self.embedding(input_seq)
# Pack padded batch of sequences for RNN module
packed = nn.utils.rnn.pack_padded_sequence(embedded, input_lengths)
# Forward pass through GRU
# output of shape (seq_len, batch, num_directions * hidden_size)
# h_n of shape (num_layers * num_directions, batch, hidden_size)
outputs, hidden = self.gru(packed, hidden)
# Unpack padding
outputs, _ = nn.utils.rnn.pad_packed_sequence(outputs)
# Sum bidirectional GRU outputs
# bidirectionalの場合、outputsのhidden_sizeは2倍の長さで出てくる
outputs = outputs[:, :, :self.hidden_size] + outputs[:, :, self.hidden_size:]
# Return output and final hidden state
return outputs, hidden
```
## Attention
```
class Attn(nn.Module):
def __init__(self, method, hidden_size):
super(Attn, self).__init__()
self.method = method
if self.method not in ['dot', 'general', 'concat']:
raise ValueError(self.method, 'is not an appropriate attention method.')
self.hidden_size = hidden_size
if self.method == 'general':
self.attn = nn.Linear(hidden_size, hidden_size)
elif self.method == 'concat':
self.attn = nn.Linear(hidden_size * 2, hidden_size)
self.v = nn.Parameter(torch.FloatTensor(hidden_size))
def dot_score(self, hidden, encoder_output):
return torch.sum(hidden * encoder_output, dim=2)
def general_score(self, hidden, encoder_output):
energy = self.attn(encoder_output)
return torch.sum(hidden * energy, dim=2)
def concat_score(self, hidden, encoder_output):
energy = self.attn(torch.cat((hidden.expand(encoder_output.size(0), -1, -1), encoder_output), 2)).tanh()
return torch.sum(self.v * energy, dim=2)
def forward(self, hidden, encoder_outputs):
# Calculate the attention weights (energies) based on the given method
if self.method == 'general':
attn_energies = self.general_score(hidden, encoder_outputs)
elif self.method == 'concat':
attn_energies = self.concat_score(hidden, encoder_outputs)
elif self.method == 'dot':
attn_energies = self.dot_score(hidden, encoder_outputs)
# Transpose max_length and batch_size dimensions
attn_energies = attn_energies.t()
# Return the softmax normalized probability scores (with added dimension)
return F.softmax(attn_energies, dim=1).unsqueeze(1)
```
## Decoder
```
class LuongAttnDecoderRNN(nn.Module):
def __init__(self, attn_model, embedding, hidden_size, output_size, n_layers=1, dropout=0.1):
super(LuongAttnDecoderRNN, self).__init__()
self.attn_model = attn_model
self.hidden_size = hidden_size
self.output_size = output_size
self.n_layers = n_layers
self.dropout = dropout
# Define layers
self.embedding = embedding
self.embedding_dropout = nn.Dropout(dropout)
self.gru = nn.GRU(hidden_size, hidden_size, n_layers, dropout=(0 if n_layers == 1 else dropout))
self.concat = nn.Linear(hidden_size * 2, hidden_size)
self.out = nn.Linear(hidden_size, output_size)
self.attn = Attn(attn_model, hidden_size)
def forward(self, input_step, last_hidden, encoder_outputs):
# Decoderは各タイムステップごとに実行される
embedded = self.embedding(input_step)
embedded = self.embedding_dropout(embedded)
# Forward through unidirectional GRU
rnn_output, hidden = self.gru(embedded, last_hidden)
# Calculate attention weights from the current GRU output
attn_weights = self.attn(rnn_output, encoder_outputs)
# Multiply attention weights to encoder outputs to get new weighted sum context vector
context = attn_weights.bmm(encoder_outputs.transpose(0, 1))
# Concatenate weighted context vector and GRU output using Luong eq. 5
rnn_output = rnn_output.squeeze(0)
context = context.squeeze(1)
concat_input = torch.cat((rnn_output, context), 1)
concat_output = torch.tanh(self.concat(concat_input))
# Predict next word using Luong eq.6
output = self.out(concat_output)
output = F.softmax(output, dim=1)
# Return output and final hidden state
return output, hidden
```
## Masked loss
```
def maskNLLLoss(inp, target, mask):
nTotal = mask.sum()
crossEntropy = -torch.log(torch.gather(inp, 1, target.view(-1, 1)).squeeze(1))
loss = crossEntropy.masked_select(mask).mean()
loss = loss.to(device)
return loss, nTotal.item()
```
## Training
```
def train(input_variable, lengths, target_variable, mask, max_target_len, encoder, decoder, embedding,
encoder_optimizer, decoder_optimizer, batch_size, clip, max_length=MAX_LENGTH):
pass
```
|
github_jupyter
|
# Work with Data
Data is the foundation on which machine learning models are built. Managing data centrally in the cloud, and making it accessible to teams of data scientists who are running experiments and training models on multiple workstations and compute targets is an important part of any professional data science solution.
In this notebook, you'll explore two Azure Machine Learning objects for working with data: *datastores*, and *datasets*.
## Connect to your workspace
To get started, connect to your workspace.
> **Note**: If you haven't already established an authenticated session with your Azure subscription, you'll be prompted to authenticate by clicking a link, entering an authentication code, and signing into Azure.
```
import azureml.core
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
```
## Work with datastores
In Azure ML, *datastores* are references to storage locations, such as Azure Storage blob containers. Every workspace has a default datastore - usually the Azure storage blob container that was created with the workspace. If you need to work with data that is stored in different locations, you can add custom datastores to your workspace and set any of them to be the default.
### View datastores
Run the following code to determine the datastores in your workspace:
```
# Get the default datastore
default_ds = ws.get_default_datastore()
# Enumerate all datastores, indicating which is the default
for ds_name in ws.datastores:
print(ds_name, "- Default =", ds_name == default_ds.name)
```
You can also view and manage datastores in your workspace on the **Datastores** page for your workspace in [Azure Machine Learning studio](https://ml.azure.com).
### Upload data to a datastore
Now that you have determined the available datastores, you can upload files from your local file system to a datastore so that it will be accessible to experiments running in the workspace, regardless of where the experiment script is actually being run.
```
default_ds.upload_files(files=['./data/diabetes.csv', './data/diabetes2.csv'], # Upload the diabetes csv files in /data
target_path='diabetes-data/', # Put it in a folder path in the datastore
overwrite=True, # Replace existing files of the same name
show_progress=True)
```
## Work with datasets
Azure Machine Learning provides an abstraction for data in the form of *datasets*. A dataset is a versioned reference to a specific set of data that you may want to use in an experiment. Datasets can be *tabular* or *file*-based.
### Create a tabular dataset
Let's create a dataset from the diabetes data you uploaded to the datastore, and view the first 20 records. In this case, the data is in a structured format in a CSV file, so we'll use a *tabular* dataset.
```
from azureml.core import Dataset
# Get the default datastore
default_ds = ws.get_default_datastore()
#Create a tabular dataset from the path on the datastore (this may take a short while)
tab_data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, 'diabetes-data/*.csv'))
# Display the first 20 rows as a Pandas dataframe
tab_data_set.take(20).to_pandas_dataframe()
```
As you can see in the code above, it's easy to convert a tabular dataset to a Pandas dataframe, enabling you to work with the data using common python techniques.
### Create a file Dataset
The dataset you created is a *tabular* dataset that can be read as a dataframe containing all of the data in the structured files that are included in the dataset definition. This works well for tabular data, but in some machine learning scenarios you might need to work with data that is unstructured; or you may simply want to handle reading the data from files in your own code. To accomplish this, you can use a *file* dataset, which creates a list of file paths in a virtual mount point, which you can use to read the data in the files.
```
#Create a file dataset from the path on the datastore (this may take a short while)
file_data_set = Dataset.File.from_files(path=(default_ds, 'diabetes-data/*.csv'))
# Get the files in the dataset
for file_path in file_data_set.to_path():
print(file_path)
```
### Register datasets
Now that you have created datasets that reference the diabetes data, you can register them to make them easily accessible to any experiment being run in the workspace.
We'll register the tabular dataset as **diabetes dataset**, and the file dataset as **diabetes files**.
```
# Register the tabular dataset
try:
tab_data_set = tab_data_set.register(workspace=ws,
name='diabetes dataset',
description='diabetes data',
tags = {'format':'CSV'},
create_new_version=True)
except Exception as ex:
print(ex)
# Register the file dataset
try:
file_data_set = file_data_set.register(workspace=ws,
name='diabetes file dataset',
description='diabetes files',
tags = {'format':'CSV'},
create_new_version=True)
except Exception as ex:
print(ex)
print('Datasets registered')
```
You can view and manage datasets on the **Datasets** page for your workspace in [Azure Machine Learning studio](https://ml.azure.com). You can also get a list of datasets from the workspace object:
```
print("Datasets:")
for dataset_name in list(ws.datasets.keys()):
dataset = Dataset.get_by_name(ws, dataset_name)
print("\t", dataset.name, 'version', dataset.version)
```
The ability to version datasets enables you to redefine datasets without breaking existing experiments or pipelines that rely on previous definitions. By default, the latest version of a named dataset is returned, but you can retrieve a specific version of a dataset by specifying the version number, like this:
```python
dataset_v1 = Dataset.get_by_name(ws, 'diabetes dataset', version = 1)
```
### Train a model from a tabular dataset
Now that you have datasets, you're ready to start training models from them. You can pass datasets to scripts as *inputs* in the estimator being used to run the script.
Run the following two code cells to create:
1. A folder named **diabetes_training_from_tab_dataset**
2. A script that trains a classification model by using a tabular dataset that is passed to it as an argument.
```
import os
# Create a folder for the experiment files
experiment_folder = 'diabetes_training_from_tab_dataset'
os.makedirs(experiment_folder, exist_ok=True)
print(experiment_folder, 'folder created')
%%writefile $experiment_folder/diabetes_training.py
# Import libraries
import os
import argparse
from azureml.core import Run, Dataset
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# Get the script arguments (regularization rate and training dataset ID)
parser = argparse.ArgumentParser()
parser.add_argument('--regularization', type=float, dest='reg_rate', default=0.01, help='regularization rate')
parser.add_argument("--input-data", type=str, dest='training_dataset_id', help='training dataset')
args = parser.parse_args()
# Set regularization hyperparameter (passed as an argument to the script)
reg = args.reg_rate
# Get the experiment run context
run = Run.get_context()
# Get the training dataset
print("Loading Data...")
diabetes = run.input_datasets['training_data'].to_pandas_dataframe()
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a logistic regression model
print('Training a logistic regression model with regularization rate of', reg)
run.log('Regularization Rate', np.float(reg))
model = LogisticRegression(C=1/reg, solver="liblinear").fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
os.makedirs('outputs', exist_ok=True)
# note file saved in the outputs folder is automatically uploaded into experiment record
joblib.dump(value=model, filename='outputs/diabetes_model.pkl')
run.complete()
```
> **Note**: In the script, the dataset is passed as a parameter (or argument). In the case of a tabular dataset, this argument will contain the ID of the registered dataset; so you could write code in the script to get the experiment's workspace from the run context, and then get the dataset using its ID; like this:
>
> ```
> run = Run.get_context()
> ws = run.experiment.workspace
> dataset = Dataset.get_by_id(ws, id=args.training_dataset_id)
> diabetes = dataset.to_pandas_dataframe()
> ```
>
> However, Azure Machine Learning runs automatically identify arguments that reference named datasets and add them to the run's **input_datasets** collection, so you can also retrieve the dataset from this collection by specifying its "friendly name" (which as you'll see shortly, is specified in the argument definition in the script run configuration for the experiment). This is the approach taken in the script above.
Now you can run a script as an experiment, defining an argument for the training dataset, which is read by the script.
> **Note**: The **Dataset** class depends on some components in the **azureml-dataprep** package, which includes optional support for **pandas** that is used by the **to_pandas_dataframe()** method. So you need to include this package in the environment where the training experiment will be run.
```
from azureml.core import Experiment, ScriptRunConfig, Environment
from azureml.core.conda_dependencies import CondaDependencies
from azureml.widgets import RunDetails
# Create a Python environment for the experiment
sklearn_env = Environment("sklearn-env")
# Ensure the required packages are installed (we need scikit-learn, Azure ML defaults, and Azure ML dataprep)
packages = CondaDependencies.create(conda_packages=['scikit-learn','pip'],
pip_packages=['azureml-defaults','azureml-dataprep[pandas]'])
sklearn_env.python.conda_dependencies = packages
# Get the training dataset
diabetes_ds = ws.datasets.get("diabetes dataset")
# Create a script config
script_config = ScriptRunConfig(source_directory=experiment_folder,
script='diabetes_training.py',
arguments = ['--regularization', 0.1, # Regularizaton rate parameter
'--input-data', diabetes_ds.as_named_input('training_data')], # Reference to dataset
environment=sklearn_env)
# submit the experiment
experiment_name = 'mslearn-train-diabetes'
experiment = Experiment(workspace=ws, name=experiment_name)
run = experiment.submit(config=script_config)
RunDetails(run).show()
run.wait_for_completion()
```
> **Note:** The **--input-data** argument passes the dataset as a *named input* that includes a *friendly name* for the dataset, which is used by the script to read it from the **input_datasets** collection in the experiment run. The string value in the **--input-data** argument is actually the registered dataset's ID. As an alternative approach, you could simply pass `diabetes_ds.id`, in which case the script can access the dataset ID from the script arguments and use it to get the dataset from the workspace, but not from the **input_datasets** collection.
The first time the experiment is run, it may take some time to set up the Python environment - subsequent runs will be quicker.
When the experiment has completed, in the widget, view the **azureml-logs/70_driver_log.txt** output log and the metrics generated by the run.
### Register the trained model
As with any training experiment, you can retrieve the trained model and register it in your Azure Machine Learning workspace.
```
from azureml.core import Model
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Tabular dataset'}, properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
```
### Train a model from a file dataset
You've seen how to train a model using training data in a *tabular* dataset; but what about a *file* dataset?
When you're using a file dataset, the dataset argument passed to the script represents a mount point containing file paths. How you read the data from these files depends on the kind of data in the files and what you want to do with it. In the case of the diabetes CSV files, you can use the Python **glob** module to create a list of files in the virtual mount point defined by the dataset, and read them all into Pandas dataframes that are concatenated into a single dataframe.
Run the following two code cells to create:
1. A folder named **diabetes_training_from_file_dataset**
2. A script that trains a classification model by using a file dataset that is passed to is as an *input*.
```
import os
# Create a folder for the experiment files
experiment_folder = 'diabetes_training_from_file_dataset'
os.makedirs(experiment_folder, exist_ok=True)
print(experiment_folder, 'folder created')
%%writefile $experiment_folder/diabetes_training.py
# Import libraries
import os
import argparse
from azureml.core import Dataset, Run
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
import glob
# Get script arguments (rgularization rate and file dataset mount point)
parser = argparse.ArgumentParser()
parser.add_argument('--regularization', type=float, dest='reg_rate', default=0.01, help='regularization rate')
parser.add_argument('--input-data', type=str, dest='dataset_folder', help='data mount point')
args = parser.parse_args()
# Set regularization hyperparameter (passed as an argument to the script)
reg = args.reg_rate
# Get the experiment run context
run = Run.get_context()
# load the diabetes dataset
print("Loading Data...")
data_path = run.input_datasets['training_files'] # Get the training data path from the input
# (You could also just use args.dataset_folder if you don't want to rely on a hard-coded friendly name)
# Read the files
all_files = glob.glob(data_path + "/*.csv")
diabetes = pd.concat((pd.read_csv(f) for f in all_files), sort=False)
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a logistic regression model
print('Training a logistic regression model with regularization rate of', reg)
run.log('Regularization Rate', np.float(reg))
model = LogisticRegression(C=1/reg, solver="liblinear").fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
os.makedirs('outputs', exist_ok=True)
# note file saved in the outputs folder is automatically uploaded into experiment record
joblib.dump(value=model, filename='outputs/diabetes_model.pkl')
run.complete()
```
Just as with tabular datasets, you can retrieve a file dataset from the **input_datasets** collection by using its friendly name. You can also retrieve it from the script argument, which in the case of a file dataset contains a mount path to the files (rather than the dataset ID passed for a tabular dataset).
Next we need to change the way we pass the dataset to the script - it needs to define a path from which the script can read the files. You can use either the **as_download** or **as_mount** method to do this. Using **as_download** causes the files in the file dataset to be downloaded to a temporary location on the compute where the script is being run, while **as_mount** creates a mount point from which the files can be streamed directly from the datasetore.
You can combine the access method with the **as_named_input** method to include the dataset in the **input_datasets** collection in the experiment run (if you omit this, for example by setting the argument to `diabetes_ds.as_mount()`, the script will be able to access the dataset mount point from the script arguments, but not from the **input_datasets** collection).
```
from azureml.core import Experiment
from azureml.widgets import RunDetails
# Get the training dataset
diabetes_ds = ws.datasets.get("diabetes file dataset")
# Create a script config
script_config = ScriptRunConfig(source_directory=experiment_folder,
script='diabetes_training.py',
arguments = ['--regularization', 0.1, # Regularizaton rate parameter
'--input-data', diabetes_ds.as_named_input('training_files').as_download()], # Reference to dataset location
environment=sklearn_env) # Use the environment created previously
# submit the experiment
experiment_name = 'mslearn-train-diabetes'
experiment = Experiment(workspace=ws, name=experiment_name)
run = experiment.submit(config=script_config)
RunDetails(run).show()
run.wait_for_completion()
```
When the experiment has completed, in the widget, view the **azureml-logs/70_driver_log.txt** output log to verify that the files in the file dataset were downloaded to a temporary folder to enable the script to read the files.
### Register the trained model
Once again, you can register the model that was trained by the experiment.
```
from azureml.core import Model
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'File dataset'}, properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
```
> **More Information**: For more information about training with datasets, see [Training with Datasets](https://docs.microsoft.com/azure/machine-learning/how-to-train-with-datasets) in the Azure ML documentation.
|
github_jupyter
|
# Part 5: Competing Journals Analysis
In this notebook we are going to
* Load the researchers impact metrics data previously extracted (see parts 1-2-3)
* Get the full publications history for these researchers
* Use this new publications dataset to determine which are the most frequent journals the researchers have also published in
* Build some visualizations in order to have a quick overview of the results
## Prerequisites: Installing the Dimensions Library and Logging in
```
# @markdown # Get the API library and login
# @markdown Click the 'play' button on the left (or shift+enter) after entering your API credentials
username = "" #@param {type: "string"}
password = "" #@param {type: "string"}
endpoint = "https://app.dimensions.ai" #@param {type: "string"}
!pip install dimcli plotly tqdm -U --quiet
import dimcli
from dimcli.shortcuts import *
dimcli.login(username, password, endpoint)
dsl = dimcli.Dsl()
#
# load common libraries
import time
import sys
import os
import json
import pandas as pd
from pandas.io.json import json_normalize
from tqdm.notebook import tqdm as progress
#
# charts libs
# import plotly_express as px
import plotly.express as px
if not 'google.colab' in sys.modules:
# make js dependecies local / needed by html exports
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
#
# create output data folder
if not(os.path.exists("data")):
os.mkdir("data")
```
## Competing Journals
From our researchers master list, we now want to extract the following:
* full list of publications for a 5 year period
* full list of journals with counts of how many publications per journal
This new dataset will let us draw up some conclusions re. which are the competing journals of the one we selected at the beginning.
### First let's reload the data obtained in previous steps
```
#
researchers = pd.read_csv("data/2.researchers_impact_metrics.csv")
#
print("Total researchers:", len(researchers))
researchers.head(5)
```
### What the query looks like
The approach we're taking consists in pulling all publications data, so that we can count journals as a second step.
This approach may take some time (as we're potentially retrieving a lot of publications data), but it will lead to precise results.
The query template to use looks like this (for a couple of researchers only):
```
%%dsldf
search publications where researchers.id in ["ur.01277776417.51", "ur.0637651205.48"]
and year >= 2015 and journal is not empty
and journal.id != "jour.1103138"
return publications[id+journal] limit 10
```
## Extracting all publications/journals information
This part may take some time to run (depending on how many years back one wants to go) so you may want to get a coffee while you wait..
```
#
journal_id = "jour.1103138" # Nature genetics
start_year = 2018
# our list of researchers
llist = list(researchers['researcher_id'])
#
# the query
q2 = """search publications
where researchers.id in {}
and year >= {} and journal is not empty and journal.id != "{}"
return publications[id+journal+year]"""
VERBOSE = False
RESEARCHER_ITERATOR_NO = 400
pubs = pd.DataFrame
for chunk in progress(list(chunks_of(llist, RESEARCHER_ITERATOR_NO))):
# get all pubs
query = q2.format(json.dumps(chunk), start_year, journal_id)
res = dsl.query_iterative(query, verbose=VERBOSE)
if pubs.empty:
# first time, init the dataframe
pubs = res.as_dataframe()
else:
pubs.append(res.as_dataframe())
# remove duplicate publications, if they have the same PUB_ID
pubs = pubs.drop_duplicates(subset="id")
# save
pubs.to_csv("data/5.journals-via-publications-RAW.csv", index=False)
# preview the data
pubs
```
Now we can create a journals-only dataset that includes counts per year, and grant total.
```
journals = pubs.copy()
# drop pub_id column
journals = journals.drop(['id'], axis=1)
#
# add total column
journals['total'] = journals.groupby('journal.id')['journal.id'].transform('count')
journals['total_year'] = journals.groupby(['journal.id', 'year'])['journal.id'].transform('count')
#
# remove multiple counts for same journal
journals = journals.drop_duplicates()
journals.reset_index(drop=True)
#
# sort by total count
journals = journals.sort_values('total', ascending=False)
# #
# # save
journals.to_csv("data/5.journals-via-publications.csv", index=False)
print("======\nDone")
# download the data
if COLAB_ENV:
files.download("data/5.journals-via-publications.csv")
#preview the data
journals.head(10)
```
# Visualizations
```
threshold = 100
temp = journals.sort_values("total", ascending=False)[:threshold]
px.bar(journals[:threshold],
x="journal.title", y="total_year",
color="year",
hover_name="journal.title",
hover_data=['journal.id', 'journal.title', 'total' ],
title=f"Top {threshold} competitors for {journal_id} (based on publications data from {start_year})")
threshold = 200
temp = journals.sort_values("year", ascending=True).groupby("year").head(threshold)
px.bar(journals[:threshold],
x="journal.title", y="total_year",
color="year",
facet_row="year",
height=900,
hover_name="journal.title",
hover_data=['journal.id', 'journal.title', 'total' ],
title=f"Top {threshold} competitors for {journal_id} - segmented by year")
```
NOTE the European Neuropsychopharmacology journal has a massive jump in 2019 cause they [published a lot of conference proceedings](https://www.sciencedirect.com/journal/european-neuropsychopharmacology/issues)! See also the journal [Dimensions page](https://app.dimensions.ai/analytics/publication/overview/timeline?and_facet_source_title=jour.1101548) for comparison..
|
github_jupyter
|
# CAMS functions
```
def get_ADS_API_key():
""" Get ADS API key to download CAMS datasets
Returns:
API_key (str): ADS API key
"""
keys_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/keys.txt'))
try:
keys_file = open(keys_path, 'r')
keys = keys_file.readlines()
environ_keys = [key.rstrip() for key in keys]
ADS_key = environ_keys[0]
except:
print('ERROR: You need to create a keys.txt file in the data folder with the ADS API key.')
print('Get your ADS API key by registering at https://ads.atmosphere.copernicus.eu/api-how-to.')
raise KeyboardInterrupt
return ADS_key
def CAMS_download(dates, start_date, end_date, component, component_nom, lat_min, lat_max, lon_min, lon_max,
area_name, model_full_name, model_level, CAMS_UID = None, CAMS_key = None):
""" Query and download the CAMS levels dataset from CDS API
Args:
dates (arr): Query dates
start_date (str): Query start date
end_date (str): Query end date
component (str): Component name
component_nom (str): Component chemical nomenclature
lat_min (int): Minimum latitude
lat_max (int): Maximum latitude
lon_min (int): Minimum longitude
lon_max (int): Maximum longitude
area_name (str): User defined area name
model_full_name (str): Full name of the CAMS model among:
- 'cams-global-atmospheric-composition-forecasts'
- 'cams-global-reanalysis-eac4-monthly'
model_level (str): Model levels:
- 'Simple' for total columns
- 'Multiple' for levels
CAMS_UID (str): ADS user ID
CAMS_key (str): ADS key
Returns:
CAMS_product_name (str): Product name of CAMS product
CAMS_type (str): Model type:
- 'Forecast'
- 'Reanalysis'
"""
# Get API key
if CAMS_UID != None and CAMS_key != None:
ADS_key = CAMS_UID + ':' + CAMS_key
else:
ADS_key = get_ADS_API_key()
# Connect to the server
c = cdsapi.Client(url = 'https://ads.atmosphere.copernicus.eu/api/v2', key = ADS_key)
# Download component concentration dataset
if model_full_name == 'cams-global-atmospheric-composition-forecasts':
CAMS_type = 'Forecast'
if model_level == 'Multiple':
CAMS_product_name = ('CAMS_FORECAST_' + component_nom + '_137_LEVELS_' + start_date + '_' + end_date +
'_' + area_name + '.grib')
CAMS_product_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/' + CAMS_product_name))
if os.path.isfile(CAMS_product_path):
print('The file exists, it will not be downloaded again.')
else:
print('The file does not exist, it will be downloaded.')
c.retrieve(
model_full_name,
{
'date': start_date + '/' + end_date,
'type': 'forecast',
'format': 'grib',
'variable': component,
'model_level': [str(x + 1) for x in range(137)],
'time': '00:00',
'leadtime_hour': [str(x) for x in range(0, 24, 3)],
'area': [lat_max, lon_min, lat_min, lon_max],
},
CAMS_product_path)
elif model_level == 'Single':
CAMS_product_name = ('CAMS_FORECAST_' + component_nom + '_TC_' + start_date + '_' + end_date +
'_' + area_name + '.grib')
CAMS_product_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/' + CAMS_product_name))
if os.path.isfile(CAMS_product_path):
print('The file exists, it will not be downloaded again.')
else:
print('The file does not exist, it will be downloaded.')
c = cdsapi.Client(url = 'https://ads.atmosphere.copernicus.eu/api/v2', key = ADS_key)
c.retrieve(
'cams-global-atmospheric-composition-forecasts',
{
'date': start_date + '/' + end_date,
'type': 'forecast',
'format': 'grib',
'variable': 'total_column_' + component,
'time': '00:00',
'leadtime_hour': [str(x) for x in range(0, 24, 3)],
'area': [lat_max, lon_min, lat_min, lon_max],
},
CAMS_product_path)
elif model_full_name == 'cams-global-reanalysis-eac4-monthly':
CAMS_type = 'Reanalysis'
if model_level == 'Single':
CAMS_product_name = ('CAMS_REANALYSIS_' + component_nom + '_TC_' + start_date + '_' + end_date +
'_' + area_name + '.grib')
CAMS_product_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/' + CAMS_product_name))
if os.path.isfile(CAMS_product_path):
print('The file exists, it will not be downloaded again.')
else:
print('The file does not exist, it will be downloaded.')
months = []
years = []
for date in dates:
year = date.split('-')[0]
month = date.split('-')[1]
if year not in years:
years.append(year)
if month not in months:
months.append(month)
c.retrieve(
model_full_name,
{
'format': 'grib',
'variable': 'total_column_' + component,
'year': years,
'month': months,
'product_type': 'monthly_mean',
'area': [lat_max, lon_min, lat_min, lon_max],
},
CAMS_product_path)
elif model_level == 'Multiple':
start_dates = pd.date_range(np.datetime64(start_date), np.datetime64(end_date), freq='MS')
start_dates = tuple(np.unique([date.strftime('%Y-%m-%d') for date in start_dates]))
end_dates = pd.date_range(np.datetime64(start_date), np.datetime64(end_date), freq='M')
end_dates = tuple(np.unique([date.strftime('%Y-%m-%d') for date in end_dates]))
# Download month by month (to avoid crashing the server)
CAMS_product_name = []
for start_date, end_date in zip(start_dates, end_dates):
CAMS_product_name_month = ('CAMS_REANALYSIS_' + component_nom + '_60_LEVELS_' + start_date + '_' + end_date +
'_' + area_name + '.grib')
CAMS_product_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/' + CAMS_product_name_month))
if os.path.isfile(CAMS_product_path):
print('The file exists, it will not be downloaded again.')
else:
print('The file does not exist, it will be downloaded.')
c.retrieve(
'cams-global-reanalysis-eac4',
{
'date': start_date + '/' + end_date,
'format': 'grib',
'variable': component,
'model_level': [str(x + 1) for x in range(60)],
'time': ['00:00', '03:00', '06:00', '09:00', '12:00', '15:00', '18:00', '21:00',],
'area': [lat_max, lon_min, lat_min, lon_max],
},
CAMS_product_path)
CAMS_product_name.append(CAMS_product_name_month)
return CAMS_product_name, CAMS_type
def CAMS_read(CAMS_product_name, component, component_nom, dates):
""" Read CAMS levels dataset as xarray dataset object
Args:
CAMS_product_name (str): Product name of CAMS product
component (str): Component name
component_nom (str): Component chemical nomenclature
dates (arr): Query dates
Returns:
CAMS_ds (xarray): CAMS levels dataset in xarray format
"""
# Read as xarray dataset object
if isinstance(CAMS_product_name, list):
CAMS_ds = xr.open_mfdataset(os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/CAMS_REANALYSIS_' + component_nom + '_60_LEVELS_*')),
concat_dim = 'time')
else:
CAMS_ds = xr.open_dataset(os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/' + CAMS_product_name)))
# Change name to component
if 'hybrid' in CAMS_ds.keys():
if component == 'ozone':
CAMS_ds = CAMS_ds.rename({'go3': 'component'})
else:
CAMS_ds = CAMS_ds.rename({component_nom.lower(): 'component'})
else:
if component == 'ozone':
CAMS_ds = CAMS_ds.rename({'gtco3': 'component'})
else:
CAMS_ds = CAMS_ds.rename({'tc' + component_nom.lower(): 'component'})
if 'REANALYSIS_' + component_nom + '_TC_' in CAMS_product_name:
# Remove data for dates that have been downloaded but not asked for (error of the CAMS API!)
all_datetimes = []
for date in dates:
year = int(date.split('-')[0])
month = int(date.split('-')[1])
time_str = np.datetime64(dt.datetime(year, month, 1, 0, 0, 0, 0))
all_datetimes.append(time_str)
# Drop datetimes
datetimes_to_delete = np.setdiff1d(CAMS_ds.time.values, np.array(all_datetimes))
if datetimes_to_delete.size != 0:
CAMS_ds = CAMS_ds.drop_sel(time = datetimes_to_delete)
# Available dates
dates_to_keep = np.intersect1d(CAMS_ds.time.values, np.array(all_datetimes))
dates = tuple(dates_to_keep.astype('datetime64[M]').astype(str))
# Remove step since there is only one
CAMS_ds = CAMS_ds.drop('step')
# Arrange coordinates
CAMS_ds = CAMS_ds.assign_coords(longitude = (((CAMS_ds.longitude + 180) % 360) - 180)).sortby('longitude')
CAMS_ds = CAMS_ds.sortby('latitude')
# Assign time as dimension (when there is only one time)
if CAMS_ds.time.values.size == 1:
CAMS_ds = CAMS_ds.expand_dims(dim = ['time'])
# Get model levels
CAMS_levels_df = CAMS_levels(CAMS_ds, CAMS_product_name)
return CAMS_ds, dates, CAMS_levels_df
def CAMS_levels(CAMS_ds, CAMS_product_name):
""" Create table with information about the CAMS model levels
Args:
CAMS_ds (xarray): CAMS levels dataset in xarray format
CAMS_product_name (str): Product name of CAMS product
Returns:
CAMS_levels_df (dataframe): Table with CAMS levels data
"""
# Read CSV table with information about the model levels
if '60_LEVELS' in CAMS_product_name:
CAMS_levels_df = pd.read_csv(os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/60-levels-definition.csv')))
else:
CAMS_levels_df = pd.read_csv(os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/137-levels-definition.csv')))
# Drop first row and set n as index hybrid
CAMS_levels_df = CAMS_levels_df.drop(0).reset_index(drop = True)
CAMS_levels_df = CAMS_levels_df.set_index('n')
CAMS_levels_df.index.names = ['hybrid']
# Change important columns to numeric
CAMS_levels_df['ph [Pa]'] = pd.to_numeric(CAMS_levels_df['ph [hPa]']) * 100
CAMS_levels_df['Geopotential Altitude [m]'] = pd.to_numeric(CAMS_levels_df['Geopotential Altitude [m]'])
CAMS_levels_df['Density [kg/m^3]'] = pd.to_numeric(CAMS_levels_df['Density [kg/m^3]'])
# Calculate difference from geopotential altitude
CAMS_levels_df['Depth [m]'] = CAMS_levels_df['Geopotential Altitude [m]'].diff(-1)
CAMS_levels_df['Depth [m]'].iloc[-1] = CAMS_levels_df['Geopotential Altitude [m]'].iloc[-1]
return CAMS_levels_df
def CAMS_pressure(CAMS_ds, CAMS_product_name, CAMS_levels_df, start_date, end_date, component_nom,
lat_min, lat_max, lon_min, lon_max, area_name, CAMS_UID = None, CAMS_key = None):
""" Download surface pressure and calculate levels pressure following the instructions given at:
https://confluence.ecmwf.int/display/OIFS/4.4+OpenIFS%3A+Vertical+Resolution+and+Configurations
Args:
CAMS_ds (xarray): CAMS levels dataset in xarray format
CAMS_product_name (str): Product name of CAMS product
CAMS_levels_df (dataframe): Table with 137 CAMS levels data
start_date (str): Query start date
end_date (str): Query end date
component_nom (str): Component chemical nomenclature
lat_min (int): Minimum latitude
lat_max (int): Maximum latitude
lon_min (int): Minimum longitude
lon_max (int): Maximum longitude
area_name (str): User defined area name
CAMS_UID (str): ADS user ID
CAMS_key (str): ADS key
Returns:
CAMS_ds (xarray): CAMS levels dataset in xarray format
"""
CAMS_pressure_product_name = ('_SURFACE_PRESSURE_' + start_date + '_' + end_date +
'_' + area_name + '.grib')
# Get API key
if CAMS_UID != None and CAMS_key != None:
ADS_key = CAMS_UID + ':' + CAMS_key
else:
ADS_key = get_ADS_API_key()
# Connect to the server
c = cdsapi.Client(url = 'https://ads.atmosphere.copernicus.eu/api/v2', key = ADS_key)
# Dowload surface pressure data
if 'FORECAST' in CAMS_product_name:
CAMS_surface_pressure_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/CAMS_FORECAST' + CAMS_pressure_product_name))
c.retrieve(
'cams-global-atmospheric-composition-forecasts',
{
'date': start_date + '/' + end_date,
'type': 'forecast',
'format': 'grib',
'variable': 'surface_pressure',
'leadtime_hour': [str(x) for x in range(0, 24, 3)],
'time': '00:00',
'area': [lat_max, lon_min, lat_min, lon_max],
},
CAMS_surface_pressure_path)
elif 'REANALYSIS' in CAMS_product_name:
CAMS_surface_pressure_path = os.path.join('/', '/'.join(
os.getcwd().split('/')[1:3]), 'adc-toolbox',
os.path.relpath('data/cams/' + component_nom + '/CAMS_REANALYSIS' + CAMS_pressure_product_name))
c.retrieve(
'cams-global-reanalysis-eac4',
{
'date': start_date + '/' + end_date,
'format': 'grib',
'variable': 'surface_pressure',
'time': ['00:00', '03:00', '06:00',
'09:00', '12:00', '15:00',
'18:00', '21:00',],
'area': [lat_max, lon_min, lat_min, lon_max],
},
CAMS_surface_pressure_path)
hybrid = CAMS_ds['hybrid'].data
time = CAMS_ds['time'].data
step = CAMS_ds['step'].data
latitude = CAMS_ds['latitude'].data
longitude = CAMS_ds['longitude'].data
# Read surface pressure
model_pressure_ds = xr.open_dataarray(CAMS_surface_pressure_path)
# Arrange coordinates
model_pressure_ds = model_pressure_ds.assign_coords(longitude = (((model_pressure_ds.longitude + 180) % 360) - 180)).sortby('longitude')
model_pressure_ds = model_pressure_ds.sortby('latitude')
# Assign time as dimension (when there is only one time)
if model_pressure_ds.time.values.size == 1:
model_pressure_ds = model_pressure_ds.expand_dims(dim = ['time'])
# Transpose dimensions
model_pressure_ds = model_pressure_ds.transpose('time', 'step', 'latitude', 'longitude')
# Subset surface pressure dataset
model_pressure_ds = subset(model_pressure_ds, bbox, sensor, component_nom, sensor_type, subset_type = 'model_subset')
sp_array = xr.DataArray(
model_pressure_ds.values,
dims = ('time', 'step', 'latitude', 'longitude'),
coords = {
'time': ('time', time),
'step': ('step', step),
'latitude': ('latitude', latitude),
'longitude': ('longitude', longitude),
},
name = 'surface_pressure'
)
a_array = xr.DataArray(
CAMS_levels_df['a [Pa]'],
dims = ('hybrid'),
coords = {'hybrid': ('hybrid', hybrid),},
name = 'a'
)
b_array = xr.DataArray(
CAMS_levels_df['b'],
dims = ('hybrid'),
coords = {'hybrid': ('hybrid', hybrid),},
name = 'b'
)
CAMS_ds['surface_pressure'] = sp_array
CAMS_ds['a'] = a_array
CAMS_ds['b'] = b_array
CAMS_ds['pressure_1/2'] = CAMS_ds['a'] + CAMS_ds['surface_pressure'] * CAMS_ds['b']
CAMS_ds['pressure_-1/2'] = CAMS_ds['pressure_1/2'].shift(hybrid = 1)
CAMS_ds['pressure_-1/2'] = CAMS_ds['pressure_-1/2'].where(~np.isnan(CAMS_ds['pressure_-1/2']), 0, drop = False)
CAMS_ds['pressure'] = 0.5 * (CAMS_ds['pressure_-1/2'] + CAMS_ds['pressure_1/2'])
CAMS_ds = CAMS_ds.drop_vars(['a', 'b', 'surface_pressure', 'pressure_1/2', 'pressure_-1/2'])
return CAMS_ds
def CAMS_get_levels_data(CAMS_ds, CAMS_product_name, CAMS_levels_df, column_type,
lat_min, lat_max, lon_min, lon_max):
""" Get the tropospheric or column model data, depending on the nature of the sensor data
Args:
CAMS_ds (xarray): CAMS levels dataset in xarray format
CAMS_product_name (str): Product name of CAMS product
CAMS_levels_df (dataframe): Table with 137 CAMS levels data
column_type (str): Tropospheric or total column
lat_min (int): Minimum latitude
lat_max (int): Maximum latitude
lon_min (int): Minimum longitude
lon_max (int): Maximum longitude
Returns:
CAMS_ds (xarray): CAMS levels dataset in xarray format
"""
# Get units and calculate tropospheric columns if needed
units = CAMS_ds.component.attrs['units']
if 'REANALYSIS' in CAMS_product_name:
if column_type == 'tropospheric':
print('The model total columns will be directly compared to the tropospheric sensor columns.')
elif column_type == 'total':
print('The model total columns will be compared to the total sensor columns.')
elif 'FORECAST' in CAMS_product_name:
if column_type == 'tropospheric':
print('The model tropospheric columns will be compared to the tropospheric sensor columns.')
print('The model tropospheric columns will be estimated (pressures above or equal to 300 hPa).')
# Calculate levels pressure
CAMS_ds = CAMS_pressure(CAMS_ds, CAMS_product_name, CAMS_levels_df, start_date, end_date, component_nom,
lat_min, lat_max, lon_min, lon_max, area_name, CAMS_UID = None, CAMS_key = None)
if apply_kernels == False:
CAMS_ds = CAMS_ds.where(CAMS_ds.pressure >= 30000, drop = True)
CAMS_ds = CAMS_ds.sum(dim = 'hybrid')
CAMS_ds['component'] = CAMS_ds.component.assign_attrs({'units': units})
if column_type == 'total':
print('The model total columns will be compared to the total sensor columns.')
return CAMS_ds
def CAMS_kg_kg_to_kg_m2(CAMS_ds, CAMS_levels_df, sensor, start_date, end_date,
component_nom, apply_kernels = False, CAMS_UID = None, CAMS_key = None):
""" Convert the units of the CAMS partial columns for any component from kg/kg to kg/m2. To do this,
calculate columns above each CAMS half level assuming it is 0 at the top of the atmosphere
Args:
CAMS_ds (xarray): CAMS levels dataset in xarray format
CAMS_levels_df (dataframe): Table with 137 CAMS levels data
sensor (str): Name of the sensor
start_date (str): Query start date
end_date (str): Query end date
component_nom (str): Component chemical nomenclature
apply_kernels (bool): Apply (True) or not (False) the averaging kernels
CAMS_UID (str): ADS user ID
CAMS_key (str): ADS key
Returns:
CAMS_ds (xarray): CAMS levels dataset in xarray format
"""
# Calculate columns above each CAMS half level
if sensor == 'tropomi' and apply_kernels == True:
print('The columns above each CAMS half level will be calculated.')
# Initialize new array
CAMS_ds_all = []
for time in CAMS_ds.time:
# Select data for each timestep
CAMS_ds_time_old = CAMS_ds.sel(time = time)
# Initialize partial columns at the top of the atmosphere (hybrid = 1) as 0
PC_hybrid_0 = CAMS_ds_time_old.sel(hybrid = 1)
PC_hybrid_0['component'] = PC_hybrid_0['component'].where(PC_hybrid_0['component'] <= 0, 0, drop = False)
PC_hybrid_0 = PC_hybrid_0.expand_dims(dim = ['hybrid'])
# Create new model dataset
PC_above_all = []
PC_above_all.append(PC_hybrid_0)
CAMS_ds_time_new = PC_hybrid_0
for hybrid in range(1, 137):
# Get current and previous partial columns and level pressures
PC_last = CAMS_ds_time_new.component.sel(hybrid = hybrid)
PC_current = CAMS_ds_time_old.component.sel(hybrid = hybrid + 1)
pressure_last = CAMS_ds_time_old.pressure.sel(hybrid = hybrid)
pressure_current = CAMS_ds_time_old.pressure.sel(hybrid = hybrid + 1)
# Calculate pressure difference
pressure_diff = pressure_current - pressure_last
# Calculate partial columns above each model level
# Units: (kg/kg * kg/m*s2) * s2/m -> kg/m2
PC_above = CAMS_ds_time_old.sel(hybrid = hybrid + 1)
PC_above['component'] = PC_last + PC_current * pressure_diff * (1/9.81)
# Append result
PC_above_all.append(PC_above)
CAMS_ds_time_new = xr.concat(PC_above_all, pd.Index(range(1, hybrid + 2), name = 'hybrid'))
CAMS_ds_all.append(CAMS_ds_time_new)
CAMS_ds = xr.concat(CAMS_ds_all, dim = 'time')
else:
# Create xarray object from CAMS model levels information
CAMS_levels_ds = CAMS_levels_df.to_xarray()
# Convert units from kg/kg to kg/m3
CAMS_ds = CAMS_ds * CAMS_levels_ds['Density [kg/m^3]']
# Convert units from kg/m3 to kg/m2
CAMS_ds = CAMS_ds * CAMS_levels_ds['Depth [m]']
return CAMS_ds
def CAMS_kg_m2_to_molecules_cm2(CAMS_ds, component_mol_weight):
""" Convert the units of the CAMS dataset for any component from kg/m2 to molecules/cm2
Args:
CAMS_ds (xarray): CAMS levels dataset in xarray format
component_mol_weight (float): Component molecular weight
Returns:
CAMS_ds (xarray): CAMS levels dataset in xarray format
"""
# Convert units from kg/m2 to molecules/cm2
NA = 6.022*10**23
CAMS_ds['component'] = (CAMS_ds['component'] * NA * 1000) / (10000 * component_mol_weight)
return CAMS_ds
def CAMS_molecules_cm2_to_DU(CAMS_ds):
""" Convert the units of the CAMS dataset for any component from molecules/cm2 to DU for ozone
Args:
CAMS_ds (xarray): CAMS levels dataset in xarray format
Returns:
CAMS_ds (xarray): CAMS levels dataset in xarray format
"""
# Convert units from molecules/cm2 to DU
CAMS_ds = CAMS_ds / (2.69*10**16)
return CAMS_ds
```
|
github_jupyter
|
# SARK-110 Time Domain and Gating Example
Example adapted from: https://scikit-rf.readthedocs.io/en/latest/examples/networktheory/Time%20Domain.html
- Measurements with a 2.8m section of rg58 coax cable not terminated at the end
This notebooks demonstrates how to use scikit-rf for time-domain analysis and gating. A quick example is given first, followed by a more detailed explanation.
S-parameters are measured in the frequency domain, but can be analyzed in time domain if you like. In many cases, measurements are not made down to DC. This implies that the time-domain transform is not complete, but it can be very useful non-theless. A major application of time-domain analysis is to use gating to isolate a single response in space. More information about the details of time domain analysis.
Please ensure that the analyzer is connected to the computer using the USB cable and in Computer Control mode.
```
from sark110 import *
import skrf as rf
rf.stylely()
from pylab import *
```
Enter frequency limits:
```
fr_start = 100000 # Frequency start in Hz
fr_stop = 230000000 # Frequency stop in Hz
points = 401 # Number of points
```
## Utility functions
```
def z2vswr(rs: float, xs: float, z0=50 + 0j) -> float:
gamma = math.sqrt((rs - z0.real) ** 2 + xs ** 2) / math.sqrt((rs + z0.real) ** 2 + xs ** 2)
if gamma > 0.980197824:
return 99.999
swr = (1 + gamma) / (1 - gamma)
return swr
def z2mag(r: float, x: float) -> float:
return math.sqrt(r ** 2 + x ** 2)
def z2gamma(rs: float, xs: float, z0=50 + 0j) -> complex:
z = complex(rs, xs)
return (z - z0) / (z + z0)
```
## Connect to the device
```
sark110 = Sark110()
sark110.open()
sark110.connect()
if not sark110.is_connected:
print("Device not connected")
exit(-1)
else:
print("Device connected")
sark110.buzzer()
print(sark110.fw_protocol, sark110.fw_version)
```
## Acquire and plot the data
```
y = []
x = []
rs = [0]
xs = [0]
for i in range(points):
fr = int(fr_start + i * (fr_stop - fr_start) / (points - 1))
sark110.measure(fr, rs, xs)
x.append(fr / 1e9) # Units in GHz
y.append(z2gamma(rs[0][0], xs[0][0]))
probe = rf.Network(frequency=x, s=y, z0=50)
probe.frequency.unit = 'mhz'
print (probe)
```
# Quick example
```
# we will focus on s11
s11 = probe.s11
# time-gate the first largest reflection
s11_gated = s11.time_gate(center=0, span=50)
s11_gated.name='gated probe'
# plot frequency and time-domain s-parameters
figure(figsize=(8,4))
subplot(121)
s11.plot_s_db()
s11_gated.plot_s_db()
title('Frequency Domain')
subplot(122)
s11.plot_s_db_time()
s11_gated.plot_s_db_time()
title('Time Domain')
tight_layout()
```
# Interpreting Time Domain
Note there are two time-domain plotting functions in scikit-rf:
- Network.plot_s_db_time()
- Network.plot_s_time_db()
The difference is that the former, plot_s_db_time(), employs windowing before plotting to enhance impluse resolution. Windowing will be discussed in a bit, but for now we just use plot_s_db_time().
Plotting all four s-parameters of the probe in both frequency and time-domain.
```
# plot frequency and time-domain s-parameters
figure(figsize=(8,4))
subplot(121)
probe.plot_s_db()
title('Frequency Domain')
subplot(122)
probe.plot_s_db_time()
title('Time Domain')
tight_layout()
```
Focusing on the reflection coefficient from the waveguide port (s11), you can see there is an interference pattern present.
```
probe.plot_s_db(0,0)
title('Reflection Coefficient From \nWaveguide Port')
```
This ripple is evidence of several discrete reflections. Plotting s11 in the time-domain allows us to see where, or when, these reflections occur.
```
probe_s11 = probe.s11
probe_s11.plot_s_db_time(0,0)
title('Reflection Coefficient From \nWaveguide Port, Time Domain')
ylim(-100,0)
```
# Gating The Reflection of Interest
To isolate the reflection from the waveguide port, we can use time-gating. This can be done by using the method Network.time_gate(), and provide it an appropriate center and span (in ns). To see the effects of the gate, both the original and gated reponse are compared.
```
probe_s11_gated = probe_s11.time_gate(center=0, span=50)
probe_s11_gated.name='gated probe'
s11.plot_s_db_time()
s11_gated.plot_s_db_time()
```
Next, compare both responses in frequency domain to see the effect of the gate.
```
s11.plot_s_db()
s11_gated.plot_s_db()
```
# Auto-gate
The time-gating method in skrf has an auto-gating feature which can also be used to gate the largest reflection. When no gate parameters are provided, time_gate() does the following:
find the two largest peaks
center the gate on the tallest peak
set span to distance between two tallest peaks
You may want to plot the gated network in time-domain to see what the determined gate shape looks like.
```
title('Waveguide Interface of Probe')
s11.plot_s_db(label='original')
s11.time_gate().plot_s_db(label='autogated') #autogate on the fly
```
# Determining Distance
To make time-domain useful as a diagnostic tool, one would like to convert the x-axis to distance. This requires knowledge of the propagation velocity in the device. skrf provides some transmission-line models in the module skrf.media, which can be used for this.
However...
For dispersive media, such as rectangular waveguide, the phase velocity is a function of frequency, and transforming time to distance is not straightforward. As an approximation, you can normalize the x-axis to the speed of light.
Alternatively, you can simulate the a known device and compare the two time domain responses. This allows you to attribute quantatative meaning to the axes. For example, you could create an ideal delayed load as shown below. Note: the magnitude of a response behind a large impulse doesn not have meaningful units.
```
from skrf.media import DistributedCircuit
# create a Media object for RG-58, based on distributed ckt values
rg58 = DistributedCircuit(
frequency = probe.frequency,
C =93.5e-12,#F/m
L =273e-9, #H/m
R =0, #53e-3, #Ohm/m
G =0, #S/m
)
# create an ideal delayed load, parameters are adjusted until the
# theoretical response agrees with the measurement
theory = rg58.delay_load(Gamma0=rf.db_2_mag(-20),
d=280, unit='cm')
probe.plot_s_db_time(0,0, label = 'Measurement')
theory.plot_s_db_time(label='-20dB @ 280cm from test-port')
ylim(-100,0)
xlim(-500,500)
```
This plot demonstrates a few important points:
the theortical delayed load is not a perfect impulse in time. This is due to the dispersion in waveguide.
the peak of the magnitude in time domain is not identical to that specified, also due to disperison (and windowing).
# What the hell is Windowing?
The 'plot_s_db_time()' function does a few things.
windows the s-parameters.
converts to time domain
takes magnitude component, convert to dB
calculates time-axis s
plots
A word about step 1: windowing. A FFT represents a signal with a basis of periodic signals (sinusoids). If your frequency response is not periodic, which in general it isnt, taking a FFT will introduces artifacts in the time-domain results. To minimize these effects, the frequency response is windowed. This makes the frequency response more periodic by tapering off the band-edges.
Windowing is just applied to improve the plot appearance,d it does not affect the original network.
In skrf this can be done explicitly using the 'windowed()' function. By default this function uses the hamming window, but can be adjusted through arguments. The result of windowing is show below.
```
probe_w = probe.windowed()
probe.plot_s_db(0,0, label = 'Original')
probe_w.plot_s_db(0,0, label = 'Windowed')
```
Comparing the two time-domain plotting functions, we can see the difference between windowed and not.
```
probe.plot_s_time_db(0,0, label = 'Original')
probe_w.plot_s_time_db(0,0, label = 'Windowed')
```
# The end!
```
sark110.close()
```
|
github_jupyter
|
# Mouse Bone Marrow - merging annotated samples from MCA
```
import scanpy as sc
import numpy as np
import scipy as sp
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import rcParams
from matplotlib import colors
import seaborn as sb
import glob
import rpy2.rinterface_lib.callbacks
import logging
from rpy2.robjects import pandas2ri
import anndata2ri
# Ignore R warning messages
#Note: this can be commented out to get more verbose R output
rpy2.rinterface_lib.callbacks.logger.setLevel(logging.ERROR)
# Automatically convert rpy2 outputs to pandas dataframes
pandas2ri.activate()
anndata2ri.activate()
%load_ext rpy2.ipython
plt.rcParams['figure.figsize']=(8,8) #rescale figures
sc.settings.verbosity = 3
#sc.set_figure_params(dpi=200, dpi_save=300)
sc.logging.print_versions()
results_file = './write/MCA_mou_BM_pp.h5ad'
%%R
# Load all the R libraries we will be using in the notebook
library(scran)
```
## Load
Here we load the pre-processed datasets (which has been annotated), and the raw matrices (which won't be filtered on the gene level).
### Raw data
```
file_paths = '../../Munich/datasets/mouse/MCA_boneMarrow/ckit/'
adatas_raw = []
for i in glob.glob(file_paths+'*.txt.gz'):
print(i)
adatas_raw.append(sc.read(i, cache=True))
samples = ['BM_1', 'BM_3', 'BM_2']
# Loop to annotate data
for i in range(len(adatas_raw)):
adata_tmp = adatas_raw[i]
adata_tmp = adata_tmp.transpose()
#Annotate data
adata_tmp.obs.index.rename('barcode', inplace=True)
adata_tmp.obs['batch'] = ['MCA_'+samples[i]]*adata_tmp.n_obs
adata_tmp.obs['study'] = ['MCA_BM']*adata_tmp.n_obs
adata_tmp.obs['chemistry'] = ['microwell-seq']*adata_tmp.n_obs
adata_tmp.obs['tissue'] = ['Bone_Marrow']*adata_tmp.n_obs
adata_tmp.obs['species'] = ['Mouse']*adata_tmp.n_obs
adata_tmp.obs['data_type'] = ['UMI']*adata_tmp.n_obs
adata_tmp.var.index.names = ['gene_symbol']
adata_tmp.var_names_make_unique()
adatas_raw[i] = adata_tmp
adatas_raw[0].obs.head()
# Concatenate to unique adata object
adata_raw = adatas_raw[0].concatenate(adatas_raw[1:], batch_key='sample_ID', index_unique=None)
adata_raw.obs.head()
adata_raw.obs.drop(columns=['sample_ID'], inplace=True)
adata_raw.obs.head()
adata_raw.shape
```
### Pre-processed data
```
file_paths = '../../Bone_Marrow_mouse/write/'
adatas_pp = []
for i in glob.glob(file_paths+'*.h5ad'):
print(i)
adatas_pp.append(sc.read(i, cache=True))
for i in range(len(adatas_pp)):
adata_tmp = adatas_pp[i]
adata_obs = adata_tmp.obs.reset_index()
adata_obs = adata_obs[['index', 'final_annotation', 'dpt_pseudotime_y', 'n_counts', 'n_genes', 'mt_frac']].rename(columns = {'index':'barcode'})
adata_obs.set_index('barcode', inplace = True)
adatas_pp[i].obs = adata_obs
# Concatenate to unique adata object
adata_pp = adatas_pp[0].concatenate(adatas_pp[1:], batch_key='sample_ID',
index_unique=None)
adata_pp.obs.drop(columns=['sample_ID'], inplace = True)
adata_pp.obs.head()
adata_raw.shape
adata_pp.shape
# Restrict to cells that passed QC and were annotated
adata_obs_raw = adata_raw.obs.reset_index()
adata_obs_pp = adata_pp.obs.reset_index()
adata_merged = adata_obs_raw.merge(adata_obs_pp, on='barcode', how='left')
adata_merged.set_index('barcode', inplace = True)
adata_raw.obs = adata_merged
adata_raw.obs.head()
adata_raw = adata_raw[~pd.isnull(adata_raw.obs['final_annotation'])]
adata_raw.shape
```
### Normalization
```
# Exclude genes that are = 0 in all cells
#Filter genes:
print('Total number of genes: {:d}'.format(adata_raw.n_vars))
# Min 20 cells - filters out 0 count genes
sc.pp.filter_genes(adata_raw, min_cells=1)
print('Number of genes after cell filter: {:d}'.format(adata_raw.n_vars))
#Perform a clustering for scran normalization in clusters
adata_pp = adata_raw.copy()
sc.pp.normalize_per_cell(adata_pp, counts_per_cell_after=1e6)
sc.pp.log1p(adata_pp)
sc.pp.pca(adata_pp, n_comps=15, svd_solver='arpack')
sc.pp.neighbors(adata_pp)
sc.tl.louvain(adata_pp, key_added='groups', resolution=0.5)
# Check if the minimum number of cells per cluster is < 21:in that case, sizes will be also passed as input to the normalization
adata_pp.obs['groups'].value_counts()
#Preprocess variables for scran normalization
input_groups = adata_pp.obs['groups']
data_mat = adata_raw.X.T
%%R -i data_mat -i input_groups -o size_factors
size_factors = computeSumFactors(data_mat, clusters=input_groups, min.mean=0.1)
#Delete adata_pp
del adata_pp
# Visualize the estimated size factors
adata_raw.obs['size_factors'] = size_factors
sc.pl.scatter(adata_raw, 'size_factors', 'n_counts')
sc.pl.scatter(adata_raw, 'size_factors', 'n_genes')
sb.distplot(size_factors, bins=50, kde=False)
plt.show()
#Keep the count data in a counts layer
adata_raw.layers["counts"] = adata_raw.X.copy()
#Normalize adata
adata_raw.X /= adata_raw.obs['size_factors'].values[:,None]
sc.pp.log1p(adata_raw)
adata_raw.write(results_file)
```
|
github_jupyter
|
```
# importing the required libraries
import os
import numpy as np
import cv2
import matplotlib.pyplot as plt
%matplotlib inline
# function for reading the image
# this image is taken from a video
# and the video is taken from a thermal camera
# converting image from BGR to RGB
def read_image(image_path):
image = cv2.imread(image_path)
return cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# thermal camera takes the heat/thermal energy
# more heat means the pixel value is closer to 255
# if it is cool then pixel value is closer to 0
# displaying the image, where white portion means that part is having more temprature
# and vice versa
image = read_image("thermal_scr_img.png")
plt.imshow(image)
# converting the image into grayscale
# changing and applying the ColorMap, black and white to black and red
heatmap_gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
heatmap = cv2.applyColorMap(heatmap_gray, cv2.COLORMAP_HOT)
heatmap = cv2.cvtColor(heatmap, cv2.COLOR_BGR2RGB)
plt.imshow(heatmap)
# now taking the heatmap_gray and converting it to black and white image
# and performing threshold operation
# the pixels having values more than 200 will become white pixels and the one having values less than 200 will become black pixels
heatmap_gray = cv2.cvtColor(heatmap, cv2.COLOR_RGB2GRAY)
ret, binary_thresh = cv2.threshold(heatmap_gray, 200, 255, cv2.THRESH_BINARY)
plt.imshow(binary_thresh, cmap='gray')
# then cleaning the small white pixels to calculate the temperature for bigger blocks/portions of the image
# doing erosion operation by taking binary threshold (it makes image pixels thinner)
# doing dilution operation by taking the image erosion (and then that's why we are removing/cleaning all small pixels)
# kernel is some kind of filter and it changes the values of these pixels
kernel = np.ones((5,5), np.uint8)
image_erosion = cv2.erode(binary_thresh, kernel, iterations=1)
image_opening = cv2.dilate(image_erosion, kernel, iterations=1)
plt.imshow(image_opening, cmap='gray')
# now creating some masks
# using function zeros_like() it will take all the structures like zero
# x, y, w, h are the coordinate for rectangle
# copying the small rectangle part from this image using mask
# and printing the avg. value of pixels to get the temperature
contours, _ = cv2.findContours(image_opening, 1, 2)
contour = contours[11]
mask = np.zeros_like(heatmap_gray)
x, y, w, h = cv2.boundingRect(contour)
mask[y:y+h, x:x+w] = image_opening[y:y+h, x:x+w]
print(cv2.mean(heatmap_gray, mask= mask))
plt.imshow(mask, cmap='gray')
# performing the bitwise and operator on heatmap
# here we have created not mask
masked = cv2.bitwise_and(heatmap, heatmap, mask=~mask)
plt.imshow(masked)
# displaying the heatmap_gray
plt.imshow(heatmap_gray)
image_with_rectangles = np.copy(heatmap)
for contour in contours:
# rectangle over each contour
x, y, w, h = cv2.boundingRect(contour)
# mask is boolean type of matrix
mask = np.zeros_like(heatmap_gray)
mask[y:y+h, x:x+w] = image_opening[y:y+h, x:x+w]
# temperature calculation
temp = round(cv2.mean(heatmap_gray, mask=mask)[0] / 2.25, 2)
# draw rectangles for visualisation
image_with_rectangles = cv2.rectangle(
image_with_rectangles, (x,y), (x+w, y+h), (0, 255, 0), 2)
# write temperature for each rectangle
cv2.putText(image_with_rectangles, f"{temp} F", (x,y),
cv2.FONT_HERSHEY_SIMPLEX, 0.7, (0, 255, 0), 2, cv2.LINE_AA)
plt.imshow(image_with_rectangles)
```
|
github_jupyter
|
### Topic Modelling Demo Code
#### Things I want to do -
- Identify a package to build / train LDA model
- Use visualization to explore Documents -> Topics Distribution -> Word distribution
```
!pip install pyLDAvis, gensim
import numpy as np
import pandas as pd
# Visualization
import matplotlib.pyplot as plt
from matplotlib.ticker import FuncFormatter
import seaborn as sns
import pyLDAvis.gensim
# Text Preprocessing and model building
from gensim.corpora import Dictionary
import nltk
from nltk.stem import WordNetLemmatizer
import re
# Iteratively read files
import glob
import os
# For displaying images in ipython
from IPython.display import HTML, display
%matplotlib inline
plt.style.use('ggplot')
plt.rcParams['figure.figsize'] = (14.0, 8.7)
#warnings.filterwarnings('ignore')
pd.options.display.float_format = '{:,.2f}'.format
```
<h2>Latent Dirichlet Allocation</h2>
<h3>From Documents -- DTM -- LDA Model</h3>
Topic modeling aims to automatically summarize large collections of documents to facilitate organization and management, as well as search and recommendations. At the same time, it can enable the understanding of documents to the extent that humans can interpret the descriptions of topics
<img src="images/lda2.png" alt="lda" style="width:60%">
<img src="images/docs_to_lda.png" alt="ldaflow" style="width:100%">
### Load Data
```
# User defined function to read and store bbc data from multipe folders
def load_data(folder_names,root_path):
fileNames = [path + '/' + 'bbc' +'/'+ folder + '/*.txt' for path,folder in zip([root_path]*len(folder_names),
folder_names )]
doc_list = []
tags = folder_names
for docs in fileNames:
#print(docs)
#print(type(docs))
doc = glob.glob(docs) # glob method iterates through the all the text documents in a folder
for text in doc:
with open(text, encoding='latin1') as f:
topic = docs.split('/')[-2]
lines = f.readlines()
heading = lines[0].strip()
body = ' '.join([l.strip() for l in lines[1:]])
doc_list.append([topic, heading, body])
print("Completed loading data from folder: %s"%topic)
print("Completed Loading entire text")
return doc_list
folder_names = ['business','entertainment','politics','sport','tech']
docs = load_data(folder_names = folder_names, root_path = os.getcwd())
docs = pd.DataFrame(docs, columns=['Category', 'Heading', 'Article'])
print(docs.head())
print('\nShape of data is {}\n'.format(docs.shape))
```
### Extract Raw Corpus
```
articles = docs.Article.tolist()
print(type(articles))
print(articles[0:2])
wordnet_lemmatizer = WordNetLemmatizer()
```
### Preprocessing of Raw Text
```
from nltk.corpus import stopwords
import nltk
# nltk.download('punkt')
# nltk.download('wordnet')
# nltk.download('stopwords')
# nltk.download('stopwords')
stopwords = stopwords.word('english')
# Method to preprocess my raw data
def preprocessText(x):
temp = x.lower()
temp = re.sub(r'[^\w]', ' ', temp)
temp = nltk.word_tokenize(temp)
temp = [wordnet_lemmatizer.lemmatize(w) for w in temp]
temp = [word for word in temp if word not in stopwords ]
return temp
articles_final = [preprocessText(article) for article in articles]
articles_final[0:2]
```
### Transformation of Preprocessed text into Vector form using Gensim
```
# Create a dictionary representation of the documents.
dictionary = Dictionary(articles_final)
# Filter out words that occur less than 20 documents, or more than 50% of the documents.
dictionary.filter_extremes(no_below=20, no_above=0.5)
print(dictionary)
# Bag-of-words representation of the documents.
corpus = [dictionary.doc2bow(doc) for doc in articles_final]
print('Number of unique tokens: %d' % len(dictionary))
print('Number of documents: %d' % len(corpus))
```
### Train LDA model using Gensim
```
dictionary
# Train LDA model.
from gensim.models import LdaModel
# Set training parameters.
num_topics = 5
chunksize = 2000
passes = 10
# iterations = 400
eval_every = None # Don't evaluate model perplexity, takes too much time.
# Make a index to word dictionary.
temp = dictionary[0] # This is only to "load" the dictionary.
id2word = dictionary.id2token
model = LdaModel(
corpus=corpus,
id2word=id2word,
chunksize=chunksize,
alpha='auto',
eta='auto',
# iterations=iterations,
num_topics=num_topics,
passes=passes,
eval_every=eval_every
)
```
### Model exploration: Top K words in each topic
```
# Print the Keyword in the 10 topics
pprint.pprint(model.print_topics(num_words= 20))
doc_lda = model[corpus]
```
### Model Visualization using PyLDAvis
```
pyLDAvis.enable_notebook()
vis = pyLDAvis.gensim.prepare(model, corpus, dictionary=dictionary)
vis
```
### Assign Topic Model Numbers to original Data Frame as Column
```
# Assigns the topics to the documents in corpus
lda_corpus = model[corpus]
topics = []
for doc in lda_corpus:
temp_id = []
temp_score = []
for doc_tuple in doc:
temp_id.append(doc_tuple[0])
temp_score.append(doc_tuple[1])
index = np.argmax(temp_score)
topics.append(temp_id[index])
docs["Topic_num"] = topics
docs.tail(n= 40)
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.