code
stringlengths 2.5k
150k
| kind
stringclasses 1
value |
|---|---|
# Problem Simulation Tutorial
```
import pyblp
import numpy as np
import pandas as pd
pyblp.options.digits = 2
pyblp.options.verbose = False
pyblp.__version__
```
Before configuring and solving a problem with real data, it may be a good idea to perform Monte Carlo analysis on simulated data to verify that it is possible to accurately estimate model parameters. For example, before configuring and solving the example problems in the prior tutorials, it may have been a good idea to simulate data according to the assumed models of supply and demand. During such Monte Carlo anaysis, the data would only be used to determine sample sizes and perhaps to choose reasonable true parameters.
Simulations are configured with the :class:`Simulation` class, which requires many of the same inputs as :class:`Problem`. The two main differences are:
1. Variables in formulations that cannot be loaded from `product_data` or `agent_data` will be drawn from independent uniform distributions.
2. True parameters and the distribution of unobserved product characteristics are specified.
First, we'll use :func:`build_id_data` to build market and firm IDs for a model in which there are $T = 50$ markets, and in each market $t$, a total of $J_t = 20$ products produced by $F = 10$ firms.
```
id_data = pyblp.build_id_data(T=50, J=20, F=10)
```
Next, we'll create an :class:`Integration` configuration to build agent data according to a Gauss-Hermite product rule that exactly integrates polynomials of degree $2 \times 9 - 1 = 17$ or less.
```
integration = pyblp.Integration('product', 9)
integration
```
We'll then pass these data to :class:`Simulation`. We'll use :class:`Formulation` configurations to create an $X_1$ that consists of a constant, prices, and an exogenous characteristic; an $X_2$ that consists only of the same exogenous characteristic; and an $X_3$ that consists of the common exogenous characteristic and a cost-shifter.
```
simulation = pyblp.Simulation(
product_formulations=(
pyblp.Formulation('1 + prices + x'),
pyblp.Formulation('0 + x'),
pyblp.Formulation('0 + x + z')
),
beta=[1, -2, 2],
sigma=1,
gamma=[1, 4],
product_data=id_data,
integration=integration,
seed=0
)
simulation
```
When :class:`Simulation` is initialized, it constructs :attr:`Simulation.agent_data` and simulates :attr:`Simulation.product_data`.
The :class:`Simulation` can be further configured with other arguments that determine how unobserved product characteristics are simulated and how marginal costs are specified.
At this stage, simulated variables are not consistent with true parameters, so we still need to solve the simulation with :meth:`Simulation.replace_endogenous`. This method replaced simulated prices and market shares with values that are consistent with the true parameters. Just like :meth:`ProblemResults.compute_prices`, to do so it iterates over the $\zeta$-markup equation from :ref:`references:Morrow and Skerlos (2011)`.
```
simulation_results = simulation.replace_endogenous()
simulation_results
```
Now, we can try to recover the true parameters by creating and solving a :class:`Problem`.
The convenience method :meth:`SimulationResults.to_problem` constructs some basic "sums of characteristics" BLP instruments that are functions of all exogenous numerical variables in the problem. In this example, excluded demand-side instruments are the cost-shifter `z` and traditional BLP instruments constructed from `x`. Excluded supply-side instruments are traditional BLP instruments constructed from `x` and `z`.
```
problem = simulation_results.to_problem()
problem
```
We'll choose starting values that are half the true parameters so that the optimization routine has to do some work. Note that since we're jointly estimating the supply side, we need to provide an initial value for the linear coefficient on prices because this parameter cannot be concentrated out of the problem (unlike linear coefficients on exogenous characteristics).
```
results = problem.solve(
sigma=0.5 * simulation.sigma,
pi=0.5 * simulation.pi,
beta=[None, 0.5 * simulation.beta[1], None],
optimization=pyblp.Optimization('l-bfgs-b', {'gtol': 1e-5})
)
results
```
The parameters seem to have been estimated reasonably well.
```
np.c_[simulation.beta, results.beta]
np.c_[simulation.gamma, results.gamma]
np.c_[simulation.sigma, results.sigma]
```
In addition to checking that the configuration for a model based on actual data makes sense, the :class:`Simulation` class can also be a helpful tool for better understanding under what general conditions BLP models can be accurately estimated. Simulations are also used extensively in pyblp's test suite.
| github_jupyter |
# Softmax exercise
*Complete and hand in this completed worksheet (including its outputs and any supporting code outside of the worksheet) with your assignment submission. For more details see the [assignments page](http://vision.stanford.edu/teaching/cs231n/assignments.html) on the course website.*
This exercise is analogous to the SVM exercise. You will:
- implement a fully-vectorized **loss function** for the Softmax classifier
- implement the fully-vectorized expression for its **analytic gradient**
- **check your implementation** with numerical gradient
- use a validation set to **tune the learning rate and regularization** strength
- **optimize** the loss function with **SGD**
- **visualize** the final learned weights
```
import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt
from __future__ import print_function
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# for auto-reloading extenrnal modules
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
def get_CIFAR10_data(num_training=49000, num_validation=1000, num_test=1000, num_dev=500):
"""
Load the CIFAR-10 dataset from disk and perform preprocessing to prepare
it for the linear classifier. These are the same steps as we used for the
SVM, but condensed to a single function.
"""
# Load the raw CIFAR-10 data
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
# subsample the data
mask = list(range(num_training, num_training + num_validation))
X_val = X_train[mask]
y_val = y_train[mask]
mask = list(range(num_training))
X_train = X_train[mask]
y_train = y_train[mask]
mask = list(range(num_test))
X_test = X_test[mask]
y_test = y_test[mask]
mask = np.random.choice(num_training, num_dev, replace=False)
X_dev = X_train[mask]
y_dev = y_train[mask]
# Preprocessing: reshape the image data into rows
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_val = np.reshape(X_val, (X_val.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
X_dev = np.reshape(X_dev, (X_dev.shape[0], -1))
# Normalize the data: subtract the mean image
mean_image = np.mean(X_train, axis = 0)
X_train -= mean_image
X_val -= mean_image
X_test -= mean_image
X_dev -= mean_image
# add bias dimension and transform into columns
X_train = np.hstack([X_train, np.ones((X_train.shape[0], 1))])
X_val = np.hstack([X_val, np.ones((X_val.shape[0], 1))])
X_test = np.hstack([X_test, np.ones((X_test.shape[0], 1))])
X_dev = np.hstack([X_dev, np.ones((X_dev.shape[0], 1))])
return X_train, y_train, X_val, y_val, X_test, y_test, X_dev, y_dev
# Invoke the above function to get our data.
X_train, y_train, X_val, y_val, X_test, y_test, X_dev, y_dev = get_CIFAR10_data()
print('Train data shape: ', X_train.shape)
print('Train labels shape: ', y_train.shape)
print('Validation data shape: ', X_val.shape)
print('Validation labels shape: ', y_val.shape)
print('Test data shape: ', X_test.shape)
print('Test labels shape: ', y_test.shape)
print('dev data shape: ', X_dev.shape)
print('dev labels shape: ', y_dev.shape)
```
## Softmax Classifier
Your code for this section will all be written inside **cs231n/classifiers/softmax.py**.
```
# First implement the naive softmax loss function with nested loops.
# Open the file cs231n/classifiers/softmax.py and implement the
# softmax_loss_naive function.
from cs231n.classifiers.softmax import softmax_loss_naive
import time
# Generate a random softmax weight matrix and use it to compute the loss.
W = np.random.randn(3073, 10) * 0.0001
loss, grad = softmax_loss_naive(W, X_dev, y_dev, 0.0)
# As a rough sanity check, our loss should be something close to -log(0.1).
print('loss: %f' % loss)
print('sanity check: %f' % (-np.log(0.1)))
```
## Inline Question 1:
Why do we expect our loss to be close to -log(0.1)? Explain briefly.**
**Your answer:** *Because it's a random classifier. Since there are 10 classes and a random classifier will correctly classify with 10% probability.*
```
# Complete the implementation of softmax_loss_naive and implement a (naive)
# version of the gradient that uses nested loops.
loss, grad = softmax_loss_naive(W, X_dev, y_dev, 0.0)
# As we did for the SVM, use numeric gradient checking as a debugging tool.
# The numeric gradient should be close to the analytic gradient.
from cs231n.gradient_check import grad_check_sparse
f = lambda w: softmax_loss_naive(w, X_dev, y_dev, 0.0)[0]
grad_numerical = grad_check_sparse(f, W, grad, 10)
# similar to SVM case, do another gradient check with regularization
loss, grad = softmax_loss_naive(W, X_dev, y_dev, 5e1)
f = lambda w: softmax_loss_naive(w, X_dev, y_dev, 5e1)[0]
grad_numerical = grad_check_sparse(f, W, grad, 10)
# Now that we have a naive implementation of the softmax loss function and its gradient,
# implement a vectorized version in softmax_loss_vectorized.
# The two versions should compute the same results, but the vectorized version should be
# much faster.
tic = time.time()
loss_naive, grad_naive = softmax_loss_naive(W, X_dev, y_dev, 0.000005)
toc = time.time()
print('naive loss: %e computed in %fs' % (loss_naive, toc - tic))
from cs231n.classifiers.softmax import softmax_loss_vectorized
tic = time.time()
loss_vectorized, grad_vectorized = softmax_loss_vectorized(W, X_dev, y_dev, 0.000005)
toc = time.time()
print('vectorized loss: %e computed in %fs' % (loss_vectorized, toc - tic))
# As we did for the SVM, we use the Frobenius norm to compare the two versions
# of the gradient.
grad_difference = np.linalg.norm(grad_naive - grad_vectorized, ord='fro')
print('Loss difference: %f' % np.abs(loss_naive - loss_vectorized))
print('Gradient difference: %f' % grad_difference)
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of over 0.35 on the validation set.
from cs231n.classifiers import Softmax
results = {}
best_val = -1
best_softmax = None
learning_rates = [5e-6, 1e-7, 5e-7]
regularization_strengths = [1e3, 2.5e4, 5e4]
################################################################################
# TODO: #
# Use the validation set to set the learning rate and regularization strength. #
# This should be identical to the validation that you did for the SVM; save #
# the best trained softmax classifer in best_softmax. #
################################################################################
for lr in learning_rates:
for reg in regularization_strengths:
softmax = Softmax()
loss_hist = softmax.train(X_train, y_train, learning_rate=lr, reg=reg,
num_iters=1500, verbose=True)
y_train_pred = softmax.predict(X_train)
y_val_pred = softmax.predict(X_val)
training_accuracy = np.mean(y_train == y_train_pred)
validation_accuracy = np.mean(y_val == y_val_pred)
#append in results
results[(lr,reg)] = (training_accuracy, validation_accuracy)
if validation_accuracy > best_val:
best_val = validation_accuracy
best_softmax = softmax
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print('best validation accuracy achieved during cross-validation: %f' % best_val)
# evaluate on test set
# Evaluate the best softmax on test set
y_test_pred = best_softmax.predict(X_test)
test_accuracy = np.mean(y_test == y_test_pred)
print('softmax on raw pixels final test set accuracy: %f' % (test_accuracy, ))
# Visualize the learned weights for each class
w = best_softmax.W[:-1,:] # strip out the bias
w = w.reshape(32, 32, 3, 10)
w_min, w_max = np.min(w), np.max(w)
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
for i in range(10):
plt.subplot(2, 5, i + 1)
# Rescale the weights to be between 0 and 255
wimg = 255.0 * (w[:, :, :, i].squeeze() - w_min) / (w_max - w_min)
plt.imshow(wimg.astype('uint8'))
plt.axis('off')
plt.title(classes[i])
```
| github_jupyter |
<a href="https://colab.research.google.com/github/ai-fast-track/timeseries/blob/master/nbs/index.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# `timeseries` package for fastai v2
> **`timeseries`** is a Timeseries Classification and Regression package for fastai v2.
> It mimics the fastai v2 vision module (fastai2.vision).
> This notebook is a tutorial that shows, and trains an end-to-end a timeseries dataset.
> The dataset example is the NATOPS dataset (see description here beow).
> First, 4 different methods of creation on how to create timeseries dataloaders are presented.
> Then, we train a model based on [Inception Time] (https://arxiv.org/pdf/1909.04939.pdf) architecture
## Credit
> timeseries for fastai v2 was inspired by by Ignacio's Oguiza timeseriesAI (https://github.com/timeseriesAI/timeseriesAI.git).
> Inception Time model definition is a modified version of [Ignacio Oguiza] (https://github.com/timeseriesAI/timeseriesAI/blob/master/torchtimeseries/models/InceptionTime.py) and [Thomas Capelle] (https://github.com/tcapelle/TimeSeries_fastai/blob/master/inception.py) implementaions
## Installing **`timeseries`** on local machine as an editable package
1- Only if you have not already installed `fastai v2`
Install [fastai2](https://dev.fast.ai/#Installing) by following the steps described there.
2- Install timeseries package by following the instructions here below:
```
git clone https://github.com/ai-fast-track/timeseries.git
cd timeseries
pip install -e .
```
# pip installing **`timeseries`** from repo either locally or in Google Colab - Start Here
## Installing fastai v2
```
!pip install git+https://github.com/fastai/fastai2.git
```
## Installing `timeseries` package from github
```
!pip install git+https://github.com/ai-fast-track/timeseries.git
```
# *pip Installing - End Here*
# `Usage`
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai2.basics import *
# hide
# Only for Windows users because symlink to `timeseries` folder is not recognized by Windows
import sys
sys.path.append("..")
from timeseries.all import *
```
# Tutorial on timeseries package for fastai v2
## Example : NATOS dataset
<img src="https://github.com/ai-fast-track/timeseries/blob/master/images/NATOPS.jpg?raw=1">
## Right Arm vs Left Arm (3: 'Not clear' Command (see picture here above))
<br>
<img src="https://github.com/ai-fast-track/timeseries/blob/master/images/ts-right-arm.png?raw=1"><img src="https://github.com/ai-fast-track/timeseries/blob/master/images/ts-left-arm.png?raw=1">
## Description
The data is generated by sensors on the hands, elbows, wrists and thumbs. The data are the x,y,z coordinates for each of the eight locations. The order of the data is as follows:
## Channels (24)
0. Hand tip left, X coordinate
1. Hand tip left, Y coordinate
2. Hand tip left, Z coordinate
3. Hand tip right, X coordinate
4. Hand tip right, Y coordinate
5. Hand tip right, Z coordinate
6. Elbow left, X coordinate
7. Elbow left, Y coordinate
8. Elbow left, Z coordinate
9. Elbow right, X coordinate
10. Elbow right, Y coordinate
11. Elbow right, Z coordinate
12. Wrist left, X coordinate
13. Wrist left, Y coordinate
14. Wrist left, Z coordinate
15. Wrist right, X coordinate
16. Wrist right, Y coordinate
17. Wrist right, Z coordinate
18. Thumb left, X coordinate
19. Thumb left, Y coordinate
20. Thumb left, Z coordinate
21. Thumb right, X coordinate
22. Thumb right, Y coordinate
23. Thumb right, Z coordinate
## Classes (6)
The six classes are separate actions, with the following meaning:
1: I have command
2: All clear
3: Not clear
4: Spread wings
5: Fold wings
6: Lock wings
## Download data using `download_unzip_data_UCR(dsname=dsname)` method
```
dsname = 'NATOPS' #'NATOPS', 'LSST', 'Wine', 'Epilepsy', 'HandMovementDirection'
# url = 'http://www.timeseriesclassification.com/Downloads/NATOPS.zip'
path = unzip_data(URLs_TS.NATOPS)
path
```
## Why do I have to concatenate train and test data?
Both Train and Train dataset contains 180 samples each. We concatenate them in order to have one big dataset and then split into train and valid dataset using our own split percentage (20%, 30%, or whatever number you see fit)
```
fname_train = f'{dsname}_TRAIN.arff'
fname_test = f'{dsname}_TEST.arff'
fnames = [path/fname_train, path/fname_test]
fnames
data = TSData.from_arff(fnames)
print(data)
items = data.get_items()
idx = 1
x1, y1 = data.x[idx], data.y[idx]
y1
# You can select any channel to display buy supplying a list of channels and pass it to `chs` argument
# LEFT ARM
# show_timeseries(x1, title=y1, chs=[0,1,2,6,7,8,12,13,14,18,19,20])
# RIGHT ARM
# show_timeseries(x1, title=y1, chs=[3,4,5,9,10,11,15,16,17,21,22,23])
# ?show_timeseries(x1, title=y1, chs=range(0,24,3)) # Only the x axis coordinates
seed = 42
splits = RandomSplitter(seed=seed)(range_of(items)) #by default 80% for train split and 20% for valid split are chosen
splits
```
# Using `Datasets` class
## Creating a Datasets object
```
tfms = [[ItemGetter(0), ToTensorTS()], [ItemGetter(1), Categorize()]]
# Create a dataset
ds = Datasets(items, tfms, splits=splits)
ax = show_at(ds, 2, figsize=(1,1))
```
# Create a `Dataloader` objects
## 1st method : using `Datasets` object
```
bs = 128
# Normalize at batch time
tfm_norm = Normalize(scale_subtype = 'per_sample_per_channel', scale_range=(0, 1)) # per_sample , per_sample_per_channel
# tfm_norm = Standardize(scale_subtype = 'per_sample')
batch_tfms = [tfm_norm]
dls1 = ds.dataloaders(bs=bs, val_bs=bs * 2, after_batch=batch_tfms, num_workers=0, device=default_device())
dls1.show_batch(max_n=9, chs=range(0,12,3))
```
# Using `DataBlock` class
## 2nd method : using `DataBlock` and `DataBlock.get_items()`
```
getters = [ItemGetter(0), ItemGetter(1)]
tsdb = DataBlock(blocks=(TSBlock, CategoryBlock),
get_items=get_ts_items,
getters=getters,
splitter=RandomSplitter(seed=seed),
batch_tfms = batch_tfms)
tsdb.summary(fnames)
# num_workers=0 is Microsoft Windows
dls2 = tsdb.dataloaders(fnames, num_workers=0, device=default_device())
dls2.show_batch(max_n=9, chs=range(0,12,3))
```
## 3rd method : using `DataBlock` and passing `items` object to the `DataBlock.dataloaders()`
```
getters = [ItemGetter(0), ItemGetter(1)]
tsdb = DataBlock(blocks=(TSBlock, CategoryBlock),
getters=getters,
splitter=RandomSplitter(seed=seed))
dls3 = tsdb.dataloaders(data.get_items(), batch_tfms=batch_tfms, num_workers=0, device=default_device())
dls3.show_batch(max_n=9, chs=range(0,12,3))
```
## 4th method : using `TSDataLoaders` class and `TSDataLoaders.from_files()`
```
dls4 = TSDataLoaders.from_files(fnames, batch_tfms=batch_tfms, num_workers=0, device=default_device())
dls4.show_batch(max_n=9, chs=range(0,12,3))
```
# Train Model
```
# Number of channels (i.e. dimensions in ARFF and TS files jargon)
c_in = get_n_channels(dls2.train) # data.n_channels
# Number of classes
c_out= dls2.c
c_in,c_out
```
## Create model
```
model = inception_time(c_in, c_out).to(device=default_device())
model
```
## Create Learner object
```
#Learner
opt_func = partial(Adam, lr=3e-3, wd=0.01)
loss_func = LabelSmoothingCrossEntropy()
learn = Learner(dls2, model, opt_func=opt_func, loss_func=loss_func, metrics=accuracy)
print(learn.summary())
```
## LR find
```
lr_min, lr_steep = learn.lr_find()
lr_min, lr_steep
```
## Train
```
#lr_max=1e-3
epochs=30; lr_max=lr_steep; pct_start=.7; moms=(0.95,0.85,0.95); wd=1e-2
learn.fit_one_cycle(epochs, lr_max=lr_max, pct_start=pct_start, moms=moms, wd=wd)
# learn.fit_one_cycle(epochs=20, lr_max=lr_steep)
```
## Plot loss function
```
learn.recorder.plot_loss()
```
## Show results
```
learn.show_results(max_n=9, chs=range(0,12,3))
#hide
from nbdev.export import notebook2script
# notebook2script()
notebook2script(fname='index.ipynb')
# #hide
# from nbdev.export2html import _notebook2html
# # notebook2script()
# _notebook2html(fname='index.ipynb')
```
# Fin
<img src="https://github.com/ai-fast-track/timeseries/blob/master/images/tree.jpg?raw=1" width="1440" height="840" alt=""/>
| github_jupyter |
<a href="https://colab.research.google.com/github/mouctarbalde/concrete-strength-prediction/blob/main/Cement_prediction_.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import r2_score
from sklearn.preprocessing import RobustScaler
from sklearn.linear_model import Lasso
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import StackingRegressor
import warnings
import random
seed = 42
random.seed(seed)
import numpy as np
np.random.seed(seed)
warnings.filterwarnings('ignore')
plt.style.use('ggplot')
df = pd.read_csv('https://raw.githubusercontent.com/mouctarbalde/concrete-strength-prediction/main/Train.csv')
df.head()
columns_name = df.columns.to_list()
columns_name =['Cement',
'Blast_Furnace_Slag',
'Fly_Ash',
'Water',
'Superplasticizer',
'Coarse Aggregate',
'Fine Aggregate',
'Age_day',
'Concrete_compressive_strength']
df.columns = columns_name
df.info()
df.shape
import missingno as ms
ms.matrix(df)
df.isna().sum()
df.describe().T
df.corr()['Concrete_compressive_strength'].sort_values().plot(kind='barh')
plt.title("Correlation based on the target variable.")
plt.show()
sns.heatmap(df.corr(),annot=True)
sns.boxplot(x='Water', y = 'Cement',data=df)
plt.figure(figsize=(15,9))
df.boxplot()
sns.regplot(x='Water', y = 'Cement',data=df)
```
As we can see from the above cell there is not correlation between **water** and our target variable.
```
sns.boxplot(x='Age_day', y = 'Cement',data=df)
sns.regplot(x='Age_day', y = 'Cement',data=df)
X = df.drop('Concrete_compressive_strength',axis=1)
y = df.Concrete_compressive_strength
X.head()
y.head()
X_train,X_test,y_train,y_test = train_test_split(X, y, test_size=.2, random_state=seed)
X_train.shape ,y_train.shape
```
In our case we notice from our analysis the presence of outliers although they are not many we are going to use Robustscaler from sklearn to scale the data.
Robust scaler is going to remove the median and put variance to 1 it will also transform the data by removing outliers(24%-75%) is considered.
```
scale = RobustScaler()
# note we have to fit_transform only on the training data. On your test data you only have to transform.
X_train = scale.fit_transform(X_train)
X_test = scale.transform(X_test)
X_train
```
# Model creation
### Linear Regression
```
lr = LinearRegression()
lr.fit(X_train,y_train)
pred_lr = lr.predict(X_test)
pred_lr[:10]
mae_lr = mean_absolute_error(y_test,pred_lr)
r2_lr = r2_score(y_test,pred_lr)
print(f'Mean absolute error of linear regression is {mae_lr}')
print(f'R2 score of Linear Regression is {r2_lr}')
```
**Graph for linear regression** the below graph is showing the relationship between the actual and the predicted values.
```
fig, ax = plt.subplots()
ax.scatter(pred_lr, y_test)
ax.plot([y_test.min(), y_test.max()],[y_test.min(), y_test.max()], color = 'red', marker = "*", markersize = 10)
```
### Decision tree Regression
```
dt = DecisionTreeRegressor(criterion='mae')
dt.fit(X_train,y_train)
pred_dt = dt.predict(X_test)
mae_dt = mean_absolute_error(y_test,pred_dt)
r2_dt = r2_score(y_test,pred_dt)
print(f'Mean absolute error of linear regression is {mae_dt}')
print(f'R2 score of Decision tree regressor is {r2_dt}')
fig, ax = plt.subplots()
plt.title('Linear relationship for decison tree')
ax.scatter(pred_dt, y_test)
ax.plot([y_test.min(), y_test.max()],[y_test.min(), y_test.max()], color = 'red', marker = "*", markersize = 10)
```
### Random Forest Regression
```
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor()
rf.fit(X_train, y_train)
# prediction
pred_rf = rf.predict(X_test)
mae_rf = mean_absolute_error(y_test,pred_rf)
r2_rf = r2_score(y_test,pred_rf)
print(f'Mean absolute error of Random forst regression is {mae_rf}')
print(f'R2 score of Random forst regressor is {r2_rf}')
fig, ax = plt.subplots()
plt.title('Linear relationship for random forest regressor')
ax.scatter(pred_rf, y_test)
ax.plot([y_test.min(), y_test.max()],[y_test.min(), y_test.max()], color = 'red', marker = "*", markersize = 10)
```
# Lasso Regression
```
laso = Lasso()
laso.fit(X_train, y_train)
pred_laso = laso.predict(X_test)
mae_laso = mean_absolute_error(y_test, pred_laso)
r2_laso = r2_score(y_test, pred_laso)
print(f'Mean absolute error of Random forst regression is {mae_laso}')
print(f'R2 score of Random forst regressor is {r2_laso}')
fig, ax = plt.subplots()
plt.title('Linear relationship for Lasso regressor')
ax.scatter(pred_laso, y_test)
ax.plot([y_test.min(), y_test.max()],[y_test.min(), y_test.max()], color = 'red', marker = "*", markersize = 10)
gb = GradientBoostingRegressor()
gb.fit(X_train, y_train)
pred_gb = gb.predict(X_test)
mae_gb = mean_absolute_error(y_test, pred_gb)
r2_gb = r2_score(y_test, pred_gb)
print(f'Mean absolute error of Random forst regression is {mae_gb}')
print(f'R2 score of Random forst regressor is {r2_gb}')
fig, ax = plt.subplots()
plt.title('Linear relationship for Lasso regressor')
ax.scatter(pred_gb, y_test)
ax.plot([y_test.min(), y_test.max()],[y_test.min(), y_test.max()], color = 'red', marker = "*", markersize = 10)
```
# Stacking Regressor:
combining multiple regression model and choosing the final model. in our case we used kfold cross validation to make sure that the model is not overfitting.
```
estimators = [('lr',LinearRegression()), ('gb',GradientBoostingRegressor()),\
('dt',DecisionTreeRegressor()), ('laso',Lasso())]
from sklearn.model_selection import KFold
kf = KFold(n_splits=10,shuffle=True, random_state=seed)
stacking = StackingRegressor(estimators=estimators, final_estimator=RandomForestRegressor(random_state=seed), cv=kf)
stacking.fit(X_train, y_train)
pred_stack = stacking.predict(X_test)
mae_stack = mean_absolute_error(y_test, pred_stack)
r2_stack = r2_score(y_test, pred_stack)
print(f'Mean absolute error of Random forst regression is {mae_stack}')
print(f'R2 score of Random forst regressor is {r2_stack}')
fig, ax = plt.subplots()
plt.title('Linear relationship for Stacking regressor')
ax.scatter(pred_stack, y_test)
ax.plot([y_test.min(), y_test.max()],[y_test.min(), y_test.max()], color = 'red', marker = "*", markersize = 10)
result = pd.DataFrame({'Model':['Linear Regression','Decison tree','Random Forest', 'Lasso',\
'Gradient Boosting Regressor', 'Stacking Regressor'],
'MAE':[mae_lr, mae_dt, mae_rf, mae_laso, mae_gb, mae_stack],
'R2 score':[r2_lr, r2_dt, r2_rf, r2_laso, r2_gb, r2_stack]
})
result
```
| github_jupyter |
# The Extended Kalman Filter
선형 칼만 필터 (Linear Kalman Filter)에 대한 이론을 바탕으로 비선형 문제에 칼만 필터를 적용해 보겠습니다. 확장칼만필터 (EKF)는 예측단계와 추정단계의 데이터를 비선형으로 가정하고 현재의 추정값에 대해 시스템을 선형화 한뒤 선형 칼만 필터를 사용하는 기법입니다.
비선형 문제에 적용되는 성능이 더 좋은 알고리즘들 (UKF, H_infinity)이 있지만 EKF 는 아직도 널리 사용되서 관련성이 높습니다.
```
%matplotlib inline
# HTML("""
# <style>
# .output_png {
# display: table-cell;
# text-align: center;
# vertical-align: middle;
# }
# </style>
# """)
```
## Linearizing the Kalman Filter
### Non-linear models
칼만 필터는 시스템이 선형일것이라는 가정을 하기 때문에 비선형 문제에는 직접적으로 사용하지 못합니다. 비선형성은 두가지 원인에서 기인될수 있는데 첫째는 프로세스 모델의 비선형성 그리고 둘째 측정 모델의 비선형성입니다. 예를 들어, 떨어지는 물체의 가속도는 속도의 제곱에 비례하는 공기저항에 의해 결정되기 때문에 비선형적인 프로세스 모델을 가지고, 레이더로 목표물의 범위와 방위 (bearing) 를 측정할때 비선형함수인 삼각함수를 사용하여 표적의 위치를 계산하기 때문에 비선형적인 측정 모델을 가지게 됩니다.
비선형문제에 기존의 칼만필터 방정식을 적용하지 못하는 이유는 비선형함수에 정규분포 (Gaussian)를 입력하면 아래와 같이 Gaussian 이 아닌 분포를 가지게 되기 때문입니다.
```
import numpy as np
import scipy.stats as stats
import matplotlib.pyplot as plt
mu, sigma = 0, 0.1
gaussian = stats.norm.pdf(x, mu, sigma)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 10000)
def nonlinearFunction(x):
return np.sin(x)
def linearFunction(x):
return 0.5*x
nonlinearOutput = nonlinearFunction(gaussian)
linearOutput = linearFunction(gaussian)
# print(x)
plt.plot(x, gaussian, label = 'Gaussian Input')
plt.plot(x, linearOutput, label = 'Linear Output')
plt.plot(x, nonlinearOutput, label = 'Nonlinear Output')
plt.grid(linestyle='dotted', linewidth=0.8)
plt.legend()
plt.show()
```
### System Equations
선형 칼만 필터의 경우 프로세스 및 측정 모델은 다음과 같이 나타낼수 있습니다.
$$\begin{aligned}\dot{\mathbf x} &= \mathbf{Ax} + w_x\\
\mathbf z &= \mathbf{Hx} + w_z
\end{aligned}$$
이때 $\mathbf A$ 는 (연속시간에서) 시스템의 역학을 묘사하는 dynamic matrix 입니다. 위의 식을 이산화(discretize)시키면 아래와 같이 나타내줄 수 있습니다.
$$\begin{aligned}\bar{\mathbf x}_k &= \mathbf{F} \mathbf{x}_{k-1} \\
\bar{\mathbf z} &= \mathbf{H} \mathbf{x}_{k-1}
\end{aligned}$$
이때 $\mathbf F$ 는 이산시간 $\Delta t$ 에 걸쳐 $\mathbf x_{k-1}$을 $\mathbf x_{k}$ 로 전환하는 상태변환행렬 또는 상태전달함수 (state transition matrix) 이고, 위에서의 $w_x$ 와 $w_z$는 각각 프로세스 노이즈 공분산 행렬 $\mathbf Q$ 과 측정 노이즈 공분산 행렬 $\mathbf R$ 에 포함됩니다.
선형 시스템에서의 $\mathbf F \mathbf x- \mathbf B \mathbf u$ 와 $\mathbf H \mathbf x$ 는 비선형 시스템에서 함수 $f(\mathbf x, \mathbf u)$ 와 $h(\mathbf x)$ 로 대체됩니다.
$$\begin{aligned}\dot{\mathbf x} &= f(\mathbf x, \mathbf u) + w_x\\
\mathbf z &= h(\mathbf x) + w_z
\end{aligned}$$
### Linearisation
선형화란 말그대로 하나의 시점에 대하여 비선형함수에 가장 가까운 선 (선형시스템) 을 찾는것이라고 볼수 있습니다. 여러가지 방법으로 선형화를 할수 있겠지만 흔히 일차 테일러 급수를 사용합니다. ($ c_0$ 과 $c_1 x$)
$$f(x) = \sum_{k=0}^\infty c_k x^k = c_0 + c_1 x + c_2 x^2 + \dotsb$$
$$c_k = \frac{f^{\left(k\right)}(0)}{k!} = \frac{1}{k!} \cdot \frac{d^k f}{dx^k}\bigg|_0 $$
행렬의 미분값을 Jacobian 이라고 하는데 이를 통해서 위와 같이 $\mathbf F$ 와 $\mathbf H$ 를 나타낼 수 있습니다.
$$
\begin{aligned}
\mathbf F
= {\frac{\partial{f(\mathbf x_t, \mathbf u_t)}}{\partial{\mathbf x}}}\biggr|_{{\mathbf x_t},{\mathbf u_t}} \;\;\;\;
\mathbf H = \frac{\partial{h(\bar{\mathbf x}_t)}}{\partial{\bar{\mathbf x}}}\biggr|_{\bar{\mathbf x}_t}
\end{aligned}
$$
$$\mathbf F = \frac{\partial f(\mathbf x, \mathbf u)}{\partial x} =\begin{bmatrix}
\frac{\partial f_1}{\partial x_1} & \frac{\partial f_1}{\partial x_2} & \dots & \frac{\partial f_1}{\partial x_n}\\
\frac{\partial f_2}{\partial x_1} & \frac{\partial f_2}{\partial x_2} & \dots & \frac{\partial f_2}{\partial x_n} \\
\\ \vdots & \vdots & \ddots & \vdots
\\
\frac{\partial f_n}{\partial x_1} & \frac{\partial f_n}{\partial x_2} & \dots & \frac{\partial f_n}{\partial x_n}
\end{bmatrix}
$$
Linear Kalman Filter 와 Extended Kalman Filter 의 식들을 아래와 같이 비교할수 있습니다.
$$\begin{array}{l|l}
\text{Linear Kalman filter} & \text{EKF} \\
\hline
& \boxed{\mathbf F = {\frac{\partial{f(\mathbf x_t, \mathbf u_t)}}{\partial{\mathbf x}}}\biggr|_{{\mathbf x_t},{\mathbf u_t}}} \\
\mathbf{\bar x} = \mathbf{Fx} + \mathbf{Bu} & \boxed{\mathbf{\bar x} = f(\mathbf x, \mathbf u)} \\
\mathbf{\bar P} = \mathbf{FPF}^\mathsf{T}+\mathbf Q & \mathbf{\bar P} = \mathbf{FPF}^\mathsf{T}+\mathbf Q \\
\hline
& \boxed{\mathbf H = \frac{\partial{h(\bar{\mathbf x}_t)}}{\partial{\bar{\mathbf x}}}\biggr|_{\bar{\mathbf x}_t}} \\
\textbf{y} = \mathbf z - \mathbf{H \bar{x}} & \textbf{y} = \mathbf z - \boxed{h(\bar{x})}\\
\mathbf{K} = \mathbf{\bar{P}H}^\mathsf{T} (\mathbf{H\bar{P}H}^\mathsf{T} + \mathbf R)^{-1} & \mathbf{K} = \mathbf{\bar{P}H}^\mathsf{T} (\mathbf{H\bar{P}H}^\mathsf{T} + \mathbf R)^{-1} \\
\mathbf x=\mathbf{\bar{x}} +\mathbf{K\textbf{y}} & \mathbf x=\mathbf{\bar{x}} +\mathbf{K\textbf{y}} \\
\mathbf P= (\mathbf{I}-\mathbf{KH})\mathbf{\bar{P}} & \mathbf P= (\mathbf{I}-\mathbf{KH})\mathbf{\bar{P}}
\end{array}$$
$\mathbf F \mathbf x_{k-1}$ 을 사용하여 $\mathbf x_{k}$의 값을 추정할수 있겠지만, 선형화 과정에서 오차가 생길수 있기 때문에 Euler 또는 Runge Kutta 수치 적분 (numerical integration) 을 통해서 사전추정값 $\mathbf{\bar{x}}$ 를 구합니다. 같은 이유로 $\mathbf y$ (innovation vector 또는 잔차(residual)) 를 구할때도 $\mathbf H \mathbf x$ 대신에 수치적인 방법으로 계산하게 됩니다.
## Example: Robot Localization
### Prediction Model (예측모델)
EKF를 4륜 로봇에 적용시켜 보겠습니다. 간단한 bicycle steering model 을 통해 아래의 시스템 모델을 나타낼 수 있습니다.
```
import kf_book.ekf_internal as ekf_internal
ekf_internal.plot_bicycle()
```
$$\begin{aligned}
\beta &= \frac d w \tan(\alpha) \\
\bar x_k &= x_{k-1} - R\sin(\theta) + R\sin(\theta + \beta) \\
\bar y_k &= y_{k-1} + R\cos(\theta) - R\cos(\theta + \beta) \\
\bar \theta_k &= \theta_{k-1} + \beta
\end{aligned}
$$
위의 식들을 토대로 상태벡터를 $\mathbf{x}=[x, y, \theta]^T$ 그리고 입력벡터를 $\mathbf{u}=[v, \alpha]^T$ 라고 정의 해주면 아래와 같이 $f(\mathbf x, \mathbf u)$ 나타내줄수 있고 $f$ 의 Jacobian $\mathbf F$를 미분하여 아래의 행렬을 구해줄수 있습니다.
$$\bar x = f(x, u) + \mathcal{N}(0, Q)$$
$$f = \begin{bmatrix}x\\y\\\theta\end{bmatrix} +
\begin{bmatrix}- R\sin(\theta) + R\sin(\theta + \beta) \\
R\cos(\theta) - R\cos(\theta + \beta) \\
\beta\end{bmatrix}$$
$$\mathbf F = \frac{\partial f(\mathbf x, \mathbf u)}{\partial \mathbf x} = \begin{bmatrix}
1 & 0 & -R\cos(\theta) + R\cos(\theta+\beta) \\
0 & 1 & -R\sin(\theta) + R\sin(\theta+\beta) \\
0 & 0 & 1
\end{bmatrix}$$
$\bar{\mathbf P}$ 을 구하기 위해 입력($\mathbf u$)에서 비롯되는 프로세스 노이즈 $\mathbf Q$ 를 아래와 같이 정의합니다.
$$\mathbf{M} = \begin{bmatrix}\sigma_{vel}^2 & 0 \\ 0 & \sigma_\alpha^2\end{bmatrix}
\;\;\;\;
\mathbf{V} = \frac{\partial f(x, u)}{\partial u} \begin{bmatrix}
\frac{\partial f_1}{\partial v} & \frac{\partial f_1}{\partial \alpha} \\
\frac{\partial f_2}{\partial v} & \frac{\partial f_2}{\partial \alpha} \\
\frac{\partial f_3}{\partial v} & \frac{\partial f_3}{\partial \alpha}
\end{bmatrix}$$
$$\mathbf{\bar P} =\mathbf{FPF}^{\mathsf T} + \mathbf{VMV}^{\mathsf T}$$
```
import sympy
from sympy.abc import alpha, x, y, v, w, R, theta
from sympy import symbols, Matrix
sympy.init_printing(use_latex="mathjax", fontsize='16pt')
time = symbols('t')
d = v*time
beta = (d/w)*sympy.tan(alpha)
r = w/sympy.tan(alpha)
fxu = Matrix([[x-r*sympy.sin(theta) + r*sympy.sin(theta+beta)],
[y+r*sympy.cos(theta)- r*sympy.cos(theta+beta)],
[theta+beta]])
F = fxu.jacobian(Matrix([x, y, theta]))
F
# reduce common expressions
B, R = symbols('beta, R')
F = F.subs((d/w)*sympy.tan(alpha), B)
F.subs(w/sympy.tan(alpha), R)
V = fxu.jacobian(Matrix([v, alpha]))
V = V.subs(sympy.tan(alpha)/w, 1/R)
V = V.subs(time*v/R, B)
V = V.subs(time*v, 'd')
V
```
### Measurement Model (측정모델)
레이더로 범위$(r)$와 방위($\phi$)를 측정할때 다음과 같은 센서모델을 사용합니다. 이때 $\mathbf p$ 는 landmark의 위치를 나타내줍니다.
$$r = \sqrt{(p_x - x)^2 + (p_y - y)^2}
\;\;\;\;
\phi = \arctan(\frac{p_y - y}{p_x - x}) - \theta
$$
$$\begin{aligned}
\mathbf z& = h(\bar{\mathbf x}, \mathbf p) &+ \mathcal{N}(0, R)\\
&= \begin{bmatrix}
\sqrt{(p_x - x)^2 + (p_y - y)^2} \\
\arctan(\frac{p_y - y}{p_x - x}) - \theta
\end{bmatrix} &+ \mathcal{N}(0, R)
\end{aligned}$$
$h$ 의 Jacobian $\mathbf H$를 미분하여 아래의 행렬을 구해줄수 있습니다.
$$\mathbf H = \frac{\partial h(\mathbf x, \mathbf u)}{\partial \mathbf x} =
\left[\begin{matrix}\frac{- p_{x} + x}{\sqrt{\left(p_{x} - x\right)^{2} + \left(p_{y} - y\right)^{2}}} & \frac{- p_{y} + y}{\sqrt{\left(p_{x} - x\right)^{2} + \left(p_{y} - y\right)^{2}}} & 0\\- \frac{- p_{y} + y}{\left(p_{x} - x\right)^{2} + \left(p_{y} - y\right)^{2}} & - \frac{p_{x} - x}{\left(p_{x} - x\right)^{2} + \left(p_{y} - y\right)^{2}} & -1\end{matrix}\right]
$$
```
import sympy
from sympy.abc import alpha, x, y, v, w, R, theta
px, py = sympy.symbols('p_x, p_y')
z = sympy.Matrix([[sympy.sqrt((px-x)**2 + (py-y)**2)],
[sympy.atan2(py-y, px-x) - theta]])
z.jacobian(sympy.Matrix([x, y, theta]))
# print(sympy.latex(z.jacobian(sympy.Matrix([x, y, theta])))
from math import sqrt
def H_of(x, landmark_pos):
""" compute Jacobian of H matrix where h(x) computes
the range and bearing to a landmark for state x """
px = landmark_pos[0]
py = landmark_pos[1]
hyp = (px - x[0, 0])**2 + (py - x[1, 0])**2
dist = sqrt(hyp)
H = array(
[[-(px - x[0, 0]) / dist, -(py - x[1, 0]) / dist, 0],
[ (py - x[1, 0]) / hyp, -(px - x[0, 0]) / hyp, -1]])
return H
from math import atan2
def Hx(x, landmark_pos):
""" takes a state variable and returns the measurement
that would correspond to that state.
"""
px = landmark_pos[0]
py = landmark_pos[1]
dist = sqrt((px - x[0, 0])**2 + (py - x[1, 0])**2)
Hx = array([[dist],
[atan2(py - x[1, 0], px - x[0, 0]) - x[2, 0]]])
return Hx
```
측정 노이즈는 다음과 같이 나타내줍니다.
$$\mathbf R=\begin{bmatrix}\sigma_{range}^2 & 0 \\ 0 & \sigma_{bearing}^2\end{bmatrix}$$
### Implementation
`FilterPy` 의 `ExtendedKalmanFilter` class 를 활용해서 EKF 를 구현해보도록 하겠습니다.
```
from filterpy.kalman import ExtendedKalmanFilter as EKF
from numpy import array, sqrt, random
import sympy
class RobotEKF(EKF):
def __init__(self, dt, wheelbase, std_vel, std_steer):
EKF.__init__(self, 3, 2, 2)
self.dt = dt
self.wheelbase = wheelbase
self.std_vel = std_vel
self.std_steer = std_steer
a, x, y, v, w, theta, time = sympy.symbols(
'a, x, y, v, w, theta, t')
d = v*time
beta = (d/w)*sympy.tan(a)
r = w/sympy.tan(a)
self.fxu = sympy.Matrix(
[[x-r*sympy.sin(theta)+r*sympy.sin(theta+beta)],
[y+r*sympy.cos(theta)-r*sympy.cos(theta+beta)],
[theta+beta]])
self.F_j = self.fxu.jacobian(sympy.Matrix([x, y, theta]))
self.V_j = self.fxu.jacobian(sympy.Matrix([v, a]))
# save dictionary and it's variables for later use
self.subs = {x: 0, y: 0, v:0, a:0,
time:dt, w:wheelbase, theta:0}
self.x_x, self.x_y, = x, y
self.v, self.a, self.theta = v, a, theta
def predict(self, u):
self.x = self.move(self.x, u, self.dt)
self.subs[self.theta] = self.x[2, 0]
self.subs[self.v] = u[0]
self.subs[self.a] = u[1]
F = array(self.F_j.evalf(subs=self.subs)).astype(float)
V = array(self.V_j.evalf(subs=self.subs)).astype(float)
# covariance of motion noise in control space
M = array([[self.std_vel*u[0]**2, 0],
[0, self.std_steer**2]])
self.P = F @ self.P @ F.T + V @ M @ V.T
def move(self, x, u, dt):
hdg = x[2, 0]
vel = u[0]
steering_angle = u[1]
dist = vel * dt
if abs(steering_angle) > 0.001: # is robot turning?
beta = (dist / self.wheelbase) * tan(steering_angle)
r = self.wheelbase / tan(steering_angle) # radius
dx = np.array([[-r*sin(hdg) + r*sin(hdg + beta)],
[r*cos(hdg) - r*cos(hdg + beta)],
[beta]])
else: # moving in straight line
dx = np.array([[dist*cos(hdg)],
[dist*sin(hdg)],
[0]])
return x + dx
```
정확한 잔차값 $y$을 구하기 방위값이 $0 \leq \phi \leq 2\pi$ 이도록 고쳐줍니다.
```
def residual(a, b):
""" compute residual (a-b) between measurements containing
[range, bearing]. Bearing is normalized to [-pi, pi)"""
y = a - b
y[1] = y[1] % (2 * np.pi) # force in range [0, 2 pi)
if y[1] > np.pi: # move to [-pi, pi)
y[1] -= 2 * np.pi
return y
from filterpy.stats import plot_covariance_ellipse
from math import sqrt, tan, cos, sin, atan2
import matplotlib.pyplot as plt
dt = 1.0
def z_landmark(lmark, sim_pos, std_rng, std_brg):
x, y = sim_pos[0, 0], sim_pos[1, 0]
d = np.sqrt((lmark[0] - x)**2 + (lmark[1] - y)**2)
a = atan2(lmark[1] - y, lmark[0] - x) - sim_pos[2, 0]
z = np.array([[d + random.randn()*std_rng],
[a + random.randn()*std_brg]])
return z
def ekf_update(ekf, z, landmark):
ekf.update(z, HJacobian = H_of, Hx = Hx,
residual=residual,
args=(landmark), hx_args=(landmark))
def run_localization(landmarks, std_vel, std_steer,
std_range, std_bearing,
step=10, ellipse_step=20, ylim=None):
ekf = RobotEKF(dt, wheelbase=0.5, std_vel=std_vel,
std_steer=std_steer)
ekf.x = array([[2, 6, .3]]).T # x, y, steer angle
ekf.P = np.diag([.1, .1, .1])
ekf.R = np.diag([std_range**2, std_bearing**2])
sim_pos = ekf.x.copy() # simulated position
# steering command (vel, steering angle radians)
u = array([1.1, .01])
plt.figure()
plt.scatter(landmarks[:, 0], landmarks[:, 1],
marker='s', s=60)
track = []
for i in range(200):
sim_pos = ekf.move(sim_pos, u, dt/10.) # simulate robot
track.append(sim_pos)
if i % step == 0:
ekf.predict(u=u)
if i % ellipse_step == 0:
plot_covariance_ellipse(
(ekf.x[0,0], ekf.x[1,0]), ekf.P[0:2, 0:2],
std=6, facecolor='k', alpha=0.3)
x, y = sim_pos[0, 0], sim_pos[1, 0]
for lmark in landmarks:
z = z_landmark(lmark, sim_pos,
std_range, std_bearing)
ekf_update(ekf, z, lmark)
if i % ellipse_step == 0:
plot_covariance_ellipse(
(ekf.x[0,0], ekf.x[1,0]), ekf.P[0:2, 0:2],
std=6, facecolor='g', alpha=0.8)
track = np.array(track)
plt.plot(track[:, 0], track[:,1], color='k', lw=2)
plt.axis('equal')
plt.title("EKF Robot localization")
if ylim is not None: plt.ylim(*ylim)
plt.show()
return ekf
landmarks = array([[5, 10], [10, 5], [15, 15]])
ekf = run_localization(
landmarks, std_vel=0.1, std_steer=np.radians(1),
std_range=0.3, std_bearing=0.1)
print('Final P:', ekf.P.diagonal())
```
## References
* Roger R Labbe, Kalman and Bayesian Filters in Python
(https://github.com/rlabbe/Kalman-and-Bayesian-Filters-in-Python/blob/master/11-Extended-Kalman-Filters.ipynb)
* https://blog.naver.com/jewdsa813/222200570774
| github_jupyter |
# Method for visualizing warping over training steps
```
import os
import imageio
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(0)
```
### Construct warping matrix
```
g = 1.02 # scaling parameter
# Matrix for rotating 45 degrees
rotate = np.array([[np.cos(np.pi/4), -np.sin(np.pi/4)],
[np.sin(np.pi/4), np.cos(np.pi/4)]])
# Matrix for scaling along x coordinate
scale_x = np.array([[g, 0],
[0, 1]])
# Matrix for scaling along y coordinate
scale_y = np.array([[1, 0],
[0, g]])
# Matrix for unrotating (-45 degrees)
unrotate = np.array([[np.cos(-np.pi/4), -np.sin(-np.pi/4)],
[np.sin(-np.pi/4), np.cos(-np.pi/4)]])
# Warping matrix
warp = rotate @ scale_x @ unrotate
# Unwarping matrix
unwarp = rotate @ scale_y @ unrotate
```
### Warp grid slowly over time
```
# Construct 4x4 grid
s = 1 # initial scale
locs = [[x,y] for x in range(4) for y in range(4)]
grid = s*np.array(locs)
# Matrix to collect data
n_steps = 50
warp_data = np.zeros([n_steps, 16, 2])
# Initial timestep has no warping
warp_data[0,:,:] = grid
# Warp slowly over time
for i in range(1,n_steps):
grid = grid @ warp
warp_data[i,:,:] = grid
fig, ax = plt.subplots(1,3, figsize=(15, 5), sharex=True, sharey=True)
ax[0].scatter(warp_data[0,:,0], warp_data[0,:,1])
ax[0].set_title("Warping: step 0")
ax[1].scatter(warp_data[n_steps//2,:,0], warp_data[n_steps//2,:,1])
ax[1].set_title("Warping: Step {}".format(n_steps//2))
ax[2].scatter(warp_data[n_steps-1,:,0], warp_data[n_steps-1,:,1])
ax[2].set_title("Warping: Step {}".format(n_steps-1))
plt.show()
```
### Unwarp grid slowly over time
```
# Matrix to collect data
unwarp_data = np.zeros([n_steps, 16, 2])
# Start with warped grid
unwarp_data[0,:,:] = grid
# Unwarp slowly over time
for i in range(1,n_steps):
grid = grid @ unwarp
unwarp_data[i,:,:] = grid
fig, ax = plt.subplots(1,3, figsize=(15, 5), sharex=True, sharey=True)
ax[0].scatter(unwarp_data[0,:,0], unwarp_data[0,:,1])
ax[0].set_title("Unwarping: Step 0")
# ax[0].set_ylim([-0.02, 0.05])
# ax[0].set_xlim([-0.02, 0.05])
ax[1].scatter(unwarp_data[n_steps//2,:,0], unwarp_data[n_steps//2,:,1])
ax[1].set_title("Unwarping: Step {}".format(n_steps//2))
ax[2].scatter(unwarp_data[n_steps-1,:,0], unwarp_data[n_steps-1,:,1])
ax[2].set_title("Unwarping: Step {}".format(n_steps-1))
plt.show()
```
### High-dimensional vectors with random projection matrix
```
# data = [warp_data, unwarp_data]
data = np.concatenate([warp_data, unwarp_data], axis=0)
# Random projection matrix
hidden_dim = 32
random_mat = np.random.randn(2, hidden_dim)
data = data @ random_mat
# Add noise to each time step
sigma = 0.2
noise = sigma*np.random.randn(2*n_steps, 16, hidden_dim)
data = data + noise
```
### Parameterize scatterplot with average "congruent" and "incongruent" distances
```
loc2idx = {i:(loc[0],loc[1]) for i,loc in enumerate(locs)}
idx2loc = {v:k for k,v in loc2idx.items()}
```
Function for computing distance matrix
```
def get_distances(M):
n,m = M.shape
D = np.zeros([n,n])
for i in range(n):
for j in range(n):
D[i,j] = np.linalg.norm(M[i,:] - M[j,:])
return D
D = get_distances(data[0])
plt.imshow(D)
plt.show()
```
Construct same-rank groups for "congruent" and "incongruent" diagonals
```
c_rank = np.array([loc[0] + loc[1] for loc in locs]) # rank along "congruent" diagonal
i_rank = np.array([3 + loc[0] - loc[1] for loc in locs]) # rank along "incongruent" diagonal
G_idxs = [] # same-rank group for "congruent" diagonal
H_idxs = [] # same-rank group for "incongruent" diagonal
for i in range(7): # total number of ranks (0 through 6)
G_set = [j for j in range(len(c_rank)) if c_rank[j] == i]
H_set = [j for j in range(len(i_rank)) if i_rank[j] == i]
G_idxs.append(G_set)
H_idxs.append(H_set)
```
Function for estimating $ \alpha $ and $ \beta $
$$ \bar{x_i} = \sum_{x \in G_i} \frac{1}{n} x $$
$$ \alpha_{i, i+1} = || \bar{x}_i - \bar{x}_{i+1} || $$
$$ \bar{y_i} = \sum_{y \in H_i} \frac{1}{n} y $$
$$ \beta_{i, i+1} = || \bar{y}_i - \bar{y}_{i+1} || $$
```
def get_parameters(M):
# M: [16, hidden_dim]
alpha = []
beta = []
for i in range(6): # total number of parameters (01,12,23,34,45,56)
# alpha_{i, i+1}
x_bar_i = np.mean(M[G_idxs[i],:], axis=0)
x_bar_ip1 = np.mean(M[G_idxs[i+1],:], axis=0)
x_dist = np.linalg.norm(x_bar_i - x_bar_ip1)
alpha.append(x_dist)
# beta_{i, i+1}
y_bar_i = np.mean(M[H_idxs[i],:], axis=0)
y_bar_ip1 = np.mean(M[H_idxs[i+1],:], axis=0)
y_dist = np.linalg.norm(y_bar_i - y_bar_ip1)
beta.append(y_dist)
return alpha, beta
alpha_data = []
beta_data = []
for t in range(len(data)):
alpha, beta = get_parameters(data[t])
alpha_data.append(alpha)
beta_data.append(beta)
plt.plot(alpha_data, color='tab:blue')
plt.plot(beta_data, color='tab:orange')
plt.show()
```
Use parameters to plot idealized 2D representations
```
idx2g = {}
for idx in range(16):
for g, group in enumerate(G_idxs):
if idx in group:
idx2g[idx] = g
idx2h = {}
for idx in range(16):
for h, group in enumerate(H_idxs):
if idx in group:
idx2h[idx] = h
def generate_grid(alpha, beta):
cum_alpha = np.zeros(7)
cum_beta = np.zeros(7)
cum_alpha[1:] = np.cumsum(alpha)
cum_beta[1:] = np.cumsum(beta)
# Get x and y coordinate in rotated basis
X = np.zeros([16, 2])
for idx in range(16):
g = idx2g[idx] # G group
h = idx2h[idx] # H group
X[idx,0] = cum_alpha[g] # x coordinate
X[idx,1] = cum_beta[h] # y coordinate
# Unrotate
unrotate = np.array([[np.cos(-np.pi/4), -np.sin(-np.pi/4)],
[np.sin(-np.pi/4), np.cos(-np.pi/4)]])
X = X @ unrotate
# Mean-center
X = X - np.mean(X, axis=0, keepdims=True)
return X
X = generate_grid(alpha, beta)
```
Get reconstructed grid for each time step
```
reconstruction = np.zeros([data.shape[0], data.shape[1], 2])
for t,M in enumerate(data):
alpha, beta = get_parameters(M)
X = generate_grid(alpha, beta)
reconstruction[t,:,:] = X
t = 50
plt.scatter(reconstruction[t,:,0], reconstruction[t,:,1])
plt.show()
```
### Make .gif
```
plt.scatter(M[:,0], M[:,1])
reconstruction.shape
xmin = np.min(reconstruction[:,:,0])
xmax = np.max(reconstruction[:,:,0])
ymin = np.min(reconstruction[:,:,1])
ymax = np.max(reconstruction[:,:,1])
for t,M in enumerate(reconstruction):
plt.scatter(M[:,0], M[:,1])
plt.title("Reconstructed grid")
plt.xlim([xmin-1.5, xmax+1.5])
plt.ylim([ymin-1.5, ymax+1.5])
plt.xticks([])
plt.yticks([])
plt.tight_layout()
plt.savefig('reconstruction_test_{}.png'.format(t), dpi=100)
plt.show()
filenames = ['reconstruction_test_{}.png'.format(i) for i in range(2*n_steps)]
with imageio.get_writer('reconstruction_test.gif', mode='I') as writer:
for filename in filenames:
image = imageio.imread(filename)
writer.append_data(image)
# remove files
for filename in filenames:
os.remove(filename)
```
<img src="reconstruction_test.gif" width="750" align="center">
| github_jupyter |
# Documenting Classes
It is almost as easy to document a class as it is to document a function. Simply add docstrings to all of the classes functions, and also below the class name itself. For example, here is a simple documented class
```
class Demo:
"""This class demonstrates how to document a class.
This class is just a demonstration, and does nothing.
However the principles of documentation are still valid!
"""
def __init__(self, name):
"""You should document the constructor, saying what it expects to
create a valid class. In this case
name -- the name of an object of this class
"""
self._name = name
def getName(self):
"""You should then document all of the member functions, just as
you do for normal functions. In this case, returns
the name of the object
"""
return self._name
d = Demo("cat")
help(d)
```
Often, when you write a class, you want to hide member data or member functions so that they are only visible within an object of the class. For example, above, the `self._name` member data should be hidden, as it should only be used by the object.
You control the visibility of member functions or member data using an underscore. If the member function or member data name starts with an underscore, then it is hidden. Otherwise, the member data or function is visible.
For example, we can hide the `getName` function by renaming it to `_getName`
```
class Demo:
"""This class demonstrates how to document a class.
This class is just a demonstration, and does nothing.
However the principles of documentation are still valid!
"""
def __init__(self, name):
"""You should document the constructor, saying what it expects to
create a valid class. In this case
name -- the name of an object of this class
"""
self._name = name
def _getName(self):
"""You should then document all of the member functions, just as
you do for normal functions. In this case, returns
the name of the object
"""
return self._name
d = Demo("cat")
help(d)
```
Member functions or data that are hidden are called "private". Member functions or data that are visible are called "public". You should document all public member functions of a class, as these are visible and designed to be used by other people. It is helpful, although not required, to document all of the private member functions of a class, as these will only really be called by you. However, in years to come, you will thank yourself if you still documented them... ;-)
While it is possible to make member data public, it is not advised. It is much better to get and set values of member data using public member functions. This makes it easier for you to add checks to ensure that the data is consistent and being used in the right way. For example, compare these two classes that represent a person, and hold their height.
```
class Person1:
"""Class that holds a person's height"""
def __init__(self):
"""Construct a person who has zero height"""
self.height = 0
class Person2:
"""Class that holds a person's height"""
def __init__(self):
"""Construct a person who has zero height"""
self._height = 0
def setHeight(self, height):
"""Set the person's height to 'height', returning whether or
not the height was set successfully
"""
if height < 0 or height > 300:
print("This is an invalid height! %s" % height)
return False
else:
self._height = height
return True
def getHeight(self):
"""Return the person's height"""
return self._height
```
The first example is quicker to write, but it does little to protect itself against a user who attempts to use the class badly.
```
p = Person1()
p.height = -50
p.height
p.height = "cat"
p.height
```
The second example takes more lines of code, but these lines are valuable as they check that the user is using the class correctly. These checks, when combined with good documentation, ensure that your classes can be safely used by others, and that incorrect use will not create difficult-to-find bugs.
```
p = Person2()
p.setHeight(-50)
p.getHeight()
p.setHeight("cat")
p.getHeight()
```
# Exercise
## Exercise 1
Below is the completed `GuessGame` class from the previous lesson. Add documentation to this class.
```
class GuessGame:
"""
This class provides a simple guessing game. You create an object
of the class with its own secret, with the aim that a user
then needs to try to guess what the secret is.
"""
def __init__(self, secret, max_guesses=5):
"""Create a new guess game
secret -- the secret that must be guessed
max_guesses -- the maximum number of guesses allowed by the user
"""
self._secret = secret
self._nguesses = 0
self._max_guesses = max_guesses
def guess(self, value):
"""Try to guess the secret. This will print out to the screen whether
or not the secret has been guessed.
value -- the user-supplied guess
"""
if (self.nGuesses() >= self.maxGuesses()):
print("Sorry, you have run out of guesses")
elif (value == self._secret):
print("Well done - you have guessed my secret")
else:
self._nguesses += 1
print("Try again...")
def nGuesses(self):
"""Return the number of incorrect guesses made so far"""
return self._nguesses
def maxGuesses(self):
"""Return the maximum number of incorrect guesses allowed"""
return self._max_guesses
help(GuessGame)
```
## Exercise 2
Below is a poorly-written class that uses public member data to store the name and age of a Person. Edit this class so that the member data is made private. Add `get` and `set` functions that allow you to safely get and set the name and age.
```
class Person:
"""Class the represents a Person, holding their name and age"""
def __init__(self, name="unknown", age=0):
"""Construct a person with unknown name and an age of 0"""
self.setName(name)
self.setAge(age)
def setName(self, name):
"""Set the person's name to 'name'"""
self._name = str(name) # str ensures the name is a string
def getName(self):
"""Return the person's name"""
return self._name
def setAge(self, age):
"""Set the person's age. This must be a number between 0 and 130"""
if (age < 0 or age > 130):
print("Cannot set the age to an invalid value: %s" % age)
self._age = age
def getAge(self):
"""Return the person's age"""
return self._age
p = Person(name="Peter Parker", age=21)
p.getName()
p.getAge()
```
## Exercise 3
Add a private member function called `_splitName` to your `Person` class that breaks the name into a surname and first name. Add new functions called `getFirstName` and `getSurname` that use this function to return the first name and surname of the person.
```
class Person:
"""Class the represents a Person, holding their name and age"""
def __init__(self, name="unknown", age=0):
"""Construct a person with unknown name and an age of 0"""
self.setName(name)
self.setAge(age)
def setName(self, name):
"""Set the person's name to 'name'"""
self._name = str(name) # str ensures the name is a string
def getName(self):
"""Return the person's name"""
return self._name
def setAge(self, age):
"""Set the person's age. This must be a number between 0 and 130"""
if (age < 0 or age > 130):
print("Cannot set the age to an invalid value: %s" % age)
self._age = age
def getAge(self):
"""Return the person's age"""
return self._age
def _splitName(self):
"""Private function that splits the name into parts"""
return self._name.split(" ")
def getFirstName(self):
"""Return the first name of the person"""
return self._splitName()[0]
def getSurname(self):
"""Return the surname of the person"""
return self._splitName()[-1]
p = Person(name="Peter Parker", age=21)
p.getFirstName()
p.getSurname()
```
| github_jupyter |
# Lab 4: EM Algorithm and Single-Cell RNA-seq Data
### Name: Your Name Here (Your netid here)
### Due April 2, 2021 11:59 PM
#### Preamble (Don't change this)
## Important Instructions -
1. Please implement all the *graded functions* in main.py file. Do not change function names in main.py.
2. Please read the description of every graded function very carefully. The description clearly states what is the expectation of each graded function.
3. After some graded functions, there is a cell which you can run and see if the expected output matches the output you are getting.
4. The expected output provided is just a way for you to assess the correctness of your code. The code will be tested on several other cases as well.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%run main.py
module = Lab4()
```
## Part 1 : Expectation-Maximization (EM) algorithm for transcript quantification
## Introduction
The EM algorithm is a very helpful tool to compute maximum likelihood estimates of parameters in models that have some latent (hidden) variables.
In the case of the transcript quantification problem, the model parameters we want to estimate are the transcript relative abundances $\rho_1,...,\rho_K$.
The latent variables are the read-to-transcript indicator variables $Z_{ik}$, which indicate whether the $i$th read comes from the $k$th transcript (in which case $Z_{ik}=1$.
In this part of the lab, you will be given the read alignment data.
For each read and transcript pair, it tells you whether the read can be mapped (i.e., aligned) to that transcript.
Using the EM algorithm, you will estimate the relative abundances of the trascripts.
### Reading read transcript data - We have 30000 reads and 30 transcripts
```
n_reads=30000
n_transcripts=30
read_mapping=[]
with open("read_mapping_data.txt",'r') as file :
lines_reads=file.readlines()
for line in lines_reads :
read_mapping.append([int(x) for x in line.split(",")])
read_mapping[:10]
```
Rather than giving you a giant binary matrix, we encoded the read mapping data in a more concise way. read_mapping is a list of lists. The $i$th list contains the indices of the transcripts that the $i$th read maps to.
### Reading true abundances and transcript lengths
```
with open("transcript_true_abundances.txt",'r') as file :
lines_gt=file.readlines()
ground_truth=[float(x) for x in lines_gt[0].split(",")]
with open("transcript_lengths.txt",'r') as file :
lines_gt=file.readlines()
tr_lengths=[float(x) for x in lines_gt[0].split(",")]
ground_truth[:5]
tr_lengths[:5]
```
## Graded Function 1 : expectation_maximization (10 marks)
Purpose : To implement the EM algorithm to obtain abundance estimates for each transcript.
E-step : In this step, we calculate the fraction of read that is assigned to each transcript (i.e., the estimate of $Z_{ik}$). For read $i$ and transicript $k$, this is calculated by dividing the current abundance estimate of transcript $k$ by the sum of abundance estimates of all transcripts that read $i$ maps to.
M-step : In this step, we update the abundance estimate of each transcript based on the fraction of all reads that is currently assigned to the transcript. First we compute the average fraction of all reads assigned to the transcript. Then, (if transcripts are of different lengths) we divide the result by the transcript length.
Finally, we normalize all abundance estimates so that they add up to 1.
Inputs - read_mapping (which is a list of lists where each sublist contains the transcripts to which a particular read belongs to. The length of this list is equal to the number of reads, i.e. 30000; tr_lengths (a list containing the length of the 30 transcripts, in order); n_iterations (the number of EM iterations to be performed)
Output - a list of lists where each sublist contains the abundance estimates for a transcript across all iterations. The length of each sublist should be equal to the number of iterations plus one (for the initialization) and the total number of sublists should be equal to the number of transcripts.
```
history=module.expectation_maximization(read_mapping,tr_lengths,20)
print(len(history))
print(len(history[0]))
print(history[0][-5:])
print(history[1][-5:])
print(history[2][-5:])
```
## Expected Output -
30
21
[0.033769639494636614, 0.03381298624783303, 0.03384568373972948, 0.0338703482393148, 0.03388895326082054]
[0.0020082674603036053, 0.0019649207071071456, 0.0019322232152109925, 0.0019075587156241912, 0.0018889536941198502]
[0.0660581789629968, 0.06606927656035864, 0.06607650126895578, 0.06608120466668756, 0.0660842666518177]
You can use the following function to visualize how the estimated relative abundances are converging with the number of iterations of the algorithm.
```
def visualize_em(history,n_iterations) :
#start code here
fig, ax = plt.subplots(figsize=(8,6))
for j in range(n_transcripts):
ax.plot([i for i in range(n_iterations+1)],[history[j][i] - ground_truth[j] for i in range(n_iterations+1)],marker='o')
#end code here
visualize_em(history,20)
```
## Part 2 : Exploring Single-Cell RNA-seq data
In a study published in 2015, Zeisel et al. used single-cell RNA-seq data to explore the cell diversity in the mouse brain.
We will explore the data used for their study.
You can read more about it [here](https://science.sciencemag.org/content/347/6226/1138).
```
#reading single-cell RNA-seq data
lines_genes=[]
with open("Zeisel_expr.txt",'r') as file :
lines_genes=file.readlines()
lines_genes[0][:300]
```
Each line in the file Zeisel_expr.txt corresponds to one gene.
The columns correspond to different cells (notice that this is the opposite of how we looked at this matrix in class).
The entries of this matrix correspond to the number of reads mapping to a given gene in the corresponding cell.
```
# reading true labels for each cell
with open("Zeisel_labels.txt",'r') as file :
true_labels = file.read().splitlines()
```
The study also provides us with true labels for each of the cells.
For each of the cells, the vector true_labels contains the name of the cell type.
There are nine different cell types in this dataset.
```
set(true_labels)
```
## Graded Function 2 : prepare_data (10 marks) :
Purpose - To create a dataframe where each row corresponds to a specific cell and each column corresponds to the expressions levels of a particular gene across all cells.
You should name the columns as "Gene_1", "Gene_2", and so on.
We will iterate through all the lines in lines_genes list created above, add 1 to each value and take log.
Each line will correspond to 1 column in the dataframe
Output - gene expression dataframe
### Note - All the values in the output dataframe should be rounded off to 5 digits after the decimal
```
data_df=module.prepare_data(lines_genes)
print(data_df.shape)
print(data_df.iloc[0:3,:5])
print(data_df.columns)
```
## Expected Output :
``(3005, 19972)``
`` Gene_0 Gene_1 Gene_2 Gene_3 Gene_4``
``0 0.0 1.38629 1.38629 0.0 0.69315``
``1 0.0 0.69315 0.69315 0.0 0.69315``
``2 0.0 0.00000 1.94591 0.0 0.69315``
## Graded Function 3 : identify_less_expressive_genes (10 marks)
Purpose : To identify genes (columns) that are expressed in less than 25 cells. We will create a list of all gene columns that have values greater than 0 for less than 25 cells.
Input - gene expression dataframe
Output - list of column names which are expressed in less than 25 cells
```
drop_columns = module.identify_less_expressive_genes(data_df)
print(len(drop_columns))
print(drop_columns[:10])
```
## Expected Output :
``5120``
``['Gene_28', 'Gene_126', 'Gene_145', 'Gene_146', 'Gene_151', 'Gene_152', 'Gene_167', 'Gene_168', 'Gene_170', 'Gene_173']``
### Filtering less expressive genes
We will now create a new dataframe in which genes which are expressed in less than 25 cells will not be present
```
df_new = data_df.drop(drop_columns, axis=1)
df_new.head()
```
## Graded Function 4 : perform_pca (10 marks)
Pupose - Perform Principal Component Analysis on the new dataframe and take the top 50 principal components
Input - df_new
Output - numpy array containing the top 50 principal components of the data.
### Note - All the values in the output should be rounded off to 5 digits after the decimal
### Note - Please use random_state=365 for the PCA object you will create
```
pca_data=module.perform_pca(df_new)
print(pca_data.shape)
print(type(pca_data))
print(pca_data[0:3,:5])
```
## Expected Output :
``(3005, 50)``
``<class 'numpy.ndarray'>``
``[[26.97148 -2.7244 0.62163 25.90148 -6.24736]``
`` [26.49135 -1.58774 -4.79315 24.01094 -7.25618]``
`` [47.82664 5.06799 2.15177 30.24367 -3.38878]]``
## (Non-graded) Function 5 : perform_tsne
Pupose - Perform t-SNE on the pca_data and obtain 2 t-SNE components
We will use TSNE class of the sklearn.manifold package. Use random_state=1000 and perplexity=50
Documenation can be found here - https://scikit-learn.org/stable/modules/generated/sklearn.manifold.TSNE.html
Input - pca_data
Output - numpy array containing the top 2 tsne components of the data.
**Note: This function will not be graded because of the random nature of t-SNE.**
```
tsne_data50 = module.perform_tsne(pca_data)
print(tsne_data50.shape)
print(tsne_data50[:3,:])
```
## Expected Output :
(These numbers can deviate a bit depending on your sklearn)
``(3005, 2)``
``[[ 15.069608 -47.535984]``
`` [ 15.251476 -47.172073]``
`` [ 13.3932 -49.909657]]``
```
fig, ax = plt.subplots(figsize=(12,8))
sns.scatterplot(tsne_data50[:,0], tsne_data50[:,1], hue=true_labels)
plt.show()
```
Notice that the different cell types form clusters (which can be easily visualized on the t-SNE space).
Zeisel et al. performed clustering on this data in order to identify and label the different cell types.
You can try using clustering methods (such as k-means and GMM) to cluster the single-cell RNA-seq data of Zeisel at al. and see if your results agree with theirs!
| github_jupyter |
# Week 2 Tasks
During this week's meeting, we have discussed about if/else statements, Loops and Lists. This notebook file will guide you through reviewing the topics discussed and assisting you to be familiarized with the concepts discussed.
## Let's first create a list
```
# Create a list that stores the multiples of 5, from 0 to 50 (inclusive)
# initialize the list using list comprehension!
# Set the list name to be 'l'
# TODO: Make the cell return 'True'
# Hint: Do you remember that you can apply arithmetic operators in the list comprehension?
# Your code goes below here
# Do not modify below
l == [0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50]
```
If you are eager to learn more about list comprehension, you can look up here -> https://www.programiz.com/python-programming/list-comprehension. You will find out how you can initialize `l` without using arithmetic operators, but using conditionals (if/else).
Now, simply run the cell below, and observe how `l` has changed.
```
l[0] = 3
print(l)
l[5]
```
As seen above, you can overwrite each elements of the list.
Using this fact, complete the task written below.
## If/elif/else practice
```
# Write a for loop such that:
# For each elements in the list l,
# If the element is divisible by 6, divide the element by 6
# Else if the element is divisible by 3, divide the element by 3 and then add 4
# Else if the element is divisible by 2, subtract 10.
# Else, square the element
# TODO: Make the cell return 'True'
l = [3, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50]
# Your code goes below here
# Do not modify below
l = [int(i) for i in l]
l == [5, 25, 0, 9, 10, 625, 5, 1225, 30, 19, 40]
```
## Limitations of a ternary operator
```
# Write a for loop that counts the number of odd number elements in the list
# and the number of even number elements in the list
# These should be stored in the variables 'odd_count' and 'even_count', which are declared below.
# Try to use the ternary operator inside the for loop and inspect why it does not work
# TODO: Make the cell return 'True'
l = [5, 25, 0, 9, 10, 625, 5, 1225, 30, 19, 40]
odd_count, even_count = 0, 0
# Your code goes below here
# Do not modify below
print("There are 7 odd numbers in the list.") if odd_count == 7 else print("Your odd_count is not correct.")
print("There are 4 even numbers in the list.") if even_count == 4 else print("Your even_count is not correct.")
print(odd_count == 7 and even_count == 4 and odd_count + even_count == len(l))
```
If you have tried using the ternary operator in the cell above, you would have found that the cell fails to compile because of a syntax error. This is because you can only write *expressions* in ternary operators, specifically **the last segment of the three segments in the operator**, not *statements*.
In other words, since your code (which last part of it would have been something like `odd_count += 1` or `even_count += 1`) is a *statement*, the code is syntactically incorrect.
To learn more about *expressions* and *statements*, please refer to this webpage -> https://runestone.academy/runestone/books/published/thinkcspy/SimplePythonData/StatementsandExpressions.html
Thus, a code like `a += 1 if <CONDITION> else b += 1` is syntactically wrong as `b += 1` is a *statement*, and we cannot use the ternary operator to achieve something like this.
In fact, ternary operators are usually used like this: `a += 1 if <CONDITION> else 0`.
The code above behaves exactly the same as this: `if <CONDITION>: a += 1 else: a = 0`.
Does this give better understanding about why statements cannot be used in ternary operators? If not, feel free to do more research on your own, or open up a discussion during the next team meeting!
## While loop and boolean practice
```
# Write a while loop that finds an index of the element in 'l' which first exceeds 1000.
# The index found should be stored in the variable 'large_index'
# If there are no element in 'l' that exceeds 1000, 'large_index' must store -1
# Use the declared 'large_not_found' as the condition for the while loop
# Use the declared 'index' to iterate through 'l'
# Do not use 'break'
# TODO: Make the cell return 'True'
l = [5, 25, 0, 9, 10, 625, 5, 1225, 30, 19, 1001]
large_not_found = True
index = 0
large_index = 0
# Your code goes below here
# Do not modify below
print(large_index == 7)
```
## Finding the minimum element
```
# For this task, you can use either for loop or while loop, depending on your preference
# Find the smallest element in 'l' and store it in the declared variable 'min_value'
# 'min_value' is initialized as a big number
# Do not use min()
# TODO: Make the cell return 'True'
import sys
min_value = sys.maxsize
min_index = 0
# Your code goes below here
# Do not modify below
print(min_value == 0)
import os
os.getpid()
```
| github_jupyter |
# launch scripts through SLURM
The script in the cell below submits SLURM jobs running the requested `script`, with all parameters specified in `param_iterators` and the folder where to dump data as last parameter.
The generated SBATCH scipts (`.job` files) are saved in the `jobs` folder and then submitted.
Output and error dumps are saved in the `out` folder.
```
import numpy as np
import os
from itertools import product
#######################
### User parameters ###
#######################
script = "TFIM-bangbang-WF.py" # name of the script to be run
data_subdir = "TFIM/bangbang/WF" # subdirectory of ´data´ where to save results
jobname_template = "BBWF-L{}JvB{}nit{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(16, 21), # L
[0.2, 1, 5], # JvB
[None], # nit
[200] # n_samples
)
time = "4-00:00" # format days-hh:mm
mem = "4GB" # can use postfixes (MB, GB, ...)
partition = "compIntel"
# insert here additional lines that should be run before the script
# (source bash scripts, load modules, activate environment, etc.)
additional_lines = [
'source ~/.bashrc\n'
]
#####################################
### Create folders, files and run ###
#####################################
current_dir = os.getcwd()
script = os.path.join(*os.path.split(current_dir)[:-1], 'scripts', script)
data_supdir = os.path.join(*os.path.split(current_dir)[:-1], 'data')
data_dir = os.path.join(data_supdir, data_subdir)
job_dir = 'jobs'
out_dir = 'out'
os.makedirs(job_dir, exist_ok=True)
os.makedirs(out_dir, exist_ok=True)
os.makedirs(data_dir, exist_ok=True)
for params in product(*param_iterators):
# ******** for BangBang ********
# redefine nit = L if it is None
if params[2] is None:
params = list(params)
params[2] = params[0]
# ******************************
job_name = jobname_template.format(*params)
job_file = os.path.join(job_dir, job_name+'.job')
with open(job_file, 'wt') as fh:
fh.writelines(
["#!/bin/bash\n",
f"#SBATCH --job-name={job_name}\n",
f"#SBATCH --output={os.path.join(out_dir, job_name+'.out')}\n",
f"#SBATCH --error={os.path.join(out_dir, job_name+'.err')}\n",
f"#SBATCH --time={time}\n",
f"#SBATCH --mem={mem}\n",
f"#SBATCH --partition={partition}\n",
f"#SBATCH --mail-type=NONE\n",
] + additional_lines + [
f"python -u {script} {' '.join(str(par) for par in params)} {data_dir}\n"]
)
os.system("sbatch %s" %job_file)
complex(1).__sizeof__() * 2**(2*15) / 1E9
```
# History of parameters that have been run
## TFIM LogSweep
### density matrix
```
script = "TFIM-logsweep-DM.py"
data_subdir = "TFIM/logsweep/DM"
param_iterators = (
[2], # L
[0.2, 1, 5], # JvB
np.arange(2, 50) # K
)
param_iterators = (
[7], # L
[0.2, 1, 5], # JvB
np.arange(2, 50) # K
)
param_iterators = (
np.arange(2, 11), # L
[0.2, 1, 5], # JvB
[2, 5, 10, 20, 40] # K
)
```
### Iterative, density matrix
```
script = "TFIM-logsweep-DM-iterative.py" # name of the script to be run
data_subdir = "TFIM/logsweep/DM/iterative" # subdirectory of ´data´ where to save results
jobname_template = "ItLS-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
[2, 7], # L
[0.2, 1, 5], # JvB
np.arange(2, 50) # K
)
```
### WF + Monte Carlo
#### old version of the script
the old version suffered from unnormalized final states due to numerical error
```
script = "TFIM-logsweep-WF.py" # name of the script to be run
data_subdir = "TFIM/logsweep/WF-raw" # subdirectory of ´data´ where to save results
jobname_template = "WF-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(2, 15), # L
[0.2, 1, 5], # JvB
[2, 3, 5, 10, 20, 40], # K
[100] # n_samples
)
```
#### new version of the script
Where normalization is forced
```
script = "TFIM-logsweep-WF.py" # name of the script to be run
data_subdir = "TFIM/logsweep/WF" # subdirectory of ´data´ where to save results
jobname_template = "WF-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(2, 10), # L
[0.2, 1, 5], # JvB
[2, 3, 5, 10], # K
[100] # n_samples
)
time = "3-00:00" # format days-hh:mm
mem = "1GB" # can use postfixes (MB, GB, ...)
partition = "compIntel"
script = "TFIM-logsweep-WF.py" # name of the script to be run
data_subdir = "TFIM/logsweep/WF" # subdirectory of ´data´ where to save results
jobname_template = "WF-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(10, 14), # L
[0.2, 1, 5], # JvB
[2, 3, 5, 10], # K
[100] # n_samples
)
time = "3-00:00" # format days-hh:mm
mem = "20GB" # can use postfixes (MB, GB, ...)
partition = "compIntel"
```
### iterative, WF + Monte Carlo
```
script = "TFIM-logsweep-WF-iterative.py" # name of the script to be run
data_subdir = "TFIM/logsweep/WF/iterative" # subdirectory of ´data´ where to save results
jobname_template = "WFiter-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(2, 14), # L
[0.2, 1, 5], # JvB
[5, 10], # K
[100] # n_samples
)
time = "3-00:00" # format days-hh:mm
mem = "20GB" # can use postfixes (MB, GB, ...)
partition = "ibIntel"
```
### continuous DM
```
script = "TFIM-logsweep-continuous-DM.py" # name of the script to be run
data_subdir = "TFIM/logsweep/continuous/DM" # subdirectory of ´data´ where to save results
jobname_template = "Rh-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(2,7), # L
[0.2, 1, 5], # JvB
[2, 3, 5, 10, 20, 40] # K
)
param_iterators = (
[7], # L
[0.2, 1, 5], # JvB
np.arange(2, 50) # K
)
param_iterators = (
np.arange(8, 15), # L
[0.2, 1, 5], # JvB
[2,3,5,10,20,40] # K
)
```
### continuous WF
```
script = "TFIM-logsweep-continuous-WF.py" # name of the script to be run
data_subdir = "TFIM/logsweep/continuous/WF" # subdirectory of ´data´ where to save results
jobname_template = "CWF-L{}JvB{}K{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(2, 12), # L
[0.2, 1, 5], # JvB
[2, 3, 5, 10, 20, 40], # K
[100] # n_samples
)
time = "3-00:00" # format days-hh:mm
mem = "1GB" # can use postfixes (MB, GB, ...)
partition = "ibIntel"
param_iterators = (
[13, 14], # L
[0.2, 1, 5], # JvB
[2, 10], # K
[100] # n_samples
)
time = "3-00:00" # format days-hh:mm
mem = "100GB" # can use postfixes (MB, GB, ...)
partition = "ibIntel"
```
## TFIM bang-bang
```
data_subdir = "TFIM/bangbang/WF" # subdirectory of ´data´ where to save results
jobname_template = "BBWF-L{}JvB{}nit{}" # job name will be created from this, inserting parameter values
param_iterators = (
np.arange(2, 21), # L
[0.2, 1, 5], # JvB
[None], # nit
[200] # n_samples
)
time = "4-00:00" # format days-hh:mm
mem = "4GB" # can use postfixes (MB, GB, ...)
partition = "compIntel"
```
| github_jupyter |
<a href="https://colab.research.google.com/github/Vanagand/DS-Unit-2-Applied-Modeling/blob/master/module1-define-ml-problems/Unit_2_Sprint_3_Module_1_LESSON.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Lambda School Data Science
*Unit 2, Sprint 3, Module 1*
---
# Define ML problems
- Choose a target to predict, and check its distribution
- Avoid leakage of information from test to train or from target to features
- Choose an appropriate evaluation metric
### Setup
```
%%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
!pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
```
# Choose a target to predict, and check its distribution
## Overview
This is the data science process at a high level:
<img src="https://image.slidesharecdn.com/becomingadatascientistadvice-pydatadc-shared-161012184823/95/becoming-a-data-scientist-advice-from-my-podcast-guests-55-638.jpg?cb=1476298295">
—Renee Teate, [Becoming a Data Scientist, PyData DC 2016 Talk](https://www.becomingadatascientist.com/2016/10/11/pydata-dc-2016-talk/)
We've focused on the 2nd arrow in the diagram, by training predictive models. Now let's zoom out and focus on the 1st arrow: defining problems, by translating business questions into code/data questions.
Last sprint, you did a Kaggle Challenge. It’s a great way to practice model validation and other technical skills. But that's just part of the modeling process. [Kaggle gets critiqued](https://speakerdeck.com/szilard/machine-learning-software-in-practice-quo-vadis-invited-talk-kdd-conference-applied-data-science-track-august-2017-halifax-canada?slide=119) because some things are done for you: Like [**defining the problem!**](https://www.linkedin.com/pulse/data-science-taught-universities-here-why-maciej-wasiak/) In today’s module, you’ll begin to practice this objective, with your dataset you’ve chosen for your personal portfolio project.
When defining a supervised machine learning problem, one of the first steps is choosing a target to predict.
Which column in your tabular dataset will you predict?
Is your problem regression or classification? You have options. Sometimes it’s not straightforward, as we'll see below.
- Discrete, ordinal, low cardinality target: Can be regression or multi-class classification.
- (In)equality comparison: Converts regression or multi-class classification to binary classification.
- Predicted probability: Seems to [blur](https://brohrer.github.io/five_questions_data_science_answers.html) the line between classification and regression.
## Follow Along
Let's reuse the [Burrito reviews dataset.](https://nbviewer.jupyter.org/github/LambdaSchool/DS-Unit-2-Linear-Models/blob/master/module4-logistic-regression/LS_DS_214_assignment.ipynb) 🌯
```
import pandas as pd
pd.options.display.max_columns = None
df = pd.read_csv(DATA_PATH+'burritos/burritos.csv')
```
### Choose your target
Which column in your tabular dataset will you predict?
```
df.head()
df['overall'].describe()
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
sns.distplot(df['overall'])
df['Great'] = df['overall'] >= 4
df['Great']
```
### How is your target distributed?
For a classification problem, determine: How many classes? Are the classes imbalanced?
```
y = df['Great']
y.unique()
y.value_counts(normalize=True)
sns.countplot(y)
y.value_counts(normalize=True).plot(kind="bar")
# Stretch: how to fix imbalanced classes
#. upsampling: randomly re-sample from the minority class to increase the sample in the minority class
#. downsampling: random re-sampling from the majority class to decrease the sample in the majority class
# Why does it matter if we have imbalanced classes?
# 1:1000 tested positive:tested negative
# 99.99% accuracy
#
```
# Avoid leakage of information from test to train or from target to features
## Overview
Overfitting is our enemy in applied machine learning, and leakage is often the cause.
> Make sure your training features do not contain data from the “future” (aka time traveling). While this might be easy and obvious in some cases, it can get tricky. … If your test metric becomes really good all of the sudden, ask yourself what you might be doing wrong. Chances are you are time travelling or overfitting in some way. — [Xavier Amatriain](https://www.quora.com/What-are-some-best-practices-for-training-machine-learning-models/answer/Xavier-Amatriain)
Choose train, validate, and test sets. Are some observations outliers? Will you exclude them? Will you do a random split or a time-based split? You can (re)read [How (and why) to create a good validation set](https://www.fast.ai/2017/11/13/validation-sets/).
## Follow Along
First, begin to **explore and clean your data.**
```
df['Burrito'].nunique()
df['Burrito'].unique()
# Combine Burrito categories
df['Burrito_rename'] = df['Burrito'].str.lower()
# All burrito types that contain 'California' are grouped into the same
#. category. Similar logic applied to asada, surf, and carnitas.
# 'California Surf and Turf'
california = df['Burrito'].str.contains('california')
asada = df['Burrito'].str.contains('asada')
surf = df['Burrito'].str.contains('surf')
carnitas = df['Burrito'].str.contains('carnitas')
df.loc[california, 'Burrito_rename'] = 'California'
df.loc[asada, 'Burrito_rename'] = 'Asada'
df.loc[surf, 'Burrito_rename'] = 'Surf & Turf'
df.loc[carnitas, 'Burrito_rename'] = 'Carnitas'
# If the burrito is not captured in one of the above categories, it is put in the
# 'Other' category.
df.loc[~california & ~asada & ~surf & ~carnitas, 'Burrito_rename'] = 'Other'
df[['Burrito', 'Burrito_rename']]
df = df.drop(columns=['Notes', 'Location', 'Reviewer', 'Address', 'URL', 'Neighborhood'])
df.info()
df.isna().sum().sort_values()
df = df.fillna('Missing')
df.info()
```
Next, do a **time-based split:**
- Train on reviews from 2016 & earlier.
- Validate on 2017.
- Test on 2018 & later.
```
df['Date'] = pd.to_datetime(df['Date'])
# create a subset of data for anything less than or equal to the year 2016, equal
#. to 2017 for validation, and test set to include >= 2018
train = df[df['Date'].dt.year <= 2016]
val = df[df['Date'].dt.year == 2017]
test = df[df['Date'].dt.year >= 2018]
train.shape, val.shape, test.shape
```
Begin to choose which features, if any, to exclude. **Would some features “leak” future information?**
What happens if we _DON’T_ drop features with leakage?
```
# Try a shallow decision tree as a fast, first model
import category_encoders as ce
from sklearn.pipeline import make_pipeline
from sklearn.tree import DecisionTreeClassifier
target = 'Great'
features = train.columns.drop([target, 'Date', 'Data'])
X_train = train[features]
y_train = train[target]
X_val = val[features]
y_val = val[target]
pipeline = make_pipeline(
ce.OrdinalEncoder(),
DecisionTreeClassifier()
)
pipeline.fit(X_train, y_train)
print('Validation Accuracy', pipeline.score(X_val, y_val))
```
Drop the column with “leakage”.
```
target = 'Great'
features = train.columns.drop([target, 'Date', 'Data', 'overall'])
X_train = train[features]
y_train = train[target]
X_val = val[features]
y_val = val[target]
pipeline = make_pipeline(
ce.OrdinalEncoder(),
DecisionTreeClassifier()
)
pipeline.fit(X_train, y_train)
print('Validation Accuracy', pipeline.score(X_val, y_val))
```
# Choose an appropriate evaluation metric
## Overview
How will you evaluate success for your predictive model? You must choose an appropriate evaluation metric, depending on the context and constraints of your problem.
**Classification & regression metrics are different!**
- Don’t use _regression_ metrics to evaluate _classification_ tasks.
- Don’t use _classification_ metrics to evaluate _regression_ tasks.
[Scikit-learn has lists of popular metrics.](https://scikit-learn.org/stable/modules/model_evaluation.html#common-cases-predefined-values)
## Follow Along
For classification problems:
As a rough rule of thumb, if your majority class frequency is >= 50% and < 70% then you can just use accuracy if you want. Outside that range, accuracy could be misleading — so what evaluation metric will you choose, in addition to or instead of accuracy? For example:
- Precision?
- Recall?
- ROC AUC?
```
# 1:3 -> 25%, 75%
y.value_counts(normalize=True)
```
### Precision & Recall
Let's review Precision & Recall. What do these metrics mean, in scenarios like these?
- Predict great burritos
- Predict fraudulent transactions
- Recommend Spotify songs
[Are false positives or false negatives more costly? Can you optimize for dollars?](https://alexgude.com/blog/machine-learning-metrics-interview/)
```
# High precision -> few false positives.
# High recall -> few false negatives.
# In lay terms, how would we translate our problem with burritos:
#. high precision- 'Great burrito'. If we make a prediction of a great burrito,
#. it probably IS a great burrito.
# Which metric would you emphasize if you were choosing a burrito place to take your first date to?
#. Precision.
# Which metric would -> feeling adventurous?
# . Recall.
# Predict Fraud:
# True negative: normal transaction
# True positive: we caught fraud!
# False Positive: normal transaction that is blocked -> annoyed customer! (low precision)
# False Negative: fraudulent transaction that was allowed -> lost money (low recall)
```
### ROC AUC
Let's also review ROC AUC (Receiver Operating Characteristic, Area Under the Curve).
[Wikipedia explains,](https://en.wikipedia.org/wiki/Receiver_operating_characteristic) "A receiver operating characteristic curve, or ROC curve, is a graphical plot that illustrates the diagnostic ability of a binary classifier system as its discrimination threshold is varied. **The ROC curve is created by plotting the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings.**"
ROC AUC is the area under the ROC curve. [It can be interpreted](https://stats.stackexchange.com/questions/132777/what-does-auc-stand-for-and-what-is-it) as "the expectation that a uniformly drawn random positive is ranked before a uniformly drawn random negative."
ROC AUC measures **how well a classifier ranks predicted probabilities.** So, when you get your classifier’s ROC AUC score, you need to **use predicted probabilities, not discrete predictions.**
ROC AUC ranges **from 0 to 1.** Higher is better. A naive majority class **baseline** will have an ROC AUC score of **0.5**, regardless of class (im)balance.
#### Scikit-Learn docs
- [User Guide: Receiver operating characteristic (ROC)](https://scikit-learn.org/stable/modules/model_evaluation.html#receiver-operating-characteristic-roc)
- [sklearn.metrics.roc_curve](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_curve.html)
- [sklearn.metrics.roc_auc_score](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_auc_score.html)
#### More links
- [StatQuest video](https://youtu.be/4jRBRDbJemM)
- [Data School article / video](https://www.dataschool.io/roc-curves-and-auc-explained/)
- [The philosophical argument for using ROC curves](https://lukeoakdenrayner.wordpress.com/2018/01/07/the-philosophical-argument-for-using-roc-curves/)
```
from sklearn.metrics import roc_auc_score
y_pred_proba = pipeline.predict_proba(X_val)[:, -1]
roc_auc_score(y_val, y_pred_proba)
from sklearn.metrics import roc_curve
fpr, tpr, thresholds = roc_curve(y_val, y_pred_proba)
(fpr, tpr, thresholds)
import matplotlib.pyplot as plt
plt.scatter(fpr, tpr)
plt.plot(fpr, tpr)
plt.title('ROC curve')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
```
### Imbalanced classes
Do you have highly imbalanced classes?
If so, you can try ideas from [Learning from Imbalanced Classes](https://www.svds.com/tbt-learning-imbalanced-classes/):
- “Adjust the class weight (misclassification costs)” — most scikit-learn classifiers have a `class_balance` parameter.
- “Adjust the decision threshold” — we did this last module. Read [Visualizing Machine Learning Thresholds to Make Better Business Decisions](https://blog.insightdatascience.com/visualizing-machine-learning-thresholds-to-make-better-business-decisions-4ab07f823415).
- “Oversample the minority class, undersample the majority class, or synthesize new minority classes” — try the the [imbalanced-learn](https://github.com/scikit-learn-contrib/imbalanced-learn) library as a stretch goal.
# BONUS: Regression example 🏘️
```
# Read our NYC apartment rental listing dataset
df = pd.read_csv(DATA_PATH+'apartments/renthop-nyc.csv')
```
### Choose your target
Which column in your tabular dataset will you predict?
```
y = df['price']
```
### How is your target distributed?
For a regression problem, determine: Is the target right-skewed?
```
# Yes, the target is right-skewed
import seaborn as sns
sns.distplot(y);
y.describe()
```
### Are some observations outliers?
Will you exclude
them?
```
# Yes! There are outliers
# Some prices are so high or low it doesn't really make sense.
# Some locations aren't even in New York City
# Remove the most extreme 1% prices,
# the most extreme .1% latitudes, &
# the most extreme .1% longitudes
import numpy as np
df = df[(df['price'] >= np.percentile(df['price'], 0.5)) &
(df['price'] <= np.percentile(df['price'], 99.5)) &
(df['latitude'] >= np.percentile(df['latitude'], 0.05)) &
(df['latitude'] < np.percentile(df['latitude'], 99.95)) &
(df['longitude'] >= np.percentile(df['longitude'], 0.05)) &
(df['longitude'] <= np.percentile(df['longitude'], 99.95))]
# The distribution has improved, but is still right-skewed
y = df['price']
sns.distplot(y);
y.describe()
```
### Log-Transform
If the target is right-skewed, you may want to “log transform” the target.
> Transforming the target variable (using the mathematical log function) into a tighter, more uniform space makes life easier for any [regression] model.
>
> The only problem is that, while easy to execute, understanding why taking the log of the target variable works and how it affects the training/testing process is intellectually challenging. You can skip this section for now, if you like, but just remember that this technique exists and check back here if needed in the future.
>
> Optimally, the distribution of prices would be a narrow “bell curve” distribution without a tail. This would make predictions based upon average prices more accurate. We need a mathematical operation that transforms the widely-distributed target prices into a new space. The “price in dollars space” has a long right tail because of outliers and we want to squeeze that space into a new space that is normally distributed. More specifically, we need to shrink large values a lot and smaller values a little. That magic operation is called the logarithm or log for short.
>
> To make actual predictions, we have to take the exp of model predictions to get prices in dollars instead of log dollars.
>
>— Terence Parr & Jeremy Howard, [The Mechanics of Machine Learning, Chapter 5.5](https://mlbook.explained.ai/prep.html#logtarget)
[Numpy has exponents and logarithms](https://docs.scipy.org/doc/numpy/reference/routines.math.html#exponents-and-logarithms). Your Python code could look like this:
```python
import numpy as np
y_train_log = np.log1p(y_train)
model.fit(X_train, y_train_log)
y_pred_log = model.predict(X_val)
y_pred = np.expm1(y_pred_log)
print(mean_absolute_error(y_val, y_pred))
```
```
import numpy as np
y_log = np.log1p(y)
sns.distplot(y_log)
sns.distplot(y)
plt.title('Original target, in the unit of US dollars');
y_log = np.log1p(y)
sns.distplot(y_log)
plt.title('Log-transformed target, in log-dollars');
y_untransformed = np.expm1(y_log)
sns.distplot(y_untransformed)
plt.title('Back to the original units');
```
## Challenge
You will use your portfolio project dataset for all assignments this sprint. (If you haven't found a dataset yet, do that today. [Review requirements for your portfolio project](https://lambdaschool.github.io/ds/unit2) and choose your dataset.)
Complete these tasks for your project, and document your decisions.
- Choose your target. Which column in your tabular dataset will you predict?
- Is your problem regression or classification?
- How is your target distributed?
- Classification: How many classes? Are the classes imbalanced?
- Regression: Is the target right-skewed? If so, you may want to log transform the target.
- Choose your evaluation metric(s).
- Classification: Is your majority class frequency >= 50% and < 70% ? If so, you can just use accuracy if you want. Outside that range, accuracy could be misleading. What evaluation metric will you choose, in addition to or instead of accuracy?
- Regression: Will you use mean absolute error, root mean squared error, R^2, or other regression metrics?
- Choose which observations you will use to train, validate, and test your model.
- Are some observations outliers? Will you exclude them?
- Will you do a random split or a time-based split?
- Begin to clean and explore your data.
- Begin to choose which features, if any, to exclude. Would some features "leak" future information?
Some students worry, ***what if my model isn't “good”?*** Then, [produce a detailed tribute to your wrongness. That is science!](https://twitter.com/nathanwpyle/status/1176860147223867393)
| github_jupyter |
<img src="images/strathsdr_banner.png" align="left">
# An RFSoC Spectrum Analyzer Dashboard with Voila
----
<div class="alert alert-box alert-info">
Please use Jupyter Labs http://board_ip_address/lab for this notebook.
</div>
The RFSoC Spectrum Analyzer is an open source tool developed by the [University of Strathclyde](https://github.com/strath-sdr/rfsoc_sam). This notebook is specifically for Voila dashboards. If you would like to see an overview of the Spectrum Analyser, see this [notebook](rfsoc_spectrum_analysis.ipynb) instead.
## Table of Contents
* [Introduction](#introduction)
* [Running this Demonstration](#running-this-demonstration)
* [The Voila Procedure](#the-voila-procedure)
* [Import Libraries](#import-libraries)
* [Initialise Overlay](#initialise-overlay)
* [Dashboard Display](#dashboard-display)
* [Conclusion](#conclusion)
## References
* [Xilinx, Inc, "USP RF Data Converter: LogiCORE IP Product Guide", PG269, v2.3, June 2020](https://www.xilinx.com/support/documentation/ip_documentation/usp_rf_data_converter/v2_3/pg269-rf-data-converter.pdf)
## Revision History
* **v1.0** | 16/02/2021 | Voila spectrum analyzer demonstration
* **v1.1** | 22/10/2021 | Voila update notes in 'running this demonstration' section
## Introduction <a class="anchor" id="introduction"></a>
You ZCU111 platform and XM500 development board is capable of quad-channel spectral analysis. The RFSoC Spectrum Analyser Module (rfsoc-sam) enables hardware accelerated analysis of signals received from the RF Analogue-to-Digital Converters (RF ADCs). This notebook is specifically for running the Spectrum Analyser using Voila dashboards. Follow the instructions outlined in [Running this Demonstration](#running-this-demonstration) to learn more.
### Hardware Setup <a class="anchor" id="hardware-setup"></a>
Your ZCU111 development board can host four Spectrum Analyzer Modules. To setup your board for this demonstration, you can connect each channel in loopback as shown in [Figure 1](#fig-1), or connect an antenna to one of the ADC channels.
Don't worry if you don't have an antenna. The default loopback configuration will still be very interesting and is connected as follows:
* Channel 0: DAC4 (Tile 229 Block 0) to ADC0 (Tile 224 Block 0)
* Channel 1: DAC5 (Tile 229 Block 1) to ADC1 (Tile 224 Block 1)
* Channel 2: DAC6 (Tile 229 Block 2) to ADC2 (Tile 225 Block 0)
* Channel 3: DAC7 (Tile 229 Block 3) to ADC3 (Tile 225 Block 1)
There has been several XM500 board revisions, and some contain different silkscreen and labels for the ADCs and DACs. Use the image below for further guidance and pay attention to the associated Tile and Block.
<a class="anchor" id="fig-1"></a>
<figure>
<img src='images/zcu111_setup.png' height='50%' width='50%'/>
<figcaption><b>Figure 1: ZCU111 and XM500 development board setup in loopback mode.</b></figcaption>
</figure>
If you have chosen to use an antenna, **do not** attach your antenna to any SMA interfaces labelled DAC.
<div class="alert alert-box alert-danger">
<b>Caution:</b>
In this demonstration, we generate tones using the RFSoC development board. Your device should be setup in loopback mode. You should understand that the RFSoC platform can also transmit RF signals wirelessly. Remember that unlicensed wireless transmission of RF signals may be illegal in your geographical location. Radio signals may also interfere with nearby devices, such as pacemakers and emergency radio equipment. Note that it is also illegal to intercept and decode particular RF signals. If you are unsure, please seek professional support.
</div>
----
## Running this Demonstration <a class="anchor" id="running-this-demonstration"></a>
Voila can be used to execute the Spectrum Analyzer Module, while ignoring all of the markdown and code cells typically found in a normal Jupyter notebook. The Voila dashboard can be launched following the instructions below:
* Click on the "Open with Voila Gridstack in a new browser tab" button at the top of the screen:
<figure>
<img src='images/open_voila.png' height='50%' width='50%'/>
</figure>
After the new tab opens the kernel will start and the notebook will run. Only the Spectrum Analyzer will be displayed. The initialisation process takes around 1 minute.
## The Voila Procedure <a class="anchor" id="the-voila-procedure"></a>
Below are the code cells that will be ran when Voila is called. The procedure is fairly straight forward. Load the rfsoc-sam library, initialise the overlay, and display the spectrum analyzer. All you have to ensure is that the above command is executed in the terminal and you have launched a browser tab using the given address. You do not need to run these code cells individually to create the voila dashboard.
### Import Libraries
```
from rfsoc_sam.overlay import Overlay
```
### Initialise Overlay
```
sam = Overlay(init_rf_clks = True)
```
### Dashboard Display
```
sam.spectrum_analyzer_application()
```
## Conclusion
This notebook has presented a hardware accelerated Spectrum Analyzer Module for the ZCU111 development board. The demonstration used Voila to enable rapid dashboarding for visualisation and control.
| github_jupyter |
# Examples I - Inferring $v_{\rm rot}$ By Minimizing the Line Width
This Notebook intends to demonstrate the method used in [Teague et al. (2018a)](https://ui.adsabs.harvard.edu/#abs/2018ApJ...860L..12T) to infer the rotation velocity as a function of radius in the disk of HD 163296. The following [Notebook](Examples%20-%20II.ipynb) demonstrates the updated method presented in Teague et al. (2018b) which relaxes many of the assumptions used in this Notebook.
## Methodology
For this method to work we make the assumption that the disk is azimuthally symmetric (note that this does not mean that the emission we observe in symmetric, but only that the underlying disk structure is). Therefore, if we were to observe the line profile at different azimuthal angles for a given radius, they should all have the same shape. What will be different is the line centre due to the line-of-sight component of the rotation,
$$v_0 = v_{\rm LSR} + v_{\rm rot} \cdot \cos \theta$$
where $i$ is the inclination of the disk, $\theta$ is the azimuthal angle measured from the red-shifted major axis and $v_{\rm LSR}$ is the systemic velocity. Note that this azimuthal angle is not the same as position angle and must be calculated accounting for the 3D structure of the disk.
It has already been shown by assuming a rotation velocity, for example from fitting a first moment map, each spectrum can be shifted back to the systemic velocity and then stacked in azimuth to boost the signal-to-noise of these lines (see [Yen et al. (2016)](https://ui.adsabs.harvard.edu/#abs/2016ApJ...832..204Y) for a thorough discussion on this and [Teague et al. (2016)](https://ui.adsabs.harvard.edu/#abs/2016A&A...592A..49T) and [Matrà et al. (2017)](https://ui.adsabs.harvard.edu/#abs/2017ApJ...842....9M) for applications of this).
---

In the above image, the left hand plot shows the typical Keplerian rotation pattern, taking into account a flared emission surface. Dotted lines show contours of constant azimuthal angle $\theta$ and radius $r$. Three spectra, shown on the right in black, are extracted at the dot locations. By shifting the velocity axis of each of this by $-v_{\rm rot} \cdot \cos \theta$ they are aligned along the systemic velocity, $v_{\rm LSR}$, and able to be stacked (shown in gray).
---
However, this only works correctly if we know the rotation velocity. If an incorrect velocity is used to deproject the spectra then the line centres will be scattered around the systemic velocity. When these lines are stacked, the resulting profile will be broader with a smaller amplitude. We can therefore assert that the correct velocity used to derproject the spectra is the one which _minimises the width of the stacked line profile_. One could make a similar argument about the line peak, however with noisy data this is a less strict constraint as this relies on one channel (the one containing the line peak) rather than the entire line profile ([Yen et al. (2018)](www.google.com), who use a similar method, use the signal-to-noise of the stacked line weighted by a Gaussian fit as their quality of fit measure).
## Python Implementation
This approach is relatively simple to code up with Python. We consider the case of very high signal-to-noise data, however it also works well with low signal-to-noise data, as we describe below. All the functions are part of the `eddy.ensemble` class which will be discussed in more detail below.
We start with an annulus of spectra which we have extracted from our data, along with their azimuthal angles and the velocity axis of the observations. We can generate model spectra through the `eddy.modelling` functions. We model an annulus of 20 spectra with a peak brightness temperature of 40K, a linewidth of 350m/s and and RMS noise of 2K. What's returned is an `ensemble` instance which containts all the deprojecting functions.
```
%matplotlib inline
from eddy.annulus import ensemble
from eddy.modelling import gaussian_ensemble
annulus = gaussian_ensemble(vrot=1500., Tb=40., dV=350., rms=2.0, N=20, plot=True, return_ensemble=True)
```
We first want to shift all the points to the systemic velocity (here at 0m/s). To do this we use the `deproject_spectra()` function which takes the rotation as its only argument. It returns the new velocity of each pixel in the annulus and it's value. Lets first deproject with the correct rotation velocity of 1500m/s to check we recover the intrinsic line profile.
```
velocity, brightness = annulus.deprojected_spectra(1500.)
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.errorbar(velocity, brightness, fmt='.k', ms=4)
ax.set_xlim(velocity[0], velocity[-1])
ax.set_xlabel(r'Velocity')
ax.set_ylabel(r'Intensity')
```
This highlights which this method can achieve such a high precision on determinations of the rotation velocity. Because we shift back all the spectra by a non-quantised amount, we end up sampling the intrinsic profile at a much higher rate (by a factor of the number of beams we have in our annulus).
We can compare this with the spectrum which is resampled backed down to the original velocity resolution using the `deprojected_spectrum()` functions.
```
fig, ax = plt.subplots()
velocity, brightness = annulus.deprojected_spectrum(1500.)
ax.errorbar(velocity, brightness, fmt='.k', ms=4)
ax.set_xlim(velocity[0], velocity[-1])
ax.set_xlabel(r'Velocity')
ax.set_ylabel(r'Intensity')
```
Now, if we projected the spectra with the incorrect velocity, we can see that the stacked spectrum becomes broader. Note also that this is symmetric about the correct velocity meaning this is a convex problem making minimization much easier.
```
import numpy as np
fig, ax = plt.subplots()
for vrot in np.arange(1100, 2100, 200):
velocity, brightness = annulus.deprojected_spectrum(vrot)
ax.plot(velocity, brightness, label='%d m/s' % vrot)
ax.legend(markerfirst=False)
ax.set_xlim(-1000, 1000)
ax.set_xlabel(r'Velocity')
ax.set_ylabel(r'Intensity')
```
We can measure the width of the stacked lines by fitting a Gaussian using the `get_deprojected_width()` function.
```
vrots = np.linspace(1300, 1700, 150)
widths = np.array([annulus.get_deprojected_width(vrot) for vrot in vrots])
fig, ax = plt.subplots()
ax.plot(vrots, widths, label='Deprojected Widths')
ax.axvline(1500., ls=':', color='k', label='Truth')
ax.set_xlabel(r'Rotation Velocity (m/s)')
ax.set_ylabel(r'Width of Stacked Line (m/s)')
ax.legend(markerfirst=False)
```
This shows that if we find the rotation velocity which minimizes the width of the stacked line we should have a pretty good idea of the rotation velocity is. The `get_vrot_dV()` function packges this all up, using the `bounded` method to search for the minimum width within a range of 0.7 to 1.3 times an initial guess. This guess can be provided (for instance if you have an idea of what the Keplerian rotation should be), otherwise it will try to guess it from the spectra based on the peaks of the spectra which are most shifted.
```
vfit = annulus.get_vrot_dV()
print("The linewidth is minimized for a rotation velocity of %.1f m/s" % vfit)
```
The power of this method is also that the fitting is performed on the stacked spectrum meaning that in the noisy regions at the edges of the disk we stack over so many independent beams that we still get a reasonable line profile to fit.
Lets try with a signal-to-noise ratio of 4.
```
annulus = gaussian_ensemble(vrot=1500., Tb=40., dV=350., rms=10.0, N=20, plot=True, return_ensemble=True)
fig, ax = plt.subplots()
velocity, brightness = annulus.deprojected_spectrum(1500.)
ax.step(velocity, brightness, color='k', where='mid', label='Shifted')
ax.legend(markerfirst=False)
ax.set_xlim(velocity[0], velocity[-1])
ax.set_xlabel(r'Velocity')
ax.set_ylabel(r'Intensity')
vfit = annulus.get_vrot_dV()
print("The linewidth is minimized for a rotation velocity of %.1f m/s" % vfit)
```
The final advtange of this method is that it is exceptionally quick. The convex nature of the problem means that a minimum width is readily found and so it can be applied very quickly, even with a large number of spectra. With 200 indiviudal beams:
```
annulus = gaussian_ensemble(vrot=1500., Tb=40., dV=350., rms=10.0, N=200, plot=True, return_ensemble=True)
%timeit annulus.get_vrot_dV()
```
This method, however, does not provide a good measure of the uncertainty on the inferred rotation velocity. Furthermore, it makes the implicit assumption that the intrinsic line profile is Gaussian which for optically thick lines is not the case. In the next [Notebook](Examples%20-%20II.ipynb) we use Gaussian Processes to model the stacked line profile and search for the smoothest model.
| github_jupyter |
## Data Extraction and load from FRED API..
```
## Import packages for the process...
import requests
import pickle
import os
import mysql.connector
import time
```
### Using pickle to wrap the database credentials and Fred API keys
```
if not os.path.exists('fred_api_secret.pk1'):
fred_key = {}
fred_key['api_key'] = ''
with open ('fred_api_secret.pk1','wb') as f:
pickle.dump(fred_key,f)
else:
fred_key=pickle.load(open('fred_api_secret.pk1','rb'))
if not os.path.exists('fred_sql.pk1'):
fred_sql = {}
fred_sql['user'] = ''
fred_sql['password'] = ''
fred_sql['database'] = ''
with open ('fred_sql.pk1','wb') as f:
pickle.dump(fred_sql,f)
else:
fred_sql=pickle.load(open('fred_sql.pk1','rb'))
```
#### testing database connection.
We have a lookup table containing the FRED series along with the value. Let's export the connection parameters and test the connection by running a select query against the lookup table.
```
cn = mysql.connector.connect(user=fred_sql['user'], password=fred_sql['password'],
host='127.0.0.1',
database=fred_sql['database'])
cursor = cn.cursor()
query = ("SELECT frd_cd,frd_val FROM frd_lkp")
cursor.execute(query)
for (frd_cd,frd_val) in cursor:
sr_list.append(frd_cd)
print(frd_cd +' - '+ frd_val)
cn.close()
```
## Helper functions..
We are doing this exercise with minimal modelling. Hence, just one target table to store the observations for all series.
Let's create few helper functions to make this process easier.
db_max_count - We are adding surrogate key to the table to make general querying operations and loads easier. COALESCE is used, to get a valid value from the database.
db_srs_count - Since we are using just one target table, we are adding the series name as part of the data. this function will help us with the count for each series present in the table.
fred_req - Helper function that sends the request to FRED API and returns the response back..
```
def db_max_count():
cn = mysql.connector.connect(user=fred_sql['user'], password=fred_sql['password'],
host='127.0.0.1',
database=fred_sql['database'])
cursor = cn.cursor()
dbquery = ("SELECT COALESCE(max(idfrd_srs),0) FROM frd_srs_data")
cursor.execute(dbquery)
for ct in cursor:
if ct is not None:
return ct[0]
cn.close()
def db_srs_count():
cn = mysql.connector.connect(user=fred_sql['user'], password=fred_sql['password'],
host='127.0.0.1',
database=fred_sql['database'])
cursor = cn.cursor()
dbquery = ("SELECT frd_srs, count(*) FROM frd_srs_data group by frd_srs")
cursor.execute(dbquery)
for ct in cursor:
print(ct)
cn.close()
def fred_req(series):
time.sleep(10)
response = requests.get('https://api.stlouisfed.org/fred/series/observations?series_id='+series+'&api_key='+fred_key['api_key']+'&file_type=json')
result = response.json()
return result
```
## Main functions..
We are creating main functions to support the process. Here are the steps
1) Get the data from FRED API. (helper function created above)
2) Validate and transform the observations data from API.
3) Create tuples according to the table structure.
4) Load the tuples into the relational database
fred_data for Step 2 & Step 3. Function dbload for Step 4.
```
def dbload(tuple_list):
try:
cn = mysql.connector.connect(user=fred_sql['user'], password=fred_sql['password'],
host='127.0.0.1',
database=fred_sql['database'])
cursor = cn.cursor()
insert_query = ("INSERT INTO frd_srs_data"
"(idfrd_srs,frd_srs,frd_srs_val_dt,frd_srs_val,frd_srs_val_yr,frd_srs_val_mth,frd_srs_val_dy,frd_srs_strt_dt,frd_srs_end_dt)"
"VALUES (%s,%s,%s,%s,%s,%s,%s,%s,%s)")
print("*** Database Connection Initialized, buckle up the seat belts..")
# Data load..
for i in range(len(tuple_list)):
data_val=tuple_list[i]
cursor.execute(insert_query, data_val)
cn.commit()
## Intended timeout before starting the next interation of load..
time.sleep(5)
print("\n *** Data load successful.. ")
db_srs_count()
# Closing database connection...
cn.close
except mysql.connector.Error as err:
cn.close
print("Something went wrong: {}".format(err))
def fred_data(series):
print("\n")
print("** Getting data for the series: " + series)
counter=db_max_count()
# Calling function to get the data from FRED API for the series.
fred_result = fred_req(series)
print("** Number of observations extracted -" '{:d}'.format(fred_result['count']))
# transforming observations and preparing for data load.
print("** Preparing data for load for series -",series)
temp_lst = fred_result['observations']
tlist = []
# from the incoming data, let's create tuple of values for data load.
for val in range(len(temp_lst)):
temp_dict = temp_lst[val]
for key,val in temp_dict.items():
if key=='date':
dt_lst = val.split("-")
yr = dt_lst[0]
mth = dt_lst[1]
dtt = dt_lst[2]
if key=='value':
if len(val.strip())>1:
out_val = val
else:
out_val = 0.00
counter+=1
tup = (counter,series,temp_dict['date'],out_val,yr,mth,dtt,temp_dict['realtime_start'],temp_dict['realtime_end'])
tlist.append(tup)
print("** Data is ready for the load.. Loading " '{:d}'.format(len(tlist)))
dbload(tlist)
```
### Starting point...
So, we have all functions created based on few assumptions (that data is all good with very minimal or no issues).
```
sr_list = ['UMCSENT', 'GDPC1', 'UNRATE']
for series in sr_list:
fred_data(series)
cn = mysql.connector.connect(user=fred_sql['user'], password=fred_sql['password'],
host='127.0.0.1',
database=fred_sql['database'])
cursor = cn.cursor()
quizquery = ("SELECT frd_srs_val_yr , avg(frd_srs_val) as avg_unrate FROM fred.frd_srs_data WHERE frd_srs='UNRATE' AND frd_srs_val_yr BETWEEN 1980 AND 2015 GROUP BY frd_srs_val_yr ORDER BY 1")
cursor.execute(quizquery)
for qz in cursor:
print(qz)
```
| github_jupyter |
```
import pathlib
import lzma
import re
import os
import datetime
import copy
import numpy as np
import pandas as pd
# Makes it so any changes in pymedphys is automatically
# propagated into the notebook without needing a kernel reset.
from IPython.lib.deepreload import reload
%load_ext autoreload
%autoreload 2
import pymedphys._utilities.filesystem
from prototyping import *
root = pathlib.Path(r'\\physics-server\iComLogFiles\patients')
compressed_files = sorted(list(root.glob('**/*.xz')))
# compressed_files
mechanical_output = root.parent.joinpath('mechanical/4299/20200116.csv')
mechanical_output.parent.mkdir(exist_ok=True)
data = b""
for path in compressed_files:
with lzma.open(path, 'r') as f:
data += f.read()
data_points = get_data_points(data)
mechanical_data = {}
for data_point in data_points:
_, result = strict_extract(data_point)
machine_id = result['Machine ID']
try:
machine_record = mechanical_data[machine_id]
except KeyError:
machine_record = {}
mechanical_data[machine_id] = machine_record
timestamp = result['Timestamp']
try:
timestamp_record = machine_record[timestamp]
except KeyError:
timestamp_record = {}
machine_record[timestamp] = timestamp_record
counter = result['Counter']
mlc = result['MLCX']
mlc_a = mlc[0::2]
mlc_b = mlc[1::2]
width_at_cra = np.mean(mlc_b[39:41] - mlc_a[39:41])
jaw = result['ASYMY']
length = np.sum(jaw)
timestamp_record[counter] = {
'Energy': result['Energy'],
'Monitor Units': result['Total MU'],
'Gantry': result['Gantry'],
'Collimator': result['Collimator'],
'Table Column': result['Table Column'],
'Table Isocentric': result['Table Isocentric'],
'Table Vertical': result['Table Vertical'],
'Table Longitudinal': result['Table Longitudinal'],
'Table Lateral': result['Table Lateral'],
'MLC distance at CRA': width_at_cra,
'Jaw distance': length
}
# pd.Timestamp('2020-01-16T17:08:45')
table_record = pd.DataFrame(
columns=[
'Timestamp', 'Counter', 'Energy', 'Monitor Units', 'Gantry', 'Collimator', 'Table Column',
'Table Isocentric', 'Table Vertical', 'Table Longitudinal',
'Table Lateral', 'MLC distance at CRA', 'Jaw distance'
]
)
for timestamp, timestamp_record in mechanical_data[4299].items():
for counter, record in timestamp_record.items():
table_record = table_record.append({
**{
'Timestamp': pd.Timestamp(timestamp),
'Counter': counter
},
**record
}, ignore_index=True)
table_record.to_csv(mechanical_output, index=False)
```
| github_jupyter |
<img src="https://storage.googleapis.com/arize-assets/arize-logo-white.jpg" width="200"/>
# Arize Tutorial: Surrogate Model Feature Importance
A surrogate model is an interpretable model trained on predicting the predictions of a black box model. The goal is to approximate the predictions of the black box model as closely as possible and generate feature importance values from the interpretable surrogate model. The benefit of this approach is that it does not require knowledge of the inner workings of the black box model.
In this tutorial we use the `MimcExplainer` from the `interpret_community` library to generate feature importance values from a surrogate model using only the prediction outputs from a black box model. Both [classification](#classification) and [regression](#regression) examples are provided below and feature importance values are logged to Arize using the Pandas [logger](https://docs.arize.com/arize/api-reference/python-sdk/arize.pandas).
# Install and import the `interpret_community` library
```
!pip install -q interpret==0.2.7 interpret-community==0.22.0
from interpret_community.mimic.mimic_explainer import (
MimicExplainer,
LGBMExplainableModel,
)
```
<a name="classification"></a>
# Classification Example
### Generate example
In this example we'll use a support vector machine (SVM) as our black box model. Only the prediction outputs of the SVM model is needed to train the surrogate model, and feature importances are generated from the surrogate model and sent to Arize.
```
import pandas as pd
import numpy as np
from sklearn.datasets import load_breast_cancer
from sklearn.svm import SVC
bc = load_breast_cancer()
feature_names = bc.feature_names
target_names = bc.target_names
data, target = bc.data, bc.target
df = pd.DataFrame(data, columns=feature_names)
model = SVC(probability=True).fit(df, target)
prediction_label = pd.Series(map(lambda v: target_names[v], model.predict(df)))
prediction_score = pd.Series(map(lambda v: v[1], model.predict_proba(df)))
actual_label = pd.Series(map(lambda v: target_names[v], target))
actual_score = pd.Series(target)
```
### Generate feature importance values
Note that the model itself is not used here. Only its prediction outputs are used.
```
def model_func(_):
return np.array(list(map(lambda p: [1 - p, p], prediction_score)))
explainer = MimicExplainer(
model_func,
df,
LGBMExplainableModel,
augment_data=False,
is_function=True,
)
feature_importance_values = pd.DataFrame(
explainer.explain_local(df).local_importance_values, columns=feature_names
)
feature_importance_values
```
### Send data to Arize
Set up Arize client. We'll be using the Pandas Logger. First copy the Arize `API_KEY` and `ORG_KEY` from your admin page linked below!
[](https://app.arize.com/admin)
```
!pip install -q arize
from arize.pandas.logger import Client, Schema
from arize.utils.types import ModelTypes, Environments
ORGANIZATION_KEY = "ORGANIZATION_KEY"
API_KEY = "API_KEY"
arize_client = Client(organization_key=ORGANIZATION_KEY, api_key=API_KEY)
if ORGANIZATION_KEY == "ORGANIZATION_KEY" or API_KEY == "API_KEY":
raise ValueError("❌ NEED TO CHANGE ORGANIZATION AND/OR API_KEY")
else:
print("✅ Import and Setup Arize Client Done! Now we can start using Arize!")
```
Helper functions to simulate prediction IDs and timestamps.
```
import uuid
from datetime import datetime, timedelta
# Prediction ID is required for logging any dataset
def generate_prediction_ids(df):
return pd.Series((str(uuid.uuid4()) for _ in range(len(df))), index=df.index)
# OPTIONAL: We can directly specify when inferences were made
def simulate_production_timestamps(df, days=30):
t = datetime.now()
current_t, earlier_t = t.timestamp(), (t - timedelta(days=days)).timestamp()
return pd.Series(np.linspace(earlier_t, current_t, num=len(df)), index=df.index)
```
Assemble Pandas DataFrame as a production dataset with prediction IDs and timestamps.
```
feature_importance_values_column_names_mapping = {
f"{feat}": f"{feat} (feature importance)" for feat in feature_names
}
production_dataset = pd.concat(
[
pd.DataFrame(
{
"prediction_id": generate_prediction_ids(df),
"prediction_ts": simulate_production_timestamps(df),
"prediction_label": prediction_label,
"actual_label": actual_label,
"prediction_score": prediction_score,
"actual_score": actual_score,
}
),
df,
feature_importance_values.rename(
columns=feature_importance_values_column_names_mapping
),
],
axis=1,
)
production_dataset
```
Send dataframe to Arize
```
# Define a Schema() object for Arize to pick up data from the correct columns for logging
production_schema = Schema(
prediction_id_column_name="prediction_id", # REQUIRED
timestamp_column_name="prediction_ts",
prediction_label_column_name="prediction_label",
prediction_score_column_name="prediction_score",
actual_label_column_name="actual_label",
actual_score_column_name="actual_score",
feature_column_names=feature_names,
shap_values_column_names=feature_importance_values_column_names_mapping,
)
# arize_client.log returns a Response object from Python's requests module
response = arize_client.log(
dataframe=production_dataset,
schema=production_schema,
model_id="surrogate_model_example_classification",
model_type=ModelTypes.SCORE_CATEGORICAL,
environment=Environments.PRODUCTION,
)
# If successful, the server will return a status_code of 200
if response.status_code != 200:
print(
f"❌ logging failed with response code {response.status_code}, {response.text}"
)
else:
print(
f"✅ You have successfully logged {len(production_dataset)} data points to Arize!"
)
```
<a name="regression"></a>
# Regression Example
### Generate example
In this example we'll use a support vector machine (SVM) as our black box model. Only the prediction outputs of the SVM model is needed to train the surrogate model, and feature importances are generated from the surrogate model and sent to Arize.
```
import pandas as pd
import numpy as np
from sklearn.datasets import fetch_california_housing
housing = fetch_california_housing()
# Use only 1,000 data point for a speedier example
data_reg = housing.data[:1000]
target_reg = housing.target[:1000]
feature_names_reg = housing.feature_names
df_reg = pd.DataFrame(data_reg, columns=feature_names_reg)
from sklearn.svm import SVR
model_reg = SVR().fit(df_reg, target_reg)
prediction_label_reg = pd.Series(model_reg.predict(df_reg))
actual_label_reg = pd.Series(target_reg)
```
### Generate feature importance values
Note that the model itself is not used here. Only its prediction outputs are used.
```
def model_func_reg(_):
return np.array(prediction_label_reg)
explainer_reg = MimicExplainer(
model_func_reg,
df_reg,
LGBMExplainableModel,
augment_data=False,
is_function=True,
)
feature_importance_values_reg = pd.DataFrame(
explainer_reg.explain_local(df_reg).local_importance_values,
columns=feature_names_reg,
)
feature_importance_values_reg
```
Assemble Pandas DataFrame as a production dataset with prediction IDs and timestamps.
```
feature_importance_values_column_names_mapping_reg = {
f"{feat}": f"{feat} (feature importance)" for feat in feature_names_reg
}
production_dataset_reg = pd.concat(
[
pd.DataFrame(
{
"prediction_id": generate_prediction_ids(df_reg),
"prediction_ts": simulate_production_timestamps(df_reg),
"prediction_label": prediction_label_reg,
"actual_label": actual_label_reg,
}
),
df_reg,
feature_importance_values_reg.rename(
columns=feature_importance_values_column_names_mapping_reg
),
],
axis=1,
)
production_dataset_reg
```
Send DataFrame to Arize.
```
# Define a Schema() object for Arize to pick up data from the correct columns for logging
production_schema_reg = Schema(
prediction_id_column_name="prediction_id", # REQUIRED
timestamp_column_name="prediction_ts",
prediction_label_column_name="prediction_label",
actual_label_column_name="actual_label",
feature_column_names=feature_names_reg,
shap_values_column_names=feature_importance_values_column_names_mapping_reg,
)
# arize_client.log returns a Response object from Python's requests module
response_reg = arize_client.log(
dataframe=production_dataset_reg,
schema=production_schema_reg,
model_id="surrogate_model_example_regression",
model_type=ModelTypes.NUMERIC,
environment=Environments.PRODUCTION,
)
# If successful, the server will return a status_code of 200
if response_reg.status_code != 200:
print(
f"❌ logging failed with response code {response_reg.status_code}, {response_reg.text}"
)
else:
print(
f"✅ You have successfully logged {len(production_dataset_reg)} data points to Arize!"
)
```
## Conclusion
You now know how to seamlessly log surrogate model feature importance values onto the Arize platform. Go to [Arize](https://app.arize.com/) in order to analyze and monitor the logged SHAP values.
### Overview
Arize is an end-to-end ML observability and model monitoring platform. The platform is designed to help ML engineers and data science practitioners surface and fix issues with ML models in production faster with:
- Automated ML monitoring and model monitoring
- Workflows to troubleshoot model performance
- Real-time visualizations for model performance monitoring, data quality monitoring, and drift monitoring
- Model prediction cohort analysis
- Pre-deployment model validation
- Integrated model explainability
### Website
Visit Us At: https://arize.com/model-monitoring/
### Additional Resources
- [What is ML observability?](https://arize.com/what-is-ml-observability/)
- [Playbook to model monitoring in production](https://arize.com/the-playbook-to-monitor-your-models-performance-in-production/)
- [Using statistical distance metrics for ML monitoring and observability](https://arize.com/using-statistical-distance-metrics-for-machine-learning-observability/)
- [ML infrastructure tools for data preparation](https://arize.com/ml-infrastructure-tools-for-data-preparation/)
- [ML infrastructure tools for model building](https://arize.com/ml-infrastructure-tools-for-model-building/)
- [ML infrastructure tools for production](https://arize.com/ml-infrastructure-tools-for-production-part-1/)
- [ML infrastructure tools for model deployment and model serving](https://arize.com/ml-infrastructure-tools-for-production-part-2-model-deployment-and-serving/)
- [ML infrastructure tools for ML monitoring and observability](https://arize.com/ml-infrastructure-tools-ml-observability/)
Visit the [Arize Blog](https://arize.com/blog) and [Resource Center](https://arize.com/resource-hub/) for more resources on ML observability and model monitoring.
| github_jupyter |
<a href="https://colab.research.google.com/github/MingSheng92/AE_denoise/blob/master/DL_Example.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!pip show tensorflow
!git clone https://github.com/MingSheng92/AE_denoise.git
from google.colab import drive
drive.mount('/content/drive')
%load /content/AE_denoise/scripts/utility.py
%load /content/AE_denoise/scripts/Denoise_NN.py
from AE_denoise.scripts.utility import load_data, faceGrid, ResultGrid, subsample, AddNoiseToMatrix, noisy
from AE_denoise.scripts.Denoise_NN import PSNRLoss, createModel
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
img_data, label, img_size = load_data('/content/drive/My Drive/FaceDataset/CroppedYaleB', 0)
#img_data, label, img_size = load_data('/content/drive/My Drive/FaceDataset/ORL', 0)
img_size
x_train, x_test, y_train, y_test = train_test_split(img_data.T, label, test_size=0.1, random_state=111)
x_train, x_val, y_train, y_val = train_test_split(x_train, y_train, test_size=0.1, random_state=111)
print("Total number of training samples: ", x_train.shape)
print("Total number of training samples: ", x_val.shape)
print("Total number of validation samples: ", x_test.shape)
x_train = x_train.astype('float32') / 255.0
x_val = x_val.astype('float32') / 255.0
x_test = x_test.astype('float32') / 255.0
#x_train = x_train.reshape(-1, img_size[0], img_size[1], 1)
#x_val = x_val.reshape(-1, img_size[0], img_size[1], 1)
x_train = np.reshape(x_train, (len(x_train), img_size[0], img_size[1], 1))
x_val = np.reshape(x_val, (len(x_val), img_size[0], img_size[1], 1))
x_test = np.reshape(x_test, (len(x_test), img_size[0], img_size[1], 1))
# add noise to the face images
noise_factor = 0.3
x_train_noisy = x_train + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_train.shape)
x_val_noisy = x_val + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_val.shape)
x_test_noisy = x_test + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=x_test.shape)
x_train_noisy = np.clip(x_train_noisy, 0., 1.)
x_val_noisy = np.clip(x_val_noisy, 0., 1.)
x_test_noisy = np.clip(x_test_noisy, 0., 1.)
faceGrid(10, x_train, img_size, 64)
faceGrid(10, x_train_noisy, img_size, 64)
model = createModel(img_size)
model.summary()
model.fit(x_train_noisy, x_train,
epochs=15,
batch_size=64,
validation_data=(x_val_noisy, x_val))
denoise_prediction = model.predict(x_test_noisy)
faceGrid(10, x_test, img_size, 5)
faceGrid(10, x_test_noisy, img_size, 5)
faceGrid(10, denoise_prediction, img_size, 5)
```
| github_jupyter |
```
!pwd
%matplotlib inline
```
PyTorch: nn
-----------
A fully-connected ReLU network with one hidden layer, trained to predict y from x
by minimizing squared Euclidean distance.
This implementation uses the nn package from PyTorch to build the network.
PyTorch autograd makes it easy to define computational graphs and take gradients,
but raw autograd can be a bit too low-level for defining complex neural networks;
this is where the nn package can help. The nn package defines a set of Modules,
which you can think of as a neural network layer that has produces output from
input and may have some trainable weights.
```
import torch
from torch.autograd import Variable
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
N, D_in, H, D_out = 64, 1000, 100, 10
# Create random Tensors to hold inputs and outputs, and wrap them in Variables.
x = Variable(torch.randn(N, D_in))
y = Variable(torch.randn(N, D_out), requires_grad=False)
# Use the nn package to define our model as a sequence of layers. nn.Sequential
# is a Module which contains other Modules, and applies them in sequence to
# produce its output. Each Linear Module computes output from input using a
# linear function, and holds internal Variables for its weight and bias.
model = torch.nn.Sequential(
torch.nn.Linear(D_in, H),
torch.nn.ReLU(),
torch.nn.Linear(H, D_out),
)
# The nn package also contains definitions of popular loss functions; in this
# case we will use Mean Squared Error (MSE) as our loss function.
loss_fn = torch.nn.MSELoss(size_average=False)
learning_rate = 1e-4
for t in range(500):
# Forward pass: compute predicted y by passing x to the model. Module objects
# override the __call__ operator so you can call them like functions. When
# doing so you pass a Variable of input data to the Module and it produces
# a Variable of output data.
y_pred = model(x)
# Compute and print loss. We pass Variables containing the predicted and true
# values of y, and the loss function returns a Variable containing the
# loss.
loss = loss_fn(y_pred, y)
print(t, loss.data[0])
# Zero the gradients before running the backward pass.
model.zero_grad()
# Backward pass: compute gradient of the loss with respect to all the learnable
# parameters of the model. Internally, the parameters of each Module are stored
# in Variables with requires_grad=True, so this call will compute gradients for
# all learnable parameters in the model.
loss.backward()
# Update the weights using gradient descent. Each parameter is a Variable, so
# we can access its data and gradients like we did before.
for param in model.parameters():
param.data -= learning_rate * param.grad.data
```
| github_jupyter |
# Understanding Data Types in Python
Effective data-driven science and computation requires understanding how data is stored and manipulated. This section outlines and contrasts how arrays of data are handled in the Python language itself, and how NumPy improves on this. Understanding this difference is fundamental to understanding much of the material throughout the rest of the course.
Python is simple to use. While a statically-typed language like C or Java requires each variable to be explicitly declared, a dynamically-typed language like Python skips this specification.
In C, the data types of each variable are explicitly declared, while in Python the types are dynamically inferred.
This means, for example, that we can assign any kind of data to any variable:
```
x = 4
x = "four"
```
This sort of flexibility is one piece that makes Python and other dynamically-typed languages convenient and easy to use.
## 1.1. Data Types
We have several data types in python:
* None
* Numeric (int, float, complex, bool)
* List
* Tuple
* Set
* String
* Range
* Dictionary (Map)
```
# NoneType
a = None
type(a)
# int
a = 1+1
print(a)
type(a)
# complex
c = 1.5 + 0.5j
type(c)
c.real
c.imag
# boolean
d = 2 > 3
print(d)
type(d)
```
## Python Lists
Let's consider now what happens when we use a Python data structure that holds many Python objects. The standard mutable multi-element container in Python is the list. We can create a list of integers as follows:
```
L = list(range(10))
L
type(L[0])
```
Or, similarly, a list of strings:
```
L2 = [str(c) for c in L]
L2
type(L2[0])
```
Because of Python's dynamic typing, we can even create heterogeneous lists:
```
L3 = [True, "2", 3.0, 4]
[type(item) for item in L3]
```
## Python Dictionaries
```
keys = [1, 2, 3, 4, 5]
values = ['monday', 'tuesday', 'wendsday', 'friday']
dictionary = dict(zip(keys, values))
dictionary
dictionary.get(1)
dictionary[1]
```
## Fixed-Type Arrays in Python
```
import numpy as np
```
First, we can use np.array to create arrays from Python lists:
```
# integer array:
np.array([1, 4, 2, 5, 3])
```
Unlike python lists, NumPy is constrained to arrays that all contain the same type.
If we want to explicitly set the data type of the resulting array, we can use the dtype keyword:
```
np.array([1, 2, 3, 4], dtype='float32')
```
### Creating Arrays from Scratch
Especially for larger arrays, it is more efficient to create arrays from scratch using routines built into NumPy. Here are several examples:
```
# Create a length-10 integer array filled with zeros
np.zeros(10, dtype=int)
# Create a 3x5 floating-point array filled with ones
np.ones((3, 5), dtype=float)
# Create a 3x5 array filled with 3.14
np.full((3, 5), 3.14)
# Create an array filled with a linear sequence
np.arange(1, 10)
# Starting at 0, ending at 20, stepping by 2
# (this is similar to the built-in range() function)
np.arange(0, 20, 2)
```
| github_jupyter |
```
import numpy as np
from keras.models import Model
from keras.layers import Input
from keras.layers.pooling import GlobalMaxPooling1D
from keras import backend as K
import json
from collections import OrderedDict
def format_decimal(arr, places=6):
return [round(x * 10**places) / 10**places for x in arr]
DATA = OrderedDict()
```
### GlobalMaxPooling1D
**[pooling.GlobalMaxPooling1D.0] input 6x6**
```
data_in_shape = (6, 6)
L = GlobalMaxPooling1D()
layer_0 = Input(shape=data_in_shape)
layer_1 = L(layer_0)
model = Model(inputs=layer_0, outputs=layer_1)
# set weights to random (use seed for reproducibility)
np.random.seed(260)
data_in = 2 * np.random.random(data_in_shape) - 1
result = model.predict(np.array([data_in]))
data_out_shape = result[0].shape
data_in_formatted = format_decimal(data_in.ravel().tolist())
data_out_formatted = format_decimal(result[0].ravel().tolist())
print('')
print('in shape:', data_in_shape)
print('in:', data_in_formatted)
print('out shape:', data_out_shape)
print('out:', data_out_formatted)
DATA['pooling.GlobalMaxPooling1D.0'] = {
'input': {'data': data_in_formatted, 'shape': data_in_shape},
'expected': {'data': data_out_formatted, 'shape': data_out_shape}
}
```
**[pooling.GlobalMaxPooling1D.1] input 3x7**
```
data_in_shape = (3, 7)
L = GlobalMaxPooling1D()
layer_0 = Input(shape=data_in_shape)
layer_1 = L(layer_0)
model = Model(inputs=layer_0, outputs=layer_1)
# set weights to random (use seed for reproducibility)
np.random.seed(261)
data_in = 2 * np.random.random(data_in_shape) - 1
result = model.predict(np.array([data_in]))
data_out_shape = result[0].shape
data_in_formatted = format_decimal(data_in.ravel().tolist())
data_out_formatted = format_decimal(result[0].ravel().tolist())
print('')
print('in shape:', data_in_shape)
print('in:', data_in_formatted)
print('out shape:', data_out_shape)
print('out:', data_out_formatted)
DATA['pooling.GlobalMaxPooling1D.1'] = {
'input': {'data': data_in_formatted, 'shape': data_in_shape},
'expected': {'data': data_out_formatted, 'shape': data_out_shape}
}
```
**[pooling.GlobalMaxPooling1D.2] input 8x4**
```
data_in_shape = (8, 4)
L = GlobalMaxPooling1D()
layer_0 = Input(shape=data_in_shape)
layer_1 = L(layer_0)
model = Model(inputs=layer_0, outputs=layer_1)
# set weights to random (use seed for reproducibility)
np.random.seed(262)
data_in = 2 * np.random.random(data_in_shape) - 1
result = model.predict(np.array([data_in]))
data_out_shape = result[0].shape
data_in_formatted = format_decimal(data_in.ravel().tolist())
data_out_formatted = format_decimal(result[0].ravel().tolist())
print('')
print('in shape:', data_in_shape)
print('in:', data_in_formatted)
print('out shape:', data_out_shape)
print('out:', data_out_formatted)
DATA['pooling.GlobalMaxPooling1D.2'] = {
'input': {'data': data_in_formatted, 'shape': data_in_shape},
'expected': {'data': data_out_formatted, 'shape': data_out_shape}
}
```
### export for Keras.js tests
```
import os
filename = '../../../test/data/layers/pooling/GlobalMaxPooling1D.json'
if not os.path.exists(os.path.dirname(filename)):
os.makedirs(os.path.dirname(filename))
with open(filename, 'w') as f:
json.dump(DATA, f)
print(json.dumps(DATA))
```
| github_jupyter |
# GWAS, PheWAS, and Mendelian Randomization
> Understanding Methods of Genetic Analysis
- categories: [jupyter]
## GWAS
Genome Wide Association Studies (GWAS) look for genetic variants across the genome in a large amount of individuals to see if any variants are associated with a specific trait such as height or disease. GWA studies typically look at single nucleotide polymorphisms (SNPs), which are germline substitutions of a single nucleotide at a specific position in the genome, meaning that these are heritable differences in the human population. A GWAS is performed by taking DNA samples from many individuals and using SNP arrays to read the different genetic variants. If a particular variant is more present in people with a specific trait, the SNP is associated with the disease. Results from a GWAS are typically shown in a Manhattan plot displaying which loci on the various chromosomes are more associated with a specific trait. In the picture below taken from Wikipedia, each dot represents a SNP, and "this example is taken from a GWA study investigating microcirculation, so the tops indicates genetic variants that more often are found in individuals with constrictions in small blood vessels."
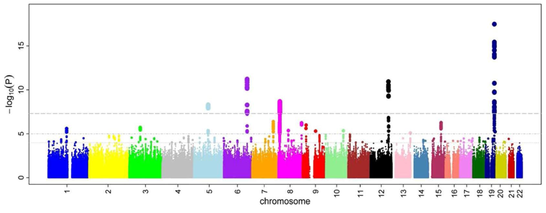
A GWAS is a non-candidate-driven approach, in that a GWAS investigate the whole genome and not specific genes. As a result, a GWAS can tell the user which genes are associated with the disease but cannot describe any causal relations between the genes identified and the trait/disease being studied.
### Methodology
Typically, two general populations are used for a GWAS, a case group with a certain disease and a control group without the disease. The individuals in the population are genotyped for the majority of known SNPs in the human genome, which surpass a million. The allele frequences at each SNP are calculated, and an odds ratio is generated in order to compare the case and control populations. An odds ratio is "the ratio of the odds of A in the presence of B and the odds of A in the absence of B", or in the case of a GWAS, it is "the odds of case for individuals having a specific allele and the odds of case for individuals who do not have that same allele".
For example, at a certain SNP, there are two main alleles, T and C. The amount of individuals in the case group having allele T is represented by A, or 4500, and the amount of individuals in the control group having allele T is represented by B, or 2000. Then the number of individuals in the case group having allele C is represented by X, or 3000, and the individuals in the control group having allele C is represented by Y, or 3500. The odds ratio for allele T is calculated as (A/B) / (X/Y) or (4500/2000) / (3000,3500).
If the allele frequency is much higher in the case group than in the control group, the odds ratio will be greater than one. Furthermore, a chi-squared test is used to calculate a p-value for the significance of the generated odds ratios. For a GWAS, typically a p-value < 5x10^-8 is required for an odds ratio to be meaningful.
Furthermore, factors such as ethnicity, geography, sex, and age must be taken into consideration and controlled for as they could confound the results.
### Imputation
Another key facet used in many studies involves imputation, or the statistical inference of unobserved genetic sequences. Since it is time-consuming and costly to do genome wide sequencing on a large population, only key areas of the genome are typically sequenced and a large portion of the genome is statistically inferred through large scale genome datasets such as the HapMap or 1000 Genomes Project. Imputation is achieved by combining the GWAS data with the reference panel of haplotypes (HapMap/1000 Genomes Project) and inferring other SNPs in the genome through shared haplotypes among individuals over short sequences. For example, if we know that a patient with an A allele at base 10 always has a G allele at base 135, we can impute this information for the entire population. This method increases the number of SNPs that can be tested for association in a GWAS as well as the power of the GWAS.
| github_jupyter |
```
import nltk
from nltk import *
emma = nltk.Text(nltk.corpus.gutenberg.words('austen-emma.txt'))
len(emma)
emma.concordance("surprise")
from nltk.corpus import gutenberg
print(gutenberg.fileids())
emma = gutenberg.words("austen-emma.txt")
type(gutenberg)
for fileid in gutenberg.fileids():
n_chars = len(gutenberg.raw(fileid))
n_words = len(gutenberg.words(fileid))
n_sents = len(gutenberg.sents(fileid))
n_vocab = len(set(w.lower() for w in gutenberg.words(fileid)))
print(f"chr: {n_chars} wor: {n_words} sen: {n_sents} voc: {n_vocab} {fileid}")
print(round(n_chars/n_words), round(n_words/n_sents), round(n_words/n_vocab), fileid)
machbeth = gutenberg.sents("shakespeare-macbeth.txt")
ls = max(len(w) for w in machbeth)
[s for s in machbeth if len(s) == ls]
from nltk.corpus import webtext
for fileid in webtext.fileids():
print(fileid, len(webtext.raw(fileid)), webtext.raw(fileid)[:20])
from nltk.corpus import brown
brown.categories()
brown.words(categories=["lore", "reviews"])
brown.words(fileids=['cg22'])
brown.sents(categories=['news', 'editorial', 'reviews'])
from nltk.corpus import reuters
print(reuters.fileids()[:10])
print(reuters.categories()[:10])
reuters.categories('training/9865')
reuters.categories(['training/9865', 'training/9880'])
reuters.fileids('barley')
reuters.fileids(['barley', 'corn'])
reuters.words('training/9865')[:14]
from nltk.corpus import inaugural
inaugural.fileids()
print([fileid[:4] for fileid in inaugural.fileids()])
import matplotlib.pyplot as plt
y = [int(fileid[:4]) for fileid in inaugural.fileids()]
x = range(0, len(y))
plt.plot(y)
cfd = nltk.ConditionalFreqDist(
(target, fileid[:4]) for fileid in inaugural.fileids() \
for w in inaugural.words(fileid) \
for target in ["america", "citizen"] if w.lower().startswith(target)
)
cfd.plot()
from nltk.corpus import udhr
languages = ['Chickasaw', 'English', 'German_Deutsch', 'Greenlandic_Inuktikut', 'Hungarian_Magyar', 'Ibibio_Efik']
cfd = nltk.ConditionalFreqDist(
(lang, len(word)) \
for lang in languages \
for word in udhr.words(lang + '-Latin1')
)
cfd.plot(cumulative=True)
cfd.tabulate(conditions=['English', 'German_Deutsch'], samples=range(10), cumulative=True)
turkish_raw = udhr.raw("Turkish_Turkce-Turkish")
nltk.FreqDist(turkish_raw).plot()
inaugural.readme()
from nltk.corpus import brown
cfd = nltk.ConditionalFreqDist(
(genre, word)\
for genre in brown.categories() \
for word in brown.words(categories=genre)
)
cfd.items()
days = ["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"]
cfd.tabulate(samples=days)
genre_word = [
(genre, word)\
for genre in ["news", "romance"] \
for word in brown.words(categories=genre)
]
len(genre_word)
genre_word[:4]
cfd = nltk.ConditionalFreqDist(genre_word)
cfd.conditions()
cfd["romance"].most_common()
sent = ['In', 'the', 'beginning', 'God', 'created', 'the', 'heaven',
... 'and', 'the', 'earth', '.']
list(nltk.bigrams(sent))
def generate_model(cfdist, word, num=15):
for i in range(num):
print(word, end=' ')
word = cfdist[word].max()
text = nltk.corpus.genesis.words('english-kjv.txt')
bigrams = nltk.bigrams(text)
cfd = nltk.ConditionalFreqDist(bigrams)
cfd['living']
generate_model(cfd, "living")
cfd['creature']
from nltk.book import *
list(nltk.bigrams(text))
vocal = sorted(set(text))
text
def unusual_words(text):
text_vocab = set(w.lower() for w in text if w.isalpha())
english_vocab = set(w.lower() for w in nltk.corpus.words.words())
unusual = text_vocab - english_vocab
return sorted(unusual)
unusual_words(nltk.corpus.gutenberg.words('austen-sense.txt'))
unusual_words(nltk.corpus.nps_chat.words())
def frac_stopwords(text):
stopwords = nltk.corpus.stopwords.words('english')
content = [w for w in text if w.lower() not in stopwords]
return len(content) / len(text)
frac_stopwords(text)
frac_stopwords(nltk.corpus.reuters.words())
puzzle_letters = nltk.FreqDist('egivrvonl')
obligatory = 'r'
wordlist = nltk.corpus.words.words()
[w for w in wordlist
if len(w) >= 6 and
obligatory in w and
nltk.FreqDist(w) <= puzzle_letters
]
nltk.FreqDist('sdfsd') <= puzzle_letters
names = nltk.corpus.names
male_names, female_names = names.words('male.txt'), names.words('female.txt')
[w for w in male_names if w in female_names]
from nltk import ConditionalFreqDist
cfd = ConditionalFreqDist(
(fileid, name[-1])
for fileid in names.fileids()
for name in names.words(fileid)
)
cfd.plot()
entries = nltk.corpus.cmudict.entries()
for entry in entries[42371:42380]:
print(entry)
for word, pron in entries:
if len(pron) == 3:
ph1, ph2, ph3 = pron
if ph1 == 'P' and ph3 == 'N':
print(word, ph2, end=' ')
syllable = ['N', 'IH0', 'K', 'S']
[word for word, pron in entries if pron[-4:] == syllable]
prondict = nltk.corpus.cmudict.dict()
prondict['fire']
from nltk.corpus import swadesh
swadesh.fileids()
swadesh.words('en')
fr2en = swadesh.entries(['fr', 'en'])
translate = dict(fr2en)
translate['chien']
translate['jeter']
languages = ['en', 'de', 'nl', 'es', 'fr', 'pt', 'la']
for i in [139, 140, 141, 142]:
print(swadesh.entries(languages)[i])
from nltk.corpus import wordnet as wn
wn.synsets('motorcar')
wn.synset('car.n.01').lemma_names()
wn.synset('car.n.01').definition()
wn.synset('car.n.01').examples()
wn.lemma('car.n.01.automobile')
wn.lemma('car.n.01.automobile').synset()
wn.synsets('car')
for synset in wn.synsets('car'):
print(synset.lemma_names())
wn.lemmas('car')
for synset in wn.synsets('dish'):
print(synset.lemma_names())
print(synset.definition())
typmtrcr = wn.synset('car.n.01').hyponyms()[0].hyponyms()
sorted(lemma.name() for synset in typmtrcr for lemma in synset.lemmas())
motorcar = wn.synset('car.n.01')
motorcar.hypernyms()
paths = motorcar.hypernym_paths()
[synset.name() for synset in paths[0]]
```
| github_jupyter |
# Water quality
## Setup software libraries
```
# Import and initialize the Earth Engine library.
import ee
ee.Initialize()
ee.__version__
# Folium setup.
import folium
print(folium.__version__)
# Skydipper library.
import Skydipper
print(Skydipper.__version__)
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import functools
import json
import uuid
import os
from pprint import pprint
import env
import time
import ee_collection_specifics
```
## Composite image
**Variables**
```
collection = 'Lake-Water-Quality-100m'
init_date = '2019-01-21'
end_date = '2019-01-31'
# Define the URL format used for Earth Engine generated map tiles.
EE_TILES = 'https://earthengine.googleapis.com/map/{mapid}/{{z}}/{{x}}/{{y}}?token={token}'
composite = ee_collection_specifics.Composite(collection)(init_date, end_date)
mapid = composite.getMapId(ee_collection_specifics.vizz_params_rgb(collection))
tiles_url = EE_TILES.format(**mapid)
map = folium.Map(location=[39.31, 0.302])
folium.TileLayer(
tiles=tiles_url,
attr='Google Earth Engine',
overlay=True,
name=str(ee_collection_specifics.ee_bands_rgb(collection))).add_to(map)
map.add_child(folium.LayerControl())
map
```
***
## Geostore
We select the areas from which we will export the training data.
**Variables**
```
def polygons_to_multipoligon(polygons):
multipoligon = []
MultiPoligon = {}
for polygon in polygons.get('features'):
multipoligon.append(polygon.get('geometry').get('coordinates'))
MultiPoligon = {
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"properties": {},
"geometry": {
"type": "MultiPolygon",
"coordinates": multipoligon
}
}
]
}
return MultiPoligon
#trainPolygons = {"type":"FeatureCollection","features":[{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-0.45043945312499994,39.142842478062505],[0.06042480468749999,39.142842478062505],[0.06042480468749999,39.55064761909318],[-0.45043945312499994,39.55064761909318],[-0.45043945312499994,39.142842478062505]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-0.2911376953125,38.659777730712534],[0.2581787109375,38.659777730712534],[0.2581787109375,39.10022600175347],[-0.2911376953125,39.10022600175347],[-0.2911376953125,38.659777730712534]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-0.3350830078125,39.56758783088905],[0.22521972656249997,39.56758783088905],[0.22521972656249997,39.757879992021756],[-0.3350830078125,39.757879992021756],[-0.3350830078125,39.56758783088905]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[0.07965087890625,39.21310328979648],[0.23345947265625,39.21310328979648],[0.23345947265625,39.54852980171147],[0.07965087890625,39.54852980171147],[0.07965087890625,39.21310328979648]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-1.0931396484375,35.7286770448517],[-0.736083984375,35.7286770448517],[-0.736083984375,35.94243575255426],[-1.0931396484375,35.94243575255426],[-1.0931396484375,35.7286770448517]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-1.7303466796874998,35.16931803601131],[-1.4666748046875,35.16931803601131],[-1.4666748046875,35.74205383068037],[-1.7303466796874998,35.74205383068037],[-1.7303466796874998,35.16931803601131]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-1.42822265625,35.285984736065764],[-1.131591796875,35.285984736065764],[-1.131591796875,35.782170703266075],[-1.42822265625,35.782170703266075],[-1.42822265625,35.285984736065764]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-1.8127441406249998,35.831174956246535],[-1.219482421875,35.831174956246535],[-1.219482421875,36.04465753921525],[-1.8127441406249998,36.04465753921525],[-1.8127441406249998,35.831174956246535]]]}}]}
trainPolygons = {"type":"FeatureCollection","features":[{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-0.406494140625,38.64476310916202],[0.27740478515625,38.64476310916202],[0.27740478515625,39.74521015328692],[-0.406494140625,39.74521015328692],[-0.406494140625,38.64476310916202]]]}},{"type":"Feature","properties":{},"geometry":{"type":"Polygon","coordinates":[[[-1.70013427734375,35.15135442846945],[-0.703125,35.15135442846945],[-0.703125,35.94688293218141],[-1.70013427734375,35.94688293218141],[-1.70013427734375,35.15135442846945]]]}}]}
trainPolys = polygons_to_multipoligon(trainPolygons)
evalPolys = None
nTrain = len(trainPolys.get('features')[0].get('geometry').get('coordinates'))
print('Number of training polygons:', nTrain)
if evalPolys:
nEval = len(evalPolys.get('features')[0].get('geometry').get('coordinates'))
print('Number of training polygons:', nEval)
```
**Display Polygons**
```
# Define the URL format used for Earth Engine generated map tiles.
EE_TILES = 'https://earthengine.googleapis.com/map/{mapid}/{{z}}/{{x}}/{{y}}?token={token}'
composite = ee_collection_specifics.Composite(collection)(init_date, end_date)
mapid = composite.getMapId(ee_collection_specifics.vizz_params_rgb(collection))
tiles_url = EE_TILES.format(**mapid)
map = folium.Map(location=[39.31, 0.302], zoom_start=6)
folium.TileLayer(
tiles=tiles_url,
attr='Google Earth Engine',
overlay=True,
name=str(ee_collection_specifics.ee_bands_rgb(collection))).add_to(map)
# Convert the GeoJSONs to feature collections
trainFeatures = ee.FeatureCollection(trainPolys.get('features'))
if evalPolys:
evalFeatures = ee.FeatureCollection(evalPolys.get('features'))
polyImage = ee.Image(0).byte().paint(trainFeatures, 1)
if evalPolys:
polyImage = ee.Image(0).byte().paint(trainFeatures, 1).paint(evalFeatures, 2)
polyImage = polyImage.updateMask(polyImage)
mapid = polyImage.getMapId({'min': 1, 'max': 2, 'palette': ['red', 'blue']})
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name='training polygons',
).add_to(map)
map.add_child(folium.LayerControl())
map
```
***
## Data pre-processing
We normalize the composite images to have values from 0 to 1.
**Variables**
```
input_dataset = 'Sentinel-2-Top-of-Atmosphere-Reflectance'
output_dataset = 'Lake-Water-Quality-100m'
init_date = '2019-01-21'
end_date = '2019-01-31'
scale = 100 #scale in meters
collections = [input_dataset, output_dataset]
```
**Normalize images**
```
def min_max_values(image, collection, scale, polygons=None):
normThreshold = ee_collection_specifics.ee_bands_normThreshold(collection)
num = 2
lon = np.linspace(-180, 180, num)
lat = np.linspace(-90, 90, num)
features = []
for i in range(len(lon)-1):
for j in range(len(lat)-1):
features.append(ee.Feature(ee.Geometry.Rectangle(lon[i], lat[j], lon[i+1], lat[j+1])))
if not polygons:
polygons = ee.FeatureCollection(features)
regReducer = {
'geometry': polygons,
'reducer': ee.Reducer.minMax(),
'maxPixels': 1e10,
'bestEffort': True,
'scale':scale,
'tileScale': 10
}
values = image.reduceRegion(**regReducer).getInfo()
print(values)
# Avoid outliers by taking into account only the normThreshold% of the data points.
regReducer = {
'geometry': polygons,
'reducer': ee.Reducer.histogram(),
'maxPixels': 1e10,
'bestEffort': True,
'scale':scale,
'tileScale': 10
}
hist = image.reduceRegion(**regReducer).getInfo()
for band in list(normThreshold.keys()):
if normThreshold[band] != 100:
count = np.array(hist.get(band).get('histogram'))
x = np.array(hist.get(band).get('bucketMeans'))
cumulative_per = np.cumsum(count/count.sum()*100)
values[band+'_max'] = x[np.where(cumulative_per < normThreshold[band])][-1]
return values
def normalize_ee_images(image, collection, values):
Bands = ee_collection_specifics.ee_bands(collection)
# Normalize [0, 1] ee images
for i, band in enumerate(Bands):
if i == 0:
image_new = image.select(band).clamp(values[band+'_min'], values[band+'_max'])\
.subtract(values[band+'_min'])\
.divide(values[band+'_max']-values[band+'_min'])
else:
image_new = image_new.addBands(image.select(band).clamp(values[band+'_min'], values[band+'_max'])\
.subtract(values[band+'_min'])\
.divide(values[band+'_max']-values[band+'_min']))
return image_new
%%time
images = []
for collection in collections:
# Create composite
image = ee_collection_specifics.Composite(collection)(init_date, end_date)
bands = ee_collection_specifics.ee_bands(collection)
image = image.select(bands)
#Create composite
if ee_collection_specifics.normalize(collection):
# Get min man values for each band
values = min_max_values(image, collection, scale, polygons=trainFeatures)
print(values)
# Normalize images
image = normalize_ee_images(image, collection, values)
else:
values = {}
images.append(image)
```
**Display composite**
```
# Define the URL format used for Earth Engine generated map tiles.
EE_TILES = 'https://earthengine.googleapis.com/map/{mapid}/{{z}}/{{x}}/{{y}}?token={token}'
map = folium.Map(location=[39.31, 0.302], zoom_start=6)
for n, collection in enumerate(collections):
for params in ee_collection_specifics.vizz_params(collection):
mapid = images[n].getMapId(params)
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name=str(params['bands']),
).add_to(map)
# Convert the GeoJSONs to feature collections
trainFeatures = ee.FeatureCollection(trainPolys.get('features'))
if evalPolys:
evalFeatures = ee.FeatureCollection(evalPolys.get('features'))
polyImage = ee.Image(0).byte().paint(trainFeatures, 1)
if evalPolys:
polyImage = ee.Image(0).byte().paint(trainFeatures, 1).paint(evalFeatures, 2)
polyImage = polyImage.updateMask(polyImage)
mapid = polyImage.getMapId({'min': 1, 'max': 2, 'palette': ['red', 'blue']})
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name='training polygons',
).add_to(map)
map.add_child(folium.LayerControl())
map
```
***
## Create TFRecords for training
### Export pixels
**Variables**
```
input_bands = ['B2','B3','B4','B5','ndvi','ndwi']
output_bands = ['turbidity_blended_mean']
bands = [input_bands, output_bands]
dataset_name = 'Sentinel2_WaterQuality'
base_names = ['training_pixels', 'eval_pixels']
bucket = env.bucket_name
folder = 'cnn-models/'+dataset_name+'/data'
```
**Select the bands**
```
# Select the bands we want
c = images[0].select(bands[0])\
.addBands(images[1].select(bands[1]))
pprint(c.getInfo())
```
**Sample pixels**
```
sr = c.sample(region = trainFeatures, scale = scale, numPixels=20000, tileScale=4, seed=999)
# Add random column
sr = sr.randomColumn(seed=999)
# Partition the sample approximately 70-30.
train_dataset = sr.filter(ee.Filter.lt('random', 0.7))
eval_dataset = sr.filter(ee.Filter.gte('random', 0.7))
# Print the first couple points to verify.
pprint({'training': train_dataset.first().getInfo()})
pprint({'testing': eval_dataset.first().getInfo()})
# Print the first couple points to verify.
from pprint import pprint
train_size=train_dataset.size().getInfo()
eval_size=eval_dataset.size().getInfo()
pprint({'training': train_size})
pprint({'testing': eval_size})
```
**Export the training and validation data**
```
def export_TFRecords_pixels(datasets, base_names, bucket, folder, selectors):
# Export all the training/evaluation data
filePaths = []
for n, dataset in enumerate(datasets):
filePaths.append(bucket+ '/' + folder + '/' + base_names[n])
# Create the tasks.
task = ee.batch.Export.table.toCloudStorage(
collection = dataset,
description = 'Export '+base_names[n],
fileNamePrefix = folder + '/' + base_names[n],
bucket = bucket,
fileFormat = 'TFRecord',
selectors = selectors)
task.start()
return filePaths
datasets = [train_dataset, eval_dataset]
selectors = input_bands + output_bands
# Export training/evaluation data
filePaths = export_TFRecords_pixels(datasets, base_names, bucket, folder, selectors)
```
***
## Inspect data
### Inspect pixels
Load the data exported from Earth Engine into a tf.data.Dataset.
**Helper functions**
```
# Tensorflow setup.
import tensorflow as tf
if tf.__version__ == '1.15.0':
tf.enable_eager_execution()
print(tf.__version__)
def parse_function(proto):
"""The parsing function.
Read a serialized example into the structure defined by FEATURES_DICT.
Args:
example_proto: a serialized Example.
Returns:
A tuple of the predictors dictionary and the labels.
"""
# Define your tfrecord
features = input_bands + output_bands
# Specify the size and shape of patches expected by the model.
columns = [
tf.io.FixedLenFeature(shape=[1,1], dtype=tf.float32) for k in features
]
features_dict = dict(zip(features, columns))
# Load one example
parsed_features = tf.io.parse_single_example(proto, features_dict)
# Convert a dictionary of tensors to a tuple of (inputs, outputs)
inputsList = [parsed_features.get(key) for key in features]
stacked = tf.stack(inputsList, axis=0)
# Convert the tensors into a stack in HWC shape
stacked = tf.transpose(stacked, [1, 2, 0])
return stacked[:,:,:len(input_bands)], stacked[:,:,len(input_bands):]
def get_dataset(glob, buffer_size, batch_size):
"""Get the dataset
Returns:
A tf.data.Dataset of training data.
"""
glob = tf.compat.v1.io.gfile.glob(glob)
dataset = tf.data.TFRecordDataset(glob, compression_type='GZIP')
dataset = dataset.map(parse_function, num_parallel_calls=5)
dataset = dataset.shuffle(buffer_size).batch(batch_size).repeat()
return dataset
```
**Variables**
```
buffer_size = 100
batch_size = 4
```
**Dataset**
```
glob = 'gs://' + bucket + '/' + folder + '/' + base_names[0] + '*'
dataset = get_dataset(glob, buffer_size, batch_size)
dataset
```
**Check the first record**
```
arr = iter(dataset.take(1)).next()
input_arr = arr[0].numpy()
print(input_arr.shape)
output_arr = arr[1].numpy()
print(output_arr.shape)
```
***
## Training the model locally
**Variables**
```
job_dir = 'gs://' + bucket + '/' + 'cnn-models/'+ dataset_name +'/trainer'
logs_dir = job_dir + '/logs'
model_dir = job_dir + '/model'
shuffle_size = 2000
batch_size = 4
epochs=50
train_size=train_size
eval_size=eval_size
output_activation=''
```
**Training/evaluation data**
The following is code to load training/evaluation data.
```
import tensorflow as tf
def parse_function(proto):
"""The parsing function.
Read a serialized example into the structure defined by FEATURES_DICT.
Args:
example_proto: a serialized Example.
Returns:
A tuple of the predictors dictionary and the labels.
"""
# Define your tfrecord
features = input_bands + output_bands
# Specify the size and shape of patches expected by the model.
columns = [
tf.io.FixedLenFeature(shape=[1,1], dtype=tf.float32) for k in features
]
features_dict = dict(zip(features, columns))
# Load one example
parsed_features = tf.io.parse_single_example(proto, features_dict)
# Convert a dictionary of tensors to a tuple of (inputs, outputs)
inputsList = [parsed_features.get(key) for key in features]
stacked = tf.stack(inputsList, axis=0)
# Convert the tensors into a stack in HWC shape
stacked = tf.transpose(stacked)
return stacked[:,:,:len(input_bands)], stacked[:,:,len(input_bands):]
def get_dataset(glob):
"""Get the dataset
Returns:
A tf.data.Dataset of training data.
"""
glob = tf.compat.v1.io.gfile.glob(glob)
dataset = tf.data.TFRecordDataset(glob, compression_type='GZIP')
dataset = dataset.map(parse_function, num_parallel_calls=5)
return dataset
def get_training_dataset():
"""Get the preprocessed training dataset
Returns:
A tf.data.Dataset of training data.
"""
glob = 'gs://' + bucket + '/' + folder + '/' + base_names[0] + '*'
dataset = get_dataset(glob)
dataset = dataset.shuffle(shuffle_size).batch(batch_size).repeat()
return dataset
def get_evaluation_dataset():
"""Get the preprocessed evaluation dataset
Returns:
A tf.data.Dataset of evaluation data.
"""
glob = 'gs://' + bucket + '/' + folder + '/' + base_names[1] + '*'
dataset = get_dataset(glob)
dataset = dataset.batch(1).repeat()
return dataset
```
**Model**
```
from tensorflow.python.keras import Model # Keras model module
from tensorflow.python.keras.layers import Input, Dense, Dropout, Activation
def create_keras_model(inputShape, nClasses, output_activation='linear'):
inputs = Input(shape=inputShape, name='vector')
x = Dense(32, input_shape=inputShape, activation='relu')(inputs)
x = Dropout(0.5)(x)
x = Dense(128, activation='relu')(x)
x = Dropout(0.5)(x)
x = Dense(nClasses)(x)
outputs = Activation(output_activation, name= 'output')(x)
model = Model(inputs=inputs, outputs=outputs, name='sequential')
return model
```
**Training task**
The following will get the training and evaluation data, train the model and save it when it's done in a Cloud Storage bucket.
```
import tensorflow as tf
import time
import os
def train_and_evaluate():
"""Trains and evaluates the Keras model.
Uses the Keras model defined in model.py and trains on data loaded and
preprocessed in util.py. Saves the trained model in TensorFlow SavedModel
format to the path defined in part by the --job-dir argument.
"""
# Create the Keras Model
if not output_activation:
keras_model = create_keras_model(inputShape = (None, None, len(input_bands)), nClasses = len(output_bands))
else:
keras_model = create_keras_model(inputShape = (None, None, len(input_bands)), nClasses = len(output_bands), output_activation = output_activation)
# Compile Keras model
keras_model.compile(loss='mse', optimizer='adam', metrics=['mse'])
# Pass a tfrecord
training_dataset = get_training_dataset()
evaluation_dataset = get_evaluation_dataset()
# Setup TensorBoard callback.
tensorboard_cb = tf.keras.callbacks.TensorBoard(logs_dir)
# Train model
keras_model.fit(
x=training_dataset,
steps_per_epoch=int(train_size / batch_size),
epochs=epochs,
validation_data=evaluation_dataset,
validation_steps=int(eval_size / batch_size),
verbose=1,
callbacks=[tensorboard_cb])
tf.keras.models.save_model(keras_model, filepath=os.path.join(model_dir, str(int(time.time()))), save_format="tf")
return keras_model
model = train_and_evaluate()
```
**Evaluate model**
```
evaluation_dataset = get_evaluation_dataset()
model.evaluate(evaluation_dataset, steps=int(eval_size / batch_size))
```
### Read pretrained model
```
job_dir = 'gs://' + env.bucket_name + '/' + 'cnn-models/' + dataset_name + '/trainer'
model_dir = job_dir + '/model'
PROJECT_ID = env.project_id
# Pick the directory with the latest timestamp, in case you've trained multiple times
exported_model_dirs = ! gsutil ls {model_dir}
saved_model_path = exported_model_dirs[-1]
model = tf.keras.models.load_model(saved_model_path)
```
***
## Predict in Earth Engine
### Prepare the model for making predictions in Earth Engine
Before we can use the model in Earth Engine, it needs to be hosted by AI Platform. But before we can host the model on AI Platform we need to *EEify* (a new word!) it. The EEification process merely appends some extra operations to the input and outputs of the model in order to accomdate the interchange format between pixels from Earth Engine (float32) and inputs to AI Platform (base64). (See [this doc](https://cloud.google.com/ml-engine/docs/online-predict#binary_data_in_prediction_input) for details.)
**`earthengine model prepare`**
The EEification process is handled for you using the Earth Engine command `earthengine model prepare`. To use that command, we need to specify the input and output model directories and the name of the input and output nodes in the TensorFlow computation graph. We can do all that programmatically:
```
dataset_name = 'Sentinel2_WaterQuality'
job_dir = 'gs://' + env.bucket_name + '/' + 'cnn-models/' + dataset_name + '/trainer'
model_dir = job_dir + '/model'
project_id = env.project_id
# Pick the directory with the latest timestamp, in case you've trained multiple times
exported_model_dirs = ! gsutil ls {model_dir}
saved_model_path = exported_model_dirs[-1]
folder_name = saved_model_path.split('/')[-2]
from tensorflow.python.tools import saved_model_utils
meta_graph_def = saved_model_utils.get_meta_graph_def(saved_model_path, 'serve')
inputs = meta_graph_def.signature_def['serving_default'].inputs
outputs = meta_graph_def.signature_def['serving_default'].outputs
# Just get the first thing(s) from the serving signature def. i.e. this
# model only has a single input and a single output.
input_name = None
for k,v in inputs.items():
input_name = v.name
break
output_name = None
for k,v in outputs.items():
output_name = v.name
break
# Make a dictionary that maps Earth Engine outputs and inputs to
# AI Platform inputs and outputs, respectively.
import json
input_dict = "'" + json.dumps({input_name: "array"}) + "'"
output_dict = "'" + json.dumps({output_name: "prediction"}) + "'"
# Put the EEified model next to the trained model directory.
EEIFIED_DIR = job_dir + '/eeified/' + folder_name
# You need to set the project before using the model prepare command.
!earthengine set_project {PROJECT_ID}
!earthengine model prepare --source_dir {saved_model_path} --dest_dir {EEIFIED_DIR} --input {input_dict} --output {output_dict}
```
### Deployed the model to AI Platform
```
from googleapiclient import discovery
from googleapiclient import errors
```
**Authenticate your GCP account**
Enter the path to your service account key as the
`GOOGLE_APPLICATION_CREDENTIALS` variable in the cell below and run the cell.
```
%env GOOGLE_APPLICATION_CREDENTIALS {env.privatekey_path}
model_name = 'water_quality_test'
version_name = 'v' + folder_name
project_id = env.project_id
```
**Create model**
```
print('Creating model: ' + model_name)
# Store your full project ID in a variable in the format the API needs.
project = 'projects/{}'.format(project_id)
# Build a representation of the Cloud ML API.
ml = discovery.build('ml', 'v1')
# Create a dictionary with the fields from the request body.
request_dict = {'name': model_name,
'description': ''}
# Create a request to call projects.models.create.
request = ml.projects().models().create(
parent=project, body=request_dict)
# Make the call.
try:
response = request.execute()
print(response)
except errors.HttpError as err:
# Something went wrong, print out some information.
print('There was an error creating the model. Check the details:')
print(err._get_reason())
```
**Create version**
```
ml = discovery.build('ml', 'v1')
request_dict = {
'name': version_name,
'deploymentUri': EEIFIED_DIR,
'runtimeVersion': '1.14',
'pythonVersion': '3.5',
'framework': 'TENSORFLOW',
'autoScaling': {
"minNodes": 10
},
'machineType': 'mls1-c4-m2'
}
request = ml.projects().models().versions().create(
parent=f'projects/{project_id}/models/{model_name}',
body=request_dict
)
# Make the call.
try:
response = request.execute()
print(response)
except errors.HttpError as err:
# Something went wrong, print out some information.
print('There was an error creating the model. Check the details:')
print(err._get_reason())
```
**Check deployment status**
```
def check_status_deployment(model_name, version_name):
desc = !gcloud ai-platform versions describe {version_name} --model={model_name}
return desc.grep('state:')[0].split(':')[1].strip()
print(check_status_deployment(model_name, version_name))
```
### Load the trained model and use it for prediction in Earth Engine
**Variables**
```
# polygon where we want to display de predictions
geometry = {
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"properties": {},
"geometry": {
"type": "Polygon",
"coordinates": [
[
[
-2.63671875,
34.56085936708384
],
[
-1.2084960937499998,
34.56085936708384
],
[
-1.2084960937499998,
36.146746777814364
],
[
-2.63671875,
36.146746777814364
],
[
-2.63671875,
34.56085936708384
]
]
]
}
}
]
}
```
**Input image**
Select bands and convert them into float
```
image = images[0].select(bands[0]).float()
```
**Output image**
```
# Load the trained model and use it for prediction.
model = ee.Model.fromAiPlatformPredictor(
projectName = project_id,
modelName = model_name,
version = version_name,
inputTileSize = [1, 1],
inputOverlapSize = [0, 0],
proj = ee.Projection('EPSG:4326').atScale(scale),
fixInputProj = True,
outputBands = {'prediction': {
'type': ee.PixelType.float(),
'dimensions': 1,
}
}
)
predictions = model.predictImage(image.toArray()).arrayFlatten([bands[1]])
predictions.getInfo()
```
Clip the prediction area with the polygon
```
# Clip the prediction area with the polygon
polygon = ee.Geometry.Polygon(geometry.get('features')[0].get('geometry').get('coordinates'))
predictions = predictions.clip(polygon)
# Get centroid
centroid = polygon.centroid().getInfo().get('coordinates')[::-1]
```
**Display**
Use folium to visualize the input imagery and the predictions.
```
# Define the URL format used for Earth Engine generated map tiles.
EE_TILES = 'https://earthengine.googleapis.com/map/{mapid}/{{z}}/{{x}}/{{y}}?token={token}'
mapid = image.getMapId({'bands': ['B4', 'B3', 'B2'], 'min': 0, 'max': 1})
map = folium.Map(location=centroid, zoom_start=8)
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name='median composite',
).add_to(map)
params = ee_collection_specifics.vizz_params(collections[1])[0]
mapid = images[1].getMapId(params)
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name=str(params['bands']),
).add_to(map)
for band in bands[1]:
mapid = predictions.getMapId({'bands': [band], 'min': 0, 'max': 1})
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name=band,
).add_to(map)
map.add_child(folium.LayerControl())
map
```
***
## Make predictions of an image outside Earth Engine
### Export the imagery
We export the imagery using TFRecord format.
**Variables**
```
#Input image
image = images[0].select(bands[0])
dataset_name = 'Sentinel2_WaterQuality'
file_name = 'image_pixel'
bucket = env.bucket_name
folder = 'cnn-models/'+dataset_name+'/data'
# polygon where we want to display de predictions
geometry = {
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"properties": {},
"geometry": {
"type": "Polygon",
"coordinates": [
[
[
-2.63671875,
34.56085936708384
],
[
-1.2084960937499998,
34.56085936708384
],
[
-1.2084960937499998,
36.146746777814364
],
[
-2.63671875,
36.146746777814364
],
[
-2.63671875,
34.56085936708384
]
]
]
}
}
]
}
# Specify patch and file dimensions.
imageExportFormatOptions = {
'patchDimensions': [256, 256],
'maxFileSize': 104857600,
'compressed': True
}
# Setup the task.
imageTask = ee.batch.Export.image.toCloudStorage(
image=image,
description='Image Export',
fileNamePrefix=folder + '/' + file_name,
bucket=bucket,
scale=scale,
fileFormat='TFRecord',
region=geometry.get('features')[0].get('geometry').get('coordinates'),
formatOptions=imageExportFormatOptions,
)
# Start the task.
imageTask.start()
```
**Read the JSON mixer file**
The mixer contains metadata and georeferencing information for the exported patches, each of which is in a different file. Read the mixer to get some information needed for prediction.
```
json_file = f'gs://{bucket}' + '/' + folder + '/' + file_name +'.json'
# Load the contents of the mixer file to a JSON object.
json_text = !gsutil cat {json_file}
# Get a single string w/ newlines from the IPython.utils.text.SList
mixer = json.loads(json_text.nlstr)
pprint(mixer)
```
**Read the image files into a dataset**
The input needs to be preprocessed differently than the training and testing. Mainly, this is because the pixels are written into records as patches, we need to read the patches in as one big tensor (one patch for each band), then flatten them into lots of little tensors.
```
# Get relevant info from the JSON mixer file.
PATCH_WIDTH = mixer['patchDimensions'][0]
PATCH_HEIGHT = mixer['patchDimensions'][1]
PATCHES = mixer['totalPatches']
PATCH_DIMENSIONS_FLAT = [PATCH_WIDTH * PATCH_HEIGHT, 1]
features = bands[0]
glob = f'gs://{bucket}' + '/' + folder + '/' + file_name +'.tfrecord.gz'
# Note that the tensors are in the shape of a patch, one patch for each band.
image_columns = [
tf.FixedLenFeature(shape=PATCH_DIMENSIONS_FLAT, dtype=tf.float32) for k in features
]
# Parsing dictionary.
features_dict = dict(zip(bands[0], image_columns))
def parse_image(proto):
return tf.io.parse_single_example(proto, features_dict)
image_dataset = tf.data.TFRecordDataset(glob, compression_type='GZIP')
image_dataset = image_dataset.map(parse_image, num_parallel_calls=5)
# Break our long tensors into many little ones.
image_dataset = image_dataset.flat_map(
lambda features: tf.data.Dataset.from_tensor_slices(features)
)
# Turn the dictionary in each record into a tuple without a label.
image_dataset = image_dataset.map(
lambda dataDict: (tf.transpose(list(dataDict.values())), )
)
# Turn each patch into a batch.
image_dataset = image_dataset.batch(PATCH_WIDTH * PATCH_HEIGHT)
image_dataset
```
**Check the first record**
```
arr = iter(image_dataset.take(1)).next()
input_arr = arr[0].numpy()
print(input_arr.shape)
```
**Display the input channels**
```
def display_channels(data, nChannels, titles = False):
if nChannels == 1:
plt.figure(figsize=(5,5))
plt.imshow(data[:,:,0])
if titles:
plt.title(titles[0])
else:
fig, axs = plt.subplots(nrows=1, ncols=nChannels, figsize=(5*nChannels,5))
for i in range(nChannels):
ax = axs[i]
ax.imshow(data[:,:,i])
if titles:
ax.set_title(titles[i])
input_arr = input_arr.reshape((PATCH_WIDTH, PATCH_HEIGHT, len(bands[0])))
input_arr.shape
display_channels(input_arr, input_arr.shape[2], titles=bands[0])
```
### Generate predictions for the image pixels
To get predictions in each pixel, run the image dataset through the trained model using model.predict(). Print the first prediction to see that the output is a list of the three class probabilities for each pixel. Running all predictions might take a while.
```
predictions = model.predict(image_dataset, steps=PATCHES, verbose=1)
output_arr = predictions.reshape((PATCHES, PATCH_WIDTH, PATCH_HEIGHT, len(bands[1])))
output_arr.shape
display_channels(output_arr[9,:,:,:], output_arr.shape[3], titles=bands[1])
```
### Write the predictions to a TFRecord file
We need to write the pixels into the file as patches in the same order they came out. The records are written as serialized `tf.train.Example` protos.
```
dataset_name = 'Sentinel2_WaterQuality'
bucket = env.bucket_name
folder = 'cnn-models/'+dataset_name+'/data'
output_file = 'gs://' + bucket + '/' + folder + '/predicted_image_pixel.TFRecord'
print('Writing to file ' + output_file)
# Instantiate the writer.
writer = tf.io.TFRecordWriter(output_file)
patch = [[]]
nPatch = 1
for prediction in predictions:
patch[0].append(prediction[0][0])
# Once we've seen a patches-worth of class_ids...
if (len(patch[0]) == PATCH_WIDTH * PATCH_HEIGHT):
print('Done with patch ' + str(nPatch) + ' of ' + str(PATCHES))
# Create an example
example = tf.train.Example(
features=tf.train.Features(
feature={
'prediction': tf.train.Feature(
float_list=tf.train.FloatList(
value=patch[0]))
}
)
)
# Write the example to the file and clear our patch array so it's ready for
# another batch of class ids
writer.write(example.SerializeToString())
patch = [[]]
nPatch += 1
writer.close()
```
**Verify the existence of the predictions file**
```
!gsutil ls -l {output_file}
```
### Upload the predicted image to an Earth Engine asset
```
asset_id = 'projects/vizzuality/skydipper-water-quality/predicted-image'
print('Writing to ' + asset_id)
# Start the upload.
!earthengine upload image --asset_id={asset_id} {output_file} {json_file}
```
### View the predicted image
```
# Get centroid
polygon = ee.Geometry.Polygon(geometry.get('features')[0].get('geometry').get('coordinates'))
centroid = polygon.centroid().getInfo().get('coordinates')[::-1]
EE_TILES = 'https://earthengine.googleapis.com/map/{mapid}/{{z}}/{{x}}/{{y}}?token={token}'
map = folium.Map(location=centroid, zoom_start=8)
for n, collection in enumerate(collections):
params = ee_collection_specifics.vizz_params(collection)[0]
mapid = images[n].getMapId(params)
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name=str(params['bands']),
).add_to(map)
# Read predicted Image
predicted_image = ee.Image(asset_id)
mapid = predicted_image.getMapId({'bands': ['prediction'], 'min': 0, 'max': 1})
folium.TileLayer(
tiles=EE_TILES.format(**mapid),
attr='Google Earth Engine',
overlay=True,
name='predicted image',
).add_to(map)
map.add_child(folium.LayerControl())
map
```
| github_jupyter |
# Calculating Area and Center Coordinates of a Polygon
```
%load_ext lab_black
%load_ext autoreload
%autoreload 2
import geopandas as gpd
import pandas as pd
%aimport src.utils
from src.utils import show_df
```
<a id="toc"></a>
## [Table of Contents](#table-of-contents)
0. [About](#about)
1. [User Inputs](#user-inputs)
2. [Load Chicago Community Areas GeoData](#load-chicago-community-areas-geodata)
3. [Calculate Area of each Community Area](#calculate-area-of-each-community-area)
4. [Calculate Coordinates of Midpoint of each Community Area](#calculate-coordinates-of-midpoint-of-each-community-area)
<a id="about"></a>
## 0. [About](#about)
We'll explore calculations of the area and central coordinates of polygons from geospatial data using the Python [`geopandas` library](https://pypi.org/project/geopandas/).
<a id="user-inputs"></a>
## 1. [User Inputs](#user-inputs)
```
ca_url = "https://data.cityofchicago.org/api/geospatial/cauq-8yn6?method=export&format=GeoJSON"
convert_sqm_to_sqft = 10.7639
```
<a id="load-chicago-community-areas-geodata"></a>
## 2. [Load Chicago Community Areas GeoData](#load-chicago-community-areas-geodata)
Load the boundaries geodata for the [Chicago community areas](https://data.cityofchicago.org/Facilities-Geographic-Boundaries/Boundaries-Community-Areas-current-/cauq-8yn6)
```
%%time
gdf_ca = gpd.read_file(ca_url)
print(gdf_ca.crs)
gdf_ca.head(2)
```
<a id="calculate-area-of-each-community-area"></a>
## 3. [Calculate Area of each Community Area](#calculate-area-of-each-community-area)
To get the area, we need to
- project the geometry into a Cylindrical Equal-Area (CEA) format, an equal area projection, with that preserves area ([1](https://learn.arcgis.com/en/projects/choose-the-right-projection/))
- calculate the area by calling the `.area()` method on the `GeoDataFrame`
- this will give area in square meters
- [convert area from square meters to square feet](https://www.metric-conversions.org/area/square-meters-to-square-feet.htm)
- through trial and error, it was found that this is the unit in which the Chicago community areas geodata gives the area (see the `shape_area` column)
```
%%time
gdf_ca["cea_area_square_feet"] = gdf_ca.to_crs({"proj": "cea"}).area * convert_sqm_to_sqft
gdf_ca["diff_sq_feet"] = gdf_ca["shape_area"].astype(float) - gdf_ca["cea_area_square_feet"]
gdf_ca["diff_pct"] = gdf_ca["diff_sq_feet"] / gdf_ca["shape_area"].astype(float) * 100
show_df(gdf_ca.drop(columns=["geometry"]))
display(gdf_ca[["diff_sq_feet", "diff_pct"]].describe())
```
**Observations**
1. It is reassuring that the CEA projection has given us areas in square feet that are within less than 0.01 percent of the areas provided with the Chicago community areas dataset. We'll use this approach to calculate shape areas.
<a id="calculate-coordinates-of-midpoint-of-each-community-area"></a>
## 4. [Calculate Coordinates of Midpoint of each Community Area](#calculate-coordinates-of-midpoint-of-each-community-area)
In order to get the centroid of a geometry, it is [recommended to first project to the CEA CRS (equal area CRS) before computing the centroid](https://gis.stackexchange.com/a/401815/135483). [Other used CRS values include 3395, 32663 or 4087](https://gis.stackexchange.com/a/390563/135483). Once the geometry is projected, we can calculate the centroid coordinates calling the `.centroid()` method on the `GeoDataFrame`'s `geometry` column
```
%%time
centroid_cea = gdf_ca["geometry"].to_crs("+proj=cea").centroid.to_crs(gdf_ca.crs)
centroid_3395 = gdf_ca["geometry"].to_crs(epsg=3395).centroid.to_crs(gdf_ca.crs)
centroid_32663 = gdf_ca["geometry"].to_crs(epsg=32663).centroid.to_crs(gdf_ca.crs)
centroid_4087 = gdf_ca["geometry"].to_crs(epsg=4087).centroid.to_crs(gdf_ca.crs)
centroid_6345 = gdf_ca["geometry"].to_crs(epsg=6345).centroid.to_crs(gdf_ca.crs)
df_centroid_coords = pd.DataFrame()
for c, centroid_coords in zip(
["cea", 3395, 32663, 4087, 6345],
[centroid_cea, centroid_3395, centroid_32663, centroid_4087, centroid_6345],
):
df_centroid_coords[f"lat_{c}"] = centroid_coords.y
df_centroid_coords[f"lon_{c}"] = centroid_coords.x
show_df(df_centroid_coords)
```
**Observations**
1. For our case, centroids computed using all projections give nearly identical co-ordinates. This is likely since each of the city's community areas cover a very small area on the surface of the Earth. Further reading will be required to understand the close agreement between these centroid locations found using the different projections. For subsequent calculation of the centroids, we'll use the equal area projection.
| github_jupyter |
# PTN Template
This notebook serves as a template for single dataset PTN experiments
It can be run on its own by setting STANDALONE to True (do a find for "STANDALONE" to see where)
But it is intended to be executed as part of a *papermill.py script. See any of the
experimentes with a papermill script to get started with that workflow.
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
import os, json, sys, time, random
import numpy as np
import torch
from torch.optim import Adam
from easydict import EasyDict
import matplotlib.pyplot as plt
from steves_models.steves_ptn import Steves_Prototypical_Network
from steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper
from steves_utils.iterable_aggregator import Iterable_Aggregator
from steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig
from steves_utils.torch_sequential_builder import build_sequential
from steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader
from steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)
from steves_utils.PTN.utils import independent_accuracy_assesment
from steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory
from steves_utils.ptn_do_report import (
get_loss_curve,
get_results_table,
get_parameters_table,
get_domain_accuracies,
)
from steves_utils.transforms import get_chained_transform
```
# Required Parameters
These are allowed parameters, not defaults
Each of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)
Papermill uses the cell tag "parameters" to inject the real parameters below this cell.
Enable tags to see what I mean
```
required_parameters = {
"experiment_name",
"lr",
"device",
"seed",
"dataset_seed",
"labels_source",
"labels_target",
"domains_source",
"domains_target",
"num_examples_per_domain_per_label_source",
"num_examples_per_domain_per_label_target",
"n_shot",
"n_way",
"n_query",
"train_k_factor",
"val_k_factor",
"test_k_factor",
"n_epoch",
"patience",
"criteria_for_best",
"x_transforms_source",
"x_transforms_target",
"episode_transforms_source",
"episode_transforms_target",
"pickle_name",
"x_net",
"NUM_LOGS_PER_EPOCH",
"BEST_MODEL_PATH",
"torch_default_dtype"
}
standalone_parameters = {}
standalone_parameters["experiment_name"] = "STANDALONE PTN"
standalone_parameters["lr"] = 0.0001
standalone_parameters["device"] = "cuda"
standalone_parameters["seed"] = 1337
standalone_parameters["dataset_seed"] = 1337
standalone_parameters["num_examples_per_domain_per_label_source"]=100
standalone_parameters["num_examples_per_domain_per_label_target"]=100
standalone_parameters["n_shot"] = 3
standalone_parameters["n_query"] = 2
standalone_parameters["train_k_factor"] = 1
standalone_parameters["val_k_factor"] = 2
standalone_parameters["test_k_factor"] = 2
standalone_parameters["n_epoch"] = 100
standalone_parameters["patience"] = 10
standalone_parameters["criteria_for_best"] = "target_accuracy"
standalone_parameters["x_transforms_source"] = ["unit_power"]
standalone_parameters["x_transforms_target"] = ["unit_power"]
standalone_parameters["episode_transforms_source"] = []
standalone_parameters["episode_transforms_target"] = []
standalone_parameters["torch_default_dtype"] = "torch.float32"
standalone_parameters["x_net"] = [
{"class": "nnReshape", "kargs": {"shape":[-1, 1, 2, 256]}},
{"class": "Conv2d", "kargs": { "in_channels":1, "out_channels":256, "kernel_size":(1,7), "bias":False, "padding":(0,3), },},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features":256}},
{"class": "Conv2d", "kargs": { "in_channels":256, "out_channels":80, "kernel_size":(2,7), "bias":True, "padding":(0,3), },},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features":80}},
{"class": "Flatten", "kargs": {}},
{"class": "Linear", "kargs": {"in_features": 80*256, "out_features": 256}}, # 80 units per IQ pair
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm1d", "kargs": {"num_features":256}},
{"class": "Linear", "kargs": {"in_features": 256, "out_features": 256}},
]
# Parameters relevant to results
# These parameters will basically never need to change
standalone_parameters["NUM_LOGS_PER_EPOCH"] = 10
standalone_parameters["BEST_MODEL_PATH"] = "./best_model.pth"
# uncomment for CORES dataset
from steves_utils.CORES.utils import (
ALL_NODES,
ALL_NODES_MINIMUM_1000_EXAMPLES,
ALL_DAYS
)
standalone_parameters["labels_source"] = ALL_NODES
standalone_parameters["labels_target"] = ALL_NODES
standalone_parameters["domains_source"] = [1]
standalone_parameters["domains_target"] = [2,3,4,5]
standalone_parameters["pickle_name"] = "cores.stratified_ds.2022A.pkl"
# Uncomment these for ORACLE dataset
# from steves_utils.ORACLE.utils_v2 import (
# ALL_DISTANCES_FEET,
# ALL_RUNS,
# ALL_SERIAL_NUMBERS,
# )
# standalone_parameters["labels_source"] = ALL_SERIAL_NUMBERS
# standalone_parameters["labels_target"] = ALL_SERIAL_NUMBERS
# standalone_parameters["domains_source"] = [8,20, 38,50]
# standalone_parameters["domains_target"] = [14, 26, 32, 44, 56]
# standalone_parameters["pickle_name"] = "oracle.frame_indexed.stratified_ds.2022A.pkl"
# standalone_parameters["num_examples_per_domain_per_label_source"]=1000
# standalone_parameters["num_examples_per_domain_per_label_target"]=1000
# Uncomment these for Metahan dataset
# standalone_parameters["labels_source"] = list(range(19))
# standalone_parameters["labels_target"] = list(range(19))
# standalone_parameters["domains_source"] = [0]
# standalone_parameters["domains_target"] = [1]
# standalone_parameters["pickle_name"] = "metehan.stratified_ds.2022A.pkl"
# standalone_parameters["n_way"] = len(standalone_parameters["labels_source"])
# standalone_parameters["num_examples_per_domain_per_label_source"]=200
# standalone_parameters["num_examples_per_domain_per_label_target"]=100
standalone_parameters["n_way"] = len(standalone_parameters["labels_source"])
# Parameters
parameters = {
"experiment_name": "tuned_1v2:oracle.run1_limited",
"device": "cuda",
"lr": 0.0001,
"labels_source": [
"3123D52",
"3123D65",
"3123D79",
"3123D80",
"3123D54",
"3123D70",
"3123D7B",
"3123D89",
"3123D58",
"3123D76",
"3123D7D",
"3123EFE",
"3123D64",
"3123D78",
"3123D7E",
"3124E4A",
],
"labels_target": [
"3123D52",
"3123D65",
"3123D79",
"3123D80",
"3123D54",
"3123D70",
"3123D7B",
"3123D89",
"3123D58",
"3123D76",
"3123D7D",
"3123EFE",
"3123D64",
"3123D78",
"3123D7E",
"3124E4A",
],
"episode_transforms_source": [],
"episode_transforms_target": [],
"domains_source": [8, 32, 50],
"domains_target": [14, 20, 26, 38, 44],
"num_examples_per_domain_per_label_source": 2000,
"num_examples_per_domain_per_label_target": 2000,
"n_shot": 3,
"n_way": 16,
"n_query": 2,
"train_k_factor": 3,
"val_k_factor": 2,
"test_k_factor": 2,
"torch_default_dtype": "torch.float32",
"n_epoch": 50,
"patience": 3,
"criteria_for_best": "target_accuracy",
"x_net": [
{"class": "nnReshape", "kargs": {"shape": [-1, 1, 2, 256]}},
{
"class": "Conv2d",
"kargs": {
"in_channels": 1,
"out_channels": 256,
"kernel_size": [1, 7],
"bias": False,
"padding": [0, 3],
},
},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features": 256}},
{
"class": "Conv2d",
"kargs": {
"in_channels": 256,
"out_channels": 80,
"kernel_size": [2, 7],
"bias": True,
"padding": [0, 3],
},
},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features": 80}},
{"class": "Flatten", "kargs": {}},
{"class": "Linear", "kargs": {"in_features": 20480, "out_features": 256}},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm1d", "kargs": {"num_features": 256}},
{"class": "Linear", "kargs": {"in_features": 256, "out_features": 256}},
],
"NUM_LOGS_PER_EPOCH": 10,
"BEST_MODEL_PATH": "./best_model.pth",
"pickle_name": "oracle.Run1_10kExamples_stratified_ds.2022A.pkl",
"x_transforms_source": ["unit_power"],
"x_transforms_target": ["unit_power"],
"dataset_seed": 1337,
"seed": 1337,
}
# Set this to True if you want to run this template directly
STANDALONE = False
if STANDALONE:
print("parameters not injected, running with standalone_parameters")
parameters = standalone_parameters
if not 'parameters' in locals() and not 'parameters' in globals():
raise Exception("Parameter injection failed")
#Use an easy dict for all the parameters
p = EasyDict(parameters)
supplied_keys = set(p.keys())
if supplied_keys != required_parameters:
print("Parameters are incorrect")
if len(supplied_keys - required_parameters)>0: print("Shouldn't have:", str(supplied_keys - required_parameters))
if len(required_parameters - supplied_keys)>0: print("Need to have:", str(required_parameters - supplied_keys))
raise RuntimeError("Parameters are incorrect")
###################################
# Set the RNGs and make it all deterministic
###################################
np.random.seed(p.seed)
random.seed(p.seed)
torch.manual_seed(p.seed)
torch.use_deterministic_algorithms(True)
###########################################
# The stratified datasets honor this
###########################################
torch.set_default_dtype(eval(p.torch_default_dtype))
###################################
# Build the network(s)
# Note: It's critical to do this AFTER setting the RNG
# (This is due to the randomized initial weights)
###################################
x_net = build_sequential(p.x_net)
start_time_secs = time.time()
###################################
# Build the dataset
###################################
if p.x_transforms_source == []: x_transform_source = None
else: x_transform_source = get_chained_transform(p.x_transforms_source)
if p.x_transforms_target == []: x_transform_target = None
else: x_transform_target = get_chained_transform(p.x_transforms_target)
if p.episode_transforms_source == []: episode_transform_source = None
else: raise Exception("episode_transform_source not implemented")
if p.episode_transforms_target == []: episode_transform_target = None
else: raise Exception("episode_transform_target not implemented")
eaf_source = Episodic_Accessor_Factory(
labels=p.labels_source,
domains=p.domains_source,
num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_source,
iterator_seed=p.seed,
dataset_seed=p.dataset_seed,
n_shot=p.n_shot,
n_way=p.n_way,
n_query=p.n_query,
train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),
pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name),
x_transform_func=x_transform_source,
example_transform_func=episode_transform_source,
)
train_original_source, val_original_source, test_original_source = eaf_source.get_train(), eaf_source.get_val(), eaf_source.get_test()
eaf_target = Episodic_Accessor_Factory(
labels=p.labels_target,
domains=p.domains_target,
num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_target,
iterator_seed=p.seed,
dataset_seed=p.dataset_seed,
n_shot=p.n_shot,
n_way=p.n_way,
n_query=p.n_query,
train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),
pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name),
x_transform_func=x_transform_target,
example_transform_func=episode_transform_target,
)
train_original_target, val_original_target, test_original_target = eaf_target.get_train(), eaf_target.get_val(), eaf_target.get_test()
transform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only
train_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)
val_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)
test_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)
train_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)
val_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)
test_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)
datasets = EasyDict({
"source": {
"original": {"train":train_original_source, "val":val_original_source, "test":test_original_source},
"processed": {"train":train_processed_source, "val":val_processed_source, "test":test_processed_source}
},
"target": {
"original": {"train":train_original_target, "val":val_original_target, "test":test_original_target},
"processed": {"train":train_processed_target, "val":val_processed_target, "test":test_processed_target}
},
})
# Some quick unit tests on the data
from steves_utils.transforms import get_average_power, get_average_magnitude
q_x, q_y, s_x, s_y, truth = next(iter(train_processed_source))
assert q_x.dtype == eval(p.torch_default_dtype)
assert s_x.dtype == eval(p.torch_default_dtype)
print("Visually inspect these to see if they line up with expected values given the transforms")
print('x_transforms_source', p.x_transforms_source)
print('x_transforms_target', p.x_transforms_target)
print("Average magnitude, source:", get_average_magnitude(q_x[0].numpy()))
print("Average power, source:", get_average_power(q_x[0].numpy()))
q_x, q_y, s_x, s_y, truth = next(iter(train_processed_target))
print("Average magnitude, target:", get_average_magnitude(q_x[0].numpy()))
print("Average power, target:", get_average_power(q_x[0].numpy()))
###################################
# Build the model
###################################
model = Steves_Prototypical_Network(x_net, device=p.device, x_shape=(2,256))
optimizer = Adam(params=model.parameters(), lr=p.lr)
###################################
# train
###################################
jig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)
jig.train(
train_iterable=datasets.source.processed.train,
source_val_iterable=datasets.source.processed.val,
target_val_iterable=datasets.target.processed.val,
num_epochs=p.n_epoch,
num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,
patience=p.patience,
optimizer=optimizer,
criteria_for_best=p.criteria_for_best,
)
total_experiment_time_secs = time.time() - start_time_secs
###################################
# Evaluate the model
###################################
source_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)
target_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)
source_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)
target_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)
history = jig.get_history()
total_epochs_trained = len(history["epoch_indices"])
val_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))
confusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)
per_domain_accuracy = per_domain_accuracy_from_confusion(confusion)
# Add a key to per_domain_accuracy for if it was a source domain
for domain, accuracy in per_domain_accuracy.items():
per_domain_accuracy[domain] = {
"accuracy": accuracy,
"source?": domain in p.domains_source
}
# Do an independent accuracy assesment JUST TO BE SURE!
# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)
# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)
# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)
# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)
# assert(_source_test_label_accuracy == source_test_label_accuracy)
# assert(_target_test_label_accuracy == target_test_label_accuracy)
# assert(_source_val_label_accuracy == source_val_label_accuracy)
# assert(_target_val_label_accuracy == target_val_label_accuracy)
experiment = {
"experiment_name": p.experiment_name,
"parameters": dict(p),
"results": {
"source_test_label_accuracy": source_test_label_accuracy,
"source_test_label_loss": source_test_label_loss,
"target_test_label_accuracy": target_test_label_accuracy,
"target_test_label_loss": target_test_label_loss,
"source_val_label_accuracy": source_val_label_accuracy,
"source_val_label_loss": source_val_label_loss,
"target_val_label_accuracy": target_val_label_accuracy,
"target_val_label_loss": target_val_label_loss,
"total_epochs_trained": total_epochs_trained,
"total_experiment_time_secs": total_experiment_time_secs,
"confusion": confusion,
"per_domain_accuracy": per_domain_accuracy,
},
"history": history,
"dataset_metrics": get_dataset_metrics(datasets, "ptn"),
}
ax = get_loss_curve(experiment)
plt.show()
get_results_table(experiment)
get_domain_accuracies(experiment)
print("Source Test Label Accuracy:", experiment["results"]["source_test_label_accuracy"], "Target Test Label Accuracy:", experiment["results"]["target_test_label_accuracy"])
print("Source Val Label Accuracy:", experiment["results"]["source_val_label_accuracy"], "Target Val Label Accuracy:", experiment["results"]["target_val_label_accuracy"])
json.dumps(experiment)
```
| github_jupyter |
# Batch Normalization – Practice
Batch normalization is most useful when building deep neural networks. To demonstrate this, we'll create a convolutional neural network with 20 convolutional layers, followed by a fully connected layer. We'll use it to classify handwritten digits in the MNIST dataset, which should be familiar to you by now.
This is **not** a good network for classfying MNIST digits. You could create a _much_ simpler network and get _better_ results. However, to give you hands-on experience with batch normalization, we had to make an example that was:
1. Complicated enough that training would benefit from batch normalization.
2. Simple enough that it would train quickly, since this is meant to be a short exercise just to give you some practice adding batch normalization.
3. Simple enough that the architecture would be easy to understand without additional resources.
This notebook includes two versions of the network that you can edit. The first uses higher level functions from the `tf.layers` package. The second is the same network, but uses only lower level functions in the `tf.nn` package.
1. [Batch Normalization with `tf.layers.batch_normalization`](#example_1)
2. [Batch Normalization with `tf.nn.batch_normalization`](#example_2)
The following cell loads TensorFlow, downloads the MNIST dataset if necessary, and loads it into an object named `mnist`. You'll need to run this cell before running anything else in the notebook.
```
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True, reshape=False)
```
# Batch Normalization using `tf.layers.batch_normalization`<a id="example_1"></a>
This version of the network uses `tf.layers` for almost everything, and expects you to implement batch normalization using [`tf.layers.batch_normalization`](https://www.tensorflow.org/api_docs/python/tf/layers/batch_normalization)
We'll use the following function to create fully connected layers in our network. We'll create them with the specified number of neurons and a ReLU activation function.
This version of the function does not include batch normalization.
```
"""
DO NOT MODIFY THIS CELL
"""
def fully_connected(prev_layer, num_units):
"""
Create a fully connectd layer with the given layer as input and the given number of neurons.
:param prev_layer: Tensor
The Tensor that acts as input into this layer
:param num_units: int
The size of the layer. That is, the number of units, nodes, or neurons.
:returns Tensor
A new fully connected layer
"""
layer = tf.layers.dense(prev_layer, num_units, activation=tf.nn.relu)
return layer
```
We'll use the following function to create convolutional layers in our network. They are very basic: we're always using a 3x3 kernel, ReLU activation functions, strides of 1x1 on layers with odd depths, and strides of 2x2 on layers with even depths. We aren't bothering with pooling layers at all in this network.
This version of the function does not include batch normalization.
```
"""
DO NOT MODIFY THIS CELL
"""
def conv_layer(prev_layer, layer_depth):
"""
Create a convolutional layer with the given layer as input.
:param prev_layer: Tensor
The Tensor that acts as input into this layer
:param layer_depth: int
We'll set the strides and number of feature maps based on the layer's depth in the network.
This is *not* a good way to make a CNN, but it helps us create this example with very little code.
:returns Tensor
A new convolutional layer
"""
strides = 2 if layer_depth % 3 == 0 else 1
conv_layer = tf.layers.conv2d(prev_layer, layer_depth*4, 3, strides, 'same', activation=tf.nn.relu)
return conv_layer
```
**Run the following cell**, along with the earlier cells (to load the dataset and define the necessary functions).
This cell builds the network **without** batch normalization, then trains it on the MNIST dataset. It displays loss and accuracy data periodically while training.
```
"""
DO NOT MODIFY THIS CELL
"""
def train(num_batches, batch_size, learning_rate):
# Build placeholders for the input samples and labels
inputs = tf.placeholder(tf.float32, [None, 28, 28, 1])
labels = tf.placeholder(tf.float32, [None, 10])
# Feed the inputs into a series of 20 convolutional layers
layer = inputs
for layer_i in range(1, 20):
layer = conv_layer(layer, layer_i)
# Flatten the output from the convolutional layers
orig_shape = layer.get_shape().as_list()
layer = tf.reshape(layer, shape=[-1, orig_shape[1] * orig_shape[2] * orig_shape[3]])
# Add one fully connected layer
layer = fully_connected(layer, 100)
# Create the output layer with 1 node for each
logits = tf.layers.dense(layer, 10)
# Define loss and training operations
model_loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=labels))
train_opt = tf.train.AdamOptimizer(learning_rate).minimize(model_loss)
# Create operations to test accuracy
correct_prediction = tf.equal(tf.argmax(logits,1), tf.argmax(labels,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# Train and test the network
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for batch_i in range(num_batches):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# train this batch
sess.run(train_opt, {inputs: batch_xs, labels: batch_ys})
# Periodically check the validation or training loss and accuracy
if batch_i % 100 == 0:
loss, acc = sess.run([model_loss, accuracy], {inputs: mnist.validation.images,
labels: mnist.validation.labels})
print('Batch: {:>2}: Validation loss: {:>3.5f}, Validation accuracy: {:>3.5f}'.format(batch_i, loss, acc))
elif batch_i % 25 == 0:
loss, acc = sess.run([model_loss, accuracy], {inputs: batch_xs, labels: batch_ys})
print('Batch: {:>2}: Training loss: {:>3.5f}, Training accuracy: {:>3.5f}'.format(batch_i, loss, acc))
# At the end, score the final accuracy for both the validation and test sets
acc = sess.run(accuracy, {inputs: mnist.validation.images,
labels: mnist.validation.labels})
print('Final validation accuracy: {:>3.5f}'.format(acc))
acc = sess.run(accuracy, {inputs: mnist.test.images,
labels: mnist.test.labels})
print('Final test accuracy: {:>3.5f}'.format(acc))
# Score the first 100 test images individually. This won't work if batch normalization isn't implemented correctly.
correct = 0
for i in range(100):
correct += sess.run(accuracy,feed_dict={inputs: [mnist.test.images[i]],
labels: [mnist.test.labels[i]]})
print("Accuracy on 100 samples:", correct/100)
num_batches = 800
batch_size = 64
learning_rate = 0.002
tf.reset_default_graph()
with tf.Graph().as_default():
train(num_batches, batch_size, learning_rate)
```
With this many layers, it's going to take a lot of iterations for this network to learn. By the time you're done training these 800 batches, your final test and validation accuracies probably won't be much better than 10%. (It will be different each time, but will most likely be less than 15%.)
Using batch normalization, you'll be able to train this same network to over 90% in that same number of batches.
# Add batch normalization
We've copied the previous three cells to get you started. **Edit these cells** to add batch normalization to the network. For this exercise, you should use [`tf.layers.batch_normalization`](https://www.tensorflow.org/api_docs/python/tf/layers/batch_normalization) to handle most of the math, but you'll need to make a few other changes to your network to integrate batch normalization. You may want to refer back to the lesson notebook to remind yourself of important things, like how your graph operations need to know whether or not you are performing training or inference.
If you get stuck, you can check out the `Batch_Normalization_Solutions` notebook to see how we did things.
**TODO:** Modify `fully_connected` to add batch normalization to the fully connected layers it creates. Feel free to change the function's parameters if it helps.
```
def fully_connected(prev_layer, num_units, is_training):
"""
Create a fully connectd layer with the given layer as input and the given number of neurons.
:param prev_layer: Tensor
The Tensor that acts as input into this layer
:param num_units: int
The size of the layer. That is, the number of units, nodes, or neurons.
:returns Tensor
A new fully connected layer
"""
layer = tf.layers.dense(prev_layer, num_units, use_bias=False, activation=None)
layer = tf.layers.batch_normalization(layer, training=is_training)
layer = tf.nn.relu(layer)
return layer
```
**TODO:** Modify `conv_layer` to add batch normalization to the convolutional layers it creates. Feel free to change the function's parameters if it helps.
```
def conv_layer(prev_layer, layer_depth, is_training):
"""
Create a convolutional layer with the given layer as input.
:param prev_layer: Tensor
The Tensor that acts as input into this layer
:param layer_depth: int
We'll set the strides and number of feature maps based on the layer's depth in the network.
This is *not* a good way to make a CNN, but it helps us create this example with very little code.
:returns Tensor
A new convolutional layer
"""
strides = 2 if layer_depth % 3 == 0 else 1
conv_layer = tf.layers.conv2d(prev_layer, layer_depth*4, 3, strides, 'same', activation=None, use_bias=False)
conv_layer = tf.layers.batch_normalization(conv_layer, training=is_training)
conv_layer = tf.nn.relu(conv_layer)
return conv_layer
```
**TODO:** Edit the `train` function to support batch normalization. You'll need to make sure the network knows whether or not it is training, and you'll need to make sure it updates and uses its population statistics correctly.
```
def train(num_batches, batch_size, learning_rate):
# Build placeholders for the input samples and labels
inputs = tf.placeholder(tf.float32, [None, 28, 28, 1])
labels = tf.placeholder(tf.float32, [None, 10])
# Add placeholder to indicate whether or not we're training the model
is_training = tf.placeholder(tf.bool)
# Feed the inputs into a series of 20 convolutional layers
layer = inputs
for layer_i in range(1, 20):
layer = conv_layer(layer, layer_i, is_training)
# Flatten the output from the convolutional layers
orig_shape = layer.get_shape().as_list()
layer = tf.reshape(layer, shape=[-1, orig_shape[1] * orig_shape[2] * orig_shape[3]])
# Add one fully connected layer
layer = fully_connected(layer, 100, is_training)
# Create the output layer with 1 node for each
logits = tf.layers.dense(layer, 10)
# Define loss and training operations
model_loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=labels))
# Tell TensorFlow to update the population statistics while training
with tf.control_dependencies(tf.get_collection(tf.GraphKeys.UPDATE_OPS)):
train_opt = tf.train.AdamOptimizer(learning_rate).minimize(model_loss)
# Create operations to test accuracy
correct_prediction = tf.equal(tf.argmax(logits,1), tf.argmax(labels,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# Train and test the network
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for batch_i in range(num_batches):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# train this batch
sess.run(train_opt, {inputs: batch_xs, labels: batch_ys, is_training: True})
# Periodically check the validation or training loss and accuracy
if batch_i % 100 == 0:
loss, acc = sess.run([model_loss, accuracy], {inputs: mnist.validation.images,
labels: mnist.validation.labels,
is_training: False})
print('Batch: {:>2}: Validation loss: {:>3.5f}, Validation accuracy: {:>3.5f}'.format(batch_i, loss, acc))
elif batch_i % 25 == 0:
loss, acc = sess.run([model_loss, accuracy], {inputs: batch_xs, labels: batch_ys, is_training: False})
print('Batch: {:>2}: Training loss: {:>3.5f}, Training accuracy: {:>3.5f}'.format(batch_i, loss, acc))
# At the end, score the final accuracy for both the validation and test sets
acc = sess.run(accuracy, {inputs: mnist.validation.images,
labels: mnist.validation.labels,
is_training: False})
print('Final validation accuracy: {:>3.5f}'.format(acc))
acc = sess.run(accuracy, {inputs: mnist.test.images,
labels: mnist.test.labels,
is_training: False})
print('Final test accuracy: {:>3.5f}'.format(acc))
# Score the first 100 test images individually, just to make sure batch normalization really worked
correct = 0
for i in range(100):
correct += sess.run(accuracy,feed_dict={inputs: [mnist.test.images[i]],
labels: [mnist.test.labels[i]],
is_training: False})
print("Accuracy on 100 samples:", correct/100)
num_batches = 800
batch_size = 64
learning_rate = 0.002
tf.reset_default_graph()
with tf.Graph().as_default():
train(num_batches, batch_size, learning_rate)
```
With batch normalization, you should now get an accuracy over 90%. Notice also the last line of the output: `Accuracy on 100 samples`. If this value is low while everything else looks good, that means you did not implement batch normalization correctly. Specifically, it means you either did not calculate the population mean and variance while training, or you are not using those values during inference.
# Batch Normalization using `tf.nn.batch_normalization`<a id="example_2"></a>
Most of the time you will be able to use higher level functions exclusively, but sometimes you may want to work at a lower level. For example, if you ever want to implement a new feature – something new enough that TensorFlow does not already include a high-level implementation of it, like batch normalization in an LSTM – then you may need to know these sorts of things.
This version of the network uses `tf.nn` for almost everything, and expects you to implement batch normalization using [`tf.nn.batch_normalization`](https://www.tensorflow.org/api_docs/python/tf/nn/batch_normalization).
**Optional TODO:** You can run the next three cells before you edit them just to see how the network performs without batch normalization. However, the results should be pretty much the same as you saw with the previous example before you added batch normalization.
**TODO:** Modify `fully_connected` to add batch normalization to the fully connected layers it creates. Feel free to change the function's parameters if it helps.
**Note:** For convenience, we continue to use `tf.layers.dense` for the `fully_connected` layer. By this point in the class, you should have no problem replacing that with matrix operations between the `prev_layer` and explicit weights and biases variables.
```
def fully_connected(prev_layer, num_units, is_training):
"""
Create a fully connectd layer with the given layer as input and the given number of neurons.
:param prev_layer: Tensor
The Tensor that acts as input into this layer
:param num_units: int
The size of the layer. That is, the number of units, nodes, or neurons.
:param is_training: bool or Tensor
Indicates whether or not the network is currently training, which tells the batch normalization
layer whether or not it should update or use its population statistics.
:returns Tensor
A new fully connected layer
"""
layer = tf.layers.dense(prev_layer, num_units, use_bias=False, activation=None)
gamma = tf.Variable(tf.ones([num_units]))
beta = tf.Variable(tf.zeros([num_units]))
pop_mean = tf.Variable(tf.zeros([num_units]), trainable=False)
pop_variance = tf.Variable(tf.ones([num_units]), trainable=False)
epsilon = 1e-3
def batch_norm_training():
batch_mean, batch_variance = tf.nn.moments(layer, [0])
decay = 0.99
train_mean = tf.assign(pop_mean, pop_mean * decay + batch_mean * (1 - decay))
train_variance = tf.assign(pop_variance, pop_variance * decay + batch_variance * (1 - decay))
with tf.control_dependencies([train_mean, train_variance]):
return tf.nn.batch_normalization(layer, batch_mean, batch_variance, beta, gamma, epsilon)
def batch_norm_inference():
return tf.nn.batch_normalization(layer, pop_mean, pop_variance, beta, gamma, epsilon)
batch_normalized_output = tf.cond(is_training, batch_norm_training, batch_norm_inference)
return tf.nn.relu(batch_normalized_output)
```
**TODO:** Modify `conv_layer` to add batch normalization to the fully connected layers it creates. Feel free to change the function's parameters if it helps.
**Note:** Unlike in the previous example that used `tf.layers`, adding batch normalization to these convolutional layers _does_ require some slight differences to what you did in `fully_connected`.
```
def conv_layer(prev_layer, layer_depth, is_training):
"""
Create a convolutional layer with the given layer as input.
:param prev_layer: Tensor
The Tensor that acts as input into this layer
:param layer_depth: int
We'll set the strides and number of feature maps based on the layer's depth in the network.
This is *not* a good way to make a CNN, but it helps us create this example with very little code.
:param is_training: bool or Tensor
Indicates whether or not the network is currently training, which tells the batch normalization
layer whether or not it should update or use its population statistics.
:returns Tensor
A new convolutional layer
"""
strides = 2 if layer_depth % 3 == 0 else 1
in_channels = prev_layer.get_shape().as_list()[3]
out_channels = layer_depth*4
weights = tf.Variable(
tf.truncated_normal([3, 3, in_channels, out_channels], stddev=0.05))
layer = tf.nn.conv2d(prev_layer, weights, strides=[1,strides, strides, 1], padding='SAME')
gamma = tf.Variable(tf.ones([out_channels]))
beta = tf.Variable(tf.zeros([out_channels]))
pop_mean = tf.Variable(tf.zeros([out_channels]), trainable=False)
pop_variance = tf.Variable(tf.ones([out_channels]), trainable=False)
epsilon = 1e-3
def batch_norm_training():
# Important to use the correct dimensions here to ensure the mean and variance are calculated
# per feature map instead of for the entire layer
batch_mean, batch_variance = tf.nn.moments(layer, [0,1,2], keep_dims=False)
decay = 0.99
train_mean = tf.assign(pop_mean, pop_mean * decay + batch_mean * (1 - decay))
train_variance = tf.assign(pop_variance, pop_variance * decay + batch_variance * (1 - decay))
with tf.control_dependencies([train_mean, train_variance]):
return tf.nn.batch_normalization(layer, batch_mean, batch_variance, beta, gamma, epsilon)
def batch_norm_inference():
return tf.nn.batch_normalization(layer, pop_mean, pop_variance, beta, gamma, epsilon)
batch_normalized_output = tf.cond(is_training, batch_norm_training, batch_norm_inference)
return tf.nn.relu(batch_normalized_output)
```
**TODO:** Edit the `train` function to support batch normalization. You'll need to make sure the network knows whether or not it is training.
```
def train(num_batches, batch_size, learning_rate):
# Build placeholders for the input samples and labels
inputs = tf.placeholder(tf.float32, [None, 28, 28, 1])
labels = tf.placeholder(tf.float32, [None, 10])
# Add placeholder to indicate whether or not we're training the model
is_training = tf.placeholder(tf.bool)
# Feed the inputs into a series of 20 convolutional layers
layer = inputs
for layer_i in range(1, 20):
layer = conv_layer(layer, layer_i, is_training)
# Flatten the output from the convolutional layers
orig_shape = layer.get_shape().as_list()
layer = tf.reshape(layer, shape=[-1, orig_shape[1] * orig_shape[2] * orig_shape[3]])
# Add one fully connected layer
layer = fully_connected(layer, 100, is_training)
# Create the output layer with 1 node for each
logits = tf.layers.dense(layer, 10)
# Define loss and training operations
model_loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=labels))
train_opt = tf.train.AdamOptimizer(learning_rate).minimize(model_loss)
# Create operations to test accuracy
correct_prediction = tf.equal(tf.argmax(logits,1), tf.argmax(labels,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# Train and test the network
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for batch_i in range(num_batches):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# train this batch
sess.run(train_opt, {inputs: batch_xs, labels: batch_ys, is_training: True})
# Periodically check the validation or training loss and accuracy
if batch_i % 100 == 0:
loss, acc = sess.run([model_loss, accuracy], {inputs: mnist.validation.images,
labels: mnist.validation.labels,
is_training: False})
print('Batch: {:>2}: Validation loss: {:>3.5f}, Validation accuracy: {:>3.5f}'.format(batch_i, loss, acc))
elif batch_i % 25 == 0:
loss, acc = sess.run([model_loss, accuracy], {inputs: batch_xs, labels: batch_ys, is_training: False})
print('Batch: {:>2}: Training loss: {:>3.5f}, Training accuracy: {:>3.5f}'.format(batch_i, loss, acc))
# At the end, score the final accuracy for both the validation and test sets
acc = sess.run(accuracy, {inputs: mnist.validation.images,
labels: mnist.validation.labels,
is_training: False})
print('Final validation accuracy: {:>3.5f}'.format(acc))
acc = sess.run(accuracy, {inputs: mnist.test.images,
labels: mnist.test.labels,
is_training: False})
print('Final test accuracy: {:>3.5f}'.format(acc))
# Score the first 100 test images individually, just to make sure batch normalization really worked
correct = 0
for i in range(100):
correct += sess.run(accuracy,feed_dict={inputs: [mnist.test.images[i]],
labels: [mnist.test.labels[i]],
is_training: False})
print("Accuracy on 100 samples:", correct/100)
num_batches = 800
batch_size = 64
learning_rate = 0.002
tf.reset_default_graph()
with tf.Graph().as_default():
train(num_batches, batch_size, learning_rate)
```
Once again, the model with batch normalization should reach an accuracy over 90%. There are plenty of details that can go wrong when implementing at this low level, so if you got it working - great job! If not, do not worry, just look at the `Batch_Normalization_Solutions` notebook to see what went wrong.
| github_jupyter |
# Country Economic Conditions for Cargo Carriers
This report is written from the point of view of a data scientist preparing a report to the Head of Analytics for a logistics company. The company needs information on economic and financial conditions is different countries, including data on their international trade, to be aware of any situations that could affect business.
## Data Summary
This dataset is taken from the International Monetary Fund (IMF) data bank. It lists country-level economic and financial statistics from all countries globally. This includes data such as gross domestic product (GDP), inflation, exports and imports, and government borrowing and revenue. The data is given in either US Dollars, or local currency depending on the country and year. Some variables, like inflation and unemployment, are given as percentages.
## Data Exploration
The initial plan for data exploration is to first model the data on country GDP and inflation, then to look further into trade statistics.
```
#Import required packages
import numpy as np
import pandas as pd
from sklearn import linear_model
from scipy import stats
import math
from sklearn import datasets, linear_model
from sklearn.linear_model import LinearRegression
import statsmodels.api as sm
#Import IMF World Economic Outlook Data from GitHub
WEO = pd.read_csv('https://raw.githubusercontent.com/jamiemfraser/machine_learning/main/WEOApr2021all.csv')
WEO=pd.DataFrame(WEO)
WEO.head()
# Print basic details of the dataset
print(WEO.shape[0])
print(WEO.columns.tolist())
print(WEO.dtypes)
#Shows that all numeric columns are type float, and string columns are type object
```
### Data Cleaning and Feature Engineering
```
#We are only interested in the most recent year for which data is available, 2019
WEO=WEO.drop(['2000', '2001', '2002', '2003', '2004', '2005', '2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015', '2016', '2017', '2018'], axis = 1)
#Reshape the data so each country is one observation
WEO=WEO.pivot_table(index=["Country"], columns='Indicator', values='2019').reset_index()
WEO.columns = ['Country', 'Current_account', 'Employment', 'Net_borrowing', 'Government_revenue', 'Government_expenditure', 'GDP_percap_constant', 'GDP_percap_current', 'GDP_constant', 'Inflation', 'Investment', 'Unemployment', 'Volume_exports', 'Volume_imports']
WEO.head()
#Describe the dataset
WEO.dropna(inplace=True)
WEO.describe()
```
### Key Findings and Insights
```
#Large differences betweeen the mean and median values could be an indication of outliers that are skewing the data
WEO.agg([np.mean, np.median])
#Create a scatterplot
import matplotlib.pyplot as plt
%matplotlib inline
ax = plt.axes()
ax.scatter(WEO.Volume_exports, WEO.Volume_imports)
# Label the axes
ax.set(xlabel='Volume Exports',
ylabel='Volume Imports',
title='Volume of Exports vs Imports');
#Create a scatterplot
import matplotlib.pyplot as plt
%matplotlib inline
ax = plt.axes()
ax.scatter(WEO.GDP_percap_constant, WEO.Volume_imports)
# Label the axes
ax.set(xlabel='GDP per capita',
ylabel='Volume Imports',
title='GDP per capita vs Volume of Imports');
#Create a scatterplot
import matplotlib.pyplot as plt
%matplotlib inline
ax = plt.axes()
ax.scatter(WEO.Investment, WEO.Volume_imports)
# Label the axes
ax.set(xlabel='Investment',
ylabel='Volume Imports',
title='Investment vs Volume of Imports');
```
### Hypotheses
Hypothesis 1: GDP per capita and the level of investment will be significant in determining the volume of goods and services imports
Hypothesis 2: There will be a strong correlation between government revenues and government expenditures
Hypothesis 3: GDP per capita and inflation will be significant in determining the unemployment rate
### Significance Test
I will conduct a formal hypothesis test on Hypothesis #1, which states that GDP per capita and the level of investment will be significant in determining the volume of goods and services imports. I will use a linear regression model because the scatterplots shown above indicate there is likely a linear relationship between both GDP per capita and investment against the volume of imports. I will take a p-value of 0.05 or less to be an indication of significance.
The null hypothesis is that there is no significant relationship between GDP per capita or the level of investment and the volume of goods and services.
The alternative hypothesis is that there is a significant relationship between either GDP per capita or the level of investment and the volume of goods and services.
```
#Set up a linear regression model for GDP per capita and evaluate
WEO=WEO.reset_index()
X = WEO['GDP_percap_constant']
X=X.values.reshape(-1,1)
y = WEO['Volume_imports']
X2 = sm.add_constant(X)
est = sm.OLS(y, X2)
est2 = est.fit()
print(est2.summary())
#Set up a linear regression model for Investment and evaluate
WEO=WEO.reset_index()
X = WEO['Investment']
X=X.values.reshape(-1,1)
y = WEO['Volume_imports']
X2 = sm.add_constant(X)
est = sm.OLS(y, X2)
est2 = est.fit()
print(est2.summary())
```
The linear regression analyses show that while GDP per capita is not significant in determining the volume of imports, investment is significant. For GDP per capita, we obtain a p-value of 0.313 which is insignificant. For Investment, we obtain a p-value of 0.000, which is significant.
## Next Steps
Next steps in analysing the data would be to see if there are any other variables that are significant in determining the volume of imports. The data scientist could also try a multiple linear regression to determine if there are variables that together produce a significant effect.
### Data Quality
The quality of this dataset is questionable. The exploratory data analysis showed several outliers that could be skewing the data. Further, there is no defined uniformity for how this data is measured. It is reported on a country-by-country basis, which leaves open the possibility that variation in definitions or methods for measuring these variables could lead to inaccurate comparison between countries.
Further data that I would request is more detailed trade data. Specifically, because this analysis finds that investment is significant in determining the volume of imports, it would be interesting to see which types of goods are more affected by investment. This could inform business decisions for a logistics company by allowing it to predict what type of cargo would need to be moved depending on investment practices in an individual country.
| github_jupyter |
```
import warnings
warnings.filterwarnings('ignore')
import os
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import pandas as pd
import seaborn as sns
sns.set(rc={'figure.figsize':(12, 6),"font.size":20,"axes.titlesize":20,"axes.labelsize":20},style="darkgrid")
```
Is there any connection with the crime and food inspection failures? May be ! For now, I am focusing on the burgalaries only. The burglary data is the chicago's crime data filtered for burgalaries only (in the same time window i.e. first 3 months of 2019).
```
burglary = pd.read_json('../data/raw/burglary.json', convert_dates=['date'])
burglary.head()
shape = burglary.shape
print(" There are %d rows and %d columns in the data" % (shape[0], shape[1]))
print(burglary.info())
```
Let's check if there are any null values in the data.
```
burglary.isna().sum()
burglary['latitude'].fillna(burglary['latitude'].mode()[0], inplace=True)
burglary['longitude'].fillna(burglary['longitude'].mode()[0], inplace=True)
ax = sns.countplot(x="ward", data=burglary)
plt.title("Burglaries by Ward")
plt.show()
plt.rcParams['figure.figsize'] = 16, 5
ax = sns.countplot(x="community_area", data=burglary)
plt.title("Burglaries by Ward")
plt.show()
```
Burglaries HeatMap
```
import gmaps
APIKEY= os.getenv('GMAPAPIKEY')
gmaps.configure(api_key=APIKEY)
def make_heatmap(locations, weights=None):
fig = gmaps.figure()
heatmap_layer = gmaps.heatmap_layer(locations)
#heatmap_layer.max_intensity = 100
heatmap_layer.point_radius = 8
fig.add_layer(heatmap_layer)
return fig
locations = zip(burglary['latitude'], burglary['longitude'])
fig = make_heatmap(locations)
fig
burglary_per_day = pd.DataFrame()
burglary_per_day = burglary[['date', 'case_number']]
burglary_per_day = burglary_per_day.set_index(
pd.to_datetime(burglary_per_day['date']))
burglary_per_day = burglary_per_day.resample('D').count()
plt.rcParams['figure.figsize'] = 12, 5
fig, ax = plt.subplots()
fig.autofmt_xdate()
#
#ax.xaxis.set_major_locator(mdates.MonthLocator())
#ax.xaxis.set_minor_locator(mdates.DayLocator())
monthFmt = mdates.DateFormatter('%Y-%b')
ax.xaxis.set_major_formatter(monthFmt)
plt.plot(burglary_per_day.index, burglary_per_day, 'r-')
plt.xlabel('Date')
plt.ylabel('Number of Cases Reported')
plt.title('Burglaries Reported')
plt.show()
burglary['event_date'] = burglary['date']
burglary = burglary.set_index('event_date')
burglary.sort_values(by='date', inplace=True)
burglary.head()
burglary.to_csv('../data/processed/burglary_data_processed.csv')
```
| github_jupyter |
# Set-up notebook environment
## NOTE: Use a QIIME2 kernel
```
import numpy as np
import pandas as pd
import seaborn as sns
import scipy
from scipy import stats
import matplotlib.pyplot as plt
import re
from pandas import *
import matplotlib.pyplot as plt
%matplotlib inline
from qiime2.plugins import feature_table
from qiime2 import Artifact
from qiime2 import Metadata
import biom
from biom.table import Table
from qiime2.plugins import diversity
from scipy.stats import ttest_ind
from scipy.stats.stats import pearsonr
%config InlineBackend.figure_formats = ['svg']
from qiime2.plugins.feature_table.methods import relative_frequency
import biom
import qiime2 as q2
import os
import math
```
# Import sample metadata
```
meta = q2.Metadata.load('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/sample_metadata/12201_metadata.txt').to_dataframe()
```
Separate round 1 and round 2 and exclude round 1 Zymo, Homebrew, and MagMAX Beta
```
meta_r1 = meta[meta['round'] == 1]
meta_clean_r1_1 = meta_r1[meta_r1['extraction_kit'] != 'Zymo MagBead']
meta_clean_r1_2 = meta_clean_r1_1[meta_clean_r1_1['extraction_kit'] != 'Homebrew']
meta_clean_r1 = meta_clean_r1_2[meta_clean_r1_2['extraction_kit'] != 'MagMax Beta']
meta_clean_r2 = meta[meta['round'] == 2]
```
Remove PowerSoil samples from each round - these samples will be used as the baseline
```
meta_clean_r1_noPS = meta_clean_r1[meta_clean_r1['extraction_kit'] != 'PowerSoil']
meta_clean_r2_noPS = meta_clean_r2[meta_clean_r2['extraction_kit'] != 'PowerSoil']
```
Create tables including only round 1 or round 2 PowerSoil samples
```
meta_clean_r1_onlyPS = meta_clean_r1[meta_clean_r1['extraction_kit'] == 'PowerSoil']
meta_clean_r2_onlyPS = meta_clean_r2[meta_clean_r2['extraction_kit'] == 'PowerSoil']
```
Merge PowerSoil samples from round 2 with other samples from round 1, and vice versa - this will allow us to get the correlations between the two rounds of PowerSoil
```
meta_clean_r1_with_r2_PS = pd.concat([meta_clean_r1_noPS, meta_clean_r2_onlyPS])
meta_clean_r2_with_r1_PS = pd.concat([meta_clean_r2_noPS, meta_clean_r1_onlyPS])
```
## Collapse feature-table to the desired level (e.g., genus)
16S
```
qiime taxa collapse \
--i-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/16S/10_filtered_data/dna_bothPS_16S_deblur_biom_lod_noChl_noMit_sepp_gg_noNTCs_noMock.qza \
--i-taxonomy /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/16S/06_taxonomy/dna_all_16S_deblur_seqs_taxonomy_silva138.qza \
--p-level 6 \
--o-collapsed-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/16S/10_filtered_data/dna_bothPS_16S_deblur_biom_lod_noChl_noMit_sepp_gg_noNTCs_noMock_taxa_collapse_genus.qza
qiime feature-table summarize \
--i-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/16S/10_filtered_data/dna_bothPS_16S_deblur_biom_lod_noChl_noMit_sepp_gg_noNTCs_noMock_taxa_collapse_genus.qza \
--o-visualization /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/16S/10_filtered_data/dna_bothPS_16S_deblur_biom_lod_noChl_noMit_sepp_gg_noNTCs_noMock_taxa_collapse_genus.qzv
# There are 846 samples and 1660 features
```
ITS
```
qiime taxa collapse \
--i-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/ITS/08_filtered_data/dna_bothPS_ITS_deblur_biom_lod_noNTCs_noMock.qza \
--i-taxonomy /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/ITS/06_taxonomy/dna_all_ITS_deblur_seqs_taxonomy_unite8.qza \
--p-level 6 \
--o-collapsed-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/ITS/08_filtered_data/dna_bothPS_ITS_deblur_biom_lod_noNTCs_noMock_taxa_collapse_genus.qza
qiime feature-table summarize \
--i-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/ITS/08_filtered_data/dna_bothPS_ITS_deblur_biom_lod_noNTCs_noMock_taxa_collapse_genus.qza \
--o-visualization /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/ITS/08_filtered_data/dna_bothPS_ITS_deblur_biom_lod_noNTCs_noMock_taxa_collapse_genus.qzv
# There are 978 samples and 791 features
```
Shotgun
```
qiime taxa collapse \
--i-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/shotgun/03_filtered_data/dna_bothPS_shotgun_woltka_wol_biom_noNTCs_noMock.qza \
--i-taxonomy /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/shotgun/wol_taxonomy.qza \
--p-level 6 \
--o-collapsed-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/shotgun/03_filtered_data/dna_bothPS_shotgun_woltka_wol_biom_noNTCs_noMock_taxa_collapse_genus.qza
qiime feature-table summarize \
--i-table /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/shotgun/03_filtered_data/dna_bothPS_shotgun_woltka_wol_biom_noNTCs_noMock_taxa_collapse_genus.qza \
--o-visualization /Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/shotgun/03_filtered_data/dna_bothPS_shotgun_woltka_wol_biom_noNTCs_noMock_taxa_collapse_genus.qzv
# There are 1044 samples and 2060 features
```
# Import feature-tables
```
dna_bothPS_16S_genus_qza = q2.Artifact.load('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/16S/10_filtered_data/dna_bothPS_16S_deblur_biom_lod_noChl_noMit_sepp_gg_noNTCs_noMock_taxa_collapse_genus.qza')
dna_bothPS_ITS_genus_qza = q2.Artifact.load('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/ITS/08_filtered_data/dna_bothPS_ITS_deblur_biom_lod_noNTCs_noMock_taxa_collapse_genus.qza')
dna_bothPS_shotgun_genus_qza = q2.Artifact.load('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/data/shotgun/03_filtered_data/dna_bothPS_shotgun_woltka_wol_biom_noNTCs_noMock_taxa_collapse_genus.qza')
```
# Convert QZA to a Pandas DataFrame
```
dna_bothPS_16S_genus_df = dna_bothPS_16S_genus_qza.view(pd.DataFrame)
dna_bothPS_ITS_genus_df = dna_bothPS_ITS_genus_qza.view(pd.DataFrame)
dna_bothPS_shotgun_genus_df = dna_bothPS_shotgun_genus_qza.view(pd.DataFrame)
```
# Melt dataframes
```
dna_bothPS_16S_genus_df_melt = dna_bothPS_16S_genus_df.unstack()
dna_bothPS_ITS_genus_df_melt = dna_bothPS_ITS_genus_df.unstack()
dna_bothPS_shotgun_genus_df_melt = dna_bothPS_shotgun_genus_df.unstack()
dna_bothPS_16S_genus = pd.DataFrame(dna_bothPS_16S_genus_df_melt)
dna_bothPS_ITS_genus = pd.DataFrame(dna_bothPS_ITS_genus_df_melt)
dna_bothPS_shotgun_genus = pd.DataFrame(dna_bothPS_shotgun_genus_df_melt)
dna_bothPS_16S_genus.reset_index(inplace=True)
dna_bothPS_16S_genus.rename(columns={'level_0':'taxa','level_1':'sample',0:'counts'}, inplace=True)
dna_bothPS_ITS_genus.reset_index(inplace=True)
dna_bothPS_ITS_genus.rename(columns={'level_0':'taxa','level_1':'sample',0:'counts'}, inplace=True)
dna_bothPS_shotgun_genus.reset_index(inplace=True)
dna_bothPS_shotgun_genus.rename(columns={'level_0':'taxa','level_1':'sample',0:'counts'}, inplace=True)
```
# Wrangle data into long form for each kit
Wrangle metadata
```
# Create empty list of extraction kit IDs
ext_kit_levels = []
# Create empty list of metadata subsets based on levels of variable of interest
ext_kit = []
# Create empty list of baseline samples for each subset
bl = []
# Populate lists with round 1 data
for ext_kit_level, ext_kit_level_df in meta_clean_r1_with_r2_PS.groupby('extraction_kit_round'):
ext_kit.append(ext_kit_level_df)
powersoil_r1_bl = meta_clean_r1_onlyPS[meta_clean_r1_onlyPS.extraction_kit_round == 'PowerSoil r1']
bl.append(powersoil_r1_bl)
ext_kit_levels.append(ext_kit_level)
print('Gathered data for',ext_kit_level)
# Populate lists with round 2 data
for ext_kit_level, ext_kit_level_df in meta_clean_r2_with_r1_PS.groupby('extraction_kit_round'):
ext_kit.append(ext_kit_level_df)
powersoil_r2_bl = meta_clean_r2_onlyPS[meta_clean_r2_onlyPS['extraction_kit_round'] == 'PowerSoil r2']
bl.append(powersoil_r2_bl)
ext_kit_levels.append(ext_kit_level)
print('Gathered data for',ext_kit_level)
# Create empty list for concatenated subset-baseline datasets
subsets_w_bl = {}
# Populate list with subset-baseline data
for ext_kit_level, ext_kit_df, ext_kit_bl in zip(ext_kit_levels, ext_kit, bl):
new_df = pd.concat([ext_kit_bl,ext_kit_df])
subsets_w_bl[ext_kit_level] = new_df
print('Merged data for',ext_kit_level)
```
16S
```
list_of_lists = []
for key, value in subsets_w_bl.items():
string = ''.join(key)
#merge metadata subsets with baseline with taxonomy
meta_16S_genera = pd.merge(value, dna_bothPS_16S_genus, left_index=True, right_on='sample')
#create new column
meta_16S_genera['taxa_subject'] = meta_16S_genera['taxa'] + meta_16S_genera['host_subject_id']
#subtract out duplicates and pivot
meta_16S_genera_clean = meta_16S_genera.drop_duplicates(subset = ['taxa_subject', 'extraction_kit_round'], keep = 'first')
meta_16S_genera_pivot = meta_16S_genera_clean.pivot(index='taxa_subject', columns='extraction_kit_round', values='counts')
meta_16S_genera_pivot_clean = meta_16S_genera_pivot.dropna()
# Export dataframe to file
meta_16S_genera_pivot_clean.to_csv('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/results/feature_abundance_correlation_images/table_correlation_16S_genera_%s.txt'%string,
sep = '\t',
index = False)
```
ITS
```
list_of_lists = []
for key, value in subsets_w_bl.items():
string = ''.join(key)
#merge metadata subsets with baseline with taxonomy
meta_ITS_genera = pd.merge(value, dna_bothPS_ITS_genus, left_index=True, right_on='sample')
#create new column
meta_ITS_genera['taxa_subject'] = meta_ITS_genera['taxa'] + meta_ITS_genera['host_subject_id']
#subtract out duplicates and pivot
meta_ITS_genera_clean = meta_ITS_genera.drop_duplicates(subset = ['taxa_subject', 'extraction_kit_round'], keep = 'first')
meta_ITS_genera_pivot = meta_ITS_genera_clean.pivot(index='taxa_subject', columns='extraction_kit_round', values='counts')
meta_ITS_genera_pivot_clean = meta_ITS_genera_pivot.dropna()
# Export dataframe to file
meta_ITS_genera_pivot_clean.to_csv('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/results/feature_abundance_correlation_images/table_correlation_ITS_genera_%s.txt'%string,
sep = '\t',
index = False)
```
Shotgun
```
list_of_lists = []
for key, value in subsets_w_bl.items():
string = ''.join(key)
#merge metadata subsets with baseline with taxonomy
meta_shotgun_genera = pd.merge(value, dna_bothPS_shotgun_genus, left_index=True, right_on='sample')
#create new column
meta_shotgun_genera['taxa_subject'] = meta_shotgun_genera['taxa'] + meta_shotgun_genera['host_subject_id']
#subtract out duplicates and pivot
meta_shotgun_genera_clean = meta_shotgun_genera.drop_duplicates(subset = ['taxa_subject', 'extraction_kit_round'], keep = 'first')
meta_shotgun_genera_pivot = meta_shotgun_genera_clean.pivot(index='taxa_subject', columns='extraction_kit_round', values='counts')
meta_shotgun_genera_pivot_clean = meta_shotgun_genera_pivot.dropna()
# Export dataframe to file
meta_shotgun_genera_pivot_clean.to_csv('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/results/feature_abundance_correlation_images/table_correlation_shotgun_genera_%s.txt'%string,
sep = '\t',
index = False)
```
# Code below is not used
## NOTE: The first cell was originally appended to the cell above
```
# check pearson correlation
x = meta_16S_genera_pivot_clean.iloc[:,1]
y = meta_16S_genera_pivot_clean[key]
corr = stats.pearsonr(x, y)
int1, int2 = corr
corr_rounded = round(int1, 2)
corr_str = str(corr_rounded)
x_key = key[0]
y_key = key[1]
list1 = []
list1.append(corr_rounded)
list1.append(key)
list_of_lists.append(list1)
list_of_lists
df = pd.DataFrame(list_of_lists, columns = ['Correlation', 'Extraction kit'])
df.to_csv('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/results/feature_abundance_correlation_images/table_correlations_16S_genera.txt',
sep = '\t',
index = False)
splot = sns.catplot(y="Correlation",
x="Extraction kit",
hue= "Extraction kit",
kind='bar',
data=df,
dodge = False)
splot.set(ylim=(0, 1))
plt.xticks(rotation=45,
horizontalalignment='right')
#new_labels = ['−20C','−20C after 1 week', '4C','Ambient','Freeze-thaw','Heat']
#for t, l in zip(splot._legend.texts, new_labels):
# t.set_text(l)
splot.savefig('correlation_16S_genera.png')
splot.savefig('correlation_16S_genera.svg', format='svg', dpi=1200)
```
### Individual correlation plots
```
for key, value in subsets_w_bl.items():
string = ''.join(key)
#merge metadata subsets with baseline with taxonomy
meta_16S_genera = pd.merge(value, dna_bothPS_16S_genus, left_index=True, right_on='sample')
#create new column
meta_16S_genera['taxa_subject'] = meta_16S_genera['taxa'] + meta_16S_genera['host_subject_id']
#subtract out duplicates and pivot
meta_16S_genera_clean = meta_16S_genera.drop_duplicates(subset = ['taxa_subject', 'extraction_kit_round'], keep = 'first')
meta_16S_genera_pivot = meta_16S_genera_clean.pivot(index='taxa_subject', columns='extraction_kit_round', values='counts')
meta_16S_genera_pivot_clean = meta_16S_genera_pivot.dropna()
# check pearson correlation
x = meta_16S_genera_pivot_clean.iloc[:,1]
y = meta_16S_genera_pivot_clean[key]
corr = stats.pearsonr(x, y)
int1, int2 = corr
corr_rounded = round(int1, 2)
corr_str = str(corr_rounded)
#make correlation plots
meta_16S_genera_pivot_clean['x1'] = meta_16S_genera_pivot_clean.iloc[:,1]
meta_16S_genera_pivot_clean['y1'] = meta_16S_genera_pivot_clean.iloc[:,0]
ax=sns.lmplot(x='x1',
y='y1',
data=meta_16S_genera_pivot_clean,
height=3.8)
ax.set(yscale='log')
ax.set(xscale='log')
ax.set(xlabel='PowerSoil', ylabel=key)
#plt.xlim(0.00001, 10000000)
#plt.ylim(0.00001, 10000000)
plt.title(string + ' (%s)' %corr_str)
ax.savefig('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/results/feature_abundance_correlation_images/figure_scatter_correlation_16S_genera_%s.png'%string)
ax.savefig('/Users/Justin/Mycelium/UCSD/00_Knight_Lab/03_Extraction_test_12201/round_02/results/feature_abundance_correlation_images/figure_scatter_correlation_16S_genera_%s.svg'%string, format='svg',dpi=1200)
```
| github_jupyter |
## Health, Wealth of Nations from 1800-2008
```
import os
import numpy as np
import pandas as pd
from pandas import Series, DataFrame
from bqplot import Figure, Tooltip, Label
from bqplot import Axis, ColorAxis
from bqplot import LogScale, LinearScale, OrdinalColorScale
from bqplot import Scatter, Lines
from bqplot import CATEGORY10
from ipywidgets import HBox, VBox, IntSlider, Play, jslink
from more_itertools import flatten
```
---
### Get Data
```
year_start = 1800
df = pd.read_json("data_files/nations.json")
df.head()
list_rows_to_drop = \
(df['income']
.apply(len)
.where(lambda i: i < 10)
.dropna()
.index
.tolist()
)
df.drop(list_rows_to_drop, inplace=True)
dict_dfs = {}
for COL in ['income', 'lifeExpectancy', 'population']:
df1 = \
DataFrame(df
.loc[:, COL]
.map(lambda l: (DataFrame(l)
.set_index(0)
.squeeze()
.reindex(range(1800, 2009))
.interpolate()
.to_dict()))
.tolist())
df1.index = df.name
dict_dfs[COL] = df1
def get_data(year):
"""
"""
income = dict_dfs['income'].loc[:, year]
lifeExpectancy = dict_dfs['lifeExpectancy'].loc[:, year]
population = dict_dfs['population'].loc[:, year]
return income, lifeExpectancy, population
get_min_max_from_df = lambda df: (df.min().min(), df.max().max())
```
---
### Create Tooltip
```
tt = Tooltip(fields=['name', 'x', 'y'],
labels=['Country', 'IncomePerCapita', 'LifeExpectancy'])
```
---
### Create Scales
```
# Income
income_min, income_max = get_min_max_from_df(dict_dfs['income'])
x_sc = LogScale(min=income_min,
max=income_max)
# Life Expectancy
life_exp_min, life_exp_max = get_min_max_from_df(dict_dfs['lifeExpectancy'])
y_sc = LinearScale(min=life_exp_min,
max=life_exp_max)
# Population
pop_min, pop_max = get_min_max_from_df(dict_dfs['population'])
size_sc = LinearScale(min=pop_min,
max=pop_max)
# Color
c_sc = OrdinalColorScale(domain=df['region'].unique().tolist(),
colors=CATEGORY10[:6])
```
---
### Create Axes
```
ax_y = Axis(label='Life Expectancy',
scale=y_sc,
orientation='vertical',
side='left',
grid_lines='solid')
ax_x = Axis(label='Income per Capita',
scale=x_sc,
grid_lines='solid')
```
---
## Create Marks
### 1. Scatter
```
cap_income, life_exp, pop = get_data(year_start)
scatter_ = Scatter(x=cap_income,
y=life_exp,
color=df['region'],
size=pop,
names=df['name'],
display_names=False,
scales={
'x': x_sc,
'y': y_sc,
'color': c_sc,
'size': size_sc
},
default_size=4112,
tooltip=tt,
animate=True,
stroke='Black',
unhovered_style={'opacity': 0.5})
```
### 2. Line
```
line_ = Lines(x=dict_dfs['income'].loc['Angola'].values,
y=dict_dfs['lifeExpectancy'].loc['Angola'].values,
colors=['Gray'],
scales={
'x': x_sc,
'y': y_sc
},
visible=False)
```
---
### Create Label
```
year_label = Label(x=[0.75],
y=[0.10],
font_size=50,
font_weight='bolder',
colors=['orange'],
text=[str(year_start)],
enable_move=True)
```
---
## Construct the Figure
```
time_interval = 10
fig_ = \
Figure(
marks=[scatter_, line_, year_label],
axes=[ax_x, ax_y],
title='Health and Wealth of Nations',
animation_duration=time_interval
)
fig_.layout.min_width = '960px'
fig_.layout.min_height = '640px'
fig_
```
---
## Add Interactivity
- Update chart when year changes
```
slider_ = IntSlider(
min=year_start,
max=2008,
step=1,
description='Year: ',
value=year_start)
def on_change_year(change):
"""
"""
scatter_.x, scatter_.y, scatter_.size = get_data(slider_.value)
year_label.text = [str(slider_.value)]
slider_.observe(on_change_year, 'value')
slider_
```
- Display line when hovered
```
def on_hover(change):
"""
"""
if change.new is not None:
display(change.new)
line_.x = dict_dfs['income'].iloc[change.new + 1]
line_.y = dict_dfs['lifeExpectancy'].iloc[change.new + 1]
line_.visible = True
else:
line_.visible = False
scatter_.observe(on_hover, 'hovered_point')
```
---
## Add Animation!
```
play_button = Play(
min=1800,
max=2008,
interval=time_interval
)
jslink(
(play_button, 'value'),
(slider_, 'value')
)
```
---
## Create the GUI
```
VBox([play_button, slider_, fig_])
```
| github_jupyter |
I want to analyze changes over time in the MOT GTFS feed.
Agenda:
1. [Get data](#Get-the-data)
3. [Tidy](#Tidy-it-up)
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import partridge as ptg
from ftplib import FTP
import datetime
import re
import zipfile
import os
%matplotlib inline
plt.rcParams['figure.figsize'] = (10, 5) # set default size of plots
sns.set_style("white")
sns.set_context("talk")
sns.set_palette('Set2', 10)
```
## Get the data
There are two options - TransitFeeds and the workshop's S3 bucket.
```
#!aws s3 cp s3://s3.obus.hasadna.org.il/2018-04-25.zip data/gtfs_feeds/2018-04-25.zip
```
## Tidy it up
Again I'm using [partridge](https://github.com/remix/partridge/tree/master/partridge) for filtering on dates, and then some tidying up and transformations.
```
from gtfs_utils import *
local_tariff_path = 'data/sample/180515_tariff.zip'
conn = ftp_connect()
get_ftp_file(conn, file_name = TARIFF_FILE_NAME, local_zip_path = local_tariff_path )
def to_timedelta(df):
'''
Turn time columns into timedelta dtype
'''
cols = ['arrival_time', 'departure_time']
numeric = df[cols].apply(pd.to_timedelta, unit='s')
df = df.copy()
df[cols] = numeric
return df
%time f2 = new_get_tidy_feed_df(feed, [zones])
f2.head()
f2.columns
def get_tidy_feed_df(feed, zones):
s = feed.stops
r = feed.routes
a = feed.agency
t = (feed.trips
# faster joins and slices with Categorical dtypes
.assign(route_id=lambda x: pd.Categorical(x['route_id'])))
f = (feed.stop_times[fields['stop_times']]
.merge(s[fields['stops']], on='stop_id')
.merge(zones, how='left')
.assign(zone_name=lambda x: pd.Categorical(x['zone_name']))
.merge(t[fields['trips']], on='trip_id', how='left')
.assign(route_id=lambda x: pd.Categorical(x['route_id']))
.merge(r[fields['routes']], on='route_id', how='left')
.assign(agency_id=lambda x: pd.Categorical(x['agency_id']))
.merge(a[fields['agency']], on='agency_id', how='left')
.assign(agency_name=lambda x: pd.Categorical(x['agency_name']))
.pipe(to_timedelta)
)
return f
LOCAL_ZIP_PATH = 'data/gtfs_feeds/2018-02-01.zip'
feed = get_partridge_feed_by_date(LOCAL_ZIP_PATH, datetime.date(2018,2 , 1))
zones = get_zones()
'route_ids' in feed.routes.columns
feed.routes.shape
f = get_tidy_feed_df(feed, zones)
f.columns
f[f.route_short_name.isin(['20', '26', '136'])].groupby('stop_name').route_short_name.nunique().sort_values(ascending=False)
```
| github_jupyter |
Write in the input space, click `Shift-Enter` or click on the `Play` button to execute.
```
(3 + 1 + 12) ** 2 + 2 * 18
```
Give a title to the notebook by clicking on `Untitled` on the very top of the page, better not to use spaces because it will be also used for the filename
Save the notebook with the `Diskette` button, check dashboard
Integer division gives integer result with truncation in Python 2, float result in Python 3:
```
5/3
1/3
```
### Quotes for strings
```
print("Hello world")
print('Hello world')
```
### Look for differences
```
"Hello world"
print("Hello world")
```
### Multiple lines in a cell
```
1 + 2
3 + 4
print(1 + 2)
print(3 + 4)
print("""This is
a multiline
Hello world""")
```
## Functions and help
```
abs(-2)
```
Write a function name followed by `?` to open the help for that function.
type in a cell and execute: `abs?`
# Heading 1
## Heading 2
Structured plain text format, it looks a lot like writing text **emails**,
you can do lists:
* like
* this
write links like <http://google.com>, or [hyperlinking words](http://www.google.com)
go to <http://markdowntutorial.com/> to learn more
$b_n=\frac{1}{\pi}\int\limits_{-\pi}^{\pi}f(x)\sin nx\,\mathrm{d}x=\\
=\frac{1}{\pi}\int\limits_{-\pi}^{\pi}x^2\sin nx\,\mathrm{d}x$
## Variables
```
weight_kg = 55
```
Once a variable has a value, we can print it:
```
print(weight_kg)
```
and do arithmetic with it:
```
print('weight in pounds:')
print(2.2 * weight_kg)
```
We can also change a variable's value by assigning it a new one:
```
weight_kg = 57.5
print('weight in kilograms is now:')
print(weight_kg)
```
As the example above shows,
we can print several things at once by separating them with commas.
If we imagine the variable as a sticky note with a name written on it,
assignment is like putting the sticky note on a particular value:
<img src="files/img/python-sticky-note-variables-01.svg" alt="Variables as Sticky Notes" />
This means that assigning a value to one variable does *not* change the values of other variables.
For example,
let's store the subject's weight in pounds in a variable:
```
weight_lb = 2.2 * weight_kg
print('weight in kilograms:')
print(weight_kg)
print('and in pounds:')
print(weight_lb)
```
<img src="files/img/python-sticky-note-variables-02.svg" alt="Creating Another Variable" />
and then change `weight_kg`:
```
weight_kg = 100.0
print('weight in kilograms is now:')
print(weight_kg)
print('and weight in pounds is still:')
print(weight_lb)
```
<img src="files/img/python-sticky-note-variables-03.svg" alt="Updating a Variable" />
Since `weight_lb` doesn't "remember" where its value came from,
it isn't automatically updated when `weight_kg` changes.
This is different from the way spreadsheets work.
### Challenge
```
x = 5
y = x
x = x**2
```
How much is `x`? how much is `y`?
### Comments
```
weight_kg = 100.0 # assigning weight
# now convert to pounds
print(2.2 * weight_kg)
```
### Strings slicing
```
my_string = "Hello world"
print(my_string)
```
Python by convention starts indexing from `0`
```
print(my_string[0:3])
print(my_string[:3])
```
Python uses intervals open on the right: $ \left[7, 9\right[ $
```
print(my_string[7:9])
```
### Challenge
What happens if you print:
```
print(my_string[4:4])
```
| github_jupyter |
# Saving and Loading Models
In this notebook, I'll show you how to save and load models with PyTorch. This is important because you'll often want to load previously trained models to use in making predictions or to continue training on new data.
```
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms
import helper
import fc_model
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
# Download and load the training data
trainset = datasets.FashionMNIST('F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)
```
Here we can see one of the images.
```
image, label = next(iter(trainloader))
helper.imshow(image[0,:]);
```
# Train a network
To make things more concise here, I moved the model architecture and training code from the last part to a file called `fc_model`. Importing this, we can easily create a fully-connected network with `fc_model.Network`, and train the network using `fc_model.train`. I'll use this model (once it's trained) to demonstrate how we can save and load models.
```
# Create the network, define the criterion and optimizer
model = fc_model.Network(784, 10, [512, 256, 128])
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.001)
fc_model.train(model, trainloader, testloader, criterion, optimizer, epochs=2)
```
## Saving and loading networks
As you can imagine, it's impractical to train a network every time you need to use it. Instead, we can save trained networks then load them later to train more or use them for predictions.
The parameters for PyTorch networks are stored in a model's `state_dict`. We can see the state dict contains the weight and bias matrices for each of our layers.
```
print("Our model: \n\n", model, '\n')
print("The state dict keys: \n\n", model.state_dict().keys())
```
The simplest thing to do is simply save the state dict with `torch.save`. For example, we can save it to a file `'checkpoint.pth'`.
```
torch.save(model.state_dict(), 'checkpoint.pth')
```
Then we can load the state dict with `torch.load`.
```
state_dict = torch.load('checkpoint.pth')
print(state_dict.keys())
```
And to load the state dict in to the network, you do `model.load_state_dict(state_dict)`.
```
model.load_state_dict(state_dict)
```
Seems pretty straightforward, but as usual it's a bit more complicated. Loading the state dict works only if the model architecture is exactly the same as the checkpoint architecture. If I create a model with a different architecture, this fails.
```
# Try this
model = fc_model.Network(784, 10, [400, 200, 100])
# This will throw an error because the tensor sizes are wrong!
model.load_state_dict(state_dict)
```
This means we need to rebuild the model exactly as it was when trained. Information about the model architecture needs to be saved in the checkpoint, along with the state dict. To do this, you build a dictionary with all the information you need to compeletely rebuild the model.
```
checkpoint = {'input_size': 784,
'output_size': 10,
'hidden_layers': [each.out_features for each in model.hidden_layers],
'state_dict': model.state_dict()}
torch.save(checkpoint, 'checkpoint.pth')
```
Now the checkpoint has all the necessary information to rebuild the trained model. You can easily make that a function if you want. Similarly, we can write a function to load checkpoints.
```
def load_checkpoint(filepath):
checkpoint = torch.load(filepath)
model = fc_model.Network(checkpoint['input_size'],
checkpoint['output_size'],
checkpoint['hidden_layers'])
model.load_state_dict(checkpoint['state_dict'])
return model
model = load_checkpoint('checkpoint.pth')
print(model)
```
| github_jupyter |
```
Function Order in a Single File¶
In the following code example, the functions are out of order, and the code will not compile. Try to fix this by rearranging the functions to be in the correct order.
#include <iostream>
using std::cout;
void OuterFunction(int i)
{
InnerFunction(i);
}
void InnerFunction(int i)
{
cout << "The value of the integer is: " << i << "\n";
}
int main()
{
int a = 5;
OuterFunction(a);
}
In the mini-project for the first half of the course, the instructions were very careful to indicate where each function should be placed, so you didn't run into the problem of functions being out of order.
Using a Header
One other way to solve the code problem above (without rearranging the functions) would have been to declare each function at the top of the file. A function declaration is much like the first line of a function definition - it contains the return type, function name, and input variable types. The details of the function definition are not needed for the declaration though.
To avoid a single file from becomming cluttered with declarations and definitions for every function, it is customary to declare the functions in another file, called the header file. In C++, the header file will have filetype .h, and the contents of the header file must be included at the top of the .cpp file. See the following example for a refactoring of the code above into a header and a cpp file.
// The header file with just the function declarations.
// When you click the "Run Code" button, this file will
// be saved as header_example.h.
#ifndef HEADER_EXAMPLE_H
#define HEADER_EXAMPLE_H
void OuterFunction(int);
void InnerFunction(int);
#endif
// The contents of header_example.h are included in
// the corresponding .cpp file using quotes:
#include "header_example.h"
#include <iostream>
using std::cout;
void OuterFunction(int i)
{
InnerFunction(i);
}
void InnerFunction(int i)
{
cout << "The value of the integer is: " << i << "\n";
}
int main()
{
int a = 5;
OuterFunction(a);
}
Notice that the code from the first example was fixed without having to rearrange the functions! In the code above, you might also have noticed several other things:
The function declarations in the header file don't need variable names, just variable types. You can put names in the declaration, however, and doing this often makes the code easier to read.
The #include statement for the header used quotes " " around the file name, and not angle brackets <>. We have stored the header in the same directory as the .cpp file, and the quotes tell the preprocessor to look for the file in the same directory as the current file - not in the usual set of directories where libraries are typically stored.
Finally, there is a preprocessor directive:
#ifndef HEADER_EXAMPLE_H
#define HEADER_EXAMPLE_H
at the top of the header, along with an #endif at the end. This is called an "include guard". Since the header will be included into another file, and #include just pastes contents into a file, the include guard prevents the same file from being pasted multiple times into another file. This might happen if multiple files include the same header, and then are all included into the same main.cpp, for example. The ifndef checks if HEADER_EXAMPLE_H has not been defined in the file already. If it has not been defined yet, then it is defined with #define HEADER_EXAMPLE_H, and the rest of the header is used. If HEADER_EXAMPLE_H has already been defined, then the preprocessor does not enter the ifndef block. Note: There are other ways to do this. Another common way is to use an #pragma oncepreprocessor directive, but we won't cover that in detail here. See this Wikipedia article for examples.
Practice
In the following two cells, there is a blank header file and a short program that won't compile due to the functions being out of order. The code should take a vector of ints, add 1 to each of the vector entries, and then print the sum over the vector entries.
Without rearranging the functions in the main .cpp file, add some function declarations to the header file to fix this problem. Don't forget to include the "header_practice.h" file in your .cpp file!
Practice
In the following two cells, there is a blank header file and a short program that won't compile due to the functions being out of order. The code should take a vector of ints, add 1 to each of the vector entries, and then print the sum over the vector entries.
Without rearranging the functions in the main .cpp file, add some function declarations to the header file to fix this problem. Don't forget to include the "header_practice.h" file in your .cpp file!
// This file will be saved as "header_practice.h"
#include <iostream>
#include <vector>
using std::vector;
using std::cout;
int IncrementAndComputeVectorSum(vector<int> v)
{
int total = 0;
AddOneToEach(v);
for (auto i: v) {
total += i;
}
return total;
}
void AddOneToEach(vector<int> &v)
{
// Note that the function passes a reference to v
// and the for loop below uses references to
// each item in v. This means the actual
// ints that v holds will be incremented.
for (auto& i: v) {
i++;
}
}
int main()
{
vector<int> v{1, 2, 3, 4};
int total = IncrementAndComputeVectorSum(v);
cout << "The total is: " << total << "\n";
}
```
| github_jupyter |
# Deep Learning & Art: Neural Style Transfer
Welcome to the second assignment of this week. In this assignment, you will learn about Neural Style Transfer. This algorithm was created by Gatys et al. (2015) (https://arxiv.org/abs/1508.06576).
**In this assignment, you will:**
- Implement the neural style transfer algorithm
- Generate novel artistic images using your algorithm
Most of the algorithms you've studied optimize a cost function to get a set of parameter values. In Neural Style Transfer, you'll optimize a cost function to get pixel values!
```
import os
import sys
import scipy.io
import scipy.misc
import matplotlib.pyplot as plt
from matplotlib.pyplot import imshow
from PIL import Image
from nst_utils import *
import numpy as np
import tensorflow as tf
%matplotlib inline
```
## 1 - Problem Statement
Neural Style Transfer (NST) is one of the most fun techniques in deep learning. As seen below, it merges two images, namely, a "content" image (C) and a "style" image (S), to create a "generated" image (G). The generated image G combines the "content" of the image C with the "style" of image S.
In this example, you are going to generate an image of the Louvre museum in Paris (content image C), mixed with a painting by Claude Monet, a leader of the impressionist movement (style image S).
<img src="images/louvre_generated.png" style="width:750px;height:200px;">
Let's see how you can do this.
## 2 - Transfer Learning
Neural Style Transfer (NST) uses a previously trained convolutional network, and builds on top of that. The idea of using a network trained on a different task and applying it to a new task is called transfer learning.
Following the original NST paper (https://arxiv.org/abs/1508.06576), we will use the VGG network. Specifically, we'll use VGG-19, a 19-layer version of the VGG network. This model has already been trained on the very large ImageNet database, and thus has learned to recognize a variety of low level features (at the earlier layers) and high level features (at the deeper layers).
Run the following code to load parameters from the VGG model. This may take a few seconds.
```
model = load_vgg_model("pretrained-model/imagenet-vgg-verydeep-19.mat")
print(model)
```
The model is stored in a python dictionary where each variable name is the key and the corresponding value is a tensor containing that variable's value. To run an image through this network, you just have to feed the image to the model. In TensorFlow, you can do so using the [tf.assign](https://www.tensorflow.org/api_docs/python/tf/assign) function. In particular, you will use the assign function like this:
```python
model["input"].assign(image)
```
This assigns the image as an input to the model. After this, if you want to access the activations of a particular layer, say layer `4_2` when the network is run on this image, you would run a TensorFlow session on the correct tensor `conv4_2`, as follows:
```python
sess.run(model["conv4_2"])
```
## 3 - Neural Style Transfer
We will build the NST algorithm in three steps:
- Build the content cost function $J_{content}(C,G)$
- Build the style cost function $J_{style}(S,G)$
- Put it together to get $J(G) = \alpha J_{content}(C,G) + \beta J_{style}(S,G)$.
### 3.1 - Computing the content cost
In our running example, the content image C will be the picture of the Louvre Museum in Paris. Run the code below to see a picture of the Louvre.
```
content_image = scipy.misc.imread("images/louvre.jpg")
imshow(content_image)
```
The content image (C) shows the Louvre museum's pyramid surrounded by old Paris buildings, against a sunny sky with a few clouds.
** 3.1.1 - How do you ensure the generated image G matches the content of the image C?**
As we saw in lecture, the earlier (shallower) layers of a ConvNet tend to detect lower-level features such as edges and simple textures, and the later (deeper) layers tend to detect higher-level features such as more complex textures as well as object classes.
We would like the "generated" image G to have similar content as the input image C. Suppose you have chosen some layer's activations to represent the content of an image. In practice, you'll get the most visually pleasing results if you choose a layer in the middle of the network--neither too shallow nor too deep. (After you have finished this exercise, feel free to come back and experiment with using different layers, to see how the results vary.)
So, suppose you have picked one particular hidden layer to use. Now, set the image C as the input to the pretrained VGG network, and run forward propagation. Let $a^{(C)}$ be the hidden layer activations in the layer you had chosen. (In lecture, we had written this as $a^{[l](C)}$, but here we'll drop the superscript $[l]$ to simplify the notation.) This will be a $n_H \times n_W \times n_C$ tensor. Repeat this process with the image G: Set G as the input, and run forward progation. Let $$a^{(G)}$$ be the corresponding hidden layer activation. We will define as the content cost function as:
$$J_{content}(C,G) = \frac{1}{4 \times n_H \times n_W \times n_C}\sum _{ \text{all entries}} (a^{(C)} - a^{(G)})^2\tag{1} $$
Here, $n_H, n_W$ and $n_C$ are the height, width and number of channels of the hidden layer you have chosen, and appear in a normalization term in the cost. For clarity, note that $a^{(C)}$ and $a^{(G)}$ are the volumes corresponding to a hidden layer's activations. In order to compute the cost $J_{content}(C,G)$, it might also be convenient to unroll these 3D volumes into a 2D matrix, as shown below. (Technically this unrolling step isn't needed to compute $J_{content}$, but it will be good practice for when you do need to carry out a similar operation later for computing the style const $J_{style}$.)
<img src="images/NST_LOSS.png" style="width:800px;height:400px;">
**Exercise:** Compute the "content cost" using TensorFlow.
**Instructions**: The 3 steps to implement this function are:
1. Retrieve dimensions from a_G:
- To retrieve dimensions from a tensor X, use: `X.get_shape().as_list()`
2. Unroll a_C and a_G as explained in the picture above
- If you are stuck, take a look at [Hint1](https://www.tensorflow.org/versions/r1.3/api_docs/python/tf/transpose) and [Hint2](https://www.tensorflow.org/versions/r1.2/api_docs/python/tf/reshape).
3. Compute the content cost:
- If you are stuck, take a look at [Hint3](https://www.tensorflow.org/api_docs/python/tf/reduce_sum), [Hint4](https://www.tensorflow.org/api_docs/python/tf/square) and [Hint5](https://www.tensorflow.org/api_docs/python/tf/subtract).
```
# GRADED FUNCTION: compute_content_cost
def compute_content_cost(a_C, a_G):
"""
Computes the content cost
Arguments:
a_C -- tensor of dimension (1, n_H, n_W, n_C),
hidden layer activations representing content of the image C
a_G -- tensor of dimension (1, n_H, n_W, n_C),
hidden layer activations representing content of the image G
Returns:
J_content -- scalar that you compute using equation 1 above.
"""
### START CODE HERE ###
# Retrieve dimensions from a_G (≈1 line)
m, n_H, n_W, n_C = a_G.get_shape().as_list()
# Reshape a_C and a_G (≈2 lines)
a_C_unrolled = tf.transpose(tf.reshape(a_C, [-1]))
a_G_unrolled = tf.transpose(tf.reshape(a_G, [-1]))
# compute the cost with tensorflow (≈1 line)
J_content = tf.reduce_sum((a_C_unrolled - a_G_unrolled)**2)
/ (4 * n_H * n_W * n_C)
#J_content = tf.reduce_sum(tf.square(tf.subtract(a_C_unrolled,
# a_G_unrolled)))/ (4*n_H*n_W*n_C)
### END CODE HERE ###
return J_content
tf.reset_default_graph()
with tf.Session() as test:
tf.set_random_seed(1)
a_C = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)
a_G = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)
J_content = compute_content_cost(a_C, a_G)
print("J_content = " + str(J_content.eval()))
```
**Expected Output**:
<table>
<tr>
<td>
**J_content**
</td>
<td>
6.76559
</td>
</tr>
</table>
<font color='blue'>
**What you should remember**:
- The content cost takes a hidden layer activation of the neural network, and measures how different $a^{(C)}$ and $a^{(G)}$ are.
- When we minimize the content cost later, this will help make sure $G$ has similar content as $C$.
### 3.2 - Computing the style cost
For our running example, we will use the following style image:
```
style_image = scipy.misc.imread("images/monet_800600.jpg")
imshow(style_image)
```
This painting was painted in the style of *[impressionism](https://en.wikipedia.org/wiki/Impressionism)*.
Lets see how you can now define a "style" const function $J_{style}(S,G)$.
### 3.2.1 - Style matrix
The style matrix is also called a "Gram matrix." In linear algebra, the Gram matrix G of a set of vectors $(v_{1},\dots ,v_{n})$ is the matrix of dot products, whose entries are ${\displaystyle G_{ij} = v_{i}^T v_{j} = np.dot(v_{i}, v_{j}) }$. In other words, $G_{ij}$ compares how similar $v_i$ is to $v_j$: If they are highly similar, you would expect them to have a large dot product, and thus for $G_{ij}$ to be large.
Note that there is an unfortunate collision in the variable names used here. We are following common terminology used in the literature, but $G$ is used to denote the Style matrix (or Gram matrix) as well as to denote the generated image $G$. We will try to make sure which $G$ we are referring to is always clear from the context.
In NST, you can compute the Style matrix by multiplying the "unrolled" filter matrix with their transpose:
<img src="images/NST_GM.png" style="width:900px;height:300px;">
The result is a matrix of dimension $(n_C,n_C)$ where $n_C$ is the number of filters. The value $G_{ij}$ measures how similar the activations of filter $i$ are to the activations of filter $j$.
One important part of the gram matrix is that the diagonal elements such as $G_{ii}$ also measures how active filter $i$ is. For example, suppose filter $i$ is detecting vertical textures in the image. Then $G_{ii}$ measures how common vertical textures are in the image as a whole: If $G_{ii}$ is large, this means that the image has a lot of vertical texture.
By capturing the prevalence of different types of features ($G_{ii}$), as well as how much different features occur together ($G_{ij}$), the Style matrix $G$ measures the style of an image.
**Exercise**:
Using TensorFlow, implement a function that computes the Gram matrix of a matrix A. The formula is: The gram matrix of A is $G_A = AA^T$. If you are stuck, take a look at [Hint 1](https://www.tensorflow.org/api_docs/python/tf/matmul) and [Hint 2](https://www.tensorflow.org/api_docs/python/tf/transpose).
```
# GRADED FUNCTION: gram_matrix
def gram_matrix(A):
"""
Argument:
A -- matrix of shape (n_C, n_H*n_W)
Returns:
GA -- Gram matrix of A, of shape (n_C, n_C)
"""
### START CODE HERE ### (≈1 line)
GA = tf.matmul(A, tf.transpose(A))
### END CODE HERE ###
return GA
tf.reset_default_graph()
with tf.Session() as test:
tf.set_random_seed(1)
A = tf.random_normal([3, 2*1], mean=1, stddev=4)
GA = gram_matrix(A)
print("GA = " + str(GA.eval()))
```
**Expected Output**:
<table>
<tr>
<td>
**GA**
</td>
<td>
[[ 6.42230511 -4.42912197 -2.09668207] <br>
[ -4.42912197 19.46583748 19.56387138] <br>
[ -2.09668207 19.56387138 20.6864624 ]]
</td>
</tr>
</table>
### 3.2.2 - Style cost
After generating the Style matrix (Gram matrix), your goal will be to minimize the distance between the Gram matrix of the "style" image S and that of the "generated" image G. For now, we are using only a single hidden layer $a^{[l]}$, and the corresponding style cost for this layer is defined as:
$$J_{style}^{[l]}(S,G) = \frac{1}{4 \times {n_C}^2 \times (n_H \times n_W)^2} \sum _{i=1}^{n_C}\sum_{j=1}^{n_C}(G^{(S)}_{ij} - G^{(G)}_{ij})^2\tag{2} $$
where $G^{(S)}$ and $G^{(G)}$ are respectively the Gram matrices of the "style" image and the "generated" image, computed using the hidden layer activations for a particular hidden layer in the network.
**Exercise**: Compute the style cost for a single layer.
**Instructions**: The 3 steps to implement this function are:
1. Retrieve dimensions from the hidden layer activations a_G:
- To retrieve dimensions from a tensor X, use: `X.get_shape().as_list()`
2. Unroll the hidden layer activations a_S and a_G into 2D matrices, as explained in the picture above.
- You may find [Hint1](https://www.tensorflow.org/versions/r1.3/api_docs/python/tf/transpose) and [Hint2](https://www.tensorflow.org/versions/r1.2/api_docs/python/tf/reshape) useful.
3. Compute the Style matrix of the images S and G. (Use the function you had previously written.)
4. Compute the Style cost:
- You may find [Hint3](https://www.tensorflow.org/api_docs/python/tf/reduce_sum), [Hint4](https://www.tensorflow.org/api_docs/python/tf/square) and [Hint5](https://www.tensorflow.org/api_docs/python/tf/subtract) useful.
```
# GRADED FUNCTION: compute_layer_style_cost
def compute_layer_style_cost(a_S, a_G):
"""
Arguments:
a_S -- tensor of dimension (1, n_H, n_W, n_C),
hidden layer activations representing style of the image S
a_G -- tensor of dimension (1, n_H, n_W, n_C),
hidden layer activations representing style of the image G
Returns:
J_style_layer -- tensor representing a scalar value,
style cost defined above by equation (2)
"""
### START CODE HERE ###
# Retrieve dimensions from a_G (≈1 line)
m, n_H, n_W, n_C = a_G.get_shape().as_list()
# Reshape the images to have them of shape (n_C, n_H*n_W) (≈2 lines)
#a_S = tf.reshape(a_S, [n_C, n_H * n_W])
#a_G = tf.reshape(a_G, [n_C, n_H * n_W])
a_S = tf.transpose(tf.reshape(a_S, [n_H*n_W, n_C]))
a_G = tf.transpose(tf.reshape(a_G, [n_H*n_W, n_C]))
# Computing gram_matrices for both images S and G (≈2 lines)
GS = gram_matrix(a_S)
GG = gram_matrix(a_G)
# Computing the loss (≈1 line)
J_style_layer = tf.reduce_sum(tf.square((GS - GG)))
/ (4 * n_C**2 * (n_W * n_H)**2)
### END CODE HERE ###
return J_style_layer
tf.reset_default_graph()
with tf.Session() as test:
tf.set_random_seed(1)
a_S = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)
a_G = tf.random_normal([1, 4, 4, 3], mean=1, stddev=4)
J_style_layer = compute_layer_style_cost(a_S, a_G)
print("J_style_layer = " + str(J_style_layer.eval()))
```
**Expected Output**:
<table>
<tr>
<td>
**J_style_layer**
</td>
<td>
9.19028
</td>
</tr>
</table>
### 3.2.3 Style Weights
So far you have captured the style from only one layer. We'll get better results if we "merge" style costs from several different layers. After completing this exercise, feel free to come back and experiment with different weights to see how it changes the generated image $G$. But for now, this is a pretty reasonable default:
```
STYLE_LAYERS = [
('conv1_1', 0.2),
('conv2_1', 0.2),
('conv3_1', 0.2),
('conv4_1', 0.2),
('conv5_1', 0.2)]
```
You can combine the style costs for different layers as follows:
$$J_{style}(S,G) = \sum_{l} \lambda^{[l]} J^{[l]}_{style}(S,G)$$
where the values for $\lambda^{[l]}$ are given in `STYLE_LAYERS`.
We've implemented a compute_style_cost(...) function. It simply calls your `compute_layer_style_cost(...)` several times, and weights their results using the values in `STYLE_LAYERS`. Read over it to make sure you understand what it's doing.
<!--
2. Loop over (layer_name, coeff) from STYLE_LAYERS:
a. Select the output tensor of the current layer. As an example, to call the tensor from the "conv1_1" layer you would do: out = model["conv1_1"]
b. Get the style of the style image from the current layer by running the session on the tensor "out"
c. Get a tensor representing the style of the generated image from the current layer. It is just "out".
d. Now that you have both styles. Use the function you've implemented above to compute the style_cost for the current layer
e. Add (style_cost x coeff) of the current layer to overall style cost (J_style)
3. Return J_style, which should now be the sum of the (style_cost x coeff) for each layer.
!-->
```
def compute_style_cost(model, STYLE_LAYERS):
"""
Computes the overall style cost from several chosen layers
Arguments:
model -- our tensorflow model
STYLE_LAYERS -- A python list containing:
- the names of the layers we would like
to extract style from
- a coefficient for each of them
Returns:
J_style -- tensor representing a scalar value, style cost
defined above by equation (2)
"""
# initialize the overall style cost
J_style = 0
for layer_name, coeff in STYLE_LAYERS:
# Select the output tensor of the currently selected layer
out = model[layer_name]
# Set a_S to be the hidden layer activation from the layer
# we have selected, by running the session on out
a_S = sess.run(out)
# Set a_G to be the hidden layer activation from same layer.
# Here, a_G references model[layer_name]
# and isn't evaluated yet. Later in the code, we'll assign
# the image G as the model input, so that
# when we run the session, this will be the activations
# drawn from the appropriate layer, with G as input.
a_G = out
# Compute style_cost for the current layer
J_style_layer = compute_layer_style_cost(a_S, a_G)
# Add coeff * J_style_layer of this layer to overall style cost
J_style += coeff * J_style_layer
return J_style
```
**Note**: In the inner-loop of the for-loop above, `a_G` is a tensor and hasn't been evaluated yet. It will be evaluated and updated at each iteration when we run the TensorFlow graph in model_nn() below.
<!--
How do you choose the coefficients for each layer? The deeper layers capture higher-level concepts, and the features in the deeper layers are less localized in the image relative to each other. So if you want the generated image to softly follow the style image, try choosing larger weights for deeper layers and smaller weights for the first layers. In contrast, if you want the generated image to strongly follow the style image, try choosing smaller weights for deeper layers and larger weights for the first layers
!-->
<font color='blue'>
**What you should remember**:
- The style of an image can be represented using the Gram matrix of a hidden layer's activations. However, we get even better results combining this representation from multiple different layers. This is in contrast to the content representation, where usually using just a single hidden layer is sufficient.
- Minimizing the style cost will cause the image $G$ to follow the style of the image $S$.
</font color='blue'>
### 3.3 - Defining the total cost to optimize
Finally, let's create a cost function that minimizes both the style and the content cost. The formula is:
$$J(G) = \alpha J_{content}(C,G) + \beta J_{style}(S,G)$$
**Exercise**: Implement the total cost function which includes both the content cost and the style cost.
```
# GRADED FUNCTION: total_cost
def total_cost(J_content, J_style, alpha = 10, beta = 40):
"""
Computes the total cost function
Arguments:
J_content -- content cost coded above
J_style -- style cost coded above
alpha -- hyperparameter weighting the importance of the content cost
beta -- hyperparameter weighting the importance of the style cost
Returns:
J -- total cost as defined by the formula above.
"""
### START CODE HERE ### (≈1 line)
J = alpha * J_content + beta * J_style
### END CODE HERE ###
return J
tf.reset_default_graph()
with tf.Session() as test:
np.random.seed(3)
J_content = np.random.randn()
J_style = np.random.randn()
J = total_cost(J_content, J_style)
print("J = " + str(J))
```
**Expected Output**:
<table>
<tr>
<td>
**J**
</td>
<td>
35.34667875478276
</td>
</tr>
</table>
<font color='blue'>
**What you should remember**:
- The total cost is a linear combination of the content cost $J_{content}(C,G)$ and the style cost $J_{style}(S,G)$
- $\alpha$ and $\beta$ are hyperparameters that control the relative weighting between content and style
## 4 - Solving the optimization problem
Finally, let's put everything together to implement Neural Style Transfer!
Here's what the program will have to do:
<font color='purple'>
1. Create an Interactive Session
2. Load the content image
3. Load the style image
4. Randomly initialize the image to be generated
5. Load the VGG16 model
7. Build the TensorFlow graph:
- Run the content image through the VGG16 model and compute the content cost
- Run the style image through the VGG16 model and compute the style cost
- Compute the total cost
- Define the optimizer and the learning rate
8. Initialize the TensorFlow graph and run it for a large number of iterations, updating the generated image at every step.
</font>
Lets go through the individual steps in detail.
You've previously implemented the overall cost $J(G)$. We'll now set up TensorFlow to optimize this with respect to $G$. To do so, your program has to reset the graph and use an "[Interactive Session](https://www.tensorflow.org/api_docs/python/tf/InteractiveSession)". Unlike a regular session, the "Interactive Session" installs itself as the default session to build a graph. This allows you to run variables without constantly needing to refer to the session object, which simplifies the code.
Lets start the interactive session.
```
# Reset the graph
tf.reset_default_graph()
# Start interactive session
sess = tf.InteractiveSession()
```
Let's load, reshape, and normalize our "content" image (the Louvre museum picture):
```
content_image = scipy.misc.imread("images/louvre_small.jpg")
content_image = reshape_and_normalize_image(content_image)
```
Let's load, reshape and normalize our "style" image (Claude Monet's painting):
```
style_image = scipy.misc.imread("images/monet.jpg")
style_image = reshape_and_normalize_image(style_image)
```
Now, we initialize the "generated" image as a noisy image created from the content_image. By initializing the pixels of the generated image to be mostly noise but still slightly correlated with the content image, this will help the content of the "generated" image more rapidly match the content of the "content" image. (Feel free to look in `nst_utils.py` to see the details of `generate_noise_image(...)`; to do so, click "File-->Open..." at the upper-left corner of this Jupyter notebook.)
```
generated_image = generate_noise_image(content_image)
imshow(generated_image[0])
```
Next, as explained in part (2), let's load the VGG16 model.
```
model = load_vgg_model("pretrained-model/imagenet-vgg-verydeep-19.mat")
```
To get the program to compute the content cost, we will now assign `a_C` and `a_G` to be the appropriate hidden layer activations. We will use layer `conv4_2` to compute the content cost. The code below does the following:
1. Assign the content image to be the input to the VGG model.
2. Set a_C to be the tensor giving the hidden layer activation for layer "conv4_2".
3. Set a_G to be the tensor giving the hidden layer activation for the same layer.
4. Compute the content cost using a_C and a_G.
```
# Assign the content image to be the input of the VGG model.
sess.run(model['input'].assign(content_image))
# Select the output tensor of layer conv4_2
out = model['conv4_2']
# Set a_C to be the hidden layer activation from the layer we have selected
a_C = sess.run(out)
# Set a_G to be the hidden layer activation from same layer.
# Here, a_G references model['conv4_2']
# and isn't evaluated yet. Later in the code, we'll assign
# the image G as the model input, so that
# when we run the session, this will be the activations
# drawn from the appropriate layer, with G as input.
a_G = out
# Compute the content cost
J_content = compute_content_cost(a_C, a_G)
```
**Note**: At this point, a_G is a tensor and hasn't been evaluated. It will be evaluated and updated at each iteration when we run the Tensorflow graph in model_nn() below.
```
# Assign the input of the model to be the "style" image
sess.run(model['input'].assign(style_image))
# Compute the style cost
J_style = compute_style_cost(model, STYLE_LAYERS)
```
**Exercise**: Now that you have J_content and J_style, compute the total cost J by calling `total_cost()`. Use `alpha = 10` and `beta = 40`.
```
### START CODE HERE ### (1 line)
J = total_cost(J_content, J_style, alpha = 10, beta = 40)
### END CODE HERE ###
```
You'd previously learned how to set up the Adam optimizer in TensorFlow. Lets do that here, using a learning rate of 2.0. [See reference](https://www.tensorflow.org/api_docs/python/tf/train/AdamOptimizer)
```
# define optimizer (1 line)
optimizer = tf.train.AdamOptimizer(2.0)
# define train_step (1 line)
train_step = optimizer.minimize(J)
```
**Exercise**: Implement the model_nn() function which initializes the variables of the tensorflow graph, assigns the input image (initial generated image) as the input of the VGG16 model and runs the train_step for a large number of steps.
```
def model_nn(sess, input_image, num_iterations = 200):
# Initialize global variables (you need to run
# the session on the initializer)
### START CODE HERE ### (1 line)
sess.run(tf.global_variables_initializer())
### END CODE HERE ###
# Run the noisy input image (initial generated image)
# through the model. Use assign().
### START CODE HERE ### (1 line)
sess.run(model['input'].assign(input_image))
### END CODE HERE ###
for i in range(num_iterations):
# Run the session on the train_step to minimize the total cost
### START CODE HERE ### (1 line)
_ = sess.run(train_step)
### END CODE HERE ###
# Compute the generated image by running the session
# on the current model['input']
### START CODE HERE ### (1 line)
generated_image = sess.run(model['input'])
### END CODE HERE ###
# Print every 20 iteration.
if i%20 == 0:
Jt, Jc, Js = sess.run([J, J_content, J_style])
print("Iteration " + str(i) + " :")
print("total cost = " + str(Jt))
print("content cost = " + str(Jc))
print("style cost = " + str(Js))
# save current generated image in the "/output" directory
save_image("output/" + str(i) + ".png", generated_image)
# save last generated image
save_image('output/generated_image.jpg', generated_image)
return generated_image
```
Run the following cell to generate an artistic image. It should take about 3min on CPU for every 20 iterations but you start observing attractive results after ≈140 iterations. Neural Style Transfer is generally trained using GPUs.
```
model_nn(sess, generated_image)
```
**Expected Output**:
<table>
<tr>
<td>
**Iteration 0 : **
</td>
<td>
total cost = 5.05035e+09 <br>
content cost = 7877.67 <br>
style cost = 1.26257e+08
</td>
</tr>
</table>
You're done! After running this, in the upper bar of the notebook click on "File" and then "Open". Go to the "/output" directory to see all the saved images. Open "generated_image" to see the generated image! :)
You should see something the image presented below on the right:
<img src="images/louvre_generated.png" style="width:800px;height:300px;">
We didn't want you to wait too long to see an initial result, and so had set the hyperparameters accordingly. To get the best looking results, running the optimization algorithm longer (and perhaps with a smaller learning rate) might work better. After completing and submitting this assignment, we encourage you to come back and play more with this notebook, and see if you can generate even better looking images.
Here are few other examples:
- The beautiful ruins of the ancient city of Persepolis (Iran) with the style of Van Gogh (The Starry Night)
<img src="images/perspolis_vangogh.png" style="width:750px;height:300px;">
- The tomb of Cyrus the great in Pasargadae with the style of a Ceramic Kashi from Ispahan.
<img src="images/pasargad_kashi.png" style="width:750px;height:300px;">
- A scientific study of a turbulent fluid with the style of a abstract blue fluid painting.
<img src="images/circle_abstract.png" style="width:750px;height:300px;">
## 5 - Test with your own image (Optional/Ungraded)
Finally, you can also rerun the algorithm on your own images!
To do so, go back to part 4 and change the content image and style image with your own pictures. In detail, here's what you should do:
1. Click on "File -> Open" in the upper tab of the notebook
2. Go to "/images" and upload your images (requirement: (WIDTH = 300, HEIGHT = 225)), rename them "my_content.png" and "my_style.png" for example.
3. Change the code in part (3.4) from :
```python
content_image = scipy.misc.imread("images/louvre.jpg")
style_image = scipy.misc.imread("images/claude-monet.jpg")
```
to:
```python
content_image = scipy.misc.imread("images/my_content.jpg")
style_image = scipy.misc.imread("images/my_style.jpg")
```
4. Rerun the cells (you may need to restart the Kernel in the upper tab of the notebook).
You can also tune your hyperparameters:
- Which layers are responsible for representing the style? STYLE_LAYERS
- How many iterations do you want to run the algorithm? num_iterations
- What is the relative weighting between content and style? alpha/beta
## 6 - Conclusion
Great job on completing this assignment! You are now able to use Neural Style Transfer to generate artistic images. This is also your first time building a model in which the optimization algorithm updates the pixel values rather than the neural network's parameters. Deep learning has many different types of models and this is only one of them!
<font color='blue'>
What you should remember:
- Neural Style Transfer is an algorithm that given a content image C and a style image S can generate an artistic image
- It uses representations (hidden layer activations) based on a pretrained ConvNet.
- The content cost function is computed using one hidden layer's activations.
- The style cost function for one layer is computed using the Gram matrix of that layer's activations. The overall style cost function is obtained using several hidden layers.
- Optimizing the total cost function results in synthesizing new images.
This was the final programming exercise of this course. Congratulations--you've finished all the programming exercises of this course on Convolutional Networks! We hope to also see you in Course 5, on Sequence models!
### References:
The Neural Style Transfer algorithm was due to Gatys et al. (2015). Harish Narayanan and Github user "log0" also have highly readable write-ups from which we drew inspiration. The pre-trained network used in this implementation is a VGG network, which is due to Simonyan and Zisserman (2015). Pre-trained weights were from the work of the MathConvNet team.
- Leon A. Gatys, Alexander S. Ecker, Matthias Bethge, (2015). A Neural Algorithm of Artistic Style (https://arxiv.org/abs/1508.06576)
- Harish Narayanan, Convolutional neural networks for artistic style transfer. https://harishnarayanan.org/writing/artistic-style-transfer/
- Log0, TensorFlow Implementation of "A Neural Algorithm of Artistic Style". http://www.chioka.in/tensorflow-implementation-neural-algorithm-of-artistic-style
- Karen Simonyan and Andrew Zisserman (2015). Very deep convolutional networks for large-scale image recognition (https://arxiv.org/pdf/1409.1556.pdf)
- MatConvNet. http://www.vlfeat.org/matconvnet/pretrained/
| github_jupyter |
# Predicting Student Admissions with Neural Networks
In this notebook, we predict student admissions to graduate school at UCLA based on three pieces of data:
- GRE Scores (Test)
- GPA Scores (Grades)
- Class rank (1-4)
The dataset originally came from here: http://www.ats.ucla.edu/
## Loading the data
To load the data and format it nicely, we will use two very useful packages called Pandas and Numpy. You can read on the documentation here:
- https://pandas.pydata.org/pandas-docs/stable/
- https://docs.scipy.org/
```
# Importing pandas and numpy
import pandas as pd
import numpy as np
# Reading the csv file into a pandas DataFrame
data = pd.read_csv('student_data.csv')
# Printing out the first 10 rows of our data
data[:10]
```
## Plotting the data
First let's make a plot of our data to see how it looks. In order to have a 2D plot, let's ingore the rank.
```
# Importing matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
# Function to help us plot
def plot_points(data):
X = np.array(data[["gre","gpa"]])
y = np.array(data["admit"])
admitted = X[np.argwhere(y==1)]
rejected = X[np.argwhere(y==0)]
plt.scatter([s[0][0] for s in rejected], [s[0][1] for s in rejected], s = 25, color = 'red', edgecolor = 'k')
plt.scatter([s[0][0] for s in admitted], [s[0][1] for s in admitted], s = 25, color = 'cyan', edgecolor = 'k')
plt.xlabel('Test (GRE)')
plt.ylabel('Grades (GPA)')
# Plotting the points
plot_points(data)
plt.show()
```
Roughly, it looks like the students with high scores in the grades and test passed, while the ones with low scores didn't, but the data is not as nicely separable as we hoped it would. Maybe it would help to take the rank into account? Let's make 4 plots, each one for each rank.
```
# Separating the ranks
data_rank1 = data[data["rank"]==1]
data_rank2 = data[data["rank"]==2]
data_rank3 = data[data["rank"]==3]
data_rank4 = data[data["rank"]==4]
# Plotting the graphs
plot_points(data_rank1)
plt.title("Rank 1")
plt.show()
plot_points(data_rank2)
plt.title("Rank 2")
plt.show()
plot_points(data_rank3)
plt.title("Rank 3")
plt.show()
plot_points(data_rank4)
plt.title("Rank 4")
plt.show()
```
This looks more promising, as it seems that the lower the rank, the higher the acceptance rate. Let's use the rank as one of our inputs. In order to do this, we should one-hot encode it.
## TODO: One-hot encoding the rank
Use the `get_dummies` function in pandas in order to one-hot encode the data.
Hint: To drop a column, it's suggested that you use `one_hot_data`[.drop( )](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.drop.html).
```
# TODO: Make dummy variables for rank
one_hot_data = pd.concat([data, pd.get_dummies(data['rank'], prefix = 'rank_')], axis = 1)
# TODO: Drop the previous rank column
one_hot_data = one_hot_data.drop(['rank'], axis = 1)
# Print the first 10 rows of our data
one_hot_data[:10]
```
## TODO: Scaling the data
The next step is to scale the data. We notice that the range for grades is 1.0-4.0, whereas the range for test scores is roughly 200-800, which is much larger. This means our data is skewed, and that makes it hard for a neural network to handle. Let's fit our two features into a range of 0-1, by dividing the grades by 4.0, and the test score by 800.
```
# Making a copy of our data
processed_data = one_hot_data[:]
# TODO: Scale the columns
processed_data['gpa'] = processed_data['gpa'] / 4.0
processed_data['gre'] = processed_data['gre'] / 800
# Printing the first 10 rows of our procesed data
processed_data[:10]
```
## Splitting the data into Training and Testing
In order to test our algorithm, we'll split the data into a Training and a Testing set. The size of the testing set will be 10% of the total data.
```
sample = np.random.choice(processed_data.index, size=int(len(processed_data)*0.9), replace=False)
train_data, test_data = processed_data.iloc[sample], processed_data.drop(sample)
print("Number of training samples is", len(train_data))
print("Number of testing samples is", len(test_data))
print(train_data[:10])
print(test_data[:10])
```
## Splitting the data into features and targets (labels)
Now, as a final step before the training, we'll split the data into features (X) and targets (y).
```
features = train_data.drop('admit', axis = 1)
targets = train_data['admit']
features_test = test_data.drop('admit', axis=1)
targets_test = test_data['admit']
print(features[:10])
print(targets[:10])
```
## Training the 2-layer Neural Network
The following function trains the 2-layer neural network. First, we'll write some helper functions.
```
# Activation (sigmoid) function
def sigmoid(x):
return 1 / (1 + np.exp(-x))
def sigmoid_prime(x):
return sigmoid(x) * (1-sigmoid(x))
def error_formula(y, output):
return - y*np.log(output) - (1 - y) * np.log(1-output)
```
# TODO: Backpropagate the error
Now it's your turn to shine. Write the error term. Remember that this is given by the equation $$ (y-\hat{y}) \sigma'(x) $$
```
# TODO: Write the error term formula
def error_term_formula(x, y, output):
return (y - output) * sigmoid_prime(x)
# Neural Network hyperparameters
epochs = 1000
learnrate = 0.5
# Training function
def train_nn(features, targets, epochs, learnrate):
# Use to same seed to make debugging easier
np.random.seed(42)
n_records, n_features = features.shape
last_loss = None
# Initialize weights
weights = np.random.normal(scale=1 / n_features**.5, size=n_features)
for e in range(epochs):
del_w = np.zeros(weights.shape)
for x, y in zip(features.values, targets):
# Loop through all records, x is the input, y is the target
# Activation of the output unit
# Notice we multiply the inputs and the weights here
# rather than storing h as a separate variable
output = sigmoid(np.dot(x, weights))
# The error, the target minus the network output
error = error_formula(y, output)
# The error term
error_term = error_term_formula(x, y, output)
# The gradient descent step, the error times the gradient times the inputs
del_w += error_term * x
# Update the weights here. The learning rate times the
# change in weights, divided by the number of records to average
weights += learnrate * del_w / n_records
# Printing out the mean square error on the training set
if e % (epochs / 10) == 0:
out = sigmoid(np.dot(features, weights))
loss = np.mean((out - targets) ** 2)
print("Epoch:", e)
if last_loss and last_loss < loss:
print("Train loss: ", loss, " WARNING - Loss Increasing")
else:
print("Train loss: ", loss)
last_loss = loss
print("=========")
print("Finished training!")
return weights
weights = train_nn(features, targets, epochs, learnrate)
```
## Calculating the Accuracy on the Test Data
```
# Calculate accuracy on test data
test_out = sigmoid(np.dot(features_test, weights))
predictions = test_out > 0.5
accuracy = np.mean(predictions == targets_test)
print("Prediction accuracy: {:.3f}".format(accuracy))
```
| github_jupyter |
<a href="https://colab.research.google.com/github/ahvblackwelltech/DS-Unit-2-Kaggle-Challenge/blob/master/module2-random-forests/Ahvi_Blackwell_LS_DS_222_assignment.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Lambda School Data Science
*Unit 2, Sprint 2, Module 2*
---
# Random Forests
## Assignment
- [ ] Read [“Adopting a Hypothesis-Driven Workflow”](https://outline.com/5S5tsB), a blog post by a Lambda DS student about the Tanzania Waterpumps challenge.
- [ ] Continue to participate in our Kaggle challenge.
- [ ] Define a function to wrangle train, validate, and test sets in the same way. Clean outliers and engineer features.
- [ ] Try Ordinal Encoding.
- [ ] Try a Random Forest Classifier.
- [ ] Submit your predictions to our Kaggle competition. (Go to our Kaggle InClass competition webpage. Use the blue **Submit Predictions** button to upload your CSV file. Or you can use the Kaggle API to submit your predictions.)
- [ ] Commit your notebook to your fork of the GitHub repo.
## Stretch Goals
### Doing
- [ ] Add your own stretch goal(s) !
- [ ] Do more exploratory data analysis, data cleaning, feature engineering, and feature selection.
- [ ] Try other [categorical encodings](https://contrib.scikit-learn.org/categorical-encoding/).
- [ ] Get and plot your feature importances.
- [ ] Make visualizations and share on Slack.
### Reading
Top recommendations in _**bold italic:**_
#### Decision Trees
- A Visual Introduction to Machine Learning, [Part 1: A Decision Tree](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/), and _**[Part 2: Bias and Variance](http://www.r2d3.us/visual-intro-to-machine-learning-part-2/)**_
- [Decision Trees: Advantages & Disadvantages](https://christophm.github.io/interpretable-ml-book/tree.html#advantages-2)
- [How a Russian mathematician constructed a decision tree — by hand — to solve a medical problem](http://fastml.com/how-a-russian-mathematician-constructed-a-decision-tree-by-hand-to-solve-a-medical-problem/)
- [How decision trees work](https://brohrer.github.io/how_decision_trees_work.html)
- [Let’s Write a Decision Tree Classifier from Scratch](https://www.youtube.com/watch?v=LDRbO9a6XPU)
#### Random Forests
- [_An Introduction to Statistical Learning_](http://www-bcf.usc.edu/~gareth/ISL/), Chapter 8: Tree-Based Methods
- [Coloring with Random Forests](http://structuringtheunstructured.blogspot.com/2017/11/coloring-with-random-forests.html)
- _**[Random Forests for Complete Beginners: The definitive guide to Random Forests and Decision Trees](https://victorzhou.com/blog/intro-to-random-forests/)**_
#### Categorical encoding for trees
- [Are categorical variables getting lost in your random forests?](https://roamanalytics.com/2016/10/28/are-categorical-variables-getting-lost-in-your-random-forests/)
- [Beyond One-Hot: An Exploration of Categorical Variables](http://www.willmcginnis.com/2015/11/29/beyond-one-hot-an-exploration-of-categorical-variables/)
- _**[Categorical Features and Encoding in Decision Trees](https://medium.com/data-design/visiting-categorical-features-and-encoding-in-decision-trees-53400fa65931)**_
- _**[Coursera — How to Win a Data Science Competition: Learn from Top Kagglers — Concept of mean encoding](https://www.coursera.org/lecture/competitive-data-science/concept-of-mean-encoding-b5Gxv)**_
- [Mean (likelihood) encodings: a comprehensive study](https://www.kaggle.com/vprokopev/mean-likelihood-encodings-a-comprehensive-study)
- [The Mechanics of Machine Learning, Chapter 6: Categorically Speaking](https://mlbook.explained.ai/catvars.html)
#### Imposter Syndrome
- [Effort Shock and Reward Shock (How The Karate Kid Ruined The Modern World)](http://www.tempobook.com/2014/07/09/effort-shock-and-reward-shock/)
- [How to manage impostor syndrome in data science](https://towardsdatascience.com/how-to-manage-impostor-syndrome-in-data-science-ad814809f068)
- ["I am not a real data scientist"](https://brohrer.github.io/imposter_syndrome.html)
- _**[Imposter Syndrome in Data Science](https://caitlinhudon.com/2018/01/19/imposter-syndrome-in-data-science/)**_
### More Categorical Encodings
**1.** The article **[Categorical Features and Encoding in Decision Trees](https://medium.com/data-design/visiting-categorical-features-and-encoding-in-decision-trees-53400fa65931)** mentions 4 encodings:
- **"Categorical Encoding":** This means using the raw categorical values as-is, not encoded. Scikit-learn doesn't support this, but some tree algorithm implementations do. For example, [Catboost](https://catboost.ai/), or R's [rpart](https://cran.r-project.org/web/packages/rpart/index.html) package.
- **Numeric Encoding:** Synonymous with Label Encoding, or "Ordinal" Encoding with random order. We can use [category_encoders.OrdinalEncoder](https://contrib.scikit-learn.org/categorical-encoding/ordinal.html).
- **One-Hot Encoding:** We can use [category_encoders.OneHotEncoder](http://contrib.scikit-learn.org/categorical-encoding/onehot.html).
- **Binary Encoding:** We can use [category_encoders.BinaryEncoder](http://contrib.scikit-learn.org/categorical-encoding/binary.html).
**2.** The short video
**[Coursera — How to Win a Data Science Competition: Learn from Top Kagglers — Concept of mean encoding](https://www.coursera.org/lecture/competitive-data-science/concept-of-mean-encoding-b5Gxv)** introduces an interesting idea: use both X _and_ y to encode categoricals.
Category Encoders has multiple implementations of this general concept:
- [CatBoost Encoder](http://contrib.scikit-learn.org/categorical-encoding/catboost.html)
- [James-Stein Encoder](http://contrib.scikit-learn.org/categorical-encoding/jamesstein.html)
- [Leave One Out](http://contrib.scikit-learn.org/categorical-encoding/leaveoneout.html)
- [M-estimate](http://contrib.scikit-learn.org/categorical-encoding/mestimate.html)
- [Target Encoder](http://contrib.scikit-learn.org/categorical-encoding/targetencoder.html)
- [Weight of Evidence](http://contrib.scikit-learn.org/categorical-encoding/woe.html)
Category Encoder's mean encoding implementations work for regression problems or binary classification problems.
For multi-class classification problems, you will need to temporarily reformulate it as binary classification. For example:
```python
encoder = ce.TargetEncoder(min_samples_leaf=..., smoothing=...) # Both parameters > 1 to avoid overfitting
X_train_encoded = encoder.fit_transform(X_train, y_train=='functional')
X_val_encoded = encoder.transform(X_train, y_val=='functional')
```
For this reason, mean encoding won't work well within pipelines for multi-class classification problems.
**3.** The **[dirty_cat](https://dirty-cat.github.io/stable/)** library has a Target Encoder implementation that works with multi-class classification.
```python
dirty_cat.TargetEncoder(clf_type='multiclass-clf')
```
It also implements an interesting idea called ["Similarity Encoder" for dirty categories](https://www.slideshare.net/GaelVaroquaux/machine-learning-on-non-curated-data-154905090).
However, it seems like dirty_cat doesn't handle missing values or unknown categories as well as category_encoders does. And you may need to use it with one column at a time, instead of with your whole dataframe.
**4. [Embeddings](https://www.kaggle.com/learn/embeddings)** can work well with sparse / high cardinality categoricals.
_**I hope it’s not too frustrating or confusing that there’s not one “canonical” way to encode categoricals. It’s an active area of research and experimentation! Maybe you can make your own contributions!**_
### Setup
You can work locally (follow the [local setup instructions](https://lambdaschool.github.io/ds/unit2/local/)) or on Colab (run the code cell below).
```
%%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge/master/data/'
!pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
import pandas as pd
from sklearn.model_selection import train_test_split
train = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'),
pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))
test = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')
sample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')
train.shape, test.shape
```
# 1. Define a function to wrangle train, validate, and test sets in the same way. Clean outliers and engineer features.
```
import pandas as pd
import numpy as np
%matplotlib inline
# Splitting the train into a train & val
train, val = train_test_split(train, train_size=0.80, test_size=0.02,
stratify=train['status_group'], random_state=42)
def wrangle(X):
X = X.copy()
X['latitude'] = X['latitude'].replace(-2e-08, 0)
cols_with_zeros = ['longitude', 'latitude', 'construction_year',
'gps_height', 'population']
for col in cols_with_zeros:
X[col] = X[col].replace(0, np.nan)
X[col+'_MISSING'] = X[col].isnull()
duplicates = ['quantity_group', 'payment_type']
X = X.drop(columns=duplicates)
unusable_variance = ['recorded_by', 'id']
X = X.drop(columns=unusable_variance)
X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)
X['year_recorded'] = X['date_recorded'].dt.year
X['month_recorded'] = X['date_recorded'].dt.month
X['day_recorded'] = X['date_recorded'].dt.day
X = X.drop(columns='date_recorded')
X['years'] = X['year_recorded'] - X['construction_year']
X['years_MISSING'] = X['years'].isnull()
return X
train = wrangle(train)
val = wrangle(val)
test = wrangle(test)
# The target is status_group
target = 'status_group'
train_features = train.drop(columns=[target])
# Getting list of the numeric features
numeric_features = train_features.select_dtypes(include='number').columns.tolist()
cardinality = train_features.select_dtypes(exclude='number').nunique()
categorical_features = cardinality[cardinality <= 50].index.tolist()
# Combined the lists
features = numeric_features + categorical_features
# Arranging the data into X features matrix & y target vector
X_train = train[features]
y_train = train[target]
X_val = val[features]
y_val = val[target]
X_test = test[features]
# 38 features
print(X_train.shape, X_val.shape)
```
# 2. Try Ordinal Encoding
```
pip install category_encoders
%%time
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.pipeline import make_pipeline
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
SimpleImputer(strategy='mean'),
RandomForestClassifier(n_jobs=-1, random_state=42)
)
pipeline.fit(X_train, y_train)
print('Validation Accuracy:', round(pipeline.score(X_val, y_val)))
encoder = pipeline.named_steps['onehotencoder']
encoded_df = encoder.transform(X_train)
print('X_train shape after encoding', encoded_df.shape)
# Now there are 182 features
%matplotlib inline
import matplotlib.pyplot as plt
# Get feature importances
rf = pipeline.named_steps['randomforestclassifier']
importances = pd.Series(rf.feature_importances_, encoded_df.columns)
n = 25
plt.figure(figsize=(10, n/2))
plt.title(f'Top {n} Features')
importances.sort_values()[-n:].plot.barh(color='grey');
# My Submission CSV
y_pred = pipeline.predict(X_test)
submission = sample_submission.copy()
submission['status_group'] = y_pred
submission.to_csv('ALB_submission_2.csv', index=False)
submission.head()
```
| github_jupyter |
```
# List all NVIDIA GPUs as avaialble in this computer (or Colab's session)
!nvidia-smi -L
import sys
print( f"Python {sys.version}\n" )
import numpy as np
print( f"NumPy {np.__version__}" )
import matplotlib.pyplot as plt
%matplotlib inline
import tensorflow as tf
print( f"TensorFlow {tf.__version__}" )
print( f"tf.keras.backend.image_data_format() = {tf.keras.backend.image_data_format()}" )
# Count the number of GPUs as detected by tensorflow
gpus = tf.config.experimental.list_physical_devices('GPU')
print( f"TensorFlow detected { len(gpus) } GPU(s):" )
for i, gpu in enumerate(gpus):
print( f".... GPU No. {i}: Name = {gpu.name} , Type = {gpu.device_type}" )
```
## 1. Load the pre-trained VGG-16 (only the feature extractor)
```
# Load the ImageNet VGG-16 model, ***excluding*** the latter part regarding the classifier
# Default of input_shape is 224x224x3 for VGG-16
img_w,img_h = 32,32
vgg_extractor = tf.keras.applications.vgg16.VGG16(weights = "imagenet", include_top=False, input_shape = (img_w, img_h, 3))
vgg_extractor.summary()
```
## 2. Extend VGG-16 to match our requirement
```
# Freeze all layers in VGG-16
for i,layer in enumerate(vgg_extractor.layers):
print( f"Layer {i}: name = {layer.name} , trainable = {layer.trainable} => {False}" )
layer.trainable = False # freeze this layer
x = vgg_extractor.output
# Add our custom layer(s) to the end of the existing model
x = tf.keras.layers.Flatten()(x)
x = tf.keras.layers.Dense(1024, activation="relu")(x)
x = tf.keras.layers.Dropout(0.5)(x)
new_outputs = tf.keras.layers.Dense(10, activation="softmax")(x)
# Create the final model
model = tf.keras.models.Model(inputs=vgg_extractor.input, outputs=new_outputs)
model.summary()
```
## 3. Prepare our own dataset
```
# Load CIFAR-10 color image dataset
(x_train , y_train), (x_test , y_test) = tf.keras.datasets.cifar10.load_data()
# Inspect the dataset
print( f"x_train: type={type(x_train)} dtype={x_train.dtype} shape={x_train.shape} max={x_train.max(axis=None)} min={x_train.min(axis=None)}" )
print( f"y_train: type={type(y_train)} dtype={y_train.dtype} shape={y_train.shape} max={max(y_train)} min={min(y_train)}" )
print( f"x_test: type={type(x_test)} dtype={x_test.dtype} shape={x_test.shape} max={x_test.max(axis=None)} min={x_test.min(axis=None)}" )
print( f"y_test: type={type(y_test)} dtype={y_test.dtype} shape={y_test.shape} max={max(y_test)} min={min(y_test)}" )
y_train[0:5]
cifar10_labels = [ 'airplane', 'automobile', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck' ]
# Visualize the first five images in x_train
plt.figure(figsize=(15,5))
for i in range(5):
plt.subplot(150 + 1 + i).set_title( f"class no. {y_train[i]}: {cifar10_labels[ int(y_train[i]) ]}" )
plt.imshow( x_train[i] )
plt.setp( plt.gcf().get_axes(), xticks=[], yticks=[]) # remove all tick marks
plt.show()
# Preprocess CIFAR-10 dataset to match VGG-16's requirements
x_train_vgg = tf.keras.applications.vgg16.preprocess_input(x_train)
x_test_vgg = tf.keras.applications.vgg16.preprocess_input(x_test)
print( x_train_vgg.dtype, x_train_vgg.shape, np.min(x_train_vgg), np.max(x_train_vgg) )
print( x_test_vgg.dtype, x_test_vgg.shape, np.min(x_test_vgg), np.max(x_test_vgg) )
```
## 4. Transfer learning
```
# Set loss function, optimizer and evaluation metric
model.compile( loss="sparse_categorical_crossentropy", optimizer="adam", metrics=["acc"] )
history = model.fit( x_train_vgg, y_train, batch_size=128, epochs=20, verbose=1, validation_data=(x_test_vgg,y_test) )
# Summarize history for accuracy
plt.figure(figsize=(15,5))
plt.plot(history.history['acc'])
plt.plot(history.history['val_acc'])
plt.title('Train accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.grid()
plt.show()
# Summarize history for loss
plt.figure(figsize=(15,5))
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('Train loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.grid()
plt.show()
```
## 5. Evaluate and test the model
```
# Evaluate the trained model on the test set
results = model.evaluate(x_test_vgg, y_test, batch_size=128)
print("test loss, test acc:", results)
# Test using the model on x_test_vgg[0]
i = 0
y_pred = model.predict( x_test_vgg[i].reshape(1,32,32,3) )
plt.imshow( x_test[i] )
plt.title( f"x_test[{i}]: predict=[{np.argmax(y_pred)}]{cifar10_labels[np.argmax(y_pred)]}, true={y_test[i]}{cifar10_labels[int(y_test[i])]}" )
plt.show()
# Test using the model on the first 20 images in x_test
for i in range(20):
y_pred = model.predict( x_test_vgg[i].reshape(1,32,32,3) )
plt.imshow( x_test[i] )
plt.title( f"x_test[{i}]: predict=[{np.argmax(y_pred)}]{cifar10_labels[np.argmax(y_pred)]}, true={y_test[i]}{cifar10_labels[int(y_test[i])]}" )
plt.show()
```
| github_jupyter |
# Imports and Paths
```
import torch
from torch import nn
import torch.nn.functional as F
import torch.optim as optim
import numpy as np
import pandas as pd
import os
import shutil
from skimage import io, transform
import torchvision
import torchvision.transforms as transforms
from torch.utils.data import Dataset, DataLoader
import matplotlib.pyplot as plt
PATH = '/data/msnow/nih_cxr/'
```
# Load the Data
```
df = pd.read_csv(f'{PATH}Data_Entry_2017.csv')
df.shape
df.head()
img_list = os.listdir(f'{PATH}images')
len(img_list)
```
## Collate the data
```
df_pa = df.loc[df.view=='PA',:]
df_pa.reset_index(drop=True, inplace=True)
trn_sz = int(df_pa.shape[0]/2)
df_pa_trn = df_pa.loc[:trn_sz,:]
df_pa_tst = df_pa.loc[trn_sz:,:]
df_pa_tst.shape
pneumo = []
for i,v in df_pa_trn.iterrows():
if "pneumo" in v['labels'].lower():
pneumo.append('pneumo')
else:
pneumo.append('no pneumo')
df_pa_trn['pneumo'] = pneumo
pneumo = []
for i,v in df_pa_tst.iterrows():
if "pneumo" in v['labels'].lower():
pneumo.append(pneumo)
else:
pneumo.append('no pneumo')
df_pa_tst['pneumo'] = pneumo
df_pa_trn.shape
```
Copy images to train and test folders
```
# dst = os.path.join(PATH,'trn')
# src = os.path.join(PATH,'images')
# for i,v in df_pa_trn.iterrows():
# src2 = os.path.join(src,v.image)
# shutil.copy2(src2,dst)
# dst = os.path.join(PATH,'tst')
# src = os.path.join(PATH,'images')
# for i,v in df_pa_tst.iterrows():
# src2 = os.path.join(src,v.image)
# shutil.copy2(src2,dst)
```
# Create the Dataset and Dataloader
```
class TDataset(Dataset):
def __init__(self, df, root_dir, transform=None):
"""
Args:
df (dataframe): df with all the annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
# self.landmarks_frame = pd.read_csv(csv_file)
self.df = df
self.root_dir = root_dir
self.transform = transform
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
img_name = os.path.join(self.root_dir,self.df.image[idx])
image = io.imread(img_name)
categ = self.df.pneumo[idx]
return image, categ
aa = trainset[0]
trainset = TDataset(df_pa_trn,f'{PATH}trn')
testset = TDataset(df_pa_tst,f'{PATH}tst')
trainloader = DataLoader(trainset, batch_size=4,shuffle=True, num_workers=4)
testloader = DataLoader(testset, batch_size=4,shuffle=False, num_workers=4)
aa[0].shape
```
# Define and train a CNN
```
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
# 1 input image channel, 6 output channels, 5x5 square convolution kernel
self.conv1 = nn.Conv2d(1, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
# 6 input image channel, 16 output channels, 5x5 square convolution kernel
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
print(f'input shape {x.shape}')
x = self.pool(F.relu(self.conv1(x)))
print(f'Lin (1,6,5) + relu + pool shape {x.shape}')
x = self.pool(F.relu(self.conv2(x)))
print(f'Lin (6,16,5) + relu + pool shape {x.shape}')
x = x.view(x.shape[0],-1)
print(f'reshape shape {x.shape}')
# x = x.view(-1, 16 * 5 * 5)
x = F.relu(self.fc1(x))
# x = F.relu(self.fc2(x))
# x = self.fc3(x)
return x
net = Net()
input = torch.randn(1, 1, 1024,1024)
out = net(input)
# print(out)
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
for i, data in enumerate(trainloader, 0):
break
inputs, labels = data
tst = inputs.view(-1,1,1024,1024)
tst = tst.type('torch.FloatTensor')
out = net(tst)
16*253*253
tst.shape
# tst = inputs[:,None,:,:]
tst.type(torch.FloatTensor)
type(tst)
list(net.parameters())[0].size()
net(tst)
conv1_tst(tst)
for epoch in range(2): # loop over the dataset multiple times
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# get the inputs
inputs, labels = data
# zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
# print statistics
running_loss += loss.item()
if i % 2000 == 1999: # print every 2000 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / 2000))
running_loss = 0.0
print('Finished Training')
```
| github_jupyter |
```
%matplotlib inline
```
# Compute LCMV inverse solution on evoked data in volume source space
Compute LCMV inverse solution on an auditory evoked dataset in a volume source
space. It stores the solution in a nifti file for visualisation e.g. with
Freeview.
```
# Author: Alexandre Gramfort <[email protected]>
#
# License: BSD (3-clause)
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.datasets import sample
from mne.beamformer import lcmv
from nilearn.plotting import plot_stat_map
from nilearn.image import index_img
print(__doc__)
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_raw-eve.fif'
fname_fwd = data_path + '/MEG/sample/sample_audvis-meg-vol-7-fwd.fif'
```
Get epochs
```
event_id, tmin, tmax = 1, -0.2, 0.5
# Setup for reading the raw data
raw = mne.io.read_raw_fif(raw_fname, preload=True, proj=True)
raw.info['bads'] = ['MEG 2443', 'EEG 053'] # 2 bads channels
events = mne.read_events(event_fname)
# Set up pick list: EEG + MEG - bad channels (modify to your needs)
left_temporal_channels = mne.read_selection('Left-temporal')
picks = mne.pick_types(raw.info, meg=True, eeg=False, stim=True, eog=True,
exclude='bads', selection=left_temporal_channels)
# Pick the channels of interest
raw.pick_channels([raw.ch_names[pick] for pick in picks])
# Re-normalize our empty-room projectors, so they are fine after subselection
raw.info.normalize_proj()
# Read epochs
proj = False # already applied
epochs = mne.Epochs(raw, events, event_id, tmin, tmax,
baseline=(None, 0), preload=True, proj=proj,
reject=dict(grad=4000e-13, mag=4e-12, eog=150e-6))
evoked = epochs.average()
forward = mne.read_forward_solution(fname_fwd)
# Read regularized noise covariance and compute regularized data covariance
noise_cov = mne.compute_covariance(epochs, tmin=tmin, tmax=0, method='shrunk')
data_cov = mne.compute_covariance(epochs, tmin=0.04, tmax=0.15,
method='shrunk')
# Run free orientation (vector) beamformer. Source orientation can be
# restricted by setting pick_ori to 'max-power' (or 'normal' but only when
# using a surface-based source space)
stc = lcmv(evoked, forward, noise_cov, data_cov, reg=0.01, pick_ori=None)
# Save result in stc files
stc.save('lcmv-vol')
stc.crop(0.0, 0.2)
# Save result in a 4D nifti file
img = mne.save_stc_as_volume('lcmv_inverse.nii.gz', stc,
forward['src'], mri_resolution=False)
t1_fname = data_path + '/subjects/sample/mri/T1.mgz'
# Plotting with nilearn ######################################################
plot_stat_map(index_img(img, 61), t1_fname, threshold=0.8,
title='LCMV (t=%.1f s.)' % stc.times[61])
# plot source time courses with the maximum peak amplitudes
plt.figure()
plt.plot(stc.times, stc.data[np.argsort(np.max(stc.data, axis=1))[-40:]].T)
plt.xlabel('Time (ms)')
plt.ylabel('LCMV value')
plt.show()
```
| github_jupyter |
```
#default_exp dataset.dataset
#export
import os
import torch
import transformers
import pandas as pd
import numpy as np
import Hasoc.config as config
#hide
df = pd.read_csv(config.DATA_PATH/'fold_df.csv')
#hide
df.head(2)
#hide
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
le.fit_transform(df.task1)
le.classes_
#hide
df['task1_encoded'] = le.transform(df.task1.values)
#hide
# TOKENIZER = transformers.BertTokenizer.from_pretrained(
# pretrained_model_name_or_path='bert-base-uncased',
# do_lower_case=True,
# # force_download = True,
# )
# MAX_LEN = 72
#export
class BertDataset(torch.utils.data.Dataset):
def __init__(self,text, target=None, is_test=False):
self.text, self.target = text, target
self.tokenizer = config.TOKENIZER
self.max_len = config.MAX_LEN
self.is_test = is_test
def __len__(self):
return len(self.target)
def __getitem__(self, i):
# sanity check
text = ' '.join(self.text[i].split())
# tokenize using Huggingface tokenizers
out = self.tokenizer.encode_plus(text, None,
add_special_tokens=True,
max_length = self.max_len,
truncation=True)
ids = out['input_ids']
mask = out['attention_mask']
token_type_ids = out['token_type_ids']
padding_length = self.max_len - len(ids)
ids = ids + ([0] * padding_length)
mask = mask + ([0] * padding_length)
token_type_ids = token_type_ids + ([0] * padding_length)
if not self.is_test:
return {
'input_ids': torch.tensor(ids, dtype=torch.long),
'attention_mask': torch.tensor(mask, dtype=torch.long),
'token_type_ids': torch.tensor(token_type_ids, dtype=torch.long),
'targets': self.onehot(len(np.unique(self.target)), self.target[i])
}
else:
return{
'input_ids': torch.tensor(ids, dtype=torch.long),
'attention_mask': torch.tensor(mask, dtype=torch.long),
'token_type_ids': torch.tensor(token_type_ids, dtype=torch.long),
}
@staticmethod
def onehot(size, target):
vec = torch.zeros(size, dtype=torch.long)
vec[target] = 1.
return vec
def get_labels(self):
return list(self.target)
#hide
d = BertDataset(df.text.values, df.task1_encoded.values)
#hide
d[10]
c = d[0]['targets']
c.argmax(dim=-1)
```
| github_jupyter |
# Speeding-up gradient-boosting
In this notebook, we present a modified version of gradient boosting which
uses a reduced number of splits when building the different trees. This
algorithm is called "histogram gradient boosting" in scikit-learn.
We previously mentioned that random-forest is an efficient algorithm since
each tree of the ensemble can be fitted at the same time independently.
Therefore, the algorithm scales efficiently with both the number of cores and
the number of samples.
In gradient-boosting, the algorithm is a sequential algorithm. It requires
the `N-1` trees to have been fit to be able to fit the tree at stage `N`.
Therefore, the algorithm is quite computationally expensive. The most
expensive part in this algorithm is the search for the best split in the
tree which is a brute-force approach: all possible split are evaluated and
the best one is picked. We explained this process in the notebook "tree in
depth", which you can refer to.
To accelerate the gradient-boosting algorithm, one could reduce the number of
splits to be evaluated. As a consequence, the statistical performance of such
a tree would be reduced. However, since we are combining several trees in a
gradient-boosting, we can add more estimators to overcome this issue.
We will make a naive implementation of such algorithm using building blocks
from scikit-learn. First, we will load the California housing dataset.
```
from sklearn.datasets import fetch_california_housing
data, target = fetch_california_housing(return_X_y=True, as_frame=True)
target *= 100 # rescale the target in k$
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
We will make a quick benchmark of the original gradient boosting.
```
from sklearn.model_selection import cross_validate
from sklearn.ensemble import GradientBoostingRegressor
gradient_boosting = GradientBoostingRegressor(n_estimators=200)
cv_results_gbdt = cross_validate(gradient_boosting, data, target, n_jobs=-1)
print("Gradient Boosting Decision Tree")
print(f"R2 score via cross-validation: "
f"{cv_results_gbdt['test_score'].mean():.3f} +/- "
f"{cv_results_gbdt['test_score'].std():.3f}")
print(f"Average fit time: "
f"{cv_results_gbdt['fit_time'].mean():.3f} seconds")
print(f"Average score time: "
f"{cv_results_gbdt['score_time'].mean():.3f} seconds")
```
We recall that a way of accelerating the gradient boosting is to reduce the
number of split considered within the tree building. One way is to bin the
data before to give them into the gradient boosting. A transformer called
`KBinsDiscretizer` is doing such transformation. Thus, we can pipeline
this preprocessing with the gradient boosting.
We can first demonstrate the transformation done by the `KBinsDiscretizer`.
```
import numpy as np
from sklearn.preprocessing import KBinsDiscretizer
discretizer = KBinsDiscretizer(
n_bins=256, encode="ordinal", strategy="quantile")
data_trans = discretizer.fit_transform(data)
data_trans
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">The code cell above will generate a couple of warnings. Indeed, for some of
the features, we requested too much bins in regard of the data dispersion
for those features. The smallest bins will be removed.</p>
</div>
We see that the discretizer transforms the original data into an integer.
This integer represents the bin index when the distribution by quantile is
performed. We can check the number of bins per feature.
```
[len(np.unique(col)) for col in data_trans.T]
```
After this transformation, we see that we have at most 256 unique values per
features. Now, we will use this transformer to discretize data before
training the gradient boosting regressor.
```
from sklearn.pipeline import make_pipeline
gradient_boosting = make_pipeline(
discretizer, GradientBoostingRegressor(n_estimators=200))
cv_results_gbdt = cross_validate(gradient_boosting, data, target, n_jobs=-1)
print("Gradient Boosting Decision Tree with KBinsDiscretizer")
print(f"R2 score via cross-validation: "
f"{cv_results_gbdt['test_score'].mean():.3f} +/- "
f"{cv_results_gbdt['test_score'].std():.3f}")
print(f"Average fit time: "
f"{cv_results_gbdt['fit_time'].mean():.3f} seconds")
print(f"Average score time: "
f"{cv_results_gbdt['score_time'].mean():.3f} seconds")
```
Here, we see that the fit time has been drastically reduced but that the
statistical performance of the model is identical. Scikit-learn provides a
specific classes which are even more optimized for large dataset, called
`HistGradientBoostingClassifier` and `HistGradientBoostingRegressor`. Each
feature in the dataset `data` is first binned by computing histograms, which
are later used to evaluate the potential splits. The number of splits to
evaluate is then much smaller. This algorithm becomes much more efficient
than gradient boosting when the dataset has over 10,000 samples.
Below we will give an example for a large dataset and we will compare
computation times with the experiment of the previous section.
```
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
histogram_gradient_boosting = HistGradientBoostingRegressor(
max_iter=200, random_state=0)
cv_results_hgbdt = cross_validate(gradient_boosting, data, target, n_jobs=-1)
print("Histogram Gradient Boosting Decision Tree")
print(f"R2 score via cross-validation: "
f"{cv_results_hgbdt['test_score'].mean():.3f} +/- "
f"{cv_results_hgbdt['test_score'].std():.3f}")
print(f"Average fit time: "
f"{cv_results_hgbdt['fit_time'].mean():.3f} seconds")
print(f"Average score time: "
f"{cv_results_hgbdt['score_time'].mean():.3f} seconds")
```
The histogram gradient-boosting is the best algorithm in terms of score.
It will also scale when the number of samples increases, while the normal
gradient-boosting will not.
| github_jupyter |
```
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import Imputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
from sklearn import metrics
import sklearn.preprocessing
train_file = "../data/train.csv"
test_file = "../data/test.csv"
train_data_raw = pd.read_csv(train_file)
test_data_raw = pd.read_csv(test_file)
target = "Survived"
### CLEAN DATA FUNC
def clean_func(train_data):
## DO IMPUTATION
# FARE
imp_fare = Imputer(missing_values="NaN", strategy="mean")
imp_fare.fit(train_data[["Fare"]])
train_data[["Fare"]]=imp_fare.transform(train_data[["Fare"]]).ravel()
# Age
imp=Imputer(missing_values="NaN", strategy="mean")
imp.fit(train_data[["Age"]])
train_data[["Age"]]=imp.transform(train_data[["Age"]]).ravel()
# Filna
train_data["Cabin"] = train_data["Cabin"].fillna("")
# one hot encoding
sex_features = pd.get_dummies(train_data["Sex"])
embarked_features = pd.get_dummies(train_data["Embarked"])
# rename embarked features
embarked_features = embarked_features.rename(columns={'C': 'embarked_cobh'
, 'Q': 'embark_queenstown'
, 'S': 'embark_southampton'})
# Concat new features
train_data_extras = pd.concat([train_data,sex_features,embarked_features],axis=1)
# HACK - REMOVE T WHICH IS NOT IN TEST LIKELY ERRROR
cabin_letters = pd.get_dummies(train_data['Cabin'].map(lambda x: "empty" if len(x)==0 or x[0]=="T" else x[0]))
# cabin_letters = pd.get_dummies(train_data['Cabin'].map(lambda x: "empty" if len(x)==0 else x[0]))
cabin_letters.columns = ["Cabin_letter_"+i for i in cabin_letters.columns]
train_data_extras = pd.concat([train_data_extras,cabin_letters],axis=1)
train_data_extras["Cabin_number"] = train_data['Cabin'].map(lambda x: -99 if len(x)==0 else x.split(" ")[0][1:])
# ONLY RETURN NUMERIC COLUMNS
num_types = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64','uint8']
train_data_numerics = train_data_extras.select_dtypes(include=num_types)
return train_data_numerics
```
## Select only numeric columns
```
train_data_raw2 = clean_func(train_data_raw)
train_data = train_data_raw2.iloc[:, train_data_raw2.columns != target]
train_data_target = train_data_raw2[target].values
X_train,X_test,Y_train,Y_test = train_test_split(train_data
,train_data_target
,test_size=0.3
,random_state=42)
```
# Models
- logreg
- random forest
### random forest naive
```
model_rf = RandomForestClassifier(
n_estimators=100
)
model_rf.fit(X_train, Y_train)
# Cross Validation RF
scores = cross_val_score(model_rf, X_train, Y_train, cv=10)
print(scores)
pred_rf = model_rf.predict(X_test)
metrics.accuracy_score(Y_test,pred_rf)
```
### Random Forest Grid Search
```
model_rf_gs = RandomForestClassifier()
# parmeter dict
param_grid = dict(
n_estimators=np.arange(60,101,20)
, min_samples_leaf=np.arange(2,4,1)
#, criterion = ["gini","entropy"]
#, max_features = np.arange(0.1,0.5,0.1)
)
print(param_grid)
grid = GridSearchCV(model_rf_gs,param_grid=param_grid,scoring = "accuracy", cv = 5)
grid.fit(train_data, train_data_target)
""
# model_rf.fit(train_data, train_data[target])
# print(grid)
# for i in ['params',"mean_train_score","mean_test_score"]:
# print(i)
# print(grid.cv_results_[i])
# grid.cv_results_
print(grid.best_params_)
print(grid.best_score_)
model_rf_gs_best = RandomForestClassifier(**grid.best_params_)
model_rf_gs_best.fit(X_train,Y_train)
## print feture importance
model = model_rf_gs_best
feature_names = X_train.columns.values
feature_importance2 = sorted(zip(map(lambda x: round(x, 4), model.feature_importances_), feature_names), reverse=True)
print(len(feature_importance2))
for feature in feature_importance2:
print('%f:%s' % feature )
###
# Recursive feature elimination
from sklearn.feature_selection import RFECV
model = model_rf_gs_best
rfecv = RFECV(estimator=model, step=1, cv=3, scoring='accuracy')
rfecv.fit(X_train,Y_train)
import matplotlib.pyplot as plt
from sklearn import base
model = model_rf_gs_best
print("Optimal number of features : %d" % rfecv.n_features_)
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
plt.title('Recursive feature elemination')
plt.xlabel('Nr of features')
plt.ylabel('Acc')
feature_short = feature_names[rfecv.support_]
print('== Feature short list ==')
print(feature_short)
model_simple = base.clone(model)
model_simple.fit(X_train[feature_short],Y_train)
```
- Converge about 16
- let us comare 16 vs full features on test set
```
Y_pred = model.predict(X_test)
model_score = metrics.accuracy_score(Y_test,Y_pred)
Y_pred_simple = model_simple.predict(X_test[feature_short])
model_simple_score = metrics.accuracy_score(Y_test,Y_pred_simple)
print("model acc: %.3f" % model_score)
print("simple model acc: %.3f" % model_simple_score)
```
ie. we sligtly increase test scores by removing extra variables with recursive feature elimination. (ie we remove extra variable that only seem to overfit on noise and don't contribute to acc)
Often an even more conservative cutoff can be used and go for 90% of max accurracy for f
| github_jupyter |
```
# Install libraries
!pip -qq install rasterio tifffile
# Import libraries
import os
import glob
import shutil
import gc
from joblib import Parallel, delayed
from tqdm import tqdm_notebook
import h5py
import pandas as pd
import numpy as np
import datetime as dt
from datetime import datetime, timedelta
import matplotlib.pyplot as plt
import rasterio
import tifffile as tiff
%matplotlib inline
pd.set_option('display.max_colwidth', None)
# Download data with a frequency of 10 days
def date_finder(start_date):
season_dates = []
m = str(start_date)[:10]
s = str(start_date)[:10]
for i in range(24):
date = datetime.strptime(s, "%Y-%m-%d")
s = str(date + timedelta(days = 10))[:10]
season_dates.append(datetime.strptime(s, "%Y-%m-%d"))
seasons_dates = [datetime.strptime(m, "%Y-%m-%d")] + season_dates
seasons_dates = [np.datetime64(x) for x in seasons_dates]
return list(seasons_dates)
# If day not in a frequency of 10 days, find the nearest date
def nearest(items, pivot):
return min(items, key=lambda x: abs(x - pivot))
%%time
# Unpack data saved in gdrive to colab
shutil.unpack_archive('/content/drive/MyDrive/CompeData/Radiant/Radiant_Data.zip', '/content/radiant')
gc.collect()
# Load files
train = pd.concat([pd.read_csv(f'/content/radiant/train{i}.csv', parse_dates=['datetime']) for i in range(1, 5)]).reset_index(drop = True)
test = pd.concat([pd.read_csv(f'/content/radiant/test{i}.csv', parse_dates=['datetime']) for i in range(1, 5)]).reset_index(drop = True)
train.file_path = train.file_path.apply(lambda x: '/'.join(['/content', 'radiant'] + x.split('/')[2:]))
test.file_path = test.file_path.apply(lambda x: '/'.join(['/content', 'radiant'] + x.split('/')[2:]))
train.datetime, test.datetime = pd.to_datetime(train.datetime.dt.date), pd.to_datetime(test.datetime.dt.date)
train['month'], test['month'] = train.datetime.dt.month, test.datetime.dt.month
train.head()
# Unique months
train.month.unique()
# Bands
bands = ['B01','B02','B03','B04','B05','B06','B07','B08','B8A','B09','B11','B12','CLM']
# Function to load tile and extract fields data into a numpy array and convert the same to a dataframe
# Train
def process_tile_train(tile):
tile_df = train[(train.tile_id == tile)].reset_index(drop = True)
y = np.expand_dims(rasterio.open(tile_df[tile_df.asset == 'labels'].file_path.values[0]).read(1).flatten(), axis = 1)
fields = np.expand_dims(rasterio.open(tile_df[tile_df.asset == 'field_ids'].file_path.values[0]).read(1).flatten(), axis = 1)
tile_df = train[(train.tile_id == tile) & (train.satellite_platform == 's2')].reset_index(drop = True)
unique_dates = list(tile_df.datetime.unique())
start_date = tile_df.datetime.unique()[0]
# Assert
diff = set([str(x)[:10] for x in date_finder(start_date)]) - set([str(x)[:10] for x in unique_dates])
if len(diff) > 0:
missing = list(set([str(x)[:10] for x in date_finder(start_date)]) - set(diff))
for d in diff:
missing.append(str(nearest(unique_dates, np.datetime64(d)))[:10])
dates = sorted([np.datetime64(x) for x in missing])
else:
dates = date_finder(start_date)
X_tile = np.empty((256 * 256, 0))
colls = []
for date, datec in zip(dates, range(25)):
for band in bands:
tif_file = tile_df[(tile_df.asset == band) & (tile_df.datetime == date)].file_path.values[0]
X_tile = np.append(X_tile, (np.expand_dims(rasterio.open(tif_file).read(1).flatten(), axis = 1)), axis = 1)
colls.append(str(datec) + '_' + band)
df = pd.DataFrame(X_tile, columns = colls)
df['y'], df['fields'] = y, fields
return df
# Preprocessing the data in chunks to avoid outofmemmory error
# Train
tiles = train.tile_id.unique()
chunks = [tiles[x:x+50] for x in range(0, len(tiles), 50)]
[len(x) for x in chunks], len(chunks)
# Preprocessing the tiles without storing them in memory but saving them as csvs in gdrive
# Train
for i in range(len(chunks)):
pd.DataFrame(np.vstack(Parallel(n_jobs=-1, verbose=1, backend="multiprocessing")(map(delayed(process_tile_train), [x for x in chunks[i]])))).to_csv(f'/content/drive/MyDrive/CompeData/Radiant/Seasonality/train/train{i}.csv', index = False)
gc.collect()
# Function to load tile and extract fields data into a numpy array and convert the same to a dataframe
# Test
def process_tile_test(tile):
tile_df = test[(test.tile_id == tile)].reset_index(drop = True)
fields = np.expand_dims(rasterio.open(tile_df[tile_df.asset == 'field_ids'].file_path.values[0]).read(1).flatten(), axis = 1)
tile_df = test[(test.tile_id == tile) & (test.satellite_platform == 's2')].reset_index(drop = True)
unique_dates = list(tile_df.datetime.unique())
start_date = tile_df.datetime.unique()[0]
# Assert
diff = set([str(x)[:10] for x in date_finder(start_date)]) - set([str(x)[:10] for x in unique_dates])
if len(diff) > 0:
missing = list(set([str(x)[:10] for x in date_finder(start_date)]) - set(diff))
for d in diff:
missing.append(str(nearest(unique_dates, np.datetime64(d)))[:10])
dates = sorted([np.datetime64(x) for x in missing])
else:
dates = date_finder(start_date)
X_tile = np.empty((256 * 256, 0))
colls = []
for date, datec in zip(dates, range(25)):
for band in bands:
tif_file = tile_df[(tile_df.asset == band) & (tile_df.datetime == date)].file_path.values[0]
X_tile = np.append(X_tile, (np.expand_dims(rasterio.open(tif_file).read(1).flatten(), axis = 1)), axis = 1)
colls.append(str(datec) + '_' + band)
df = pd.DataFrame(X_tile, columns = colls)
df['fields'] = fields
return df
# Preprocessing the data in chunks to avoid outofmemmory error
# Train
tiles = test.tile_id.unique()
chunks = [tiles[x:x+50] for x in range(0, len(tiles), 50)]
[len(x) for x in chunks], len(chunks)
# Preprocessing the tiles without storing them in memory but saving them as csvs in gdrive
# Train
for i in range(len(chunks)):
pd.DataFrame(np.vstack(Parallel(n_jobs=-1, verbose=1, backend="multiprocessing")(map(delayed(process_tile_test), [x for x in chunks[i]])))).to_csv(f'/content/drive/MyDrive/CompeData/Radiant/Seasonality/test/test{i}.csv', index = False)
gc.collect()
```
| github_jupyter |
# Explore endangered languages from UNESCO Atlas of the World's Languages in Danger
### Input
Endangered languages
- https://www.kaggle.com/the-guardian/extinct-languages/version/1 (updated in 2016)
- original data: http://www.unesco.org/languages-atlas/index.php?hl=en&page=atlasmap (published in 2010)
Countries of the world
- https://www.ethnologue.com/sites/default/files/CountryCodes.tab
### Output
- `endangered_languages_europe.csv`
## Imports
```
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
```
## Load data
```
df = pd.read_csv("../../data/endangerment/extinct_languages.csv")
print(df.shape)
print(df.dtypes)
df.head()
df.columns
ENDANGERMENT_MAP = {
"Vulnerable": 1,
"Definitely endangered": 2,
"Severely endangered": 3,
"Critically endangered": 4,
"Extinct": 5,
}
df["Endangerment code"] = df["Degree of endangerment"].apply(lambda x: ENDANGERMENT_MAP[x])
df[["Degree of endangerment", "Endangerment code"]]
```
## Distribution of the degree of endangerment
```
plt.xticks(fontsize=16)
plt.yticks(fontsize=16)
df["Degree of endangerment"].hist(figsize=(15,5)).get_figure().savefig('endangered_hist.png', format="png")
```
## Show distribution on map
```
countries_map = gpd.read_file(gpd.datasets.get_path("naturalearth_lowres"))
countries_map.head()
# Plot Europe
fig, ax = plt.subplots(figsize=(20, 10))
countries_map.plot(color='lightgrey', ax=ax)
plt.xlim([-30, 50])
plt.ylim([30, 75])
df.plot(
x="Longitude",
y="Latitude",
kind="scatter",
title="Endangered languages in Europe (1=Vulnerable, 5=Extinct)",
c="Endangerment code",
colormap="YlOrRd",
ax=ax,
)
plt.show()
```
## Get endangered languages only for Europe
```
countries = pd.read_csv("../../data/general/country_codes.tsv", sep="\t")
europe = countries[countries["Area"] == "Europe"]
europe
europe_countries = set(europe["Name"].to_list())
europe_countries
df[df["Countries"].isna()]
df = df[df["Countries"].notna()]
df[df["Countries"].isna()]
df["In Europe"] = df["Countries"].apply(lambda x: len(europe_countries.intersection(set(x.split(",")))) > 0)
df_europe = df.loc[df["In Europe"] == True]
print(df_europe.shape)
df_europe.head(20)
# Plot only European endangered languages
fig, ax = plt.subplots(figsize=(20, 10))
countries_map.plot(color='lightgrey', ax=ax)
plt.xlim([-30, 50])
plt.ylim([30, 75])
df_europe.plot(
x="Longitude",
y="Latitude",
kind="scatter",
title="Endangered languages in Europe",
c="Endangerment code",
colormap="YlOrRd",
ax=ax,
)
plt.xticks(fontsize=16)
plt.yticks(fontsize=16)
plt.xlabel('Longitude', fontsize=18)
plt.ylabel('Latitude', fontsize=18)
plt.title("Endangered languages in Europe (1=Vulnerable, 5=Extinct)", fontsize=18)
plt.show()
fig.savefig("endangered_languages_in_europe.png", format="png", bbox_inches="tight")
```
## Save output
```
df_europe.to_csv("../../data/endangerment/endangered_languages_europe.csv", index=False)
```
| github_jupyter |
# Polynomials Class
```
from sympy import *
import numpy as np
x = Symbol('x')
class polinomio:
def __init__(self, coefficienti: list):
self.coefficienti = coefficienti
self.grado = 0 if len(self.coefficienti) == 0 else len(
self.coefficienti) - 1
i = 0
while i < len(self.coefficienti):
if self.coefficienti[0] == 0:
self.coefficienti.pop(0)
i += 1
# scrittura del polinomio:
def __str__(self):
output = ""
for i in range(0, len(self.coefficienti)):
# and x[grado_polinomio]!=0):
if (((self.coefficienti[i] == 1 or self.coefficienti[i] == 1.0) and self.grado-i == 1)):
output += "x "
if self.grado-i == 1 and (self.coefficienti[i] != 0 and self.coefficienti[i] != 1 and self.coefficienti[i] != -1 and self.coefficienti[i] != 1.0 and self.coefficienti[i] != -1.0):
output += "{}x ".format(self.coefficienti[i])
if self.coefficienti[i] == 0:
pass
# continue
if self.grado-i != 0 and self.grado-i != 1 and (self.coefficienti[i] != 0 and self.coefficienti[i] != 1 and self.coefficienti[i] != -1 and self.coefficienti[i] != 1.0 and self.coefficienti[i] != -1.0):
output += "{}x^{} ".format(
self.coefficienti[i], self.grado-i)
# continue
#print(x[i], "$x^", grado_polinomio-i, "$ + ")
if (self.coefficienti[i] == 1 or self.coefficienti[i] == 1.0) and self.grado-i != 1 and self.grado-i != 0:
output += "x^{} ".format(self.grado-i)
# continue
elif (self.coefficienti[i] == -1 or self.coefficienti[i] == -1.0) and self.grado-i != 1 and self.grado-i != 0:
output += "- x^{} ".format(self.grado-i)
# continue
elif self.coefficienti[i] != 0 and self.grado-i == 0 and (self.coefficienti[i] != 1 or self.coefficienti[i] != 1.0):
output += "{} ".format(self.coefficienti[i])
elif self.coefficienti[i] != 0 and self.grado-i == 0 and (self.coefficienti[i] == 1 or self.coefficienti[i] == 1.0):
output += "1 "
if ((self.coefficienti[i] == -1 or self.coefficienti[i] == -1.0) and self.grado-i == 1):
output += "- x "
if (i != self.grado and self.grado-i != 0) and self.coefficienti[i+1] > 0:
output += "+ "
continue
return output
def latex(self):
latex_polinomio = 0
for i in range(0, len(self.coefficienti)):
# and x[grado_polinomio]!=0):
if (((self.coefficienti[i] == 1 or self.coefficienti[i] == 1.0) and self.grado-i == 1)):
latex_polinomio += x
if self.grado-i == 1 and (self.coefficienti[i] != 0 and self.coefficienti[i] != 1 and self.coefficienti[i] != -1 and self.coefficienti[i] != 1.0 and self.coefficienti[i] != -1.0):
latex_polinomio += self.coefficienti[i]*x
if self.coefficienti[i] == 0:
pass
# continue
if self.grado-i != 0 and self.grado-i != 1 and (self.coefficienti[i] != 0 and self.coefficienti[i] != 1 and self.coefficienti[i] != -1 and self.coefficienti[i] != 1.0 and self.coefficienti[i] != -1.0):
latex_polinomio += self.coefficienti[i]*x**(self.grado-i)
# continue
#print(x[i], "$x^", grado_polinomio-i, "$ + ")
if (self.coefficienti[i] == 1 or self.coefficienti[i] == 1.0) and self.grado-i != 1 and self.grado-i != 0:
latex_polinomio += x**(self.grado-i)
# continue
elif (self.coefficienti[i] == -1 or self.coefficienti[i] == -1.0) and self.grado-i != 1 and self.grado-i != 0:
latex_polinomio += -x**(self.grado-i)
# continue
elif self.coefficienti[i] != 0 and self.grado-i == 0 and (self.coefficienti[i] != 1 or self.coefficienti[i] != 1.0):
latex_polinomio += self.coefficienti[i]
elif self.coefficienti[i] != 0 and self.grado-i == 0 and (self.coefficienti[i] == 1 or self.coefficienti[i] == 1.0):
latex_polinomio += 1
if ((self.coefficienti[i] == -1 or self.coefficienti[i] == -1.0) and self.grado-i == 1):
latex_polinomio += -x
# if (i != self.grado and self.grado-i != 0) and self.coefficienti[i+1] > 0:
# latex_polinomio += +
# continue
return latex_polinomio
def __add__(self, y):
if type(y).__name__ != "polinomio":
raise Exception(
f"You are trying to sum a polinomio with a {type(y).__name__}")
c = []
n = min(len(self.coefficienti), len(y.coefficienti))
m = max(len(self.coefficienti), len(y.coefficienti))
d = []
if m == len(self.coefficienti):
d = self.coefficienti
else:
d = y.coefficienti
for i in range(0, m-n):
c.append(d[i])
if m == len(self.coefficienti):
for j in range(m-n, m):
z = self.coefficienti[j] + y.coefficienti[j-m+n]
c.append(z)
else:
for j in range(m-n, m):
z = self.coefficienti[j-m+n] + y.coefficienti[j]
c.append(z)
i = 0
while i < len(c):
if c[0] == 0:
c.pop(0)
i += 1
d = polinomio(c)
return d
def __sub__(self, y):
c = []
for i in y.coefficienti:
c.append(-i)
f = self + polinomio(c)
return f
def __mul__(self, y):
grado_prodotto = self.grado + y.grado
d = [[], []]
for i in range(len(self.coefficienti)):
for j in range(len(y.coefficienti)):
d[0].append(self.coefficienti[i]*y.coefficienti[j])
d[1].append(i+j) # grado del monomio
d[1] = d[1][::-1]
# print(d)
for i in range(grado_prodotto+1):
if d[1].count(grado_prodotto-i) > 1:
j = d[1].index(grado_prodotto - i)
#print("j vale: ", j)
z = j+1
while z < len(d[1]):
if d[1][z] == d[1][j]:
#print("z vale:", z)
d[0][j] = d[0][j]+d[0][z]
d[1].pop(z)
d[0].pop(z)
# print(d)
z += 1
i = 0
while i < len(d[0]):
if d[0][0] == 0:
d[0].pop(0)
i += 1
return polinomio(d[0])
def __pow__(self, var: int):
p = self
i = 0
while i < var-1:
p *= self
i += 1
return p
def __truediv__(self, y, c=[]):
d = []
s = self.grado
v = y.grado
grado_polinomio_risultante = s-v
output = 0
if grado_polinomio_risultante > 0:
d.append(self.coefficienti[0]/y.coefficienti[0])
i = 0
while i < grado_polinomio_risultante:
d.append(0)
i += 1
c.append(d[0])
a = polinomio(d)
g = a*y
f = self - g
if (f.grado - y.grado) == 0 and (len(f.coefficienti)-len(c)) > 1:
c.append(0)
if (f.grado-y.grado) < 0 and f.grado != 0:
j = 0
while j < y.grado-f.grado:
c.append(0)
self = f
return f.__truediv__(y, c)
elif grado_polinomio_risultante == 0:
d.append(self.coefficienti[0]/y.coefficienti[0])
c.append(d[0])
a = polinomio(d)
g = a*y
f = self - g
if f.grado == 0 and (f.coefficienti == [] or f.coefficienti[0] == 0):
return polinomio(c).latex()
elif f.grado >= 0:
self = f
return f.__truediv__(y, c)
elif grado_polinomio_risultante < 0:
output += polinomio(c).latex() + self.latex()/y.latex()
# output += self.latex()/y.latex()
# output += y.latex()
# if polinomio(c).grado != 0:
# output += "+"
# output += "(" + str(self) + ")/("
# output += str(y) + ")"
return output
elif s == 0:
return polinomio(c).latex()
def __eq__(self, y):
equality = 0
if len(self.coefficienti) != len(y.coefficienti):
return False
for i in range(len(self.coefficienti)):
if self.coefficienti[i] == y.coefficienti[i]:
equality += 1
if equality == len(self.coefficienti):
return True
else:
return False
def __ne__(self, y):
inequality = 0
if len(self.coefficienti) != len(y.coefficienti):
return True
else:
for i in range(len(self.coefficienti)):
if self.coefficienti[i] != y.coefficienti[i]:
inequality += 1
if inequality == len(self.coefficienti):
return True
else:
return False
a = [1, 1, 2, 1, 1]
b = [1, 1, 2, 1, 1]
c = polinomio(a)
d = polinomio(b)
(c+d).latex()
# a = [1, 0, 2, 0, 1]
# b = [1, 0, 1]
# c = polinomio(a)
# d = polinomio(b)
# c/d
a = [1,1,1]
b = [1,0]
c = polinomio(a)
d = polinomio(b)
(c*d).latex()
a = [1]
b = [1,1]
c = polinomio(a)
d = polinomio(b)
c/d
# a = [3,3,3]
# b = [3]
# c = polinomio(a)
# d = polinomio(b)
# c/d
a = [1, 1, 2, 1, 1]
b = [1, 1, 2, 1, 1]
c = polinomio(a)
d = polinomio(b)
print(c+d)
```
| github_jupyter |
# Function to list overlapping Landsat 8 scenes
This function is based on the following tutorial: http://geologyandpython.com/get-landsat-8.html
This function uses the area of interest (AOI) to retrieve overlapping Landsat 8 scenes. It will also output on the scenes with the largest portion of overlap and with less than 5% cloud cover.
```
def landsat_scene_list(aoi, start_date, end_date):
'''Creates a list of Landsat 8, level 1, tier 1
scenes that overlap with an aoi and are captured
within a specified date range.
Parameters
----------
aoi : str
The path to a shape file of an aoi with geometry.
start-date : str
The first date from which to start looking for
Landsat image capture in the format yyyy-mm,dd,
e.g. '2017-10-01'.
end-date : str
The last date from which to looking for
Landsat image capture in the format yyyy-mm,dd,
e.g. '2017-10-31'.
Returns
-------
wrs : shapefile
A catalog of Landsat 8 scenes.
scenes : geopandas.geodataframe.GeoDataFrame
A dataframe containing the information
of Landsat 8, Level 1, Tier 1 scenes that
overlap with the aoi.
'''
# Download Landsat 8 catalog from USGS (get_data auto unzips)
USGS_url = 'https://landsat.usgs.gov/sites/default/files/documents/WRS2_descending.zip'
et.data.get_data(url=USGS_url, replace=True)
# Open Landsat catalog
wrs = gpd.GeoDataFrame.from_file(os.path.join('data', 'earthpy-downloads',
'WRS2_descending',
'WRS2_descending.shp'))
# Find polygons that intersect Landsat catalog and aoi
wrs_intersection = wrs[wrs.intersects(aoi.geometry[0])]
# Calculated paths and rows
paths, rows = wrs_intersection['PATH'].values, wrs_intersection['ROW'].values
# Iterate through each Polygon of paths and rows intersecting the area
for i, row in wrs_intersection.iterrows():
# Create a string for the name containing the path and row of this Polygon
name = 'path: %03d, row: %03d' % (row.PATH, row.ROW)
# Removing scenes with small amounts of overlap using threshold of intersection area
b = (paths > 23) & (paths < 26)
paths = paths[b]
rows = rows[b]
# # Path(s) and row(s) covering the intersection
# ############################ WHY NOT PRINTING? ###################################
# for i, (path, row) in enumerate(zip(paths, rows)):
# print('Image', i+1, ' - path:', path, 'row:', row)
# Check scene availability in Amazon S3 bucket list of Landsat scenes
s3_scenes = pd.read_csv('http://landsat-pds.s3.amazonaws.com/c1/L8/scene_list.gz',
compression='gzip', parse_dates=['acquisitionDate'],
index_col=['acquisitionDate'])
# Capture only Landsat T1 scenes within dates of interest
scene_mask = (s3_scenes.index > start_date) & (s3_scenes.index <= end_date)
scene_dates = s3_scenes.loc[scene_mask]
scene_product = scene_dates[scene_dates['productId'].str.contains("_T1")]
# Geodataframe of scenes with <5% cloud cover, the url to retrieve them
#############################row.ROW and row.PATH will need to be fixed##################
scenes = scene_product[(scene_product.path == row.PATH) &
(scene_product.row == row.ROW) &
(scene_product.cloudCover <= 5)]
return wrs, scenes
```
# TEST
**Can DELETE everything below once tested and approved!**
```
# WILL DELETE WHEN FUNCTIONS ARE SEPARATED OUT
def NEON_site_extent(path_to_NEON_boundaries, site):
'''Extracts a NEON site extent from an individual site as
long as the original NEON site extent shape file contains
a column named 'siteID'.
Parameters
----------
path_to_NEON_boundaries : str
The path to a shape file that contains the list
of all NEON site extents, also known as field
sampling boundaries (can be found at NEON and
ESRI sites)
site : str
One siteID contains 4 capital letters,
e.g. CPER, HARV, ONAQ or SJER.
Returns
-------
site_boundary : geopandas.geodataframe.GeoDataFrame
A vector containing a single polygon
per the site specified.
'''
NEON_boundaries = gpd.read_file(path_to_NEON_boundaries)
boundaries_indexed = NEON_boundaries.set_index(['siteID'])
site_boundary = boundaries_indexed.loc[[site]]
site_boundary.reset_index(inplace=True)
return site_boundary
# Import packages
import os
from glob import glob
import requests
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import folium
import geopandas as gpd
import rasterio as rio
#from bs4 import BeautifulSoup
import shutil
import earthpy as et
# Set working directory
os.chdir(os.path.join(et.io.HOME, 'earth-analytics'))
# Download shapefile of all NEON site boundaries
url = 'https://www.neonscience.org/sites/default/files/Field_Sampling_Boundaries_2020.zip'
et.data.get_data(url=url, replace=True)
# Create path to shapefile
terrestrial_sites = os.path.join(
'data', 'earthpy-downloads',
'Field_Sampling_Boundaries_2020',
'terrestrialSamplingBoundaries.shp')
# Retrieving the boundaries of CPER
aoi = NEON_site_extent(terrestrial_sites, 'ONAQ')
# Test out new landsat retrieval process
scene_catalog, scene_df = landsat_scene_list(aoi, '2017-10-01', '2017-10-31')
# Visualize the catalog
scene_catalog.head(3)
# Visualize the scenes of interest based on the input parameters
scene_df
```
| github_jupyter |
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#@title MIT License
#
# Copyright (c) 2017 François Chollet
#
# Permission is hereby granted, free of charge, to any person obtaining a
# copy of this software and associated documentation files (the "Software"),
# to deal in the Software without restriction, including without limitation
# the rights to use, copy, modify, merge, publish, distribute, sublicense,
# and/or sell copies of the Software, and to permit persons to whom the
# Software is furnished to do so, subject to the following conditions:
#
# The above copyright notice and this permission notice shall be included in
# all copies or substantial portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
# DEALINGS IN THE SOFTWARE.
```
# Menyimpan dan memuat model
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/tutorials/keras/save_and_load"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />Liht di TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/id/tutorials/keras/save_and_load.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Jalankan di Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs-l10n/blob/master/site/id/tutorials/keras/save_and_load.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />Lihat kode di GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/id/tutorials/keras/save_and_load.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Unduh notebook</a>
</td>
</table>
Progres dari model dapat disimpan ketika proses training dan setelah training. Ini berarti sebuah model dapat melanjutkan proses training dengan kondisi yang sama dengan ketika proses training sebelumnya dihentikan dan dapat menghindari waktu training yang panjng. Menyimpan juga berarti Anda dapat membagikan model Anda dan orang lain dapat membuat ulang proyek Anda. Ketika mempublikasikan hasil riset dan teknik dari suatu model, kebanyakan praktisi *machine learning* membagikan:
* kode untuk membuat model, dan
* berat, atau parameter, dari sebuah model
Membagikan data ini akan membantu orang lain untuk memahami bagaimana model bekerja dan mencoba model tersebut dengan data yang baru.
Perhatian: Hati-hati dengan kode yang tidak dapat dipercaya—model-model TensorFlow adalah kode. Lihat [Menggunakan TensorFlow dengan aman](https://github.com/tensorflow/tensorflow/blob/master/SECURITY.md) untuk lebih detail.
### Opsi
Terdapat beberapa cara untuk menyimpan model TensorFlow—bergantung kepada API yang Anda gunakan. Panduan ini menggunakan [tf.keras](https://www.tensorflow.org/guide/keras), sebuah API tingkat tinggi yang digunakan untuk membangun dan melatih model di TensorFlow. Untuk pendekatan lainnya, lihat panduan Tensorflow [Simpan dan Restorasi](https://www.tensorflow.org/guide/saved_model) atau [Simpan sesuai keinginan](https://www.tensorflow.org/guide/eager#object-based_saving).
## Pengaturan
### Instal dan import
Install dan import TensorFlow dan beberapa *dependency*:
```
try:
# %tensorflow_version only exists in Colab.
%tensorflow_version 2.x
except Exception:
pass
!pip install pyyaml h5py # Required to save models in HDF5 format
from __future__ import absolute_import, division, print_function, unicode_literals
import os
import tensorflow as tf
from tensorflow import keras
print(tf.version.VERSION)
```
### Memperoleh dataset
Untuk menunjukan bagaimana cara untuk menyimpan dan memuat berat dari model, Anda akan menggunakan [Dataset MNIST](http://yann.lecun.com/exdb/mnist/). Untuk mempercepat operasi ini, gunakan hanya 1000 data pertama:
```
(train_images, train_labels), (test_images, test_labels) = tf.keras.datasets.mnist.load_data()
train_labels = train_labels[:1000]
test_labels = test_labels[:1000]
train_images = train_images[:1000].reshape(-1, 28 * 28) / 255.0
test_images = test_images[:1000].reshape(-1, 28 * 28) / 255.0
```
### Mendefinisikan sebuah model
Mulai dengan membangun sebuah model sekuensial sederhana:
```
# Define a simple sequential model
def create_model():
model = tf.keras.models.Sequential([
keras.layers.Dense(512, activation='relu', input_shape=(784,)),
keras.layers.Dropout(0.2),
keras.layers.Dense(10, activation='softmax')
])
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
return model
# Create a basic model instance
model = create_model()
# Display the model's architecture
model.summary()
```
## Menyimpan cek poin ketika proses training
You can use a trained model without having to retrain it, or pick-up training where you left off—in case the training process was interrupted. The `tf.keras.callbacks.ModelCheckpoint` callback allows to continually save the model both *during* and at *the end* of training.
Anda dapat menggunakan model terlatih tanpa harus melatihnya kembali, atau melanjutkan proses training di titik di mana proses training sebelumnya berhenti. *Callback* `tf.keras.callbacks.ModelCheckpoint` memungkinkan sebuah model untuk disimpan ketika dan setelah proses training dilakukan.
### Penggunaan *callback* cek poin
Buat sebuah callback `tf.keras.callbacks.ModelCheckpoint` yang menyimpan berat hanya ketika proses training berlangsung:
```
checkpoint_path = "training_1/cp.ckpt"
checkpoint_dir = os.path.dirname(checkpoint_path)
# Create a callback that saves the model's weights
cp_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True,
verbose=1)
# Train the model with the new callback
model.fit(train_images,
train_labels,
epochs=10,
validation_data=(test_images,test_labels),
callbacks=[cp_callback]) # Pass callback to training
# This may generate warnings related to saving the state of the optimizer.
# These warnings (and similar warnings throughout this notebook)
# are in place to discourage outdated usage, and can be ignored.
```
This creates a single collection of TensorFlow checkpoint files that are updated at the end of each epoch:
```
!ls {checkpoint_dir}
```
Create a new, untrained model. When restoring a model from weights-only, you must have a model with the same architecture as the original model. Since it's the same model architecture, you can share weights despite that it's a different *instance* of the model.
Now rebuild a fresh, untrained model, and evaluate it on the test set. An untrained model will perform at chance levels (~10% accuracy):
```
# Create a basic model instance
model = create_model()
# Evaluate the model
loss, acc = model.evaluate(test_images, test_labels, verbose=2)
print("Untrained model, accuracy: {:5.2f}%".format(100*acc))
```
Then load the weights from the checkpoint and re-evaluate:
```
# Loads the weights
model.load_weights(checkpoint_path)
# Re-evaluate the model
loss,acc = model.evaluate(test_images, test_labels, verbose=2)
print("Restored model, accuracy: {:5.2f}%".format(100*acc))
```
### Checkpoint callback options
The callback provides several options to provide unique names for checkpoints and adjust the checkpointing frequency.
Train a new model, and save uniquely named checkpoints once every five epochs:
```
# Include the epoch in the file name (uses `str.format`)
checkpoint_path = "training_2/cp-{epoch:04d}.ckpt"
checkpoint_dir = os.path.dirname(checkpoint_path)
# Create a callback that saves the model's weights every 5 epochs
cp_callback = tf.keras.callbacks.ModelCheckpoint(
filepath=checkpoint_path,
verbose=1,
save_weights_only=True,
period=5)
# Create a new model instance
model = create_model()
# Save the weights using the `checkpoint_path` format
model.save_weights(checkpoint_path.format(epoch=0))
# Train the model with the new callback
model.fit(train_images,
train_labels,
epochs=50,
callbacks=[cp_callback],
validation_data=(test_images,test_labels),
verbose=0)
```
Sekarang, lihat hasil cek poin dan pilih yang terbaru:
```
!ls {checkpoint_dir}
latest = tf.train.latest_checkpoint(checkpoint_dir)
latest
```
Catatan: secara default format tensorflow hanya menyimpan 5 cek poin terbaru.
Untuk tes, reset model dan muat cek poin terakhir:
```
# Create a new model instance
model = create_model()
# Load the previously saved weights
model.load_weights(latest)
# Re-evaluate the model
loss, acc = model.evaluate(test_images, test_labels, verbose=2)
print("Restored model, accuracy: {:5.2f}%".format(100*acc))
```
## Apa sajakah file-file ini?
Kode di atas menyimpan berat dari model ke sebuah kumpulan [cek poin](https://www.tensorflow.org/guide/saved_model#save_and_restore_variables)-file yang hanya berisikan berat dari model yan sudah dilatih dalam format biner. Cek poin terdiri atas:
* Satu atau lebih bagian (*shard*) yang berisi berat dari model Anda.
* Sebuah file index yang mengindikasikan suatu berat disimpan pada *shard* yang mana.
Jika Anda hanya melakukan proses training dari sebuah model pada sebuah komputer, Anda akan hanya memiliki satu *shard* dengan sufiks `.data-00000-of-00001`
## Menyimpan berat secara manual
Anda telah melihat bagaimana caranya memuat berat yang telah disimpan sebelumnya menjadi model. Menyimpannya secara manual dapat dilakukan dengan mudah dengan *method* `Model.save_weights`. Secara default, `tf.keras`—dan `save_weights` menggunakan format TensorFlow [cek poin](../../guide/keras/checkpoints) dengan ekstensi `.ckpt` (menyimpan dalam format [HDF5](https://js.tensorflow.org/tutorials/import-keras.html) dengan ekstensi `.h5` dijelaskan dalam panduan ini [Menyimpan dan serialisasi model](../../guide/keras/save_and_serialize#weights-only_saving_in_savedmodel_format)):
```
# Save the weights
model.save_weights('./checkpoints/my_checkpoint')
# Create a new model instance
model = create_model()
# Restore the weights
model.load_weights('./checkpoints/my_checkpoint')
# Evaluate the model
loss,acc = model.evaluate(test_images, test_labels, verbose=2)
print("Restored model, accuracy: {:5.2f}%".format(100*acc))
```
## Menyimpan keseluruhan model
Gunakan [`model.save`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#save) untuk menyimpan arsitektur dari model, berat, dan konfigurasi training dalam satu file/folder. Hal ini menyebabkan Anda dapat melakukan ekspor dari suatu model sehingga model tersebut dapat digunakan tanpa harus mengakses kode Python secara langsung*. Karena kondisi optimizer dipulihkan, Anda dapat melanjutkan proses training tepat ketika proses training sebelumnya ditinggalkan.
Meneyimpan sebuah model fungsional sangat berguna—Anda dapat memuatnya di TensorFlow.js [HDF5](https://js.tensorflow.org/tutorials/import-keras.html), [Saved Model](https://js.tensorflow.org/tutorials/import-saved-model.html)) dan kemudian melatih dan menggunakan model tersebut di web browser, atau mengubahnya sehingga dapat beroperasi di perangkat *mobile* menggunakan TensorFlw Lite [HDF5](https://www.tensorflow.org/lite/convert/python_api#exporting_a_tfkeras_file_), [Saved Model](https://www.tensorflow.org/lite/convert/python_api#exporting_a_savedmodel_))
\*Objek-objek custom (model subkelas atau layer) membutuhkan perhatian lebih ketika proses disimpan atau dimuat. Lihat bagian **Penyimpanan objek custom** di bawah.
### Format HDF5
Keras menyediakan format penyimpanan menggunakan menggunakan [HDF5](https://en.wikipedia.org/wiki/Hierarchical_Data_Format)
```
# Create and train a new model instance.
model = create_model()
model.fit(train_images, train_labels, epochs=5)
# Save the entire model to a HDF5 file.
# The '.h5' extension indicates that the model shuold be saved to HDF5.
model.save('my_model.h5')
```
Sekarang, buat ulang model dari file tersebut:
```
# Recreate the exact same model, including its weights and the optimizer
new_model = tf.keras.models.load_model('my_model.h5')
# Show the model architecture
new_model.summary()
```
Cek akurasi dari model:
```
loss, acc = new_model.evaluate(test_images, test_labels, verbose=2)
print('Restored model, accuracy: {:5.2f}%'.format(100*acc))
```
Teknik ini menyimpan semuanya:
* Nilai berat
* Konfigurasi model (arsitektur)
* konfigurasi dari optimizer
Keras menyimpan model dengan cara menginspeksi arsitekturnya. Saat ini, belum bisa menyimpan optimizer TensorFlow (dari `tf.train`). Ketika menggunakannya, Anda harus mengkompilasi kembali model setelah dimuat, dan Anda akan kehilangan kondisi dari optimizer.
### Format SavedModel
Format SavedModel adalah suatu cara lainnya untuk melakukan serialisasi model. Model yang disimpan dalam format ini dapat direstorasi menggunakan `tf.keras.models.load_model` dan kompatibel dengan TensorFlow Serving. [Panduan SavedModel](https://www.tensorflow.org/guide/saved_model) menjelaskan detail bagaimana untuk menyediakan/memeriksa SavedModel. Kode di bawah ini mengilustrasikan langkah-langkah yang dilakukan untuk menyimpan dan memuat kembali model.
```
# Create and train a new model instance.
model = create_model()
model.fit(train_images, train_labels, epochs=5)
# Save the entire model as a SavedModel.
!mkdir -p saved_model
model.save('saved_model/my_model')
```
Format SavedModel merupakan direktori yang berisi sebuah *protobuf binary* dan sebuah cek poin TensorFlow. Mememiksa direktori dari model tersimpan:
```
# my_model directory
!ls saved_model
# Contains an assets folder, saved_model.pb, and variables folder.
!ls saved_model/my_model
```
Muat ulang Keras model yang baru dari model tersimpan:
```
new_model = tf.keras.models.load_model('saved_model/my_model')
# Check its architecture
new_model.summary()
```
Model yang sudah terestorasi dikompilasi dengan argument yang sama dengan model asli. Coba lakukan evaluasi dan prediksi menggunakan model tersebut:
```
# Evaluate the restored model
loss, acc = new_model.evaluate(test_images, test_labels, verbose=2)
print('Restored model, accuracy: {:5.2f}%'.format(100*acc))
print(new_model.predict(test_images).shape)
```
### Menyimpan objek custom
Apabila Anda menggunakan format SavedModel, Anda dapat melewati bagian ini. Perbedaan utama antara HDF5 dan SavedModel adalah HDF5 menggunakan konfigurasi objek untuk menyimpan arsitektur dari model, sementara SavedModel menyimpan *execution graph*. Sehingga, SavedModel dapat menyimpan objek custom seperti model subkelas dan layer custom tanpa membutuhkan kode yang asli.
Untuk menyimpan objek custom dalam bentuk HDF5, Anda harus melakukan hal sebagai berikut:
1. Mendefinisikan sebuah *method* `get_config` pada objek Anda, dan mendefinisikan *classmethod* (opsional) `from_config`.
* `get_config(self)` mengembalikan *JSON-serializable dictionary* dari parameter-parameter yang dibutuhkan untuk membuat kembali objek.
* `from_config(cls, config)` menggunakan dan mengembalikan konfigurasi dari `get_config` untuk membuat objek baru. Secara default, fungsi ini menggunakan konfigurasi teresbut untuk menginisialisasi kwargs (`return cls(**config)`).
2. Gunakan objek tersebut sebagai argumen dari `custom_objects` ketika memuat model. Argumen tersebut harus merupakan sebuah *dictionary* yang memetakan string dari nama kelas ke class Python. Misalkan `tf.keras.models.load_model(path, custom_objects={'CustomLayer': CustomLayer})`
Lihat tutorial [Menulis layers and models from awal](https://www.tensorflow.org/guide/keras/custom_layers_and_models) untuk contoh dari objek custom dan `get_config`.
| github_jupyter |
# 증식을 통한 데이터셋 크기 확장
## 1. Google Drive와 연동
```
from google.colab import drive
drive.mount("/content/gdrive")
path = "gdrive/'My Drive'/'Colab Notebooks'/CNN"
!ls gdrive/'My Drive'/'Colab Notebooks'/CNN/datasets
```
## 2. 모델 생성
```
from tensorflow.keras import layers, models, optimizers
```
0. Sequential 객체 생성
1. conv layer(filter32, kernel size(3,3), activation 'relu', input_shape()
2. pooling layer(pool_size(2,2))
3. conv layer(filter 64, kernel size(3,3), activation 'relu'
4. pooling layer(pool_size(2,2))
5. conv layer(filter 128, kernel size(3,3), activation 'relu'
6. pooling layer(pool_size(2,2))
7. conv layer(filter 128, kernel size(3,3), activation 'relu'
8. pooling layer(pool_size(2,2))
-------
9. flatten layer
10. Dense layer 512, relu
11. Dense layer 1, sigmoid
```
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.summary()
from tensorflow.keras import optimizers
model.compile(loss='binary_crossentropy', optimizer=optimizers.RMSprop(lr=1e-4), metrics=['accuracy'])
from tensorflow.keras.preprocessing.image import ImageDataGenerator
```
## 3. 데이터 전처리
```
import os
base_dir = '/content/gdrive/My Drive/Colab Notebooks/CNN/datasets/cats_and_dogs_small'
train_dir = os.path.join(base_dir,'train')
validation_dir = os.path.join(base_dir,'validation')
test_dir=os.path.join(base_dir,'test')
# [코드작성]
# train_datagen이라는 ImageDataGenerator 객체 생성
# train_datagen의 증식 옵션
# 1. scale : 0~1
# 2. 회전 각도 범위 : -40~+40
# 3. 수평이동 범위 : 전체 너비의 20% 비율
# 4. 수직이동 범위 : 전체 높이의 20% 비율
# 5. 전단 변환(shearing) 각도 범위 : 10%
# 6. 사진 확대 범위 : 20%
# 7. 이미지를 수평으로 뒤집기 : True
# 8. 회전이나 가로/세로 이동으로 인해 새롭게 생성해야 할 픽셀을 채울 전략 : 'nearest'
train_datagen = ImageDataGenerator(rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.1,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
validation_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
train_dir, target_size=(150,150), batch_size=20,class_mode='binary')
validation_generator = validation_datagen.flow_from_directory(
validation_dir, target_size=(150,150), batch_size=20,class_mode='binary')
test_generator = test_datagen.flow_from_directory(
test_dir, target_size=(150,150), batch_size=20,class_mode='binary')
```
## 4. 모델 훈련
```
history = model.fit_generator(train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50)
```
## 5. 성능 시각화
```
import matplotlib.pyplot as plt
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(1, len(acc) +1)
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.show()
```
* acc와 val_acc 모두 증가하는 경향을 보아 과적합이 발생하지 않았음
```
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
```
## 6. 모델 평가하기
```
test_loss, test_accuracy = model.evaluate_generator(test_generator, steps=50)
print(test_loss)
print(test_accuracy)
```
## 7. 모델 저장
```
model.save('/content/gdrive/My Drive/Colab Notebooks/CNN/datasets/cats_and_dogs_small_augmentation.h5')
```
| github_jupyter |
<a href="https://colab.research.google.com/github/coenarrow/MNistTests/blob/main/MNIST.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
### Start Julia evironment
```
# Install any required python packages here
# !pip install <packages>
# Here we install Julia
%%capture
%%shell
if ! command -v julia 3>&1 > /dev/null
then
wget -q 'https://julialang-s3.julialang.org/bin/linux/x64/1.6/julia-1.6.2-linux-x86_64.tar.gz' \
-O /tmp/julia.tar.gz
tar -x -f /tmp/julia.tar.gz -C /usr/local --strip-components 1
rm /tmp/julia.tar.gz
fi
julia -e 'using Pkg; pkg"add IJulia; precompile;"'
echo 'Done'
```
After you run the first cell (the the cell directly above this text), go to Colab's menu bar and select **Edit** and select **Notebook settings** from the drop down. Select *Julia 1.6* in Runtime type. You can also select your prefered harwdware acceleration (defaults to GPU).
<br/>You should see something like this:
> 
<br/>Click on SAVE
<br/>**We are ready to get going**
```
VERSION
```
**The next three cells are for GPU benchmarking. If you are using this notebook for the first time and have GPU enabled, you can give it a try.**
### Import all the Julia Packages
Here, we first import all the required packages. CUDA is used to offload some of the processing to the gpu, Flux is the package for putting together the NN. MLDatasets contains the MNIST dataset which we will use in this example. Images contains some functionality to actually view images. Makie, and CairoMakie are used for plotting.
```
import Pkg
Pkg.add(["CUDA","Flux","MLDatasets","Images","Makie","CairoMakie","ImageMagick"])
using CUDA, Flux, MLDatasets, Images, Makie, Statistics, CairoMakie,ImageMagick
using Base.Iterators: partition
```
Let's look at the functions we can call from the MNIST set itself
```
names(MNIST)
```
Let's assume we want to get the training data from the MNIST package. Now, let's see what we get returned if we call that function
```
Base.return_types(MNIST.traindata)
```
This does not mean a heck of a lot to me initially, but we can basically see we get 2 tuples returned. So let's go ahead and assign some x and y to each of the tuples so we can probe further.
```
x, y = MNIST.traindata();
```
Let's now further investigate the x
```
size(x)
```
We know from the MNIST dataset, that the set contains 60000 images, each of size 28x28. So clearly we are looking at the images themselves. So this is our input. Let's plot an example to make sure.
```
i = rand(1:60000)
heatmap(x[:,:,i],colormap = :grays)
```
Similarly, let's have a quick look at the size of y. I expect this is the label associated with the images
```
y[i]
```
And then let's check that the image above is labelled as what we expect.
```
y[7]
show(names(Images))
?imshow
names(ImageShow)
```
| github_jupyter |
# 컴파일러에서 변수, 조건문 다루기
변수를 다루기 위해서는 기계상태에 메모리를 추가하고 메모리 연산을 위한 저급언어 명령을 추가한다.
조건문을 다루기 위해서는 실행코드를 순차적으로만 실행하는 것이 아니라 특정 코드 위치로 이동하여 실행하는 저급언어 명령을 추가한다.
```
data Expr = Var Name -- x
| Val Value -- n
| Add Expr Expr -- e1 + e2
-- | Sub Expr Expr
-- | Mul Expr Expr
-- | Div Expr Expr
| If Expr Expr Expr -- if e then e1 else e0
deriving Show
type Name = String -- 변수의 이름은 문자열로 표현
type Value = Int -- 상수값은 정수
type Stack = [Value]
data Inst = ADD | PUSH Value -- 스택 명령
| GOTO Code | JMPZ Code -- 실행코드 명령
| READ Addr -- 메모리 명령
deriving Show
type Code = [Inst]
-- type Env = [ (Name, Value) ] 라는 인터프리터 실행 환경을
-- 두 단계로 아래와 같이 나눈다
type SymTbl = [ (Name, Addr) ] -- 컴파일 단계에서 사용하는 심볼 테이블
type Memory = [ (Addr, Value) ] -- 기계(가상머신) 실행 단계에서 사용하는 메모리
type Addr = Int -- 주소는 정수로 표현
-- 이제 Kont는 스택만이 아니라 세 요소로 이루어진 기계상태를 변화시키는 함수 타입이다
type Kont = (Stack,Memory,Code) -> (Stack,Memory,Code)
-- 더 이상 실행할 코드가 없는 기계상태로 변화시키는 함수
haltK :: Kont
haltK (s, mem, _) = (s, mem, [])
-- 스택 명령을 실행시키기 위한 기계상태변화 함수들
pushK :: Int -> Kont
pushK n (s, mem, code) = (n:s, mem, code)
addK :: Kont
addK (n2:n1:s, mem, code) = ((n1+n2):s, mem, code)
-- 실행코드 명령을 실행시키기 위한 기계상태변화 함수들
jmpzK :: Code -> Kont
jmpzK code (0:s, mem, _) = (s, mem, code) -- 스택 맨 위 값이 0이면 새로운 code 위치로 점프
jmpzK _ (_:s, mem, c) = (s, mem, c) -- 스택 맨 위가 0이 아니면 원래 실행하던 코드 c실행
gotoK :: Code -> Kont
gotoK code (s, mem, _) = (s, mem, code) -- 무조건 새로운 code 위치로 이동
-- 메모리 명령을 실행시키기 위한 기계상태변화 함수
-- (메모리에서 값을 읽어 스택 맨 위에 쌓는다)
readK a (s, mem, code) = case lookup a mem of
Nothing -> error (show a ++ " uninitialized memory address")
Just v -> (v:s, mem, code)
compile :: SymTbl -> Expr -> Code
compile tbl (Var x) = case lookup x tbl of
Nothing -> error (x ++ " not found")
Just a -> [READ a]
compile tbl (Val n) = [PUSH n]
compile tbl (Add e1 e2) = compile tbl e1 ++ compile tbl e2 ++ [ADD]
compile tbl (If e e1 e0) =
compile tbl e ++ [JMPZ c0] ++ c1 ++ [GOTO []] ++ c0
where
c1 = compile tbl e1
c0 = compile tbl e0
step :: Inst -> Kont
step (PUSH n) = pushK n
step ADD = addK
step (GOTO c) = gotoK c
step (JMPZ c) = jmpzK c
step (READ a) = readK a
run :: Kont
run (s, mem, []) = (s, mem, [])
run (s, mem, c:cs) = run (step c (s, mem, cs))
import Data.List (union)
vars (Var x) = [x]
vars (Val _) = []
vars (Add e1 e2) = vars e1 `union` vars e2
vars (If e e1 e0) = vars e `union` vars e1 `union` vars e0
-- 인터프리터에서는 아래 식을 실행하려면 [("x",2),("y",3)]와 같은
-- 실행환경을 만들어 한방에 실행하면 되지만 컴파일러에는 두 단계
e0 = Add (Add (Var "x") (Var "y")) (Val 100)
e0
-- 컴파일할 때는 변수를 메모리 주소에 대응시키는 심볼테이블이 필요
code0 = compile [("x",102),("y",103)] e0
code0
-- 실행할 때는 해당 주소에 적절한 값을 할당한 메모리가 필요
vm0 = ([], [(102,7), (103,3)], code0)
run vm0
{- b = 2, x = 12, y = 123 -}
-- if b then (x + 3) else y
e1 = If (Var "b") (Add (Var "x") (Val 3)) (Var "y")
-- (if b then (x + 3) else y) + 1000
e2 = e1 `Add` Val 1000
tbl0 = [("b",101),("x",102),("y",103)]
tbl0
mem0 = [(101,2), (102,12), (103,123)]
mem0
code1 = compile tbl0 e1
code1
code2 = compile tbl0 e2
code2
{-
import GHC.HeapView
putStr =<< ppHeapGraph <$> buildHeapGraph 15 code2 (asBox code2)
-}
-- 예상대로 e1의 계산 결과 스택 맨 위에 15가 나온다
run ([], mem0, code1)
-- e2의 계산 결과는 1015이어야 하는데 e1과 마찬가지로 15가 되어버린다
run ([], mem0, code2)
```
아래는 e2를 컴파일한 code2를 실행한 결과가 왜 원하는 대로 나오지 않는지 좀더 자세히 살펴보기 위해
step 함수를 한단계씩 호출해 가며 각각의 명령 실행 전후의 기계상태 vm0,...,vm6를 알아본 내용이다.
```
vm0@(s0, _,c0:cs0) = ([], mem0, code2)
vm0
vm1@(s1,mem1,c1:cs1) = step c0 (s0,mem0,cs0)
vm1
vm2@(s2,mem2,c2:cs2) = step c1 (s1,mem1,cs1)
vm2
vm3@(s3,mem3,c3:cs3) = step c2 (s2,mem2,cs2)
vm3
vm4@(s4,mem4,c4:cs4) = step c3 (s3,mem3,cs3)
vm4
vm5@(s5,mem5,c5:cs5) = step c4 (s4,mem4,cs4)
vm5
vm6 = step c5 (s5,mem5,cs5)
vm6
```
----
# HW02-compiler2019fall (10점)
지금까지 살펴본 `compile` 함수의 문제점을 해결하여
제대로 된 컴파일러를 정의하려면 다음과 같은 개념으로 접근하면 된다.
> 지금 내가 주목해서 컴파일하는 부분의 목적 코드를 생성해서
> 그 다음에 뒤이어 할 일의 코드 앞에다 이어붙이는
> 코드변환함수를 만드는 것이 컴파일러다.
그러니까 다음과 같은 타입으로 `compile` 함수를 재작성해야 한다.
```haskell
type Control = Code -> Code -- 코드변환함수 타입
compile :: SymTbl -> Expr -> Control
```
(테스트 코드 작성조건 안내 추가예정)
과제에 도움이 될만한 내용 (아래에 추가예정)
| github_jupyter |
# <span style="color: #B40486">BASIC PYTHON FOR RESEARCHERS</span>
_by_ [**_Megat Harun Al Rashid bin Megat Ahmad_**](https://www.researchgate.net/profile/Megat_Harun_Megat_Ahmad)
last updated: April 14, 2016
-------
## _<span style="color: #29088A">8. Database and Data Analysis</span>_
---
<span style="color: #0000FF">$Pandas$</span> is an open source library for data analysis in _Python_. It gives _Python_ similar capabilities to _R_ programming language and even though it is possible to run _R_ in _Jupyter Notebook_, it would be more practical to do data analysis with a _Python_ friendly syntax. Similar to other libraries, the first step to use <span style="color: #0000FF">$Pandas$</span> is to import the library and usually together with the <span style="color: #0000FF">$Numpy$</span> library.
```
import pandas as pd
import numpy as np
```
***
### **_8.1 Data Structures_**
Data structures (similar to _Sequence_ in _Python_) of <span style="color: #0000FF">$Pandas$</span> revolves around the **_Series_** and **_DataFrame_** structures. Both are fast as they are built on top of <span style="color: #0000FF">$Numpy$</span>.
A **_Series_** is a one-dimensional object with a lot of similar properties similar to a list or dictionary in _Python_'s _Sequence_. Each element or item in a **_Series_** will be assigned by default an index label from _0_ to _N-1_ (where _N_ is the length of the **_Series_**) and it can contains the various type of _Python_'s data.
```
# Creating a series (with different type of data)
s1 = pd.Series([34, 'Material', 4*np.pi, 'Reactor', [100,250,500,750], 'kW'])
s1
```
The index of a **_Series_** can be specified during its creation and giving it a similar function to a dictionary.
```
# Creating a series with specified index
lt = [34, 'Material', 4*np.pi, 'Reactor', [100,250,500,750], 'kW']
s2 = pd.Series(lt, index = ['b1', 'r1', 'solid angle', 18, 'reactor power', 'unit'])
s2
```
Data can be extracted by specifying the element position or index (similar to list/dictionary).
```
s1[3], s2['solid angle']
```
**_Series_** can also be constructed from a dictionary.
```
pop_cities = {'Kuala Lumpur':1588750, 'Seberang Perai':818197, 'Kajang':795522,
'Klang':744062, 'Subang Jaya':708296}
cities = pd.Series(pop_cities)
cities
```
The elements can be sort using the <span style="color: #0000FF">$Series.order()$</span> function. This will not change the structure of the original variable.
```
cities.order(ascending=False)
cities
```
Another sorting function is the <span style="color: #0000FF">$sort()$</span> function but this will change the structure of the **_Series_** variable.
```
# Sorting with descending values
cities.sort(ascending=False)
cities
cities
```
Conditions can be applied to the elements.
```
# cities with population less than 800,000
cities[cities<800000]
# cities with population between 750,000 and 800,000
cities[cities<800000][cities>750000]
```
---
A **_DataFrame_** is a 2-dimensional data structure with named rows and columns. It is similar to _R_'s _data.frame_ object and function like a spreadsheet. **_DataFrame_** can be considered to be made of series of **_Series_** data according to the column names. **_DataFrame_** can be created by passing a 2-dimensional array of data and specifying the rows and columns names.
```
# Creating a DataFrame by passing a 2-D numpy array of random number
# Creating first the date-time index using date_range function
# and checking it.
dates = pd.date_range('20140801', periods = 8, freq = 'D')
dates
# Creating the column names as list
Kedai = ['Kedai A', 'Kedai B', 'Kedai C', 'Kedai D', 'Kedai E']
# Creating the DataFrame with specified rows and columns
df = pd.DataFrame(np.random.randn(8,5),index=dates,columns=Kedai)
df
```
---
Some of the useful functions that can be applied to a **_DataFrame_** include:
```
df.head() # Displaying the first five (default) rows
df.head(3) # Displaying the first three (specified) rows
df.tail(2) # Displaying the last two (specified) rows
df.index # Showing the index of rows
df.columns # Showing the fields of columns
df.values # Showing the data only in its original 2-D array
df.describe() # Simple statistical data for each column
df.T # Transposing the DataFrame (index becomes column and vice versa)
df.sort_index(axis=1,ascending=False) # Sorting with descending column
df.sort(columns='Kedai D') # Sorting according to ascending specific column
df['Kedai A'] # Extract specific column (using python list syntax)
df['Kedai A'][2:4] # Slicing specific column (using python list syntax)
df[2:4] # Slicing specific row data (using python list syntax)
# Slicing specific index range
df['2014-08-03':'2014-08-05']
# Slicing specific index range for a particular column
df['2014-08-03':'2014-08-05']['Kedai B']
# Using the loc() function
# Slicing specific index and column ranges
df.loc['2014-08-03':'2014-08-05','Kedai B':'Kedai D']
# Slicing specific index range with specific column names
df.loc['2014-08-03':'2014-08-05',['Kedai B','Kedai D']]
# Possibly not yet to have something like this
df.loc[['2014-08-01','2014-08-03':'2014-08-05'],['Kedai B','Kedai D']]
# Using the iloc() function
df.iloc[3] # Specific row location
df.iloc[:,3] # Specific column location (all rows)
df.iloc[2:4,1:3] # Python like slicing for range
df.iloc[[2,4],[1,3]] # Slicing with python like list
# Conditionals on the data
df>0 # Array values > 0 OR
df[df>0] # Directly getting the value
```
**_NaN_** means empty, missing data or unavailable.
```
df[df['Kedai B']<0] # With reference to specific value in a column (e.g. Kedai B)
df2 = df.copy() # Made a copy of a database
df2
# Adding column
df2['Tambah'] = ['satu','satu','dua','tiga','empat','tiga','lima','enam']
df2
# Adding row using append() function. The previous loc() is possibly deprecated.
# Assign a new name to the new row (with the same format)
new_row_name = pd.date_range('20140809', periods = 1, freq = 'D')
# Appending new row with new data
df2.append(list(np.random.randn(5))+['sembilan'])
# Renaming the new row (here actually is a reassignment)
df2 = df2.rename(index={10: new_row_name[0]})
df2
# Assigning new data to a row
df2.loc['2014-08-05'] = list(np.random.randn(5))+['tujuh']
df2
# Assigning new data to a specific element
df2.loc['2014-08-05','Tambah'] = 'lapan'
df2
# Using the isin() function (returns boolean data frame)
df2.isin(['satu','tiga'])
# Select specific row based on additonal column
df2[df2['Tambah'].isin(['satu','tiga'])]
# Use previous command - select certain column based on selected additional column
df2[df2['Tambah'].isin(['satu','tiga'])].loc[:,'Kedai B':'Kedai D']
# Select > 0 from previous cell...
(df2[df2['Tambah'].isin(['satu','tiga'])].loc[:,'Kedai B':'Kedai D']>0)
```
***
### **_8.2 Data Operations_**
We have seen few operations previously on **_Series_** and **_DataFrame_** and here this will be explored further.
```
df.mean() # Statistical mean (column) - same as df.mean(0), 0 means column
df.mean(1) # Statistical mean (row) - 1 means row
df.mean()['Kedai C':'Kedai E'] # Statistical mean (range of columns)
df.max() # Statistical max (column)
df.max()['Kedai C'] # Statistical max (specific column)
df.max(1)['2014-08-04':'2014-08-07'] # Statistical max (specific row)
df.max(1)[dates[3]] # Statistical max (specific row by variable)
```
---
Other statistical functions can be checked by typing df._< TAB >_.
The data in a **_DataFrame_** can be represented by a variable declared using the <span style="color: #0000FF">$lambda$</span> operator.
```
df.apply(lambda x: x.max() - x.min()) # Operating array values with function
df.apply(lambda z: np.log(z)) # Operating array values with function
```
Replacing, rearranging and operations of data between columns can be done much like spreadsheet.
```
df3 = df.copy()
df3[r'Kedai A^2/Kedai E'] = df3['Kedai A']**2/df3['Kedai E']
df3
```
Tables can be split, rearranged and combined.
```
df4 = df.copy()
df4
pieces = [df4[6:], df4[3:6], df4[:3]] # split row 2+3+3
pieces
df5 = pd.concat(pieces) # concantenate (rearrange/combine)
df5
df4+df5 # Operation between tables with original index sequence
df0 = df.loc[:,'Kedai A':'Kedai C'] # Slicing and extracting columns
pd.concat([df4, df0], axis = 1) # Concatenating columns (axis = 1 -> refers to column)
```
***
### **_8.3 Plotting Functions_**
---
Let us look on some of the simple plotting function on <span style="color: #0000FF">$Pandas$</span> (requires <span style="color: #0000FF">$Matplotlib$</span> library).
```
df_add = df.copy()
# Simple auto plotting
%matplotlib inline
df_add.cumsum().plot()
# Reposition the legend
import matplotlib.pyplot as plt
df_add.cumsum().plot()
plt.legend(bbox_to_anchor=[1.3, 1])
```
In the above example, repositioning the legend requires the legend function in <span style="color: #0000FF">$Matplotlib$</span> library. Therefore, the <span style="color: #0000FF">$Matplotlib$</span> library must be explicitly imported.
```
df_add.cumsum().plot(kind='bar')
plt.legend(bbox_to_anchor=[1.3, 1])
df_add.cumsum().plot(kind='barh', stacked=True)
df_add.cumsum().plot(kind='hist', alpha=0.5)
df_add.cumsum().plot(kind='area', alpha=0.4, stacked=False)
plt.legend(bbox_to_anchor=[1.3, 1])
```
A 3-dimensional plot can be projected on a canvas but requires the <span style="color: #0000FF">$Axes3D$</span> library with slightly complicated settings.
```
# Plotting a 3D bar plot
from mpl_toolkits.mplot3d import Axes3D
import numpy as np
# Convert the time format into ordinary strings
time_series = pd.Series(df.index.format())
fig = plt.figure(figsize=(8,6))
ax = fig.add_subplot(111, projection='3d')
# Plotting the bar graph column by column
for c, z in zip(['r', 'g', 'b', 'y','m'], np.arange(len(df.columns))):
xs = df.index
ys = df.values[:,z]
ax.bar(xs, ys, zs=z, zdir='y', color=c, alpha=0.5)
ax.set_zlabel('Z')
ax.set_xticklabels(time_series, va = 'baseline', ha = 'right', rotation = 15)
ax.set_yticks(np.arange(len(df.columns)))
ax.set_yticklabels(df.columns, va = 'center', ha = 'left', rotation = -42)
ax.view_init(30, -30)
fig.tight_layout()
```
***
### **_8.4 Reading And Writing Data To File_**
Data in **_DataFrame_** can be exported into **_csv_** (comma separated value) and **_Excel_** file. The users can also create a **_DataFrame_** from data in **_csv_** and **_Excel_** file, the data can then be processed.
```
# Export data to a csv file but separated with < TAB > rather than comma
# the default separation is with comma
df.to_csv('Tutorial8/Kedai.txt', sep='\t')
# Export to Excel file
df.to_excel('Tutorial8/Kedai.xlsx', sheet_name = 'Tarikh', index = True)
# Importing data from csv file (without header)
from_file = pd.read_csv('Tutorial8/Malaysian_Town.txt',sep='\t',header=None)
from_file.head()
# Importing data from Excel file (with header (the first row) that became the column names)
from_excel = pd.read_excel('Tutorial8/Malaysian_Town.xlsx','Sheet1')
from_excel.head()
```
---
Further <span style="color: #0000FF">$Pandas$</span> features can be found in http://pandas.pydata.org/.
| github_jupyter |
# Investigation of No-show Appointments Data
## Table of Contents
<ul>
<li><a href="#intro">Introduction</a></li>
<li><a href="#wrangling">Data Wrangling</a></li>
<li><a href="#eda">Exploratory Data Analysis</a></li>
<li><a href="#conclusions">Conclusions</a></li>
</ul>
<a id='intro'></a>
## Introduction
The data includes some information about more than 100,000 Braxzilian medical appointments. It gives if the patient shows up or not for the appointment as well as some characteristics of patients and appointments. When we calculate overall no-show rate for all records, we see that it is pretty high; above 20%. It means more than one out of 5 patients does not show up at all. In this project, we specifically look at the associatons between no show-up rate and other variables and try to understand why the rate is at the level it is.
```
import pandas as pd
import seaborn as sb
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
```
<a id='wrangling'></a>
## Data Wrangling
```
# Load your data and print out a few lines. Perform operations to inspect data
# types and look for instances of missing or possibly errant data.
filename = 'noshowappointments-kagglev2-may-2016.csv'
df= pd.read_csv(filename)
df.head()
df.info() # no missing values
```
The data gives information about gender and age of the patient, neighbourhood of the hospital, if the patient has hypertension, diabetes, alcoholism or not, date and time of appointment and schedule, if the patient is registered in scholarship or not, and if SMS received or not as a reminder.
When I look at the data types of columns, it is realized that AppointmentDay and ScheduledDay are recorded as object, or string to be more specific. And also, PatientId is recorded as float instead of integer. But I most probably will not make use of this information since it is very specific to the patient.
The data seems pretty clear. There is no missing value or duplicated rows.
First, I start with creating a dummy variable for No-show variable. So it makes easier for us to look at no show rate across different groups.
```
df.describe()
df.isnull().any().sum() # no missing value
df.duplicated().any()
```
### Data Cleaning
A dummy variable named no_showup is created. It takes the value 1 if the patient did not show up, and 0 otherwise. I omitted PatientId, AppointmentID and No-show columns.
There are some rows with Age value of -1 which does not make much sense. So I dropped these rows.
Other than that, the data seems pretty clean; no missing values, no duplicated rows.
```
df['No-show'].unique()
df['no_showup'] = np.where(df['No-show'] == 'Yes', 1, 0)
df.drop(['PatientId', 'AppointmentID', 'No-show'], axis = 1, inplace = True)
noshow = df.no_showup == 1
show = df.no_showup == 0
index = df[df.Age == -1].index
df.drop(index, inplace = True)
```
<a id='eda'></a>
## Exploratory Data Analysis
### What factors are important in predicting no-show rate?
```
plt.figure(figsize = (10,6))
df.Age[noshow].plot(kind = 'hist', alpha= 0.5, color= 'green', bins =20, label = 'no-show');
df.Age[show].plot(kind = 'hist', alpha= 0.4, color= 'orange', bins =20, label = 'show');
plt.legend();
plt.xlabel('Age');
plt.ylabel('Number of Patients');
```
I started exploratory data analysis by first looking at the relationship between age and no_showup. By looking age distributions for patients who showed up and not showed up, we can not say much. There is a spike at around age 0, and no show up number is not that high compared to other ages. We can infer that adults are careful about babies' appointments. As age increases, the number of patients in both groups decreases, which is plausible taking general demographics into account. To be able to say more about showup rate across different age groups we need to look at ratio of one group to another.
First, I created age bins which are equally spaced from age 0 to the maximum age which is 115. It is called age_bins. Basically, it shows which bin the age of patient falls in. So I can look at the no_showup rate across different age bins.
```
bin_edges = np.arange(0, df.Age.max()+3, 3)
df['age_bins'] = pd.cut(df.Age, bin_edges)
base_color = sb.color_palette()[0]
age_order = df.age_bins.unique().sort_values()
g= sb.FacetGrid(data= df, row= 'Gender', row_order = ['M', 'F'], height=4, aspect = 2);
g = g.map(sb.barplot, 'age_bins', 'no_showup', color = base_color, ci = None, order = age_order);
g.axes[0,0].set_ylabel('No-show Rate');
g.axes[1,0].set_ylabel('No-show Rate');
plt.xlabel('Age Intervals')
plt.xticks(rotation = 90);
```
No-show rate is smaller than average for babies ((0-3] interval). Then it increases as age get larger and it reaches peak at around 15-18 depending on gender. After that point, as age gets larger the no-show rate gets smaller. So middle-aged and old people are much more careful about their doctor appointments which is understandable as you get older, your health might not be in a good condition, you become more concerned about your health and do not miss your appointments. Or another explanation might be that as person ages, it is more probable to have a health condition which requires close doctor watch which incentivizes you attend to your scheduled appointments.
There are spikes at the end of graphs, I suspect this happens due to small number of patients in corresponding bins.
There are only 5 people in (114,117] bin which proves my suspicion right.
```
df.groupby('age_bins').size().sort_values().head(8)
df.groupby('Gender').no_showup.mean()
```
There is no much difference across genders. No-show rates are close.
```
order_scholar = [0, 1]
g= sb.FacetGrid(data= df, col= 'Gender', col_order = ['M', 'F'], height=4);
g = g.map(sb.barplot, 'Scholarship', 'no_showup', order = order_scholar, color = base_color, ci = None,);
g.axes[0,0].set_ylabel('No-show Rate');
g.axes[0,1].set_ylabel('No-show Rate');
```
If the patient is in Brazilian welfare program, then the probability of her not showing up for the appointment is larger than the probablity of a patient which is not registered in welfare program. There is no significant difference between males and females.
```
order_hyper = [0, 1]
g= sb.FacetGrid(data= df, col= 'Gender', col_order = ['M', 'F'], height=4);
g = g.map(sb.barplot, 'Hipertension', 'no_showup', order = order_hyper, color = base_color, ci = None,);
g.axes[0,0].set_ylabel('No-show Rate');
g.axes[0,1].set_ylabel('No-show Rate');
```
When the patient has hypertension or diabetes, she would not want to miss doctor appointments. So having a disease to be watched closely incentivizes you to show up for your appointments. Again, being male or female does not make a significant difference in no-show rate.
```
order_diabetes = [0, 1]
sb.barplot(data =df, x = 'Diabetes', y = 'no_showup', hue = 'Gender', ci = None, order = order_diabetes);
sb.despine();
plt.ylabel('No-show Rate');
plt.legend(loc = 'lower right');
order_alcol = [0, 1]
sb.barplot(data =df, x = 'Alcoholism', y = 'no_showup', hue = 'Gender', ci = None, order = order_alcol);
sb.despine();
plt.ylabel('No-show Rate');
plt.legend(loc = 'lower right');
```
The story for alcoholism is a bit different. If the patient is a male with alcoholism, the probability of his no showing up is smaller than the one of male with no alcoholism. On the other hand, having alcoholism makes a female patient's probability of not showing up larger. Here I suspect if the number of females having alcoholism is very small or not, but I see below that the numbers in both groups are comparable.
```
df.groupby(['Gender', 'Alcoholism']).size()
order_handcap = [0, 1, 2, 3, 4]
sb.barplot(data =df, x = 'Handcap', y = 'no_showup', hue = 'Gender', ci = None, order = order_handcap);
sb.despine();
plt.ylabel('No-show Rate');
plt.legend(loc = 'lower right');
df.groupby(['Handcap', 'Gender']).size()
```
We cannot see a significant difference across levels of Handcap variable. Label 4 for females is 1 but I do not pay attention to this since there are only 2 data points in this group. So being in different Handcap levels does not say much when predicting if a patient will show up.
```
plt.figure(figsize = (16,6))
sb.barplot(data = df, x='Neighbourhood', y='no_showup', color =base_color, ci = None);
plt.xticks(rotation = 90);
plt.ylabel('No-show Rate');
df.groupby('Neighbourhood').size().sort_values(ascending = True).head(10)
```
I want to see no-show rate in different neighborhoods. There is no significant difference across neighborhoods except ILHAS OCEÂNICAS DE TRINDADE. There are only 2 data points from this place in the dataset. The exceptions can occur with only 2 data points.
Lastly, I want to look at how sending SMS to patients to remind their appointments effects no-show rate.
```
plt.figure(figsize = (5,5))
sb.barplot(data = df, x='SMS_received', y='no_showup', color =base_color, ci = None);
plt.title('No-show Rate vs SMS received');
plt.ylabel('No-show Rate');
```
The association between SMS_received variable and no-show rate is very counterintuitive. I expect that when the patient receives SMS as a reminder, she is more likely to go to the appointment. Here the graph says exact opposite thing; when no SMS, the rate is around 16% whereas when SMS received it is more than 27%. It needs further and deeper examination.
### Understanding Negative Association between No-show Rate and SMS_received Variable
```
sb.barplot(data = df, x = 'SMS_received', y = 'no_showup', hue = 'Gender', ci = None);
plt.title('No-show Rate vs SMS received');
plt.ylabel('No-show Rate');
plt.legend(loc ='lower right');
```
Gender does not make a significant impact on the rate with SMS and no SMS.
Below I try to look at how no-show rate changes with time to appointment day. I convert ScheduledDay and AppointmentDay to datetime. There is no information about hour in AppointmentDay variable. It includes 00:00:00 for all rows whereas ScheduledDay column includes hour information.
New variable named time_to_app represent time difference between AppointmentDay and ScheduledDay. It is supposed to be positive but because AppointmentDay includes 00:00:00 as hour for all appointments, time_to_app value is negative if both variables are on the same day. For example, if the patient schedules at 10 am for the appointment at 3pm the same day, time_to_app value for this appointment is (-1 days + 10 hours) since instead of 3 pm, midnight is recorded in AppointmentDay variable.
```
df['ScheduledDay'] = pd.to_datetime(df['ScheduledDay'])
df['AppointmentDay'] = pd.to_datetime(df['AppointmentDay'])
df['time_to_app']= df['AppointmentDay'] - df['ScheduledDay']
import datetime as dt
rows_to_drop = df[df.time_to_app < dt.timedelta(days = -1)].index
df.drop(rows_to_drop, inplace = True)
```
All time_to_app values smaller than 1 day are omitted since it points another error.
```
time_bins = [dt.timedelta(days=-1, hours= 0), dt.timedelta(days=-1, hours= 6), dt.timedelta(days=-1, hours= 12), dt.timedelta(days=-1, hours= 15),
dt.timedelta(days=-1, hours = 18),
dt.timedelta(days=1), dt.timedelta(days=2), dt.timedelta(days=3), dt.timedelta(days=7), dt.timedelta(days=15),
dt.timedelta(days=30), dt.timedelta(days=90), dt.timedelta(days=180)]
df['time_bins'] = pd.cut(df['time_to_app'], time_bins)
df.groupby('time_bins').size()
```
I created bins for time_to_app variable. They are not equally spaced. I notice that there are significant number of patients in (-1 days, 0 days] bin. I partitioned it into smaller time bins to see the picture. The number of points in each bin is given above.
I group the data by time_bins and look at no-show rate.
```
plt.figure(figsize =(9,6))
sb.barplot(data= df, y ='time_bins', x = 'no_showup', hue = 'SMS_received', ci = None);
plt.xlabel('No-show Rate');
plt.ylabel('Time to Appointment');
```
When patient schedules an appointment for the same day which represented by the first 4 upper rows in the graaph above, no-show rate is pretty smaller than the average rate higher than 20%. If patients schedule an appointment for the same day (meaning patients make a schedule several hours before the appointment hour), with more than 95% probability they show up in the appointment. And unless there is more than 2 days to the appointment at the time the patient schedules the appointment, he does not receive SMS as a reminder. This explains why we see counterintuitive negative association between no-show rate and SMS_received variable. All patients schedule an appointment for the same day fall in no SMS received group with very low no-show rate and high number of patients and they pull down averall no-show rate of the group substantially. At the end, the rate for no SMS ends up much smaller than the rate for SMS getting patients.
We can see the effect of SMS on grouped data in the graph. SMS lowers no-show rate in every group including both 0 and 1 values for SMS_received variable. For instance, no-show rate when no SMS is a bit higher than 27% whereas it is 24% when SMS is sent for (3 days, 7 days) group. As time to appointment gets larger, SMS is being more effective. For example, SMS improves no-show rate by 3%, 5.5% and 7.7% when there are 3-7, 7-15, 30-90 days to appointment when it is scheduled, respectively.
We can see the overall effect of SMS on no-show rate by taking only those groups which have both SMS sent and no SMS sent patients. Excluding time bins smaller than 2 days, it is found that the rate is 0.33% with no SMS, and 28% with SMS sent.
But it is pretty interesting that patient attends appointment with high probability if it is the same day, and no-show rate jumps abruptly from below 5% to above 20% even if schedule day and appointment day are only 1 day apart.
```
sms_sent = df[( df.AppointmentDay - df.ScheduledDay) >= dt.timedelta(days = 2) ]
sms_sent.groupby('SMS_received').no_showup.mean()
```
<a id='conclusions'></a>
## Conclusions
- If schedule day and appointment day are on the same day, the patient will show up with very high probability (higher than 95%).
- This probability drops abruptly below 80% even when scheduled appointment is as early as tomorrow.
- The probability of no showing up increases as appointment is scheduled for distant future than near future.
- No-show rate does not show significant difference across neighbourhoods.
- Having hypertension and diabetes, being old, non-being registered in Brazilian welfare program and receving SMS as a reminder of appointment increases the probability of patient showing up for her scheduled appointment.
- The effect of receiving SMS increases as the appointment is scheduled more in advance.
| github_jupyter |
# Test for Embedding, to later move it into a layer
```
import numpy as np
# Set-up numpy generator for random numbers
random_number_generator = np.random.default_rng()
# First tokenize the protein sequence (or any sequence) in kmers.
def tokenize(protein_seqs, kmer_sz):
kmers = set()
# Loop over protein sequences
for protein_seq in protein_seqs:
# Loop over the whole sequence
for i in range(len(protein_seq) - (kmer_sz - 1)):
# Add kmers to the set, thus only unique kmers will remain
kmers.add(protein_seq[i: i + kmer_sz])
# Map kmers for one hot-encoding
kmer_to_id = dict()
id_to_kmer = dict()
for ind, kmer in enumerate(kmers):
kmer_to_id[kmer] = ind
id_to_kmer[ind] = kmer
vocab_sz = len(kmers)
assert vocab_sz == len(kmer_to_id.keys())
# Tokenize the protein sequence to integers
tokenized = []
for protein_seq in protein_seqs:
sequence = []
for i in range(len(protein_seq) - (kmer_sz -1)):
# Convert kmer to integer
kmer = protein_seq[i: i + kmer_sz]
sequence.append(kmer_to_id[kmer])
tokenized.append(sequence)
return tokenized, vocab_sz, kmer_to_id, id_to_kmer
# Embedding dictionary to embed the tokenized sequence
def embed(EMBEDDING_DIM, vocab_sz, rng):
embedding = {}
for i in range(vocab_sz):
# Use random number generator to fill the embedding with embedding_dimension random numbers
embedding[i] = rng.random(size=(embedding_dim, 1))
return embedding
if __name__ == '__main__':
# Globals
KMER_SIZE = 3 # Choose a Kmer_size (this is a hyperparameter which can be optimized)
EMBEDDING_DIM = 10 # Also a hyperparameter
# Store myoglobin protein sequence in a list of protein sequences
protein_seqs = ['MGLSDGEWQLVLNVWGKVEADIPGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASEDLKKHGATVLTALGGILKKKGHHEAEIKPLAQSHATKHKIPVKYLEFISECIIQVLQSKHPGDFGADAQGAMNKALELFRKDMASNYKELGFQG']
# Tokenize the protein sequence
tokenized_seqs, vocab_sz, kmer_to_id, id_to_kmer = tokenize(protein_seqs, KMER_SIZE)
embedding = embed(embedding_dim, vocab_sz, random_number_generator)
assert vocab_sz == len(embedding)
# Embed the tokenized protein sequence
for protein_seq in tokenized_seqs:
for token in protein_seq:
print(embedding[token])
break
# Embedding matrix to embed the tokenized sequence
def embed(embedding_dim, vocab_sz, rng):
embedding = rng.random(size=(embedding_dim, vocab_sz))
return embedding
emb = embed(EMBEDDING_DIM, vocab_sz, random_number_generator)
emb.shape
# First tokenize the protein sequence (or any sequence) in kmers.
def tokenize(protein_seqs, kmer_sz):
kmers = set()
# Loop over protein sequences
for protein_seq in protein_seqs:
# Loop over the whole sequence
for i in range(len(protein_seq) - (kmer_sz - 1)):
# Add kmers to the set, thus only unique kmers will remain
kmers.add(protein_seq[i: i + kmer_sz])
# Map kmers for one hot-encoding
kmer_to_id = dict()
id_to_kmer = dict()
for ind, kmer in enumerate(kmers):
kmer_to_id[kmer] = ind
id_to_kmer[ind] = kmer
vocab_sz = len(kmers)
assert vocab_sz == len(kmer_to_id.keys())
# Tokenize the protein sequence to a one-hot-encoded matrix
tokenized = []
for protein_seq in protein_seqs:
sequence = []
for i in range(len(protein_seq) - (kmer_sz -1)):
# Convert kmer to integer
kmer = protein_seq[i: i + kmer_sz]
# One hot encode the kmer
x = kmer_to_id[kmer]
x_vec = np.zeros((vocab_sz, 1))
x_vec[x] = 1
sequence.append(x_vec)
tokenized.append(sequence)
return tokenized, vocab_sz, kmer_to_id, id_to_kmer
# Tokenize the protein sequence
tokenized_seqs, vocab_sz, kmer_to_id, id_to_kmer = tokenize(protein_seqs, KMER_SIZE)
for tokenized_seq in tokenized_seqs:
y = np.dot(emb, tokenized_seq)
y.shape
np.array()
```
| github_jupyter |
# Spectral encoding of categorical features
About a year ago I was working on a regression model, which had over a million features. Needless to say, the training was super slow, and the model was overfitting a lot. After investigating this issue, I realized that most of the features were created using 1-hot encoding of the categorical features, and some of them had tens of thousands of unique values.
The problem of mapping categorical features to lower-dimensional space is not new. Recently one of the popular way to deal with it is using entity embedding layers of a neural network. However that method assumes that neural networks are used. What if we decided to use tree-based algorithms instead? In tis case we can use Spectral Graph Theory methods to create low dimensional embedding of the categorical features.
The idea came from spectral word embedding, spectral clustering and spectral dimensionality reduction algorithms.
If you can define a similarity measure between different values of the categorical features, we can use spectral analysis methods to find the low dimensional representation of the categorical feature.
From the similarity function (or kernel function) we can construct an Adjacency matrix, which is a symmetric matrix, where the ij element is the value of the kernel function between category values i and j:
$$ A_{ij} = K(i,j) \tag{1}$$
It is very important that I only need a Kernel function, not a high-dimensional representation. This means that 1-hot encoding step is not necessary here. Also for the kernel-base machine learning methods, the categorical variable encoding step is not necessary as well, because what matters is the kernel function between two points, which can be constructed using the individual kernel functions.
Once the adjacency matrix is constructed, we can construct a degree matrix:
$$ D_{ij} = \delta_{ij} \sum_{k}{A_{ik}} \tag{2} $$
Here $\delta$ is the Kronecker delta symbol. The Laplacian matrix is the difference between the two:
$$ L = D - A \tag{3} $$
And the normalize Laplacian matrix is defined as:
$$ \mathscr{L} = D^{-\frac{1}{2}} L D^{-\frac{1}{2}} \tag{4} $$
Following the Spectral Graph theory, we proceed with eigendecomposition of the normalized Laplacian matrix. The number of zero eigenvalues correspond to the number of connected components. In our case, let's assume that our categorical feature has two sets of values that are completely dissimilar. This means that the kernel function $K(i,j)$ is zero if $i$ and $j$ belong to different groups. In this case we will have two zero eigenvalues of the normalized Laplacian matrix.
If there is only one connected component, we will have only one zero eigenvalue. Normally it is uninformative and is dropped to prevent multicollinearity of features. However we can keep it if we are planning to use tree-based models.
The lower eigenvalues correspond to "smooth" eigenvectors (or modes), that are following the similarity function more closely. We want to keep only these eigenvectors and drop the eigenvectors with higher eigenvalues, because they are more likely represent noise. It is very common to look for a gap in the matrix spectrum and pick the eigenvalues below the gap. The resulting truncated eigenvectors can be normalized and represent embeddings of the categorical feature values.
As an example, let's consider the Day of Week. 1-hot encoding assumes every day is similar to any other day ($K(i,j) = 1$). This is not a likely assumption, because we know that days of the week are different. For example, the bar attendance spikes on Fridays and Saturdays (at least in USA) because the following day is a weekend. Label encoding is also incorrect, because it will make the "distance" between Monday and Wednesday twice higher than between Monday and Tuesday. And the "distance" between Sunday and Monday will be six times higher, even though the days are next to each other. By the way, the label encoding corresponds to the kernel $K(i, j) = exp(-\gamma |i-j|)$
```
import numpy as np
import pandas as pd
np.set_printoptions(linewidth=130)
def normalized_laplacian(A):
'Compute normalized Laplacian matrix given the adjacency matrix'
d = A.sum(axis=0)
D = np.diag(d)
L = D-A
D_rev_sqrt = np.diag(1/np.sqrt(d))
return D_rev_sqrt @ L @ D_rev_sqrt
```
We will consider an example, where weekdays are similar to each other, but differ a lot from the weekends.
```
#The adjacency matrix for days of the week
A_dw = np.array([[0,10,9,8,5,2,1],
[0,0,10,9,5,2,1],
[0,0,0,10,8,2,1],
[0,0,0,0,10,2,1],
[0,0,0,0,0,5,3],
[0,0,0,0,0,0,10],
[0,0,0,0,0,0,0]])
A_dw = A_dw + A_dw.T
A_dw
#The normalized Laplacian matrix for days of the week
L_dw_noem = normalized_laplacian(A_dw)
L_dw_noem
#The eigendecomposition of the normalized Laplacian matrix
sz, sv = np.linalg.eig(L_dw_noem)
sz
```
Notice, that the eigenvalues are not ordered here. Let's plot the eigenvalues, ignoring the uninformative zero.
```
%matplotlib inline
from matplotlib import pyplot as plt
import seaborn as sns
sns.stripplot(data=sz[1:], jitter=False, );
```
We can see a pretty substantial gap between the first eigenvalue and the rest of the eigenvalues. If this does not give enough model performance, you can include the second eigenvalue, because the gap between it and the higher eigenvalues is also quite substantial.
Let's print all eigenvectors:
```
sv
```
Look at the second eigenvector. The weekend values have a different size than the weekdays and Friday is close to zero. This proves the transitional role of Friday, that, being a day of the week, is also the beginning of the weekend.
If we are going to pick two lowest non-zero eigenvalues, our categorical feature encoding will result in these category vectors:
```
#Picking only two eigenvectors
category_vectors = sv[:,[1,3]]
category_vectors
category_vector_frame=pd.DataFrame(category_vectors, index=['mon', 'tue', 'wed', 'thu', 'fri', 'sat', 'sun'],
columns=['col1', 'col2']).reset_index()
sns.scatterplot(data=category_vector_frame, x='col1', y='col2', hue='index');
```
In the plot above we see that Monday and Tuesday, and also Saturday and Sunday are clustered close together, while Wednesday, Thursday and Friday are far apart.
## Learning the kernel function
In the previous example we assumed that the similarity function is given. Sometimes this is the case, where it can be defined based on the business rules. However it may be possible to learn it from data.
One of the ways to compute the Kernel is using [Wasserstein distance](https://en.wikipedia.org/wiki/Wasserstein_metric). It is a good way to tell how far apart two distributions are.
The idea is to estimate the data distribution (including the target variable, but excluding the categorical variable) for each value of the categorical variable. If for two values the distributions are similar, then the divergence will be small and the similarity value will be large. As a measure of similarity I choose the RBF kernel (Gaussian radial basis function):
$$ A_{ij} = exp(-\gamma W(i, j)^2) \tag{5}$$
Where $W(i,j)$ is the Wasserstein distance between the data distributions for the categories i and j, and $\gamma$ is a hyperparameter that has to be tuned
To try this approach will will use [liquor sales data set](https://www.kaggle.com/residentmario/iowa-liquor-sales/downloads/iowa-liquor-sales.zip/1). To keep the file small I removed some columns and aggregated the data.
```
liq = pd.read_csv('Iowa_Liquor_agg.csv', dtype={'Date': 'str', 'Store Number': 'str', 'Category': 'str', 'orders': 'int', 'sales': 'float'},
parse_dates=True)
liq.Date = pd.to_datetime(liq.Date)
liq.head()
```
Since we care about sales, let's encode the day of week using the information from the sales column
Let's check the histogram first:
```
sns.distplot(liq.sales, kde=False);
```
We see that the distribution is very skewed, so let's try to use log of sales columns instead
```
sns.distplot(np.log10(1+liq.sales), kde=False);
```
This is much better. So we will use a log for our distribution
```
liq["log_sales"] = np.log10(1+liq.sales)
```
Here we will follow [this blog](https://amethix.com/entropy-in-machine-learning/) for computation of the Kullback-Leibler divergence.
Also note, that since there are no liquor sales on Sunday, we consider only six days in a week
```
from scipy.stats import wasserstein_distance
from numpy import histogram
from scipy.stats import iqr
def dw_data(i):
return liq[liq.Date.dt.dayofweek == i].log_sales
def wass_from_data(i,j):
return wasserstein_distance(dw_data(i), dw_data(j)) if i > j else 0.0
distance_matrix = np.fromfunction(np.vectorize(wass_from_data), (6,6))
distance_matrix += distance_matrix.T
distance_matrix
```
As we already mentioned, the hyperparameter $\gamma$ has to be tuned. Here we just pick the value that will give a plausible result
```
gamma = 100
kernel = np.exp(-gamma * distance_matrix**2)
np.fill_diagonal(kernel, 0)
kernel
norm_lap = normalized_laplacian(kernel)
sz, sv = np.linalg.eig(norm_lap)
sz
sns.stripplot(data=sz[1:], jitter=False, );
```
Ignoring the zero eigenvalue, we can see that there is a bigger gap between the first eigenvalue and the rest of the eigenvalues, even though the values are all in the range between 1 and 1.3. Looking at the eigenvectors,
```
sv
```
Ultimately the number of eigenvectors to use is another hyperparameter, that should be optimized on a supervised learning task. The Category field is another candidate to do spectral analysis, and is, probably, a better choice since it has more unique values
```
len(liq.Category.unique())
unique_categories = liq.Category.unique()
def dw_data_c(i):
return liq[liq.Category == unique_categories[int(i)]].log_sales
def wass_from_data_c(i,j):
return wasserstein_distance(dw_data_c(i), dw_data_c(j)) if i > j else 0.0
#WARNING: THIS WILL TAKE A LONG TIME
distance_matrix = np.fromfunction(np.vectorize(wass_from_data_c), (107,107))
distance_matrix += distance_matrix.T
distance_matrix
def plot_eigenvalues(gamma):
"Eigendecomposition of the kernel and plot of the eigenvalues"
kernel = np.exp(-gamma * distance_matrix**2)
np.fill_diagonal(kernel, 0)
norm_lap = normalized_laplacian(kernel)
sz, sv = np.linalg.eig(norm_lap)
sns.stripplot(data=sz[1:], jitter=True, );
plot_eigenvalues(100);
```
We can see, that a lot of eigenvalues are grouped around the 1.1 mark. The eigenvalues that are below that cluster can be used for encoding the Category feature.
Please also note that this method is highly sensitive on selection of hyperparameter $\gamma$. For illustration let me pick a higher and a lower gamma
```
plot_eigenvalues(500);
plot_eigenvalues(10)
```
## Conclusion and next steps
We presented a way to encode the categorical features as a low dimensional vector that preserves most of the feature similarity information. For this we use methods of Spectral analysis on the values of the categorical feature. In order to find the kernel function we can either use heuristics, or learn it using a variety of methods, for example, using Kullback–Leibler divergence of the data distribution conditional on the category value. To select the subset of the eigenvectors we used gap analysis, but what we really need is to validate this methods by analyzing a variety of datasets and both classification and regression problems. We also need to compare it with other encoding methods, for example, entity embedding using Neural Networks. The kernel function we used can also include the information about category frequency, which will help us deal with high information, but low frequency values.
| github_jupyter |
# Multivariate SuSiE and ENLOC model
## Aim
This notebook aims to demonstrate a workflow of generating posterior inclusion probabilities (PIPs) from GWAS summary statistics using SuSiE regression and construsting SNP signal clusters from global eQTL analysis data obtained from multivariate SuSiE models.
## Methods overview
This procedure assumes that molecular phenotype summary statistics and GWAS summary statistics are aligned and harmonized to have consistent allele coding (see [this module](../../misc/summary_stats_merger.html) for implementation details). Both molecular phenotype QTL and GWAS should be fine-mapped beforehand using mvSusiE or SuSiE. We further assume (and require) that molecular phenotype and GWAS data come from the same population ancestry. Violations from this assumption may not cause an error in the analysis computational workflow but the results obtained may not be valid.
## Input
1) GWAS Summary Statistics with the following columns:
- chr: chromosome number
- bp: base pair position
- a1: effect allele
- a2: other allele
- beta: effect size
- se: standard error of beta
- z: z score
2) eQTL data from multivariate SuSiE model with the following columns:
- chr: chromosome number
- bp: base pair position
- a1: effect allele
- a2: other allele
- pip: posterior inclusion probability
3) LD correlation matrix
## Output
Intermediate files:
1) GWAS PIP file with the following columns
- var_id
- ld_block
- snp_pip
- block_pip
2) eQTL annotation file with the following columns
- chr
- bp
- var_id
- a1
- a2
- annotations, in the format: `gene:cs_num@tissue=snp_pip[cs_pip:cs_total_snps]`
Final Outputs:
1) Enrichment analysis result prefix.enloc.enrich.rst: estimated enrichment parameters and standard errors.
2) Signal-level colocalization result prefix.enloc.sig.out: the main output from the colocalization analysis with the following format
- column 1: signal cluster name (from eQTL analysis)
- column 2: number of member SNPs
- column 3: cluster PIP of eQTLs
- column 4: cluster PIP of GWAS hits (without eQTL prior)
- column 5: cluster PIP of GWAS hits (with eQTL prior)
- column 6: regional colocalization probability (RCP)
3) SNP-level colocalization result prefix.enloc.snp.out: SNP-level colocalization output with the following form at
- column 1: signal cluster name
- column 2: SNP name
- column 3: SNP-level PIP of eQTLs
- column 4: SNP-level PIP of GWAS (without eQTL prior)
- column 5: SNP-level PIP of GWAS (with eQTL prior)
- column 6: SNP-level colocalization probability
4) Sorted list of colocalization signals
Takes into consideration 3 situations:
1) "Major" and "minor" alleles flipped
2) Different strand but same variant
3) Remove variants with A/T and C/G alleles due to ambiguity
## Minimal working example
```
sos run mvenloc.ipynb merge \
--cwd output \
--eqtl-sumstats .. \
--gwas-sumstats ..
sos run mvenloc.ipynb eqtl \
--cwd output \
--sumstats-file .. \
--ld-region ..
sos run mvenloc.ipynb gwas \
--cwd output \
--sumstats-file .. \
--ld-region ..
sos run mvenloc.ipynb enloc \
--cwd output \
--eqtl-pip .. \
--gwas-pip ..
```
### Summary
```
head enloc.enrich.out
head enloc.sig.out
head enloc.snp.out
```
## Command interface
```
sos run mvenloc.ipynb -h
```
## Implementation
```
[global]
parameter: cwd = path
parameter: container = ""
```
### Step 0: data formatting
#### Extract common SNPS between the GWAS summary statistics and eQTL data
```
[merger]
# eQTL summary statistics as a list of RData
parameter: eqtl_sumstats = path
# GWAS summary stats in gz format
parameter: gwas_sumstats = path
input: eqtl_sumstats, gwas_sumstats
output: f"{cwd:a}/{eqtl_sumstats:bn}.standardized.gz", f"{cwd:a}/{gwas_sumstats:bn}.standardized.gz"
R: expand = "${ }"
###
# functions
###
allele.qc = function(a1,a2,ref1,ref2) {
# a1 and a2 are the first data-set
# ref1 and ref2 are the 2nd data-set
# Make all the alleles into upper-case, as A,T,C,G:
a1 = toupper(a1)
a2 = toupper(a2)
ref1 = toupper(ref1)
ref2 = toupper(ref2)
# Strand flip, to change the allele representation in the 2nd data-set
strand_flip = function(ref) {
flip = ref
flip[ref == "A"] = "T"
flip[ref == "T"] = "A"
flip[ref == "G"] = "C"
flip[ref == "C"] = "G"
flip
}
flip1 = strand_flip(ref1)
flip2 = strand_flip(ref2)
snp = list()
# Remove strand ambiguous SNPs (scenario 3)
snp[["keep"]] = !((a1=="A" & a2=="T") | (a1=="T" & a2=="A") | (a1=="C" & a2=="G") | (a1=="G" & a2=="C"))
# Remove non-ATCG coding
snp[["keep"]][ a1 != "A" & a1 != "T" & a1 != "G" & a1 != "C" ] = F
snp[["keep"]][ a2 != "A" & a2 != "T" & a2 != "G" & a2 != "C" ] = F
# as long as scenario 1 is involved, sign_flip will return TRUE
snp[["sign_flip"]] = (a1 == ref2 & a2 == ref1) | (a1 == flip2 & a2 == flip1)
# as long as scenario 2 is involved, strand_flip will return TRUE
snp[["strand_flip"]] = (a1 == flip1 & a2 == flip2) | (a1 == flip2 & a2 == flip1)
# remove other cases, eg, tri-allelic, one dataset is A C, the other is A G, for example.
exact_match = (a1 == ref1 & a2 == ref2)
snp[["keep"]][!(exact_match | snp[["sign_flip"]] | snp[["strand_flip"]])] = F
return(snp)
}
# Extract information from RData
eqtl.split = function(eqtl){
rows = length(eqtl)
chr = vector(length = rows)
pos = vector(length = rows)
a1 = vector(length = rows)
a2 = vector(length = rows)
for (i in 1:rows){
split1 = str_split(eqtl[i], ":")
split2 = str_split(split1[[1]][2], "_")
chr[i]= split1[[1]][1]
pos[i] = split2[[1]][1]
a1[i] = split2[[1]][2]
a2[i] = split2[[1]][3]
}
eqtl.df = data.frame(eqtl,chr,pos,a1,a2)
}
remove.dup = function(df){
df = df %>% arrange(PosGRCh37, -N)
df = df[!duplicated(df$PosGRCh37),]
return(df)
}
###
# Code
###
# gene regions:
# 1 = ENSG00000203710
# 2 = ENSG00000064687
# 3 = ENSG00000203710
# eqtl
gene.name = scan(${_input[0]:r}, what='character')
# initial filter of gwas variants that are in eqtl
gwas = gwas_sumstats
gwas_filter = gwas[which(gwas$id %in% var),]
# create eqtl df
eqtl.df = eqtl.split(eqtl$var)
# allele flip
f_gwas = gwas %>% filter(chr %in% eqtl.df$chr & PosGRCh37 %in% eqtl.df$pos)
eqtl.df.f = eqtl.df %>% filter(pos %in% f_gwas$PosGRCh37)
# check if there are duplicate pos
length(unique(f_gwas$PosGRCh37))
# multiple snps with same pos
dup.pos = f_gwas %>% group_by(PosGRCh37) %>% filter(n() > 1)
f_gwas = remove.dup(f_gwas)
qc = allele.qc(f_gwas$testedAllele, f_gwas$otherAllele, eqtl.df.f$a1, eqtl.df.f$a2)
keep = as.data.frame(qc$keep)
sign = as.data.frame(qc$sign_flip)
strand = as.data.frame(qc$strand_flip)
# sign flip
f_gwas$z[qc$sign_flip] = -1 * f_gwas$z[qc$sign_flip]
f_gwas$testedAllele[qc$sign_flip] = eqtl.df.f$a1[qc$sign_flip]
f_gwas$otherAllele[qc$sign_flip] = eqtl.df.f$a2[qc$sign_flip]
f_gwas$testedAllele[qc$strand_flip] = eqtl.df.f$a1[qc$strand_flip]
f_gwas$otherAllele[qc$strand_flip] = eqtl.df.f$a2[qc$strand_flip]
# remove ambigiuous
if ( sum(!qc$keep) > 0 ) {
eqtl.df.f = eqtl.df.f[qc$keep,]
f_gwas = f_gwas[qc$keep,]
}
```
#### Extract common SNPS between the summary statistics and LD
```
[eqtl_1, gwas_1 (filter LD file and sumstat file)]
parameter: sumstat_file = path
# LD and region information: chr, start, end, LD file
paramter: ld_region = path
input: sumstat_file, for_each = 'ld_region'
output: f"{cwd:a}/{sumstat_file:bn}_{region[0]}_{region[1]}_{region[2]}.z.rds",
f"{cwd:a}/{sumstat_file:bn}_{region[0]}_{region[1]}_{region[2]}.ld.rds"
R:
# FIXME: need to filter both ways for sumstats and for LD
# lds filtered
eqtl_id = which(var %in% eqtl.df.f$eqtl)
ld_f = ld[eqtl_id, eqtl_id]
# ld missing
miss = which(is.na(ld_f), arr.ind=TRUE)
miss_r = unique(as.data.frame(miss)$row)
miss_c = unique(as.data.frame(miss)$col)
total_miss = unique(union(miss_r,miss_c))
# FIXME: LD should not have missing data if properly processed by our pipeline
# In the future we should throw an error when it happens
if (length(total_miss)!=0){
ld_f2 = ld_f[-total_miss,]
ld_f2 = ld_f2[,-total_miss]
dim(ld_f2)
}else{ld_f2 = ld_f}
f_gwas.f = f_gwas %>% filter(id %in% eqtl_id.f$eqtl)
```
### Step 1: fine-mapping
```
[eqtl_2, gwas_2 (finemapping)]
# FIXME: RDS file should have included region information
output: f"{_input[0]:nn}.susieR.rds", f"{_input[0]:nn}.susieR_plot.rds"
R:
susie_results = susieR::susie_rss(z = f_gwas.f$z,R = ld_f2, check_prior = F)
susieR::susie_plot(susie_results,"PIP")
susie_results$z = f_gwas.f$z
susieR::susie_plot(susie_results,"z_original")
```
### Step 2: fine-mapping results processing
#### Construct eQTL annotation file using eQTL SNP PIPs and credible sets
```
[eqtl_3 (create signal cluster using CS)]
output: f"{_input[0]:nn}.enloc_annot.gz"
R:
cs = eqtl[["sets"]][["cs"]][["L1"]]
o_id = which(var %in% eqtl_id.f$eqtl)
pip = eqtl$pip[o_id]
eqtl_annot = cbind(eqtl_id.f, pip) %>% mutate(gene = gene.name,cluster = -1, cluster_pip = 0, total_snps = 0)
for(snp in cs){
eqtl_annot$cluster[snp] = 1
eqtl_annot$cluster_pip[snp] = eqtl[["sets"]][["coverage"]]
eqtl_annot$total_snps[snp] = length(cs)
}
eqtl_annot1 = eqtl_annot %>% filter(cluster != -1)%>%
mutate(annot = sprintf("%s:%d@=%e[%e:%d]",gene,cluster,pip,cluster_pip,total_snps)) %>%
select(c(chr,pos,eqtl,a1,a2,annot))
# FIXME: repeats whole process (extracting+fine-mapping+cs creation) 3 times before this next step
eqtl_annot_comb = rbind(eqtl_annot3, eqtl_annot1, eqtl_annot2)
# FIXME: write to a zip file
write.table(eqtl_annot_comb, file = "eqtl.annot.txt", col.names = T, row.names = F, quote = F)
```
#### Export GWAS PIP
```
[gwas_3 (format PIP into enloc GWAS input)]
output: f"{_input[0]:nn}.enloc_gwas.gz"
R:
gwas_annot1 = f_gwas.f %>% mutate(pip = susie_results$pip)
# FIXME: repeat whole process (extracting common snps + fine-mapping) 3 times before the next steps
gwas_annot_comb = rbind(gwas_annot3, gwas_annot1, gwas_annot2)
gwas_loc_annot = gwas_annot_comb %>% select(id, chr, PosGRCh37,z)
write.table(gwas_loc_annot, file = "loc.gwas.txt", col.names = F, row.names = F, quote = F)
bash:
perl format2torus.pl loc.gwas.txt > loc2.gwas.txt
R:
loc = data.table::fread("loc2.gwas.txt")
loc = loc[["V2"]]
gwas_annot_comb2 = gwas_annot_comb %>% select(id, chr, PosGRCh37,pip)
gwas_annot_comb2 = cbind(gwas_annot_comb2, loc) %>% select(id, loc, pip)
write.table(gwas_annot_comb2, file = "gwas.pip.txt", col.names = F, row.names = F, quote = F)
bash:
perl format2torus.pl gwas.pip.txt | gzip --best > gwas.pip.gz
```
### Step 3: Colocalization with FastEnloc
```
[enloc]
# eQTL summary statistics as a list of RData
# FIXME: to replace later
parameter: eqtl_pip = path
# GWAS summary stats in gz format
parameter: gwas_pip = path
input: eqtl_pip, gwas_pip
output: f"{cwd:a}/{eqtl_pip:bnn}.{gwas_pip:bnn}.xx.gz"
bash:
fastenloc -eqtl eqtl.annot.txt.gz -gwas gwas.pip.txt.gz
sort -grk6 prefix.enloc.sig.out | gzip --best > prefix.enloc.sig.sorted.gz
rm -f prefix.enloc.sig.out
```
| github_jupyter |
```
from utils import config, parse_midas_data, sample_utils as su, temporal_changes_utils, stats_utils, midas_db_utils, parse_patric
from collections import defaultdict
import math, random, numpy as np
import pickle, sys, bz2
import matplotlib.pyplot as plt
# Cohort list
cohorts = ['backhed', 'ferretti', 'yassour', 'shao', 'olm', 'hmp']
# Plot directory
plot_dir = "%s/" % (config.analysis_directory)
# Species list
good_species_list = parse_midas_data.load_pickled_good_species_list()
# Sample-subject-order maps
sys.stderr.write("Loading sample metadata...\n")
subject_sample_map = su.parse_subject_sample_map()
sample_order_map = su.parse_sample_order_map()
sample_subject_map = su.parse_sample_subject_map()
same_mi_pair_dict = su.get_same_mi_pair_dict(subject_sample_map)
sys.stderr.write("Done!\n")
# Timepoint pair types
tp_pair_names = ['MM', 'MI', 'II', 'AA']
# Cohorts
cohorts = ['backhed', 'ferretti', 'yassour', 'shao', 'hmp']
mi_cohorts = ['backhed', 'ferretti', 'yassour', 'shao']
# Samples for each cohort
samples = {cohort: su.get_sample_names(cohort) for cohort in cohorts}
hmp_samples = su.get_sample_names('hmp')
mother_samples = su.get_sample_names('mother')
infant_samples = su.get_sample_names('infant')
olm_samples = su.get_sample_names('olm')
infant_samples_no_olm = [sample for sample in infant_samples if sample not in olm_samples]
mi_samples_no_olm = [sample for sample in (mother_samples + infant_samples) if sample not in olm_samples]
# Sample-cohort map
sample_cohort_map = su.parse_sample_cohort_map()
# Sample-timepoint map
mi_sample_day_dict = su.get_mi_sample_day_dict(exclude_cohorts=['olm'])
mi_tp_sample_dict = su.get_mi_tp_sample_dict(exclude_cohorts=['olm']) # no binning
mi_tp_sample_dict_binned, mi_tp_binned_labels = su.get_mi_tp_sample_dict(exclude_cohorts=['olm'], binned=True)
# ======================================================================
# Load pickled data
# ======================================================================
# Parameters
sweep_type = 'full' # assume full for now
pp_prev_cohort = 'all'
min_coverage = 0
ddir = config.data_directory
pdir = "%s/pickles/cov%i_prev_%s/" % (ddir, min_coverage, pp_prev_cohort)
snp_changes = pickle.load(open('%s/big_snp_changes_%s.pkl' % (pdir, sweep_type), 'rb'))
gene_changes = pickle.load(open('%s/big_gene_changes_%s.pkl' % (pdir, sweep_type), 'rb'))
snp_change_freqs = pickle.load(open('%s/snp_change_freqs_%s.pkl' % (pdir, sweep_type), 'rb'))
snp_change_null_freqs = pickle.load(open('%s/snp_change_null_freqs_%s.pkl' % (pdir, sweep_type), 'rb'))
gene_gain_freqs = pickle.load(open('%s/gene_gain_freqs_%s.pkl' % (pdir, sweep_type), 'rb'))
gene_loss_freqs = pickle.load(open('%s/gene_loss_freqs_%s.pkl' % (pdir, sweep_type), 'rb'))
gene_loss_null_freqs = pickle.load(open('%s/gene_loss_null_freqs_%s.pkl' % (pdir, sweep_type), 'rb'))
between_snp_change_counts = pickle.load(open('%s/between_snp_change_counts_%s.pkl' % (pdir, sweep_type), 'rb'))
between_gene_change_counts = pickle.load(open('%s/between_gene_change_counts_%s.pkl' % (pdir, sweep_type), 'rb'))
# Relative abundance file
relab_fpath = "%s/species/relative_abundance.txt.bz2" % (config.data_directory)
relab_file = open(relab_fpath, 'r')
decompressor = bz2.BZ2Decompressor()
raw = decompressor.decompress(relab_file.read())
data = [row.split('\t') for row in raw.split('\n')]
data.pop() # Get rid of extra element due to terminal newline
header = su.parse_merged_sample_names(data[0]) # species_id, samples...
# Load species presence/absence information
sample_species_dict = defaultdict(set)
for row in data[1:]:
species = row[0]
for relab_str, sample in zip(row[1:], header[1:]):
relab = float(relab_str)
if relab > 0:
sample_species_dict[sample].add(species)
# Custom sample pair cohorts [not just sample!]
is_mi = lambda sample_i, sample_j: ((sample_i in mother_samples and sample_j in infant_samples_no_olm) and mi_sample_day_dict[sample_i] >= 0 and mi_sample_day_dict[sample_i] <= 7 and mi_sample_day_dict[sample_j] <= 7)
num_transmission = 0 # Number of MI QP pairs which are strain transmissions
num_transmission_shared_species = []
num_replacement = 0 # Number of MI QP pairs which are strain replacements
num_replacement_shared_species = []
num_total = 0 # Total number of MI QP pairs (sanity check)
num_shared_species_per_dyad = {}
shared_highcov_species_per_dyad = defaultdict(set)
existing_hosts = set()
# For every mother-infant QP pair, also count number of shared species
for species in snp_changes:
for sample_i, sample_j in snp_changes[species]:
# Only consider mother-infant QP pairs
if not is_mi(sample_i, sample_j):
continue
# Make sure only one sample pair per host
host = sample_order_map[sample_i][0][:-2]
if host in existing_hosts:
continue
existing_hosts.add(host)
num_total += 1
# Get number of shared species
shared_species = sample_species_dict[sample_i].intersection(sample_species_dict[sample_j])
num_shared_species = len(shared_species)
num_shared_species_per_dyad[(sample_i, sample_j)] = num_shared_species
shared_highcov_species_per_dyad[(sample_i, sample_j)].add(species)
# Get number of SNP differences
val = snp_changes[species][(sample_i, sample_j)]
if (type(val) == type(1)): # Replacement
num_replacement += 1
num_replacement_shared_species.append(num_shared_species)
else: # Modification or no change
num_transmission += 1
num_transmission_shared_species.append(num_shared_species)
hosts = defaultdict(int)
for s1, s2 in num_shared_species_per_dyad:
if sample_order_map[s1][0][:-2] != sample_order_map[s2][0][:-2]:
print("Weird")
hosts[sample_order_map[s1][0][:-2]] += 1
print("%i transmissions" % num_transmission)
print("%i shared species aggregated over transmissions" % sum([nss for nss in num_transmission_shared_species]))
print("%i replacements" % num_replacement)
print("%i shared species aggregated over replacements" % sum([nss for nss in num_replacement_shared_species]))
print("%i total mother-infant QP pairs" % num_total)
print("%i total shared species aggregated over dyads" % sum(num_shared_species_per_dyad.values()))
print("%i dyads" % len(shared_highcov_species_per_dyad))
print("%i total shared highcov species aggregated over dyads" % sum([len(shared_highcov_species_per_dyad[dyad]) for dyad in shared_highcov_species_per_dyad]))
float(num_transmission)/(sum([len(nss) for nss in num_transmission_shared_species])*2)
```
| github_jupyter |
```
import keras
from keras.applications import VGG16
from keras.models import Model
from keras.layers import Dense, Dropout, Input
from keras.regularizers import l2, activity_l2,l1
from keras.utils import np_utils
from keras.preprocessing.image import array_to_img, img_to_array, load_img
from keras.applications.vgg16 import preprocess_input
from PIL import Image
from scipy import misc
from keras.optimizers import SGD
# from keras.utils.visualize_util import plot
from os import listdir
import numpy as np
import matplotlib.pyplot as plt
import scipy
temperature=1
def softmaxTemp(x):
return K.softmax(x/temperature)
def getModel( output_dim):
# output_dim: the number of classes (int)
# return: compiled model (keras.engine.training.Model)
vgg_model = VGG16( weights='imagenet', include_top=True )
vgg_out = vgg_model.layers[-1].output
out = Dense( output_dim, activation='softmax')( vgg_out )
tl_model = Model( input=vgg_model.input, output=out)
tl_model.layers[-2].activation=softmaxTemp
for layer in tl_model.layers[0:-1]:
layer.trainable = False
tl_model.compile(loss= "categorical_crossentropy", optimizer="adagrad", metrics=["accuracy"])
tl_model.summary()
return tl_model
# define functions to laod images
def loadBatchImages(path,s, nVal = 2):
# return array of images
catList = listdir(path)
loadedImagesTrain = []
loadedLabelsTrain = []
loadedImagesVal = []
loadedLabelsVal = []
for cat in catList[0:256]:
deepPath = path+cat+"/"
# if cat == ".DS_Store": continue
imageList = listdir(deepPath)
indx = 0
for images in imageList[0:s + nVal]:
img = load_img(deepPath + images)
img = img_to_array(img)
img = misc.imresize(img, (224,224))
img = scipy.misc.imrotate(img,180)
if indx < s:
loadedLabelsTrain.append(int(images[0:3])-1)
loadedImagesTrain.append(img)
else:
loadedLabelsVal.append(int(images[0:3])-1)
loadedImagesVal.append(img)
indx += 1
# return np.asarray(loadedImages), np.asarray(loadedLabels)
return loadedImagesTrain, np_utils.to_categorical(loadedLabelsTrain), loadedImagesVal, np_utils.to_categorical(loadedLabelsVal)
def shuffledSet(a, b):
# shuffle the entire dataset
assert np.shape(a)[0] == np.shape(b)[0]
p = np.random.permutation(np.shape(a)[0])
return (a[p], b[p])
path = "/mnt/cube/VGG_/256_ObjectCategories/"
samCat = 8 # number of samples per category
data, labels, dataVal, labelsVal = loadBatchImages(path,samCat, nVal = 2)
data = preprocess_input(np.float64(data))
data = data.swapaxes(1, 3).swapaxes(2, 3)
dataVal = preprocess_input(np.float64(dataVal))
dataVal = dataVal.swapaxes(1, 3).swapaxes(2, 3)
train = shuffledSet(np.asarray(data),labels)
val = shuffledSet(np.asarray(dataVal),labelsVal)
# plt.imshow(train[0][0][0])
# plt.show()
print train[0].shape, val[0].shape
output_dim = 256
tl_model = getModel(output_dim)
nb_epoch = 20
history = tl_model.fit(train[0], train[1], batch_size = 16, nb_epoch = nb_epoch, validation_data = val,
shuffle = True)
keras.callbacks.EarlyStopping(monitor='val_loss', min_delta = 0, patience = 2, verbose = 0, mode='auto')
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('Model loss for %d samples per category' % samCat)
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='right left')
plt.show()
plt.plot(history.history['val_acc'])
plt.title('model accuracy for %d samples per category' % samCat)
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.show()
1 22.07
2 19.82
4 25.20
8 18.36
16 18.75
X=[2, 4, 8, 16, 64]
Y=[19.82, 25.20, 18.36, 18.75]
plt.plot(X,Y)
plt.show()
```
| github_jupyter |
# Guided Investigation - Anomaly Lookup
__Notebook Version:__ 1.0<br>
__Python Version:__ Python 3.6 (including Python 3.6 - AzureML)<br>
__Required Packages:__ azure 4.0.0, azure-cli-profile 2.1.4<br>
__Platforms Supported:__<br>
- Azure Notebooks Free Compute
- Azure Notebook on DSVM
__Data Source Required:__<br>
- Log Analytics tables
### Description
Gain insights into the possible root cause of an alert by searching for related anomalies on the corresponding entities around the alert’s time. This notebook will provide valuable leads for an alert’s investigation, listing all suspicious increase in event counts or their properties around the time of the alert, and linking to the corresponding raw records in Log Analytics for the investigator to focus on and interpret.
<font>When you switch between Azure Notebooks Free Compute and Data Science Virtual Machine (DSVM), you may need to select Python version: please select Python 3.6 for Free Compute, and Python 3.6 - AzureML for DSVM.</font>
## Table of Contents
1. Initialize Azure Resource Management Clients
2. Looking up for anomaly entities
## 1. Initialize Azure Resource Management Clients
```
# only run once
!pip install --upgrade Azure-Sentinel-Utilities
!pip install azure-cli-core
# User Input and Save to Environmental store
import os
from SentinelWidgets import WidgetViewHelper
env_dir = %env
helper = WidgetViewHelper()
# Enter Tenant Domain
helper.set_env(env_dir, 'tenant_domain')
# Enter Azure Subscription Id
helper.set_env(env_dir, 'subscription_id')
# Enter Azure Resource Group
helper.set_env(env_dir, 'resource_group')
env_dir = %env
if 'tenant_domain' in env_dir:
tenant_domain = env_dir['tenant_domain']
if 'subscription_id' in env_dir:
subscription_id = env_dir['subscription_id']
if 'resource_group' in env_dir:
resource_group = env_dir['resource_group']
from azure.loganalytics import LogAnalyticsDataClient
from azure.loganalytics.models import QueryBody
from azure.mgmt.loganalytics import LogAnalyticsManagementClient
import SentinelAzure
from SentinelAnomalyLookup import AnomalyFinder, AnomalyLookupViewHelper
from pandas.io.json import json_normalize
import sys
import timeit
import datetime as dt
import pandas as pd
import copy
from IPython.display import HTML
# Authentication to Log Analytics
from azure.common.client_factory import get_client_from_cli_profile
from azure.common.credentials import get_azure_cli_credentials
# please enter your tenant domain below, for Microsoft, using: microsoft.onmicrosoft.com
!az login --tenant $tenant_domain
la_client = get_client_from_cli_profile(LogAnalyticsManagementClient, subscription_id = subscription_id)
la = SentinelAzure.azure_loganalytics_helper.LogAnalyticsHelper(la_client)
creds, _ = get_azure_cli_credentials(resource="https://api.loganalytics.io")
la_data_client = LogAnalyticsDataClient(creds)
```
## 2. Looking up for anomaly entities
```
# Select a workspace
selected_workspace = WidgetViewHelper.select_log_analytics_workspace(la)
display(selected_workspace)
import ipywidgets as widgets
workspace_id = la.get_workspace_id(selected_workspace.value)
#DateTime format: 2019-07-15T07:05:20.000
q_timestamp = widgets.Text(value='2019-09-15',description='DateTime: ')
display(q_timestamp)
#Entity format: computer
q_entity = widgets.Text(value='computer',description='Entity for search: ')
display(q_entity)
anomaly_lookup = AnomalyFinder(workspace_id, la_data_client)
selected_tables = WidgetViewHelper.select_multiple_tables(anomaly_lookup)
display(selected_tables)
# This action may take a few minutes or more, please be patient.
start = timeit.default_timer()
anomalies, queries = anomaly_lookup.run(q_timestamp.value, q_entity.value, list(selected_tables.value))
display(anomalies)
if queries is not None:
url = WidgetViewHelper.construct_url_for_log_analytics_logs(tenant_domain, subscription_id, resource_group, selected_workspace.value)
WidgetViewHelper.display_html(WidgetViewHelper.copy_to_clipboard(url, queries, 'Add queries to clipboard and go to Log Analytics'))
print('==================')
print('Elapsed time: ', timeit.default_timer() - start, ' seconds')
```
| github_jupyter |
# 4 - Train models and make predictions
## Motivation
- **`tf.keras`** API offers built-in functions for training, validation and prediction.
- Those functions are easy to use and enable you to train any ML model.
- They also give you a high level of customizability.
## Objectives
- Understand the common training workflow in TensorFlow.
- Set an optimizer, a loss functions, and metrics with `Model.compile()`
- Create custom losses and metrics from scratch
- Train your model with `Model.fit()`
- Evaluate your model with `Model.evaluate()`
- Make predictions with `Model.predict()`
- Discover useful callbacks during training like checkpointing and learning rate scheduling
- Create custom callbacks to get 100% control on your training
- Practise what you learned on a concrete example
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
tf.__version__
```
# Table of contents:
* ### [Overview](#overview)
* ### [Part 1: Setting an optimizer, a loss function, and metrics](#part-1)
* ### [Part 2: Training models and make predictions](#part-2)
* ### [Part 3: Using callbacks](#part-3)
* ### [Part 4: Exercise](#part-4)
* ### [Summary](#summary)
* ### [Where to go next](#next)
# Overview <a class="anchor" id="overview"></a>
- Model training and evaluation works exactly the same whether your model is a Sequential model, a model built with the Functional API, or a model written from scratch via model subclassing.
- Here's what the typical end-to-end workflow looks like:
- Define optimizer, training loss, and evaluation metrics (via `Model.compile()`)
- Train your model on your training data (via `Model.fit()`)
- Validate on a holdout set generated from the original training data
- Evaluate the model on the test data (via `Model.evaluate()`)
In the next sections we will use the **MNIST dataset** to explained in details how to train a model with the `keras.Model`'s methods listed above. As a reminder from chapter 2, the **MNIST dataset** is a large dataset of handwritten digits. Each image is a 28x28 matrix having values between 0 and 255.

The following code cells build a `tf.data` pipeline for the MNIST dataset, splitting the dataset into a training set, a validation set and a test set (resp. 60%, 20%, and 20%), and build a simple artificial neural network model (ANN) for classification.
```
# Load the MNIST dataset
train, test = tf.keras.datasets.mnist.load_data()
# Overview of the dataset:
images, labels = train
print(type(images), type(labels))
print(images.shape, labels.shape)
# First 9 images of the training set:
plt.figure(figsize=(3,3))
for i in range(9):
plt.subplot(3,3,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(images[i], cmap=plt.cm.binary)
plt.show()
# creates tf.data.Dataset
train_ds = tf.data.Dataset.from_tensor_slices(train)
test_ds = tf.data.Dataset.from_tensor_slices(test)
# split train into train and validation
num_val = train_ds.cardinality().numpy() * 0.2
train_ds = train_ds.skip(num_val)
val_ds = train_ds.take(num_val)
def configure_dataset(ds, is_training=True):
if is_training:
ds = ds.shuffle(48000).repeat()
ds = ds.batch(64)
ds = ds.map(lambda image, label: (image/255, label), num_parallel_calls=tf.data.AUTOTUNE)
ds = ds.prefetch(tf.data.AUTOTUNE)
return ds
train_ds = configure_dataset(train_ds, is_training=True)
val_ds = configure_dataset(val_ds, is_training=False)
# Build the model:
model = keras.Sequential([
keras.Input(shape=(28, 28), name="digits"),
layers.Flatten(),
layers.Dense(64, activation="relu", name="dense_1"),
layers.Dense(64, activation="relu", name="dense_2"),
layers.Dense(10, activation="softmax", name="predictions"),
])
model.summary()
```
# Part 1: Setting an optimizer, a loss function, and metrics <a class="anchor" id="part-1"></a>
## 1.1 The `compile()` method <a class="anchor" id="1.1"></a>
## 1.2 The `compile()` method <a class="anchor" id="1.2"></a>
## 1.3 The `compile()` method <a class="anchor" id="1.3"></a>
## 1.4 The `compile()` method <a class="anchor" id="1.4"></a>
## 1.5 The `compile()` method <a class="anchor" id="1.5"></a>
# Part 2: Training models and make predictions <a class="anchor" id="part-2"></a>
# Part 3: Using callbacks <a class="anchor" id="part-3"></a>
# Part 4: Exercise <a class="anchor" id="part-4"></a>
# Summary <a class="anchor" id="summary"></a>
# Where to go next <a class="anchor" id="next"></a>
| github_jupyter |
<a href="https://colab.research.google.com/github/dribnet/clipit/blob/future/demos/CLIP_GradCAM_Visualization.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# CLIP GradCAM Colab
This Colab notebook uses [GradCAM](https://arxiv.org/abs/1610.02391) on OpenAI's [CLIP](https://openai.com/blog/clip/) model to produce a heatmap highlighting which regions in an image activate the most to a given caption.
**Note:** Currently only works with the ResNet variants of CLIP. ViT support coming soon.
```
#@title Install dependencies
#@markdown Please execute this cell by pressing the _Play_ button
#@markdown on the left.
#@markdown **Note**: This installs the software on the Colab
#@markdown notebook in the cloud and not on your computer.
%%capture
!pip install ftfy regex tqdm matplotlib opencv-python scipy scikit-image
!pip install git+https://github.com/openai/CLIP.git
import numpy as np
import torch
import os
import torch.nn as nn
import torch.nn.functional as F
import cv2
import urllib.request
import matplotlib.pyplot as plt
import clip
from PIL import Image
from skimage import transform as skimage_transform
from scipy.ndimage import filters
#@title Helper functions
#@markdown Some helper functions for overlaying heatmaps on top
#@markdown of images and visualizing with matplotlib.
def normalize(x: np.ndarray) -> np.ndarray:
# Normalize to [0, 1].
x = x - x.min()
if x.max() > 0:
x = x / x.max()
return x
# Modified from: https://github.com/salesforce/ALBEF/blob/main/visualization.ipynb
def getAttMap(img, attn_map, blur=True):
if blur:
attn_map = filters.gaussian_filter(attn_map, 0.02*max(img.shape[:2]))
attn_map = normalize(attn_map)
cmap = plt.get_cmap('jet')
attn_map_c = np.delete(cmap(attn_map), 3, 2)
attn_map = 1*(1-attn_map**0.7).reshape(attn_map.shape + (1,))*img + \
(attn_map**0.7).reshape(attn_map.shape+(1,)) * attn_map_c
return attn_map
def viz_attn(img, attn_map, blur=True):
fig, axes = plt.subplots(1, 2, figsize=(10, 5))
axes[0].imshow(img)
axes[1].imshow(getAttMap(img, attn_map, blur))
for ax in axes:
ax.axis("off")
plt.show()
def load_image(img_path, resize=None):
image = Image.open(image_path).convert("RGB")
if resize is not None:
image = image.resize((resize, resize))
return np.asarray(image).astype(np.float32) / 255.
#@title GradCAM: Gradient-weighted Class Activation Mapping
#@markdown Our gradCAM implementation registers a forward hook
#@markdown on the model at the specified layer. This allows us
#@markdown to save the intermediate activations and gradients
#@markdown at that layer.
#@markdown To visualize which parts of the image activate for
#@markdown a given caption, we use the caption as the target
#@markdown label and backprop through the network using the
#@markdown image as the input.
#@markdown In the case of CLIP models with resnet encoders,
#@markdown we save the activation and gradients at the
#@markdown layer before the attention pool, i.e., layer4.
class Hook:
"""Attaches to a module and records its activations and gradients."""
def __init__(self, module: nn.Module):
self.data = None
self.hook = module.register_forward_hook(self.save_grad)
def save_grad(self, module, input, output):
self.data = output
output.requires_grad_(True)
output.retain_grad()
def __enter__(self):
return self
def __exit__(self, exc_type, exc_value, exc_traceback):
self.hook.remove()
@property
def activation(self) -> torch.Tensor:
return self.data
@property
def gradient(self) -> torch.Tensor:
return self.data.grad
# Reference: https://arxiv.org/abs/1610.02391
def gradCAM(
model: nn.Module,
input: torch.Tensor,
target: torch.Tensor,
layer: nn.Module
) -> torch.Tensor:
# Zero out any gradients at the input.
if input.grad is not None:
input.grad.data.zero_()
# Disable gradient settings.
requires_grad = {}
for name, param in model.named_parameters():
requires_grad[name] = param.requires_grad
param.requires_grad_(False)
# Attach a hook to the model at the desired layer.
assert isinstance(layer, nn.Module)
with Hook(layer) as hook:
# Do a forward and backward pass.
output = model(input)
output.backward(target)
grad = hook.gradient.float()
act = hook.activation.float()
# Global average pool gradient across spatial dimension
# to obtain importance weights.
alpha = grad.mean(dim=(2, 3), keepdim=True)
# Weighted combination of activation maps over channel
# dimension.
gradcam = torch.sum(act * alpha, dim=1, keepdim=True)
# We only want neurons with positive influence so we
# clamp any negative ones.
gradcam = torch.clamp(gradcam, min=0)
# Resize gradcam to input resolution.
gradcam = F.interpolate(
gradcam,
input.shape[2:],
mode='bicubic',
align_corners=False)
# Restore gradient settings.
for name, param in model.named_parameters():
param.requires_grad_(requires_grad[name])
return gradcam
#@title Run
#@markdown #### Image & Caption settings
image_url = 'https://images2.minutemediacdn.com/image/upload/c_crop,h_706,w_1256,x_0,y_64/f_auto,q_auto,w_1100/v1554995050/shape/mentalfloss/516438-istock-637689912.jpg' #@param {type:"string"}
image_caption = 'the cat' #@param {type:"string"}
#@markdown ---
#@markdown #### CLIP model settings
clip_model = "RN50" #@param ["RN50", "RN101", "RN50x4", "RN50x16"]
saliency_layer = "layer4" #@param ["layer4", "layer3", "layer2", "layer1"]
#@markdown ---
#@markdown #### Visualization settings
blur = True #@param {type:"boolean"}
device = "cuda" if torch.cuda.is_available() else "cpu"
model, preprocess = clip.load(clip_model, device=device, jit=False)
# Download the image from the web.
image_path = 'image.png'
urllib.request.urlretrieve(image_url, image_path)
image_input = preprocess(Image.open(image_path)).unsqueeze(0).to(device)
image_np = load_image(image_path, model.visual.input_resolution)
text_input = clip.tokenize([image_caption]).to(device)
attn_map = gradCAM(
model.visual,
image_input,
model.encode_text(text_input).float(),
getattr(model.visual, saliency_layer)
)
attn_map = attn_map.squeeze().detach().cpu().numpy()
viz_attn(image_np, attn_map, blur)
```
| github_jupyter |
# Capstone Project - Flight Delays
# Does weather events have impact the delay of flights (Brazil)?
### It is important to see this notebook with the step-by-step of the dataset cleaning process:
[https://github.com/davicsilva/dsintensive/blob/master/notebooks/flightDelayPrepData_v2.ipynb](https://github.com/davicsilva/dsintensive/blob/master/notebooks/flightDelayPrepData_v2.ipynb)
```
from datetime import datetime
# Pandas and NumPy
import pandas as pd
import numpy as np
# Matplotlib for additional customization
from matplotlib import pyplot as plt
%matplotlib inline
# Seaborn for plotting and styling
import seaborn as sns
# 1. Flight delay: any flight with (real_departure - planned_departure >= 15 minutes)
# 2. The Brazilian Federal Agency for Civil Aviation (ANAC) does not define exactly what is a "flight delay" (in minutes)
# 3. Anyway, the ANAC has a resolution for this subject: https://goo.gl/YBwbMy (last access: nov, 15th, 2017)
# ---
# DELAY, for this analysis, is defined as greater than 15 minutes (local flights only)
DELAY = 15
```
### 1 - Local flights dataset. For now, only flights from January to September, 2017
**A note about date columns on this dataset**
* In the original dataset (CSV file from ANAC), the date was not in ISO8601 format (e.g. '2017-10-31 09:03:00')
* To fix this I used regex (regular expression) to transform this column directly on CSV file
* The original date was "31/10/2017 09:03" (october, 31, 2017 09:03)
```
#[flights] dataset_01 => all "Active Regular Flights" from 2017, from january to september
#source: http://www.anac.gov.br/assuntos/dados-e-estatisticas/historico-de-voos
#Last access this website: nov, 14th, 2017
flights = pd.read_csv('data/arf2017ISO.csv', sep = ';', dtype = str)
flights['departure-est'] = flights[['departure-est']].apply(lambda row: row.str.replace("(?P<day>\d{2})/(?P<month>\d{2})/(?P<year>\d{4}) (?P<HOUR>\d{2}):(?P<MIN>\d{2})", "\g<year>/\g<month>/\g<day> \g<HOUR>:\g<MIN>:00"), axis=1)
flights['departure-real'] = flights[['departure-real']].apply(lambda row: row.str.replace("(?P<day>\d{2})/(?P<month>\d{2})/(?P<year>\d{4}) (?P<HOUR>\d{2}):(?P<MIN>\d{2})", "\g<year>/\g<month>/\g<day> \g<HOUR>:\g<MIN>:00"), axis=1)
flights['arrival-est'] = flights[['arrival-est']].apply(lambda row: row.str.replace("(?P<day>\d{2})/(?P<month>\d{2})/(?P<year>\d{4}) (?P<HOUR>\d{2}):(?P<MIN>\d{2})", "\g<year>/\g<month>/\g<day> \g<HOUR>:\g<MIN>:00"), axis=1)
flights['arrival-real'] = flights[['arrival-real']].apply(lambda row: row.str.replace("(?P<day>\d{2})/(?P<month>\d{2})/(?P<year>\d{4}) (?P<HOUR>\d{2}):(?P<MIN>\d{2})", "\g<year>/\g<month>/\g<day> \g<HOUR>:\g<MIN>:00"), axis=1)
# Departure and Arrival columns: from 'object' to 'date' format
flights['departure-est'] = pd.to_datetime(flights['departure-est'], errors='ignore')
flights['departure-real'] = pd.to_datetime(flights['departure-real'], errors='ignore')
flights['arrival-est'] = pd.to_datetime(flights['arrival-est'], errors='ignore')
flights['arrival-real'] = pd.to_datetime(flights['arrival-real'], errors='ignore')
# translate the flight status from portuguese to english
flights['flight-status'] = flights[['flight-status']].apply(lambda row: row.str.replace("REALIZADO", "ACCOMPLISHED"), axis=1)
flights['flight-status'] = flights[['flight-status']].apply(lambda row: row.str.replace("CANCELADO", "CANCELED"), axis=1)
flights.head()
flights.size
flights.to_csv("flights_csv.csv")
```
## Some EDA's tasks
```
# See: https://stackoverflow.com/questions/37287938/sort-pandas-dataframe-by-value
#
df_departures = flights.groupby(['airport-A']).size().reset_index(name='number_departures')
df_departures.sort_values(by=['number_departures'], ascending=False, inplace=True)
df_departures
```
### 2 - Local airports (list with all the ~600 brazilian public airports)
Source: https://goo.gl/mNFuPt (a XLS spreadsheet in portuguese; last access on nov, 15th, 2017)
```
# Airports dataset: all brazilian public airports (updated until october, 2017)
airports = pd.read_csv('data/brazilianPublicAirports-out2017.csv', sep = ';', dtype= str)
airports.head()
# Merge "flights" dataset with "airports" in order to identify
# local flights (origin and destination are in Brazil)
flights = pd.merge(flights, airports, left_on="airport-A", right_on="airport", how='left')
flights = pd.merge(flights, airports, left_on="airport-B", right_on="airport", how='left')
flights.tail()
```
### 3 - List of codes (two letters) used when there was a flight delay (departure)
I have found two lists that define two-letter codes used by the aircraft crew to justify the delay of the flights: a short and a long one.
Source: https://goo.gl/vUC8BX (last access: nov, 15th, 2017)
```
# ------------------------------------------------------------------
# List of codes (two letters) used to justify a delay on the flight
# - delayCodesShortlist.csv: list with YYY codes
# - delayCodesLongList.csv: list with XXX codes
# ------------------------------------------------------------------
delaycodes = pd.read_csv('data/delayCodesShortlist.csv', sep = ';', dtype = str)
delaycodesLongList = pd.read_csv('data/delayCodesLonglist.csv', sep = ';', dtype = str)
delaycodes.head()
```
### 4 - The Weather data from https://www.wunderground.com/history
From this website I captured a sample data from local airport (Campinas, SP, Brazil): January to September, 2017.
The website presents data like this (see [https://goo.gl/oKwzyH](https://goo.gl/oKwzyH)):
```
# Weather sample: load the CSV with weather historical data (from Campinas, SP, Brazil, 2017)
weather = pd.read_csv('data/DataScience-Intensive-weatherAtCampinasAirport-2017-Campinas_Airport_2017Weather.csv', \
sep = ',', dtype = str)
weather["date"] = weather["year"].map(str) + "-" + weather["month"].map(str) + "-" + weather["day"].map(str)
weather["date"] = pd.to_datetime(weather['date'],errors='ignore')
weather.head()
```
| github_jupyter |
```
# Reload when code changed:
%load_ext autoreload
%reload_ext autoreload
%autoreload 2
%pwd
import sys
import os
path = "../"
sys.path.append(path)
#os.path.abspath("../")
print(os.path.abspath(path))
import os
import core
import logging
import importlib
importlib.reload(core)
try:
logging.shutdown()
importlib.reload(logging)
except:
pass
import pandas as pd
import numpy as np
import json
import time
from event_handler import EventHandler
from event_handler import get_list_from_interval
print(core.__file__)
pd.__version__
user_id_1 = 'user_1'
user_id_2 = 'user_2'
user_1_ws_1 = 'mw1'
print(path)
paths = {'user_id': user_id_1,
'workspace_directory': 'D:/git/ekostat_calculator/workspaces',
'resource_directory': path + '/resources',
'log_directory': path + '/log',
'test_data_directory': path + '/test_data',
'temp_directory': path + '/temp',
'cache_directory': path + '/cache'}
ekos = EventHandler(**paths)
ekos.test_timer()
ekos.mapping_objects['quality_element'].get_indicator_list_for_quality_element('secchi')
def update_workspace_uuid_in_test_requests(workspace_alias='New test workspace'):
ekos = EventHandler(**paths)
workspace_uuid = ekos.get_unique_id_for_alias(workspace_alias=workspace_alias)
if workspace_uuid:
print('Updating user {} with uuid: {}'.format(user_id_1, workspace_uuid))
print('-'*70)
ekos.update_workspace_uuid_in_test_requests(workspace_uuid)
else:
print('No workspaces for user: {}'.format(user_id_1))
def update_subset_uuid_in_test_requests(workspace_alias='New test workspace',
subset_alias=False):
ekos = EventHandler(**paths)
workspace_uuid = ekos.get_unique_id_for_alias(workspace_alias=workspace_alias)
if workspace_uuid:
ekos.load_workspace(workspace_uuid)
subset_uuid = ekos.get_unique_id_for_alias(workspace_alias=workspace_alias, subset_alias=subset_alias)
print('Updating user {} with workspace_uuid {} and subset_uuid {}'.format(user_id_1, workspace_uuid, subset_uuid))
print(workspace_uuid, subset_uuid)
print('-'*70)
ekos.update_subset_uuid_in_test_requests(subset_uuid=subset_uuid)
else:
print('No workspaces for user: {}'.format(user_id_1))
def print_boolean_structure(workspace_uuid):
workspace_object = ekos.get_workspace(unique_id=workspace_uuid)
workspace_object.index_handler.print_boolean_keys()
# update_workspace_uuid_in_test_requests()
```
### Request workspace add
```
t0 = time.time()
ekos = EventHandler(**paths)
request = ekos.test_requests['request_workspace_add_1']
response_workspace_add = ekos.request_workspace_add(request)
ekos.write_test_response('request_workspace_add_1', response_workspace_add)
# request = ekos.test_requests['request_workspace_add_2']
# response_workspace_add = ekos.request_workspace_add(request)
# ekos.write_test_response('request_workspace_add_2', response_workspace_add)
print('-'*50)
print('Time for request: {}'.format(time.time()-t0))
```
#### Update workspace uuid in test requests
```
update_workspace_uuid_in_test_requests()
```
### Request workspace import default data
```
# ekos = EventHandler(**paths)
# # When copying data the first time all sources has status=0, i.e. no data will be loaded.
# request = ekos.test_requests['request_workspace_import_default_data']
# response_import_data = ekos.request_workspace_import_default_data(request)
# ekos.write_test_response('request_workspace_import_default_data', response_import_data)
```
### Import data from sharkweb
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_sharkweb_import']
response_sharkweb_import = ekos.request_sharkweb_import(request)
ekos.write_test_response('request_sharkweb_import', response_sharkweb_import)
ekos.data_params
ekos.selection_dicts
# ekos = EventHandler(**paths)
# ekos.mapping_objects['sharkweb_mapping'].df
```
### Request data source list/edit
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_workspace_data_sources_list']
response = ekos.request_workspace_data_sources_list(request)
ekos.write_test_response('request_workspace_data_sources_list', response)
request = response
request['data_sources'][0]['status'] = False
request['data_sources'][1]['status'] = False
request['data_sources'][2]['status'] = False
request['data_sources'][3]['status'] = False
# request['data_sources'][4]['status'] = True
# Edit data source
response = ekos.request_workspace_data_sources_edit(request)
ekos.write_test_response('request_workspace_data_sources_edit', response)
```
### Request subset add
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_subset_add_1']
response_subset_add = ekos.request_subset_add(request)
ekos.write_test_response('request_subset_add_1', response_subset_add)
update_subset_uuid_in_test_requests(subset_alias='mw_subset')
```
### Request subset get data filter
```
ekos = EventHandler(**paths)
update_subset_uuid_in_test_requests(subset_alias='mw_subset')
request = ekos.test_requests['request_subset_get_data_filter']
response_subset_get_data_filter = ekos.request_subset_get_data_filter(request)
ekos.write_test_response('request_subset_get_data_filter', response_subset_get_data_filter)
# import re
# string = """{
# "workspace_uuid": "52725df4-b4a0-431c-a186-5e542fc6a3a4",
# "data_sources": [
# {
# "status": true,
# "loaded": false,
# "filename": "physicalchemical_sharkweb_data_all_2013-2014_20180916.txt",
# "datatype": "physicalchemical"
# }
# ]
# }"""
# r = re.sub('"workspace_uuid": ".{36}"', '"workspace_uuid": "new"', string)
```
### Request subset set data filter
```
ekos = EventHandler(**paths)
update_subset_uuid_in_test_requests(subset_alias='mw_subset')
request = ekos.test_requests['request_subset_set_data_filter']
response_subset_set_data_filter = ekos.request_subset_set_data_filter(request)
ekos.write_test_response('request_subset_set_data_filter', response_subset_set_data_filter)
```
### Request subset get indicator settings
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_subset_get_indicator_settings']
# request = ekos.test_requests['request_subset_get_indicator_settings_no_areas']
# print(request['subset']['subset_uuid'])
# request['subset']['subset_uuid'] = 'fel'
# print(request['subset']['subset_uuid'])
response_subset_get_indicator_settings = ekos.request_subset_get_indicator_settings(request)
ekos.write_test_response('request_subset_get_indicator_settings', response_subset_get_indicator_settings)
```
### Request subset set indicator settings
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_subset_set_indicator_settings']
response_subset_set_indicator_settings = ekos.request_subset_set_indicator_settings(request)
ekos.write_test_response('request_subset_set_indicator_settings', response_subset_set_indicator_settings)
```
### Request subset calculate status
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_subset_calculate_status']
response = ekos.request_subset_calculate_status(request)
ekos.write_test_response('request_subset_calculate_status', response)
```
### Request subset result get
```
ekos = EventHandler(**paths)
request = ekos.test_requests['request_workspace_result']
response_workspace_result = ekos.request_workspace_result(request)
ekos.write_test_response('request_workspace_result', response_workspace_result)
response_workspace_result['subset']['a4e53080-2c68-40d5-957f-8cc4dbf77815']['result']['SE552170-130626']['result']['indicator_din_winter']['data']
workspace_uuid = 'fccc7645-8501-4541-975b-bdcfb40a5092'
subset_uuid = 'a4e53080-2c68-40d5-957f-8cc4dbf77815'
result = ekos.dict_data_timeseries(workspace_uuid=workspace_uuid,
subset_uuid=subset_uuid,
viss_eu_cd='SE575150-162700',
element_id='indicator_din_winter')
print(result['datasets'][0]['x'])
print()
print(result['y'])
for k in range(len(result['datasets'])):
print(result['datasets'][k]['x'])
import datetime
# Extend date list
start_year = all_dates[0].year
end_year = all_dates[-1].year+1
date_intervall = []
for year in range(start_year, end_year+1):
for month in range(1, 13):
d = datetime.datetime(year, month, 1)
if d >= all_dates[0] and d <= all_dates[-1]:
date_intervall.append(d)
extended_dates = sorted(set(all_dates + date_intervall))
# Loop dates and add/remove values
new_x = []
new_y = dict((item, []) for item in date_to_y)
for date in extended_dates:
if date in date_intervall:
new_x.append(date.strftime('%y-%b'))
else:
new_x.append('')
for i in new_y:
new_y[i].append(date_to_y[i].get(date, None))
# new_y = {}
# for i in date_to_y:
# new_y[i] = []
# for date in all_dates:
# d = date_to_y[i].get(date)
# if d:
# new_y[i].append(d)
# else:
# new_y[i].append(None)
new_y[0]
import datetime
year_list = range(2011, 2013+1)
month_list = range(1, 13)
date_list = []
for year in year_list:
for month in month_list:
date_list.append(datetime.datetime(year, month, 1))
date_list
a
y[3][i]
sorted(pd.to_datetime(df['SDATE']))
```
| github_jupyter |
Notebook prirejen s strani http://www.pieriandata.com
# NumPy Indexing and Selection
In this lecture we will discuss how to select elements or groups of elements from an array.
```
import numpy as np
#Creating sample array
arr = np.arange(0,11)
#Show
arr
```
## Bracket Indexing and Selection
The simplest way to pick one or some elements of an array looks very similar to python lists:
```
#Get a value at an index
arr[8]
#Get values in a range
arr[1:5]
#Get values in a range
arr[0:5]
# l = ['a', 'b', 'c']
# l[0:2]
```
## Broadcasting
NumPy arrays differ from normal Python lists because of their ability to broadcast. With lists, you can only reassign parts of a list with new parts of the same size and shape. That is, if you wanted to replace the first 5 elements in a list with a new value, you would have to pass in a new 5 element list. With NumPy arrays, you can broadcast a single value across a larger set of values:
```
l = list(range(10))
l
l[0:5] = [100,100,100,100,100]
l
#Setting a value with index range (Broadcasting)
arr[0:5]=100
#Show
arr
# Reset array, we'll see why I had to reset in a moment
arr = np.arange(0,11)
#Show
arr
#Important notes on Slices
slice_of_arr = arr[0:6]
#Show slice
slice_of_arr
#Change Slice
slice_of_arr[:]=99
#Show Slice again
slice_of_arr
```
Now note the changes also occur in our original array!
```
arr
```
Data is not copied, it's a view of the original array! This avoids memory problems!
```
#To get a copy, need to be explicit
arr_copy = arr.copy()
arr_copy
```
## Indexing a 2D array (matrices)
The general format is **arr_2d[row][col]** or **arr_2d[row,col]**. I recommend using the comma notation for clarity.
```
arr_2d = np.array(([5,10,15],[20,25,30],[35,40,45]))
#Show
arr_2d
#Indexing row
arr_2d[1]
# Format is arr_2d[row][col] or arr_2d[row,col]
# Getting individual element value
arr_2d[1][0]
# Getting individual element value
arr_2d[1,0]
# 2D array slicing
#Shape (2,2) from top right corner
arr_2d[:2,1:]
#Shape bottom row
arr_2d[2]
#Shape bottom row
arr_2d[2,:]
```
## More Indexing Help
Indexing a 2D matrix can be a bit confusing at first, especially when you start to add in step size. Try google image searching *NumPy indexing* to find useful images, like this one:
<img src= 'numpy_indexing.png' width=500/> Image source: http://www.scipy-lectures.org/intro/numpy/numpy.html
## Conditional Selection
This is a very fundamental concept that will directly translate to pandas later on, make sure you understand this part!
Let's briefly go over how to use brackets for selection based off of comparison operators.
```
arr = np.arange(1,11)
arr
arr > 4
bool_arr = arr>4
bool_arr
arr[bool_arr]
arr[arr>2]
x = 2
arr[arr>x]
```
# Great Job!
| github_jupyter |
# Chapter 4: Linear models
[Link to outline](https://docs.google.com/document/d/1fwep23-95U-w1QMPU31nOvUnUXE2X3s_Dbk5JuLlKAY/edit#heading=h.9etj7aw4al9w)
Concept map:

#### Notebook setup
```
import numpy as np
import pandas as pd
import scipy as sp
import seaborn as sns
from scipy.stats import uniform, norm
# notebooks figs setup
%matplotlib inline
import matplotlib.pyplot as plt
sns.set(rc={'figure.figsize':(8,5)})
blue, orange = sns.color_palette()[0], sns.color_palette()[1]
# silence annoying warnings
import warnings
warnings.filterwarnings('ignore')
```
## 4.1 Linear models for relationship between two numeric variables
- def'n linear model: **y ~ m*x + b**, a.k.a. linear regression
- Amy has collected a new dataset:
- Instead of receiving a fixed amount of stats training (100 hours),
**each employee now receives a variable amount of stats training (anywhere from 0 hours to 100 hours)**
- Amy has collected ELV values after one year as previously
- Goal find best fit line for relationship $\textrm{ELV} \sim \beta_0 + \beta_1\!*\!\textrm{hours}$
- Limitation: **we assume the change in ELV is proportional to number of hours** (i.e. linear relationship).
Other types of hours-ELV relationship possible, but we will not be able to model them correctly (see figure below).
### New dataset
- The `hours` column contains the `x` values (how many hours of statistics training did the employee receive),
- The `ELV` column contains the `y` values (the employee ELV after one year)

```
# Load data into a pandas dataframe
df2 = pd.read_excel("data/ELV_vs_hours.ods", sheet_name="Data")
# df2
df2.describe()
# plot ELV vs. hours data
sns.scatterplot(x='hours', y='ELV', data=df2)
# linear model plot (preview)
# sns.lmplot(x='hours', y='ELV', data=df2, ci=False)
```
#### Types of linear relationship between input and output
Different possible relationships between the number of hours of stats training and ELV gains:

## 4.2 Fitting linear models
- Main idea: use `fit` method from `statsmodels.ols` and a formula (approach 1)
- Visual inspection
- Results of linear model fit are:
- `beta0` = $\beta_0$ = baseline ELV (y-intercept)
- `beta1` = $\beta_1$ = increase in ELV for each additional hour of stats training (slope)
- Five more alternative fitting methods (bonus material):
2. fit using statsmodels `OLS`
3. solution using `linregress` from `scipy`
4. solution using `optimize` from `scipy`
5. linear algebra solution using `numpy`
6. solution using `LinearRegression` model from scikit-learn
### Using statsmodels formula API
The `statsmodels` Python library offers a convenient way to specify statistics model as a "formula" that describes the relationship we're looking for.
Mathematically, the linear model is written:
$\large \textrm{ELV} \ \ \sim \ \ \beta_0\cdot 1 \ + \ \beta_1\cdot\textrm{hours}$
and the formula is:
`ELV ~ 1 + hours`
Note the variables $\beta_0$ and $\beta_1$ are omitted, since the whole point of fitting a linear model is to find these coefficients. The parameters of the model are:
- Instead of $\beta_0$, the constant parameter will be called `Intercept`
- Instead of a new name $\beta_1$, we'll call it `hours` coefficient (i.e. the coefficient associated with the `hours` variable in the model)
```
import statsmodels.formula.api as smf
model = smf.ols('ELV ~ 1 + hours', data=df2)
result = model.fit()
# extact the best-fit model parameters
beta0, beta1 = result.params
beta0, beta1
# data points
sns.scatterplot(x='hours', y='ELV', data=df2)
# linear model for data
x = df2['hours'].values # input = hours
ymodel = beta0 + beta1*x # output = ELV
sns.lineplot(x, ymodel)
result.summary()
```
### Alternative model fitting methods
2. fit using statsmodels [`OLS`](https://www.statsmodels.org/stable/generated/statsmodels.regression.linear_model.OLS.html)
3. solution using [`linregress`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.linregress.html) from `scipy`
4. solution using [`minimize`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html) from `scipy`
5. [linear algebra](https://numpy.org/doc/stable/reference/routines.linalg.html) solution using `numpy`
6. solution using [`LinearRegression`](https://scikit-learn.org/stable/modules/linear_model.html#ordinary-least-squares) model from scikit-learn
#### Data pre-processing
The `statsmodels` formula `ols` approach we used above was able to get the data
directly from the dataframe `df2`, but some of the other model fitting methods
require data to be provided as regular arrays: the x-values and the y-values.
```
# extract hours and ELV data from df2
x = df2['hours'].values # hours data as an array
y = df2['ELV'].values # ELV data as an array
x.shape, y.shape
# x
```
Two of the approaches required "packaging" the x-values along with a column of ones,
to form a matrix (called a design matrix). Luckily `statsmodels` provides a convenient function for this:
```
import statsmodels.api as sm
# add a column of ones to the x data
X = sm.add_constant(x)
X.shape
# X
```
____
#### 2. fit using statsmodels OLS
```
model2 = sm.OLS(y, X)
result2 = model2.fit()
# result2.summary()
result2.params
```
____
#### 3. solution using `linregress` from `scipy`
```
from scipy.stats import linregress
result3 = linregress(x, y)
result3.intercept, result3.slope
```
____
#### 4. Using an optimization approach
```
from scipy.optimize import minimize
def sse(beta, x=x, y=y):
"""Compute the sum-of-squared-errors objective function."""
sumse = 0.0
for xi, yi in zip(x, y):
yi_pred = beta[0] + beta[1]*xi
ei = (yi_pred-yi)**2
sumse += ei
return sumse
result4 = minimize(sse, x0=[0,0])
beta0, beta1 = result4.x
beta0, beta1
```
____
#### 5. Linear algebra solution
We obtain the least squares solution using the Moore–Penrose inverse formula:
$$ \large
\vec{\beta} = (X^{\sf T} X)^{-1}X^{\sf T}\; \vec{y}
$$
```
# 5. linear algebra solution using `numpy`
import numpy as np
result5 = np.linalg.inv(X.T.dot(X)).dot(X.T).dot(y)
beta0, beta1 = result5
beta0, beta1
```
_____
#### Using scikit-learn
```
# 6. solution using `LinearRegression` from scikit-learn
from sklearn import linear_model
model6 = linear_model.LinearRegression()
model6.fit(x[:,np.newaxis], y)
model6.intercept_, model6.coef_
```
## 4.3 Interpreting linear models
- model fit checks
- $R^2$ [coefficient of determination](https://en.wikipedia.org/wiki/Coefficient_of_determination)
= the proportion of the variation in the dependent variable that is predictable from the independent variable
- plot of residuals
- many other: see [scikit docs](https://scikit-learn.org/stable/modules/model_evaluation.html#regression-metrics)
- hypothesis tests
- is slope zero or nonzero? (and CI interval)
- caution: cannot make any cause-and-effect claims; only a correlation
- Predictions
- given best-fir model obtained from data, we can make predictions (interpolations),
e.g., what is the expected ELV after 50 hours of stats training?
### Interpreting the results
Let's review some of the other data included in the `results.summary()` report for the linear model fit we did earlier.
```
result.summary()
```
### Model parameters
```
beta0, beta1 = result.params
result.params
```
### The $R^2$ coefficient of determination
$R^2 = 1$ corresponds to perfect prediction
```
result.rsquared
```
### Hypothesis testing for slope coefficient
Is there a non-zero slope coefficient?
- **null hypothesis $H_0$**: `hours` has no effect on `ELV`,
which is equivalent to $\beta_1 = 0$:
$$ \large
H_0: \qquad \textrm{ELV} \sim \mathcal{N}(\color{red}{\beta_0}, \sigma^2) \qquad \qquad \qquad
$$
- **alternative hypothesis $H_A$**: `hours` has an effect on `ELV`,
and the slope is not zero, $\beta_1 \neq 0$:
$$ \large
H_A: \qquad \textrm{ELV}
\sim
\mathcal{N}\left(
\color{blue}{\beta_0 + \beta_1\!\cdot\!\textrm{hours}},
\ \sigma^2
\right)
$$
```
# p-value under the null hypotheis of zero slope or "no effect of `hours` on `ELV`"
result.pvalues.loc['hours']
# 95% confidence interval for the hours-slope parameter
# result.conf_int()
CI_hours = list(result.conf_int().loc['hours'])
CI_hours
```
### Predictions using the model
We can use the model we obtained to predict (interpolate) the ELV for future employees.
```
sns.scatterplot(x='hours', y='ELV', data=df2)
ymodel = beta0 + beta1*x
sns.lineplot(x, ymodel)
```
What ELV can we expect from a new employee that takes 50 hours of stats training?
```
result.predict({'hours':[50]})
result.predict({'hours':[100]})
```
**WARNING**: it's not OK to extrapolate the validity of the model outside of the range of values where we have observed data.
For example, there is no reason to believe in the model's predictions about ELV for 200 or 2000 hours of stats training:
```
result.predict({'hours':[200]})
```
## Discussion
Further topics that will be covered in the book:
- Generalized linear models, e.g., [Logistic regression](https://en.wikipedia.org/wiki/Logistic_regression)
- [Everything is a linear model](https://www.eigenfoo.xyz/tests-as-linear/) article
- The verbs `fit` and `predict` will come up A LOT in machine learning,
so it's worth learning linear models in detail to be prepared for further studies.
____
Congratulations on completing this overview of statistics! We covered a lot of topics and core ideas from the book. I know some parts seemed kind of complicated at first, but if you think about them a little you'll see there is nothing too difficult to learn. The good news is that the examples in these notebooks contain all the core ideas, and you won't be exposed to anything more complicated that what you saw here!
If you were able to handle these notebooks, you'll be able to handle the **No Bullshit Guide to Statistics** too! In fact the book will cover the topics in a much smoother way, and with better explanations. You'll have a lot of exercises and problems to help you practice statistical analysis.
### Next steps
- I encourage you to check out the [book outline shared gdoc](https://docs.google.com/document/d/1fwep23-95U-w1QMPU31nOvUnUXE2X3s_Dbk5JuLlKAY/edit) if you haven't seen it already. Please leave me a comment in the google document if you see something you don't like in the outline, or if you think some important statistics topics are missing. You can also read the [book proposal blog post](https://minireference.com/blog/no-bullshit-guide-to-statistics-progress-update/) for more info about the book.
- Check out also the [concept map](https://minireference.com/static/excerpts/noBSstats/conceptmaps/BookSubjectsOverview.pdf). You can print it out and annotate with the concepts you heard about in these notebooks.
- If you want to be involved in the stats book in the coming months, sign up to the [stats reviewers mailing list](https://confirmsubscription.com/h/t/A17516BF2FCB41B2) to receive chapter drafts as they are being prepared (Nov+Dec 2021). I'll appreciate your feedback on the text. The goal is to have the book finished in the Spring 2022, and feedback and "user testing" will be very helpful.
| github_jupyter |
```
try:
import saspy
except ImportError as e:
print('Installing saspy')
%pip install saspy
import pandas as pd
# The following imports are only necessary for automated sascfg_personal.py creation
from pathlib import Path
import os
from shutil import copyfile
import getpass
# Imports without the setup check codes
import saspy
import pandas as pd
```
# Set up your connection
The next cell contains code to check if you already have a sascfg_personal.py file in your current conda environment. If you do not one is created for you.
Next [choose your access method](https://sassoftware.github.io/saspy/install.html#choosing-an-access-method) and the read through the configuration properties in sascfg_personal.py
```
# Setup for the configuration file - for running inside of a conda environment
saspyPfad = f"C:\\Users\\{getpass.getuser()}\\.conda\\envs\\{os.environ['CONDA_DEFAULT_ENV']}\\Lib\\site-packages\\saspy\\"
saspycfg_personal = Path(f'{saspyPfad}sascfg_personal.py')
if saspycfg_personal.is_file():
print('All setup and ready to go')
else:
copyfile(f'{saspyPfad}sascfg.py', f'{saspyPfad}sascfg_personal.py')
print('The configuration file was created for you, please setup your connection method')
print(f'Find sascfg_personal.py here: {saspyPfad}')
```
# Configuration
prod = {
'iomhost': 'rfnk01-0068.exnet.sas.com', <-- SAS Host Name
'iomport': 8591, <-- SAS Workspace Server Port
'class_id': '440196d4-90f0-11d0-9f41-00a024bb830c', <-- static, if the value is wrong use proc iomoperate
'provider': 'sas.iomprovider', <-- static
'encoding': 'windows-1252' <-- Python encoding for SAS session encoding
}
```
# If no configuration name is specified, you get a list of the configured ones
# sas = saspy.SASsession(cfgname='prod')
sas = saspy.SASsession()
```
# Explore some interactions with SAS
Getting a feeling for what SASPy can do.
```
# Let's take a quick look at all the different methods and variables provided by SASSession object
dir(sas)
# Get a list of all tables inside of the library sashelp
table_df = sas.list_tables(libref='sashelp', results='pandas')
# Search for a table containing a capital C in its name
table_df[table_df['MEMNAME'].str.contains('C')]
# If teach_me_sas is true instead of executing the code, we get the generated code returned
sas.teach_me_SAS(True)
sas.list_tables(libref='sashelp', results='pandas')
# Let's turn it off again to actually run the code
sas.teach_me_SAS(False)
# Create a sasdata object, based on the table cars in the sashelp library
cars = sas.sasdata('cars', 'sashelp')
# Get information about the columns in the table
cars.columnInfo()
# Creating a simple heat map
cars.heatmap('Horsepower', 'EngineSize')
# Clean up for this section
del cars, table_df
```
# Reading in data from local disc with Pandas and uploading it to SAS
1. First we are going to read in a local csv file
2. Creating a copy of the base data file in SAS
3. Append the local data to the data stored in SAS and sort it
The Opel data set:
Make,Model,Type,Origin,DriveTrain,MSRP,Invoice,EngineSize,Cylinders,Horsepower,MPG_City,MPG_Highway,Weight,Wheelbase,Length
Opel,Astra Edition,Sedan,Europe,Rear,28495,26155,3,6,22.5,16,23,4023,110,180
Opel,Astra Design & Tech,Sedan,Europe,Rear,30795,28245,4.4,8,32.5,16,22,4824,111,184
Opel,Astra Elegance,Sedan,Europe,Rear,37995,34800,2.5,6,18.4,20,29,3219,107,176
Opel,Astra Ultimate,Sedan,Europe,Rear,42795,38245,2.5,6,18.4,20,29,3197,107,177
Opel,Astra Business Edition,Sedan,Europe,Rear,28495,24800,2.5,6,18.4,19,27,3560,107,177
Opel,Astra Elegance,Sedan,Europe,Rear,30245,27745,2.5,6,18.4,19,27,3461,107,176
```
# Read a local csv file with pandas and take a look
opel = pd.read_csv('cars_opel.csv')
opel.describe()
# Looks like the horsepower isn't right, let's fix that
opel.loc[:, 'Horsepower'] *= 10
opel.describe()
# Create a working copy of the cars data set
sas.submitLOG('''data work.cars; set sashelp.cars; run;''')
# Append the panda dataframe to the working copy of the cars data set in SAS
cars = sas.sasdata('cars', 'work')
# The pandas data frame is appended to the SAS data set
cars.append(opel)
cars.tail()
# Sort the data set in SAS to restore the old order
cars.sort('make model type')
cars.tail()
# Confirm that Opel has been added
cars.bar('Make')
```
# Reading in data from SAS and manipulating it with Pandas
```
# Short form is sd2df()
df = sas.sasdata2dataframe('cars', 'sashelp', dsopts={'where': 'make="BMW"'})
type(df)
```
Now that the data set is available as a Pandas DataFrame you can use it in e.g. a sklearn pipeline
```
df
```
# Creating a model
The data can be found [here](https://www.kaggle.com/gsr9099/best-model-for-credit-card-approval)
```
# Read two local csv files
df_applications = pd.read_csv('application_record.csv')
df_credit = pd.read_csv('credit_record.csv')
# Get a feel for the data
print(df_applications.columns)
print(df_applications.head(5))
df_applications.describe()
# Join the two data sets together
df_application_credit = df_applications.join(df_credit, lsuffix='_applications', rsuffix='_credit')
print(df_application_credit.head())
df_application_credit.columns
# Upload the data to the SAS server
# Here just a small sample, as the data set is quite large and the data is pre-loaded on SAS server
sas.df2sd(df_application_credit[:10], table='application_credit_sample', libref='saspy')
# Create a training data set and test data set in SAS
application_credit_sas = sas.sasdata('application_credit', 'saspy')
application_credit_part = application_credit_sas.partition(fraction=.7, var='status')
application_credit_part.info()
# Creating a SAS/STAT object
stat = sas.sasstat()
dir(stat)
# Target
target = 'status'
# Class Variables
var_class = ['FLAG_OWN_CAR','FLAG_OWN_REALTY', 'OCCUPATION_TYPE', 'STATUS']
```
The HPSPLIT procedure is a high-performance procedure that builds tree-based statistical models for classification and regression. The procedure produces classification trees, which model a categorical response, and regression trees, which model a continuous response. Both types of trees are referred to as decision trees because the model is expressed as a series of if-then statements - [documentation](https://support.sas.com/documentation/onlinedoc/stat/141/hpsplit.pdf)
```
hpsplit_model = stat.hpsplit(data=application_credit_part,
cls=var_class,
model="status(event='N')= FLAG_OWN_CAR FLAG_OWN_REALTY OCCUPATION_TYPE MONTHS_BALANCE AMT_INCOME_TOTAL",
code='trescore.sas',
procopts='assignmissing=similar',
out = 'work.dt_score',
id = "ID",
partition="rolevar=_partind_(TRAIN='1' VALIDATE='0');")
dir(hpsplit_model)
hpsplit_model.ROCPLOT
hpsplit_model.varimportance
sas.set_results('HTML')
hpsplit_model.wholetreeplot
```
| github_jupyter |
# VacationPy
----
#### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
import json
# Import API key
from config import g_key
```
### Store Part I results into DataFrame
* Load the csv exported in Part I to a DataFrame
```
#read in weather data
weather_data = pd.read_csv('../cities.csv')
weather_data.head()
```
### Humidity Heatmap
* Configure gmaps.
* Use the Lat and Lng as locations and Humidity as the weight.
* Add Heatmap layer to map.
```
#Filter columns to be used in weather dataframe
cols = ["City", "Cloudiness", "Country", "Date", "Humidity", "Lat", "Lng", "Temp", "Wind Speed"]
weather_data = weather_data[cols]
#configure gmaps
gmaps.configure(api_key=g_key)
#create coordinates
locations = weather_data[["Lat", "Lng"]].astype(float)
humidity = weather_data["Humidity"].astype(float)
fig = gmaps.figure()
#create heatmap to display humidity across globe
heat_layer = gmaps.heatmap_layer(locations, weights=humidity,
dissipating=False, max_intensity=100,
point_radius = 1)
#add heatmap layer
fig.add_layer(heat_layer)
#display heatmap
fig
```
### Create new DataFrame fitting weather criteria
* Narrow down the cities to fit weather conditions.
* Drop any rows will null values.
```
weather_data = weather_data[weather_data["Temp"].between(70,80,inclusive=True)]
weather_data = weather_data[weather_data["Temp"] > 70]
weather_data = weather_data[weather_data["Wind Speed"] < 10]
weather_data = weather_data[weather_data["Cloudiness"] == 0]
weather_data.head()
```
### Hotel Map
* Store into variable named `hotel_df`.
* Add a "Hotel Name" column to the DataFrame.
* Set parameters to search for hotels with 5000 meters.
* Hit the Google Places API for each city's coordinates.
* Store the first Hotel result into the DataFrame.
* Plot markers on top of the heatmap.
```
hotel_df = weather_data
hotel_df["Hotel Name"]= ''
hotel_df
params = {
"types": "lodging",
"radius":5000,
"key": g_key
}
# Use the lat/lng we recovered to identify airports
for index, row in hotel_df.iterrows():
# get lat, lng from df
lat = row["Lat"]
lng = row["Lng"]
# change location each iteration while leaving original params in place
params["location"] = f"{lat},{lng}"
# Use the search term: "International Airport" and our lat/lng
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
# make request and print url
name_address = requests.get(base_url, params=params).json()
print(json.dumps(name_address, indent=4, sort_keys=True))
try:
hotel_df.loc[index, "Hotel Name"] = name_address["results"][0]["name"]
except (KeyError, IndexError):
print("Missing field/result... skipping.")
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
marker_layer = gmaps.marker_layer(locations, info_box_content=hotel_info)
# Display Map
fig.add_layer(marker_layer)
fig
```
| github_jupyter |
# Support Vector Machine (SVM) Tutorial
Follow from: [link](https://towardsdatascience.com/support-vector-machine-introduction-to-machine-learning-algorithms-934a444fca47)
- SVM can be used for both regression and classification problems.
- The goal of SVM models is to find a hyperplane in an N-dimensional space that distinctly classifies the data points.
- The hyperplane must be the one with the maximum margin.
- Support vectors are data points that are closer to the hyperplane and influence the position and orientation of the hyperplane.
- In SVM, if the output of the model is greater than 1, it is identified with one class and if it is -1, it is identified with the other class $[-1, 1]$.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
%matplotlib inline
plt.style.use('seaborn')
df = pd.read_csv('data/Iris.csv')
df.head()
df = df.drop(['Id'], axis=1)
df.head()
target = df['Species']
s = list(set(target))
rows = list(range(100, 150))
# Since the Iris dataset has three classes, we remove the third class. This then results in a binary classification problem
df = df.drop(df.index[rows])
x, y = df['SepalLengthCm'], df['PetalLengthCm']
setosa_x, setosa_y = x[:50], y[:50]
versicolor_x, versicolor_y = x[50:], y[50:]
plt.figure(figsize=(8,6))
plt.scatter(setosa_x, setosa_y, marker='.', color='green')
plt.scatter(versicolor_x, versicolor_y, marker='*', color='red')
plt.show()
df = df.drop(['SepalWidthCm', 'PetalWidthCm'], axis=1)
Y = []
target = df['Species']
for val in target:
if(val == 'Iris-setosa'):
Y.append(-1)
else:
Y.append(1)
df = df.drop(['Species'], axis=1)
X = df.values.tolist()
# Shuffle and split the data
X, Y = shuffle(X, Y)
x_train, x_test, y_train, y_test = train_test_split(X, Y, train_size=0.9)
x_train = np.array(x_train)
y_train = np.array(y_train).reshape(90, 1)
x_test = np.array(x_test)
y_test = np.array(y_test).reshape(10, 1)
```
## SVM implementation with Numpy
```
train_f1 = x_train[:, 0].reshape(90, 1)
train_f2 = x_train[:, 1].reshape(90, 1)
w1, w2 = np.zeros((90, 1)), np.zeros((90, 1))
epochs = 1
alpha = 1e-4
while epochs < 10000:
y = w1 * train_f1 + w2 * train_f2
prod = y * y_train
count = 0
for val in prod:
if val >= 1:
cost = 0
w1 = w1 - alpha * (2 * 1/epochs * w1)
w2 = w2 - alpha * (2 * 1/epochs * w2)
else:
cost = 1 - val
w1 = w1 + alpha * (train_f1[count] * y_train[count] - 2 * 1/epochs * w1)
w2 = w2 + alpha * (train_f2[count] * y_train[count] - 2 * 1/epochs * w2)
count += 1
epochs += 1
```
### Evaluation
```
index = list(range(10, 90))
w1 = np.delete(w1, index).reshape(10, 1)
w2 = np.delete(w2, index).reshape(10, 1)
## Extract the test data features
test_f1 = x_test[:,0].reshape(10, 1)
test_f2 = x_test[:,1].reshape(10, 1)
## Predict
y_pred = w1 * test_f1 + w2 * test_f2
predictions = []
for val in y_pred:
if val > 1:
predictions.append(1)
else:
predictions.append(-1)
print(accuracy_score(y_test,predictions))
```
## SVM via sklearn
```
from sklearn.svm import SVC
clf = SVC(kernel='linear')
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(accuracy_score(y_test, y_pred))
```
| github_jupyter |
<small><i>This notebook was put together by [Jake Vanderplas](http://www.vanderplas.com). Source and license info is on [GitHub](https://github.com/jakevdp/sklearn_tutorial/).</i></small>
# Supervised Learning In-Depth: Random Forests
Previously we saw a powerful discriminative classifier, **Support Vector Machines**.
Here we'll take a look at motivating another powerful algorithm. This one is a *non-parametric* algorithm called **Random Forests**.
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
plt.style.use('seaborn')
```
## Motivating Random Forests: Decision Trees
Random forests are an example of an *ensemble learner* built on decision trees.
For this reason we'll start by discussing decision trees themselves.
Decision trees are extremely intuitive ways to classify or label objects: you simply ask a series of questions designed to zero-in on the classification:
```
import fig_code
fig_code.plot_example_decision_tree()
```
The binary splitting makes this extremely efficient.
As always, though, the trick is to *ask the right questions*.
This is where the algorithmic process comes in: in training a decision tree classifier, the algorithm looks at the features and decides which questions (or "splits") contain the most information.
### Creating a Decision Tree
Here's an example of a decision tree classifier in scikit-learn. We'll start by defining some two-dimensional labeled data:
```
from sklearn.datasets import make_blobs
X, y = make_blobs(n_samples=300, centers=4,
random_state=0, cluster_std=1.0)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='rainbow');
```
We have some convenience functions in the repository that help
```
from fig_code import visualize_tree, plot_tree_interactive
```
Now using IPython's ``interact`` (available in IPython 2.0+, and requires a live kernel) we can view the decision tree splits:
```
plot_tree_interactive(X, y);
```
Notice that at each increase in depth, every node is split in two **except** those nodes which contain only a single class.
The result is a very fast **non-parametric** classification, and can be extremely useful in practice.
**Question: Do you see any problems with this?**
### Decision Trees and over-fitting
One issue with decision trees is that it is very easy to create trees which **over-fit** the data. That is, they are flexible enough that they can learn the structure of the noise in the data rather than the signal! For example, take a look at two trees built on two subsets of this dataset:
```
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier()
plt.figure()
visualize_tree(clf, X[:200], y[:200], boundaries=False)
plt.figure()
visualize_tree(clf, X[-200:], y[-200:], boundaries=False)
```
The details of the classifications are completely different! That is an indication of **over-fitting**: when you predict the value for a new point, the result is more reflective of the noise in the model rather than the signal.
## Ensembles of Estimators: Random Forests
One possible way to address over-fitting is to use an **Ensemble Method**: this is a meta-estimator which essentially averages the results of many individual estimators which over-fit the data. Somewhat surprisingly, the resulting estimates are much more robust and accurate than the individual estimates which make them up!
One of the most common ensemble methods is the **Random Forest**, in which the ensemble is made up of many decision trees which are in some way perturbed.
There are volumes of theory and precedent about how to randomize these trees, but as an example, let's imagine an ensemble of estimators fit on subsets of the data. We can get an idea of what these might look like as follows:
```
def fit_randomized_tree(random_state=0):
X, y = make_blobs(n_samples=300, centers=4,
random_state=0, cluster_std=2.0)
clf = DecisionTreeClassifier(max_depth=15)
rng = np.random.RandomState(random_state)
i = np.arange(len(y))
rng.shuffle(i)
visualize_tree(clf, X[i[:250]], y[i[:250]], boundaries=False,
xlim=(X[:, 0].min(), X[:, 0].max()),
ylim=(X[:, 1].min(), X[:, 1].max()))
from ipywidgets import interact
interact(fit_randomized_tree, random_state=(0, 100));
```
See how the details of the model change as a function of the sample, while the larger characteristics remain the same!
The random forest classifier will do something similar to this, but use a combined version of all these trees to arrive at a final answer:
```
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=100, random_state=0)
visualize_tree(clf, X, y, boundaries=False);
```
By averaging over 100 randomly perturbed models, we end up with an overall model which is a much better fit to our data!
*(Note: above we randomized the model through sub-sampling... Random Forests use more sophisticated means of randomization, which you can read about in, e.g. the [scikit-learn documentation](http://scikit-learn.org/stable/modules/ensemble.html#forest)*)
## Quick Example: Moving to Regression
Above we were considering random forests within the context of classification.
Random forests can also be made to work in the case of regression (that is, continuous rather than categorical variables). The estimator to use for this is ``sklearn.ensemble.RandomForestRegressor``.
Let's quickly demonstrate how this can be used:
```
from sklearn.ensemble import RandomForestRegressor
x = 10 * np.random.rand(100)
def model(x, sigma=0.3):
fast_oscillation = np.sin(5 * x)
slow_oscillation = np.sin(0.5 * x)
noise = sigma * np.random.randn(len(x))
return slow_oscillation + fast_oscillation + noise
y = model(x)
plt.errorbar(x, y, 0.3, fmt='o');
xfit = np.linspace(0, 10, 1000)
yfit = RandomForestRegressor(100).fit(x[:, None], y).predict(xfit[:, None])
ytrue = model(xfit, 0)
plt.errorbar(x, y, 0.3, fmt='o')
plt.plot(xfit, yfit, '-r');
plt.plot(xfit, ytrue, '-k', alpha=0.5);
```
As you can see, the non-parametric random forest model is flexible enough to fit the multi-period data, without us even specifying a multi-period model!
## Example: Random Forest for Classifying Digits
We previously saw the **hand-written digits** data. Let's use that here to test the efficacy of the SVM and Random Forest classifiers.
```
from sklearn.datasets import load_digits
digits = load_digits()
digits.keys()
X = digits.data
y = digits.target
print(X.shape)
print(y.shape)
```
To remind us what we're looking at, we'll visualize the first few data points:
```
# set up the figure
fig = plt.figure(figsize=(6, 6)) # figure size in inches
fig.subplots_adjust(left=0, right=1, bottom=0, top=1, hspace=0.05, wspace=0.05)
# plot the digits: each image is 8x8 pixels
for i in range(64):
ax = fig.add_subplot(8, 8, i + 1, xticks=[], yticks=[])
ax.imshow(digits.images[i], cmap=plt.cm.binary, interpolation='nearest')
# label the image with the target value
ax.text(0, 7, str(digits.target[i]))
```
We can quickly classify the digits using a decision tree as follows:
```
from sklearn.model_selection import train_test_split
from sklearn import metrics
Xtrain, Xtest, ytrain, ytest = train_test_split(X, y, random_state=0)
clf = DecisionTreeClassifier(max_depth=11)
clf.fit(Xtrain, ytrain)
ypred = clf.predict(Xtest)
```
We can check the accuracy of this classifier:
```
metrics.accuracy_score(ypred, ytest)
```
and for good measure, plot the confusion matrix:
```
plt.imshow(metrics.confusion_matrix(ypred, ytest),
interpolation='nearest', cmap=plt.cm.binary)
plt.grid(False)
plt.colorbar()
plt.xlabel("predicted label")
plt.ylabel("true label");
```
### Exercise
1. Repeat this classification task with ``sklearn.ensemble.RandomForestClassifier``. How does the ``max_depth``, ``max_features``, and ``n_estimators`` affect the results?
2. Try this classification with ``sklearn.svm.SVC``, adjusting ``kernel``, ``C``, and ``gamma``. Which classifier performs optimally?
3. Try a few sets of parameters for each model and check the F1 score (``sklearn.metrics.f1_score``) on your results. What's the best F1 score you can reach?
| github_jupyter |
# Talks markdown generator for academicpages
Takes a TSV of talks with metadata and converts them for use with [academicpages.github.io](academicpages.github.io). This is an interactive Jupyter notebook ([see more info here](http://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/what_is_jupyter.html)). The core python code is also in `talks.py`. Run either from the `markdown_generator` folder after replacing `talks.tsv` with one containing your data.
TODO: Make this work with BibTex and other databases, rather than Stuart's non-standard TSV format and citation style.
```
import pandas as pd
import os
```
## Data format
The TSV needs to have the following columns: title, type, url_slug, venue, date, location, talk_url, description, with a header at the top. Many of these fields can be blank, but the columns must be in the TSV.
- Fields that cannot be blank: `title`, `url_slug`, `date`. All else can be blank. `type` defaults to "Talk"
- `date` must be formatted as YYYY-MM-DD.
- `url_slug` will be the descriptive part of the .md file and the permalink URL for the page about the paper.
- The .md file will be `YYYY-MM-DD-[url_slug].md` and the permalink will be `https://[yourdomain]/talks/YYYY-MM-DD-[url_slug]`
- The combination of `url_slug` and `date` must be unique, as it will be the basis for your filenames
This is how the raw file looks (it doesn't look pretty, use a spreadsheet or other program to edit and create).
```
!type talks.tsv
```
## Import TSV
Pandas makes this easy with the read_csv function. We are using a TSV, so we specify the separator as a tab, or `\t`.
I found it important to put this data in a tab-separated values format, because there are a lot of commas in this kind of data and comma-separated values can get messed up. However, you can modify the import statement, as pandas also has read_excel(), read_json(), and others.
```
talks = pd.read_csv("talks.tsv", sep="\t", header=0)
talks
```
## Escape special characters
YAML is very picky about how it takes a valid string, so we are replacing single and double quotes (and ampersands) with their HTML encoded equivilents. This makes them look not so readable in raw format, but they are parsed and rendered nicely.
```
html_escape_table = {
"&": "&",
'"': """,
"'": "'"
}
def html_escape(text):
if type(text) is str:
return "".join(html_escape_table.get(c,c) for c in text)
else:
return "False"
```
## Creating the markdown files
This is where the heavy lifting is done. This loops through all the rows in the TSV dataframe, then starts to concatentate a big string (```md```) that contains the markdown for each type. It does the YAML metadata first, then does the description for the individual page.
```
loc_dict = {}
for row, item in talks.iterrows():
md_filename = str(item.date) + "-" + item.url_slug + ".md"
html_filename = str(item.date) + "-" + item.url_slug
year = item.date[:4]
md = "---\ntitle: \"" + item.title + '"\n'
md += "collection: talks" + "\n"
if len(str(item.type)) > 3:
md += 'type: "' + item.type + '"\n'
else:
md += 'type: "Talk"\n'
md += "permalink: /talks/" + html_filename + "\n"
if len(str(item.venue)) > 3:
md += 'venue: "' + item.venue + '"\n'
if len(str(item.location)) > 3:
md += "date: " + str(item.date) + "\n"
if len(str(item.location)) > 3:
md += 'location: "' + str(item.location) + '"\n'
md += "---\n"
if len(str(item.talk_url)) > 3:
md += "\n[More information here](" + item.talk_url + ")\n"
if len(str(item.description)) > 3:
md += "\n" + html_escape(item.description) + "\n"
md_filename = os.path.basename(md_filename)
#print(md)
with open("../_talks/" + md_filename, 'w') as f:
f.write(md)
```
These files are in the talks directory, one directory below where we're working from.
```
!ls ../_talks
!cat ../_talks/2013-03-01-tutorial-1.md
```
| github_jupyter |
<a href="https://colab.research.google.com/github/isb-cgc/Community-Notebooks/blob/master/MachineLearning/How_to_build_an_RNAseq_logistic_regression_classifier_with_BigQuery_ML.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# How to build an RNA-seq logistic regression classifier with BigQuery ML
Check out other notebooks at our [Community Notebooks Repository](https://github.com/isb-cgc/Community-Notebooks)!
- **Title:** How to build an RNA-seq logistic regression classifier with BigQuery ML
- **Author:** John Phan
- **Created:** 2021-07-19
- **Purpose:** Demonstrate use of BigQuery ML to predict a cancer endpoint using gene expression data.
- **URL:** https://github.com/isb-cgc/Community-Notebooks/blob/master/MachineLearning/How_to_build_an_RNAseq_logistic_regression_classifier_with_BigQuery_ML.ipynb
- **Note:** This example is based on the work published by [Bosquet et al.](https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-016-0548-9)
This notebook builds upon the [scikit-learn notebook](https://github.com/isb-cgc/Community-Notebooks/blob/master/MachineLearning/How_to_build_an_RNAseq_logistic_regression_classifier.ipynb) and demonstrates how to build a machine learning model using BigQuery ML to predict ovarian cancer treatment outcome. BigQuery is used to create a temporary data table that contains both training and testing data. These datasets are then used to fit and evaluate a Logistic Regression classifier.
# Import Dependencies
```
# GCP libraries
from google.cloud import bigquery
from google.colab import auth
```
## Authenticate
Before using BigQuery, we need to get authorization for access to BigQuery and the Google Cloud. For more information see ['Quick Start Guide to ISB-CGC'](https://isb-cancer-genomics-cloud.readthedocs.io/en/latest/sections/HowToGetStartedonISB-CGC.html). Alternative authentication methods can be found [here](https://googleapis.dev/python/google-api-core/latest/auth.html)
```
# if you're using Google Colab, authenticate to gcloud with the following
auth.authenticate_user()
# alternatively, use the gcloud SDK
#!gcloud auth application-default login
```
## Parameters
Customize the following parameters based on your notebook, execution environment, or project. BigQuery ML must create and store classification models, so be sure that you have write access to the locations stored in the "bq_dataset" and "bq_project" variables.
```
# set the google project that will be billed for this notebook's computations
google_project = 'google-project' ## CHANGE ME
# bq project for storing ML model
bq_project = 'bq-project' ## CHANGE ME
# bq dataset for storing ML model
bq_dataset = 'scratch' ## CHANGE ME
# name of temporary table for data
bq_tmp_table = 'tmp_data'
# name of ML model
bq_ml_model = 'tcga_ov_therapy_ml_lr_model'
# in this example, we'll be using the Ovarian cancer TCGA dataset
cancer_type = 'TCGA-OV'
# genes used for prediction model, taken from Bosquet et al.
genes = "'RHOT1','MYO7A','ZBTB10','MATK','ST18','RPS23','GCNT1','DROSHA','NUAK1','CCPG1',\
'PDGFD','KLRAP1','MTAP','RNF13','THBS1','MLX','FAP','TIMP3','PRSS1','SLC7A11',\
'OLFML3','RPS20','MCM5','POLE','STEAP4','LRRC8D','WBP1L','ENTPD5','SYNE1','DPT',\
'COPZ2','TRIO','PDPR'"
# clinical data table
clinical_table = 'isb-cgc-bq.TCGA_versioned.clinical_gdc_2019_06'
# RNA seq data table
rnaseq_table = 'isb-cgc-bq.TCGA.RNAseq_hg38_gdc_current'
```
## BigQuery Client
Create the BigQuery client.
```
# Create a client to access the data within BigQuery
client = bigquery.Client(google_project)
```
## Create a Table with a Subset of the Gene Expression Data
Pull RNA-seq gene expression data from the TCGA RNA-seq BigQuery table, join it with clinical labels, and pivot the table so that it can be used with BigQuery ML. In this example, we will label the samples based on therapy outcome. "Complete Remission/Response" will be labeled as "1" while all other therapy outcomes will be labeled as "0". This prepares the data for binary classification.
Prediction modeling with RNA-seq data typically requires a feature selection step to reduce the dimensionality of the data before training a classifier. However, to simplify this example, we will use a pre-identified set of 33 genes (Bosquet et al. identified 34 genes, but PRSS2 and its aliases are not available in the hg38 RNA-seq data).
Creation of a BQ table with only the data of interest reduces the size of the data passed to BQ ML and can significantly reduce the cost of running BQ ML queries. This query also randomly splits the dataset into "training" and "testing" sets using the "FARM_FINGERPRINT" hash function in BigQuery. "FARM_FINGERPRINT" generates an integer from the input string. More information can be found [here](https://cloud.google.com/bigquery/docs/reference/standard-sql/hash_functions).
```
tmp_table_query = client.query(("""
BEGIN
CREATE OR REPLACE TABLE `{bq_project}.{bq_dataset}.{bq_tmp_table}` AS
SELECT * FROM (
SELECT
labels.case_barcode as sample,
labels.data_partition as data_partition,
labels.response_label AS label,
ge.gene_name AS gene_name,
-- Multiple samples may exist per case, take the max value
MAX(LOG(ge.HTSeq__FPKM_UQ+1)) AS gene_expression
FROM `{rnaseq_table}` AS ge
INNER JOIN (
SELECT
*
FROM (
SELECT
case_barcode,
primary_therapy_outcome_success,
CASE
-- Complete Reponse --> label as 1
-- All other responses --> label as 0
WHEN primary_therapy_outcome_success = 'Complete Remission/Response' THEN 1
WHEN (primary_therapy_outcome_success IN (
'Partial Remission/Response','Progressive Disease','Stable Disease'
)) THEN 0
END AS response_label,
CASE
WHEN MOD(ABS(FARM_FINGERPRINT(case_barcode)), 10) < 5 THEN 'training'
WHEN MOD(ABS(FARM_FINGERPRINT(case_barcode)), 10) >= 5 THEN 'testing'
END AS data_partition
FROM `{clinical_table}`
WHERE
project_short_name = '{cancer_type}'
AND primary_therapy_outcome_success IS NOT NULL
)
) labels
ON labels.case_barcode = ge.case_barcode
WHERE gene_name IN ({genes})
GROUP BY sample, label, data_partition, gene_name
)
PIVOT (
MAX(gene_expression) FOR gene_name IN ({genes})
);
END;
""").format(
bq_project=bq_project,
bq_dataset=bq_dataset,
bq_tmp_table=bq_tmp_table,
rnaseq_table=rnaseq_table,
clinical_table=clinical_table,
cancer_type=cancer_type,
genes=genes
)).result()
print(tmp_table_query)
```
Let's take a look at this subset table. The data has been pivoted such that each of the 33 genes is available as a column that can be "SELECTED" in a query. In addition, the "label" and "data_partition" columns simplify data handling for classifier training and evaluation.
```
tmp_table_data = client.query(("""
SELECT
* --usually not recommended to use *, but in this case, we want to see all of the 33 genes
FROM `{bq_project}.{bq_dataset}.{bq_tmp_table}`
""").format(
bq_project=bq_project,
bq_dataset=bq_dataset,
bq_tmp_table=bq_tmp_table
)).result().to_dataframe()
print(tmp_table_data.info())
tmp_table_data
```
# Train the Machine Learning Model
Now we can train a classifier using BigQuery ML with the data stored in the subset table. This model will be stored in the location specified by the "bq_ml_model" variable, and can be reused to predict samples in the future.
We pass three options to the BQ ML model: model_type, auto_class_weights, and input_label_cols. Model_type specifies the classifier model type. In this case, we use "LOGISTIC_REG" to train a logistic regression classifier. Other classifier options are documented [here](https://cloud.google.com/bigquery-ml/docs/reference/standard-sql/bigqueryml-syntax-create). Auto_class_weights indicates whether samples should be weighted to balance the classes. For example, if the dataset happens to have more samples labeled as "Complete Response", those samples would be less weighted to ensure that the model is not biased towards predicting those samples. Input_label_cols tells BigQuery that the "label" column should be used to determine each sample's label.
**Warning**: BigQuery ML models can be very time-consuming and expensive to train. Please check your data size before running BigQuery ML commands. Information about BigQuery ML costs can be found [here](https://cloud.google.com/bigquery-ml/pricing).
```
# create ML model using BigQuery
ml_model_query = client.query(("""
CREATE OR REPLACE MODEL `{bq_project}.{bq_dataset}.{bq_ml_model}`
OPTIONS
(
model_type='LOGISTIC_REG',
auto_class_weights=TRUE,
input_label_cols=['label']
) AS
SELECT * EXCEPT(sample, data_partition) -- when training, we only the labels and feature columns
FROM `{bq_project}.{bq_dataset}.{bq_tmp_table}`
WHERE data_partition = 'training' -- using training data only
""").format(
bq_project=bq_project,
bq_dataset=bq_dataset,
bq_ml_model=bq_ml_model,
bq_tmp_table=bq_tmp_table
)).result()
print(ml_model_query)
# now get the model metadata
ml_model = client.get_model('{}.{}.{}'.format(bq_project, bq_dataset, bq_ml_model))
print(ml_model)
```
# Evaluate the Machine Learning Model
Once the model has been trained and stored, we can evaluate the model's performance using the "testing" dataset from our subset table. Evaluating a BQ ML model is generally less expensive than training.
Use the following query to evaluate the BQ ML model. Note that we're using the "data_partition = 'testing'" clause to ensure that we're only evaluating the model with test samples from the subset table.
BigQuery's ML.EVALUATE function returns several performance metrics: precision, recall, accuracy, f1_score, log_loss, and roc_auc. More details about these performance metrics are available from [Google's ML Crash Course](https://developers.google.com/machine-learning/crash-course/classification/video-lecture). Specific topics can be found at the following URLs: [precision and recall](https://developers.google.com/machine-learning/crash-course/classification/precision-and-recall), [accuracy](https://developers.google.com/machine-learning/crash-course/classification/accuracy), [ROC and AUC](https://developers.google.com/machine-learning/crash-course/classification/roc-and-auc).
```
ml_eval = client.query(("""
SELECT * FROM ML.EVALUATE (MODEL `{bq_project}.{bq_dataset}.{bq_ml_model}`,
(
SELECT * EXCEPT(sample, data_partition)
FROM `{bq_project}.{bq_dataset}.{bq_tmp_table}`
WHERE data_partition = 'testing'
)
)
""").format(
bq_project=bq_project,
bq_dataset=bq_dataset,
bq_ml_model=bq_ml_model,
bq_tmp_table=bq_tmp_table
)).result().to_dataframe()
# Display the table of evaluation results
ml_eval
```
# Predict Outcome for One or More Samples
ML.EVALUATE evaluates a model's performance, but does not produce actual predictions for each sample. In order to do that, we need to use the ML.PREDICT function. The syntax is similar to that of the ML.EVALUATE function and returns "label", "predicted_label", "predicted_label_probs", and all feature columns. Since the feature columns are unchanged from the input dataset, we select only the original label, predicted label, and probabilities for each sample.
Note that the input dataset can include one or more samples, and must include the same set of features as the training dataset.
```
ml_predict = client.query(("""
SELECT
label,
predicted_label,
predicted_label_probs
FROM ML.PREDICT (MODEL `{bq_project}.{bq_dataset}.{bq_ml_model}`,
(
SELECT * EXCEPT(sample, data_partition)
FROM `{bq_project}.{bq_dataset}.{bq_tmp_table}`
WHERE data_partition = 'testing' -- Use the testing dataset
)
)
""").format(
bq_project=bq_project,
bq_dataset=bq_dataset,
bq_ml_model=bq_ml_model,
bq_tmp_table=bq_tmp_table
)).result().to_dataframe()
# Display the table of prediction results
ml_predict
# Calculate the accuracy of prediction, which should match the result of ML.EVALUATE
accuracy = 1-sum(abs(ml_predict['label']-ml_predict['predicted_label']))/len(ml_predict)
print('Accuracy: ', accuracy)
```
# Next Steps
The BigQuery ML logistic regression model trained in this notebook is comparable to the scikit-learn model developed in our [companion notebook](https://github.com/isb-cgc/Community-Notebooks/blob/master/MachineLearning/How_to_build_an_RNAseq_logistic_regression_classifier.ipynb). BigQuery ML simplifies the model building and evaluation process by enabling bioinformaticians to use machine learning within the BigQuery ecosystem. However, it is often necessary to optimize performance by evaluating several types of models (i.e., other than logistic regression), and tuning model parameters. Due to the cost of BigQuery ML for training, such iterative model fine-tuning may be cost prohibitive. In such cases, a combination of scikit-learn (or other libraries such as Keras and TensorFlow) and BigQuery ML may be appropriate. E.g., models can be fine-tuned using scikit-learn and published as a BigQuery ML model for production applications. In future notebooks, we will explore methods for model selection, optimization, and publication with BigQuery ML.
| github_jupyter |
# FireCARES ops management notebook
### Using this notebook
In order to use this notebook, a single production/test web node will need to be bootstrapped w/ ipython and django-shell-plus python libraries. After bootstrapping is complete and while forwarding a local port to the port that the ipython notebook server will be running on the node, you can open the ipython notebook using the token provided in the SSH session after ipython notebook server start.
#### Bootstrapping a prod/test node
To bootstrap a specific node for use of this notebook, you'll need to ssh into the node and forward a local port # to localhost:8888 on the node.
e.g. `ssh firecares-prod -L 8890:localhost:8888` to forward the local port 8890 to 8888 on the web node, assumes that the "firecares-prod" SSH config is listed w/ the correct webserver IP in your `~/.ssh/config`
- `sudo chown -R firecares: /run/user/1000` as the `ubuntu` user
- `sudo su firecares`
- `workon firecares`
- `pip install -r dev_requirements.txt`
- `python manage.py shell_plus --notebook --no-browser --settings=firecares.settings.local`
At this point, there will be a mention of "The jupyter notebook is running at: http://localhost:8888/?token=XXXX". Copy the URL, but be sure to use the local port that you're forwarding instead for the connection vs the default of 8888 if necessary.
Since the ipython notebook server supports django-shell-plus, all of the FireCARES models will automatically be imported. From here any command that you execute in the notebook will run on the remote web node immediately.
## Fire department management
### Re-generate performance score for a specific fire department
Useful for when a department's FDID has been corrected. Will do the following:
1. Pull NFIRS counts for the department (cached in FireCARES database)
1. Generate fires heatmap
1. Update department owned census tracts geom
1. Regenerate structure hazard counts in jurisdiction
1. Regenerate population_quartiles materialized view to get safe grades for department
1. Re-run performance score for the department
```
import psycopg2
from firecares.tasks import update
from firecares.utils import dictfetchall
from django.db import connections
from django.conf import settings
from django.core.management import call_command
from IPython.display import display
import pandas as pd
fd = {'fdid': '18M04', 'state': 'WA'}
nfirs = connections['nfirs']
department = FireDepartment.objects.filter(**fd).first()
fid = department.id
print 'FireCARES id: %s' % fid
print 'https://firecares.org/departments/%s' % fid
%%time
# Get raw fire incident counts (prior to intersection with )
with nfirs.cursor() as cur:
cur.execute("""
select count(1), fdid, state, extract(year from inc_date) as year
from fireincident where fdid=%(fdid)s and state=%(state)s
group by fdid, state, year
order by year""", fd)
fire_years = dictfetchall(cur)
display(fire_years)
print 'Total fires: %s\n' % sum([x['count'] for x in fire_years])
%%time
# Get building fire counts after structure hazard level calculations
sql = update.STRUCTURE_FIRES
print sql
with nfirs.cursor() as cur:
cur.execute(sql, dict(fd, years=tuple([x['year'] for x in fire_years])))
fires_by_hazard_level = dictfetchall(cur)
display(fires_by_hazard_level)
print 'Total geocoded fires: %s\n' % sum([x['count'] for x in fires_by_hazard_level])
sql = """
select alarm, a.inc_type, alarms,ff_death, oth_death, ST_X(geom) as x, st_y(geom) as y, COALESCE(y.risk_category, 'Unknown') as risk_category
from buildingfires a
LEFT JOIN (
SELECT state, fdid, inc_date, inc_no, exp_no, x.geom, x.parcel_id, x.risk_category
FROM (
SELECT * FROM incidentaddress a
LEFT JOIN parcel_risk_category_local using (parcel_id)
) AS x
) AS y
USING (state, fdid, inc_date, inc_no, exp_no)
WHERE a.state = %(state)s and a.fdid = %(fdid)s"""
with nfirs.cursor() as cur:
cur.execute(sql, fd)
rows = dictfetchall(cur)
out_name = '{id}-building-fires.csv'.format(id=fid)
full_path = '/tmp/' + out_name
with open(full_path, 'w') as f:
writer = csv.DictWriter(f, fieldnames=[x.name for x in cur.description])
writer.writeheader()
writer.writerows(rows)
# Push building fires to S3
!aws s3 cp $full_path s3://firecares-test/$out_name --acl="public-read"
update.update_nfirs_counts(fid)
update.calculate_department_census_geom(fid)
# Fire counts by hazard level over all years, keep in mind that the performance score model will currently ONLY work
# hazard levels w/
display(pd.DataFrame(fires_by_hazard_level).groupby(['risk_level']).sum()['count'])
update.update_performance_score(fid)
```
## User management
### Whitelist
| github_jupyter |
# Solving vertex cover with a quantum annealer
The problem of vertex cover is, given an undirected graph $G = (V, E)$, colour the smallest amount of vertices such that each edge $e \in E$ is connected to a coloured vertex.
This notebooks works through the process of creating a random graph, translating to an optimization problem, and eventually finding the ground state using a quantum annealer.
### Graph setup
The first thing we will do is create an instance of the problem, by constructing a small, random undirected graph. We are going to use the `networkx` package, which should already be installed if you have installed if you are using Anaconda.
```
import dimod
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
n_vertices = 5
n_edges = 6
small_graph = nx.gnm_random_graph(n_vertices, n_edges)
nx.draw(small_graph, with_labels=True)
```
### Constructing the Hamiltonian
I showed in class that the objective function for vertex cover looks like this:
\begin{equation}
\sum_{(u,v) \in E} (1 - x_u) (1 - x_v) + \gamma \sum_{v \in V} x_v
\end{equation}
We want to find an assignment of the $x_u$ of 1 (coloured) or 0 (uncoloured) that _minimizes_ this function. The first sum tries to force us to choose an assignment that makes sure every edge gets attached to a coloured vertex. The second sum is essentially just counting the number of coloured vertices.
**Task**: Expand out the QUBO above to see how you can convert it to a more 'traditional' looking QUBO:
\begin{equation}
\sum_{(u,v) \in E} x_u x_v + \sum_{v \in V} (\gamma - \hbox{deg}(x_v)) x_v
\end{equation}
where deg($x_v$) indicates the degree of vertex $x_v$ in the graph.
```
γ = 0.8
Q = {x : 1 for x in small_graph.edges()}
r = {x : (γ - small_graph.degree[x]) for x in small_graph.nodes}
```
Let's convert it to the appropriate data structure, and solve using the exact solver.
```
bqm = dimod.BinaryQuadraticModel(r, Q, 0, dimod.BINARY)
response = dimod.ExactSolver().sample(bqm)
print(f"Sample energy = {next(response.data(['energy']))[0]}")
```
Let's print the graph with proper colours included
```
colour_assignments = next(response.data(['sample']))[0]
colours = ['grey' if colour_assignments[x] == 0 else 'red' for x in range(len(colour_assignments))]
nx.draw(small_graph, with_labels=True, node_color=colours)
```
### Scaling up...
That one was easy enough to solve by hand. Let's try a much larger instance...
```
n_vertices = 20
n_edges = 60
large_graph = nx.gnm_random_graph(n_vertices, n_edges)
nx.draw(large_graph, with_labels=True)
# Create h, J and put it into the exact solver
γ = 0.8
Q = {x : 1 for x in large_graph.edges()}
r = {x : (γ - large_graph.degree[x]) for x in large_graph.nodes}
bqm = dimod.BinaryQuadraticModel(r, Q, 0, dimod.BINARY)
response = dimod.ExactSolver().sample(bqm)
print(f"Sample energy = {next(response.data(['energy']))[0]}")
colour_assignments = next(response.data(['sample']))[0]
colours = ['grey' if colour_assignments[x] == 0 else 'red' for x in range(len(colour_assignments))]
nx.draw(large_graph, with_labels=True, node_color=colours)
print(f"Coloured {list(colour_assignments.values()).count(1)}/{n_vertices} vertices.")
```
### Running on the D-Wave
You'll only be able to run the next few cells if you have D-Wave access. We will send the same graph as before to the D-Wave QPU and see what kind of results we get back!
```
from dwave.system.samplers import DWaveSampler
from dwave.system.composites import EmbeddingComposite
sampler = EmbeddingComposite(DWaveSampler())
ising_conversion = bqm.to_ising()
h, J = ising_conversion[0], ising_conversion[1]
response = sampler.sample_ising(h, J, num_reads = 1000)
best_solution =np.sort(response.record, order='energy')[0]
print(f"Sample energy = {best_solution['energy']}")
colour_assignments_qpu = {x : best_solution['sample'][x] for x in range(n_vertices)}
for x in range(n_vertices):
if colour_assignments_qpu[x] == -1:
colour_assignments_qpu[x] = 0
colours = ['grey' if colour_assignments_qpu[x] == 0 else 'red' for x in range(len(colour_assignments_qpu))]
nx.draw(large_graph, with_labels=True, node_color=colours)
print(f"Coloured {list(colour_assignments_qpu.values()).count(1)}/{n_vertices} vertices.")
print("Node\tExact\tQPU")
for x in range(n_vertices):
print(f"{x}\t{colour_assignments[x]}\t{colour_assignments_qpu[x]}")
```
Here is a scatter plot of all the different energies we got out, against the number of times each solution occurred.
```
plt.scatter(response.record['energy'], response.record['num_occurrences'])
response.record['num_occurrences']
```
| github_jupyter |
```
import sys
sys.path.append("/Users/sgkang/Projects/DamGeophysics/codes/")
from Readfiles import getFnames
from DCdata import readReservoirDC
%pylab inline
from SimPEG.EM.Static import DC
from SimPEG import EM
from SimPEG import Mesh
```
Read DC data
```
fname = "../data/ChungCheonDC/20150101000000.apr"
survey = readReservoirDC(fname)
dobsAppres = survey.dobs
fig, ax = plt.subplots(1,1, figsize = (10, 2))
dat = EM.Static.Utils.StaticUtils.plot_pseudoSection(survey, ax, dtype='volt', sameratio=False)
cb = dat[2]
cb.set_label("Apprent resistivity (ohm-m)")
geom = np.hstack(dat[3])
dobsDC = dobsAppres * geom
# problem = DC.Problem2D_CC(mesh)
cs = 2.5
npad = 6
hx = [(cs,npad, -1.3),(cs,160),(cs,npad, 1.3)]
hy = [(cs,npad, -1.3),(cs,20)]
mesh = Mesh.TensorMesh([hx, hy])
mesh = Mesh.TensorMesh([hx, hy],x0=[-mesh.hx[:6].sum()-0.25, -mesh.hy.sum()])
def from3Dto2Dsurvey(survey):
srcLists2D = []
nSrc = len(survey.srcList)
for iSrc in range (nSrc):
src = survey.srcList[iSrc]
locsM = np.c_[src.rxList[0].locs[0][:,0], np.ones_like(src.rxList[0].locs[0][:,0])*-0.75]
locsN = np.c_[src.rxList[0].locs[1][:,0], np.ones_like(src.rxList[0].locs[1][:,0])*-0.75]
rx = DC.Rx.Dipole_ky(locsM, locsN)
locA = np.r_[src.loc[0][0], -0.75]
locB = np.r_[src.loc[1][0], -0.75]
src = DC.Src.Dipole([rx], locA, locB)
srcLists2D.append(src)
survey2D = DC.Survey_ky(srcLists2D)
return survey2D
from SimPEG import (Mesh, Maps, Utils, DataMisfit, Regularization,
Optimization, Inversion, InvProblem, Directives)
mapping = Maps.ExpMap(mesh)
survey2D = from3Dto2Dsurvey(survey)
problem = DC.Problem2D_N(mesh, mapping=mapping)
problem.pair(survey2D)
m0 = np.ones(mesh.nC)*np.log(1e-2)
from ipywidgets import interact
nSrc = len(survey2D.srcList)
def foo(isrc):
figsize(10, 5)
mesh.plotImage(np.ones(mesh.nC)*np.nan, gridOpts={"color":"k", "alpha":0.5}, grid=True)
# isrc=0
src = survey2D.srcList[isrc]
plt.plot(src.loc[0][0], src.loc[0][1], 'bo')
plt.plot(src.loc[1][0], src.loc[1][1], 'ro')
locsM = src.rxList[0].locs[0]
locsN = src.rxList[0].locs[1]
plt.plot(locsM[:,0], locsM[:,1], 'ko')
plt.plot(locsN[:,0], locsN[:,1], 'go')
plt.gca().set_aspect('equal', adjustable='box')
interact(foo, isrc=(0, nSrc-1, 1))
pred = survey2D.dpred(m0)
# data_anal = []
# nSrc = len(survey.srcList)
# for isrc in range(nSrc):
# src = survey.srcList[isrc]
# locA = src.loc[0]
# locB = src.loc[1]
# locsM = src.rxList[0].locs[0]
# locsN = src.rxList[0].locs[1]
# rxloc=[locsM, locsN]
# a = EM.Analytics.DCAnalyticHalf(locA, rxloc, 1e-3, earth_type="halfspace")
# b = EM.Analytics.DCAnalyticHalf(locB, rxloc, 1e-3, earth_type="halfspace")
# data_anal.append(a-b)
# data_anal = np.hstack(data_anal)
survey.dobs = pred
fig, ax = plt.subplots(1,1, figsize = (10, 2))
dat = EM.Static.Utils.StaticUtils.plot_pseudoSection(survey, ax, dtype='appr', sameratio=False, scale="linear", clim=(0, 200))
out = hist(np.log10(abs(dobsDC)), bins = 100)
weight = 1./abs(mesh.gridCC[:,1])**1.5
mesh.plotImage(np.log10(weight))
survey2D.dobs = dobsDC
survey2D.eps = 10**(-2.3)
survey2D.std = 0.02
dmisfit = DataMisfit.l2_DataMisfit(survey2D)
regmap = Maps.IdentityMap(nP=int(mesh.nC))
reg = Regularization.Simple(mesh,mapping=regmap,cell_weights=weight)
opt = Optimization.InexactGaussNewton(maxIter=5)
invProb = InvProblem.BaseInvProblem(dmisfit, reg, opt)
# Create an inversion object
beta = Directives.BetaSchedule(coolingFactor=5, coolingRate=2)
betaest = Directives.BetaEstimate_ByEig(beta0_ratio=1e0)
inv = Inversion.BaseInversion(invProb, directiveList=[beta, betaest])
problem.counter = opt.counter = Utils.Counter()
opt.LSshorten = 0.5
opt.remember('xc')
mopt = inv.run(m0)
xc = opt.recall("xc")
fig, ax = plt.subplots(1,1, figsize = (10, 1.5))
sigma = mapping*mopt
dat = mesh.plotImage(1./sigma, clim=(10, 150),grid=False, ax=ax, pcolorOpts={"cmap":"jet"})
ax.set_ylim(-50, 0)
ax.set_xlim(-10, 290)
print np.log10(sigma).min(), np.log10(sigma).max()
survey.dobs = invProb.dpred
fig, ax = plt.subplots(1,1, figsize = (10, 2))
dat = EM.Static.Utils.StaticUtils.plot_pseudoSection(survey, ax, dtype='appr', sameratio=False, clim=(40, 170))
survey.dobs = dobsDC
fig, ax = plt.subplots(1,1, figsize = (10, 2))
dat = EM.Static.Utils.StaticUtils.plot_pseudoSection(survey, ax, dtype='appr', sameratio=False, clim=(40, 170))
survey.dobs = abs(dmisfit.Wd*(dobsDC-invProb.dpred))
fig, ax = plt.subplots(1,1, figsize = (10, 2))
dat = EM.Static.Utils.StaticUtils.plot_pseudoSection(survey, ax, dtype='volt', sameratio=False, clim=(0, 2))
# sigma = np.ones(mesh.nC)
modelname = "sigma0101.npy"
np.save(modelname, sigma)
```
| github_jupyter |
# FloPy
## Using FloPy to simplify the use of the MT3DMS ```SSM``` package
A multi-component transport demonstration
```
import os
import sys
import numpy as np
# run installed version of flopy or add local path
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
print(sys.version)
print('numpy version: {}'.format(np.__version__))
print('flopy version: {}'.format(flopy.__version__))
```
First, we will create a simple model structure
```
nlay, nrow, ncol = 10, 10, 10
perlen = np.zeros((10), dtype=float) + 10
nper = len(perlen)
ibound = np.ones((nlay,nrow,ncol), dtype=int)
botm = np.arange(-1,-11,-1)
top = 0.
```
## Create the ```MODFLOW``` packages
```
model_ws = 'data'
modelname = 'ssmex'
mf = flopy.modflow.Modflow(modelname, model_ws=model_ws)
dis = flopy.modflow.ModflowDis(mf, nlay=nlay, nrow=nrow, ncol=ncol,
perlen=perlen, nper=nper, botm=botm, top=top,
steady=False)
bas = flopy.modflow.ModflowBas(mf, ibound=ibound, strt=top)
lpf = flopy.modflow.ModflowLpf(mf, hk=100, vka=100, ss=0.00001, sy=0.1)
oc = flopy.modflow.ModflowOc(mf)
pcg = flopy.modflow.ModflowPcg(mf)
rch = flopy.modflow.ModflowRch(mf)
```
We'll track the cell locations for the ```SSM``` data using the ```MODFLOW``` boundary conditions.
Get a dictionary (```dict```) that has the ```SSM``` ```itype``` for each of the boundary types.
```
itype = flopy.mt3d.Mt3dSsm.itype_dict()
print(itype)
print(flopy.mt3d.Mt3dSsm.get_default_dtype())
ssm_data = {}
```
Add a general head boundary (```ghb```). The general head boundary head (```bhead```) is 0.1 for the first 5 stress periods with a component 1 (comp_1) concentration of 1.0 and a component 2 (comp_2) concentration of 100.0. Then ```bhead``` is increased to 0.25 and comp_1 concentration is reduced to 0.5 and comp_2 concentration is increased to 200.0
```
ghb_data = {}
print(flopy.modflow.ModflowGhb.get_default_dtype())
ghb_data[0] = [(4, 4, 4, 0.1, 1.5)]
ssm_data[0] = [(4, 4, 4, 1.0, itype['GHB'], 1.0, 100.0)]
ghb_data[5] = [(4, 4, 4, 0.25, 1.5)]
ssm_data[5] = [(4, 4, 4, 0.5, itype['GHB'], 0.5, 200.0)]
for k in range(nlay):
for i in range(nrow):
ghb_data[0].append((k, i, 0, 0.0, 100.0))
ssm_data[0].append((k, i, 0, 0.0, itype['GHB'], 0.0, 0.0))
ghb_data[5] = [(4, 4, 4, 0.25, 1.5)]
ssm_data[5] = [(4, 4, 4, 0.5, itype['GHB'], 0.5, 200.0)]
for k in range(nlay):
for i in range(nrow):
ghb_data[5].append((k, i, 0, -0.5, 100.0))
ssm_data[5].append((k, i, 0, 0.0, itype['GHB'], 0.0, 0.0))
```
Add an injection ```well```. The injection rate (```flux```) is 10.0 with a comp_1 concentration of 10.0 and a comp_2 concentration of 0.0 for all stress periods. WARNING: since we changed the ```SSM``` data in stress period 6, we need to add the well to the ssm_data for stress period 6.
```
wel_data = {}
print(flopy.modflow.ModflowWel.get_default_dtype())
wel_data[0] = [(0, 4, 8, 10.0)]
ssm_data[0].append((0, 4, 8, 10.0, itype['WEL'], 10.0, 0.0))
ssm_data[5].append((0, 4, 8, 10.0, itype['WEL'], 10.0, 0.0))
```
Add the ```GHB``` and ```WEL``` packages to the ```mf``` ```MODFLOW``` object instance.
```
ghb = flopy.modflow.ModflowGhb(mf, stress_period_data=ghb_data)
wel = flopy.modflow.ModflowWel(mf, stress_period_data=wel_data)
```
## Create the ```MT3DMS``` packages
```
mt = flopy.mt3d.Mt3dms(modflowmodel=mf, modelname=modelname, model_ws=model_ws)
btn = flopy.mt3d.Mt3dBtn(mt, sconc=0, ncomp=2, sconc2=50.0)
adv = flopy.mt3d.Mt3dAdv(mt)
ssm = flopy.mt3d.Mt3dSsm(mt, stress_period_data=ssm_data)
gcg = flopy.mt3d.Mt3dGcg(mt)
```
Let's verify that ```stress_period_data``` has the right ```dtype```
```
print(ssm.stress_period_data.dtype)
```
## Create the ```SEAWAT``` packages
```
swt = flopy.seawat.Seawat(modflowmodel=mf, mt3dmodel=mt,
modelname=modelname, namefile_ext='nam_swt', model_ws=model_ws)
vdf = flopy.seawat.SeawatVdf(swt, mtdnconc=0, iwtable=0, indense=-1)
mf.write_input()
mt.write_input()
swt.write_input()
```
And finally, modify the ```vdf``` package to fix ```indense```.
```
fname = modelname + '.vdf'
f = open(os.path.join(model_ws, fname),'r')
lines = f.readlines()
f.close()
f = open(os.path.join(model_ws, fname),'w')
for line in lines:
f.write(line)
for kper in range(nper):
f.write("-1\n")
f.close()
```
| github_jupyter |
# Home2
Your home away from home <br>
The best location for your needs, anywhere in the world <br>
### Inputs:
Addresses (eg. 'Pune, Maharashtra')
Category List (eg. 'Food', 'Restaurant', 'Gym', 'Trails', 'School', 'Train Station')
Limit of Results to return (eg. 75)
Radius of search in metres (eg. 10,000)
Radiums of hotels to search
### Outputs:
Cluster of venues coded by criteria
map of cluster
Centroid latitude and longitude for these venues
Address near centroid
Hotels near centroid
## User Input
```
# Addresses to analyze venues around and obtain best location
addresses=['Bend, Oregon']
# 4square Venue categories of interest (https://developer.foursquare.com/docs/build-with-foursquare/categories)
categories=['4bf58dd8d48988d181941735','4bf58dd8d48988d149941735','4bf58dd8d48988d176941735','53e510b7498ebcb1801b55d4',
'52e81612bcbc57f1066b7a21','5bae9231bedf3950379f89d0','4bf58dd8d48988d159941735']
# Limit of search results
LIMIT=500
# Radius of search in metres (maximum 100000)
radius=20000
# Radius in metres to search hotels around from final optimum location (centroid). This list is also sorted by likes
hotel_radius=20000
# Remove Outliers? 'Y' or 'N'
remove_outliers='Y'
```
## Import Libraries
```
import numpy as np # library to handle data in a vectorized manner
import pandas as pd # library for data analsysis
pd.set_option('display.max_columns', None)
pd.set_option('display.max_rows', None)
import math
import json # library to handle JSON files
!conda install -c conda-forge geopy --yes # uncomment this line if you haven't completed the Foursquare API lab
from geopy.geocoders import Nominatim # convert an address into latitude and longitude values
import requests # library to handle requests
from pandas.io.json import json_normalize # tranform JSON file into a pandas dataframe
# Matplotlib and associated plotting modules
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
# import k-means from clustering stage
from sklearn.cluster import KMeans
#!conda install -c conda-forge folium=0.5.0 --yes # uncomment this line if you haven't completed the Foursquare API lab
import folium # map rendering library
! pip install lxml
print('Libraries imported.')
```
## Obtain Location and Venue information in a Dataframe
### Create Geolocator using Nominatim and Obtain Location info. for the addresses
```
geoagent="explorer"
lat=[]
long=[]
for address in addresses:
geolocator = Nominatim(user_agent=geoagent)
loc = geolocator.geocode(address)
lat.append(loc.latitude)
long.append(loc.longitude)
print('The geograpical coordinate of '+address +' are {}, {}.'.format(lat[-1], long[-1]))
df_loc=pd.DataFrame({'Name': addresses,'Latitude': lat, 'Longitude':long})
df_loc
```
### Foursquare Credentials
```
CLIENT_ID = '' # your Foursquare ID
CLIENT_SECRET = '' # your Foursquare Secret
VERSION = '' # Foursquare API version
print('Your credentails:')
print('CLIENT_ID: ' + CLIENT_ID)
print('CLIENT_SECRET:' + CLIENT_SECRET)
```
### Make list of category names by ID
```
categories_url='https://api.foursquare.com/v2/venues/categories?&client_id={}&client_secret={}&v={}'.format(CLIENT_ID,
CLIENT_SECRET,VERSION)
result=requests.get(categories_url).json()
cat_id_list=[]
cat_name_list=[]
try:
len_major_cat=len(result['response']['categories'])
for i in range(len_major_cat-1):
cat_id_list.append(result['response']['categories'][i]['id'])
cat_name_list.append(result['response']['categories'][i]['name'])
len_sub_cat=len(result['response']['categories'][i]['categories'])
for j in range(len_sub_cat-1):
cat_id_list.append(result['response']['categories'][i]['categories'][j]['id'])
cat_name_list.append(result['response']['categories'][i]['categories'][j]['name'])
len_sub_sub_cat=len(result['response']['categories'][i]['categories'][j]['categories'])
for k in range(len_sub_sub_cat-1):
cat_id_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['id'])
cat_name_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['name'])
len_sub_sub_sub_cat=len(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'])
for l in range(len_sub_sub_sub_cat-1):
cat_id_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['id'])
cat_name_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['name'])
len_sub_sub_sub_sub_cat=len(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'])
for m in range(len_sub_sub_sub_sub_cat-1):
cat_id_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['id'])
cat_name_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['name'])
len_sub_sub_sub_sub_sub_cat=len(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m])
for n in range(len_sub_sub_sub_sub_sub_cat-1):
cat_id_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['categories'][n]['id'])
cat_name_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['categories'][n]['name'])
len_sub_sub_sub_sub_sub_sub_cat=len(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['categories'][n])
for o in range(len_sub_sub_sub_sub_sub_sub_cat-1):
cat_id_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['categories'][n]['categories'][o]['id'])
cat_name_list.append(result['response']['categories'][i]['categories'][j]['categories'][k]['categories'][l]['categories'][m]['categories'][n]['categories'][o]['name'])
except:
pass
cat_dict={}
for i in range (len(cat_name_list)):
cat_dict[cat_id_list[i]] = cat_name_list[i]
for cat in categories:
print(cat_dict[cat])
# General Search URL string
url_str='https://api.foursquare.com/v2/venues/search?categoryId={}&client_id={}&client_secret={}&ll={},{}&v={}&radius={}&limit={}'
# Zipcode Search URL string
url_str_zip='https://api.foursquare.com/v2/venues/search?&client_id={}&client_secret={}&ll={},{}&v={}&radius={}&limit={}'
```
### Explore nearby venues
<b> Function to get nearby venues matching the "categories" given address information </b>
```
def getNearbyVenues(names, latitudes, longitudes,url_link,categories, radius):
''' Create the venue search url and lookup nearby venues and return as dataframe'''
venues_list=[]
for i in range(0,len(names)):
name=names[i]
lat=latitudes[i]
lng = longitudes[i]
for category in categories:
# create the API request URL
url = url_link.format(
category,
CLIENT_ID,
CLIENT_SECRET,
lat,
lng,
VERSION,
radius,
LIMIT)
# make the GET request
results = requests.get(url).json()
# return only relevant information for each nearby venue
for j in range(0,len(results['response']['venues'])):
venues_list.append([
name,
results['response']['venues'][j]['name'],
results['response']['venues'][j]['id'],
results['response']['venues'][j]['location']['lat'],
results['response']['venues'][j]['location']['lng'],
results['response']['venues'][j]['categories'][0]['name'],
category,
results['response']['venues'][j]['location']['distance'],
])
nearby_venues = pd.DataFrame(venues_list)
nearby_venues.columns = ['Address',
'Venue',
'Venue_id',
'Venue Latitude',
'Venue Longitude',
'Venue Category',
'Category ID',
'Distance [m]']
return(nearby_venues)
def getVenueLikes(venue_ids):
''' Obtain the list of number of likes for the venues in "venue_ids"'''
likes_list=[]
for i in range(0,len(venue_ids)):
# create the API request URL
url_link='https://api.foursquare.com/v2/venues/{}?&client_id={}&client_secret={}&v={}'
url = url_link.format(venue_ids[i],
CLIENT_ID,
CLIENT_SECRET,
VERSION)
# make the GET request
results = requests.get(url).json()
likes_list.append(results['response']['venue']['likes']['count'])
return(likes_list)
```
Dataframe of Venues for each address, matching the "categories"
```
# Implement 'getNearbyVenues'
loc_venues=getNearbyVenues(df_loc['Name'], df_loc['Latitude'],df_loc['Longitude'],url_str,categories,radius)
loc_venues.head()
```
## Pre-Processing
Drop NaN values, set 'Venue' as index column since we are dealing with venues.
```
loc_venues.set_index('Venue',inplace=True)
loc_venues.dropna(inplace=True)
loc_venues.head()
loc_venues.shape
```
Print number of categories for each address
```
for i in range(0,len(addresses)):
print('There are {} uniques categories for '
.format(len(loc_venues.loc[loc_venues['Address']==addresses[i],'Venue Category'].unique()))+addresses[i])
```
## Exploratory Data Analysis
### Make Folium plot to show venues
```
maps={}
loc_results_lat=[]
loc_results_long=[]
zip_results={}
i=0
for address in addresses:
# create map
clustered=loc_venues[loc_venues['Address']==address]
lat_array=clustered['Venue Latitude']
long_array=clustered['Venue Longitude']
venue_name=clustered.index
# Calculate mean latitude and longitude
latitude=lat_array.mean()
longitude=long_array.mean()
# Update results latitude and longitude arrays
loc_results_lat.append(latitude)
loc_results_long.append(longitude)
# Obtain Zipcode
url_zip=url_str_zip.format(CLIENT_ID, CLIENT_SECRET, latitude, longitude, VERSION,500,1)
zip_result=requests.get(url_zip).json()
try:
zip_results[address]=zip_result['response']['venues'][0]['location']['formattedAddress']
except:
zip_results[address]='0'
print('Centroid for '+str(address)+' at: '+str(round(latitude,5))+', '+str(round(longitude,5))
+', Address:',zip_results[address][0])
map_clusters = folium.Map(location=[latitude, longitude],zoom_start=10)
# add markers to the map
markers_colors = []
for lat, lon, name in zip(lat_array, long_array,
venue_name ):
label = folium.Popup(name, parse_html=True)
folium.CircleMarker(
[lat, lon],
radius=5,
popup=label,
color='blue',
fill=True,
fill_color='blue',
fill_opacity=0.7,
).add_to(map_clusters)
folium.RegularPolygonMarker(location=[latitude, longitude], popup='Centroid',
fill_color='yellow', radius=10).add_to(map_clusters)
maps[address]=map_clusters
i=i+1
lat1=latitude
long1=longitude
maps[addresses[0]]
```
### Make Venue Longitude and Latitude box plots
```
fnum=1
unique_cat=len(loc_venues['Category ID'].unique())+1
bp={} # Box plot object dict.
for i in range(0,unique_cat):
plt.figure()
plt.subplot(1,5,1)
Y=loc_venues.loc[loc_venues['Category ID']==categories[i],'Venue Latitude']
bp[categories[i]+'.Latitude']=(plt.boxplot(Y))
plt.xlabel('Latitude')
plt.xticks([])
plt.title(str(cat_dict[categories[i]]))
plt.subplot(1,5,5)
Y=loc_venues.loc[loc_venues['Category ID']==categories[i],'Venue Longitude']
bp[categories[i]+'.Longitude']=(plt.boxplot(Y))
plt.xlabel('Longitude')
plt.xticks([])
plt.title(str(cat_dict[categories[i]]))
fnum=fnum+1
```
Remove the outliers from data, by referencing the category ID and latitude/longitude values
```
if remove_outliers=='Y':
flag=1
while flag==1:
flag=0
for category in categories:
Y=loc_venues.loc[loc_venues['Category ID']==category,'Venue Latitude']
bp[category+'.Latitude']=plt.boxplot(Y)
Y=loc_venues.loc[loc_venues['Category ID']==category,'Venue Longitude']
bp[category+'.Longitude']=plt.boxplot(Y)
outliers_lat=bp[category+'.Latitude']['fliers'][0].get_data()[1]
outliers_long=bp[category+'.Longitude']['fliers'][0].get_data()[1]
if len(outliers_lat)>0 or len(outliers_long)>0:
flag=1
for outlier_lat in outliers_lat:
idx1=loc_venues['Category ID']==category
idx2=loc_venues['Venue Latitude']==outlier_lat
idx1=idx1[idx1==True].index
idx2=idx2[idx2==True].index
loc_venues.drop(idx1.intersection(idx2),axis=0,inplace=True)
for outlier_long in outliers_long:
idx1=loc_venues['Category ID']==category
idx2=loc_venues['Venue Longitude']==outlier_long
idx1=idx1[idx1==True].index
idx2=idx2[idx2==True].index
loc_venues.drop(idx1.intersection(idx2),axis=0,inplace=True)
loc_venues.shape
```
Re-Plot the box plots to check that there are no outliers remaining
```
fnum=1
unique_cat=len(loc_venues['Category ID'].unique())+1
bp={} # Box plot object dict.
for i in range(0,unique_cat):
plt.figure()
plt.subplot(1,5,1)
Y=loc_venues.loc[loc_venues['Category ID']==categories[i],'Venue Latitude']
bp[categories[i]+'.Latitude']=(plt.boxplot(Y))
plt.xlabel('Latitude')
plt.xticks([])
plt.title(str(cat_dict[categories[i]]))
plt.subplot(1,5,5)
Y=loc_venues.loc[loc_venues['Category ID']==categories[i],'Venue Longitude']
bp[categories[i]+'.Longitude']=(plt.boxplot(Y))
plt.xlabel('Longitude')
plt.xticks([])
plt.title(str(cat_dict[categories[i]]))
fnum=fnum+1
```
Re-plot the folium plot
```
maps={}
loc_results_lat=[]
loc_results_long=[]
zip_results={}
i=0
for address in addresses:
# create map
clustered=loc_venues[loc_venues['Address']==address]
lat_array=clustered['Venue Latitude']
long_array=clustered['Venue Longitude']
venue_name=clustered.index
# Calculate mean latitude and longitude
latitude=lat_array.mean()
longitude=long_array.mean()
# Update results latitude and longitude arrays
loc_results_lat.append(latitude)
loc_results_long.append(longitude)
# Obtain Zipcode
url_zip=url_str_zip.format(CLIENT_ID, CLIENT_SECRET, latitude, longitude, VERSION,500,1)
zip_result=requests.get(url_zip).json()
try:
zip_results[address]=zip_result['response']['venues'][0]['location']['formattedAddress']
except:
zip_results[address]='0'
print('Centroid for '+str(address)+' at: '+str(round(latitude,5))+', '+str(round(longitude,5))
+', Address:',zip_results[address][0])
map_clusters = folium.Map(location=[latitude, longitude],zoom_start=10)
# add markers to the map
markers_colors = []
for lat, lon, name in zip(lat_array, long_array,
venue_name ):
label = folium.Popup(name, parse_html=True)
folium.CircleMarker(
[lat, lon],
radius=5,
popup=label,
color='blue',
fill=True,
fill_color='blue',
fill_opacity=0.7,
).add_to(map_clusters)
folium.RegularPolygonMarker(location=[latitude, longitude], popup='Centroid',
fill_color='yellow', radius=10).add_to(map_clusters)
maps[address]=map_clusters
i=i+1
lat2=latitude
long2=longitude
maps[addresses[-1]]
```
### New dataframe for the venues with outliers removed and distance calculated from new centroid
```
loc_venues_new=getNearbyVenues(df_loc['Name'], loc_results_lat,loc_results_long,url_str,categories,radius)
loc_venues_new.head()
# Find common venues between initial and outlier adjusted venue list
set_loc_venues=set(loc_venues['Venue_id'].tolist())
set_loc_venues_new=set(loc_venues_new['Venue_id'].tolist())
list_common_venues=list(set_loc_venues.intersection(set_loc_venues_new))
#Filter loc_venues_new to common venue ids
loc_venues_new=loc_venues_new.set_index(['Venue_id']).loc[list_common_venues,:].reset_index()
loc_venues_new.head()
#Sum of distances between centroid before and after removing outliers
dist_1=loc_venues['Distance [m]'].mean()/1000
print('Mean distance between venues and centroid is {} km.'.format(dist_1))
if remove_outliers=='Y':
dist_2=loc_venues.set_index(['Venue_id']).loc[list_common_venues,:]['Distance [m]'].mean()/1000
dist_3=loc_venues_new['Distance [m]'].mean()/1000
# Print values
print('Mean distance between non-outlier venues and centroid before removing outliers is {} km.'.format(dist_1))
print('Mean distance between non-outlier venues and centroid after removing outliers is {} km.'.format(dist_2))
print('Difference in mean distance between non-outlier venues and centroid before and after removing outliers is {} km'.
format(dist_2-dist_3))
print('Difference in mean distance between venues and centroid before and after removing outliers is {} km'.
format(dist_1-dist_3))
```
### Geo-distance shift in coordinates
Define function to calculate distance based on geo-coordinates
```
def geodistance (coord1,coord2):
R = 6373.0 ## Radius of Earth in kms.
latd1 = math.radians(coord1[0]) # Latitude of coord1 calculated in radians
lon1 = math.radians(coord1[1]) # Longitude of coord1 calculated in radians
latd2 = math.radians(coord2[0]) # Latitude of coord2 calculated in radians
lon2 = math.radians(coord2[1]) # Longitude of coord2 calculated in radians
dlon = lon2 - lon1
dlat = latd2 - latd1
a = math.sin(dlat / 2)**2 + math.cos(lat1) * math.cos(lat2) * math.sin(dlon / 2)**2
c = 2 * math.atan2(math.sqrt(a), math.sqrt(1 - a))
distance = R * c * 1000 # Geo-distance in kms.
return distance
```
Distance shift in centroid after omitting the outliers:
```
shift_outlier=geodistance([lat1, long1],[lat2,long2])
print('Geo-Distance shift in Centroid before and after removing outliers is: '+str(round(shift_outlier/1000,2))+'kms.' )
for i in range(0,len(addresses)):
shift_original=geodistance([df_loc.loc[i,'Latitude'],df_loc.loc[i,'Longitude']],[lat2,long2])
print('Geo-Distance shift in Centroid for {} before and after processing is: {}'
.format(addresses[i],str(round(shift_original/1000,2))+'kms.'))
```
## Encoding and Clustering
### Encoding by user specified category
```
# one hot encoding
loc_encoded=pd.get_dummies(loc_venues_new[['Category ID']], prefix="", prefix_sep="") # Dataframe of encoding
loc_venues_encoded=pd.concat([loc_venues_new,loc_encoded],axis=1) # Encoded dataframe with venue details
loc_venues_encoded.head()
```
### Clustering by category encoding
```
# set number of clusters
kclusters = len(categories)
# run k-means clustering
kmeans = KMeans(n_clusters=kclusters, random_state=0).fit(loc_encoded)
# check cluster labels generated for each row in the dataframe
(kmeans.labels_)
# add clustering labels
loc_venues_clustered=loc_venues_encoded
loc_venues_clustered['Cluster Labels']= kmeans.labels_
# Display clustered dataframe
loc_venues_clustered.head(2)
```
## Display Results
### Generate analyzed map array for each address using folium, and display centroid location for each address
```
maps={}
loc_results_lat=[]
loc_results_long=[]
zip_results={}
i=0
for address in addresses:
# create map
clustered=loc_venues_clustered[loc_venues_clustered['Address']==address]
lat_array=clustered['Venue Latitude']
long_array=clustered['Venue Longitude']
# Calculate mean latitude and longitude
latitude=lat_array.mean()
longitude=long_array.mean()
# Update results latitude and longitude arrays
loc_results_lat.append(latitude)
loc_results_long.append(longitude)
# Obtain Zipcode
url_zip=url_str_zip.format(CLIENT_ID, CLIENT_SECRET, latitude, longitude, VERSION,500,1)
zip_result=requests.get(url_zip).json()
try:
zip_results[address]=zip_result['response']['venues'][0]['location']['formattedAddress']
except:
zip_results[address]='0'
print('Centroid for '+str(address)+' at: '+str(round(latitude,5))+', '+str(round(longitude,5))
+', Address:',zip_results[address][0])
map_clusters = folium.Map(location=[latitude, longitude],zoom_start=10)
# set color scheme for the clusters
x = np.arange(kclusters)
ys = [j + x + (j*x)**2 for j in range(kclusters)]
colors_array = cm.rainbow(np.linspace(0, 1, len(ys)))
rainbow = [colors.rgb2hex(j) for j in colors_array]
# add markers to the map
markers_colors = []
for lat, lon, poi, cluster, category in zip(lat_array, long_array,
clustered.index, clustered['Cluster Labels'], clustered['Category ID']):
label = folium.Popup(str(poi) + ' Cluster: ' + str(cat_dict[category]), parse_html=True)
folium.CircleMarker(
[lat, lon],
radius=5,
popup=label,
color=rainbow[cluster],
fill=True,
fill_color=rainbow[cluster],
fill_opacity=0.7,
).add_to(map_clusters)
folium.RegularPolygonMarker(location=[latitude, longitude], popup='Centroid',
fill_color='yellow', radius=10).add_to(map_clusters)
maps[address]=map_clusters
i=i+1
```
### Plot map
```
maps[addresses[-1]]
hotel_category=['4bf58dd8d48988d1fa931735'] # Category for Hotels
# Enter Hotel Search URL string
url_str_hotels='https://api.foursquare.com/v2/venues/search?categoryId={}&client_id={}&client_secret={}&ll={},{}&v={}&radius={}&limit={}'
df_hotels=df_loc.copy()
df_hotels['Latitude']=latitude
df_hotels['Longitude']=longitude
# Radius in metres
hotels=getNearbyVenues(names=df_hotels['Name'], latitudes=df_hotels['Latitude'],longitudes=df_hotels['Longitude'],
url_link=url_str_hotels,categories=hotel_category,radius=hotel_radius)
hotel_ids=hotels['Venue_id'].tolist()
likes_list=getVenueLikes(hotel_ids)
hotels['Likes']=likes_list
hotels.drop(columns=['Category ID','Address','Venue_id'],inplace=True)
hotels.sort_values(by=['Likes','Distance [m]'],ascending=[False,True],inplace=True)
hotels.reset_index(inplace=True,drop=True)
hotels
```
| github_jupyter |
# Introduction
In a prior notebook, documents were partitioned by assigning them to the domain with the highest Dice similarity of their term and structure occurrences. The occurrences of terms and structures in each domain is what we refer to as the domain "archetype." Here, we'll assess whether the observed similarity between documents and the archetype is greater than expected by chance. This would indicate that information in the framework generalizes well to individual documents.
# Load the data
```
import os
import pandas as pd
import numpy as np
import sys
sys.path.append("..")
import utilities
from ontology import ontology
from style import style
version = 190325 # Document-term matrix version
clf = "lr" # Classifier used to generate the framework
suffix = "_" + clf # Suffix for term lists
n_iter = 1000 # Iterations for null distribution
circuit_counts = range(2, 51) # Range of k values
```
## Brain activation coordinates
```
act_bin = utilities.load_coordinates()
print("Document N={}, Structure N={}".format(
act_bin.shape[0], act_bin.shape[1]))
```
## Document-term matrix
```
dtm_bin = utilities.load_doc_term_matrix(version=version, binarize=True)
print("Document N={}, Term N={}".format(
dtm_bin.shape[0], dtm_bin.shape[1]))
```
## Document splits
```
splits = {}
# splits["train"] = [int(pmid.strip()) for pmid in open("../data/splits/train.txt")]
splits["validation"] = [int(pmid.strip()) for pmid in open("../data/splits/validation.txt")]
splits["test"] = [int(pmid.strip()) for pmid in open("../data/splits/test.txt")]
for split, split_pmids in splits.items():
print("{:12s} N={}".format(split.title(), len(split_pmids)))
pmids = dtm_bin.index.intersection(act_bin.index)
```
## Document assignments and distances
Indexing by min:max will be faster in subsequent computations
```
from collections import OrderedDict
from scipy.spatial.distance import cdist
def load_doc2dom(k, clf="lr"):
doc2dom_df = pd.read_csv("../partition/data/doc2dom_k{:02d}_{}.csv".format(k, clf),
header=None, index_col=0)
doc2dom = {int(pmid): str(dom.values[0]) for pmid, dom in doc2dom_df.iterrows()}
return doc2dom
def load_dom2docs(k, domains, splits, clf="lr"):
doc2dom = load_doc2dom(k, clf=clf)
dom2docs = {dom: {split: [] for split, _ in splits.items()} for dom in domains}
for doc, dom in doc2dom.items():
for split, split_pmids in splits.items():
if doc in splits[split]:
dom2docs[dom][split].append(doc)
return dom2docs
sorted_pmids, doc_dists, dom_idx = {}, {}, {}
for k in circuit_counts:
print("Processing k={:02d}".format(k))
sorted_pmids[k], doc_dists[k], dom_idx[k] = {}, {}, {}
for split, split_pmids in splits.items():
lists, circuits = ontology.load_ontology(k, path="../ontology/", suffix=suffix)
words = sorted(list(set(lists["TOKEN"])))
structures = sorted(list(set(act_bin.columns)))
domains = list(OrderedDict.fromkeys(lists["DOMAIN"]))
dtm_words = dtm_bin.loc[pmids, words]
act_structs = act_bin.loc[pmids, structures]
docs = dtm_words.copy()
docs[structures] = act_structs.copy()
doc2dom = load_doc2dom(k, clf=clf)
dom2docs = load_dom2docs(k, domains, splits, clf=clf)
ids = []
for dom in domains:
ids += [pmid for pmid, sys in doc2dom.items() if sys == dom and pmid in split_pmids]
sorted_pmids[k][split] = ids
doc_dists[k][split] = pd.DataFrame(cdist(docs.loc[ids], docs.loc[ids], metric="dice"),
index=ids, columns=ids)
dom_idx[k][split] = {}
for dom in domains:
dom_idx[k][split][dom] = {}
dom_pmids = dom2docs[dom][split]
if len(dom_pmids) > 0:
dom_idx[k][split][dom]["min"] = sorted_pmids[k][split].index(dom_pmids[0])
dom_idx[k][split][dom]["max"] = sorted_pmids[k][split].index(dom_pmids[-1]) + 1
else:
dom_idx[k][split][dom]["min"] = 0
dom_idx[k][split][dom]["max"] = 0
```
# Index by PMID and sort by structure
```
structures = sorted(list(set(act_bin.columns)))
act_structs = act_bin.loc[pmids, structures]
```
# Compute domain modularity
## Observed values
## Distances internal and external to articles in each domain
```
dists_int, dists_ext = {}, {}
for k in circuit_counts:
dists_int[k], dists_ext[k] = {}, {}
lists, circuits = ontology.load_ontology(k, path="../ontology/", suffix=suffix)
domains = list(OrderedDict.fromkeys(lists["DOMAIN"]))
for split, split_pmids in splits.items():
dists_int[k][split], dists_ext[k][split] = {}, {}
for dom in domains:
dom_min, dom_max = dom_idx[k][split][dom]["min"], dom_idx[k][split][dom]["max"]
dom_dists = doc_dists[k][split].values[:,dom_min:dom_max][dom_min:dom_max,:]
dists_int[k][split][dom] = dom_dists
other_dists_lower = doc_dists[k][split].values[:,dom_min:dom_max][:dom_min,:]
other_dists_upper = doc_dists[k][split].values[:,dom_min:dom_max][dom_max:,:]
other_dists = np.concatenate((other_dists_lower, other_dists_upper))
dists_ext[k][split][dom] = other_dists
```
## Domain-averaged ratio of external to internal distances
```
means = {split: np.empty((len(circuit_counts),)) for split in splits.keys()}
for k_i, k in enumerate(circuit_counts):
file_obs = "data/kvals/mod_obs_k{:02d}_{}_{}.csv".format(k, clf, split)
if not os.path.isfile(file_obs):
print("Processing k={:02d}".format(k))
lists, circuits = ontology.load_ontology(k, path="../ontology/", suffix=suffix)
domains = list(OrderedDict.fromkeys(lists["DOMAIN"]))
dom2docs = load_dom2docs(k, domains, splits, clf=clf)
pmid_list, split_list, dom_list, obs_list = [], [], [], []
for split, split_pmids in splits.items():
for dom in domains:
n_dom_docs = dists_int[k][split][dom].shape[0]
if n_dom_docs > 0:
mean_dist_int = np.nanmean(dists_int[k][split][dom], axis=0)
mean_dist_ext = np.nanmean(dists_ext[k][split][dom], axis=0)
ratio = mean_dist_ext / mean_dist_int
ratio[ratio == np.inf] = np.nan
pmid_list += dom2docs[dom][split]
dom_list += [dom] * len(ratio)
split_list += [split] * len(ratio)
obs_list += list(ratio)
df_obs = pd.DataFrame({"PMID": pmid_list, "SPLIT": split_list,
"DOMAIN": dom_list, "OBSERVED": obs_list})
df_obs.to_csv(file_obs, index=None)
else:
df_obs = pd.read_csv(file_obs)
for split, split_pmids in splits.items():
dom_means = []
for dom in set(df_obs["DOMAIN"]):
dom_vals = df_obs.loc[(df_obs["SPLIT"] == split) & (df_obs["DOMAIN"] == dom), "OBSERVED"]
dom_means.append(np.nanmean(dom_vals))
means[split][k_i] = np.nanmean(dom_means)
```
## Null distributions
```
nulls = {split: np.empty((len(circuit_counts),n_iter)) for split in splits.keys()}
for split, split_pmids in splits.items():
for k_i, k in enumerate(circuit_counts):
file_null = "data/kvals/mod_null_k{:02d}_{}_{}iter.csv".format(k, split, n_iter)
if not os.path.isfile(file_null):
print("Processing k={:02d}".format(k))
lists, circuits = ontology.load_ontology(k, path="../ontology/", suffix=suffix)
domains = list(OrderedDict.fromkeys(lists["DOMAIN"]))
n_docs = len(split_pmids)
df_null = np.empty((len(domains), n_iter))
for i, dom in enumerate(domains):
n_dom_docs = dists_int[k][split][dom].shape[0]
if n_dom_docs > 0:
dist_int_ext = np.concatenate((dists_int[k][split][dom], dists_ext[k][split][dom]))
for n in range(n_iter):
null = np.random.choice(range(n_docs), size=n_docs, replace=False)
dist_int_ext_null = dist_int_ext[null,:]
mean_dist_int = np.nanmean(dist_int_ext_null[:n_dom_docs,:], axis=0)
mean_dist_ext = np.nanmean(dist_int_ext_null[n_dom_docs:,:], axis=0)
ratio = mean_dist_ext / mean_dist_int
ratio[ratio == np.inf] = np.nan
df_null[i,n] = np.nanmean(ratio)
else:
df_null[i,:] = np.nan
df_null = pd.DataFrame(df_null, index=domains, columns=range(n_iter))
df_null.to_csv(file_null)
else:
df_null = pd.read_csv(file_null, index_col=0, header=0)
nulls[split][k_i,:] = np.nanmean(df_null, axis=0)
```
## Bootstrap distributions
```
boots = {split: np.empty((len(circuit_counts),n_iter)) for split in splits.keys()}
for split, split_pmids in splits.items():
for k_i, k in enumerate(circuit_counts):
file_boot = "data/kvals/mod_boot_k{:02d}_{}_{}iter.csv".format(k, split, n_iter)
if not os.path.isfile(file_boot):
print("Processing k={:02d}".format(k))
lists, circuits = ontology.load_ontology(k, path="../ontology/", suffix=suffix)
domains = list(OrderedDict.fromkeys(lists["DOMAIN"]))
df_boot = np.empty((len(domains), n_iter))
for i, dom in enumerate(domains):
n_dom_docs = dists_int[k][split][dom].shape[0]
if n_dom_docs > 0:
for n in range(n_iter):
boot = np.random.choice(range(n_dom_docs), size=n_dom_docs, replace=True)
mean_dist_int = np.nanmean(dists_int[k][split][dom][:,boot], axis=0)
mean_dist_ext = np.nanmean(dists_ext[k][split][dom][:,boot], axis=0)
ratio = mean_dist_ext / mean_dist_int
ratio[ratio == np.inf] = np.nan
df_boot[i,n] = np.nanmean(ratio)
else:
df_boot[i,:] = np.nan
df_boot = pd.DataFrame(df_boot, index=domains, columns=range(n_iter))
df_boot.to_csv(file_boot)
else:
df_boot = pd.read_csv(file_boot, index_col=0, header=0)
boots[split][k_i,:] = np.nanmean(df_boot, axis=0)
```
# Plot results over k
```
from matplotlib import rcParams
%matplotlib inline
rcParams["axes.linewidth"] = 1.5
for split in splits.keys():
print(split.upper())
utilities.plot_stats_by_k(means, nulls, boots, circuit_counts, metric="mod",
split=split, op_k=6, clf=clf, interval=0.999,
ylim=[0.8,1.4], yticks=[0.8, 0.9,1,1.1,1.2,1.3,1.4])
```
| github_jupyter |
```
import pandas as pd
import praw
import re
import datetime as dt
import seaborn as sns
import requests
import json
import sys
import time
## acknowledgements
'''
https://stackoverflow.com/questions/48358837/pulling-reddit-comments-using-python-praw-and-creating-a-dataframe-with-the-resu
https://www.reddit.com/r/redditdev/comments/2e2q2l/praw_downvote_count_always_zero/
https://towardsdatascience.com/an-easy-tutorial-about-sentiment-analysis-with-deep-learning-and-keras-2bf52b9cba91
For navigating pushshift: https://github.com/Watchful1/Sketchpad/blob/master/postDownloader.py
# traffic = reddit.subreddit(subreddit).traffic() is not available to us, sadly.
'''
with open("../API.env") as file:
exec(file.read())
reddit = praw.Reddit(
client_id = client_id,
client_secret = client_secret,
user_agent = user_agent
)
'''
Some helper functions for the reddit API.
'''
def extract_num_rewards(awardings_data):
return sum( x["count"] for x in awardings_data)
def extract_data(submission, comments = False):
postlist = []
# extracts top level comments
if comments:
submission.comments.replace_more(limit=0)
for comment in submission.comments:
post = vars(comment)
postlist.append(post)
content = vars(submission)
content["total_awards"] = extract_num_rewards(content["all_awardings"])
return content
'''
Sample num_samples random submissions, and get the top num_samples submissions, and put them into dataframes.
Opted instead to scrape the entire thing.
'''
def random_sample(num_samples, subreddit):
sample = []
for i in range(num_samples):
submission = reddit.subreddit(subreddit).random()
sample.append(extract_data(submission))
return(pd.DataFrame(sample))
def sample(source):
submissions = []
for submission in source:
submissions.append(extract_data(submission))
print(f"Got {len(submissions)} submissions. (This can be less than num_samples.)")
return(pd.DataFrame(submissions))
def top_sample(num_samples, subreddit):
return sample(reddit.subreddit(subreddit).top(limit=num_samples) )
def rising_sample(num_samples, subreddit):
return sample(reddit.subreddit(subreddit).rising(limit=num_samples))
def controversial_sample(num_samples, subreddit):
return sample(reddit.subreddit(subreddit).controversial(limit=num_samples) )
num_samples = 10
subreddit ='wallstreetbets'
#random_wsb = random_sample(num_samples, subreddit)
#top_wsb = top_sample(num_samples,subreddit)
#rising_wsb = rising_sample(num_samples, subreddit)
#controversial_wsb = controversial_sample(num_samples, subreddit)
#random_wsb.to_pickle("random_wsb.pkl")
#top_wsb.to_pickle("top_wsb.pkl")
#rising_wsb.to_pickle("rising_wsb.pkl")
#controversial_wsb.to_pickle("controversial_wsb.pkl")
# other commands here: https://praw.readthedocs.io/en/latest/code_overview/models/subreddit.html#praw.models.Subreddit.rising
# NB: The subreddit stream option seems useful.
# NB: There is also rising_random
submission = reddit.subreddit(subreddit).random()
submission.approved_at_utc
vars(submission)
str(submission.flair)
```
| github_jupyter |
```
#pip install sklearn numpy scipy matplotlib
from sklearn import datasets
iris = datasets.load_iris()
digits = datasets.load_digits()
print(digits.data)
digits.target
digits.images[0]
```
create a support vector classifier and manually set the gamma
```
from sklearn import svm, metrics
clf = svm.SVC(gamma=0.001, C=100.)
```
fit the classifier to the model and use all the images in our dataset except the last one
```
clf.fit(digits.data[:-1], digits.target[:-1])
svm.SVC(C=100.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma=0.001, kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)
clf.predict(digits.data[-1:])
```
reshape the image data into a 8x8 array prior to rendering it
```
import matplotlib.pyplot as plt
plt.imshow(digits.data[-1:].reshape(8,8), cmap=plt.cm.gray_r)
plt.show()
```
persist the model using pickle and load it again to ensure it works
```
import pickle
s = pickle.dumps(clf)
with open(b"digits.model.obj", "wb") as f:
pickle.dump(clf, f)
clf2 = pickle.loads(s)
clf2.predict(digits.data[0:1])
plt.imshow(digits.data[0:1].reshape(8,8), cmap=plt.cm.gray_r)
plt.show()
```
alternately use joblib.dump
```
from sklearn.externals import joblib
joblib.dump(clf, 'digits.model.pkl')
```
example from http://scikit-learn.org/stable/auto_examples/classification/plot_digits_classification.html#sphx-glr-auto-examples-classification-plot-digits-classification-py
```
images_and_labels = list(zip(digits.images, digits.target))
for index, (image, label) in enumerate(images_and_labels[:4]):
plt.subplot(2, 4, index + 1)
plt.axis('off')
plt.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
plt.title('Training: %i' % label)
# To apply a classifier on this data, we need to flatten the image, to
# turn the data in a (samples, feature) matrix:
n_samples = len(digits.images)
data = digits.images.reshape((n_samples, -1))
# Create a classifier: a support vector classifier
classifier = svm.SVC(gamma=0.001)
# We learn the digits on the first half of the digits
classifier.fit(data[:n_samples // 2], digits.target[:n_samples // 2])
# Now predict the value of the digit on the second half:
expected = digits.target[n_samples // 2:]
predicted = classifier.predict(data[n_samples // 2:])
print("Classification report for classifier %s:\n%s\n"
% (classifier, metrics.classification_report(expected, predicted)))
print("Confusion matrix:\n%s" % metrics.confusion_matrix(expected, predicted))
images_and_predictions = list(zip(digits.images[n_samples // 2:], predicted))
for index, (image, prediction) in enumerate(images_and_predictions[:4]):
plt.subplot(2, 4, index + 5)
plt.axis('off')
plt.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
plt.title('Prediction: %i' % prediction)
plt.show()
```
| github_jupyter |
# Residual Networks
Welcome to the second assignment of this week! You will learn how to build very deep convolutional networks, using Residual Networks (ResNets). In theory, very deep networks can represent very complex functions; but in practice, they are hard to train. Residual Networks, introduced by [He et al.](https://arxiv.org/pdf/1512.03385.pdf), allow you to train much deeper networks than were previously practically feasible.
**In this assignment, you will:**
- Implement the basic building blocks of ResNets.
- Put together these building blocks to implement and train a state-of-the-art neural network for image classification.
## <font color='darkblue'>Updates</font>
#### If you were working on the notebook before this update...
* The current notebook is version "2a".
* You can find your original work saved in the notebook with the previous version name ("v2")
* To view the file directory, go to the menu "File->Open", and this will open a new tab that shows the file directory.
#### List of updates
* For testing on an image, replaced `preprocess_input(x)` with `x=x/255.0` to normalize the input image in the same way that the model's training data was normalized.
* Refers to "shallower" layers as those layers closer to the input, and "deeper" layers as those closer to the output (Using "shallower" layers instead of "lower" or "earlier").
* Added/updated instructions.
This assignment will be done in Keras.
Before jumping into the problem, let's run the cell below to load the required packages.
```
import numpy as np
from keras import layers
from keras.layers import Input, Add, Dense, Activation, ZeroPadding2D, BatchNormalization, Flatten, Conv2D, AveragePooling2D, MaxPooling2D, GlobalMaxPooling2D
from keras.models import Model, load_model
from keras.preprocessing import image
from keras.utils import layer_utils
from keras.utils.data_utils import get_file
from keras.applications.imagenet_utils import preprocess_input
import pydot
from IPython.display import SVG
from keras.utils.vis_utils import model_to_dot
from keras.utils import plot_model
from resnets_utils import *
from keras.initializers import glorot_uniform
import scipy.misc
from matplotlib.pyplot import imshow
%matplotlib inline
import keras.backend as K
K.set_image_data_format('channels_last')
K.set_learning_phase(1)
```
## 1 - The problem of very deep neural networks
Last week, you built your first convolutional neural network. In recent years, neural networks have become deeper, with state-of-the-art networks going from just a few layers (e.g., AlexNet) to over a hundred layers.
* The main benefit of a very deep network is that it can represent very complex functions. It can also learn features at many different levels of abstraction, from edges (at the shallower layers, closer to the input) to very complex features (at the deeper layers, closer to the output).
* However, using a deeper network doesn't always help. A huge barrier to training them is vanishing gradients: very deep networks often have a gradient signal that goes to zero quickly, thus making gradient descent prohibitively slow.
* More specifically, during gradient descent, as you backprop from the final layer back to the first layer, you are multiplying by the weight matrix on each step, and thus the gradient can decrease exponentially quickly to zero (or, in rare cases, grow exponentially quickly and "explode" to take very large values).
* During training, you might therefore see the magnitude (or norm) of the gradient for the shallower layers decrease to zero very rapidly as training proceeds:
<img src="images/vanishing_grad_kiank.png" style="width:450px;height:220px;">
<caption><center> <u> <font color='purple'> **Figure 1** </u><font color='purple'> : **Vanishing gradient** <br> The speed of learning decreases very rapidly for the shallower layers as the network trains </center></caption>
You are now going to solve this problem by building a Residual Network!
## 2 - Building a Residual Network
In ResNets, a "shortcut" or a "skip connection" allows the model to skip layers:
<img src="images/skip_connection_kiank.png" style="width:650px;height:200px;">
<caption><center> <u> <font color='purple'> **Figure 2** </u><font color='purple'> : A ResNet block showing a **skip-connection** <br> </center></caption>
The image on the left shows the "main path" through the network. The image on the right adds a shortcut to the main path. By stacking these ResNet blocks on top of each other, you can form a very deep network.
We also saw in lecture that having ResNet blocks with the shortcut also makes it very easy for one of the blocks to learn an identity function. This means that you can stack on additional ResNet blocks with little risk of harming training set performance.
(There is also some evidence that the ease of learning an identity function accounts for ResNets' remarkable performance even more so than skip connections helping with vanishing gradients).
Two main types of blocks are used in a ResNet, depending mainly on whether the input/output dimensions are same or different. You are going to implement both of them: the "identity block" and the "convolutional block."
### 2.1 - The identity block
The identity block is the standard block used in ResNets, and corresponds to the case where the input activation (say $a^{[l]}$) has the same dimension as the output activation (say $a^{[l+2]}$). To flesh out the different steps of what happens in a ResNet's identity block, here is an alternative diagram showing the individual steps:
<img src="images/idblock2_kiank.png" style="width:650px;height:150px;">
<caption><center> <u> <font color='purple'> **Figure 3** </u><font color='purple'> : **Identity block.** Skip connection "skips over" 2 layers. </center></caption>
The upper path is the "shortcut path." The lower path is the "main path." In this diagram, we have also made explicit the CONV2D and ReLU steps in each layer. To speed up training we have also added a BatchNorm step. Don't worry about this being complicated to implement--you'll see that BatchNorm is just one line of code in Keras!
In this exercise, you'll actually implement a slightly more powerful version of this identity block, in which the skip connection "skips over" 3 hidden layers rather than 2 layers. It looks like this:
<img src="images/idblock3_kiank.png" style="width:650px;height:150px;">
<caption><center> <u> <font color='purple'> **Figure 4** </u><font color='purple'> : **Identity block.** Skip connection "skips over" 3 layers.</center></caption>
Here are the individual steps.
First component of main path:
- The first CONV2D has $F_1$ filters of shape (1,1) and a stride of (1,1). Its padding is "valid" and its name should be `conv_name_base + '2a'`. Use 0 as the seed for the random initialization.
- The first BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2a'`.
- Then apply the ReLU activation function. This has no name and no hyperparameters.
Second component of main path:
- The second CONV2D has $F_2$ filters of shape $(f,f)$ and a stride of (1,1). Its padding is "same" and its name should be `conv_name_base + '2b'`. Use 0 as the seed for the random initialization.
- The second BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2b'`.
- Then apply the ReLU activation function. This has no name and no hyperparameters.
Third component of main path:
- The third CONV2D has $F_3$ filters of shape (1,1) and a stride of (1,1). Its padding is "valid" and its name should be `conv_name_base + '2c'`. Use 0 as the seed for the random initialization.
- The third BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2c'`.
- Note that there is **no** ReLU activation function in this component.
Final step:
- The `X_shortcut` and the output from the 3rd layer `X` are added together.
- **Hint**: The syntax will look something like `Add()([var1,var2])`
- Then apply the ReLU activation function. This has no name and no hyperparameters.
**Exercise**: Implement the ResNet identity block. We have implemented the first component of the main path. Please read this carefully to make sure you understand what it is doing. You should implement the rest.
- To implement the Conv2D step: [Conv2D](https://keras.io/layers/convolutional/#conv2d)
- To implement BatchNorm: [BatchNormalization](https://faroit.github.io/keras-docs/1.2.2/layers/normalization/) (axis: Integer, the axis that should be normalized (typically the 'channels' axis))
- For the activation, use: `Activation('relu')(X)`
- To add the value passed forward by the shortcut: [Add](https://keras.io/layers/merge/#add)
```
# GRADED FUNCTION: identity_block
def identity_block(X, f, filters, stage, block):
"""
Implementation of the identity block as defined in Figure 4
Arguments:
X -- input tensor of shape (m, n_H_prev, n_W_prev, n_C_prev)
f -- integer, specifying the shape of the middle CONV's window for the main path
filters -- python list of integers, defining the number of filters in the CONV layers of the main path
stage -- integer, used to name the layers, depending on their position in the network
block -- string/character, used to name the layers, depending on their position in the network
Returns:
X -- output of the identity block, tensor of shape (n_H, n_W, n_C)
"""
# defining name basis
conv_name_base = 'res' + str(stage) + block + '_branch'
bn_name_base = 'bn' + str(stage) + block + '_branch'
# Retrieve Filters
F1, F2, F3 = filters
# Save the input value. You'll need this later to add back to the main path.
X_shortcut = X
# First component of main path
X = Conv2D(filters = F1, kernel_size = (1, 1), strides = (1,1), padding = 'valid', name = conv_name_base + '2a', kernel_initializer = glorot_uniform(seed=0))(X)
X = BatchNormalization(axis = 3, name = bn_name_base + '2a')(X)
X = Activation('relu')(X)
### START CODE HERE ###
# Second component of main path (≈3 lines)
X = Conv2D(filters = F2, kernel_size = (f, f), strides = (1,1), padding = 'same', name = conv_name_base + '2b', kernel_initializer = glorot_uniform(seed=0))(X)
X = BatchNormalization(axis = 3, name = bn_name_base + '2b')(X)
X = Activation('relu')(X)
# Third component of main path (≈2 lines)
X = Conv2D(filters = F3, kernel_size = (1, 1), strides = (1,1), padding = 'valid', name = conv_name_base + '2c', kernel_initializer = glorot_uniform(seed=0))(X)
X = BatchNormalization(axis = 3, name = bn_name_base + '2c')(X)
# Final step: Add shortcut value to main path, and pass it through a RELU activation (≈2 lines)
X = Add()([X, X_shortcut])
X = Activation('relu')(X)
### END CODE HERE ###
return X
tf.reset_default_graph()
with tf.Session() as test:
np.random.seed(1)
A_prev = tf.placeholder("float", [3, 4, 4, 6])
X = np.random.randn(3, 4, 4, 6)
A = identity_block(A_prev, f = 2, filters = [2, 4, 6], stage = 1, block = 'a')
test.run(tf.global_variables_initializer())
out = test.run([A], feed_dict={A_prev: X, K.learning_phase(): 0})
print("out = " + str(out[0][1][1][0]))
```
**Expected Output**:
<table>
<tr>
<td>
**out**
</td>
<td>
[ 0.94822985 0. 1.16101444 2.747859 0. 1.36677003]
</td>
</tr>
</table>
## 2.2 - The convolutional block
The ResNet "convolutional block" is the second block type. You can use this type of block when the input and output dimensions don't match up. The difference with the identity block is that there is a CONV2D layer in the shortcut path:
<img src="images/convblock_kiank.png" style="width:650px;height:150px;">
<caption><center> <u> <font color='purple'> **Figure 4** </u><font color='purple'> : **Convolutional block** </center></caption>
* The CONV2D layer in the shortcut path is used to resize the input $x$ to a different dimension, so that the dimensions match up in the final addition needed to add the shortcut value back to the main path. (This plays a similar role as the matrix $W_s$ discussed in lecture.)
* For example, to reduce the activation dimensions's height and width by a factor of 2, you can use a 1x1 convolution with a stride of 2.
* The CONV2D layer on the shortcut path does not use any non-linear activation function. Its main role is to just apply a (learned) linear function that reduces the dimension of the input, so that the dimensions match up for the later addition step.
The details of the convolutional block are as follows.
First component of main path:
- The first CONV2D has $F_1$ filters of shape (1,1) and a stride of (s,s). Its padding is "valid" and its name should be `conv_name_base + '2a'`. Use 0 as the `glorot_uniform` seed.
- The first BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2a'`.
- Then apply the ReLU activation function. This has no name and no hyperparameters.
Second component of main path:
- The second CONV2D has $F_2$ filters of shape (f,f) and a stride of (1,1). Its padding is "same" and it's name should be `conv_name_base + '2b'`. Use 0 as the `glorot_uniform` seed.
- The second BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2b'`.
- Then apply the ReLU activation function. This has no name and no hyperparameters.
Third component of main path:
- The third CONV2D has $F_3$ filters of shape (1,1) and a stride of (1,1). Its padding is "valid" and it's name should be `conv_name_base + '2c'`. Use 0 as the `glorot_uniform` seed.
- The third BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '2c'`. Note that there is no ReLU activation function in this component.
Shortcut path:
- The CONV2D has $F_3$ filters of shape (1,1) and a stride of (s,s). Its padding is "valid" and its name should be `conv_name_base + '1'`. Use 0 as the `glorot_uniform` seed.
- The BatchNorm is normalizing the 'channels' axis. Its name should be `bn_name_base + '1'`.
Final step:
- The shortcut and the main path values are added together.
- Then apply the ReLU activation function. This has no name and no hyperparameters.
**Exercise**: Implement the convolutional block. We have implemented the first component of the main path; you should implement the rest. As before, always use 0 as the seed for the random initialization, to ensure consistency with our grader.
- [Conv2D](https://keras.io/layers/convolutional/#conv2d)
- [BatchNormalization](https://keras.io/layers/normalization/#batchnormalization) (axis: Integer, the axis that should be normalized (typically the features axis))
- For the activation, use: `Activation('relu')(X)`
- [Add](https://keras.io/layers/merge/#add)
```
# GRADED FUNCTION: convolutional_block
def convolutional_block(X, f, filters, stage, block, s = 2):
"""
Implementation of the convolutional block as defined in Figure 4
Arguments:
X -- input tensor of shape (m, n_H_prev, n_W_prev, n_C_prev)
f -- integer, specifying the shape of the middle CONV's window for the main path
filters -- python list of integers, defining the number of filters in the CONV layers of the main path
stage -- integer, used to name the layers, depending on their position in the network
block -- string/character, used to name the layers, depending on their position in the network
s -- Integer, specifying the stride to be used
Returns:
X -- output of the convolutional block, tensor of shape (n_H, n_W, n_C)
"""
# defining name basis
conv_name_base = 'res' + str(stage) + block + '_branch'
bn_name_base = 'bn' + str(stage) + block + '_branch'
# Retrieve Filters
F1, F2, F3 = filters
# Save the input value
X_shortcut = X
##### MAIN PATH #####
# First component of main path
X = Conv2D(F1, (1, 1), strides = (s,s), padding='valid' , name = conv_name_base + '2a', kernel_initializer = glorot_uniform(seed=0))(X)
X = BatchNormalization(axis = 3, name = bn_name_base + '2a')(X)
X = Activation('relu')(X)
### START CODE HERE ###
X = Conv2D(filters=F2, kernel_size=(f, f), strides=(1, 1), padding='same', name=conv_name_base + '2b', kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3, name=bn_name_base + '2b')(X)
X = Activation('relu')(X)
# Third component of main path (≈2 lines)
X = Conv2D(filters=F3, kernel_size=(1, 1), strides=(1, 1), padding='valid', name=conv_name_base + '2c', kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3, name=bn_name_base + '2c')(X)
##### SHORTCUT PATH #### (≈2 lines)
X_shortcut = Conv2D(filters=F3, kernel_size=(1, 1), strides=(s, s), padding='valid', name=conv_name_base + '1', kernel_initializer=glorot_uniform(seed=0))(X_shortcut)
X_shortcut = BatchNormalization(axis=3, name=bn_name_base + '1')(X_shortcut)
# Final step: Add shortcut value to main path, and pass it through a RELU activation (≈2 lines)
X = Add()([X, X_shortcut])
X = Activation('relu')(X)
### END CODE HERE ###
return X
tf.reset_default_graph()
with tf.Session() as test:
np.random.seed(1)
A_prev = tf.placeholder("float", [3, 4, 4, 6])
X = np.random.randn(3, 4, 4, 6)
A = convolutional_block(A_prev, f = 2, filters = [2, 4, 6], stage = 1, block = 'a')
test.run(tf.global_variables_initializer())
out = test.run([A], feed_dict={A_prev: X, K.learning_phase(): 0})
print("out = " + str(out[0][1][1][0]))
```
**Expected Output**:
<table>
<tr>
<td>
**out**
</td>
<td>
[ 0.09018463 1.23489773 0.46822017 0.0367176 0. 0.65516603]
</td>
</tr>
</table>
## 3 - Building your first ResNet model (50 layers)
You now have the necessary blocks to build a very deep ResNet. The following figure describes in detail the architecture of this neural network. "ID BLOCK" in the diagram stands for "Identity block," and "ID BLOCK x3" means you should stack 3 identity blocks together.
<img src="images/resnet_kiank.png" style="width:850px;height:150px;">
<caption><center> <u> <font color='purple'> **Figure 5** </u><font color='purple'> : **ResNet-50 model** </center></caption>
The details of this ResNet-50 model are:
- Zero-padding pads the input with a pad of (3,3)
- Stage 1:
- The 2D Convolution has 64 filters of shape (7,7) and uses a stride of (2,2). Its name is "conv1".
- BatchNorm is applied to the 'channels' axis of the input.
- MaxPooling uses a (3,3) window and a (2,2) stride.
- Stage 2:
- The convolutional block uses three sets of filters of size [64,64,256], "f" is 3, "s" is 1 and the block is "a".
- The 2 identity blocks use three sets of filters of size [64,64,256], "f" is 3 and the blocks are "b" and "c".
- Stage 3:
- The convolutional block uses three sets of filters of size [128,128,512], "f" is 3, "s" is 2 and the block is "a".
- The 3 identity blocks use three sets of filters of size [128,128,512], "f" is 3 and the blocks are "b", "c" and "d".
- Stage 4:
- The convolutional block uses three sets of filters of size [256, 256, 1024], "f" is 3, "s" is 2 and the block is "a".
- The 5 identity blocks use three sets of filters of size [256, 256, 1024], "f" is 3 and the blocks are "b", "c", "d", "e" and "f".
- Stage 5:
- The convolutional block uses three sets of filters of size [512, 512, 2048], "f" is 3, "s" is 2 and the block is "a".
- The 2 identity blocks use three sets of filters of size [512, 512, 2048], "f" is 3 and the blocks are "b" and "c".
- The 2D Average Pooling uses a window of shape (2,2) and its name is "avg_pool".
- The 'flatten' layer doesn't have any hyperparameters or name.
- The Fully Connected (Dense) layer reduces its input to the number of classes using a softmax activation. Its name should be `'fc' + str(classes)`.
**Exercise**: Implement the ResNet with 50 layers described in the figure above. We have implemented Stages 1 and 2. Please implement the rest. (The syntax for implementing Stages 3-5 should be quite similar to that of Stage 2.) Make sure you follow the naming convention in the text above.
You'll need to use this function:
- Average pooling [see reference](https://keras.io/layers/pooling/#averagepooling2d)
Here are some other functions we used in the code below:
- Conv2D: [See reference](https://keras.io/layers/convolutional/#conv2d)
- BatchNorm: [See reference](https://keras.io/layers/normalization/#batchnormalization) (axis: Integer, the axis that should be normalized (typically the features axis))
- Zero padding: [See reference](https://keras.io/layers/convolutional/#zeropadding2d)
- Max pooling: [See reference](https://keras.io/layers/pooling/#maxpooling2d)
- Fully connected layer: [See reference](https://keras.io/layers/core/#dense)
- Addition: [See reference](https://keras.io/layers/merge/#add)
```
# GRADED FUNCTION: ResNet50
def ResNet50(input_shape = (64, 64, 3), classes = 6):
"""
Implementation of the popular ResNet50 the following architecture:
CONV2D -> BATCHNORM -> RELU -> MAXPOOL -> CONVBLOCK -> IDBLOCK*2 -> CONVBLOCK -> IDBLOCK*3
-> CONVBLOCK -> IDBLOCK*5 -> CONVBLOCK -> IDBLOCK*2 -> AVGPOOL -> TOPLAYER
Arguments:
input_shape -- shape of the images of the dataset
classes -- integer, number of classes
Returns:
model -- a Model() instance in Keras
"""
# Define the input as a tensor with shape input_shape
X_input = Input(input_shape)
# Zero-Padding
X = ZeroPadding2D((3, 3))(X_input)
# Stage 1
X = Conv2D(64, (7, 7), strides = (2, 2), name = 'conv1', kernel_initializer = glorot_uniform(seed=0))(X)
X = BatchNormalization(axis = 3, name = 'bn_conv1')(X)
X = Activation('relu')(X)
X = MaxPooling2D((3, 3), strides=(2, 2))(X)
# Stage 2
X = convolutional_block(X, f = 3, filters = [64, 64, 256], stage = 2, block='a', s = 1)
X = identity_block(X, 3, [64, 64, 256], stage=2, block='b')
X = identity_block(X, 3, [64, 64, 256], stage=2, block='c')
### START CODE HERE ###
# Stage 3 (≈4 lines)
X = convolutional_block(X, f=3, filters=[128, 128, 512], stage=3, block='a', s=2)
X = identity_block(X, 3, [128, 128, 512], stage=3, block='b')
X = identity_block(X, 3, [128, 128, 512], stage=3, block='c')
X = identity_block(X, 3, [128, 128, 512], stage=3, block='d')
# Stage 4 (≈6 lines)
X = convolutional_block(X, f=3, filters=[256, 256, 1024], stage=4, block='a', s=2)
X = identity_block(X, 3, [256, 256, 1024], stage=4, block='b')
X = identity_block(X, 3, [256, 256, 1024], stage=4, block='c')
X = identity_block(X, 3, [256, 256, 1024], stage=4, block='d')
X = identity_block(X, 3, [256, 256, 1024], stage=4, block='e')
X = identity_block(X, 3, [256, 256, 1024], stage=4, block='f')
# Stage 5 (≈3 lines)
X = X = convolutional_block(X, f=3, filters=[512, 512, 2048], stage=5, block='a', s=2)
X = identity_block(X, 3, [512, 512, 2048], stage=5, block='b')
X = identity_block(X, 3, [512, 512, 2048], stage=5, block='c')
# AVGPOOL (≈1 line). Use "X = AveragePooling2D(...)(X)"
X = AveragePooling2D(pool_size=(2, 2))(X)
### END CODE HERE ###
# output layer
X = Flatten()(X)
X = Dense(classes, activation='softmax', name='fc' + str(classes), kernel_initializer = glorot_uniform(seed=0))(X)
# Create model
model = Model(inputs = X_input, outputs = X, name='ResNet50')
return model
```
Run the following code to build the model's graph. If your implementation is not correct you will know it by checking your accuracy when running `model.fit(...)` below.
```
model = ResNet50(input_shape = (64, 64, 3), classes = 6)
```
As seen in the Keras Tutorial Notebook, prior training a model, you need to configure the learning process by compiling the model.
```
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
```
The model is now ready to be trained. The only thing you need is a dataset.
Let's load the SIGNS Dataset.
<img src="images/signs_data_kiank.png" style="width:450px;height:250px;">
<caption><center> <u> <font color='purple'> **Figure 6** </u><font color='purple'> : **SIGNS dataset** </center></caption>
```
X_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()
# Normalize image vectors
X_train = X_train_orig/255.
X_test = X_test_orig/255.
# Convert training and test labels to one hot matrices
Y_train = convert_to_one_hot(Y_train_orig, 6).T
Y_test = convert_to_one_hot(Y_test_orig, 6).T
print ("number of training examples = " + str(X_train.shape[0]))
print ("number of test examples = " + str(X_test.shape[0]))
print ("X_train shape: " + str(X_train.shape))
print ("Y_train shape: " + str(Y_train.shape))
print ("X_test shape: " + str(X_test.shape))
print ("Y_test shape: " + str(Y_test.shape))
```
Run the following cell to train your model on 2 epochs with a batch size of 32. On a CPU it should take you around 5min per epoch.
```
model.fit(X_train, Y_train, epochs = 2, batch_size = 32)
```
**Expected Output**:
<table>
<tr>
<td>
** Epoch 1/2**
</td>
<td>
loss: between 1 and 5, acc: between 0.2 and 0.5, although your results can be different from ours.
</td>
</tr>
<tr>
<td>
** Epoch 2/2**
</td>
<td>
loss: between 1 and 5, acc: between 0.2 and 0.5, you should see your loss decreasing and the accuracy increasing.
</td>
</tr>
</table>
Let's see how this model (trained on only two epochs) performs on the test set.
```
preds = model.evaluate(X_test, Y_test)
print ("Loss = " + str(preds[0]))
print ("Test Accuracy = " + str(preds[1]))
```
**Expected Output**:
<table>
<tr>
<td>
**Test Accuracy**
</td>
<td>
between 0.16 and 0.25
</td>
</tr>
</table>
For the purpose of this assignment, we've asked you to train the model for just two epochs. You can see that it achieves poor performances. Please go ahead and submit your assignment; to check correctness, the online grader will run your code only for a small number of epochs as well.
After you have finished this official (graded) part of this assignment, you can also optionally train the ResNet for more iterations, if you want. We get a lot better performance when we train for ~20 epochs, but this will take more than an hour when training on a CPU.
Using a GPU, we've trained our own ResNet50 model's weights on the SIGNS dataset. You can load and run our trained model on the test set in the cells below. It may take ≈1min to load the model.
```
model = load_model('ResNet50.h5')
preds = model.evaluate(X_test, Y_test)
print ("Loss = " + str(preds[0]))
print ("Test Accuracy = " + str(preds[1]))
```
ResNet50 is a powerful model for image classification when it is trained for an adequate number of iterations. We hope you can use what you've learnt and apply it to your own classification problem to perform state-of-the-art accuracy.
Congratulations on finishing this assignment! You've now implemented a state-of-the-art image classification system!
## 4 - Test on your own image (Optional/Ungraded)
If you wish, you can also take a picture of your own hand and see the output of the model. To do this:
1. Click on "File" in the upper bar of this notebook, then click "Open" to go on your Coursera Hub.
2. Add your image to this Jupyter Notebook's directory, in the "images" folder
3. Write your image's name in the following code
4. Run the code and check if the algorithm is right!
```
img_path = 'images/my_image.jpg'
img = image.load_img(img_path, target_size=(64, 64))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = x/255.0
print('Input image shape:', x.shape)
my_image = scipy.misc.imread(img_path)
imshow(my_image)
print("class prediction vector [p(0), p(1), p(2), p(3), p(4), p(5)] = ")
print(model.predict(x))
```
You can also print a summary of your model by running the following code.
```
model.summary()
```
Finally, run the code below to visualize your ResNet50. You can also download a .png picture of your model by going to "File -> Open...-> model.png".
```
plot_model(model, to_file='model.png')
SVG(model_to_dot(model).create(prog='dot', format='svg'))
```
## What you should remember
- Very deep "plain" networks don't work in practice because they are hard to train due to vanishing gradients.
- The skip-connections help to address the Vanishing Gradient problem. They also make it easy for a ResNet block to learn an identity function.
- There are two main types of blocks: The identity block and the convolutional block.
- Very deep Residual Networks are built by stacking these blocks together.
### References
This notebook presents the ResNet algorithm due to He et al. (2015). The implementation here also took significant inspiration and follows the structure given in the GitHub repository of Francois Chollet:
- Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun - [Deep Residual Learning for Image Recognition (2015)](https://arxiv.org/abs/1512.03385)
- Francois Chollet's GitHub repository: https://github.com/fchollet/deep-learning-models/blob/master/resnet50.py
| github_jupyter |
# Project 3 Sandbox-Blue-O, NLP using webscraping to create the dataset
## Objective: Determine if posts are in the SpaceX Subreddit or the Blue Origin Subreddit
We'll utilize the RESTful API from pushshift.io to scrape subreddit posts from r/blueorigin and r/spacex and see if we cannot use the Bag-of-words algorithm to predict which posts are from where.
Author: Matt Paterson, [email protected]
This notebook is the SANDBOX and should be used to play around. The formal presentation will be in a different notebook
```
import requests
from bs4 import BeautifulSoup
import pandas as pd
import lebowski as dude
from sklearn.feature_extraction.text import CountVectorizer
import re, regex
# Establish a connection to the API and search for a specific keyword. Maybe we'll add this function to the
# lebowski library? Or maybe make a new and slicker Library called spaceman or something
# CREDIT: code below adapted from Riley Dallas Lesson on webscraping
# keyword = 'propulsion'
# url_boeing = 'https://api.pushshift.io/reddit/search/comment/?q=' + keyword + '&subreddit=boeing'
# res = requests.get(url_boeing)
# res.status_code
# instantiate a Beautiful Soup object for Boeing
#boeing = BeautifulSoup(res.content, 'lxml')
#boeing.find("body")
spacex = dude.create_lexicon('spacex', 5000)
blueorigin = dude.create_lexicon('blueorigin', 5000)
spacex.head()
blueorigin.head()
spacex[['subreddit', 'selftext', 'title']].head() # predict the subreddit column
blueorigin[['subreddit', 'selftext', 'title']].head() # predict the subreddit column
print('Soux City Sarsparilla?') # silly print statement to check progress of long print
spacex_comments = dude.create_lexicon('spacex', 5000, post_type='comment')
spacex_comments.head()
spacex_comments[['subreddit', 'body']].head() # predict the subreddit column
blueorigin_comments = dude.create_lexicon('blueorigin', 5000, post_type='comment')
blueorigin_comments[['subreddit', 'body']].head() # predict the subreddit column
blueorigin_comments.columns
```
There's not a "title" column in the comments dataframe, so how is the comment tied to the original post?
```
# View the first entry in the dataframe and see if you can find that answer
# permalink?
blueorigin_comments.iloc[0]
```
IN EDA below, we find: "We have empty rows in 'body' in many columns. It's likely that all of those are postings, not comments, and we should actually map the postings to the body for those before merging the datafrmes."
```
def strip_and_rep(word):
if len(str(word).strip().replace(" ", "")) < 1:
return 'replace_me'
else:
return word
blueorigin['selftext'] = blueorigin['selftext'].map(strip_and_rep)
spacex['selftext'] = spacex['selftext'].map(strip_and_rep)
spacex.selftext.isna().sum()
blueorigin.selftext.isna().sum()
blueorigin.selftext.head()
spacex.iloc[2300:2320]
blo_coms = blueorigin_comments[['subreddit', 'body', 'permalink']]
blo_posts = blueorigin[['subreddit', 'selftext', 'permalink']].copy()
spx_coms = spacex_comments[['subreddit', 'body', 'permalink']]
spx_posts = spacex[['subreddit', 'selftext', 'permalink']].copy()
#blueorigin['selftext'][len(blueorigin['selftext'])>0]
type(blueorigin.selftext.iloc[1])
blo_posts.rename(columns={'selftext': 'body'}, inplace=True)
spx_posts.rename(columns={'selftext': 'body'}, inplace=True)
# result = pd.concat(frames)
space_wars_2 = pd.concat([blo_coms, blo_posts, spx_coms, spx_posts])
space_wars_2.shape
space_wars_2.head()
dude.show_details(space_wars_2)
```
We have empty rows in 'body' in many columns. It's likely that all of those are postings, not comments, and we should actually map the postings to the body for those before merging the datafrmes.
However, when trying that above, we ended up with more null values. Mapping 'replace_me' in to empty fileds kept the number of null values low. We'll add that token to our stop_words dictionary when creating the BOW from this corpus.
```
space_wars_2.dropna(inplace=True)
space_wars_2.isna().sum()
space_wars.to_csv('./data/betaset.csv', index=False)
```
# Before we split up the training and testing sets, establish our X and y. If you need to reset the dataframe, run the next cell FIRST
keyword = RESET
```
space_wars_2 = pd.read_csv('./data/betaset.csv')
space_wars_2.columns
```
I believe that the 'permalink' will be almost as indicative as the 'subreddit' that we are trying to predict, so the X will only include the words...
```
space_wars_2.head()
```
## Convert target column to binary before moving forward
We want to predict whether this post is Spacex, 1, or is not Spacex, 0
```
space_wars_2['subreddit'].value_counts()
space_wars_2['subreddit'] = space_wars_2['subreddit'].map({'spacex': 1, 'BlueOrigin': 0})
space_wars_2['subreddit'].value_counts()
X = space_wars_2.body
y = space_wars_2.subreddit
```
Calculate our baseline split
```
space_wars_2.subreddit.value_counts(normalize=True)
base_set = space_wars_2.subreddit.value_counts(normalize=True)
baseline = 0.0
if base_set[0] > base_set[1]:
baseline = base_set[0]
else:
baseline = base_set[1]
baseline
```
Before we sift out stopwords, etc, let's just run a logistic regression on the words, as well as a decision tree:
```
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import GridSearchCV, train_test_split, cross_val_score
```
## Before we can fit the models we need to convert the data to numbers...we can use CountVectorizer or TF-IDF for this
```
# from https://stackoverflow.com/questions/5511708/adding-words-to-nltk-stoplist
# add certain words to the stop_words library
import nltk
stopwords = nltk.corpus.stopwords.words('english')
new_words=('replace_me', 'removed', 'deleted', '0','1', '2', '3', '4', '5', '6', '7', '8','9', '00', '000')
for i in new_words:
stopwords.append(i)
print(stopwords)
space_wars_2.isna().sum()
space_wars_2.dropna(inplace=True)
# This section, next number of cells, borrowed from Noelle's lesson on NLP EDA
# Instantiate the "CountVectorizer" object, which is sklearn's
# bag of words tool.
cnt_vec = CountVectorizer(analyzer = "word",
tokenizer = None,
preprocessor = None,
stop_words = stopwords,
max_features = 5000)
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=.20,
random_state=42,
stratify=y)
```
Keyword = CHANGELING
```
y_test
# This section, next number of cells, borrowed from Noelle's lesson on NLP EDA
# fit_transform() does two things: First, it fits the model and
# learns the vocabulary; second, it transforms our training data
# into feature vectors. The input to fit_transform should be a
# list of strings.
train_data_features = cnt_vec.fit_transform(X_train, y_train)
test_data_features = cnt_vec.transform(X_test)
train_data_features.shape
train_data_df = pd.DataFrame(train_data_features)
test_data_features.shape
test_data_df = pd.DataFrame(test_data_features)
test_data_df['subreddit']
lr = LogisticRegression( max_iter = 10_000)
lr.fit(train_data_features, y_train)
train_data_features.shape
dt = DecisionTreeClassifier()
dt.fit(train_data_features, y_train)
print('Logistic Regression without doing anything, really:', lr.score(train_data_features, y_train))
print('Decision Tree without doing anything, really:', dt.score(train_data_features, y_train))
print('*'*80)
print('Logistic Regression Test Score without doing anything, really:', lr.score(test_data_features, y_test))
print('Decision Tree Test Score without doing anything, really:', dt.score(test_data_features, y_test))
print('*'*80)
print(f'The baseline split is {baseline}')
```
So we see that we are above our baseline of 57% accuracy by only guessing a single subreddit without trying to predict. We also see that our initial runs without any GridSearch or HPO tuning gives us a fairly overfit model for either mode.
**Let's see next what happens when we sift through our data with stopwords, etc, to really clean up the dataset and also let's do some comparative EDA including comparing lengths of posts, etc. Finally we can create a sepatate dataframe with engineered features and try running a Logistic Regression model using only descriptors in the dataset such as post lenth, word length, most common words, etc.**
## Deep EDA of our words
```
space_wars.shape
space_wars.describe()
```
## Feature Engineering
Map word count and character length funcitons on to the 'body' column to see a difference in each.
```
def word_count(string):
'''
returns the number of words or tokens in a string literal, splitting on spaces,
regardless of word lenth. This function will include space-separated
punctuation as a word, such as " : " where the colon would be counted
string, a string
'''
str_list = string.split()
return len(str_list)
def count_chars(string):
'''
returns the total number of characters including spaces in a string literal
string, a string
'''
count=0
for s in string:
count+=1
return count
import lebowski as dude
space_wars['word_count'] = space_wars['body'].map(word_count)
space_wars['word_count'].value_counts().head()
# code from https://stackoverflow.com/questions/39132742/groupby-value-counts-on-the-dataframe-pandas
#df.groupby(['id', 'group', 'term']).size().unstack(fill_value=0)
space_wars.groupby(['subreddit', 'word_count']).size().head()
space_wars['post_length'] = space_wars['body'].map(count_chars)
space_wars['post_length'].value_counts().head()
space_wars.columns
import seaborn as sns
import matplotlib.pyplot as plt
sns.distplot(space_wars['word_count'])
# Borrowing from Noelle's nlp II lesson, import the following,
# and think about what you want to use in the presentation
# imports
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import confusion_matrix, plot_confusion_matrix
# Import CountVectorizer and TFIDFVectorizer from feature_extraction.text.
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
```
## Text Feature Extraction
## Follow along in the NLP EDA II video and do some analysis
```
X_train_df = pd.DataFrame(train_data_features.toarray(),
columns=cntv.get_feature_names())
X_train_df
X_train_df['subreddit']
# get count of top-occurring words
# empty dictionary
top_words = {}
# loop through columns
for i in X_train_df.columns:
# save sum of each column in dictionary
top_words[i] = X_train_df[i].sum()
# top_words to dataframe sorted by highest occurance
most_freq = pd.DataFrame(sorted(top_words.items(), key = lambda x: x[1], reverse = True))
most_freq.head()
# Make a different CountVectorizer
count_v = CountVectorizer(analyzer='word',
stop_words = stopwords,
max_features = 1_000,
min_df = 50,
max_df = .80,
ngram_range=(2,3),
)
# Redefine the training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y,
test_size = .1,
stratify = y,
random_state=42)
baseline
```
## Implement Naive Bayes because it's in the project instructions
Multinomial Naive Bayes often outperforms other models despite text data being non-independent data
```
pipe = Pipeline([
('count_v', CountVectorizer()),
('nb', MultinomialNB())
])
pipe_params = {
'count_v__max_features': [2000, 5000, 9000],
'count_v__stop_words': [stopwords],
'count_v__min_df': [2, 3, 10],
'count_v__max_df': [.9, .8, .7],
'count_v__ngram_range': [(1, 1), (1, 2)]
}
gs = GridSearchCV(pipe,
pipe_params,
cv = 5,
n_jobs=6
)
%%time
gs.fit(X_train, y_train)
gs.best_params_
print(gs.best_score_)
gs.score(X_train, y_train)
gs.score(X_test, y_test)
```
So far, the Multinomial Naive Bayes Algorithm is the top function at 79.28% Accuracy. The confusion matrix below is very simiar to that of other models
```
# Get predictions
preds = gs.predict(X_test)
# Save confusion matrix values
tn, fp, fn, tp = confusion_matrix(y_test, preds).ravel()
# View confusion matrix
plot_confusion_matrix(gs, X_test, y_test, cmap='Blues', values_format='d');
# Calculate the specificity
spec = tn / (tn + fp)
print('Specificity:', spec)
```
None of the 1620 different models we tried in this pipeline performed noticibly better than the thrown-together Logistic Regression Classifier that we started out with. Let's try TF-IDF, then Random Cut Forest, and finally Vector Machines. Our last run brought the best accuracy score to 79.3%
# TF-IDF
```
# Redefine the training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y,
test_size = .1,
stratify = y,
random_state=42)
tvec = TfidfVectorizer(stop_words=stopwords)
df = pd.DataFrame(tvec.fit_transform(X_train).toarray(),
columns=tvec.get_feature_names())
df.head()
# get count of top-occurring words
top_words_tf = {}
for i in df.columns:
top_words_tf[i] = df[i].sum()
# top_words to dataframe sorted by highest occurance
most_freq_tf = pd.DataFrame(sorted(top_words_tf.items(), key = lambda x: x[1], reverse = True))
plt.figure(figsize = (10, 5))
# visualize top 10 words
plt.bar(most_freq_tf[0][:10], most_freq_tf[1][:10]);
pipe_tvec = Pipeline([
('tvec', TfidfVectorizer()),
('nb', MultinomialNB())
])
pipe_params_tvec = {
'tvec__max_features': [2000, 9000],
'tvec__stop_words' : [None, stopwords],
'tvec__ngram_range': [(1, 1), (1, 2)]
}
gs_tvec = GridSearchCV(pipe_tvec, pipe_params_tvec, cv = 5)
%%time
gs_tvec.fit(X_train, y_train)
gs_tvec.best_params_
gs_tvec.score(X_train, y_train)
gs_tvec.score(X_test, y_test)
# Get predictions
preds = gs_tvec.predict(X_test)
# Save confusion matrix values
tn, fp, fn, tp = confusion_matrix(y_test, preds).ravel()
# View confusion matrix
plot_confusion_matrix(gs_tvec, X_test, y_test, cmap='Blues', values_format='d');
# Calculate the specificity
spec = tn / (tn + fp)
print('Specificity:', spec)
```
## Random Cut Forest, Bagging, and Support Vector Machines
```
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier
```
Before we run the decision tree model or RandomForestClassifier(), we need to convert all of the data to numeric data
```
rf = RandomForestClassifier()
et = ExtraTreesClassifier()
cross_val_score(rf, train_data_features, X_train_df['subreddit']).mean()
cross_val_score(et, train_data_features, X_train_df['subreddit']).mean()
#cross_val_score(rf, test_data_features, y_test).mean()
```
## Make sure that we are using X and y data that are completely numeric and free of nulls
```
space_wars.head(1)
space_wars.shape
pipe_rf = Pipeline([
('count_v', CountVectorizer()),
('rf', RandomForestClassifier()),
])
pipe_ef = Pipeline([
('count_v', CountVectorizer()),
('ef', ExtraTreesClassifier()),
])
pipe_params =
'count_v__max_features': [2000, 5000, 9000],
'count_v__stop_words': [stopwords],
'count_v__min_df': [2, 3, 10],
'count_v__max_df': [.9, .8, .7],
'count_v__ngram_range': [(1, 1), (1, 2)]
}
%%time
gs_rf = GridSearchCV(pipe_rf,
pipe_params,
cv = 5,
n_jobs=6)
gs_rf.fit(X_train, y_train)
print(gs_rf.best_score_)
gs_rf.best_params_
gs_rf.score(X_train, y_train)
gs_rf.score(X_test, y_test)
# %%time
# gs_ef = GridSearchCV(pipe_ef,
# pipe_params,
# cv = 5,
# n_jobs=6)
# gs_ef.fit(X_train, y_train)
# print(gs_ef.best_score_)
# gs_ef.best_params_
#gs_ef.score(X_train, y_train)
#gs_ef.score(X_test, y_test)
```
## Now run through Gradient Boosting and SVM
```
from sklearn.ensemble import GradientBoostingClassifier, AdaBoostClassifier, VotingClassifier
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsClassifier
```
Using samples from Riley's Lessons:
```
AdaBoostClassifier()
GradientBoostingClassifier()
```
Use the CountVectorizer to convert the data to numeric data prior to running it through the below VotingClassifier
```
'count_v__max_df': 0.9,
'count_v__max_features': 9000,
'count_v__min_df': 2,
'count_v__ngram_range': (1, 1),
knn_pipe = Pipeline([
('ss', StandardScaler()),
('knn', KNeighborsClassifier())
])
%%time
vote = VotingClassifier([
('ada', AdaBoostClassifier(base_estimator=DecisionTreeClassifier())),
('grad_boost', GradientBoostingClassifier()),
('tree', DecisionTreeClassifier()),
('knn_pipe', knn_pipe)
])
params = {}
# 'ada__n_estimators': [50, 51],
# 'grad_boost__n_estimators': [10, 11],
# 'knn_pipe__knn__n_neighbors': [5],
# 'ada__base_estimator__max_depth': [1, 2],
# 'weights': [[.25] * 4, [.3, .3, .3, .1]]
# }
gs = GridSearchCV(vote, param_grid=params, cv=3)
gs.fit(X_train, y_train)
print(gs.best_score_)
gs.best_params_
```
| github_jupyter |
Uses Fine-Tuned BERT network to classify biomechanics papers from PubMed
```
# Check date
!rm /etc/localtime
!ln -s /usr/share/zoneinfo/America/Los_Angeles /etc/localtime
!date
# might need to restart runtime if timezone didn't change
## Install & load libraries
!pip install tensorflow==2.7.0
try:
from official.nlp import optimization
except:
!pip install -q -U tf-models-official==2.4.0
from official.nlp import optimization
try:
from Bio import Entrez
except:
!pip install -q -U biopython
from Bio import Entrez
try:
import tensorflow_text as text
except:
!pip install -q -U tensorflow_text==2.7.3
import tensorflow_text as text
import pandas as pd
import numpy as np
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
import tensorflow as tf # probably have to lock version
import string
import datetime
from bs4 import BeautifulSoup
from sklearn.preprocessing import LabelEncoder
from tensorflow.keras.models import load_model
import tensorflow_hub as hub
from google.colab import drive
import datetime as dt
#Define date range
today = dt.date.today()
yesterday = today - dt.timedelta(days=1)
week_ago = yesterday - dt.timedelta(days=7) # ensure overlap in pubmed search
days_ago_6 = yesterday - dt.timedelta(days=6) # for text output
# Mount Google Drive for model and csv up/download
drive.mount('/content/gdrive')
print(today)
# Define Search Criteria ----
def search(query):
Entrez.email = '[email protected]'
handle = Entrez.esearch(db='pubmed',
sort='most recent',
retmax='5000',
retmode='xml',
datetype='pdat', # pdat is published date, edat is entrez date.
# reldate=7, # only within n days from now
mindate= min_date,
maxdate= max_date, # for searching date range
term=query)
results = Entrez.read(handle)
return results
# Perform Search and Pull Paper Titles ----
def fetch_details(ids):
Entrez.email = '[email protected]'
handle = Entrez.efetch(db='pubmed',
retmode='xml',
id=ids)
results = Entrez.read(handle)
return results
# Make the stop words for string cleaning ----
def html_strip(text):
text = BeautifulSoup(text, 'lxml').text
text = text.replace('[','').replace(']','')
return text
def clean_str(text, stops):
text = BeautifulSoup(text, 'lxml').text
text = text.split()
return ' '.join([word for word in text if word not in stops])
stop = list(stopwords.words('english'))
stop_c = [string.capwords(word) for word in stop]
for word in stop_c:
stop.append(word)
new_stop = ['The', 'An', 'A', 'Do', 'Is', 'In', 'StringElement',
'NlmCategory', 'Label', 'attributes', 'INTRODUCTION',
'METHODS', 'BACKGROUND', 'RESULTS', 'CONCLUSIONS']
for s in new_stop:
stop.append(s)
# Search terms (can test string with Pubmed Advanced Search) ----
# search_results = search('(Biomech*[Title/Abstract] OR locomot*[Title/Abstract])')
min_date = week_ago.strftime('%m/%d/%Y')
max_date = yesterday.strftime('%m/%d/%Y')
search_results = search('(biomech*[Title/Abstract] OR locomot*[Title/Abstract] NOT opiod*[Title/Abstract] NOT pharm*[Journal] NOT mouse[Title/Abstract] NOT drosophil*[Title/Abstract] NOT mice[Title/Abstract] NOT rats*[Title/Abstract] NOT elegans[Title/Abstract])')
id_list = search_results['IdList']
papers = fetch_details(id_list)
print(len(papers['PubmedArticle']), 'Papers found')
titles, full_titles, keywords, authors, links, journals, abstracts = ([] for i in range(7))
for paper in papers['PubmedArticle']:
# clean and store titles, abstracts, and links
t = clean_str(paper['MedlineCitation']['Article']['ArticleTitle'],
stop).replace('[','').replace(']','').capitalize() # rm brackets that survived beautifulsoup, sentence case
titles.append(t)
full_titles.append(paper['MedlineCitation']['Article']['ArticleTitle'])
pmid = paper['MedlineCitation']['PMID']
links.append('[URL="https://www.ncbi.nlm.nih.gov/pubmed/{0}"]{1}[/URL]'.format(pmid, html_strip(paper['MedlineCitation']['Article']['ArticleTitle'])))
try:
abstracts.append(clean_str(paper['MedlineCitation']['Article']['Abstract']['AbstractText'][0],
stop).replace('[','').replace(']','').capitalize()) # rm brackets that survived beautifulsoup, sentence case
except:
abstracts.append('')
# clean and store authors
auths = []
try:
for auth in paper['MedlineCitation']['Article']['AuthorList']:
try: # see if there is a last name and initials
auth_name = [auth['LastName'], auth['Initials'] + ',']
auth_name = ' '.join(auth_name)
auths.append(auth_name)
except:
if 'LastName' in auth.keys(): # maybe they don't have initials
auths.append(auth['LastName'] + ',')
else: # no last name
auths.append('')
print(paper['MedlineCitation']['Article']['ArticleTitle'],
'has an issue with an author name:')
except:
auths.append('AUTHOR NAMES ERROR')
print(paper['MedlineCitation']['Article']['ArticleTitle'], 'has no author list?')
# compile authors
authors.append(' '.join(auths).replace('[','').replace(']','')) # rm brackets in names
# journal names
journals.append(paper['MedlineCitation']['Article']['Journal']['Title'].replace('[','').replace(']','')) # rm brackets
# store keywords
if paper['MedlineCitation']['KeywordList'] != []:
kwds = []
for kw in paper['MedlineCitation']['KeywordList'][0]:
kwds.append(kw[:])
keywords.append(', '.join(kwds).lower())
else:
keywords.append('')
# Put Titles, Abstracts, Authors, Journal, and Keywords into dataframe
papers_df = pd.DataFrame({'title': titles,
'keywords': keywords,
'abstract': abstracts,
'authors': authors,
'journal': journals,
'links': links,
'raw_title': full_titles,
'mindate': min_date,
'maxdate': max_date})
# remove papers with no title or no authors
for index, row in papers_df.iterrows():
if row['title'] == '' or row['authors'] == 'AUTHOR NAMES ERROR':
papers_df.drop(index, inplace=True)
papers_df.reset_index(drop=True, inplace=True)
# join titles and abstract
papers_df['BERT_input'] = pd.DataFrame(papers_df['title'] + ' ' + papers_df['abstract'])
# Load Fine-Tuned BERT Network ----
model = tf.saved_model.load('/content/gdrive/My Drive/BiomchBERT/Data/BiomchBERT/')
print('Loaded model from disk')
# Load Label Encoder ----
le = LabelEncoder()
le.classes_ = np.load('/content/gdrive/My Drive/BiomchBERT/Data/BERT_label_encoder.npy')
print('Loaded Label Encoder')
# Predict Paper Topic ----
predicted_topic = model(papers_df['BERT_input'], training=False) # will run out of GPU memory (14GB) if predicting more than ~2000 title+abstracts at once
# Determine Publications that BiomchBERT is unsure about ----
topics, pred_val_str = ([] for i in range(2))
for pred_prob in predicted_topic:
pred_val = np.max(pred_prob)
if pred_val > 1.5 * np.sort(pred_prob)[-2]: # Is top confidence score more than 1.5x the second best confidence score?
topics.append(le.inverse_transform([np.argmax(pred_prob)])[0])
top1 = le.inverse_transform([np.argmax(pred_prob)])[0]
top2 = le.inverse_transform([list(pred_prob).index([np.sort(pred_prob)[-2]])])[0]
# pred_val_str.append(pred_val * 100) # just report top category
pred_val_str.append(str(np.round(pred_val * 100, 1)) + '% ' + str(top1) + '; ' + str(
np.round(np.sort(pred_prob)[-2] * 100, 1)) + '% ' + str(top2)) # report top 2 categories
else:
topics.append('UNKNOWN')
top1 = le.inverse_transform([np.argmax(pred_prob)])[0]
top2 = le.inverse_transform([list(pred_prob).index([np.sort(pred_prob)[-2]])])[0]
pred_val_str.append(str(np.round(pred_val * 100, 1)) + '% ' + str(top1) + '; ' + str(
np.round(np.sort(pred_prob)[-2] * 100, 1)) + '% ' + str(top2))
papers_df['topic'] = topics
papers_df['pred_val'] = pred_val_str
print('BiomchBERT is unsure about {0} papers\n'.format(len(papers_df[papers_df['topic'] == 'UNKNOWN'])))
# Prompt User to decide for BiomchBERT ----
unknown_papers = papers_df[papers_df['topic'] == 'UNKNOWN']
for indx, paper in unknown_papers.iterrows():
print(paper['raw_title'])
print(paper['journal'])
print(paper['pred_val'])
print()
splt_str = paper['pred_val'].split(';')
options = [str for pred_cls in splt_str for str in le.classes_ if (str in pred_cls)]
choice = input('(1)st topic, (2)nd topic, (o)ther topic, or (r)emove paper? ')
print()
if choice == '1':
papers_df.iloc[indx]['topic'] = str(options[0])
elif choice == '2':
papers_df.iloc[indx]['topic'] = str(options[1])
elif choice == 'o':
# print all categories so you can select
for i in zip(range(len(le.classes_)),le.classes_):
print(i)
new_cat = input('Enter number of new class or type "r" to remove paper: ')
print()
if new_cat == 'r':
papers_df.iloc[indx]['topic'] = '_REMOVE_' # not deleted, but withheld from text file output
else:
papers_df.iloc[indx]['topic'] = le.classes_[int(new_cat)]
elif choice == 'r':
papers_df.iloc[indx]['topic'] = '_REMOVE_' # not deleted, but withheld from text file output
print('Removing {0} papers\n'.format(len(papers_df[papers_df['topic'] == '_REMOVE_'])))
# Double check that none of these papers were included in past literature updates ----
# load prior papers
# papers_df.to_csv('/content/gdrive/My Drive/BiomchBERT/Updates/prior_papers.csv', index=False) # run ONLY if there are no prior papers
prior_papers = pd.read_csv('/content/gdrive/My Drive/BiomchBERT/Updates/prior_papers.csv')
prior_papers.dropna(subset=['title'], inplace=True)
prior_papers.reset_index(drop=True, inplace=True)
# NEED TO DO: find matching papers between current week and prior papers using Pubmed ID since titles can change from ahead of print to final version.
# match = papers_df['links'].split(']')[0].isin(prior_papers['links'].split(']')[0])
match = papers_df['title'].isin(prior_papers['title']) # boolean
print('Removing {0} papers found in prior literature updates\n'.format(sum(match)))
# filter and check if everything accidentally was removed
filtered_papers_df = papers_df.drop(papers_df[match].index)
if filtered_papers_df.shape[0] < 1:
raise ValueError('might have removed all the papers for some reason. ')
else:
papers_df = filtered_papers_df
papers_df.reset_index(drop=True, inplace=True)
updated_prior_papers = pd.concat([prior_papers, papers_df], axis=0)
updated_prior_papers.reset_index(drop=True, inplace=True)
updated_prior_papers.to_csv('/content/gdrive/My Drive/BiomchBERT/Updates/prior_papers.csv', index=False)
# Create Text File for Biomch-L ----
# Compile papers grouped by topic
txtname = '/content/gdrive/My Drive/BiomchBERT/Updates/' + today.strftime("%Y-%m-%d") + '-litupdate.txt'
txt = open(txtname, 'w', encoding='utf-8')
txt.write('[SIZE=16px][B]LITERATURE UPDATE[/B][/SIZE]\n')
txt.write(days_ago_6.strftime("%b %d, %Y") + ' - '+ yesterday.strftime("%b %d, %Y")+'\n') # a week ago from yesterday.
txt.write(
"""
Literature search terms: biomech* & locomot*
Publications are classified by [URL="https://www.ryan-alcantara.com/projects/p88_BiomchBERT/"]BiomchBERT[/URL], a neural network trained on past Biomch-L Literature Updates. BiomchBERT is managed by [URL="https://jouterleys.github.io"]Jereme Outerleys[/URL], a Doctoral Student at Queen's University. Each publication has a score (out of 100%) reflecting how confident BiomchBERT is that the publication belongs in a particular category (top 2 shown). If something doesn't look right, email jereme.outerleys[at]queensu.ca.
Twitter: [URL="https://www.twitter.com/jouterleys"]@jouterleys[/URL].
"""
)
# Write papers to text file grouped by topic ----
topic_list = np.unique(papers_df.sort_values('topic')['topic'])
for topic in topic_list:
papers_subset = pd.DataFrame(papers_df[papers_df.topic == topic].reset_index(drop=True))
txt.write('\n')
# TOPIC NAME (with some cleaning)
if topic == '_REMOVE_':
continue
elif topic == 'UNKNOWN':
txt.write('[SIZE=16px][B]*Papers BiomchBERT is unsure how to classify*[/B][/SIZE]\n')
elif topic == 'CARDIOVASCULAR/CARDIOPULMONARY':
topic = 'CARDIOVASCULAR/PULMONARY'
txt.write('[SIZE=16px][B]*%s*[/B][/SIZE]\n' % topic)
elif topic == 'CELLULAR/SUBCELLULAR':
topic = 'CELLULAR'
txt.write('[SIZE=16px][B]*%s*[/B][/SIZE]\n' % topic)
elif topic == 'ORTHOPAEDICS/SURGERY':
topic = 'ORTHOPAEDICS (SURGERY)'
txt.write('[SIZE=16px][B]*%s*[/B][/SIZE]\n' % topic)
elif topic == 'ORTHOPAEDICS/SPINE':
topic = 'ORTHOPAEDICS (SPINE)'
txt.write('[SIZE=16px][B]*%s*[/B][/SIZE]\n' % topic)
else:
txt.write('[SIZE=16px][B]*%s*[/B][/SIZE]\n' % topic)
# HYPERLINKED PAPERS, AUTHORS, JOURNAL NAME
for i, paper in enumerate(papers_subset['links']):
txt.write('[B]%s[/B] ' % paper)
txt.write('%s ' % papers_subset['authors'][i])
txt.write('[I]%s[/I]. ' % papers_subset['journal'][i])
# CONFIDENCE SCORE (BERT softmax categorical crossentropy)
try:
txt.write('(%.1f%%) \n\n' % papers_subset['pred_val'][i])
except:
txt.write('(%s)\n\n' % papers_subset['pred_val'][i])
txt.write('[SIZE=16px][B]*PICK OF THE WEEK*[/B][/SIZE]\n')
txt.close()
print('Literature Update Exported for Biomch-L')
print('Location:', txtname)
```
| github_jupyter |
# Wilcoxon and Chi Squared
```
import numpy as np
import pandas as pd
df = pd.read_csv("prepared_neuror2_data.csv")
def stats_for_neuror2_range(lo, hi):
admissions = df[df.NR2_Score.between(lo, hi)]
total_patients = admissions.shape[0]
readmits = admissions[admissions.UnplannedReadmission]
total_readmits = readmits.shape[0]
return (total_readmits, total_patients, "%.1f" % (total_readmits/total_patients*100,))
mayo_davis = []
for (expected, (lo, hi)) in [(1.4, (0, 0)),
(4, (1, 4)),
(5.6, (5, 8)),
(14.2, (9, 13)),
(33.0, (14, 19)),
(0.0, (20, 22))]:
(total_readmits, total_patients, readmit_percent) = stats_for_neuror2_range(lo, hi)
mayo_davis.append([lo, hi, expected, readmit_percent, total_readmits, total_patients])
title="Davis and Mayo Populations by NeuroR2 Score"
print(title)
print("-" * len(title))
print(pd.DataFrame(mayo_davis, columns=["Low", "High", "Mayo %", "Davis %",
"Readmits", "Total"]).to_string(index=False))
# Continuous variables were compared using wilcoxon
from scipy.stats import ranksums as wilcoxon
def create_samples(col_name):
unplanned = df[df.UnplannedReadmission][col_name].values
planned = df[~df.UnplannedReadmission][col_name].values
return (unplanned, planned)
continous_vars = ["AdmissionAgeYears", "LengthOfStay", "NR2_Score"]#, "MsDrgWeight"]
for var in continous_vars:
(unplanned, planned) = create_samples(var)
(stat, p) = wilcoxon(unplanned, planned)
print ("%30s" % (var,), "p-value %f" % (p,))
unplanned, planned = create_samples("LengthOfStay")
print(pd.DataFrame(unplanned, columns=["Unplanned Readmission"]).describe())
print(pd.DataFrame(planned, columns=[" Index Only Admission"]).describe())
# Categorical variables were compared using chi squared
from scipy.stats import chi2, chi2_contingency
from IPython.core.display import display, HTML
# Collect all the categorical features
cols = sorted([col for col in df.columns if "_" in col])
for var in continous_vars:
try:
cols.remove(var)
except:
pass
index_only = df[~df.UnplannedReadmission].shape[0]
unplanned_readmit = df[df.UnplannedReadmission].shape[0]
html = "<table><tr>"
for th in ["Characteristic", "Index admission only</br>(n=%d)" % (index_only,),
"Unplanned readmission</br>(n = %d)" % (unplanned_readmit,),"<i>p</i> Value"]:
html += "<th>%s</th>" % (th,)
html += "</tr>"
start_row = "<tr><td>%s</td>"
end_row = "<td>%d (%.1f)</td><td>%d (%.1f)</td><td></td></tr>"
pval_str = lambda p: "<0.001" if p<0.001 else "%.3f" % p
col_str = lambda col, p: "<b><i>%s</i></b>" % (col,) if p < 0.05 else col
for col in sorted(cols):
table = pd.crosstab(df[col], df.UnplannedReadmission)
stat, p, dof, expected = chi2_contingency(table)
html += "<tr><td>%s</td><td></td><td></td><td>%s</td></tr>" % (col_str(col,p), pval_str(p))
html += start_row % ("No",)
html += end_row % (table.values[0][0], expected[0][0],
table.values[0][1], expected[0][1])
try:
html += start_row % ("Yes",)
html += end_row % (table.values[1][0], expected[1,0],
table.values[1][1], expected[1][1])
except IndexError:
html += "<td>-</td><td>-</td><td></td></tr>"
html += "</table>"
display(HTML(html))
```
| github_jupyter |
Note:
This notebook was executed on google colab pro.
```
!pip3 install pytorch-lightning --quiet
from google.colab import drive
drive.mount('/content/drive')
import os
os.chdir('/content/drive/MyDrive/Colab Notebooks/atmacup11/experiments')
```
# Settings
```
EXP_NO = 27
SEED = 1
N_SPLITS = 5
TARGET = 'target'
GROUP = 'art_series_id'
REGRESSION = False
assert((TARGET, REGRESSION) in (('target', True), ('target', False), ('sorting_date', True)))
MODEL_NAME = 'resnet'
BATCH_SIZE = 512
NUM_EPOCHS = 500
```
# Library
```
from collections import defaultdict
from functools import partial
import gc
import glob
import json
from logging import getLogger, StreamHandler, FileHandler, DEBUG, Formatter
import pickle
import os
import sys
import time
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from sklearn.metrics import confusion_matrix, mean_squared_error, cohen_kappa_score
# from sklearnex import patch_sklearn
from pytorch_lightning import seed_everything
import torch
import torch.nn as nn
import torch.optim
from torch.utils.data import DataLoader
from torchvision import transforms
SCRIPTS_DIR = os.path.join('..', 'scripts')
assert(os.path.isdir(SCRIPTS_DIR))
if SCRIPTS_DIR not in sys.path: sys.path.append(SCRIPTS_DIR)
from cross_validation import load_cv_object_ids
from dataset import load_csvfiles, load_photofile,load_photofiles, AtmaImageDatasetV02
from folder import experiment_dir_of
from models import initialize_model
from utils import train_model, predict_by_model
pd.options.display.float_format = '{:.5f}'.format
DEVICE = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
DEVICE
```
# Prepare directory
```
output_dir = experiment_dir_of(EXP_NO)
output_dir
```
# Prepare logger
```
logger = getLogger(__name__)
'''Refference
https://docs.python.org/ja/3/howto/logging-cookbook.html
'''
logger.setLevel(DEBUG)
# create file handler which logs even debug messages
fh = FileHandler(os.path.join(output_dir, 'log.log'))
fh.setLevel(DEBUG)
# create console handler with a higher log level
ch = StreamHandler()
ch.setLevel(DEBUG)
# create formatter and add it to the handlers
formatter = Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
fh.setFormatter(formatter)
ch.setFormatter(formatter)
# add the handlers to the logger
logger.addHandler(fh)
logger.addHandler(ch)
len(logger.handlers)
logger.info('Experiment no: {}'.format(EXP_NO))
logger.info('CV: StratifiedGroupKFold')
logger.info('SEED: {}'.format(SEED))
logger.info('REGRESSION: {}'.format(REGRESSION))
```
# Load csv files
```
SINCE = time.time()
logger.debug('Start loading csv files ({:.3f} seconds passed)'.format(time.time() - SINCE))
train, test, materials, techniques, sample_submission = load_csvfiles()
logger.debug('Complete loading csv files ({:.3f} seconds passed)'.format(time.time() - SINCE))
train
test
```
# Cross validation
```
seed_everything(SEED)
train.set_index('object_id', inplace=True)
fold_object_ids = load_cv_object_ids()
for i, (train_object_ids, valid_object_ids) in enumerate(zip(fold_object_ids[0], fold_object_ids[1])):
assert(set(train_object_ids) & set(valid_object_ids) == set())
num_fold = i + 1
logger.debug('Start fold {} ({:.3f} seconds passed)'.format(num_fold, time.time() - SINCE))
# Separate dataset into training/validation fold
y_train = train.loc[train_object_ids, TARGET].values
y_valid = train.loc[valid_object_ids, TARGET].values
torch.cuda.empty_cache()
# Training
logger.debug('Start training model ({:.3f} seconds passed)'.format(time.time() - SINCE))
## Prepare model
num_classes = len(set(list(y_train)))
model, input_size = initialize_model(MODEL_NAME, num_classes)
model.to(DEVICE)
## Prepare transformers
train_transformer = transforms.Compose([
transforms.RandomResizedCrop(input_size),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
val_transformer = transforms.Compose([
transforms.Resize(input_size),
transforms.CenterCrop(input_size),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
# Prepare dataset
if not REGRESSION:
# label should be one-hot style
y_train = np.identity(num_classes)[y_train].astype('int')
y_valid = np.identity(num_classes)[y_valid].astype('int')
train_dataset = AtmaImageDatasetV02(train_object_ids, train_transformer, y_train)
val_dataset = AtmaImageDatasetV02(valid_object_ids, val_transformer, y_valid)
# Prepare dataloader
dataloaders = {
'train': DataLoader(dataset=train_dataset, batch_size=BATCH_SIZE, shuffle=True, num_workers=os.cpu_count()),
'val': DataLoader(dataset=val_dataset, batch_size=BATCH_SIZE, shuffle=False, num_workers=os.cpu_count()),
}
## train estimator
estimator, train_losses, valid_losses = train_model(
model, dataloaders, criterion=nn.BCEWithLogitsLoss(), num_epochs=NUM_EPOCHS, device=DEVICE,
optimizer=torch.optim.Adam(model.parameters()), log_func=logger.debug,
is_inception=MODEL_NAME == 'inception')
logger.debug('Complete training ({:.3f} seconds passed)'.format(time.time() - SINCE))
## Visualize training loss
plt.plot(train_losses, label='train')
plt.plot(valid_losses, label='valid')
plt.legend(loc='upper left', bbox_to_anchor=[1., 1.])
plt.title(f'Fold{num_fold}')
plt.show()
# Save model and prediction
## Prediction
predictions = {}
for fold_, object_ids_ in zip(['train', 'val', 'test'],
[train_object_ids, valid_object_ids, test['object_id']]):
# Prepare transformer
transformer_ = transforms.Compose([
transforms.Resize(input_size),
transforms.CenterCrop(input_size),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
# Prepare dataset
dataset_ = AtmaImageDatasetV02(object_ids_, transformer_)
# Prepare dataloader
dataloader_ = DataLoader(dataset=dataset_, batch_size=BATCH_SIZE, shuffle=False,
num_workers=os.cpu_count())
# Prediction
predictions[fold_] = predict_by_model(estimator, dataloader_, DEVICE)
logger.debug('Complete prediction for {} fold ({:.3f} seconds passed)' \
.format(fold_, time.time() - SINCE))
if REGRESSION:
pred_train = pd.DataFrame(data=predictions['train'], columns=['pred'])
pred_valid = pd.DataFrame(data=predictions['val'], columns=['pred'])
pred_test = pd.DataFrame(data=predictions['test'], columns=['pred'])
else:
columns = list(range(num_classes))
pred_train = pd.DataFrame(data=predictions['train'], columns=columns)
pred_valid = pd.DataFrame(data=predictions['val'], columns=columns)
pred_test = pd.DataFrame(data=predictions['test'], columns=columns)
# else: # Do not come here!
# raise NotImplemented
# try:
# pred_train = pd.DataFrame(data=estimator.predict_proba(X_train),
# columns=estimator.classes_)
# pred_valid = pd.DataFrame(data=estimator.predict_proba(X_valid),
# columns=estimator.classes_)
# pred_test = pd.DataFrame(data=estimator.predict_proba(X_test),
# columns=estimator.classes_)
# except AttributeError:
# pred_train = pd.DataFrame(data=estimator.decision_function(X_train),
# columns=estimator.classes_)
# pred_valid = pd.DataFrame(data=estimator.decision_function(X_valid),
# columns=estimator.classes_)
# pred_test = pd.DataFrame(data=estimator.decision_function(X_test),
# columns=estimator.classes_)
## Training set
pred_train['object_id'] = train_object_ids
filepath_fold_train = os.path.join(output_dir, f'cv_fold{num_fold}_training.csv')
pred_train.to_csv(filepath_fold_train, index=False)
logger.debug('Save training fold to {} ({:.3f} seconds passed)' \
.format(filepath_fold_train, time.time() - SINCE))
## Validation set
pred_valid['object_id'] = valid_object_ids
filepath_fold_valid = os.path.join(output_dir, f'cv_fold{num_fold}_validation.csv')
pred_valid.to_csv(filepath_fold_valid, index=False)
logger.debug('Save validation fold to {} ({:.3f} seconds passed)' \
.format(filepath_fold_valid, time.time() - SINCE))
## Test set
pred_test['object_id'] = test['object_id'].values
filepath_fold_test = os.path.join(output_dir, f'cv_fold{num_fold}_test.csv')
pred_test.to_csv(filepath_fold_test, index=False)
logger.debug('Save test result {} ({:.3f} seconds passed)' \
.format(filepath_fold_test, time.time() - SINCE))
## Model
filepath_fold_model = os.path.join(output_dir, f'cv_fold{num_fold}_model.torch')
torch.save(estimator.state_dict(), filepath_fold_model)
# with open(filepath_fold_model, 'wb') as f:
# pickle.dump(estimator, f)
logger.debug('Save model {} ({:.3f} seconds passed)'.format(filepath_fold_model, time.time() - SINCE))
# Save memory
del (estimator, y_train, y_valid, pred_train, pred_valid, pred_test)
gc.collect()
logger.debug('Complete fold {} ({:.3f} seconds passed)'.format(num_fold, time.time() - SINCE))
```
# Evaluation
```
rmse = partial(mean_squared_error, squared=False)
# qwk = partial(cohen_kappa_score, labels=np.sort(train['target'].unique()), weights='quadratic')
@np.vectorize
def predict(proba_0: float, proba_1: float, proba_2: float, proba_3: float) -> int:
return np.argmax((proba_0, proba_1, proba_2, proba_3))
metrics = defaultdict(list)
```
## Training set
```
pred_train_dfs = []
for i in range(N_SPLITS):
num_fold = i + 1
logger.debug('Evaluate cv result (training set) Fold {}'.format(num_fold))
# Read cv result
filepath_fold_train = os.path.join(output_dir, f'cv_fold{num_fold}_training.csv')
pred_train_df = pd.read_csv(filepath_fold_train)
pred_train_df['actual'] = train.loc[pred_train_df['object_id'], TARGET].values
if REGRESSION:
if TARGET == 'target':
pred_train_df['pred'].clip(lower=0, upper=3, inplace=True)
else:
pred_train_df['pred'] = np.vectorize(soring_date2target)(pred_train_df['pred'])
pred_train_df['actual'] = np.vectorize(soring_date2target)(pred_train_df['actual'])
else:
pred_train_df['pred'] = predict(pred_train_df['0'], pred_train_df['1'],
pred_train_df['2'], pred_train_df['3'])
if not (REGRESSION and TARGET == 'target'):
print(confusion_matrix(pred_train_df['actual'], pred_train_df['pred'],
labels=np.sort(train['target'].unique())))
loss = rmse(pred_train_df['actual'], pred_train_df['pred'])
# score = qwk(pred_train_df['actual'], pred_train_df['pred'])
logger.debug('Loss: {}'.format(loss))
# logger.debug('Score: {}'.format(score))
metrics['train_losses'].append(loss)
# metrics['train_scores'].append(score)
pred_train_dfs.append(pred_train_df)
metrics['train_losses_avg'] = np.mean(metrics['train_losses'])
metrics['train_losses_std'] = np.std(metrics['train_losses'])
# metrics['train_scores_avg'] = np.mean(metrics['train_scores'])
# metrics['train_scores_std'] = np.std(metrics['train_scores'])
pred_train = pd.concat(pred_train_dfs).groupby('object_id').sum()
pred_train = pred_train / N_SPLITS
if not REGRESSION:
pred_train['pred'] = predict(pred_train['0'], pred_train['1'], pred_train['2'], pred_train['3'])
pred_train['actual'] = train.loc[pred_train.index, TARGET].values
if REGRESSION and TARGET == 'sorting_date':
pred_train['actual'] = np.vectorize(soring_date2target)(pred_train['actual'])
# for c in ('pred', 'actual'):
# pred_train[c] = pred_train[c].astype('int')
pred_train
if not (REGRESSION and TARGET == 'target'):
print(confusion_matrix(pred_train['actual'], pred_train['pred'], labels=np.sort(train['target'].unique())))
loss = rmse(pred_train['actual'], pred_train['pred'])
# score = qwk(pred_train['actual'], pred_train['pred'])
metrics['train_loss'] = loss
# metrics['train_score'] = score
logger.info('Training loss: {}'.format(loss))
# logger.info('Training score: {}'.format(score))
pred_train.to_csv(os.path.join(output_dir, 'prediction_train.csv'))
logger.debug('Write cv result to {}'.format(os.path.join(output_dir, 'prediction_train.csv')))
```
## Validation set
```
pred_valid_dfs = []
for i in range(N_SPLITS):
num_fold = i + 1
logger.debug('Evaluate cv result (validation set) Fold {}'.format(num_fold))
# Read cv result
filepath_fold_valid = os.path.join(output_dir, f'cv_fold{num_fold}_validation.csv')
pred_valid_df = pd.read_csv(filepath_fold_valid)
pred_valid_df['actual'] = train.loc[pred_valid_df['object_id'], TARGET].values
if REGRESSION:
if TARGET == 'target':
pred_valid_df['pred'].clip(lower=0, upper=3, inplace=True)
else:
pred_valid_df['pred'] = np.vectorize(soring_date2target)(pred_valid_df['pred'])
pred_valid_df['actual'] = np.vectorize(soring_date2target)(pred_valid_df['actual'])
else:
pred_valid_df['pred'] = predict(pred_valid_df['0'], pred_valid_df['1'],
pred_valid_df['2'], pred_valid_df['3'])
if not (REGRESSION and TARGET == 'target'):
print(confusion_matrix(pred_valid_df['actual'], pred_valid_df['pred'],
labels=np.sort(train['target'].unique())))
loss = rmse(pred_valid_df['actual'], pred_valid_df['pred'])
# score = qwk(pred_valid_df['actual'], pred_valid_df['pred'])
logger.debug('Loss: {}'.format(loss))
# logger.debug('Score: {}'.format(score))
metrics['valid_losses'].append(loss)
# metrics['valid_scores'].append(score)
pred_valid_dfs.append(pred_valid_df)
metrics['valid_losses_avg'] = np.mean(metrics['valid_losses'])
metrics['valid_losses_std'] = np.std(metrics['valid_losses'])
# metrics['valid_scores_avg'] = np.mean(metrics['valid_scores'])
# metrics['valid_scores_std'] = np.std(metrics['valid_scores'])
pred_valid = pd.concat(pred_valid_dfs).groupby('object_id').sum()
pred_valid = pred_valid / N_SPLITS
if not REGRESSION:
pred_valid['pred'] = predict(pred_valid['0'], pred_valid['1'], pred_valid['2'], pred_valid['3'])
pred_valid['actual'] = train.loc[pred_valid.index, TARGET].values
if REGRESSION and TARGET == 'sorting_date':
pred_valid['actual'] = np.vectorize(soring_date2target)(pred_valid['actual'])
# for c in ('pred', 'actual'):
# pred_valid[c] = pred_valid[c].astype('int')
pred_valid
if not REGRESSION:
print(confusion_matrix(pred_valid['actual'], pred_valid['pred'], labels=np.sort(train['target'].unique())))
loss = rmse(pred_valid['actual'], pred_valid['pred'])
# score = qwk(pred_valid['actual'], pred_valid['pred'])
metrics['valid_loss'] = loss
# metrics['valid_score'] = score
logger.info('Validatino loss: {}'.format(loss))
# logger.info('Validatino score: {}'.format(score))
pred_valid.to_csv(os.path.join(output_dir, 'prediction_valid.csv'))
logger.debug('Write cv result to {}'.format(os.path.join(output_dir, 'prediction_valid.csv')))
with open(os.path.join(output_dir, 'metrics.json'), 'w') as f:
json.dump(dict(metrics), f)
logger.debug('Write metrics to {}'.format(os.path.join(output_dir, 'metrics.json')))
```
# Prediction
```
pred_test_dfs = []
for i in range(N_SPLITS):
num_fold = i + 1
# Read cv result
filepath_fold_test = os.path.join(output_dir, f'cv_fold{num_fold}_test.csv')
pred_test_df = pd.read_csv(filepath_fold_test)
pred_test_dfs.append(pred_test_df)
pred_test = pd.concat(pred_test_dfs).groupby('object_id').sum()
pred_test = pred_test / N_SPLITS
if REGRESSION:
if TARGET == 'target':
pred_test['pred'].clip(lower=0, upper=3, inplace=True)
else:
pred_test['pred'] = np.vectorize(soring_date2target)(pred_test['pred'])
else:
pred_test['pred'] = predict(pred_test['0'], pred_test['1'], pred_test['2'], pred_test['3'])
pred_test
test['target'] = pred_test.loc[test['object_id'], 'pred'].values
test = test[['target']]
test
sample_submission
test.to_csv(os.path.join(output_dir, f'{str(EXP_NO).zfill(3)}_submission.csv'), index=False)
logger.debug('Write submission to {}'.format(os.path.join(output_dir, f'{str(EXP_NO).zfill(3)}_submission.csv')))
fig = plt.figure()
if not (REGRESSION and TARGET == 'target'):
sns.countplot(data=test, x='target')
else:
sns.histplot(data=test, x='target')
sns.despine()
fig.savefig(os.path.join(output_dir, 'prediction.png'))
logger.debug('Write figure to {}'.format(os.path.join(output_dir, 'prediction.png')))
logger.debug('Complete ({:.3f} seconds passed)'.format(time.time() - SINCE))
```
| github_jupyter |
```
%reset -f
# libraries used
# https://stats.stackexchange.com/questions/181/how-to-choose-the-number-of-hidden-layers-and-nodes-in-a-feedforward-neural-netw
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, classification_report
from sklearn import preprocessing
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
import keras
from keras.models import Sequential
from keras.layers import Dense, Dropout
import itertools
emotions = pd.read_csv("drive/MyDrive/EEG/emotions.csv")
emotions.replace(['NEGATIVE', 'POSITIVE', 'NEUTRAL'], [2, 1, 0], inplace=True)
emotions['label'].unique()
X = emotions.drop('label', axis=1).copy()
y = (emotions['label'].copy())
# Splitting data into training and testing as 80-20
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)
x = X_train #returns a numpy array
min_max_scaler = preprocessing.MinMaxScaler()
x_scaled = min_max_scaler.fit_transform(x)
df = pd.DataFrame(x_scaled)
# resetting the data - https://www.tensorflow.org/api_docs/python/tf/keras/backend/clear_session
tf.keras.backend.clear_session()
model = Sequential()
model.add(Dense((2*X_train.shape[1]/3), input_dim=X_train.shape[1], activation='relu'))
model.add(Dense((2*X_train.shape[1]/3), activation='relu'))
model.add(Dense((1*X_train.shape[1]/3), activation='relu'))
model.add(Dense((1*X_train.shape[1]/3), activation='relu'))
model.add(Dense(3, activation='softmax'))
#model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
print(model.summary())
# for categorical entropy
# https://stackoverflow.com/questions/63211181/error-while-using-categorical-crossentropy
from tensorflow.keras.utils import to_categorical
Y_one_hot=to_categorical(y_train) # convert Y into an one-hot vector
# https://stackoverflow.com/questions/59737875/keras-change-learning-rate
#optimizer = tf.keras.optimizers.Adam(0.001)
#optimizer.learning_rate.assign(0.01)
opt = keras.optimizers.SGD(learning_rate=0.001)
model.compile(
optimizer=opt,
loss='sparse_categorical_crossentropy',
metrics=['accuracy']
)
# to be run for categorical cross entropy
# model.compile(loss='categorical_crossentropy', optimizer=tf.keras.optimizers.Adam(lr=0.01), metrics=['accuracy'])
# make sure that the input data is shuffled before hand so that the model doesn't notice patterns and generalizes well
# change y_train to y_hot_encoded when using categorical cross entorpy
import time
start_time = time.time()
history = model.fit(
df,
y_train,
validation_split=0.2,
batch_size=32,
epochs=75)
history.history
print("--- %s seconds ---" % (time.time() - start_time))
x_test = X_test #returns a numpy array
min_max_scaler = preprocessing.MinMaxScaler()
x_scaled_test = min_max_scaler.fit_transform(x_test)
df_test = pd.DataFrame(x_scaled_test)
predictions = model.predict(x=df_test, batch_size=32)
rounded_predictions = np.argmax(predictions, axis=-1)
cm = confusion_matrix(y_true=y_test, y_pred=rounded_predictions)
label_mapping = {'NEGATIVE': 0, 'NEUTRAL': 1, 'POSITIVE': 2}
# for diff dataset
# label_mapping = {'NEGATIVE': 0, 'POSITIVE': 1}
plt.figure(figsize=(8, 8))
sns.heatmap(cm, annot=True, vmin=0, fmt='g', cbar=False, cmap='Blues')
clr = classification_report(y_test, rounded_predictions, target_names=label_mapping.keys())
plt.xticks(np.arange(3) + 0.5, label_mapping.keys())
plt.yticks(np.arange(3) + 0.5, label_mapping.keys())
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title("Confusion Matrix")
plt.show()
print("Classification Report:\n----------------------\n", clr)
# https://stackoverflow.com/questions/26413185/how-to-recover-matplotlib-defaults-after-setting-stylesheet
import matplotlib as mpl
mpl.rcParams.update(mpl.rcParamsDefault)
training_acc = history.history['accuracy']
validation_acc = history.history['val_accuracy']
training_loss = history.history['loss']
validation_loss = history.history['val_loss']
epochs = history.epoch
plt.plot(epochs, training_acc, color = '#17e6e6', label='Training Accuracy')
plt.plot(epochs, validation_acc,color = '#e61771', label='Validation Accuracy')
plt.title('Accuracy vs Epochs')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.savefig('AccuracyVsEpochs.png')
plt.show()
from google.colab import files
files.download('AccuracyVsEpochs.png')
```
| github_jupyter |
# Essential Objects
This tutorial covers several object types that are foundational to much of what pyGSTi does: [circuits](#circuits), [processor specifications](#pspecs), [models](#models), and [data sets](#datasets). Our objective is to explain what these objects are and how they relate to one another at a high level while providing links to other notebooks that cover details we skip over here.
```
import pygsti
from pygsti.circuits import Circuit
from pygsti.models import Model
from pygsti.data import DataSet
```
<a id="circuits"></a>
## Circuits
The `Circuit` object encapsulates a quantum circuit as a sequence of *layers*, each of which contains zero or more non-identity *gates*. A `Circuit` has some number of labeled *lines* and each gate label is assigned to one or more lines. Line labels can be integers or strings. Gate labels have two parts: a `str`-type name and a tuple of line labels. A gate name typically begins with 'G' because this is expected when we parse circuits from text files.
For example, `('Gx',0)` is a gate label that means "do the Gx gate on qubit 0", and `('Gcnot',(2,3))` means "do the Gcnot gate on qubits 2 and 3".
A `Circuit` can be created from a list of gate labels:
```
c = Circuit( [('Gx',0),('Gcnot',0,1),(),('Gy',3)], line_labels=[0,1,2,3])
print(c)
```
If you want multiple gates in a single layer, just put those gate labels in their own nested list:
```
c = Circuit( [('Gx',0),[('Gcnot',0,1),('Gy',3)],()] , line_labels=[0,1,2,3])
print(c)
```
We distinguish three basic types of circuit layers. We call layers containing quantum gates *operation layers*. All the circuits we've seen so far just have operation layers. It's also possible to have a *preparation layer* at the beginning of a circuit and a *measurement layer* at the end of a circuit. There can also be a fourth type of layer called an *instrument layer* which we dicuss in a separate [tutorial on Instruments](objects/advanced/Instruments.ipynb). Assuming that `'rho'` labels a (n-qubit) state preparation and `'Mz'` labels a (n-qubit) measurement, here's a circuit with all three types of layers:
```
c = Circuit( ['rho',('Gz',1),[('Gswap',0,1),('Gy',2)],'Mz'] , line_labels=[0,1,2])
print(c)
```
Finally, when dealing with small systems (e.g. 1 or 2 qubits), we typically just use a `str`-type label (without any line-labels) to denote every possible layer. In this case, all the labels operate on the entire state space so we don't need the notion of 'lines' in a `Circuit`. When there are no line-labels, a `Circuit` assumes a single default **'\*'-label**, which you can usually just ignore:
```
c = Circuit( ['Gx','Gy','Gi'] )
print(c)
```
Pretty simple, right? The `Circuit` object allows you to easily manipulate its labels (similar to a NumPy array) and even perform some basic operations like depth reduction and simple compiling. For lots more details on how to create, modify, and use circuit objects see the [circuit tutorial](objects/Circuit.ipynb).
<a id="models"></a>
<a id="pspecs"></a>
## Processor Specifications
A processor specification describes the interface that a quantum processor exposes to the outside world. Actual quantum processors often have a "native" interface associated with them, but can also be viewed as implementing various other derived interfaces. For example, while a 1-qubit quantum processor may natively implement the $X(\pi/2)$ and $Z(\pi/2)$ gates, it can also implement the set of all 1-qubit Clifford gates. Both of these interfaces would correspond to a processor specification in pyGSTi.
Currently pyGSTi only supports processor specifications having an integral number of qubits. The `QubitProcessorSpec` object describes the number of qubits and what gates are available on them. For example,
```
pspec = pygsti.processors.QubitProcessorSpec(num_qubits=2, gate_names=['Gxpi2', 'Gypi2', 'Gcnot'],
geometry="line")
print("Qubit labels are", pspec.qubit_labels)
print("X(pi/2) gates on qubits: ", pspec.resolved_availability('Gxpi2'))
print("CNOT gates on qubits: ", pspec.resolved_availability('Gcnot'))
```
creates a processor specification for a 2-qubits with $X(\pi/2)$, $Y(\pi/2)$, and CNOT gates. Setting the geometry to `"line"` causes 1-qubit gates to be available on each qubit and the CNOT between the two qubits (in either control/target direction). Processor specifications are used to build experiment designs and models, and so defining or importing an appropriate processor specification is often the first step in many analyses. To learn more about processor specification objects, see the [processor specification tutorial](objects/ProcessorSpec.ipynb).
## Models
An instance of the `Model` class represents something that can predict the outcome probabilities of quantum circuits. We define any such thing to be a "QIP model", or just a "model", as these probabilities define the behavior of some real or virtual QIP. Because there are so many types of models, the `Model` class in pyGSTi is just a base class and is never instaniated directly. Classes `ExplicitOpModel` and `ImplicitOpModel` (subclasses of `Model`) define two broad categories of models, both of which sequentially operate on circuit *layers* (the "Op" in the class names is short for "layer operation").
#### Explicit layer-operation models
An `ExplicitOpModel` is a container object. Its `.preps`, `.povms`, and `.operations` members are essentially dictionaires of state preparation, measurement, and layer-operation objects, respectively. How to create these objects and build up explicit models from scratch is a central capability of pyGSTi and a topic of the [explicit-model tutorial](objects/ExplicitModel.ipynb). Presently, we'll create a 2-qubit model using the processor specification above via the `create_explicit_model` function:
```
mdl = pygsti.models.create_explicit_model(pspec)
```
This creates an `ExplicitOpModel` with a default preparation (prepares all qubits in the zero-state) labeled `'rho0'`, a default measurement labeled `'Mdefault'` in the Z-basis and with 5 layer-operations given by the labels in the 2nd argument (the first argument is akin to a circuit's line labels and the third argument contains special strings that the function understands):
```
print("Preparations: ", ', '.join(map(str,mdl.preps.keys())))
print("Measurements: ", ', '.join(map(str,mdl.povms.keys())))
print("Layer Ops: ", ', '.join(map(str,mdl.operations.keys())))
```
We can now use this model to do what models were made to do: compute the outcome probabilities of circuits.
```
c = Circuit( [('Gxpi2',0),('Gcnot',0,1),('Gypi2',1)] , line_labels=[0,1])
print(c)
mdl.probabilities(c) # Compute the outcome probabilities of circuit `c`
```
An `ExplictOpModel` only "knows" how to operate on circuit layers it explicitly contains in its dictionaries,
so, for example, a circuit layer with two X gates in parallel (layer-label = `[('Gxpi2',0),('Gxpi2',1)]`) cannot be used with our model until we explicitly associate an operation with the layer-label `[('Gxpi2',0),('Gxpi2',1)]`:
```
import numpy as np
c = Circuit( [[('Gxpi2',0),('Gxpi2',1)],('Gxpi2',1)] , line_labels=[0,1])
print(c)
try:
p = mdl.probabilities(c)
except KeyError as e:
print("!!KeyError: ",str(e))
#Create an operation for two parallel X-gates & rerun (now it works!)
mdl.operations[ [('Gxpi2',0),('Gxpi2',1)] ] = np.dot(mdl.operations[('Gxpi2',0)].to_dense(),
mdl.operations[('Gxpi2',1)].to_dense())
p = mdl.probabilities(c)
print("Probability_of_outcome(00) = ", p['00']) # p is like a dictionary of outcomes
mdl.probabilities((('Gxpi2',0),('Gcnot',0,1)))
```
#### Implicit layer-operation models
In the above example, you saw how it is possible to manually add a layer-operation to an `ExplicitOpModel` based on its other, more primitive layer operations. This often works fine for a few qubits, but can quickly become tedious as the number of qubits increases (since the number of potential layers that involve a given set of gates grows exponentially with qubit number). This is where `ImplicitOpModel` objects come into play: these models contain rules for building up arbitrary layer-operations based on more primitive operations. PyGSTi offers several "built-in" types of implicit models and a rich set of tools for building your own custom ones. See the [tutorial on implicit models](objects/ImplicitModel.ipynb) for details.
<a id="datasets"></a>
## Data Sets
The `DataSet` object is a container for tabulated outcome counts. It behaves like a dictionary whose keys are `Circuit` objects and whose values are dictionaries that associate *outcome labels* with (usually) integer counts. There are two primary ways you go about getting a `DataSet`. The first is by reading in a simply formatted text file:
```
dataset_txt = \
"""## Columns = 00 count, 01 count, 10 count, 11 count
{} 100 0 0 0
Gxpi2:0 55 5 40 0
Gxpi2:0Gypi2:1 20 27 23 30
Gxpi2:0^4 85 3 10 2
Gxpi2:0Gcnot:0:1 45 1 4 50
[Gxpi2:0Gxpi2:1]Gypi2:0 25 32 17 26
"""
with open("tutorial_files/Example_Short_Dataset.txt","w") as f:
f.write(dataset_txt)
ds = pygsti.io.read_dataset("tutorial_files/Example_Short_Dataset.txt")
```
The second is by simulating a `Model` and thereby generating "fake data". This essentially calls `mdl.probabilities(c)` for each circuit in a given list, and samples from the output probability distribution to obtain outcome counts:
```
circuit_list = pygsti.circuits.to_circuits([ (),
(('Gxpi2',0),),
(('Gxpi2',0),('Gypi2',1)),
(('Gxpi2',0),)*4,
(('Gxpi2',0),('Gcnot',0,1)),
((('Gxpi2',0),('Gxpi2',1)),('Gxpi2',0)) ], line_labels=(0,1))
ds_fake = pygsti.data.simulate_data(mdl, circuit_list, num_samples=100,
sample_error='multinomial', seed=8675309)
```
Outcome counts are accessible by indexing a `DataSet` as if it were a dictionary with `Circuit` keys:
```
c = Circuit( (('Gxpi2',0),('Gypi2',1)), line_labels=(0,1) )
print(ds[c]) # index using a Circuit
print(ds[ [('Gxpi2',0),('Gypi2',1)] ]) # or with something that can be converted to a Circuit
```
Because `DataSet` object can also store *timestamped* data (see the [time-dependent data tutorial](objects/advanced/TimestampedDataSets.ipynb), the values or "rows" of a `DataSet` aren't simple dictionary objects. When you'd like a `dict` of counts use the `.counts` member of a data set row:
```
row = ds[c]
row['00'] # this is ok
for outlbl, cnt in row.counts.items(): # Note: `row` doesn't have .items(), need ".counts"
print(outlbl, cnt)
```
Another thing to note is that `DataSet` objects are "sparse" in that 0-counts are not typically stored:
```
c = Circuit([('Gxpi2',0)], line_labels=(0,1))
print("No 01 or 11 outcomes here: ",ds_fake[c])
for outlbl, cnt in ds_fake[c].counts.items():
print("Item: ",outlbl, cnt) # Note: this loop never loops over 01 or 11!
```
You can manipulate `DataSets` in a variety of ways, including:
- adding and removing rows
- "trucating" a `DataSet` to include only a subset of it's string
- "filtering" a $n$-qubit `DataSet` to a $m < n$-qubit dataset
To find out more about these and other operations, see our [data set tutorial](objects/DataSet.ipynb).
## What's next?
You've learned about the three main object types in pyGSTi! The next step is to learn about how these objects are used within pyGSTi, which is the topic of the next [overview tutorial on applications](02-Applications.ipynb). Alternatively, if you're interested in learning more about the above-described or other objects, here are some links to relevant tutorials:
- [Circuit](objects/Circuit.ipynb) - how to build circuits ([GST circuits](objects/advanced/GSTCircuitConstruction.ipynb) in particular)
- [ExplicitModel](objects/ExplicitModel.ipynb) - constructing explicit layer-operation models
- [ImplicitModel](objects/ImplicitModel.ipynb) - constructing implicit layer-operation models
- [DataSet](objects/DataSet.ipynb) - constructing data sets ([timestamped data](objects/advanced/TimestampedDataSets.ipynb) in particular)
- [Basis](objects/advanced/MatrixBases.ipynb) - defining matrix and vector bases
- [Results](objects/advanced/Results.ipynb) - the container object for model-based results
- [QubitProcessorSpec](objects/advanced/QubitProcessorSpec.ipynb) - represents a QIP as a collection of models and meta information.
- [Instrument](objects/advanced/Instruments.ipynb) - allows for circuits with intermediate measurements
- [Operation Factories](objects/advanced/OperationFactories.ipynb) - allows continuously parameterized gates
| github_jupyter |
<img src="Techzooka.png">
## Hacker Factory Cyber Hackathon Solution
### by Team Jugaad (Abhiraj Singh Rajput, Deepanshu Gupta, Manuj Mehrotra)
We are a team of members, that are NOT moved by the buzzwords like Machine Learning, Data Science, AI etc. However we are a team of people who get adrenaline rush for seeking the solution to a problem. And the approach to solve the problem is never a constraint for us. Keeping our heads down we tried out bit to solve the problem of <i><b>“Preventive analytics with AI – How to use AI to predict probability of occurrence of a crime.”</b></i>
Formally our team members: -
<ul>
<i><b>
<li>Abhiraj Singh Rajput(BI Engineer, Emp ID -1052530)</li>
<li>Deepanshu Gupta(Performance Test Engineer, Emp ID - 1048606)</li>
<li>Manuj Mehrotra(Analyst-Data Science, Emp ID - 1061322)</li>
</b></i>
</ul>
### Preventive analytics with AI – How to use AI to predict probability of occurrence of a crime
<ul>
<li><b>Context</b>:- We tried to create a classification ML model to analyze the data points and using that we tried to Forecast the occurrence of the Malware attack. For the study we have taken two separate datasets i.e. bifurcated datasets, on the basis of Static and Dynamic features (Sources: Ref[3]).</li><br>
<li><b>Scope of the solution covered</b> :- Since the Model that we have created considers nearly 350 Features (i.e. 331 statics features and 13 dynamic Features) for predicting the attack, so the model is very Robust and is scalable very easily. The objective behind building this predictive model was to forecast the attack of a malicious app by capturing these features and hence preventing them from attacking the device.</li>
</ul>
## Soultion Archicture
<img src="Solution Architecture.png">
# Additional Information – How can it enhance further
<ul>
<li>The data set that we used for Static Analysis has just 398 data points that is comparatively very less to generalize a statistical model (to population).</li>
<li>We haven’t tuned the all the hyper parameter of ML models, however we have considered the important hyper parameters while model building ex: - tuning K in K-NN.</li>
<li>We have analyses the Static and Dynamic Analysis separately. However a more robust model will be that, analyzes both the features together, provided we have sufficient number of data points.</li>
<li>Stacking or ensembling of the ML models from both the data sets could be done to make the model more Robust, provided we capture both the static and dynamic feature of the application.</li>
<li>Dynamic features likes duracion , avg_local_pkt_rate and avg_remote_pkt_rate were not captured which would have degraded the model quality by some amount.</li>
</ul>
# Proof Of Concept
### Static Analysis
Includes analysing the application that we want to analyse without executing it, like the study of resources, app permission etc.
```
import pandas as pd
df = pd.read_csv("train.csv", sep=";")
df.head()
df.columns
df.shape
```
### Let's get the top 10 of permissions that are used for our malware samples
#### Malicious
```
series = pd.Series.sort_values(df[df.type==1].sum(axis=0), ascending=False)[1:11]
series
pd.Series.sort_values(df[df.type==0].sum(axis=0), ascending=False)[:10]
import matplotlib.pyplot as plt
fig, axs = plt.subplots(nrows=2, sharex=True)
pd.Series.sort_values(df[df.type==0].sum(axis=0), ascending=False)[:10].plot.bar(ax=axs[0], color="green")
pd.Series.sort_values(df[df.type==1].sum(axis=0), ascending=False)[1:11].plot.bar(ax=axs[1], color="red")
```
## Now will try to predict with the exsisting data set, i.e. model creation
### Machine Learning Models
```
from sklearn.naive_bayes import GaussianNB, BernoulliNB
from sklearn.metrics import accuracy_score, classification_report,roc_auc_score
from sklearn.ensemble import BaggingClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import SGDClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import cohen_kappa_score
from sklearn.metrics import confusion_matrix
from sklearn.ensemble import RandomForestClassifier
from sklearn import preprocessing
#import torch
from sklearn import svm
from sklearn import tree
import pandas as pd
from sklearn.externals import joblib
import pickle
import numpy as np
import seaborn as sns
y = df["type"]
X = df.drop("type", axis=1)
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.33,random_state=7)
# Naive Bayes algorithm
gnb = GaussianNB()
gnb.fit(X_train, y_train)
# pred
pred = gnb.predict(X_test)
# accuracy
accuracy = accuracy_score(pred, y_test)
print("naive_bayes")
print(accuracy)
print(classification_report(pred, y_test, labels=None))
for i in range(3,15,3):
neigh = KNeighborsClassifier(n_neighbors=i)
neigh.fit(X_train, y_train)
pred = neigh.predict(X_test)
# accuracy
accuracy = accuracy_score(pred, y_test)
print("kneighbors {}".format(i))
print(accuracy)
print(classification_report(pred, y_test, labels=None))
print("")
clf = tree.DecisionTreeClassifier()
clf.fit(X_train, y_train)
# Read the csv test file
pred = clf.predict(X_test)
# accuracy
accuracy = accuracy_score(pred, y_test)
print(clf)
print(accuracy)
print(classification_report(pred, y_test, labels=None))
```
### Dynamic Analysis
For this approach, we used a set of pcap files from the DroidCollector project integrated by 4705 benign and 7846 malicious applications. All of the files were processed by our feature extractor script (a result from [4]), the idea of this analysis is to answer the next question, according to the static analysis previously seen a lot of applications use a network connection, in other words, they are trying to communicate or transmit information, so.. is it possible to distinguish between malware and benign application using network traffic?
```
import pandas as pd
data = pd.read_csv("android_traffic.csv", sep=";")
data.head()
data.columns
data.shape
data.type.value_counts()
data.isna().sum()
data = data.drop(['duracion','avg_local_pkt_rate','avg_remote_pkt_rate'], axis=1).copy()
data.describe()
sns.pairplot(data)
data.loc[data.tcp_urg_packet > 0].shape[0]
data = data.drop(columns=["tcp_urg_packet"], axis=1).copy()
data.shape
data=data[data.tcp_packets<20000].copy()
data=data[data.dist_port_tcp<1400].copy()
data=data[data.external_ips<35].copy()
data=data[data.vulume_bytes<2000000].copy()
data=data[data.udp_packets<40].copy()
data=data[data.remote_app_packets<15000].copy()
data[data.duplicated()].sum()
data=data.drop('source_app_packets.1',axis=1).copy()
scaler = preprocessing.RobustScaler()
scaledData = scaler.fit_transform(data.iloc[:,1:11])
scaledData = pd.DataFrame(scaledData, columns=['tcp_packets','dist_port_tcp','external_ips','vulume_bytes','udp_packets','source_app_packets','remote_app_packets',' source_app_bytes','remote_app_bytes','dns_query_times'])
X_train, X_test, y_train, y_test = train_test_split(scaledData.iloc[:,0:10], data.type.astype("str"), test_size=0.25, random_state=45)
gnb = GaussianNB()
gnb.fit(X_train, y_train)
pred = gnb.predict(X_test)
## accuracy
accuracy = accuracy_score(y_test,pred)
print("naive_bayes")
print(accuracy)
print(classification_report(y_test,pred, labels=None))
print("cohen kappa score")
print(cohen_kappa_score(y_test, pred))
for i in range(3,15,3):
neigh = KNeighborsClassifier(n_neighbors=i)
neigh.fit(X_train, y_train)
pred = neigh.predict(X_test)
# accuracy
accuracy = accuracy_score(pred, y_test)
print("kneighbors {}".format(i))
print(accuracy)
print(classification_report(pred, y_test, labels=None))
print("cohen kappa score")
print(cohen_kappa_score(y_test, pred))
print("")
rdF=RandomForestClassifier(n_estimators=250, max_depth=50,random_state=45)
rdF.fit(X_train,y_train)
pred=rdF.predict(X_test)
cm=confusion_matrix(y_test, pred)
accuracy = accuracy_score(y_test,pred)
print(rdF)
print(accuracy)
print(classification_report(y_test,pred, labels=None))
print("cohen kappa score")
print(cohen_kappa_score(y_test, pred))
print(cm)
from lightgbm import LGBMClassifier
rdF=LGBMClassifier(n_estimators=250, max_depth=50,random_state=45)
rdF.fit(X_train,y_train)
pred=rdF.predict(X_test)
cm=confusion_matrix(y_test, pred)
accuracy = accuracy_score(y_test,pred)
print(rdF)
print(accuracy)
print(classification_report(y_test,pred, labels=None))
print("cohen kappa score")
print(cohen_kappa_score(y_test, pred))
print(cm)
import pandas as pd
feature_importances = pd.DataFrame(rdF.feature_importances_,index = X_train.columns,columns=['importance']).sort_values('importance',ascending=False)
feature_importances
x= feature_importances.index
y=feature_importances["importance"]
plt.figure(figsize=(6,4))
sns.barplot(x=y,y=x)
```
# Refrence
<ul>
<li>Christian Camilo Urcuqui López(https://github.com/urcuqui/)</li>
<li>Android Genome Project (MalGenome)</li>
<li>Data Set Source (https://www.kaggle.com/xwolf12/datasetandroidpermissions , https://www.kaggle.com/xwolf12/network-traffic-android-malware)</li>
<li> [1] López, U., Camilo, C., García Peña, M., Osorio Quintero, J. L., & Navarro Cadavid, A. (2018). Ciberseguridad: un enfoque desde la ciencia de datos-Primera edición.</li>
<li>[2] Navarro Cadavid, A., Londoño, S., Urcuqui López, C. C., & Gomez, J. (2014, June). Análisis y caracterización de frameworks para detección de aplicaciones maliciosas en Android. In Conference: XIV Jornada Internacional de Seguridad Informática ACIS-2014 (Vol. 14). ResearchGate.</li>
<li>[3] Urcuqui-López, C., & Cadavid, A. N. (2016). Framework for malware analysis in Android.</li>
<li>[4] Urcuqui, C., Navarro, A., Osorio, J., & Garcıa, M. (2017). Machine Learning Classifiers to Detect Malicious Websites. CEUR Workshop Proceedings. Vol 1950, 14-17.</li>
<li>[5] López, C. C. U., Villarreal, J. S. D., Belalcazar, A. F. P., Cadavid, A. N., & Cely, J. G. D. (2018, May). Features to Detect Android Malware. In 2018 IEEE Colombian Conference on Communications and Computing (COLCOM) (pp. 1-6). IEEE</li>
</ul>
| github_jupyter |
# Day and Night Image Classifier
---
The day/night image dataset consists of 200 RGB color images in two categories: day and night. There are equal numbers of each example: 100 day images and 100 night images.
We'd like to build a classifier that can accurately label these images as day or night, and that relies on finding distinguishing features between the two types of images!
*Note: All images come from the [AMOS dataset](http://cs.uky.edu/~jacobs/datasets/amos/) (Archive of Many Outdoor Scenes).*
### Import resources
Before you get started on the project code, import the libraries and resources that you'll need.
```
import cv2 # computer vision library
import helpers
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
%matplotlib inline
```
## Training and Testing Data
The 200 day/night images are separated into training and testing datasets.
* 60% of these images are training images, for you to use as you create a classifier.
* 40% are test images, which will be used to test the accuracy of your classifier.
First, we set some variables to keep track of some where our images are stored:
image_dir_training: the directory where our training image data is stored
image_dir_test: the directory where our test image data is stored
```
# Image data directories
image_dir_training = "day_night_images/training/"
image_dir_test = "day_night_images/test/"
```
## Load the datasets
These first few lines of code will load the training day/night images and store all of them in a variable, `IMAGE_LIST`. This list contains the images and their associated label ("day" or "night").
For example, the first image-label pair in `IMAGE_LIST` can be accessed by index:
``` IMAGE_LIST[0][:]```.
```
# Using the load_dataset function in helpers.py
# Load training data
IMAGE_LIST = helpers.load_dataset(image_dir_training)
```
---
# 1. Visualize the input images
```
# Print out 1. The shape of the image and 2. The image's label
# Select an image and its label by list index
image_index = 0
selected_image = IMAGE_LIST[image_index][0]
selected_label = IMAGE_LIST[image_index][1]
# Display image and data about it
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label: " + str(selected_label))
```
# 2. Pre-process the Data
After loading in each image, you have to standardize the input and output.
#### Solution code
You are encouraged to try to complete this code on your own, but if you are struggling or want to make sure your code is correct, there i solution code in the `helpers.py` file in this directory. You can look at that python file to see complete `standardize_input` and `encode` function code. For this day and night challenge, you can often jump one notebook ahead to see the solution code for a previous notebook!
---
### Input
It's important to make all your images the same size so that they can be sent through the same pipeline of classification steps! Every input image should be in the same format, of the same size, and so on.
#### TODO: Standardize the input images
* Resize each image to the desired input size: 600x1100px (hxw).
```
# This function should take in an RGB image and return a new, standardized version
def standardize_input(image):
## TODO: Resize image so that all "standard" images are the same size 600x1100 (hxw)
standard_im = image[0:600, 0:1100]
return standard_im
```
### TODO: Standardize the output
With each loaded image, you also need to specify the expected output. For this, use binary numerical values 0/1 = night/day.
```
# Examples:
# encode("day") should return: 1
# encode("night") should return: 0
def encode(label):
numerical_val = 0
## TODO: complete the code to produce a numerical label
if label == "day":
numerical_val = 1
return numerical_val
```
## Construct a `STANDARDIZED_LIST` of input images and output labels.
This function takes in a list of image-label pairs and outputs a **standardized** list of resized images and numerical labels.
This uses the functions you defined above to standardize the input and output, so those functions must be complete for this standardization to work!
```
def standardize(image_list):
# Empty image data array
standard_list = []
# Iterate through all the image-label pairs
for item in image_list:
image = item[0]
label = item[1]
# Standardize the image
standardized_im = standardize_input(image)
# Create a numerical label
binary_label = encode(label)
# Append the image, and it's one hot encoded label to the full, processed list of image data
standard_list.append((standardized_im, binary_label))
return standard_list
# Standardize all training images
STANDARDIZED_LIST = standardize(IMAGE_LIST)
```
## Visualize the standardized data
Display a standardized image from STANDARDIZED_LIST.
```
# Display a standardized image and its label
# Select an image by index
image_num = 0
selected_image = STANDARDIZED_LIST[image_num][0]
selected_label = STANDARDIZED_LIST[image_num][1]
# Display image and data about it
## TODO: Make sure the images have numerical labels and are of the same size
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label [1 = day, 0 = night]: " + str(selected_label))
```
| github_jupyter |
```
from plot import *
from gen import *
# from load_data import *
from func_tools import *
from AGM import *
from GM import *
from BFGS import *
from LBFGS import *
from sklearn import metrics
import warnings
warnings.filterwarnings('ignore')
def purity_score(y_true, y_pred):
contingency_matrix = metrics.cluster.contingency_matrix(y_true, y_pred)
return np.sum(np.amax(contingency_matrix, axis=0)) / np.sum(contingency_matrix)
```
### 生成随机数
```
# 这里使用默认的参数,按照均匀分布的中心点
# TODO: task 上说可以尝试有趣的pattern,我们可以手动给定centroid再生成周围点,详见 gen.py 的文档
centroids, points, N = gen_data()
y_true = np.repeat(np.arange(len(N)),N)
len(y_true)
len(points)
# 简单画个图
plt.figure(figsize=(10,10))
plot_generated_data(centroids, points, N)
len(points)
```
## AGM Sample
```
lbd = 0.05
delta = 1e-3
n = len(points)
step = step_size(n,lbd,delta)
grad = lambda X,B,D: grad_hub_matrix(X,delta,points,lbd,B,D)
ans,AGM_loss = AGM(grad,points,step,0.005)
groups = get_group(ans, tol=1.5)
groups
purity_score(y_true,groups)
plt.figure(figsize=(10,10))
plot_res_data(points,ans,groups)
plt.figure(figsize=(10,10))
plot_res_data(points,ans,groups,way='ans')
plt.figure(figsize=(10,10))
plot_res_data(points,ans,groups,way='points')
plt.plot(np.log(AGM_loss))
```
## GM Sample
```
lbd = 0.05
delta = 1e-3
func = lambda X,B: loss_func(X,points,lbd,delta,B)
grad = lambda X,B,D: grad_hub_matrix(X,delta,points,lbd,B,D)
ans2,GM_loss = GM(points,func,grad,1e-2)
len(GM_loss)
groups = get_group(ans2, tol=2)
plt.figure(figsize=(10,10))
plot_res_data(points,ans2,groups,way='points')
plt.rc_context({'axes.edgecolor':'orange', 'xtick.color':'green', 'ytick.color':'green', 'figure.facecolor':'white'})
plt.plot(np.log(GM_loss - GM_loss[len(GM_loss)-1]))
plt.ylabel("Loss: |f(xk) - f(x*)|")
plt.xlabel("Iters")
```
## GM_BB Sample
```
lbd = 0.05
delta = 1e-3
func = lambda X,B: loss_func(X,points,lbd,delta,B)
grad = lambda X,B,D: grad_hub_matrix(X,delta,points,lbd,B,D)
ans_BB,GM_BB_loss = GM_BB(points,func,grad,1e-5)
len(GM_BB_loss)
groups = get_group(ans_BB, tol=2)
plt.figure(figsize=(10,10))
plot_res_data(points,ans_BB,groups,way='points')
plt.rc_context({'axes.edgecolor':'black', 'xtick.color':'black', 'ytick.color':'black', 'figure.facecolor':'white'})
plt.figure(figsize=(8,8))
plt.ylabel("Loss: log(|f(xk) - f(x*)|)")
plt.plot(np.log(GM_BB_loss - GM_BB_loss[len(GM_BB_loss)-1]),label="GM_BB")
plt.plot(np.log(GM_loss - GM_loss[len(GM_loss)-1]),color="green",label="GM")
plt.legend()
plt.savefig("D:\Study\MDS\Term 1\Optimization\Final\Figure\BB_GM_Loss")
plt.show()
```
## BFGS
tol=0.03 is quite almost minimum, if smaller s.y is too small 1/s.y=nan
```
lbd = 0.05
delta = 1e-3
func = lambda X,B: loss_func(X,points,lbd,delta,B)
grad = lambda X,B,D: grad_hub_matrix(X,delta,points,lbd,B,D)
ans_BFGS,BFGS_loss = BFGS(points,func,grad,0.003)
groups = get_group(ans_BFGS, tol=2)
plt.figure(figsize=(10,10))
plot_res_data(points,ans_BFGS,groups)
plt.rc_context({'axes.edgecolor':'orange', 'xtick.color':'green', 'ytick.color':'green', 'figure.facecolor':'white'})
plt.figure(figsize=(5,5))
plt.ylabel("Loss")
plt.plot(np.log(BFGS_loss - BFGS_loss[len(BFGS_loss)-1]))
plt.show()
```
## LBFGS
tol=0.03 is quite almost minimum, if smaller s.y is too small 1/s.y=nan
```
lbd = 0.05
delta = 1e-3
func = lambda X,B: loss_func(X,points,lbd,delta,B)
grad = lambda X,B,D: grad_hub_matrix(X,delta,points,lbd,B,D)
ans_LBFGS,LBFGS_loss = LBFGS(points,func,grad,0.003,1,5)
groups = get_group(ans_LBFGS, tol=2)
plt.figure(figsize=(10,10))
plot_res_data(points,ans_LBFGS,groups)
plt.rc_context({'axes.edgecolor':'orange', 'xtick.color':'green', 'ytick.color':'green', 'figure.facecolor':'white'})
plt.figure(figsize=(5,5))
plt.ylabel("Loss")
plt.plot(np.log(LBFGS_loss - LBFGS_loss[len(LBFGS_loss)-1]))
plt.show()
```
## 计算Hessian
```
from itertools import combinations
def huber(x, delta):
'''
Args:
x: input that has been norm2ed (n*(n-1)/2,)
delta: threshold
Output:
(n*(n-1)/2,)
'''
return np.where(x > delta ** 2, np.sqrt(x) - delta / 2, x / (2 * delta))
def pair_col_diff_norm2(x, idx):
'''
compute norm2 of pairwise column difference
Args:
x: (d, n)
idx: (n*(n - 1)/2, 2), used to indexing pairwise column combinations
Output:
(n*(n-1)/2,)
'''
x = x[:, idx] # (d, n*(n - 1)/2, 2)
x = np.diff(x, axis=-1).squeeze() # (d, n*(n-1)/2)
x = np.sum(x ** 2, axis=0) # (n*(n-1)/2,)
return x
def pair_col_diff_sum(x, t, idx):
'''
compute sum of pairwise column difference
Args:
x: (d, n)
t: (d, n)
idx: (n*(n - 1)/2, 2), used to indexing pairwise column combinations
Output:
(n*(n-1)/2,)
'''
x = np.diff(x[:, idx], axis=-1).squeeze() # (d, n*(n-1)/2)
t = np.diff(t[:, idx], axis=-1).squeeze() # (d, n*(n-1)/2)
return np.sum(x * t, axis=0) # (n*(n-1)/2,)
class OBJ:
def __init__(self, d, n, delta):
'''
a: training data samples of shape (d, n)
'''
self.d = d
self.n = n
self.delta = delta
self.idx = np.array(list(combinations(list(range(n)), 2)))
self.triu_idx = np.triu_indices(self.n, 1)
def __call__(self, x, a, lamb):
'''
Args:
x: (d, n)
a: (d, n)
lamb: control effect of regularization
Output:
scalar
'''
v = np.sum((x - a) ** 2) / 2
v += lamb * np.sum(huber(pair_col_diff_norm2(x, self.idx), self.delta))
return v
def grad(self, x, a, lamb):
'''
gradient
Output:
(d, n)
'''
g = x - a
diff_norm2 = pair_col_diff_norm2(x, self.idx) # (n*(n-1)/2,)
tmp = np.zeros((self.n, self.n))
tmp[self.triu_idx] = diff_norm2
tmp += tmp.T # (n, n)
mask = (tmp > self.delta ** 2)
tmp = np.where(mask,
np.divide(1, np.sqrt(tmp), where=mask),
0)
x = x.T
g = g + lamb * (tmp.sum(axis=1, keepdims=True) * x - tmp @ x).T
tmp = 1 - mask
g = g + lamb * (tmp.sum(axis=1, keepdims=True) * x - tmp @ x).T / self.delta
return g.flatten()
def hessiant(self, x, t, lamb):
'''
returns the result of hessian matrix dot product a vector t
Args:
t: (d, n)
Output:
(d, n)
'''
ht = 0
ht += t
diff_norm2 = pair_col_diff_norm2(x, self.idx) # (n*(n-1)/2,)
diff_sum = pair_col_diff_sum(x, t, self.idx)
tmp = np.zeros((self.n, self.n))
tmp[self.triu_idx] = diff_norm2
tmp += tmp.T
mask = (tmp > self.delta ** 2)
tmp = np.where(mask,
np.divide(1, np.sqrt(tmp), where=mask),
0)
t = t.T
x = x.T
ht += (lamb * (tmp.sum(axis=1, keepdims=True) * t - tmp @ t).T)
# tmp1 = np.where(tmp1 > 0, tmp1 ** 3, 0)
tmp = tmp ** 3
tmp[self.triu_idx] *= diff_sum
tmp[(self.triu_idx[1], self.triu_idx[0])] *= diff_sum
ht -= lamb * (tmp.sum(axis=1, keepdims=True) * x - tmp @ x).T
tmp = 1 - mask
ht += (lamb * (tmp.sum(axis=1, keepdims=True) * t - tmp @ t).T / self.delta)
return ht.flatten()
import numpy as np
# from numpy.lib.function_base import _delete_dispatcher
def Hessian_hub(X, p, delta, B):
n = X.shape[0]; d = X.shape[1]
res = np.zeros(n*d).reshape((n*d,1))
for i in range(n):
H_tmp = Hessian_rows(i,n,d,delta,B,X)
res[i*d:(i+1)*d] = H_tmp.dot(p).reshape((d,1))
return np.array(res)
def Hessian_rows(i,n,d,delta,B,X):#i从0开始
I = np.identity(d)
DF = np.tile((-1/delta) * np.identity(d), n)
choose_BX = (B.T[i] != 0) #choose material Xi-Xk, n-1 in total
DBX = B.dot(X)[choose_BX]
DBX[:i,:] = -DBX[:i,:]
mask = np.linalg.norm(DBX, axis=1) > delta #find ||Xi-Xk|| which is greater than delta
mask2 = np.tile(mask,(d,1)).T.reshape(1,-1)[0] #will be further use
norm = np.linalg.norm(DBX[mask], axis=1) #Calculate the norm which is greater than delta (prepare for the left part of tmp)
#prepare for the right part of tmp
row = np.repeat(np.arange(n-1),d)
col = np.arange((n-1)*d)
DBX_trans = np.array(sps.csr_matrix((DBX.flatten(),(row,col)),shape=((n-1),(n-1)*d)).todense())
tmp = -(np.tile(I,(1,len(norm)))/np.repeat(norm,d)) + \
(DBX[mask].T.dot(DBX_trans[:,mask2]))/np.repeat(norm**3,d)
#change the values of items whose norm are greater than delta
DF[:,:i*d][:, mask2[:i*d]] = tmp[:,:i*d]
DF[:,(i+1)*d:][:,mask2[i*d:]] = tmp[:,i*d:]
z = np.zeros((d,d))
DF[:,i*d:(i+1)*d] = z
I_tmp = np.tile(I,(n,1))
iblock_tmp = -DF.dot(I_tmp)
DF[:,i*d:(i+1)*d] = iblock_tmp
return np.array(DF)
i=1
n, d = 5,3
delta = 0.1
B = gen_B(n, sparse=False)
X = np.arange(n*d).reshape(n,d)
DBX = B.T.dot(B.dot(X))
p = np.arange(n*d).reshape((n*d,1))
# Hessian_hub(X, p, 0.1, B)
Hdn = Hessian_rows(i,n,d,delta,B,X)
Hdn[0].dot(p)
```
## 测试Hessian
```
n, d = 4,2
test = OBJ(d,n,0.1)
X = np.arange(n*d).reshape(n,d)
t = np.arange(n*d).reshape((d,n)).astype(float)
test.hessiant(X.T, t, 0.1)
test.hessiant(X.T, t, 0.1)
n, d = 4,2
delta = 0.1
X = np.arange(n*d).reshape(n,d)
B = gen_B(n, sparse=False)
p = np.arange(n*d).reshape((n*d,1))
Hessian_hub(X, p, delta, B)
i=1
B = gen_B(n, sparse=False)
X = np.random.randn(n,d)
DBX = B.T.dot(B.dot(X))
i = 0
n, d = 5,3
delta = 0.1
B = gen_B(n, sparse=False)
X = np.random.randn(n,d)
DBX = B.T.dot(B.dot(X))
I = np.identity(d)
DF = np.tile((-1/delta) * np.identity(d), n)
choose_BX = (B.T[i] != 0) #choose material Xi-Xk, n-1 in total
DBX = B.dot(X)[choose_BX]
mask = np.linalg.norm(DBX, axis=1) > delta #find ||Xi-Xk|| which is greater than delta
mask2 = np.tile(mask,(d,1)).T.reshape(1,-1)[0] #will be further use
norm = np.linalg.norm(DBX[mask], axis=1) #Calculate the norm which is greater than delta (prepare for the left part of tmp)
#prepare for the right part of tmp
row = np.repeat(np.arange(n-1),d)
col = np.arange((n-1)*d)
DBX_trans = np.array(sps.csr_matrix((DBX.flatten(),(row,col)),shape=((n-1),(n-1)*d)).todense())
tmp = -(np.tile(I,(1,len(norm)))/np.repeat(norm,d)) + \
(DBX[mask].T.dot(DBX_trans[:,mask2]))/np.repeat(norm**3,d)
#change the values of items whose norm are greater than delta
DF[:,:i*d][:, mask2[:i*d]] = tmp[:,:i*d]
DF[:,(i+1)*d:][:,mask2[i*d:]] = tmp[:,i*d:]
z = np.zeros((d,d))
DF[:,i*d:(i+1)*d] = z
I_tmp = np.tile(-I,(n,1))
iblock_tmp = DF.dot(I_tmp)
DF[:,i*d:(i+1)*d] = iblock_tmp
DF
```
## 使用原数据集转sparse后操作
| github_jupyter |
##### Copyright 2018 The AdaNet Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Customizing AdaNet
Often times, as a researcher or machine learning practitioner, you will have
some prior knowledge about a dataset. Ideally you should be able to encode that
knowledge into your machine learning algorithm. With `adanet`, you can do so by
defining the *neural architecture search space* that the AdaNet algorithm should
explore.
In this tutorial, we will explore the flexibility of the `adanet` framework, and
create a custom search space for an image-classificatio dataset using high-level
TensorFlow libraries like `tf.layers`.
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import functools
import adanet
from adanet.examples import simple_dnn
import tensorflow as tf
# The random seed to use.
RANDOM_SEED = 42
```
## Fashion MNIST dataset
In this example, we will use the Fashion MNIST dataset
[[Xiao et al., 2017](https://arxiv.org/abs/1708.07747)] for classifying fashion
apparel images into one of ten categories:
1. T-shirt/top
2. Trouser
3. Pullover
4. Dress
5. Coat
6. Sandal
7. Shirt
8. Sneaker
9. Bag
10. Ankle boot

## Download the data
Conveniently, the data is available via Keras:
```
(x_train, y_train), (x_test, y_test) = (
tf.keras.datasets.fashion_mnist.load_data())
```
## Supply the data in TensorFlow
Our first task is to supply the data in TensorFlow. Using the
tf.estimator.Estimator covention, we will define a function that returns an
`input_fn` which returns feature and label `Tensors`.
We will also use the `tf.data.Dataset` API to feed the data into our models.
```
FEATURES_KEY = "images"
def generator(images, labels):
"""Returns a generator that returns image-label pairs."""
def _gen():
for image, label in zip(images, labels):
yield image, label
return _gen
def preprocess_image(image, label):
"""Preprocesses an image for an `Estimator`."""
# First let's scale the pixel values to be between 0 and 1.
image = image / 255.
# Next we reshape the image so that we can apply a 2D convolution to it.
image = tf.reshape(image, [28, 28, 1])
# Finally the features need to be supplied as a dictionary.
features = {FEATURES_KEY: image}
return features, label
def input_fn(partition, training, batch_size):
"""Generate an input_fn for the Estimator."""
def _input_fn():
if partition == "train":
dataset = tf.data.Dataset.from_generator(
generator(x_train, y_train), (tf.float32, tf.int32), ((28, 28), ()))
else:
dataset = tf.data.Dataset.from_generator(
generator(x_test, y_test), (tf.float32, tf.int32), ((28, 28), ()))
# We call repeat after shuffling, rather than before, to prevent separate
# epochs from blending together.
if training:
dataset = dataset.shuffle(10 * batch_size, seed=RANDOM_SEED).repeat()
dataset = dataset.map(preprocess_image).batch(batch_size)
iterator = dataset.make_one_shot_iterator()
features, labels = iterator.get_next()
return features, labels
return _input_fn
```
## Establish baselines
The next task should be to get somes baselines to see how our model performs on
this dataset.
Let's define some information to share with all our `tf.estimator.Estimators`:
```
# The number of classes.
NUM_CLASSES = 10
# We will average the losses in each mini-batch when computing gradients.
loss_reduction = tf.losses.Reduction.SUM_OVER_BATCH_SIZE
# A `Head` instance defines the loss function and metrics for `Estimators`.
head = tf.contrib.estimator.multi_class_head(
NUM_CLASSES, loss_reduction=loss_reduction)
# Some `Estimators` use feature columns for understanding their input features.
feature_columns = [
tf.feature_column.numeric_column(FEATURES_KEY, shape=[28, 28, 1])
]
# Estimator configuration.
config = tf.estimator.RunConfig(
save_checkpoints_steps=50000,
save_summary_steps=50000,
tf_random_seed=RANDOM_SEED)
```
Let's start simple, and train a linear model:
```
#@test {"skip": true}
#@title Parameters
LEARNING_RATE = 0.001 #@param {type:"number"}
TRAIN_STEPS = 5000 #@param {type:"integer"}
BATCH_SIZE = 64 #@param {type:"integer"}
estimator = tf.estimator.LinearClassifier(
feature_columns=feature_columns,
n_classes=NUM_CLASSES,
optimizer=tf.train.RMSPropOptimizer(learning_rate=LEARNING_RATE),
loss_reduction=loss_reduction,
config=config)
results, _ = tf.estimator.train_and_evaluate(
estimator,
train_spec=tf.estimator.TrainSpec(
input_fn=input_fn("train", training=True, batch_size=BATCH_SIZE),
max_steps=TRAIN_STEPS),
eval_spec=tf.estimator.EvalSpec(
input_fn=input_fn("test", training=False, batch_size=BATCH_SIZE),
steps=None))
print("Accuracy:", results["accuracy"])
print("Loss:", results["average_loss"])
```
The linear model with default parameters achieves about **84.13% accuracy**.
Let's see if we can do better with the `simple_dnn` AdaNet:
```
#@test {"skip": true}
#@title Parameters
LEARNING_RATE = 0.003 #@param {type:"number"}
TRAIN_STEPS = 5000 #@param {type:"integer"}
BATCH_SIZE = 64 #@param {type:"integer"}
ADANET_ITERATIONS = 2 #@param {type:"integer"}
estimator = adanet.Estimator(
head=head,
subnetwork_generator=simple_dnn.Generator(
feature_columns=feature_columns,
optimizer=tf.train.RMSPropOptimizer(learning_rate=LEARNING_RATE),
seed=RANDOM_SEED),
max_iteration_steps=TRAIN_STEPS // ADANET_ITERATIONS,
evaluator=adanet.Evaluator(
input_fn=input_fn("train", training=False, batch_size=BATCH_SIZE),
steps=None),
config=config)
results, _ = tf.estimator.train_and_evaluate(
estimator,
train_spec=tf.estimator.TrainSpec(
input_fn=input_fn("train", training=True, batch_size=BATCH_SIZE),
max_steps=TRAIN_STEPS),
eval_spec=tf.estimator.EvalSpec(
input_fn=input_fn("test", training=False, batch_size=BATCH_SIZE),
steps=None))
print("Accuracy:", results["accuracy"])
print("Loss:", results["average_loss"])
```
The `simple_dnn` AdaNet model with default parameters achieves about **85.66%
accuracy**.
This improvement can be attributed to `simple_dnn` searching over
fully-connected neural networks which have more expressive power than the linear
model due to their non-linear activations.
Fully-connected layers are permutation invariant to their inputs, meaning that
if we consistently swapped two pixels before training, the final model would
perform identically. However, there is spatial and locality information in
images that we should try to capture. Applying a few convolutions to our inputs
will allow us to do so, and that will require defining a custom
`adanet.subnetwork.Builder` and `adanet.subnetwork.Generator`.
## Define a convolutional AdaNet model
Creating a new search space for AdaNet to explore is straightforward. There are
two abstract classes you need to extend:
1. `adanet.subnetwork.Builder`
2. `adanet.subnetwork.Generator`
Similar to the tf.estimator.Estimator `model_fn`, `adanet.subnetwork.Builder`
allows you to define your own TensorFlow graph for creating a neural network,
and specify the training operations.
Below we define one that applies a 2D convolution, max-pooling, and then a
fully-connected layer to the images:
```
class SimpleCNNBuilder(adanet.subnetwork.Builder):
"""Builds a CNN subnetwork for AdaNet."""
def __init__(self, learning_rate, max_iteration_steps, seed):
"""Initializes a `SimpleCNNBuilder`.
Args:
learning_rate: The float learning rate to use.
max_iteration_steps: The number of steps per iteration.
seed: The random seed.
Returns:
An instance of `SimpleCNNBuilder`.
"""
self._learning_rate = learning_rate
self._max_iteration_steps = max_iteration_steps
self._seed = seed
def build_subnetwork(self,
features,
logits_dimension,
training,
iteration_step,
summary,
previous_ensemble=None):
"""See `adanet.subnetwork.Builder`."""
images = features.values()[0]
kernel_initializer = tf.keras.initializers.he_normal(seed=self._seed)
x = tf.layers.conv2d(
images,
filters=16,
kernel_size=3,
padding="same",
activation="relu",
kernel_initializer=kernel_initializer)
x = tf.layers.max_pooling2d(x, pool_size=2, strides=2)
x = tf.layers.flatten(x)
x = tf.layers.dense(
x, units=64, activation="relu", kernel_initializer=kernel_initializer)
# The `Head` passed to adanet.Estimator will apply the softmax activation.
logits = tf.layers.dense(
x, units=10, activation=None, kernel_initializer=kernel_initializer)
# Use a constant complexity measure, since all subnetworks have the same
# architecture and hyperparameters.
complexity = tf.constant(1)
return adanet.Subnetwork(
last_layer=x,
logits=logits,
complexity=complexity,
persisted_tensors={})
def build_subnetwork_train_op(self,
subnetwork,
loss,
var_list,
labels,
iteration_step,
summary,
previous_ensemble=None):
"""See `adanet.subnetwork.Builder`."""
# Momentum optimizer with cosine learning rate decay works well with CNNs.
learning_rate = tf.train.cosine_decay(
learning_rate=self._learning_rate,
global_step=iteration_step,
decay_steps=self._max_iteration_steps)
optimizer = tf.train.MomentumOptimizer(learning_rate, .9)
# NOTE: The `adanet.Estimator` increments the global step.
return optimizer.minimize(loss=loss, var_list=var_list)
def build_mixture_weights_train_op(self, loss, var_list, logits, labels,
iteration_step, summary):
"""See `adanet.subnetwork.Builder`."""
return tf.no_op("mixture_weights_train_op")
@property
def name(self):
"""See `adanet.subnetwork.Builder`."""
return "simple_cnn"
```
Next, we extend a `adanet.subnetwork.Generator`, which defines the search
space of candidate `SimpleCNNBuilders` to consider including the final network.
It can create one or more at each iteration with different parameters, and the
AdaNet algorithm will select the candidate that best improves the overall neural
network's `adanet_loss` on the training set.
The one below is very simple: it always creates the same architecture, but gives
it a different random seed at each iteration:
```
class SimpleCNNGenerator(adanet.subnetwork.Generator):
"""Generates a `SimpleCNN` at each iteration.
"""
def __init__(self, learning_rate, max_iteration_steps, seed=None):
"""Initializes a `Generator` that builds `SimpleCNNs`.
Args:
learning_rate: The float learning rate to use.
max_iteration_steps: The number of steps per iteration.
seed: The random seed.
Returns:
An instance of `Generator`.
"""
self._seed = seed
self._dnn_builder_fn = functools.partial(
SimpleCNNBuilder,
learning_rate=learning_rate,
max_iteration_steps=max_iteration_steps)
def generate_candidates(self, previous_ensemble, iteration_number,
previous_ensemble_reports, all_reports):
"""See `adanet.subnetwork.Generator`."""
seed = self._seed
# Change the seed according to the iteration so that each subnetwork
# learns something different.
if seed is not None:
seed += iteration_number
return [self._dnn_builder_fn(seed=seed)]
```
With these defined, we pass them into a new `adanet.Estimator`:
```
#@title Parameters
LEARNING_RATE = 0.05 #@param {type:"number"}
TRAIN_STEPS = 5000 #@param {type:"integer"}
BATCH_SIZE = 64 #@param {type:"integer"}
ADANET_ITERATIONS = 2 #@param {type:"integer"}
max_iteration_steps = TRAIN_STEPS // ADANET_ITERATIONS
estimator = adanet.Estimator(
head=head,
subnetwork_generator=SimpleCNNGenerator(
learning_rate=LEARNING_RATE,
max_iteration_steps=max_iteration_steps,
seed=RANDOM_SEED),
max_iteration_steps=max_iteration_steps,
evaluator=adanet.Evaluator(
input_fn=input_fn("train", training=False, batch_size=BATCH_SIZE),
steps=None),
adanet_loss_decay=.99,
config=config)
results, _ = tf.estimator.train_and_evaluate(
estimator,
train_spec=tf.estimator.TrainSpec(
input_fn=input_fn("train", training=True, batch_size=BATCH_SIZE),
max_steps=TRAIN_STEPS),
eval_spec=tf.estimator.EvalSpec(
input_fn=input_fn("test", training=False, batch_size=BATCH_SIZE),
steps=None))
print("Accuracy:", results["accuracy"])
print("Loss:", results["average_loss"])
```
Our `SimpleCNNGenerator` code achieves **90.41% accuracy**.
## Conclusion and next steps
In this tutorial, you learned how to customize `adanet` to encode your
understanding of a particular dataset, and explore novel search spaces with
AdaNet.
One use-case that has worked for us at Google, has been to take a production
model's TensorFlow code, convert it to into an `adanet.subnetwork.Builder`, and
adaptively grow it into an ensemble. In many cases, this has given significant
performance improvements.
As an exercise, you can swap out the FASHION-MNIST with the MNIST handwritten
digits dataset in this notebook using `tf.keras.datasets.mnist.load_data()`, and
see how `SimpleCNN` performs.
| github_jupyter |
# CW Attack Example
TJ Kim <br />
1.28.21
### Summary:
Implement CW attack on toy network example given in the readme of the github. <br />
https://github.com/tj-kim/pytorch-cw2?organization=tj-kim&organization=tj-kim
A dummy network is made using CIFAR example. <br />
https://pytorch.org/tutorials/beginner/blitz/cifar10_tutorial.html
### Build Dummy Pytorch Network
```
import torch
import torchvision
import torchvision.transforms as transforms
```
Download a few classes from the dataset.
```
batch_size = 10
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data', train=True,
download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=batch_size,
shuffle=True, num_workers=2)
testset = torchvision.datasets.CIFAR10(root='./data', train=False,
download=True, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=batch_size,
shuffle=False, num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
```
Show a few images from the dataset.
```
import matplotlib.pyplot as plt
import numpy as np
# functions to show an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
# get some random training images
dataiter = iter(trainloader)
images, labels = dataiter.next()
# show images
imshow(torchvision.utils.make_grid(images))
# print labels
print(' '.join('%5s' % classes[labels[j]] for j in range(4)))
```
Define a NN.
```
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(-1, 16 * 5 * 5)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
```
Define loss and optimizer
```
import torch.optim as optim
net = Net()
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
```
Train the Network
```
train_flag = False
PATH = './cifar_net.pth'
if train_flag:
for epoch in range(2): # loop over the dataset multiple times
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# get the inputs; data is a list of [inputs, labels]
inputs, labels = data
# zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
# print statistics
running_loss += loss.item()
if i % 2000 == 1999: # print every 2000 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / 2000))
running_loss = 0.0
print('Finished Training')
else:
net.load_state_dict(torch.load(PATH))
```
Save Existing Network.
```
if train_flag:
torch.save(net.state_dict(), PATH)
```
Test Acc.
```
correct = 0
total = 0
with torch.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' % (
100 * correct / total))
```
### C&W Attack
Perform attack on toy network.
Before running the example code, we have to set the following parameters:
- dataloader
- mean
- std
The mean and std are one value per each channel of input
```
dataloader = trainloader
mean = (0.5,0.5,0.5)
std = (0.5,0.5,0.5)
import torch
import cw
inputs_box = (min((0 - m) / s for m, s in zip(mean, std)),
max((1 - m) / s for m, s in zip(mean, std)))
"""
# an untargeted adversary
adversary = cw.L2Adversary(targeted=False,
confidence=0.0,
search_steps=10,
box=inputs_box,
optimizer_lr=5e-4)
inputs, targets = next(iter(dataloader))
adversarial_examples = adversary(net, inputs, targets, to_numpy=False)
assert isinstance(adversarial_examples, torch.FloatTensor)
assert adversarial_examples.size() == inputs.size()
"""
# a targeted adversary
adversary = cw.L2Adversary(targeted=True,
confidence=0.0,
search_steps=10,
box=inputs_box,
optimizer_lr=5e-4)
inputs, orig_label = next(iter(dataloader))
# a batch of any attack targets
attack_targets = torch.ones(inputs.size(0), dtype = torch.long) * 3
adversarial_examples = adversary(net, inputs, attack_targets, to_numpy=False)
assert isinstance(adversarial_examples, torch.FloatTensor)
assert adversarial_examples.size() == inputs.size()
# Obtain the outputs of the adversarial perturbations vs. original
print("attacked:", torch.argmax(net(adversarial_examples),dim=1))
print("original:", orig_label)
```
| github_jupyter |
# Simple RNN
In this notebook, we're going to train a simple RNN to do **time-series prediction**. Given some set of input data, it should be able to generate a prediction for the next time step!
<img src='assets/time_prediction.png' width=40% />
> * First, we'll create our data
* Then, define an RNN in PyTorch
* Finally, we'll train our network and see how it performs
### Import resources and create data
```
import torch
from torch import nn
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=(8,5))
# how many time steps/data pts are in one batch of data
seq_length = 20
# generate evenly spaced data pts
time_steps = np.linspace(0, np.pi, seq_length + 1)
data = np.sin(time_steps)
data.resize((seq_length + 1, 1)) # size becomes (seq_length+1, 1), adds an input_size dimension
x = data[:-1] # all but the last piece of data
y = data[1:] # all but the first
# display the data
plt.plot(time_steps[1:], x, 'r.', label='input, x') # x
plt.plot(time_steps[1:], y, 'b.', label='target, y') # y
plt.legend(loc='best')
plt.show()
```
---
## Define the RNN
Next, we define an RNN in PyTorch. We'll use `nn.RNN` to create an RNN layer, then we'll add a last, fully-connected layer to get the output size that we want. An RNN takes in a number of parameters:
* **input_size** - the size of the input
* **hidden_dim** - the number of features in the RNN output and in the hidden state
* **n_layers** - the number of layers that make up the RNN, typically 1-3; greater than 1 means that you'll create a stacked RNN
* **batch_first** - whether or not the input/output of the RNN will have the batch_size as the first dimension (batch_size, seq_length, hidden_dim)
Take a look at the [RNN documentation](https://pytorch.org/docs/stable/nn.html#rnn) to read more about recurrent layers.
```
class RNN(nn.Module):
def __init__(self, input_size, output_size, hidden_dim, n_layers):
super(RNN, self).__init__()
self.hidden_dim=hidden_dim
# define an RNN with specified parameters
# batch_first means that the first dim of the input and output will be the batch_size
self.rnn = nn.RNN(input_size, hidden_dim, n_layers, batch_first=True)
# last, fully-connected layer
self.fc = nn.Linear(hidden_dim, output_size)
def forward(self, x, hidden):
# x (batch_size, seq_length, input_size)
# hidden (n_layers, batch_size, hidden_dim)
# r_out (batch_size, seq_length, hidden_dim)
batch_size = x.size(0)
# get RNN outputs
r_out, hidden = self.rnn(x, hidden)
# shape output to be (batch_size*seq_length, hidden_dim)
r_out = r_out.view(-1, self.hidden_dim)
# get final output
output = self.fc(r_out)
return output, hidden
```
### Check the input and output dimensions
As a check that your model is working as expected, test out how it responds to input data.
```
# test that dimensions are as expected
test_rnn = RNN(input_size=1, output_size=1, hidden_dim=10, n_layers=2)
# generate evenly spaced, test data pts
time_steps = np.linspace(0, np.pi, seq_length)
data = np.sin(time_steps)
data.resize((seq_length, 1))
test_input = torch.Tensor(data).unsqueeze(0) # give it a batch_size of 1 as first dimension
print('Input size: ', test_input.size())
# test out rnn sizes
test_out, test_h = test_rnn(test_input, None)
print('Output size: ', test_out.size())
print('Hidden state size: ', test_h.size())
```
---
## Training the RNN
Next, we'll instantiate an RNN with some specified hyperparameters. Then train it over a series of steps, and see how it performs.
```
# decide on hyperparameters
input_size=1
output_size=1
hidden_dim=32
n_layers=1
# instantiate an RNN
rnn = RNN(input_size, output_size, hidden_dim, n_layers)
print(rnn)
```
### Loss and Optimization
This is a regression problem: can we train an RNN to accurately predict the next data point, given a current data point?
>* The data points are coordinate values, so to compare a predicted and ground_truth point, we'll use a regression loss: the mean squared error.
* It's typical to use an Adam optimizer for recurrent models.
```
# MSE loss and Adam optimizer with a learning rate of 0.01
criterion = nn.MSELoss()
optimizer = torch.optim.Adam(rnn.parameters(), lr=0.01)
```
### Defining the training function
This function takes in an rnn, a number of steps to train for, and returns a trained rnn. This function is also responsible for displaying the loss and the predictions, every so often.
#### Hidden State
Pay close attention to the hidden state, here:
* Before looping over a batch of training data, the hidden state is initialized
* After a new hidden state is generated by the rnn, we get the latest hidden state, and use that as input to the rnn for the following steps
```
# train the RNN
def train(rnn, n_steps, print_every):
# initialize the hidden state
hidden = None
for batch_i, step in enumerate(range(n_steps)):
# defining the training data
time_steps = np.linspace(step * np.pi, (step+1)*np.pi, seq_length + 1)
data = np.sin(time_steps)
data.resize((seq_length + 1, 1)) # input_size=1
x = data[:-1]
y = data[1:]
# convert data into Tensors
x_tensor = torch.Tensor(x).unsqueeze(0) # unsqueeze gives a 1, batch_size dimension
y_tensor = torch.Tensor(y)
# outputs from the rnn
prediction, hidden = rnn(x_tensor, hidden)
## Representing Memory ##
# make a new variable for hidden and detach the hidden state from its history
# this way, we don't backpropagate through the entire history
hidden = hidden.data
# calculate the loss
loss = criterion(prediction, y_tensor)
# zero gradients
optimizer.zero_grad()
# perform backprop and update weights
loss.backward()
optimizer.step()
# display loss and predictions
if batch_i%print_every == 0:
print('Loss: ', loss.item())
plt.plot(time_steps[1:], x, 'r.') # input
plt.plot(time_steps[1:], prediction.data.numpy().flatten(), 'b.') # predictions
plt.show()
return rnn
# train the rnn and monitor results
n_steps = 75
print_every = 15
trained_rnn = train(rnn, n_steps, print_every)
```
### Time-Series Prediction
Time-series prediction can be applied to many tasks. Think about weather forecasting or predicting the ebb and flow of stock market prices. You can even try to generate predictions much further in the future than just one time step!
| github_jupyter |
```
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib import rc,rcParams
rc('text', usetex=True)
rcParams['figure.figsize'] = (8, 6.5)
rcParams['ytick.labelsize'],rcParams['xtick.labelsize'] = 17.,17.
rcParams['axes.labelsize']=19.
rcParams['legend.fontsize']=17.
rcParams['axes.titlesize']=20.
rcParams['text.latex.preamble'] = ['\\usepackage{siunitx}']
import seaborn
seaborn.despine()
seaborn.set_style('white', {'xes.linewidth': 0.5, 'axes.edgecolor':'black'})
seaborn.despine(left=True)
%load_ext autoreload
from astroML.linear_model import TLS_logL
from astroML.plotting.mcmc import convert_to_stdev
import astroML.datasets as adata
from astropy.table import Table
from scipy import polyfit,linalg
from scipy.optimize import curve_fit
```
Εστω οτι παρατηρούμε εναν αστέρα στον ουρανό και μετράμε τη ροή φωτονίων. Θεωρώντας ότι η ροή είναι σταθερή με το χρόνο ίση με $F_{\mathtt{true}}$.
Παίρνουμε $N$ παρατηρήσεις, μετρώντας τη ροή $F_i$ και το σφάλμα $e_i$.
Η ανίχνευση ενός φωτονίου είναι ενα ανεξάρτητο γεγονός που ακολουθεί μια τυχαία κατανομή Poisson. Από τη διακύμανση της κατανομής Poisson υπολογίζουμε το σφάλμα $e_i=\sqrt{F_i}$
```
N=100
F_true=1000.
F=np.random.poisson(F_true*np.ones(N))
e=np.sqrt(F)
plt.errorbar(np.arange(N),F,yerr=e, fmt='ok', ecolor='gray', alpha=0.5)
plt.hlines(np.mean(F),0,N,linestyles='--')
plt.hlines(F_true,0,N)
print np.mean(F),np.mean(F)-F_true,np.std(F)
ax=seaborn.distplot(F,bins=N/3)
xx=np.linspace(F.min(),F.max())
gaus=np.exp(-0.5*((xx-F_true)/np.std(F))**2)/np.sqrt(2.*np.pi*np.std(F)**2)
ax.plot(xx,gaus)
```
Η αρχική προσέγγιση μας είναι μέσω της μεγιστοποιήσης της πιθανοφάνειας. Με βάση τα δεδομένωα $D_i=(F_i,e_i)$ μπορούμε να υπολογίσουμε τη πιθανότητα να τα έχουμε παρατηρήσει δεδομένου της αληθινής τιμής $F_{\mathtt{true}}$ υποθέτωντας ότι τα σφάλματα είναι gaussian
$$
P(D_i|F_{\mathtt{true}})=\frac{1}{\sqrt{2\pi e_i^2}}e^{-\frac{(F_i-F_{\mathtt{true}})^2}{2e_i^2}}
$$
Ορίζουμε τη συνάρτηση πιθανοφάνειας σαν το σύνολο των πιθανοτήτων για κάθε σημείο
$$
L(D|F_{\mathtt{true}})=\prod _{i=1}^N P(D_i|F_{\mathtt{true}})
$$
Επειδή η τιμή της συνάρτηση πιθανοφάνειας μπορεί να γίνει πολύ μικρή, είναι πιο έυκολο να χρησιμοποιήσουμε το λογάριθμο της
$$
\log L = -\frac{1}{2} \sum _{i=0}^N \big[ \log(2\pi e_i^2) + \frac{(F_i-F_\mathtt{true})^2}{e_i^2} \big]
$$
```
#xx=np.linspace(0,10,5000)
xx=np.ones(1000)
#seaborn.distplot(np.random.poisson(xx),kde=False)
plt.hist(np.random.poisson(xx))
w = 1. / e ** 2
print("""
F_true = {0}
F_est = {1:.0f} +/- {2:.0f} (based on {3} measurements)
""".format(F_true, (w * F).sum() / w.sum(), w.sum() ** -0.5, N))
np.sum(((F-F.mean())/F.std())**2)/(N-1)
def log_prior(theta):
return 1 # flat prior
def log_likelihood(theta, F, e):
return -0.5 * np.sum(np.log(2 * np.pi * e ** 2)
+ (F - theta[0]) ** 2 / e ** 2)
def log_posterior(theta, F, e):
return log_prior(theta) + log_likelihood(theta, F, e)
ndim = 1 # number of parameters in the model
nwalkers = 100 # number of MCMC walkers
nburn = 1000 # "burn-in" period to let chains stabilize
nsteps = 5000 # number of MCMC steps to take
# we'll start at random locations between 0 and 2000
starting_guesses = 20 * np.random.rand(nwalkers, ndim)
import emcee
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_posterior, args=[F, e])
sampler.run_mcmc(starting_guesses, nsteps)
sample = sampler.chain # shape = (nwalkers, nsteps, ndim)
sample = sampler.chain[:, nburn:, :].ravel() # discard burn-in points
sampler.chain[0]
# plot a histogram of the sample
plt.hist(sampler.chain, bins=50, histtype="stepfilled", alpha=0.3, normed=True)
# plot a best-fit Gaussian
F_fit = np.linspace(F.min(),F.max(),500)
pdf = stats.norm(np.mean(sample), np.std(sample)).pdf(F_fit)
#plt.plot(F_fit, pdf, '-k')
plt.xlabel("F"); plt.ylabel("P(F)")
print("""
F_true = {0}
F_est = {1:.0f} +/- {2:.0f} (based on {3} measurements)
""".format(F_true, np.mean(sample), np.std(sample), N))
```
| github_jupyter |
# Load MNIST Data
```
# MNIST dataset downloaded from Kaggle :
#https://www.kaggle.com/c/digit-recognizer/data
# Functions to read and show images.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
d0 = pd.read_csv('./mnist_train.csv')
print(d0.head(5)) # print first five rows of d0.
# save the labels into a variable l.
l = d0['label']
# Drop the label feature and store the pixel data in d.
d = d0.drop("label",axis=1)
print(d.shape)
print(l.shape)
# display or plot a number.
plt.figure(figsize=(7,7))
idx = 1
grid_data = d.iloc[idx].as_matrix().reshape(28,28) # reshape from 1d to 2d pixel array
plt.imshow(grid_data, interpolation = "none", cmap = "gray")
plt.show()
print(l[idx])
```
# 2D Visualization using PCA
```
# Pick first 15K data-points to work on for time-effeciency.
#Excercise: Perform the same analysis on all of 42K data-points.
labels = l.head(15000)
data = d.head(15000)
print("the shape of sample data = ", data.shape)
# Data-preprocessing: Standardizing the data
from sklearn.preprocessing import StandardScaler
standardized_data = StandardScaler().fit_transform(data)
print(standardized_data.shape)
#find the co-variance matrix which is : A^T * A
sample_data = standardized_data
# matrix multiplication using numpy
covar_matrix = np.matmul(sample_data.T , sample_data)
print ( "The shape of variance matrix = ", covar_matrix.shape)
# finding the top two eigen-values and corresponding eigen-vectors
# for projecting onto a 2-Dim space.
from scipy.linalg import eigh
# the parameter 'eigvals' is defined (low value to heigh value)
# eigh function will return the eigen values in asending order
# this code generates only the top 2 (782 and 783) eigenvalues.
values, vectors = eigh(covar_matrix, eigvals=(782,783))
print("Shape of eigen vectors = ",vectors.shape)
# converting the eigen vectors into (2,d) shape for easyness of further computations
vectors = vectors.T
print("Updated shape of eigen vectors = ",vectors.shape)
# here the vectors[1] represent the eigen vector corresponding 1st principal eigen vector
# here the vectors[0] represent the eigen vector corresponding 2nd principal eigen vector
# projecting the original data sample on the plane
#formed by two principal eigen vectors by vector-vector multiplication.
import matplotlib.pyplot as plt
new_coordinates = np.matmul(vectors, sample_data.T)
print (" resultanat new data points' shape ", vectors.shape, "X", sample_data.T.shape," = ", new_coordinates.shape)
import pandas as pd
# appending label to the 2d projected data
new_coordinates = np.vstack((new_coordinates, labels)).T
# creating a new data frame for ploting the labeled points.
dataframe = pd.DataFrame(data=new_coordinates, columns=("1st_principal", "2nd_principal", "label"))
print(dataframe.head())
# ploting the 2d data points with seaborn
import seaborn as sn
sn.FacetGrid(dataframe, hue="label", size=6).map(plt.scatter, '1st_principal', '2nd_principal').add_legend()
plt.show()
```
# PCA using Scikit-Learn
```
# initializing the pca
from sklearn import decomposition
pca = decomposition.PCA()
# configuring the parameteres
# the number of components = 2
pca.n_components = 2
pca_data = pca.fit_transform(sample_data)
# pca_reduced will contain the 2-d projects of simple data
print("shape of pca_reduced.shape = ", pca_data.shape)
# attaching the label for each 2-d data point
pca_data = np.vstack((pca_data.T, labels)).T
# creating a new data fram which help us in ploting the result data
pca_df = pd.DataFrame(data=pca_data, columns=("1st_principal", "2nd_principal", "label"))
sn.FacetGrid(pca_df, hue="label", size=6).map(plt.scatter, '1st_principal', '2nd_principal').add_legend()
plt.show()
```
# PCA for dimensionality redcution (not for visualization)
```
# PCA for dimensionality redcution (non-visualization)
pca.n_components = 784
pca_data = pca.fit_transform(sample_data)
percentage_var_explained = pca.explained_variance_ / np.sum(pca.explained_variance_);
cum_var_explained = np.cumsum(percentage_var_explained)
# Plot the PCA spectrum
plt.figure(1, figsize=(6, 4))
plt.clf()
plt.plot(cum_var_explained, linewidth=2)
plt.axis('tight')
plt.grid()
plt.xlabel('n_components')
plt.ylabel('Cumulative_explained_variance')
plt.show()
# If we take 200-dimensions, approx. 90% of variance is expalined.
```
# t-SNE using Scikit-Learn
```
# TSNE
from sklearn.manifold import TSNE
# Picking the top 1000 points as TSNE takes a lot of time for 15K points
data_1000 = standardized_data[0:1000,:]
labels_1000 = labels[0:1000]
model = TSNE(n_components=2, random_state=0)
# configuring the parameteres
# the number of components = 2
# default perplexity = 30
# default learning rate = 200
# default Maximum number of iterations for the optimization = 1000
tsne_data = model.fit_transform(data_1000)
# creating a new data frame which help us in ploting the result data
tsne_data = np.vstack((tsne_data.T, labels_1000)).T
tsne_df = pd.DataFrame(data=tsne_data, columns=("Dim_1", "Dim_2", "label"))
# Ploting the result of tsne
sn.FacetGrid(tsne_df, hue="label", size=6).map(plt.scatter, 'Dim_1', 'Dim_2').add_legend()
plt.show()
model = TSNE(n_components=2, random_state=0, perplexity=50)
tsne_data = model.fit_transform(data_1000)
# creating a new data fram which help us in ploting the result data
tsne_data = np.vstack((tsne_data.T, labels_1000)).T
tsne_df = pd.DataFrame(data=tsne_data, columns=("Dim_1", "Dim_2", "label"))
# Ploting the result of tsne
sn.FacetGrid(tsne_df, hue="label", size=6).map(plt.scatter, 'Dim_1', 'Dim_2').add_legend()
plt.title('With perplexity = 50')
plt.show()
model = TSNE(n_components=2, random_state=0, perplexity=50, n_iter=5000)
tsne_data = model.fit_transform(data_1000)
# creating a new data fram which help us in ploting the result data
tsne_data = np.vstack((tsne_data.T, labels_1000)).T
tsne_df = pd.DataFrame(data=tsne_data, columns=("Dim_1", "Dim_2", "label"))
# Ploting the result of tsne
sn.FacetGrid(tsne_df, hue="label", size=6).map(plt.scatter, 'Dim_1', 'Dim_2').add_legend()
plt.title('With perplexity = 50, n_iter=5000')
plt.show()
model = TSNE(n_components=2, random_state=0, perplexity=2)
tsne_data = model.fit_transform(data_1000)
# creating a new data fram which help us in ploting the result data
tsne_data = np.vstack((tsne_data.T, labels_1000)).T
tsne_df = pd.DataFrame(data=tsne_data, columns=("Dim_1", "Dim_2", "label"))
# Ploting the result of tsne
sn.FacetGrid(tsne_df, hue="label", size=6).map(plt.scatter, 'Dim_1', 'Dim_2').add_legend()
plt.title('With perplexity = 2')
plt.show()
#Excercise: Run the same analysis using 42K points with various
#values of perplexity and iterations.
# If you use all of the points, you can expect plots like this blog below:
# http://colah.github.io/posts/2014-10-Visualizing-MNIST/
```
| github_jupyter |
```
!pip install vcrpy
import vcr
offline = vcr.VCR(
record_mode='new_episodes',
)
```
# APIs and data
Catherine Devlin (@catherinedevlin)
Innovation Specialist, 18F
Oakwood High School, Feb 16 2017
# Who am I?
(hint: not Jean Valjean)

# Cool things I've done
- Chemical engineer in college
- Oops, became a programmer
- Created IPython `%sql` magic
# [Dayton Dynamic Languages](http://d8ndl.org)
# PyOhio

# [18F](18f.gsa.gov)
<a href="https://18f.gsa.gov"><img src="https://18f.gsa.gov/assets/img/logos/18f-logo.svg"
alt="18F logo" width="30%" /></a>
"Digital startup" within the Federal government
It's like college!
Much of what I'm teaching you I've learned the last couple years... some of it last week
# Federal Election Commission
[Old site](http://www.fec.gov/)
[New site](https://beta.fec.gov/)
User research & best practices
Let's look up our Representative
# API

# Webpage vs. API
# FEC API
https://api.open.fec.gov/developers/
Every API works differently.
Let's find the committee ID for our Congressional representative.
C00373001
https://api.open.fec.gov/v1/committee/C00373001/totals/?page=1&api_key=DEMO_KEY&sort=-cycle&per_page=20
- Knife
- Cheese grater
- Vegetable peeler
- Apple corer
- Food processor

# `requests` library
First, we install. That's like buying it.
```
!pip install requests
```
Then, we import. That's like getting it out of the cupboard.
```
import requests
```
# Oakwood High School
```
with offline.use_cassette('offline.vcr'):
response = requests.get('http://ohs.oakwoodschools.org/pages/Oakwood_High_School')
response.ok
response.status_code
print(response.text)
```
We have backed our semi up to the front door.
OK, back to checking out politicians.
```
url = 'https://api.open.fec.gov/v1/committee/C00373001/totals/?page=1&api_key=DEMO_KEY&sort=-cycle&per_page=20'
with offline.use_cassette('offline.vcr'):
response = requests.get(url)
response.ok
response.status_code
response.json()
response.json()['results']
results = response.json()['results']
results[0]['cycle']
results[0]['disbursements']
for result in results:
print(result['cycle'])
for result in results:
year = result['cycle']
spent = result['disbursements']
print('year: {}\t spent:{}'.format(year, spent))
```
# [Pandas](http://pandas.pydata.org/)
```
!pip install pandas
import pandas as pd
data = pd.DataFrame(response.json()['results'])
data
data = data.set_index('cycle')
data
data['disbursements']
data[data['disbursements'] < 1000000 ]
```
# [Bokeh](http://bokeh.pydata.org/en/latest/)
```
!pip install bokeh
from bokeh.charts import Bar, show, output_notebook
by_year = Bar(data, values='disbursements')
output_notebook()
show(by_year)
```
# Playtime
[so many options](http://bokeh.pydata.org/en/latest/docs/user_guide/charts.html)
- Which column to map?
- Colors or styles?
- Scatter
- Better y-axis label?
- Some other candidate committee?
- Portman C00458463, Brown C00264697
- Filter it
# Where's it coming from?
https://api.open.fec.gov/v1/committee/C00373001/schedules/schedule_a/by_state/?per_page=20&api_key=DEMO_KEY&page=1&cycle=2016
```
url = 'https://api.open.fec.gov/v1/committee/C00373001/schedules/schedule_a/by_state/?per_page=20&api_key=DEMO_KEY&page=1&cycle=2016'
with offline.use_cassette('offline.vcr'):
response = requests.get(url)
results = response.json()['results']
data = pd.DataFrame(results)
data
data = data.set_index('state')
by_state = Bar(data, values='total')
show(by_state)
```
# More data
[data.gov](https://www.data.gov/)
Websearch on anything + "API"
# What can you *really* do?
[Moneyfollower](http://moneyfollower.us/)
# Learning more
- [PyVideo](http://pyvideo.org/)
- [Beginner's Guide](https://wiki.python.org/moin/BeginnersGuide)
- [Hitchhiker's Guide](http://docs.python-guide.org/en/latest/)
- [DDL](http://d8ndl.org/), [PyOhio](http://pyohio.org/)
- @catherinedevlin
| github_jupyter |
# Random Forest
Aplicação do random forest em uma mão de poker
***Dataset:*** https://archive.ics.uci.edu/ml/datasets/Poker+Hand
***Apresentação:*** https://docs.google.com/presentation/d/1zFS4cTf9xwvcVPiCOA-sV_RFx_UeoNX2dTthHkY9Am4/edit
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.utils import column_or_1d
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.preprocessing import OneHotEncoder
from sklearn.ensemble import AdaBoostClassifier
from sklearn.metrics import confusion_matrix
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import cross_val_score
import seaborn as sn
import timeit
from format import format_poker_data
train_data = pd.read_csv('train.csv')
test_data = pd.read_csv('test.csv')
X_train, y_train = np.split(train_data,[-1],axis=1)
test_data = test_data.dropna()
X_test, y_test = np.split(test_data,[-1],axis=1)
start_time = timeit.default_timer()
X_train , equal_suit_train = format_poker_data(X_train)
elapsed = timeit.default_timer() - start_time
print(str(elapsed)+" ns")
X_test , equal_suit_test = format_poker_data(X_test)
rf = RandomForestClassifier(n_estimators=50,random_state=42)
rf2 = RandomForestClassifier(n_estimators=50,random_state=42)
y_train = column_or_1d(y_train)
y_test = column_or_1d(y_test)
rf.fit(X_train,y_train)
rf.score(X_train,y_train)
rf.score(X_test,y_test)
n_data_train = pd.DataFrame()
n_data_train['predict'] = rf.predict(X_train)
n_data_train['is_the_same'] = equal_suit_train
n_data_train.shape
n_data_test = pd.DataFrame()
n_data_test['predict'] = rf.predict(X_test)
n_data_test['is_the_same'] = equal_suit_test
n_data_train.head()
n_data_train = pd.get_dummies(n_data_train,columns=['predict']).astype('bool')
n_data_test = pd.get_dummies(n_data_test,columns=['predict']).astype('bool')
rf2.fit(n_data_train,y_train)
rf2.score(n_data_train,y_train)
rf2.score(n_data_test,y_test)
#Confusion Matrixfor Test Data
conf_array_test = confusion_matrix(y_test,rf2.predict(n_data_test))
conf_array_test = conf_array_test / conf_array_test.astype(np.float).sum(axis=1)
df_class_test = pd.DataFrame(conf_array_test, range(10),range(10))
sn.set(font_scale=0.7)#for label size
sn.heatmap(df_class_test,annot=True)# font size
```
| github_jupyter |
```
import numpy as np
import matplotlib.pylab as plt
def Weight(phi,A=5, phi_o=0):
return 1-(0.5*np.tanh(A*((np.abs(phi)-phi_o)))+0.5)
def annot_max(x,y, ax=None):
x=np.array(x)
y=np.array(y)
xmax = x[np.argmax(y)]
ymax = y.max()
text= "x={:.3f}, y={:.3f}".format(xmax, ymax)
if not ax:
ax=plt.gca()
bbox_props = dict(boxstyle="square,pad=0.3", fc="w", ec="k", lw=0.72)
arrowprops=dict(arrowstyle="->",connectionstyle="angle,angleA=0,angleB=60")
kw = dict(xycoords='data',textcoords="axes fraction",
arrowprops=arrowprops, bbox=bbox_props, ha="right", va="top")
ax.annotate(text, xy=(xmax, ymax), xytext=(0.94,0.96), **kw)
def plotweighting(philist, A, p, delta, ctephi_o, enumeration):
label=enumeration+r" $w(\phi,\phi_o=$"+"$\delta$"+r"$ \cdot $"+"{cte}".format(cte=ctephi_o)+r"$,A = $"+"{A}".format(A=A)+r"$\frac{p}{\delta})$"+"\np = {p}, $\delta$ = {delta}m".format(p=p,delta=delta)
plt.plot(philist,[Weight(phi, A = A*p/delta, phi_o = phi_o) for phi in philist], label = label)
plt.figure(figsize= [10, 4],dpi=100)
delta = 50.05;
philist=np.arange(-(delta+10),(delta+10),.5).tolist()
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = 0.65; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(A)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 12; p = 2; ctephi_o = 0.85; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(B)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 8; p = 2; ctephi_o = 0.80; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(C)")
plt.axvline([delta],c="k");plt.axvline([-delta],c="k")
plt.xlabel("$\phi$")
plt.title(r"$w(\phi,\phi_o,A)=1-(0.5\ tanh(A(|\phi|-\phi_o))+0.5)$")
plt.grid()
plt.legend()
plt.show()
plt.figure(figsize= [10, 4],dpi=100)
delta = 50.05;
philist=np.arange(-(delta+10),(delta+10),.5).tolist()
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = .999; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(D)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = .65; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(A)")
plt.axvline([delta],c="k");plt.axvline([-delta],c="k")
plt.xlabel("$\phi$")
plt.title(r"$w(\phi,\phi_o,A)=1-(0.5\ tanh(A(|\phi|-\phi_o))+0.5)$")
plt.grid()
plt.legend()
plt.show()
plt.figure(figsize= [10, 4],dpi=100)
delta = 50.05;
philist=np.arange(-(delta+10),(delta+10),.5).tolist()
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = .65; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(A)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = .75; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(.75)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = .85; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(.85)")
plt.axvline([delta],c="k");plt.axvline([-delta],c="k")
plt.xlabel("$\phi$")
plt.title(r"$w(\phi,\phi_o,A)=1-(0.5\ tanh(A(|\phi|-\phi_o))+0.5)$")
plt.grid()
plt.legend()
plt.show()
plt.figure(figsize= [10, 4],dpi=100)
delta = 50.05;
philist=np.arange(-(delta+10),(delta+10),.5).tolist()
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 2; ctephi_o = .85; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(A)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 6; p = 2; ctephi_o = .75; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(?)")
plt.axvline([delta],c="k");plt.axvline([-delta],c="k")
plt.xlabel("$\phi$")
plt.title(r"$w(\phi,\phi_o,A)=1-(0.5\ tanh(A(|\phi|-\phi_o))+0.5)$")
plt.grid()
plt.legend()
plt.show()
plt.figure(figsize= [10, 4],dpi=100)
delta = 50.05;
philist=np.arange(-(delta+10),(delta+10),.5).tolist()
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 6.5; p = 2; ctephi_o = .85; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(A)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 6; p = 3; ctephi_o = .65; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(B)")
#%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
A = 4; p = 3; ctephi_o = .65; phi_o = delta*ctephi_o;
plotweighting(philist, A, p, delta, ctephi_o,"(C)")
plt.axvline([delta],c="k");plt.axvline([-delta],c="k")
plt.xlabel("$\phi$")
plt.title(r"$w(\phi,\phi_o,A)=1-(0.5\ tanh(A(|\phi|-\phi_o))+0.5)$")
plt.grid()
plt.legend()
plt.show()
```
| github_jupyter |
# Description
This task is to do an exploratory data analysis on the balance-scale dataset
## Data Set Information
This data set was generated to model psychological experimental results. Each example is classified as having the balance scale tip to the right, tip to the left, or be balanced. The attributes are the left weight, the left distance, the right weight, and the right distance. The correct way to find the class is the greater of (left-distance left-weight) and (right-distance right-weight). If they are equal, it is balanced.
### Attribute Information:-
1. Class Name: 3 (L, B, R)
2. Left-Weight: 5 (1, 2, 3, 4, 5)
3. Left-Distance: 5 (1, 2, 3, 4, 5)
4. Right-Weight: 5 (1, 2, 3, 4, 5)
5. Right-Distance: 5 (1, 2, 3, 4, 5)
```
#importing libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
#reading the data
data=pd.read_csv('balance-scale.data')
#shape of the data
data.shape
#first five rows of the data
data.head()
#Generating the x values
x=data.drop(['Class'],axis=1)
x.head()
#Generating the y values
y=data['Class']
y.head()
#Checking for any null data in x
x.isnull().any()
#Checking for any null data in y
y.isnull().any()
#Adding left and right torque as a new data frame
x1=pd.DataFrame()
x1['LT']=x['LW']*x['LD']
x1['RT']=x['RW']*x['RD']
x1.head()
#Converting the results of "Classs" attribute ,i.e., Balanced(B), Left(L) and Right(R) to numerical values for computation in sklearn
y=y.map(dict(B=0,L=1,R=2))
y.head()
```
### Using the Weight and Distance parameters
Splitting the data set into a ratio of 70:30 by the built in 'train_test_split' function in sklearn to get a better idea of accuracy of the model
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(x,y,stratify=y, test_size=0.3, random_state=2)
X_train.describe()
#Importing decision tree classifier and creating it's object
from sklearn.tree import DecisionTreeClassifier
clf= DecisionTreeClassifier()
clf.fit(X_train,y_train)
y_pred=clf.predict(X_test)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred)
```
We observe that the accuracy score is pretty low. Thus, we need to find optimal parameters to get the best accuracy. We do that by using GridSearchCV
```
#Using GridSearchCV to find the maximun optimal depth
from sklearn.model_selection import GridSearchCV
tree_para={"criterion":["gini","entropy"], "max_depth":[3,4,5,6,7,8,9,10,11,12]}
dt_model_grid= GridSearchCV(DecisionTreeClassifier(random_state=3),tree_para, cv=10)
dt_model_grid.fit(X_train,y_train)
# To print the optimum parameters computed by GridSearchCV required for best accuracy score
dt_model=dt_model_grid.best_estimator_
print(dt_model)
#To find the best accuracy score for all possible combinations of parameters provided
dt_model_grid.best_score_
dt_model_grid.best_params_
#Scoring the model
from sklearn.metrics import classification_report
y_pred1=dt_model.predict(X_test)
print(classification_report(y_test,y_pred1,target_names=["Balanced","Left","Right"]))
from sklearn import tree
!pip install graphviz
#Plotting the Tree
from sklearn.tree import export_graphviz
export_graphviz(
dt_model,
out_file=("model1.dot"),
feature_names=["Left Weight","Left Distance","Right Weight","Right Distance"],
class_names=["Balanced","Left","Right"],
filled=True)
#Run this to print png
# !dot -Tpng model1.dot -o model1.png
```
## Using the created Torque
```
dt_model2 = DecisionTreeClassifier(random_state=31)
X_train, X_test, y_train, y_test= train_test_split(x1,y, stratify=y, test_size=0.3, random_state=8)
X_train.head(
)
X_train.shape
dt_model2.fit(X_train, y_train)
y_pred2= dt_model2.predict(X_test)
print(classification_report(y_test, y_pred2, target_names=["Balanced","Left","Right"]))
#Plotting the Tree
from sklearn import export_graphviz
export_graphviz(
dt_model2,
out_file=("model2.dot"),
feature_names=["Left Torque", "Right Torque"],
class_names=["Balanced","Left","Right"],
filled=True)
# run this to make png
# dot -Tpng model2.dot -o model2.png
```
## Increasing the optimization
After observing the trees, we conclude that differences are not being taken into account. Hence, we add the differences attribute to try and increase the accuracy.
```
x1['Diff']= x1['LT']- x1['RT']
x1.head()
X_train, X_test, y_train, y_test =train_test_split(x1,y, stratify=y, test_size=0.3,random_state=40)
dt_model3= DecisionTreeClassifier(random_state=40)
dt_model3.fit(X_train, y_train)
#Create Classification Report
y_pred3= dt_model3.predict(X_test)
print(classification_report(y_test, y_pred3, target_names=["Balanced", "Left", "Right"]))
#Plotting the tree
from sklearn.tree import export_graphviz
export_graphviz(
dt_model3
out_file=("model3.dot"),
feature_names=["Left Torque","Right Torque","Difference"],
class_names=["Balanced","Left","Right"]
filled=True)
# run this to make png
# dot -Tpng model3.dot -o model3.png
from sklearn.metrics import accuracy_score
accuracy_score(y_pred3,y_test)
```
## Final Conclusion
The model returns a perfect accuracy score as desired.
```
!pip install seaborn
```
| github_jupyter |
# Summarizing Emails using Machine Learning: Data Wrangling
## Table of Contents
1. Imports & Initalization <br>
2. Data Input <br>
A. Enron Email Dataset <br>
B. BC3 Corpus <br>
3. Preprocessing <br>
A. Data Cleaning. <br>
B. Sentence Cleaning <br>
C. Tokenizing <br>
4. Store Data <br>
A. Locally as pickle <br>
B. Into database <br>
5. Data Exploration <br>
A. Enron Emails <br>
B. BC3 Corpus <br>
The goal of this notebook is to clean both the Enron Email and BC3 Corpus data sets to perform email text summarization. The BC3 Corpus contains human summarizations that can be used to calculate ROUGE metrics to better understand how accurate the summarizations are. The Enron dataset is far more comprehensive, but lacks summaries to test against.
You can find the text summarization notebook that uses the preprocessed data [here.](https://github.com/dailykirt/ML_Enron_email_summary/blob/master/notebooks/Text_rank_summarization.ipynb)
A visual summary of the preprocessing steps are in the figure below.
<img src="./images/Preprocess_Flow.jpg">
## 1. Imports & Initalization
```
import sys
from os import listdir
from os.path import isfile, join
import configparser
from sqlalchemy import create_engine
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import email
import mailparser
import xml.etree.ElementTree as ET
from talon.signature.bruteforce import extract_signature
from nltk.tokenize import word_tokenize, sent_tokenize
from nltk.corpus import stopwords
import re
import dask.dataframe as dd
from distributed import Client
import multiprocessing as mp
#Set local location of emails.
mail_dir = '../data/maildir/'
#mail_dir = '../data/testdir/'
```
## 2. Data Input
### A. Enron Email Dataset
The raw enron email dataset contains a maildir directory that contains folders seperated by employee which contain the emails. The following processes the raw text of each email into a dask dataframe with the following columns:
Employee: The username of the email owner. <br>
Body: Cleaned body of the email. <br>
Subject: The title of the email. <br>
From: The original sender of the email <br>
Message-ID: Used to remove duplicate emails, as each email has a unique ID. <br>
Chain: The parsed out email chain from a email that was forwarded. <br>
Signature: The extracted signature from the body.<br>
Date: Time the email was sent. <br>
All of the Enron emails were sent using the Multipurpose Internet Mail Extensions 1.0 (MIME) format. Keeping this in mind helps find the correct libraries and methods to clean the emails in a standardized fashion.
```
def process_email(index):
'''
This function splits a raw email into constituent parts that can be used as features.
'''
email_path = index[0]
employee = index[1]
folder = index[2]
mail = mailparser.parse_from_file(email_path)
full_body = email.message_from_string(mail.body)
#Only retrieve the body of the email.
if full_body.is_multipart():
return
else:
mail_body = full_body.get_payload()
split_body = clean_body(mail_body)
headers = mail.headers
#Reformating date to be more pandas readable
date_time = process_date(headers.get('Date'))
email_dict = {
"employee" : employee,
"email_folder": folder,
"message_id": headers.get('Message-ID'),
"date" : date_time,
"from" : headers.get('From'),
"subject": headers.get('Subject'),
"body" : split_body['body'],
"chain" : split_body['chain'],
"signature": split_body['signature'],
"full_email_path" : email_path #for debug purposes.
}
#Append row to dataframe.
return email_dict
def clean_body(mail_body):
'''
This extracts both the email signature, and the forwarding email chain if it exists.
'''
delimiters = ["-----Original Message-----","To:","From"]
#Trying to split string by biggest delimiter.
old_len = sys.maxsize
for delimiter in delimiters:
split_body = mail_body.split(delimiter,1)
new_len = len(split_body[0])
if new_len <= old_len:
old_len = new_len
final_split = split_body
#Then pull chain message
if (len(final_split) == 1):
mail_chain = None
else:
mail_chain = final_split[1]
#The following uses Talon to try to get a clean body, and seperate out the rest of the email.
clean_body, sig = extract_signature(final_split[0])
return {'body': clean_body, 'chain' : mail_chain, 'signature': sig}
def process_date(date_time):
'''
Converts the MIME date format to a more pandas friendly type.
'''
try:
date_time = email.utils.format_datetime(email.utils.parsedate_to_datetime(date_time))
except:
date_time = None
return date_time
def generate_email_paths(mail_dir):
'''
Given a mail directory, this will generate the file paths to each email in each inbox.
'''
mailboxes = listdir(mail_dir)
for mailbox in mailboxes:
inbox = listdir(mail_dir + mailbox)
for folder in inbox:
path = mail_dir + mailbox + "/" + folder
emails = listdir(path)
for single_email in emails:
full_path = path + "/" + single_email
if isfile(full_path): #Skip directories.
yield (full_path, mailbox, folder)
#Use multiprocessing to speed up initial data load and processing. Also helps partition DASK dataframe.
try:
cpus = mp.cpu_count()
except NotImplementedError:
cpus = 2
pool = mp.Pool(processes=cpus)
print("CPUS: " + str(cpus))
indexes = generate_email_paths(mail_dir)
enron_email_df = pool.map(process_email,indexes)
#Remove Nones from the list
enron_email_df = [i for i in enron_email_df if i]
enron_email_df = pd.DataFrame(enron_email_df)
enron_email_df.describe()
```
### B. BC3 Corpus
This dataset is split into two xml files. One contains the original emails split line by line, and the other contains the summarizations created by the annotators. Each email may contain several summarizations from different annotators and summarizations may also be over several emails. This will create a data frame for both xml files, then join them together using the thread number in combination of the email number for a single final dataframe.
The first dataframe will contain the wrangled original emails containing the following information:
Listno: Thread identifier <br>
Email_num: Email in thread sequence <br>
From: The original sender of the email <br>
To: The recipient of the email. <br>
Recieved: Time email was recieved. <br>
Subject: Title of email. <br>
Body: Original body. <br>
```
def parse_bc3_emails(root):
'''
This adds every BC3 email to a newly created dataframe.
'''
BC3_email_list = []
#The emails are seperated by threads.
for thread in root:
email_num = 0
#Iterate through the thread elements <name, listno, Doc>
for thread_element in thread:
#Getting the listno allows us to link the summaries to the correct emails
if thread_element.tag == "listno":
listno = thread_element.text
#Each Doc element is a single email
if thread_element.tag == "DOC":
email_num += 1
email_metadata = []
for email_attribute in thread_element:
#If the email_attri is text, then each child contains a line from the body of the email
if email_attribute.tag == "Text":
email_body = ""
for sentence in email_attribute:
email_body += sentence.text
else:
#The attributes of the Email <Recieved, From, To, Subject, Text> appends in this order.
email_metadata.append(email_attribute.text)
#Use same enron cleaning methods on the body of the email
split_body = clean_body(email_body)
email_dict = {
"listno" : listno,
"date" : process_date(email_metadata[0]),
"from" : email_metadata[1],
"to" : email_metadata[2],
"subject" : email_metadata[3],
"body" : split_body['body'],
"email_num": email_num
}
BC3_email_list.append(email_dict)
return pd.DataFrame(BC3_email_list)
#load BC3 Email Corpus. Much smaller dataset has no need for parallel processing.
parsedXML = ET.parse( "../data/BC3_Email_Corpus/corpus.xml" )
root = parsedXML.getroot()
#Clean up BC3 emails the same way as the Enron emails.
bc3_email_df = parse_bc3_emails(root)
bc3_email_df.info()
bc3_email_df.head(3)
```
The second dataframe contains the summarizations of each email:
Annotator: Person who created summarization. <br>
Email_num: Email in thread sequence. <br>
Listno: Thread identifier. <br>
Summary: Human summarization of the email. <br>
```
def parse_bc3_summaries(root):
'''
This parses every BC3 Human summary that is contained in the dataset.
'''
BC3_summary_list = []
for thread in root:
#Iterate through the thread elements <listno, name, annotation>
for thread_element in thread:
if thread_element.tag == "listno":
listno = thread_element.text
#Each Doc element is a single email
if thread_element.tag == "annotation":
for annotation in thread_element:
#If the email_attri is summary, then each child contains a summarization line
if annotation.tag == "summary":
summary_dict = {}
for summary in annotation:
#Generate the set of emails the summary sentence belongs to (often a single email)
email_nums = summary.attrib['link'].split(',')
s = set()
for num in email_nums:
s.add(num.split('.')[0].strip())
#Remove empty strings, since they summarize whole threads instead of emails.
s = [x for x in set(s) if x]
for email_num in s:
if email_num in summary_dict:
summary_dict[email_num] += ' ' + summary.text
else:
summary_dict[email_num] = summary.text
#get annotator description
elif annotation.tag == "desc":
annotator = annotation.text
#For each email summarizaiton create an entry
for email_num, summary in summary_dict.items():
email_dict = {
"listno" : listno,
"annotator" : annotator,
"email_num" : email_num,
"summary" : summary
}
BC3_summary_list.append(email_dict)
return pd.DataFrame(BC3_summary_list)
#Load summaries and process
parsedXML = ET.parse( "../data/BC3_Email_Corpus/annotation.xml" )
root = parsedXML.getroot()
bc3_summary_df = parse_bc3_summaries(root)
bc3_summary_df['email_num'] = bc3_summary_df['email_num'].astype(int)
bc3_summary_df.info()
#merge the dataframes together
bc3_df = pd.merge(bc3_email_df,
bc3_summary_df[['annotator', 'email_num', 'listno', 'summary']],
on=['email_num', 'listno'])
bc3_df.head()
```
## 3. Preprocessing
### A. Data Cleaning
```
#Convert date to pandas datetime.
enron_email_df['date'] = pd.to_datetime(enron_email_df['date'], utc=True)
bc3_df['date'] = pd.to_datetime(bc3_df.date, utc=True)
#Look at the timeframe
start_date = str(enron_email_df.date.min())
end_date = str(enron_email_df.date.max())
print("Start Date: " + start_date)
print("End Date: " + end_date)
```
Since the Enron data was collected in May 2002 according to wikipedia its a bit strange to see emails past that date. Reading some of the emails seem to suggest it's mostly spam.
```
enron_email_df[(enron_email_df.date > '2003-01-01')].head()
#Quick look at emails before 1999,
enron_email_df[(enron_email_df.date < '1999-01-01')].date.value_counts().head()
enron_email_df[(enron_email_df.date == '1980-01-01')].head()
```
There seems to be a glut of emails dated exactly on 1980-01-01. The emails seem legitimate, but these should be droped since without the true date we won't be able to figure out where the email fits in the context of a batch of summaries. Keep emails between Jan 1st 1999 and June 1st 2002.
```
enron_email_df = enron_email_df[(enron_email_df.date > '1998-01-01') & (enron_email_df.date < '2002-06-01')]
```
### B. Sentence Cleaning
The raw enron email Corpus tends to have a large amount of unneeded characters that can interfere with tokenizaiton. It's best to do a bit more cleaning.
```
def clean_email_df(df):
'''
These remove symbols and character patterns that don't aid in producing a good summary.
'''
#Removing strings related to attatchments and certain non numerical characters.
patterns = ["\[IMAGE\]","-", "_", "\*", "+","\".\""]
for pattern in patterns:
df['body'] = pd.Series(df['body']).str.replace(pattern, "")
#Remove multiple spaces.
df['body'] = df['body'].replace('\s+', ' ', regex=True)
#Blanks are replaced with NaN in the whole dataframe. Then rows with a 'NaN' in the body will be dropped.
df = df.replace('',np.NaN)
df = df.dropna(subset=['body'])
#Remove all Duplicate emails
#df = df.drop_duplicates(subset='body')
return df
#Apply clean to both datasets.
enron_email_df = clean_email_df(enron_email_df)
bc3_df = clean_email_df(bc3_df)
```
### C. Tokenizing
It's important to split up sentences into it's constituent parts for the ML algorithim that will be used for text summarization. This will aid in further processing like removing extra whitespace. We can also remove stopwords, which are very commonly used words that don't provide additional sentence meaning like 'and' 'or' and 'the'. This will be applied to both the Enron and BC3 datasets.
```
def remove_stopwords(sen):
'''
This function removes stopwords
'''
stop_words = stopwords.words('english')
sen_new = " ".join([i for i in sen if i not in stop_words])
return sen_new
def tokenize_email(text):
'''
This function splits up the body into sentence tokens and removes stop words.
'''
clean_sentences = sent_tokenize(text, language='english')
#removing punctuation, numbers and special characters. Then lowercasing.
clean_sentences = [re.sub('[^a-zA-Z ]', '',s) for s in clean_sentences]
clean_sentences = [s.lower() for s in clean_sentences]
clean_sentences = [remove_stopwords(r.split()) for r in clean_sentences]
return clean_sentences
```
Starting with the Enron dataset.
```
#This tokenizing will be the extracted sentences that may be chosen to form the email summaries.
enron_email_df['extractive_sentences'] = enron_email_df['body'].apply(sent_tokenize)
#Splitting the text in emails into cleaned sentences
enron_email_df['tokenized_body'] = enron_email_df['body'].apply(tokenize_email)
#Tokenizing the bodies might have revealed more duplicate emails that should be droped.
enron_email_df = enron_email_df.loc[enron_email_df.astype(str).drop_duplicates(subset='tokenized_body').index]
```
Now working on the BC3 Dataset.
```
bc3_df['extractive_sentences'] = bc3_df['body'].apply(sent_tokenize)
bc3_df['tokenized_body'] = bc3_df['body'].apply(tokenize_email)
#bc3_email_df = bc3_email_df.loc[bc3_email_df.astype(str).drop_duplicates(subset='tokenized_body').index]
```
## Store Data
### A. Locally as pickle
After all the preprocessing is finished its best to store the the data so it can be quickly and easily retrieved by other software. Pickles are best used if you are working locally and want a simple way to store and load data. You can also use a cloud database that can be accessed by other production services such as Heroku to retrieve the data. In this case, I load the data up into a AWS postgres database.
```
#Local locations for pickle files.
ENRON_PICKLE_LOC = "../data/dataframes/wrangled_enron_full_df.pkl"
BC3_PICKLE_LOC = "../data/dataframes/wrangled_BC3_df.pkl"
#Store dataframes to disk
enron_email_df.to_pickle(ENRON_PICKLE_LOC)
bc3_df.head()
bc3_df.to_pickle(BC3_PICKLE_LOC)
```
### B. Into database
I used a Postgres database with the DB configurations stored in a config_notebook.ini file. This allows me to easily switch between local and AWS configurations.
```
#Configure postgres database
config = configparser.ConfigParser()
config.read('config_notebook.ini')
#database_config = 'LOCAL_POSTGRES'
database_config = 'AWS_POSTGRES'
POSTGRES_ADDRESS = config[database_config]['POSTGRES_ADDRESS']
POSTGRES_USERNAME = config[database_config]['POSTGRES_USERNAME']
POSTGRES_PASSWORD = config[database_config]['POSTGRES_PASSWORD']
POSTGRES_DBNAME = config[database_config]['POSTGRES_DBNAME']
#now create database connection
postgres_str = ('postgresql+psycopg2://{username}:{password}@{ipaddress}/{dbname}'
.format(username=POSTGRES_USERNAME,
password=POSTGRES_PASSWORD,
ipaddress=POSTGRES_ADDRESS,
dbname=POSTGRES_DBNAME))
cnx = create_engine(postgres_str)
#Store data.
enron_email_df.to_sql('full_enron_emails', cnx)
```
## 5. Data Exploration
Exploring the dataset can go a long way to building more accurate machine learning models and spotting any possible issues with the dataset. Since the Enron dataset is quite large, we can speed up some of our computations by using Dask. While not strictly necessary, iterating on this dataset should be much faster.
### A. Enron Emails
```
client = Client(processes = True)
client.cluster
#Make into dask dataframe.
enron_email_df = dd.from_pandas(enron_email_df, npartitions=cpus)
enron_email_df.columns
#Used to create a describe summary of the dataset. Ignoring tokenized columns.
enron_email_df[['body', 'chain', 'date', 'email_folder', 'employee', 'from', 'full_email_path', 'message_id', 'signature', 'subject']].describe().compute()
#Get word frequencies from tokenized word lists
def get_word_freq(df):
freq_words=dict()
for tokens in df.tokenized_words.compute():
for token in tokens:
if token in freq_words:
freq_words[token] += 1
else:
freq_words[token] = 1
return freq_words
def tokenize_word(sentences):
tokens = []
for sentence in sentences:
tokens = word_tokenize(sentence)
return tokens
#Tokenize the sentences
enron_email_df['tokenized_words'] = enron_email_df['tokenized_body'].apply(tokenize_word).compute()
#Creating word dictionary to understand word frequencies.
freq_words = get_word_freq(enron_email_df)
print('Unique words: {:,}'.format(len(freq_words)))
word_data = []
#Sort dictionary by highest word frequency.
for key, value in sorted(freq_words.items(), key=lambda item: item[1], reverse=True):
word_data.append([key, freq_words[key]])
#Prepare to plot bar graph of top words.
#Create dataframe with Word and Frequency, then sort in Descending order.
freq_words_df = pd.DataFrame.from_dict(freq_words, orient='index').reset_index()
freq_words_df = freq_words_df.rename(columns={"index": "Word", 0: "Frequency"})
freq_words_df = freq_words_df.sort_values(by=['Frequency'],ascending = False)
freq_words_df.reset_index(drop = True, inplace=True)
freq_words_df.head(30).plot(x='Word', kind='bar', figsize=(20,10))
```
### B. BC3 Corpus
```
bc3_df.head()
bc3_df['to'].value_counts().head()
```
| github_jupyter |
```
# Load essential libraries
import csv
import numpy as np
import matplotlib.pyplot as plt
import statistics
import numpy as np
from scipy.signal import butter, lfilter, freqz
from IPython.display import Image
from datetime import datetime
# Time and robot egomotion
time = []
standardized_time = []
standardized_time2 = []
compass_heading = []
speed = []
# sonde data
temp = []
PH = []
cond = [] # ms
chlorophyll = []
ODO = [] # mg/L
sonar = []
angular_z = []
# wp data
wp_time = []
wp_seq = []
initial_time = None
time_crop = 4000
time_crop1 = 580
time_crop2 = 800
# File loading from relative path
file = '../../../Data/ISER2021/Sunapee-20200715-path-1.csv'
# File loading from relative path
file2 = '../../../Data/ISER2021/Sunapee-20200715-path-1-mavros.csv'
# original data
with open(file, 'r') as csvfile:
csvreader= csv.reader(csvfile, delimiter=',')
header = next(csvreader)
for row in csvreader:
# robot data
if initial_time is None:
initial_time = float(row[0])
current_time = float(row[0])
if current_time - initial_time >= time_crop1 and current_time - initial_time < time_crop2:
#if current_time - initial_time <= time_crop:
time.append(float(row[0]))
compass_heading.append(float(row[4]))
speed.append(float(row[10]))
angular_z.append(float(row[18]))
# sonde data
temp.append(float(row[23]))
PH.append(float(row[26]))
cond.append(float(row[25]))
chlorophyll.append(float(row[29]))
ODO.append(float(row[30]))
sonar.append(float(row[8]))
minimum_time = min(time)
for time_stamp in time:
standardized_time.append(time_stamp - minimum_time)
# wp data
with open(file2, 'r') as csvfile2:
csvreader2 = csv.reader(csvfile2, delimiter=',')
header = next(csvreader2)
for row in csvreader2:
current_time = float(row[0])
#if current_time - initial_time <= time_crop:
if current_time - initial_time >= time_crop1 and current_time - initial_time < time_crop2:
wp_time.append(float(row[0]))
wp_seq.append(float(row[1]))
for time_stamp in wp_time:
standardized_time2.append(time_stamp - minimum_time)
# collision time around 790
```
### Compass heading
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('Heading [degree]', fontsize=16)
ax1.plot(standardized_time, compass_heading, label='compass heading')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('ground_speed_x [m/s]', fontsize=16)
ax1.plot(standardized_time, speed, label='ground_speed_x', color='m')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('angular_z [rad/s]', fontsize=16)
ax1.plot(standardized_time, angular_z, label='angular_z', color='r')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
```
### Temperature
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('Temperature [degree]', fontsize=16)
ax1.plot(standardized_time, temp, label='temp', color='k')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
print("Standard Deviation of the temp is % s " %(statistics.stdev(temp)))
print("Mean of the temp is % s " %(statistics.mean(temp)))
```
### PH
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('PH', fontsize=16)
ax1.plot(standardized_time, PH, label='PH', color='r')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
print("Standard Deviation of the temp is % s " %(statistics.stdev(PH)))
print("Mean of the temp is % s " %(statistics.mean(PH)))
```
### Conductivity
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('Conductivity [ms]', fontsize=16)
ax1.plot(standardized_time, cond, label='conductivity', color='b')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
print("Standard Deviation of the chlorophyll is % s " %(statistics.stdev(cond)))
print("Mean of the chlorophyll is % s " %(statistics.mean(cond)))
```
### Chlorophyll
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('chlorophyll [RFU]', fontsize=16)
ax1.plot(standardized_time, chlorophyll, label='chlorophyll', color='g')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
print("Standard Deviation of the chlorophyll is % s " %(statistics.stdev(chlorophyll)))
print("Mean of the chlorophyll is % s " %(statistics.mean(chlorophyll)))
```
### ODO
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('ODO [%sat]', fontsize=16)
ax1.plot(standardized_time, ODO, label='ODO', color='m')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
print("Standard Deviation of the DO is % s " %(statistics.stdev(ODO)))
print("Mean of the DO is % s " %(statistics.mean(ODO)))
```
### Sonar depth
```
# Figure initialization
fig, ax1 = plt.subplots()
ax1.set_xlabel('Time [sec]', fontsize=16)
ax1.set_ylabel('sonar [m]', fontsize=16)
ax1.plot(standardized_time, sonar, label='sonar', color='c')
ax1.legend()
for wp in standardized_time2:
plt.axvline(x=wp, color='gray', linestyle='--')
plt.show()
```
| github_jupyter |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.