code
stringlengths 2.5k
150k
| kind
stringclasses 1
value |
|---|---|
```
!git clone https://github.com/ninomiyalab/Memory_Less_Momentum_Quasi_Newton
import tensorflow as tf
import tensorflow.keras
from tensorflow.keras.models import Model, load_model
from tensorflow.keras.layers import Input, Dense, Activation, Conv2D, Flatten
from tensorflow.keras import optimizers
from Memory_Less_Momentum_Quasi_Newton.MLQN import *
from Memory_Less_Momentum_Quasi_Newton.MLMoQ import *
import matplotlib.pyplot as plt
import numpy as np
import csv
def compare_MNIST(i = 0):
np.random.seed(i)
mnist = tf.keras.datasets.mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train, x_test = x_train[..., np.newaxis]/255.0, x_test[..., np.newaxis]/255.0
y_train, y_test = tf.keras.utils.to_categorical(y_train), tf.keras.utils.to_categorical(y_test)
#defined Neural Network Model
def My_Model(input_shape, Output_dim):
inputs = Input(shape = (input_shape))
x = Flatten()(inputs)
x = Dense(10, activation="sigmoid")(x)
outputs = Dense(Output_dim, activation="softmax")(x)
model = Model(inputs = [inputs], outputs = [outputs])
return model
model = My_Model(x_train.shape[1:], Output_dim=10)
model.save("model.h5")
loss_fn = tf.keras.losses.CategoricalCrossentropy()
epochs = 2000
#if verbose is True, the results of each iteration will be printed.
verbose = True
#if graph is True, the results of all algorithms will be plotted in the graph.
graph = True
# MLQN Training
# --------------------------------------------------------------------------------------
model = load_model("model.h5")
optimizer = MLQN( )
model.compile(loss=loss_fn, optimizer=optimizer, metrics=['accuracy'])
MLQN_history = model.fit(x_train, y_train, epochs = epochs, verbose = verbose, batch_size = x_train.shape[0], validation_data = (x_test, y_test))
# --------------------------------------------------------------------------------------
# MLMoQ Training
# --------------------------------------------------------------------------------------
model = load_model("model.h5")
optimizer = MLMoQ()
model.compile(loss=loss_fn, optimizer=optimizer, metrics=['accuracy'])
MLMoQ_history = model.fit(x_train, y_train, epochs = epochs, verbose = verbose, batch_size = x_train.shape[0], validation_data = (x_test, y_test))
# --------------------------------------------------------------------------------------
# Adam Training
# --------------------------------------------------------------------------------------
model = load_model("model.h5")
optimizer = tf.keras.optimizers.Adam()
model.compile(loss=loss_fn, optimizer=optimizer, metrics=['accuracy'])
Adam_history = model.fit(x_train, y_train, epochs = epochs, verbose = verbose, batch_size = x_train.shape[0], validation_data = (x_test, y_test))
# --------------------------------------------------------------------------------------
if graph:
fig, (axL, axR) = plt.subplots(ncols=2, figsize=(10,4))
#Train Loss vs. Iteration graph
axL.set_title("Train_Loss")
axL.plot(MLQN_history.history['loss'],color="blue", label="MLQN")
axL.plot(MLMoQ_history.history['loss'], color="m",label="MLMoQ")
axL.plot(Adam_history.history['loss'], color="orange",label="Adam")
axL.set_xlabel('Iterations')
axL.set_ylabel('Train Loss')
axL.legend(bbox_to_anchor=(0, -0.2), loc='upper left', borderaxespad=0)
axL.legend()
#Train Accuracy vs. Iteration graph
axR.set_title("Train_Accuracy")
axR.plot(MLQN_history.history['accuracy'],color="blue", label="MLQN")
axR.plot(MLMoQ_history.history['accuracy'], color="m",label="MLMoQ")
axR.plot(Adam_history.history['accuracy'],color="orange", label="Adam")
axR.set_xlabel('Iterations')
axR.set_ylabel('Train Accuracy')
axR.legend(bbox_to_anchor=(0, -0.2), loc='upper left', borderaxespad=0)
axR.legend()
plt.show()
fig, (axL, axR) = plt.subplots(ncols=2, figsize=(10,4))
#Test Loss vs. Iteration graph
axL.set_title("Test_Loss")
axL.plot(MLQN_history.history['val_loss'],color="blue", label="MLQN")
axL.plot(MLMoQ_history.history['val_loss'],color="m", label="MLMoQ")
axL.plot(Adam_history.history['val_loss'],color="orange",label="Adam")
axL.set_xlabel('Iterations')
axL.set_ylabel('Test Loss')
axL.legend(bbox_to_anchor=(0, -0.2), loc='upper left', borderaxespad=0)
axL.legend()
#Test Accuracy vs. Iteration graph
axR.set_title("Test_Accuracy")
axR.plot(MLQN_history.history['val_accuracy'],color="blue",label="MLQN")
axR.plot(MLMoQ_history.history['val_accuracy'],color="m", label="MLMoQ")
axR.plot(Adam_history.history['val_accuracy'],color="orange", label="Adam")
axR.set_xlabel('Iterations')
axR.set_ylabel('Test Accuracy')
axR.legend(bbox_to_anchor=(0, -0.2), loc='upper left', borderaxespad=0)
axR.legend()
plt.show()
for i in range(10):
print(i + 1)
compare_MNIST(i)
```
| github_jupyter |
<a href="https://colab.research.google.com/github/zevan07/DS-Unit-1-Sprint-3-Statistical-Tests-and-Experiments/blob/master/Copy_of_LS_DS_142_Sampling_Confidence_Intervals_and_Hypothesis_Testing.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Lambda School Data Science Module 142
## Sampling, Confidence Intervals, and Hypothesis Testing
## Prepare - examine other available hypothesis tests
If you had to pick a single hypothesis test in your toolbox, t-test would probably be the best choice - but the good news is you don't have to pick just one! Here's some of the others to be aware of:
```
import numpy as np
from scipy.stats import chisquare # One-way chi square test
# Chi square can take any crosstab/table and test the independence of rows/cols
# The null hypothesis is that the rows/cols are independent -> low chi square
# The alternative is that there is a dependence -> high chi square
# Be aware! Chi square does *not* tell you direction/causation
ind_obs = np.array([[1, 1], [2, 2]]).T
print(ind_obs)
print(chisquare(ind_obs, axis=None))
dep_obs = np.array([[16, 18, 16, 14, 12, 12], [32, 24, 16, 28, 20, 24]]).T
print(dep_obs)
print(chisquare(dep_obs, axis=None))
# Distribution tests:
# We often assume that something is normal, but it can be important to *check*
# For example, later on with predictive modeling, a typical assumption is that
# residuals (prediction errors) are normal - checking is a good diagnostic
from scipy.stats import normaltest
# Poisson models arrival times and is related to the binomial (coinflip)
sample = np.random.poisson(5, 1000)
print(normaltest(sample)) # Pretty clearly not normal
# Kruskal-Wallis H-test - compare the median rank between 2+ groups
# Can be applied to ranking decisions/outcomes/recommendations
# The underlying math comes from chi-square distribution, and is best for n>5
from scipy.stats import kruskal
x1 = [1, 3, 5, 7, 9]
y1 = [2, 4, 6, 8, 10]
print(kruskal(x1, y1)) # x1 is a little better, but not "significantly" so
x2 = [1, 1, 1]
y2 = [2, 2, 2]
z = [2, 2] # Hey, a third group, and of different size!
print(kruskal(x2, y2, z)) # x clearly dominates
```
And there's many more! `scipy.stats` is fairly comprehensive, though there are even more available if you delve into the extended world of statistics packages. As tests get increasingly obscure and specialized, the importance of knowing them by heart becomes small - but being able to look them up and figure them out when they *are* relevant is still important.
## Live Lecture - let's explore some more of scipy.stats
Candidate topics to explore:
- `scipy.stats.chi2` - the Chi-squared distribution, which we can use to reproduce the Chi-squared test
- Calculate the Chi-Squared test statistic "by hand" (with code), and feed it into `chi2`
- Build a confidence interval with `stats.t.ppf`, the t-distribution percentile point function (the inverse of the CDF) - we can write a function to return a tuple of `(mean, lower bound, upper bound)` that you can then use for the assignment (visualizing confidence intervals)
```
# Taking requests! Come to lecture with a topic or problem and we'll try it.
```
## Assignment - Build a confidence interval
A confidence interval refers to a neighborhood around some point estimate, the size of which is determined by the desired p-value. For instance, we might say that 52% of Americans prefer tacos to burritos, with a 95% confidence interval of +/- 5%.
52% (0.52) is the point estimate, and +/- 5% (the interval $[0.47, 0.57]$) is the confidence interval. "95% confidence" means a p-value $\leq 1 - 0.95 = 0.05$.
In this case, the confidence interval includes $0.5$ - which is the natural null hypothesis (that half of Americans prefer tacos and half burritos, thus there is no clear favorite). So in this case, we could use the confidence interval to report that we've failed to reject the null hypothesis.
But providing the full analysis with a confidence interval, including a graphical representation of it, can be a helpful and powerful way to tell your story. Done well, it is also more intuitive to a layperson than simply saying "fail to reject the null hypothesis" - it shows that in fact the data does *not* give a single clear result (the point estimate) but a whole range of possibilities.
How is a confidence interval built, and how should it be interpreted? It does *not* mean that 95% of the data lies in that interval - instead, the frequentist interpretation is "if we were to repeat this experiment 100 times, we would expect the average result to lie in this interval ~95 times."
For a 95% confidence interval and a normal(-ish) distribution, you can simply remember that +/-2 standard deviations contains 95% of the probability mass, and so the 95% confidence interval based on a given sample is centered at the mean (point estimate) and has a range of +/- 2 (or technically 1.96) standard deviations.
Different distributions/assumptions (90% confidence, 99% confidence) will require different math, but the overall process and interpretation (with a frequentist approach) will be the same.
Your assignment - using the data from the prior module ([congressional voting records](https://archive.ics.uci.edu/ml/datasets/Congressional+Voting+Records)):
1. Generate and numerically represent a confidence interval
2. Graphically (with a plot) represent the confidence interval
3. Interpret the confidence interval - what does it tell you about the data and its distribution?
Stretch goals:
1. Write a summary of your findings, mixing prose and math/code/results. *Note* - yes, this is by definition a political topic. It is challenging but important to keep your writing voice *neutral* and stick to the facts of the data. Data science often involves considering controversial issues, so it's important to be sensitive about them (especially if you want to publish).
2. Apply the techniques you learned today to your project data or other data of your choice, and write/discuss your findings here.
3. Refactor your code so it is elegant, readable, and can be easily run for all issues.
```
import pandas as pd
import numpy as np
from scipy import stats
from scipy.stats import normaltest
from scipy.stats import kruskal
from random import randint
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
# the data file does not have a header so we'll need to create one
# attribute info copy and pasted from name file
attribute_info = '''1. Class-Name: 2 (democrat, republican)
2. handicapped-infants: 2 (y,n)
3. water-project-cost-sharing: 2 (y,n)
4. adoption-of-the-budget-resolution: 2 (y,n)
5. physician-fee-freeze: 2 (y,n)
6. el-salvador-aid: 2 (y,n)
7. religious-groups-in-schools: 2 (y,n)
8. anti-satellite-test-ban: 2 (y,n)
9. aid-to-nicaraguan-contras: 2 (y,n)
10. mx-missile: 2 (y,n)
11. immigration: 2 (y,n)
12. synfuels-corporation-cutback: 2 (y,n)
13. education-spending: 2 (y,n)
14. superfund-right-to-sue: 2 (y,n)
15. crime: 2 (y,n)
16. duty-free-exports: 2 (y,n)
17. export-administration-act-south-africa: 2 (y,n)'''
# clean up attribute info to use for column headers
names = (attribute_info.replace(': 2 (y,n)', ' ')
.replace(': 2 (democrat, republican)', ' ')
.replace('.', ' ')
.split())
# finish cleaning by getting rid of numbers
for x in names:
nums = [str(x) for x in range(0, 18)]
if x in nums:
names.remove(x)
# import the csv without the first row as a header
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/voting-records/house-votes-84.data', header=None)
# add header (names)
df.columns = names
# replace all 'y', 'n', and '?' values with python friendly values
# replaced '?' with random numbers to avoid NaNs
df = df.replace({'y': 1, 'n': 0, '?': randint(0,1)})
print(df.shape)
# create dataframes for each party
rep = df[df['Class-Name'] == 'republican']
dem = df[df['Class-Name'] == 'democrat']
# create a function to get mean, confidence interval, and the interval (for use in graphing)
def confidence_interval(data, confidence = 0.95):
data = np.array(data)
mean = np.mean(data)
n = len(data)
stderr = stats.sem(data)
interval = stderr * stats.t.ppf((1 + confidence) / 2.0, n - 1)
return (mean, mean - interval, mean + interval, interval)
# create a reporter for all of the values calculated with the above function
def report_confidence_interval(confidence_interval):
print('Mean: {}'.format(confidence_interval[0]))
print('Lower bound: {}'.format(confidence_interval[1]))
print('Upper bound: {}'.format(confidence_interval[2]))
s = "our mean lies in the interval [{:.5}, {:.5}]".format(confidence_interval[1], confidence_interval[2])
return s, confidence_interval[0]
dem_means = []
rep_means = []
dem_er = []
rep_er = []
for name in names[1:]:
print(name)
print('Democrats')
dem_means.append(confidence_interval(dem[name])[0])
dem_er.append(confidence_interval(dem[name])[3])
print(report_confidence_interval(confidence_interval(dem[name])))
print('Republicans')
rep_means.append(confidence_interval(rep[name])[0])
rep_er.append(confidence_interval(rep[name])[3])
print(report_confidence_interval(confidence_interval(rep[name])))
print(' ')
# bar heights (with a subset of the data)
part_dem_means = dem_means[:5]
part_rep_means = rep_means[:5]
# we need to cut down the names to fit
part_names = names [1:6]
# error bars (with a subset of the data)
part_dem_ers = dem_er[:5]
part_rep_ers = rep_er[:5]
# plot a bar graph
plt.style.use('fivethirtyeight')
barWidth = 0.4
r1 = np.arange(len(part_dem_means))
r2 = [x + barWidth for x in r1]
plt.bar(r1, part_dem_means, width = barWidth, color = 'blue', edgecolor = 'black', yerr = part_dem_ers, capsize = 4, label = 'Democrats')
plt.bar(r2, part_rep_means, width = barWidth, color = 'red', edgecolor = 'black', yerr = part_rep_ers, capsize = 4, label = 'Republicans')
plt.title('Support for bills by party')
plt.legend()
plt.xticks([r + barWidth for r in range(len(part_dem_means))], names[1:6], rotation = 45, ha="right");
```
## Interpretation
Most of the confidence intervals are pretty large. If you were trying to extrapolate this data to a population (sort of a nonsensical situation, because congress is the population), you might find a value much different from what you predicted.
Using the handicapped infants bill as an example, the predicted outcome would be ~62%, but because the confidence interval is ~6%, the actual value could be expected to be anywhere between ~56% and ~68%.
```
print(dem_means[0])
print(dem_er[0])
```
## Resources
- [Interactive visualize the Chi-Squared test](https://homepage.divms.uiowa.edu/~mbognar/applets/chisq.html)
- [Calculation of Chi-Squared test statistic](https://en.wikipedia.org/wiki/Pearson%27s_chi-squared_test)
- [Visualization of a confidence interval generated by R code](https://commons.wikimedia.org/wiki/File:Confidence-interval.svg)
- [Expected value of a squared standard normal](https://math.stackexchange.com/questions/264061/expected-value-calculation-for-squared-normal-distribution) (it's 1 - which is why the expected value of a Chi-Squared with $n$ degrees of freedom is $n$, as it's the sum of $n$ squared standard normals)
| github_jupyter |
# Fitting distribution with R
```
x.norm <- rnorm(n=200,m=10,sd=2)
hist(x.norm,main="Histogram of observed data")
plot(density(x.norm),main="Density estimate of data")
plot(ecdf(x.norm),main="Empirical cumulative distribution function")
z.norm <- (x.norm-mean(x.norm))/sd(x.norm) # standardize data
qqnorm(z.norm) ## drawing the QQplot
abline(0,1) ## drawing a 45-degree reference line
```
If data differ from a normal distribution (i.e. data belonging from a Weibull pdf) we cna use `qqplot()` in this way:
```
x.wei <- rweibull(n=200,shape=2.1,scale=1.1) ## sampling from a Weibull
x.teo <- rweibull(n=200,shape=2,scale=1) ## theoretical quantiles from a Weibull population
qqplot(x.teo,x.wei,main="QQ-plot distr. Weibull")
abline(0,1)
```
# Model choice
Dealing with discrete data we can refer to Poisson's distribution with probability mass function:
$$ f(x,\lambda)=e^{-\lambda\dfrac{\lambda^x}{x!}} \quad \text{where } x=0,1,2,\ldots$$
```
x.poi <- rpois(n=200, lambda=2.5)
hist(x.poi, main="Poisson distribution")
```
As concern continuous data we have the normal (gaussian) dsitrubition:
$$ f(x,\lambda,\sigma)=\dfrac{1}{\sqrt{2\pi}\sigma} e^{\dfrac{1(x-\mu)^2}{2\sigma^2}} $$
with $x \in \mathbb{R}$.
```
curve(dnorm(x,m=10,sd=2),from=0,to=20,main="Normal distribution")
```
Gamma distribution:
$$ f(x,\alpha,\lambda)=\dfrac{\lambda^\alpha}{\gamma(\alpha)}x^{\alpha-1}e^{-\lambda x} $$
with $x \in \mathbb{R}^+$.
```
curve(dgamma(x, scale=1.5, shape=2), from=0, to=15, main="Gamma distribution")
```
Weibull distribition:
$$ f(x,\alpha,\beta)=\alpha\beta^{-\alpha}x^{\alpha-1}e^{-\left[\left(\dfrac{x}{\beta}\right)^\alpha\right]} $$
```
curve(dweibull(x, scale=2.5, shape=1.5), from=0, to=15, main="Weibull distribution")
h<-hist(x.norm,breaks=15)
xhist<-c(min(h$breaks),h$breaks)
yhist<-c(0,h$density,0)
xfit<-seq(min(x.norm),max(x.norm),length=40)
yfit<-dnorm(xfit,mean=mean(x.norm),sd=sd(x.norm))
plot(xhist,yhist,type="s",ylim=c(0,max(yhist,yfit)), main="Normal pdf and histogram")
lines(xfit,yfit, col="red")
yfit
yhist
ks.test(yfit,yhist)
```
# StackOverflow example
The following is from this StackOverflow example: https://stats.stackexchange.com/questions/132652/how-to-determine-which-distribution-fits-my-data-best
This requires you to install the following packages with the R package manager: `fitdistrplus` and `logspline`.
```
library(fitdistrplus)
library(logspline)
x <- c(37.50,46.79,48.30,46.04,43.40,39.25,38.49,49.51,40.38,36.98,40.00,
38.49,37.74,47.92,44.53,44.91,44.91,40.00,41.51,47.92,36.98,43.40,
42.26,41.89,38.87,43.02,39.25,40.38,42.64,36.98,44.15,44.91,43.40,
49.81,38.87,40.00,52.45,53.13,47.92,52.45,44.91,29.54,27.13,35.60,
45.34,43.37,54.15,42.77,42.88,44.26,27.14,39.31,24.80,16.62,30.30,
36.39,28.60,28.53,35.84,31.10,34.55,52.65,48.81,43.42,52.49,38.00,
38.65,34.54,37.70,38.11,43.05,29.95,32.48,24.63,35.33,41.34)
descdist(x, discrete = FALSE)
fit.weibull <- fitdist(x, "weibull")
fit.norm <- fitdist(x, "norm")
plot(fit.norm)
plot(fit.weibull)
fit.weibull$aic
fit.norm$aic
```
## Kolmogorov-Smirnov test simulation
```
n.sims <- 5e4
stats <- replicate(n.sims, {
r <- rweibull(n = length(x)
, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"]
)
as.numeric(ks.test(r
, "pweibull"
, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"])$statistic
)
})
plot(ecdf(stats), las = 1, main = "KS-test statistic simulation (CDF)", col = "darkorange", lwd = 1.7)
grid()
fit <- logspline(stats)
1 - plogspline(ks.test(x
, "pweibull"
, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"])$statistic
, fit
)
xs <- seq(10, 65, len=500)
true.weibull <- rweibull(1e6, shape= fit.weibull$estimate["shape"]
, scale = fit.weibull$estimate["scale"])
boot.pdf <- sapply(1:1000, function(i) {
xi <- sample(x, size=length(x), replace=TRUE)
MLE.est <- suppressWarnings(fitdist(xi, distr="weibull"))
dweibull(xs, shape=MLE.est$estimate["shape"], scale = MLE.est$estimate["scale"])
}
)
boot.cdf <- sapply(1:1000, function(i) {
xi <- sample(x, size=length(x), replace=TRUE)
MLE.est <- suppressWarnings(fitdist(xi, distr="weibull"))
pweibull(xs, shape= MLE.est$estimate["shape"], scale = MLE.est$estimate["scale"])
}
)
#-----------------------------------------------------------------------------
# Plot PDF
#-----------------------------------------------------------------------------
par(bg="white", las=1, cex=1.2)
plot(xs, boot.pdf[, 1], type="l", col=rgb(.6, .6, .6, .1), ylim=range(boot.pdf),
xlab="x", ylab="Probability density")
for(i in 2:ncol(boot.pdf)) lines(xs, boot.pdf[, i], col=rgb(.6, .6, .6, .1))
# Add pointwise confidence bands
quants <- apply(boot.pdf, 1, quantile, c(0.025, 0.5, 0.975))
min.point <- apply(boot.pdf, 1, min, na.rm=TRUE)
max.point <- apply(boot.pdf, 1, max, na.rm=TRUE)
lines(xs, quants[1, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[3, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[2, ], col="darkred", lwd=2)
#-----------------------------------------------------------------------------
# Plot CDF
#-----------------------------------------------------------------------------
par(bg="white", las=1, cex=1.2)
plot(xs, boot.cdf[, 1], type="l", col=rgb(.6, .6, .6, .1), ylim=range(boot.cdf),
xlab="x", ylab="F(x)")
for(i in 2:ncol(boot.cdf)) lines(xs, boot.cdf[, i], col=rgb(.6, .6, .6, .1))
# Add pointwise confidence bands
quants <- apply(boot.cdf, 1, quantile, c(0.025, 0.5, 0.975))
min.point <- apply(boot.cdf, 1, min, na.rm=TRUE)
max.point <- apply(boot.cdf, 1, max, na.rm=TRUE)
lines(xs, quants[1, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[3, ], col="red", lwd=1.5, lty=2)
lines(xs, quants[2, ], col="darkred", lwd=2)
#lines(xs, min.point, col="purple")
#lines(xs, max.point, col="purple")
```
| github_jupyter |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv('dataset-of-10s.csv')
data.head()
```
# checking basic integrity
```
data.shape
data.info()
```
# no. of rows = non null values for each column -> no null value
```
data.head()
```
# checking unique records using uri
```
# extracting exact id
def extract(x):
splited_list = x.split(':') # spliting text at colons
return splited_list[2] # returning third element
data['uri'] = data['uri'].apply(extract)
data.head() #successfully extracted the id
```
# checking for duplicate rows
```
data['uri'].nunique(),
data['uri'].value_counts()
data['uri'].value_counts().unique()
dupe_mask = data['uri'].value_counts()==2
dupe_ids = dupe_mask[dupe_mask]
dupe_ids.value_counts, dupe_ids.shape
#converting duplicate ids into a list
dupe_ids = dupe_ids.index
dupe_ids = dupe_ids.tolist()
dupe_ids
duplicate_index = data.loc[data['uri'].isin(dupe_ids),:].index # all the duplicted records
duplicate_index = duplicate_index.tolist()
```
# We will be removing all the duplication as they are few compared to data
```
data.drop(duplicate_index,axis=0,inplace=True)
data.shape
data.info()
print("shape of data",data.shape )
print("no. of unique rows",data['uri'].nunique()) # no duplicates
data.head()
```
# now we will be dropping all the unnecessary columns which contain string which cant be eficiently converted into numerics
```
data.drop(['track','artist','uri'],axis=1,inplace=True)
data.head()
```
# Univariate analysis
```
#analysing class imbalance
sns.countplot(data=data,x='target')
data.columns
# checking appropriate data type
data[['danceability', 'energy', 'key', 'loudness']].info() # every feature have appropriate datatype
# checking range of first 4 features
data[['danceability', 'energy', 'key', 'loudness']].describe()
plt.figure(figsize=(10,10))
plt.subplot(2,2,1)
data['danceability'].plot()
plt.subplot(2,2,2)
plt.plot(data['energy'],color='red')
plt.subplot(2,2,3)
plt.plot(data[['key','loudness']])
```
# danceabilty is well inside the range(0,1)
# energy is well inside the range(0,1)
# there's no -1 for keys-> every track has been assigned respective keys
# loudness values are out of range(0,-60)db
```
loudness_error_idnex = data[data['loudness']>0].index
loudness_error_idnex
# removing rows with out of range values in loudness column
data.drop(loudness_error_idnex,axis=0, inplace=True)
data.shape # record is removed
# checking appropriate datatype for next 5 columns
data[['mode', 'speechiness',
'acousticness', 'instrumentalness', 'liveness',]].info() # datatypes are in acoordance with provided info
data[['mode', 'speechiness',
'acousticness', 'instrumentalness', 'liveness',]].describe() # every feautre is within range
sns.countplot(x=data['mode']) # have only two possible values 0 and 1, no noise in the feature
data[['valence', 'tempo',
'duration_ms', 'time_signature', 'chorus_hit', 'sections']].info() # data type is in accordance with provided info
data[['valence', 'tempo',
'duration_ms', 'time_signature', 'chorus_hit', 'sections']].describe() # all the data are in specified range
```
# Performing F-test to know the relation between every feature and target
```
data.head()
x = data.iloc[:,:-1].values
y = data.iloc[:,-1].values
x.shape,y.shape
from sklearn.feature_selection import f_classif
f_stat,p_value = f_classif(x,y)
feat_list = data.iloc[:,:-1].columns.tolist()
# making a dataframe
dict = {'Features':feat_list,'f_statistics':f_stat,'p_value':p_value}
relation = pd.DataFrame(dict)
relation.sort_values(by='p_value')
```
# Multivariate analysis
```
correlation = data.corr()
plt.figure(figsize=(15,12))
sns.heatmap(correlation, annot=True)
plt.tight_layout
```
# strong features(accordance with f-test) -->
danceability, loudness, acousticness, instrumentalness, valence
# less imortant feature(accordance with f-test)-->
duration, section, mode, time_signature, chorus hit
# least imortant-->
energy,key,speecheness,liveliness,tempo
| github_jupyter |
# Python Language Basics, IPython, and Jupyter Notebooks
```
import numpy as np
np.random.seed(12345)
np.set_printoptions(precision=4, suppress=True)
```
## The Python Interpreter
```python
$ python
Python 3.6.0 | packaged by conda-forge | (default, Jan 13 2017, 23:17:12)
[GCC 4.8.2 20140120 (Red Hat 4.8.2-15)] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>> a = 5
>>> print(a)
5
```
```python
print('Hello world')
```
```python
$ python hello_world.py
Hello world
```
```shell
$ ipython
Python 3.6.0 | packaged by conda-forge | (default, Jan 13 2017, 23:17:12)
Type "copyright", "credits" or "license" for more information.
IPython 5.1.0 -- An enhanced Interactive Python.
? -> Introduction and overview of IPython's features.
%quickref -> Quick reference.
help -> Python's own help system.
object? -> Details about 'object', use 'object??' for extra details.
In [1]: %run hello_world.py
Hello world
In [2]:
```
## IPython Basics
### Running the IPython Shell
$
```
import numpy as np
data = {i : np.random.randn() for i in range(7)}
data
```
>>> from numpy.random import randn
>>> data = {i : randn() for i in range(7)}
>>> print(data)
{0: -1.5948255432744511, 1: 0.10569006472787983, 2: 1.972367135977295,
3: 0.15455217573074576, 4: -0.24058577449429575, 5: -1.2904897053651216,
6: 0.3308507317325902}
### Running the Jupyter Notebook
```shell
$ jupyter notebook
[I 15:20:52.739 NotebookApp] Serving notebooks from local directory:
/home/wesm/code/pydata-book
[I 15:20:52.739 NotebookApp] 0 active kernels
[I 15:20:52.739 NotebookApp] The Jupyter Notebook is running at:
http://localhost:8888/
[I 15:20:52.740 NotebookApp] Use Control-C to stop this server and shut down
all kernels (twice to skip confirmation).
Created new window in existing browser session.
```
### Tab Completion
```
In [1]: an_apple = 27
In [2]: an_example = 42
In [3]: an
```
```
In [3]: b = [1, 2, 3]
In [4]: b.
```
```
In [1]: import datetime
In [2]: datetime.
```
```
In [7]: datasets/movielens/
```
### Introspection
```
In [8]: b = [1, 2, 3]
In [9]: b?
Type: list
String Form:[1, 2, 3]
Length: 3
Docstring:
list() -> new empty list
list(iterable) -> new list initialized from iterable's items
In [10]: print?
Docstring:
print(value, ..., sep=' ', end='\n', file=sys.stdout, flush=False)
Prints the values to a stream, or to sys.stdout by default.
Optional keyword arguments:
file: a file-like object (stream); defaults to the current sys.stdout.
sep: string inserted between values, default a space.
end: string appended after the last value, default a newline.
flush: whether to forcibly flush the stream.
Type: builtin_function_or_method
```
```python
def add_numbers(a, b):
"""
Add two numbers together
Returns
-------
the_sum : type of arguments
"""
return a + b
```
```python
In [11]: add_numbers?
Signature: add_numbers(a, b)
Docstring:
Add two numbers together
Returns
-------
the_sum : type of arguments
File: <ipython-input-9-6a548a216e27>
Type: function
```
```python
In [12]: add_numbers??
Signature: add_numbers(a, b)
Source:
def add_numbers(a, b):
"""
Add two numbers together
Returns
-------
the_sum : type of arguments
"""
return a + b
File: <ipython-input-9-6a548a216e27>
Type: function
```
```python
In [13]: np.*load*?
np.__loader__
np.load
np.loads
np.loadtxt
np.pkgload
```
### The %run Command
```python
def f(x, y, z):
return (x + y) / z
a = 5
b = 6
c = 7.5
result = f(a, b, c)
```
```python
In [14]: %run ipython_script_test.py
```
```python
In [15]: c
Out [15]: 7.5
In [16]: result
Out[16]: 1.4666666666666666
```
```python
>>> %load ipython_script_test.py
def f(x, y, z):
return (x + y) / z
a = 5
b = 6
c = 7.5
result = f(a, b, c)
```
#### Interrupting running code
### Executing Code from the Clipboard
```python
x = 5
y = 7
if x > 5:
x += 1
y = 8
```
```python
In [17]: %paste
x = 5
y = 7
if x > 5:
x += 1
y = 8
## -- End pasted text --
```
```python
In [18]: %cpaste
Pasting code; enter '--' alone on the line to stop or use Ctrl-D.
:x = 5
:y = 7
:if x > 5:
: x += 1
:
: y = 8
:--
```
### Terminal Keyboard Shortcuts
### About Magic Commands
```python
In [20]: a = np.random.randn(100, 100)
In [20]: %timeit np.dot(a, a)
10000 loops, best of 3: 20.9 µs per loop
```
```python
In [21]: %debug?
Docstring:
::
%debug [--breakpoint FILE:LINE] [statement [statement ...]]
Activate the interactive debugger.
This magic command support two ways of activating debugger.
One is to activate debugger before executing code. This way, you
can set a break point, to step through the code from the point.
You can use this mode by giving statements to execute and optionally
a breakpoint.
The other one is to activate debugger in post-mortem mode. You can
activate this mode simply running %debug without any argument.
If an exception has just occurred, this lets you inspect its stack
frames interactively. Note that this will always work only on the last
traceback that occurred, so you must call this quickly after an
exception that you wish to inspect has fired, because if another one
occurs, it clobbers the previous one.
If you want IPython to automatically do this on every exception, see
the %pdb magic for more details.
positional arguments:
statement Code to run in debugger. You can omit this in cell
magic mode.
optional arguments:
--breakpoint <FILE:LINE>, -b <FILE:LINE>
Set break point at LINE in FILE.
```
```python
In [22]: %pwd
Out[22]: '/home/wesm/code/pydata-book
In [23]: foo = %pwd
In [24]: foo
Out[24]: '/home/wesm/code/pydata-book'
```
### Matplotlib Integration
```python
In [26]: %matplotlib
Using matplotlib backend: Qt4Agg
```
```python
In [26]: %matplotlib inline
```
## Python Language Basics
### Language Semantics
#### Indentation, not braces
```python
for x in array:
if x < pivot:
less.append(x)
else:
greater.append(x)
```
```python
a = 5; b = 6; c = 7
```
#### Everything is an object
#### Comments
```python
results = []
for line in file_handle:
# keep the empty lines for now
# if len(line) == 0:
# continue
results.append(line.replace('foo', 'bar'))
```
```python
print("Reached this line") # Simple status report
```
#### Function and object method calls
```
result = f(x, y, z)
g()
```
```
obj.some_method(x, y, z)
```
```python
result = f(a, b, c, d=5, e='foo')
```
#### Variables and argument passing
```
a = [1, 2, 3]
b = a
a.append(4)
b
```
```python
def append_element(some_list, element):
some_list.append(element)
```
```python
In [27]: data = [1, 2, 3]
In [28]: append_element(data, 4)
In [29]: data
Out[29]: [1, 2, 3, 4]
```
#### Dynamic references, strong types
```
a = 5
type(a)
a = 'foo'
type(a)
'5' + 5
a = 4.5
b = 2
# String formatting, to be visited later
print('a is {0}, b is {1}'.format(type(a), type(b)))
a / b
a = 5
isinstance(a, int)
a = 5; b = 4.5
isinstance(a, (int, float))
isinstance(b, (int, float))
```
#### Attributes and methods
```python
In [1]: a = 'foo'
In [2]: a.<Press Tab>
a.capitalize a.format a.isupper a.rindex a.strip
a.center a.index a.join a.rjust a.swapcase
a.count a.isalnum a.ljust a.rpartition a.title
a.decode a.isalpha a.lower a.rsplit a.translate
a.encode a.isdigit a.lstrip a.rstrip a.upper
a.endswith a.islower a.partition a.split a.zfill
a.expandtabs a.isspace a.replace a.splitlines
a.find a.istitle a.rfind a.startswith
```
```
a = 'foo'
getattr(a, 'split')
```
#### Duck typing
```
def isiterable(obj):
try:
iter(obj)
return True
except TypeError: # not iterable
return False
isiterable('a string')
isiterable([1, 2, 3])
isiterable(5)
```
if not isinstance(x, list) and isiterable(x):
x = list(x)
#### Imports
```python
# some_module.py
PI = 3.14159
def f(x):
return x + 2
def g(a, b):
return a + b
```
import some_module
result = some_module.f(5)
pi = some_module.PI
from some_module import f, g, PI
result = g(5, PI)
import some_module as sm
from some_module import PI as pi, g as gf
r1 = sm.f(pi)
r2 = gf(6, pi)
#### Binary operators and comparisons
```
5 - 7
12 + 21.5
5 <= 2
a = [1, 2, 3]
b = a
c = list(a)
a is b
a is not c
a == c
a = None
a is None
```
#### Mutable and immutable objects
```
a_list = ['foo', 2, [4, 5]]
a_list[2] = (3, 4)
a_list
a_tuple = (3, 5, (4, 5))
a_tuple[1] = 'four'
```
### Scalar Types
#### Numeric types
```
ival = 17239871
ival ** 6
fval = 7.243
fval2 = 6.78e-5
3 / 2
3 // 2
```
#### Strings
a = 'one way of writing a string'
b = "another way"
```
c = """
This is a longer string that
spans multiple lines
"""
c.count('\n')
a = 'this is a string'
a[10] = 'f'
b = a.replace('string', 'longer string')
b
a
a = 5.6
s = str(a)
print(s)
s = 'python'
list(s)
s[:3]
s = '12\\34'
print(s)
s = r'this\has\no\special\characters'
s
a = 'this is the first half '
b = 'and this is the second half'
a + b
template = '{0:.2f} {1:s} are worth US${2:d}'
template.format(4.5560, 'Argentine Pesos', 1)
```
#### Bytes and Unicode
```
val = "español"
val
val_utf8 = val.encode('utf-8')
val_utf8
type(val_utf8)
val_utf8.decode('utf-8')
val.encode('latin1')
val.encode('utf-16')
val.encode('utf-16le')
bytes_val = b'this is bytes'
bytes_val
decoded = bytes_val.decode('utf8')
decoded # this is str (Unicode) now
```
#### Booleans
```
True and True
False or True
```
#### Type casting
```
s = '3.14159'
fval = float(s)
type(fval)
int(fval)
bool(fval)
bool(0)
```
#### None
```
a = None
a is None
b = 5
b is not None
```
def add_and_maybe_multiply(a, b, c=None):
result = a + b
if c is not None:
result = result * c
return result
```
type(None)
```
#### Dates and times
```
from datetime import datetime, date, time
dt = datetime(2011, 10, 29, 20, 30, 21)
dt.day
dt.minute
dt.date()
dt.time()
dt.strftime('%m/%d/%Y %H:%M')
datetime.strptime('20091031', '%Y%m%d')
dt.replace(minute=0, second=0)
dt2 = datetime(2011, 11, 15, 22, 30)
delta = dt2 - dt
delta
type(delta)
dt
dt + delta
```
### Control Flow
#### if, elif, and else
if x < 0:
print('It's negative')
if x < 0:
print('It's negative')
elif x == 0:
print('Equal to zero')
elif 0 < x < 5:
print('Positive but smaller than 5')
else:
print('Positive and larger than or equal to 5')
```
a = 5; b = 7
c = 8; d = 4
if a < b or c > d:
print('Made it')
4 > 3 > 2 > 1
```
#### for loops
for value in collection:
# do something with value
sequence = [1, 2, None, 4, None, 5]
total = 0
for value in sequence:
if value is None:
continue
total += value
sequence = [1, 2, 0, 4, 6, 5, 2, 1]
total_until_5 = 0
for value in sequence:
if value == 5:
break
total_until_5 += value
```
for i in range(4):
for j in range(4):
if j > i:
break
print((i, j))
```
for a, b, c in iterator:
# do something
#### while loops
x = 256
total = 0
while x > 0:
if total > 500:
break
total += x
x = x // 2
#### pass
if x < 0:
print('negative!')
elif x == 0:
# TODO: put something smart here
pass
else:
print('positive!')
#### range
```
range(10)
list(range(10))
list(range(0, 20, 2))
list(range(5, 0, -1))
```
seq = [1, 2, 3, 4]
for i in range(len(seq)):
val = seq[i]
sum = 0
for i in range(100000):
# % is the modulo operator
if i % 3 == 0 or i % 5 == 0:
sum += i
#### Ternary expressions
value =
if
```
x = 5
'Non-negative' if x >= 0 else 'Negative'
```
| github_jupyter |
```
# last edited Apr 4, 2021, by GO.
# to do:
################################################################################
# script uses 'seagrid' E grid (500 m) and a bathymetric data file to generate
# new bathymetric .nc file at coarser resolutions (multiples of 500 m).
# Based on original code provided by M Dunphy.
# Assumes input bathymetric file is on exact grid as 'seagrid'.
# Output can be used by NEMO to generate a 'mesh mask'
# in:
# coordinates_seagrid_SalishSea2.nc - 500m coordinates, same region
# bathymetry_201702.nc - 500m bathymetry, same region
#
# out:
# coordinates_seagrid_SalishSea_1500m.nc
# bathymetry_201702.nc
# change log:
# - Apr 3 2021 -
# issue with np.nan vs np.NaN or similar causing issues with XIOS.
# Set all land values to zero instead of nan.
# - Feb 5, 2021, by GO (previously called 'Working Grid Generator')
# - fixes to the Fraser river extension and southeast corner
#
# - mike made improvements - Dec 29, 2020
# - to 'decimante' a 500 m Arakawa E grid to 1 km, 1.5 km, 2 km etc grids
# (factors of 500 m) we can extract by skipping n cells and taking coords
# from previous 500 m Arakawa E grid.
# However, the point we extract from the 500 m grid depends on the new grid
# - it's either an 'f' point (even number of skipped cells, n; e.g., 1 km =
# 2 x 500 m = even) or a 't' point (if n is odd).
################################################################################
%matplotlib notebook
import netCDF4 as nc
import numpy as np
from helpers import writebathy, expandf # custom helper fns from MD, MEOPAR
from helpers import gete1, gete2, writecoords, t2u, t2v, t2f
import matplotlib.pyplot as plt
res = "1500m"
km = "1500m"
gridfilename = "..//data//grid//coordinates_salishsea_{}.nc".format(res) # in
n = 3 # e.g., 500m x n = new res
datetag = "20210406"
bathyout_filename = "..//data//bathymetry//bathy_salishsea_{}_{}.nc".format(res,datetag)
bathyout_filename_preedits = "..//data//bathymetry//bathy_salishsea_{}_before_manual_edits.nc".format(res)
def loadreduce_md(pt, n):
c0 = '..//data//grid//etc//coordinates_seagrid_SalishSea2.nc'
with nc.Dataset(c0) as ncid:
if pt=='t':
glam = ncid.variables["glamt"][0, 1::n, 1::n].filled()
gphi = ncid.variables["gphit"][0, 1::n, 1::n].filled()
if pt=='u':
glam = ncid.variables["glamu"][0, 1::n, 2::n].filled()
gphi = ncid.variables["gphiu"][0, 1::n, 2::n].filled()
if pt=='v':
glam = ncid.variables["glamv"][0, 2::n, 1::n].filled()
gphi = ncid.variables["gphiv"][0, 2::n, 1::n].filled()
if pt=='f':
glam = ncid.variables["glamf"][0, 2::n, 2::n].filled()
gphi = ncid.variables["gphif"][0, 2::n, 2::n].filled()
return glam, gphi
#########################################################
######### DECIMATE GRID FROM 500m to 1500m ##############
# Since we're doing a 3-way reduction, we can re-use the original points and not calculate new ones
glamt, gphit = loadreduce_md('t', n)
glamu, gphiu = loadreduce_md('u', n)
glamv, gphiv = loadreduce_md('v', n)
glamf, gphif = loadreduce_md('f', n)
# Compute scaling factors (with extrapolation for the left/bottom most scaling factor)
e1t = gete1(glamu,gphiu,expandleft=True) # Need a left u point
e1u = gete1(glamt,gphit)
e1v = gete1(glamf,gphif,expandleft=True) # Need a left f point
e1f = gete1(glamv,gphiv)
#
e2t = gete2(glamv,gphiv,expanddown=True) # Need a lower v point
e2u = gete2(glamf,gphif,expanddown=True) # Need a lower f point
e2v = gete2(glamt,gphit)
e2f = gete2(glamu,gphiu)
# Output slices
NY,NX = glamt.shape
J,I = slice(0,NY), slice(0,NX-1)
writecoords(gridfilename,
glamt[J,I],glamu[J,I],glamv[J,I],glamf[J,I],
gphit[J,I],gphiu[J,I],gphiv[J,I],gphif[J,I],
e1t[J,I],e1u[J,I],e1v[J,I],e1f[J,I],
e2t[J,I],e2u[J,I],e2v[J,I],e2f[J,I])
##########################################################
############### DECIMATE AND EDIT BATHY ##################
# --------------------------------------------------------------------------------
# 1) get the grid centres (t points) for new grid
with nc.Dataset(gridfilename) as ncid:
glamt = ncid.variables["glamt"][0, :, :].filled()
gphit = ncid.variables["gphit"][0, :, :].filled()
# --------------------------------------------------------------------------------
# 2) get depths from 500 m bathy file
with nc.Dataset('..//data//bathymetry//etc//bathymetry_201702.nc') as nc_b_file:
a = nc_b_file.variables["Bathymetry"][:, :].filled()
# --------------------------------------------------------------------------------
# 3) 'land mask' from 500 m bathy
mask = a.copy()
mask[mask > 0] = 1
# --------------------------------------------------------------------------------
# 4) create new grid taking mean of surrounding cells
a2 = np.zeros(glamt.shape)
m2 = np.zeros(glamt.shape)
for j in range(a2.shape[0]):
for i in range(a2.shape[1]):
i1, i2 = 3*i, 3*i+3
j1, j2 = 3*j, 3*j+3
bvals = a[j1:j2, i1:i2] # extract 3x3 box of bathy values
a2[j,i] = np.mean(bvals)
mvals = mask[j1:j2, i1:i2]
m2[j,i] = np.mean(mvals)
# --------------------------------------------------------------------------------
# 5) filter new bathy grid based on % land
# (m2 is the % of the new 1500m cell that was land in 500m version)
a2[m2 < 0.5] = 0
# --------------------------------------------------------------------------------
# 6) set min depth
a2[(a2 > 0) & (a2 < 4)] = 4
# --------------------------------------------------------------------------------
# 6a) write to file pre-edits
writebathy(bathyout_filename_preedits,glamt,gphit,a2)
def manualedits(a, n):
# a = array of depths, 1.5 km grid
# manual edits for 1.5 km bathy
if n == 3:
# north to south
a[296,57] = 40 #northern fjord
a[296,54] = 60 #northern fjord
a[296,53] = 60 #northern fjord
a[295,52] = 150 #northern fjord
a[286,44] = 200 #northern fjord
a[289,32] = 20 #hardwick
a[289,33] = 20 #hardwick
a[286,33] = 20 #hardwick
a[285,30] = 20 #hardwick
a[284,41] = 20 #west thurlow
a[284,42] = 20 #west thurlow
a[283,43] = 20 #west thurlow
a[282,43] = 20 #west thurlow
a[281,44] = 20 #west thurlow
a[279,43] = 20 #west thurlow
a[279,45] = 20 #west thurlow
a[269,59] = 20 #sonora
a[265,54] = 10 #maurelle
a[265,57] = 20 #maurelle
a[266,52] = 20 #quadra
a[268,46] = 20 #quadra
a[259,53] = 20 #quadra
a[260,57] = 20 #read
a[254,63] = 20 #cortes
a[254,62] = 20 #cortes
a[255,62] = 20 #cortes
a[254,72] = 6 #redonda
a[254,73] = 20 #redonda
a[252,72] = 20 #redonda
a[252,71] = 20 #redonda
a[251,71] = 20 #redonda
a[197,82] = 30 #nelson
a[197,84] = 60 #nelson
a[199,86] = 60 #nelson
a[200,86] = 60 #nelson
a[156,73] = 30 #gabriola
a[132,72] = 100 #salt spring
a[128,71] = 50 #salt spring
a[123,86] = 30 #mayne
a[146,112] = 0.0 #north fraser
a[146,114] = 0.0 #north fraser
a[146,113] = 0.0 #north fraser
a[146,108] = 6 #north fraser
a[146,109] = 6 #north fraser
a[145,112] = 0.0 #north fraser
a[145,115] = 0.0 #north fraser
#a[145,108] = 10 #north fraser
a[144,108] = 6 #north fraser
a[144,109] = 6 #north fraser
a[144,110] = 6 #north fraser
a[144,111] = 6 #north fraser
a[144,112] = 6 #north fraser
a[144,115] = 0.0 #north fraser
a[145,110] = 0.0 #north fraser
a[145,111] = 0.0 #north fraser
a[145,114] = 0.0 #north fraser
a[144,107] = 6 #north fraser
a[143,112] = 6 #north fraser
a[143,113] = 6 #north fraser
a[143,115] = 0.0 #north fraser
a[143,116] = 0.0 #north fraser
a[142,113] = 6 #north fraser
a[142,114] = 6 #north fraser
a[142,116] = 0.0 #north fraser
a[141,116] = 0.0 #north fraser
a[141,118] = 6 #north fraser
a[141,120] = 6 #north fraser
a[142,120] = 6 #north fraser
a[136,103] = 6 #south fraser
a[137,104] = 6 #south fraser
a[138,109] = 10 #south fraser
a[138,112] = 12 #south fraser
a[139,104] = 10 #south fraser
a[139,113] = 10 #south fraser
a[139,114] = 10 #south fraser
a[138,113] = 0.0 #south fraser
a[138,117] = 0.0 #south fraser
a[137,107] = 0.0 #south fraser
a[137,109] = 0.0 #south fraser
a[137,110] = 0.0 #south fraser
a[137,113] = 0.0 #south fraser
a[137,114] = 0.0 #south fraser
a[137,115] = 0.0 #south fraser
a[137,116] = 10 #south fraser
a[137,117] = 10 #south fraser
a[137,118] = 12 #south fraser
a[136,116] = 0.0 #south fraser
a[136,117] = 0.0 #south fraser
a[136,118] = 0.0 #south fraser
a[136,119] = 0.0 #south fraser
a[140,119] = 6 # fraser
a[140,125] = 0.0 # fraser
a[141,117] = 6 # fraser
a[141,118] = 0.0 # fraser
a[142,118] = 0.0 # fraser
a[142,119] = 0.0 # fraser
a[142,120] = 0.0 # fraser
a[141,120] = 0.0 # fraser
a[140,120] = 0.0 # fraser
a[140,121] = 0.0 # fraser
a[141,121] = 0.0 # fraser
a[140,123] = 0.0 # fraser
a[141,123] = 0.0 # fraser
a[142,123] = 0.0 # fraser
a[142,122] = 0.0 # fraser
a[141,122] = 0.0 # fraser
a[141,124] = 0.0 # fraser
a[140,124] = 0.0 # fraser
a[140,125] = 0.0 # fraser
a[141,115] = 6 # fraser
a[141,116] = 6 # fraser
a[142,115] = 6 # fraser
a[140,115] = 0.0 # fraser
a[140,116] = 0.0 # fraser
a[140,117] = 6 # fraser
a[140,118] = 6 # fraser
a[138,114] = 10 # fraser
a[138,115] = 10 # fraser
a[138,116] = 10 # fraser
a[138,118] = 6 # fraser
a[138,127] = 10 # fraser
a[138,128] = 10 # fraser
a[138,129] = 10 # fraser
a[138,130] = 0.0 # fraser
a[137,129] = 10 # fraser
a[136,129] = 10 # fraser
a[135,129] = 0 # fraser
a[135,130] = 10 # fraser
a[135,131] = 10 # fraser
a[136,131] = 10 # fraser
a[137,131] = 10 # fraser
a[139,112] = 10 # fraser
a[139,117] = 0.0 # fraser
a[139,118] = 0.0 # fraser
a[139,121] = 10 # fraser
a[139,122] = 10 # fraser
a[139,123] = 10 # fraser
a[139,124] = 10 # fraser
a[139,125] = 10 # fraser
a[139,126] = 10 # fraser
a[139,130] = 6 #artificial frsr riv extension
a[140,130] = 6 #artificial frsr riv extension
a[141,130] = 6 #artificial frsr riv extension
a[142,130] = 6 #artificial frsr riv extension
a[143,130] = 6 #artificial frsr riv extension
a[144,130] = 6 #artificial frsr riv extension
a[145,130] = 6 #artificial frsr riv extension
a[146,130] = 6 #artificial frsr riv extension
a[147,130] = 6 #artificial frsr riv extension
a[148,130] = 6 #artificial frsr riv extension
a[149,130] = 6 #artificial frsr riv extension
a[150,130] = 6 #artificial frsr riv extension
a[151,130] = 6 #artificial frsr riv extension
a[140,129] = 6 #artificial frsr riv extension
a[139,129] = 6 #artificial frsr riv extension
a[136,129] = 0.0 #artificial frsr riv extension
a[135,131] = 0.0 #artificial frsr riv extension
a[136,131] = 0.0 #artificial frsr riv extension
a[137,131] = 0.0 #artificial frsr riv extension
a[138,131] = 0.0 #artificial frsr riv extension
a[135,130] = 0.0 #artificial frsr riv extension
a[137,129] = 0.0 #artificial frsr riv extension
a[100,93] = 15 #shaw is
a[99,94] = 15 #shaw is
a[102,81] = 15 #san juan is
a[102,82] = 10 #san juan is
a[103,82] = 10 #san juan is
a[104,82] = 10 #san juan is
a[104,83] = 10 #san juan is
a[94,98] = 30 #lopez is
a[82,101] = 20 #rosario b
a[81,101] = 20 #rosario b
a[81,102] = 10 #rosario b
a[33,43] = 200 #hood cnl
a[34,44] = 200 #hood cnl
a[24,39] = 30 #hood cnl
a[23,41] = 30 #hood cnl
a[23,48] = 30 #hood cnl
a[7,59] = 30 #tacoma
a[5,58] = 30 #tacoma
a[8,54] = 20 #fox is
a[28,71] = 10 #bremerton
a[28,72] = 10 #bremerton
a[30,72] = 10 #bremerton
a[27,68] = 20 #bremerton
a[26,68] = 30 #bremerton
a[26,66] = 30 #bremerton
a[4,31] = 30 #southwest
a[4,32] = 30 #southwest
a[4,33] = 30 #southwest
a[9,31] = 10 #southwest
a[11,39] = 10 #southwest
a[12,39] = 10 #southwest
a[13,40] = 10 #southwest
a[14,41] = 10 #southwest
a[6,35] = 10 #southwest
a[8,35] = 10 #southwest
a[19,50] = 10 #southwest
a[19,49] = 10 #southwest
a[10,68] = 10 #southwest
a[8,30] = 10 #southwest
a[8,29] = 10 #southwest
a[1,46] = 10 #southwest - MD fix 20210208
return a
# --------------------------------------------------------------------------------
# 7) apply manual edits
a3 = manualedits(a2, n)
# Apr 6 --------------------------------------------------------------------------
# 8) replace all np.nan and nan with 0.0
# --------------------------------------------------------------------------------
# 9) write to file
writebathy(bathyout_filename,glamt,gphit,a3)
print("success")
```
# Plots to check channels etc
- took from old checkbathy and plotgrids files
```
import scipy.io as sio
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:90% !important; }</style>"))
from helpers import expandf, grid_angle
# grid
def load1(f):
with nc.Dataset(f) as ncid:
glamt = ncid.variables["glamt"][0, :, :].filled()
gphit = ncid.variables["gphit"][0, :, :].filled()
glamu = ncid.variables["glamu"][0, :, :].filled()
gphiu = ncid.variables["gphiu"][0, :, :].filled()
glamv = ncid.variables["glamv"][0, :, :].filled()
gphiv = ncid.variables["gphiv"][0, :, :].filled()
glamf = ncid.variables["glamf"][0, :, :].filled()
gphif = ncid.variables["gphif"][0, :, :].filled()
return glamt, glamu, glamv, glamf, gphit, gphiu, gphiv, gphif
#
def load2(f):
with nc.Dataset(f) as ncid:
e1t = ncid.variables["e1t"][0, :, :].filled()
e1u = ncid.variables["e1u"][0, :, :].filled()
e1v = ncid.variables["e1v"][0, :, :].filled()
e1f = ncid.variables["e1f"][0, :, :].filled()
e2t = ncid.variables["e2t"][0, :, :].filled()
e2u = ncid.variables["e2u"][0, :, :].filled()
e2v = ncid.variables["e2v"][0, :, :].filled()
e2f = ncid.variables["e2f"][0, :, :].filled()
return e1t,e1u,e1v,e1f,e2t,e2u,e2v,e2f
def load3(f):
with nc.Dataset(f) as ncid:
depth = ncid.variables["Bathymetry"][:, :].filled()
latt = ncid.variables["nav_lat"][:, :].filled()
lont = ncid.variables["nav_lon"][:, :].filled()
return depth, latt, lont
# for rivers - GO
def load4(f):
with nc.Dataset(f) as ncid:
rorunoff = ncid.variables["rorunoff"][6, :, :].filled()
latt = ncid.variables["nav_lat"][:, :].filled()
lont = ncid.variables["nav_lon"][:, :].filled()
return rorunoff, latt, lont
# grid
def plotgrid1(f):
glamt, glamu, glamv, glamf, gphit, gphiu, gphiv, gphif = load1(f)
plt.figure(figsize=(7,5)); plt.clf()
# Draw sides of every box
glamfe, gphife = expandf(glamf, gphif)
NY,NX = glamfe.shape
print(glamt.shape)
print(glamu.shape)
print(glamf.shape)
for j in range(NY):
plt.plot(glamfe[j,:],gphife[j,:], 'k')
for i in range(NX):
plt.plot(glamfe[:,i],gphife[:,i], 'k')
# Plot t, u, v, f points in red, green, blue, magenta
plt.plot(glamt, gphit, 'r.')
plt.plot(glamu, gphiu, 'g.')
plt.plot(glamv, gphiv, 'b.')
plt.plot(glamf, gphif, 'm.')
plt.tight_layout()
plt.xlim([-123.5,-123.3])
plt.ylim([46.84,46.95])
#plt.savefig(f.replace(".nc","_gridpts.png"))
# grid
def plotgrid2(f):
glamt, glamu, glamv, glamf, gphit, gphiu, gphiv, gphif = load1(f)
e1t,e1u,e1v,e1f,e2t,e2u,e2v,e2f = load2(f)
glamfe, gphife = expandf(glamf, gphif)
A = grid_angle(f)
plt.figure(figsize=(12,4))
plt.subplot(1,3,1)
plt.pcolormesh(glamfe,gphife,e1t); plt.colorbar(); plt.title("e1t (m)")
plt.subplot(1,3,2)
plt.pcolormesh(glamfe,gphife,e2t); plt.colorbar(); plt.title("e2t (m)")
plt.subplot(1,3,3)
plt.pcolormesh(glamf,gphif,A); plt.colorbar(); plt.title("angle (deg)")
plt.tight_layout()
plt.savefig(f.replace(".nc","_resolution_angle.png"))
# bathy
def plotgrid3(f):
depth, latt, lont = load3(f)
depth[depth==0]=np.nan
depth[depth>0]=1
#print(depth.shape)
# can do edits below
# made permanent in the main create bathy above
# north to south
#depth[178,128] = 400 #northern fjord
# depth[296,54] = 60 #northern fjord
# depth[296,53] = 60 #northern fjord
plt.figure(figsize=(8,8))
plt.subplot(1,1,1)
plt.pcolormesh(depth, cmap=plt.plasma()); plt.colorbar(); plt.title("depth")
#plt.pcolormesh(depth); plt.colorbar(); plt.title("depth")
#plt.pcolormesh(ma_rorunoff, cmap=plt.pink()); plt.title("rodepth")
plt.tight_layout()
plt.savefig(f.replace(".nc","_bathycheck.png"))
# runoff / rivers
def plotgrid4(f):
depth, latt, lont = load3(f)
# added for river runoff overlay
rorunoff, latt2, lontt2 = load4('c:/temp/runofftools/rivers_month_202101GO.nc')
#rorunoff[rorunoff==0]=np.nan
#print(rorunoff.shape)
ma_rorunoff = np.ma.masked_array(rorunoff, rorunoff == 0)
depth[depth==0]=np.nan
depth[depth>0]=1
#print(depth.shape)
plt.figure(figsize=(8,8))
plt.subplot(1,1,1)
plt.pcolormesh(depth, cmap=plt.plasma()); plt.colorbar(); plt.title("depth")
#plt.pcolormesh(depth); plt.colorbar(); plt.title("depth")
#plt.pcolormesh(ma_rorunoff, cmap=plt.pink()); plt.title("rodepth")
plt.tight_layout()
plt.savefig("C:/temp/runofftools/runoffcheck2.png")
# #################################################################
# #################### BASIC PLOT OF BATHY ########################
gridfilename = '..//data//grid//coordinates_salishsea_1500m.nc'
#bathyfilename = 'bathy_salishsea_1500m_before_manual_edits.nc'
#bathyfilename = '..//data//bathymetry//bathy_salishsea_1500m_Dec30.nc'
with nc.Dataset(gridfilename) as ncid:
glamt = ncid.variables["glamt"][0, :, :].filled()
gphit = ncid.variables["gphit"][0, :, :].filled()
glamf = ncid.variables["glamf"][0, :, :].filled()
gphif = ncid.variables["gphif"][0, :, :].filled()
glamfe,gphife=expandf(glamf,gphif)
with nc.Dataset(bathyout_filename) as nc_b_file:
bathy = nc_b_file.variables["Bathymetry"][:, :].filled()
bb=np.copy(bathy); bb[bb==0]=np.nan
plt.figure(figsize=(8,8))
plt.subplot(1,1,1)
plt.pcolormesh(glamfe,gphife,bb); plt.colorbar()
# Coastlines
mfile = sio.loadmat('..//data//reference//PNW.mat')
ncst = mfile['ncst']
plt.plot(ncst[:,0],ncst[:,1],'k')
mfile2 = sio.loadmat('..//data//reference//PNWrivers.mat')
ncst2 = mfile2['ncst']
plt.plot(ncst2[:,0],ncst2[:,1],'k')
##########################################################
############### PLOTS TO CHECK BATHY ETC #################
# plotgrid1('coordinates_seagrid_SalishSea2.nc')
#plotgrid1('coordinates_salishsea_1km.nc')
#plotgrid1('coordinates_salishsea_1500m.nc')
#plotgrid1('coordinates_salishsea_2km.nc')
#plotgrid2('coordinates_seagrid_SalishSea2.nc')
# plotgrid2('coordinates_salishsea_1km.nc')
#plotgrid2('coordinates_salishsea_2km.nc')
#plotgrid2('coordinates_salishsea_1p5km.nc')
#plotgrid3('bathy_salishsea_1500m_Dec21.nc')
plotgrid3(bathyout_filename)
#plotgrid3('bathy_salishsea_2km.nc')
# junk code below
a = range(24)
b = a[::3]
list(b)
my_list[0] = [_ for _ in 'abcdefghi']
my_list[1] = [_ for _ in 'abcdefghi']
my_list[0:-1]
glamu.shape
a[296,10]
############################################################
### EXPLORE TWO MESHES - NEMO ORAS5 and SS1500 #############
### Apr 2021
import sys
# load mask (tmask)
def loadmask(f):
with nc.Dataset(f) as ncid:
tmaskutil = ncid.variables["tmaskutil"][0,:, :].filled()
latt = ncid.variables["nav_lat"][:, :].filled()
lont = ncid.variables["nav_lon"][:, :].filled()
e1t = ncid.variables["e1t"][0,:, :].filled()
e2t = ncid.variables["e2t"][0,:, :].filled()
return tmaskutil, latt, lont, e1t, e2t
def plot_two_grids(f,g):
# load ss1500mask
tmask, latt, lont, e1t, e2t = loadmask(f)
# load ORAS5
tmask2, latt2, lont2, e1t2, e2t2 = loadmask(g)
#print(tmask[:,])
#plt.subplot(1,1,1)
#plt.figure(figsize=(7,5)); plt.clf()
plt.scatter(lont, latt, tmask)
plt.scatter(lont2, latt2, tmask2)
# Draw sides of every box
#glamfe, gphife = expandf(glamf, gphif)
#NY,NX = glamfe.shape
#for j in range(NY):
# plt.plot(glamfe[j,:],gphife[j,:], 'k')
#for i in range(NX):
# plt.plot(glamfe[:,i],gphife[:,i], 'k')
# Plot t, u, v, f points in red, green, blue, magenta
#plt.plot(glamt, gphit, 'r.')
#plt.plot(glamu, gphiu, 'g.')
#plt.plot(glamv, gphiv, 'b.')
#plt.plot(glamf, gphif, 'm.')
#plt.plot(glamt_2, gphit_2, 'b.')
#plt.plot(glamu, gphiu, 'g.')
#plt.plot(glamv, gphiv, 'b.')
#plt.plot(glamf, gphif, 'm.')
plt.tight_layout()
plt.xlim([-126.2,-122.1])
plt.ylim([46.84,52])
#plt.savefig(f.replace(".nc","_gridpts.png"))
res = "1500m"
ss1500grid = "..//data//grid//coordinates_salishsea_{}.nc".format(res) # in
datetag = "20210406"
oras5grid = "..//data//reference//ORAS5 Mask and Bathy//mesh_mask.nc"
ss1500meshmask = "..//data//mesh mask//mesh_mask_20210406.nc"
np.set_printoptions(threshold=sys.maxsize)
plot_two_grids(ss1500meshmask, oras5grid)
tmask, latt, lont, e1t, e2t = load2(f)
plt.figure(figsize=(8,8))
plt.subplot(1,1,1)
plt.pcolormesh(tmask[:,:], cmap=plt.pink()); plt.title("model_mask")
plt.tight_layout()
plt.figure(figsize=(7,5)); plt.clf()
plt.plot(tmaskutil[0,:],tmaskutil[:,0], 'r.')
with nc.Dataset(ss1500meshmask) as ncid:
print(tmaskutil[:,0])
```
| github_jupyter |
# PART 3 - Metadata Knowledge Graph creation in Amazon Neptune.
Amazon Neptune is a fast, reliable, fully managed graph database service that makes it easy to build and run applications that work with highly connected datasets. The core of Neptune is a purpose-built, high-performance graph database engine. This engine is optimized for storing billions of relationships and querying the graph with milliseconds latency. Neptune supports the popular graph query languages Apache TinkerPop Gremlin and W3C’s SPARQL, enabling you to build queries that efficiently navigate highly connected datasets.
https://docs.aws.amazon.com/neptune/latest/userguide/feature-overview.html
In that section we're going to use TinkerPop Gremlin as the language to create and query our graph.
### Important
We need to downgrade the tornado library for the gremlin libraries to work in our notebook.
Without doing this, you'll most likely run into the following error when executing some gremlin queries:
"RuntimeError: Cannot run the event loop while another loop is running"
```
!pip install --upgrade tornado==4.5.3
```
### Restart your kernel
Because the notebook itself has some dependencies with the tornado library, we need to restart the kernel before proceeding.
To do so, go to the top menu > Kernel > Restart Kernel.. > Restart
Then proceed and execute the following cells.
```
!pip install pandas
!pip install jsonlines
!pip install gremlinpython
!pip install networkx
!pip install matplotlib
import os
import jsonlines
import networkx as nx
import matplotlib.pyplot as plt
import pandas as pd
#load stored variable from previous notebooks
%store -r
```
Loading the Gremlin libraries and connecting to our Neptune instance
```
from gremlin_python import statics
from gremlin_python.process.anonymous_traversal import traversal
from gremlin_python.process.graph_traversal import __
from gremlin_python.process.strategies import *
from gremlin_python.driver.driver_remote_connection import DriverRemoteConnection
from gremlin_python.process.traversal import T
from gremlin_python.process.traversal import Order
from gremlin_python.process.traversal import Cardinality
from gremlin_python.process.traversal import Column
from gremlin_python.process.traversal import Direction
from gremlin_python.process.traversal import Operator
from gremlin_python.process.traversal import P
from gremlin_python.process.traversal import Pop
from gremlin_python.process.traversal import Scope
from gremlin_python.process.traversal import Barrier
from gremlin_python.process.traversal import Bindings
from gremlin_python.process.traversal import WithOptions
from gremlin_python.structure.graph import Graph
graph = Graph()
def start_remote_connection_neptune():
remoteConn = DriverRemoteConnection(your_neptune_endpoint_url,'g')
g = graph.traversal().withRemote(remoteConn)
return g
# g is the traversal source to use to query the graph
g = start_remote_connection_neptune()
```
<b>IMPORTANT:</b>
- Note that the remote connection will time out after few minutes if unused so if you're encountering exceptions after having paused the notebook execution for a while, please re-run the above cell.
- <b>Make sure your Neptune DB is created for the sole purpose of this labs as we'll be cleaning it before starting.</b>
```
#CAREFUL - the below line of code empties your graph. Again, make sure you're using a dedicated instance for this workshop
g.V().drop().iterate()
```
## A note on Gremlin
Gremlin is a functional, data-flow language that enables users to succinctly express complex traversals on (or queries of) their application's property graph. Every Gremlin traversal is composed of a sequence of (potentially nested) steps. A step performs an atomic operation on the data stream. Every step is either a map-step (transforming the objects in the stream), a filter-step (removing objects from the stream), or a sideEffect-step (computing statistics about the stream).
More info here: https://tinkerpop.apache.org/gremlin.html
The image below is an extract from:
https://tinkerpop.apache.org/docs/3.5.1/tutorials/getting-started/#_the_next_fifteen_minutes
I highly recommend you to be familiar with the concepts of Vertex and Edges at the very minimum before proceeding with the notebook.

## Vertices and Edges names
See below the variables containing the labels for our vertices and edges that we'll create across the notebook.
```
#Vertex representing a Video
V_VIDEO = "video"
#Vertex representing a "scene" e.g. SHOT, TECHNICAL_CUE
V_VIDEO_SCENE = "video_scene"
#Vertex representing a Video segment. we arbitrary split our video into 1min segments and attach metadata to the segments itselves
V_VIDEO_SEGMENT = 'video_segment'
#Edge between VIDEO and SEGMENT
E_HAS_SEGMENT = 'contains_segment'
#Edge between VIDEO and SCENE
E_HAS_SCENE = 'contains_scene'
#Edge between Scene and Segment
E_BELONG_TO_SEGMENT = 'belong_to_segment'
#Vertex representing a label extracted by Rekognition from the video
V_LABEL = 'label'
#Edge between SEGMENT and LABEL
E_HAS_LABEL = 'has_label'
#Edge between parent LABEL and child LABEL e.g. construction -> bulldozer
E_HAS_CHILD_LABEL = 'has_child_label'
#Vertex representing the NER
V_ENTITY = 'entities'
#Vertex representing the type of NER
V_ENTITY_TYPE = 'entity_type'
#Edge between ENTITY and ENTITY_TYPE
E_IS_OF_ENTITY_TYPE = 'is_of_entity_type'
#Edge between SEGMENT and ENTITY
E_HAS_ENTITY = 'has_entity'
#Vertex representing a TOPIC
V_TOPIC = 'topic'
#Vertex representing a TOPIC_TERM
V_TOPIC_TERM = 'topic_term'
#Edge between a VIDEO_SEGMENT and a TOPIC
E_HAS_TOPIC = 'has_topic'
#Edge between a TOPIC and a TOPIC_TERM
E_HAS_TERM = 'has_term'
#Vertex representing a TERM
V_TERM = 'term'
```
## We start by adding our video to the Graph
Note how I start with g, our traversal graph, then call the addV (V for Vertex) method and then attach properties to the new vertex. I end the line with ".next()" which will return the newly created node (similar to how an iterator would work). all method are "chained" together in one expression.
```
sample_video_vertex = g.addV(V_VIDEO).property("name", video_name).property("filename", video_file) .property('description', 'description of the video').next()
```
[QUERY] We're listing all the vertices in the graph with their metadata. At this stage, we only have one.
Explanation: g.V() gets us all vertices in the graph, the .hasLabel() filters the vertices based on the vertex label(=type), the .valueMap() returns all properties for all vertices and the .toList() returns the full list. Note that you can use .next() instead of toList() to just return the next element in the list.
```
g.V().hasLabel(V_VIDEO).valueMap().toList()
```
[QUERY] Below is a different way to precisely return a vertex based on its name.
Explanation: g.V() gives us all the vertices, .has() allows us to filter based on the name of the vertex and .next() returns the first (and only) item from the iterator. note that we haven't used .valueMap() so what is returned is the ID of the vertex.
```
g.V().has('name', video_name).next()
```
## Creating 1min segments vertices in Neptune
As mentioned in the previous notebook, we are creating metadata segments that we'll use to store labels and other information related to those 1min video segments.
This will give us a more fine grained view of the video's topics and metadata.
```
print(segment_size_ms)
#get the video duration by looking at the end of the last segment.
def get_video_duration_in_ms(segment_detection_output):
return segment_detection_output['Segments'][-1]['EndTimestampMillis']
#create a new segment vertex and connect it to the video
def add_segment_vertex(video_name, start, end, g):
#retrieving the video vertex
video_vertex = g.V().has(V_VIDEO, 'name', video_name).next()
#generating a segment ID
segment_id = video_name + '-' + str(start) + '-' + str(end)
#creating a new vertex for the segment
new_segment_vert = g.addV(V_VIDEO_SEGMENT).property("name", segment_id).property('StartTimestampMillis', start).property('EndTimestampMillis', end).next()
#connecting the video vertex to the segment vertex
g.V(video_vertex).addE(E_HAS_SEGMENT).to(new_segment_vert).iterate()
#generate segment vertices of a specific duration (default 60s) for a specific video
def generate_segment_vertices(video_name, g, duration_in_millisecs, segment_size_in_millisecs=60000):
#retrieve the mod
modulo = duration_in_millisecs % segment_size_in_millisecs
#counter that we'll increment by segment_size_in_millisecs steps
counter = 0
while ((counter + segment_size_in_millisecs) < duration_in_millisecs) :
start = counter
end = counter + segment_size_in_millisecs
add_segment_vertex(video_name, start, end, g)
counter += segment_size_in_millisecs
#adding the segment vertex to the video vertex
add_segment_vertex(video_name, duration_in_millisecs - modulo, duration_in_millisecs, g)
#add a vertex if it doesn't already exist
def add_vertex(vertex_label, vertex_name, g):
g.V().has(vertex_label,'name', vertex_name).fold().coalesce(__.unfold(), __.addV(vertex_label).property('name',vertex_name)).iterate()
#add an edge between 2 vertices
def add_edge(vertex_label_from, vertex_label_to, vertex_name_from, vertex_name_to, edge_name, g, weight=None):
if weight == None:
g.V().has(vertex_label_to, 'name', vertex_name_to).as_('v1').V().has(vertex_label_from, 'name', vertex_name_from).coalesce(__.outE(edge_name).where(__.inV().as_('v1')), __.addE(edge_name).to('v1')).iterate()
else:
g.V().has(vertex_label_to, 'name', vertex_name_to).as_('v1').V().has(vertex_label_from, 'name', vertex_name_from).coalesce(__.outE(edge_name).where(__.inV().as_('v1')), __.addE(edge_name).property('weight', weight).to('v1')).iterate()
```
Note: remember, the SegmentDetectionOutput object contains the output of the Amazon Rekognition segment (=scene) detection job
```
duration = get_video_duration_in_ms(SegmentDetectionOutput)
generate_segment_vertices(video_name, g, duration, segment_size_ms)
```
[QUERY] Let's retrieve the segments that are connected to the video vertex via an edge, ordered by StartTimestampMillis. In that case we limit the result set to 5 items.
Explanation: g.V() get us all vertices, .has(V_VIDEO, 'name', video_name) filters on the video vertices with name=video_name, .out() gives us all vertices connected to this vertex by an outgoing edge, .hasLabel(V_VIDEO_SEGMENT) filters the vertices to video segments only, .order().by() orders the vertices by StartTimestampMillis, .valueMap() gives us all properties for those vertices, .limit(5) reduces the results to 5 items, .toList() gives us the list of items.
```
list_of_segments = g.V().has(V_VIDEO, 'name', video_name).out().hasLabel(V_VIDEO_SEGMENT) \
.order().by('StartTimestampMillis', Order.asc).valueMap().limit(5).toList()
list_of_segments
```
## Graph Visualisation
The networkx library alongside with matplotlib allows us to draw visually the graph.
Let's draw our vertex video and the 1min segments we just created.
```
#Function printing the graph from a start vertex and a list of edges that will be traversed/displayed.
def print_graph(start_vertex_label, start_vertex_name, list_edges, displayLabels=True, node_size=2000, node_limit=200):
#getting the paths between vertices
paths = g.V().has(start_vertex_label, 'name', start_vertex_name)
#adding the edges that we want to traverse
for edge in list_edges:
paths = paths.out(edge)
paths = paths.path().toList()
#creating graph object
G=nx.DiGraph()
#counters to limit the number of nodes being displayed.
limit_nodes_counter = 0
#creating the graph by iterating over the paths
for p in paths:
#depth of the graph
depth = len(p)
#we build our graph
for i in range(0, depth -1):
label1 = g.V(p[i]).valueMap().next()['name'][0]
label2 = g.V(p[i+1]).valueMap().next()['name'][0]
if limit_nodes_counter < node_limit:
G.add_edge(label1, label2)
limit_nodes_counter += 1
plt.figure(figsize=(12,7))
nx.draw(G, node_size=node_size, with_labels=displayLabels)
plt.show()
#please note that we limit the number of nodes being displayed
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT], node_limit=15)
```
# Add the scenes into our graph
In the below steps we're connecting the scenes to the video itself and not the segments as we want to be able to search and list the different types of scenes at the video level. However, note that we're not going to attach any specific metadata at the scene level, only at the segment level.
```
def store_video_segment(original_video_name, json_segment_detection_output, orig_video_vertex):
shot_counter = 0
tech_cue_counter = 0
for technicalCue in json_segment_detection_output['Segments']:
#start
frameStartValue = technicalCue['StartTimestampMillis'] / 1000
#end
frameEndValue = technicalCue['EndTimestampMillis'] / 1000
#SHOT or TECHNICAL_CUE
segment_type = technicalCue['Type']
counter = -1
if (segment_type == 'SHOT'):
shot_counter += 1
counter = shot_counter
elif (segment_type == 'TECHNICAL_CUE'):
tech_cue_counter += 1
counter = tech_cue_counter
segment_id = original_video_name + '-' + segment_type + '-' + str(counter)
#creating the vertex for the video segment with all the metadata extracted from the segment generation job
new_vert = g.addV(V_VIDEO_SCENE).property("name", segment_id).property("type", segment_type) \
.property('StartTimestampMillis', technicalCue['StartTimestampMillis']).property('EndTimestampMillis', technicalCue['EndTimestampMillis']) \
.property('StartFrameNumber', technicalCue['StartFrameNumber']).property('EndFrameNumber', technicalCue['EndFrameNumber']) \
.property('DurationFrames', technicalCue['DurationFrames']).next()
#creating the edge between the original video vertex and the segment vertex with the type as a property of the relationship
g.V(orig_video_vertex).addE(E_HAS_SCENE).to(new_vert).properties("type", segment_type).iterate()
store_video_segment(video_name, SegmentDetectionOutput, sample_video_vertex)
```
[QUERY] We're retrieving the list of edges/branches created between the video and the scenes.
Explanation: g.V() returns all vertices, .has(V_VIDEO, 'name', video_name) returns the V_VIDEO vertex with name=video_name, .out(E_HAS_SCENE) returns the list of vertices that are connected to the V_VIDEO vertex by a E_HAS_SCENE edge, toList() returns the list of items.
```
list_of_edges = g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SCENE).toList()
print(f"the sample video vertex has now {len(list_of_edges)} edges connecting to the scenes vertices")
```
[QUERY] Let's search for the technical cues (black and fix screens) at the end of the video.
Explanation: g.V() returns all vertices, .has(V_VIDEO, 'name', video_name) returns the V_VIDEO vertex with name=video_name, .out(E_HAS_SCENE) returns the list of vertices that are connected to the V_VIDEO vertex by a E_HAS_SCENE edge, .has('type', 'TECHNICAL_CUE') filters the list on type=TECHNICAL_CUE, the rest was seen above already.
```
g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SCENE) \
.has('type', 'TECHNICAL_CUE') \
.order().by('EndTimestampMillis', Order.desc) \
.limit(5).valueMap().toList()
```
</br>
Let's print the graph for those newly created SCENE vertices
```
#please note that we limit the number of nodes being displayed
print_graph(V_VIDEO, video_name, [E_HAS_SCENE], node_limit=15)
```
## Create the labels vertices and link them to the segments
We're now going to create vertices to represent the labels in our graph and connect them to the 1min segments
```
def create_label_vertices(LabelDetectionOutput, video_name, g, confidence_threshold=80):
labels = LabelDetectionOutput['Labels']
for instance in labels:
#keeping only the labels with high confidence
label_details_obj = instance['Label']
confidence = label_details_obj['Confidence']
if confidence > confidence_threshold:
#adding then main label name to the list
label_name = str(label_details_obj['Name']).lower()
#adding the label vertex
add_vertex(V_LABEL, label_name, g)
#adding the link between video and label
add_edge(V_VIDEO, V_LABEL, video_name, label_name, E_HAS_LABEL, g, weight=None)
#adding parent labels too
parents = label_details_obj['Parents']
if len(parents) > 0:
for parent in parents:
#create parent vertex if it doesn't exist
parent_label_name = str(parent['Name']).lower()
add_vertex(V_LABEL, parent_label_name, g)
#create the relationship between parent and children if it doesn't already exist
add_edge(V_LABEL, V_LABEL, parent_label_name, label_name, E_HAS_CHILD_LABEL, g, weight=None)
create_label_vertices(LabelDetectionOutput, video_name, g, 80)
```
[QUERY] Let's list the labels vertices to see what was created above.
Explanation: g.V() returns all vertices, .hasLabel(V_LABEL) returns only the vertices of label/type V_LABEL, .valueMap().limit(20).toList() gives us the list with properties for the first 20 items.
```
#retrieving a list of the first 20 labels
label_list = g.V().hasLabel(V_LABEL).valueMap().limit(20).toList()
label_list
```
Let's display a graph with our video's labels and the child labels relationships in between labels.
```
print_graph(V_VIDEO, video_name, [E_HAS_LABEL, E_HAS_CHILD_LABEL], node_limit=15)
```
[QUERY] A typical query would be to search for videos who have a specific label.
Explanation: g.V().has(V_LABEL, 'name', ..) returns the first label vertex from the previous computed list, .in_(E_HAS_LABEL) returns all vertices who have an incoming edge (inE) pointing to this label vertex, .valueMap().toList() returns the list with properties.
note that in_(E_HAS_LABEL) is equivalent to .inE(E_HAS_LABEL).outV() where .inE(E_HAS_LABEL) returns all incoming edges with the specified label and .outV() will traverse to the vertices attached to that edge.
Obviously we only have the one result as we've only processed one video so far.
```
g.V().has(V_LABEL, 'name', label_list[0]['name'][0]).in_(E_HAS_LABEL).valueMap().toList()
```
## Create the topics and associated topic terms vertices
We are going to re-arrange a bit the raw results from the topic modeling job to make it more readable
```
comprehend_topics_df.head()
```
We extract the segment id/number from the docname column in a separate column, cast it to numeric values, drop the docname column and sort by segment_id
```
comprehend_topics_df['segment_id'] = comprehend_topics_df['docname'].apply(lambda x: x.split(':')[-1])
comprehend_topics_df['segment_id'] = pd.to_numeric(comprehend_topics_df['segment_id'], errors='coerce')
comprehend_topics_df = comprehend_topics_df.drop('docname', axis=1)
comprehend_topics_df = comprehend_topics_df.sort_values(by='segment_id')
comprehend_topics_df.head(5)
```
Looks better!
Note that:
- a segment_id can belong to several topics
- proportion = the proportion of the document that is concerned with the topic
Let's now create our topic vertices
```
def create_topic_vertices(topics_df, terms_df, video_name, g):
#retrieve all segments for the video
segments_vertex_list = g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SEGMENT).order().by('StartTimestampMillis', Order.asc).valueMap().toList()
for index, row in topics_df.iterrows():
topic = row['topic']
segment_id = int(row['segment_id'])
#string formating to use as name for our vertices
topic_str = str(int(row['topic']))
#adding terms vertices that are associated with that topic and create the topic -> term edge
list_of_terms = terms_df[comprehend_terms_df['topic'] == topic]
#getting the segment name
segment_name = segments_vertex_list[segment_id]['name'][0]
#adding the topic vertex
add_vertex(V_TOPIC, topic_str, g)
#adding the link between entity and entity_type
add_edge(V_VIDEO_SEGMENT, V_TOPIC, segment_name, topic_str, E_HAS_TOPIC, g, weight=None)
#looping across all
for index2, row2 in list_of_terms.iterrows():
term = row2['term']
weight = row2['weight']
add_vertex(V_TERM, term, g)
add_edge(V_TOPIC, V_TERM, topic_str, term, E_HAS_TERM, g, weight=weight)
create_topic_vertices(comprehend_topics_df, comprehend_terms_df, video_name, g)
```
Let's display our video, few segments and their associated topics
```
#please note that we limit the number of nodes being displayed
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT, E_HAS_TOPIC], node_limit=10)
```
Let's display a partial graph showing relationships between the video -> segment -> topic -> term
```
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT, E_HAS_TOPIC, E_HAS_TERM], node_limit=20)
```
[QUERY] We're now listing all the segments that are in topic 2 (try different topic numbers if you want)
Explanation: g.V().has(V_TOPIC, 'name', '2') returns the topic vertex with name=2, .in_(E_HAS_TOPIC) returns all vertices that have a edge pointing into that topic vertex, .valueMap().toList() returns the list of items with their properties
```
g.V().has(V_TOPIC, 'name', '2').in_(E_HAS_TOPIC).valueMap().toList()
```
## Create the NER vertices and link them to the segments
```
#create the entity and entity_type vertices including the related edges
def create_ner_vertices(ner_job_data, video_name, g, score_threshold=0.8):
#retrieve all segments for the video
segments_vertex_list = g.V().has(V_VIDEO, 'name', video_name).out(E_HAS_SEGMENT).order().by('StartTimestampMillis', Order.asc).valueMap().toList()
counter_vertex = 0
for doc in ner_job_data:
#each jsonline from the ner job is already segmented by 1min chunks, so we're just matching them to our ordered segments list.
segment_vertex_name = segments_vertex_list[counter_vertex]['name'][0]
for entity in doc:
text = entity['Text']
type_ = entity['Type']
score = entity['Score']
if score > score_threshold:
#adding the entity type vertex
entity_type_vertex = g.V().has(V_ENTITY_TYPE,'name', type_).fold().coalesce(__.unfold(), __.addV(V_ENTITY_TYPE).property('name',type_)).iterate()
#adding the entity type vertex
entity_vertex = g.V().has(V_ENTITY,'name', text).fold().coalesce(__.unfold(), __.addV(V_ENTITY).property('name',text)).iterate()
#adding the link between entity and entity_type
entity_entity_type_edge = g.V().has(V_ENTITY_TYPE, 'name', type_).as_('v1').V().has(V_ENTITY, 'name', text).coalesce(__.outE(E_IS_OF_ENTITY_TYPE).where(__.inV().as_('v1')), __.addE(E_IS_OF_ENTITY_TYPE).to('v1')).iterate()
#adding the edge between entity and segment
segment_entity_edge = g.V().has(V_ENTITY,'name', text).as_('v1').V().has(V_VIDEO_SEGMENT, 'name', segment_vertex_name).coalesce(__.outE(E_HAS_ENTITY).where(__.inV().as_('v1')), __.addE(E_HAS_ENTITY).to('v1')).iterate()
#print(f"attaching entity: {text} to segment: {segment_vertex_name}")
counter_vertex += 1
create_ner_vertices(ner_job_data, video_name, g, 0.8)
```
[QUERY] Let's get a list of the first 20 entities
Explanation: g.V().hasLabel(V_ENTITY) returns all vertices of label/type V_ENTITY, .valueMap().limit(20).toList() returns the list of the first 20 items with their properties (just name in that case).
```
entities_list = g.V().hasLabel(V_ENTITY).valueMap().limit(20).toList()
entities_list
```
[QUERY] Let's now look up the first entity of the previous entities_list and check its type
Explanation: g.V().has(V_ENTITY, 'name', ...) return the first V_ENTITY vertex of the entities_list list, .out(E_IS_OF_ENTITY_TYPE) returns vertices connected to this V_ENTITY vertex by a E_IS_OF_ENTITY_TYPE edge.
```
g.V().has(V_ENTITY, 'name', entities_list[0]['name'][0]).out(E_IS_OF_ENTITY_TYPE).valueMap().toList()
```
[QUERY] Let's see now which video segments contains that entity
Explanation: g.V().has(V_ENTITY, 'name', ...) return the first V_ENTITY vertex of the entities_list list, .in_(E_HAS_ENTITY) returns all vertices that have an incoming edge into that V_ENTITY vertex and .valueMap().toList() returns the list with properties.
```
g.V().has(V_ENTITY, 'name', entities_list[0]['name'][0]).in_(E_HAS_ENTITY).valueMap().toList()
```
[QUERY] Similar query but this time we traverse further the graph and only return the list of videos which have this specific entity.
Explanation: g.V().has(V_ENTITY, 'name', ...) return the first V_ENTITY vertex of the entities_list list, .in_(E_HAS_ENTITY) returns the V_VIDEO_SEGMENT vertices that have an incoming edge into that V_ENTITY vertex, .in_(E_HAS_SEGMENT) returns the V_VIDEO vertices that have an incoming edge into those V_VIDEO_SEGMENT vertices and .valueMap().toList() returns the list with properties.
Note how by chaining the .in_() methods we are able to traverse the graph from one type of vertex to the other.
```
g.V().has(V_ENTITY, 'name', entities_list[0]['name'][0]).in_(E_HAS_ENTITY).in_(E_HAS_SEGMENT).dedup().valueMap().toList()
```
</br>
Let's now display a graph showing the relationship between Video -> Segment -> Entity
```
print_graph(V_VIDEO, video_name, [E_HAS_SEGMENT, E_HAS_ENTITY], node_size=800, node_limit=30)
```
# Summary
This notebook only touched the surface of what you can do with Graph databases but it should give you an idea of how powerful they are at modeling highly dimensional relationships between entities. This specific architecture allows them to be especially scalable and performing even with billions of vertices and edges.
Gremlin is the most widely used query language for graph DB and provides quite an intuitive way to traverse/query those graphs by chaining those instructions but if you want a more traditional SQL language, you can also look into SPARQL as an alternative.
https://graphdb.ontotext.com/documentation/free/devhub/sparql.html#using-sparql-in-graphdb
| github_jupyter |
```
import os
import pickle
import sys
import numpy as np
import torch
import torch.utils.data
from skimage.color import lab2rgb, rgb2lab, rgb2gray
from torchvision import datasets, transforms
import matplotlib.pyplot as plt
%matplotlib inline
class CIFAR10ImageDataSet(torch.utils.data.Dataset):
def __init__(self, data, transforms=None):
self.data = data
self.transforms = transforms
def __getitem__(self, index):
img = self.data[index]
img_original = transforms.functional.to_pil_image(torch.from_numpy(img.astype(np.uint8)))
if self.transforms is not None:
img_original = self.transforms(img_original)
img_original = np.asarray(img_original)
img_original = img_original / 255
img_lab = rgb2lab(img_original)
img_lab = (img_lab + 128) / 255
img_ab = img_lab[:, :, 1:3]
img_ab = torch.from_numpy(img_ab.transpose((2, 0, 1))).float()
img_gray = rgb2gray(img_original)
img_gray = torch.from_numpy(img_gray).unsqueeze(0).float()
return img_gray, img_ab, img_original
def __len__(self):
return self.data.shape[0]
def unpickle_cifar10(file):
with open(file, 'rb') as fo:
dict = pickle.load(fo, encoding='bytes')
return dict[b"data"]
def get_cifar10_loaders(dataset_path, batch_size):
"""
Get CIFAR-10 data set loaders
"""
'''
Process training data into a DataLoader object
'''
train_transforms = transforms.Compose([
transforms.RandomHorizontalFlip()
])
train_set = datasets.CIFAR10(root=dataset_path, train=True, download=True)
num_training_points = train_set.__len__()
num_points_training_batch = int(num_training_points / batch_size)
train_data = np.array([]).reshape(0, 3, 32, 32)
data_batch_name = 'cifar-10-batches-py/data_batch_{}'
for batch_num in range(1, 6):
data_batch = data_batch_name.format(batch_num)
batch_dir = os.path.join(dataset_path, data_batch)
train_data = np.append(train_data, np.reshape(unpickle_cifar10(batch_dir),
(num_points_training_batch, 3, 32, 32)), 0)
train_lab_data = CIFAR10ImageDataSet(train_data, transforms=train_transforms)
train_loader = torch.utils.data.DataLoader(train_lab_data, batch_size=batch_size, shuffle=True, num_workers=1)
'''
Process validation data into a DataLoader object
'''
val_transforms = transforms.Compose([
transforms.Scale(32)
])
val_set_name = 'cifar-10-batches-py/test_batch'
val_dir = os.path.join(dataset_path, val_set_name)
val_data = unpickle_cifar10(val_dir)
num_points_val_batch = val_data.shape[0]
val_data = np.reshape(val_data, (num_points_val_batch, 3, 32, 32))
val_lab_data = CIFAR10ImageDataSet(val_data, transforms=val_transforms)
val_loader = torch.utils.data.DataLoader(val_lab_data, batch_size=1, shuffle=False, num_workers=1)
return train_loader, val_loader
train_loader, val_loader = get_cifar10_loaders('../data/cifar10', 5)
def to_rgb(grayscale_input, ab_input, colour):
plt.clf() # clear matplotlib
color_image = torch.cat((grayscale_input, ab_input), 0).numpy() # combine channels
color_image = color_image.transpose((1, 2, 0)) # rescale for matplotlib
color_image[:, :, 0:1] = color_image[:, :, 0:1] * 100
color_image[:, :, 1:3] = color_image[:, :, 1:3] * 255 - 128
color_image = lab2rgb(color_image.astype(np.float64))
grayscale_input = grayscale_input.squeeze().numpy()
f, axarr = plt.subplots(1, 3)
axarr[0].imshow(grayscale_input, cmap='gray')
axarr[1].imshow(color_image)
axarr[2].imshow(colour)
axarr[0].axis('off'), axarr[1].axis('off'), axarr[2].axis('off')
plt.show();
for i, (input_gray, input_ab, colour) in enumerate(val_loader):
for j in range(1):
to_rgb(input_gray[j].cpu(), input_ab[j].cpu(), colour[j].cpu())
if i == 10: break
```
| github_jupyter |
```
import pandas as pd
import numpy as np
df = pd.DataFrame({'Map': [0,0,0,1,1,2,2], 'Values': [1,2,3,5,4,2,5]})
df['S'] = df.groupby('Map')['Values'].transform(np.sum)
df['M'] = df.groupby('Map')['Values'].transform(np.mean)
df['V'] = df.groupby('Map')['Values'].transform(np.var)
print (df)
import numpy as np
import pandas as pd
df = pd.DataFrame({'A': [2,3,1], 'B': [1,2,3], 'C': [5,3,4]})
df = df.drop(df.index[[1]])
print (df)
df = df.drop('B', 1)
print (df)
import pandas as pd
df = pd.DataFrame({'A': [0,0,0,0,0,1,1], 'B': [1,2,3,5,4,2,5],
'C': [5,3,4,1,1,2,3]})
a_group_desc = df.groupby('A').describe()
print (a_group_desc)
unstacked = a_group_desc.unstack()
print (unstacked)
import pandas as pd
import numpy as np
s = pd.Series([1, 2, 3, np.NaN, 5, 6, None])
print (s.isnull())
print (s[s.isnull()])
import pandas as pd
import numpy as np
s = pd.Series([1, 2, 3, np.NaN, 5, 6, None])
print (s.fillna(int(s.mean())))
print (s.dropna())
x = np.array([[[1, 2, 3], [4, 5, 6], [7, 8, 9],], [[11,12,13], [14,15,16], [17,18,19],],
[[21,22,23], [24,25,26], [27,28,29]]])
print(x[[[0]]])
values = [1, 5, 8, 9, 2, 0, 3, 10, 4, 7]
import matplotlib.pyplot as plt
plt.plot(range(1,11), values)
plt.savefig('Image.jpeg', format='jpeg')
values = [1, 5, 8, 9, 2, 0, 3, 10, 4, 7]
import matplotlib.pyplot as plt
plt.plot(range(1,11), values)
plt.savefig('MySamplePlot.png', format='png')
values = [1, 5, 8, 9, 2, 0, 3, 10, 4, 7]
import matplotlib.pyplot as plt
plt.plot(range(1,11), values)
plt.savefig('plt.pdf', format='pdf')
import numpy as np
import matplotlib.pyplot as plt
x1 = 50 * np.random.rand(40)
x2 = 25 * np.random.rand(40) + 25
x = np.concatenate((x1, x2))
y1 = 25 * np.random.rand(40)
y2 = 50 * np.random.rand(40) + 25
y = np.concatenate((y1, y2))
plt.scatter(x, y, s=[100], marker='^', c='m')
plt.show()
pip install matplotlib
pip install --upgrade matplotlib
pip install mpl_toolkits
pip install basemap
from mpl_toolkits.basemap import Basemap
import matplotlib.pyplot as plt
m = Basemap(projection='mill')
m.drawcoastlines()
plt.show()
conda install basemap
pip install mpltoolkits.basemap
per = (11438/500)*100
x = 'result = {r:3.2f}%'.format(r=per)
x
coords = {'lat':'37.25N','long':'-115.45W'}
'Coords : {long}, {lat}'.format(**coords)
l = list(x for x in range(1,20))
l
x = [2,4,8,6,3,1,7,9]
x.sort()
x.reverse()
x
l = [(1,2,3), (4,5,6), (7,8,9)]
for x in l:
for y in x:
print(y)
import numpy as np
import matplotlib.pyplot as plt
x = 20 * np.random.randint(1,10,10000)
plt.hist(x, 25,histtype='stepfilled', align='mid', color='g',label='TestData')
plt.legend()
plt.title('Step Filled Histogram')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
data = 50 * np.random.rand(100) - 25
plt.boxplot(data)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x1 = 5 * np.random.rand(40)
x2 = 5 * np.random.rand(40) + 25
x3 = 25 * np.random.rand(20)
x = np.concatenate((x1, x2, x3))
y1 = 5 * np.random.rand(40)
y2 = 5 * np.random.rand(40) + 25
y3 = 25 * np.random.rand(20)
y = np.concatenate((y1, y2, y3))
plt.scatter(x, y, s=[100], marker='^', c='m')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x1 = 5 * np.random.rand(50)
x2 = 5 * np.random.rand(50) + 25
x3 = 30 * np.random.rand(25)
x = np.concatenate((x1, x2, x3))
y1 = 5 * np.random.rand(50)
y2 = 5 * np.random.rand(50) + 25
y3 = 30 * np.random.rand(25)
y = np.concatenate((y1, y2, y3))
color_array = ['b'] * 50 + ['g'] * 50 + ['r'] * 25
plt.scatter(x, y, s=[50], marker='D', c=color_array)
plt.show()
import networkx as nx
g = nx.Graph()
g.add_node(1)
g.add_nodes_from([2,7])
g.add_edge(1,2)
g.add_edges_from([(2,3),(4,5),(6,7),(3,7),(2,5),(4,6)])
nx.draw_networkx(g)
nx.info(g)
import pandas as pd
df = pd.DataFrame({'A': [0,0,0,0,0,1,1], 'B': [1,2,3,5,4,2,5],
'C': [5,3,4,1,1,2,3]})
a_group_desc = df.groupby('A').describe()
print (a_group_desc)
unstacked = a_group_desc.unstack()
print (unstacked)
import nltk
nltk.download()
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
example_sent = "This is a sample sentence, showing off the stop words filtration."
stop_words = set(stopwords.words('english'))
word_tokens = word_tokenize(example_sent)
filtered_sentence = [w for w in word_tokens if not w in stop_words]
print(word_tokens)
print(filtered_sentence)
import networkx as nx
G = nx.cycle_graph(10)
A = nx.adjacency_matrix(G)
print(A.todense())
import numpy as np
import pandas as pd
c = pd.Series(["a", "b", "d", "a", "d"], dtype ="category")
print ("\nCategorical without pandas.Categorical() : \n", c)
c1 = pd.Categorical([1, 2, 3, 1, 2, 3])
print ("\n\nc1 : ", c1)
c2 = pd.Categorical(['e', 'm', 'f', 'i', 'f', 'e', 'h', 'm' ])
print ("\nc2 : ", c2)
import sys
sys.getdefaultencoding( )
from scipy.sparse import csc_matrix
print (csc_matrix([1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0]))
sklearn_hashing_trick = txt.HashingVectorizer( n_features=20, binary=True,norm=None)
text_vector = sklearn_hashing_trick.transform( ['Python for data science','Python for machine learning'])
text_vector
from sklearn.feature_extraction.text import CountVectorizer
document = ["One Geek helps Two Geeks",
"Two Geeks help Four Geeks",
"Each Geek helps many other Geeks at GeeksforGeeks"]
# Create a Vectorizer Object
vectorizer = CountVectorizer()
vectorizer.fit(document)
# Printing the identified Unique words along with their indices
print("Vocabulary: ", vectorizer.vocabulary_)
# Encode the Document
vector = vectorizer.transform(document)
# Summarizing the Encoded Texts
print("Encoded Document is:")
print(vector.toarray())
from sklearn.feature_extraction.text import HashingVectorizer
document = ["One Geek helps Two Geeks",
"Two Geeks help Four Geeks",
"Each Geek helps many other Geeks at GeeksforGeeks"]
# Create a Vectorizer Object
vectorizer = HashingVectorizer()
vectorizer.fit(document)
# Encode the Document
vector = vectorizer.transform(document)
# Summarizing the Encoded Texts
print("Encoded Document is:")
print(vector.toarray())
from sklearn.datasets import load_digits
digits = load_digits()
X, y = digits.data,digits.target
from sklearn.svm import SVC
from sklearn.model_selection import cross_val_score
%timeit single_core_learning = cross_val_score(SVC(), X,y, cv=20, n_jobs=1)
%timeit multi_core_learning = cross_val_score(SVC(), X, y, cv=20, n_jobs=-1)from sklearn.datasets import load_iris
iris = load_iris()
from sklearn.datasets import load_iris
iris = load_iris()
import pandas as pd
import numpy as np
iris_nparray = iris.data
iris_dataframe = pd.DataFrame(iris.data, columns=iris.feature_names)
iris_dataframe['group'] = pd.Series([iris.target_names[k] for k in iris.target],dtype="category")
print (iris_dataframe.mean(numeric_only=True))
print (iris_dataframe.median(numeric_only=True))
print (iris_dataframe.std())
print (iris_dataframe.max(numeric_only=True)-iris_dataframe.min(numeric_only=True) )
print (iris_dataframe.quantile(np.array([0,.25,.50,.75,1])))
pip install scipy
from scipy.stats import kurtosis, kurtosistest
k = kurtosis(iris_dataframe['petal length (cm)'])
zscore, pvalue = kurtosistest(iris_dataframe['petal length (cm)'])
print ('Kurtosis %0.3f\nz-score %0.3f\np-value %0.3f' % (k, zscore, pvalue) )
from scipy.stats import skew, skewtest
s = skew(iris_dataframe['petal length (cm)'])
zscore, pvalue = skewtest(iris_dataframe['petal length (cm)'])
print ('Skewness %0.3f\nz-score %0.3f\np-value %0.3f' % (s, zscore, pvalue))
iris_binned = pd.concat([
pd.qcut(iris_dataframe.iloc[:,0], [0, .25, .5, .75, 1]),
pd.qcut(iris_dataframe.iloc[:,1], [0, .25, .5, .75, 1]),
pd.qcut(iris_dataframe.iloc[:,2], [0, .25, .5, .75, 1]),
pd.qcut(iris_dataframe.iloc[:,3], [0, .25, .5, .75, 1]),
], join='outer', axis = 1)
print(iris_dataframe['group'].value_counts())
print(iris_binned['petal length (cm)'].value_counts())
print(iris_binned.describe())
print (pd.crosstab(iris_dataframe['group'], iris_binned['petal length (cm)']) )
boxplots = iris_dataframe.boxplot(return_type='axes')
from scipy.stats import ttest_ind
group0 = iris_dataframe['group'] == 'setosa'
group1 = iris_dataframe['group'] == 'versicolor'
group2 = iris_dataframe['group'] == 'virginica'
print('var1 %0.3f var2 %03f' % (iris_dataframe['petal length (cm)'][group1].var(),iris_dataframe['petal length (cm)'][group2].var()))
t, pvalue = ttest_ind(iris_dataframe['sepal width (cm)'][group1], iris_dataframe['sepal width (cm)'][group2], axis=0, equal_var=False)
print('t statistic %0.3f p-value %0.3f' % (t, pvalue))
from scipy.stats import f_oneway
f, pvalue = f_oneway(iris_dataframe['sepal width (cm)'][group0],iris_dataframe['sepal width (cm)'][group1],iris_dataframe['sepal width (cm)'][group2])
print("One-way ANOVA F-value %0.3f p-value %0.3f" % (f,pvalue))
from pandas.plotting import parallel_coordinates
iris_dataframe['labels'] = [iris.target_names[k] for k in iris_dataframe['group']]
pll = parallel_coordinates(iris_dataframe,'labels')
densityplot = iris_dataframe[iris_dataframe.columns[:4]].plot(kind='density’)
single_distribution = iris_dataframe['petal length (cm)'].plot(kind='hist')
simple_scatterplot = iris_dataframe.plot(kind='scatter', x='petal length (cm)', y='petal width (cm)')
from pandas import scatter_matrix
matrix_of_scatterplots = scatter_matrix(iris_dataframe, figsize=(6, 6),diagonal='kde')
from sklearn.datasets import load_iris
iris = load_iris()
import pandas as pd
import numpy as np
iris_nparray = iris.data
iris_dataframe = pd.DataFrame(iris.data, columns=iris.feature_names)
iris_dataframe['group'] = pd.Series([iris.target_names[k] for k in iris.target],
dtype="category")
print(iris_dataframe['group'])
from scipy.stats import spearmanr
from scipy.stats.stats import pearsonr
spearmanr_coef, spearmanr_p = spearmanr(iris_dataframe['sepal length (cm)'],iris_dataframe['sepal width (cm)'])
pearsonr_coef, pearsonr_p = pearsonr(iris_dataframe['sepal length (cm)'],iris_dataframe['sepal width (cm)'])
print ('Pearson correlation %0.3f | Spearman correlation %0.3f' % (pearsonr_coef,spearmanr_coef))
from scipy.stats import chi2_contingency
table = pd.crosstab(iris_dataframe['group'], iris_binned['petal length (cm)'])
chi2, p, dof, expected = chi2_contingency(table.values)
print('Chi-square %0.2f p-value %0.3f' % (chi2, p))
from sklearn.preprocessing import scale
stand_sepal_width = scale(iris_dataframe['sepal width (cm)'])
import matplotlib.pyplot as plt
values = [5, 8, 9, 10, 4, 7]
colors = ['b', 'g', 'r', 'c', 'm', 'y']
labels = ['A', 'B', 'C', 'D', 'E', 'F']
explode = (0, 0.2, 0, 0, 0, 0)
plt.pie(values, colors=colors, labels=labels, explode=explode, shadow=True, autopct='%1.2f%%')
plt.title('Values')
plt.show()
import matplotlib.pyplot as plt
values = [5, 8, 9, 10, 4, 7]
widths = [0.7, 0.8, 0.7, 0.7, 0.7, 0.7]
colors = ['b', 'r', 'b', 'b', 'b', 'b']
plt.bar(range(0, 6), values, width=widths,color=colors, align='center')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x = 100 * np.random.randn(10000)
plt.hist(x, histtype='stepfilled', color='g',label='TestData')
plt.legend()
plt.title('Step Filled Histogram')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
x = 100 * np.random.randn(1000)
plt.boxplot(x)
plt.title('Step Filled Histogram')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.pylab as plb
x1 = 15 * np.random.rand(50)
x2 = 15 * np.random.rand(50) + 15
x3 = 30 * np.random.rand(30)
x = np.concatenate((x1, x2, x3))
y1 = 15 * np.random.rand(50)
y2 = 15 * np.random.rand(50) + 15
y3 = 30 * np.random.rand(30)
y = np.concatenate((y1, y2, y3))
color_array = ['b'] * 50 + ['g'] * 50 + ['r'] * 30
plt.scatter(x, y, s=[90], marker='*', c=color_array)
z = np.polyfit(x, y, 1)
p = np.poly1d(z)
plb.plot(x, p(x), 'm-')
plt.show()
import matplotlib.pyplot as plt
import datetime
import numpy as np
x = np.array([datetime.datetime(2021, 1, 1, i, 0) for i in range(24)])
y = np.random.randint(100, size=x.shape)
plt.plot(x,y)
plt.show()
from sklearn.datasets import fetch_20newsgroups
import sklearn.feature_extraction.text as ext
categories = ['sci.space']
twenty_train = fetch_20newsgroups(subset='train',categories=categories,remove=('headers', 'footers', 'quotes’), shuffle=True,random_state=42)
import pandas as pd
import numpy as np
df = pd.DataFrame({'A': [2,1,2,3,3,5,4], 'B': [1,2,3,5,4,2,5], 'C': [5,3,4,1,1,2,3]})
df = df.sort_index(by=['A', 'B'], ascending=[True, True])
df = df.reset_index(drop=True)
df
```
| github_jupyter |
<a href="https://colab.research.google.com/github/allanstar-byte/ESTRELLA/blob/master/SQL_WORLD_SUICIDE_ANALYTICS.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# **SQL DATA CLEANING, OUTLIERS AND ANALYTICS**
# **1. Connecting to our Database**
```
#loading the sql extension into our environment
%load_ext sql
# Then connect to our in memory sqlite database
%sql sqlite://
```
# **2. Importing Data from CSV files**
The dataset we will use contains suicide cases from different countries in the world with different generations, age groups and other factors as outlined below.
```
# Importing the pandas library
# We will use a function read_csv from pandas to read our datasets as shown
#
import pandas as pd
# Loading our table from the respective CSV files
with open('/content/Suicide.csv','r') as f:
Suicide = pd.read_csv(f, index_col=0, encoding='utf-8')
%sql DROP TABLE if EXISTS Suicide
%sql PERSIST Suicide;
%sql SELECT * FROM Suicide LIMIT 5;
```
# **3. Analytics**
```
#1. identifying top 5 countries with the highest suicide cases in the world
%%sql
SELECT Country,
SUM (Suicides_no)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_no) DESC
limit 5;
#2. identifying top 5 countries with the lowest suicide cases in the world
%%sql
SELECT Country,
SUM (Suicides_no)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_no) ASC
limit 5;
#3. identifying the generation with the highest suicide cases
%%sql
SELECT Generation,
SUM (Suicide_rate)
FROM Suicide
GROUP BY Generation
ORDER BY SUM (Suicide_rate) DESC
limit 5;
#4. identifying the generations with the lowest suicide cases
%%sql
SELECT Generation,
SUM (Suicide_rate)
FROM Suicide
GROUP BY Generation
ORDER BY SUM (Suicide_rate) ASC
limit 5;
#5 Investigating which gender has more suicide rates compared to the other one
%%sql
SELECT Sex,
SUM (Suicides_no)
FROM Suicide
GROUP BY Sex
ORDER BY SUM (Suicides_no) DESC
limit 5;
#6. Knowing the age group which most people commit suicide
%%sql
SELECT Age,
SUM (Suicides_no)
FROM Suicide
GROUP BY Age
ORDER BY SUM (Suicide_rate) DESC
limit 5;
#7. Finding out the year where people committed suicide the most
%%sql
SELECT Year,
SUM (Suicides_no)
FROM Suicide
GROUP BY Year
ORDER BY SUM (Suicides_no) DESC
limit 5;
#8. Finding which countries has the most suicides comited at every 100,000
%%sql
SELECT Country,
SUM (Suicides_per_hundred_thousand_pop)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_per_hundred_thousand_pop) DESC
limit 5;
#9. Finding which countries has the leas suicides comited at every 100,000
%%sql
SELECT Country,
SUM (Suicides_per_hundred_thousand_pop)
FROM Suicide
GROUP BY Country
ORDER BY SUM (Suicides_per_hundred_thousand_pop) ASC
limit 7;
#10. Finding which Age groups has the most suicides commited at every 100,000
%%sql
SELECT Age,
SUM (Suicides_per_hundred_thousand_pop)
FROM Suicide
GROUP BY Age
ORDER BY SUM (Suicides_per_hundred_thousand_pop) DESC
limit 5;
```
| github_jupyter |
# Introduction to Python
##***Welcome to your first iPython Notebook.***

## **About iPython Notebooks**
iPython Notebooks are interactive coding environments embedded in a webpage. You will be using iPython notebooks in this class. You only need to write code between the ### START CODE HERE ### and ### END CODE HERE ### comments. After writing your code, you can run the cell by either pressing "SHIFT"+"ENTER" or by clicking on "Run Cell" (denoted by a play symbol) in the left bar of the cell.
**In this notebook you will learn -**
* Basic Syntax
* Variables
* Numbers
* Casting
* String
#Your First Program
**Printing statements and numbers.**
We can use **print** function to display a string , integers, float, complex numbers.
**Example:**
```
print("Hello Friend")
print(30)
```
**Exercise 1.1:**
Display "Batman is the best superhero" using print function.
```
### START CODE HERE ### (1 line of code)
print("Batman is the best superhero")
### END CODE HERE ###
```
**Expected Output: **"Batman is the best superhero"
#Python Variables
**Creating Variables :**
Unlike other programming languages, Python has no command for declaring a variable.
A variable is created the moment you first assign a value to it.
**Example:**
```
x = 5
y = "Python"
print(x)
print(y)
```
Variables do not need to be declared with any particular type and can even change type after they have been set.
```
x = 4 # x is of type int
y = "python" # x is now of type str
print(x)
print(y)
```
**Variable Names :**
A variable can have a short name (like x and y) or a more descriptive name (age, carname, total_volume). Rules for Python variables:
* A variable name must start with a letter or the underscore character.
* A variable name cannot start with a number.
* A variable name can only contain alpha-numeric characters and underscores (A-z, 0-9, and _ ).
* Variable names are case-sensitive (age, Age and AGE are three different variables).
**NOTE:** Remember that variables are case-sensitive.
**Exercise 1.2:**
Create a variable **x** and assign value 10 to it. Create another variable **y** and assign the string **Hello there**. Print both variables.
```
### START CODE HERE ### (4 line of code)
x = 10
y = "Hello there"
print(x)
print(y)
### END CODE HERE ###
```
**Expected Output:**
10
Hello there
**Exercise 1.3:**
Create a variable called **z**, assign** x + y** to it, and display the result.
```
### START CODE HERE ###
x = 5
y = 15
z = x + y
print(z)
### END CODE HERE ###
```
**Expected output: **
20
# Python Numbers
**There are three numeric types in Python:**
* ** int**
* ** float**
* **complex**
**Int **:
Int, or integer, is a whole number, positive or negative, without decimals, of unlimited length.
```
x = 1
y = 35656222554887711
z = -3255522
print(type(x)) # To verify the type of any object in Python, use the type() function
print(type(y))
print(type(z))
```
**Float :**
Float, or "floating point number" is a number, positive or negative, containing one or more decimals.
```
x = 1.10
y = 1.0
z = -35.59
print(type(x))
print(type(y))
print(type(z))
```
**Complex :**
Complex numbers are written with a "j" as the imaginary part.
```
x = 3+5j
y = 5j
z = -5j
print(type(x))
print(type(y))
print(type(z))
```
**Exercise 1.4:**
Find whether E=3.4j is integer, float or complex.
```
### START CODE HERE ### (1 line of code)
E=3.4j
print(type(E))
### END CODE HERE ###
```
**Expected output:** class 'complex'
# Python Casting
**Specify a Variable Type :**
There may be times when you want to specify a type on to a variable. This can be done with casting. Python is an object-orientated language, and as such it uses classes to define data types, including its primitive types.
Casting in python is therefore done using constructor functions:
A **literal** is a notation for representing a fixed value in source code.
* **int()** - constructs an integer number from an integer literal, a float literal (by rounding down to the previous whole number), or a string literal (providing the string represents a whole number)
* **float()** - constructs a float number from an integer literal, a float literal or a string literal (providing the string represents a float or an integer)
* **str()** - constructs a string from a wide variety of data types, including strings, integer literals and float literals
**Integers**:
```
x = int(1) # x will be 1
y = int(2.8) # y will be 2
z = int("3") # z will be 3
```
**Floats:**
```
x = float(1) # x will be 1.0
y = float(2.8) # y will be 2.8
z = float("3") # z will be 3.0
w = float("4.2") # w will be 4.2
```
**Strings:**
```
x = str("s1") # x will be 's1'
y = str(2) # y will be '2'
z = str(3.0) # z will be '3.0'
```
Main advantage of type casting is you can print and integer and a string in the same line.
**Example: **
```
a = " kingdoms in Westeros"
b = str(7)
print (b + a)
```
**Excercise 1.5:**
Create a variable **x** and assign the integer 3 to it. Create another variable **y** and assign string '4' to it. Add both variables using **int** function.
```
### START CODE HERE ### (≈ 4 lines of code)
x = 3
y = "4"
z = x + int(y)
print(z)
### END CODE HERE ###
```
**Expected Output:**
7
#Python Strings
**String literals**:
String literals in python are surrounded by either single quotation marks, or double quotation marks.
'hello' is the same as "hello".
Like many other popular programming languages, strings in Python are arrays of bytes representing unicode characters. Square brackets can be used to access elements of the string.
```
a = "Hello, World!"
print(a[1]) # Gets the character at position 1 (remember that the first character has the position 0)
b = "Hello, World!"
print(b[2:5]) #Gets the characters from position 2 to position 5 (not included)
```
**The strip() method:**
The strip() method removes any whitespace from the beginning or the end:
```
a = " Hello, World! "
print(a.strip()) # returns "Hello, World!"
```
**The len() method:**
The len() method returns the length of a string
```
a = "Hello, World!"
print(len(a))
```
**The lower() method:**
The lower() method returns the string in lower case
```
a = "Hello, World!"
print(a.lower())
print(a) #Orignal value of a is not changed
a=a.lower() #Orignal value of a is changed
print(a)
```
**The upper() method:**
The upper() method returns the string in upper case
```
a = "Hello, World!"
print(a.upper())
```
**The replace() method :**
The replace() method replaces a string with another string.
```
a = "Hello, World!"
print(a.replace("H", "J"))
```
**The split() method :**
The split() method splits the string into substrings if it finds instances of the separator
```
a = "Hello, World!"
print(a.split(",")) # returns ['Hello', ' World!']
```
**Exercise 1.6:**
Get the first character of the string **str** and print it.
```
### START CODE HERE ### (≈ 3 lines of code)
str="Learning python"
x = str[0]
print(x)
### END CODE HERE ###
```
**Expected Output:**
L
**Exercise 1.7:**
Get the characters from position 3 to position 8 (not included) using strinf slicing method and print it.
```
### START CODE HERE ### (≈ 3 lines of code)
str="Learning python"
x = str[3:8]
print(x)
### END CODE HERE ###
```
**Expected Output:**
rning
**Exercise 1.8:**
For E="HELLO FRIENS" make the string lowercase, print, replace **s** by **d** and return the length of the string.
```
### START CODE HERE ### (≈ 4-5 lines of code)
E = "HELLO FRIENS"
E = E.lower()
print(E)
E = E.replace("s","d")
print(E)
print(len(E))
### END CODE HERE ###
```
**Expected Output:**
hello friens
hello friend
12
# Great Job!
| github_jupyter |
<a href="https://colab.research.google.com/github/AngieCat26/MujeresDigitales/blob/main/Taller_semana_7.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Introducción
**Contexto comercial.** Usted es un analista en una entidad bancaria, y se le proporciona un conjunto de datos de los clientes. Su jefe le pide que analice la información para determinar si existen similaridades entre grupos de clientes para lanzar una campaña de mercadeo.
**Problema comercial.** Su tarea es **crear un modelo de clusterización para determinar si existen grupos de clientes similares**.
**Contexto analítico.** Como científico de datos, se le pide realizar una clusterización de los clientes para identificar
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import scipy
import seaborn as sns
import sklearn # Paquete base de ML
from scipy.stats import norm
from sklearn.cluster import KMeans
from sklearn.preprocessing import MinMaxScaler, MaxAbsScaler, RobustScaler, StandardScaler
%matplotlib inline
url = 'https://raw.githubusercontent.com/AngieCat26/MujeresDigitales/main/Lending_club_cleaned_2.csv'
df = pd.read_csv(url)
df.head()
```
## Ejercicio 1:
Realice una normalización de los datos numéricos es decir que los valores oscilen entre 0 y 1 en las columnas annual_inc y loan_amnt.
Consejo: antes de realizar la normalización asegúrese de que el tipo de dichas columnas si sea numérico.
```
# Escriba aquí su codigo
def normalize(df):
resultado = df.copy()
for normalizado in df.columns:
max = df[normalizado].max()
min = df[normalizado].min()
resultado[normalizado] = (df[normalizado] - min) / (max - min)
return resultado
df_normalizado = normalize(df[['annual_inc', 'loan_amnt']])
df_normalizado.head(11)
```
## Ejercicio 2:
Emplee el algoritmo de k-means para agrupar a los clientes usando un número de clusters de 4.
```
# Escriba aquí su codigo
k = 4
kmeans = KMeans(n_clusters = k, init='k-means++')
kmeans.fit(df_normalizado)
labels = kmeans.predict(df_normalizado)
centroids = kmeans.cluster_centers_
centroids
```
## Ejercicio 3 (Opcional):
Realice un gráfico de dispersión (scatter) para vizualizar los cluster que descubrió en el punto anterior (ejercicio 2). Usando colores diferentes para identificar los 4 cluster.
```
# Escriba aquí su codigo
plt.figure(figsize=(6, 6))
color_map = {1:'r', 2:'g', 3:'b' , 4:'c'}
colors = [color_map[x+1] for x in labels]
plt.scatter(df_normalizado['annual_inc'], df_normalizado['loan_amnt'], color=colors, alpha=0.4, edgecolor='k')
for idx, centroid in enumerate(centroids):
plt.scatter(*centroid, marker='*', edgecolor='k')
plt.xlim(-0.25, 1.25)
plt.xlabel('annual_inc', fontsize=12)
plt.xticks(fontsize=12)
plt.ylim(-0.25, 1.25)
plt.ylabel('loan_amnt', fontsize=12)
plt.yticks(fontsize=12)
plt.title('annual_inc VS loan_amnt', fontsize=16)
plt.show()
```
## Ejercicio 4 (Opcional):
Use el método del codo para verificar cual es el número de clusters óptimo. Revise desde 1 clúster hasta 11 para realizar esta validación.
```
# Escriba aquí su codigo
sum_sq_d = []
K = range(1, 11)
for k in K:
km = KMeans(n_clusters = k)
km = km.fit(df_normalizado[['annual_inc', 'loan_amnt']])
sum_sq_d.append(km.inertia_)
plt.figure(figsize=(8,6))
plt.plot(K, sum_sq_d, 'rx-.')
plt.xlabel('Numero de Clusters, k', fontsize=12)
plt.xticks(range(1,11), fontsize=12)
plt.ylabel('suma de Distancias al Cuadradp', fontsize=12)
plt.xticks(fontsize=12)
plt.title('Metodo del Codo', fontsize=16)
plt.show()
```
| github_jupyter |
# Hands On: Seleksi Fitur
Seleksi fitur (feature selection) adalah proses memilih feature yang tepat untuk melatih model ML.
Untuk melakukan feature selection, kita perlu memahami hubungan antara variables.
Hubungan antar dua random variables disebut correlation dan dapat dihitung dengan menggunakan correlation coefficient.
Range nilai correlation coeficient adalah:
Positif maks +1, korelasi positif, artinya kedua variable akan bergerak searah.
Negatif maks -1, korelasi negatif, artinya kedua variable akan bergerak berlawanan.
Nol, menunjukan antara kedua variable tidak ada correlation.
Teknik perhitungan correlation cukup banyak, berikut yang umum digunakan: Pearson, Kendall dan Spearman.
A. Pearson
* Paling umum digunakan.
* Digunakan untuk numerical data.
* Tidak bisa digunakan untuk ordinal data.
* Mengukur linear data dengan asumsi data terdistribusi normal.
B. Kendall
* Rank correlation measure.
* Dapat digunakan untuk numerical dan ordinal data, namun tidak untuk nominal data.
* Tidak diperlukan linear relationship antar variable.
* Digunakan untuk mengukur kemiripan ranked ordering data.
* Untuk kondisi normal lebih baik menggunakan Kendall dibandingkan Spearman.
C. Spearman
* Rank correlation measure
* Dapat digunakan untuk numerical dan ordinal data, namun tidak untuk nominal data.
* Tidak diperlukan linear relationship antar variable.
* Monotonic relationship
Ada beberapa metoda feature selection yang umum digunakan, yaitu Filter, Embedded dan Wrapper.
**Filter Method**
Umumnya digunakan pada tahap preprocessing. Pemilihan features tidak tergantung kepada algoritma ML yang akan digunakan . Features dipilih berdasarkan score test statistik kolerasi.
**Embedded Method**
Feature dipilih saat proses model training. Menggunakan learning algorithm untuk melakukan variable selection dan feature selection and classification secara simultan. Harus memilih algoritma machine learning yang sesuai.
**Wrapper Method**
Menggunakan subset of features untuk melatih model. Berdasarkan hasil yang dihasilkan dari model sebelumnya, kita tentukan untuk menambah atau membuang features dari subset. Kelemahannya membutuhkan resource besar dalam melakukan komputasi.
Ada jenis seleksi fitur lainnya, seperti dalam slide modul 8 ini, diantaranya:
1. Seleksi Univariat (Univariate Selection)
2. Pentingnya Fitur (Feature Importance)
3. Matriks Korelasi (Correlation Matrix) dengan Heatmap
Teknik pemilihan fitur yang perlu kita ketahui, untuk mendapatkan performa terbaik dari model Anda.
1. SelectKBest
2. Regresi linier
3. Random Forest
4. XGBoost
5. Penghapusan Fitur Rekursif
6. Boruta
### Berikut ini adalah sebagian kecil dari metode/teknik dalam Seleksi Fitur
#### Sumber dataset:
---
https://www.kaggle.com/iabhishekofficial/mobile-price-classification#train.csv
### 1. Seleksi Unvariate
---
Metode paling sederhana dan tercepat didasarkan pada uji statistik univariat. Untuk setiap fitur, ukur seberapa kuat target bergantung pada fitur menggunakan uji statistik seperti χ2 (chi-square) or ANOVA.
Uji statistik dapat digunakan untuk memilih fitur-fitur tersebut yang memiliki relasi paling kuat dengan variabel output/target.
Library scikit-learn menyediakan class *SelectKBest* yang digunakan untuk serangkaian uji statistik berbeda untuk memilih angka spesifik dari fitur. Berikut ini adalah uji statistik chi-square utk fitur non-negatif untuk memilih 10 fitur terbaik dari dataset *Mobile Price Range Prediction*.
```
import pandas as pd
import numpy as np
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
data = pd.read_csv("train.csv")
X = data.iloc[:,0:20] #independent colums
y = data.iloc[:,-1] # target colum i.e price range
# apply SelectKBest class to extract
bestfeatures = SelectKBest(score_func=chi2, k=10)
fit = bestfeatures.fit(X,y)
dfscores = pd.DataFrame(fit.scores_)
dfcolumns = pd.DataFrame(X.columns)
#concat two dataframes for better visualization
featureScores = pd.concat([dfcolumns,dfscores],axis=1)
featureScores.columns = ['Specs','Score'] #naming the dataframe columns
print(featureScores.nlargest(10,'Score')) #print 10 best features
```
### 2. Feature Importance
---
*Feature importance* mengacu pada kelas teknik untuk menetapkan skor ke fitur input ke model prediktif yang menunjukkan *importance* relatif dari setiap fitur saat membuat prediksi.
Skor *Feature importance* dapat dihitung untuk masalah yang melibatkan prediksi nilai numerik, yang disebut regresi, dan masalah yang melibatkan prediksi label kelas, yang disebut klasifikasi.
Skor berguna dan dapat digunakan dalam berbagai situasi dalam masalah pemodelan prediktif, seperti:
* Lebih memahami data.
* Lebih memahami model.
* Mengurangi jumlah fitur input.
* memberi skor untuk setiap fitur data, semakin tinggi skor semakin penting atau relevan fitur tersebut terhadap variabel output
inbuilt yang dilengkapi dengan Pengklasifikasi Berbasis Pohon (Tree Based Classifier), kami akan menggunakan Pengklasifikasi Pohon Ekstra untuk mengekstraksi 10 fitur teratas untuk kumpulan data
```
import pandas as pd
import numpy as np
data = pd.read_csv("train.csv")
X = data.iloc[:,0:20] #independent columns
y = data.iloc[:,-1] #target column i.e price range
from sklearn.ensemble import ExtraTreesClassifier
import matplotlib.pyplot as plt
model = ExtraTreesClassifier()
model.fit(X,y)
print(model.feature_importances_) #use inbuilt class feature_importances of tree based classifiers
#plot graph of feature importances for better visualization
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(10).plot(kind='barh')
plt.show()
```
### 3. Matriks Korelasi dengan Heatmap
---
* Korelasi menyatakan bagaimana fitur terkait satu sama lain atau variabel target.
* Korelasi bisa positif (kenaikan satu nilai fitur meningkatkan nilai variabel target) atau negatif (kenaikan satu nilai fitur menurunkan nilai variabel target)
* Heatmap memudahkan untuk mengidentifikasi fitur mana yang paling terkait dengan variabel target, kami akan memplot peta panas fitur yang berkorelasi menggunakan seaborn library
```
import pandas as pd
import numpy as np
import seaborn as sns
data = pd.read_csv("train.csv")
X = data.iloc[:,0:20] #independent columns
y = data.iloc[:,-1] #target column i.e price range
#get correlations of each features in dataset
corrmat = data.corr()
top_corr_features = corrmat.index
plt.figure(figsize=(20,20))
#plot heat map
g=sns.heatmap(data[top_corr_features].corr(),annot=True,cmap="RdYlGn")
```
| github_jupyter |
# Your first neural network
In this project, you'll build your first neural network and use it to predict daily bike rental ridership. We've provided some of the code, but left the implementation of the neural network up to you (for the most part). After you've submitted this project, feel free to explore the data and the model more.
```
%matplotlib inline
%load_ext autoreload
%autoreload 2
%config InlineBackend.figure_format = 'retina'
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
```
## Load and prepare the data
A critical step in working with neural networks is preparing the data correctly. Variables on different scales make it difficult for the network to efficiently learn the correct weights. Below, we've written the code to load and prepare the data. You'll learn more about this soon!
```
data_path = 'Bike-Sharing-Dataset/hour.csv'
rides = pd.read_csv(data_path)
rides.head()
```
## Checking out the data
This dataset has the number of riders for each hour of each day from January 1 2011 to December 31 2012. The number of riders is split between casual and registered, summed up in the `cnt` column. You can see the first few rows of the data above.
Below is a plot showing the number of bike riders over the first 10 days or so in the data set. (Some days don't have exactly 24 entries in the data set, so it's not exactly 10 days.) You can see the hourly rentals here. This data is pretty complicated! The weekends have lower over all ridership and there are spikes when people are biking to and from work during the week. Looking at the data above, we also have information about temperature, humidity, and windspeed, all of these likely affecting the number of riders. You'll be trying to capture all this with your model.
```
rides[:24*10].plot(x='dteday', y='cnt')
```
### Dummy variables
Here we have some categorical variables like season, weather, month. To include these in our model, we'll need to make binary dummy variables. This is simple to do with Pandas thanks to `get_dummies()`.
```
dummy_fields = ['season', 'weathersit', 'mnth', 'hr', 'weekday']
for each in dummy_fields:
dummies = pd.get_dummies(rides[each], prefix=each, drop_first=False)
rides = pd.concat([rides, dummies], axis=1)
fields_to_drop = ['instant', 'dteday', 'season', 'weathersit',
'weekday', 'atemp', 'mnth', 'workingday', 'hr']
data = rides.drop(fields_to_drop, axis=1)
data.head()
```
### Scaling target variables
To make training the network easier, we'll standardize each of the continuous variables. That is, we'll shift and scale the variables such that they have zero mean and a standard deviation of 1.
The scaling factors are saved so we can go backwards when we use the network for predictions.
```
quant_features = ['casual', 'registered', 'cnt', 'temp', 'hum', 'windspeed']
# Store scalings in a dictionary so we can convert back later
scaled_features = {}
for each in quant_features:
mean, std = data[each].mean(), data[each].std()
scaled_features[each] = [mean, std]
data.loc[:, each] = (data[each] - mean)/std
```
### Splitting the data into training, testing, and validation sets
We'll save the data for the last approximately 21 days to use as a test set after we've trained the network. We'll use this set to make predictions and compare them with the actual number of riders.
```
# Save data for approximately the last 21 days
test_data = data[-21*24:]
# Now remove the test data from the data set
data = data[:-21*24]
# Separate the data into features and targets
target_fields = ['cnt', 'casual', 'registered']
features, targets = data.drop(target_fields, axis=1), data[target_fields]
test_features, test_targets = test_data.drop(target_fields, axis=1), test_data[target_fields]
```
We'll split the data into two sets, one for training and one for validating as the network is being trained. Since this is time series data, we'll train on historical data, then try to predict on future data (the validation set).
```
# Hold out the last 60 days or so of the remaining data as a validation set
train_features, train_targets = features[:-60*24], targets[:-60*24]
val_features, val_targets = features[-60*24:], targets[-60*24:]
```
## Time to build the network
Below you'll build your network. We've built out the structure. You'll implement both the forward pass and backwards pass through the network. You'll also set the hyperparameters: the learning rate, the number of hidden units, and the number of training passes.
<img src="assets/neural_network.png" width=300px>
The network has two layers, a hidden layer and an output layer. The hidden layer will use the sigmoid function for activations. The output layer has only one node and is used for the regression, the output of the node is the same as the input of the node. That is, the activation function is $f(x)=x$. A function that takes the input signal and generates an output signal, but takes into account the threshold, is called an activation function. We work through each layer of our network calculating the outputs for each neuron. All of the outputs from one layer become inputs to the neurons on the next layer. This process is called *forward propagation*.
We use the weights to propagate signals forward from the input to the output layers in a neural network. We use the weights to also propagate error backwards from the output back into the network to update our weights. This is called *backpropagation*.
> **Hint:** You'll need the derivative of the output activation function ($f(x) = x$) for the backpropagation implementation. If you aren't familiar with calculus, this function is equivalent to the equation $y = x$. What is the slope of that equation? That is the derivative of $f(x)$.
Below, you have these tasks:
1. Implement the sigmoid function to use as the activation function. Set `self.activation_function` in `__init__` to your sigmoid function.
2. Implement the forward pass in the `train` method.
3. Implement the backpropagation algorithm in the `train` method, including calculating the output error.
4. Implement the forward pass in the `run` method.
```
#############
# In the my_answers.py file, fill out the TODO sections as specified
#############
def MSE(y, Y):
return np.mean((y-Y)**2)
```
## Unit tests
Run these unit tests to check the correctness of your network implementation. This will help you be sure your network was implemented correctly befor you starting trying to train it. These tests must all be successful to pass the project.
```
from my_answers import NeuralNetwork
import unittest
inputs = np.array([[0.5, -0.2, 0.1]])
targets = np.array([[0.4]])
test_w_i_h = np.array([[0.1, -0.2],
[0.4, 0.5],
[-0.3, 0.2]])
test_w_h_o = np.array([[0.3],
[-0.1]])
class TestMethods(unittest.TestCase):
##########
# Unit tests for data loading
##########
def test_data_path(self):
# Test that file path to dataset has been unaltered
self.assertTrue(data_path.lower() == 'bike-sharing-dataset/hour.csv')
def test_data_loaded(self):
# Test that data frame loaded
self.assertTrue(isinstance(rides, pd.DataFrame))
##########
# Unit tests for network functionality
##########
def test_activation(self):
network = NeuralNetwork(3, 2, 1, 0.5)
# Test that the activation function is a sigmoid
self.assertTrue(np.all(network.activation_function(0.5) == 1/(1+np.exp(-0.5))))
def test_train(self):
# Test that weights are updated correctly on training
network = NeuralNetwork(3, 2, 1, 0.5)
network.weights_input_to_hidden = test_w_i_h.copy()
network.weights_hidden_to_output = test_w_h_o.copy()
network.train(inputs, targets)
self.assertTrue(np.allclose(network.weights_hidden_to_output,
np.array([[ 0.37275328],
[-0.03172939]])))
self.assertTrue(np.allclose(network.weights_input_to_hidden,
np.array([[ 0.10562014, -0.20185996],
[0.39775194, 0.50074398],
[-0.29887597, 0.19962801]])))
def test_run(self):
# Test correctness of run method
network = NeuralNetwork(3, 2, 1, 0.5)
network.weights_input_to_hidden = test_w_i_h.copy()
network.weights_hidden_to_output = test_w_h_o.copy()
self.assertTrue(np.allclose(network.run(inputs), 0.09998924))
suite = unittest.TestLoader().loadTestsFromModule(TestMethods())
unittest.TextTestRunner().run(suite)
```
## Training the network
Here you'll set the hyperparameters for the network. The strategy here is to find hyperparameters such that the error on the training set is low, but you're not overfitting to the data. If you train the network too long or have too many hidden nodes, it can become overly specific to the training set and will fail to generalize to the validation set. That is, the loss on the validation set will start increasing as the training set loss drops.
You'll also be using a method know as Stochastic Gradient Descent (SGD) to train the network. The idea is that for each training pass, you grab a random sample of the data instead of using the whole data set. You use many more training passes than with normal gradient descent, but each pass is much faster. This ends up training the network more efficiently. You'll learn more about SGD later.
### Choose the number of iterations
This is the number of batches of samples from the training data we'll use to train the network. The more iterations you use, the better the model will fit the data. However, this process can have sharply diminishing returns and can waste computational resources if you use too many iterations. You want to find a number here where the network has a low training loss, and the validation loss is at a minimum. The ideal number of iterations would be a level that stops shortly after the validation loss is no longer decreasing.
### Choose the learning rate
This scales the size of weight updates. If this is too big, the weights tend to explode and the network fails to fit the data. Normally a good choice to start at is 0.1; however, if you effectively divide the learning rate by n_records, try starting out with a learning rate of 1. In either case, if the network has problems fitting the data, try reducing the learning rate. Note that the lower the learning rate, the smaller the steps are in the weight updates and the longer it takes for the neural network to converge.
### Choose the number of hidden nodes
In a model where all the weights are optimized, the more hidden nodes you have, the more accurate the predictions of the model will be. (A fully optimized model could have weights of zero, after all.) However, the more hidden nodes you have, the harder it will be to optimize the weights of the model, and the more likely it will be that suboptimal weights will lead to overfitting. With overfitting, the model will memorize the training data instead of learning the true pattern, and won't generalize well to unseen data.
Try a few different numbers and see how it affects the performance. You can look at the losses dictionary for a metric of the network performance. If the number of hidden units is too low, then the model won't have enough space to learn and if it is too high there are too many options for the direction that the learning can take. The trick here is to find the right balance in number of hidden units you choose. You'll generally find that the best number of hidden nodes to use ends up being between the number of input and output nodes.
```
import sys
####################
### Set the hyperparameters in you myanswers.py file ###
####################
from my_answers import iterations, learning_rate, hidden_nodes, output_nodes
N_i = train_features.shape[1]
network = NeuralNetwork(N_i, hidden_nodes, output_nodes, learning_rate)
losses = {'train':[], 'validation':[]}
for ii in range(iterations):
# Go through a random batch of 128 records from the training data set
batch = np.random.choice(train_features.index, size=128)
X, y = train_features.loc[batch].values, train_targets.loc[batch]['cnt']
network.train(X, y)
# Printing out the training progress
train_loss = MSE(network.run(train_features).T, train_targets['cnt'].values)
val_loss = MSE(network.run(val_features).T, val_targets['cnt'].values)
sys.stdout.write("\rProgress: {:2.1f}".format(100 * ii/float(iterations)) \
+ "% ... Training loss: " + str(train_loss)[:5] \
+ " ... Validation loss: " + str(val_loss)[:5])
sys.stdout.flush()
losses['train'].append(train_loss)
losses['validation'].append(val_loss)
plt.plot(losses['train'], label='Training loss')
plt.plot(losses['validation'], label='Validation loss')
plt.legend()
_ = plt.ylim()
```
## Check out your predictions
Here, use the test data to view how well your network is modeling the data. If something is completely wrong here, make sure each step in your network is implemented correctly.
```
fig, ax = plt.subplots(figsize=(8,4))
mean, std = scaled_features['cnt']
predictions = network.run(test_features).T*std + mean
ax.plot(predictions[0], label='Prediction')
ax.plot((test_targets['cnt']*std + mean).values, label='Data')
ax.set_xlim(right=len(predictions))
ax.legend()
dates = pd.to_datetime(rides.loc[test_data.index]['dteday'])
dates = dates.apply(lambda d: d.strftime('%b %d'))
ax.set_xticks(np.arange(len(dates))[12::24])
_ = ax.set_xticklabels(dates[12::24], rotation=45)
```
## OPTIONAL: Thinking about your results(this question will not be evaluated in the rubric).
Answer these questions about your results. How well does the model predict the data? Where does it fail? Why does it fail where it does?
> **Note:** You can edit the text in this cell by double clicking on it. When you want to render the text, press control + enter
#### Your answer below
The prediction of the model is almost on point for the timespan before the 21.12.
After the 21.12 the model predicts the demain of bike too high.
In my option the reason for that can befound in th
* that the model doenst take into account that most people are on holiday between 21.12 and New Year (the Witching Week)
* To less data: The whole data set consist of 1 year. In order for the model to learn that the 21.12-31.12 is a special time of the year it would need a data set which consist of several years.
| github_jupyter |
<a href="https://colab.research.google.com/github/dyjdlopez/linearAlgebra2021/blob/main/Week%202%20-%20Intro%20to%20Vectors%20and%20Numpy/LinAlg_Lab_2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Lab 2 - Plotting Vector using NumPy and MatPlotLib
In this laboratory we will be disucssin the basics of numerical and scientific programming by working with Vectors using NumPy and MatPlotLib.
### Objectives
At the end of this activity you will be able to:
1. Be familiar with the libraries in Python for numerical and scientific programming.
2. Visualize vectors through Python programming.
3. Perform simple vector operations through code.
## Discussion
### *NumPy*
NumPy or Numerical Python, is mainly used for matrix and vector operations. It is capable of declaring computing and representing matrices. Most Python scienitifc programming libraries uses NumPy as the basic code.
### Defining Vectors, Matrices, and Tensors
Vectors, Matrices, and Tensors are the fundamental objects in Linear Algebra programming. We'll be defining each of these objects specifically in the Computer Science/Engineering perspective since it would be much confusing if we consider their Physics and Pure Mathematics definitions.
#### <i>Scalars</i>
Scalars are numerical entities that are represented by a single value.
```
import numpy as np
x = np.array(-0.5)
x
```
#### *Vectors*
Vectors are array of numerical values or scalars that would represent any feature space. Feature spaces or simply dimensions or the parameters of an equation or a function.
#### *Representing Vectors*
Now that you know how to represent vectors using their component and matrix form we can now hard-code them in Python. Let's say that you have the vectors:
$$ A = 4\hat{x} + 3\hat{y} \\
B = 2\hat{x} - 5\hat{y}$$
In which it's matrix equivalent is:
$$ A = \begin{bmatrix} 4 \\ 3\end{bmatrix} , B = \begin{bmatrix} 2 \\ -5\end{bmatrix}\\
A = \begin{bmatrix} 4 & 3\end{bmatrix} \\
B = \begin{bmatrix} 2 & -5\end{bmatrix}
$$
We can then start doing numpy code with this by:
```
A = np.array([4,3])
B = np.array([2, -5])
print('Vector A is ', A)
print('Vector B is ', B)
```
#### Describing vectors in NumPy
Describing vectors is very important if we want to perform basic to advanced operations with them. The fundamental ways in describing vectors are knowing their shape, size and dimensions.
```
### Checking shapes
### Shapes tells us how many rows and columns are there
ball1 = np.array([1,2,3])
ball2 = np.array([0,1,-1])
pool = np.array([J,K]) ## Matrix
pool.shape
U = np.array([
[1, 2],
[2, 3]
])
U.shape
### Checking size
### Array/Vector sizes tells us many total number of elements are there in the vector
U.size
### Checking dimensions
### The dimensions or rank of a vector tells us how many dimensions are there for the vector.
A.ndim
pool.ndim
```
Great! Now let's try to explore in performing operations with these vectors.
#### Addition
The addition rule is simple, the we just need to add the elements of the matrices according to their index. So in this case if we add vector $A$ and vector $B$ we will have a resulting vector:
$$R = 6\hat{x}-2\hat{y} \\ \\or \\ \\ R = \begin{bmatrix} 6 \\ -2\end{bmatrix} $$
So let's try to do that in NumPy in several number of ways:
```
position1 = np.array([0, 0, 0])
position2 = np.array([1, 1, 0])
position3 = np.array([-1, 2, 0])
position4 = np.array([2, 5, 3])
R = position1 + position2 + position3 + position4 #Eager execution
R
R1 = np.add(position1,position2) #functional method
R2 = np.add(R1,position3)
R3 = np.add(R2,position4)
R3
Rm = np.multiply(position3, position4)
Rm
Rm = position3 * position4
Rm
```
##### Try for yourself!
Try to implement subtraction and division with vectors $A$ and $B$!
```
### Try out you code here! Don't forget to take a screenshot or a selfie!
```
$$
W = \hat{x} + \hat{y}\\
T = -2\hat{x} -3\hat{y}\\
R3 = W + (T*-W)
$$
```
W = np.array([1, 1])
T = np.array([-2, -3])
# R3 = np.add(W,np.multiply(T,np.multiply(-1,W)))
R3 = W + (T*(-1*W))
R3
```
### Scaling
Scaling or scalar multiplication takes a scalar value and performs multiplication with a vector. Let's take the example below:
$$S = 5 \cdot A$$
We can do this in numpy through:
```
A = np.array([1,5,8,9])
S = 5*A
S
S = np.multiply(5,A)
S
```
$$R = 3X - Y\\X = \hat{x} + \hat{y} , Y = 2\hat{x} - 3\hat{y}$$
```
X = np.array([1, 1])
Y = np.array([2, -3])
R = np.subtract(np.multiply(3,X),Y) ## functional method
# R = 3*X - Y
R
```
### MatPlotLib
MatPlotLib or MATLab Plotting library is Python's take on MATLabs plotting feature. MatPlotLib can be used vastly from graping values to visualizing several dimensions of data.
#### Visualizing Data
It's not enough just sloving these vectors so might need to visualize them. So we'll use MatPlotLib for that. We'll need to import it first.
```
import matplotlib.pyplot as plt ## use this one if not in jupyterlab/notebook
# from matplotlib import pyplot as plt
import matplotlib
```

```
A = [2,-1]
B = [5,2]
plt.scatter(A[0],A[1], label='A', c='magenta')
plt.scatter(B[0],B[1], label='B', c='mediumspringgreen')
plt.grid()
plt.legend()
plt.show()
A = np.array([-5,0])
B = np.array([0,5])
plt.title("Resultant Vector\nMagnitude:{:.2f}".format(R_mag))
plt.xlim(-15, 15)
plt.ylim(-15, 15)
# print(B)
plt.quiver(0,0, A[0], A[1], angles='xy', scale_units='xy',scale=1, color='red') # Red --> A
plt.quiver(A[0], A[1], B[0], B[1],angles='xy', scale_units='xy',scale=1, color='green')
R = A+B
plt.quiver(0, 0, R[0], R[1],angles='xy', scale_units='xy',scale=1, color='orange')
plt.grid()
plt.show()
```
$\sqrt{A^2+B^2+C^2}$
```
R
R_mag = np.sqrt(np.sum(A**2+B**2)) ##Euclidean Distance / Euclidean Norm
rise = R[1]
run = R[0]
slope = rise/run
slope
## angle of the vector? arctan(rise/run)
```
| github_jupyter |
```
import json
import joblib
import pickle
import pandas as pd
from lightgbm import LGBMClassifier
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline, Pipeline
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.metrics import precision_score, recall_score
import numpy as np
from sklearn.metrics import precision_recall_curve
import matplotlib.pyplot as plt
df = pd.read_csv("data/train_searched.csv")
df.head()
# lowercaes departments and location names
df['Department Name'] = df['Department Name'].apply(lambda x: str(x).lower())
df['InterventionLocationName'] = df['InterventionLocationName'].apply(lambda x: str(x).lower())
train_features = df.columns.drop(['VehicleSearchedIndicator', 'ContrabandIndicator'])
categorical_features = train_features.drop(['InterventionDateTime', 'SubjectAge'])
numerical_features = ['SubjectAge']
target = 'ContrabandIndicator'
# show the most common feature values for all the categorical features
for feature in categorical_features:
display(df[feature].value_counts())
# I'm going to remove less common features.
# Let's create a dictionary with the minimum required number of appearences
min_frequency = {
"Department Name": 50,
"InterventionLocationName": 50,
"ReportingOfficerIdentificationID": 30,
"StatuteReason": 10
}
def filter_values(df: pd.DataFrame, column_name: str, threshold: int):
value_counts = df[column_name].value_counts()
to_keep = value_counts[value_counts > threshold].index
filtered = df[df[column_name].isin(to_keep)]
return filtered
df.shape
for feature, threshold in min_frequency.items():
df = filter_values(df, feature, threshold)
df.shape
X = df[train_features]
y = df[target]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=42)
categorical_transformer = Pipeline(steps=[
('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('onehot', OneHotEncoder(handle_unknown='ignore'))])
preprocessor = ColumnTransformer(
transformers=[('cat', categorical_transformer, categorical_features)])
pipeline = make_pipeline(
preprocessor,
LGBMClassifier(n_jobs=-1, random_state=42),
)
pipeline.fit(X_train, y_train)
preds = pipeline.predict(X_test)
def verify_success_rate_above(y_true, y_pred, min_success_rate=0.5):
"""
Verifies the success rate on a test set is above a provided minimum
"""
precision = precision_score(y_true, y_pred, pos_label=True)
is_satisfied = (precision >= min_success_rate)
return is_satisfied, precision
def verify_amount_found(y_true, y_pred):
"""
Verifies the amout of contraband found in the test dataset - a.k.a the recall in our test set
"""
recall = recall_score(y_true, y_pred)
return recall
verify_success_rate_above(y_test, preds)
verify_amount_found(y_test, preds)
```
Now let's find the best threshold for our requirements.
Precision needs to be at least 0.5, and recall has to be as max as possible.
It's usually true that the bigger is precision, the lower is the recall.
So we need to find the threshold that coresponds to precision = 0.5
```
proba = pipeline.predict_proba(X_test)
precision, recall, thresholds = precision_recall_curve(y_test, proba[:, 1])
print(len(precision), len(recall), len(thresholds))
# according to documentation, precision and recall
# have 1 and 0 at the end, so we should remove them before plotting.
precision = precision[:-1]
recall = recall[:-1]
fig=plt.figure()
ax1 = plt.subplot(211)
ax2 = plt.subplot(212)
ax1.hlines(y=0.5,xmin=0, xmax=1, colors='red')
ax1.plot(thresholds,precision)
ax2.plot(thresholds,recall)
ax1.get_shared_x_axes().join(ax1, ax2)
ax1.set_xticklabels([])
ax1.set_title('Precision')
ax2.set_title('Recall')
plt.xlabel('Threshold')
plt.show()
```
Red line shows the point where precision is equal 0.5.
It looks like the biggest recall for precision >= 0.5 is around 0.2
Let's find the exact value.
```
min_index = [i for i, prec in enumerate(precision) if prec >= 0.5][0]
print(min_index)
precision[min_index]
recall[min_index]
thresholds[min_index]
best_preds = [1 if pred > thresholds[min_index] else 0 for pred in proba[:, 1]]
verify_success_rate_above(y_test, best_preds)
verify_amount_found(y_test, best_preds)
with open('columns.json', 'w') as fh:
json.dump(X_train.columns.tolist(), fh)
with open('dtypes.pickle', 'wb') as fh:
pickle.dump(X_train.dtypes, fh)
joblib.dump(pipeline, 'pipeline.pickle');
```
| github_jupyter |
# Parsing Inputs
In the chapter on [Grammars](Grammars.ipynb), we discussed how grammars can be
used to represent various languages. We also saw how grammars can be used to
generate strings of the corresponding language. Grammars can also perform the
reverse. That is, given a string, one can decompose the string into its
constituent parts that correspond to the parts of grammar used to generate it
– the _derivation tree_ of that string. These parts (and parts from other similar
strings) can later be recombined using the same grammar to produce new strings.
In this chapter, we use grammars to parse and decompose a given set of valid seed inputs into their corresponding derivation trees. This structural representation allows us to mutate, crossover, and recombine their parts in order to generate new valid, slightly changed inputs (i.e., fuzz)
```
from bookutils import YouTubeVideo
YouTubeVideo('2yS9EfBEirE')
```
**Prerequisites**
* You should have read the [chapter on grammars](Grammars.ipynb).
* An understanding of derivation trees from the [chapter on grammar fuzzer](GrammarFuzzer.ipynb)
is also required.
## Synopsis
<!-- Automatically generated. Do not edit. -->
To [use the code provided in this chapter](Importing.ipynb), write
```python
>>> from fuzzingbook.Parser import <identifier>
```
and then make use of the following features.
This chapter introduces `Parser` classes, parsing a string into a _derivation tree_ as introduced in the [chapter on efficient grammar fuzzing](GrammarFuzzer.ipynb). Two important parser classes are provided:
* [Parsing Expression Grammar parsers](#Parsing-Expression-Grammars) (`PEGParser`). These are very efficient, but limited to specific grammar structure. Notably, the alternatives represent *ordered choice*. That is, rather than choosing all rules that can potentially match, we stop at the first match that succeed.
* [Earley parsers](#Parsing-Context-Free-Grammars) (`EarleyParser`). These accept any kind of context-free grammars, and explore all parsing alternatives (if any).
Using any of these is fairly easy, though. First, instantiate them with a grammar:
```python
>>> from Grammars import US_PHONE_GRAMMAR
>>> us_phone_parser = EarleyParser(US_PHONE_GRAMMAR)
```
Then, use the `parse()` method to retrieve a list of possible derivation trees:
```python
>>> trees = us_phone_parser.parse("(555)987-6543")
>>> tree = list(trees)[0]
>>> display_tree(tree)
```

These derivation trees can then be used for test generation, notably for mutating and recombining existing inputs.

```
import bookutils
from typing import Dict, List, Tuple, Collection, Set, Iterable, Generator, cast
from Fuzzer import Fuzzer # minor dependendcy
from Grammars import EXPR_GRAMMAR, START_SYMBOL, RE_NONTERMINAL
from Grammars import is_valid_grammar, syntax_diagram, Grammar
from GrammarFuzzer import GrammarFuzzer, display_tree, tree_to_string, dot_escape
from GrammarFuzzer import DerivationTree
from ExpectError import ExpectError
from IPython.display import display
from Timer import Timer
```
## Why Parsing for Fuzzing?
Why would one want to parse existing inputs in order to fuzz? Let us illustrate the problem with an example. Here is a simple program that accepts a CSV file of vehicle details and processes this information.
```
def process_inventory(inventory):
res = []
for vehicle in inventory.split('\n'):
ret = process_vehicle(vehicle)
res.extend(ret)
return '\n'.join(res)
```
The CSV file contains details of one vehicle per line. Each row is processed in `process_vehicle()`.
```
def process_vehicle(vehicle):
year, kind, company, model, *_ = vehicle.split(',')
if kind == 'van':
return process_van(year, company, model)
elif kind == 'car':
return process_car(year, company, model)
else:
raise Exception('Invalid entry')
```
Depending on the kind of vehicle, the processing changes.
```
def process_van(year, company, model):
res = ["We have a %s %s van from %s vintage." % (company, model, year)]
iyear = int(year)
if iyear > 2010:
res.append("It is a recent model!")
else:
res.append("It is an old but reliable model!")
return res
def process_car(year, company, model):
res = ["We have a %s %s car from %s vintage." % (company, model, year)]
iyear = int(year)
if iyear > 2016:
res.append("It is a recent model!")
else:
res.append("It is an old but reliable model!")
return res
```
Here is a sample of inputs that the `process_inventory()` accepts.
```
mystring = """\
1997,van,Ford,E350
2000,car,Mercury,Cougar\
"""
print(process_inventory(mystring))
```
Let us try to fuzz this program. Given that the `process_inventory()` takes a CSV file, we can write a simple grammar for generating comma separated values, and generate the required CSV rows. For convenience, we fuzz `process_vehicle()` directly.
```
import string
CSV_GRAMMAR: Grammar = {
'<start>': ['<csvline>'],
'<csvline>': ['<items>'],
'<items>': ['<item>,<items>', '<item>'],
'<item>': ['<letters>'],
'<letters>': ['<letter><letters>', '<letter>'],
'<letter>': list(string.ascii_letters + string.digits + string.punctuation + ' \t\n')
}
```
We need some infrastructure first for viewing the grammar.
```
syntax_diagram(CSV_GRAMMAR)
```
We generate `1000` values, and evaluate the `process_vehicle()` with each.
```
gf = GrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
trials = 1000
valid: List[str] = []
time = 0
for i in range(trials):
with Timer() as t:
vehicle_info = gf.fuzz()
try:
process_vehicle(vehicle_info)
valid.append(vehicle_info)
except:
pass
time += t.elapsed_time()
print("%d valid strings, that is GrammarFuzzer generated %f%% valid entries from %d inputs" %
(len(valid), len(valid) * 100.0 / trials, trials))
print("Total time of %f seconds" % time)
```
This is obviously not working. But why?
```
gf = GrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
trials = 10
time = 0
for i in range(trials):
vehicle_info = gf.fuzz()
try:
print(repr(vehicle_info), end="")
process_vehicle(vehicle_info)
except Exception as e:
print("\t", e)
else:
print()
```
None of the entries will get through unless the fuzzer can produce either `van` or `car`.
Indeed, the reason is that the grammar itself does not capture the complete information about the format. So here is another idea. We modify the `GrammarFuzzer` to know a bit about our format.
```
import copy
import random
class PooledGrammarFuzzer(GrammarFuzzer):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self._node_cache = {}
def update_cache(self, key, values):
self._node_cache[key] = values
def expand_node_randomly(self, node):
(symbol, children) = node
assert children is None
if symbol in self._node_cache:
if random.randint(0, 1) == 1:
return super().expand_node_randomly(node)
return copy.deepcopy(random.choice(self._node_cache[symbol]))
return super().expand_node_randomly(node)
```
Let us try again!
```
gf = PooledGrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
gf.update_cache('<item>', [
('<item>', [('car', [])]),
('<item>', [('van', [])]),
])
trials = 10
time = 0
for i in range(trials):
vehicle_info = gf.fuzz()
try:
print(repr(vehicle_info), end="")
process_vehicle(vehicle_info)
except Exception as e:
print("\t", e)
else:
print()
```
At least we are getting somewhere! It would be really nice if _we could incorporate what we know about the sample data in our fuzzer._ In fact, it would be nice if we could _extract_ the template and valid values from samples, and use them in our fuzzing. How do we do that? The quick answer to this question is: Use a *parser*.
## Using a Parser
Generally speaking, a _parser_ is the part of a a program that processes (structured) input. The parsers we discuss in this chapter transform an input string into a _derivation tree_ (discussed in the [chapter on efficient grammar fuzzing](GrammarFuzzer.ipynb)). From a user's perspective, all it takes to parse an input is two steps:
1. Initialize the parser with a grammar, as in
```
parser = Parser(grammar)
```
2. Using the parser to retrieve a list of derivation trees:
```python
trees = parser.parse(input)
```
Once we have parsed a tree, we can use it just as the derivation trees produced from grammar fuzzing.
We discuss a number of such parsers, in particular
* [parsing expression grammar parsers](#Parsing-Expression-Grammars) (`PEGParser`), which are very efficient, but limited to specific grammar structure; and
* [Earley parsers](#Parsing-Context-Free-Grammars) (`EarleyParser`), which accept any kind of context-free grammars.
If you just want to _use_ parsers (say, because your main focus is testing), you can just stop here and move on [to the next chapter](LangFuzzer.ipynb), where we learn how to make use of parsed inputs to mutate and recombine them. If you want to _understand_ how parsers work, though, this chapter is right for you.
## An Ad Hoc Parser
As we saw in the previous section, programmers often have to extract parts of data that obey certain rules. For example, for *CSV* files, each element in a row is separated by *commas*, and multiple raws are used to store the data.
To extract the information, we write an ad hoc parser `simple_parse_csv()`.
```
def simple_parse_csv(mystring: str) -> DerivationTree:
children: List[DerivationTree] = []
tree = (START_SYMBOL, children)
for i, line in enumerate(mystring.split('\n')):
children.append(("record %d" % i, [(cell, [])
for cell in line.split(',')]))
return tree
```
We also change the default orientation of the graph to *left to right* rather than *top to bottom* for easier viewing using `lr_graph()`.
```
def lr_graph(dot):
dot.attr('node', shape='plain')
dot.graph_attr['rankdir'] = 'LR'
```
The `display_tree()` shows the structure of our CSV file after parsing.
```
tree = simple_parse_csv(mystring)
display_tree(tree, graph_attr=lr_graph)
```
This is of course simple. What if we encounter slightly more complexity? Again, another example from the Wikipedia.
```
mystring = '''\
1997,Ford,E350,"ac, abs, moon",3000.00\
'''
print(mystring)
```
We define a new annotation method `highlight_node()` to mark the nodes that are interesting.
```
def highlight_node(predicate):
def hl_node(dot, nid, symbol, ann):
if predicate(dot, nid, symbol, ann):
dot.node(repr(nid), dot_escape(symbol), fontcolor='red')
else:
dot.node(repr(nid), dot_escape(symbol))
return hl_node
```
Using `highlight_node()` we can highlight particular nodes that we were wrongly parsed.
```
tree = simple_parse_csv(mystring)
bad_nodes = {5, 6, 7, 12, 13, 20, 22, 23, 24, 25}
def hl_predicate(_d, nid, _s, _a): return nid in bad_nodes
highlight_err_node = highlight_node(hl_predicate)
display_tree(tree, log=False, node_attr=highlight_err_node,
graph_attr=lr_graph)
```
The marked nodes indicate where our parsing went wrong. We can of course extend our parser to understand quotes. First we define some of the helper functions `parse_quote()`, `find_comma()` and `comma_split()`
```
def parse_quote(string, i):
v = string[i + 1:].find('"')
return v + i + 1 if v >= 0 else -1
def find_comma(string, i):
slen = len(string)
while i < slen:
if string[i] == '"':
i = parse_quote(string, i)
if i == -1:
return -1
if string[i] == ',':
return i
i += 1
return -1
def comma_split(string):
slen = len(string)
i = 0
while i < slen:
c = find_comma(string, i)
if c == -1:
yield string[i:]
return
else:
yield string[i:c]
i = c + 1
```
We can update our `parse_csv()` procedure to use our advanced quote parser.
```
def parse_csv(mystring):
children = []
tree = (START_SYMBOL, children)
for i, line in enumerate(mystring.split('\n')):
children.append(("record %d" % i, [(cell, [])
for cell in comma_split(line)]))
return tree
```
Our new `parse_csv()` can now handle quotes correctly.
```
tree = parse_csv(mystring)
display_tree(tree, graph_attr=lr_graph)
```
That of course does not survive long:
```
mystring = '''\
1999,Chevy,"Venture \\"Extended Edition, Very Large\\"",,5000.00\
'''
print(mystring)
```
A few embedded quotes are sufficient to confuse our parser again.
```
tree = parse_csv(mystring)
bad_nodes = {4, 5}
display_tree(tree, node_attr=highlight_err_node, graph_attr=lr_graph)
```
Here is another record from that CSV file:
```
mystring = '''\
1996,Jeep,Grand Cherokee,"MUST SELL!
air, moon roof, loaded",4799.00
'''
print(mystring)
tree = parse_csv(mystring)
bad_nodes = {5, 6, 7, 8, 9, 10}
display_tree(tree, node_attr=highlight_err_node, graph_attr=lr_graph)
```
Fixing this would require modifying both inner `parse_quote()` and the outer `parse_csv()` procedures. We note that each of these features actually documented in the CSV [RFC 4180](https://tools.ietf.org/html/rfc4180)
Indeed, each additional improvement falls apart even with a little extra complexity. The problem becomes severe when one encounters recursive expressions. For example, JSON is a common alternative to CSV files for saving data. Similarly, one may have to parse data from an HTML table instead of a CSV file if one is getting the data from the web.
One might be tempted to fix it with a little more ad hoc parsing, with a bit of *regular expressions* thrown in. However, that is the [path to insanity](https://stackoverflow.com/a/1732454).
It is here that _formal parsers_ shine. The main idea is that, any given set of strings belong to a language, and these languages can be specified by their grammars (as we saw in the [chapter on grammars](Grammars.ipynb)). The great thing about grammars is that they can be _composed_. That is, one can introduce finer and finer details into an internal structure without affecting the external structure, and similarly, one can change the external structure without much impact on the internal structure.
## Grammars in Parsing
We briefly describe grammars in the context of parsing.
### Excursion: Grammars and Derivation Trees
A grammar, as you have read from the [chapter on grammars](Grammars.ipynb) is a set of _rules_ that explain how the start symbol can be expanded. Each rule has a name, also called a _nonterminal_, and a set of _alternative choices_ in how the nonterminal can be expanded.
```
A1_GRAMMAR: Grammar = {
"<start>": ["<expr>"],
"<expr>": ["<expr>+<expr>", "<expr>-<expr>", "<integer>"],
"<integer>": ["<digit><integer>", "<digit>"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
syntax_diagram(A1_GRAMMAR)
```
In the above expression, the rule `<expr> : [<expr>+<expr>,<expr>-<expr>,<integer>]` corresponds to how the nonterminal `<expr>` might be expanded. The expression `<expr>+<expr>` corresponds to one of the alternative choices. We call this an _alternative_ expansion for the nonterminal `<expr>`. Finally, in an expression `<expr>+<expr>`, each of `<expr>`, `+`, and `<expr>` are _symbols_ in that expansion. A symbol could be either a nonterminal or a terminal symbol based on whether its expansion is available in the grammar.
Here is a string that represents an arithmetic expression that we would like to parse, which is specified by the grammar above:
```
mystring = '1+2'
```
The _derivation tree_ for our expression from this grammar is given by:
```
tree = ('<start>', [('<expr>',
[('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>', [('<integer>', [('<digit>', [('2',
[])])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
```
While a grammar can be used to specify a given language, there could be multiple
grammars that correspond to the same language. For example, here is another
grammar to describe the same addition expression.
```
A2_GRAMMAR: Grammar = {
"<start>": ["<expr>"],
"<expr>": ["<integer><expr_>"],
"<expr_>": ["+<expr>", "-<expr>", ""],
"<integer>": ["<digit><integer_>"],
"<integer_>": ["<integer>", ""],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
syntax_diagram(A2_GRAMMAR)
```
The corresponding derivation tree is given by:
```
tree = ('<start>', [('<expr>', [('<integer>', [('<digit>', [('1', [])]),
('<integer_>', [])]),
('<expr_>', [('+', []),
('<expr>',
[('<integer>',
[('<digit>', [('2', [])]),
('<integer_>', [])]),
('<expr_>', [])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
```
Indeed, there could be different classes of grammars that
describe the same language. For example, the first grammar `A1_GRAMMAR`
is a grammar that sports both _right_ and _left_ recursion, while the
second grammar `A2_GRAMMAR` does not have left recursion in the
nonterminals in any of its productions, but contains _epsilon_ productions.
(An epsilon production is a production that has empty string in its right
hand side.)
### End of Excursion
### Excursion: Recursion
You would have noticed that we reuse the term `<expr>` in its own definition. Using the same nonterminal in its own definition is called *recursion*. There are two specific kinds of recursion one should be aware of in parsing, as we see in the next section.
#### Recursion
A grammar is _left recursive_ if any of its nonterminals are left recursive,
and a nonterminal is directly left-recursive if the left-most symbol of
any of its productions is itself.
```
LR_GRAMMAR: Grammar = {
'<start>': ['<A>'],
'<A>': ['<A>a', ''],
}
syntax_diagram(LR_GRAMMAR)
mystring = 'aaaaaa'
display_tree(
('<start>', [('<A>', [('<A>', [('<A>', []), ('a', [])]), ('a', [])]),
('a', [])]))
```
A grammar is indirectly left-recursive if any
of the left-most symbols can be expanded using their definitions to
produce the nonterminal as the left-most symbol of the expansion. The left
recursion is called a _hidden-left-recursion_ if during the series of
expansions of a nonterminal, one reaches a rule where the rule contains
the same nonterminal after a prefix of other symbols, and these symbols can
derive the empty string. For example, in `A1_GRAMMAR`, `<integer>` will be
considered hidden-left recursive if `<digit>` could derive an empty string.
Right recursive grammars are defined similarly.
Below is the derivation tree for the right recursive grammar that represents the same
language as that of `LR_GRAMMAR`.
```
RR_GRAMMAR: Grammar = {
'<start>': ['<A>'],
'<A>': ['a<A>', ''],
}
syntax_diagram(RR_GRAMMAR)
display_tree(('<start>', [('<A>', [
('a', []), ('<A>', [('a', []), ('<A>', [('a', []), ('<A>', [])])])])]
))
```
#### Ambiguity
To complicate matters further, there could be
multiple derivation trees – also called _parses_ – corresponding to the
same string from the same grammar. For example, a string `1+2+3` can be parsed
in two ways as we see below using the `A1_GRAMMAR`
```
mystring = '1+2+3'
tree = ('<start>',
[('<expr>',
[('<expr>', [('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>', [('<integer>',
[('<digit>', [('2', [])])])])]), ('+', []),
('<expr>', [('<integer>', [('<digit>', [('3', [])])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
tree = ('<start>',
[('<expr>', [('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>',
[('<expr>', [('<integer>', [('<digit>', [('2', [])])])]),
('+', []),
('<expr>', [('<integer>', [('<digit>', [('3',
[])])])])])])])
assert tree_to_string(tree) == mystring
display_tree(tree)
```
There are many ways to resolve ambiguities. One approach taken by *Parsing Expression Grammars* explained in the next section is to specify a particular order of resolution, and choose the first one. Another approach is to simply return all possible derivation trees, which is the approach taken by *Earley parser* we develop later.
### End of Excursion
## A Parser Class
Next, we develop different parsers. To do that, we define a minimal interface for parsing that is obeyed by all parsers. There are two approaches to parsing a string using a grammar.
1. The traditional approach is to use a *lexer* (also called a *tokenizer* or a *scanner*) to first tokenize the incoming string, and feed the grammar one token at a time. The lexer is typically a smaller parser that accepts a *regular language*. The advantage of this approach is that the grammar used by the parser can eschew the details of tokenization. Further, one gets a shallow derivation tree at the end of the parsing which can be directly used for generating the *Abstract Syntax Tree*.
2. The second approach is to use a tree pruner after the complete parse. With this approach, one uses a grammar that incorporates complete details of the syntax. Next, the nodes corresponding to tokens are pruned and replaced with their corresponding strings as leaf nodes. The utility of this approach is that the parser is more powerful, and further there is no artificial distinction between *lexing* and *parsing*.
In this chapter, we use the second approach. This approach is implemented in the `prune_tree` method.
The *Parser* class we define below provides the minimal interface. The main methods that need to be implemented by the classes implementing this interface are `parse_prefix` and `parse`. The `parse_prefix` returns a tuple, which contains the index until which parsing was completed successfully, and the parse forest until that index. The method `parse` returns a list of derivation trees if the parse was successful.
```
class Parser:
"""Base class for parsing."""
def __init__(self, grammar: Grammar, *,
start_symbol: str = START_SYMBOL,
log: bool = False,
coalesce: bool = True,
tokens: Set[str] = set()) -> None:
"""Constructor.
`grammar` is the grammar to be used for parsing.
Keyword arguments:
`start_symbol` is the start symbol (default: '<start>').
`log` enables logging (default: False).
`coalesce` defines if tokens should be coalesced (default: True).
`tokens`, if set, is a set of tokens to be used."""
self._grammar = grammar
self._start_symbol = start_symbol
self.log = log
self.coalesce_tokens = coalesce
self.tokens = tokens
def grammar(self) -> Grammar:
"""Return the grammar of this parser."""
return self._grammar
def start_symbol(self) -> str:
"""Return the start symbol of this parser."""
return self._start_symbol
def parse_prefix(self, text: str) -> Tuple[int, Iterable[DerivationTree]]:
"""Return pair (cursor, forest) for longest prefix of text.
To be defined in subclasses."""
raise NotImplementedError
def parse(self, text: str) -> Iterable[DerivationTree]:
"""Parse `text` using the grammar.
Return an iterable of parse trees."""
cursor, forest = self.parse_prefix(text)
if cursor < len(text):
raise SyntaxError("at " + repr(text[cursor:]))
return [self.prune_tree(tree) for tree in forest]
def parse_on(self, text: str, start_symbol: str) -> Generator:
old_start = self._start_symbol
try:
self._start_symbol = start_symbol
yield from self.parse(text)
finally:
self._start_symbol = old_start
def coalesce(self, children: List[DerivationTree]) -> List[DerivationTree]:
last = ''
new_lst: List[DerivationTree] = []
for cn, cc in children:
if cn not in self._grammar:
last += cn
else:
if last:
new_lst.append((last, []))
last = ''
new_lst.append((cn, cc))
if last:
new_lst.append((last, []))
return new_lst
def prune_tree(self, tree: DerivationTree) -> DerivationTree:
name, children = tree
assert isinstance(children, list)
if self.coalesce_tokens:
children = self.coalesce(cast(List[DerivationTree], children))
if name in self.tokens:
return (name, [(tree_to_string(tree), [])])
else:
return (name, [self.prune_tree(c) for c in children])
```
### Excursion: Canonical Grammars
The `EXPR_GRAMMAR` we import from the [chapter on grammars](Grammars.ipynb) is oriented towards generation. In particular, the production rules are stored as strings. We need to massage this representation a little to conform to a _canonical representation_ where each token in a rule is represented separately. The `canonical` format uses separate tokens to represent each symbol in an expansion.
```
CanonicalGrammar = Dict[str, List[List[str]]]
import re
def single_char_tokens(grammar: Grammar) -> Dict[str, List[List[Collection[str]]]]:
g_ = {}
for key in grammar:
rules_ = []
for rule in grammar[key]:
rule_ = []
for token in rule:
if token in grammar:
rule_.append(token)
else:
rule_.extend(token)
rules_.append(rule_)
g_[key] = rules_
return g_
def canonical(grammar: Grammar) -> CanonicalGrammar:
def split(expansion):
if isinstance(expansion, tuple):
expansion = expansion[0]
return [token for token in re.split(
RE_NONTERMINAL, expansion) if token]
return {
k: [split(expression) for expression in alternatives]
for k, alternatives in grammar.items()
}
CE_GRAMMAR: CanonicalGrammar = canonical(EXPR_GRAMMAR)
CE_GRAMMAR
```
We also provide a convenience method for easier display of canonical grammars.
```
def recurse_grammar(grammar, key, order):
rules = sorted(grammar[key])
old_len = len(order)
for rule in rules:
for token in rule:
if token not in grammar: continue
if token not in order:
order.append(token)
new = order[old_len:]
for ckey in new:
recurse_grammar(grammar, ckey, order)
def show_grammar(grammar, start_symbol=START_SYMBOL):
order = [start_symbol]
recurse_grammar(grammar, start_symbol, order)
return {k: sorted(grammar[k]) for k in order}
show_grammar(CE_GRAMMAR)
```
We provide a way to revert a canonical expression.
```
def non_canonical(grammar):
new_grammar = {}
for k in grammar:
rules = grammar[k]
new_rules = []
for rule in rules:
new_rules.append(''.join(rule))
new_grammar[k] = new_rules
return new_grammar
non_canonical(CE_GRAMMAR)
```
It is easier to work with the `canonical` representation during parsing. Hence, we update our parser class to store the `canonical` representation also.
```
class Parser(Parser):
def __init__(self, grammar, **kwargs):
self._start_symbol = kwargs.get('start_symbol', START_SYMBOL)
self.log = kwargs.get('log', False)
self.tokens = kwargs.get('tokens', set())
self.coalesce_tokens = kwargs.get('coalesce', True)
canonical_grammar = kwargs.get('canonical', False)
if canonical_grammar:
self.cgrammar = single_char_tokens(grammar)
self._grammar = non_canonical(grammar)
else:
self._grammar = dict(grammar)
self.cgrammar = single_char_tokens(canonical(grammar))
# we do not require a single rule for the start symbol
if len(grammar.get(self._start_symbol, [])) != 1:
self.cgrammar['<>'] = [[self._start_symbol]]
```
We update the `prune_tree()` to account for the phony start symbol if it was insserted.
```
class Parser(Parser):
def prune_tree(self, tree):
name, children = tree
if name == '<>':
assert len(children) == 1
return self.prune_tree(children[0])
if self.coalesce_tokens:
children = self.coalesce(children)
if name in self.tokens:
return (name, [(tree_to_string(tree), [])])
else:
return (name, [self.prune_tree(c) for c in children])
```
### End of Excursion
## Parsing Expression Grammars
A _[Parsing Expression Grammar](http://bford.info/pub/lang/peg)_ (*PEG*) \cite{Ford2004} is a type of _recognition based formal grammar_ that specifies the sequence of steps to take to parse a given string.
A _parsing expression grammar_ is very similar to a _context-free grammar_ (*CFG*) such as the ones we saw in the [chapter on grammars](Grammars.ipynb). As in a CFG, a parsing expression grammar is represented by a set of nonterminals and corresponding alternatives representing how to match each. For example, here is a PEG that matches `a` or `b`.
```
PEG1 = {
'<start>': ['a', 'b']
}
```
However, unlike the _CFG_, the alternatives represent *ordered choice*. That is, rather than choosing all rules that can potentially match, we stop at the first match that succeed. For example, the below _PEG_ can match `ab` but not `abc` unlike a _CFG_ which will match both. (We call the sequence of ordered choice expressions *choice expressions* rather than alternatives to make the distinction from _CFG_ clear.)
```
PEG2 = {
'<start>': ['ab', 'abc']
}
```
Each choice in a _choice expression_ represents a rule on how to satisfy that particular choice. The choice is a sequence of symbols (terminals and nonterminals) that are matched against a given text as in a _CFG_.
Beyond the syntax of grammar definitions we have seen so far, a _PEG_ can also contain a few additional elements. See the exercises at the end of the chapter for additional information.
The PEGs model the typical practice in handwritten recursive descent parsers, and hence it may be considered more intuitive to understand.
### The Packrat Parser for Predicate Expression Grammars
Short of hand rolling a parser, _Packrat_ parsing is one of the simplest parsing techniques, and is one of the techniques for parsing PEGs.
The _Packrat_ parser is so named because it tries to cache all results from simpler problems in the hope that these solutions can be used to avoid re-computation later. We develop a minimal _Packrat_ parser next.
We derive from the `Parser` base class first, and we accept the text to be parsed in the `parse()` method, which in turn calls `unify_key()` with the `start_symbol`.
__Note.__ While our PEG parser can produce only a single unambiguous parse tree, other parsers can produce multiple parses for ambiguous grammars. Hence, we return a list of trees (in this case with a single element).
```
class PEGParser(Parser):
def parse_prefix(self, text):
cursor, tree = self.unify_key(self.start_symbol(), text, 0)
return cursor, [tree]
```
### Excursion: Implementing `PEGParser`
#### Unify Key
The `unify_key()` algorithm is simple. If given a terminal symbol, it tries to match the symbol with the current position in the text. If the symbol and text match, it returns successfully with the new parse index `at`.
If on the other hand, it was given a nonterminal, it retrieves the choice expression corresponding to the key, and tries to match each choice *in order* using `unify_rule()`. If **any** of the rules succeed in being unified with the given text, the parse is considered a success, and we return with the new parse index returned by `unify_rule()`.
```
class PEGParser(PEGParser):
"""Packrat parser for Parsing Expression Grammars (PEGs)."""
def unify_key(self, key, text, at=0):
if self.log:
print("unify_key: %s with %s" % (repr(key), repr(text[at:])))
if key not in self.cgrammar:
if text[at:].startswith(key):
return at + len(key), (key, [])
else:
return at, None
for rule in self.cgrammar[key]:
to, res = self.unify_rule(rule, text, at)
if res is not None:
return (to, (key, res))
return 0, None
mystring = "1"
peg = PEGParser(EXPR_GRAMMAR, log=True)
peg.unify_key('1', mystring)
mystring = "2"
peg.unify_key('1', mystring)
```
#### Unify Rule
The `unify_rule()` method is similar. It retrieves the tokens corresponding to the rule that it needs to unify with the text, and calls `unify_key()` on them in sequence. If **all** tokens are successfully unified with the text, the parse is a success.
```
class PEGParser(PEGParser):
def unify_rule(self, rule, text, at):
if self.log:
print('unify_rule: %s with %s' % (repr(rule), repr(text[at:])))
results = []
for token in rule:
at, res = self.unify_key(token, text, at)
if res is None:
return at, None
results.append(res)
return at, results
mystring = "0"
peg = PEGParser(EXPR_GRAMMAR, log=True)
peg.unify_rule(peg.cgrammar['<digit>'][0], mystring, 0)
mystring = "12"
peg.unify_rule(peg.cgrammar['<integer>'][0], mystring, 0)
mystring = "1 + 2"
peg = PEGParser(EXPR_GRAMMAR, log=False)
peg.parse(mystring)
```
The two methods are mutually recursive, and given that `unify_key()` tries each alternative until it succeeds, `unify_key` can be called multiple times with the same arguments. Hence, it is important to memoize the results of `unify_key`. Python provides a simple decorator `lru_cache` for memoizing any function call that has hashable arguments. We add that to our implementation so that repeated calls to `unify_key()` with the same argument get cached results.
This memoization gives the algorithm its name – _Packrat_.
```
from functools import lru_cache
class PEGParser(PEGParser):
@lru_cache(maxsize=None)
def unify_key(self, key, text, at=0):
if key not in self.cgrammar:
if text[at:].startswith(key):
return at + len(key), (key, [])
else:
return at, None
for rule in self.cgrammar[key]:
to, res = self.unify_rule(rule, text, at)
if res is not None:
return (to, (key, res))
return 0, None
```
We wrap initialization and calling of `PEGParser` in a method `parse()` already implemented in the `Parser` base class that accepts the text to be parsed along with the grammar.
### End of Excursion
Here are a few examples of our parser in action.
```
mystring = "1 + (2 * 3)"
peg = PEGParser(EXPR_GRAMMAR)
for tree in peg.parse(mystring):
assert tree_to_string(tree) == mystring
display(display_tree(tree))
mystring = "1 * (2 + 3.35)"
for tree in peg.parse(mystring):
assert tree_to_string(tree) == mystring
display(display_tree(tree))
```
One should be aware that while the grammar looks like a *CFG*, the language described by a *PEG* may be different. Indeed, only *LL(1)* grammars are guaranteed to represent the same language for both PEGs and other parsers. Behavior of PEGs for other classes of grammars could be surprising \cite{redziejowski2008}.
## Parsing Context-Free Grammars
### Problems with PEG
While _PEGs_ are simple at first sight, their behavior in some cases might be a bit unintuitive. For example, here is an example \cite{redziejowski2008}:
```
PEG_SURPRISE: Grammar = {
"<A>": ["a<A>a", "aa"]
}
```
When interpreted as a *CFG* and used as a string generator, it will produce strings of the form `aa, aaaa, aaaaaa` that is, it produces strings where the number of `a` is $ 2*n $ where $ n > 0 $.
```
strings = []
for nn in range(4):
f = GrammarFuzzer(PEG_SURPRISE, start_symbol='<A>')
tree = ('<A>', None)
for _ in range(nn):
tree = f.expand_tree_once(tree)
tree = f.expand_tree_with_strategy(tree, f.expand_node_min_cost)
strings.append(tree_to_string(tree))
display_tree(tree)
strings
```
However, the _PEG_ parser can only recognize strings of the form $2^n$
```
peg = PEGParser(PEG_SURPRISE, start_symbol='<A>')
for s in strings:
with ExpectError():
for tree in peg.parse(s):
display_tree(tree)
print(s)
```
This is not the only problem with _Parsing Expression Grammars_. While *PEGs* are expressive and the *packrat* parser for parsing them is simple and intuitive, *PEGs* suffer from a major deficiency for our purposes. *PEGs* are oriented towards language recognition, and it is not clear how to translate an arbitrary *PEG* to a *CFG*. As we mentioned earlier, a naive re-interpretation of a *PEG* as a *CFG* does not work very well. Further, it is not clear what is the exact relation between the class of languages represented by *PEG* and the class of languages represented by *CFG*. Since our primary focus is *fuzzing* – that is _generation_ of strings – , we next look at _parsers that can accept context-free grammars_.
The general idea of *CFG* parser is the following: Peek at the input text for the allowed number of characters, and use these, and our parser state to determine which rules can be applied to complete parsing. We next look at a typical *CFG* parsing algorithm, the Earley Parser.
### The Earley Parser
The Earley parser is a general parser that is able to parse any arbitrary *CFG*. It was invented by Jay Earley \cite{Earley1970} for use in computational linguistics. While its computational complexity is $O(n^3)$ for parsing strings with arbitrary grammars, it can parse strings with unambiguous grammars in $O(n^2)$ time, and all *[LR(k)](https://en.wikipedia.org/wiki/LR_parser)* grammars in linear time ($O(n)$ \cite{Leo1991}). Further improvements – notably handling epsilon rules – were invented by Aycock et al. \cite{Aycock2002}.
Note that one restriction of our implementation is that the start symbol can have only one alternative in its alternative expressions. This is not a restriction in practice because any grammar with multiple alternatives for its start symbol can be extended with a new start symbol that has the original start symbol as its only choice. That is, given a grammar as below,
```
grammar = {
'<start>': ['<A>', '<B>'],
...
}
```
one may rewrite it as below to conform to the *single-alternative* rule.
```
grammar = {
'<start>': ['<start_>'],
'<start_>': ['<A>', '<B>'],
...
}
```
Let us implement a class `EarleyParser`, again derived from `Parser` which implements an Earley parser.
### Excursion: Implementing `EarleyParser`
We first implement a simpler parser that is a parser for nearly all *CFGs*, but not quite. In particular, our parser does not understand _epsilon rules_ – rules that derive empty string. We show later how the parser can be extended to handle these.
We use the following grammar in our examples below.
```
SAMPLE_GRAMMAR: Grammar = {
'<start>': ['<A><B>'],
'<A>': ['a<B>c', 'a<A>'],
'<B>': ['b<C>', '<D>'],
'<C>': ['c'],
'<D>': ['d']
}
C_SAMPLE_GRAMMAR = canonical(SAMPLE_GRAMMAR)
syntax_diagram(SAMPLE_GRAMMAR)
```
The basic idea of Earley parsing is the following:
* Start with the alternative expressions corresponding to the START_SYMBOL. These represent the possible ways to parse the string from a high level. Essentially each expression represents a parsing path. Queue each expression in our set of possible parses of the string. The parsed index of an expression is the part of expression that has already been recognized. In the beginning of parse, the parsed index of all expressions is at the beginning. Further, each letter gets a queue of expressions that recognizes that letter at that point in our parse.
* Examine our queue of possible parses and check if any of them start with a nonterminal. If it does, then that nonterminal needs to be recognized from the input before the given rule can be parsed. Hence, add the alternative expressions corresponding to the nonterminal to the queue. Do this recursively.
* At this point, we are ready to advance. Examine the current letter in the input, and select all expressions that have that particular letter at the parsed index. These expressions can now advance one step. Advance these selected expressions by incrementing their parsed index and add them to the queue of expressions in line for recognizing the next input letter.
* If while doing these things, we find that any of the expressions have finished parsing, we fetch its corresponding nonterminal, and advance all expressions that have that nonterminal at their parsed index.
* Continue this procedure recursively until all expressions that we have queued for the current letter have been processed. Then start processing the queue for the next letter.
We explain each step in detail with examples in the coming sections.
The parser uses dynamic programming to generate a table containing a _forest of possible parses_ at each letter index – the table contains as many columns as there are letters in the input, and each column contains different parsing rules at various stages of the parse.
For example, given an input `adcd`, the Column 0 would contain the following:
```
<start> : ● <A> <B>
```
which is the starting rule that indicates that we are currently parsing the rule `<start>`, and the parsing state is just before identifying the symbol `<A>`. It would also contain the following which are two alternative paths it could take to complete the parsing.
```
<A> : ● a <B> c
<A> : ● a <A>
```
Column 1 would contain the following, which represents the possible completion after reading `a`.
```
<A> : a ● <B> c
<A> : a ● <A>
<B> : ● b <C>
<B> : ● <D>
<A> : ● a <B> c
<A> : ● a <A>
<D> : ● d
```
Column 2 would contain the following after reading `d`
```
<D> : d ●
<B> : <D> ●
<A> : a <B> ● c
```
Similarly, Column 3 would contain the following after reading `c`
```
<A> : a <B> c ●
<start> : <A> ● <B>
<B> : ● b <C>
<B> : ● <D>
<D> : ● d
```
Finally, Column 4 would contain the following after reading `d`, with the `●` at the end of the `<start>` rule indicating that the parse was successful.
```
<D> : d ●
<B> : <D> ●
<start> : <A> <B> ●
```
As you can see from above, we are essentially filling a table (a table is also called a **chart**) of entries based on each letter we read, and the grammar rules that can be applied. This chart gives the parser its other name -- Chart parsing.
#### Columns
We define the `Column` first. The `Column` is initialized by its own `index` in the input string, and the `letter` at that index. Internally, we also keep track of the states that are added to the column as the parsing progresses.
```
class Column:
def __init__(self, index, letter):
self.index, self.letter = index, letter
self.states, self._unique = [], {}
def __str__(self):
return "%s chart[%d]\n%s" % (self.letter, self.index, "\n".join(
str(state) for state in self.states if state.finished()))
```
The `Column` only stores unique `states`. Hence, when a new `state` is `added` to our `Column`, we check whether it is already known.
```
class Column(Column):
def add(self, state):
if state in self._unique:
return self._unique[state]
self._unique[state] = state
self.states.append(state)
state.e_col = self
return self._unique[state]
```
#### Items
An item represents a _parse in progress for a specific rule._ Hence the item contains the name of the nonterminal, and the corresponding alternative expression (`expr`) which together form the rule, and the current position of parsing in this expression -- `dot`.
**Note.** If you are familiar with [LR parsing](https://en.wikipedia.org/wiki/LR_parser), you will notice that an item is simply an `LR0` item.
```
class Item:
def __init__(self, name, expr, dot):
self.name, self.expr, self.dot = name, expr, dot
```
We also provide a few convenience methods. The method `finished()` checks if the `dot` has moved beyond the last element in `expr`. The method `advance()` produces a new `Item` with the `dot` advanced one token, and represents an advance of the parsing. The method `at_dot()` returns the current symbol being parsed.
```
class Item(Item):
def finished(self):
return self.dot >= len(self.expr)
def advance(self):
return Item(self.name, self.expr, self.dot + 1)
def at_dot(self):
return self.expr[self.dot] if self.dot < len(self.expr) else None
```
Here is how an item could be used. We first define our item
```
item_name = '<B>'
item_expr = C_SAMPLE_GRAMMAR[item_name][1]
an_item = Item(item_name, tuple(item_expr), 0)
```
To determine where the status of parsing, we use `at_dot()`
```
an_item.at_dot()
```
That is, the next symbol to be parsed is `<D>`
If we advance the item, we get another item that represents the finished parsing rule `<B>`.
```
another_item = an_item.advance()
another_item.finished()
```
#### States
For `Earley` parsing, the state of the parsing is simply one `Item` along with some meta information such as the starting `s_col` and ending column `e_col` for each state. Hence we inherit from `Item` to create a `State`.
Since we are interested in comparing states, we define `hash()` and `eq()` with the corresponding methods.
```
class State(Item):
def __init__(self, name, expr, dot, s_col, e_col=None):
super().__init__(name, expr, dot)
self.s_col, self.e_col = s_col, e_col
def __str__(self):
def idx(var):
return var.index if var else -1
return self.name + ':= ' + ' '.join([
str(p)
for p in [*self.expr[:self.dot], '|', *self.expr[self.dot:]]
]) + "(%d,%d)" % (idx(self.s_col), idx(self.e_col))
def copy(self):
return State(self.name, self.expr, self.dot, self.s_col, self.e_col)
def _t(self):
return (self.name, self.expr, self.dot, self.s_col.index)
def __hash__(self):
return hash(self._t())
def __eq__(self, other):
return self._t() == other._t()
def advance(self):
return State(self.name, self.expr, self.dot + 1, self.s_col)
```
The usage of `State` is similar to that of `Item`. The only difference is that it is used along with the `Column` to track the parsing state. For example, we initialize the first column as follows:
```
col_0 = Column(0, None)
item_tuple = tuple(*C_SAMPLE_GRAMMAR[START_SYMBOL])
start_state = State(START_SYMBOL, item_tuple, 0, col_0)
col_0.add(start_state)
start_state.at_dot()
```
The first column is then updated by using `add()` method of `Column`
```
sym = start_state.at_dot()
for alt in C_SAMPLE_GRAMMAR[sym]:
col_0.add(State(sym, tuple(alt), 0, col_0))
for s in col_0.states:
print(s)
```
#### The Parsing Algorithm
The _Earley_ algorithm starts by initializing the chart with columns (as many as there are letters in the input). We also seed the first column with a state representing the expression corresponding to the start symbol. In our case, the state corresponds to the start symbol with the `dot` at `0` is represented as below. The `●` symbol represents the parsing status. In this case, we have not parsed anything.
```
<start>: ● <A> <B>
```
We pass this partial chart to a method for filling the rest of the parse chart.
Before starting to parse, we seed the chart with the state representing the ongoing parse of the start symbol.
```
class EarleyParser(Parser):
"""Earley Parser. This parser can parse any context-free grammar."""
def __init__(self, grammar: Grammar, **kwargs) -> None:
super().__init__(grammar, **kwargs)
self.chart: List = [] # for type checking
def chart_parse(self, words, start):
alt = tuple(*self.cgrammar[start])
chart = [Column(i, tok) for i, tok in enumerate([None, *words])]
chart[0].add(State(start, alt, 0, chart[0]))
return self.fill_chart(chart)
```
The main parsing loop in `fill_chart()` has three fundamental operations. `predict()`, `scan()`, and `complete()`. We discuss `predict` next.
#### Predicting States
We have already seeded `chart[0]` with a state `[<A>,<B>]` with `dot` at `0`. Next, given that `<A>` is a nonterminal, we `predict` the possible parse continuations of this state. That is, it could be either `a <B> c` or `A <A>`.
The general idea of `predict()` is as follows: Say you have a state with name `<A>` from the above grammar, and expression containing `[a,<B>,c]`. Imagine that you have seen `a` already, which means that the `dot` will be on `<B>`. Below, is a representation of our parse status. The left hand side of ● represents the portion already parsed (`a`), and the right hand side represents the portion yet to be parsed (`<B> c`).
```
<A>: a ● <B> c
```
To recognize `<B>`, we look at the definition of `<B>`, which has different alternative expressions. The `predict()` step adds each of these alternatives to the set of states, with `dot` at `0`.
```
<A>: a ● <B> c
<B>: ● b c
<B>: ● <D>
```
In essence, the `predict()` method, when called with the current nonterminal, fetches the alternative expressions corresponding to this nonterminal, and adds these as predicted _child_ states to the _current_ column.
```
class EarleyParser(EarleyParser):
def predict(self, col, sym, state):
for alt in self.cgrammar[sym]:
col.add(State(sym, tuple(alt), 0, col))
```
To see how to use `predict`, we first construct the 0th column as before, and we assign the constructed column to an instance of the EarleyParser.
```
col_0 = Column(0, None)
col_0.add(start_state)
ep = EarleyParser(SAMPLE_GRAMMAR)
ep.chart = [col_0]
```
It should contain a single state -- `<start> at 0`
```
for s in ep.chart[0].states:
print(s)
```
We apply predict to fill out the 0th column, and the column should contain the possible parse paths.
```
ep.predict(col_0, '<A>', s)
for s in ep.chart[0].states:
print(s)
```
#### Scanning Tokens
What if rather than a nonterminal, the state contained a terminal symbol such as a letter? In that case, we are ready to make some progress. For example, consider the second state:
```
<B>: ● b c
```
We `scan` the next column's letter. Say the next token is `b`.
If the letter matches what we have, then create a new state by advancing the current state by one letter.
```
<B>: b ● c
```
This new state is added to the next column (i.e the column that has the matched letter).
```
class EarleyParser(EarleyParser):
def scan(self, col, state, letter):
if letter == col.letter:
col.add(state.advance())
```
As before, we construct the partial parse first, this time adding a new column so that we can observe the effects of `scan()`
```
ep = EarleyParser(SAMPLE_GRAMMAR)
col_1 = Column(1, 'a')
ep.chart = [col_0, col_1]
new_state = ep.chart[0].states[1]
print(new_state)
ep.scan(col_1, new_state, 'a')
for s in ep.chart[1].states:
print(s)
```
#### Completing Processing
When we advance, what if we actually `complete()` the processing of the current rule? If so, we want to update not just this state, but also all the _parent_ states from which this state was derived.
For example, say we have states as below.
```
<A>: a ● <B> c
<B>: b c ●
```
The state `<B>: b c ●` is now complete. So, we need to advance `<A>: a ● <B> c` one step forward.
How do we determine the parent states? Note from `predict` that we added the predicted child states to the _same_ column as that of the inspected state. Hence, we look at the starting column of the current state, with the same symbol `at_dot` as that of the name of the completed state.
For each such parent found, we advance that parent (because we have just finished parsing that non terminal for their `at_dot`) and add the new states to the current column.
```
class EarleyParser(EarleyParser):
def complete(self, col, state):
return self.earley_complete(col, state)
def earley_complete(self, col, state):
parent_states = [
st for st in state.s_col.states if st.at_dot() == state.name
]
for st in parent_states:
col.add(st.advance())
```
Here is an example of completed processing. First we complete the Column 0
```
ep = EarleyParser(SAMPLE_GRAMMAR)
col_1 = Column(1, 'a')
col_2 = Column(2, 'd')
ep.chart = [col_0, col_1, col_2]
ep.predict(col_0, '<A>', s)
for s in ep.chart[0].states:
print(s)
```
Then we use `scan()` to populate Column 1
```
for state in ep.chart[0].states:
if state.at_dot() not in SAMPLE_GRAMMAR:
ep.scan(col_1, state, 'a')
for s in ep.chart[1].states:
print(s)
for state in ep.chart[1].states:
if state.at_dot() in SAMPLE_GRAMMAR:
ep.predict(col_1, state.at_dot(), state)
for s in ep.chart[1].states:
print(s)
```
Then we use `scan()` again to populate Column 2
```
for state in ep.chart[1].states:
if state.at_dot() not in SAMPLE_GRAMMAR:
ep.scan(col_2, state, state.at_dot())
for s in ep.chart[2].states:
print(s)
```
Now, we can use `complete()`:
```
for state in ep.chart[2].states:
if state.finished():
ep.complete(col_2, state)
for s in ep.chart[2].states:
print(s)
```
#### Filling the Chart
The main driving loop in `fill_chart()` essentially calls these operations in order. We loop over each column in order.
* For each column, fetch one state in the column at a time, and check if the state is `finished`.
* If it is, then we `complete()` all the parent states depending on this state.
* If the state was not finished, we check to see if the state's current symbol `at_dot` is a nonterminal.
* If it is a nonterminal, we `predict()` possible continuations, and update the current column with these states.
* If it was not, we `scan()` the next column and advance the current state if it matches the next letter.
```
class EarleyParser(EarleyParser):
def fill_chart(self, chart):
for i, col in enumerate(chart):
for state in col.states:
if state.finished():
self.complete(col, state)
else:
sym = state.at_dot()
if sym in self.cgrammar:
self.predict(col, sym, state)
else:
if i + 1 >= len(chart):
continue
self.scan(chart[i + 1], state, sym)
if self.log:
print(col, '\n')
return chart
```
We now can recognize a given string as belonging to a language represented by a grammar.
```
ep = EarleyParser(SAMPLE_GRAMMAR, log=True)
columns = ep.chart_parse('adcd', START_SYMBOL)
```
The chart we printed above only shows completed entries at each index. The parenthesized expression indicates the column just before the first character was recognized, and the ending column.
Notice how the `<start>` nonterminal shows fully parsed status.
```
last_col = columns[-1]
for state in last_col.states:
if state.name == '<start>':
print(state)
```
Since `chart_parse()` returns the completed table, we now need to extract the derivation trees.
#### The Parse Method
For determining how far we have managed to parse, we simply look for the last index from `chart_parse()` where the `start_symbol` was found.
```
class EarleyParser(EarleyParser):
def parse_prefix(self, text):
self.table = self.chart_parse(text, self.start_symbol())
for col in reversed(self.table):
states = [
st for st in col.states if st.name == self.start_symbol()
]
if states:
return col.index, states
return -1, []
```
Here is the `parse_prefix()` in action.
```
ep = EarleyParser(SAMPLE_GRAMMAR)
cursor, last_states = ep.parse_prefix('adcd')
print(cursor, [str(s) for s in last_states])
```
The following is adapted from the excellent reference on Earley parsing by [Loup Vaillant](http://loup-vaillant.fr/tutorials/earley-parsing/).
Our `parse()` method is as follows. It depends on two methods `parse_forest()` and `extract_trees()` that will be defined next.
```
class EarleyParser(EarleyParser):
def parse(self, text):
cursor, states = self.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(text[cursor:]))
forest = self.parse_forest(self.table, start)
for tree in self.extract_trees(forest):
yield self.prune_tree(tree)
```
#### Parsing Paths
The `parse_paths()` method tries to unify the given expression in `named_expr` with the parsed string. For that, it extracts the last symbol in `named_expr` and checks if it is a terminal symbol. If it is, then it checks the chart at `til` to see if the letter corresponding to the position matches the terminal symbol. If it does, extend our start index by the length of the symbol.
If the symbol was a nonterminal symbol, then we retrieve the parsed states at the current end column index (`til`) that correspond to the nonterminal symbol, and collect the start index. These are the end column indexes for the remaining expression.
Given our list of start indexes, we obtain the parse paths from the remaining expression. If we can obtain any, then we return the parse paths. If not, we return an empty list.
```
class EarleyParser(EarleyParser):
def parse_paths(self, named_expr, chart, frm, til):
def paths(state, start, k, e):
if not e:
return [[(state, k)]] if start == frm else []
else:
return [[(state, k)] + r
for r in self.parse_paths(e, chart, frm, start)]
*expr, var = named_expr
starts = None
if var not in self.cgrammar:
starts = ([(var, til - len(var),
't')] if til > 0 and chart[til].letter == var else [])
else:
starts = [(s, s.s_col.index, 'n') for s in chart[til].states
if s.finished() and s.name == var]
return [p for s, start, k in starts for p in paths(s, start, k, expr)]
```
Here is the `parse_paths()` in action
```
print(SAMPLE_GRAMMAR['<start>'])
ep = EarleyParser(SAMPLE_GRAMMAR)
completed_start = last_states[0]
paths = ep.parse_paths(completed_start.expr, columns, 0, 4)
for path in paths:
print([list(str(s_) for s_ in s) for s in path])
```
That is, the parse path for `<start>` given the input `adcd` included recognizing the expression `<A><B>`. This was recognized by the two states: `<A>` from input(0) to input(2) which further involved recognizing the rule `a<B>c`, and the next state `<B>` from input(3) which involved recognizing the rule `<D>`.
#### Parsing Forests
The `parse_forest()` method takes the state which represents the completed parse, and determines the possible ways that its expressions corresponded to the parsed expression. For example, say we are parsing `1+2+3`, and the state has `[<expr>,+,<expr>]` in `expr`. It could have been parsed as either `[{<expr>:1+2},+,{<expr>:3}]` or `[{<expr>:1},+,{<expr>:2+3}]`.
```
class EarleyParser(EarleyParser):
def forest(self, s, kind, chart):
return self.parse_forest(chart, s) if kind == 'n' else (s, [])
def parse_forest(self, chart, state):
pathexprs = self.parse_paths(state.expr, chart, state.s_col.index,
state.e_col.index) if state.expr else []
return state.name, [[(v, k, chart) for v, k in reversed(pathexpr)]
for pathexpr in pathexprs]
ep = EarleyParser(SAMPLE_GRAMMAR)
result = ep.parse_forest(columns, last_states[0])
result
```
#### Extracting Trees
What we have from `parse_forest()` is a forest of trees. We need to extract a single tree from that forest. That is accomplished as follows.
(For now, we return the first available derivation tree. To do that, we need to extract the parse forest from the state corresponding to `start`.)
```
class EarleyParser(EarleyParser):
def extract_a_tree(self, forest_node):
name, paths = forest_node
if not paths:
return (name, [])
return (name, [self.extract_a_tree(self.forest(*p)) for p in paths[0]])
def extract_trees(self, forest):
yield self.extract_a_tree(forest)
```
We now verify that our parser can parse a given expression.
```
A3_GRAMMAR: Grammar = {
"<start>": ["<bexpr>"],
"<bexpr>": [
"<aexpr><gt><aexpr>", "<aexpr><lt><aexpr>", "<aexpr>=<aexpr>",
"<bexpr>=<bexpr>", "<bexpr>&<bexpr>", "<bexpr>|<bexpr>", "(<bexrp>)"
],
"<aexpr>":
["<aexpr>+<aexpr>", "<aexpr>-<aexpr>", "(<aexpr>)", "<integer>"],
"<integer>": ["<digit><integer>", "<digit>"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
"<lt>": ['<'],
"<gt>": ['>']
}
syntax_diagram(A3_GRAMMAR)
mystring = '(1+24)=33'
parser = EarleyParser(A3_GRAMMAR)
for tree in parser.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
```
We now have a complete parser that can parse almost arbitrary *CFG*. There remains a small corner to fix -- the case of epsilon rules as we will see later.
#### Ambiguous Parsing
Ambiguous grammars are grammars that can produce multiple derivation trees for some given string. For example, the `A3_GRAMMAR` can parse `1+2+3` in two different ways – `[1+2]+3` and `1+[2+3]`.
Extracting a single tree might be reasonable for unambiguous parses. However, what if the given grammar produces ambiguity when given a string? We need to extract all derivation trees in that case. We enhance our `extract_trees()` method to extract multiple derivation trees.
```
import itertools as I
class EarleyParser(EarleyParser):
def extract_trees(self, forest_node):
name, paths = forest_node
if not paths:
yield (name, [])
for path in paths:
ptrees = [self.extract_trees(self.forest(*p)) for p in path]
for p in I.product(*ptrees):
yield (name, p)
```
As before, we verify that everything works.
```
mystring = '1+2'
parser = EarleyParser(A1_GRAMMAR)
for tree in parser.parse(mystring):
assert mystring == tree_to_string(tree)
display_tree(tree)
```
One can also use a `GrammarFuzzer` to verify that everything works.
```
gf = GrammarFuzzer(A1_GRAMMAR)
for i in range(5):
s = gf.fuzz()
print(i, s)
for tree in parser.parse(s):
assert tree_to_string(tree) == s
```
#### The Aycock Epsilon Fix
While parsing, one often requires to know whether a given nonterminal can derive an empty string. For example, in the following grammar A can derive an empty string, while B can't. The nonterminals that can derive an empty string are called _nullable_ nonterminals. For example, in the below grammar `E_GRAMMAR_1`, `<A>` is _nullable_, and since `<A>` is one of the alternatives of `<start>`, `<start>` is also _nullable_. But `<B>` is not _nullable_.
```
E_GRAMMAR_1: Grammar = {
'<start>': ['<A>', '<B>'],
'<A>': ['a', ''],
'<B>': ['b']
}
```
One of the problems with the original Earley implementation is that it does not handle rules that can derive empty strings very well. For example, the given grammar should match `a`
```
EPSILON = ''
E_GRAMMAR: Grammar = {
'<start>': ['<S>'],
'<S>': ['<A><A><A><A>'],
'<A>': ['a', '<E>'],
'<E>': [EPSILON]
}
syntax_diagram(E_GRAMMAR)
mystring = 'a'
parser = EarleyParser(E_GRAMMAR)
with ExpectError():
trees = parser.parse(mystring)
```
Aycock et al.\cite{Aycock2002} suggests a simple fix. Their idea is to pre-compute the `nullable` set and use it to advance the `nullable` states. However, before we do that, we need to compute the `nullable` set. The `nullable` set consists of all nonterminals that can derive an empty string.
Computing the `nullable` set requires expanding each production rule in the grammar iteratively and inspecting whether a given rule can derive the empty string. Each iteration needs to take into account new terminals that have been found to be `nullable`. The procedure stops when we obtain a stable result. This procedure can be abstracted into a more general method `fixpoint`.
##### Fixpoint
A `fixpoint` of a function is an element in the function's domain such that it is mapped to itself. For example, 1 is a `fixpoint` of square root because `squareroot(1) == 1`.
(We use `str` rather than `hash` to check for equality in `fixpoint` because the data structure `set`, which we would like to use as an argument has a good string representation but is not hashable).
```
def fixpoint(f):
def helper(arg):
while True:
sarg = str(arg)
arg_ = f(arg)
if str(arg_) == sarg:
return arg
arg = arg_
return helper
```
Remember `my_sqrt()` from [the first chapter](Intro_Testing.ipynb)? We can define `my_sqrt()` using fixpoint.
```
def my_sqrt(x):
@fixpoint
def _my_sqrt(approx):
return (approx + x / approx) / 2
return _my_sqrt(1)
my_sqrt(2)
```
##### Nullable
Similarly, we can define `nullable` using `fixpoint`. We essentially provide the definition of a single intermediate step. That is, assuming that `nullables` contain the current `nullable` nonterminals, we iterate over the grammar looking for productions which are `nullable` -- that is, productions where the entire sequence can yield an empty string on some expansion.
We need to iterate over the different alternative expressions and their corresponding nonterminals. Hence we define a `rules()` method converts our dictionary representation to this pair format.
```
def rules(grammar):
return [(key, choice)
for key, choices in grammar.items()
for choice in choices]
```
The `terminals()` method extracts all terminal symbols from a `canonical` grammar representation.
```
def terminals(grammar):
return set(token
for key, choice in rules(grammar)
for token in choice if token not in grammar)
def nullable_expr(expr, nullables):
return all(token in nullables for token in expr)
def nullable(grammar):
productions = rules(grammar)
@fixpoint
def nullable_(nullables):
for A, expr in productions:
if nullable_expr(expr, nullables):
nullables |= {A}
return (nullables)
return nullable_({EPSILON})
for key, grammar in {
'E_GRAMMAR': E_GRAMMAR,
'E_GRAMMAR_1': E_GRAMMAR_1
}.items():
print(key, nullable(canonical(grammar)))
```
So, once we have the `nullable` set, all that we need to do is, after we have called `predict` on a state corresponding to a nonterminal, check if it is `nullable` and if it is, advance and add the state to the current column.
```
class EarleyParser(EarleyParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.epsilon = nullable(self.cgrammar)
def predict(self, col, sym, state):
for alt in self.cgrammar[sym]:
col.add(State(sym, tuple(alt), 0, col))
if sym in self.epsilon:
col.add(state.advance())
mystring = 'a'
parser = EarleyParser(E_GRAMMAR)
for tree in parser.parse(mystring):
display_tree(tree)
```
To ensure that our parser does parse all kinds of grammars, let us try two more test cases.
```
DIRECTLY_SELF_REFERRING: Grammar = {
'<start>': ['<query>'],
'<query>': ['select <expr> from a'],
"<expr>": ["<expr>", "a"],
}
INDIRECTLY_SELF_REFERRING: Grammar = {
'<start>': ['<query>'],
'<query>': ['select <expr> from a'],
"<expr>": ["<aexpr>", "a"],
"<aexpr>": ["<expr>"],
}
mystring = 'select a from a'
for grammar in [DIRECTLY_SELF_REFERRING, INDIRECTLY_SELF_REFERRING]:
forest = EarleyParser(grammar).parse(mystring)
print('recognized', mystring)
try:
for tree in forest:
print(tree_to_string(tree))
except RecursionError as e:
print("Recursion error", e)
```
Why do we get recursion error here? The reason is that, our implementation of `extract_trees()` is eager. That is, it attempts to extract _all_ inner parse trees before it can construct the outer parse tree. When there is a self reference, this results in recursion. Here is a simple extractor that avoids this problem. The idea here is that we randomly and lazily choose a node to expand, which avoids the infinite recursion.
#### Tree Extractor
As you saw above, one of the problems with attempting to extract all trees is that the parse forest can consist of an infinite number of trees. So, here, we solve that problem by extracting one tree at a time.
```
class SimpleExtractor:
def __init__(self, parser, text):
self.parser = parser
cursor, states = parser.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(cursor))
self.my_forest = parser.parse_forest(parser.table, start)
def extract_a_node(self, forest_node):
name, paths = forest_node
if not paths:
return ((name, 0, 1), []), (name, [])
cur_path, i, length = self.choose_path(paths)
child_nodes = []
pos_nodes = []
for s, kind, chart in cur_path:
f = self.parser.forest(s, kind, chart)
postree, ntree = self.extract_a_node(f)
child_nodes.append(ntree)
pos_nodes.append(postree)
return ((name, i, length), pos_nodes), (name, child_nodes)
def choose_path(self, arr):
length = len(arr)
i = random.randrange(length)
return arr[i], i, length
def extract_a_tree(self):
pos_tree, parse_tree = self.extract_a_node(self.my_forest)
return self.parser.prune_tree(parse_tree)
```
Using it is as folows:
```
de = SimpleExtractor(EarleyParser(DIRECTLY_SELF_REFERRING), mystring)
for i in range(5):
tree = de.extract_a_tree()
print(tree_to_string(tree))
```
On the indirect reference:
```
ie = SimpleExtractor(EarleyParser(INDIRECTLY_SELF_REFERRING), mystring)
for i in range(5):
tree = ie.extract_a_tree()
print(tree_to_string(tree))
```
Note that the `SimpleExtractor` gives no guarantee of the uniqueness of the returned trees. This can however be fixed by keeping track of the particular nodes that were expanded from `pos_tree` variable, and hence, avoiding exploration of the same paths.
For implementing this, we extract the random stream passing into the `SimpleExtractor`, and use it to control which nodes are explored. Different exploration paths can then form a tree of nodes.
We start with the node definition for a single choice. The `self._chosen` is the current choice made, `self.next` holds the next choice done using `self._chosen`. The `self.total` holds the total number of choices that one can have in this node.
```
class ChoiceNode:
def __init__(self, parent, total):
self._p, self._chosen = parent, 0
self._total, self.next = total, None
def chosen(self):
assert not self.finished()
return self._chosen
def __str__(self):
return '%d(%s/%s %s)' % (self._i, str(self._chosen),
str(self._total), str(self.next))
def __repr__(self):
return repr((self._i, self._chosen, self._total))
def increment(self):
# as soon as we increment, next becomes invalid
self.next = None
self._chosen += 1
if self.finished():
if self._p is None:
return None
return self._p.increment()
return self
def finished(self):
return self._chosen >= self._total
```
Now we come to the enhanced `EnhancedExtractor()`.
```
class EnhancedExtractor(SimpleExtractor):
def __init__(self, parser, text):
super().__init__(parser, text)
self.choices = ChoiceNode(None, 1)
```
First we define `choose_path()` that given an array and a choice node, returns the element in array corresponding to the next choice node if it exists, or produces a new choice nodes, and returns that element.
```
class EnhancedExtractor(EnhancedExtractor):
def choose_path(self, arr, choices):
arr_len = len(arr)
if choices.next is not None:
if choices.next.finished():
return None, None, None, choices.next
else:
choices.next = ChoiceNode(choices, arr_len)
next_choice = choices.next.chosen()
choices = choices.next
return arr[next_choice], next_choice, arr_len, choices
```
We define `extract_a_node()` here. While extracting, we have a choice. Should we allow infinite forests, or should we have a finite number of trees with no direct recursion? A direct recursion is when there exists a parent node with the same nonterminal that parsed the same span. We choose here not to extract such trees. They can be added back after parsing.
This is a recursive procedure that inspects a node, extracts the path required to complete that node. A single path (corresponding to a nonterminal) may again be composed of a sequence of smaller paths. Such paths are again extracted using another call to `extract_a_node()` recursively.
What happens when we hit on one of the node recursions we want to avoid? In that case, we return the current choice node, which bubbles up to `extract_a_tree()`. That procedure increments the last choice, which in turn increments up the parents until we reach a choice node that still has options to explore.
What if we hit the end of choices for a particular choice node(i.e, we have exhausted paths that can be taken from a node)? In this case also, we return the current choice node, which bubbles up to `extract_a_tree()`.
That procedure increments the last choice, which bubbles up to the next choice that has some unexplored paths.
```
class EnhancedExtractor(EnhancedExtractor):
def extract_a_node(self, forest_node, seen, choices):
name, paths = forest_node
if not paths:
return (name, []), choices
cur_path, _i, _l, new_choices = self.choose_path(paths, choices)
if cur_path is None:
return None, new_choices
child_nodes = []
for s, kind, chart in cur_path:
if kind == 't':
child_nodes.append((s, []))
continue
nid = (s.name, s.s_col.index, s.e_col.index)
if nid in seen:
return None, new_choices
f = self.parser.forest(s, kind, chart)
ntree, newer_choices = self.extract_a_node(f, seen | {nid}, new_choices)
if ntree is None:
return None, newer_choices
child_nodes.append(ntree)
new_choices = newer_choices
return (name, child_nodes), new_choices
```
The `extract_a_tree()` is a depth first extractor of a single tree. It tries to extract a tree, and if the extraction returns `None`, it means that a particular choice was exhausted, or we hit on a recursion. In that case, we increment the choice, and explore a new path.
```
class EnhancedExtractor(EnhancedExtractor):
def extract_a_tree(self):
while not self.choices.finished():
parse_tree, choices = self.extract_a_node(self.my_forest, set(), self.choices)
choices.increment()
if parse_tree is not None:
return self.parser.prune_tree(parse_tree)
return None
```
Note that the `EnhancedExtractor` only extracts nodes that are not directly recursive. That is, if it finds a node with a nonterminal that covers the same span as that of a parent node with the same nonterminal, it skips the node.
```
ee = EnhancedExtractor(EarleyParser(INDIRECTLY_SELF_REFERRING), mystring)
i = 0
while True:
i += 1
t = ee.extract_a_tree()
if t is None: break
print(i, t)
s = tree_to_string(t)
assert s == mystring
istring = '1+2+3+4'
ee = EnhancedExtractor(EarleyParser(A1_GRAMMAR), istring)
i = 0
while True:
i += 1
t = ee.extract_a_tree()
if t is None: break
print(i, t)
s = tree_to_string(t)
assert s == istring
```
#### More Earley Parsing
A number of other optimizations exist for Earley parsers. A fast industrial strength Earley parser implementation is the [Marpa parser](https://jeffreykegler.github.io/Marpa-web-site/). Further, Earley parsing need not be restricted to character data. One may also parse streams (audio and video streams) \cite{qi2018generalized} using a generalized Earley parser.
### End of Excursion
Here are a few examples of the Earley parser in action.
```
mystring = "1 + (2 * 3)"
earley = EarleyParser(EXPR_GRAMMAR)
for tree in earley.parse(mystring):
assert tree_to_string(tree) == mystring
display(display_tree(tree))
mystring = "1 * (2 + 3.35)"
for tree in earley.parse(mystring):
assert tree_to_string(tree) == mystring
display(display_tree(tree))
```
In contrast to the `PEGParser`, above, the `EarleyParser` can handle arbitrary context-free grammars.
### Excursion: Testing the Parsers
While we have defined two parser variants, it would be nice to have some confirmation that our parses work well. While it is possible to formally prove that they work, it is much more satisfying to generate random grammars, their corresponding strings, and parse them using the same grammar.
```
def prod_line_grammar(nonterminals, terminals):
g = {
'<start>': ['<symbols>'],
'<symbols>': ['<symbol><symbols>', '<symbol>'],
'<symbol>': ['<nonterminals>', '<terminals>'],
'<nonterminals>': ['<lt><alpha><gt>'],
'<lt>': ['<'],
'<gt>': ['>'],
'<alpha>': nonterminals,
'<terminals>': terminals
}
if not nonterminals:
g['<nonterminals>'] = ['']
del g['<lt>']
del g['<alpha>']
del g['<gt>']
return g
syntax_diagram(prod_line_grammar(["A", "B", "C"], ["1", "2", "3"]))
def make_rule(nonterminals, terminals, num_alts):
prod_grammar = prod_line_grammar(nonterminals, terminals)
gf = GrammarFuzzer(prod_grammar, min_nonterminals=3, max_nonterminals=5)
name = "<%s>" % ''.join(random.choices(string.ascii_uppercase, k=3))
return (name, [gf.fuzz() for _ in range(num_alts)])
make_rule(["A", "B", "C"], ["1", "2", "3"], 3)
from Grammars import unreachable_nonterminals
def make_grammar(num_symbols=3, num_alts=3):
terminals = list(string.ascii_lowercase)
grammar = {}
name = None
for _ in range(num_symbols):
nonterminals = [k[1:-1] for k in grammar.keys()]
name, expansions = \
make_rule(nonterminals, terminals, num_alts)
grammar[name] = expansions
grammar[START_SYMBOL] = [name]
# Remove unused parts
for nonterminal in unreachable_nonterminals(grammar):
del grammar[nonterminal]
assert is_valid_grammar(grammar)
return grammar
make_grammar()
```
Now we verify if our arbitrary grammars can be used by the Earley parser.
```
for i in range(5):
my_grammar = make_grammar()
print(my_grammar)
parser = EarleyParser(my_grammar)
mygf = GrammarFuzzer(my_grammar)
s = mygf.fuzz()
print(s)
for tree in parser.parse(s):
assert tree_to_string(tree) == s
display_tree(tree)
```
With this, we have completed both implementation and testing of *arbitrary* CFG, which can now be used along with `LangFuzzer` to generate better fuzzing inputs.
### End of Excursion
## Background
Numerous parsing techniques exist that can parse a given string using a
given grammar, and produce corresponding derivation tree or trees. However,
some of these techniques work only on specific classes of grammars.
These classes of grammars are named after the specific kind of parser
that can accept grammars of that category. That is, the upper bound for
the capabilities of the parser defines the grammar class named after that
parser.
The *LL* and *LR* parsing are the main traditions in parsing. Here, *LL* means left-to-right, leftmost derivation, and it represents a top-down approach. On the other hand, and LR (left-to-right, rightmost derivation) represents a bottom-up approach. Another way to look at it is that LL parsers compute the derivation tree incrementally in *pre-order* while LR parsers compute the derivation tree in *post-order* \cite{pingali2015graphical}).
Different classes of grammars differ in the features that are available to
the user for writing a grammar of that class. That is, the corresponding
kind of parser will be unable to parse a grammar that makes use of more
features than allowed. For example, the `A2_GRAMMAR` is an *LL*
grammar because it lacks left recursion, while `A1_GRAMMAR` is not an
*LL* grammar. This is because an *LL* parser parses
its input from left to right, and constructs the leftmost derivation of its
input by expanding the nonterminals it encounters. If there is a left
recursion in one of these rules, an *LL* parser will enter an infinite loop.
Similarly, a grammar is LL(k) if it can be parsed by an LL parser with k lookahead token, and LR(k) grammar can only be parsed with LR parser with at least k lookahead tokens. These grammars are interesting because both LL(k) and LR(k) grammars have $O(n)$ parsers, and can be used with relatively restricted computational budget compared to other grammars.
The languages for which one can provide an *LL(k)* grammar is called *LL(k)* languages (where k is the minimum lookahead required). Similarly, *LR(k)* is defined as the set of languages that have an *LR(k)* grammar. In terms of languages, LL(k) $\subset$ LL(k+1) and LL(k) $\subset$ LR(k), and *LR(k)* $=$ *LR(1)*. All deterministic *CFLs* have an *LR(1)* grammar. However, there exist *CFLs* that are inherently ambiguous \cite{ogden1968helpful}, and for these, one can't provide an *LR(1)* grammar.
The other main parsing algorithms for *CFGs* are GLL \cite{scott2010gll}, GLR \cite{tomita1987efficient,tomita2012generalized}, and CYK \cite{grune2008parsing}.
The ALL(\*) (used by ANTLR) on the other hand is a grammar representation that uses *Regular Expression* like predicates (similar to advanced PEGs – see [Exercise](#Exercise-3:-PEG-Predicates)) rather than a fixed lookahead. Hence, ALL(\*) can accept a larger class of grammars than CFGs.
In terms of computational limits of parsing, the main CFG parsers have a complexity of $O(n^3)$ for arbitrary grammars. However, parsing with arbitrary *CFG* is reducible to boolean matrix multiplication \cite{Valiant1975} (and the reverse \cite{Lee2002}). This is at present bounded by $O(2^{23728639}$) \cite{LeGall2014}. Hence, worse case complexity for parsing arbitrary CFG is likely to remain close to cubic.
Regarding PEGs, the actual class of languages that is expressible in *PEG* is currently unknown. In particular, we know that *PEGs* can express certain languages such as $a^n b^n c^n$. However, we do not know if there exist *CFLs* that are not expressible with *PEGs*. In Section 2.3, we provided an instance of a counter-intuitive PEG grammar. While important for our purposes (we use grammars for generation of inputs) this is not a criticism of parsing with PEGs. PEG focuses on writing grammars for recognizing a given language, and not necessarily in interpreting what language an arbitrary PEG might yield. Given a Context-Free Language to parse, it is almost always possible to write a grammar for it in PEG, and given that 1) a PEG can parse any string in $O(n)$ time, and 2) at present we know of no CFL that can't be expressed as a PEG, and 3) compared with *LR* grammars, a PEG is often more intuitive because it allows top-down interpretation, when writing a parser for a language, PEGs should be under serious consideration.
## Synopsis
This chapter introduces `Parser` classes, parsing a string into a _derivation tree_ as introduced in the [chapter on efficient grammar fuzzing](GrammarFuzzer.ipynb). Two important parser classes are provided:
* [Parsing Expression Grammar parsers](#Parsing-Expression-Grammars) (`PEGParser`). These are very efficient, but limited to specific grammar structure. Notably, the alternatives represent *ordered choice*. That is, rather than choosing all rules that can potentially match, we stop at the first match that succeed.
* [Earley parsers](#Parsing-Context-Free-Grammars) (`EarleyParser`). These accept any kind of context-free grammars, and explore all parsing alternatives (if any).
Using any of these is fairly easy, though. First, instantiate them with a grammar:
```
from Grammars import US_PHONE_GRAMMAR
us_phone_parser = EarleyParser(US_PHONE_GRAMMAR)
```
Then, use the `parse()` method to retrieve a list of possible derivation trees:
```
trees = us_phone_parser.parse("(555)987-6543")
tree = list(trees)[0]
display_tree(tree)
```
These derivation trees can then be used for test generation, notably for mutating and recombining existing inputs.
```
# ignore
from ClassDiagram import display_class_hierarchy
# ignore
display_class_hierarchy([PEGParser, EarleyParser],
public_methods=[
Parser.parse,
Parser.__init__,
Parser.grammar,
Parser.start_symbol
],
types={
'DerivationTree': DerivationTree,
'Grammar': Grammar
},
project='fuzzingbook')
```
## Lessons Learned
* Grammars can be used to generate derivation trees for a given string.
* Parsing Expression Grammars are intuitive, and easy to implement, but require care to write.
* Earley Parsers can parse arbitrary Context Free Grammars.
## Next Steps
* Use parsed inputs to [recombine existing inputs](LangFuzzer.ipynb)
## Exercises
### Exercise 1: An Alternative Packrat
In the _Packrat_ parser, we showed how one could implement a simple _PEG_ parser. That parser kept track of the current location in the text using an index. Can you modify the parser so that it simply uses the current substring rather than tracking the index? That is, it should no longer have the `at` parameter.
**Solution.** Here is a possible solution:
```
class PackratParser(Parser):
def parse_prefix(self, text):
txt, res = self.unify_key(self.start_symbol(), text)
return len(txt), [res]
def parse(self, text):
remain, res = self.parse_prefix(text)
if remain:
raise SyntaxError("at " + res)
return res
def unify_rule(self, rule, text):
results = []
for token in rule:
text, res = self.unify_key(token, text)
if res is None:
return text, None
results.append(res)
return text, results
def unify_key(self, key, text):
if key not in self.cgrammar:
if text.startswith(key):
return text[len(key):], (key, [])
else:
return text, None
for rule in self.cgrammar[key]:
text_, res = self.unify_rule(rule, text)
if res:
return (text_, (key, res))
return text, None
mystring = "1 + (2 * 3)"
for tree in PackratParser(EXPR_GRAMMAR).parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
```
### Exercise 2: More PEG Syntax
The _PEG_ syntax provides a few notational conveniences reminiscent of regular expressions. For example, it supports the following operators (letters `T` and `A` represents tokens that can be either terminal or nonterminal. `ε` is an empty string, and `/` is the ordered choice operator similar to the non-ordered choice operator `|`):
* `T?` represents an optional greedy match of T and `A := T?` is equivalent to `A := T/ε`.
* `T*` represents zero or more greedy matches of `T` and `A := T*` is equivalent to `A := T A/ε`.
* `T+` represents one or more greedy matches – equivalent to `TT*`
If you look at the three notations above, each can be represented in the grammar in terms of basic syntax.
Remember the exercise from [the chapter on grammars](Grammars.ipynb) that developed `define_ex_grammar()` that can represent grammars as Python code? extend `define_ex_grammar()` to `define_peg()` to support the above notational conveniences. The decorator should rewrite a given grammar that contains these notations to an equivalent grammar in basic syntax.
### Exercise 3: PEG Predicates
Beyond these notational conveniences, it also supports two predicates that can provide a powerful lookahead facility that does not consume any input.
* `T&A` represents an _And-predicate_ that matches `T` if `T` is matched, and it is immediately followed by `A`
* `T!A` represents a _Not-predicate_ that matches `T` if `T` is matched, and it is *not* immediately followed by `A`
Implement these predicates in our _PEG_ parser.
### Exercise 4: Earley Fill Chart
In the `Earley Parser`, `Column` class, we keep the states both as a `list` and also as a `dict` even though `dict` is ordered. Can you explain why?
**Hint**: see the `fill_chart` method.
**Solution.** Python allows us to append to a list in flight, while a dict, eventhough it is ordered does not allow that facility.
That is, the following will work
```python
values = [1]
for v in values:
values.append(v*2)
```
However, the following will result in an error
```python
values = {1:1}
for v in values:
values[v*2] = v*2
```
In the `fill_chart`, we make use of this facility to modify the set of states we are iterating on, on the fly.
### Exercise 5: Leo Parser
One of the problems with the original Earley parser is that while it can parse strings using arbitrary _Context Free Gramamrs_, its performance on right-recursive grammars is quadratic. That is, it takes $O(n^2)$ runtime and space for parsing with right-recursive grammars. For example, consider the parsing of the following string by two different grammars `LR_GRAMMAR` and `RR_GRAMMAR`.
```
mystring = 'aaaaaa'
```
To see the problem, we need to enable logging. Here is the logged version of parsing with the `LR_GRAMMAR`
```
result = EarleyParser(LR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass # consume the generator so that we can see the logs
```
Compare that to the parsing of `RR_GRAMMAR` as seen below:
```
result = EarleyParser(RR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
```
As can be seen from the parsing log for each letter, the number of states with representation `<A>: a <A> ● (i, j)` increases at each stage, and these are simply a left over from the previous letter. They do not contribute anything more to the parse other than to simply complete these entries. However, they take up space, and require resources for inspection, contributing a factor of `n` in analysis.
Joop Leo \cite{Leo1991} found that this inefficiency can be avoided by detecting right recursion. The idea is that before starting the `completion` step, check whether the current item has a _deterministic reduction path_. If such a path exists, add a copy of the topmost element of the _deteministic reduction path_ to the current column, and return. If not, perform the original `completion` step.
**Definition 2.1**: An item is said to be on the deterministic reduction path above $[A \rightarrow \gamma., i]$ if it is $[B \rightarrow \alpha A ., k]$ with $[B \rightarrow \alpha . A, k]$ being the only item in $ I_i $ with the dot in front of A, or if it is on the deterministic reduction path above $[B \rightarrow \alpha A ., k]$. An item on such a path is called *topmost* one if there is no item on the deterministic reduction path above it\cite{Leo1991}.
Finding a _deterministic reduction path_ is as follows:
Given a complete state, represented by `<A> : seq_1 ● (s, e)` where `s` is the starting column for this rule, and `e` the current column, there is a _deterministic reduction path_ **above** it if two constraints are satisfied.
1. There exist a *single* item in the form `<B> : seq_2 ● <A> (k, s)` in column `s`.
2. That should be the *single* item in s with dot in front of `<A>`
The resulting item is of the form `<B> : seq_2 <A> ● (k, e)`, which is simply item from (1) advanced, and is considered above `<A>:.. (s, e)` in the deterministic reduction path.
The `seq_1` and `seq_2` are arbitrary symbol sequences.
This forms the following chain of links, with `<A>:.. (s_1, e)` being the child of `<B>:.. (s_2, e)` etc.
Here is one way to visualize the chain:
```
<C> : seq_3 <B> ● (s_3, e)
| constraints satisfied by <C> : seq_3 ● <B> (s_3, s_2)
<B> : seq_2 <A> ● (s_2, e)
| constraints satisfied by <B> : seq_2 ● <A> (s_2, s_1)
<A> : seq_1 ● (s_1, e)
```
Essentially, what we want to do is to identify potential deterministic right recursion candidates, perform completion on them, and *throw away the result*. We do this until we reach the top. See Grune et al.~\cite{grune2008parsing} for further information.
Note that the completions are in the same column (`e`), with each candidates with constraints satisfied
in further and further earlier columns (as shown below):
```
<C> : seq_3 ● <B> (s_3, s_2) --> <C> : seq_3 <B> ● (s_3, e)
|
<B> : seq_2 ● <A> (s_2, s_1) --> <B> : seq_2 <A> ● (s_2, e)
|
<A> : seq_1 ● (s_1, e)
```
Following this chain, the topmost item is the item `<C>:.. (s_3, e)` that does not have a parent. The topmost item needs to be saved is called a *transitive* item by Leo, and it is associated with the non-terminal symbol that started the lookup. The transitive item needs to be added to each column we inspect.
Here is the skeleton for the parser `LeoParser`.
```
class LeoParser(EarleyParser):
def complete(self, col, state):
return self.leo_complete(col, state)
def leo_complete(self, col, state):
detred = self.deterministic_reduction(state)
if detred:
col.add(detred.copy())
else:
self.earley_complete(col, state)
def deterministic_reduction(self, state):
raise NotImplementedError
```
Can you implement the `deterministic_reduction()` method to obtain the topmost element?
**Solution.** Here is a possible solution:
First, we update our `Column` class with the ability to add transitive items. Note that, while Leo asks the transitive to be added to the set $ I_k $ there is no actual requirement for the transitive states to be added to the `states` list. The transitive items are only intended for memoization and not for the `fill_chart()` method. Hence, we track them separately.
```
class Column(Column):
def __init__(self, index, letter):
self.index, self.letter = index, letter
self.states, self._unique, self.transitives = [], {}, {}
def add_transitive(self, key, state):
assert key not in self.transitives
self.transitives[key] = state
return self.transitives[key]
```
Remember the picture we drew of the deterministic path?
```
<C> : seq_3 <B> ● (s_3, e)
| constraints satisfied by <C> : seq_3 ● <B> (s_3, s_2)
<B> : seq_2 <A> ● (s_2, e)
| constraints satisfied by <B> : seq_2 ● <A> (s_2, s_1)
<A> : seq_1 ● (s_1, e)
```
We define a function `uniq_postdot()` that given the item `<A> := seq_1 ● (s_1, e)`, returns a `<B> : seq_2 ● <A> (s_2, s_1)` that satisfies the constraints mentioned in the above picture.
```
class LeoParser(LeoParser):
def uniq_postdot(self, st_A):
col_s1 = st_A.s_col
parent_states = [
s for s in col_s1.states if s.expr and s.at_dot() == st_A.name
]
if len(parent_states) > 1:
return None
matching_st_B = [s for s in parent_states if s.dot == len(s.expr) - 1]
return matching_st_B[0] if matching_st_B else None
lp = LeoParser(RR_GRAMMAR)
[(str(s), str(lp.uniq_postdot(s))) for s in columns[-1].states]
```
We next define the function `get_top()` that is the core of deterministic reduction which gets the topmost state above the current state (`A`).
```
class LeoParser(LeoParser):
def get_top(self, state_A):
st_B_inc = self.uniq_postdot(state_A)
if not st_B_inc:
return None
t_name = st_B_inc.name
if t_name in st_B_inc.e_col.transitives:
return st_B_inc.e_col.transitives[t_name]
st_B = st_B_inc.advance()
top = self.get_top(st_B) or st_B
return st_B_inc.e_col.add_transitive(t_name, top)
```
Once we have the machinery in place, `deterministic_reduction()` itself is simply a wrapper to call `get_top()`
```
class LeoParser(LeoParser):
def deterministic_reduction(self, state):
return self.get_top(state)
lp = LeoParser(RR_GRAMMAR)
columns = lp.chart_parse(mystring, lp.start_symbol())
[(str(s), str(lp.get_top(s))) for s in columns[-1].states]
```
Now, both LR and RR grammars should work within $O(n)$ bounds.
```
result = LeoParser(RR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
```
We verify the Leo parser with a few more right recursive grammars.
```
RR_GRAMMAR2 = {
'<start>': ['<A>'],
'<A>': ['ab<A>', ''],
}
mystring2 = 'ababababab'
result = LeoParser(RR_GRAMMAR2, log=True).parse(mystring2)
for _ in result: pass
RR_GRAMMAR3 = {
'<start>': ['c<A>'],
'<A>': ['ab<A>', ''],
}
mystring3 = 'cababababab'
result = LeoParser(RR_GRAMMAR3, log=True).parse(mystring3)
for _ in result: pass
RR_GRAMMAR4 = {
'<start>': ['<A>c'],
'<A>': ['ab<A>', ''],
}
mystring4 = 'ababababc'
result = LeoParser(RR_GRAMMAR4, log=True).parse(mystring4)
for _ in result: pass
RR_GRAMMAR5 = {
'<start>': ['<A>'],
'<A>': ['ab<B>', ''],
'<B>': ['<A>'],
}
mystring5 = 'abababab'
result = LeoParser(RR_GRAMMAR5, log=True).parse(mystring5)
for _ in result: pass
RR_GRAMMAR6 = {
'<start>': ['<A>'],
'<A>': ['a<B>', ''],
'<B>': ['b<A>'],
}
mystring6 = 'abababab'
result = LeoParser(RR_GRAMMAR6, log=True).parse(mystring6)
for _ in result: pass
RR_GRAMMAR7 = {
'<start>': ['<A>'],
'<A>': ['a<A>', 'a'],
}
mystring7 = 'aaaaaaaa'
result = LeoParser(RR_GRAMMAR7, log=True).parse(mystring7)
for _ in result: pass
```
We verify that our parser works correctly on `LR_GRAMMAR` too.
```
result = LeoParser(LR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
```
__Advanced:__ We have fixed the complexity bounds. However, because we are saving only the topmost item of a right recursion, we need to fix our parser to be aware of our fix while extracting parse trees. Can you fix it?
__Hint:__ Leo suggests simply transforming the Leo item sets to normal Earley sets, with the results from deterministic reduction expanded to their originals. For that, keep in mind the picture of constraint chain we drew earlier.
**Solution.** Here is a possible solution.
We first change the definition of `add_transitive()` so that results of deterministic reduction can be identified later.
```
class Column(Column):
def add_transitive(self, key, state):
assert key not in self.transitives
self.transitives[key] = TState(state.name, state.expr, state.dot,
state.s_col, state.e_col)
return self.transitives[key]
```
We also need a `back()` method to create the constraints.
```
class State(State):
def back(self):
return TState(self.name, self.expr, self.dot - 1, self.s_col, self.e_col)
```
We update `copy()` to make `TState` items instead.
```
class TState(State):
def copy(self):
return TState(self.name, self.expr, self.dot, self.s_col, self.e_col)
```
We now modify the `LeoParser` to keep track of the chain of constrains that we mentioned earlier.
```
class LeoParser(LeoParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self._postdots = {}
```
Next, we update the `uniq_postdot()` so that it tracks the chain of links.
```
class LeoParser(LeoParser):
def uniq_postdot(self, st_A):
col_s1 = st_A.s_col
parent_states = [
s for s in col_s1.states if s.expr and s.at_dot() == st_A.name
]
if len(parent_states) > 1:
return None
matching_st_B = [s for s in parent_states if s.dot == len(s.expr) - 1]
if matching_st_B:
self._postdots[matching_st_B[0]._t()] = st_A
return matching_st_B[0]
return None
```
We next define a method `expand_tstate()` that, when given a `TState`, generates all the intermediate links that we threw away earlier for a given end column.
```
class LeoParser(LeoParser):
def expand_tstate(self, state, e):
if state._t() not in self._postdots:
return
c_C = self._postdots[state._t()]
e.add(c_C.advance())
self.expand_tstate(c_C.back(), e)
```
We define a `rearrange()` method to generate a reversed table where each column contains states that start at that column.
```
class LeoParser(LeoParser):
def rearrange(self, table):
f_table = [Column(c.index, c.letter) for c in table]
for col in table:
for s in col.states:
f_table[s.s_col.index].states.append(s)
return f_table
```
Here is the rearranged table. (Can you explain why the Column 0 has a large number of `<start>` items?)
```
ep = LeoParser(RR_GRAMMAR)
columns = ep.chart_parse(mystring, ep.start_symbol())
r_table = ep.rearrange(columns)
for col in r_table:
print(col, "\n")
```
We save the result of rearrange before going into `parse_forest()`.
```
class LeoParser(LeoParser):
def parse(self, text):
cursor, states = self.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(text[cursor:]))
self.r_table = self.rearrange(self.table)
forest = self.extract_trees(self.parse_forest(self.table, start))
for tree in forest:
yield self.prune_tree(tree)
```
Finally, during `parse_forest()`, we first check to see if it is a transitive state, and if it is, expand it to the original sequence of states using `traverse_constraints()`.
```
class LeoParser(LeoParser):
def parse_forest(self, chart, state):
if isinstance(state, TState):
self.expand_tstate(state.back(), state.e_col)
return super().parse_forest(chart, state)
```
This completes our implementation of `LeoParser`.
We check whether the previously defined right recursive grammars parse and return the correct parse trees.
```
result = LeoParser(RR_GRAMMAR).parse(mystring)
for tree in result:
assert mystring == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR2).parse(mystring2)
for tree in result:
assert mystring2 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR3).parse(mystring3)
for tree in result:
assert mystring3 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR4).parse(mystring4)
for tree in result:
assert mystring4 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR5).parse(mystring5)
for tree in result:
assert mystring5 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR6).parse(mystring6)
for tree in result:
assert mystring6 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR7).parse(mystring7)
for tree in result:
assert mystring7 == tree_to_string(tree)
result = LeoParser(LR_GRAMMAR).parse(mystring)
for tree in result:
assert mystring == tree_to_string(tree)
RR_GRAMMAR8 = {
'<start>': ['<A>'],
'<A>': ['a<A>', 'a']
}
mystring8 = 'aa'
RR_GRAMMAR9 = {
'<start>': ['<A>'],
'<A>': ['<B><A>', '<B>'],
'<B>': ['b']
}
mystring9 = 'bbbbbbb'
result = LeoParser(RR_GRAMMAR8).parse(mystring8)
for tree in result:
print(repr(tree_to_string(tree)))
assert mystring8 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR9).parse(mystring9)
for tree in result:
print(repr(tree_to_string(tree)))
assert mystring9 == tree_to_string(tree)
```
### Exercise 6: Filtered Earley Parser
One of the problems with our Earley and Leo Parsers is that it can get stuck in infinite loops when parsing with grammars that contain token repetitions in alternatives. For example, consider the grammar below.
```
RECURSION_GRAMMAR: Grammar = {
"<start>": ["<A>"],
"<A>": ["<A>", "<A>aa", "AA", "<B>"],
"<B>": ["<C>", "<C>cc", "CC"],
"<C>": ["<B>", "<B>bb", "BB"]
}
```
With this grammar, one can produce an infinite chain of derivations of `<A>`, (direct recursion) or an infinite chain of derivations of `<B> -> <C> -> <B> ...` (indirect recursion). The problem is that, our implementation can get stuck trying to derive one of these infinite chains. One possibility is to use the `LazyExtractor`. Another, is to simply avoid generating such chains.
```
from ExpectError import ExpectTimeout
with ExpectTimeout(1, print_traceback=False):
mystring = 'AA'
parser = LeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
```
Can you implement a solution such that any tree that contains such a chain is discarded?
**Solution.** Here is a possible solution.
```
class FilteredLeoParser(LeoParser):
def forest(self, s, kind, seen, chart):
return self.parse_forest(chart, s, seen) if kind == 'n' else (s, [])
def parse_forest(self, chart, state, seen=None):
if isinstance(state, TState):
self.expand_tstate(state.back(), state.e_col)
def was_seen(chain, s):
if isinstance(s, str):
return False
if len(s.expr) > 1:
return False
return s in chain
if len(state.expr) > 1: # things get reset if we have a non loop
seen = set()
elif seen is None: # initialization
seen = {state}
pathexprs = self.parse_paths(state.expr, chart, state.s_col.index,
state.e_col.index) if state.expr else []
return state.name, [[(s, k, seen | {s}, chart)
for s, k in reversed(pathexpr)
if not was_seen(seen, s)] for pathexpr in pathexprs]
```
With the `FilteredLeoParser`, we should be able to recover minimal parse trees in reasonable time.
```
mystring = 'AA'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'AAaa'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'AAaaaa'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'CC'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BBcc'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BB'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BBccbb'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
```
As can be seen, we are able to recover minimal parse trees without hitting on infinite chains.
### Exercise 7: Iterative Earley Parser
Recursive algorithms are quite handy in some cases but sometimes we might want to have iteration instead of recursion due to memory or speed problems.
Can you implement an iterative version of the `EarleyParser`?
__Hint:__ In general, you can use a stack to replace a recursive algorithm with an iterative one. An easy way to do this is pushing the parameters onto a stack instead of passing them to the recursive function.
**Solution.** Here is a possible solution.
First, we define `parse_paths()` that extract paths from a parsed expression, which is very similar to the original.
```
class IterativeEarleyParser(EarleyParser):
def parse_paths(self, named_expr_, chart, frm, til_):
return_paths = []
path_build_stack = [(named_expr_, til_, [])]
def iter_paths(path_prefix, path, start, k, e):
x = path_prefix + [(path, k)]
if not e:
return_paths.extend([x] if start == frm else [])
else:
path_build_stack.append((e, start, x))
while path_build_stack:
named_expr, til, path_prefix = path_build_stack.pop()
*expr, var = named_expr
starts = None
if var not in self.cgrammar:
starts = ([(var, til - len(var), 't')]
if til > 0 and chart[til].letter == var else [])
else:
starts = [(s, s.s_col.index, 'n') for s in chart[til].states
if s.finished() and s.name == var]
for s, start, k in starts:
iter_paths(path_prefix, s, start, k, expr)
return return_paths
```
Next we used these paths to recover the forest data structure using `parse_forest()`. Since `parse_forest()` does not recurse, we reuse the original definition. Next, we define `extract_a_tree()`
Now we are ready to extract trees from the forest using `extract_a_tree()`
```
class IterativeEarleyParser(IterativeEarleyParser):
def choose_a_node_to_explore(self, node_paths, level_count):
first, *rest = node_paths
return first
def extract_a_tree(self, forest_node_):
start_node = (forest_node_[0], [])
tree_build_stack = [(forest_node_, start_node[-1], 0)]
while tree_build_stack:
forest_node, tree, level_count = tree_build_stack.pop()
name, paths = forest_node
if not paths:
tree.append((name, []))
else:
new_tree = []
current_node = self.choose_a_node_to_explore(paths, level_count)
for p in reversed(current_node):
new_forest_node = self.forest(*p)
tree_build_stack.append((new_forest_node, new_tree, level_count + 1))
tree.append((name, new_tree))
return start_node
```
For now, we simply extract the first tree found.
```
class IterativeEarleyParser(IterativeEarleyParser):
def extract_trees(self, forest):
yield self.extract_a_tree(forest)
```
Let's see if it works with some of the grammars we have seen so far.
```
test_cases: List[Tuple[Grammar, str]] = [
(A1_GRAMMAR, '1-2-3+4-5'),
(A2_GRAMMAR, '1+2'),
(A3_GRAMMAR, '1+2+3-6=6-1-2-3'),
(LR_GRAMMAR, 'aaaaa'),
(RR_GRAMMAR, 'aa'),
(DIRECTLY_SELF_REFERRING, 'select a from a'),
(INDIRECTLY_SELF_REFERRING, 'select a from a'),
(RECURSION_GRAMMAR, 'AA'),
(RECURSION_GRAMMAR, 'AAaaaa'),
(RECURSION_GRAMMAR, 'BBccbb')
]
for i, (grammar, text) in enumerate(test_cases):
print(i, text)
tree, *_ = IterativeEarleyParser(grammar).parse(text)
assert text == tree_to_string(tree)
```
As can be seen, our `IterativeEarleyParser` is able to handle recursive grammars. However, it can only extract the first tree found. What should one do to get all possible parses? What we can do, is to keep track of options to explore at each `choose_a_node_to_explore()`. Next, capture in the nodes explored in a tree data structure, adding new paths each time a new leaf is expanded. See the `TraceTree` datastructure in the [chapter on Concolic fuzzing](ConcolicFuzzer.ipynb) for an example.
### Exercise 8: First Set of a Nonterminal
We previously gave a way to extract a the `nullable` (epsilon) set, which is often used for parsing.
Along with `nullable`, parsing algorithms often use two other sets [`first` and `follow`](https://en.wikipedia.org/wiki/Canonical_LR_parser#FIRST_and_FOLLOW_sets).
The first set of a terminal symbol is itself, and the first set of a nonterminal is composed of terminal symbols that can come at the beginning of any derivation
of that nonterminal. The first set of any nonterminal that can derive the empty string should contain `EPSILON`. For example, using our `A1_GRAMMAR`, the first set of both `<expr>` and `<start>` is `{0,1,2,3,4,5,6,7,8,9}`. The extraction first set for any self-recursive nonterminal is simple enough. One simply has to recursively compute the first set of the first element of its choice expressions. The computation of `first` set for a self-recursive nonterminal is tricky. One has to recursively compute the first set until one is sure that no more terminals can be added to the first set.
Can you implement the `first` set using our `fixpoint()` decorator?
**Solution.** The first set of all terminals is the set containing just themselves. So we initialize that first. Then we update the first set with rules that derive empty strings.
```
def firstset(grammar, nullable):
first = {i: {i} for i in terminals(grammar)}
for k in grammar:
first[k] = {EPSILON} if k in nullable else set()
return firstset_((rules(grammar), first, nullable))[1]
```
Finally, we rely on the `fixpoint` to update the first set with the contents of the current first set until the first set stops changing.
```
def first_expr(expr, first, nullable):
tokens = set()
for token in expr:
tokens |= first[token]
if token not in nullable:
break
return tokens
@fixpoint
def firstset_(arg):
(rules, first, epsilon) = arg
for A, expression in rules:
first[A] |= first_expr(expression, first, epsilon)
return (rules, first, epsilon)
firstset(canonical(A1_GRAMMAR), EPSILON)
```
### Exercise 9: Follow Set of a Nonterminal
The follow set definition is similar to the first set. The follow set of a nonterminal is the set of terminals that can occur just after that nonterminal is used in any derivation. The follow set of the start symbol is `EOF`, and the follow set of any nonterminal is the super set of first sets of all symbols that come after it in any choice expression.
For example, the follow set of `<expr>` in `A1_GRAMMAR` is the set `{EOF, +, -}`.
As in the previous exercise, implement the `followset()` using the `fixpoint()` decorator.
**Solution.** The implementation of `followset()` is similar to `firstset()`. We first initialize the follow set with `EOF`, get the epsilon and first sets, and use the `fixpoint()` decorator to iteratively compute the follow set until nothing changes.
```
EOF = '\0'
def followset(grammar, start):
follow = {i: set() for i in grammar}
follow[start] = {EOF}
epsilon = nullable(grammar)
first = firstset(grammar, epsilon)
return followset_((grammar, epsilon, first, follow))[-1]
```
Given the current follow set, one can update the follow set as follows:
```
@fixpoint
def followset_(arg):
grammar, epsilon, first, follow = arg
for A, expression in rules(grammar):
f_B = follow[A]
for t in reversed(expression):
if t in grammar:
follow[t] |= f_B
f_B = f_B | first[t] if t in epsilon else (first[t] - {EPSILON})
return (grammar, epsilon, first, follow)
followset(canonical(A1_GRAMMAR), START_SYMBOL)
```
### Exercise 10: A LL(1) Parser
As we mentioned previously, there exist other kinds of parsers that operate left-to-right with right most derivation (*LR(k)*) or left-to-right with left most derivation (*LL(k)*) with _k_ signifying the amount of lookahead the parser is permitted to use.
What should one do with the lookahead? That lookahead can be used to determine which rule to apply. In the case of an *LL(1)* parser, the rule to apply is determined by looking at the _first_ set of the different rules. We previously implemented `first_expr()` that takes a an expression, the set of `nullables`, and computes the first set of that rule.
If a rule can derive an empty set, then that rule may also be applicable if of sees the `follow()` set of the corresponding nonterminal.
#### Part 1: A LL(1) Parsing Table
The first part of this exercise is to implement the _parse table_ that describes what action to take for an *LL(1)* parser on seeing a terminal symbol on lookahead. The table should be in the form of a _dictionary_ such that the keys represent the nonterminal symbol, and the value should contain another dictionary with keys as terminal symbols and the particular rule to continue parsing as the value.
Let us illustrate this table with an example. The `parse_table()` method populates a `self.table` data structure that should conform to the following requirements:
```
class LL1Parser(Parser):
def parse_table(self):
self.my_rules = rules(self.cgrammar)
self.table = ... # fill in here to produce
def rules(self):
for i, rule in enumerate(self.my_rules):
print(i, rule)
def show_table(self):
ts = list(sorted(terminals(self.cgrammar)))
print('Rule Name\t| %s' % ' | '.join(t for t in ts))
for k in self.table:
pr = self.table[k]
actions = list(str(pr[t]) if t in pr else ' ' for t in ts)
print('%s \t| %s' % (k, ' | '.join(actions)))
```
On invocation of `LL1Parser(A2_GRAMMAR).show_table()`
It should result in the following table:
```
for i, r in enumerate(rules(canonical(A2_GRAMMAR))):
print("%d\t %s := %s" % (i, r[0], r[1]))
```
|Rule Name || + | - | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9|
|-----------||---|---|---|---|---|---|---|---|---|---|---|--|
|start || | | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0|
|expr || | | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1|
|expr_ || 2 | 3 | | | | | | | | | | |
|integer || | | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5|
|integer_ || 7 | 7 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6|
|digit || | | 8 | 9 |10 |11 |12 |13 |14 |15 |16 |17|
**Solution.** We define `predict()` as we explained before. Then we use the predicted rules to populate the parse table.
```
class LL1Parser(LL1Parser):
def predict(self, rulepair, first, follow, epsilon):
A, rule = rulepair
rf = first_expr(rule, first, epsilon)
if nullable_expr(rule, epsilon):
rf |= follow[A]
return rf
def parse_table(self):
self.my_rules = rules(self.cgrammar)
epsilon = nullable(self.cgrammar)
first = firstset(self.cgrammar, epsilon)
# inefficient, can combine the three.
follow = followset(self.cgrammar, self.start_symbol())
ptable = [(i, self.predict(rule, first, follow, epsilon))
for i, rule in enumerate(self.my_rules)]
parse_tbl = {k: {} for k in self.cgrammar}
for i, pvals in ptable:
(k, expr) = self.my_rules[i]
parse_tbl[k].update({v: i for v in pvals})
self.table = parse_tbl
ll1parser = LL1Parser(A2_GRAMMAR)
ll1parser.parse_table()
ll1parser.show_table()
```
#### Part 2: The Parser
Once we have the parse table, implementing the parser is as follows: Consider the first item from the sequence of tokens to parse, and seed the stack with the start symbol.
While the stack is not empty, extract the first symbol from the stack, and if the symbol is a terminal, verify that the symbol matches the item from the input stream. If the symbol is a nonterminal, use the symbol and input item to lookup the next rule from the parse table. Insert the rule thus found to the top of the stack. Keep track of the expressions being parsed to build up the parse table.
Use the parse table defined previously to implement the complete LL(1) parser.
**Solution.** Here is the complete parser:
```
class LL1Parser(LL1Parser):
def parse_helper(self, stack, inplst):
inp, *inplst = inplst
exprs = []
while stack:
val, *stack = stack
if isinstance(val, tuple):
exprs.append(val)
elif val not in self.cgrammar: # terminal
assert val == inp
exprs.append(val)
inp, *inplst = inplst or [None]
else:
if inp is not None:
i = self.table[val][inp]
_, rhs = self.my_rules[i]
stack = rhs + [(val, len(rhs))] + stack
return self.linear_to_tree(exprs)
def parse(self, inp):
self.parse_table()
k, _ = self.my_rules[0]
stack = [k]
return self.parse_helper(stack, inp)
def linear_to_tree(self, arr):
stack = []
while arr:
elt = arr.pop(0)
if not isinstance(elt, tuple):
stack.append((elt, []))
else:
# get the last n
sym, n = elt
elts = stack[-n:] if n > 0 else []
stack = stack[0:len(stack) - n]
stack.append((sym, elts))
assert len(stack) == 1
return stack[0]
ll1parser = LL1Parser(A2_GRAMMAR)
tree = ll1parser.parse('1+2')
display_tree(tree)
```
| github_jupyter |
# 100 numpy exercises
This is a collection of exercises that have been collected in the numpy mailing list, on stack overflow
and in the numpy documentation. The goal of this collection is to offer a quick reference for both old
and new users but also to provide a set of exercises for those who teach.
If you find an error or think you've a better way to solve some of them, feel
free to open an issue at <https://github.com/rougier/numpy-100>.
File automatically generated. See the documentation to update questions/answers/hints programmatically.
Run the `initialize.py` module, then for each question you can query the
answer or an hint with `hint(n)` or `answer(n)` for `n` question number.
```
import initialise as ini
```
#### 1. Import the numpy package under the name `np` (★☆☆)
```
import numpy as np
```
#### 2. Print the numpy version and the configuration (★☆☆)
```
np.__version__
```
#### 3. Create a null vector of size 10 (★☆☆)
```
np.zeros(10)
```
#### 4. How to find the memory size of any array (★☆☆)
```
random_array = np.array(45)
random_array.nbytes
```
#### 5. How to get the documentation of the numpy add function from the command line? (★☆☆)
```
np.info('add')
```
#### 6. Create a null vector of size 10 but the fifth value which is 1 (★☆☆)
```
x = np.zeros(10)
x[4] = 1
```
#### 7. Create a vector with values ranging from 10 to 49 (★☆☆)
```
a = np.array(range(10,50))
```
#### 8. Reverse a vector (first element becomes last) (★☆☆)
```
a[::-1]
```
#### 9. Create a 3x3 matrix with values ranging from 0 to 8 (★☆☆)
```
np.reshape(np.array(range(0,9)), (3, 3))
```
#### 10. Find indices of non-zero elements from [1,2,0,0,4,0] (★☆☆)
```
a = [1,2,0,0,4,0]
np.nonzero(a)
```
#### 11. Create a 3x3 identity matrix (★☆☆)
```
np.eye(3)
```
#### 12. Create a 3x3x3 array with random values (★☆☆)
```
from numpy import random
random.randint(10, size=(3,3))
```
#### 13. Create a 10x10 array with random values and find the minimum and maximum values (★☆☆)
```
from numpy import random
x = random.randint(100, size=(10,10))
x, np.amax(x), np.amin(x)
```
#### 14. Create a random vector of size 30 and find the mean value (★☆☆)
```
from numpy import random
np.mean(random.randint(10, size=(30)))
```
#### 15. Create a 2d array with 1 on the border and 0 inside (★☆☆)
```
a = np.ones((4,4))
a[1: -1, 1:-1] = 0
a
```
#### 16. How to add a border (filled with 0's) around an existing array? (★☆☆)
```
a = np.ones((3,3))
np.pad(a, 1)
```
#### 17. What is the result of the following expression? (★☆☆)
```python
0 * np.nan
np.nan == np.nan
np.inf > np.nan
np.nan - np.nan
np.nan in set([np.nan])
0.3 == 3 * 0.1
```
#### 18. Create a 5x5 matrix with values 1,2,3,4 just below the diagonal (★☆☆)
```
x = random.randint(5, size=(5, 5))
np.diag(x) = np.array([1, 2, 3, 4, 5])
```
#### 19. Create a 8x8 matrix and fill it with a checkerboard pattern (★☆☆)
#### 20. Consider a (6,7,8) shape array, what is the index (x,y,z) of the 100th element?
#### 21. Create a checkerboard 8x8 matrix using the tile function (★☆☆)
#### 22. Normalize a 5x5 random matrix (★☆☆)
#### 23. Create a custom dtype that describes a color as four unsigned bytes (RGBA) (★☆☆)
#### 24. Multiply a 5x3 matrix by a 3x2 matrix (real matrix product) (★☆☆)
#### 25. Given a 1D array, negate all elements which are between 3 and 8, in place. (★☆☆)
#### 26. What is the output of the following script? (★☆☆)
```python
# Author: Jake VanderPlas
print(sum(range(5),-1))
from numpy import *
print(sum(range(5),-1))
```
#### 27. Consider an integer vector Z, which of these expressions are legal? (★☆☆)
```python
Z**Z
2 << Z >> 2
Z <- Z
1j*Z
Z/1/1
Z<Z>Z
```
#### 28. What are the result of the following expressions?
```python
np.array(0) / np.array(0)
np.array(0) // np.array(0)
np.array([np.nan]).astype(int).astype(float)
```
#### 29. How to round away from zero a float array ? (★☆☆)
#### 30. How to find common values between two arrays? (★☆☆)
#### 31. How to ignore all numpy warnings (not recommended)? (★☆☆)
#### 32. Is the following expressions true? (★☆☆)
```python
np.sqrt(-1) == np.emath.sqrt(-1)
```
#### 33. How to get the dates of yesterday, today and tomorrow? (★☆☆)
#### 34. How to get all the dates corresponding to the month of July 2016? (★★☆)
#### 35. How to compute ((A+B)*(-A/2)) in place (without copy)? (★★☆)
#### 36. Extract the integer part of a random array of positive numbers using 4 different methods (★★☆)
#### 37. Create a 5x5 matrix with row values ranging from 0 to 4 (★★☆)
#### 38. Consider a generator function that generates 10 integers and use it to build an array (★☆☆)
#### 39. Create a vector of size 10 with values ranging from 0 to 1, both excluded (★★☆)
#### 40. Create a random vector of size 10 and sort it (★★☆)
#### 41. How to sum a small array faster than np.sum? (★★☆)
#### 42. Consider two random array A and B, check if they are equal (★★☆)
#### 43. Make an array immutable (read-only) (★★☆)
#### 44. Consider a random 10x2 matrix representing cartesian coordinates, convert them to polar coordinates (★★☆)
#### 45. Create random vector of size 10 and replace the maximum value by 0 (★★☆)
#### 46. Create a structured array with `x` and `y` coordinates covering the [0,1]x[0,1] area (★★☆)
#### 47. Given two arrays, X and Y, construct the Cauchy matrix C (Cij =1/(xi - yj))
#### 48. Print the minimum and maximum representable value for each numpy scalar type (★★☆)
#### 49. How to print all the values of an array? (★★☆)
#### 50. How to find the closest value (to a given scalar) in a vector? (★★☆)
#### 51. Create a structured array representing a position (x,y) and a color (r,g,b) (★★☆)
#### 52. Consider a random vector with shape (100,2) representing coordinates, find point by point distances (★★☆)
#### 53. How to convert a float (32 bits) array into an integer (32 bits) in place?
#### 54. How to read the following file? (★★☆)
```
1, 2, 3, 4, 5
6, , , 7, 8
, , 9,10,11
```
#### 55. What is the equivalent of enumerate for numpy arrays? (★★☆)
#### 56. Generate a generic 2D Gaussian-like array (★★☆)
#### 57. How to randomly place p elements in a 2D array? (★★☆)
#### 58. Subtract the mean of each row of a matrix (★★☆)
#### 59. How to sort an array by the nth column? (★★☆)
#### 60. How to tell if a given 2D array has null columns? (★★☆)
#### 61. Find the nearest value from a given value in an array (★★☆)
#### 62. Considering two arrays with shape (1,3) and (3,1), how to compute their sum using an iterator? (★★☆)
#### 63. Create an array class that has a name attribute (★★☆)
#### 64. Consider a given vector, how to add 1 to each element indexed by a second vector (be careful with repeated indices)? (★★★)
#### 65. How to accumulate elements of a vector (X) to an array (F) based on an index list (I)? (★★★)
#### 66. Considering a (w,h,3) image of (dtype=ubyte), compute the number of unique colors (★★☆)
#### 67. Considering a four dimensions array, how to get sum over the last two axis at once? (★★★)
#### 68. Considering a one-dimensional vector D, how to compute means of subsets of D using a vector S of same size describing subset indices? (★★★)
#### 69. How to get the diagonal of a dot product? (★★★)
#### 70. Consider the vector [1, 2, 3, 4, 5], how to build a new vector with 3 consecutive zeros interleaved between each value? (★★★)
#### 71. Consider an array of dimension (5,5,3), how to mulitply it by an array with dimensions (5,5)? (★★★)
#### 72. How to swap two rows of an array? (★★★)
#### 73. Consider a set of 10 triplets describing 10 triangles (with shared vertices), find the set of unique line segments composing all the triangles (★★★)
#### 74. Given a sorted array C that corresponds to a bincount, how to produce an array A such that np.bincount(A) == C? (★★★)
#### 75. How to compute averages using a sliding window over an array? (★★★)
#### 76. Consider a one-dimensional array Z, build a two-dimensional array whose first row is (Z[0],Z[1],Z[2]) and each subsequent row is shifted by 1 (last row should be (Z[-3],Z[-2],Z[-1]) (★★★)
#### 77. How to negate a boolean, or to change the sign of a float inplace? (★★★)
#### 78. Consider 2 sets of points P0,P1 describing lines (2d) and a point p, how to compute distance from p to each line i (P0[i],P1[i])? (★★★)
#### 79. Consider 2 sets of points P0,P1 describing lines (2d) and a set of points P, how to compute distance from each point j (P[j]) to each line i (P0[i],P1[i])? (★★★)
#### 80. Consider an arbitrary array, write a function that extract a subpart with a fixed shape and centered on a given element (pad with a `fill` value when necessary) (★★★)
#### 81. Consider an array Z = [1,2,3,4,5,6,7,8,9,10,11,12,13,14], how to generate an array R = [[1,2,3,4], [2,3,4,5], [3,4,5,6], ..., [11,12,13,14]]? (★★★)
#### 82. Compute a matrix rank (★★★)
#### 83. How to find the most frequent value in an array?
#### 84. Extract all the contiguous 3x3 blocks from a random 10x10 matrix (★★★)
#### 85. Create a 2D array subclass such that Z[i,j] == Z[j,i] (★★★)
#### 86. Consider a set of p matrices wich shape (n,n) and a set of p vectors with shape (n,1). How to compute the sum of of the p matrix products at once? (result has shape (n,1)) (★★★)
#### 87. Consider a 16x16 array, how to get the block-sum (block size is 4x4)? (★★★)
#### 88. How to implement the Game of Life using numpy arrays? (★★★)
#### 89. How to get the n largest values of an array (★★★)
#### 90. Given an arbitrary number of vectors, build the cartesian product (every combinations of every item) (★★★)
#### 91. How to create a record array from a regular array? (★★★)
#### 92. Consider a large vector Z, compute Z to the power of 3 using 3 different methods (★★★)
#### 93. Consider two arrays A and B of shape (8,3) and (2,2). How to find rows of A that contain elements of each row of B regardless of the order of the elements in B? (★★★)
#### 94. Considering a 10x3 matrix, extract rows with unequal values (e.g. [2,2,3]) (★★★)
#### 95. Convert a vector of ints into a matrix binary representation (★★★)
#### 96. Given a two dimensional array, how to extract unique rows? (★★★)
#### 97. Considering 2 vectors A & B, write the einsum equivalent of inner, outer, sum, and mul function (★★★)
#### 98. Considering a path described by two vectors (X,Y), how to sample it using equidistant samples (★★★)?
#### 99. Given an integer n and a 2D array X, select from X the rows which can be interpreted as draws from a multinomial distribution with n degrees, i.e., the rows which only contain integers and which sum to n. (★★★)
#### 100. Compute bootstrapped 95% confidence intervals for the mean of a 1D array X (i.e., resample the elements of an array with replacement N times, compute the mean of each sample, and then compute percentiles over the means). (★★★)
| github_jupyter |
# Predicting Boston Housing Prices
## Using XGBoost in SageMaker (Batch Transform)
_Deep Learning Nanodegree Program | Deployment_
---
As an introduction to using SageMaker's Low Level Python API we will look at a relatively simple problem. Namely, we will use the [Boston Housing Dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html) to predict the median value of a home in the area of Boston Mass.
The documentation reference for the API used in this notebook is the [SageMaker Developer's Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/)
## General Outline
Typically, when using a notebook instance with SageMaker, you will proceed through the following steps. Of course, not every step will need to be done with each project. Also, there is quite a lot of room for variation in many of the steps, as you will see throughout these lessons.
1. Download or otherwise retrieve the data.
2. Process / Prepare the data.
3. Upload the processed data to S3.
4. Train a chosen model.
5. Test the trained model (typically using a batch transform job).
6. Deploy the trained model.
7. Use the deployed model.
In this notebook we will only be covering steps 1 through 5 as we just want to get a feel for using SageMaker. In later notebooks we will talk about deploying a trained model in much more detail.
```
# Make sure that we use SageMaker 1.x
!pip install sagemaker==1.72.0
```
## Step 0: Setting up the notebook
We begin by setting up all of the necessary bits required to run our notebook. To start that means loading all of the Python modules we will need.
```
%matplotlib inline
import os
import time
from time import gmtime, strftime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_boston
import sklearn.model_selection
```
In addition to the modules above, we need to import the various bits of SageMaker that we will be using.
```
import sagemaker
from sagemaker import get_execution_role
from sagemaker.amazon.amazon_estimator import get_image_uri
# This is an object that represents the SageMaker session that we are currently operating in. This
# object contains some useful information that we will need to access later such as our region.
session = sagemaker.Session()
# This is an object that represents the IAM role that we are currently assigned. When we construct
# and launch the training job later we will need to tell it what IAM role it should have. Since our
# use case is relatively simple we will simply assign the training job the role we currently have.
role = get_execution_role()
```
## Step 1: Downloading the data
Fortunately, this dataset can be retrieved using sklearn and so this step is relatively straightforward.
```
boston = load_boston()
```
## Step 2: Preparing and splitting the data
Given that this is clean tabular data, we don't need to do any processing. However, we do need to split the rows in the dataset up into train, test and validation sets.
```
# First we package up the input data and the target variable (the median value) as pandas dataframes. This
# will make saving the data to a file a little easier later on.
X_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)
Y_bos_pd = pd.DataFrame(boston.target)
# We split the dataset into 2/3 training and 1/3 testing sets.
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)
# Then we split the training set further into 2/3 training and 1/3 validation sets.
X_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)
```
## Step 3: Uploading the data files to S3
When a training job is constructed using SageMaker, a container is executed which performs the training operation. This container is given access to data that is stored in S3. This means that we need to upload the data we want to use for training to S3. In addition, when we perform a batch transform job, SageMaker expects the input data to be stored on S3. We can use the SageMaker API to do this and hide some of the details.
### Save the data locally
First we need to create the test, train and validation csv files which we will then upload to S3.
```
# This is our local data directory. We need to make sure that it exists.
data_dir = '../data/boston'
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# We use pandas to save our test, train and validation data to csv files. Note that we make sure not to include header
# information or an index as this is required by the built in algorithms provided by Amazon. Also, for the train and
# validation data, it is assumed that the first entry in each row is the target variable.
X_test.to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)
pd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)
pd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)
```
### Upload to S3
Since we are currently running inside of a SageMaker session, we can use the object which represents this session to upload our data to the 'default' S3 bucket. Note that it is good practice to provide a custom prefix (essentially an S3 folder) to make sure that you don't accidentally interfere with data uploaded from some other notebook or project.
```
prefix = 'boston-xgboost-LL'
test_location = session.upload_data(os.path.join(data_dir, 'test.csv'), key_prefix=prefix)
val_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)
train_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)
```
## Step 4: Train and construct the XGBoost model
Now that we have the training and validation data uploaded to S3, we can construct a training job for our XGBoost model and build the model itself.
### Set up the training job
First, we will set up and execute a training job for our model. To do this we need to specify some information that SageMaker will use to set up and properly execute the computation. For additional documentation on constructing a training job, see the [CreateTrainingJob API](https://docs.aws.amazon.com/sagemaker/latest/dg/API_CreateTrainingJob.html) reference.
```
# We will need to know the name of the container that we want to use for training. SageMaker provides
# a nice utility method to construct this for us.
container = get_image_uri(session.boto_region_name, 'xgboost')
# We now specify the parameters we wish to use for our training job
training_params = {}
# We need to specify the permissions that this training job will have. For our purposes we can use
# the same permissions that our current SageMaker session has.
training_params['RoleArn'] = role
# Here we describe the algorithm we wish to use. The most important part is the container which
# contains the training code.
training_params['AlgorithmSpecification'] = {
"TrainingImage": container,
"TrainingInputMode": "File"
}
# We also need to say where we would like the resulting model artifacts stored.
training_params['OutputDataConfig'] = {
"S3OutputPath": "s3://" + session.default_bucket() + "/" + prefix + "/output"
}
# We also need to set some parameters for the training job itself. Namely we need to describe what sort of
# compute instance we wish to use along with a stopping condition to handle the case that there is
# some sort of error and the training script doesn't terminate.
training_params['ResourceConfig'] = {
"InstanceCount": 1,
"InstanceType": "ml.m4.xlarge",
"VolumeSizeInGB": 5
}
training_params['StoppingCondition'] = {
"MaxRuntimeInSeconds": 86400
}
# Next we set the algorithm specific hyperparameters. You may wish to change these to see what effect
# there is on the resulting model.
training_params['HyperParameters'] = {
"max_depth": "5",
"eta": "0.2",
"gamma": "4",
"min_child_weight": "6",
"subsample": "0.8",
"objective": "reg:linear",
"early_stopping_rounds": "10",
"num_round": "200"
}
# Now we need to tell SageMaker where the data should be retrieved from.
training_params['InputDataConfig'] = [
{
"ChannelName": "train",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": train_location,
"S3DataDistributionType": "FullyReplicated"
}
},
"ContentType": "csv",
"CompressionType": "None"
},
{
"ChannelName": "validation",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": val_location,
"S3DataDistributionType": "FullyReplicated"
}
},
"ContentType": "csv",
"CompressionType": "None"
}
]
```
### Execute the training job
Now that we've built the dictionary object containing the training job parameters, we can ask SageMaker to execute the job.
```
# First we need to choose a training job name. This is useful for if we want to recall information about our
# training job at a later date. Note that SageMaker requires a training job name and that the name needs to
# be unique, which we accomplish by appending the current timestamp.
training_job_name = "boston-xgboost-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
training_params['TrainingJobName'] = training_job_name
# And now we ask SageMaker to create (and execute) the training job
training_job = session.sagemaker_client.create_training_job(**training_params)
```
The training job has now been created by SageMaker and is currently running. Since we need the output of the training job, we may wish to wait until it has finished. We can do so by asking SageMaker to output the logs generated by the training job and continue doing so until the training job terminates.
```
session.logs_for_job(training_job_name, wait=True)
```
### Build the model
Now that the training job has completed, we have some model artifacts which we can use to build a model. Note that here we mean SageMaker's definition of a model, which is a collection of information about a specific algorithm along with the artifacts which result from a training job.
```
# We begin by asking SageMaker to describe for us the results of the training job. The data structure
# returned contains a lot more information than we currently need, try checking it out yourself in
# more detail.
training_job_info = session.sagemaker_client.describe_training_job(TrainingJobName=training_job_name)
model_artifacts = training_job_info['ModelArtifacts']['S3ModelArtifacts']
# Just like when we created a training job, the model name must be unique
model_name = training_job_name + "-model"
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the xgboost container that we used for training
# can also be used for inference.
primary_container = {
"Image": container,
"ModelDataUrl": model_artifacts
}
# And lastly we construct the SageMaker model
model_info = session.sagemaker_client.create_model(
ModelName = model_name,
ExecutionRoleArn = role,
PrimaryContainer = primary_container)
```
## Step 5: Testing the model
Now that we have fit our model to the training data, using the validation data to avoid overfitting, we can test our model. To do this we will make use of SageMaker's Batch Transform functionality. In other words, we need to set up and execute a batch transform job, similar to the way that we constructed the training job earlier.
### Set up the batch transform job
Just like when we were training our model, we first need to provide some information in the form of a data structure that describes the batch transform job which we wish to execute.
We will only be using some of the options available here but to see some of the additional options please see the SageMaker documentation for [creating a batch transform job](https://docs.aws.amazon.com/sagemaker/latest/dg/API_CreateTransformJob.html).
```
# Just like in each of the previous steps, we need to make sure to name our job and the name should be unique.
transform_job_name = 'boston-xgboost-batch-transform-' + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# Now we construct the data structure which will describe the batch transform job.
transform_request = \
{
"TransformJobName": transform_job_name,
# This is the name of the model that we created earlier.
"ModelName": model_name,
# This describes how many compute instances should be used at once. If you happen to be doing a very large
# batch transform job it may be worth running multiple compute instances at once.
"MaxConcurrentTransforms": 1,
# This says how big each individual request sent to the model should be, at most. One of the things that
# SageMaker does in the background is to split our data up into chunks so that each chunks stays under
# this size limit.
"MaxPayloadInMB": 6,
# Sometimes we may want to send only a single sample to our endpoint at a time, however in this case each of
# the chunks that we send should contain multiple samples of our input data.
"BatchStrategy": "MultiRecord",
# This next object describes where the output data should be stored. Some of the more advanced options which
# we don't cover here also describe how SageMaker should collect output from various batches.
"TransformOutput": {
"S3OutputPath": "s3://{}/{}/batch-bransform/".format(session.default_bucket(),prefix)
},
# Here we describe our input data. Of course, we need to tell SageMaker where on S3 our input data is stored, in
# addition we need to detail the characteristics of our input data. In particular, since SageMaker may need to
# split our data up into chunks, it needs to know how the individual samples in our data file appear. In our
# case each line is its own sample and so we set the split type to 'line'. We also need to tell SageMaker what
# type of data is being sent, in this case csv, so that it can properly serialize the data.
"TransformInput": {
"ContentType": "text/csv",
"SplitType": "Line",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": test_location,
}
}
},
# And lastly we tell SageMaker what sort of compute instance we would like it to use.
"TransformResources": {
"InstanceType": "ml.m4.xlarge",
"InstanceCount": 1
}
}
```
### Execute the batch transform job
Now that we have created the request data structure, it is time to ask SageMaker to set up and run our batch transform job. Just like in the previous steps, SageMaker performs these tasks in the background so that if we want to wait for the transform job to terminate (and ensure the job is progressing) we can ask SageMaker to wait of the transform job to complete.
```
transform_response = session.sagemaker_client.create_transform_job(**transform_request)
transform_desc = session.wait_for_transform_job(transform_job_name)
```
### Analyze the results
Now that the transform job has completed, the results are stored on S3 as we requested. Since we'd like to do a bit of analysis in the notebook we can use some notebook magic to copy the resulting output from S3 and save it locally.
```
transform_output = "s3://{}/{}/batch-bransform/".format(session.default_bucket(),prefix)
!aws s3 cp --recursive $transform_output $data_dir
```
To see how well our model works we can create a simple scatter plot between the predicted and actual values. If the model was completely accurate the resulting scatter plot would look like the line $x=y$. As we can see, our model seems to have done okay but there is room for improvement.
```
Y_pred = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)
plt.scatter(Y_test, Y_pred)
plt.xlabel("Median Price")
plt.ylabel("Predicted Price")
plt.title("Median Price vs Predicted Price")
```
## Optional: Clean up
The default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.
```
# First we will remove all of the files contained in the data_dir directory
!rm $data_dir/*
# And then we delete the directory itself
!rmdir $data_dir
```
| github_jupyter |
# Problema do negócio
A empresa deseja uma análise dos preços dos produtos das lojas concorrentes para precificar melhor o próprio produto no mercado, neste caso, calças.
### Saída (Produto final)
1. Descobrir a reposta para a pergunta calculando a mediana dos preços dos concorrentes
2. Formato da entrega: tabela ou gráfico
3. Aplicação de entrega: Streamlit
### Processo
1. Realizar o calculo da mediana sobre o produto, tipo e cor
2. Gerar gráfico de barraas com a mediana dos preços dos produtos, por tipo e cr dos últimos 10 dias
3. Tabela com: id | product_name | product_type | product_color | product_price
4. Definiçãao do schema: coluna e seu tipo
5. Infraestrutura de armazenamento (SQLite)
6. Design do ETL
7. Planejamento de agendamento dos scripts (dependencias entre os scripts)
8. Fazer as visualizações
9. Entrega do produto final
### Entrada (Fonte de dados)
1. Fonte de dados: site da H&M e Macy's (lojas de e-commerce)
2. Ferramentas: Python 3.8.0, Biblioteca de Webscraping (BeautifulSoup e Selenium), Jupyter Notebook, Vs Code, Scheduler e Streamlit
# Métricas de um E-commerce
## Crescimento
1. Porcentagem do Market Share
1. Quantidade de clientes novos
## Faturamento
1. Quantidade de vendas
2. Ticket médio *
3. LTV
4. Recência média
5. Basket Size
6. Markup Médio *
## Custo
1. CAC (Aquisição de clientes)
2. Desconto médio
3. Custo de produção *
4. Taxa de devolução
5. Custos fixos (folha de pagamento, escritório, ferramentas)
6. Impostos
# Extração dos dados em HTML
## Beautiful Soup
```
import requests
import pandas as pd
import numpy as np
from bs4 import BeautifulSoup
from datetime import datetime
html_doc = """
<html><head><title>The Dormouse's story</title></head>
<body>
<p class="title"><b>The Dormouse's story</b></p>
<p class="story">Once upon a time there were three little sisters; and their names were
<a href="http://example.com/elsie" class="sister" id="link1">Elsie</a>,
<a href="http://example.com/lacie" class="sister" id="link2">Lacie</a> and
<a href="http://example.com/tillie" class="sister" id="link3">Tillie</a>;
and they lived at the bottom of a well.</p>
<p class="story">...</p>
"""
soup = BeautifulSoup(html_doc, 'html.parser')
soup
print(soup.body.p)
soup.find_all('p')[1] #vai buscar todos os paragrafos cuja a classe é title
#busco dentre todos os "sisters", apenas o que tem id=link1 = Elsie
soup.find_all('a', id='link1')
#busco dentre todos os "sisters", apenas o que tem id=link1 = Elsie e EXTRAIR APENAS O NOME
soup.find_all('a', id='link1')[0].get_text()
# ou [0].string
url= 'https://www2.hm.com/en_us/men/products/jeans.html'
headers={'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36'}
page = requests.get(url, headers=headers) #requisição provinda de um navegador para o site
soup = BeautifulSoup(page.text, 'html.parser')
soup #cria o objeto que contem todo o html da pagina
#busca todos os objetos da lista não ordenanada 'ul' product listing small = vitrine de calças
products = soup.find('ul', class_='products-listing small')
products
#busca apenas um produto/item da vitrine
products_list = soup.find_all('article', class_="hm-product-item")
products_list
#Extraindo com um loop, todos os codigos dos items da vitrine
product_id = [p.get('data-articlecode') for p in products_list]
product_id
#extraindo as categorias de cada produto
product_category = [p.get('data-category') for p in products_list]
product_category
product_list = products.find_all('a', class_='link')
product_name=[p.get_text() for p in product_list]
product_name
product_list = products.find_all('span', class_='price regular')
product_price=[p.get_text() for p in product_list]
product_price
data = pd.DataFrame( [product_id, product_category, product_name, product_price] ).T
data.columns=['product_id', 'product_category','product_name', 'product_price']
data['scrapy_datetime'] = datetime.now().strftime( '%Y-%m-%d %H:%M:%S' )
data.head()
#Buscar os produtos para as proximas paginas. Paginação serve para melhorar a navegação do usuário no site
url= 'https://www2.hm.com/en_us/men/products/jeans.html'
headers={'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36'}
page = requests.get(url, headers=headers) #requisição provinda de um navegador para o site
soup = BeautifulSoup(page.text, 'html.parser')
#extraindo todos os itens de todas as páginas , no total são 94 itens
total_item = soup.find_all('h2', class_='load-more-heading')[0].get('data-total')
total_item
#3 paginas de 36 items
page_number = int(total_item)/36
page_number
url02 = url + '?page-size='+str(int(page_number*36))
url02 #variavel com o link de toda a vitrine de produtos do tipo calça
```
### Buscar os detalhes de cada produto em suas paginas individuais: para extrair cor, tipo do tecido e codigo do produto
```
#Realizando a request da API
url= 'https://www2.hm.com/en_us/productpage.0985159001.html'
headers={'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36'}
page = requests.get(url, headers=headers) #requisição provinda de um navegador para o site
soup = BeautifulSoup(page.text, 'html.parser')
#Extraindo as cores disponiveis para apenas uma calça da vitrine
product_list = soup.find_all('a', role = 'radio') #tem que descobrir a classe em comum
color_name = [p.get('data-color') for p in product_list]
color_name
product_id = [p.get('data-articlecode') for p in product_list]
product_id
df_color = pd.DataFrame([product_id, color_name]).T
df_color.columns = ['product_id', 'color_name']
#gerar style_id + color_id
df_color['style_id'] = df_color['product_id'].apply(lambda x: x[:-3])
df_color['color_id'] = df_color['product_id'].apply(lambda x: x[-3:])
df_color
#Extraindo o tipo de tecido das calças
product_composition_list = soup.find_all('div', class_='pdp-description-list-item')
product_composition = [list(filter(None, p.get_text().split('\n'))) for p in product_composition_list]
product_composition
pd.DataFrame(product_composition).T
#É necessário promover a primeira linha para o titulo das colunas e substituir os valores 'None'
#renomeando o dataframe
df_composition = pd.DataFrame(product_composition).T
df_composition.columns = df_composition.iloc[0]
#deletando a primeira linha
df_composition = df_composition.iloc[1:].fillna(method='ffill') # preenchendo os valores nulos com os valores da linha acima
#gerar as colunas style id + color id para fazer o merge dos dois dataframes
df_composition['style_id'] = df_composition['Art. No.'].apply(lambda x: x[:-3]) #cria a coluna style_id e pega o codigo até os ultimos 3 numeros
df_composition['color_id'] = df_composition['Art. No.'].apply(lambda x: x[-3:])
del df_composition['Size']
df_composition
#explicado In[105]
data_sku = pd.merge(df_color, df_composition[['style_id','Fit','Composition']], how='left', on='style_id')
df_color
#Unidos os dois dataframes, com left join, considerando style_id como parametro de união
#left join = todos os dados que estao na df_composition sao adicionados na df_color
pd.merge(df_color, df_composition[['style_id','Fit','Composition']], how='left', on='style_id')
```
### Codigo único para extração dos detalhes de um produto
```
#Realizando a request da API
url= 'https://www2.hm.com/en_us/productpage.0985159001.html'
headers={'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36'}
page = requests.get(url, headers=headers) #requisição provinda de um navegador para o site
soup = BeautifulSoup(page.text, 'html.parser')
#==================== color_name ===========================
#Extraindo as cores disponiveis para apenas uma calça da vitrine
product_list = soup.find_all('a', role = 'radio') #tem que descobrir a classe em comum
color_name = [p.get('data-color') for p in product_list]
color_name
product_id = [p.get('data-articlecode') for p in product_list]
product_id
df_color = pd.DataFrame([product_id, color_name]).T
df_color.columns = ['product_id', 'color_name']
#gerar style_id + color_id
df_color['style_id'] = df_color['product_id'].apply(lambda x: x[:-3])
df_color['color_id'] = df_color['product_id'].apply(lambda x: x[-3:])
df_color
#==================== composition ===========================
#Extraindo o tipo de tecido das calças
product_composition_list = soup.find_all('div', class_='pdp-description-list-item')
product_composition = [list(filter(None, p.get_text().split('\n'))) for p in product_composition_list]
product_composition
pd.DataFrame(product_composition).T
#É necessário promover a primeira linha para o titulo das colunas e substituir os valores 'None'
#renomeando o dataframe
df_composition = pd.DataFrame(product_composition).T
df_composition.columns = df_composition.iloc[0]
#deletando a primeira linha
df_composition = df_composition.iloc[1:].fillna(method='ffill') # preenchendo os valores nulos com os valores da linha acima
#gerar as colunas style id + color id para fazer o merge dos dois dataframes
df_composition['style_id'] = df_composition['Art. No.'].apply(lambda x: x[:-3]) #cria a coluna style_id e pega o codigo até os ultimos 3 numeros
df_composition['color_id'] = df_composition['Art. No.'].apply(lambda x: x[-3:])
del df_composition['Size']
df_composition
#explicado In[105]
data_sku = pd.merge(df_color, df_composition[['style_id','Fit','Composition']], how='left', on='style_id')
data_sku
```
### Para vários produtos
```
headers={'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36'}
#criando um dataframe vazio
df_details = pd.DataFrame()
aux=[]
cols=['Art. No.','Composition','Fit','More sustainable materials','color_id','style_id','Size']
df_pattern = pd.DataFrame(columns=cols)
for i in range(len(data)):
#Realizando a request da API para percorrer todas as paginas de detalhes dos produtos da vitrine
url = 'https://www2.hm.com/en_us/productpage.' +data.loc[i, 'product_id'] + '.html'
page = requests.get(url, headers=headers) #requisição provinda de um navegador para o site
soup = BeautifulSoup(page.text, 'html.parser')
#==================== color_name ===========================
#Extraindo as cores disponiveis para apenas uma calça da vitrine
product_list = soup.find_all('a', role = 'radio') #tem que descobrir a classe em comum
color_name = [p.get('data-color') for p in product_list]
color_name
product_id = [p.get('data-articlecode') for p in product_list]
product_id
df_color = pd.DataFrame([product_id, color_name]).T
df_color.columns = ['product_id', 'color_name']
#gerar style_id + color_id
df_color['style_id'] = df_color['product_id'].apply(lambda x: x[:-3])
df_color['color_id'] = df_color['product_id'].apply(lambda x: x[-3:])
df_color
#==================== composition ===========================
#Extraindo o tipo de tecido das calças
product_composition_list = soup.find_all('div', class_='pdp-description-list-item')
product_composition = [list(filter(None, p.get_text().split('\n'))) for p in product_composition_list]
product_composition
pd.DataFrame(product_composition).T
#É necessário promover a primeira linha para o titulo das colunas e substituir os valores 'None'
#renomeando o dataframe
df_composition = pd.DataFrame(product_composition).T
df_composition.columns = df_composition.iloc[0]
#deletando a primeira linha
df_composition = df_composition.iloc[1:].fillna(method='ffill') # preenchendo os valores nulos com os valores da linha acima
#garantir a mesma quantidade de colunas
df_composition = pd.concat([df_pattern, df_composition], axis=0)
#gerar as colunas style id + color id para fazer o merge dos dois dataframes
df_composition['style_id'] = df_composition['Art. No.'].apply(lambda x: x[:-3]) #cria a coluna style_id e pega o codigo até os ultimos 3 numeros
df_composition['color_id'] = df_composition['Art. No.'].apply(lambda x: x[-3:])
#buscar todos os detalhes de todos os items (pagina de detalhes) da vitrine
aux = aux + df_composition.columns.tolist()
#explicado In[105]
data_sku = pd.merge(df_color, df_composition[['style_id','Fit','Composition','More sustainable materials','Size']], how='left', on='style_id')
df_details = pd.concat([df_details, data_sku], axis=0)
#União dos produtos da vitrine + detalhes de cada item
data['style_id'] = data['product_id'].apply(lambda x: x[:-3])
data['color_id'] = data['product_id'].apply(lambda x: x[-3:])
data_raw = pd.merge(data, df_details[['style_id','color_name','Fit', 'Composition', 'Size', 'More sustainable materials']],
how='left', on='style_id')
data_raw.head()
```
| github_jupyter |
# Project: Investigate Children Out of School
## Table of Contents
<ul>
<li><a href="#intro">Introduction</a></li>
<li><a href="#wrangling">Data Wrangling</a></li>
<li><a href="#eda">Exploratory Data Analysis</a></li>
<li><a href="#conclusions">Conclusions</a></li>
</ul>
<a id='intro'></a>
## Introduction
> **Key notes**: "Gapminder has collected a lot of information about how people live their lives in different countries, tracked across the years, and on a number of different indicators.
> **Questions to explore**:
><ul>
><li><a href="#q1"> 1. Research Question 1: What is the total numbers of children out of primary school over years, indicate the male and female numbers as well?</a></li>
><li><a href="#q2"> 2. Research Question 2: What is distribution of female children who was out of primary school from 1980 to 1995?</a></li>
><li><a href="#q3"> 3. Research Question 3: What are numbers of children out of school in total, by male and female in China, 1985?</a></li>
><li><a href="#q4"> 4. What are relationship of children out of school of female in China in russian and usa over time? Which has a better trend?</a></li>
><li><a href="#q5"> 5. Research Question 5: What is the overall trend for children out of primary school over the years?</a></li>
```
# Set up import statements for all of the packages that are planed to use;
# Include a 'magic word' so that visualizations are plotted;
# call on dataframe to display the first 5 rows.
import pandas as pd
import numpy as np
import datetime
from statistics import mode
% matplotlib inline
import matplotlib.pyplot as plt
%config InlineBackend.figure_format = 'retina'
import seaborn as sns
sns.set_style('darkgrid')
# Reading an Excel file in python using pandas
# call on dataframe to display the first 5 rows
xl = pd.ExcelFile('Child out of school primary.xlsx')
xl.sheet_names
[u'Data']
df_tot = xl.parse("Data")
df_tot.head()
x2 = pd.ExcelFile('Child out of school primiary female.xlsx')
x2.sheet_names
[u'Data']
df_f = x2.parse("Data")
df_f.head()
x3 = pd.ExcelFile('Child out of school primiary male.xlsx')
x3.sheet_names
[u'Data']
df_m = x3.parse("Data")
df_m.head()
# Check if the three dataframe have the same shape
df_tot.shape, df_m.shape, df_f.shape
# Check if the first columns from the 3 dataframe are exactly the same
assert (df_tot['Children out of school, primary'].tolist() == df_m['Children out of school, primary, male'].tolist()\
== df_f['Children out of school, primary, female'].tolist())
# Merge the 3 dataframe
df1 = df_tot.merge(df_f, how='outer', left_index = True, right_index = True)
df1 = df1.merge(df_m, how='outer', left_index = True, right_index = True)
# Confirm changes
df1.shape
```
<a id='wrangling'></a>
## Data Wrangling
> **Key notes**: In this section of the report, the following work will be done: load the data; check for cleanliness; trim and clean dataset for analysis.
### General Properties
```
# return the datatypes of the columns.
df1.dtypes
# check for duplicates in the data.
sum(df1.duplicated())
# check if any value is NaN in DataFrame and in how many columns
df1.isnull().any().any(), sum(df1.isnull().any())
# Generates descriptive statistics, excluding NaN values.
df1.describe()
```
### Data Cleaning
```
# Locate the columns whose NaN values needs to be treated
col = df1.drop(['Children out of school, primary', 'Children out of school, primary, female'\
, 'Children out of school, primary, male'], axis=1)
# Replace NaN with mean
for c in col:
c_mean = df1[c].mean()
df1[c].fillna(c_mean, inplace = True)
# Confirm changes
df1.isnull().any().any()
# Rename column for simplification
df1.rename(columns = {'Children out of school, primary':'country'}, inplace = True)
# check the new dataframe
df1.head()
```
<a id='eda'></a>
## Exploratory Data Analysis
<a id='q1'></a>
### Research Question 1: What is the total numbers of children out of primary school over years, indicate the male and female numbers as well?
```
# Get the sum for each group
sum_tot = df1.iloc[:, 1:43]
m_tot = df1.iloc[:, 44:86]
f_tot = df1.iloc[:, 87:]
tot = []
for t in sum_tot.columns:
tot.append(sum_tot[t].sum())
m = []
for ma in m_tot.columns:
m.append(m_tot[ma].sum())
f = []
for fa in f_tot.columns:
f.append(f_tot[fa].sum())
# Plot
x = ['total number', 'male number', 'female number']
y = [sum(tot), sum(m), sum(f)]
plt.subplots(figsize=(10,6))
sns.barplot(x,y, alpha = 0.8);
```
<a id='q2'></a>
### Research Question 2: What is distribution of female children who was out of primary school from 1980 to 1995?
```
# Target the year and plot
sum_tot1 = sum_tot.iloc[:, 10:26]
new_col = []
for ele in sum_tot1.columns:
new_col.append(ele.split('_x')[0])
sum_tot1.columns = new_col
plt.figure(figsize=(20,15))
sns.boxplot(data = sum_tot1);
```
<a id='q3'></a>
### Research Question 3: What are numbers of children out of school in total, by male and female in China, 1985?
```
china = df1.copy()
china = china.set_index('country')
tot_chi = china.loc['China', '1985_x']
f_chi = china.loc['China', '1985_y']
m_chi = china.loc['China', '1985']
print('The numbers of children out of school in total, by male and female in China were \
{0:.0f}, {1:.0f} and {2:.0f} in 1985, respectively.'.format(tot_chi, f_chi, m_chi))
```
<a id='q4'></a>
### Research Question 4: What are relationship of children out of school of female in China in russian and usa over time? Which has a better trend?
```
rus_us = df1.iloc[:, 0:42].copy()
new_col1 = []
for ele in rus_us:
new_col1.append(ele.split('_x')[0])
rus_us.columns = new_col1
rus_us = rus_us.set_index('country')
rus_us_df = pd.DataFrame(columns=['USA','Russia'])
rus_us_df['USA'] = rus_us.loc['United States'].values
rus_us_df['Russia'] = rus_us.loc['Russia'].values
sns.lmplot(x = 'USA', y = 'Russia', data = rus_us_df);
sns.boxplot(data=rus_us_df);
rus_us_df['year'] = rus_us.columns
rus_us_df.index = rus_us_df.year
rus_us_df.plot();
plt.ylabel('Numers')
plt.xlabel('Country')
plt.title('Numbers of children out of primary school from 1970 to 2011');
```
> There is a positive correlation between children droped out of primary school in Russia and USA. The estimated linear regression is shown as the blue line, the estimates varies in the light blue shade with 95% confident level. The trend of children out of school in USA is much higher than that of Russia over that past 40 years.
<a id='q5'></a>
### Research Question 5: What is the overall trend for children out of primary school over the years?
```
overall_df = pd.DataFrame(columns=['year','numbers'])
overall_df['year'] = rus_us.columns
n_list =[]
for n in rus_us.columns:
n_list.append(rus_us[n].mean())
overall_df['numbers'] = np.array(n_list)
overall_df.index = overall_df.year
overall_df.plot();
plt.ylabel('Numers')
plt.xlabel('Year')
plt.title('Numbers of children out of primary school from 1970 to 2011');
```
> From the analysis we can conclude that the overall trend of children out of primary school had been descreasing starting between 1970 and 1975 at which point of time the numbers fell down dramatically
<a id='conclusions'></a>
## Conclusions
> In current study, a good amount of profound analysis has been carried out. Prior to each step, deailed instructions was given and interpretions was also provided afterwards. The dataset across 41 years from 1970 to 2011.
> The limitations of current study was that the structure is only 275*42 in shape, thus the analysis would not be much reliable due to small scale samples.
> In addition, the parameters in the dataset is very simple, it only focus on the number of children out of school.
```
from subprocess import call
call(['python', '-m', 'nbconvert', 'Investigate_Children_Out_of_School_20180108.ipynb'])
```
| github_jupyter |
# Apache Kafka Integration + Preprocessing / Interactive Analysis with KSQL
This notebook uses the combination of Python, Apache Kafka, KSQL for Machine Learning infrastructures.
It includes code examples using ksql-python and other widespread components from Python’s machine learning ecosystem, like Numpy, pandas, TensorFlow and Keras.
The use case is fraud detection for credit card payments. We use a test data set from Kaggle as foundation to train an unsupervised autoencoder to detect anomalies and potential fraud in payments. Focus of this example is not just model training, but the whole Machine Learning infrastructure including data ingestion, data preprocessing, model training, model deployment and monitoring. All of this needs to be scalable, reliable and performant.
If you want to learn more about the relation between the Apache Kafka open source ecosystem and Machine Learning, please check out these two blog posts:
- [How to Build and Deploy Scalable Machine Learning in Production with Apache Kafka](https://www.confluent.io/blog/build-deploy-scalable-machine-learning-production-apache-kafka/)
- [Using Apache Kafka to Drive Cutting-Edge Machine Learning](https://www.confluent.io/blog/using-apache-kafka-drive-cutting-edge-machine-learning)
##### This notebook is not meant to be perfect using all coding and ML best practices, but just a simple guide how to build your own notebooks where you can combine Python APIs with Kafka and KSQL
### Start Backend Services (Zookeeper, Kafka, KSQL)
The only server requirement is a local KSQL server running (with Kafka broker ZK node). If you don't have it running, just use Confluent CLI:
```
# Shows correct startup but does not work 100% yet. Better run this command from outside Jupyter if you have any problems (e.g. from Terminal)!
! confluent start ksql-server
```
## Data Integration and Preprocessing with Python and KSQL
First of all, create the Kafka Topic 'creditcardfraud_source' if it does not exist already:
```
! kafka-topics --zookeeper localhost:2181 --create --topic creditcardfraud_source --partitions 3 --replication-factor 1
```
Then load KSQL library and initiate connection to KSQL server:
```
from ksql import KSQLAPI
client = KSQLAPI('http://localhost:8088')
```
Consume source data from Kafka Topic "creditcardfraud_source":
```
client.create_stream(table_name='creditcardfraud_source',
columns_type=['Id bigint', 'Timestamp varchar', 'User varchar', 'Time int', 'V1 double', 'V2 double', 'V3 double', 'V4 double', 'V5 double', 'V6 double', 'V7 double', 'V8 double', 'V9 double', 'V10 double', 'V11 double', 'V12 double', 'V13 double', 'V14 double', 'V15 double', 'V16 double', 'V17 double', 'V18 double', 'V19 double', 'V20 double', 'V21 double', 'V22 double', 'V23 double', 'V24 double', 'V25 double', 'V26 double', 'V27 double', 'V28 double', 'Amount double', 'Class string'],
topic='creditcardfraud_source',
value_format='DELIMITED')
```
Preprocessing:
- Filter columns which are not needed
- Filter messages where column 'class' is empty
- Change data format to Avro for more convenient further processing
```
client.create_stream_as(table_name='creditcardfraud_preprocessed_avro',
select_columns=['Time', 'V1', 'V2', 'V3', 'V4', 'V5', 'V6', 'V7', 'V8', 'V9', 'V10', 'V11', 'V12', 'V13', 'V14', 'V15', 'V16', 'V17', 'V18', 'V19', 'V20', 'V21', 'V22', 'V23', 'V24', 'V25', 'V26', 'V27', 'V28', 'Amount', 'Class'],
src_table='creditcardfraud_source',
conditions='Class IS NOT NULL',
kafka_topic='creditcardfraud_preprocessed_avro',
value_format='AVRO')
```
Take a look at the creates KSQL Streams:
```
client.ksql('show streams')
```
Take a look at the metadata of the KSQL Stream:
```
client.ksql('describe CREDITCARDFRAUD_PREPROCESSED_AVRO')
```
Interactive query statement:
```
query = client.query('SELECT * FROM CREDITCARDFRAUD_PREPROCESSED_AVRO LIMIT 1')
for item in query:
print(item)
```
Produce single test data manually (if you did not connect to a real data stream which produces data continuously), e.g. from terminal:
confluent produce creditcardfraud_source
1,"2018-12- 18T12:00:00Z","Hans",0,-1.3598071336738,-0.0727811733098497,2.53634673796914,1.37815522427443,-0.338320769942518,0.462387777762292,0.239598554061257,0.0986979012610507,0.363786969611213,0.0907941719789316,-0.551599533260813,-0.617800855762348,-0.991389847235408,-0.311169353699879,1.46817697209427,-0.470400525259478,0.207971241929242,0.0257905801985591,0.403992960255733,0.251412098239705,-0.018306777944153,0.277837575558899,-0.110473910188767,0.0669280749146731,0.128539358273528,-0.189114843888824,0.133558376740387,-0.0210530534538215,149.62,"0"
*BE AWARE: The KSQL Python API does a REST call. This only waits a few seconds by default and then throws a timeout exception. You need to get data into the query before the timeout (e.g. by using above command).*
```
# TODO How to embed ' ' in Python ???
# See https://github.com/bryanyang0528/ksql-python/issues/54
# client.ksql('SET \'auto.offset.reset\'=\'earliest\'');
```
### Additional (optional) analysis and preprocessing examples
Some more examples for possible data wrangling and preprocessing with KSQL:
- Anonymization
- Augmentation
- Merge / Join data frames
```
query = client.query('SELECT Id, MASK_LEFT(User, 2) FROM creditcardfraud_source LIMIT 1')
for item in query:
print(item)
query = client.query('SELECT Id, IFNULL(Class, \'-1\') FROM creditcardfraud_source LIMIT 1')
for item in query:
print(item)
```
#### Stream-Table-Join
For the STREAM-TABLE-JOIN, you first need to create a Kafka Topic 'Users' (for the corresponding KSQL TABLE 'Users):
```
! kafka-topics --zookeeper localhost:2181 --create --topic users --partitions 3 --replication-factor 1
```
Then create the KSQL Table:
```
client.create_table(table_name='users',
columns_type=['userid varchar', 'gender varchar', 'regionid varchar'],
topic='users',
key='userid',
value_format='AVRO')
client.ksql("CREATE STREAM creditcardfraud_per_user WITH (VALUE_FORMAT='AVRO', KAFKA_TOPIC='creditcardfraud_per_user') AS SELECT Time, Amount, Class FROM creditcardfraud_source c INNER JOIN USERS u on c.user = u.userid WHERE u.USERID = 1")
```
# Mapping from KSQL to NumPy / pandas for Machine Learning tasks
```
import numpy as np
import pandas as pd
import json
```
The query below command returns a Python generator. It can be printed e.g. by reading its values via next(query) or a for loop.
Due to a current [bug in ksql-python library](https://github.com/bryanyang0528/ksql-python/issues/57), we need to to an additional line of Python code to strip out unnecessary info and change to 2D array
```
query = client.query('SELECT * FROM CREDITCARDFRAUD_PREPROCESSED_AVRO LIMIT 8') # Returns a Python generator object
#items = [item for item in query][:-1] # -1 to remove last record that is a dummy msg for "Limit Reached"
#one_record = json.loads(''.join(items)) # Join two records as one as ksql-python is splitting it into two?
#data = [one_record['row']['columns'][2:-1]] # Strip out unnecessary info and change to 2D array
#df = pd.DataFrame(data=data)
records = [json.loads(r) for r in ''.join(query).strip().replace('\n\n\n\n', '').split('\n')]
data = [r['row']['columns'][2:] for r in records[:-1]]
#data = r['row']['columns'][2] for r in records
df = pd.DataFrame(data=data, columns=['Time', 'V1' , 'V2' , 'V3' , 'V4' , 'V5' , 'V6' , 'V7' , 'V8' , 'V9' , 'V10' , 'V11' , 'V12' , 'V13' , 'V14' , 'V15' , 'V16' , 'V17' , 'V18' , 'V19' , 'V20' , 'V21' , 'V22' , 'V23' , 'V24' , 'V25' , 'V26' , 'V27' , 'V28' , 'Amount' , 'Class'])
df
```
### Generate some test data
As discussed in the step-by-step guide, you have various options. Here we - ironically - read messages from a CSV file. This is for simple demo purposes so that you don't have to set up a real continuous Kafka stream.
In real world or more advanced examples, you should connect to a real Kafka data stream (for instance using the Kafka data generator or Kafka Connect).
Here we just consume a few messages for demo purposes so that they get mapped into a pandas dataframe:
cat /Users/kai.waehner/git-projects/python-jupyter-apache-kafka-ksql-tensorflow-keras/data/creditcard_extended.csv | kafka-console-producer --broker-list localhost:9092 --topic creditcardfraud_source
You need to do this from command line because Jupyter cannot execute this in parallel to above KSQL query.
# Preprocessing with Pandas + Model Training with TensorFlow / Keras
#### BE AWARE: You need enough messages in the pandas data frame to train the model in the below cells (if you just play around with ksql-python and just add a few Kafka events, it is not a sufficient number of rows to continue. You can simply change to df = pd.read_csv("data/creditcard.csv") as shown below in this case to get a bigger data set...
This part only includes the steps required for model training of the Autoencoder with Keras and TensorFlow.
If you want to get a better understanding of the model, take a look at the other notebook [Python Tensorflow Keras Fraud Detection Autoencoder.ipynb](http://localhost:8888/notebooks/Python%20Tensorflow%20Keras%20Fraud%20Detection%20Autoencoder.ipynb) which includes many more details, plots and explanations.
[Kudos to David Ellison](https://www.datascience.com/blog/fraud-detection-with-tensorflow).
[The credit card fraud data set is available at Kaggle](https://www.kaggle.com/mlg-ulb/creditcardfraud/data).
```
# import packages
# matplotlib inline
#import pandas as pd
#import numpy as np
from scipy import stats
import tensorflow as tf
import matplotlib.pyplot as plt
import seaborn as sns
import pickle
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, precision_recall_curve
from sklearn.metrics import recall_score, classification_report, auc, roc_curve
from sklearn.metrics import precision_recall_fscore_support, f1_score
from sklearn.preprocessing import StandardScaler
from pylab import rcParams
from keras.models import Model, load_model
from keras.layers import Input, Dense
from keras.callbacks import ModelCheckpoint, TensorBoard
from keras import regularizers
# Use the dataframe from above (imported and preprocessed with KSQL)
# As alternative directly import from a CSV file ("the normal approach without Kafka and streaming data")
# "data/creditcard_small.csv" is a very small data set (just for quick demo purpose to get a model binary)
# => replace with "data/creditcard.csv" to use a real data set to train a model with good accuracy
#df = pd.read_csv("data/creditcard.csv")
df.head(n=5) #just to check you imported the dataset properly
#set random seed and percentage of test data
RANDOM_SEED = 314 #used to help randomly select the data points
TEST_PCT = 0.2 # 20% of the data
#set up graphic style in this case I am using the color scheme from xkcd.com
rcParams['figure.figsize'] = 14, 8.7 # Golden Mean
LABELS = ["Normal","Fraud"]
#col_list = ["cerulean","scarlet"]# https://xkcd.com/color/rgb/
#sns.set(style='white', font_scale=1.75, palette=sns.xkcd_palette(col_list))
normal_df = [df.Class == 0] #save normal_df observations into a separate df
fraud_df = [df.Class == 1] #do the same for frauds
#data = df.drop(['Time'], axis=1) #if you think the var is unimportant
df_norm = df
df_norm['Time'] = StandardScaler().fit_transform(df_norm['Time'].values.reshape(-1, 1))
df_norm['Amount'] = StandardScaler().fit_transform(df_norm['Amount'].values.reshape(-1, 1))
train_x, test_x = train_test_split(df_norm, test_size=TEST_PCT, random_state=RANDOM_SEED)
train_x = train_x[train_x.Class == 0] #where normal transactions
train_x = train_x.drop(['Class'], axis=1) #drop the class column
test_y = test_x['Class'] #save the class column for the test set
test_x = test_x.drop(['Class'], axis=1) #drop the class column
train_x = train_x.values #transform to ndarray
test_x = test_x.values
```
### My Jupyter Notebook crashed sometimes in the next step 'model training' (probably memory issues):
```
# Reduce number of epochs and batch_size if your Jupyter crashes (due to memory issues)
# nb_epoch = 100
# batch_size = 128
nb_epoch = 5
batch_size = 32
input_dim = train_x.shape[1] #num of columns, 30
encoding_dim = 14
hidden_dim = int(encoding_dim / 2) #i.e. 7
learning_rate = 1e-7
input_layer = Input(shape=(input_dim, ))
encoder = Dense(encoding_dim, activation="tanh", activity_regularizer=regularizers.l1(learning_rate))(input_layer)
encoder = Dense(hidden_dim, activation="relu")(encoder)
decoder = Dense(hidden_dim, activation='tanh')(encoder)
decoder = Dense(input_dim, activation='relu')(decoder)
autoencoder = Model(inputs=input_layer, outputs=decoder)
autoencoder.compile(metrics=['accuracy'],
loss='mean_squared_error',
optimizer='adam')
cp = ModelCheckpoint(filepath="models/autoencoder_fraud.h5",
save_best_only=True,
verbose=0)
tb = TensorBoard(log_dir='./logs',
histogram_freq=0,
write_graph=True,
write_images=True)
history = autoencoder.fit(train_x, train_x,
epochs=nb_epoch,
batch_size=batch_size,
shuffle=True,
validation_data=(test_x, test_x),
verbose=1,
callbacks=[cp, tb]).history
autoencoder = load_model('models/autoencoder_fraud.h5')
test_x_predictions = autoencoder.predict(test_x)
mse = np.mean(np.power(test_x - test_x_predictions, 2), axis=1)
error_df = pd.DataFrame({'Reconstruction_error': mse,
'True_class': test_y})
error_df.describe()
```
The binary 'models/autoencoder_fraud.h5' is the trained model which can then be deployed anywhere to do prediction on new incoming events in real time.
# Model Deployment
This demo focuses on the combination of Python and KSQL for data preprocessing and model training. If you want to understand the relation between Apache Kafka, KSQL and Python-related Machine Learning tools like TensorFlow for model deployment and monitoring, please check out my other Github projects:
Some examples of model deployment in Kafka environments:
- [Analytic models (TensorFlow, Keras, H2O and Deeplearning4j) embedded in Kafka Streams microservices](https://github.com/kaiwaehner/kafka-streams-machine-learning-examples)
- [Anomaly detection of IoT sensor data with a model embedded into a KSQL UDF](https://github.com/kaiwaehner/ksql-udf-deep-learning-mqtt-iot)
- [RPC communication between Kafka Streams application and model server (TensorFlow Serving)](https://github.com/kaiwaehner/tensorflow-serving-java-grpc-kafka-streams)
# Appendix: Pandas analysis with above Fraud Detection Data
```
df = pd.read_csv("data/creditcard.csv")
df.head()
df.shape
df.index
df.columns
df.values
df.describe()
df['Amount']
df[0:3]
df.iloc[1,1]
# Takes a minute or two (big CSV file)...
#df.plot()
```
| github_jupyter |
# Multi-Layer Perceptron, MNIST
---
In this notebook, we will train an MLP to classify images from the [MNIST database](http://yann.lecun.com/exdb/mnist/) hand-written digit database.
The process will be broken down into the following steps:
>1. Load and visualize the data
2. Define a neural network
3. Train the model
4. Evaluate the performance of our trained model on a test dataset!
Before we begin, we have to import the necessary libraries for working with data and PyTorch.
```
# import libraries
import torch
import numpy as np
```
---
## Load and Visualize the [Data](http://pytorch.org/docs/stable/torchvision/datasets.html)
Downloading may take a few moments, and you should see your progress as the data is loading. You may also choose to change the `batch_size` if you want to load more data at a time.
This cell will create DataLoaders for each of our datasets.
```
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 0
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# convert data to torch.FloatTensor
transform = transforms.ToTensor()
# choose the training and test datasets
train_data = datasets.MNIST(root='data', train=True,
download=True, transform=transform)
test_data = datasets.MNIST(root='data', train=False,
download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=train_sampler, num_workers=num_workers)
valid_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
sampler=valid_sampler, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size,
num_workers=num_workers)
```
### Visualize a Batch of Training Data
The first step in a classification task is to take a look at the data, make sure it is loaded in correctly, then make any initial observations about patterns in that data.
```
import matplotlib.pyplot as plt
%matplotlib inline
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(20):
ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
# print out the correct label for each image
# .item() gets the value contained in a Tensor
ax.set_title(str(labels[idx].item()))
```
### View an Image in More Detail
```
img = np.squeeze(images[1])
fig = plt.figure(figsize = (12,12))
ax = fig.add_subplot(111)
ax.imshow(img, cmap='gray')
width, height = img.shape
thresh = img.max()/2.5
for x in range(width):
for y in range(height):
val = round(img[x][y],2) if img[x][y] !=0 else 0
ax.annotate(str(val), xy=(y,x),
horizontalalignment='center',
verticalalignment='center',
color='white' if img[x][y]<thresh else 'black')
```
---
## Define the Network [Architecture](http://pytorch.org/docs/stable/nn.html)
The architecture will be responsible for seeing as input a 784-dim Tensor of pixel values for each image, and producing a Tensor of length 10 (our number of classes) that indicates the class scores for an input image. This particular example uses two hidden layers and dropout to avoid overfitting.
```
import torch.nn as nn
import torch.nn.functional as F
# define the NN architecture
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
# number of hidden nodes in each layer (512)
hidden_1 = 512
hidden_2 = 512
# linear layer (784 -> hidden_1)
self.fc1 = nn.Linear(28 * 28, hidden_1)
# linear layer (n_hidden -> hidden_2)
self.fc2 = nn.Linear(hidden_1, hidden_2)
# linear layer (n_hidden -> 10)
self.fc3 = nn.Linear(hidden_2, 10)
# dropout layer (p=0.2)
# dropout prevents overfitting of data
self.dropout = nn.Dropout(0.2)
def forward(self, x):
# flatten image input
x = x.view(-1, 28 * 28)
# add hidden layer, with relu activation function
x = F.relu(self.fc1(x))
# add dropout layer
x = self.dropout(x)
# add hidden layer, with relu activation function
x = F.relu(self.fc2(x))
# add dropout layer
x = self.dropout(x)
# add output layer
x = self.fc3(x)
return x
# initialize the NN
model = Net()
print(model)
```
### Specify [Loss Function](http://pytorch.org/docs/stable/nn.html#loss-functions) and [Optimizer](http://pytorch.org/docs/stable/optim.html)
It's recommended that you use cross-entropy loss for classification. If you look at the documentation (linked above), you can see that PyTorch's cross entropy function applies a softmax funtion to the output layer *and* then calculates the log loss.
```
# specify loss function (categorical cross-entropy)
criterion = nn.CrossEntropyLoss()
# specify optimizer (stochastic gradient descent) and learning rate = 0.01
optimizer = torch.optim.SGD(model.parameters(), lr=0.01)
```
---
## Train the Network
The steps for training/learning from a batch of data are described in the comments below:
1. Clear the gradients of all optimized variables
2. Forward pass: compute predicted outputs by passing inputs to the model
3. Calculate the loss
4. Backward pass: compute gradient of the loss with respect to model parameters
5. Perform a single optimization step (parameter update)
6. Update average training loss
The following loop trains for 50 epochs; take a look at how the values for the training loss decrease over time. We want it to decrease while also avoiding overfitting the training data.
```
model.to('cuda')
# number of epochs to train the model
n_epochs = 50
# initialize tracker for minimum validation loss
valid_loss_min = np.Inf # set initial "min" to infinity
for epoch in range(n_epochs):
# monitor training loss
train_loss = 0.0
valid_loss = 0.0
###################
# train the model #
###################
model.train() # prep model for training
for data, target in train_loader:
data, target = data.to('cuda'), target.to('cuda')
# clear the gradients of all optimized variables
optimizer.zero_grad()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# backward pass: compute gradient of the loss with respect to model parameters
loss.backward()
# perform a single optimization step (parameter update)
optimizer.step()
# update running training loss
train_loss += loss.item()*data.size(0)
######################
# validate the model #
######################
model.eval() # prep model for evaluation
for data, target in valid_loader:
data, target = data.to('cuda'), target.to('cuda')
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update running validation loss
valid_loss += loss.item()*data.size(0)
# print training/validation statistics
# calculate average loss over an epoch
train_loss = train_loss/len(train_loader.sampler)
valid_loss = valid_loss/len(valid_loader.sampler)
print('Epoch: {} \tTraining Loss: {:.6f} \tValidation Loss: {:.6f}'.format(
epoch+1,
train_loss,
valid_loss
))
# save model if validation loss has decreased
if valid_loss <= valid_loss_min:
print('Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...'.format(
valid_loss_min,
valid_loss))
torch.save(model.state_dict(), 'model.pt')
valid_loss_min = valid_loss
```
### Load the Model with the Lowest Validation Loss
```
model.load_state_dict(torch.load('model.pt'))
```
---
## Test the Trained Network
Finally, we test our best model on previously unseen **test data** and evaluate it's performance. Testing on unseen data is a good way to check that our model generalizes well. It may also be useful to be granular in this analysis and take a look at how this model performs on each class as well as looking at its overall loss and accuracy.
```
model.to('cpu')
# initialize lists to monitor test loss and accuracy
test_loss = 0.0
class_correct = list(0. for i in range(10))
class_total = list(0. for i in range(10))
model.eval() # prep model for evaluation
for data, target in test_loader:
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update test loss
test_loss += loss.item()*data.size(0)
# convert output probabilities to predicted class
_, pred = torch.max(output, 1)
# compare predictions to true label
correct = np.squeeze(pred.eq(target.data.view_as(pred)))
# calculate test accuracy for each object class
for i in range(len(target)):
label = target.data[i]
class_correct[label] += correct[i].item()
class_total[label] += 1
# calculate and print avg test loss
test_loss = test_loss/len(test_loader.sampler)
print('Test Loss: {:.6f}\n'.format(test_loss))
for i in range(10):
if class_total[i] > 0:
print('Test Accuracy of %5s: %2d%% (%2d/%2d)' % (
str(i), 100 * class_correct[i] / class_total[i],
np.sum(class_correct[i]), np.sum(class_total[i])))
else:
print('Test Accuracy of %5s: N/A (no training examples)' % (classes[i]))
print('\nTest Accuracy (Overall): %2d%% (%2d/%2d)' % (
100. * np.sum(class_correct) / np.sum(class_total),
np.sum(class_correct), np.sum(class_total)))
```
### Visualize Sample Test Results
This cell displays test images and their labels in this format: `predicted (ground-truth)`. The text will be green for accurately classified examples and red for incorrect predictions.
```
# obtain one batch of test images
dataiter = iter(test_loader)
images, labels = dataiter.next()
# get sample outputs
output = model(images)
# convert output probabilities to predicted class
_, preds = torch.max(output, 1)
# prep images for display
images = images.numpy()
# plot the images in the batch, along with predicted and true labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(20):
ax = fig.add_subplot(2, 20/2, idx+1, xticks=[], yticks=[])
ax.imshow(np.squeeze(images[idx]), cmap='gray')
ax.set_title("{} ({})".format(str(preds[idx].item()), str(labels[idx].item())),
color=("green" if preds[idx]==labels[idx] else "red"))
```
| github_jupyter |
These notebook is used for initial training. Only necessary preprocessing is done, mainly categorical features encoding and Nans replacement.
It should show the main problems with observations, show main model difficulties, and feaures importances. It should also guide the way of validation Therefore we have:
- data preparation
- cross-validation and modeling
- features and error analysis
```
import os
import pandas as pd
import numpy as np
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import Ridge
from sklearn.ensemble import RandomForestRegressor
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.model_selection import KFold
import mlflow
import IPython.display as ipd
import seaborn as sns
import matplotlib.pylab as plt
data = pd.read_csv(os.path.join('..', 'data', 'raw', 'ubaar-competition', 'train.csv'), encoding="utf-8", index_col="ID")
data['distanceKM'].fillna(data['distanceKM'].median(), inplace=True)
data['taxiDurationMin'].fillna(data['taxiDurationMin'].median(), inplace=True)
data.head()
data.columns
columns_countinous = ['date', 'sourceLatitude', 'sourceLongitude', 'destinationLatitude', 'destinationLongitude', 'distanceKM', 'taxiDurationMin', 'weight']
columns_cat = ['vehicleType', 'vehicleOption']
data_oh = pd.get_dummies(data, columns=columns_cat, drop_first=True)
data_oh = data_oh.drop(columns=['SourceState', 'destinationState'])
data_oh.head()
features_columns = data_oh.columns[data_oh.columns != 'price'].values
remote_server_uri = "http://18.185.244.61:5050"
mlflow.set_tracking_uri(remote_server_uri)
mlflow.set_experiment("UbaarCVinitial")
mlflow.start_run(run_name='')
mlflow.log_param('features', features_columns)
y_full = data_oh['price'].values
x_full = data_oh[features_columns].values
kfold = KFold(n_splits=5, shuffle=True, random_state=42)
train_mapes = []
dev_mapes = []
dev_preds = []
dev_refs = []
dev_inds = []
for train_ind, dev_ind in kfold.split(x_full):
model = RandomForestRegressor(n_estimators=20, max_depth=None, min_samples_leaf=8, random_state=42)
# model = Ridge(alpha=10, solver='auto')
mlflow.log_param('features', features_columns)
mlflow.log_param('model', model.__dict__)
x_train = x_full[train_ind]
y_train = y_full[train_ind]
x_dev = x_full[dev_ind]
y_dev = y_full[dev_ind]
# scaler = StandardScaler()
# scaler.fit(x_train)
# x_train = scaler.transform(x_train)
# x_dev = scaler.transform(x_dev)
model.fit(x_train, y_train)
preds_train = model.predict(x_train)
preds_dev = model.predict(x_dev)
train_mape = mean_absolute_percentage_error(y_train, preds_train)
dev_mape = mean_absolute_percentage_error(y_dev, preds_dev)
train_mapes.append(train_mape)
dev_mapes.append(dev_mape)
dev_preds.extend(list(preds_dev))
dev_refs.extend(list(y_dev))
dev_inds.extend(list(dev_ind))
print(f"Train MAPE: {train_mape}")
print(f"Dev MAPE: {dev_mape}")
print("================")
print(f"Mean MAPE: {np.mean(dev_mapes)}")
print(f"Std MAPE: {np.std(dev_mapes)}")
mlflow.log_metric("Mean dev MAPE", np.mean(dev_mapes))
mlflow.log_metric("Std dev MAPE", np.std(dev_mapes))
mlflow.end_run()
results = pd.DataFrame(list(zip(dev_refs, dev_preds, dev_inds)), columns=['refs', 'preds', 'inds'])
results = results.sort_values('inds')
results.head()
sorted_idx = model.feature_importances_.argsort()
plt.figure(figsize=(10,20))
plt.barh(features_columns[sorted_idx], model.feature_importances_[sorted_idx])
plt.xlabel("Random Forest Feature Importance")
plt.figure(figsize=(10,10))
sns.histplot(data=results, x='refs', y='preds')
plt.plot([0, 50000000], [0, 50000000], linewidth=1, c='r')
plt.xlim([0, 50000000])
plt.ylim([0, 50000000])
```
# Error analysis
Error analysis is a crucial step in working on a model. We can check the performance of the model according to specific features
to find the weakest aspect of the model.
```
data['refs'] = results['refs'].values
data['preds'] = results['preds'].values
column = 'vehicleOption'
for vehicle_type in data[column].unique():
tmp_data = data[data[column] == vehicle_type]
title = f"{vehicle_type}, MPAE:{mean_absolute_percentage_error(tmp_data['refs'], tmp_data['preds']):.2f} #:{len(tmp_data)}"
plt.title(title)
sns.scatterplot(data=tmp_data, x='refs', y='preds')
plt.xlim([0, 50000000])
plt.ylim([0, 50000000])
plt.show()
column = 'SourceState'
for vehicle_type in data[column].unique():
tmp_data = data[data[column] == vehicle_type]
title = f"{vehicle_type}, MPAE:{mean_absolute_percentage_error(tmp_data['refs'], tmp_data['preds']):.2f} #:{len(tmp_data)}"
plt.title(title)
sns.scatterplot(data=tmp_data, x='refs', y='preds')
plt.xlim([0, 50000000])
plt.ylim([0, 50000000])
plt.show()
```
| github_jupyter |
1. Crie uma classe Bola cujos atributos são cor e raio. Crie um método que imprime a cor da bola. Crie um método para calcular a área dessa bola. Crie um método para calcular o volume da bola. Crie um objeto dessa classe e calcule a área e o volume, imprimindo ambos em seguida.
Obs.:
Área da esfera = 4 * 3.14 * r * r;
Volume da esfera = 4 * 3.14 * r* r * r/3
```
class Bola:
'''
Cria a representação de uma bola
'''
def __init__(self, cor, raio):
'''
Construtor
Parâmetros
----------
cor : str
cor associada a bola
raio : float
raio da bola
'''
self.cor = cor
self.raio = raio
self.area = None
self.volume = None
def calcula_area(self):
self.area = 4*3.14*(self.raio)**2
return self.area
def calcula_volume(self):
self.volume = 4/3*3.14*(self.raio)**3
return self.volume
def __repr__(self):
if self.area == None and self.volume == None:
return f'A cor da bola é: {self.cor}, O raio da bola é: {self.raio}'
elif self.volume == None:
return f'A cor da bola é: {self.cor}, O raio da bola é: {self.raio}, A area da bola é {self.area}'
else:
return f'A cor da bola é: {self.cor}, O raio da bola é: {self.raio}, A area da bola é {self.area}, O volume da bola é {self.volume}'
Bola_teste = Bola(cor = 'azul',
raio = 4)
Are = Bola_teste.calcula_area()
Vol = Bola_teste.calcula_volume()
repr(Bola_teste)
Are = Bola_teste.calcula_area()
Vol = Bola_teste.calcula_volume()
print(Are)
print(Vol)
Bola_123 = Bola(cor = 'verde',
raio = 10)
Bola_123.calcula_area()
Bola_123.area
```
2. Crie uma classe Retângulo cujos atributos são lado_a e lado_b. Crie um método para calcular a área desse retângulo. Crie um objeto dessa classe e calcule a área e a imprima em seguida.
```
class Retângulo:
'''
Cria a representação de um retângulo
'''
def __init__(self, lado_a, lado_b):
'''
Construtor
Parâmetros
----------
lado_a : float
medida do primeiro lado do retângulo
lado_b : float
medida do segundo lado do retângulo
'''
self.lado_a = lado_a
self.lado_b = lado_b
self.area = None
def calcula_area(self):
self.area = self.lado_a*self.lado_b
return self.area
def __repr__(self):
if self.area == None:
return f'O lado a vale {self.lado_a} e o lado b vale {self.lado_b}'
else:
return f'O lado a vale {self.lado_a} e o lado b vale {self.lado_b} e a área do retangulo vale {self.area}'
Teste = Retângulo (lado_a = 10,
lado_b = 2)
Teste.calcula_area()
repr(Teste)
```
3. Crie uma classe Funcionario cujos atributos são nome e e-mail. Guarde as horas trabalhadas em um dicionário cujas chaves são o mês em questão e, em outro dicionário, guarde o salário por hora relativo ao mês em questão. Crie um método que retorna o salário mensal do funcionário.
```
class Funcionario:
'''
Cria uma representação do funcionário
'''
def __init__(self,nome,email):
'''
Construtor
Parâmetros
----------
nome : str
nome do funcionário
email : str
email do funcionário
'''
self.nome = nome
self.email = email
self.horas_mes = {}
self.salario_hora = {}
def define_horas_mes(self, mes, horas):
'''
Define a quantidade de horas trabalhadas em determinado mês
Parâmetros
---------
mes : str
mes no formato: 'nov/2021'
quantidade_horas : int
quantidade de horas trabalhadas no mês
'''
self.horas_mes[mes] = horas
def define_valor_hora(self, mes, salario_hora):
'''
Define o valor a ser recebido por hora naquele mês
Parâmetros
----------
mes : str
mes no formato: 'nov/2021'
valor_salario : float
valor da hora no mês
'''
self.salario_hora[mes] = salario_hora
def calcula_salario_mes(self, mes):
'''
Calcula o salário a ser recebido no mês pelo funcionário
Parâmetros
---------
mes : str
mes no formato: 'nov/2021'
'''
if mes in self.horas_mes and mes in self.salario_hora:
self.salario = self.salario_hora[mes]*self.horas_mes[mes]
return self.salario
else:
print('O mês desejado não possui horas ou valor por hora cadastrado!')
def __repr__(self):
return f'Nome: {self.nome}\nEmail: {self.email}'
Augusto = Funcionario(nome = 'Augusto', email = '[email protected]')
Augusto.define_valor_hora('nov/2021',20)
Augusto.define_valor_hora('dez/2021',30)
Augusto
```
4. Crie uma classe Televisor cujos atributos são:
a. fabricante;
b. modelo;
c. canal atual;
d. lista de canais; e
e. volume.
<p style='text-align: justify;'> Faça métodos para aumentar/diminuir volume, trocar o canal e sintonizar um novo canal, que adiciona um novo canal à lista de canais (somente se esse canal não estiver nessa lista). No atributo lista de canais, devem estar armazenados todos os canais já sintonizados dessa TV.
Obs.: O volume não pode ser menor que zero e maior que cem; só se pode trocar para um canal que já esteja na lista de canais. </p>
```
class Televisor:
'''
Cria uma representação de Televisor
'''
def __init__(self, fabricante, modelo):
'''
Construtor
Parâmetros
---------
fabricante : str
nome do fabricante do televisor
modelo : str
modelo do televisor
'''
self.fabricante = fabricante
self.modelo = modelo
self.lista_canais = [2,5,7,10,13] # Canais sintonizados de fábrica
self.volume = 20 # Valor padrão
self.canal_atual = 2 # Canal atual padrão
def aumentar_volume(self, quantidade):
'''
Aumenta o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja aumentar o volume do televisor
'''
volume = self.volume + quantidade
while volume > 100:
print('Erro! O volume não pode superar 100!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume + quantidade
self.volume = volume
# return True
def diminuir_volume(self, quantidade):
'''
Diminui o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja diminuir o volume do televisor
'''
volume = self.volume - quantidade
while volume < 0:
print('Erro! O volume não pode ser menor que 0!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume - quantidade
self.volume = volume
# return True
def sintonizar_canal(self, canal):
'''
Adicional um novo canal a lista de canais padrão do televisor
Parâmetros
--------------
canal : int
canal que deseja incluir na lista de canais
'''
self.lista_canais.append(canal)
print(f'Canal {canal} sintonizado com sucesso.')
def trocar_canal(self, canal):
'''
Troca de canal
Parâmetros
----------
canal : int
canal para o qual deseja mudar
'''
if canal in self.lista_canais:
self.canal_atual = canal
else:
print('Canal não alterado, pois não está na lista de canais.')
return print(f'O canal atual é: {self.canal_atual}')
def __repr__(self):
return f'Fabricante: {self.fabricante}\nModelo: {self.modelo}\nLista de Canais: {self.lista_canais}\nVolume: {self.volume}\nCanal atual: {self.canal_atual}'
Tv_sala = Televisor('TCL','ABC123')
Tv_sala.sintonizar_canal(35)
Tv_sala
Tv_sala.trocar_canal(35)
Tv_sala.aumentar_volume(85)
Tv_sala.diminuir_volume(90)
Tv_sala.lista_canais
```
<p style='text-align: justify;'> 5. Crie uma classe ControleRemoto cujo atributo é televisão (isso é, recebe um objeto da classe do exercício 4). Crie métodos para aumentar/diminuir volume, trocar o canal e sintonizar um novo canal, que adiciona um novo canal à lista de canais (somente se esse canal não estiver nessa lista). </p>
```
class ControleRemoto:
'''
Cria uma representação do controle remoto
'''
def __init__(self, Televisor):
'''
Construtor
Parâmetros
----------
Televisor : objeto da classe Televisor
'''
self.fabricante = Televisor.fabricante
self.modelo = Televisor.modelo
self.lista_canais = Televisor.lista_canais
self.volume = Televisor.volume
self.canal_atual = Televisor.canal_atual
def aumentar_volume(self, quantidade, Televisor):
'''
Aumenta o volume do televisor
Parâmetros
----------
Televisor : objeto televisor
televisor do qual se deseja realizar a ação
quantidade : int
quantidade da qual se deseja aumentar o volume do televisor
'''
volume = self.volume + quantidade
while volume > 100:
print('Erro! O volume não pode superar 100!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume + quantidade
self.volume = volume
Televisor.volume = self.volume
def diminuir_volume(self, quantidade, Televisor):
'''
Diminui o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja diminuir o volume do televisor
'''
volume = self.volume - quantidade
while volume < 0:
print('Erro! O volume não pode ser menor que 0!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume - quantidade
self.volume = volume
Televisor.volume = self.volume
# return True
def sintonizar_canal(self, canal, Televisor):
'''
Adicional um novo canal a lista de canais padrão do televisor
Parâmetros
--------------
canal : int
canal que deseja incluir na lista de canais
'''
self.lista_canais.append(canal)
Televisor.lista_canais.append(canal)
print(f'Canal {canal} sintonizado com sucesso.')
def trocar_canal(self, canal, Televisor):
'''
Troca de canal
Parâmetros
----------
canal : int
canal para o qual deseja mudar
'''
if canal in self.lista_canais:
self.canal_atual = canal
Televisor.canal_atual = canal
else:
print('Canal não alterado, pois não está na lista de canais.')
return print(f'O canal atual é: {self.canal_atual}')
class Televisor:
'''
Cria uma representação de Televisor
'''
def __init__(self, fabricante, modelo):
'''
Construtor
Parâmetros
----------
fabricante : str
nome do fabricante do televisor
modelo : str
modelo do televisor
'''
self.fabricante = fabricante
self.modelo = modelo
self.lista_canais = [2,5,7,10,13] # Canais sintonizados de fábrica
self.volume = 20 # Valor padrão
self.canal_atual = 2 # Canal atual padrão
def aumentar_volume(self, quantidade):
'''
Aumenta o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja aumentar o volume do televisor
'''
volume = self.volume + quantidade
while volume > 100:
print('Erro! O volume não pode superar 100!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume + quantidade
self.volume = volume
# return True
def diminuir_volume(self, quantidade):
'''
Diminui o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja diminuir o volume do televisor
'''
volume = self.volume - quantidade
while volume < 0:
print('Erro! O volume não pode ser menor que 0!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume - quantidade
self.volume = volume
# return True
def sintonizar_canal(self, canal):
'''
Adicional um novo canal a lista de canais padrão do televisor
Parâmetros
--------------
canal : int
canal que deseja incluir na lista de canais
'''
self.lista_canais.append(canal)
print(f'Canal {canal} sintonizado com sucesso.')
def trocar_canal(self, canal):
'''
Troca de canal
Parâmetros
----------
canal : int
canal para o qual deseja mudar
'''
if canal in self.lista_canais:
self.canal_atual = canal
else:
print('Canal não alterado, pois não está na lista de canais.')
return print(f'O canal atual é: {self.canal_atual}')
Tv1 = Televisor('TLC','123')
C1 = ControleRemoto(Tv1)
C1.diminuir_volume(5,Tv1)
C1.sintonizar_canal(99,Tv1)
C1.trocar_canal(99,Tv1)
Tv1.volume
```
<p style='text-align: justify;'> 6. O módulo time possui a função time.sleep(x), que faz seu programa “dormir” por x segundos. Utilizando essa função, crie uma classe Cronômetro e faça um programa que cronometre o tempo.
</p>
```
from time import sleep
class cronometro:
def __init__(self):
self.hora = 0
self.minuto = 0
self.segundo = 0
def contagem_progressiva(self):
flag = True
while flag == True:
if self.segundo < 60:
self.segundo = self.segundo + 1
print(f'{self.hora}:{self.minuto}:{self.segundo}')
sleep(1)
if self.segundo == 60:
self.segundo = 0
self.minuto = self.minuto + 1
if self.minuto == 60:
self.minuto = 0
self.hora = self.hora + 1
def timer(self, hora, minuto, segundo):
self.hora = hora
self.minuto = minuto
self.segundo = segundo
flag = True
print(f'{self.hora}:{self.minuto}:{self.segundo}')
while flag == True:
if self.segundo <= 60 and self.segundo != 0:
self.segundo = self.segundo - 1
print(f'{self.hora}:{self.minuto}:{self.segundo}')
sleep(1)
if self.segundo == 0 and self.minuto ==0 and self.hora == 0:
flag = False
if self.segundo == 0 and self.hora == 0:
self.segundo = 60
self.minuto = self.minuto - 1
if self.minuto == 0:
self.minuto = 60
self.hora = self.hora - 1
cron1 = cronometro()
cron1.timer(1,0,0)
cron1.contagem_progressiva()
```
<p style='text-align: justify;'> 7. Crie uma modelagem de classes para uma agenda capaz de armazenar contatos. Através dessa agenda é possível incluir, remover, buscar e listar contatos já cadastrados.
</p>
```
class Contato:
def __init__(self, nome, email, telefone, endereco):
'''
Construtor
Parâmetros
----------
nome : str
nome do contato
email : str
email do contato
telefone : str
telefone do contato
endereço : str
endereço do contato
'''
self.nome = nome
self.email = email
self.telefone = telefone
self.endereco = endereco
def ver_contato(self):
print(f'Nome: {self.nome}')
print(f'Email: {self.email}')
print(f'Telefone: {self.telefone}')
print(f'Endereço: {self.endereco}')
class Agenda:
def __init__(self):
self.contatos = {}
def adicionar_cadastro(self):
'''
Adiciona novos contatos na agenda
'''
nome = input('Insira o nome do contato: ')
nome = nome.title()
email = input('Insira o email do contato: ')
telefone = int(input('Insira o telefone do contato: '))
endereco = input('Insira o endereço do contato: ')
novo_contato = Contato(nome, email, telefone, endereco)
self.contatos[nome] = novo_contato
def visualizar_cadastros(self):
'''
Mostra a lista de cadastrados
'''
if len(self.contatos) == 0:
print('Lista Vazia!')
else:
for nome in self.contatos:
self.contatos[nome].ver_contato()
print('_________________________')
def remover_cadastro(self):
'''
Remove cadastro do usuário
'''
nome = input('Digite o nome do contato que deseja excluir: ')
self.contatos.pop(nome)
agenda1 = Agenda()
agenda1.adicionar_cadastro()
agenda1.visualizar_cadastros()
agenda1.buscar_cadastro('Augusto')
agenda1.remover_cadastro()
```
<p style='text-align: justify;'> 8. Crie uma classe Cliente cujos atributos são nome, idade e e-mail. Construa um método que imprima as informações tal como abaixo:
Nome: Fulano de Tal
Idade: 40
E-mail: [email protected]
</p>
```
class Cliente:
'''
Cria a representação de um cliente
'''
def __init__(self, nome, idade, email):
'''
Construtor
Parâmetros
----------
nome : str
nome do cliente
idade : int
idade do cliente
email : str
email do cliente
'''
nome = nome.title()
self.nome = nome
self.idade = idade
self.email = email
def imprimir_info(self):
print(f'Nome: {self.nome}')
print(f'Idade: {self.idade}')
print(f'Email: {self.email}')
Augusto = Cliente('augusto', 29, '[email protected]' )
Augusto.imprimir_info()
```
<p style='text-align: justify;'> 9. Com base no exercício anterior, crie um sistema de cadastro e a classe Cliente. Seu programa deve perguntar se o usuário quer cadastrar um novo cliente, alterar um cadastro ou sair.
Dica: Você pode fazer esse exercício criando uma classe Sistema, que irá controlar o sistema de cadastros. Essa classe deve ter o atributo cadastro e os métodos para imprimir os cadastrados, cadastrar um novo cliente, alterar um cadastro ou sair.
</p>
```
class Cliente:
'''
Cria a representação de um cliente
'''
def __init__(self, nome, idade, email):
'''
Construtor
Parâmetros
----------
nome : str
nome do cliente
idade : int
idade do cliente
email : str
email do cliente
'''
nome = nome.title()
self.nome = nome
self.idade = idade
self.email = email
def ver_cliente(self):
print(f'Nome: {self.nome}')
print(f'Idade: {self.idade}')
print(f'Email: {self.email}')
class Sistema_Cadastro:
'''
Cria a representação do sistema de cadastro
'''
def __init__(self):
'''
Construtor
'''
self.cadastrados = {}
def adicionar_cadastrados(self):
'''
Adiciona cadastros ao sistema por meio de inputs pedidos ao usuário
'''
nome = input('Insira o nome do cadastrado: ')
nome = nome.title()
email = input('Insira o email do cadastrado: ')
idade = input('Insira a idade do cadastrado: ')
while not idade.isdigit():
print('Idade inválida!')
idade = input('Insira uma idade válida: ')
if email in self.cadastrados:
print('Cliente já cadastrado!\n')
else:
novo_cadastrado = Cliente(nome, idade, email)
self.cadastrados[email] = novo_cadastrado
print('Cadastro realizado com sucesso!\n')
def ver_cadastrados(self):
'''
Visualiza a lista de cadastrados no sistema
'''
for email in self.cadastrados:
self.cadastrados[email].ver_cliente()
print('________________________________')
def alterar_cadastro(self, email):
'''
Altera o cadastro de alguém que já está no sistema
Parâmetros
----------
email : str
email de quem se deseja alterar o cadastro
'''
alterar = input('Insira 1 para alterar nome, 2 para alterar email e 3 para alterar idade')
while (alterar != '1') and (alterar != '2') and (alterar != '3'):
print('Erro! Opção não reconhecida. Tente novamente.')
alterar = input('Insira 1 para alterar nome, 2 para alterar email e 3 para alterar idade')
if alterar == '1':
novo_nome = input('Digite o novo nome: ')
novo_nome = novo_nome.title()
self.cadastrados[email].nome = novo_nome
print('Nome alterado com sucesso!\n')
elif alterar == '2':
novo_email = input('Digite o novo email: ')
nome = self.cadastrados[email].nome
idade = self.cadastrados[email].idade
novo_cadastrado = Cliente(nome, idade, novo_email)
self.cadastrados[novo_email] = novo_cadastrado
self.cadastrados.pop(email)
print('Email alterado com sucesso!\n')
elif alterar == '3':
nova_idade = int(input('Digite a nova idade: '))
while not nova_idade.isdigit():
print('Idade inválida!')
nova_idade = input('Insira uma idade válida: ')
self.cadastrados[email].idade = nova_idade
print('Idade alterada com sucesso!\n')
def rodar(self):
'''
Roda o sistema de cadastro
'''
flag = True
print('Olá! Bem vindo ao seu sistema de Cadastro!')
while flag == True:
print('O que deseja fazer?')
print('Digite 1 para adicionar um cliente ao seu sistema de cadastro.')
print('Digite 2 para ver a lista de clientes do seu sistema de cadastro.')
print('Digite 3 para alterar os dados de um cliente do seu sistema de cadastro.')
print('Digite 0 para sair do seus sistema de Cadastro.')
opcao = input('Digite a opção desejada: ')
if opcao == '1':
self.adicionar_cadastrados()
elif opcao == '2':
if len(self.cadastrados) == 0:
print('Cadastro vazio!\n')
else:
self.ver_cadastrados()
elif opcao == '3':
email = input('Digite o email do cliente que deseja alterar alguma informação: ')
if email in self.cadastrados:
self.alterar_cadastro(email)
else:
print('Cliente não cadastrado!\n')
elif opcao == '0':
flag = False
else:
print('Erro! Opção não reconhecida!\nEscolha uma opção válida.')
print('Saida do sistema realizada com sucesso.')
sistema_1 = Sistema_Cadastro()
sistema_1.rodar()
```
<p style='text-align: justify;'> 10. Crie uma classe ContaCorrente com os atributos cliente (que deve ser um objeto da classe Cliente) e saldo. Crie métodos para depósito, saque e transferência. Os métodos de saque e transferência devem verificar se é possível realizar a transação.
</p>
```
class Cliente:
'''
Cria representação de um cliente
'''
def __init__(self, nome, cpf):
'''
Construtor
Parâmetro
---------
nome : str
nome do cliente
cpf : str
cpf do cliente
'''
self.nome = nome.capitalize()
self.cpf = cpf
class ContaCorrente:
'''
Cria a representação da conta corrente
'''
def __init__(self, Cliente):
'''
Construtor
Parâmetro
---------
Cliente : objeto da classe Cliente
Objeto criado a partir de cliente
'''
self.cliente = Cliente.nome
self.saldo = 0
def depositar(self, deposito):
self.saldo = self.saldo + deposito
def sacar(self, saque):
if saque > self.saldo:
print('Saldo insuficiente!')
else:
self.saldo = self.saldo - saque
def transferir(self, other, transferencia):
if transferencia > self.saldo:
print('Saldo insuficiente para realizar operação!')
else:
self.saldo = self.saldo - transferencia
other.saldo = other.saldo + transferencia
# Exemplo de uso
augusto = Cliente('augusto', '12345678900')
joeise = Cliente('joeise','98765432100')
contaAugusto = ContaCorrente(augusto)
contaJoeise = ContaCorrente(joeise)
contaAugusto.depositar(1000)
contaAugusto.transferir(contaJoeise,600)
contaAugusto.saldo
contaJoeise.saldo
```
<p style='text-align: justify;'> 11. Crie uma classe Fração cujos atributos são numerador (número de cima) e denominador (número de baixo). <br/><br/>
Implemente os métodos de adição, subtração, multiplicação, divisão que retornam objetos do tipo Fração.<br/><br/>
Implemente também o método _ repr _.<br/><br/>
Implemente métodos para comparação: igualdade (==) e desigualdades (!=, <=, >=, < e >).
</p>
```
class Fracao:
'''
Cria a representação de uma fração
'''
def __init__(self, numerador, denominador):
'''
Construtor
Parâmetros
---------
numerador : int
numerador da fração
denominador : int
denominador da fração
'''
if (denominador == 0):
raise ValueError('Denominador deve ser diferente de zero.')
else:
self.numerador = numerador
self.denominador = denominador
def __repr__(self):
return f'{self.numerador}/{self.denominador}'
def __add__(self, other):
# numerador = self.numerador*other.denominador + self.denominador*other.numerador
# denominador = self.denominador*other.denominador
numerador_1 = self.numerador
numerador_2 = other.numerador
denominador_1 = self.denominador
denominador_2 = other.denominador
#mmc
if denominador_1 > denominador_2:
maior = denominador_1
else:
maior = denominador_2
for i in range(maior):
aux = denominador_1 * i
if (aux % denominador_2) == 0:
mmc = aux
#Calculo
numerador_resultante = ((mmc/denominador_1)*numerador_1 + (mmc/denominador_2)*numerador_2)
denominador_resultante = mmc
return Fracao(numerador_resultante,denominador_resultante)
def __sub__(self,other):
# numerador = self.numerador*other.denominador - self.denominador*other.numerador
# denominador = self.denominador*other.denominador
numerador_1 = self.numerador
numerador_2 = other.numerador
denominador_1 = self.denominador
denominador_2 = other.denominador
#mmc
if denominador_1 > denominador_2:
maior = denominador_1
else:
maior = denominador_2
for i in range(maior):
aux = denominador_1 * i
if (aux % denominador_2) == 0:
mmc = aux
#Calculo
numerador_resultante = ((mmc/denominador_1)*numerador_1 - (mmc/denominador_2)*numerador_2)
denominador_resultante = mmc
return Fracao(numerador_resultante,denominador_resultante)
def __mul__(self,other):
numerador_1 = self.numerador
numerador_2 = other.numerador
denominador_1 = self.denominador
denominador_2 = other.denominador
numerador_resultante = numerador_1*numerador_2
denominador_resultante = denominador_1*denominador_2
return Fracao(numerador_resultante,denominador_resultante)
def __truediv__(self, other):
numerador_1 = self.numerador
numerador_2 = other.denominador
denominador_1 = self.denominador
denominador_2 = other.numerador
numerador_resultante = numerador_1*numerador_2
denominador_resultante = denominador_1*denominador_2
return Fracao(numerador_resultante,denominador_resultante)
def __eq__(self, other):
return self.numerador/self.denominador == other.numerador/other.denominador
def __lt__(self, other):
return self.numerador/self.denominador < other.numerador/other.denominador
def __le__(self, other):
return self.numerador/self.denominador <= other.numerador/other.denominador
def __gt__(self, other):
return self.numerador/self.denominador > other.numerador/other.denominador
def __ge__(self, other):
return self.numerador/self.denominador >= other.numerador/other.denominador
fr1 = Fracao(6,8)
fr2 = Fracao(7,6)
# repr(fr1)
fr1/fr2
fr1<fr2
```
<p style='text-align: justify;'> 12. Crie uma classe Data cujos atributos são dia, mês e ano. Implemente métodos _ repr _ e para comparação: igualdade (==) e desigualdades (!=, <=, >=, < e >).
</p>
```
class Data:
def __init__(self, dia, mes, ano):
self.dia = dia
self.mes = mes
self.ano = ano
def __repr__(self):
return f'{self.dia}/{self.mes}/{self.ano}'
def __eq__(self, other):
if (self.dia == other.dia) and (self.mes == other.mes) and (self.ano == other.ano):
return True
else:
return False
def __lt__(self, other):
if (self.ano < other.ano):
return True
elif (self.ano > other.ano):
return False
elif (self.mes < other.mes):
return True
elif (self.mes > other.mes):
return False
elif (self.dia < other.dia):
return True
elif (self.dia > other.dia):
return False
def __gt__(self, other):
if (self.ano > other.ano):
return True
elif (self.ano < other.ano):
return False
elif (self.mes > other.mes):
return True
elif (self.mes < other.mes):
return False
elif (self.dia > other.dia):
return True
elif (self.dia < other.dia):
return False
def __ne__(self, other):
if (self.dia != other.dia) or (self.mes != other.mes) or (self.ano != other.ano):
return True
else:
return False
data1 = Data(24,9,2023)
data2 = Data(30,1,2022)
data1 == data2
```
<p style='text-align: justify;'> 13. Nos exercícios 1, 2, 3, 4 e 6, implemente o método _ repr _ para exibir as informações desejadas de cada uma das classes.
</p>
```
# Questão 1
class Bola:
'''
Cria a representação de uma bola
'''
def __init__(self, cor, raio):
'''
Construtor
Parâmetros
----------
cor : str
cor associada a bola
raio : float
raio da bola
'''
self.cor = cor
self.raio = raio
self.area = None
self.volume = None
def calcula_area(self):
self.area = 4*3.14*(self.raio)**2
return self.area
def calcula_volume(self):
self.volume = 4/3*3.14*(self.raio)**3
return self.volume
def __repr__(self):
if self.area == None and self.volume == None:
return f'A cor da bola é: {self.cor}, O raio da bola é: {self.raio}'
elif self.volume == None:
return f'A cor da bola é: {self.cor}, O raio da bola é: {self.raio}, A area da bola é {self.area}'
else:
return f'A cor da bola é: {self.cor}, O raio da bola é: {self.raio}, A area da bola é {self.area}, O volume da bola é {self.volume}'
#Questão 2
class Retângulo:
'''
Cria a representação de um retângulo
'''
def __init__(self, lado_a, lado_b):
'''
Construtor
Parâmetros
----------
lado_a : float
medida do primeiro lado do retângulo
lado_b : float
medida do segundo lado do retângulo
'''
self.lado_a = lado_a
self.lado_b = lado_b
self.area = None
def calcula_area(self):
self.area = self.lado_a*self.lado_b
return self.area
def __repr__(self):
if self.area == None:
return f'O lado a vale {self.lado_a} e o lado b vale {self.lado_b}'
else:
return f'O lado a vale {self.lado_a} e o lado b vale {self.lado_b} e a área do retangulo vale {self.area}'
#Questão 3
class Funcionario:
'''
Cria uma representação do funcionário
'''
def __init__(self,nome,email):
'''
Construtor
Parâmetros
----------
nome : str
nome do funcionário
email : str
email do funcionário
'''
self.nome = nome
self.email = email
self.horas_mes = {}
self.salario_hora = {}
def define_horas_mes(self, mes, horas):
'''
Define a quantidade de horas trabalhadas em determinado mês
Parâmetros
---------
mes : str
mes no formato: 'nov/2021'
quantidade_horas : int
quantidade de horas trabalhadas no mês
'''
self.horas_mes[mes] = horas
def define_valor_hora(self, mes, salario_hora):
'''
Define o valor a ser recebido por hora naquele mês
Parâmetros
----------
mes : str
mes no formato: 'nov/2021'
valor_salario : float
valor da hora no mês
'''
self.salario_hora[mes] = salario_hora
def calcula_salario_mes(self, mes):
'''
Calcula o salário a ser recebido no mês pelo funcionário
Parâmetros
---------
mes : str
mes no formato: 'nov/2021'
'''
if mes in self.horas_mes and mes in self.salario_hora:
self.salario = self.salario_hora[mes]*self.horas_mes[mes]
return self.salario
else:
print('O mês desejado não possui horas ou valor por hora cadastrado!')
def __repr__(self):
return f'Nome: {self.nome}\nEmail: {self.email}'
#Questão 4
class Televisor:
'''
Cria uma representação de Televisor
'''
def __init__(self, fabricante, modelo):
'''
Construtor
Parâmetros
---------
fabricante : str
nome do fabricante do televisor
modelo : str
modelo do televisor
'''
self.fabricante = fabricante
self.modelo = modelo
self.lista_canais = [2,5,7,10,13] # Canais sintonizados de fábrica
self.volume = 20 # Valor padrão
self.canal_atual = 2 # Canal atual padrão
def aumentar_volume(self, quantidade):
'''
Aumenta o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja aumentar o volume do televisor
'''
volume = self.volume + quantidade
while volume > 100:
print('Erro! O volume não pode superar 100!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume + quantidade
self.volume = volume
# return True
def diminuir_volume(self, quantidade):
'''
Diminui o volume do televisor
Parâmetros
----------
quantidade : int
quantidade da qual se deseja diminuir o volume do televisor
'''
volume = self.volume - quantidade
while volume < 0:
print('Erro! O volume não pode ser menor que 0!\n')
quantidade = int(input('Escolha outra fator para aumentar o volume: '))
volume = self.volume - quantidade
self.volume = volume
# return True
def sintonizar_canal(self, canal):
'''
Adicional um novo canal a lista de canais padrão do televisor
Parâmetros
--------------
canal : int
canal que deseja incluir na lista de canais
'''
self.lista_canais.append(canal)
print(f'Canal {canal} sintonizado com sucesso.')
def trocar_canal(self, canal):
'''
Troca de canal
Parâmetros
----------
canal : int
canal para o qual deseja mudar
'''
if canal in self.lista_canais:
self.canal_atual = canal
else:
print('Canal não alterado, pois não está na lista de canais.')
return print(f'O canal atual é: {self.canal_atual}')
def __repr__(self):
return f'Fabricante: {self.fabricante}\nModelo: {self.modelo}\nLista de Canais: {self.lista_canais}\nVolume: {self.volume}\nCanal atual: {self.canal_atual}'
#Questão 6
# Não coloquei __rep__ pois não consegui enxergar aonde poderia coloccá-lo no cronometro que montei.
```
<p style='text-align: justify;'> 14. Faça uma classe ContaVip que difere da ContaCorrente por ter cheque especial (novo atributo) e é filha da classe ContaCorrente. Você precisa implementar os métodos para saque, transferência ou depósito?
</p>
```
class Cliente:
'''
Cria representação de um cliente
'''
def __init__(self, nome, cpf):
'''
Construtor
Parâmetro
---------
nome : str
nome do cliente
cpf : str
cpf do cliente
'''
self.nome = nome.capitalize()
self.cpf = cpf
class ContaCorrente:
'''
Cria a representação da conta corrente
'''
def __init__(self, cliente, saldo=0):
'''
Construtor
Parâmetro
---------
Cliente : str
Nome do cliente
'''
self.cliente = cliente
self.saldo = saldo
def depositar(self, deposito):
self.saldo = self.saldo + deposito
def sacar(self, saque):
if saque > self.saldo:
print('Saldo insuficiente!')
else:
self.saldo = self.saldo - saque
def transferir(self, other, transferencia):
if transferencia > self.saldo:
print('Saldo insuficiente para realizar operação!')
else:
self.saldo = self.saldo - transferencia
other.saldo = other.saldo + transferencia
class ContaVip(ContaCorrente):
'''
Cria a representação de uma conta vip que tem o atributo cheque especial como diferencial
'''
def __init__(self, cliente, saldo = 0, cheque_especial=0):
super().__init__(cliente, saldo)
self.cheque_especial = cheque_especial
def sacar(self, saque):
if (self.saldo + self.cheque_especial) - saque < 0:
print('Saldo insuficiente!')
else:
self.depositar(-saque)
def depositar (self, deposito):
self.saldo = self.saldo + deposito
def transferir(self, other, transferencia):
if transferencia > self.saldo + self.cheque_especial:
print('Saldo insuficiente para realizar operação!')
else:
self.saldo = self.saldo - transferencia
other.saldo = other.saldo + transferencia
augusto = Cliente('augusto', '12345678900')
joeise = Cliente('joeise','98765432100')
contaAugusto = ContaCorrente('augusto')
contaJoeise = ContaCorrente('joeise')
contaAugusto.depositar(1000)
contaAugusto.saldo
ContaVipAugusto = ContaVip(contaAugusto, contaAugusto.saldo, cheque_especial = 200)
ContaVipAugusto.sacar(1100)
```
15. Crie uma classe Quadrado, filha da classe Retângulo do exercício 2.
```
class Retângulo:
'''
Cria a representação de um retângulo
'''
def __init__(self, lado_a, lado_b):
'''
Construtor
Parâmetros
----------
lado_a : float
medida do primeiro lado do retângulo
lado_b : float
medida do segundo lado do retângulo
'''
self.lado_a = lado_a
self.lado_b = lado_b
def calcula_area(self):
self.area = self.lado_a*self.lado_b
return self.area
class Quadrado(Retângulo):
'''
Cria a representação de um quadrado
'''
def __init__(self, lado):
super().__init__(lado, lado)
quad1 = Quadrado(5)
quad1.calcula_area()
import time
def countdown(num_of_secs):
while num_of_secs:
m, s = divmod(num_of_secs, 60)
min_sec_format = '{:02d}:{:02d}'.format(m, s)
print(min_sec_format, end='\n')
time.sleep(1)
num_of_secs -= 1
print('Countdown finished.')
inp = int(input('Input number of seconds to countdown: '))
countdown(inp)
```
| github_jupyter |
# US Production Data for RBC Modeling
```
import pandas as pd
import numpy as np
import fredpy as fp
import matplotlib.pyplot as plt
plt.style.use('classic')
%matplotlib inline
pd.plotting.register_matplotlib_converters()
# Load API key
fp.api_key = fp.load_api_key('fred_api_key.txt')
# Download nominal GDP, nominal personal consumption expenditures, nominal
# gross private domestic investment, the GDP deflator, and an index of hours
# worked in the nonfarm business sector produced by the BLS. All data are
# from FRED and are quarterly.
gdp = fp.series('GDP')
cons = fp.series('PCEC')
invest = fp.series('GPDI')
hours = fp.series('HOANBS')
defl = fp.series('GDPDEF')
pcec = fp.series('PCEC')
m2 = fp.series('M2SL')
tb3mo = fp.series('TB3MS')
unemp = fp.series('UNRATE')
# Convert monthly M2, 3-mo T-Bill, and unemployment to quarterly
m2 = m2.as_frequency('Q')
tb3mo = tb3mo.as_frequency('Q')
unemp = unemp.as_frequency('Q')
# Convert unemployment and t-bill data to decimals instead of percents
unemp.data = unemp.data/100
tb3mo.data = tb3mo.data/100
# pcec inflation as pecent change over past year
pcec = pcec.apc()
pcec.data = pcec.data/100
# Make sure that all of the downloaded series have the same data ranges
gdp,cons,invest,hours,defl,pcec,m2,tb3mo,unemp = fp.window_equalize([gdp,cons,invest,hours,defl,pcec,m2,tb3mo,unemp])
# Compute real GDP, real consumption, real investment
gdp.data = gdp.data/defl.data*100
cons.data = cons.data/defl.data*100
invest.data = invest.data/defl.data*100
m2.data = m2.data/defl.data*100
# Print units
print('Hours units: ',hours.units)
print('Deflator units:',defl.units)
```
Next, compute the quarterly capital stock series for the US using the perpetual inventory method. The discrete-time Solow growth model is given by:
\begin{align}
Y_t & = A_tK_t^{\alpha}L_t^{1-\alpha} \tag{1}\\
C_t & = (1-s)Y_t \tag{2}\\
Y_t & = C_t + I_t \tag{3}\\
K_{t+1} & = I_t + (1-\delta)K_t \tag{4}\\
A_{t+1} & = (1+g)A_t \tag{5}\\
L_{t+1} & = (1+n)L_t \tag{6}.
\end{align}
Here the model is assumed to be quarterly so $n$ is the *quarterly* growth rate of labor hours, $g$ is the *quarterly* growth rate of TFP, and $\delta$ is the *quarterly* rate of depreciation of the capital stock. Given a value of the quarterly depreciation rate $\delta$, an investment series $I_t$, and an initial capital stock $K_0$, the law of motion for the capital stock, Equation (4), can be used to compute an implied capital series. But we don't know $K_0$ or $\delta$ so we'll have to *calibrate* these values using statistics computed from the data that we've already obtained.
Let lowercase letters denote a variable that's been divided by $A_t^{1/(1-\alpha)}L_t$. E.g.,
\begin{align}
y_t = \frac{Y_t}{A_t^{1/(1-\alpha)}L_t}\tag{7}
\end{align}
Then (after substituting consumption from the model), the scaled version of the model can be written as:
\begin{align}
y_t & = k_t^{\alpha} \tag{8}\\
i_t & = sy_t \tag{9}\\
k_{t+1} & = i_t + (1-\delta-n-g')k_t,\tag{10}
\end{align}
where $g' = g/(1-\alpha)$ is the growth rate of $A_t^{1/(1-\alpha)}$. In the steady state:
\begin{align}
k & = \left(\frac{s}{\delta+n+g'}\right)^{\frac{1}{1-\alpha}} \tag{11}
\end{align}
which means that the ratio of capital to output is constant:
\begin{align}
\frac{k}{y} & = \frac{s}{\delta+n+g'} \tag{12}
\end{align}
and therefore the steady state ratio of depreciation to output is:
\begin{align}
\overline{\delta K/ Y} & = \frac{\delta s}{\delta + n + g'} \tag{13}
\end{align}
where $\overline{\delta K/ Y}$ is the long-run average ratio of depreciation to output. We can use Equation (13) to calibrate $\delta$ given $\overline{\delta K/ Y}$, $s$, $n$, and $g'$.
Furthermore, in the steady state, the growth rate of output is constant:
\begin{align}
\frac{\Delta Y}{Y} & = n + g' \tag{14}
\end{align}
1. Assume $\alpha = 0.35$.
2. Calibrate $s$ as the average of ratio of investment to GDP.
3. Calibrate $n$ as the average quarterly growth rate of labor hours.
4. Calibrate $g'$ as the average quarterly growth rate of real GDP minus n.
5. Calculate the average ratio of depreciation to GDP $\overline{\delta K/ Y}$ and use the result to calibrate $\delta$. That is, find the average ratio of Current-Cost Depreciation of Fixed Assets (FRED series ID: M1TTOTL1ES000) to GDP (FRED series ID: GDPA). Then calibrate $\delta$ from the following steady state relationship:
\begin{align}
\delta & = \frac{\left( \overline{\delta K/ Y} \right)\left(n + g' \right)}{s - \left( \overline{\delta K/ Y} \right)} \tag{15}
\end{align}
6. Calibrate $K_0$ by asusming that the capital stock is initially equal to its steady state value:
\begin{align}
K_0 & = \left(\frac{s}{\delta + n + g'}\right) Y_0 \tag{16}
\end{align}
Then, armed with calibrated values for $K_0$ and $\delta$, compute $K_1, K_2, \ldots$ recursively. See Timothy Kehoe's notes for more information on the perpetual inventory method:
http://users.econ.umn.edu/~tkehoe/classes/GrowthAccountingNotes.pdf
```
# Set the capital share of income
alpha = 0.35
# Average saving rate
s = np.mean(invest.data/gdp.data)
# Average quarterly labor hours growth rate
n = (hours.data[-1]/hours.data[0])**(1/(len(hours.data)-1)) - 1
# Average quarterly real GDP growth rate
g = ((gdp.data[-1]/gdp.data[0])**(1/(len(gdp.data)-1)) - 1) - n
# Compute annual depreciation rate
depA = fp.series('M1TTOTL1ES000')
gdpA = fp.series('gdpa')
gdpA = gdpA.window([gdp.data.index[0],gdp.data.index[-1]])
gdpA,depA = fp.window_equalize([gdpA,depA])
deltaKY = np.mean(depA.data/gdpA.data)
delta = (n+g)*deltaKY/(s-deltaKY)
# print calibrated values:
print('Avg saving rate: ',round(s,5))
print('Avg annual labor growth:',round(4*n,5))
print('Avg annual gdp growth: ',round(4*g,5))
print('Avg annual dep rate: ',round(4*delta,5))
# Construct the capital series. Note that the GPD and investment data are reported on an annualized basis
# so divide by 4 to get quarterly data.
capital = np.zeros(len(gdp.data))
capital[0] = gdp.data[0]/4*s/(n+g+delta)
for t in range(len(gdp.data)-1):
capital[t+1] = invest.data[t]/4 + (1-delta)*capital[t]
# Save in a fredpy series
capital = fp.to_fred_series(data = capital,dates =gdp.data.index,units = gdp.units,title='Capital stock of the US',frequency='Quarterly')
# plot the computed capital series
plt.plot(capital.data.index,capital.data,'-',lw=3,alpha = 0.7)
plt.ylabel(capital.units)
plt.title(capital.title)
plt.grid()
# Compute TFP
tfp = gdp.data/capital.data**alpha/hours.data**(1-alpha)
tfp = fp.to_fred_series(data = tfp,dates =gdp.data.index,units = gdp.units,title='TFP of the US',frequency='Quarterly')
# Plot the computed capital series
plt.plot(tfp.data.index,tfp.data,'-',lw=3,alpha = 0.7)
plt.ylabel(tfp.units)
plt.title(tfp.title)
plt.grid()
# Convert each series into per capita using civilian pop 16 and over
gdp = gdp.per_capita(civ_pop=True)
cons = cons.per_capita(civ_pop=True)
invest = invest.per_capita(civ_pop=True)
hours = hours.per_capita(civ_pop=True)
capital = capital.per_capita(civ_pop=True)
m2 = m2.per_capita(civ_pop=True)
# Put GDP, consumption, investment, and M2 in units of thousands of dollars per person
gdp.data = gdp.data*1000
cons.data = cons.data*1000
invest.data = invest.data*1000
capital.data = capital.data*1000
m2.data = m2.data/1000
# Scale hours per person to equal 100 on October (Quarter III) of 2012
hours.data = hours.data/hours.data.loc['2012-10-01']*100
# Compute and plot log real GDP, log consumption, log investment, log hours
gdp_log = gdp.log()
cons_log = cons.log()
invest_log = invest.log()
hours_log = hours.log()
capital_log = capital.log()
tfp_log = tfp.log()
m2_log = m2.log()
m2_log = m2.log()
# HP filter to isolate trend and cyclical components
gdp_log_cycle,gdp_log_trend = gdp_log.hp_filter()
cons_log_cycle,cons_log_trend = cons_log.hp_filter()
invest_log_cycle,invest_log_trend = invest_log.hp_filter()
hours_log_cycle,hours_log_trend = hours_log.hp_filter()
capital_log_cycle,capital_log_trend = capital_log.hp_filter()
tfp_log_cycle,tfp_log_trend = tfp_log.hp_filter()
m2_log_cycle,m2_log_trend = m2_log.hp_filter()
tb3mo_cycle,tb3mo_trend = tb3mo.hp_filter()
unemp_cycle,unemp_trend = unemp.hp_filter()
pcec_cycle,pcec_trend = pcec.hp_filter()
# Create a DataFrame with actual and trend data
data = pd.DataFrame({
'gdp':gdp.data,
'gdp_trend':np.exp(gdp_log_trend.data),
'gdp_cycle':gdp_log_cycle.data,
'consumption':cons.data,
'consumption_trend':np.exp(cons_log_trend.data),
'consumption_cycle':cons_log_cycle.data,
'investment':invest.data,
'investment_trend':np.exp(invest_log_trend.data),
'investment_cycle':invest_log_cycle.data,
'hours':hours.data,
'hours_trend':np.exp(hours_log_trend.data),
'hours_cycle':hours_log_cycle.data,
'capital':capital.data,
'capital_trend':np.exp(capital_log_trend.data),
'capital_cycle':capital_log_cycle.data,
'tfp':tfp.data,
'tfp_trend':np.exp(tfp_log_trend.data),
'tfp_cycle':tfp_log_cycle.data,
'real_m2':m2.data,
'real_m2_trend':np.exp(m2_log_trend.data),
'real_m2_cycle':m2_log_cycle.data,
't_bill_3mo':tb3mo.data,
't_bill_3mo_trend':tb3mo_trend.data,
't_bill_3mo_cycle':tb3mo_cycle.data,
'pce_inflation':pcec.data,
'pce_inflation_trend':pcec_trend.data,
'pce_inflation_cycle':pcec_cycle.data,
'unemployment':unemp.data,
'unemployment_trend':unemp_trend.data,
'unemployment_cycle':unemp_cycle.data,
},index = gdp.data.index)
# # RBC Data
# columns_ordered =[]
# names = ['gdp','consumption','investment','hours','capital','tfp']
# for name in names:
# columns_ordered.append(name)
# columns_ordered.append(name+'_trend')
# data[columns_ordered].to_csv('../Csv/rbc_data_actual_trend.csv')
# # Create a DataFrame with actual, trend, and cycle data
# columns_ordered =[]
# names = ['gdp','consumption','investment','hours','capital','tfp']
# for name in names:
# columns_ordered.append(name)
# columns_ordered.append(name+'_trend')
# columns_ordered.append(name+'_cycle')
# data[columns_ordered].to_csv('../Csv/rbc_data_actual_trend_cycle.csv')
# Business Cycle Data
columns_ordered =[]
names = ['gdp','consumption','investment','hours','capital','tfp','real_m2','t_bill_3mo','pce_inflation','unemployment']
for name in names:
columns_ordered.append(name)
columns_ordered.append(name+'_trend')
data[columns_ordered].to_csv('../Csv/business_cycle_data_actual_trend.csv')
# Create a DataFrame with actual, trend, and cycle data
columns_ordered =[]
names = ['gdp','consumption','investment','hours','capital','tfp','real_m2','t_bill_3mo','pce_inflation','unemployment']
for name in names:
columns_ordered.append(name)
columns_ordered.append(name+'_trend')
columns_ordered.append(name+'_cycle')
data[columns_ordered].to_csv('../Csv/business_cycle_data_actual_trend_cycle.csv')
```
| github_jupyter |
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
```
# Reading QoS analysis raw info
Temporarily, this info is saved in a CSV file but it will be in the database
**qos_analysis_13112018.csv**
- columns = ['url','protocol','code','start','end','duration','runid']
- First try of qos analysis.
- It was obtained from 50 repetitions of each of 3921 eepsites gathered.
- Just one i2router (UCA desktop host)
- Time gap between each eepsite request 3921*0.3sec=1176sec/60sec ~ 19 mins
- Total experiment elapsed time 50rep X 19 mins ~ 16 hours
**qos_analysis_29112018_local.csv**
- columns = ['url','code','duration','runid']
- 100 repetions of the first 10 eepsite from the list. Just for testing.
- local i2prouter from my laptop
- Time gap between each eepsite 10*5sec=50sec ~ 60s
- Total experiment elapsed time 100rep x 1min ~ 100 mins
```
# File for processing it
qos_file = 'qos_analysis_13112018.csv'
path_to_file = 'data/' + qos_file
columns = ['url','protocol','code','start','end','duration','runid']
df_qos = pd.read_csv(path_to_file,names=columns,delimiter="|")
# File for processing it - local router
qos_file = 'qos_analysis_29112018_local.csv'
path_to_file = 'data/' + qos_file
columns = ['url','code','duration','runid']
df_qos_local = pd.read_csv(path_to_file,names=columns,delimiter="|")
# File for processing it - local router
qos_file = 'qos_analysis_29112018_remote.csv'
path_to_file = 'data/' + qos_file
columns = ['url','code','duration','runid']
df_qos_remote = pd.read_csv(path_to_file,names=columns,delimiter="|")
# File for testing - to be removed
qos_file = 'analitica.csv'
path_to_file = 'data/' + qos_file
columns = ['url','code','duration','runid','intervals']
df_qos_testing = pd.read_csv(path_to_file,names=columns,delimiter="|")
# DF to analize
df_qos = df_qos_testing.copy()
# Removing not valid rounds
df_qos['runid'] = pd.to_numeric(df_qos['runid'], errors='coerce').dropna()
df_qos.head()
# Duration distribution by http response
fig, ax1 = plt.subplots(figsize=(10, 6))
# http code
code = 200
df_to_plot = df_qos[(df_qos['code']==code)]['duration']
#df_qos[(df_qos['code']==500)]['duration'].hist(bins=100)
df_to_plot.plot(kind='hist',bins=100, ax=ax1, color={'r','g'}, alpha=0.7)
ax1.set_ylabel('Frequency')
ax1.set_xlabel('Duration (seconds)')
ax1.set_title('HTTP ' + str(code))
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=75)
df_qos['code'].hist(bins=100)
df_qos['code'].unique()
df_qos.code.value_counts()
df_qos.describe()
# Average duration by error code
df = pd.DataFrame({
'code': df_qos['code'],
'duration': df_qos['duration'],
})
df = df.sort_values(by='code')
fig, ax1 = plt.subplots(figsize=(12, 8))
to_drop = []
df = df[~df['code'].isin(to_drop)]
means = df.groupby('code').mean()
std = df.groupby('code').std()
means.plot(kind='bar',yerr=std, ax=ax1, color={'r','g'}, alpha=0.7)
ax1.set_ylabel('Duration average (seconds)')
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=75)
df.groupby('code').describe()
# Duration by error code
df = pd.DataFrame({
'code': df_qos['code'],
'duration': df_qos['duration'],
})
to_drop = [504]
df = df[~df['code'].isin(to_drop)]
fig, ax1 = plt.subplots(figsize=(12, 8))
ax = sns.boxplot(x="code", y="duration", data=df, ax=ax1)
ax1.set_ylabel('Duration (seconds)')
ax1.set_xticklabels(set(df.code))
plt.sca(ax1)# matplotlib
# Average duration by eepsite
df = pd.DataFrame({
'url': df_qos['url'],
'duration': df_qos['duration'],
})
fig, ax1 = plt.subplots(figsize=(15, 8),)
df = df.sort_values(by='url')
means = df.groupby('url').mean()
std = df.groupby('url').std()
means = means[0:50]
std = std[0:50]
means.plot(kind='bar',yerr=std, ax=ax1, color={'r'}, alpha=0.7)
ax1.set_ylabel('Duration average (seconds)')
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=90)
# Average duration by eepsite
df = pd.DataFrame({
'url': df_qos['url'],
'duration': df_qos['duration'],
'code': df_qos['code']
})
fig, ax1 = plt.subplots(figsize=(15, 8),)
df = df.sort_values(by='duration',ascending=False)
eepsites = list(df[0:10000].groupby('url').groups.keys())[0:20]
df = df[df['url'].isin(eepsites)]
ax = sns.boxplot(x="url", y="duration", data=df, hue='code', ax=ax1)
ax1.set_ylabel('Duration (seconds)')
#ax1.set_ylim((0,3))
plt.sca(ax1)# matplotlib only acts over the current axis
plt.xticks(rotation=90)
```
# Availability study
```
HTTP_RESPONSE_CODES = {200:'OK',
301:'Moved Permanently',
302:'Found (Previously "Moved temporarily")',
400:'Bad Request',
401:'Unauthorized',
403:'Forbidden',
429:'Too Many Requests',
500:'Internal Server Error',
502:'Bad Gateway',
503:'Service Unavailable',
504:'Gateway Timeout'}
df_qos
```
| github_jupyter |
```
import pandas as pd
df = pd.read_csv('data/Consumer_Complaints.csv')
df.info()
feature_col = ['Consumer complaint narrative']
res_col = ['Product', 'Issue']
df.dropna(subset= feature_col + res_col, inplace=True)
df.drop_duplicates(subset=feature_col, inplace=True)
df.info()
#print(df['Product'].unique())
df_cat = None
for col in res_col:
temp = df[col].astype('category')
df_cat = pd.concat([df_cat, temp], axis=1)
df.drop(columns=res_col, inplace=True)
df = pd.concat([df, df_cat], axis=1)
df.info()
# print(df['Issue'].unique())
# randomly select train/test data
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(df[feature_col[0]], df[res_col[0]], test_size=0.2, random_state=42)
y_train.head()
#X_train.head()
import nltk
nltk.download('wordnet')
from nltk.stem.wordnet import WordNetLemmatizer
import gensim
from gensim.utils import simple_preprocess
import gensim.corpora as corpora
from nltk.corpus import stopwords
stop_words = stopwords.words('english')
stop_words.extend(['xxxx', 'xx'])
stop_words = set(stop_words)
def tokenize(doc):
# doc is a string
# return an array of words
return simple_preprocess(doc, deacc=True, min_len=2, max_len=15)
def rm_stopwords_and_lemmatize(token_array, flag_rm_stop=True, flag_lemmatize=True):
out = []
for token in token_array:
if flag_lemmatize:
token = WordNetLemmatizer().lemmatize(token)
if flag_rm_stop:
if token not in stop_words:
out.append(token)
else:
out.append(token)
return out
def my_tokenizer(doc, flag_rm_stop=True, flag_lemmatize=True):
return rm_stopwords_and_lemmatize(tokenize(doc), flag_rm_stop, flag_lemmatize)
#text = 'I struggled so much with the settings.'
#tokens = tokenize(text)
#print(tokens)
#print(rm_stopwords_and_lemmatize(tokens))
#print(rm_stopwords_and_lemmatize(tokens, flag_rm_stop=False))
#print(rm_stopwords_and_lemmatize(tokens, flag_lemmatize=False))
#print(rm_stopwords_and_lemmatize(tokens, flag_rm_stop=False, flag_lemmatize=False))
#print(my_tokenizer(text))
#define vectorizer parameters
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf_vectorizer = TfidfVectorizer(max_features=1000,
min_df=5,
stop_words='english',
use_idf=True,
tokenizer=my_tokenizer,
token_pattern=r"\b\w[\w']+\b",
ngram_range=(1,2))
tfidf_matrix = tfidf_vectorizer.fit_transform(X_train) #fit the vectorizer to corpus (min = 0.0, max = 1.0)
print (tfidf_matrix.shape)
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
clf = LogisticRegression(random_state=0, solver='lbfgs', multi_class='multinomial')
#clf = RandomForestClassifier()
from sklearn.model_selection import cross_val_score
acc = cross_val_score(clf, tfidf_matrix, y_train, scoring='accuracy', cv=5)
print(acc)
#model = clf.fit(tfidf_matrix, y_train)
from sklearn.decomposition import LatentDirichletAllocation
n_topics = 20
lda = LatentDirichletAllocation(n_components=n_topics,
learning_method='online')
tfidf_matrix_lda = (tfidf_matrix * 100)
tfidf_matrix_lda = tfidf_matrix_lda.astype(int)
lda.fit(tfidf_matrix_lda)
print(lda.components_.shape)
import pyLDAvis
import pyLDAvis.sklearn
pyLDAvis.enable_notebook()
pyLDAvis.sklearn.prepare(lda, tfidf_matrix_lda, tfidf_vectorizer)
```
| github_jupyter |
# Distributed Training of Mask-RCNN in Amazon SageMaker using EFS
This notebook is a step-by-step tutorial on distributed training of [Mask R-CNN](https://arxiv.org/abs/1703.06870) implemented in [TensorFlow](https://www.tensorflow.org/) framework. Mask R-CNN is also referred to as heavy weight object detection model and it is part of [MLPerf](https://www.mlperf.org/training-results-0-6/).
Concretely, we will describe the steps for training [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) and [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) in [Amazon SageMaker](https://aws.amazon.com/sagemaker/) using [Amazon EFS](https://aws.amazon.com/efs/) file-system as data source.
The outline of steps is as follows:
1. Stage COCO 2017 dataset in [Amazon S3](https://aws.amazon.com/s3/)
2. Copy COCO 2017 dataset from S3 to Amazon EFS file-system mounted on this notebook instance
3. Build Docker training image and push it to [Amazon ECR](https://aws.amazon.com/ecr/)
4. Configure data input channels
5. Configure hyper-prarameters
6. Define training metrics
7. Define training job and start training
Before we get started, let us initialize two python variables ```aws_region``` and ```s3_bucket``` that we will use throughout the notebook. The ```s3_bucket``` must be located in the region of this notebook instance.
```
import boto3
session = boto3.session.Session()
aws_region = session.region_name
s3_bucket = # your-s3-bucket-name
try:
s3_client = boto3.client('s3')
response = s3_client.get_bucket_location(Bucket=s3_bucket)
print(f"Bucket region: {response['LocationConstraint']}")
except:
print(f"Access Error: Check if '{s3_bucket}' S3 bucket is in '{aws_region}' region")
```
## Stage COCO 2017 dataset in Amazon S3
We use [COCO 2017 dataset](http://cocodataset.org/#home) for training. We download COCO 2017 training and validation dataset to this notebook instance, extract the files from the dataset archives, and upload the extracted files to your Amazon [S3 bucket](https://docs.aws.amazon.com/AmazonS3/latest/gsg/CreatingABucket.html). The ```prepare-s3-bucket.sh``` script executes this step.
```
!cat ./prepare-s3-bucket.sh
```
Using your *Amazon S3 bucket* as argument, run the cell below. If you have already uploaded COCO 2017 dataset to your Amazon S3 bucket, you may skip this step.
```
%%time
!./prepare-s3-bucket.sh {s3_bucket}
```
## Copy COCO 2017 dataset from S3 to Amazon EFS
Next, we copy COCO 2017 dataset from S3 to EFS file-system. The ```prepare-efs.sh``` script executes this step.
```
!cat ./prepare-efs.sh
```
If you have already copied COCO 2017 dataset from S3 to your EFS file-system, skip this step.
```
%%time
!./prepare-efs.sh {s3_bucket}
```
## Build and push SageMaker training images
For this step, the [IAM Role](https://docs.aws.amazon.com/IAM/latest/UserGuide/id_roles.html) attached to this notebook instance needs full access to Amazon ECR service. If you created this notebook instance using the ```./stack-sm.sh``` script in this repository, the IAM Role attached to this notebook instance is already setup with full access to ECR service.
Below, we have a choice of two different implementations:
1. [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) implementation supports a maximum per-GPU batch size of 1, and does not support mixed precision. It can be used with mainstream TensorFlow releases.
2. [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) is an optimized implementation that supports a maximum batch size of 4 and supports mixed precision. This implementation uses custom TensorFlow ops. The required custom TensorFlow ops are available in [AWS Deep Learning Container](https://github.com/aws/deep-learning-containers/blob/master/available_images.md) images in ```tensorflow-training``` repository with image tag ```1.15.2-gpu-py36-cu100-ubuntu18.04```, or later.
It is recommended that you build and push both SageMaker training images and use either image for training later.
### TensorPack Faster-RCNN/Mask-RCNN
Use ```./container-script-mode/build_tools/build_and_push.sh``` script to build and push the TensorPack Faster-RCNN/Mask-RCNN training image to Amazon ECR.
```
!cat ./container-script-mode/build_tools/build_and_push.sh
%%time
! ./container-script-mode/build_tools/build_and_push.sh {aws_region}
```
Set ```tensorpack_image``` below to Amazon ECR URI of the image you pushed above.
```
tensorpack_image = #<amazon-ecr-uri>
```
### AWS Samples Mask R-CNN
Use ```./container-optimized-script-mode/build_tools/build_and_push.sh``` script to build and push the AWS Samples Mask R-CNN training image to Amazon ECR.
```
!cat ./container-optimized-script-mode/build_tools/build_and_push.sh
```
Using your *AWS region* as argument, run the cell below.
```
%%time
! ./container-optimized-script-mode/build_tools/build_and_push.sh {aws_region}
```
Set ```aws_samples_image``` below to Amazon ECR URI of the image you pushed above.
```
aws_samples_image = #<amazon-ecr-uri>
```
### Upgrade SageMaker Python SDK
If needed, upgrade SageMaker Python SDK.
```
!pip install --upgrade pip
!pip install sagemaker
```
## SageMaker Initialization
We have staged the data and we have built and pushed the training docker image to Amazon ECR. Now we are ready to start using Amazon SageMaker.
```
%%time
import os
import time
import sagemaker
from sagemaker import get_execution_role
from sagemaker.tensorflow.estimator import TensorFlow
role = get_execution_role() # provide a pre-existing role ARN as an alternative to creating a new role
print(f'SageMaker Execution Role:{role}')
client = boto3.client('sts')
account = client.get_caller_identity()['Account']
print(f'AWS account:{account}')
```
Next, we set the Amazon ECR image URI used for training. You saved this URI in a previous step.
```
training_image = # set to tensorpack_image or aws_samples_image
print(f'Training image: {training_image}')
```
## Define SageMaker Data Channels
Next, we define the *train* and *log* data channels using EFS file-system. To do so, we need to specify the EFS file-system id, which is shown in the output of the command below.
```
notebook_attached_efs=!df -kh | grep 'fs-' | sed 's/\(fs-[0-9a-z]*\).*/\1/'
print(f"SageMaker notebook attached EFS: {notebook_attached_efs}")
```
In the cell below, we define the `train` data input channel.
```
from sagemaker.inputs import FileSystemInput
# Specify EFS file system id.
file_system_id = notebook_attached_efs[0]
print(f"EFS file-system-id: {file_system_id}")
# Specify directory path for input data on the file system.
# You need to provide normalized and absolute path below.
file_system_directory_path = '/mask-rcnn/sagemaker/input/train'
print(f'EFS file-system data input path: {file_system_directory_path}')
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'ro'(read-only).
file_system_access_mode = 'ro'
# Specify your file system type
file_system_type = 'EFS'
train = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=file_system_directory_path,
file_system_access_mode=file_system_access_mode)
```
Below we create the log output directory and define the `log` data output channel.
```
# Specify directory path for log output on the EFS file system.
# You need to provide normalized and absolute path below.
# For example, '/mask-rcnn/sagemaker/output/log'
# Log output directory must not exist
file_system_directory_path = f'/mask-rcnn/sagemaker/output/log-{int(time.time())}'
# Create the log output directory.
# EFS file-system is mounted on '$HOME/efs' mount point for this notebook.
home_dir=os.environ['HOME']
local_efs_path = os.path.join(home_dir,'efs', file_system_directory_path[1:])
print(f"Creating log directory on EFS: {local_efs_path}")
assert not os.path.isdir(local_efs_path)
! sudo mkdir -p -m a=rw {local_efs_path}
assert os.path.isdir(local_efs_path)
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'rw'(read-write).
file_system_access_mode = 'rw'
log = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=file_system_directory_path,
file_system_access_mode=file_system_access_mode)
data_channels = {'train': train, 'log': log}
```
Next, we define the model output location in S3. Set ```s3_bucket``` to your S3 bucket name prior to running the cell below.
The model checkpoints, logs and Tensorboard events will be written to the log output directory on the EFS file system you created above. At the end of the model training, they will be copied from the log output directory to the `s3_output_location` defined below.
```
prefix = "mask-rcnn/sagemaker" #prefix in your bucket
s3_output_location = f's3://{s3_bucket}/{prefix}/output'
print(f'S3 model output location: {s3_output_location}')
```
## Configure Hyper-parameters
Next we define the hyper-parameters.
Note, some hyper-parameters are different between the two implementations. The batch size per GPU in TensorPack Faster-RCNN/Mask-RCNN is fixed at 1, but is configurable in AWS Samples Mask-RCNN. The learning rate schedule is specified in units of steps in TensorPack Faster-RCNN/Mask-RCNN, but in epochs in AWS Samples Mask-RCNN.
The detault learning rate schedule values shown below correspond to training for a total of 24 epochs, at 120,000 images per epoch.
<table align='left'>
<caption>TensorPack Faster-RCNN/Mask-RCNN Hyper-parameters</caption>
<tr>
<th style="text-align:center">Hyper-parameter</th>
<th style="text-align:center">Description</th>
<th style="text-align:center">Default</th>
</tr>
<tr>
<td style="text-align:center">mode_fpn</td>
<td style="text-align:left">Flag to indicate use of Feature Pyramid Network (FPN) in the Mask R-CNN model backbone</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">mode_mask</td>
<td style="text-align:left">A value of "False" means Faster-RCNN model, "True" means Mask R-CNN moodel</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">eval_period</td>
<td style="text-align:left">Number of epochs period for evaluation during training</td>
<td style="text-align:center">1</td>
</tr>
<tr>
<td style="text-align:center">lr_schedule</td>
<td style="text-align:left">Learning rate schedule in training steps</td>
<td style="text-align:center">'[240000, 320000, 360000]'</td>
</tr>
<tr>
<td style="text-align:center">batch_norm</td>
<td style="text-align:left">Batch normalization option ('FreezeBN', 'SyncBN', 'GN', 'None') </td>
<td style="text-align:center">'FreezeBN'</td>
</tr>
<tr>
<td style="text-align:center">images_per_epoch</td>
<td style="text-align:left">Images per epoch </td>
<td style="text-align:center">120000</td>
</tr>
<tr>
<td style="text-align:center">data_train</td>
<td style="text-align:left">Training data under data directory</td>
<td style="text-align:center">'coco_train2017'</td>
</tr>
<tr>
<td style="text-align:center">data_val</td>
<td style="text-align:left">Validation data under data directory</td>
<td style="text-align:center">'coco_val2017'</td>
</tr>
<tr>
<td style="text-align:center">resnet_arch</td>
<td style="text-align:left">Must be 'resnet50' or 'resnet101'</td>
<td style="text-align:center">'resnet50'</td>
</tr>
<tr>
<td style="text-align:center">backbone_weights</td>
<td style="text-align:left">ResNet backbone weights</td>
<td style="text-align:center">'ImageNet-R50-AlignPadding.npz'</td>
</tr>
<tr>
<td style="text-align:center">load_model</td>
<td style="text-align:left">Pre-trained model to load</td>
<td style="text-align:center"></td>
</tr>
<tr>
<td style="text-align:center">config:</td>
<td style="text-align:left">Any hyperparamter prefixed with <b>config:</b> is set as a model config parameter</td>
<td style="text-align:center"></td>
</tr>
</table>
<table align='left'>
<caption>AWS Samples Mask-RCNN Hyper-parameters</caption>
<tr>
<th style="text-align:center">Hyper-parameter</th>
<th style="text-align:center">Description</th>
<th style="text-align:center">Default</th>
</tr>
<tr>
<td style="text-align:center">mode_fpn</td>
<td style="text-align:left">Flag to indicate use of Feature Pyramid Network (FPN) in the Mask R-CNN model backbone</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">mode_mask</td>
<td style="text-align:left">A value of "False" means Faster-RCNN model, "True" means Mask R-CNN moodel</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">eval_period</td>
<td style="text-align:left">Number of epochs period for evaluation during training</td>
<td style="text-align:center">1</td>
</tr>
<tr>
<td style="text-align:center">lr_epoch_schedule</td>
<td style="text-align:left">Learning rate schedule in epochs</td>
<td style="text-align:center">'[(16, 0.1), (20, 0.01), (24, None)]'</td>
</tr>
<tr>
<td style="text-align:center">batch_size_per_gpu</td>
<td style="text-align:left">Batch size per gpu ( Minimum 1, Maximum 4)</td>
<td style="text-align:center">4</td>
</tr>
<tr>
<td style="text-align:center">batch_norm</td>
<td style="text-align:left">Batch normalization option ('FreezeBN', 'SyncBN', 'GN', 'None') </td>
<td style="text-align:center">'FreezeBN'</td>
</tr>
<tr>
<td style="text-align:center">images_per_epoch</td>
<td style="text-align:left">Images per epoch </td>
<td style="text-align:center">120000</td>
</tr>
<tr>
<td style="text-align:center">data_train</td>
<td style="text-align:left">Training data under data directory</td>
<td style="text-align:center">'train2017'</td>
</tr>
<tr>
<td style="text-align:center">backbone_weights</td>
<td style="text-align:left">ResNet backbone weights</td>
<td style="text-align:center">'ImageNet-R50-AlignPadding.npz'</td>
</tr>
<tr>
<td style="text-align:center">load_model</td>
<td style="text-align:left">Pre-trained model to load</td>
<td style="text-align:center"></td>
</tr>
<tr>
<td style="text-align:center">config:</td>
<td style="text-align:left">Any hyperparamter prefixed with <b>config:</b> is set as a model config parameter</td>
<td style="text-align:center"></td>
</tr>
</table>
```
hyperparameters = {
"mode_fpn": "True",
"mode_mask": "True",
"eval_period": 1,
"batch_norm": "FreezeBN"
}
```
## Define Training Metrics
Next, we define the regular expressions that SageMaker uses to extract algorithm metrics from training logs and send them to [AWS CloudWatch metrics](https://docs.aws.amazon.com/en_pv/AmazonCloudWatch/latest/monitoring/working_with_metrics.html). These algorithm metrics are visualized in SageMaker console.
```
metric_definitions=[
{
"Name": "fastrcnn_losses/box_loss",
"Regex": ".*fastrcnn_losses/box_loss:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_loss",
"Regex": ".*fastrcnn_losses/label_loss:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/accuracy",
"Regex": ".*fastrcnn_losses/label_metrics/accuracy:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/false_negative",
"Regex": ".*fastrcnn_losses/label_metrics/false_negative:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/fg_accuracy",
"Regex": ".*fastrcnn_losses/label_metrics/fg_accuracy:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/num_fg_label",
"Regex": ".*fastrcnn_losses/num_fg_label:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/accuracy",
"Regex": ".*maskrcnn_loss/accuracy:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/fg_pixel_ratio",
"Regex": ".*maskrcnn_loss/fg_pixel_ratio:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/maskrcnn_loss",
"Regex": ".*maskrcnn_loss/maskrcnn_loss:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/pos_accuracy",
"Regex": ".*maskrcnn_loss/pos_accuracy:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.5",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.5:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.5:0.95",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.5:0\\.95:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.75",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.75:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/large",
"Regex": ".*mAP\\(bbox\\)/large:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/medium",
"Regex": ".*mAP\\(bbox\\)/medium:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/small",
"Regex": ".*mAP\\(bbox\\)/small:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.5",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.5:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.5:0.95",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.5:0\\.95:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.75",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.75:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/large",
"Regex": ".*mAP\\(segm\\)/large:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/medium",
"Regex": ".*mAP\\(segm\\)/medium:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/small",
"Regex": ".*mAP\\(segm\\)/small:\\s*(\\S+).*"
}
]
```
## Define SageMaker Training Job
Next, we use SageMaker [Tensorflow](https://sagemaker.readthedocs.io/en/stable/frameworks/tensorflow/sagemaker.tensorflow.html) API to define a SageMaker Training Job that uses SageMaker script mode.
### Select script
In script-mode, first we have to select an entry point script that acts as interface with SageMaker and launches the training job. For training [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) model, set ```script``` to ```"tensorpack-mask-rcnn.py"```. For training [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) model, set ```script``` to ```"aws-mask-rcnn.py"```.
```
script= # "tensorpack-mask-rcnn.py" or "aws-mask-rcnn.py"
```
### Select distribution mode
We use Message Passing Interface (MPI) to distribute the training job across multiple hosts. The ```custom_mpi_options``` below is only used by [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) model, and can be safely commented out for [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) model.
```
mpi_distribution={'mpi':
{
'enabled': True,
"custom_mpi_options" : "-x TENSORPACK_FP16=1 "
}
}
```
### Define SageMaker Tensorflow Estimator
We recommned using 32 GPUs, so we set ```instance_count=4``` and ```instance_type='ml.p3.16xlarge'```, because there are 8 Tesla V100 GPUs per ```ml.p3.16xlarge``` instance. We recommend using 100 GB [Amazon EBS](https://aws.amazon.com/ebs/) storage volume with each training instance, so we set ```volume_size = 100```.
We run the training job in your private VPC, so we need to set the ```subnets``` and ```security_group_ids``` prior to running the cell below. You may specify multiple subnet ids in the ```subnets``` list. The subnets included in the ```sunbets``` list must be part of the output of ```./stack-sm.sh``` CloudFormation stack script used to create this notebook instance. Specify only one security group id in ```security_group_ids``` list. The security group id must be part of the output of ```./stack-sm.sh``` script.
For ```instance_type``` below, you have the option to use ```ml.p3.16xlarge``` with 16 GB per-GPU memory and 25 Gbs network interconnectivity, or ```ml.p3dn.24xlarge``` with 32 GB per-GPU memory and 100 Gbs network interconnectivity. The ```ml.p3dn.24xlarge``` instance type offers significantly better performance than ```ml.p3.16xlarge``` for Mask R-CNN distributed TensorFlow training.
We use MPI to distribute the training job across multiple hosts.
```
# Give Amazon SageMaker Training Jobs Access to FileSystem Resources in Your Amazon VPC.
security_group_ids = # ['sg-xxxxxxxx']
subnets = # [ 'subnet-xxxxxxx']
sagemaker_session = sagemaker.session.Session(boto_session=session)
mask_rcnn_estimator = TensorFlow(image_uri=training_image,
role=role,
py_version='py3',
instance_count=4,
instance_type='ml.p3.16xlarge',
distribution=mpi_distribution,
entry_point=script,
volume_size = 100,
max_run = 400000,
output_path=s3_output_location,
sagemaker_session=sagemaker_session,
hyperparameters = hyperparameters,
metric_definitions = metric_definitions,
subnets=subnets,
security_group_ids=security_group_ids)
```
### Launch training job
Finally, we launch the SageMaker training job. See ```Training Jobs``` in SageMaker console to monitor the training job.
```
import time
job_name=f'mask-rcnn-efs-script-mode-{int(time.time())}'
print(f"Launching Training Job: {job_name}")
# set wait=True below if you want to print logs in cell output
mask_rcnn_estimator.fit(inputs=data_channels, job_name=job_name, logs="All", wait=False)
```
| github_jupyter |
# Exercise 03 - Booleans and Conditionals
## 1. Simple Function with Conditionals
Many programming languages have [sign](https://en.wikipedia.org/wiki/Sign_function) available as a built-in function. Python does not, but we can define our own!
In the cell below, define a function called `sign` which takes a numerical argument and returns -1 if it's negative, 1 if it's positive, and 0 if it's 0.
```
# Your code goes here. Define a function called 'sign'
def sign(num) {
return num % 1
}
```
## 2. Singular vs Plural Nouns
We've decided to add "print" to our `to_smash` function from Exercise 02
```
def to_smash(total_candies):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
print("Splitting", total_candies, "candies")
return total_candies % 3
to_smash(91)
```
What happens if we call it with `total_candies = 1`?
```
to_smash(1)
```
**Wrong grammar there!**
Modify the definition in the cell below to correct the grammar of our print statement.
**Your Task:**
> If there's only one candy, we should use the singular "candy" instead of the plural "candies"
```
def to_smash(total_candies):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
print("Splitting", total_candies, "candies")
return total_candies % 3
to_smash(91)
to_smash(1)
```
## 3. Checking weather again
In the main lesson we talked about deciding whether we're prepared for the weather. I said that I'm safe from today's weather if...
- I have an umbrella...
- or if the rain isn't too heavy and I have a hood...
- otherwise, I'm still fine unless it's raining *and* it's a workday
The function below uses our first attempt at turning this logic into a Python expression. I claimed that there was a bug in that code. Can you find it?
To prove that `prepared_for_weather` is buggy, come up with a set of inputs where it returns the wrong answer.
```
def prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday):
# Don't change this code. Our goal is just to find the bug, not fix it!
return have_umbrella or rain_level < 5 and have_hood or not rain_level > 0 and is_workday
# Change the values of these inputs so they represent a case where prepared_for_weather
# returns the wrong answer.
have_umbrella = True
rain_level = 0.0
have_hood = True
is_workday = True
# Check what the function returns given the current values of the variables above
actual = prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday)
print(actual)
```
## 4. Start being lazy...
The function `is_negative` below is implemented correctly
- It returns True if the given number is negative and False otherwise.
However, it's more verbose than it needs to be. We can actually reduce the number of lines of code in this function by *75%* while keeping the same behaviour.
**Your task:**
> See if you can come up with an equivalent body that uses just **one line** of code, and put it in the function `concise_is_negative`. (HINT: you don't even need Python's ternary syntax)
```
def is_negative(number):
if number < 0:
return True
else:
return False
def concise_is_negative(number):
pass # Your code goes here (try to keep it to one line!)
```
## 5. Adding Toppings
The boolean variables `ketchup`, `mustard` and `onion` represent whether a customer wants a particular topping on their hot dog. We want to implement a number of boolean functions that correspond to some yes-or-no questions about the customer's order. For example:
```
def onionless(ketchup, mustard, onion):
"""Return whether the customer doesn't want onions.
"""
return not onion
```
**Your task:**
> For each of the remaining functions, fill in the body to match the English description in the docstring.
```
def wants_all_toppings(ketchup, mustard, onion):
"""Return whether the customer wants "the works" (all 3 toppings)
"""
pass
def wants_plain_hotdog(ketchup, mustard, onion):
"""Return whether the customer wants a plain hot dog with no toppings.
"""
pass
def exactly_one_sauce(ketchup, mustard, onion):
"""Return whether the customer wants either ketchup or mustard, but not both.
(You may be familiar with this operation under the name "exclusive or")
"""
pass
```
## 6. <span title="A bit spicy" style="color: darkgreen ">🌶️</span>
We’ve seen that calling `bool()` on an integer returns `False` if it’s equal to 0 and `True` otherwise. What happens if we call `int()` on a bool? Try it out in the notebook cell below.
Can you take advantage of this to write a succinct function that corresponds to the English sentence "*Does the customer want exactly one topping?*"?
> *HINT*: You may have already found that `int(True)` is `1`, and `int(False)` is `0`. Think about what kinds of basic arithmetic operations you might want to perform on ketchup, mustard, and onion after converting them to integers.
```
def exactly_one_topping(ketchup, mustard, onion):
"""Return whether the customer wants exactly one of the three available toppings
on their hot dog.
"""
pass
```
# Keep Going 💪
| github_jupyter |
<a href="https://colab.research.google.com/github/neurorishika/PSST/blob/master/Tutorial/Day%205%20Optimal%20Mind%20Control/Day%205.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a> <a href="https://kaggle.com/kernels/welcome?src=https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/Day%205.ipynb" target="_parent"><img src="https://kaggle.com/static/images/open-in-kaggle.svg" alt="Open in Kaggle"/></a>
## Day 5: Optimal Mind Control
Welcome to Day 5! Now that we can simulate a model network of conductance-based neurons, we discuss the limitations of our approach and attempts to work around these issues.
### Memory Management
Using Python and TensorFlow allowed us to write code that is readable, parallizable and scalable across a variety of computational devices. However, our implementation is very memory intensive. The iterators in TensorFlow do not follow the normal process of memory allocation and garbage collection. Since, TensorFlow is designed to work on diverse hardware like GPUs, TPUs and distributed platforms, memory allocation is done adaptively during the TensorFlow session and not cleared until the Python kernel has stopped execution. The memory used increases linearly with time as the state matrix is computed recursively by the tf.scan function. The maximum memory used by the computational graph is 2 times the total state matrix size at the point when the computation finishes and copies the final data into the memory. The larger the network and longer the simulation, the larger the solution matrix. Each run is limited by the total available memory. For a system with a limited memory of K bytes, The length of a given simulation (L timesteps) of a given network (N differential equations) with 64-bit floating-point precision will follow:
$$2\times64\times L\times N=K$$
That is, for any given network, our maximum simulation length is limited. One way to improve our maximum length is to divide the simulation into smaller batches. There will be a small queuing time between batches, which will slow down our code by a small amount but we will be able to simulate longer times. Thus, if we split the simulation into K sequential batches, the maximum memory for the simulation becomes $(1+\frac{1}{K})$ times the total matrix size. Thus the memory relation becomes:
$$\Big(1+\frac{1}{K}\Big)\times64\times L\times N=K$$
This way, we can maximize the length of out simulation that we can run in a single python kernel.
Let us implement this batch system for our 3 neuron feed-forward model.
### Implementing the Model
To improve the readability of our code we separate the integrator into a independent import module. The integrator code was placed in a file called tf integrator.py. The file must be present in the same directory as the implementation of the model.
Note: If you are using Jupyter Notebook, remember to remove the %matplotlib inline command as it is specific to jupyter.
#### Importing tf_integrator and other requirements
Once the Integrator is saved in tf_integrator.py in the same directory as the Notebook, we can start importing the essentials including the integrator.
**WARNING: If you are running this notebook using Kaggle, make sure you have logged in to your verified Kaggle account and enabled Internet Access for the kernel. For instructions on enabling Internet on Kaggle Kernels, visit: https://www.kaggle.com/product-feedback/63544**
```
#@markdown Import required files and code from previous tutorials
!wget --no-check-certificate \
"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/tf_integrator.py" \
-O "tf_integrator.py"
!wget --no-check-certificate \
"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/call.py" \
-O "call.py"
!wget --no-check-certificate \
"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/run.py" \
-O "run.py"
import numpy as np
import tf_integrator as tf_int
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow.compat.v1 as tf
tf.disable_eager_execution()
```
### Recall the Model
For implementing a Batch system, we do not need to change how we construct our model only how we execute it.
#### Step 1: Initialize Parameters and Dynamical Equations; Define Input
```
n_n = 3 # Number of simultaneous neurons to simulate
sim_res = 0.01 # Time Resolution of the Simulation
sim_time = 700 # Length of the Simulation
t = np.arange(0,sim_time,sim_res) # Time points at which to simulate the network
# Acetylcholine
ach_mat = np.zeros((n_n,n_n)) # Ach Synapse Connectivity Matrix
ach_mat[1,0]=1
## PARAMETERS FOR ACETYLCHLOLINE SYNAPSES ##
n_ach = int(np.sum(ach_mat)) # Number of Acetylcholine (Ach) Synapses
alp_ach = [10.0]*n_ach # Alpha for Ach Synapse
bet_ach = [0.2]*n_ach # Beta for Ach Synapse
t_max = 0.3 # Maximum Time for Synapse
t_delay = 0 # Axonal Transmission Delay
A = [0.5]*n_n # Synaptic Response Strength
g_ach = [0.35]*n_n # Ach Conductance
E_ach = [0.0]*n_n # Ach Potential
# GABAa
gaba_mat = np.zeros((n_n,n_n)) # GABAa Synapse Connectivity Matrix
gaba_mat[2,1] = 1
## PARAMETERS FOR GABAa SYNAPSES ##
n_gaba = int(np.sum(gaba_mat)) # Number of GABAa Synapses
alp_gaba = [10.0]*n_gaba # Alpha for GABAa Synapse
bet_gaba = [0.16]*n_gaba # Beta for GABAa Synapse
V0 = [-20.0]*n_n # Decay Potential
sigma = [1.5]*n_n # Decay Time Constant
g_gaba = [0.8]*n_n # fGABA Conductance
E_gaba = [-70.0]*n_n # fGABA Potential
## Storing Firing Thresholds ##
F_b = [0.0]*n_n # Fire threshold
def I_inj_t(t):
"""
This function returns the external current to be injected into the network at any time step from the current_input matrix.
Parameters:
-----------
t: float
The time at which the current injection is being performed.
"""
# Turn indices to integer and extract from matrix
index = tf.cast(t/sim_res,tf.int32)
return tf.constant(current_input.T,dtype=tf.float64)[index]
## Acetylcholine Synaptic Current ##
def I_ach(o,V):
"""
This function returns the synaptic current for the Acetylcholine (Ach) synapses for each neuron.
Parameters:
-----------
o: float
The fraction of open acetylcholine channels for each synapse.
V: float
The membrane potential of the postsynaptic neuron.
"""
o_ = tf.constant([0.0]*n_n**2,dtype=tf.float64) # Initialize the flattened matrix to store the synaptic open fractions
ind = tf.boolean_mask(tf.range(n_n**2),ach_mat.reshape(-1) == 1) # Get the indices of the synapses that exist
o_ = tf.tensor_scatter_nd_update(o_,tf.reshape(ind,[-1,1]),o) # Update the flattened open fraction matrix
o_ = tf.transpose(tf.reshape(o_,(n_n,n_n))) # Reshape and Transpose the matrix to be able to multiply it with the conductance matrix
return tf.reduce_sum(tf.transpose((o_*(V-E_ach))*g_ach),1) # Calculate the synaptic current
## GABAa Synaptic Current ##
def I_gaba(o,V):
"""
This function returns the synaptic current for the GABA synapses for each neuron.
Parameters:
-----------
o: float
The fraction of open GABA channels for each synapse.
V: float
The membrane potential of the postsynaptic neuron.
"""
o_ = tf.constant([0.0]*n_n**2,dtype=tf.float64) # Initialize the flattened matrix to store the synaptic open fractions
ind = tf.boolean_mask(tf.range(n_n**2),gaba_mat.reshape(-1) == 1) # Get the indices of the synapses that exist
o_ = tf.tensor_scatter_nd_update(o_,tf.reshape(ind,[-1,1]),o) # Update the flattened open fraction matrix
o_ = tf.transpose(tf.reshape(o_,(n_n,n_n))) # Reshape and Transpose the matrix to be able to multiply it with the conductance matrix
return tf.reduce_sum(tf.transpose((o_*(V-E_gaba))*g_gaba),1) # Calculate the synaptic current
## Other Currents ##
def I_K(V, n):
"""
This function determines the K-channel current.
Parameters:
-----------
V: float
The membrane potential.
n: float
The K-channel gating variable n.
"""
return g_K * n**4 * (V - E_K)
def I_Na(V, m, h):
"""
This function determines the Na-channel current.
Parameters:
-----------
V: float
The membrane potential.
m: float
The Na-channel gating variable m.
h: float
The Na-channel gating variable h.
"""
return g_Na * m**3 * h * (V - E_Na)
def I_L(V):
"""
This function determines the leak current.
Parameters:
-----------
V: float
The membrane potential.
"""
return g_L * (V - E_L)
def dXdt(X, t):
"""
This function determines the derivatives of the membrane voltage and gating variables for n_n neurons.
Parameters:
-----------
X: float
The state vector given by the [V1,V2,...,Vn_n,m1,m2,...,mn_n,h1,h2,...,hn_n,n1,n2,...,nn_n] where
Vx is the membrane potential for neuron x
mx is the Na-channel gating variable for neuron x
hx is the Na-channel gating variable for neuron x
nx is the K-channel gating variable for neuron x.
t: float
The time points at which the derivatives are being evaluated.
"""
V = X[:1*n_n] # First n_n values are Membrane Voltage
m = X[1*n_n:2*n_n] # Next n_n values are Sodium Activation Gating Variables
h = X[2*n_n:3*n_n] # Next n_n values are Sodium Inactivation Gating Variables
n = X[3*n_n:4*n_n] # Next n_n values are Potassium Gating Variables
o_ach = X[4*n_n : 4*n_n + n_ach] # Next n_ach values are Acetylcholine Synapse Open Fractions
o_gaba = X[4*n_n + n_ach : 4*n_n + n_ach + n_gaba] # Next n_gaba values are GABAa Synapse Open Fractions
fire_t = X[-n_n:] # Last n_n values are the last fire times as updated by the modified integrator
dVdt = (I_inj_t(t) - I_Na(V, m, h) - I_K(V, n) - I_L(V) - I_ach(o_ach,V) - I_gaba(o_gaba,V)) / C_m # The derivative of the membrane potential
## Updation for gating variables ##
m0,tm,h0,th = Na_prop(V) # Calculate the dynamics of the Na-channel gating variables for all n_n neurons
n0,tn = K_prop(V) # Calculate the dynamics of the K-channel gating variables for all n_n neurons
dmdt = - (1.0/tm)*(m-m0) # The derivative of the Na-channel gating variable m for all n_n neurons
dhdt = - (1.0/th)*(h-h0) # The derivative of the Na-channel gating variable h for all n_n neurons
dndt = - (1.0/tn)*(n-n0) # The derivative of the K-channel gating variable n for all n_n neurons
## Updation for o_ach ##
A_ = tf.constant(A,dtype=tf.float64) # Get the synaptic response strengths of the pre-synaptic neurons
Z_ = tf.zeros(tf.shape(A_),dtype=tf.float64) # Create a zero matrix of the same size as A_
T_ach = tf.where(tf.logical_and(tf.greater(t,fire_t+t_delay),tf.less(t,fire_t+t_max+t_delay)),A_,Z_) # Find which synapses would have received an presynaptic spike in the past window and assign them the corresponding synaptic response strength
T_ach = tf.multiply(tf.constant(ach_mat,dtype=tf.float64),T_ach) # Find the postsynaptic neurons that would have received an presynaptic spike in the past window
T_ach = tf.boolean_mask(tf.reshape(T_ach,(-1,)),ach_mat.reshape(-1) == 1) # Get the pre-synaptic activation function for only the existing synapses
do_achdt = alp_ach*(1.0-o_ach)*T_ach - bet_ach*o_ach # Calculate the derivative of the open fraction of the acetylcholine synapses
## Updation for o_gaba ##
T_gaba = 1.0/(1.0+tf.exp(-(V-V0)/sigma)) # Calculate the presynaptic activation function for all n_n neurons
T_gaba = tf.multiply(tf.constant(gaba_mat,dtype=tf.float64),T_gaba) # Find the postsynaptic neurons that would have received an presynaptic spike in the past window
T_gaba = tf.boolean_mask(tf.reshape(T_gaba,(-1,)),gaba_mat.reshape(-1) == 1) # Get the pre-synaptic activation function for only the existing synapses
do_gabadt = alp_gaba*(1.0-o_gaba)*T_gaba - bet_gaba*o_gaba # Calculate the derivative of the open fraction of the GABAa synapses
## Updation for fire times ##
dfdt = tf.zeros(tf.shape(fire_t),dtype=fire_t.dtype) # zero change in fire_t as it will be updated by the modified integrator
out = tf.concat([dVdt,dmdt,dhdt,dndt,do_achdt,do_gabadt,dfdt],0) # Concatenate the derivatives of the membrane potential, gating variables, and open fractions
return out
def K_prop(V):
"""
This function determines the K-channel gating dynamics.
Parameters:
-----------
V: float
The membrane potential.
"""
T = 22 # Temperature
phi = 3.0**((T-36.0)/10) # Temperature-correction factor
V_ = V-(-50) # Voltage baseline shift
alpha_n = 0.02*(15.0 - V_)/(tf.exp((15.0 - V_)/5.0) - 1.0) # Alpha for the K-channel gating variable n
beta_n = 0.5*tf.exp((10.0 - V_)/40.0) # Beta for the K-channel gating variable n
t_n = 1.0/((alpha_n+beta_n)*phi) # Time constant for the K-channel gating variable n
n_0 = alpha_n/(alpha_n+beta_n) # Steady-state value for the K-channel gating variable n
return n_0, t_n
def Na_prop(V):
"""
This function determines the Na-channel gating dynamics.
Parameters:
-----------
V: float
The membrane potential.
"""
T = 22 # Temperature
phi = 3.0**((T-36)/10) # Temperature-correction factor
V_ = V-(-50) # Voltage baseline shift
alpha_m = 0.32*(13.0 - V_)/(tf.exp((13.0 - V_)/4.0) - 1.0) # Alpha for the Na-channel gating variable m
beta_m = 0.28*(V_ - 40.0)/(tf.exp((V_ - 40.0)/5.0) - 1.0) # Beta for the Na-channel gating variable m
alpha_h = 0.128*tf.exp((17.0 - V_)/18.0) # Alpha for the Na-channel gating variable h
beta_h = 4.0/(tf.exp((40.0 - V_)/5.0) + 1.0) # Beta for the Na-channel gating variable h
t_m = 1.0/((alpha_m+beta_m)*phi) # Time constant for the Na-channel gating variable m
t_h = 1.0/((alpha_h+beta_h)*phi) # Time constant for the Na-channel gating variable h
m_0 = alpha_m/(alpha_m+beta_m) # Steady-state value for the Na-channel gating variable m
h_0 = alpha_h/(alpha_h+beta_h) # Steady-state value for the Na-channel gating variable h
return m_0, t_m, h_0, t_h
# Initializing the Parameters
C_m = [1.0]*n_n # Membrane capacitances
g_K = [10.0]*n_n # K-channel conductances
E_K = [-95.0]*n_n # K-channel reversal potentials
g_Na = [100]*n_n # Na-channel conductances
E_Na = [50]*n_n # Na-channel reversal potentials
g_L = [0.15]*n_n # Leak conductances
E_L = [-55.0]*n_n # Leak reversal potentials
# Creating the Current Input
current_input= np.zeros((n_n,t.shape[0])) # The current input to the network
current_input[0,int(100/sim_res):int(200/sim_res)] = 2.5
current_input[0,int(300/sim_res):int(400/sim_res)] = 5.0
current_input[0,int(500/sim_res):int(600/sim_res)] = 7.5
```
#### Step 2: Define the Initial Condition of the Network and Add some Noise to the initial conditions
```
# Initializing the State Vector and adding 1% noise
state_vector = [-71]*n_n+[0,0,0]*n_n+[0]*n_ach+[0]*n_gaba+[-9999999]*n_n
state_vector = np.array(state_vector)
state_vector = state_vector + 0.01*state_vector*np.random.normal(size=state_vector.shape)
```
#### Step 3: Splitting Time Series into independent batches and Run Each Batch Sequentially
Since we will be dividing the computation into batches, we have to split the time array such that for each new call, the final state vector of the last batch will be the initial condition for the current batch. The function $np.array\_split()$ splits the array into non-overlapping vectors. Therefore, we append the last time of the previous batch to the beginning of the current time array batch.
```
# Define the Number of Batches
n_batch = 2
# Split t array into batches using numpy
t_batch = np.array_split(t,n_batch)
# Iterate over the batches of time array
for n,i in enumerate(t_batch):
# Inform start of Batch Computation
print("Batch",(n+1),"Running...",end="")
# In np.array_split(), the split edges are present in only one array and since
# our initial vector to successive calls is corresposnding to the last output
# our first element in the later time array should be the last element of the
# previous output series, Thus, we append the last time to the beginning of
# the current time array batch.
if n>0:
i = np.append(i[0]-sim_res,i)
# Set state_vector as the initial condition
init_state = tf.constant(state_vector, dtype=tf.float64)
# Create the Integrator computation graph over the current batch of t array
tensor_state = tf_int.odeint(dXdt, init_state, i, n_n, F_b)
# Initialize variables and run session
with tf.Session() as sess:
tf.global_variables_initializer().run()
state = sess.run(tensor_state)
sess.close()
# Reset state_vector as the last element of output
state_vector = state[-1,:]
# Save the output of the simulation to a binary file
np.save("part_"+str(n+1),state)
# Clear output
state=None
print("Finished")
```
#### Putting the Output Together
The output from our batch implementation is a set of binary files that store parts of our total simulation. To get the overall output we have to stitch them back together.
```
overall_state = []
# Iterate over the generated output files
for n,i in enumerate(["part_"+str(n+1)+".npy" for n in range(n_batch)]):
# Since the first element in the series was the last output, we remove them
if n>0:
overall_state.append(np.load(i)[1:,:])
else:
overall_state.append(np.load(i))
# Concatenate all the matrix to get a single state matrix
overall_state = np.concatenate(overall_state)
```
#### Visualizing the Overall Data
Finally, we plot the same voltage traces of the 3 neurons from Day 4 as a Voltage vs Time heatmap. While this visualization may seem unnecessary for just 3 neurons, it becomes an useful tool when on visualizes the dynamics of a large network of neurons as illustrated in the Example Implementation of the Locust Antennal Lobe.
```
# Plot the voltage traces of the three neurons
plt.figure(figsize=(12,6))
sns.heatmap(overall_state[::100,:3].T,xticklabels=100,yticklabels=5,cmap='RdBu_r')
plt.xlabel("Time (in ms)")
plt.ylabel("Neuron Number")
plt.title("Voltage vs Time Heatmap for Projection Neurons (PNs)")
plt.tight_layout()
plt.show()
```
By this method, we have maximized the usage of our available memory but we can go further and develop a method to allow indefinitely long simulation. The issue behind this entire algorithm is that the memory is not cleared until the python kernel finishes. One way to overcome this is to save the parameters of the model (such as connectivity matrix) and the state vector in a file, and start a new python kernel from a python script to compute successive batches. This way after each large batch, the memory gets cleaned. By combining the previous batch implementation and this system, we can maximize our computability.
### Implementing a Runner and a Caller
Firstly, we have to create an implementation of the model that takes in previous input as current parameters. Thus, we create a file, which we call "run.py" that takes an argument ie. the current batch number. The implementation for "run.py" is mostly same as the above model but there is a small difference.
When the batch number is 0, we initialize all variable parameters and save them, but otherwise we use the saved values. The parameters we save include: Acetylcholine Matrix, GABAa Matrix and Final/Initial State Vector. It will also save the files with both batch number and sub-batch number listed.
The time series will be created and split initially by the caller, which we call "call.py", and stored in a file. Each execution of the Runner will extract its relevant time series and compute on it.
#### Implementing the Runner code
"run.py" is essentially identical to the batch-implemented model we developed above with the changes described below:
```
# Additional Imports #
import sys
# Duration of Simulation #
# t = np.arange(0,sim_time,sim_res)
t = np.load("time.npy",allow_pickle=True)[int(sys.argv[1])] # get first argument to run.py
# Connectivity Matrix Definitions #
if sys.argv[1] == '0':
ach_mat = np.zeros((n_n,n_n)) # Ach Synapse Connectivity Matrix
ach_mat[1,0]=1 # If connectivity is random, once initialized it will be the same.
np.save("ach_mat",ach_mat)
else:
ach_mat = np.load("ach_mat.npy")
if sys.argv[1] == '0':
gaba_mat = np.zeros((n_n,n_n)) # GABAa Synapse Connectivity Matrix
gaba_mat[2,1] = 1 # If connectivity is random, once initialized it will be the same.
np.save("gaba_mat",gaba_mat)
else:
gaba_mat = np.load("gaba_mat.npy")
# Current Input Definition #
if sys.argv[1] == '0':
current_input= np.zeros((n_n,int(sim_time/sim_res)))
current_input[0,int(100/sim_res):int(200/sim_res)] = 2.5
current_input[0,int(300/sim_res):int(400/sim_res)] = 5.0
current_input[0,int(500/sim_res):int(600/sim_res)] = 7.5
np.save("current_input",current_input)
else:
current_input = np.load("current_input.npy")
# State Vector Definition #
if sys.argv[1] == '0':
state_vector = [-71]*n_n+[0,0,0]*n_n+[0]*n_ach+[0]*n_gaba+[-9999999]*n_n
state_vector = np.array(state_vector)
state_vector = state_vector + 0.01*state_vector*np.random.normal(size=state_vector.shape)
np.save("state_vector",state_vector)
else:
state_vector = np.load("state_vector.npy")
# Saving of Output #
# np.save("part_"+str(n+1),state)
np.save("batch"+str(int(sys.argv[1])+1)+"_part_"+str(n+1),state)
```
#### Implementing the Caller code
The caller will create the time series, split it and use python subprocess module to call "run.py" with appropriate arguments. The code for "call.py" is given below.
```
from subprocess import call
import numpy as np
total_time = 700
n_splits = 2
time = np.split(np.arange(0,total_time,0.01),n_splits)
# Append the last time point to the beginning of the next batch
for n,i in enumerate(time):
if n>0:
time[n] = np.append(i[0]-0.01,i)
np.save("time",time)
# call successive batches with a new python subprocess and pass the batch number
for i in range(n_splits):
call(['python','run.py',str(i)])
print("Simulation Completed.")
```
#### Using call.py
```
!python call.py
```
#### Combining all Data
Just like we merged all the batches, we merge all the sub-batches and batches.
```
n_splits = 2
n_batch = 2
overall_state = []
# Iterate over the generated output files
for n,i in enumerate(["batch"+str(x+1) for x in range(n_splits)]):
for m,j in enumerate(["_part_"+str(x+1)+".npy" for x in range(n_batch)]):
# Since the first element in the series was the last output, we remove them
if n>0 and m>0:
overall_state.append(np.load(i+j)[1:,:])
else:
overall_state.append(np.load(i+j))
# Concatenate all the matrix to get a single state matrix
overall_state = np.concatenate(overall_state)
# Plot the simulation results
plt.figure(figsize=(12,6))
sns.heatmap(overall_state[::100,:3].T,xticklabels=100,yticklabels=5,cmap='RdBu_r')
plt.xlabel("Time (in ms)")
plt.ylabel("Neuron Number")
plt.title("Voltage vs Time Heatmap for Projection Neurons (PNs)")
plt.tight_layout()
plt.show()
```
| github_jupyter |
# II - Wavefronts and optical systems
First let's import HCIPy, and a few supporting libraries:
```
from hcipy import *
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
```
Wavefronts in HCIPy are monochromatic. They consist of an electric field (as an HCIP `Field`), and a wavelength. If broadband images are needed, multiple `Wavefront`s must be constructed and propagated through the optical system, sampling the required wavelength range. Let us construct a `Wavefront`.
```
pupil_grid = make_pupil_grid(1024)
aperture = circular_aperture(1)(pupil_grid)
wavefront = Wavefront(aperture, 1)
```
A note must be made at this point regarding units. HCIPy is averse w.r.t. the used units. If the user fills in all quantities in SI, then all calculations in HCIPy will be returned in SI units. This allows the user to use any unit he/she wants, while still being able to seamlessly use dimensionless quantities. Ie. the convention that is used in this document, is that if the diameter of the aperture is 1, the wavelength is 1, and the focal length is 1 as well, then the focal-plane will be given in $\lambda/D$.
To propagate this wavefront to the focal plane, we first need to construct a grid on which the focal plane will be sampled:
```
focal_grid = make_focal_grid(pupil_grid, 8, 16)
```
This constructs a `Grid` with 8 samples per $\lambda/D$ and a 16 $\lambda/D$ radius field of view (so 32 $\lambda/D$ diameter field of view). Now we can construct a Fraunhofer propagator that can actually propagate the light to the focal plane.
```
prop = FraunhoferPropagator(pupil_grid, focal_grid)
img = prop.forward(wavefront)
imshow_field(np.log10(img.intensity / img.intensity.max()), vmin=-5)
plt.colorbar()
plt.show()
```
All Fourier transforms concerning the propagation are done internally. In this case a Matrix Fourier transform was used, as it was deemed quicker than a Fast Fourier transform in this case. Also note that when defining the propagator, we didn't pass the wavelength of the wavefront. This wavelength is taken from the `Wavefront` object during the propagation.
Also note that a `Wavefront` supports many properties to make it easier to use. One that we used above is `Wavefront.intensity`, but others exist as well: for example `Wavefront.phase` and `Wavefront.amplitude`, which yield the phase and amplitude of the electric field respectively. All these properties are returned as `Field`s, and can therefore be shown using `imshow_field()`.
For a more interesting results, let's do a propagation with physical quantities. We calculate the intensity pattern of a circular aperture with a diameter of 1cm, after a free-space propagation of 2m, at a wavelength of 500nm.
```
pupil_grid_2 = make_pupil_grid(1024, 0.015)
aperture_2 = circular_aperture(0.01)(pupil_grid_2)
fresnel_prop = FresnelPropagator(pupil_grid_2, 2)
wf = Wavefront(aperture_2, 500e-9)
img = fresnel_prop(wf)
imshow_field(img.intensity)
plt.show()
```
The propagators shown previously are part of a larger group of optical elements. All `OpticalElement`s can propagate a `Wavefront` through them. Examples include simple `Apodizer`s, which act as an infinitely-thin screen with a (complex) transmission. A little more complicated example is `SurfaceAberration`, which simulates a surface error with a power-law PSD (power spectral density). Optical elements can be linked to represent more complicated optical systems.
```
aberration = SurfaceAberration(pupil_grid, 0.25, 1)
wf = Wavefront(aperture)
img = prop(aberration(wf))
imshow_field(np.log10(img.intensity / img.intensity.max()), vmin=-5)
plt.colorbar()
plt.show()
```
These simple optical elements can be combined into more complicated optical systems. These include full wavefront sensor implementations and coronagraphs. Both of these will be handled in later sections.
To convert a `Wavefront` into an observed image, one can simply use the `Wavefront.power` attribute, which is the `Wavefront.intensity` multiplied by the weight at each pixel. If one wants to use a more complicated detector model, HCIPy supplies a `Detector` class. and its derivatives. A detector uses an integration/readout scheme:
```
flat_field = 0.01
dark = 10
detector = NoisyDetector(focal_grid, dark_current_rate=dark, flat_field=flat_field)
wf.total_power = 5000
img = prop(aberration(wf))
detector.integrate(img, 0.5)
image = detector.read_out()
imshow_field(np.log10(image), vmax=np.log10(image).max(), vmin=0)
plt.colorbar()
plt.show()
```
| github_jupyter |
# 📝 Exercise M6.03
This exercise aims at verifying if AdaBoost can over-fit.
We will make a grid-search and check the scores by varying the
number of estimators.
We will first load the California housing dataset and split it into a
training and a testing set.
```
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
data, target = fetch_california_housing(return_X_y=True, as_frame=True)
target *= 100 # rescale the target in k$
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=0, test_size=0.5)
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
Then, create an `AdaBoostRegressor`. Use the function
`sklearn.model_selection.validation_curve` to get training and test scores
by varying the number of estimators. Use the mean absolute error as a metric
by passing `scoring="neg_mean_absolute_error"`.
*Hint: vary the number of estimators between 1 and 60.*
```
# Write your code here.
from sklearn.ensemble import AdaBoostRegressor
from sklearn.model_selection import validation_curve
import pandas as pd
import numpy as np
n_estimators = np.arange(1, 100, 2)
n_estimators
model = AdaBoostRegressor()
model.get_params()
train_scores, test_scores = validation_curve(
model, data, target, param_name="n_estimators", param_range=n_estimators,
cv=5, scoring="neg_mean_absolute_error", n_jobs=2)
train_errors, test_errors = -train_scores, -test_scores
```
Plot both the mean training and test errors. You can also plot the
standard deviation of the errors.
*Hint: you can use `plt.errorbar`.*
```
# Write your code here.
from matplotlib import pyplot as plt
plt.errorbar(n_estimators, train_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Training error")
plt.errorbar(n_estimators, test_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Testing error")
```
Plotting the validation curve, we can see that AdaBoost is not immune against
overfitting. Indeed, there is an optimal number of estimators to be found.
Adding too many estimators is detrimental for the statistical performance of
the model.
Repeat the experiment using a random forest instead of an AdaBoost regressor.
```
# Write your code here.
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor()
train_scores, test_scores = validation_curve(
model, data, target, param_name="n_estimators", param_range=n_estimators,
cv=5, scoring="neg_mean_absolute_error", n_jobs=2)
# PLOT
plt.errorbar(n_estimators, train_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Training error")
plt.errorbar(n_estimators, test_errors.mean(axis=1),
yerr=train_errors.std(axis=1), label="Testing error")
```
| github_jupyter |
# SmallPebble
[](https://github.com/sradc/smallpebble/commits/)
**Project status: unstable.**
<br><p align="center"><img src="https://raw.githubusercontent.com/sradc/SmallPebble/master/pebbles.jpg"/></p><br>
SmallPebble is a minimal automatic differentiation and deep learning library written from scratch in [Python](https://www.python.org/), using [NumPy](https://numpy.org/)/[CuPy](https://cupy.dev/).
The implementation is relatively small, and mainly in the file: [smallpebble.py](https://github.com/sradc/SmallPebble/blob/master/smallpebble/smallpebble.py). To help understand it, check out [this](https://sidsite.com/posts/autodiff/) introduction to autodiff, which presents an autodiff framework that works in the same way as SmallPebble (except using scalars instead of NumPy arrays).
SmallPebble's *raison d'etre* is to be a simplified deep learning implementation,
for those who want to learn what’s under the hood of deep learning frameworks.
However, because it is written in terms of vectorised NumPy/CuPy operations,
it performs well enough for non-trivial models to be trained using it.
**Highlights**
- Relatively simple implementation.
- Can run on GPU, using CuPy.
- Various operations, such as matmul, conv2d, maxpool2d.
- Array broadcasting support.
- Eager or lazy execution.
- Powerful API for creating models.
- It's easy to add new SmallPebble functions.
**Notes**
Graphs are built implicitly via Python objects referencing Python objects.
When `get_gradients` is called, autodiff is carried out on the whole sub-graph. The default array library is NumPy.
---
**Read on to see:**
- Example models created and trained using SmallPebble.
- A brief guide to using SmallPebble.
```
import matplotlib.pyplot as plt
import numpy as np
from tqdm.notebook import tqdm
import smallpebble as sp
from smallpebble.misc import load_data
```
## Training a neural network to classify handwritten digits (MNIST)
```
"Load the dataset, and create a validation set."
X_train, y_train, _, _ = load_data('mnist') # load / download from openml.org
X_train = X_train/255 # normalize
# Seperate out data for validation.
X = X_train[:50_000, ...]
y = y_train[:50_000]
X_eval = X_train[50_000:60_000, ...]
y_eval = y_train[50_000:60_000]
"Plot, to check we have the right data."
plt.figure(figsize=(5,5))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(X_train[i,:].reshape(28,28), cmap='gray', vmin=0, vmax=1)
plt.show()
"Create a model, with two fully connected hidden layers."
X_in = sp.Placeholder()
y_true = sp.Placeholder()
h = sp.linearlayer(28*28, 100)(X_in)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.linearlayer(100, 100)(h)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.linearlayer(100, 10)(h)
y_pred = sp.Lazy(sp.softmax)(h)
loss = sp.Lazy(sp.cross_entropy)(y_pred, y_true)
learnables = sp.get_learnables(y_pred)
loss_vals = []
validation_acc = []
"Train model, while measuring performance on the validation dataset."
NUM_ITERS = 300
BATCH_SIZE = 200
eval_batch = sp.batch(X_eval, y_eval, BATCH_SIZE)
adam = sp.Adam() # Adam optimization
for i, (xbatch, ybatch) in tqdm(enumerate(sp.batch(X, y, BATCH_SIZE)), total=NUM_ITERS):
if i >= NUM_ITERS: break
X_in.assign_value(sp.Variable(xbatch))
y_true.assign_value(ybatch)
loss_val = loss.run() # run the graph
if np.isnan(loss_val.array):
print("loss is nan, aborting.")
break
loss_vals.append(loss_val.array)
# Compute gradients, and use to carry out learning step:
gradients = sp.get_gradients(loss_val)
adam.training_step(learnables, gradients)
# Compute validation accuracy:
x_eval_batch, y_eval_batch = next(eval_batch)
X_in.assign_value(sp.Variable(x_eval_batch))
predictions = y_pred.run()
predictions = np.argmax(predictions.array, axis=1)
accuracy = (y_eval_batch == predictions).mean()
validation_acc.append(accuracy)
# Plot results:
print(f'Final validation accuracy: {np.mean(validation_acc[-10:])}')
plt.figure(figsize=(14, 4))
plt.subplot(1, 2, 1)
plt.ylabel('Loss')
plt.xlabel('Iteration')
plt.plot(loss_vals)
plt.subplot(1, 2, 2)
plt.ylabel('Validation accuracy')
plt.xlabel('Iteration')
plt.suptitle('Neural network trained on MNIST, using SmallPebble.')
plt.ylim([0, 1])
plt.plot(validation_acc)
plt.show()
```
## Training a convolutional neural network on CIFAR-10, using CuPy
This was run on [Google Colab](https://colab.research.google.com/), with a GPU.
```
"Load the CIFAR dataset."
X_train, y_train, _, _ = load_data('cifar') # load/download from openml.org
X_train = X_train/255 # normalize
"""Plot, to check it's the right data.
(This cell's code is from: https://www.tensorflow.org/tutorials/images/cnn#verify_the_data)
"""
class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
plt.figure(figsize=(8,8))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(X_train[i,:].reshape(32,32,3))
plt.xlabel(class_names[y_train[i]])
plt.show()
"Switch array library to CuPy, so can use GPU."
import cupy
sp.use(cupy)
print(sp.array_library.library.__name__) # should be 'cupy'
"Convert data to CuPy arrays"
X_train = cupy.array(X_train)
y_train = cupy.array(y_train)
# Seperate out data for validation as before.
X = X_train[:45_000, ...]
y = y_train[:45_000]
X_eval = X_train[45_000:50_000, ...]
y_eval = y_train[45_000:50_000]
"""Define a model."""
X_in = sp.Placeholder()
y_true = sp.Placeholder()
h = sp.convlayer(height=3, width=3, depth=3, n_kernels=32)(X_in)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)
h = sp.convlayer(3, 3, 32, 128, padding='VALID')(h)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)
h = sp.convlayer(3, 3, 128, 128, padding='VALID')(h)
h = sp.Lazy(sp.leaky_relu)(h)
h = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)
h = sp.Lazy(lambda x: sp.reshape(x, [-1, 3*3*128]))(h)
h = sp.linearlayer(3*3*128, 10)(h)
h = sp.Lazy(sp.softmax)(h)
y_pred = h
loss = sp.Lazy(sp.cross_entropy)(y_pred, y_true)
learnables = sp.get_learnables(y_pred)
loss_vals = []
validation_acc = []
# Check we get the expected dimensions
X_in.assign_value(sp.Variable(X[0:3, :].reshape([-1, 32, 32, 3])))
h.run().shape
```
Train the model.
```
NUM_ITERS = 3000
BATCH_SIZE = 128
eval_batch = sp.batch(X_eval, y_eval, BATCH_SIZE)
adam = sp.Adam()
for i, (xbatch, ybatch) in tqdm(enumerate(sp.batch(X, y, BATCH_SIZE)), total=NUM_ITERS):
if i >= NUM_ITERS: break
xbatch_images = xbatch.reshape([-1, 32, 32, 3])
X_in.assign_value(sp.Variable(xbatch_images))
y_true.assign_value(ybatch)
loss_val = loss.run()
if np.isnan(loss_val.array):
print("Aborting, loss is nan.")
break
loss_vals.append(loss_val.array)
# Compute gradients, and carry out learning step.
gradients = sp.get_gradients(loss_val)
adam.training_step(learnables, gradients)
# Compute validation accuracy:
x_eval_batch, y_eval_batch = next(eval_batch)
X_in.assign_value(sp.Variable(x_eval_batch.reshape([-1, 32, 32, 3])))
predictions = y_pred.run()
predictions = np.argmax(predictions.array, axis=1)
accuracy = (y_eval_batch == predictions).mean()
validation_acc.append(accuracy)
print(f'Final validation accuracy: {np.mean(validation_acc[-10:])}')
plt.figure(figsize=(14, 4))
plt.subplot(1, 2, 1)
plt.ylabel('Loss')
plt.xlabel('Iteration')
plt.plot(loss_vals)
plt.subplot(1, 2, 2)
plt.ylabel('Validation accuracy')
plt.xlabel('Iteration')
plt.suptitle('CNN trained on CIFAR-10, using SmallPebble.')
plt.ylim([0, 1])
plt.plot(validation_acc)
plt.show()
```
It looks like we could improve our results by training for longer (and we could improve our model architecture).
---
# Brief guide to using SmallPebble
SmallPebble provides the following building blocks to make models with:
- `sp.Variable`
- Operations, such as `sp.add`, `sp.mul`, etc.
- `sp.get_gradients`
- `sp.Lazy`
- `sp.Placeholder` (this is really just `sp.Lazy` on the identity function)
- `sp.learnable`
- `sp.get_learnables`
The following examples show how these are used.
## Switching between NumPy and CuPy
We can dynamically switch between NumPy and CuPy. (Assuming you have a CuPy compatible GPU and CuPy set up. Note, CuPy is available on Google Colab, if you change the runtime to GPU.)
```
import cupy
import numpy
import smallpebble as sp
# Switch to CuPy
sp.use(cupy)
print(sp.array_library.library.__name__) # should be 'cupy'
# Switch back to NumPy:
sp.use(numpy)
print(sp.array_library.library.__name__) # should be 'numpy'
```
## sp.Variable & sp.get_gradients
With SmallPebble, you can:
- Wrap NumPy arrays in `sp.Variable`
- Apply SmallPebble operations (e.g. `sp.matmul`, `sp.add`, etc.)
- Compute gradients with `sp.get_gradients`
```
a = sp.Variable(np.random.random([2, 2]))
b = sp.Variable(np.random.random([2, 2]))
c = sp.Variable(np.random.random([2]))
y = sp.mul(a, b) + c
print('y.array:\n', y.array)
gradients = sp.get_gradients(y)
grad_a = gradients[a]
grad_b = gradients[b]
grad_c = gradients[c]
print('grad_a:\n', grad_a)
print('grad_b:\n', grad_b)
print('grad_c:\n', grad_c)
```
Note that `y` is computed straight away, i.e. the (forward) computation happens immediately.
Also note that `y` is a sp.Variable and we could continue to carry out SmallPebble operations on it.
## sp.Lazy & sp.Placeholder
Lazy graphs are constructed using `sp.Lazy` and `sp.Placeholder`.
```
lazy_node = sp.Lazy(lambda a, b: a + b)(1, 2)
print(lazy_node)
print(lazy_node.run())
a = sp.Lazy(lambda a: a)(2)
y = sp.Lazy(lambda a, b, c: a * b + c)(a, 3, 4)
print(y)
print(y.run())
```
Forward computation does not happen immediately - only when .run() is called.
```
a = sp.Placeholder()
b = sp.Variable(np.random.random([2, 2]))
y = sp.Lazy(sp.matmul)(a, b)
a.assign_value(sp.Variable(np.array([[1,2], [3,4]])))
result = y.run()
print('result.array:\n', result.array)
```
You can use .run() as many times as you like.
Let's change the placeholder value and re-run the graph:
```
a.assign_value(sp.Variable(np.array([[10,20], [30,40]])))
result = y.run()
print('result.array:\n', result.array)
```
Finally, let's compute gradients:
```
gradients = sp.get_gradients(result)
```
Note that `sp.get_gradients` is called on `result`,
which is a `sp.Variable`,
not on `y`, which is a `sp.Lazy` instance.
## sp.learnable & sp.get_learnables
Use `sp.learnable` to flag parameters as learnable,
allowing them to be extracted from a lazy graph with `sp.get_learnables`.
This enables a workflow of: building a model, while flagging parameters as learnable, and then extracting all the parameters in one go at the end.
```
a = sp.Placeholder()
b = sp.learnable(sp.Variable(np.random.random([2, 1])))
y = sp.Lazy(sp.matmul)(a, b)
y = sp.Lazy(sp.add)(y, sp.learnable(sp.Variable(np.array([5]))))
learnables = sp.get_learnables(y)
for learnable in learnables:
print(learnable)
```
| github_jupyter |
```
import rioxarray as rio
import xarray as xr
import glob
import os
import numpy as np
import requests
import geopandas as gpd
from pathlib import Path
from datetime import datetime
from rasterio.enums import Resampling
import matplotlib.pyplot as plt
%matplotlib inline
site = "BRC"
# Change site name
chirps_seas_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_season_precip_data_brc')
eeflux_seas_int_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_growing_season_integrated_brc')
chirps_wy_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/wy_total_chirps_brc')
eeflux_seas_mean_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_season_mean_brc')
all_scenes_f_precip = Path('/scratch/waves/rhone-ecostress/rasters/chirps-clipped')
all_scenes_f_et = Path('/home/serdp/rhone/rhone-ecostress/rasters/eeflux/BRC') # Change file path based on site
all_precip_paths = list(all_scenes_f_precip.glob("*"))
all_et_paths = list(all_scenes_f_et.glob("*.tif")) # Variable name agnostic to site?
# for some reason the fll value is not correct. this is the correct bad value to mask by
testf = all_precip_paths[0]
x = rio.open_rasterio(testf)
badvalue = np.unique(x.where(x != x._FillValue).sel(band=1))[0]
def chirps_path_date(path):
_, _, year, month, day, _ = path.name.split(".")
day = day.split("-")[0]
return datetime(int(year), int(month), int(day))
def open_chirps(path):
data_array = rio.open_rasterio(path) #chunks makes i lazyily executed
data_array = data_array.sel(band=1).drop("band") # gets rid of old coordinate dimension since we need bands to have unique coord ids
data_array["date"] = chirps_path_date(path) # makes a new coordinate
return data_array.expand_dims({"date":1}) # makes this coordinate a dimension
### data is not tiled so not a good idea to use chunking
#https://github.com/pydata/xarray/issues/2314
import rasterio
with rasterio.open(testf) as src:
print(src.profile)
len(all_precip_paths) * 41.7 / 10e3 # convert from in to mm
%timeit open_chirps(testf)
all_daily_precip_path = "/home/serdp/ravery/rhone-ecostress/netcdfs/all_chirps_daily_i.nc"
if Path(all_daily_precip_path).exists():
all_chirps_arr = xr.open_dataarray(all_daily_precip_path)
all_chirps_arr = all_chirps_arr.sortby("date")
else:
daily_chirps_arrs = []
for path in all_precip_paths:
daily_chirps_arrs.append(open_chirps(path))
all_chirps_arr = xr.concat(daily_chirps_arrs, dim="date")
all_chirps_arr = all_chirps_arr.sortby("date")
all_chirps_arr.to_netcdf(all_daily_precip_path)
def eeflux_path_date(path):
year, month, day, _, _ = path.name.split("-") # Change this line accordingly based on format of eeflux dates
return datetime(int(year), int(month), int(day))
def open_eeflux(path, da_for_match):
data_array = rio.open_rasterio(path) #chunks makes i lazyily executed
data_array.rio.reproject_match(da_for_match)
data_array = data_array.sel(band=1).drop("band") # gets rid of old coordinate dimension since we need bands to have unique coord ids
data_array["date"] = eeflux_path_date(path) # makes a new coordinate
return data_array.expand_dims({"date":1}) # makes this coordinate a dimension
# The following lines seem to write the lists of rasters to netcdf files? Do we need to replicate for chirps?
da_for_match = rio.open_rasterio(all_et_paths[0])
daily_eeflux_arrs = [open_eeflux(path, da_for_match) for path in all_et_paths]
all_eeflux_arr = xr.concat(daily_eeflux_arrs, dim="date")
all_daily_eeflux_path = "/home/serdp/ravery/rhone-ecostress/netcdfs/all_eeflux_daily_i.nc"
all_eeflux_arr.to_netcdf(all_daily_eeflux_path)
all_eeflux_arr[-3,:,:].plot.imshow()
all_eeflux_arr = all_eeflux_arr.sortby("date")
ey = max(all_eeflux_arr['date.year'].values)
ey
sy = min(all_eeflux_arr['date.year'].values)
sy
all_eeflux_arr['date.dayofyear'].values
# THIS IS IMPORTANT
def years_list(all_arr):
ey = max(all_arr['date.year'].values)
sy = min(all_arr['date.year'].values)
start_years = range(sy, ey)
end_years = range(sy+1, ey+1) # Change to sy+1, ey+1 for across-calendar-year (e.g. winter) calculations
return list(zip(start_years, end_years))
def group_by_custom_doy(all_arr, doy_start, doy_end):
start_end_years = years_list(all_arr)
water_year_arrs = []
for water_year in start_end_years:
start_mask = ((all_arr['date.dayofyear'].values > doy_start) & (all_arr['date.year'].values == water_year[0]))
end_mask = ((all_arr['date.dayofyear'].values < doy_end) & (all_arr['date.year'].values == water_year[1]))
water_year_arrs.append(all_arr[start_mask | end_mask]) # | = or, & = and
return water_year_arrs
def group_by_season(all_arr, doy_start, doy_end):
yrs = np.unique(all_arr['date.year'])
season_arrs = []
for yr in yrs:
start_mask = ((all_arr['date.dayofyear'].values >= doy_start) & (all_arr['date.year'].values == yr))
end_mask = ((all_arr['date.dayofyear'].values <= doy_end) & (all_arr['date.year'].values == yr))
season_arrs.append(all_arr[start_mask & end_mask])
return season_arrs
# THIS IS IMPORTANT
doystart = 125 # Edit these variables to change doy length of year
doyend = 275
# eeflux_water_year_arrs = group_by_custom_doy(all_eeflux_arr, doystart, doyend) # Replaced by eeflux_seas_arrs below
chirps_water_year_arrs = group_by_custom_doy(all_chirps_arr, doyend, doystart)
eeflux_seas_arrs = group_by_season(all_eeflux_arr, doystart, doyend)
eeflux_seas_arrs
chirps_water_year_arrs[-1]
fig = plt.figure()
plt.plot(wy_list,[arr.mean() for arr in chirps_wy_sums],'.')
plt.ylabel('WY Precipitation (mm)')
# Creates figure of ET availability
group_counts = list(map(lambda x: len(x['date']), water_year_arrs))
year_tuples = years_list(all_eeflux_arr)
indexes = np.arange(len(year_tuples))
plt.bar(indexes, group_counts)
degrees = 80
plt.xticks(indexes, year_tuples, rotation=degrees, ha="center")
plt.title("Availability of EEFLUX between DOY 125 and 275")
plt.savefig("eeflux_availability.png")
# Figure below shows empty years in 85, 88, 92, 93, 96; no winter precip rasters generated for these years b/c no ET data w/in winter window
def sum_seasonal_precip(precip_arr, eeflux_group_arr):
return precip_arr.sel(date=slice(eeflux_group_arr.date.min(), eeflux_group_arr.date.max())).sum(dim="date")
# This is matching up precip w/ available ET window for each year
for index, eeflux_group in enumerate(eeflux_seas_arrs):
if len(eeflux_group['date']) > 0:
seasonal_precip = sum_seasonal_precip(all_chirps_arr, eeflux_group) # Variable/array name matters here
seasonal_et = eeflux_group.integrate(coord="date", datetime_unit="D")
year = eeflux_group['date.year'].values[0]
et_doystart = eeflux_group['date.dayofyear'].values[0]
et_doyend = eeflux_group['date.dayofyear'].values[-1]
pname = os.path.join(chirps_seas_out_dir,f"seas_chirps_{site}_{year}_{et_doystart}_{et_doyend}.tif") #Edit output raster labels
eename = os.path.join(eeflux_seas_int_out_dir, f"seasonal_eeflux_integrated_{site}_{year}_{et_doystart}_{et_doyend}.tif")
seasonal_precip.rio.to_raster(pname)
seasonal_et.rio.to_raster(eename)
# This chunk actually outputs the rasters
## Elmera Additions for winter precip:
for index, (eeflux_group,chirps_group) in enumerate(zip(eeflux_seas_arrs,chirps_water_year_arrs[3:])): #changed eeflux_group to eeflux_seas_arrs & changed from water_year_arrs to season_arrs
if len(eeflux_group['date']) > 0: # eeflux_group to eeflux_seas_arrs
mean_seas_et = eeflux_group.mean(dim='date',skipna=False)
chirps_wy_sum = chirps_group.sum(dim='date',skipna=False)
# seasonal_precip = sum_seasonal_precip(chirps_water_year_arrs, eeflux_seas_arr) # Here's where above fxn is applied to rasters, need to replace eeflux_group
year = eeflux_group['date.year'].values[0]
pname = os.path.join(chirps_wy_out_dir,f"wy_total_chirps_{site}_{year}.tif") #Edit output raster labels
eename = os.path.join(eeflux_seas_mean_out_dir,f"mean_daily_seas_et_{site}_{year}.tif")
chirps_wy_sum.rio.to_raster(pname)
mean_seas_et.rio.to_raster(eename)
# This chunk actually outputs the rasters, ET lines removed - including seasonal_precip line?
[arr['date.year'] for arr in chirps_water_year_arrs]
seasonal_precip # This just shows the array - corner cells have empty values b/c of projection mismatch @ edge of raster
water_year_arrs[0][0].plot.imshow()
water_year_arrs[0].integrate(dim="date", datetime_unit="D").plot.imshow()
# This chunk does the actual integration
all_eeflux_arr.integrate(dim="date", datetime_unit="D")
import pandas as pd
import numpy as np
labels = ['<=2', '3-9', '>=10']
bins = [0,2,9, np.inf]
pd.cut(all_eeflux_arr, bins, labels=labels)
all_eeflux_arr
import pandas as pd
all_scene_ids = [str(i) for i in list(all_scenes_f.glob("L*"))]
df = pd.DataFrame({"scene_id":all_scene_ids}).reindex()
split_vals_series = df.scene_id.str.split("/")
dff = pd.DataFrame(split_vals_series.to_list(), columns=['_', '__', '___', '____', '_____', '______', 'fname'])
df['date'] = dff['fname'].str.slice(10,18)
df['pathrow'] = dff['fname'].str.slice(4,10)
df['sensor'] = dff['fname'].str.slice(0,4)
df['datetime'] = pd.to_datetime(df['date'])
df = df.set_index("datetime").sort_index()
marc_df = df['2014-01-01':'2019-12-31']
marc_df = marc_df[marc_df['sensor']=="LC08"]
x.where(x != badvalue).sel(band=1).plot.imshow()
# Evan additions
year_tuples = years_list(all_eeflux_arr)
year_tuples
# Winter precip calculations
year_tuples_p = years_list(all_chirps_arr)
year_tuples_p
def group_p_by_custom_doy(all_chirps_arr, doy_start, doy_end):
start_end_years = years_list(all_chirps_arr)
water_year_arrs = []
for water_year in start_end_years:
start_mask = ((all_chirps_arr['date.dayofyear'].values > doy_start) & (all_chirps_arr['date.year'].values == water_year[0]))
end_mask = ((all_chirps_arr['date.dayofyear'].values < doy_end) & (all_chirps_arr['date.year'].values == water_year[0]))
water_year_arrs.append(all_chirps_arr[start_mask | end_mask])
return water_year_arrs
doystart = 275 # Edit these variables to change doy length of year
doyend = 125
water_year_arrs = group_p_by_custom_doy(all_chirps_arr, doystart, doyend)
water_year_arrs
def sum_seasonal_precip(precip_arr, eeflux_group_arr):
return precip_arr.sel(date=slice(eeflux_group_arr.date.min(), eeflux_group_arr.date.max())).sum(dim="date")
# This is matching up precip w/ available ET window for each year, need to figure out what to feed in for 2nd variable
for index, eeflux_group in enumerate(water_year_arrs):
if len(eeflux_group['date']) > 0:
seasonal_precip = sum_seasonal_precip(all_chirps_arr, eeflux_group) # Here's where above fxn is applied to rasters, need to replace eeflux_group
year_range = year_tuples_p[index]
pname = f"winter_chirps_{year_range[0]}_{year_range[1]}_{doystart}_{doyend}.tif" #Edit output raster labels
seasonal_precip.rio.to_raster(pname)
# This chunk actually outputs the rasters, ET lines removed
```
| github_jupyter |
# ChainerRL Quickstart Guide
This is a quickstart guide for users who just want to try ChainerRL for the first time.
If you have not yet installed ChainerRL, run the command below to install it:
```
%%bash
pip install chainerrl
```
If you have already installed ChainerRL, let's begin!
First, you need to import necessary modules. The module name of ChainerRL is `chainerrl`. Let's import `gym` and `numpy` as well since they are used later.
```
import chainer
import chainer.functions as F
import chainer.links as L
import chainerrl
import gym
import numpy as np
```
ChainerRL can be used for any problems if they are modeled as "environments". [OpenAI Gym](https://github.com/openai/gym) provides various kinds of benchmark environments and defines the common interface among them. ChainerRL uses a subset of the interface. Specifically, an environment must define its observation space and action space and have at least two methods: `reset` and `step`.
- `env.reset` will reset the environment to the initial state and return the initial observation.
- `env.step` will execute a given action, move to the next state and return four values:
- a next observation
- a scalar reward
- a boolean value indicating whether the current state is terminal or not
- additional information
- `env.render` will render the current state.
Let's try 'CartPole-v0', which is a classic control problem. You can see below that its observation space consists of four real numbers while its action space consists of two discrete actions.
```
env = gym.make('CartPole-v0')
print('observation space:', env.observation_space)
print('action space:', env.action_space)
obs = env.reset()
env.render(close=True)
print('initial observation:', obs)
action = env.action_space.sample()
obs, r, done, info = env.step(action)
print('next observation:', obs)
print('reward:', r)
print('done:', done)
print('info:', info)
```
Now you have defined your environment. Next, you need to define an agent, which will learn through interactions with the environment.
ChainerRL provides various agents, each of which implements a deep reinforcement learning algorithm.
To use [DQN (Deep Q-Network)](http://dx.doi.org/10.1038/nature14236), you need to define a Q-function that receives an observation and returns an expected future return for each action the agent can take. In ChainerRL, you can define your Q-function as `chainer.Link` as below. Note that the outputs are wrapped by `chainerrl.action_value.DiscreteActionValue`, which implements `chainerrl.action_value.ActionValue`. By wrapping the outputs of Q-functions, ChainerRL can treat discrete-action Q-functions like this and [NAFs (Normalized Advantage Functions)](https://arxiv.org/abs/1603.00748) in the same way.
```
class QFunction(chainer.Chain):
def __init__(self, obs_size, n_actions, n_hidden_channels=50):
super().__init__(
l0=L.Linear(obs_size, n_hidden_channels),
l1=L.Linear(n_hidden_channels, n_hidden_channels),
l2=L.Linear(n_hidden_channels, n_actions))
def __call__(self, x, test=False):
"""
Args:
x (ndarray or chainer.Variable): An observation
test (bool): a flag indicating whether it is in test mode
"""
h = F.tanh(self.l0(x))
h = F.tanh(self.l1(h))
return chainerrl.action_value.DiscreteActionValue(self.l2(h))
obs_size = env.observation_space.shape[0]
n_actions = env.action_space.n
q_func = QFunction(obs_size, n_actions)
```
If you want to use CUDA for computation, as usual as in Chainer, call `to_gpu`.
```
# Uncomment to use CUDA
# q_func.to_gpu(0)
```
You can also use ChainerRL's predefined Q-functions.
```
_q_func = chainerrl.q_functions.FCStateQFunctionWithDiscreteAction(
obs_size, n_actions,
n_hidden_layers=2, n_hidden_channels=50)
```
As in Chainer, `chainer.Optimizer` is used to update models.
```
# Use Adam to optimize q_func. eps=1e-2 is for stability.
optimizer = chainer.optimizers.Adam(eps=1e-2)
optimizer.setup(q_func)
```
A Q-function and its optimizer are used by a DQN agent. To create a DQN agent, you need to specify a bit more parameters and configurations.
```
# Set the discount factor that discounts future rewards.
gamma = 0.95
# Use epsilon-greedy for exploration
explorer = chainerrl.explorers.ConstantEpsilonGreedy(
epsilon=0.3, random_action_func=env.action_space.sample)
# DQN uses Experience Replay.
# Specify a replay buffer and its capacity.
replay_buffer = chainerrl.replay_buffer.ReplayBuffer(capacity=10 ** 6)
# Since observations from CartPole-v0 is numpy.float64 while
# Chainer only accepts numpy.float32 by default, specify
# a converter as a feature extractor function phi.
phi = lambda x: x.astype(np.float32, copy=False)
# Now create an agent that will interact with the environment.
agent = chainerrl.agents.DoubleDQN(
q_func, optimizer, replay_buffer, gamma, explorer,
replay_start_size=500, update_interval=1,
target_update_interval=100, phi=phi)
```
Now you have an agent and an environment. It's time to start reinforcement learning!
In training, use `agent.act_and_train` to select exploratory actions. `agent.stop_episode_and_train` must be called after finishing an episode. You can get training statistics of the agent via `agent.get_statistics`.
```
n_episodes = 200
max_episode_len = 200
for i in range(1, n_episodes + 1):
obs = env.reset()
reward = 0
done = False
R = 0 # return (sum of rewards)
t = 0 # time step
while not done and t < max_episode_len:
# Uncomment to watch the behaviour
# env.render()
action = agent.act_and_train(obs, reward)
obs, reward, done, _ = env.step(action)
R += reward
t += 1
if i % 10 == 0:
print('episode:', i,
'R:', R,
'statistics:', agent.get_statistics())
agent.stop_episode_and_train(obs, reward, done)
print('Finished.')
```
Now you finished training the agent. How good is the agent now? You can test it by using `agent.act` and `agent.stop_episode` instead. Exploration such as epsilon-greedy is not used anymore.
```
for i in range(10):
obs = env.reset()
done = False
R = 0
t = 0
while not done and t < 200:
env.render(close=True)
action = agent.act(obs)
obs, r, done, _ = env.step(action)
R += r
t += 1
print('test episode:', i, 'R:', R)
agent.stop_episode()
```
If test scores are good enough, the only remaining task is to save the agent so that you can reuse it. What you need to do is to simply call `agent.save` to save the agent, then `agent.load` to load the saved agent.
```
# Save an agent to the 'agent' directory
agent.save('agent')
# Uncomment to load an agent from the 'agent' directory
# agent.load('agent')
```
RL completed!
But writing code like this every time you use RL might be boring. So, ChainerRL has utility functions that do these things.
```
# Set up the logger to print info messages for understandability.
import logging
import sys
gym.undo_logger_setup() # Turn off gym's default logger settings
logging.basicConfig(level=logging.INFO, stream=sys.stdout, format='')
chainerrl.experiments.train_agent_with_evaluation(
agent, env,
steps=2000, # Train the agent for 2000 steps
eval_n_runs=10, # 10 episodes are sampled for each evaluation
max_episode_len=200, # Maximum length of each episodes
eval_interval=1000, # Evaluate the agent after every 1000 steps
outdir='result') # Save everything to 'result' directory
```
That's all of the ChainerRL quickstart guide. To know more about ChainerRL, please look into the `examples` directory and read and run the examples. Thank you!
| github_jupyter |
# Introduction to Kubernetes
**Learning Objectives**
* Create GKE cluster from command line
* Deploy an application to your cluster
* Cleanup, delete the cluster
## Overview
Kubernetes is an open source project (available on [kubernetes.io](kubernetes.io)) which can run on many different environments, from laptops to high-availability multi-node clusters; from public clouds to on-premise deployments; from virtual machines to bare metal.
The goal of this lab is to provide a short introduction to Kubernetes (k8s) and some basic functionality.
## Create a GKE cluster
A cluster consists of at least one cluster master machine and multiple worker machines called nodes. Nodes are Compute Engine virtual machine (VM) instances that run the Kubernetes processes necessary to make them part of the cluster.
**Note**: Cluster names must start with a letter and end with an alphanumeric, and cannot be longer than 40 characters.
We'll call our cluster `asl-cluster`.
```
import os
CLUSTER_NAME = "asl-cluster"
ZONE = "us-central1-a"
os.environ["CLUSTER_NAME"] = CLUSTER_NAME
os.environ["ZONE"] = ZONE
```
We'll set our default compute zone to `us-central1-a` and use `gcloud container clusters create ...` to create the GKE cluster. Let's first look at all the clusters we currently have.
```
!gcloud container clusters list
```
**Exercise**
Use `gcloud container clusters create` to create a new cluster using the `CLUSTER_NAME` we set above. This takes a few minutes...
```
%%bash
gcloud container clusters create $CLUSTER_NAME --zone $ZONE
```
Now when we list our clusters again, we should see the cluster we created.
```
!gcloud container clusters list
```
## Get authentication credentials and deploy and application
After creating your cluster, you need authentication credentials to interact with it. Use `get-credentials` to authenticate the cluster.
**Exercise**
Use `gcloud container clusters get-credentials` to authenticate the cluster you created.
```
%%bash
gcloud container clusters get-credentials asl-cluster --zone $ZONE
```
You can now deploy a containerized application to the cluster. For this lab, you'll run `hello-app` in your cluster.
GKE uses Kubernetes objects to create and manage your cluster's resources. Kubernetes provides the [Deployment](https://kubernetes.io/docs/concepts/workloads/controllers/deployment/) object for deploying stateless applications like web servers. [Service](https://kubernetes.io/docs/concepts/services-networking/service/) objects define rules and load balancing for accessing your application from the internet.
**Exercise**
Use the `kubectl create` command to create a new Deployment `hello-server` from the `hello-app` container image. The `--image` flag to specify a container image to deploy. The `kubectl create` command pulls the example image from a Container Registry bucket. Here, use [gcr.io/google-samples/hello-app:1.0](gcr.io/google-samples/hello-app:1.0) to indicate the specific image version to pull. If a version is not specified, the latest version is used.
```
%%bash
kubectl create deployment hello-server --image=gcr.io/google-samples/hello-app:1.0
```
This Kubernetes command creates a Deployment object that represents `hello-server`. To create a Kubernetes Service, which is a Kubernetes resource that lets you expose your application to external traffic, run the `kubectl expose` command.
**Exercise**
Use the `kubectl expose` to expose the application. In this command,
* `--port` specifies the port that the container exposes.
* `type="LoadBalancer"` creates a Compute Engine load balancer for your container.
```
%%bash
kubectl expose deployment hello-server --type=LoadBalancer --port 8080
```
Use the `kubectl get service` command to inspect the `hello-server` Service.
**Note**: It might take a minute for an external IP address to be generated. Run the previous command again if the `EXTERNAL-IP` column for `hello-server` status is pending.
```
!kubectl get service hello-server
```
You can now view the application from your web browser, open a new tab and enter the following address, replacing `EXTERNAL IP` with the EXTERNAL-IP for `hello-server`:
```bash
http://[EXTERNAL_IP]:8080
```
You should see a simple page which displays
```bash
Hello, world!
Version: 1.0.0
Hostname: hello-server-5bfd595c65-7jqkn
```
## Cleanup
Delete the cluster using `gcloud` to free up those resources. Use the `--quiet` flag if you are executing this in a notebook. Deleting the cluster can take a few minutes.
**Exercise**
Delete the cluster. Use the `--quiet` flag since we're executing in a notebook.
```
%%bash
kubectl delete deployment hello-server
```
Copyright 2020 Google LLC Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at https://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
| github_jupyter |
# Advanced Usage Exampes for Seldon Client
## Istio Gateway Request with token over HTTPS - no SSL verification
Test against a current kubeflow cluster with Dex token authentication.
1. Install kubeflow with Dex authentication
```
INGRESS_HOST=!kubectl -n istio-system get service istio-ingressgateway -o jsonpath='{.status.loadBalancer.ingress[0].ip}'
ISTIO_GATEWAY=INGRESS_HOST[0]
ISTIO_GATEWAY
```
Get a token from the Dex gateway. At present as Dex does not support curl password credentials you will need to get it from your browser logged into the cluster. Open up a browser console and run `document.cookie`
```
TOKEN="eyJhbGciOiJIUzUxMiIsInR5cCI6IkpXVCJ9.eyJleHAiOjE1NjM2MjA0ODYsImlhdCI6MTU2MzUzNDA4NiwiaXNzIjoiMzQuNjUuNzMuMjU1IiwianRpIjoiYjllNDQxOGQtZjNmNC00NTIyLTg5ODEtNDcxOTY0ODNmODg3IiwidWlmIjoiZXlKcGMzTWlPaUpvZEhSd2N6b3ZMek0wTGpZMUxqY3pMakkxTlRvMU5UVTJMMlJsZUNJc0luTjFZaUk2SWtOcFVYZFBSMFUwVG1wbk1GbHBNV3RaYW1jMFRGUlNhVTU2VFhSUFZFSm9UMU13ZWxreVVYaE9hbGw0V21wVk1FNXFXVk5DVjNoMldUSkdjeUlzSW1GMVpDSTZJbXQxWW1WbWJHOTNMV0YxZEdoelpYSjJhV05sTFc5cFpHTWlMQ0psZUhBaU9qRTFOak0yTWpBME9EWXNJbWxoZENJNk1UVTJNelV6TkRBNE5pd2lZWFJmYUdGemFDSTZJbE5OWlZWRGJUQmFOVkZoUTNCdVNHTndRMWgwTVZFaUxDSmxiV0ZwYkNJNkltRmtiV2x1UUhObGJHUnZiaTVwYnlJc0ltVnRZV2xzWDNabGNtbG1hV1ZrSWpwMGNuVmxMQ0p1WVcxbElqb2lZV1J0YVc0aWZRPT0ifQ.7CQIz4A1s9m6lJeWTqpz_JKGArGX4e_zpRCOXXjVRJgguB3z48rSfei_KL7niMCWpruhU11c8UIw9E79PwHNNw"
```
## Start Seldon Core
Use the setup notebook to [Install Seldon Core](seldon_core_setup.ipynb#Install-Seldon-Core) with [Istio Ingress](seldon_core_setup.ipynb#Istio). Instructions [also online](./seldon_core_setup.html).
**Note** When running helm install for this example you will need to set the istio.gateway flag to kubeflow-gateway (```--set istio.gateway=kubeflow-gateway```).
```
deployment_name="test1"
namespace="default"
from seldon_core.seldon_client import SeldonClient, SeldonChannelCredentials, SeldonCallCredentials
sc = SeldonClient(deployment_name=deployment_name,namespace=namespace,gateway_endpoint=ISTIO_GATEWAY,debug=True,
channel_credentials=SeldonChannelCredentials(verify=False),
call_credentials=SeldonCallCredentials(token=TOKEN))
r = sc.predict(gateway="istio",transport="rest",shape=(1,4))
print(r)
```
Its not presently possible to use gRPC without getting access to the certificates. We will update this once its clear how to obtain them from a Kubeflow cluser setup.
## Istio - SSL Endpoint - Client Side Verification - No Authentication
1. First run through the [Istio Secure Gateway SDS example](https://istio.io/docs/tasks/traffic-management/ingress/secure-ingress-sds/) and make sure this works for you.
* This will create certificates for `httpbin.example.com` and test them out.
1. Update your `/etc/hosts` file to include an entry for the ingress gateway for `httpbin.example.com` e.g. add a line like: `10.107.247.132 httpbin.example.com` replacing the ip address with your ingress gateway ip address.
```
# Set to folder where the httpbin certificates are
ISTIO_HTTPBIN_CERT_FOLDER='/home/clive/work/istio/httpbin.example.com'
```
## Start Seldon Core
Use the setup notebook to [Install Seldon Core](seldon_core_setup.ipynb#Install-Seldon-Core) with [Istio Ingress](seldon_core_setup.ipynb#Istio). Instructions [also online](./seldon_core_setup.html).
**Note** When running ```helm install``` for this example you will need to set the ```istio.gateway``` flag to ```mygateway``` (```--set istio.gateway=mygateway```) used in the example.
```
deployment_name="mymodel"
namespace="default"
from seldon_core.seldon_client import SeldonClient, SeldonChannelCredentials, SeldonCallCredentials
sc = SeldonClient(deployment_name=deployment_name,namespace=namespace,gateway_endpoint="httpbin.example.com",debug=True,
channel_credentials=SeldonChannelCredentials(certificate_chain_file=ISTIO_HTTPBIN_CERT_FOLDER+'/2_intermediate/certs/ca-chain.cert.pem',
root_certificates_file=ISTIO_HTTPBIN_CERT_FOLDER+'/4_client/certs/httpbin.example.com.cert.pem',
private_key_file=ISTIO_HTTPBIN_CERT_FOLDER+'/4_client/private/httpbin.example.com.key.pem'
))
r = sc.predict(gateway="istio",transport="rest",shape=(1,4))
print(r)
r = sc.predict(gateway="istio",transport="grpc",shape=(1,4))
print(r)
```
| github_jupyter |
<table width="100%">
<tr style="border-bottom:solid 2pt #009EE3">
<td style="text-align:left" width="10%">
<a href="prepare_anaconda.dwipynb" download><img src="../../images/icons/download.png"></a>
</td>
<td style="text-align:left" width="10%">
<a href="https://mybinder.org/v2/gh/biosignalsnotebooks/biosignalsnotebooks/biosignalsnotebooks_binder?filepath=biosignalsnotebooks_environment%2Fcategories%2FInstall%2Fprepare_anaconda.dwipynb" target="_blank"><img src="../../images/icons/program.png" title="Be creative and test your solutions !"></a>
</td>
<td></td>
<td style="text-align:left" width="5%">
<a href="../MainFiles/biosignalsnotebooks.ipynb"><img src="../../images/icons/home.png"></a>
</td>
<td style="text-align:left" width="5%">
<a href="../MainFiles/contacts.ipynb"><img src="../../images/icons/contacts.png"></a>
</td>
<td style="text-align:left" width="5%">
<a href="https://github.com/biosignalsnotebooks/biosignalsnotebooks" target="_blank"><img src="../../images/icons/github.png"></a>
</td>
<td style="border-left:solid 2pt #009EE3" width="15%">
<img src="../../images/ost_logo.png">
</td>
</tr>
</table>
<link rel="stylesheet" href="../../styles/theme_style.css">
<!--link rel="stylesheet" href="../../styles/header_style.css"-->
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css">
<table width="100%">
<tr>
<td id="image_td" width="15%" class="header_image_color_13"><div id="image_img"
class="header_image_13"></div></td>
<td class="header_text"> Download, Install and Execute Anaconda </td>
</tr>
</table>
<div id="flex-container">
<div id="diff_level" class="flex-item">
<strong>Difficulty Level:</strong> <span class="fa fa-star checked"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
</div>
<div id="tag" class="flex-item-tag">
<span id="tag_list">
<table id="tag_list_table">
<tr>
<td class="shield_left">Tags</td>
<td class="shield_right" id="tags">install☁jupyter☁notebook☁anaconda☁download</td>
</tr>
</table>
</span>
<!-- [OR] Visit https://img.shields.io in order to create a tag badge-->
</div>
</div>
In every journey we always need to prepare our toolbox with the needed resources !
With <strong><span class="color1">biosignalsnotebooks</span></strong> happens the same, being <strong><span class="color4">Jupyter Notebook</span></strong> environment the most relevant application (that supports <strong><span class="color1">biosignalsnotebooks</span></strong>) to take the maximum advantage during your learning process.
In the following sequence of instruction it will be presented the operations that should be completed in order to have <strong><span class="color4">Jupyter Notebook</span></strong> ready to use and to open our <strong>ipynb</strong> files on local server.
<table width="100%">
<tr>
<td style="text-align:left;font-size:12pt;border-top:dotted 2px #62C3EE">
<span class="color1">☌</span> The current <span class="color4"><strong>Jupyter Notebook</strong></span> is focused on a complete Python toolbox called <a href="https://www.anaconda.com/distribution/"><span class="color4"><strong>Anaconda <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></span></a>.
However, there is an alternative approach to get all things ready for starting our journey, which is described on <a href="../Install/prepare_jupyter.ipynb"><span class="color1"><strong>"Download, Install and Execute Jypyter Notebook Environment" <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></span></a>
</td>
</tr>
</table>
<hr>
<hr>
<p class="steps">1 - Access the <strong><span class="color4">Anaconda</span></strong> official page at <a href="https://www.anaconda.com/distribution/">https://www.anaconda.com/distribution/</a></p>
<img src="../../images/other/anaconda_page.png">
<p class="steps">2 - Click on "Download" button, giving a first but strong step into our final objective</p>
<img src="../../images/other/anaconda_download.gif">
<p class="steps">3 - Specify the operating system of your local machine</p>
<img src="../../images/other/anaconda_download_os.gif">
<p class="steps">4 - Select the version of <span class="color1">Python</span> compiler to be included on <span class="color4">Anaconda</span></p>
It is strongly advisable that you chose version <strong>3.-</strong> to ensure that all functionalities of packages like <strong><span class="color1">biosignalsnotebooks</span></strong> are fully operational.
<img src="../../images/other/anaconda_download_version.gif">
<p class="steps">5 - After defining the directory where the downloaded file will be stored, please, wait a few minutes for the end of transfer</p>
<span class="color13" style="font-size:30px">⚠</span>
The waiting time will depend on the quality of the Internet connection !
<p class="steps">6 - When download is finished navigate through your directory tree until reaching the folder where the downloaded file is located</p>
In our case the destination folder was <img src="../../images/other/anaconda_download_location.png" style="display:inline;margin-top:0px">
<p class="steps">7 - Execute <span class="color4">Anaconda</span> installer file with a double-click</p>
<img src="../../images/other/anaconda_download_installer.gif">
<p class="steps">8 - Follow the sequential instructions presented on the <span class="color4">Anaconda</span> installer</p>
<img src="../../images/other/anaconda_download_install_steps.gif">
<p class="steps">9 - <span class="color4">Jupyter Notebook</span> environment is included on the previous installation. For starting your first Notebook execute <span class="color4">Jupyter Notebook</span></p>
Launch from "Anaconda Navigator" or through a command window, like described on the following steps.
<p class="steps">9.1 - For executing <span class="color4">Jupyter Notebook</span> environment you should open a <strong>console</strong> (in your operating system).</p>
<i>If you are a Microsoft Windows native, just type click on Windows logo (bottom-left corner of the screen) and type "cmd". Then press "Enter".</i>
<p class="steps">9.2 - Type <strong>"jupyter notebook"</strong> inside the opened console. A local <span class="color4"><strong>Jupyter Notebook</strong></span> server will be launched.</p>
<img src="../../images/other/open_jupyter.gif">
<p class="steps">10 - Create a blank Notebook</p>
<p class="steps">10.1 - Now, you should navigate through your directories until reaching the folder where you want to create or open a Notebook (as demonstrated in the following video)</p>
<span class="color13" style="font-size:30px">⚠</span>
<p style="margin-top:0px">You should note that your folder hierarchy is unique, so, the steps followed in the next image, will depend on your folder organisation, being merely illustrative </p>
<img src="../../images/other/create_notebook_part1.gif">
<p class="steps">10.2 - For creating a new Notebook, "New" button (top-right zone of Jupyter Notebook interface) should be pressed and <span class="color1"><strong>Python 3</strong></span> option selected.</p>
<i>A blank Notebook will arise and now you just need to be creative and expand your thoughts to others persons!!!</i>
<img src="../../images/other/create_notebook_part2.gif">
This can be the start of something great. Now you have all the software conditions to create and develop interactive tutorials, combining Python with HTML !
<span class="color4"><strong>Anaconda</strong></span> contains lots of additional functionalities, namely <a href="https://anaconda.org/anaconda/spyder"><span class="color7"><strong>Spyder <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></span></a>, which is an intuitive Python editor for creating and testing your own scripts.
<strong><span class="color7">We hope that you have enjoyed this guide. </span><span class="color2">biosignalsnotebooks</span><span class="color4"> is an environment in continuous expansion, so don't stop your journey and learn more with the remaining <a href="../MainFiles/biosignalsnotebooks.ipynb">Notebooks <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></a></span></strong> !
<hr>
<table width="100%">
<tr>
<td style="border-right:solid 3px #009EE3" width="20%">
<img src="../../images/ost_logo.png">
</td>
<td width="40%" style="text-align:left">
<a href="../MainFiles/aux_files/biosignalsnotebooks_presentation.pdf" target="_blank">☌ Project Presentation</a>
<br>
<a href="https://github.com/biosignalsnotebooks/biosignalsnotebooks" target="_blank">☌ GitHub Repository</a>
<br>
<a href="https://pypi.org/project/biosignalsnotebooks/" target="_blank">☌ How to install biosignalsnotebooks Python package ?</a>
<br>
<a href="../MainFiles/signal_samples.ipynb">☌ Signal Library</a>
</td>
<td width="40%" style="text-align:left">
<a href="../MainFiles/biosignalsnotebooks.ipynb">☌ Notebook Categories</a>
<br>
<a href="../MainFiles/by_diff.ipynb">☌ Notebooks by Difficulty</a>
<br>
<a href="../MainFiles/by_signal_type.ipynb">☌ Notebooks by Signal Type</a>
<br>
<a href="../MainFiles/by_tag.ipynb">☌ Notebooks by Tag</a>
</td>
</tr>
</table>
```
from biosignalsnotebooks.__notebook_support__ import css_style_apply
css_style_apply()
%%html
<script>
// AUTORUN ALL CELLS ON NOTEBOOK-LOAD!
require(
['base/js/namespace', 'jquery'],
function(jupyter, $) {
$(jupyter.events).on("kernel_ready.Kernel", function () {
console.log("Auto-running all cells-below...");
jupyter.actions.call('jupyter-notebook:run-all-cells-below');
jupyter.actions.call('jupyter-notebook:save-notebook');
});
}
);
</script>
```
| github_jupyter |
# Testing cosmogan
April 19, 2021
Borrowing pieces of code from :
- https://github.com/pytorch/tutorials/blob/11569e0db3599ac214b03e01956c2971b02c64ce/beginner_source/dcgan_faces_tutorial.py
- https://github.com/exalearn/epiCorvid/tree/master/cGAN
```
import os
import random
import logging
import sys
import torch
import torch.nn as nn
import torch.nn.parallel
import torch.backends.cudnn as cudnn
import torch.optim as optim
import torch.utils.data
from torchsummary import summary
from torch.utils.data import DataLoader, TensorDataset
import torch.distributed as dist
from torch.nn.parallel import DistributedDataParallel
# import torch.fft
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.animation as animation
# from IPython.display import HTML
import argparse
import time
from datetime import datetime
import glob
import pickle
import yaml
import collections
import socket
import shutil
# # Import modules from other files
# from utils import *
# from spec_loss import *
%matplotlib widget
```
## Modules
```
### Transformation functions for image pixel values
def f_transform(x):
return 2.*x/(x + 4.) - 1.
def f_invtransform(s):
return 4.*(1. + s)/(1. - s)
# Generator Code
class View(nn.Module):
def __init__(self, shape):
super(View, self).__init__()
self.shape = shape
def forward(self, x):
return x.view(*self.shape)
def f_get_model(gdict):
''' Module to define Generator and Discriminator'''
if gdict['image_size']==64:
class Generator(nn.Module):
def __init__(self, gdict):
super(Generator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']
ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# nn.ConvTranspose3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Linear(nz,nc*ngf*8**3),# 262144
nn.BatchNorm3d(nc,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
View(shape=[-1,ngf*8,4,4,4]),
nn.ConvTranspose3d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),
nn.BatchNorm3d(ngf*4,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*4) x 8 x 8
nn.ConvTranspose3d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf*2,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*2) x 16 x 16
nn.ConvTranspose3d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf) x 32 x 32
nn.ConvTranspose3d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),
nn.Tanh()
)
def forward(self, ip):
return self.main(ip)
class Discriminator(nn.Module):
def __init__(self, gdict):
super(Discriminator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']
ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# input is (nc) x 64 x 64 x 64
# nn.Conv3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Conv3d(nc, ndf,kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf) x 32 x 32
nn.Conv3d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*2) x 16 x 16
nn.Conv3d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*4) x 8 x 8
nn.Conv3d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*8) x 4 x 4
nn.Flatten(),
nn.Linear(nc*ndf*8*8*8, 1)
# nn.Sigmoid()
)
def forward(self, ip):
# print(ip.shape)
results=[ip]
lst_idx=[]
for i,submodel in enumerate(self.main.children()):
mid_output=submodel(results[-1])
results.append(mid_output)
## Select indices in list corresponding to output of Conv layers
if submodel.__class__.__name__.startswith('Conv'):
# print(submodel.__class__.__name__)
# print(mid_output.shape)
lst_idx.append(i)
FMloss=True
if FMloss:
ans=[results[1:][i] for i in lst_idx + [-1]]
else :
ans=results[-1]
return ans
elif gdict['image_size']==128:
class Generator(nn.Module):
def __init__(self, gdict):
super(Generator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']
ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# nn.ConvTranspose3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Linear(nz,nc*ngf*8**3*8),# 262144
nn.BatchNorm3d(nc,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
View(shape=[-1,ngf*8,8,8,8]),
nn.ConvTranspose3d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),
nn.BatchNorm3d(ngf*4,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*4) x 8 x 8
nn.ConvTranspose3d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf*2,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf*2) x 16 x 16
nn.ConvTranspose3d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),
nn.BatchNorm3d(ngf,eps=1e-05, momentum=0.9, affine=True),
nn.ReLU(inplace=True),
# state size. (ngf) x 32 x 32
nn.ConvTranspose3d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),
nn.Tanh()
)
def forward(self, ip):
return self.main(ip)
class Discriminator(nn.Module):
def __init__(self, gdict):
super(Discriminator, self).__init__()
## Define new variables from dict
keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']
ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
self.main = nn.Sequential(
# input is (nc) x 64 x 64 x 64
# nn.Conv3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
nn.Conv3d(nc, ndf,kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf) x 32 x 32
nn.Conv3d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*2) x 16 x 16
nn.Conv3d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*4) x 8 x 8
nn.Conv3d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),
nn.BatchNorm3d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),
nn.LeakyReLU(0.2, inplace=True),
# state size. (ndf*8) x 4 x 4
nn.Flatten(),
nn.Linear(nc*ndf*8*8*8*8, 1)
# nn.Sigmoid()
)
def forward(self, ip):
results=[ip]
lst_idx=[]
for i,submodel in enumerate(self.main.children()):
mid_output=submodel(results[-1])
results.append(mid_output)
## Select indices in list corresponding to output of Conv layers
if submodel.__class__.__name__.startswith('Conv'):
# print(submodel.__class__.__name__)
# print(mid_output.shape)
lst_idx.append(i)
FMloss=True
if FMloss:
ans=[results[1:][i] for i in lst_idx + [-1]]
else :
ans=results[-1]
return ans
return Generator, Discriminator
def f_gen_images(gdict,netG,optimizerG,ip_fname,op_loc,op_strg='inf_img_',op_size=500):
'''Generate images for best saved models
Arguments: gdict, netG, optimizerG,
ip_fname: name of input file
op_strg: [string name for output file]
op_size: Number of images to generate
'''
nz,device=gdict['nz'],gdict['device']
try:# handling cpu vs gpu
if torch.cuda.is_available(): checkpoint=torch.load(ip_fname)
else: checkpoint=torch.load(ip_fname,map_location=torch.device('cpu'))
except Exception as e:
print(e)
print("skipping generation of images for ",ip_fname)
return
## Load checkpoint
if gdict['multi-gpu']:
netG.module.load_state_dict(checkpoint['G_state'])
else:
netG.load_state_dict(checkpoint['G_state'])
## Load other stuff
iters=checkpoint['iters']
epoch=checkpoint['epoch']
optimizerG.load_state_dict(checkpoint['optimizerG_state_dict'])
# Generate batch of latent vectors
noise = torch.randn(op_size, 1, 1, 1, nz, device=device)
# Generate fake image batch with G
netG.eval() ## This is required before running inference
with torch.no_grad(): ## This is important. fails without it for multi-gpu
gen = netG(noise)
gen_images=gen.detach().cpu().numpy()
print(gen_images.shape)
op_fname='%s_epoch-%s_step-%s.npy'%(op_strg,epoch,iters)
np.save(op_loc+op_fname,gen_images)
print("Image saved in ",op_fname)
def f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,netG,netD,optimizerG,optimizerD,save_loc):
''' Checkpoint model '''
if gdict['multi-gpu']: ## Dataparallel
torch.save({'epoch':epoch,'iters':iters,'best_chi1':best_chi1,'best_chi2':best_chi2,
'G_state':netG.module.state_dict(),'D_state':netD.module.state_dict(),'optimizerG_state_dict':optimizerG.state_dict(),
'optimizerD_state_dict':optimizerD.state_dict()}, save_loc)
else :
torch.save({'epoch':epoch,'iters':iters,'best_chi1':best_chi1,'best_chi2':best_chi2,
'G_state':netG.state_dict(),'D_state':netD.state_dict(),'optimizerG_state_dict':optimizerG.state_dict(),
'optimizerD_state_dict':optimizerD.state_dict()}, save_loc)
def f_load_checkpoint(ip_fname,netG,netD,optimizerG,optimizerD,gdict):
''' Load saved checkpoint
Also loads step, epoch, best_chi1, best_chi2'''
print("torch device",torch.device('cuda',torch.cuda.current_device()))
try:
checkpoint=torch.load(ip_fname,map_location=torch.device('cuda',torch.cuda.current_device()))
except Exception as e:
print("Error loading saved checkpoint",ip_fname)
print(e)
raise SystemError
## Load checkpoint
if gdict['multi-gpu']:
netG.module.load_state_dict(checkpoint['G_state'])
netD.module.load_state_dict(checkpoint['D_state'])
else:
netG.load_state_dict(checkpoint['G_state'])
netD.load_state_dict(checkpoint['D_state'])
optimizerD.load_state_dict(checkpoint['optimizerD_state_dict'])
optimizerG.load_state_dict(checkpoint['optimizerG_state_dict'])
iters=checkpoint['iters']
epoch=checkpoint['epoch']
best_chi1=checkpoint['best_chi1']
best_chi2=checkpoint['best_chi2']
netG.train()
netD.train()
return iters,epoch,best_chi1,best_chi2,netD,optimizerD,netG,optimizerG
####################
### Pytorch code ###
####################
def f_get_rad(img):
''' Get the radial tensor for use in f_torch_get_azimuthalAverage '''
height,width,depth=img.shape[-3:]
# Create a grid of points with x and y and z coordinates
z,y,x = np.indices([height,width,depth])
center=[]
if not center:
center = np.array([(x.max()-x.min())/2.0, (y.max()-y.min())/2.0, (z.max()-z.min())/2.0])
# Get the radial coordinate for every grid point. Array has the shape of image
r= torch.tensor(np.sqrt((x-center[0])**2 + (y-center[1])**2 + (z-center[2])**2))
# Get sorted radii
ind = torch.argsort(torch.reshape(r, (-1,)))
return r.detach(),ind.detach()
def f_torch_get_azimuthalAverage(image,r,ind):
"""
Calculate the azimuthally averaged radial profile.
image - The 2D image
center - The [x,y] pixel coordinates used as the center. The default is
None, which then uses the center of the image (including
fracitonal pixels).
source: https://www.astrobetter.com/blog/2010/03/03/fourier-transforms-of-images-in-python/
"""
# height, width = image.shape
# # Create a grid of points with x and y coordinates
# y, x = np.indices([height,width])
# if not center:
# center = np.array([(x.max()-x.min())/2.0, (y.max()-y.min())/2.0])
# # Get the radial coordinate for every grid point. Array has the shape of image
# r = torch.tensor(np.hypot(x - center[0], y - center[1]))
# # Get sorted radii
# ind = torch.argsort(torch.reshape(r, (-1,)))
r_sorted = torch.gather(torch.reshape(r, ( -1,)),0, ind)
i_sorted = torch.gather(torch.reshape(image, ( -1,)),0, ind)
# Get the integer part of the radii (bin size = 1)
r_int=r_sorted.to(torch.int32)
# Find all pixels that fall within each radial bin.
deltar = r_int[1:] - r_int[:-1] # Assumes all radii represented
rind = torch.reshape(torch.where(deltar)[0], (-1,)) # location of changes in radius
nr = (rind[1:] - rind[:-1]).type(torch.float) # number of radius bin
# Cumulative sum to figure out sums for each radius bin
csum = torch.cumsum(i_sorted, axis=-1)
tbin = torch.gather(csum, 0, rind[1:]) - torch.gather(csum, 0, rind[:-1])
radial_prof = tbin / nr
return radial_prof
def f_torch_fftshift(real, imag):
for dim in range(0, len(real.size())):
real = torch.roll(real, dims=dim, shifts=real.size(dim)//2)
imag = torch.roll(imag, dims=dim, shifts=imag.size(dim)//2)
return real, imag
def f_torch_compute_spectrum(arr,r,ind):
GLOBAL_MEAN=1.0
arr=(arr-GLOBAL_MEAN)/(GLOBAL_MEAN)
y1=torch.rfft(arr,signal_ndim=3,onesided=False)
real,imag=f_torch_fftshift(y1[:,:,:,0],y1[:,:,:,1]) ## last index is real/imag part ## Mod for 3D
# # For pytorch 1.8
# y1=torch.fft.fftn(arr,dim=(-3,-2,-1))
# real,imag=f_torch_fftshift(y1.real,y1.imag)
y2=real**2+imag**2 ## Absolute value of each complex number
z1=f_torch_get_azimuthalAverage(y2,r,ind) ## Compute radial profile
return z1
def f_torch_compute_batch_spectrum(arr,r,ind):
batch_pk=torch.stack([f_torch_compute_spectrum(i,r,ind) for i in arr])
return batch_pk
def f_torch_image_spectrum(x,num_channels,r,ind):
'''
Data has to be in the form (batch,channel,x,y)
'''
mean=[[] for i in range(num_channels)]
var=[[] for i in range(num_channels)]
for i in range(num_channels):
arr=x[:,i,:,:,:] # Mod for 3D
batch_pk=f_torch_compute_batch_spectrum(arr,r,ind)
mean[i]=torch.mean(batch_pk,axis=0)
# var[i]=torch.std(batch_pk,axis=0)/np.sqrt(batch_pk.shape[0])
# var[i]=torch.std(batch_pk,axis=0)
var[i]=torch.var(batch_pk,axis=0)
mean=torch.stack(mean)
var=torch.stack(var)
if (torch.isnan(mean).any() or torch.isnan(var).any()):
print("Nans in spectrum",mean,var)
if torch.isnan(x).any():
print("Nans in Input image")
return mean,var
def f_compute_hist(data,bins):
try:
hist_data=torch.histc(data,bins=bins)
## A kind of normalization of histograms: divide by total sum
hist_data=(hist_data*bins)/torch.sum(hist_data)
except Exception as e:
print(e)
hist_data=torch.zeros(bins)
return hist_data
### Losses
def loss_spectrum(spec_mean,spec_mean_ref,spec_var,spec_var_ref,image_size,lambda_spec_mean,lambda_spec_var):
''' Loss function for the spectrum : mean + variance
Log(sum( batch value - expect value) ^ 2 )) '''
if (torch.isnan(spec_mean).any() or torch.isnan(spec_var).any()):
ans=torch.tensor(float("inf"))
return ans
idx=int(image_size/2) ### For the spectrum, use only N/2 indices for loss calc.
### Warning: the first index is the channel number.For multiple channels, you are averaging over them, which is fine.
loss_mean=torch.log(torch.mean(torch.pow(spec_mean[:,:idx]-spec_mean_ref[:,:idx],2)))
loss_var=torch.log(torch.mean(torch.pow(spec_var[:,:idx]-spec_var_ref[:,:idx],2)))
ans=lambda_spec_mean*loss_mean+lambda_spec_var*loss_var
if (torch.isnan(ans).any()) :
print("loss spec mean %s, loss spec var %s"%(loss_mean,loss_var))
print("spec mean %s, ref %s"%(spec_mean, spec_mean_ref))
print("spec var %s, ref %s"%(spec_var, spec_var_ref))
# raise SystemExit
return ans
def loss_hist(hist_sample,hist_ref):
lambda1=1.0
return lambda1*torch.log(torch.mean(torch.pow(hist_sample-hist_ref,2)))
def f_FM_loss(real_output,fake_output,lambda_fm,gdict):
'''
Module to implement Feature-Matching loss. Reads all but last elements of Discriminator ouput
'''
FM=torch.Tensor([0.0]).to(gdict['device'])
for i,j in zip(real_output[:-1],fake_output[:-1]):
# print(i.shape,j.shape)
real_mean=torch.mean(i)
fake_mean=torch.mean(j)
# print(real_mean,fake_mean)
FM=FM.clone()+torch.sum(torch.square(real_mean-fake_mean))
return lambda_fm*FM
def f_gp_loss(grads,l=1.0):
'''
Module to implement gradient penalty loss.
'''
loss=torch.mean(torch.sum(torch.square(grads),dim=[1,2,3]))
return l*loss
```
## Train loop
```
### Train code ###
def f_train_loop(gan_model,Dset,metrics_df,gdict,fixed_noise):
''' Train epochs '''
## Define new variables from dict
keys=['image_size','start_epoch','epochs','iters','best_chi1','best_chi2','save_dir','device','flip_prob','nz','batch_size','bns']
image_size,start_epoch,epochs,iters,best_chi1,best_chi2,save_dir,device,flip_prob,nz,batchsize,bns=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
for epoch in range(start_epoch,epochs):
t_epoch_start=time.time()
for count, data in enumerate(Dset.train_dataloader):
####### Train GAN ########
gan_model.netG.train(); gan_model.netD.train(); ### Need to add these after inference and before training
tme1=time.time()
### Update D network: maximize log(D(x)) + log(1 - D(G(z)))
gan_model.netD.zero_grad()
real_cpu = data[0].to(device)
real_cpu.requires_grad=True
b_size = real_cpu.size(0)
real_label = torch.full((b_size,), 1, device=device,dtype=float)
fake_label = torch.full((b_size,), 0, device=device,dtype=float)
g_label = torch.full((b_size,), 1, device=device,dtype=float) ## No flipping for Generator labels
# Flip labels with probability flip_prob
for idx in np.random.choice(np.arange(b_size),size=int(np.ceil(b_size*flip_prob))):
real_label[idx]=0; fake_label[idx]=1
# Generate fake image batch with G
noise = torch.randn(b_size, 1, 1, 1, nz, device=device) ### Mod for 3D
fake = gan_model.netG(noise)
# Forward pass real batch through D
real_output = gan_model.netD(real_cpu)
errD_real = gan_model.criterion(real_output[-1].view(-1), real_label.float())
errD_real.backward(retain_graph=True)
D_x = real_output[-1].mean().item()
# Forward pass fake batch through D
fake_output = gan_model.netD(fake.detach()) # The detach is important
errD_fake = gan_model.criterion(fake_output[-1].view(-1), fake_label.float())
errD_fake.backward(retain_graph=True)
D_G_z1 = fake_output[-1].mean().item()
errD = errD_real + errD_fake
if gdict['lambda_gp']: ## Add gradient - penalty loss
grads=torch.autograd.grad(outputs=real_output[-1],inputs=real_cpu,grad_outputs=torch.ones_like(real_output[-1]),allow_unused=False,create_graph=True)[0]
gp_loss=f_gp_loss(grads,gdict['lambda_gp'])
gp_loss.backward(retain_graph=True)
errD = errD + gp_loss
else:
gp_loss=torch.Tensor([np.nan])
if gdict['grad_clip']:
nn.utils.clip_grad_norm_(gan_model.netD.parameters(),gdict['grad_clip'])
gan_model.optimizerD.step()
lr_d=gan_model.optimizerD.param_groups[0]['lr']
gan_model.schedulerD.step()
# dict_keys(['train_data_loader', 'r', 'ind', 'train_spec_mean', 'train_spec_var', 'train_hist', 'val_spec_mean', 'val_spec_var', 'val_hist'])
###Update G network: maximize log(D(G(z)))
gan_model.netG.zero_grad()
output = gan_model.netD(fake)
errG_adv = gan_model.criterion(output[-1].view(-1), g_label.float())
# errG_adv.backward(retain_graph=True)
# Histogram pixel intensity loss
hist_gen=f_compute_hist(fake,bins=bns)
hist_loss=loss_hist(hist_gen,Dset.train_hist.to(device))
# Add spectral loss
mean,var=f_torch_image_spectrum(f_invtransform(fake),1,Dset.r.to(device),Dset.ind.to(device))
spec_loss=loss_spectrum(mean,Dset.train_spec_mean.to(device),var,Dset.train_spec_var.to(device),image_size,gdict['lambda_spec_mean'],gdict['lambda_spec_var'])
errG=errG_adv
if gdict['lambda_spec_mean']:
# spec_loss.backward(retain_graph=True)
errG = errG+ spec_loss
if gdict['lambda_fm']:## Add feature matching loss
fm_loss=f_FM_loss([i.detach() for i in real_output],output,gdict['lambda_fm'],gdict)
# fm_loss.backward(retain_graph=True)
errG= errG+ fm_loss
else:
fm_loss=torch.Tensor([np.nan])
if torch.isnan(errG).any():
logging.info(errG)
raise SystemError
# Calculate gradients for G
errG.backward()
D_G_z2 = output[-1].mean().item()
### Implement Gradient clipping
if gdict['grad_clip']:
nn.utils.clip_grad_norm_(gan_model.netG.parameters(),gdict['grad_clip'])
gan_model.optimizerG.step()
lr_g=gan_model.optimizerG.param_groups[0]['lr']
gan_model.schedulerG.step()
tme2=time.time()
####### Store metrics ########
# Output training stats
if gdict['world_rank']==0:
if ((count % gdict['checkpoint_size'] == 0)):
logging.info('[%d/%d][%d/%d]\tLoss_D: %.4f\tLoss_adv: %.4f\tLoss_G: %.4f\tD(x): %.4f\tD(G(z)): %.4f / %.4f'
% (epoch, epochs, count, len(Dset.train_dataloader), errD.item(), errG_adv.item(),errG.item(), D_x, D_G_z1, D_G_z2)),
logging.info("Spec loss: %s,\t hist loss: %s"%(spec_loss.item(),hist_loss.item())),
logging.info("Training time for step %s : %s"%(iters, tme2-tme1))
# Save metrics
cols=['step','epoch','Dreal','Dfake','Dfull','G_adv','G_full','spec_loss','hist_loss','fm_loss','gp_loss','D(x)','D_G_z1','D_G_z2','lr_d','lr_g','time']
vals=[iters,epoch,errD_real.item(),errD_fake.item(),errD.item(),errG_adv.item(),errG.item(),spec_loss.item(),hist_loss.item(),fm_loss.item(),gp_loss.item(),D_x,D_G_z1,D_G_z2,lr_d,lr_g,tme2-tme1]
for col,val in zip(cols,vals): metrics_df.loc[iters,col]=val
### Checkpoint the best model
checkpoint=True
iters += 1 ### Model has been updated, so update iters before saving metrics and model.
### Compute validation metrics for updated model
gan_model.netG.eval()
with torch.no_grad():
fake = gan_model.netG(fixed_noise)
hist_gen=f_compute_hist(fake,bins=bns)
hist_chi=loss_hist(hist_gen,Dset.val_hist.to(device))
mean,var=f_torch_image_spectrum(f_invtransform(fake),1,Dset.r.to(device),Dset.ind.to(device))
spec_chi=loss_spectrum(mean,Dset.val_spec_mean.to(device),var,Dset.val_spec_var.to(device),image_size,gdict['lambda_spec_mean'],gdict['lambda_spec_var'])
# Storing chi for next step
for col,val in zip(['spec_chi','hist_chi'],[spec_chi.item(),hist_chi.item()]): metrics_df.loc[iters,col]=val
# Checkpoint model for continuing run
if count == len(Dset.train_dataloader)-1: ## Check point at last step of epoch
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_last.tar')
if (checkpoint and (epoch > 1)): # Choose best models by metric
if hist_chi< best_chi1:
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_best_hist.tar')
best_chi1=hist_chi.item()
logging.info("Saving best hist model at epoch %s, step %s."%(epoch,iters))
if spec_chi< best_chi2:
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_best_spec.tar')
best_chi2=spec_chi.item()
logging.info("Saving best spec model at epoch %s, step %s"%(epoch,iters))
# if (iters in gdict['save_steps_list']) :
if ((gdict['save_steps_list']=='all') and (iters % gdict['checkpoint_size'] == 0)):
f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_{0}.tar'.format(iters))
logging.info("Saving given-step at epoch %s, step %s."%(epoch,iters))
# Save G's output on fixed_noise
if ((iters % gdict['checkpoint_size'] == 0) or ((epoch == epochs-1) and (count == len(Dset.train_dataloader)-1))):
gan_model.netG.eval()
with torch.no_grad():
fake = gan_model.netG(fixed_noise).detach().cpu()
img_arr=np.array(fake)
fname='gen_img_epoch-%s_step-%s'%(epoch,iters)
np.save(save_dir+'/images/'+fname,img_arr)
t_epoch_end=time.time()
if gdict['world_rank']==0:
logging.info("Time taken for epoch %s, count %s: %s for rank %s"%(epoch,count,t_epoch_end-t_epoch_start,gdict['world_rank']))
# Save Metrics to file after each epoch
metrics_df.to_pickle(save_dir+'/df_metrics.pkle')
logging.info("best chis: {0}, {1}".format(best_chi1,best_chi2))
```
## Start
```
### Setup modules ###
def f_manual_add_argparse():
''' use only in jpt notebook'''
args=argparse.Namespace()
args.config='config_3dgan_128_cori.yaml'
args.mode='fresh'
args.local_rank=0
args.facility='cori'
args.distributed=False
# args.mode='continue'
return args
def f_parse_args():
"""Parse command line arguments.Only for .py file"""
parser = argparse.ArgumentParser(description="Run script to train GAN using pytorch", formatter_class=argparse.ArgumentDefaultsHelpFormatter)
add_arg = parser.add_argument
add_arg('--config','-cfile', type=str, default='config_3d_Cgan.yaml', help='Name of config file')
add_arg('--mode','-m', type=str, choices=['fresh','continue','fresh_load'],default='fresh', help='Whether to start fresh run or continue previous run or fresh run loading a config file.')
add_arg("--local_rank", default=0, type=int,help='Local rank of GPU on node. Using for pytorch DDP. ')
add_arg("--facility", default='cori', choices=['cori','summit'],type=str,help='Facility: cori or summit ')
add_arg("--ddp", dest='distributed' ,default=False,action='store_true',help='use Distributed DataParallel for Pytorch or DataParallel')
return parser.parse_args()
def try_barrier(rank):
"""
Used in Distributed data parallel
Attempt a barrier but ignore any exceptions
"""
print('BAR %d'%rank)
try:
dist.barrier()
except:
pass
def f_init_gdict(args,gdict):
''' Create global dictionary gdict from args and config file'''
## read config file
config_file=args.config
with open(config_file) as f:
config_dict= yaml.load(f, Loader=yaml.SafeLoader)
gdict=config_dict['parameters']
args_dict=vars(args)
## Add args variables to gdict
for key in args_dict.keys():
gdict[key]=args_dict[key]
if gdict['distributed']:
assert not gdict['lambda_gp'],"GP couplings is %s. Cannot use Gradient penalty loss in pytorch DDP"%(gdict['lambda_gp'])
else : print("Not using DDP")
return gdict
def f_get_img_samples(ip_arr,rank=0,num_ranks=1):
'''
Module to get part of the numpy image file
'''
data_size=ip_arr.shape[0]
size=data_size//num_ranks
if gdict['batch_size']>size:
print("Caution: batchsize %s is greater than samples per GPU %s"%(gdict['batch_size'],size))
raise SystemExit
### Get a set of random indices from numpy array
random=False
if random:
idxs=np.arange(ip_arr.shape[0])
np.random.shuffle(idxs)
rnd_idxs=idxs[rank*(size):(rank+1)*size]
arr=ip_arr[rnd_idxs].copy()
else: arr=ip_arr[rank*(size):(rank+1)*size].copy()
return arr
def f_setup(gdict,metrics_df,log):
'''
Set up directories, Initialize random seeds, add GPU info, add logging info.
'''
torch.backends.cudnn.benchmark=True
# torch.autograd.set_detect_anomaly(True)
## New additions. Code taken from Jan B.
os.environ['MASTER_PORT'] = "8885"
if gdict['facility']=='summit':
get_master = "echo $(cat {} | sort | uniq | grep -v batch | grep -v login | head -1)".format(os.environ['LSB_DJOB_HOSTFILE'])
os.environ['MASTER_ADDR'] = str(subprocess.check_output(get_master, shell=True))[2:-3]
os.environ['WORLD_SIZE'] = os.environ['OMPI_COMM_WORLD_SIZE']
os.environ['RANK'] = os.environ['OMPI_COMM_WORLD_RANK']
gdict['local_rank'] = int(os.environ['OMPI_COMM_WORLD_LOCAL_RANK'])
else:
if gdict['distributed']:
os.environ['WORLD_SIZE'] = os.environ['SLURM_NTASKS']
os.environ['RANK'] = os.environ['SLURM_PROCID']
gdict['local_rank'] = int(os.environ['SLURM_LOCALID'])
## Special declarations
gdict['ngpu']=torch.cuda.device_count()
gdict['device']=torch.device("cuda" if (torch.cuda.is_available()) else "cpu")
gdict['multi-gpu']=True if (gdict['device'].type == 'cuda') and (gdict['ngpu'] > 1) else False
########################
###### Set up Distributed Data parallel ######
if gdict['distributed']:
# gdict['local_rank']=args.local_rank ## This is needed when using pytorch -m torch.distributed.launch
gdict['world_size']=int(os.environ['WORLD_SIZE'])
torch.cuda.set_device(gdict['local_rank']) ## Very important
dist.init_process_group(backend='nccl', init_method="env://")
gdict['world_rank']= dist.get_rank()
device = torch.cuda.current_device()
logging.info("World size %s, world rank %s, local rank %s device %s, hostname %s, GPUs on node %s\n"%(gdict['world_size'],gdict['world_rank'],gdict['local_rank'],device,socket.gethostname(),gdict['ngpu']))
# Divide batch size by number of GPUs
# gdict['batch_size']=gdict['batch_size']//gdict['world_size']
else:
gdict['world_size'],gdict['world_rank'],gdict['local_rank']=1,0,0
########################
###### Set up directories #######
### sync up so that time is the same for each GPU for DDP
if gdict['mode'] in ['fresh','fresh_load']:
### Create prefix for foldername
if gdict['world_rank']==0: ### For rank=0, create directory name string and make directories
dt_strg=datetime.now().strftime('%Y%m%d_%H%M%S') ## time format
dt_lst=[int(i) for i in dt_strg.split('_')] # List storing day and time
dt_tnsr=torch.LongTensor(dt_lst).to(gdict['device']) ## Create list to pass to other GPUs
else: dt_tnsr=torch.Tensor([0,0]).long().to(gdict['device'])
### Pass directory name to other ranks
if gdict['distributed']: dist.broadcast(dt_tnsr, src=0)
gdict['save_dir']=gdict['op_loc']+str(int(dt_tnsr[0]))+'_'+str(int(dt_tnsr[1]))+'_'+gdict['run_suffix']
if gdict['world_rank']==0: # Create directories for rank 0
### Create directories
if not os.path.exists(gdict['save_dir']):
os.makedirs(gdict['save_dir']+'/models')
os.makedirs(gdict['save_dir']+'/images')
shutil.copy(gdict['config'],gdict['save_dir'])
elif gdict['mode']=='continue': ## For checkpointed runs
gdict['save_dir']=gdict['ip_fldr']
### Read loss data
metrics_df=pd.read_pickle(gdict['save_dir']+'/df_metrics.pkle').astype(np.float64)
########################
### Initialize random seed
manualSeed = np.random.randint(1, 10000) if gdict['seed']=='random' else int(gdict['seed'])
# print("Seed",manualSeed,gdict['world_rank'])
random.seed(manualSeed)
np.random.seed(manualSeed)
torch.manual_seed(manualSeed)
torch.cuda.manual_seed_all(manualSeed)
if gdict['deterministic']:
logging.info("Running with deterministic sequence. Performance will be slower")
torch.backends.cudnn.deterministic=True
# torch.backends.cudnn.enabled = False
torch.backends.cudnn.benchmark = False
########################
if log:
### Write all logging.info statements to stdout and log file
logfile=gdict['save_dir']+'/log.log'
if gdict['world_rank']==0:
logging.basicConfig(level=logging.DEBUG, filename=logfile, filemode="a+", format="%(asctime)-15s %(levelname)-8s %(message)s")
Lg = logging.getLogger()
Lg.setLevel(logging.DEBUG)
lg_handler_file = logging.FileHandler(logfile)
lg_handler_stdout = logging.StreamHandler(sys.stdout)
Lg.addHandler(lg_handler_file)
Lg.addHandler(lg_handler_stdout)
logging.info('Args: {0}'.format(args))
logging.info('Start: %s'%(datetime.now().strftime('%Y-%m-%d %H:%M:%S')))
if gdict['distributed']: try_barrier(gdict['world_rank'])
if gdict['world_rank']!=0:
logging.basicConfig(level=logging.DEBUG, filename=logfile, filemode="a+", format="%(asctime)-15s %(levelname)-8s %(message)s")
return metrics_df
class Dataset:
def __init__(self,gdict):
'''
Load training dataset and compute spectrum and histogram for a small batch of training and validation dataset.
'''
## Load training dataset
t0a=time.time()
img=np.load(gdict['ip_fname'],mmap_mode='r')[:gdict['num_imgs']]
# print("Shape of input file",img.shape)
img=f_get_img_samples(img,gdict['world_rank'],gdict['world_size'])
t_img=torch.from_numpy(img)
dataset=TensorDataset(t_img)
self.train_dataloader=DataLoader(dataset,batch_size=gdict['batch_size'],shuffle=True,num_workers=0,drop_last=True)
logging.info("Size of dataset for GPU %s : %s"%(gdict['world_rank'],len(self.train_dataloader.dataset)))
t0b=time.time()
logging.info("Time for creating dataloader",t0b-t0a,gdict['world_rank'])
# Precompute spectrum and histogram for small training and validation data for computing losses
with torch.no_grad():
val_img=np.load(gdict['ip_fname'],mmap_mode='r')[-100:].copy()
t_val_img=torch.from_numpy(val_img).to(gdict['device'])
# Precompute radial coordinates
r,ind=f_get_rad(val_img)
self.r,self.ind=r.to(gdict['device']),ind.to(gdict['device'])
# Compute
self.train_spec_mean,self.train_spec_var=f_torch_image_spectrum(f_invtransform(t_val_img),1,self.r,self.ind)
self.train_hist=f_compute_hist(t_val_img,bins=gdict['bns'])
# Repeat for validation dataset
val_img=np.load(gdict['ip_fname'],mmap_mode='r')[-200:-100].copy()
t_val_img=torch.from_numpy(val_img).to(gdict['device'])
# Compute
self.val_spec_mean,self.val_spec_var=f_torch_image_spectrum(f_invtransform(t_val_img),1,self.r,self.ind)
self.val_hist=f_compute_hist(t_val_img,bins=gdict['bns'])
del val_img; del t_val_img; del img; del t_img;
class GAN_model():
def __init__(self,gdict,print_model=False):
def weights_init(m):
'''custom weights initialization called on netG and netD '''
classname = m.__class__.__name__
if classname.find('Conv') != -1:
nn.init.normal_(m.weight.data, 0.0, 0.02)
elif classname.find('BatchNorm') != -1:
nn.init.normal_(m.weight.data, 1.0, 0.02)
nn.init.constant_(m.bias.data, 0)
## Choose model
Generator, Discriminator=f_get_model(gdict) ## Mod for cGAN
# Create Generator
self.netG = Generator(gdict).to(gdict['device'])
self.netG.apply(weights_init)
# Create Discriminator
self.netD = Discriminator(gdict).to(gdict['device'])
self.netD.apply(weights_init)
if print_model:
if gdict['world_rank']==0:
print(self.netG)
# summary(netG,(1,1,64))
print(self.netD)
# summary(netD,(1,128,128))
print("Number of GPUs used %s"%(gdict['ngpu']))
if (gdict['multi-gpu']):
if not gdict['distributed']:
self.netG = nn.DataParallel(self.netG, list(range(gdict['ngpu'])))
self.netD = nn.DataParallel(self.netD, list(range(gdict['ngpu'])))
else:
self.netG=DistributedDataParallel(self.netG,device_ids=[gdict['local_rank']],output_device=[gdict['local_rank']])
self.netD=DistributedDataParallel(self.netD,device_ids=[gdict['local_rank']],output_device=[gdict['local_rank']])
#### Initialize networks ####
# self.criterion = nn.BCELoss()
self.criterion = nn.BCEWithLogitsLoss()
self.optimizerD = optim.Adam(self.netD.parameters(), lr=gdict['learn_rate_d'], betas=(gdict['beta1'], 0.999),eps=1e-7)
self.optimizerG = optim.Adam(self.netG.parameters(), lr=gdict['learn_rate_g'], betas=(gdict['beta1'], 0.999),eps=1e-7)
if gdict['distributed']: try_barrier(gdict['world_rank'])
if gdict['mode']=='fresh':
iters,start_epoch,best_chi1,best_chi2=0,0,1e10,1e10
elif gdict['mode']=='continue':
iters,start_epoch,best_chi1,best_chi2,self.netD,self.optimizerD,self.netG,self.optimizerG=f_load_checkpoint(gdict['save_dir']+'/models/checkpoint_last.tar',\
self.netG,self.netD,self.optimizerG,self.optimizerD,gdict)
if gdict['world_rank']==0: logging.info("\nContinuing existing run. Loading checkpoint with epoch {0} and step {1}\n".format(start_epoch,iters))
if gdict['distributed']: try_barrier(gdict['world_rank'])
start_epoch+=1 ## Start with the next epoch
elif gdict['mode']=='fresh_load':
iters,start_epoch,best_chi1,best_chi2,self.netD,self.optimizerD,self.netG,self.optimizerG=f_load_checkpoint(gdict['chkpt_file'],\
self.netG,self.netD,self.optimizerG,self.optimizerD,gdict)
if gdict['world_rank']==0: logging.info("Fresh run loading checkpoint file {0}".format(gdict['chkpt_file']))
# if gdict['distributed']: try_barrier(gdict['world_rank'])
iters,start_epoch,best_chi1,best_chi2=0,0,1e10,1e10
## Add to gdict
for key,val in zip(['best_chi1','best_chi2','iters','start_epoch'],[best_chi1,best_chi2,iters,start_epoch]): gdict[key]=val
## Set up learn rate scheduler
lr_stepsize=int((gdict['num_imgs'])/(gdict['batch_size']*gdict['world_size'])) # convert epoch number to step
lr_d_epochs=[i*lr_stepsize for i in gdict['lr_d_epochs']]
lr_g_epochs=[i*lr_stepsize for i in gdict['lr_g_epochs']]
self.schedulerD = optim.lr_scheduler.MultiStepLR(self.optimizerD, milestones=lr_d_epochs,gamma=gdict['lr_d_gamma'])
self.schedulerG = optim.lr_scheduler.MultiStepLR(self.optimizerG, milestones=lr_g_epochs,gamma=gdict['lr_g_gamma'])
```
## Main
```
#########################
### Main code #######
#########################
if __name__=="__main__":
jpt=False
jpt=True ##(different for jupyter notebook)
t0=time.time()
t0=time.time()
args=f_parse_args() if not jpt else f_manual_add_argparse()
#################################
### Set up global dictionary###
gdict={}
gdict=f_init_gdict(args,gdict)
# gdict['num_imgs']=200
if jpt: ## override for jpt nbks
gdict['num_imgs']=400
gdict['run_suffix']='nb_test'
### Set up metrics dataframe
cols=['step','epoch','Dreal','Dfake','Dfull','G_adv','G_full','spec_loss','hist_loss','spec_chi','hist_chi','gp_loss','fm_loss','D(x)','D_G_z1','D_G_z2','time']
metrics_df=pd.DataFrame(columns=cols)
# Setup
metrics_df=f_setup(gdict,metrics_df,log=(not jpt))
## Build GAN
gan_model=GAN_model(gdict,False)
fixed_noise = torch.randn(gdict['op_size'], 1, 1, 1, gdict['nz'], device=gdict['device']) #Latent vectors to view G progress # Mod for 3D
if gdict['distributed']: try_barrier(gdict['world_rank'])
## Load data and precompute
Dset=Dataset(gdict)
#################################
########## Train loop and save metrics and images ######
if gdict['distributed']: try_barrier(gdict['world_rank'])
if gdict['world_rank']==0:
logging.info(gdict)
logging.info("Starting Training Loop...")
f_train_loop(gan_model,Dset,metrics_df,gdict,fixed_noise)
if gdict['world_rank']==0: ## Generate images for best saved models ######
op_loc=gdict['save_dir']+'/images/'
ip_fname=gdict['save_dir']+'/models/checkpoint_best_spec.tar'
f_gen_images(gdict,gan_model.netG,gan_model.optimizerG,ip_fname,op_loc,op_strg='best_spec',op_size=32)
ip_fname=gdict['save_dir']+'/models/checkpoint_best_hist.tar'
f_gen_images(gdict,gan_model.netG,gan_model.optimizerG,ip_fname,op_loc,op_strg='best_hist',op_size=32)
tf=time.time()
logging.info("Total time %s"%(tf-t0))
logging.info('End: %s'%(datetime.now().strftime('%Y-%m-%d %H:%M:%S')))
# metrics_df.plot('step','time')
metrics_df
gan_model.optimizerG.param_groups[0]['lr']
# metrics_df['lr_d']
# summary(gan_model.netG,(1,1,64))
summary(gan_model.netD,(1,128,128,128))
# gdict
```
### Debug
```
# class Generator(nn.Module):
# def __init__(self, gdict):
# super(Generator, self).__init__()
# ## Define new variables from dict
# keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']
# ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
# self.main = nn.Sequential(
# # nn.ConvTranspose2d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
# nn.Linear(nz,nc*ngf*8*8*8),# 32768
# nn.BatchNorm2d(nc,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# View(shape=[-1,ngf*8,8,8]),
# nn.ConvTranspose2d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),
# nn.BatchNorm2d(ngf*4,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# # state size. (ngf*4) x 8 x 8
# nn.ConvTranspose2d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),
# nn.BatchNorm2d(ngf*2,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# # state size. (ngf*2) x 16 x 16
# nn.ConvTranspose2d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),
# nn.BatchNorm2d(ngf,eps=1e-05, momentum=0.9, affine=True),
# nn.ReLU(inplace=True),
# # state size. (ngf) x 32 x 32
# nn.ConvTranspose2d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),
# nn.Tanh()
# )
# def forward(self, ip):
# return self.main(ip)
# class Discriminator(nn.Module):
# def __init__(self, gdict):
# super(Discriminator, self).__init__()
# ## Define new variables from dict
# keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']
# ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())
# self.main = nn.Sequential(
# # input is (nc) x 64 x 64
# # nn.Conv2d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)
# nn.Conv2d(nc, ndf,kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf) x 32 x 32
# nn.Conv2d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf*2) x 16 x 16
# nn.Conv2d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf*4) x 8 x 8
# nn.Conv2d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),
# nn.BatchNorm2d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),
# nn.LeakyReLU(0.2, inplace=True),
# # state size. (ndf*8) x 4 x 4
# nn.Flatten(),
# nn.Linear(nc*ndf*8*8*8, 1)
# # nn.Sigmoid()
# )
# def forward(self, ip):
# # print(ip.shape)
# results=[ip]
# lst_idx=[]
# for i,submodel in enumerate(self.main.children()):
# mid_output=submodel(results[-1])
# results.append(mid_output)
# ## Select indices in list corresponding to output of Conv layers
# if submodel.__class__.__name__.startswith('Conv'):
# # print(submodel.__class__.__name__)
# # print(mid_output.shape)
# lst_idx.append(i)
# FMloss=True
# if FMloss:
# ans=[results[1:][i] for i in lst_idx + [-1]]
# else :
# ans=results[-1]
# return ans
# netG = Generator(gdict).to(gdict['device'])
# netG.apply(weights_init)
# # # # print(netG)
# # summary(netG,(1,1,64))
# # Create Discriminator
# netD = Discriminator(gdict).to(gdict['device'])
# netD.apply(weights_init)
# # print(netD)
# summary(netD,(1,128,128))
# noise = torch.randn(gdict['batchsize'], 1, 1, gdict['nz'], device=gdict['device'])
# fake = netG(noise)
# # Forward pass real batch through D
# output = netD(fake)
# print([i.shape for i in output])
0.5**10
70000/(8*6*8)
gdict.keys()
for key in ['batch_size','num_imgs','ngpu']:
print(key,gdict[key])
gdict['world_size']
```
| github_jupyter |
# Exploring Clustering Results
The file containing the clustering results is stored in the processed data folder with the suffix clean. The index is set to the first __Product group key__.
As a reminder the file is organized in three columns: _Product Group Key_, _Cluster Number_ and the corresponding _Centroid_ of the cluster.
```
import os
import sys
# add the 'src' directory as one where we can import modules
root_dir = os.path.join(os.getcwd(),os.pardir,os.pardir)
import pandas as pd
import math
import numpy as np
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import copy as cp
import seaborn as sns
import statsmodels.api as sm
from IPython.display import display
raw_path = os.path.join(root_dir,"data\\raw\\")
interim_path = os.path.join(root_dir,"data\\interim\\")
processed_path = os.path.join(root_dir,"data\\processed\\")
reports_path = os.path.join(root_dir,"reports\\")
models_path = os.path.join(root_dir,"models\\")
file_name = "euc_p2_clustering_clean_mois_v2.csv"
df_prd_cluster = pd.read_csv(models_path+file_name, sep=';', encoding='utf-8').drop('Unnamed: 0',axis=1).set_index('Product')
print(df_prd_cluster.shape)
df_prd_cluster.head()
```
## Get clients description
## Get Products description
In order to get the product features description, an inner join on the product group key is operated on the cluster result with the products description file.
Since the clustering was calculated on the second level group, some columns of the description file must be dropped in order to avoid duplicates of the first level products (mainly Promo and Standard version of the products)
```
file_name1 = "bnd_products_desc.csv"
file_name2 = "bnd_products_desc2.csv"
non_unique_features=["Key","Description","CONFEZIONE",\
"CONFEZIONE (Description)","IMBALLO","STD/PROMO","IMBALLO (Description)","STD/PROMO (Description)",\
"TIPO ARTICOLO","TIPO ARTICOLO (Description)"]
code_features = ["FAM DETTAGLIATA","FAM AGGREGATA","MARCHIO","GRUPPO MARCHIO","PACKAGING","SOTTO-TECNO",\
"PRODOTTO","CANALE DISTRIB","CLASSE COGE","FAM MARKETING","BIOLOGICO","GRUPPO MARCA COGE","Product Group key6"]
unbalanced = ["CANALE DISTRIB (Description)","CLASSE COGE (Description)","BIOLOGICO (Description)"]
df_produit1 = pd.read_csv(raw_path+file_name1, sep=';', encoding='iso8859_2')\
.drop(non_unique_features,axis=1)\
.drop(code_features,axis=1)\
.drop_duplicates()\
.dropna().reset_index(drop=True).apply(lambda x:x.astype(str).str.upper())
df_produit2 = pd.read_csv(raw_path+file_name2, sep=';', encoding='iso8859_2')\
.drop(non_unique_features,axis=1)\
.drop(code_features,axis=1)\
.drop_duplicates()\
.dropna().reset_index(drop=True).apply(lambda x:x.astype(str).str.upper())
m = pd.merge(df_produit1.iloc[:,:1],df_produit2.iloc[:,:1],how='outer',on=['Product Group key'],indicator='both').drop_duplicates()
dif = m[m['both']!='both'].reset_index(drop=True)
# df_produit = pd.concat([df_produit1,df_produit2], axis=0, ignore_index=True, copy=True)\
# .drop_duplicates(["Product Group key","Product Group key2"])
#df_produit1.to_csv(interim_path+"\\unique\\bnd_products_desc1.csv",sep=';',encoding='iso8859_2',index=False)
#df_produit2.to_csv(interim_path+"\\unique\\bnd_products_desc2.csv",sep=';',encoding='iso8859_2',index=False)
#df_produit.to_csv(interim_path+"\\unique\\bnd_products_desc.csv",sep=';',encoding='iso8859_2',index=False)
df_produit = df_produit2.drop_duplicates(["Product Group key"])
#Remove XX products weird
mask_XX = df_produit["Product Group key3"].str.endswith("XXX")
df_produit = df_produit[~mask_XX]
#Join with clusters
product_cluster = df_produit.join(df_prd_cluster,on='Product Group key',how='inner').reset_index(drop = True)
print(product_cluster.shape)
product_cluster.head()
#product_cluster.to_csv(interim_path+"\\unique\\bnd_product_cluster.csv",sep=';',encoding='iso8859_2',index=False)
display(df[["Product Group key2"]].drop_duplicates())
display(df[["Product Group key3"]].drop_duplicates())
display(df[["Product Group key4"]].drop_duplicates())
```
## Merge Products and Clients tables
```
# produit_client_cluster = pd.merge(product_cluster,client_df,how='inner', left_on=["Client"],right_on =["Key_lvl4"] )#.drop(["Key_lvl4"],axis=1)
# clusters = produit_client_cluster["Cluster"]
# centroids = produit_client_cluster["Centroid"]
# produit_client_cluster = produit_client_cluster.drop(["Cluster","Centroid"],axis=1)
# pos = len(produit_client_cluster.columns)
# produit_client_cluster.insert(pos,"Cluster",clusters)
# produit_client_cluster.insert(pos+1,"Centroid",centroids)
# all_features = produit_client_cluster.columns[:-2].drop(unbalanced)
# print(produit_client_cluster.shape)
# produit_client_cluster.tail()
# absent = produit_client_cluster[produit_client_cluster["Key_lvl5"].isnull()]["Client"].drop_duplicates()
# absent.tail()
all_features = product_cluster.columns[:-2]
```
Save the final result into a csv file for further exploration
```
filename = 'bnd_product_cluster_clean.csv'
file_name = "p2_clustering_clean_mois.csv"
product_cluster.to_csv(processed_path+filename,sep=';',encoding='iso8859_2')
```
# Homogeneity Test
In order to detect specific caraterstics for each resulted cluster we perform a statistic test based on Pearsons chi-square score with the hypothesis of a uniform distribution.
Features with the pvalues lower than 0.1 are displayed for analysis
```
def cramer_v(chisq,n,k,r=1):
return math.sqrt(chisq/(n * min(k-1,r-1) ))
```
## Calculate modalities frequency through clusters
As a first step, all the distrubtions of modalities across features and clusters are calculated and stored in one array structered as follows:
One array for each cluster which contains a dictionnary of features. Each feature is again a dictionary of modalities and their occurence in that cluster
```
#get the features
features = all_features
#get the clusters (actually its a range(1,nb_cluster))
clusters = set(product_cluster['Cluster'].values)
#array to store each cluster and freq for all the features
clusters_feature_dist = [0] #to shift the indices to clusters
#loop trhough features
for c in clusters:
feature_dist = dict()
for feature in features:
freq = product_cluster[product_cluster['Cluster']==c].groupby(feature)[feature].count()
feature_dist[feature]=freq.to_dict()
clusters_feature_dist.append(feature_dist)
```
## Chi-square test over clusters
```
from scipy.stats import chisquare
pthreashold = 0.2
#get the features
features = all_features
clusters = [6]
res_features_over_cluster = [0]
for c in clusters:
#align each feature with its distrubtion in this cluster c
cluster_feature_dist = clusters_feature_dist[c]
dist = [len(x) for x in list(cluster_feature_dist.values())]
keys = list(cluster_feature_dist.keys())
#plot the dist of number of elements by feature in this clust
plt.title("Feature distribution in the cluster %d"%c)
plt.bar(range(len(keys)),dist)
plt.xticks(range(len(keys)),keys,rotation=70)
#for each feature display its distribution over modalities
for feature in features:
#get information from the previous array
cluster_feature_dist = clusters_feature_dist[c]
feature_distribution = list(cluster_feature_dist[feature].values())
feature_keys = list(cluster_feature_dist[feature].keys())
nftrs = len(feature_keys)
chisq, p = chisquare(feature_distribution)
if p<pthreashold:
plt.figure()
plt.title("%s modalities distribution - pvalue = %.9f"%(feature,p))
plt.bar(np.arange(nftrs),feature_distribution)
plt.xticks(np.arange(nftrs)+(1.0/nftrs),feature_keys,rotation=70 if nftrs>5 else 0)
plt.show(block = True)
```
## Calculate modalities frequency through features
```
#get the features
features = all_features
#get the clusters (actually its a range(1,nb_cluster))
clusters = set(product_cluster['Cluster'].values)
#dict to store each feater and freq for all the clusters
features_clust_dist = dict()
#invert the dict and get it by feature
for f in features:
freq = dict()
for c in clusters:
freq[c] = clusters_feature_dist[c][f]
features_clust_dist[f] = freq
```
## Chi-square test over features
```
pthreashold = 0.2
clusters = set(product_cluster['Cluster'].values)
features = all_features
features = ["FAM MARKETING (Description)"]
for f in features:
for c in clusters:
#get information from the previous array
feature_clust_dist = features_clust_dist[f]
feature_distribution = list(feature_clust_dist[c].values())
feature_keys = list(feature_clust_dist[c].keys())
nftrs = len(feature_keys)
chisq, p = chisquare(feature_distribution)
if p<pthreashold:
plt.figure()
plt.title("%s: Cluster %d distribution - pvalue = %.9f"%(f,c,p))
plt.bar(np.arange(nftrs),feature_distribution)
plt.xticks(np.arange(nftrs)+(1.0/nftrs),feature_keys,rotation=70 if nftrs>5 else 0)
plt.show(block = True)
```
## Modalities distribution
```
clusters = set(product_cluster['Cluster'].values)
nclusters = len(clusters)
#get the features
features = product_cluster.columns[0:-2]
features = features.drop(unbalanced)
modalities_clust_dist = dict()
for f in features:
feature_sum=[]
modalities = set(product_cluster[f].values)
modalities_distribution=dict()
for m in modalities:
modality_distribution = np.zeros((nclusters+1))
for c in clusters:
#get information from the previous array
feature_clust_dist = features_clust_dist[f]
modality_distribution[c] +=(feature_clust_dist[c][m] if m in feature_clust_dist[c] else 0)
modalities_distribution[m] = modality_distribution
modalities_clust_dist[f] = modalities_distribution
```
## Chi-square test for modalities over clusters
```
%matplotlib inline
clusters = set(product_cluster['Cluster'].values)
nclusters = len(clusters)
pthreashold = 0.2
n_min_dist = 1
min_members = 5
min_dust = True
for f in features:
modalities = set(product_cluster[f].values)
r = len(modalities)
for m in modalities:
modality_dist = modalities_clust_dist[f][m]
md = np.count_nonzero(modality_dist)<=n_min_dist and np.max(modality_dist)>min_members
chisq, p = chisquare(modality_dist)
if p<pthreashold and (md and min_dust):
plt.figure()
plt.title("%s: %s Distribution - pvalue = %.9f"%(f,m,p))
plt.bar(np.arange(nclusters)+1,modality_dist[1:])
plt.xticks(np.arange(nclusters)+(1.0/nclusters)+1,np.arange(nclusters)+1,rotation=90,size=8)
if np.max(modality_dist[1:])<10: plt.ylim(0,10)
plt.show(block = True)
```
## MCA Analysis
### Remove unbalanced columns
```
features_df = product_cluster.iloc[:,3:-2]
plt.figure(figsize=(16,20))
features = features_df.columns
for i,f in enumerate(features):
counts = features_df.groupby([f])[f].count().to_dict()
dist = list(counts.values())
keys = list(counts.keys())
chisq, p = chisquare(dist)
plt.subplot(6,3,i+1)
plt.title("%s"%(f))
plt.bar(range(len(keys)),dist)
plt.xticks(range(len(keys)),keys,rotation=70)
if len(keys)>10: plt.tick_params(axis='x', which='both', bottom=False, top=False, labelbottom=False)
plt.subplots_adjust(wspace=0.5, hspace=0.5)
plt.show()
```
### Apply MCA on Products
```
import prince
unbalanced = ["CANALE DISTRIB (Description)","CLASSE COGE (Description)","BIOLOGICO (Description)"]
features_df = product_cluster.iloc[:,1:-2].drop(unbalanced,axis=1)
features_df = df_produit
mca = prince.MCA(features_df)
mca.plot_relationship_square()
plt.show()
```
### Apply MCA on Clients
```
import prince
features_df = client_df.astype(str).fillna("NA")
mca = prince.MCA(features_df)
mca.plot_relationship_square()
plt.show()
```
## Classification Tree
```
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
import subprocess
from sklearn.tree import export_graphviz
from sklearn.preprocessing import OneHotEncoder,LabelBinarizer,LabelEncoder
def visualize_tree(tree, feature_names,class_names=None):
with open(reports_path+"dt.dot", 'w') as f:
export_graphviz(tree, out_file=f, feature_names=feature_names, filled=True, rounded=True, class_names=class_names )
command = ["C:\\Program Files (x86)\\Graphviz2.38\\bin\\dot.exe", "-Tpng", reports_path+"dt.dot", "-o", "dt.png"]
try:
subprocess.check_call(command)
except:
exit("Could not run dot, ie graphviz, to "
"produce visualization")
drop = ["Product Group key","Centroid","PRODOTTO (Description)","PACKAGING (Description)"]
keep = ["FAM DETTAGLIATA (Description)","FAM AGGREGATA (Description)","MARCHIO (Description)","GRUPPO MARCHIO (Description)","SOTTO-TECNO (Description)","CANALE DISTRIB (Description)","CLASSE COGE (Description)","FAM MARKETING (Description)","Cluster"]
#data = product_cluster.drop(drop,axis=1)
data = product_cluster[keep]
# cat_data = []
# i=0
# for label,col in data.iteritems():
# cat_data.append(col.astype('category'))
# df = pd.DataFrame(np.array(cat_data).T,columns = data.columns)
lb = LabelBinarizer()
X = pd.get_dummies(data.drop(["Cluster"],axis=1).iloc[:,:])
print(X.shape)
display(data.head())
features = X.columns
y = lb.fit_transform(data.values[:,-1].astype(int).T)
y = data.values[:,-1].astype(int)
x_data = data.drop(["Cluster"],axis=1).iloc[:,:]
y_data = data.values[:,-1].astype(int)
from sklearn.feature_extraction import DictVectorizer
X_dict = x_data.T.to_dict().values()
vect = DictVectorizer(sparse=False)
X_vector = vect.fit_transform(X_dict)
print(X_vector.shape)
y = lb.fit_transform(y_data.T)
y = y_data.T
X = X_vector
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
classifier = DecisionTreeClassifier(criterion = "gini", max_depth=None, min_samples_leaf=1)
classifier.fit(X_train, y_train)
# from sklearn.svm import SVC
# classifier = SVC()
# classifier.fit(X_train, y_train)
y_pred = classifier.predict(X_test)
```
## Evaluation the algorithm
```
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.metrics import precision_recall_fscore_support as report
#print(confusion_matrix(y_test, y_pred))
precision,recall,fscore,support = report(y_test, y_pred,warn_for=())
print(precision.mean(),recall.mean(),fscore.mean(),support.sum())
#print(classification_report(y_test, y_pred))
# imp = np.array(classifier.feature_importances_)
# imp_ft = features[np.argsort(imp)[::-1]]
# print(imp_ft.values)
visualize_tree(classifier, features,class_names=True)
from graphviz import Graph,Source
from IPython.display import SVG
graph = Source(export_graphviz(classifier, out_file=None
, feature_names=features, class_names=True
, filled = True))
display(SVG(graph.pipe(format='svg')))
print(classifier.tree_)
```
| github_jupyter |
```
%matplotlib inline
```
# Decoding in time-frequency space using Common Spatial Patterns (CSP)
The time-frequency decomposition is estimated by iterating over raw data that
has been band-passed at different frequencies. This is used to compute a
covariance matrix over each epoch or a rolling time-window and extract the CSP
filtered signals. A linear discriminant classifier is then applied to these
signals.
```
# Authors: Laura Gwilliams <[email protected]>
# Jean-Remi King <[email protected]>
# Alex Barachant <[email protected]>
# Alexandre Gramfort <[email protected]>
#
# License: BSD (3-clause)
import numpy as np
import matplotlib.pyplot as plt
from mne import Epochs, create_info, events_from_annotations
from mne.io import concatenate_raws, read_raw_edf
from mne.datasets import eegbci
from mne.decoding import CSP
from mne.time_frequency import AverageTFR
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import StratifiedKFold, cross_val_score
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import LabelEncoder
```
Set parameters and read data
```
event_id = dict(hands=2, feet=3) # motor imagery: hands vs feet
subject = 1
runs = [6, 10, 14]
raw_fnames = eegbci.load_data(subject, runs)
raw = concatenate_raws([read_raw_edf(f) for f in raw_fnames])
# Extract information from the raw file
sfreq = raw.info['sfreq']
events, _ = events_from_annotations(raw, event_id=dict(T1=2, T2=3))
raw.pick_types(meg=False, eeg=True, stim=False, eog=False, exclude='bads')
raw.load_data()
# Assemble the classifier using scikit-learn pipeline
clf = make_pipeline(CSP(n_components=4, reg=None, log=True, norm_trace=False),
LinearDiscriminantAnalysis())
n_splits = 5 # how many folds to use for cross-validation
cv = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=42)
# Classification & time-frequency parameters
tmin, tmax = -.200, 2.000
n_cycles = 10. # how many complete cycles: used to define window size
min_freq = 5.
max_freq = 25.
n_freqs = 8 # how many frequency bins to use
# Assemble list of frequency range tuples
freqs = np.linspace(min_freq, max_freq, n_freqs) # assemble frequencies
freq_ranges = list(zip(freqs[:-1], freqs[1:])) # make freqs list of tuples
# Infer window spacing from the max freq and number of cycles to avoid gaps
window_spacing = (n_cycles / np.max(freqs) / 2.)
centered_w_times = np.arange(tmin, tmax, window_spacing)[1:]
n_windows = len(centered_w_times)
# Instantiate label encoder
le = LabelEncoder()
```
Loop through frequencies, apply classifier and save scores
```
# init scores
freq_scores = np.zeros((n_freqs - 1,))
# Loop through each frequency range of interest
for freq, (fmin, fmax) in enumerate(freq_ranges):
# Infer window size based on the frequency being used
w_size = n_cycles / ((fmax + fmin) / 2.) # in seconds
# Apply band-pass filter to isolate the specified frequencies
raw_filter = raw.copy().filter(fmin, fmax, n_jobs=1, fir_design='firwin',
skip_by_annotation='edge')
# Extract epochs from filtered data, padded by window size
epochs = Epochs(raw_filter, events, event_id, tmin - w_size, tmax + w_size,
proj=False, baseline=None, preload=True)
epochs.drop_bad()
y = le.fit_transform(epochs.events[:, 2])
X = epochs.get_data()
# Save mean scores over folds for each frequency and time window
freq_scores[freq] = np.mean(cross_val_score(estimator=clf, X=X, y=y,
scoring='roc_auc', cv=cv,
n_jobs=1), axis=0)
```
Plot frequency results
```
plt.bar(freqs[:-1], freq_scores, width=np.diff(freqs)[0],
align='edge', edgecolor='black')
plt.xticks(freqs)
plt.ylim([0, 1])
plt.axhline(len(epochs['feet']) / len(epochs), color='k', linestyle='--',
label='chance level')
plt.legend()
plt.xlabel('Frequency (Hz)')
plt.ylabel('Decoding Scores')
plt.title('Frequency Decoding Scores')
```
Loop through frequencies and time, apply classifier and save scores
```
# init scores
tf_scores = np.zeros((n_freqs - 1, n_windows))
# Loop through each frequency range of interest
for freq, (fmin, fmax) in enumerate(freq_ranges):
# Infer window size based on the frequency being used
w_size = n_cycles / ((fmax + fmin) / 2.) # in seconds
# Apply band-pass filter to isolate the specified frequencies
raw_filter = raw.copy().filter(fmin, fmax, n_jobs=1, fir_design='firwin',
skip_by_annotation='edge')
# Extract epochs from filtered data, padded by window size
epochs = Epochs(raw_filter, events, event_id, tmin - w_size, tmax + w_size,
proj=False, baseline=None, preload=True)
epochs.drop_bad()
y = le.fit_transform(epochs.events[:, 2])
# Roll covariance, csp and lda over time
for t, w_time in enumerate(centered_w_times):
# Center the min and max of the window
w_tmin = w_time - w_size / 2.
w_tmax = w_time + w_size / 2.
# Crop data into time-window of interest
X = epochs.copy().crop(w_tmin, w_tmax).get_data()
# Save mean scores over folds for each frequency and time window
tf_scores[freq, t] = np.mean(cross_val_score(estimator=clf, X=X, y=y,
scoring='roc_auc', cv=cv,
n_jobs=1), axis=0)
```
Plot time-frequency results
```
# Set up time frequency object
av_tfr = AverageTFR(create_info(['freq'], sfreq), tf_scores[np.newaxis, :],
centered_w_times, freqs[1:], 1)
chance = np.mean(y) # set chance level to white in the plot
av_tfr.plot([0], vmin=chance, title="Time-Frequency Decoding Scores",
cmap=plt.cm.Reds)
```
| github_jupyter |
## week03: Логистическая регрессия и анализ изображений
В этом ноутбуке предлагается построить классификатор изображений на основе логистической регрессии.
*Забегая вперед, мы попробуем решить задачу классификации изображений используя лишь простые методы. В третьей части нашего курса мы вернемся к этой задаче.*
```
import numpy as np
import matplotlib.pyplot as plt
import h5py
%matplotlib inline
```
## 1. Постановка задачи ##
**Задача**: Есть датасет [прямая ссылка](https://drive.google.com/file/d/15tOimf2QYWsMtPJXTUCwgZaOTF8Nxcsm/view?usp=sharing) ("catvnoncat.h5") состоящий из:
- обучающей выборки из m_train изображений, помеченных "cat" (y=1) или "non-cat" (y=0)
- тестовой выборки m_test изображений, помеченных "cat" или "non-cat"
- каждое цветное изображение имеет размер (src_size, src_size, 3), где 3 - число каналов (RGB).
Таким образом, каждый слой - квадрат размера src_size x src_size$.
Давайте построим простой алгоритм классификации изображений на классы "cat"/"non-cat".
Автоматическая загрузка доступна ниже.
<img src="img/LogReg_kiank.png" style="width:650px;height:400px;">
**Recap**:
Для каждого примера $x^{(i)}$:
$$z^{(i)} = w^T x^{(i)} + b \tag{1}$$
$$\hat{y}^{(i)} = a^{(i)} = sigmoid(z^{(i)})\tag{2}$$
$$ \mathcal{L}(a^{(i)}, y^{(i)}) = - y^{(i)} \log(a^{(i)}) - (1-y^{(i)} ) \log(1-a^{(i)})\tag{3}$$
Функция потерь:
$$ J = \frac{1}{m} \sum_{i=1}^m \mathcal{L}(a^{(i)}, y^{(i)})\tag{6}$$
```
# Uncomment this cell to download the data
# !wget "https://downloader.disk.yandex.ru/disk/7ef1d1e30e23740a4a30799a825319154815ddc85bf689542add0a3d11ccb91c/5d7fdcb0/3dcxK38Q0fG3ui0g2gMZgKkLls8ULwVpoYNkWpBm9d24EceJ6mIoH5l3_wKkFv3PfZ0WMGYjfJULynuJkuGaug%3D%3D?uid=76549735&filename=data.zip&disposition=attachment&hash=&limit=0&content_type=application%2Fzip&owner_uid=76549735&fsize=2815580&hid=084389255415f71a92d0f1024ab741d4&media_type=compressed&tknv=v2&etag=2b348ac8eca72d223108e36b2a671210" -O data.zip
# !unzip data.zip
```
### 1.1 Загрузка данных и визуализация ###
```
def load_dataset():
train_data = h5py.File("data/train_catvnoncat.h5", "r")
train_set_x_orig = np.array(train_data["train_set_x"][:]) # признаки
train_set_y_orig = np.array(train_data["train_set_y"][:]) # метки классов
test_data = h5py.File("data/test_catvnoncat.h5", "r")
test_set_x_orig = np.array(test_data["test_set_x"][:]) # признаки
test_set_y_orig = np.array(test_data["test_set_y"][:]) # метки классов
classes = np.array(test_data["list_classes"][:]) # the list of classes
classes = np.array(list(map(lambda x: x.decode('utf-8'), classes)))
train_set_y = train_set_y_orig.reshape(train_set_y_orig.shape[0])
test_set_y = test_set_y_orig.reshape(test_set_y_orig.shape[0])
return train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes
train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes = load_dataset()
```
Цветные изображения в формате RGB представлены в виде трёхмерных numpy.array.
Порядок измерений $H \times W \times C$: $H$ - высота, $W$ - ширина и $C$ - число каналов.
Значение каждого пиксела находится в интервале $[0;255]$.
```
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
def show_image_interact(i=0):
f, ax = plt.subplots(1,4, figsize=(15,20), sharey=True)
ax[0].imshow(train_set_x_orig[i])
ax[0].set_title('RGB image')
ax[1].imshow(train_set_x_orig[i][:,:,0], cmap='gray')
ax[1].set_title('R channel')
ax[2].imshow(train_set_x_orig[i][:,:,1], cmap='gray')
ax[2].set_title('G channel')
ax[3].imshow(train_set_x_orig[i][:,:,2], cmap='gray')
ax[3].set_title('B channel')
print("y = {} belongs to '{}' class.".format(str(train_set_y[i]),classes[np.squeeze(train_set_y[i])]))
interact(show_image_interact,
i=widgets.IntSlider(min=0, max=len(train_set_y)-1, step=1))
```
При работе с данными полезно будет сохранить размерности входных изображений для дальнейшей обработки.
```
m_train = train_set_x_orig.shape[0]
m_test = test_set_x_orig.shape[0]
src_size = train_set_x_orig.shape[1]
print ("Размер обучающей выборки: m_train = " + str(m_train))
print ("Размер тестовой выборки: m_test = " + str(m_test))
print ("Ширина/Высота каждого изображения: src_size = " + str(src_size))
print ("Размерны трёхмерной матрицы для каждого изображения: (" + str(src_size) + ", " + str(src_size) + ", 3)")
print ("Размерность train_set_x: " + str(train_set_x_orig.shape))
print ("Размерность train_set_y: " + str(train_set_y.shape))
print ("Размерность test_set_x: " + str(test_set_x_orig.shape))
print ("Размерность test_set_y: " + str(test_set_y.shape))
```
## 2. Предварительная обработка
Преобразуем входные изображения размера (num_px, num_px, 3) в вектор признаков размера (num_px $*$ num_px $*$ 3, 1), чтобы сформировать матрицы объект-признак в виде numpy-array для обучающей и тестовой выборок.
Каждой строке матрицы объект-признак соответствует входное развёрнутое в вектор-строку изображение.
Помимо этого, для предварительной обработки (препроцессинга) изображений применяют центрирование значений: из значения каждого пиксела вычитается среднее и делят полученное значение на среднеквадратичное отклонение значений пикселей всего изображения.
Однако, на практике обычно просто делят значения пикселей на 255 (максимальное значение пикселя).
Оформим эти шаги в функцию предварительной обработки
```
def image_preprocessing_simple(data):
assert type(data) == np.ndarray
assert data.ndim == 4
n,h,w,c = data.shape
data_vectorized = <ваш код>
data_normalized = <ваш код>
return data_normalized
# Изменить размеры входных данных
train_set_x_vectorized = image_preprocessing_simple(train_set_x_orig)
test_set_x_vectorized = image_preprocessing_simple(test_set_x_orig)
print('Train set:')
print("Размеры train_set_x_vectorized: {}".format(str(train_set_x_vectorized.shape)))
print("Размеры train_set_y: {}".format(str(train_set_y.shape)))
print("Размеры классов 'cat'/'non-cat': {} / {}".format(sum(train_set_y==1), sum(train_set_y==0)))
print('Test set:')
print("Размеры test_set_x_vectorized: {}".format(str(test_set_x_vectorized.shape)))
print("Размеры test_set_y: {}".format(str(test_set_y.shape)))
print("Размеры классов 'cat'/'non-cat': {} / {}".format(sum(test_set_y==1), sum(test_set_y==0)))
```
## 3. Классификация
```
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
import warnings
warnings.filterwarnings('ignore')
```
**Вопрос**: Какую метрику качества стоит использовать?
### 3.1 Построение модели
Построим модель с параметрами по умолчанию и посмотрим, как хорошо она справится с задачей.
```
clf = # <ваш код>
score = <ваш код>
print('Точность для простой модели с параметрами по умолчанию: {:.4f}'.format(score))
# from sklearn.metrics import f1_score
y_predicted = clf.predict(test_set_x_vectorized)
correct_score = <ваш код>
print('<Имя метрики> для простой модели: {:.4f}'.format(correct_score))
```
Попробуем подобрать параметры регуляризации в надежде, что это повысит точность предсказаний.
```
<ваш код>
print('Оптимальные параметры: {}'.format(<ваш код>))
print('Наилучшее значение метрики качества: {}'.format(<ваш код>))
```
Обучим модель с оптимальными параметрами на всей обучающей выборке и посмотрим на метрики качества:
```
best_clf = <ваш код>
best_clf.fit(train_set_x_vectorized, train_set_y)
y_predicted = best_clf.predict(test_set_x_vectorized)
metric_score = <ваш код>(y_predicted, test_set_y)
print('Optimal model hyperparameters accuracy score: {:.4f}'.format(metric_score))
```
### 3.2 Анализ ошибок
```
is_outlier = (y_predicted != test_set_y)
test_outliers_x, test_outliers_y, predicted_y = test_set_x_orig[is_outlier], test_set_y[is_outlier], y_predicted[is_outlier]
def show_image_outliers(i=0):
f = plt.figure(figsize=(5,5))
plt.imshow(test_outliers_x[i])
plt.title('RGB image')
fmt_string = "Sample belongs to '{}' class, but '{}' is predicted'"
print(fmt_string.format(classes[test_outliers_y[i]], classes[predicted_y[i]]))
interact(show_image_outliers,
i=widgets.IntSlider(min=0, max=len(test_outliers_y)-1, step=1))
```
**Вопрос**: Как по-вашему можно повысить точность? Каким недостатком обладает данный подход к классификации?
### 3.3 Модель с аугментациями
Как можно увеличить количество данных для обучения?
Сформировать новые примеры из уже имеющихся!
Например, можно пополнить class 'cat' обучающей выборки [зеркально отображёнными](https://docs.scipy.org/doc/numpy/reference/generated/numpy.fliplr.html) изображениями котов.
```
def augment_sample(src, label):
<ваш код>
def image_preprocessing_augment(data, labels):
assert type(data) == np.ndarray
assert data.ndim == 4
## ВАШ КОД ##
data_augmented =
labels_augmented =
## ВАШ КОД ЗАКАНЧИВАЕТСЯ ЗДЕСЬ ##
n,h,w,c = data_augmented.shape
data_vectorized = data_augmented.reshape(n, -1) # <ваш код>
data_normalized = data_vectorized / 255
return data_normalized, labels_augmented
train_set_x_augmented, train_set_y_augmented = image_preprocessing_augment(train_set_x_orig, train_set_y)
clf = LogisticRegression(solver='liblinear')
clf.fit(train_set_x_augmented, train_set_y_augmented)
y_pred = clf.predict(test_set_x_vectorized)
print('F-мера для модели с аугментациями: {:.4f}'.format(f1_score(y_pred, test_set_y)))
```
## 4. Проверьте работу классификатора на своей картинке
Библиотека [OpenCV](https://opencv.org) для работы с изображениями для [python](https://pypi.org/project/opencv-python/):
`pip install opencv-python`
Вместе с contrib-модулями:
`pip install opencv-contrib-python`
```
import cv2
# Путь к картинке на вашем ПК
fname = "cat-non-cat.jpg"
# Считываем картинку через scipy
src = cv2.cvtColor(cv2.imread((fname)), cv2.COLOR_BGR2RGB)
src_resized = cv2.resize(src, (src_size,src_size), interpolation=cv2.INTER_LINEAR).reshape(1, src_size*src_size*3)
my_image_predict = clf.predict(src_resized)[0]
plt.imshow(src)
print("Алгоритм говорит, что это '{}': {}".format(my_image_predict, classes[my_image_predict]))
```
| github_jupyter |
# Using results
Since json is a dictionary, you can pull out a single datapoint using the key.
```
{
"source": "ensembl_havana",
"object_type": "Gene",
"logic_name": "ensembl_havana_gene",
"version": 12,
"species": "homo_sapiens",
"description": "B-Raf proto-oncogene, serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1097]",
"display_name": "BRAF",
"assembly_name": "GRCh38",
"biotype": "protein_coding",
"end": 140924764,
"seq_region_name": "7",
"db_type": "core",
"strand": -1,
"id": "ENSG00000157764",
"start": 140719327
}
```
We can add this to our previous script:
```
import requests, json
from pprint import pprint
def fetch_endpoint(server, request, content_type):
r = requests.get(server+request, headers={ "Accept" : content_type})
if not r.ok:
r.raise_for_status()
sys.exit()
if content_type == 'application/json':
return r.json()
else:
return r.text
server = "http://rest.ensembl.org/"
ext = "lookup/id/ENSG00000157764?"
con = "application/json"
get_gene = fetch_endpoint(server, ext, con)
symbol = get_gene['display_name']
print (symbol)
```
## Exercises 3
1\. Write a script to lookup the gene called *ESPN* in human and print the stable ID of this gene.
```
# Exercise 3.1
#!/usr/bin/env python
# Get modules needed for script
import sys, requests, json
from pprint import pprint
def fetch_endpoint(server, request, content_type):
r = requests.get(server+request, headers={ "Accept" : content_type})
if not r.ok:
r.raise_for_status()
sys.exit()
if content_type == 'application/json':
return r.json()
else:
return r.text
# define the gene name
gene_name = "ESPN"
# define the general URL parameters
server = "http://rest.ensembl.org/"
# define REST query to get the gene ID from the gene name
ext_get_lookup = "lookup/symbol/homo_sapiens/" + gene_name + "?"
# define the content type
con = "application/json"
# submit the query
get_lookup = fetch_endpoint(server, ext_get_lookup, con)
#pprint(get_lookup)
pprint(get_lookup['id'])
```
2\. Get all variants that are associated with the phenotype 'Coffee consumption'. For each variant print
a. the p-value for the association
b. the PMID for the publication which describes the association between that variant and ‘Coffee consumption’
c. the risk allele and the associated gene.
```
# Exercise 3.2
pprint(get_lookup)
```
3\. Get the mouse homologue of the human BRCA2 and print the ID and sequence of both.
Note that the JSON for the endpoint you need is several layers deep, containing nested lists (appear as square brackets [ ] in the JSON) and key value sets (dictionary; appear as curly brackets { } in the JSON). Pretty print (pprint) comes in very useful here for the intermediate stage when you're trying to work out the json.
```
# Exercise 3.3
```
[Next page: Exercises 3 – answers](3_Using_results_answers.ipynb)
| github_jupyter |
```
%%html
<style>div.run_this_cell{display:block;}</style>
<style>table {float:left;width:100%;}</style>
```
<img style="float:right;margin-left:50px;margin-right:50px;" width="300" src="images/discovercoding.png">
# 1. Welcome to the Hour of Callysto!
Lesson created and taught by [Discover Coding](https://discovercoding.ca).
With support and funding from [Callysto](https://callysto.ca/) and the [Pacific Institute for Mathematical Sciences](https://www.pims.math.ca/)
<img style="float:right;padding-left:50px;padding-right:50px;" width="400" src="images/library.jpg">
## 2. What is Callysto?
<div style="padding-top:20px;padding-left:50px;font-size:large;">
- **Callysto** is a free and online collection of special textbooks for Canadian students.
- It's your own personal school library of the future.
</div>
<img style="float:left;padding-left:50px;padding-right:10px;margin-right:50px;" width="400px" src="images/notebook.jpg">
<div style="padding-left:50px;padding-top:20px;font-size:large;">
- Callysto uses **Jupyter** notebooks to display text, images, videos, and even code!
- It's a notebook where you **WRITE** and **RUN** code!
- It's the same tool used in universities by programmers and data scientists!
</div>
<img style="float:right;padding-right:100px;margin-right:100px;" width="200px" src="images/python.png">
<div style="padding-left:50px;font-size:large;">
- We are going to use this notebook to learn *coding* with **Python**.
- Python *code* are instructions in a *language* that a computer can understand.
- When we are *coding*, we are telling the computer what to do!
</div>
## 3. Getting Started with Callysto
You've already made it here! To use Callysto:
1. Log into Callysto: https://hub.callysto.ca
1. Find an interesting notebook [from here](https://callysto.ca/learning_modules/)
1. Click on it to get your own copy of it
1. Open, read, run it from the hub (look for the .ipynb file)
<img src="images/hub.png">
# 4. Python
What makes these notebooks cool is that we can write and run code, such as *Python*, directly inside the notebook! We're not just *READING* a textbook anymore; we can now use it to solve problems for us!
### *Example 4.1*
Let's run our first program using [Turtle graphics](https://en.wikipedia.org/wiki/Turtle_graphics)! Select the code block below and click <img src="images/run-button.png" style="display:inline-block;"> or press `CTRL+ENTER`
```
# My First Turtle Program!
from mobilechelonian import Turtle
ted = Turtle()
ted.forward(50)
```
Let's break down what we each line of code that we see:
1. `# My First Turtle Program!` - Any line starting with a `#` is a comment (or notes). It is NOT CODE!
1. `from mobilechelonian import Turtle` - This lets use some `Turtle` code that someone else wrote. We won't worry about it for this class.
1. `ted = Turtle()` - This creates our turtle, named `ted`.
1. `ted.forward(50)` - This tells our turtle `ted` to move `forward` by 50 spaces.
### *ACTIVITY 4.2*
Let's experiment with the turtle. Change the code above, and re-run it.
Can you:
1. Change how far `forward` turtle moves. How far can it go?
1. Can you make turtle move a *negative* number?
1. Instead of moving `forward`, can you tell turtle to move `backward`?
1. CHALLENGE 1: Can you rename the turtle from `ted` to a better name?
1. CHALLENGE 2: Can you tell your turtle to touch BOTH edges of the screen, by only going `backward`?
## 5. More Turtle *Functions*
`forward()` and `backward()` are called *Functions*. They are commands that a `Turtle()`, like `ted`, understands.
There are more *functions* that we can use to make turtles do more interesting things.
| Function | What it does |Example|
|:---|:---|:---|
| `Turtle.speed(number)` | Set the speed of our turtle, between 1-10 | `t.speed(7)` |
| `Turtle.right(degrees)` | Turn the turtle `number` of degrees to the right | `t.right(90)` |
| `Turtle.left(degrees)` | Turn the turtle `number` of degrees to the left | `t.left(90)` |
| `Turtle.pencolor('color')` | Sets the color of the turtle’s line. <br> The color can be a [color name from this list](https://www.w3schools.com/tags/ref_colornames.asp) | `t.pencolor('Blue')` |
||||
**OBSERVE!!** The functions are applied to `Turtle` objects using the names you gave them.
The examples above use a Turtle named `t`. You can create more than one turtle, and give them different names!
**Now we can draw more shapes with turtle, and do it faster!**
### *Example 5.1*
Let's make a 2D shape, like a triangle!
Run the following code:
```
# My Turtle is now 2D!
from mobilechelonian import Turtle
ted = Turtle()
ted.speed(5)
ted.forward(100)
ted.left(120)
ted.forward(100)
ted.left(120)
ted.forward(100)
```
### *ACTIVITY 5.2*
Can you change the color of the triangle?
Try inserting this line of code into Example 5.1:
`ted.pencolor('red')`
Try other [colors named here](https://www.w3schools.com/tags/ref_colornames.asp)
**Observe**
1. What happens if you write this BEFORE the first move `forward` (on line 5)?
1. What happens if you write this AFTER the last move `forward` (on line 9)?
<img style="float:right;margin-right:100px;" width="300px" src="images/turtle-house.png">
### *ACTIVITY 5.3*
Can you make a program where the turtle draws a house that looks like this one?
- **Hint 1:** Start by copy-and-paste from example 5.1
- **Hint 2:** After drawing the triangle, turtle should turn 30 degrees
- **Hint 3:** Draw a line (100 is a good length), turn, draw a line, turn, draw a line...
```
# My Turtle is now 2D!
from mobilechelonian import Turtle
ted = Turtle()
ted.speed(1)
# hint 1 - copy and paste the previous code
# hint 2 - the next turn should be 30 degrees
# hint 3 - forward, turn, forward, turn, forward...
```
# 6. LOOPS
<img style="float:right;" width="30%" src="images/loop.gif">
Did you notice that drawing shapes used the same lines of code over and over?
Instead of typing the same things over and over, we can use a *loop* to run the same code multiple times.
Loops run code __*OVER and OVER and OVER and . . .*__
Loops have two parts:
1. A line starting with the special keyword `for` or `while`
1. *indented* lines of code which are run each time
### *Example 6.1*
Below is an example of a loop using the special keyword `for`. This code says:
- `for` each number called `index`, in the `range` from 0 up to (but not including) 10, move forward, turn right
*Can you predict what it will draw?*
RUN the following code and see!
```
from mobilechelonian import Turtle
ted = Turtle()
ted.speed(5)
# This loop runs (how many?) times
for index in range(10):
ted.forward(50)
ted.right(80)
```
### *ACTIVITY 6.2*
Experiment with the code in Example 6.1, and try different values.
Can you:
1. Make turtle draw faster?
1. Change how BIG the shape is? (Hint: change the distance turtle moves `forward`)
1. Change how much the turtle turns each time?
1. What happens when turtle turns less than 90?
1. What happens when turtle turns 90?
1. What happens when turtle turns more than 90?
1. Can you add some color?
# 7. RANDOM
<img style="float:left;margin-right:30px;" width="400px" src="https://media.giphy.com/media/H4uFElBB9Nt7zq3RZ9/source.gif">
So far, we've only been drawing with one color.
Let's make it a little more interesting with using RANDOM colors!
We'll do 3 things:
1. First, use `import random` in our code.
1. Next, use `random.randint(0,255)` to pick random numbers from 0 to 255
1. Last, use the 3 numbers to set a new color using `RGB( number, number, number )`
Don't worry about too much about how this code works.
### *Example 7.1*
Let's try it out!
Run the following code:
```
from mobilechelonian import Turtle
import random
ted = Turtle()
ted.speed(10)
# This loop runs 10 times
for index in range(10):
red = random.randint(0,255)
green = random.randint(0,255)
blue = random.randint(0,255)
random_color = "RGB(%d,%d,%d)" % (red,green,blue)
ted.pencolor(random_color)
ted.forward(50)
ted.right(80)
```
### *ACTIVITY 7.2*
Let's experiment with the code in Example 7.1
1. On line 9, we pick a random number between 0 to 255 for `red`. What happens if we pick a random number between 200 to 255?
1. What happens if we pick a random number between 0 and 10 for the color `red`?
1. Try to change the range of random numbers for `green` and `blue`
### *Example 7.3*
Let's try a loop where we draw a SQUARE each time, but do an extra *small* turn before we draw the next square...
```
from mobilechelonian import Turtle
import random
ted = Turtle()
ted.speed(10)
# Now we'll run a loop.
for index in range(5):
red = random.randint(0,255)
green = random.randint(0,255)
blue = random.randint(0,255)
random_color = "RGB(%d,%d,%d)" % (red,green,blue)
ted.pencolor(random_color)
ted.forward(100)
ted.right(90)
ted.forward(100)
ted.right(90)
ted.forward(100)
ted.right(90)
ted.forward(100)
ted.right(90)
ted.right(20) # Additional small turn before we draw the next square to make a pattern
```
### *ACTIVITY 7.4*
The picture in Example 7.3 looks incomplete....
1. Can you increase the number of times the loop will run so it'll look better?
1. Can you make a different pattern just by changing the amount you `right()` turn on line 23?
1. Can you create a SECOND pattern using another LOOP after the first one completes?
- HINT 1: COPY and PASTE the ENTIRE LOOP
- HINT 2: Make sure the second `for` is NOT indented (but the rest of the code IS indented)
- HINT 3: Change the distance you move `forward()`, and the last `right()` turn
# 8. SUMMARY
CONGRATULATIONS for making it to the end of this notebook!
Today, you learned:
1. What is Callysto, Jupyter Notebooks, and Python
1. Wrote a program to draw line art using `turtle`
1. Used *LOOPS* and *RANDOM* to draw colorful patterns
***
### GREAT JOB!
### Continue modifying the examples or re-do the activities to make more cool art!
| github_jupyter |
```
import numpy as np
import matplotlib.pyplot as plt
from scipy.io import loadmat
from scipy.interpolate import RectBivariateSpline as rbs
from scipy.integrate import romb
import scipy.sparse as sp
import os
import pywt
wvt = 'db12'
%matplotlib inline
import matplotlib as mpl
norm = mpl.colors.Normalize(vmin=0.0,vmax=1.5)
nx = ny = 32
t = np.linspace(0,320,nx+1)
s = np.linspace(0,320,17)
x = y = (t[:-1]+t[1:]) / 2
x = y = (t[:-1]+t[1:]) / 2
xst = yst = (s[:-1]+s[1:]) / 2
xs, ys = np.meshgrid(xst,yst)
xs = xs.flatten()
ys = ys.flatten()
from scipy.signal import butter, lfilter
def butter_bandpass(lowcut, highcut, fs, order=5):
nyq = 0.5 * fs
low = lowcut / nyq
high = highcut / nyq
b, a = butter(order, [low, high], btype='band')
return b, a
def butter_bandpass_filter(data, lowcut, highcut, fs, order=5, axis=0):
b, a = butter_bandpass(lowcut, highcut, fs, order=order)
y = lfilter(b, a, data, axis=axis)
return y
def butter_lowpass(lowcut, fs, order=5):
nyq = 0.5 * fs
low = lowcut / nyq
b, a = butter(order, [low], btype='low')
return b, a
def butter_low_filter(data, lowcut, fs, order=5, axis=0):
b, a = butter_lowpass(lowcut, fs, order=order)
y = lfilter(b, a, data, axis=axis)
return y
shot = np.reshape(np.fromfile("Testing/TestData/shot1.dat", dtype=np.float32), (4001,64,64))
t = np.linspace(0, 0.5, 4001)
shotf = butter_low_filter(shot, 10, 8000)
tf = t[::20]
shotf = shotf[::20,:,:]
tf_freq = 1/(tf[1]-tf[0])
xc = np.linspace(0,320,65)
xc = (xc[:-1]+xc[1:])/2
yc = xc
shotf_itps = [rbs(xc, yc, s) for s in shotf[:-1]]
def reconstruction(w, wvt_lens, wvt):
starts = np.hstack([0,np.cumsum(wvt_lens)])
wcoef = [w[starts[i]:starts[i+1]] for i in range(len(wvt_lens))]
return pywt.waverec(wcoef, wvt)
```
# ZigZag
```
das_template_x = np.array([2.5*np.sqrt(2)*i for i in range(24)])
das_template_y = np.array([2.5*np.sqrt(2)*i for i in range(24)])
das_template_x2 = np.hstack([das_template_x,das_template_x[::-1],das_template_x,das_template_x[::-1]])
das_template_y2 = np.hstack([das_template_y,das_template_y+das_template_y[-1],das_template_y+2*das_template_y[-1],das_template_y+3*das_template_y[-1]])
das_x = np.hstack([das_template_x2+i*das_template_x[-1] for i in range(4)])
das_y = np.hstack([das_template_y2 for i in range(4)])
offset = (320-np.max(das_x))/2
das_x += offset
das_y += offset
azimuth_template_1 = np.array([[[45 for i in range(24)], [-45 for i in range(24)]] for i in range(2)]).flatten()
azimuth_template_2 = np.array([[[135 for i in range(24)], [215 for i in range(24)]] for i in range(2)]).flatten()
das_az = np.hstack([azimuth_template_1, azimuth_template_2,
azimuth_template_1, azimuth_template_2])
raz = np.deg2rad(das_az)
cscale = 2
generate_kernels = True
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
G3 = np.zeros((len(xs), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G3[:,j] = frame.ev(xs, ys) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
# Gn=1
G_zigzag = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
plt.plot(np.sort(np.diag(G @ np.linalg.solve(G.T@G + 1e-10*np.eye(G.shape[1]), G.T))))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
cuxr = np.array([s.ev(xs, ys, dx=1) for s in shotf_itps])
cuyr = np.array([s.ev(xs, ys, dy=1) for s in shotf_itps])
np.save("Testing/zigzag.npy", das_wvt_data)
wvt_tmp = pywt.wavedec(edasr.T[0], wvt)
wvt_lens = [len(wc) for wc in wvt_tmp]
resi = np.load(f"Testing/zigzag_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
plt.plot(np.std(resi, axis=1))
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.colorbar()
res
plt.plot(cuxr.T[100])
plt.plot(txpredi[100])
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
np.sqrt(np.square(das_x[1:]-das_x[:-1])+np.square(das_y[1:]-das_y[:-1]))
```
# Spiral
```
das_theta2 = np.linspace(0,(360*4)**2, 192*2)
das_theta = np.deg2rad(np.sqrt(das_theta2))
a = 0
b = 1
das_r = b*das_theta
das_x = das_r * np.cos(das_theta)
das_y = das_r * np.sin(das_theta)
raz = np.pi/2-np.arctan2(b*np.tan(das_theta)+(a+b*das_theta), b-(a+b*das_theta)*np.tan(das_theta))
das_az = np.rad2deg(raz)
xwidth = np.max(das_x)-np.min(das_x)
das_x = das_x / xwidth * 320
das_y = das_y / xwidth * 320
x_offset = 320 - np.max(das_x)
das_x = das_x + x_offset
y_offset = np.min(das_y)
das_y = das_y - y_offset
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
G_spiral = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
np.save("Testing/spiral.npy", das_wvt_data)
resi = np.load(f"Testing/spiral_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
res
plt.plot(cuxr.T[100])
plt.plot(txpredi[100])
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.colorbar()
np.hstack([cuxr, cuyr]).shape
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
np.sqrt(np.square(das_x[1:]-das_x[:-1])+np.square(das_y[1:]-das_y[:-1]))
```
# Crossing
```
template = np.linspace(0,320, 65)
template = (template[1:]+template[:-1])/2
das_x = np.hstack([template, template, template,[80 for i in range(len(template))], [160 for i in range(len(template))], [240 for i in range(len(template))]])
das_y = np.hstack([[80 for i in range(len(template))], [160 for i in range(len(template))], [240 for i in range(len(template))],template,template,template])
das_az = np.hstack([[90 for i in range(len(template))], [270 for i in range(len(template))], [90 for i in range(len(template))],[0 for i in range(len(template))], [180 for i in range(len(template))], [0 for i in range(len(template))]])
raz = np.deg2rad(das_az)
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
G_cross = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
np.save("Testing/crossing.npy", das_wvt_data )
resi = np.load(f"Testing/crossing_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
res
plt.plot(cuxr.T[150])
plt.plot(txpredi[150])
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.scatter(xs[150], ys[150], color='r')
plt.colorbar()
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
```
# Random
```
template = np.linspace(0,320, 65)
template = (template[1:]+template[:-1])/2
np.random.seed(94899109)
das_x = np.random.uniform(5,315,384)
das_y = np.random.uniform(5,315,384)
das_az = np.random.uniform(0,360,384)
raz = np.deg2rad(das_az)
L = 10 #gauge length
ll = np.linspace(-L/2, L/2, 2**5+1)
dl = ll[1]-ll[0]
p1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]
p2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]
if generate_kernels:
os.makedirs("Kernels", exist_ok=True)
crv = loadmat(f"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat")
G_mat = np.reshape(crv["G_mat"].T, (crv["G_mat"].shape[1], nx, ny))
crvscales = crv["scales"].flatten()
cvtscaler = 2.0**(cscale*crvscales)
G1 = np.zeros((len(raz), G_mat.shape[0]))
G2 = np.zeros((len(raz), G_mat.shape[0]))
for j in range(G_mat.shape[0]):
frame = rbs(x,y,G_mat[j])
#average derivatives of frame along gauge length
fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L
fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L
G1[:,j] = (np.sin(raz)**2*fd1 +
np.sin(2*raz)*fd2/2) / cvtscaler[j]
G2[:,j] = (np.cos(raz)**2*fd2 +
np.sin(2*raz)*fd1/2) / cvtscaler[j]
G = np.hstack([G1, G2])
Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))
G = G / Gn
G_random = G
np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))
exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])
eyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])
exyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])
edasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr)
das_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])
np.save("Testing/random.npy", das_wvt_data)
resi = np.load(f"Testing/random_res.npy")
Gs = np.std(G)
resxi = resi[:G3.shape[1], :]
resyi = resi[G3.shape[1]:, :]
xpredi = (G3/Gn/Gs) @ resxi
ypredi = (G3/Gn/Gs) @ resyi
txpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))
typredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))
res = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))
plt.plot(np.std(resi, axis=1))
res
plt.plot(cuxr.T[100])
plt.plot(txpredi[100])
cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)
plt.xlim(0,320)
plt.ylim(0,320)
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
plt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))
, norm=norm)
plt.colorbar()
cax = plt.scatter(das_x, das_y,c=das_az)
plt.xlim(0,320)
plt.ylim(0,320)
plt.colorbar(cax, label="Cable Azimuth")
plt.xlabel("Easting (m)")
plt.ylabel("Northing (m)")
plt.gca().set_aspect("equal")
np.save("Kernels/G_zigzag.npy", G_zigzag)
np.save("Kernels/G_spiral.npy", G_spiral)
np.save("Kernels/G_cross.npy", G_cross)
np.save("Kernels/G_random.npy", G_random)
```
# Eigenvalue Spectrum
```
G_full = np.vstack([np.hstack([G3, np.zeros(G3.shape)]), np.hstack([np.zeros(G3.shape), G3])])
idet = 1e-10*np.eye(G_zigzag.shape[1])
ezig = np.sort(np.linalg.eigvals(G_zigzag.T@G_zigzag+idet))[::-1]
espi = np.sort(np.linalg.eigvals(G_spiral.T@G_spiral+idet))[::-1]
ecro = np.sort(np.linalg.eigvals(G_cross.T@G_cross+idet))[::-1]
eran = np.sort(np.linalg.eigvals(G_random.T@G_random+idet))[::-1]
# efull= np.sort(np.linalg.eigvals(G_full.T@G_full+idet))[::-1]
ezign = ezig / ezig[0]
espin = espi / espi[0]
ecron = ecro / ecro[0]
erann = eran / eran[0]
# efulln = efull / efull[0]
plt.plot(np.sort(np.diag(G_zigzag @ np.linalg.solve(G_zigzag.T@G_zigzag + 1e-0*np.eye(G_zigzag.shape[1]), G_zigzag.T))), label="ZigZag")
plt.plot(np.sort(np.diag(G_spiral @ np.linalg.solve(G_spiral.T@G_spiral + 1e-0*np.eye(G_spiral.shape[1]), G_spiral.T))), label="Spiral")
plt.plot(np.sort(np.diag(G_cross @ np.linalg.solve(G_cross.T@G_cross + 1e-0*np.eye(G_cross.shape[1]), G_cross.T))), label="Crossing")
plt.plot(np.sort(np.diag(G_random @ np.linalg.solve(G_random.T@G_random + 1e-0*np.eye(G_random.shape[1]), G_random.T))), label="Random")
plt.xlim(0,384)
plt.ylabel("Coherence")
plt.xlabel("Sorted Diagonal")
plt.legend(loc="lower right")
plt.plot(np.log10(np.real(ezig)), label="ZigZag")
plt.plot(np.log10(np.real(espi)), label="Spiral")
plt.plot(np.log10(np.real(ecro)), label="Crossing")
plt.plot(np.log10(np.real(eran)), label="Random")
plt.xlim(0,384)
plt.ylabel("Log10 Normalized Eigenvalues")
plt.xlabel("Eigenvalue Index")
plt.legend(loc="upper right")
```
| github_jupyter |
$\newcommand{\xv}{\mathbf{x}}
\newcommand{\wv}{\mathbf{w}}
\newcommand{\yv}{\mathbf{y}}
\newcommand{\zv}{\mathbf{z}}
\newcommand{\Chi}{\mathcal{X}}
\newcommand{\R}{\rm I\!R}
\newcommand{\sign}{\text{sign}}
\newcommand{\Tm}{\mathbf{T}}
\newcommand{\Xm}{\mathbf{X}}
\newcommand{\Xlm}{\mathbf{X1}}
\newcommand{\Wm}{\mathbf{W}}
\newcommand{\Vm}{\mathbf{V}}
\newcommand{\Ym}{\mathbf{Y}}
\newcommand{\Zm}{\mathbf{Z}}
\newcommand{\Zlm}{\mathbf{Z1}}
\newcommand{\I}{\mathbf{I}}
\newcommand{\muv}{\boldsymbol\mu}
\newcommand{\Sigmav}{\boldsymbol\Sigma}
\newcommand{\Phiv}{\boldsymbol\Phi}
$
# Neural Networks
Neural networks, or artificial neural networks, are the computational models inspired by the brain. Mimicing the neurons' synaptic connecions (Figure 1), we build or stack multiple neuron-like hidden units to map data into nonlinear space for rich representation.
<img src="https://upload.wikimedia.org/wikipedia/commons/1/10/Blausen_0657_MultipolarNeuron.png" width=500/>
<center>Figure 1. Anatomy of a neuron (wikipedia) </center>
Now, let us review the perceptron model.
<img src="http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/perceptron.png" width=600 />
In perceptron, passing the output of linear model to the step function, we get discrete outputs.
Now, you can think a perceptron as a neuron. With a threshold zero, when the linear model outputs are over it, it passes the signal to next neuron.
By connecting the perceptrons, we can actually build synaptic connections.
We call this model as *multi-layer perceptron* (MLP).
**Q:** For inputs $x \in \{-1, +1 \}$, think about what the following picture represents and answer for it.
1)
<img src="http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_q1.png" width=300/>
2)
<img src="http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_q2.png" width=300/>
3)
<img src="http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_q3.png" width=700/>
Answer:
1) +1
2) -1
3) -1
## Feed Forward Neural Networks
Fitting the data with MLP is a combinatorial optimization problem with non-smooth step function.
So, we can consider smooth step function, a s-shaped sigmoid function.
We call this smooth function as **activation function**.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
fig, ax = plt.subplots()
# x - y axis
ax.axhline(y=0, color='k', linewidth=1)
ax.axvline(x=0, color='k', linewidth=1)
# step function in blue
plt.plot([0, 6], [1, 1], 'b-', linewidth=3)
plt.plot([-6, 0], [-1, -1], 'b-', linewidth=3)
plt.plot([0, 0], [-1, 1], 'b-', linewidth=3)
# tanh in red
x = np.linspace(-6, 6, 100)
plt.plot(x, np.tanh(x), 'r-', linewidth=3)
```
## Non-linear Extension of Linear Model
As we discussed, feed forward neural networks have a rich representation. Thus, it can represent the linear model with single layer.
<img src="http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_linear.png" width=400/>
Considering the multiple outputs, we formulated this in matrix:
$$
\begin{align}
E &= \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{k=1}^{K} (t_{nk} - y_{nk})^2 \\
\\
\Ym &= \Xlm \cdot \Wm
\end{align}
$$
Here, we assume the first column of $\Xlm$ is the bias column with 1's.
Thus, the weight matrix $\Wm$ is $(D+1) \times K$ with the bias row in the first row.
From this model, we can convert the raw data $\Xm$ to $\Phiv$, which is a nonlinear mapping.
$$
\phi: \Xm \rightarrow \Phiv
$$
Then, we can rewrite the linear model with as follows:
$$
\begin{align}
E &= \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{k=1}^{K} (t_{nk} - y_{nk})^2
\\
\Ym &= \Phiv \Wm \\
\\
\Ym_{nk} &= \Phiv_n^\top \Wm_k
\end{align}
$$
Now, let $\phi(\xv) = h(\xv)$ where $h$ is the *activation function*.
$$
\begin{align}
\Zm &= h(\Xlm \cdot \Vm) \\
\\
\Ym & = \Zlm \cdot \Wm
\end{align}
$$
Figure below depics this model.
<img src="http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/nn.png" width=500/>
The size of each matrix is listed:
- $\Xm: N \times D$
- $\Xlm: N \times (D+1)$
- $\Vm: (D+1) \times G$
- $\Zm: N \times G$
- $\Zlm: N \times (G+1)$
- $\Wm: (G+1) \times K$
- $\Ym: N \times K$
For this two-layer network, we call the blue circle layer with the activation functions as **hidden layer** and the organge layer with summation as **output layer**.
# Why Sigmoid?
The resemblance to the step function can be good reason. But is there any other reason for choosing a sigmoid function as activation?
Let us take a look at a polinomial function and the sigmoid.
$$
y = x^4 + 3 x^2 + 7 x + 3 \quad\quad\text{vs.}\quad\quad y = tanh(x)
$$
```
# polinomial function
def h_poly(x):
return x**4 + 3 * x**2 + 7 * x + 3
# sigmoid function
def h_sigmoid(x):
return np.tanh(x)
##### Gradient functions
# polinomial function
def dh_poly(x):
return 4 * x**3 + 6 * x + 7
# polinomial function
def dh_sigmoid(x):
h = h_sigmoid(x)
return 1 - h ** 2
x = np.linspace(-6, 6, 100)
plt.figure(figsize=(16,8))
plt.subplot(121)
plt.plot(x, h_poly(x), label="$y = x^4 + 3 x^2 + 7 x + 3$")
plt.plot(x, dh_poly(x), label="$dy$")
plt.legend()
plt.subplot(122)
plt.plot(x, h_sigmoid(x), label="$y = tanh(x)$")
plt.plot(x, dh_sigmoid(x), label="$dy$")
plt.legend()
```
Here, we can see the polinomial gradients are very huge when $x$ is moving away from 0. A gradient descent procedure takes this huge step for the large positive or negative $x$ values, which can make learning divergent and unstable.
In the right figure, we can see the gradient is nearly turned off for large $x$ values. Only on the nonlinear region of sigmoid function, small gradient is applied for stable learning.
# Gradient Descent
From the error function $E$,
$$
E = \frac{1}{N} \frac{1}{K}\sum_{n=1}^{N} \sum_{k=1}^{K} (t_{nk} - y_{nk})^2,
$$
we can derive the gradient to update the weights for each layer.
Since we can change the output and eventually the error by changing the weights $\Vm$ and $\Wm$,
$$
\begin{align}
v_{dg} &\leftarrow v_{dg} - \alpha_h \frac{\partial{E}} {\partial{v_{dg}}} \\
\\
w_{gk} &\leftarrow w_{gk} - \alpha_o \frac{\partial{E}} {\partial{w_{gk}}},
\end{align}
$$
where $\alpha_h$ and $\alpha_o$ are the learning rate for hidden and output layer respectively.
$$
\begin{align}
\frac{\partial{E}}{\partial{w_{gk}}} &= \frac{\partial{\Big( \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl})^2} \Big)}{\partial{w_{gk}}} \\
&= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N}(t_{nk} - y_{nk}) \frac{\partial{y_{nl}}}{\partial{w_{gk}}}
\end{align}
$$
where
$$
y_{nl} = z1_{n}^\top w_{*l} = \sum_{g=0}^{G} z1_{ng} w_{gl} .
$$
The gradient for the output layer can be computed as follows:
$$
\begin{align}
\frac{\partial{E}}{\partial{w_{gk}}} &= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} (t_{nk} - y_{nk}) z1_{nk} \\
&= -2 \frac{1}{N} \frac{1}{K} \Zlm^\top (\Tm - \Ym).
\end{align}
$$
For the hidden layer,
$$
\begin{align}
\frac{\partial{E}}{\partial{v_{dg}}} &= \frac{\partial{\Big( \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl})^2} \Big)}{\partial{v_{dg}}} \\
&= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl}) \frac{\partial{y_{nl}}}{\partial{v_{dg}}}
\end{align}
$$
where
$$
y_{nl} = \sum_{g=0}^{G} z1_{ng} w_{gl} = \sum_{g=0}^G w_{gl} h (\sum_{d=0}^D v_{dg} x1_{nd}) .
$$
Let $a_{ng} = \sum_{d=0}^D x1_{nd} v_{dg}$. Then, we can use a chain rule for the derivation.
$$
\begin{align}
\frac{\partial{E}}{\partial{v_{dg}}} &= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl}) \frac{\partial{\Big( \sum_{q=0}^G w_{ql} h (\sum_{p=0}^D v_{pq} x1_{np}) \Big)}}{\partial{v_{dg}}} \\
&= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl}) \sum_{q=0}^G w_{ql} \frac{\partial{\Big( h (\sum_{p=0}^D v_{pq} x1_{np}) \Big)}}{\partial{v_{dg}}} \\
&= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl}) \sum_{q=0}^G w_{ql} \frac{\partial{h(a_{ng})}}{\partial{a_{ng}}} \frac{\partial{a_{ng}}}{\partial{v_{dg}}} \\
&= -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nl} - y_{nl}) \sum_{q=0}^G w_{ql} \frac{\partial{h(a_{ng})}}{\partial{a_{ng}}} x1_{nd}.
\end{align}
$$
When $h = tanh$,
$$
\frac{\partial{h(a_{ng})}}{\partial{a_{ng}}} = \frac{z_{ng}}{\partial{a_{ng}}} = (1 - z_{ng}^2).
$$
Thus,
$$
\frac{\partial{E}}{\partial{v_{dg}}} = -2 \frac{1}{N} \frac{1}{K} \sum_{n=1}^{N} \sum_{l=1}^{K} (t_{nk} - y_{nl}) \sum_{g=0}^G w_{gl} (1 - z_{ng}^2) x1_{nd}.
$$
Rewriting this in matrix form,
$$
\frac{\partial{E}}{\partial{v_{dg}}} = -2 \frac{1}{N} \frac{1}{K} \Xlm^\top \Big( (\Tm - \Ym) \Wm^\top \odot (1 - \Zm^2) \Big).
$$
Here, $\odot$ denotes the element-wise multiplication.
To summarize, the backpropagation performs the this weight updates iteratively:
$$
\begin{align}
\Vm &\leftarrow \Vm + \rho_h \frac{1}{N} \frac{1}{K} \Xlm^\top \Big( (\Tm - \Ym) \Wm^\top \odot (1 - \Zm^2) \Big), \\
\Wm &\leftarrow \Wm + \rho_o \frac{1}{N} \frac{1}{K} \Zlm^\top \Big( \Tm - \Ym \Big)
\end{align}
$$
where $\rho_h$ and $\rho_o$ are the learning rate for hidden and output layer weights.
Implemented iteration follows.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import IPython.display as ipd # for display and clear_output
import time # for sleep
# Make some training data
n = 20
X = np.linspace(0.,20.0,n).reshape((n,1)) - 10
T = 0.2 + 0.05 * (X+10) + 0.4 * np.sin(X+10) + 0.2 * np.random.normal(size=(n,1))
# Make some testing data
Xtest = X + 0.1*np.random.normal(size=(n,1))
Ttest = 0.2 + 0.05 * (Xtest+10) + 0.4 * np.sin(Xtest+10) + 0.2 * np.random.normal(size=(n,1))
nSamples = X.shape[0]
nOutputs = T.shape[1]
# Set parameters of neural network
nHiddens = 10
rhoh = 0.5
rhoo = 0.1
rh = rhoh / (nSamples*nOutputs)
ro = rhoo / (nSamples*nOutputs)
# Initialize weights to uniformly distributed values between small normally-distributed between -0.1 and 0.1
V = 0.1*2*(np.random.uniform(size=(1+1,nHiddens))-0.5)
W = 0.1*2*(np.random.uniform(size=(1+nHiddens,nOutputs))-0.5)
# Add constant column of 1's
def addOnes(A):
return np.insert(A, 0, 1, axis=1)
X1 = addOnes(X)
Xtest1 = addOnes(Xtest)
# Take nReps steepest descent steps in gradient descent search in mean-squared-error function
nReps = 30000
# collect training and testing errors for plotting
errorTrace = np.zeros((nReps,2))
N_ = X1.shape[0]
K_ = W.shape[1]
fig = plt.figure(figsize=(10,8))
for reps in range(nReps):
# Forward pass on training data
Z = np.tanh(X1 @ V)
Z1 = addOnes(Z)
Y = Z1 @ W
# Error in output
error = T - Y
print("V:", V.shape)
print("X1:", X1.T.shape)
print("error:", error.shape)
print("W.T:", W.T.shape)
print("Z:", Z.shape)
print(np.square(Z).shape)
# TODO: Backward pass - the backpropagation and weight update steps
V = V + ((rh / (N_ * K_)) * X1.T * ((error * W.T) @ (1 - np.square(Z))))
W = W + (ro / (N_ * K_)) * Z1.T * error
# error traces for plotting
errorTrace[reps,0] = np.sqrt(np.mean((error**2)))
Ytest = addOnes(np.tanh(Xtest1 @ V)) @ W #!! Forward pass in one line
errorTrace[reps,1] = np.sqrt(np.mean((Ytest-Ttest)**2))
if reps % 1000 == 0 or reps == nReps-1:
plt.clf()
plt.subplot(3,1,1)
plt.plot(errorTrace[:reps,:])
plt.ylim(0,0.7)
plt.xlabel('Epochs')
plt.ylabel('RMSE')
plt.legend(('Train','Test'),loc='upper left')
plt.subplot(3,1,2)
plt.plot(X,T,'o-',Xtest,Ttest,'o-',Xtest,Ytest,'o-')
plt.xlim(-10,10)
plt.legend(('Training','Testing','Model'),loc='upper left')
plt.xlabel('$x$')
plt.ylabel('Actual and Predicted $f(x)$')
plt.subplot(3,1,3)
plt.plot(X,Z)
plt.ylim(-1.1,1.1)
plt.xlabel('$x$')
plt.ylabel('Hidden Unit Outputs ($z$)');
ipd.clear_output(wait=True)
ipd.display(fig)
ipd.clear_output(wait=True)
```
$\newcommand{\xv}{\mathbf{x}}
\newcommand{\wv}{\mathbf{w}}
\newcommand{\yv}{\mathbf{y}}
\newcommand{\zv}{\mathbf{z}}
\newcommand{\av}{\mathbf{a}}
\newcommand{\Chi}{\mathcal{X}}
\newcommand{\R}{\rm I\!R}
\newcommand{\sign}{\text{sign}}
\newcommand{\Tm}{\mathbf{T}}
\newcommand{\Xm}{\mathbf{X}}
\newcommand{\Xlm}{\mathbf{X1}}
\newcommand{\Wm}{\mathbf{W}}
\newcommand{\Vm}{\mathbf{V}}
\newcommand{\Ym}{\mathbf{Y}}
\newcommand{\Zm}{\mathbf{Z}}
\newcommand{\Zlm}{\mathbf{Z1}}
\newcommand{\I}{\mathbf{I}}
\newcommand{\muv}{\boldsymbol\mu}
\newcommand{\Sigmav}{\boldsymbol\Sigma}
\newcommand{\Phiv}{\boldsymbol\Phi}
$
# Optimization
So far, we have been using gradient descent to find minimum or maximum values in our error function.
In general, we call this maximization or minimization problem as an **optimization problem**.
In optimization problems, we look for the largest or the smallest value that a function can take. By systematically choosing input vales within the constraint set, optimization problem seeks for the best available values of an objective function.
So, for a given function $f(x)$ that maps $f: \Xm \rightarrow \Ym $ where $\Ym \subset \R$,
we are looking for a $x^* \in \Xm$ that satisfies
$$
\begin{cases}
f(x^*) \le f(x) &\forall x & \quad \text{if } \text{ minimization}\\
f(x^*) \ge f(x) &\forall x & \quad \text{if } \text{ maximization}.
\end{cases}
$$
The optimization problems are often expressed in following notation.
$$
\begin{equation*}
\begin{aligned}
& \underset{x}{\text{minimize}}
& & f(x) \\
& \text{subject to}
& & x \le b_i, \; i = 1, \ldots, m,\\
&&& x \ge 0.
\end{aligned}
\end{equation*}
$$
## Least Squares
$$
\begin{equation*}
\begin{aligned}
& \underset{\wv}{\text{minimize}}
& & \Vert \Xm \wv - t\Vert^2
\end{aligned}
\end{equation*}
$$

As we discussed, least-squares problems can be solved analytically, $\wv = (\Xm^\top \Xm)^{-1} \Xm^\top t$.
We easily formulate least-sqaures and solve very efficiently.
## Linear Programming
$$
\begin{equation*}
\begin{aligned}
& \underset{\xv}{\text{minimize}}
& & \wv^\top \xv \\
& \text{subject to}
& & \av_i^\top \xv \le b_i, \; i = 1, \ldots, m.
\end{aligned}
\end{equation*}
$$

Linear programming or linear optimization finds a maximum or minimum from a mathematical model that is represented by linear relationships.
There is no analytical formular for a solution, but there are reliable algorithms that solve LP efficiently.
## Convex Optimization
$$
\begin{equation*}
\begin{aligned}
& \underset{x}{\text{minimize}}
& & f_0(x) \\
& \text{subject to}
& & f_i(x) \leq b_i, \; i = 1, \ldots, m.
\end{aligned}
\end{equation*}
$$

Convex condition:
$$
f_i(\alpha x_1 + (1-\alpha) x_2) \le \alpha f_i(x_1) + (1-\alpha) f_i(x_2)
$$
Convex optimization generalizes the linear programming problems. A convex optimization problem has the constraint set that forms convex functions. As a general model of LP, convex optimization problems do not have analytical solution but they also have reliable and efficient algorithms for it. Thus, it can be solved very quickly and reliably up to very large problems.
However, it is difficulty to recognize if it is convex or not.
## Nonlinear Optimization
For non-convex problems, we can apply local optimization methods, which is called nonlinear programming.
Starting from initial guess, it searchs for a minimal point near neighborhood. It can be fast and can be applicable large problems. However, there is no guarantee for discovery of global optimum.
## Newton's method
Newton's method approximates the curve with quadratic function repeatedly to find a temporary point or stationary point of $f$.
If we assume that for each measurement point $x^{(k)}$, we can compute $f(x^{(k)})$, $f^{\prime}(x^{(k)})$, and $f^{\prime\prime}(x^{(k)})$.
Using second order Taylor expansion, we can approximate $q(x)$ for $f(x + \Delta x)$:
$$
q(x) = f(x^{(k)}) + f^{\prime}(x^{(k)}) \Delta x + \frac{1}{2} f^{\prime\prime}(x^{(k)}) \Delta x^2
$$
where $\Delta x = (x - x^{(k)})$.
Minimizing this quadratic function,
$$
0 = q^\prime(x) = f^{\prime}(x^{(k)}) + f^{\prime\prime}(x^{(k)}) \Delta x.
$$
Setting $x = x^{(k+1)}$, we can get
$$
x^{(k+1)} = x^{(k)} - \frac{f^{\prime}(x^{(k)})}{f^{\prime\prime}(x^{(k)})}.
$$
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import scipy.optimize as opt
from scipy.optimize import rosen, minimize
# examples are from http://people.duke.edu/~ccc14/sta-663-2017/14C_Optimization_In_Python.html
x = np.linspace(-5, 5, 1000)
y = np.linspace(-5, 5, 1000)
xs, ys = np.meshgrid(x, y)
zs = rosen(np.vstack([xs.ravel(), ys.ravel()])).reshape(xs.shape)
plt.figure(figsize=(8,8))
plt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')
plt.text(1, 1, 'x', va='center', ha='center', color='k', fontsize=30);
from scipy.optimize import rosen_der, rosen_hess
def reporter(p):
"""record the points visited"""
global ps
ps.append(p)
# starting position
x0 = np.array([4,-4.1])
ps = [x0]
minimize(rosen, x0, method="Newton-CG", jac=rosen_der, hess=rosen_hess, callback=reporter)
ps = np.array(ps)
plt.figure(figsize=(16, 8))
plt.subplot(121)
plt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')
plt.plot(ps[:, 0], ps[:, 1], '-ro')
plt.subplot(122)
plt.semilogy(range(len(ps)), rosen(ps.T));
```
## Vs. others?
Now, let us take a look at other optimization tools including naive steepest descent and scaled conjugate gradient ([Moller, 1997](http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.50.8063&rep=rep1&type=pdf)).
To run this properly, you need to download [grad.py](https://webpages.uncc.edu/mlee173/teach/itcs4156online/notes/grad.py) under your current work folder.
```
from grad import steepest
res = steepest(np.array(x0), rosen_der, rosen, stepsize=0.0001, wtracep=True, ftracep=True)
ps = np.array(res['wtrace'])
plt.figure(figsize=(16, 8))
plt.subplot(121)
plt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')
plt.plot(ps[:, 0], ps[:, 1], '-ro')
plt.subplot(122)
plt.semilogy(range(len(ps)), res['ftrace']);
from grad import scg
res = scg(np.array(x0), rosen_der, rosen, wtracep=True, ftracep=True)
res1 = scg(np.array([-4, 4]), rosen_der, rosen, wtracep=True, ftracep=True)
ps = np.array(res['wtrace'])
ps1 = np.array(res1['wtrace'])
plt.figure(figsize=(16, 8))
plt.subplot(121)
plt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')
plt.plot(ps[:, 0], ps[:, 1], '-ro')
plt.plot(ps1[:, 0], ps1[:, 1], '-bo')
plt.subplot(122)
plt.semilogy(range(len(ps)), res['ftrace']);
x0 = [-4, 4]
ps = [x0]
minimize(rosen, x0, method="Newton-CG", jac=rosen_der, hess=rosen_hess, callback=reporter)
ps = np.array(ps)
plt.figure(figsize=(16, 8))
plt.subplot(121)
plt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')
plt.plot(ps[:, 0], ps[:, 1], '-ro')
plt.subplot(122)
plt.semilogy(range(len(ps)), rosen(ps.T));
```
# Neural Network!
Now, let us use this optimization trick for our neural networks.
```
# standardization class
class Standardizer:
""" class version of standardization """
def __init__(self, X, explore=False):
self._mu = np.mean(X,0)
self._sigma = np.std(X,0)
if explore:
print ("mean: ", self._mu)
print ("sigma: ", self._sigma)
print ("min: ", np.min(X,0))
print ("max: ", np.max(X,0))
def set_sigma(self, s):
self._sigma[:] = s
def standardize(self,X):
return (X - self._mu) / self._sigma
def unstandardize(self,X):
return (X * self._sigma) + self._mu
""" Neural Network
referenced NN code by Chuck Anderson in R and C++
by Jake Lee (lemin)
example usage:
X = numpy.array([0,0,1,0,0,1,1,1]).reshape(4,2)
T = numpy.array([0,1,1,0,1,0,0,1]).reshape(4,2)
nn = nnet.NeuralNet([2,3,2])
nn.train(X,T, wprecision=1e-20, fprecision=1e-2)
Y = nn.use(X)
"""
from grad import scg, steepest
from copy import copy
class NeuralNet:
""" neural network class for regression
Parameters
----------
nunits: list
the number of inputs, hidden units, and outputs
Methods
-------
set_hunit
update/initiate weights
pack
pack multiple weights of each layer into one vector
forward
forward processing of neural network
backward
back-propagation of neural network
train
train the neural network
use
appply the trained network for prediction
Attributes
----------
_nLayers
the number of hidden unit layers
rho
learning rate
_W
weights
_weights
weights in one dimension (_W is referencing _weight)
stdX
standardization class for data
stdT
standardization class for target
Notes
-----
"""
# TODO: Try to implement Neural Network class with the member variables and methods described above
X = np.array([0,0,1,0,0,1,1,1]).reshape(4,2)
T = np.array([0,1,1,0,1,0,0,1]).reshape(4,2)
nn = NeuralNet([2,3,2])
nn.train(X, T)
Y = nn.use(X)
Y
T
X
# repeating the previous example
# Make some training data
n = 20
X = np.linspace(0.,20.0,n).reshape((n,1)) - 10
T = 0.2 + 0.05 * (X+10) + 0.4 * np.sin(X+10) + 0.2 * np.random.normal(size=(n,1))
# Make some testing data
Xtest = X + 0.1*np.random.normal(size=(n,1))
Ttest = 0.2 + 0.05 * (Xtest+10) + 0.4 * np.sin(Xtest+10) + 0.2 * np.random.normal(size=(n,1))
nSamples = X.shape[0]
nOutputs = T.shape[1]
nn = NeuralNet([1,3,1])
nn.train(X, T, ftracep=True)
Ytest, Z = nn.use(Xtest, retZ=True)
plt.figure(figsize=(10,8))
plt.subplot(3,1,1)
plt.plot(nn.ftrace)
plt.ylim(0,0.7)
plt.xlabel('Epochs')
plt.ylabel('RMSE')
plt.legend(('Train','Test'),loc='upper left')
plt.subplot(3,1,2)
plt.plot(X,T,'o-',Xtest,Ttest,'o-',Xtest,Ytest,'o-')
plt.xlim(-10,10)
plt.legend(('Training','Testing','Model'),loc='upper left')
plt.xlabel('$x$')
plt.ylabel('Actual and Predicted $f(x)$')
plt.subplot(3,1,3)
plt.plot(X, Z[1])
plt.ylim(-1.1,1.1)
plt.xlabel('$x$')
plt.ylabel('Hidden Unit Outputs ($z$)');
```
| github_jupyter |
## Reinforcement Learning for seq2seq
This time we'll solve a problem of transribing hebrew words in english, also known as g2p (grapheme2phoneme)
* word (sequence of letters in source language) -> translation (sequence of letters in target language)
Unlike what most deep learning practicioners do, we won't only train it to maximize likelihood of correct translation, but also employ reinforcement learning to actually teach it to translate with as few errors as possible.
### About the task
One notable property of Hebrew is that it's consonant language. That is, there are no wovels in the written language. One could represent wovels with diacritics above consonants, but you don't expect people to do that in everyay life.
Therefore, some hebrew characters will correspond to several english letters and others - to none, so we should use encoder-decoder architecture to figure that out.
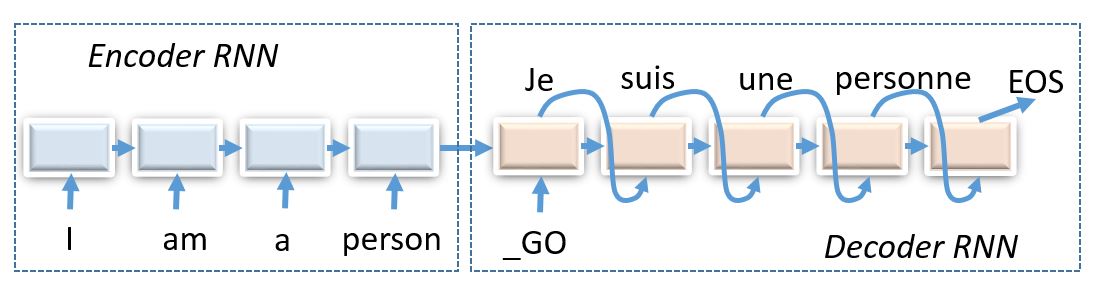
_(img: esciencegroup.files.wordpress.com)_
Encoder-decoder architectures are about converting anything to anything, including
* Machine translation and spoken dialogue systems
* [Image captioning](http://mscoco.org/dataset/#captions-challenge2015) and [image2latex](https://htmlpreview.github.io/?https://github.com/openai/requests-for-research/blob/master/_requests_for_research/im2latex.html) (convolutional encoder, recurrent decoder)
* Generating [images by captions](https://arxiv.org/abs/1511.02793) (recurrent encoder, convolutional decoder)
* Grapheme2phoneme - convert words to transcripts
We chose simplified __Hebrew->English__ machine translation for words and short phrases (character-level), as it is relatively quick to train even without a gpu cluster.
```
import sys
if 'google.colab' in sys.modules:
!wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/basic_model_torch.py -O basic_model_torch.py
!wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/main_dataset.txt -O main_dataset.txt
!wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/voc.py -O voc.py
!pip3 install torch==1.0.0 nltk editdistance
# If True, only translates phrases shorter than 20 characters (way easier).
EASY_MODE = True
# Useful for initial coding.
# If false, works with all phrases (please switch to this mode for homework assignment)
# way we translate. Either "he-to-en" or "en-to-he"
MODE = "he-to-en"
# maximal length of _generated_ output, does not affect training
MAX_OUTPUT_LENGTH = 50 if not EASY_MODE else 20
REPORT_FREQ = 100 # how often to evaluate validation score
```
### Step 1: preprocessing
We shall store dataset as a dictionary
`{ word1:[translation1,translation2,...], word2:[...],...}`.
This is mostly due to the fact that many words have several correct translations.
We have implemented this thing for you so that you can focus on more interesting parts.
__Attention python2 users!__ You may want to cast everything to unicode later during homework phase, just make sure you do it _everywhere_.
```
import numpy as np
from collections import defaultdict
word_to_translation = defaultdict(list) # our dictionary
bos = '_'
eos = ';'
with open("main_dataset.txt", encoding="utf-8") as fin:
for line in fin:
en, he = line[:-1].lower().replace(bos, ' ').replace(eos,
' ').split('\t')
word, trans = (he, en) if MODE == 'he-to-en' else (en, he)
if len(word) < 3:
continue
if EASY_MODE:
if max(len(word), len(trans)) > 20:
continue
word_to_translation[word].append(trans)
print("size = ", len(word_to_translation))
# get all unique lines in source language
all_words = np.array(list(word_to_translation.keys()))
# get all unique lines in translation language
all_translations = np.array(list(set(
[ts for all_ts in word_to_translation.values() for ts in all_ts])))
```
### split the dataset
We hold out 10% of all words to be used for validation.
```
from sklearn.model_selection import train_test_split
train_words, test_words = train_test_split(
all_words, test_size=0.1, random_state=42)
```
### Building vocabularies
We now need to build vocabularies that map strings to token ids and vice versa. We're gonna need these fellas when we feed training data into model or convert output matrices into english words.
```
from voc import Vocab
inp_voc = Vocab.from_lines(''.join(all_words), bos=bos, eos=eos, sep='')
out_voc = Vocab.from_lines(''.join(all_translations), bos=bos, eos=eos, sep='')
# Here's how you cast lines into ids and backwards.
batch_lines = all_words[:5]
batch_ids = inp_voc.to_matrix(batch_lines)
batch_lines_restored = inp_voc.to_lines(batch_ids)
print("lines")
print(batch_lines)
print("\nwords to ids (0 = bos, 1 = eos):")
print(batch_ids)
print("\nback to words")
print(batch_lines_restored)
```
Draw word/translation length distributions to estimate the scope of the task.
```
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=[8, 4])
plt.subplot(1, 2, 1)
plt.title("words")
plt.hist(list(map(len, all_words)), bins=20)
plt.subplot(1, 2, 2)
plt.title('translations')
plt.hist(list(map(len, all_translations)), bins=20)
```
### Step 3: deploy encoder-decoder (1 point)
__assignment starts here__
Our architecture consists of two main blocks:
* Encoder reads words character by character and outputs code vector (usually a function of last RNN state)
* Decoder takes that code vector and produces translations character by character
Than it gets fed into a model that follows this simple interface:
* __`model(inp, out, **flags) -> logp`__ - takes symbolic int32 matrices of hebrew words and their english translations. Computes the log-probabilities of all possible english characters given english prefices and hebrew word.
* __`model.translate(inp, **flags) -> out, logp`__ - takes symbolic int32 matrix of hebrew words, produces output tokens sampled from the model and output log-probabilities for all possible tokens at each tick.
* if given flag __`greedy=True`__, takes most likely next token at each iteration. Otherwise samples with next token probabilities predicted by model.
That's all! It's as hard as it gets. With those two methods alone you can implement all kinds of prediction and training.
```
import torch
import torch.nn as nn
import torch.nn.functional as F
from basic_model_torch import BasicTranslationModel
model = BasicTranslationModel(inp_voc, out_voc,
emb_size=64, hid_size=256)
# Play around with symbolic_translate and symbolic_score
inp = torch.tensor(np.random.randint(0, 10, [3, 5]), dtype=torch.int64)
out = torch.tensor(np.random.randint(0, 10, [3, 5]), dtype=torch.int64)
# translate inp (with untrained model)
sampled_out, logp = model.translate(inp, greedy=False)
print("Sample translations:\n", sampled_out)
print("Log-probabilities at each step:\n", logp)
# score logp(out | inp) with untrained input
logp = model(inp, out)
print("Symbolic_score output:\n", logp)
print("Log-probabilities of output tokens:\n",
torch.gather(logp, dim=2, index=out[:, :, None]))
def translate(lines, max_len=MAX_OUTPUT_LENGTH):
"""
You are given a list of input lines.
Make your neural network translate them.
:return: a list of output lines
"""
# Convert lines to a matrix of indices
lines_ix = inp_voc.to_matrix(lines)
lines_ix = torch.tensor(lines_ix, dtype=torch.int64)
# Compute translations in form of indices
trans_ix = <YOUR CODE>
# Convert translations back into strings
return out_voc.to_lines(trans_ix.data.numpy())
print("Sample inputs:", all_words[:3])
print("Dummy translations:", translate(all_words[:3]))
trans = translate(all_words[:3])
assert translate(all_words[:3]) == translate(
all_words[:3]), "make sure translation is deterministic (use greedy=True and disable any noise layers)"
assert type(translate(all_words[:3])) is list and (type(translate(all_words[:1])[0]) is str or type(
translate(all_words[:1])[0]) is unicode), "translate(lines) must return a sequence of strings!"
# note: if translation freezes, make sure you used max_len parameter
print("Tests passed!")
```
### Scoring function
LogLikelihood is a poor estimator of model performance.
* If we predict zero probability once, it shouldn't ruin entire model.
* It is enough to learn just one translation if there are several correct ones.
* What matters is how many mistakes model's gonna make when it translates!
Therefore, we will use minimal Levenshtein distance. It measures how many characters do we need to add/remove/replace from model translation to make it perfect. Alternatively, one could use character-level BLEU/RougeL or other similar metrics.
The catch here is that Levenshtein distance is not differentiable: it isn't even continuous. We can't train our neural network to maximize it by gradient descent.
```
import editdistance # !pip install editdistance
def get_distance(word, trans):
"""
A function that takes word and predicted translation
and evaluates (Levenshtein's) edit distance to closest correct translation
"""
references = word_to_translation[word]
assert len(references) != 0, "wrong/unknown word"
return min(editdistance.eval(trans, ref) for ref in references)
def score(words, bsize=100):
"""a function that computes levenshtein distance for bsize random samples"""
assert isinstance(words, np.ndarray)
batch_words = np.random.choice(words, size=bsize, replace=False)
batch_trans = translate(batch_words)
distances = list(map(get_distance, batch_words, batch_trans))
return np.array(distances, dtype='float32')
# should be around 5-50 and decrease rapidly after training :)
[score(test_words, 10).mean() for _ in range(5)]
```
## Step 2: Supervised pre-training (2 points)
Here we define a function that trains our model through maximizing log-likelihood a.k.a. minimizing crossentropy.
```
import random
def sample_batch(words, word_to_translation, batch_size):
"""
sample random batch of words and random correct translation for each word
example usage:
batch_x,batch_y = sample_batch(train_words, word_to_translations,10)
"""
# choose words
batch_words = np.random.choice(words, size=batch_size)
# choose translations
batch_trans_candidates = list(map(word_to_translation.get, batch_words))
batch_trans = list(map(random.choice, batch_trans_candidates))
return batch_words, batch_trans
bx, by = sample_batch(train_words, word_to_translation, batch_size=3)
print("Source:")
print(bx)
print("Target:")
print(by)
from basic_model_torch import infer_length, infer_mask, to_one_hot
def compute_loss_on_batch(input_sequence, reference_answers):
""" Compute crossentropy loss given a batch of sources and translations """
input_sequence = torch.tensor(inp_voc.to_matrix(input_sequence), dtype=torch.int64)
reference_answers = torch.tensor(out_voc.to_matrix(reference_answers), dtype=torch.int64)
# Compute log-probabilities of all possible tokens at each step. Use model interface.
logprobs_seq = <YOUR CODE>
# compute elementwise crossentropy as negative log-probabilities of reference_answers.
crossentropy = - \
torch.sum(logprobs_seq *
to_one_hot(reference_answers, len(out_voc)), dim=-1)
assert crossentropy.dim(
) == 2, "please return elementwise crossentropy, don't compute mean just yet"
# average with mask
mask = infer_mask(reference_answers, out_voc.eos_ix)
loss = torch.sum(crossentropy * mask) / torch.sum(mask)
return loss
# test it
loss = compute_loss_on_batch(*sample_batch(train_words, word_to_translation, 3))
print('loss = ', loss)
assert loss.item() > 0.0
loss.backward()
for w in model.parameters():
assert w.grad is not None and torch.max(torch.abs(w.grad)).item() != 0, \
"Loss is not differentiable w.r.t. a weight with shape %s. Check comput_loss_on_batch." % (
w.size(),)
```
##### Actually train the model
Minibatches and stuff...
```
from IPython.display import clear_output
from tqdm import tqdm, trange # or use tqdm_notebook,tnrange
loss_history = []
editdist_history = []
entropy_history = []
opt = torch.optim.Adam(model.parameters())
for i in trange(25000):
loss = compute_loss_on_batch(*sample_batch(train_words, word_to_translation, 32))
# train with backprop
loss.backward()
opt.step()
opt.zero_grad()
loss_history.append(loss.item())
if (i+1) % REPORT_FREQ == 0:
clear_output(True)
current_scores = score(test_words)
editdist_history.append(current_scores.mean())
print("llh=%.3f, mean score=%.3f" %
(np.mean(loss_history[-10:]), np.mean(editdist_history[-10:])))
plt.figure(figsize=(12, 4))
plt.subplot(131)
plt.title('train loss / traning time')
plt.plot(loss_history)
plt.grid()
plt.subplot(132)
plt.title('val score distribution')
plt.hist(current_scores, bins=20)
plt.subplot(133)
plt.title('val score / traning time (lower is better)')
plt.plot(editdist_history)
plt.grid()
plt.show()
```
__How to interpret the plots:__
* __Train loss__ - that's your model's crossentropy over minibatches. It should go down steadily. Most importantly, it shouldn't be NaN :)
* __Val score distribution__ - distribution of translation edit distance (score) within batch. It should move to the left over time.
* __Val score / training time__ - it's your current mean edit distance. This plot is much whimsier than loss, but make sure it goes below 8 by 2500 steps.
If it doesn't, first try to re-create both model and opt. You may have changed it's weight too much while debugging. If that doesn't help, it's debugging time.
```
for word in train_words[:10]:
print("%s -> %s" % (word, translate([word])[0]))
test_scores = []
for start_i in trange(0, len(test_words), 32):
batch_words = test_words[start_i:start_i+32]
batch_trans = translate(batch_words)
distances = list(map(get_distance, batch_words, batch_trans))
test_scores.extend(distances)
print("Supervised test score:", np.mean(test_scores))
```
## Self-critical policy gradient (2 points)
In this section you'll implement algorithm called self-critical sequence training (here's an [article](https://arxiv.org/abs/1612.00563)).
The algorithm is a vanilla policy gradient with a special baseline.
$$ \nabla J = E_{x \sim p(s)} E_{y \sim \pi(y|x)} \nabla log \pi(y|x) \cdot (R(x,y) - b(x)) $$
Here reward R(x,y) is a __negative levenshtein distance__ (since we minimize it). The baseline __b(x)__ represents how well model fares on word __x__.
In practice, this means that we compute baseline as a score of greedy translation, $b(x) = R(x,y_{greedy}(x)) $.

Luckily, we already obtained the required outputs: `model.greedy_translations, model.greedy_mask` and we only need to compute levenshtein using `compute_levenshtein` function.
```
def compute_reward(input_sequence, translations):
""" computes sample-wise reward given token ids for inputs and translations """
distances = list(map(get_distance,
inp_voc.to_lines(input_sequence.data.numpy()),
out_voc.to_lines(translations.data.numpy())))
# use negative levenshtein distance so that larger reward means better policy
return - torch.tensor(distances, dtype=torch.int64)
def scst_objective_on_batch(input_sequence, max_len=MAX_OUTPUT_LENGTH):
""" Compute pseudo-loss for policy gradient given a batch of sources """
input_sequence = torch.tensor(inp_voc.to_matrix(input_sequence), dtype=torch.int64)
# use model to __sample__ symbolic translations given input_sequence
sample_translations, sample_logp = <YOUR CODE>
# use model to __greedy__ symbolic translations given input_sequence
greedy_translations, greedy_logp = <YOUR CODE>
# compute rewards and advantage
rewards = compute_reward(input_sequence, sample_translations)
baseline = <YOUR CODE: compute __negative__ levenshtein for greedy mode>
# compute advantage using rewards and baseline
advantage = <YOUR CODE>
# compute log_pi(a_t|s_t), shape = [batch, seq_length]
logp_sample = <YOUR CODE>
# ^-- hint: look at how crossentropy is implemented in supervised learning loss above
# mind the sign - this one should not be multiplied by -1 :)
# policy gradient pseudo-loss. Gradient of J is exactly policy gradient.
J = logp_sample * advantage[:, None]
assert J.dim() == 2, "please return elementwise objective, don't compute mean just yet"
# average with mask
mask = infer_mask(sample_translations, out_voc.eos_ix)
loss = - torch.sum(J * mask) / torch.sum(mask)
# regularize with negative entropy. Don't forget the sign!
# note: for entropy you need probabilities for all tokens (sample_logp), not just logp_sample
entropy = <YOUR CODE: compute entropy matrix of shape[batch, seq_length], H = -sum(p*log_p), don't forget the sign!>
# hint: you can get sample probabilities from sample_logp using math :)
assert entropy.dim(
) == 2, "please make sure elementwise entropy is of shape [batch,time]"
reg = - 0.01 * torch.sum(entropy * mask) / torch.sum(mask)
return loss + reg, torch.sum(entropy * mask) / torch.sum(mask)
```
# Policy gradient training
```
entropy_history = [np.nan] * len(loss_history)
opt = torch.optim.Adam(model.parameters(), lr=1e-5)
for i in trange(100000):
loss, ent = scst_objective_on_batch(
sample_batch(train_words, word_to_translation, 32)[0]) # [0] = only source sentence
# train with backprop
loss.backward()
opt.step()
opt.zero_grad()
loss_history.append(loss.item())
entropy_history.append(ent.item())
if (i+1) % REPORT_FREQ == 0:
clear_output(True)
current_scores = score(test_words)
editdist_history.append(current_scores.mean())
plt.figure(figsize=(12, 4))
plt.subplot(131)
plt.title('val score distribution')
plt.hist(current_scores, bins=20)
plt.subplot(132)
plt.title('val score / traning time')
plt.plot(editdist_history)
plt.grid()
plt.subplot(133)
plt.title('policy entropy / traning time')
plt.plot(entropy_history)
plt.grid()
plt.show()
print("J=%.3f, mean score=%.3f" %
(np.mean(loss_history[-10:]), np.mean(editdist_history[-10:])))
```
__Debugging tips:__
<img src=https://github.com/yandexdataschool/Practical_RL/raw/master/yet_another_week/_resource/do_something_scst.png width=400>
* As usual, don't expect improvements right away, but in general the model should be able to show some positive changes by 5k steps.
* Entropy is a good indicator of many problems.
* If it reaches zero, you may need greater entropy regularizer.
* If it has rapid changes time to time, you may need gradient clipping.
* If it oscillates up and down in an erratic manner... it's perfectly okay for entropy to do so. But it should decrease at the end.
* We don't show loss_history cuz it's uninformative for pseudo-losses in policy gradient. However, if something goes wrong you can check it to see if everything isn't a constant zero.
### Results
```
for word in train_words[:10]:
print("%s -> %s" % (word, translate([word])[0]))
test_scores = []
for start_i in trange(0, len(test_words), 32):
batch_words = test_words[start_i:start_i+32]
batch_trans = translate(batch_words)
distances = list(map(get_distance, batch_words, batch_trans))
test_scores.extend(distances)
print("Supervised test score:", np.mean(test_scores))
# ^^ If you get Out Of MemoryError, please replace this with batched computation
```
## Step 6: Make it actually work (5++ pts)
In this section we want you to finally __restart with EASY_MODE=False__ and experiment to find a good model/curriculum for that task.
We recommend you to start with the following architecture
```
encoder---decoder
P(y|h)
^
LSTM -> LSTM
^ ^
biLSTM -> LSTM
^ ^
input y_prev
```
__Note:__ you can fit all 4 state tensors of both LSTMs into a in a single state - just assume that it contains, for example, [h0, c0, h1, c1] - pack it in encode and update in decode.
Here are some cool ideas on what you can do then.
__General tips & tricks:__
* You will likely need to adjust pre-training time for such a network.
* Supervised pre-training may benefit from clipping gradients somehow.
* SCST may indulge a higher learning rate in some cases and changing entropy regularizer over time.
* It's often useful to save pre-trained model parameters to not re-train it every time you want new policy gradient parameters.
* When leaving training for nighttime, try setting REPORT_FREQ to a larger value (e.g. 500) not to waste time on it.
__Formal criteria:__
To get 5 points we want you to build an architecture that:
* _doesn't consist of single GRU_
* _works better_ than single GRU baseline.
* We also want you to provide either learning curve or trained model, preferably both
* ... and write a brief report or experiment log describing what you did and how it fared.
### Attention
There's more than one way to connect decoder to encoder
* __Vanilla:__ layer_i of encoder last state goes to layer_i of decoder initial state
* __Every tick:__ feed encoder last state _on every iteration_ of decoder.
* __Attention:__ allow decoder to "peek" at one (or several) positions of encoded sequence on every tick.
The most effective (and cool) of those is, of course, attention.
You can read more about attention [in this nice blog post](https://distill.pub/2016/augmented-rnns/). The easiest way to begin is to use "soft" attention with "additive" or "dot-product" intermediate layers.
__Tips__
* Model usually generalizes better if you no longer allow decoder to see final encoder state
* Once your model made it through several epochs, it is a good idea to visualize attention maps to understand what your model has actually learned
* There's more stuff [here](https://github.com/yandexdataschool/Practical_RL/blob/master/week8_scst/bonus.ipynb)
* If you opted for hard attention, we recommend [gumbel-softmax](https://blog.evjang.com/2016/11/tutorial-categorical-variational.html) instead of sampling. Also please make sure soft attention works fine before you switch to hard.
### UREX
* This is a way to improve exploration in policy-based settings. The main idea is that you find and upweight under-appreciated actions.
* Here's [video](https://www.youtube.com/watch?v=fZNyHoXgV7M&feature=youtu.be&t=3444)
and an [article](https://arxiv.org/abs/1611.09321).
* You may want to reduce batch size 'cuz UREX requires you to sample multiple times per source sentence.
* Once you got it working, try using experience replay with importance sampling instead of (in addition to) basic UREX.
### Some additional ideas:
* (advanced deep learning) It may be a good idea to first train on small phrases and then adapt to larger ones (a.k.a. training curriculum).
* (advanced nlp) You may want to switch from raw utf8 to something like unicode or even syllables to make task easier.
* (advanced nlp) Since hebrew words are written __with vowels omitted__, you may want to use a small Hebrew vowel markup dataset at `he-pron-wiktionary.txt`.
```
assert not EASY_MODE, "make sure you set EASY_MODE = False at the top of the notebook."
```
`[your report/log here or anywhere you please]`
__Contributions:__ This notebook is brought to you by
* Yandex [MT team](https://tech.yandex.com/translate/)
* Denis Mazur ([DeniskaMazur](https://github.com/DeniskaMazur)), Oleg Vasilev ([Omrigan](https://github.com/Omrigan/)), Dmitry Emelyanenko ([TixFeniks](https://github.com/tixfeniks)) and Fedor Ratnikov ([justheuristic](https://github.com/justheuristic/))
* Dataset is parsed from [Wiktionary](https://en.wiktionary.org), which is under CC-BY-SA and GFDL licenses.
| github_jupyter |
# DEAP
DEAP is a novel evolutionary computation framework for rapid prototyping and testing of ideas. It seeks to make algorithms explicit and data structures transparent. It works in perfect harmony with parallelisation mechanism such as multiprocessing and SCOOP. The following documentation presents the key concepts and many features to build your own evolutions.
Library documentation: <a>http://deap.readthedocs.org/en/master/</a>
## One Max Problem (GA)
This problem is very simple, we search for a 1 filled list individual. This problem is widely used in the evolutionary computation community since it is very simple and it illustrates well the potential of evolutionary algorithms.
```
import random
from deap import base
from deap import creator
from deap import tools
# creator is a class factory that can build new classes at run-time
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
# a toolbox stores functions and their arguments
toolbox = base.Toolbox()
# attribute generator
toolbox.register("attr_bool", random.randint, 0, 1)
# structure initializers
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_bool, 100)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
# evaluation function
def evalOneMax(individual):
return sum(individual),
# register the required genetic operators
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
random.seed(64)
# instantiate a population
pop = toolbox.population(n=300)
CXPB, MUTPB, NGEN = 0.5, 0.2, 40
# evaluate the entire population
fitnesses = list(map(toolbox.evaluate, pop))
for ind, fit in zip(pop, fitnesses):
ind.fitness.values = fit
print(" Evaluated %i individuals" % len(pop))
# begin the evolution
for g in range(NGEN):
print("-- Generation %i --" % g)
# select the next generation individuals
offspring = toolbox.select(pop, len(pop))
# clone the selected individuals
offspring = list(map(toolbox.clone, offspring))
# apply crossover and mutation on the offspring
for child1, child2 in zip(offspring[::2], offspring[1::2]):
if random.random() < CXPB:
toolbox.mate(child1, child2)
del child1.fitness.values
del child2.fitness.values
for mutant in offspring:
if random.random() < MUTPB:
toolbox.mutate(mutant)
del mutant.fitness.values
# evaluate the individuals with an invalid fitness
invalid_ind = [ind for ind in offspring if not ind.fitness.valid]
fitnesses = map(toolbox.evaluate, invalid_ind)
for ind, fit in zip(invalid_ind, fitnesses):
ind.fitness.values = fit
print(" Evaluated %i individuals" % len(invalid_ind))
# the population is entirely replaced by the offspring
pop[:] = offspring
# gather all the fitnesses in one list and print the stats
fits = [ind.fitness.values[0] for ind in pop]
length = len(pop)
mean = sum(fits) / length
sum2 = sum(x*x for x in fits)
std = abs(sum2 / length - mean**2)**0.5
print(" Min %s" % min(fits))
print(" Max %s" % max(fits))
print(" Avg %s" % mean)
print(" Std %s" % std)
best_ind = tools.selBest(pop, 1)[0]
print("Best individual is %s, %s" % (best_ind, best_ind.fitness.values))
```
## Symbolic Regression (GP)
Symbolic regression is one of the best known problems in GP. It is commonly used as a tuning problem for new algorithms, but is also widely used with real-life distributions, where other regression methods may not work.
All symbolic regression problems use an arbitrary data distribution, and try to fit the most accurately the data with a symbolic formula. Usually, a measure like the RMSE (Root Mean Square Error) is used to measure an individual’s fitness.
In this example, we use a classical distribution, the quartic polynomial (x^4 + x^3 + x^2 + x), a one-dimension distribution. 20 equidistant points are generated in the range [-1, 1], and are used to evaluate the fitness.
```
import operator
import math
import random
import numpy
from deap import algorithms
from deap import base
from deap import creator
from deap import tools
from deap import gp
# define a new function for divison that guards against divide by 0
def protectedDiv(left, right):
try:
return left / right
except ZeroDivisionError:
return 1
# add aritmetic primitives
pset = gp.PrimitiveSet("MAIN", 1)
pset.addPrimitive(operator.add, 2)
pset.addPrimitive(operator.sub, 2)
pset.addPrimitive(operator.mul, 2)
pset.addPrimitive(protectedDiv, 2)
pset.addPrimitive(operator.neg, 1)
pset.addPrimitive(math.cos, 1)
pset.addPrimitive(math.sin, 1)
# constant terminal
pset.addEphemeralConstant("rand101", lambda: random.randint(-1,1))
# define number of inputs
pset.renameArguments(ARG0='x')
# create fitness and individual objects
creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", gp.PrimitiveTree, fitness=creator.FitnessMin)
# register evolution process parameters through the toolbox
toolbox = base.Toolbox()
toolbox.register("expr", gp.genHalfAndHalf, pset=pset, min_=1, max_=2)
toolbox.register("individual", tools.initIterate, creator.Individual, toolbox.expr)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("compile", gp.compile, pset=pset)
# evaluation function
def evalSymbReg(individual, points):
# transform the tree expression in a callable function
func = toolbox.compile(expr=individual)
# evaluate the mean squared error between the expression
# and the real function : x**4 + x**3 + x**2 + x
sqerrors = ((func(x) - x**4 - x**3 - x**2 - x)**2 for x in points)
return math.fsum(sqerrors) / len(points),
toolbox.register("evaluate", evalSymbReg, points=[x/10. for x in range(-10,10)])
toolbox.register("select", tools.selTournament, tournsize=3)
toolbox.register("mate", gp.cxOnePoint)
toolbox.register("expr_mut", gp.genFull, min_=0, max_=2)
toolbox.register("mutate", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)
# prevent functions from getting too deep/complex
toolbox.decorate("mate", gp.staticLimit(key=operator.attrgetter("height"), max_value=17))
toolbox.decorate("mutate", gp.staticLimit(key=operator.attrgetter("height"), max_value=17))
# compute some statistics about the population
stats_fit = tools.Statistics(lambda ind: ind.fitness.values)
stats_size = tools.Statistics(len)
mstats = tools.MultiStatistics(fitness=stats_fit, size=stats_size)
mstats.register("avg", numpy.mean)
mstats.register("std", numpy.std)
mstats.register("min", numpy.min)
mstats.register("max", numpy.max)
random.seed(318)
pop = toolbox.population(n=300)
hof = tools.HallOfFame(1)
# run the algorithm
pop, log = algorithms.eaSimple(pop, toolbox, 0.5, 0.1, 40, stats=mstats,
halloffame=hof, verbose=True)
```
| github_jupyter |
```
# !wget https://malaya-dataset.s3-ap-southeast-1.amazonaws.com/crawler/academia/academia-pdf.json
import json
import cleaning
from tqdm import tqdm
with open('../academia/academia-pdf.json') as fopen:
pdf = json.load(fopen)
len(pdf)
import os
os.path.split(pdf[0]['file'])
import malaya
fast_text = malaya.language_detection.fasttext()
fast_text.predict(['Prosiding_Kolokium_Siswazah_JUF_2017.pdf'])
from unidecode import unidecode
def clean(string):
string = [cleaning.cleaning(s) for s in string]
string = [s.strip() for s in string if 'tarikh' not in s.lower() and 'soalan no' not in s.lower()]
string = [s for s in string if not ''.join(s.split()[:1]).isdigit() and '.soalan' not in s.lower() and 'jum ' not in s.lower()]
string = [s for s in string if not s[:3].isdigit() and not s[-3:].isdigit()]
return string
outer = []
for k in tqdm(range(len(pdf))):
c = clean(pdf[k]['content']['content'].split('\n'))
t, last = [], 0
i = 0
while i < len(c):
text = c[i]
if len(text) > 5:
if len(text.split()) > 1:
t.append(text)
last = i
else:
if len(t) and (i - last) > 2:
t.append('')
outer.extend(t)
t = []
last = i
elif not len(t):
last = i
i += 1
if len(t):
t.append('')
outer.extend(t)
len(outer)
%%time
temp_vocab = list(set(cleaning.multiprocessing(outer, cleaning.unique_words)))
%%time
# important
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.duplicate_dots_marks_exclamations, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
# important
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_underscore, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
# important
temp_dict = cleaning.multiprocessing(outer, cleaning.isolate_spamchars, list_mode = False)
print(len(temp_dict))
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.break_short_words, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.break_long_words, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_ending_underscore, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_starting_underscore, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.end_punct, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.start_punct, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
%%time
temp_dict = cleaning.multiprocessing(temp_vocab, cleaning.join_dashes, list_mode = False)
print(len(temp_dict))
outer = cleaning.string_dict_cleaning(outer, temp_dict)
results, result = [], []
for i in tqdm(outer):
if not len(i) and len(result):
results.append(result)
result = []
else:
result.append(i)
if len(result):
results.append(result)
import re
alphabets = '([A-Za-z])'
prefixes = (
'(Mr|St|Mrs|Ms|Dr|Prof|Capt|Cpt|Lt|Mt|Puan|puan|Tuan|tuan|sir|Sir)[.]'
)
suffixes = '(Inc|Ltd|Jr|Sr|Co|Mo)'
starters = '(Mr|Mrs|Ms|Dr|He\s|She\s|It\s|They\s|Their\s|Our\s|We\s|But\s|However\s|That\s|This\s|Wherever|Dia|Mereka|Tetapi|Kita|Itu|Ini|Dan|Kami|Beliau|Seri|Datuk|Dato|Datin|Tuan|Puan)'
acronyms = '([A-Z][.][A-Z][.](?:[A-Z][.])?)'
websites = '[.](com|net|org|io|gov|me|edu|my)'
another_websites = '(www|http|https)[.]'
digits = '([0-9])'
before_digits = '([Nn]o|[Nn]ombor|[Nn]umber|[Kk]e|=|al)'
month = '([Jj]an(?:uari)?|[Ff]eb(?:ruari)?|[Mm]a(?:c)?|[Aa]pr(?:il)?|Mei|[Jj]u(?:n)?|[Jj]ula(?:i)?|[Aa]ug(?:ust)?|[Ss]ept?(?:ember)?|[Oo]kt(?:ober)?|[Nn]ov(?:ember)?|[Dd]is(?:ember)?)'
def split_into_sentences(text, minimum_length = 5):
text = text.replace('\x97', '\n')
text = '. '.join([s for s in text.split('\n') if len(s)])
text = text + '.'
text = unidecode(text)
text = ' ' + text + ' '
text = text.replace('\n', ' ')
text = re.sub(prefixes, '\\1<prd>', text)
text = re.sub(websites, '<prd>\\1', text)
text = re.sub(another_websites, '\\1<prd>', text)
text = re.sub('[,][.]+', '<prd>', text)
if '...' in text:
text = text.replace('...', '<prd><prd><prd>')
if 'Ph.D' in text:
text = text.replace('Ph.D.', 'Ph<prd>D<prd>')
text = re.sub('[.]\s*[,]', '<prd>,', text)
text = re.sub(before_digits + '\s*[.]\s*' + digits, '\\1<prd>\\2', text)
text = re.sub(month + '[.]\s*' + digits, '\\1<prd>\\2', text)
text = re.sub('\s' + alphabets + '[.][ ]+', ' \\1<prd> ', text)
text = re.sub(acronyms + ' ' + starters, '\\1<stop> \\2', text)
text = re.sub(
alphabets + '[.]' + alphabets + '[.]' + alphabets + '[.]',
'\\1<prd>\\2<prd>\\3<prd>',
text,
)
text = re.sub(
alphabets + '[.]' + alphabets + '[.]', '\\1<prd>\\2<prd>', text
)
text = re.sub(' ' + suffixes + '[.][ ]+' + starters, ' \\1<stop> \\2', text)
text = re.sub(' ' + suffixes + '[.]', ' \\1<prd>', text)
text = re.sub(' ' + alphabets + '[.]', ' \\1<prd>', text)
text = re.sub(digits + '[.]' + digits, '\\1<prd>\\2', text)
if '”' in text:
text = text.replace('.”', '”.')
if '"' in text:
text = text.replace('."', '".')
if '!' in text:
text = text.replace('!"', '"!')
if '?' in text:
text = text.replace('?"', '"?')
text = text.replace('.', '.<stop>')
text = text.replace('?', '?<stop>')
text = text.replace('!', '!<stop>')
text = text.replace('<prd>', '.')
sentences = text.split('<stop>')
sentences = sentences[:-1]
sentences = [s.strip() for s in sentences if len(s) > minimum_length]
return sentences
split_into_sentences('733 ke . 633 , berlaku penurunan akibat kesan program PMI .')
import malaya
import re
def strip(string):
string = ' '.join(string)
string = re.sub(r'[ ]+', ' ', string.replace('\n', ' ').replace('\t', ' ')).strip()
return split_into_sentences(string)
output = []
for r in tqdm(results):
output.extend(strip(r) + [''])
len(output)
output[10000:11000]
with open('dumping-academia.txt', 'w') as fopen:
fopen.write('\n'.join(output))
```
| github_jupyter |
<a href="https://colab.research.google.com/github/CarlosNeto2804/imersao-dados-2/blob/main/imersao_dados_aula_1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Introdução
```
import pandas as pd;
dados_enem = pd.read_csv('https://github.com/alura-cursos/imersao-dados-2-2020/blob/master/MICRODADOS_ENEM_2019_SAMPLE_43278.csv?raw=true')
# retorna as 5 primeiras linhas da colecao
dados_enem.head()
# para acessar apenas uma coluna -> dados_enem['nome_da_coluna']
dados_enem['SG_UF_RESIDENCIA']
# para verificar quais colunas existem no data frame -> dados_enem.colums.values
dados_enem.columns.values
# acessar mais de um valor no DataFrame data_frame[["cabeçalho1","cabeçalho2"]]
dados_enem[["SG_UF_RESIDENCIA","Q025"]]
# retornar os valores sem repeticao de uma coluna
dados_enem["SG_UF_RESIDENCIA"].unique()
# soma dos elementos de uma derminada chave
# ordena pelo valor
#ESTADOS
dados_enem["SG_UF_RESIDENCIA"].value_counts()
#IDADES
dados_enem['NU_IDADE'].value_counts()
# ordenacao pelo index
dados_enem['NU_IDADE'].value_counts().sort_index()
dados_enem['NU_IDADE'].describe()
```
# Continuação
```
# visualização em histograma
#dados_enem['NU_IDADE'].hist()
dados_enem['NU_IDADE'].hist(bins=100,figsize=(11,9),legend=True)
treineiros = dados_enem.query('IN_TREINEIRO == 1')
treineiros['NU_IDADE'].value_counts()
# Notas Redacao
dados_enem['NU_NOTA_REDACAO'].hist(bins=20)
# analise geral
provas = ["NU_NOTA_CN","NU_NOTA_CH","NU_NOTA_MT","NU_NOTA_LC","NU_NOTA_REDACAO"]
dados_enem[provas].describe()
```
# Desafios
- 01 : Informar a proporção de inscritos por idades
```
# desafio 1
def proporcao(total_itens):
def funcao_calculo(x):
res = x * 100 / total_itens
return round(res, 6);
return funcao_calculo
total = len(dados_enem)
inscritos_por_idade = dados_enem['NU_IDADE'].value_counts()
inscritos_por_idade.apply(proporcao(total))
```
- 02: Descobrir de quais estados são os inscritos com 13 anos
```
# desafio 02
cabecalhos= ['SG_UF_RESIDENCIA','NU_IDADE']
inscritos=dados_enem[cabecalhos]
inscritos.query('NU_IDADE==13')
```
- 03: Qual a proporcao dos alunos com 18 anos por estado
```
# desafio 3
cabecalhos = ['SG_UF_RESIDENCIA','NU_IDADE']
novo_df = dados_enem[cabecalhos]
inscritos = novo_df.query('NU_IDADE==18')
total_inscritos = len(inscritos)
inscritos.value_counts().apply(proporcao(total_inscritos))
```
- 04: Plotar Histogramas das idades de treineiros e não treineiros
```
# desafio 4
inscritos_treineiros = dados_enem.query('IN_TREINEIRO == 1')['NU_IDADE'].value_counts();
inscritos_treineiros.hist(bins=30,figsize=(10,7),legend=True)
inscritos_nao_treineiros = dados_enem.query('IN_TREINEIRO == 0')['NU_IDADE'].value_counts();
inscritos_nao_treineiros.hist(bins=30,figsize=(10,7),legend=True)
```
- 05: Comparar as distribuições das provas em ingles e espanhol
```
# desafio 5
# TP_LINGUA==0 -> Ingles
# TP_LINGUA==1 -> Espanhol
dados_enem.query('TP_LINGUA==1')['TP_LINGUA'].hist(bins=20,figsize=(10,7),legend=True)
dados_enem.query('TP_LINGUA==0')['TP_LINGUA'].hist(bins=20,figsize=(10,7),legend=True)
```
| github_jupyter |
```
import numpy as np
import pandas as pd
import xarray as xr
import geopandas as gpd
from shapely.geometry import Point
import sys
import os
sys.path.insert(0, os.path.dirname(os.getcwd()))
from time_space_reductions.match_ups_over_polygons import get_zonal_match_up
def make_fake_data(N=200):
# creating example GeoDataframe for match-ups in EPSG 4326
xx = np.random.randint(low=-60, high=-33, size=N)*1.105
yy = np.random.randint(low=-4, high=20, size=N)*1.105
df = pd.DataFrame({'lon':xx, 'lat':yy})
df['geometry'] = df.apply(lambda x: Point(x['lon'], x['lat']), axis=1)
gdf = gpd.GeoDataFrame(df, geometry='geometry', crs={'init':'epsg:4326'})
gdf['Datetime'] = pd.date_range('2010-05-19', '2010-06-24', periods=gdf.shape[0])
gdf.crs = {'init' :'epsg:4326'}
return gdf
def get_netcdf_example():
import glob
cpath = r'C:\Users\Philipe Leal\Dropbox\Profissao\Python\OSGEO\Matrizes\NetCDF\Time_Space_Concatenations\time_space_reductions\tests\data'
path_file = glob.glob(cpath + '/*.nc' )
return xr.open_mfdataset(path_file[0])
```
from time_space_reductions.netcdf_gdf_setter import Base_class_space_time_netcdf_gdf
class Space_Time_Agg_over_polygons(Base_class_space_time_netcdf_gdf):
def __init__(self, gdf, xarray_dataset=None,
netcdf_temporal_coord_name='time',
geo_series_temporal_attribute_name = 'Datetime',
longitude_dimension='lon',
latitude_dimension='lat',
):
'''
Class description:
------------------
This class is a base class for ensuring that the given netcdf is in conformity with the algorithm.
Ex: the Netcdf has to be sorted in ascending order for all dimensions (ex: time ,longitude, latitude).
Otherwise, the returned algorithm would return Nan values for all slices
Also, it is mandatory for the user to define the longitude and latitude dimension Names (ex: 'lon', 'lat'),
since there is no stadardization for defining these properties in the Netcdf files worldwide.
Attributes:
gdf (geodataframe):
-----------------------
The geodataframe object containing geometries to be analyzed.
xarray_dataset (None):
-----------------------
the Xarry Netcdf object to be analyzed
netcdf_temporal_coord_name (str = 'time'):
-----------------------------------
the name of the time dimension in the netcdf file
geo_series_temporal_attribute_name(str = 'Datetime'):
-----------------------------------
the name of the time dimension in the geoseries file
longitude_dimension (str = 'lon'):
----------------------------------
the name of the longitude/horizontal dimension in the netcdf file
latitude_dimension (str = 'lat'):
----------------------------------
the name of the latitude/vertical dimension in the netcdf file
'''
Base_class_space_time_netcdf_gdf.__init__(self, xarray_dataset=xarray_dataset,
netcdf_temporal_coord_name=netcdf_temporal_coord_name,
geo_series_temporal_attribute_name = geo_series_temporal_attribute_name,
longitude_dimension=longitude_dimension,
latitude_dimension=latitude_dimension,
)
self.__netcdf_ds = xarray_dataset
self.__gdf = gdf
self.__geo_series_temporal_attribute_name = geo_series_temporal_attribute_name
self.__netcdf_ds = self.netcdf_ds.sortby([self._temporal_coords,
longitude_dimension,
latitude_dimension])
self.netcdf_ds = self._slice_bounding_box()
@ property
def netcdf_ds(self):
return self.__netcdf_ds
@ netcdf_ds.setter
def netcdf_ds(self, new_netcdf_ds):
'''
This property-setter alters the former netcdf_ds for the new gdf provided
'''
self.__netcdf_ds = new_netcdf_ds
@ property
def gdf(self):
return self.__gdf
@ gdf.setter
def gdf(self, new_gdf):
'''
This property-setter alters the former GDF for the new gdf provided
'''
self.__gdf = new_gdf
def _slice_bounding_box(self):
xmin, ymin, xmax, ymax = self.gdf.geometry.total_bounds
dx = float(self.coord_resolution(self.spatial_coords['x']))
dy = float(self.coord_resolution(self.spatial_coords['y']))
xmin -= dx # to ensure full pixel slicing
xmax += dx # to ensure full pixel slicing
ymin -= dy # to ensure full pixel slicing
ymax += dy # to ensure full pixel slicing
result = self.netcdf_ds.sel({self.spatial_coords['x']:slice(xmin, xmax),
self.spatial_coords['y']:slice(ymin, ymax)})
return result
def _slice_time_interval(self, time_init, final_time):
result = self.netcdf_ds.sel({self._temporal_coords:slice(time_init, final_time)})
return result
def _make_time_space_aggregations(self,
geoDataFrame,
date_offset,
netcdf_varnames,
agg_functions):
Tmin = geoDataFrame[self.__geo_series_temporal_attribute_name].min()
Tmax = geoDataFrame[self.__geo_series_temporal_attribute_name].max()
time_init = Tmin - date_offset
final_time = Tmax + date_offset
netcdf_sliced = self._slice_time_interval(time_init, final_time)
netcdf_sliced_as_gpd_geodataframe = self.netcdf_to_gdf(netcdf_sliced)
if not netcdf_sliced_as_gpd_geodataframe.empty:
sjoined = gpd.sjoin(geoDataFrame, netcdf_sliced_as_gpd_geodataframe, how="left", op='contains')
sjoined_agg = sjoined[netcdf_varnames].agg(agg_functions)
else:
sjoined = geoDataFrame
for key in netcdf_varnames:
sjoined[key] = np.nan
sjoined_agg = sjoined_agg.T
sjoined_agg['period_sliced'] = time_init.strftime("%Y/%m/%d %H:%M:%S") + ' <-> ' + final_time.strftime("%Y/%m/%d %H:%M:%S")
sjoined_agg.index.name = 'Variables'
print(sjoined_agg)
#sjoined_agg.index = geodataframe.index ?
return sjoined_agg
def _evaluate_space_time_agg(self,
netcdf_varnames=['adg_443_qaa'],
dict_of_windows=dict(time_window='1D'),
agg_functions=['nanmean','nansum','nanstd'],
verbose=True):
date_offset = pd.tseries.frequencies.to_offset(dict_of_windows['time_window'])
self.gdf2 = self.gdf.groupby(self.__geo_series_temporal_attribute_name).apply(lambda group:
self._make_time_space_aggregations(group,
date_offset=date_offset,
netcdf_varnames=netcdf_varnames,
agg_functions=agg_functions)
)
if self.gdf.index.name == None:
self.gdf.index.name = 'index'
idx_name = 'index'
else:
idx_name = self.gdf.index.name
T = self.gdf2
T[idx_name] = list(self.gdf.index) * (len(self.gdf2) // len(self.gdf))
T = T.set_index('index', append=True, inplace=False).swaplevel(2, 0)
self.gdf2 = self.gdf.merge(T, on=idx_name)
def _base(gdf,
netcdf,
netcdf_varnames =['adg_443_qaa'],
netcdf_temporal_coord_name='time',
geo_series_temporal_attribute_name = 'Datetime',
longitude_dimension='lon',
latitude_dimension='lat',
dict_of_windows=dict(time_window='M'),
agg_functions=['mean', 'max', 'min', 'std'],
verbose=True):
Match_Upper = Space_Time_Agg_over_polygons( gdf=gdf,
xarray_dataset=netcdf,
netcdf_temporal_coord_name=netcdf_temporal_coord_name,
geo_series_temporal_attribute_name = geo_series_temporal_attribute_name,
longitude_dimension=longitude_dimension,
latitude_dimension=latitude_dimension)
Match_Upper._evaluate_space_time_agg(netcdf_varnames=netcdf_varnames,
dict_of_windows=dict_of_windows,
agg_functions=agg_functions,
verbose=verbose)
return Match_Upper.gdf2
def get_zonal_match_up(netcdf,
gdf,
netcdf_varnames =['adg_443_qaa'],
dict_of_windows=dict(time_window='5D'),
agg_functions=['mean', 'max', 'min', 'std'],
netcdf_temporal_coord_name='time',
geo_series_temporal_attribute_name = 'Datetime',
longitude_dimension='lon',
latitude_dimension='lat',
verbose=True):
"""
This function does Match - Up operations from centroids of Geoseries or GeoDataFrames over Netcdfs.
Attributes:
netcdf (xarray Dataset/Dataarray):
--------------------------------------------------------------------------
gdf (geopandas GeoDataFrame):
--------------------------------------------------------------------------
netcdf_varnames (list): a list containing the netcdf variable names to apply the aggregation.
Example: netcdf_varnames=['adg_443_qaa']
--------------------------------------------------------------------------
dict_of_windows(dictionary)
Example: dict_of_windows=dict(time_window='5D') # for 5 day window integration
Other time integration options, follow pandas pattern (e.g.: 'Q', '3Y',...etc.)
--------------------------------------------------------------------------
agg_functions(list):
Example: agg_functions = ['mean', 'max', 'min', 'std']
--------------------------------------------------------------------------
verbose (bool): it sets the function to verbose (or not).
Example verbose=True
--------------------------------------------------------------------------
Returns:
(geopandas GeoDataFrame)
"""
if isinstance(gdf.index, pd.MultiIndex):
gdf = gdf.reset_index()
return _base(gdf=gdf.copy(),
netcdf=netcdf.copy(),
netcdf_varnames=netcdf_varnames,
dict_of_windows=dict_of_windows,
agg_functions=agg_functions,
verbose=verbose,
netcdf_temporal_coord_name=netcdf_temporal_coord_name,
geo_series_temporal_attribute_name = geo_series_temporal_attribute_name,
longitude_dimension=longitude_dimension,
latitude_dimension=latitude_dimension,
)
# Getting data
```
gdf = make_fake_data(3)
gdf.geometry = gdf.geometry.buffer(1.15) # in degrees
xnetcdf = get_netcdf_example()
```
# Using the algorithm
```
xnetcdf['new_data'] = xnetcdf['adg_443_qaa'] * 5 - 15
Match_Upper = get_zonal_match_up(gdf=gdf,
netcdf=xnetcdf,
netcdf_varnames =['adg_443_qaa', 'new_data'],
dict_of_windows=dict(time_window='1M'),
agg_functions=['mean', 'max', 'min', 'std']
)
Match_Upper
Match_Upper
```
| github_jupyter |
<a href="https://colab.research.google.com/github/csy99/dna-nn-theory/blob/master/supervised_UCI_adam256_save_embedding.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
from itertools import product
import re
import time
from sklearn.model_selection import train_test_split
from sklearn.manifold import TSNE
import tensorflow as tf
from tensorflow import keras
```
# Read Data
```
!pip install PyDrive
from google.colab import drive
drive.mount('/content/gdrive')
def convert_label(row):
if row["Classes"] == 'EI':
return 0
if row["Classes"] == 'IE':
return 1
if row["Classes"] == 'N':
return 2
data_path = '/content/gdrive/My Drive/Colab Notebooks/UCI/'
splice_df = pd.read_csv(data_path + 'splice.data', header=None)
splice_df.columns = ['Classes', 'Name', 'Seq']
splice_df["Seq"] = splice_df["Seq"].str.replace(' ', '').str.replace('N', 'A').str.replace('D', 'T').str.replace('S', 'C').str.replace('R', 'G')
splice_df["Label"] = splice_df.apply(lambda row: convert_label(row), axis=1)
print('The shape of the datasize is', splice_df.shape)
splice_df.head()
seq_num = 0
for seq in splice_df["Seq"]:
char_num = 0
for char in seq:
if char != 'A' and char != 'C' and char != 'T' and char != 'G':
print("seq", seq_num, 'char', char_num, 'is', char)
char_num += 1
seq_num += 1
# check if the length of the sequence is the same
seq_len = len(splice_df.Seq[0])
print("The length of the sequence is", seq_len)
for seq in splice_df.Seq[:200]:
assert len(seq) == seq_len
xtrain_full, xtest, ytrain_full, ytest = train_test_split(splice_df, splice_df.Label, test_size=0.2, random_state=100, stratify=splice_df.Label)
xtrain, xval, ytrain, yval = train_test_split(xtrain_full, ytrain_full, test_size=0.2, random_state=100, stratify=ytrain_full)
print("shape of training, validation, test set\n", xtrain.shape, xval.shape, xtest.shape, ytrain.shape, yval.shape, ytest.shape)
word_size = 1
vocab = [''.join(p) for p in product('ACGT', repeat=word_size)]
word_to_idx = {word: i for i, word in enumerate(vocab)}
vocab_size = len(word_to_idx)
print('vocab_size:', vocab_size)
create1gram = keras.layers.experimental.preprocessing.TextVectorization(
standardize=lambda x: tf.strings.regex_replace(x, '(.)', '\\1 '), ngrams=1
)
create1gram.adapt(vocab)
def ds_preprocess(x, y):
x_index = tf.subtract(create1gram(x), 2)
return x_index, y
# not sure the correct way to get mapping from word to its index
create1gram('A C G T') - 2
BATCH_SIZE = 256
xtrain_ds = tf.data.Dataset.from_tensor_slices((xtrain['Seq'], ytrain)).map(ds_preprocess).batch(BATCH_SIZE)
xval_ds = tf.data.Dataset.from_tensor_slices((xval['Seq'], yval)).map(ds_preprocess).batch(BATCH_SIZE)
xtest_ds = tf.data.Dataset.from_tensor_slices((xtest['Seq'], ytest)).map(ds_preprocess).batch(BATCH_SIZE)
latent_size = 30
model = keras.Sequential([
keras.Input(shape=(seq_len,)),
keras.layers.Embedding(seq_len, latent_size),
keras.layers.LSTM(latent_size, return_sequences=False),
keras.layers.Dense(128, activation="relu", input_shape=[latent_size]),
keras.layers.Dropout(0.2),
keras.layers.Dense(64, activation="relu"),
keras.layers.Dropout(0.2),
keras.layers.Dense(32, activation="relu"),
keras.layers.Dropout(0.2),
keras.layers.Dense(16, activation="relu"),
keras.layers.Dropout(0.2),
keras.layers.Dense(3, activation="softmax")
])
model.summary()
es_cb = keras.callbacks.EarlyStopping(patience=100, restore_best_weights=True)
model.compile(keras.optimizers.Adam(), loss=keras.losses.SparseCategoricalCrossentropy(), metrics=['accuracy'])
hist = model.fit(xtrain_ds, validation_data=xval_ds, epochs=4000, callbacks=[es_cb])
def save_hist():
filename = data_path + "baseline_uci_adam256_history.csv"
hist_df = pd.DataFrame(hist.history)
with open(filename, mode='w') as f:
hist_df.to_csv(f)
save_hist()
fig, axes = plt.subplots(1, 2, figsize=(10, 5))
for i in range(1):
ax1 = axes[0]
ax2 = axes[1]
ax1.plot(hist.history['loss'], label='training')
ax1.plot(hist.history['val_loss'], label='validation')
ax1.set_ylim((0.2, 1.2))
ax1.set_title('lstm autoencoder loss')
ax1.set_xlabel('epoch')
ax1.set_ylabel('loss')
ax1.legend(['train', 'validation'], loc='upper left')
ax2.plot(hist.history['accuracy'], label='training')
ax2.plot(hist.history['val_accuracy'], label='validation')
ax2.set_ylim((0.5, 1.0))
ax2.set_title('lstm autoencoder accuracy')
ax2.set_xlabel('epoch')
ax2.set_ylabel('accuracy')
ax2.legend(['train', 'validation'], loc='upper left')
fig.tight_layout()
def eval_model(model, ds, ds_name="Training"):
loss, acc = model.evaluate(ds, verbose=0)
print("{} Dataset: loss = {} and acccuracy = {}%".format(ds_name, np.round(loss, 3), np.round(acc*100, 2)))
eval_model(model, xtrain_ds, "Training")
eval_model(model, xval_ds, "Validation")
eval_model(model, xtest_ds, "Test")
```
| github_jupyter |
```
import os
import plaid
import requests
import datetime
import json
import pandas as pd
%matplotlib inline
def pretty_print_response(response):
print(json.dumps(response, indent=4, sort_keys=True))
PLAID_CLIENT_ID = ('PLAID_CLIENT_ID')
PLAID_SBX_SECRET_KEY = ('PLAID_SBX_SECRET_KEY')
PLAID_PUBLIC_KEY = ('PLAID_PUBLIC_KEY')
PLAID_ENV = os.getenv('PLAID_ENV', 'sandbox')
PLAID_PRODUCTS = os.getenv('PLAID_PRODUCTS', 'transactions')
```
# Plaid Access Token
In this section, you will use the plaid-python api to generate the correct authentication tokens to access data in the free developer Sandbox. This mimics how you might connect to your own account or a customer account, but due to privacy issues, this homework will only require connecting to and analyzing the fake data from the developer sandbox that Plaid provides.
Complete the following steps to generate an access token:
1. Create a client to connect to plaid
2. Use the client to generate a public token and request the following items:
['transactions', 'income', 'assets']
3. Exchange the public token for an access token
4. Test the access token by requesting and printing the available test accounts
### 1. Create a client to connect to plaid
```
INSTITUTION_ID = "ins_109508"
client = plaid.Client(client_id=PLAID_CLIENT_ID,
secret=PLAID_SBX_SECRET_KEY,
public_key=PLAID_PUBLIC_KEY,
environment= PLAID_ENV )
```
### 2. Generate a public token
```
generate_tkn_response = client.Sandbox.public_token.create(INSTITUTION_ID,['transactions','income', 'assets'])
generate_tkn_response
```
### 3. Exchange the public token for an access token
```
exchange_tkn_response = client.Item.public_token.exchange(generate_tkn_response['public_token'])
access_token = exchange_tkn_response['access_token']
```
### 4. Fetch Accounts
```
client.Accounts.get(access_token)
```
---
# Account Transactions with Plaid
In this section, you will use the Plaid Python SDK to connect to the Developer Sandbox account and grab a list of transactions. You will need to complete the following steps:
1. Use the access token to fetch the transactions for the last 90 days
2. Print the categories for each transaction type
3. Create a new DataFrame using the following fields from the JSON transaction data: `date, name, amount, category`. (For categories with more than one label, just use the first category label in the list)
4. Convert the data types to the appropriate types (i.e. datetimeindex for the date and float for the amount)
### 1. Fetch the Transactions for the last 90 days
```
start_date = '{:%Y-%m-%d}'.format(datetime.datetime.now() + datetime.timedelta(-90))
end_date = '{:%Y-%m-%d}'.format(datetime.datetime.now())
transaction_response = client.Transactions.get(access_token,start_date,end_date)
pretty_print_response(transaction_response['transactions'][:1])
```
### 2. Print the categories for each transaction
```
for transaction in transaction_response['transactions']:
print(transaction['category'])
```
### 3. Create a new DataFrame using the following fields from the JSON transaction data: date, name, amount, category.
(For categories with more than one label, just use the first category label in the list)
```
transaction_df = pd.DataFrame(columns=['Date','Name','Amount','Category'])
date = []
name = []
amount = []
category = []
for i in transactions:
date.append(i['date'])
name.append(i['name'])
amount.append(i['amount'])
category.append(i['category'][0])
transaction_df['Date'] = date
transaction_df['Name'] = name
transaction_df['Amount'] = amount
transaction_df['Category'] = category
transaction_df.head()
```
### 4. Convert the data types to the appropriate types
(i.e. datetimeindex for the date and float for the amount)
```
transaction_df.dtypes
transaction_df['Date'] = pd.to_datetime(transaction_df['Date'])
transaction_df = transaction_df.set_index(['Date'])
transaction_df.dtypes
```
---
# Income Analysis with Plaid
In this section, you will use the Plaid Sandbox to complete the following:
1. Determine the previous year's gross income and print the results
2. Determine the current monthly income and print the results
3. Determine the projected yearly income and print the results
```
income_response = client.Income.get(access_token)
pretty_print_response(income_response)
print(f"Last Year's income: {income_response['income']['last_year_income_before_tax']}")
print(f"Current monthly income: {income_response['income']['income_streams'][0]['monthly_income']}")
print(f"Projected Year's income: {income_response['income']['projected_yearly_income_before_tax']}")
```
---
# Budget Analysis
In this section, you will use the transactions DataFrame to analyze the customer's budget
1. Calculate the total spending per category and print the results (Hint: groupby or count transactions per category)
2. Generate a bar chart with the number of transactions for each category
3. Calculate the expenses per month
4. Plot the total expenses per month
### Calculate the expenses per category
```
# YOUR CODE HERE
expenses_by_category = transaction_df.groupby('Category').sum()["Amount"]
expenses_by_category
expenses_by_category.plot(kind = "pie", title = "Expenses by Category",subplots=True, figsize = (10,10))
```
### Calculate the expenses per month
```
transaction_df.reset_index(inplace=True)
transaction_df['month'] = pd.DatetimeIndex(transaction_df['Date']).month
transaction_df.head()
transactions_per_month = transaction_df.groupby('month').sum()
transactions_per_month
transactions_per_month.plot(kind = 'bar', title = "Transactions per Month", rot=45)
```
| github_jupyter |
```
import numpy as np
import Cluster_Ensembles as CE
from functools import reduce
# require(data.table)
# require(bit64)
# require(dbscan)
# require(doParallel)
# require(rBayesianOptimization)
# path='../input/train_1/'
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import os
from trackml.dataset import load_event, load_dataset
from trackml.score import score_event
from trackml.randomize import shuffle_hits
from sklearn.preprocessing import StandardScaler
import hdbscan as _hdbscan
from scipy import stats
from tqdm import tqdm
import time
from sklearn.cluster.dbscan_ import dbscan
from sklearn.cluster import DBSCAN
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KDTree
import hdbscan
from bayes_opt import BayesianOptimization
# https://www.ellicium.com/python-multiprocessing-pool-process/
# http://sebastianraschka.com/Articles/2014_multiprocessing.html
from multiprocessing import Pool
import os
import time
import hdbscan as _hdbscan
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
def create_one_event_submission(event_id, hits, labels):
sub_data = np.column_stack(([event_id]*len(hits), hits.hit_id.values, labels))
submission = pd.DataFrame(data=sub_data, columns=["event_id", "hit_id", "track_id"]).astype(int)
return submission
def preprocess(hits):
x = hits.x.values
y = hits.y.values
z = hits.z.values
r = np.sqrt(x**2 + y**2 + z**2)
hits['x2'] = x/r
hits['y2'] = y/r
r = np.sqrt(x**2 + y**2)
hits['z2'] = z/r
ss = StandardScaler()
X = ss.fit_transform(hits[['x2', 'y2', 'z2']].values)
# for i, rz_scale in enumerate(self.rz_scales):
# X[:,i] = X[:,i] * rz_scale
return X
def _eliminate_outliers(clusters,M):
my_labels = np.unique(clusters)
norms=np.zeros((len(my_labels)),np.float32)
indices=np.zeros((len(my_labels)),np.float32)
for i, cluster in tqdm(enumerate(my_labels),total=len(my_labels)):
if cluster == 0:
continue
index = np.argwhere(clusters==cluster)
index = np.reshape(index,(index.shape[0]))
indices[i] = len(index)
x = M[index]
norms[i] = self._test_quadric(x)
threshold1 = np.percentile(norms,90)*5
threshold2 = 25
threshold3 = 6
for i, cluster in enumerate(my_labels):
if norms[i] > threshold1 or indices[i] > threshold2 or indices[i] < threshold3:
clusters[clusters==cluster]=0
def _test_quadric(x):
if x.size == 0 or len(x.shape)<2:
return 0
Z = np.zeros((x.shape[0],10), np.float32)
Z[:,0] = x[:,0]**2
Z[:,1] = 2*x[:,0]*x[:,1]
Z[:,2] = 2*x[:,0]*x[:,2]
Z[:,3] = 2*x[:,0]
Z[:,4] = x[:,1]**2
Z[:,5] = 2*x[:,1]*x[:,2]
Z[:,6] = 2*x[:,1]
Z[:,7] = x[:,2]**2
Z[:,8] = 2*x[:,2]
Z[:,9] = 1
v, s, t = np.linalg.svd(Z,full_matrices=False)
smallest_index = np.argmin(np.array(s))
T = np.array(t)
T = T[smallest_index,:]
norm = np.linalg.norm(np.dot(Z,T), ord=2)**2
return norm
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
#------------------------------------------------------
def make_counts(labels):
_,reverse,count = np.unique(labels,return_counts=True,return_inverse=True)
counts = count[reverse]
counts[labels==0]=0
return counts
def one_loop(param):
# <todo> tune your parameters or design your own features here!
i,m, x,y,z, d,r, a, a_start,a_step = param
#print('\r %3d %+0.8f '%(i,da), end='', flush=True)
da = m*(a_start - (i*a_step))
aa = a + np.sign(z)*z*da
zr = z/r
X = StandardScaler().fit_transform(np.column_stack([aa, aa/zr, zr, 1/zr, aa/zr + 1/zr]))
_,l = dbscan(X, eps=0.0035, min_samples=1,)
return l
def one_loop1(param):
# <todo> tune your parameters or design your own features here!
i,m, x,y,z, d,r,r2,z2,a, a_start,a_step = param
#print('\r %3d %+0.8f '%(i,da), end='', flush=True)
da = m*(a_start - (i*a_step))
aa = a + np.sign(z)*z*da
# if m == 1:
# print(da)
zr = z/r # this is cot(theta), 1/zr is tan(theta)
theta = np.arctan2(r, z)
ct = np.cos(theta)
st = np.sin(theta)
tt = np.tan(theta)
# ctt = np.cot(theta)
z2r = z2/r
z2r2 = z2/r2
# X = StandardScaler().fit_transform(df[['r2', 'theta_1', 'dip_angle', 'z2', 'z2_1', 'z2_2']].values)
caa = np.cos(aa)
saa = np.sin(aa)
taa = np.tan(aa)
ctaa = 1/taa
# 0.000005
deps = 0.0000025
X = StandardScaler().fit_transform(np.column_stack([caa, saa, tt, 1/tt]))
l= DBSCAN(eps=0.0035+i*deps,min_samples=1,metric='euclidean',n_jobs=8).fit(X).labels_
# _,l = dbscan(X, eps=0.0035, min_samples=1,algorithm='auto')
return l
def one_loop2(param):
# <todo> tune your parameters or design your own features here!
i,m, x,y,z, d,r,r2,z2,a, a_start,a_step = param
#print('\r %3d %+0.8f '%(i,da), end='', flush=True)
da = m*(a_start - (i*a_step))
aa = a + np.sign(z)*z*da
# if m == 1:
# print(da)
zr = z/r # this is cot(theta), 1/zr is tan(theta)
theta = np.arctan2(r, z)
ct = np.cos(theta)
st = np.sin(theta)
tt = np.tan(theta)
# ctt = np.cot(theta)
z2r = z2/r
z2r2 = z2/r2
# X = StandardScaler().fit_transform(df[['r2', 'theta_1', 'dip_angle', 'z2', 'z2_1', 'z2_2']].values)
caa = np.cos(aa)
saa = np.sin(aa)
taa = np.tan(aa)
ctaa = 1/taa
# 0.000005
deps = 0.0000025
X = StandardScaler().fit_transform(np.column_stack([caa, saa, tt, 1/tt]))
l= DBSCAN(eps=0.0035+i*deps,min_samples=1,metric='euclidean',n_jobs=8).fit(X).labels_
# _,l = dbscan(X, eps=0.0035, min_samples=1,algorithm='auto')
return l
def do_dbscan_predict(df):
x = df.x.values
y = df.y.values
z = df.z.values
r = np.sqrt(x**2+y**2)
d = np.sqrt(x**2+y**2+z**2)
a = np.arctan2(y,x)
x2 = df['x']/d
y2 = df['y']/d
z2 = df['z']/r
r2 = np.sqrt(x2**2 + y2**2)
phi = np.arctan2(y, x)
phi_deg= np.degrees(np.arctan2(y, x))
phi2 = np.arctan2(y2, x2)
phi2_deg = np.degrees(np.arctan2(y2, x2))
for angle in range(-180,180,1):
df1 = df.loc[(df.phi_deg>(angle-1.0)) & (df.phi_deg<(angle+1.0))]
x = df1.x.values
y = df1.y.values
z = df1.z.values
r = np.sqrt(x**2+y**2)
d = np.sqrt(x**2+y**2+z**2)
a = np.arctan2(y,x)
x2 = df1['x']/d
y2 = df1['y']/d
z2 = df1['z']/r
r2 = np.sqrt(x2**2 + y2**2)
theta= np.arctan2(r, z)
theta1 = np.arctan2(r2, z2)
tan_dip = phi/theta
tan_dip1 = phi/z2
z2_1 = 1/z2
z2_2 = phi/z2 + 1/z2
dip_angle = np.arctan2(z2, (np.sqrt(x2**2 +y2**2)) * np.arccos(x2/np.sqrt(x2**2 + y2**2)))
dip_angle1 = np.arctan2(z, (np.sqrt(x**2 +y**2)) * np.arccos(x2/np.sqrt(x**2 + y**2)))
scores = []
a_start,a_step,a_num = 0.00100,0.0000095,150
params = [(i,m, x,y,z,d,r,r2,z2, a, a_start,a_step) for i in range(a_num) for m in [-1,1]]
if 1:
pool = Pool(processes=1)
ls = pool.map( one_loop1, params )
if 0:
ls = [ one_loop(param) for param in params ]
##------------------------------------------------
num_hits=len(df)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for l in ls:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
# cl = hdbscan.HDBSCAN(min_samples=1,min_cluster_size=7,
# metric='braycurtis',cluster_selection_method='leaf',algorithm='best',
# leaf_size=50)
# X = preprocess(df)
# l1 = pd.Series(labels)
# labels = np.unique(l1)
# # print(X.shape)
# # print(len(labels_org))
# # print(len(labels_org[labels_org ==0]))
# # print(len(labels_org[labels_org ==-1]))
# n_labels = 0
# while n_labels < len(labels):
# n_labels = len(labels)
# max_len = np.max(l1)
# s = list(l1[l1 == 0].keys())
# X = X[s]
# print(X.shape)
# if X.shape[0] <= 1:
# break
# l = cl.fit_predict(X)+max_len
# # print(len(l))
# l1[l1 == 0] = l
# labels = np.unique(l1)
return labels
## reference----------------------------------------------
def do_dbscan0_predict(df):
x = df.x.values
y = df.y.values
z = df.z.values
r = np.sqrt(x**2+y**2)
d = np.sqrt(x**2+y**2+z**2)
X = StandardScaler().fit_transform(np.column_stack([
x/d, y/d, z/r]))
_,labels = dbscan(X,
eps=0.0075,
min_samples=1,
algorithm='auto',
n_jobs=-1)
#labels = hdbscan(X, min_samples=1, min_cluster_size=5, cluster_selection_method='eom')
return labels
## reference----------------------------------------------
def do_dbscan0_predict(df):
x = df.x.values
y = df.y.values
z = df.z.values
r = np.sqrt(x**2+y**2)
d = np.sqrt(x**2+y**2+z**2)
X = StandardScaler().fit_transform(np.column_stack([
x/d, y/d, z/r]))
_,labels = dbscan(X,
eps=0.0075,
min_samples=1,
algorithm='auto',
n_jobs=-1)
#labels = hdbscan(X, min_samples=1, min_cluster_size=5, cluster_selection_method='eom')
return labels
def extend(submission,hits):
df = submission.merge(hits, on=['hit_id'], how='left')
# df = submission.append(hits)
# print(df.head())
df = df.assign(d = np.sqrt( df.x**2 + df.y**2 + df.z**2 ))
df = df.assign(r = np.sqrt( df.x**2 + df.y**2))
df = df.assign(arctan2 = np.arctan2(df.z, df.r))
for angle in range(-180,180,1):
print ('\r %f'%angle, end='',flush=True)
#df1 = df.loc[(df.arctan2>(angle-0.5)/180*np.pi) & (df.arctan2<(angle+0.5)/180*np.pi)]
df1 = df.loc[(df.arctan2>(angle-1.0)/180*np.pi) & (df.arctan2<(angle+1.0)/180*np.pi)]
min_num_neighbours = len(df1)
if min_num_neighbours<4: continue
hit_ids = df1.hit_id.values
x,y,z = df1.as_matrix(columns=['x', 'y', 'z']).T
r = (x**2 + y**2)**0.5
r = r/1000
a = np.arctan2(y,x)
tree = KDTree(np.column_stack([a,r]), metric='euclidean')
track_ids = list(df1.track_id.unique())
num_track_ids = len(track_ids)
min_length=3
for i in range(num_track_ids):
p = track_ids[i]
if p==0: continue
idx = np.where(df1.track_id==p)[0]
if len(idx)<min_length: continue
if angle>0:
idx = idx[np.argsort( z[idx])]
else:
idx = idx[np.argsort(-z[idx])]
## start and end points ##
idx0,idx1 = idx[0],idx[-1]
a0 = a[idx0]
a1 = a[idx1]
r0 = r[idx0]
r1 = r[idx1]
da0 = a[idx[1]] - a[idx[0]] #direction
dr0 = r[idx[1]] - r[idx[0]]
direction0 = np.arctan2(dr0,da0)
da1 = a[idx[-1]] - a[idx[-2]]
dr1 = r[idx[-1]] - r[idx[-2]]
direction1 = np.arctan2(dr1,da1)
## extend start point
ns = tree.query([[a0,r0]], k=min(20,min_num_neighbours), return_distance=False)
ns = np.concatenate(ns)
direction = np.arctan2(r0-r[ns],a0-a[ns])
ns = ns[(r0-r[ns]>0.01) &(np.fabs(direction-direction0)<0.04)]
for n in ns:
df.loc[ df.hit_id==hit_ids[n],'track_id' ] = p
## extend end point
ns = tree.query([[a1,r1]], k=min(20,min_num_neighbours), return_distance=False)
ns = np.concatenate(ns)
direction = np.arctan2(r[ns]-r1,a[ns]-a1)
ns = ns[(r[ns]-r1>0.01) &(np.fabs(direction-direction1)<0.04)]
for n in ns:
df.loc[ df.hit_id==hit_ids[n],'track_id' ] = p
#print ('\r')
# df = df[['particle_id', 'weight', 'event_id', 'hit_id', 'track_id']]
df = df[['event_id', 'hit_id', 'track_id']]
return df
import hdbscan
import math
seed = 123
np.random.seed(seed)
def shift(l, n):
return l[n:] + l[:n]
# https://stackoverflow.com/questions/29246455/python-setting-decimal-place-range-without-rounding
def truncate(f, n):
return math.floor(f * 10 ** n) / 10 ** n
def trackML31(df, w1, w2, w3, w4, w5, w6, w7, w8, w9, w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_shift, r0Inv_nu):
x = df.x.values
y = df.y.values
z = df.z.values
# dz = 0
z = z + z_shift
rt = np.sqrt(x**2+y**2)
r = np.sqrt(x**2+y**2+z**2)
a0 = np.arctan2(y,x)
x2 = x/r
y2 = y/r
phi = np.arctan2(y, x)
phi_deg= np.degrees(np.arctan2(y, x))
z1 = z/rt
z2 = z/r
z3 = np.log1p(abs(z/r))*np.sign(z)
x1 = x/rt
y1 = y/rt
y3 = np.log1p(abs(y/r))*np.sign(y)
theta = np.arctan2(rt, z)
theta_deg = np.degrees(np.arctan2(rt, z))
tt = np.tan(theta_deg)
z4 = np.sqrt(abs(z/rt))
x4 = np.sqrt(abs(x/r))
y4 = np.sqrt(abs(y/r))
mm = 1
ls = []
# def f(x):
# return a0+mm*(rt+ 0.0000145*rt**2)/1000*(x/2)/180*np.pi
for ii in range(Niter):
mm = mm * (-1)
a1 = a0+mm*(rt+ 0.0000145*rt**2)/1000*(ii/2)/180*np.pi
da1 = mm*(1 + (2 * 0.0000145 * rt))/1000*(ii/2)/180*np.pi
ia1 = a0*rt + mm*(((rt**2)/2) + (0.0000145*rt**3)/3)/1000*(ii/2)/180*np.pi
saa = np.sin(a1)
caa = np.cos(a1)
raa = x*caa + y*saa
t1 = theta+mm*(rt+ 0.8435*rt**2)/1000*(ii/2)/180*np.pi
ctt = np.cos(t1)
stt = np.sin(t1)
# mom = np.sqrt(1 + (z1 **2))
# mom2 = r0Inv * np.sqrt(1 + (z2 **2)).round(2)
mom2 = [truncate(np.sqrt(1 + (i**2)),4) for i in z2]
# theta0= np.arcsin[np.sqrt(x**2+y**2)/(2*R)]- a0
r0_list = list(np.linspace(30, 3100, 100))
for r0 in r0_list:
r0Inv = 1./r0
theta0= np.nan_to_num(np.arcsin(rt*0.5*r0Inv))- a0
# r0Inv = 2. * np.cos(a0 - theta0) / rt
# r0Inv = 2. * caa / r
# r0Inv2 = 2. * np.cos(a0 - theta0) / rt
# r0Inv_d = -2. * np.sin(a1-t1) * da1 /r
# https://www.kaggle.com/okhlopkov/attempts-to-struggle-with-clustering
# fundu = np.arcsin((y * np.sin(theta0) - x * np.cos(theta0)) / rt) / (z - z0)
# fundu = np.arcsin((y * np.sin(theta0) - x * np.cos(theta0)) / rt) / (z-z_shift)
# fundu2 = np.arcsin((y * np.sin(theta0) - x * np.cos(theta0)) / r) / (z-z_shift)
X = StandardScaler().fit_transform(np.column_stack([caa, saa, z1, z2, rt/r, x/r, y/r, z3, y1, y3,
z4, x4, y4, raa, mom2,
da1, ia1, theta0]))
# X = StandardScaler().fit_transform(np.column_stack([caa, saa, z1, z2, fundu2]))
# print(X.shape)
# X = StandardScaler().fit_transform(np.column_stack([caa,saa,z1,z2]))
cx = [w1,w1,w2,w3, w4, w5, w6, w7, w8, w9, w11, w12, w13, w14, w15, w16, w17, 1.]
# cx = [w1,w1,w2,w3, w15]
X = np.multiply(X, cx)
l= DBSCAN(eps=0.004,min_samples=1,metric='euclidean',n_jobs=4).fit(X).labels_
ls.append(l)
num_hits=len(df)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
lss = []
for l in ls:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
# lss.append(labels)
# for i in range(Niter):
# labels1 = np.zeros(num_hits,np.int32)
# counts1 = np.zeros(num_hits,np.int32)
# ls1 = ls.copy()
# ls1 = shift(ls1, 1)
# np.random.shuffle(ls1)
# for l in ls1:
# c = make_counts(l)
# idx = np.where((c-counts>0) & (c<20))[0]
# labels1[idx] = l[idx] + labels1.max()
# counts1 = make_counts(labels1)
# l1 = labels1.copy()
# lss.append(l1)
# labels = np.zeros(num_hits,np.int32)
# counts = np.zeros(num_hits,np.int32)
# for l in lss:
# c = make_counts(l)
# idx = np.where((c-counts>0) & (c<20))[0]
# labels[idx] = l[idx] + labels.max()
# counts = make_counts(labels)
# df = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id', 'vlm'],
# data=np.column_stack(([int(0),]*len(hits), hits.hit_id.values, labels, hits.vlm.values))
# )
# df = pd.DataFrame()
# df['hit_id']=hits.hit_id.values
# df['vlm'] = hits.vlm.values
# df['track_id'] = labels
# for l in np.unique(labels):
# df_l = df[df.track_id == l]
# df_l['vlm_count'] =df_l.groupby('vlm')['vlm'].transform('count')
# same_vlm_multiple_hits = np.any(df_l.vlm_count > 1)
# if same_vlm_multiple_hits == True:
# print(l)
# which_vlm_multiple_hits = list(df_l[df_l.vlm_count > 1].index)
# which_vlm_multiple_hits.pop(0)
# df.loc[which_vlm_multiple_hits, 'track_id'] = 9999999999
# return df.track_id.values
# sub = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
# data=np.column_stack(([int(0),]*len(df), df.hit_id.values, labels))
# )
# sub['track_count'] = sub.groupby('track_id')['track_id'].transform('count')
# # sub.loc[sub.track_count < 5, 'track_id'] = 0
# sub1 = sub[sub.track_id < 0]
# sub2 = sub[sub.track_id >= 0]
# L_neg = sub1.track_id.values
# L_pos = sub2.track_id.values
# a = 1
# for l in L_neg:
# for l1 in range(a, np.iinfo(np.int32).max):
# if l1 in L_pos:
# continue
# sub.loc[sub.track_id == l, 'track_id'] = l1
# a = l1 +1
# break
# L = list(sub.track_id.values)
# labels = np.zeros(num_hits,np.int32)
# for ii in range(num_hits):
# labels[ii] = L[ii]
# print(np.any(labels < 0))
return labels
%%time
# def run_dbscan():
data_dir = '../data/train'
# event_ids = [
# '000001030',##
# '000001025','000001026','000001027','000001028','000001029',
# ]
event_ids = [
'000001030',##
]
sum=0
sum_score=0
for i,event_id in enumerate(event_ids):
particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)
hits = pd.read_csv(data_dir + '/event%s-hits.csv'%event_id)
cells = pd.read_csv(data_dir + '/event%s-cells.csv'%event_id)
truth = pd.read_csv(data_dir + '/event%s-truth.csv'%event_id)
particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)
truth = pd.merge(truth, particles, how='left', on='particle_id')
hits = pd.merge(hits, truth, how='left', on='hit_id')
%%time
w1 = 1.1932215111905984
w2 = 0.39740553885387364
w3 = 0.3512647720585538
w4 = 0.1470
w5 = 0.01201
w6 = 0.0003864
w7 = 0.0205
w8 = 0.0049
w9 = 0.00121
w10 = 1.4930496676654575e-05
w11 = 0.0318
w12 = 0.000435
w13 = 0.00038
w14 = 0.00072
w15 = 5.5e-05
# w15 = 0.000265
w16 = 0.0031
w17 = 0.00021
w18 = 7.5e-05
Niter=247
print(w18)
z_shift = 0
# ls = []
track_id = trackML31(hits, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_shift)
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(event_id),]*len(hits), hits.hit_id.values, track_id))
).astype(int)
for i in range(8):
submission = extend(submission,hits)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
# caa, saa: 5 mins score 0
# caa, saa, z1: 0.3942327679531816, 6 min 14s
# z1: 5.99028402551861e-05, 11 mins
# caa,saa,z1,z2: 7 mins, 0.5315668141457246
num_hits = len(hits)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for i in range(len(ls)):
labels1 = np.zeros(num_hits,np.int32)
counts1 = np.zeros(num_hits,np.int32)
ls1 = ls.copy()
ls1 = shift(ls1, 1)
np.random.shuffle(ls1)
for l in ls1:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels1[idx] = l[idx] + labels1.max()
counts1 = make_counts(labels1)
l1 = labels1.copy()
lss.append(l1)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for l in lss:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(event_id),]*len(hits), hits.hit_id.values, labels))
).astype(int)
for i in range(8):
submission = extend(submission,hits)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
# 179.0000000[ 0] score : 0.63363358
# 179.0000000[ 1] score : 0.63765912
# 179.0000000[ 2] score : 0.63883962
# 179.0000000[ 3] score : 0.64030808
# 179.0000000[ 4] score : 0.64120567
# 179.0000000[ 5] score : 0.64168075
# 179.0000000[ 6] score : 0.64064708
# 179.0000000[ 7] score : 0.64116239
# --------------------------------------
# 0.63939203643381
hits.head()
def Fun4BO22221(params):
tic = t = time.time()
df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, z_s, t = params
l = trackML31(df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_s, t)
toc = time.time()
print('\r z_s : %0.6f, t: %0.6f , %0.0f min'%(z_s, t, (toc-tic)/60))
return l
%%time
ls = []
lss = []
def Fun4BO21(df, truth):
w1 = 1.1932215111905984
w2 = 0.39740553885387364
w3 = 0.3512647720585538
w4 = 0.1470
w5 = 0.01201
w6 = 0.0003864
w7 = 0.0205
w8 = 0.0049
w9 = 0.00121
w10 = 1.4930496676654575e-05
w11 = 0.0318
w12 = 0.000435
w13 = 0.00038
w14 = 0.00072
w15 = 0.01
# w15 = 0.00109
# w15 = 0.001
# w15 = 5.5e-05
# w15 = 1.
w16 = 0.0031
w17 = 0.00021
w18 = 7.5e-05
Niter=247
# print(w18)
# z_shift = 0
T = []
L = []
params = []
if 1:
L = ['r0Inv']
params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
0, 0))
pool = Pool(processes=1)
ls1 = pool.map(Fun4BO22221, params, chunksize=1)
pool.close()
j = 0
ls = ls1
num_hits = len(df)
# lss = []
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
ls2 = []
if 1:
for l in ls:
print(L[j])
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))
).astype(int)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(j, score))
for i in range(8):
submission = extend(submission,df)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
# l2 = submission.track_id.values
# ls2.append(l2)
j += 1
if 0:
L = [1., 0.1,0.01, 0.001, 0.0001]
for w15 in L:
params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
0, 0))
pool = Pool(processes=len(L))
ls1 = pool.map(Fun4BO22221, params, chunksize=1)
pool.close()
j = 0
ls = ls1
num_hits = len(df)
# lss = []
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
ls2 = []
if 1:
for l in ls:
print(L[j])
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))
).astype(int)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(j, score))
for i in range(8):
submission = extend(submission,df)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
# l2 = submission.track_id.values
# ls2.append(l2)
j += 1
if 0:
params = []
r0Inv = [1]
r0Inv_list = list(np.linspace(-180, 180, 10))
theta0 = theta0 + theta_list
z_shifts = [0]
z_list = list(np.linspace(-5.5, 5.5, 10))
z_shifts = z_shifts + z_list
for z_s in z_shifts:
t = 0
if z_s != 0:
np.random.shuffle(theta0)
t = theta0.pop()
T.append(t)
L.append(z_s)
params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_s, t))
pool = Pool(processes=len(z_shifts))
ls1 = pool.map(Fun4BO22221, params, chunksize=1)
pool.close()
j = 0
ls = ls1
num_hits = len(df)
# lss = []
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
ls2 = []
if 0:
for l in ls:
print(L[j], T[j])
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))
).astype(int)
score = score_event(truth, submission)
print('[%2d, %2d] score : %0.8f'%(L[j], T[j], score))
for i in range(8):
submission = extend(submission,df)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
l2 = submission.track_id.values
ls2.append(l2)
j += 1
# labels = np.zeros(num_hits,np.int32)
# counts = np.zeros(num_hits,np.int32)
for i in range(len(ls2)):
labels1 = np.zeros(num_hits,np.int32)
counts1 = np.zeros(num_hits,np.int32)
ls1 = ls.copy()
ls1 = shift(ls1, 1)
np.random.shuffle(ls1)
for l in ls1:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels1[idx] = l[idx] + labels1.max()
counts1 = make_counts(labels1)
l1 = labels1.copy()
lss.append(l1)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for l in lss:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(0),]*len(df), df.hit_id.values, labels))
).astype(int)
for i in range(8):
submission = extend(submission,df)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
if 0:
params = []
theta0 = [0]
theta_list = list(np.linspace(-180, 180, 10))
theta0 = theta0 + theta_list
z_shifts = [0]
z_list = list(np.linspace(-5.5, 5.5, 10))
z_shifts = z_shifts + z_list
for z_s in z_shifts:
t = 0
if z_s != 0:
np.random.shuffle(theta0)
t = theta0.pop()
T.append(t)
L.append(z_s)
params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_s, t))
pool = Pool(processes=len(z_shifts))
ls1 = pool.map(Fun4BO22221, params, chunksize=1)
pool.close()
j = 0
ls = ls1
num_hits = len(df)
# lss = []
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
ls2 = []
if 0:
for l in ls:
print(L[j], T[j])
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))
).astype(int)
score = score_event(truth, submission)
print('[%2d, %2d] score : %0.8f'%(L[j], T[j], score))
for i in range(8):
submission = extend(submission,df)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
l2 = submission.track_id.values
ls2.append(l2)
j += 1
# labels = np.zeros(num_hits,np.int32)
# counts = np.zeros(num_hits,np.int32)
for i in range(len(ls2)):
labels1 = np.zeros(num_hits,np.int32)
counts1 = np.zeros(num_hits,np.int32)
ls1 = ls.copy()
ls1 = shift(ls1, 1)
np.random.shuffle(ls1)
for l in ls1:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels1[idx] = l[idx] + labels1.max()
counts1 = make_counts(labels1)
l1 = labels1.copy()
lss.append(l1)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for l in lss:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(0),]*len(df), df.hit_id.values, labels))
).astype(int)
for i in range(8):
submission = extend(submission,df)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
if 0:
theta0 = [0]
theta_list = list(np.linspace(-np.pi, np.pi, 50))
theta0 = theta0 + theta_list
params = []
for t in theta0:
params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
0, t))
pool = Pool(processes=20)
ls1 = pool.map(Fun4BO22221, params, chunksize=1)
pool.close()
ls = ls + ls1
num_hits = len(df)
# lss = []
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for i in range(len(ls)):
labels1 = np.zeros(num_hits,np.int32)
counts1 = np.zeros(num_hits,np.int32)
ls1 = ls.copy()
ls1 = shift(ls1, 1)
np.random.shuffle(ls1)
for l in ls1:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels1[idx] = l[idx] + labels1.max()
counts1 = make_counts(labels1)
l1 = labels1.copy()
lss.append(l1)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for l in lss:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(event_id),]*len(df), df.hit_id.values, labels))
).astype(int)
for i in range(8):
submission = extend(submission,hits)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
return sc
return 0
# return labels
def Fun4BO2222(params):
df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, z_s = params
l = trackML31(df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_s)
return l
%%time
def Fun4BO2(df):
w1 = 1.1932215111905984
w2 = 0.39740553885387364
w3 = 0.3512647720585538
w4 = 0.1470
w5 = 0.01201
w6 = 0.0003864
w7 = 0.0205
w8 = 0.0049
w9 = 0.00121
w10 = 1.4930496676654575e-05
w11 = 0.0318
w12 = 0.000435
w13 = 0.00038
w14 = 0.00072
w15 = 0.00109
# w15 = 5.5e-05
# w15 = 0.000265
w16 = 0.0031
w17 = 0.00021
w18 = 7.5e-05
Niter=247
# print(w18)
ls = []
lss = []
z_shift = 0
z_shifts = [0]
z_shift_list = list(np.linspace(-5.5, 5.5, 5))
z_shifts = z_shifts + z_shift_list
params = []
for z_s in z_shifts:
params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter,
z_s))
pool = Pool(processes=6)
ls1 = pool.map(Fun4BO2222, params, chunksize=1)
pool.close()
ls = ls + ls1
num_hits = len(df)
# lss = []
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for i in range(len(ls)):
labels1 = np.zeros(num_hits,np.int32)
counts1 = np.zeros(num_hits,np.int32)
ls1 = ls.copy()
ls1 = shift(ls1, 1)
np.random.shuffle(ls1)
for l in ls1:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels1[idx] = l[idx] + labels1.max()
counts1 = make_counts(labels1)
l1 = labels1.copy()
lss.append(l1)
labels = np.zeros(num_hits,np.int32)
counts = np.zeros(num_hits,np.int32)
for l in lss:
c = make_counts(l)
idx = np.where((c-counts>0) & (c<20))[0]
labels[idx] = l[idx] + labels.max()
counts = make_counts(labels)
sum_score=0
sum = 0
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([int(event_id),]*len(df), df.hit_id.values, labels))
).astype(int)
for i in range(8):
submission = extend(submission,hits)
score = score_event(truth, submission)
print('[%2d] score : %0.8f'%(i, score))
sum_score += score
sum += 1
print('--------------------------------------')
sc = sum_score/sum
print(sc)
return sc
# return labels
%%time
# def run_dbscan():
data_dir = '../data/train'
# event_ids = [
# '000001030',##
# '000001025','000001026','000001027','000001028','000001029',
# ]
event_ids = [
'000001030',##
]
sum=0
sum_score=0
for i,event_id in enumerate(event_ids):
particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)
hits = pd.read_csv(data_dir + '/event%s-hits.csv'%event_id)
cells = pd.read_csv(data_dir + '/event%s-cells.csv'%event_id)
truth = pd.read_csv(data_dir + '/event%s-truth.csv'%event_id)
particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)
truth = pd.merge(truth, particles, how='left', on='particle_id')
hits = pd.merge(hits, truth, how='left', on='hit_id')
# bo = BayesianOptimization(Fun4BO,pbounds = {'w1':w1,'w2':w2,'w3':w3,'Niter':Niter})
# bo.maximize(init_points = 3, n_iter = 20, acq = "ucb", kappa = 2.576)
# w1 = 1.1932215111905984
# w2 = 0.39740553885387364
# w3 = 0.3512647720585538
# w4 = [0.1, 0.2] # 0.1470 -> 0.55690
# w4 = 0.1470
# w5 = [0.001, 1.2] # 0.7781 -> 0.55646, 0.7235 + N = 247 => 0.56025
# Niter = 179
# Niter = 247
# w5 = 0.01
# for w6 in [0.012, 0.01201, 0.01202, 0.01203, 0.01204, 0.01205, 0.01206, 0.01207, 0.01208, 0.01209, 0.0121]:
# EPS = 1e-12
# w6 = [0.001, 1.2]
# w6 = 0.0205
# w18 = [0.00001, 0.05]
# w13 = 0.00038
# w14 = 0.0007133505234834969
# for w8 in np.arange(0.00008, 0.00015, 0.000005):
# print(w8)
# Fun4BO2(1)
# for w18 in np.arange(1.0e-05, 9.0e-05, 5.0e-06):
# print(w18)
Fun4BO21(hits, truth)
# Niter = [240, 480]
# w18 = [0.00001, 0.0003]
# bo = BayesianOptimization(Fun4BO2,pbounds = {'w18':w18})
# bo.maximize(init_points = 20, n_iter = 5, acq = "ucb", kappa = 2.576)
# x/y: 7 | 06m30s | 0.55302 | 0.0100 |
# x/y: 0.001: 0.55949
# x/y: 0.0001: 0.55949
# x/y: 0.002: 0.55959
# x/y: 0.003: 0.55915
# x/y: 0.0025: 0.55925
# x/y: 0.0015: 0.55953
# x/r: 0.0015: 0.56186
# x/r: 0.002: 0.56334
# x/r: 0.0025: 0.563989
# x/r: 0.003: 0.56447
# x/r: 0.01: 0.569822
# x/r: 0.015: 0.56940
# x/r: 0.012: 0.5719
# x/r: 0.01201: 0.57192
# 1.4499999999999993e-05 * rt**2: 0.5720702851970194
# 0.0000145
# z3: 10 | 07m12s | 0.57208 | 0.0205 |
# count: 19: 0.572567, 17: 0.57263
# ctt, stt after change: 2 | 07m56s | 0.57345 | 0.0001 | (0.00010567777727496665)
# x4: 25 | 09m42s | 0.57359 | 0.0002 | (0.000206214286412982)
# x4: 0.000435 (0.5737387485278771) (x4 = np.sqrt(abs(x/r)))
# w13: 00038 (ctt,stt): 0.5737528800479372
# ensemble of 10: 0.5772859116242378
# ensemble of Niter=247 (random shuffle+ shift): 0.5787580886742594
# ensemble of Niter=247 (shift only): 0.5743461440542145
# ensemble of Niter=247 (random shuffle+ shift+ eps=0.004+vlm): 0.5865991424251623
# 14 + ensemble: (0.0007133505234834969) 0.58787
# w14 + ensemble: 1 | 30m13s | 0.58787 | 0.0007 | (0.0007133505234834969)
# w14: 0.00027 (0.5873896523922799)
# test w14, raa = x*caa + y*saa(0.00072: 0.5878990304956998)
# test w16: r0Inv1 (21 | 21m40s | 0.58735 | 0.0000 | (1.0002801729384074e-05))
# test w16: r0Inv1 (5.5e-06: 0.5881860039044223)
# test r0Inv1 (5.5e-06, Niter=246, 0.5867403075395137)
# test r0Inv1 (5.5e-06, Niter=247, 0.5872846547180826)
# Niter = 247 (0.5880986018552999):
# X = StandardScaler().fit_transform(np.column_stack([caa, saa, z1, z2, rt/r, x/r, y/r, z3, y1, y3,
# ctt, stt, z4, x4, y4, raa, r0Inv]))
# cx = [w1,w1,w2,w3, w4, w5, w6, w7, w8, w9, w10, w10, w11, w12, w13, w14, w15]
# w15: 5.0611615056082495e-05 (17 | 21m37s | 0.58790 | 0.0001 | )
# w15 test (5.5e-05: 0.5881768870518835)
# w15 alone: 5.5e-05: 0.5870504337495849
# w15 again: 5.5e-05 0.5864220587506578 (strange)
# w15 again: 5.5e-05 (0.5880689577051738)
# w16: 5.5e-06: 0.587602145623185 (bad since w16 was not being used)
# after reset: w16 not being used - 0.5880689577051738
# a2: 0.0206: 0.58157
# org (no shift + ensemble): 0.5859135274547416
# org (with shift + ensemble + ia1): 0.5901965251117371
# org (with shift + ensemble + no ia1): 0.5901656684266057
# r0Inv_d1: 7.401866174854672e-05, 0.58592 (7.5e-05: 0.5892)
# multiprocessing - 0.6377253549867099 ( 25 mins: i7)
# multiprocessing - z_shift (10 linspace + 1)- 0.637725354987 (32 mins)
# z_shift (20 linspace + 1) - 0.639556619038 (44 mins)
# z_shift ( 5 linspace + 1) - (0.62971260568) (19 mins)
# theta0 - w15= 1.0, 1 hr 40 mins, 0.397164972826)
# theta:0 , r0Inv using r, w15: (0.1: 0.263406003077, 0.01: 0.555401034484, 0.001:0.58657067095, 0.0001: 0.585995954153,
# 1e-05: 0.585960618134 )
# w15: 0.0009: 0.586267891213, 0.015: 0.586183567052, 0.002: 0.585828179853)
# theta:0 , r0Inv using rt, w15: (0.1: 0.145904897094, 0.01: 0.536585229252, 0.001:0.585387197245, 0.0001: 0.585950889554,
# 1e-05 to 7e-05: 0.585960618134, 40 min, 8e-05, 9e-05: 0.585950889554 )
# theta: 0, r0Inv using r, 0.00109:0.58714214929
# Niter : 10 , 1 min
# Niter : 50 , 4 min
# Niter : 75 , 5 min
# Niter : 100 , 7 min
# Niter : 125 , 8 min
# Niter : 150 , 9 min (0.567703152923)
# Niter : 200 , 11 min (0.58431499816)
# z = 10 items, theta0, 10 items: 2h 3min 2s, 0.625977081867
# r0Inv with rt, a1: w15 = 0.0001: 0.585950889554
# rt/z: w15 = 1e-05: 0.585902687253
# rt * ln(|z|): w15, 1e-05 (same for 9e-06 and 2e-05): 0.585960618134
# ln |z|: w15: 1e-05: 0.585960618134
# fundu: w15: 0.001: 0.58599381397
# fundu2: same as above: w15: 0.001: 0.58599381397
# mom: 0.01:0.585854225793, 0.001, 0.0001, 0.00001: 0.585960618134
# mom2: 0.01: 0.585960872497, 0.001 onwards: 0.585960618134, 0.011: 0.585972357309
# mom2 (round2): 0.001 onwards decreasing: 0.585960618134
# mom2 trunc 4: 0.01: 0.585988797098
cluster_runs = np.random.randint(0, 50, (50, 15000))
cluster_runs.shape
consensus_clustering_labels = CE.cluster_ensembles(cluster_runs, verbose = True, N_clusters_max = 50)
cluster_runs.shape
consensus_clustering_labels.shape
w1 = 1.1932215111905984
w2 = 0.39740553885387364
w3 = 0.3512647720585538
w4 = 0.1470
w5 = 0.01201
w6 = 0.0003864
w7 = 0.0205
w8 = 0.0049
w9 = 0.00121
w10 = 1.4930496676654575e-05
w11 = 0.0318
w12 = 0.000435
w13 = 0.00038
w14 = 0.00072
w15 = 5.5e-05
# w15 = 0.000265
w16 = 0.0031
w17 = 0.00021
w18 = 7.5e-05
Niter=247
def run_make_submission():
data_dir = '../data/test'
tic = t = time.time()
event_ids = [ '%09d'%i for i in range(0,125) ] #(0,125)
if 1:
submissions = []
for i,event_id in enumerate(event_ids):
hits = pd.read_csv(data_dir + '/event%s-hits.csv'%event_id)
cells = pd.read_csv(data_dir + '/event%s-cells.csv'%event_id)
labels = Fun4BO2(hits)
toc = time.time()
print('\revent_id : %s , %0.0f min'%(event_id, (toc-tic)/60))
# Prepare submission for an event
submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],
data=np.column_stack(([event_id,]*len(hits), hits.hit_id.values, labels))
).astype(int)
submissions.append(submission)
for i in range(8):
submission = extend(submission,hits)
submission.to_csv('../cache/sub2/%s.csv.gz'%event_id,
index=False, compression='gzip')
#------------------------------------------------------
if 1:
event_ids = [ '%09d'%i for i in range(0,125) ] #(0,125)
submissions = []
for i,event_id in enumerate(event_ids):
submission = pd.read_csv('../cache/sub2/%s.csv.gz'%event_id, compression='gzip')
submissions.append(submission)
# Create submission file
submission = pd.concat(submissions, axis=0)
submission.to_csv('../submissions/sub2/submission-0029.csv.gz',
index=False, compression='gzip')
print(len(submission))
run_make_submission()
```
| github_jupyter |
```
import pandas as pd
import numpy as np
import os
import math
import graphlab
import graphlab as gl
import graphlab.aggregate as agg
from graphlab import SArray
'''钢炮'''
path = '/home/zongyi/bimbo_data/'
train = gl.SFrame.read_csv(path + 'train_lag5.csv', verbose=False)
town = gl.SFrame.read_csv(path + 'towns.csv', verbose=False)
train = train.join(town, on=['Agencia_ID','Producto_ID'], how='left')
train = train.fillna('t_c',1)
train = train.fillna('tcc',0)
train = train.fillna('tp_sum',0)
del train['Town']
del train['id']
del train['Venta_uni_hoy']
del train['Venta_hoy']
del train['Dev_uni_proxima']
del train['Dev_proxima']
del train['Demanda_uni_equil']
# relag_train = gl.SFrame.read_csv(path + 're_lag_train.csv', verbose=False)
# train = train.join(relag_train, on=['Cliente_ID','Producto_ID','Semana'], how='left')
# train = train.fillna('re_lag1',0)
# train = train.fillna('re_lag2',0)
# train = train.fillna('re_lag3',0)
# train = train.fillna('re_lag4',0)
# train = train.fillna('re_lag5',0)
# del relag_train
# pd = gl.SFrame.read_csv(path + 'products.csv', verbose=False)
# train = train.join(pd, on=['Producto_ID'], how='left')
# train = train.fillna('prom',0)
# train = train.fillna('weight',0)
# train = train.fillna('pieces',1)
# train = train.fillna('w_per_piece',0)
# train = train.fillna('healthy',0)
# train = train.fillna('drink',0)
# del train['brand']
# del train['NombreProducto']
# del pd
# client = gl.SFrame.read_csv(path + 'clients.csv', verbose=False)
# train = train.join(client, on=['Cliente_ID'], how='left')
# del client
# cluster = gl.SFrame.read_csv(path + 'prod_cluster.csv', verbose=False)
# cluster = cluster[['Producto_ID','cluster']]
# train = train.join(cluster, on=['Producto_ID'], how='left')
train
# Make a train-test split
train_data, test_data = train.random_split(0.999)
# Create a model.
model = gl.boosted_trees_regression.create(train_data, target='Demada_log',
step_size=0.1,
max_iterations=500,
max_depth = 10,
metric='rmse',
random_seed=395,
column_subsample=0.7,
row_subsample=0.85,
validation_set=test_data,
model_checkpoint_path=path,
model_checkpoint_interval=500)
model1 = gl.boosted_trees_regression.create(train, target='Demada_log',
step_size=0.1,
max_iterations=4,
max_depth = 10,
metric='rmse',
random_seed=395,
column_subsample=0.7,
row_subsample=0.85,
validation_set=None,
resume_from_checkpoint=path+'model_checkpoint_4',
model_checkpoint_path=path,
model_checkpoint_interval=2)
model
w = model.get_feature_importance()
w = w.add_row_number()
w
from IPython.core.pylabtools import figsize
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
sns.set_style('darkgrid', {'grid.color': '.8','grid.linestyle': u'--'})
%matplotlib inline
figsize(12, 6)
plt.bar(w['id'], w['count'], tick_label=w['name'])
plt.xticks(rotation=45)
# Save predictions to an SArray
predictions = model.predict(train)
# Evaluate the model and save the results into a dictionary
results = model.evaluate(train)
print results
model.summary()
test = gl.SFrame.read_csv(path + 'test_lag5.csv', verbose=False)
test = test.join(town, on=['Agencia_ID','Producto_ID'], how='left')
del test['Town']
test = test.fillna('t_c',1)
test = test.fillna('tcc',0)
test = test.fillna('tp_sum',0)
test
ids = test['id']
del test['id']
demand_log = model.predict(test)
sub = gl.SFrame({'id':ids,'Demanda_uni_equil':demand_log})
import math
sub['Demanda_uni_equil'] = sub['Demanda_uni_equil'].apply(lambda x: math.expm1(max(0, x)))
sub
sub.save(path+'gbrt_sub3.csv',format='csv')
math.expm1(math.log1p(2))
```
| github_jupyter |
## Multi-label classification
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai.conv_learner import *
PATH = 'data/planet/'
# Data preparation steps if you are using Crestle:
os.makedirs('data/planet/models', exist_ok=True)
os.makedirs('/cache/planet/tmp', exist_ok=True)
!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train-jpg {PATH}
!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/test-jpg {PATH}
!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train_v2.csv {PATH}
!ln -s /cache/planet/tmp {PATH}
ls {PATH}
```
## Multi-label versus single-label classification
```
from fastai.plots import *
def get_1st(path): return glob(f'{path}/*.*')[0]
dc_path = "data/dogscats/valid/"
list_paths = [get_1st(f"{dc_path}cats"), get_1st(f"{dc_path}dogs")]
plots_from_files(list_paths, titles=["cat", "dog"], maintitle="Single-label classification")
```
In single-label classification each sample belongs to one class. In the previous example, each image is either a *dog* or a *cat*.
```
list_paths = [f"{PATH}train-jpg/train_0.jpg", f"{PATH}train-jpg/train_1.jpg"]
titles=["haze primary", "agriculture clear primary water"]
plots_from_files(list_paths, titles=titles, maintitle="Multi-label classification")
```
In multi-label classification each sample can belong to one or more clases. In the previous example, the first images belongs to two clases: *haze* and *primary*. The second image belongs to four clases: *agriculture*, *clear*, *primary* and *water*.
## Multi-label models for Planet dataset
```
from planet import f2
metrics=[f2]
f_model = resnet34
label_csv = f'{PATH}train_v2.csv'
n = len(list(open(label_csv)))-1
val_idxs = get_cv_idxs(n)
```
We use a different set of data augmentations for this dataset - we also allow vertical flips, since we don't expect vertical orientation of satellite images to change our classifications.
```
def get_data(sz):
tfms = tfms_from_model(f_model, sz, aug_tfms=transforms_top_down, max_zoom=1.05)
return ImageClassifierData.from_csv(PATH, 'train-jpg', label_csv, tfms=tfms,
suffix='.jpg', val_idxs=val_idxs, test_name='test-jpg')
data = get_data(256)
x,y = next(iter(data.val_dl))
y
list(zip(data.classes, y[0]))
plt.imshow(data.val_ds.denorm(to_np(x))[0]*1.4);
sz=64
data = get_data(sz)
data = data.resize(int(sz*1.3), 'tmp')
learn = ConvLearner.pretrained(f_model, data, metrics=metrics)
lrf=learn.lr_find()
learn.sched.plot()
lr = 0.05
learn.fit(lr, 3, cycle_len=1, cycle_mult=2)
lrs = np.array([lr/9,lr/3,lr])
learn.unfreeze()
learn.fit(lrs, 3, cycle_len=1, cycle_mult=2)
learn.save(f'{sz}')
learn.sched.plot_loss()
sz=128
learn.set_data(get_data(sz))
learn.freeze()
learn.fit(lr, 3, cycle_len=1, cycle_mult=2)
learn.unfreeze()
learn.fit(lrs, 3, cycle_len=1, cycle_mult=2)
learn.save(f'{sz}')
sz=256
learn.set_data(get_data(sz))
learn.freeze()
learn.fit(lr, 3, cycle_len=1, cycle_mult=2)
learn.unfreeze()
learn.fit(lrs, 3, cycle_len=1, cycle_mult=2)
learn.save(f'{sz}')
multi_preds, y = learn.TTA()
preds = np.mean(multi_preds, 0)
files = !ls {PATH}test/
```
### End
| github_jupyter |
```
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.layers import Embedding, LSTM, Dense, Dropout, Bidirectional
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
from tensorflow.keras import regularizers
import tensorflow.keras.utils as ku
import numpy as np
tokenizer = Tokenizer()
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/sonnets.txt \
-O /tmp/sonnets.txt
data = open('/tmp/sonnets.txt').read()
corpus = data.lower().split("\n")
tokenizer.fit_on_texts(corpus)
total_words = len(tokenizer.word_index) + 1
# create input sequences using list of tokens
input_sequences = []
for line in corpus:
token_list = tokenizer.texts_to_sequences([line])[0]
for i in range(1, len(token_list)):
n_gram_sequence = token_list[:i+1]
input_sequences.append(n_gram_sequence)
# pad sequences
max_sequence_len = max([len(x) for x in input_sequences])
input_sequences = np.array(pad_sequences(input_sequences, maxlen=max_sequence_len, padding='pre'))
# create predictors and label
predictors, label = input_sequences[:,:-1],input_sequences[:,-1]
label = ku.to_categorical(label, num_classes=total_words)
model = Sequential()
model.add(Embedding(total_words, 100, input_length=max_sequence_len-1))
model.add(Bidirectional(LSTM(150, return_sequences = True)))
model.add(Dropout(0.2))
model.add(LSTM(100))
model.add(Dense(total_words/2, activation='relu', kernel_regularizer=regularizers.l2(0.01)))
model.add(Dense(total_words, activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
print(model.summary())
history = model.fit(predictors, label, epochs=100, verbose=1)
import matplotlib.pyplot as plt
acc = history.history['accuracy']
loss = history.history['loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'b', label='Training accuracy')
plt.title('Training accuracy')
plt.figure()
plt.plot(epochs, loss, 'b', label='Training Loss')
plt.title('Training loss')
plt.legend()
plt.show()
seed_text = "I Love you"
next_words = 100
for _ in range(next_words):
token_list = tokenizer.texts_to_sequences([seed_text])[0]
token_list = pad_sequences([token_list], maxlen=max_sequence_len-1, padding='pre')
predicted = model.predict_classes(token_list, verbose=0)
output_word = ""
for word, index in tokenizer.word_index.items():
if index == predicted:
output_word = word
break
seed_text += " " + output_word
print(seed_text)
```
| github_jupyter |
# sentinelRequest
sentinelRequest can be used to colocate a geodataframe (ie areas, trajectories, buoys, etc ...) with sentinel (1, but also 2 , 3 : all known by scihub)
## Install
```
conda install -c conda-forge lxml numpy geopandas shapely requests fiona matplotlib jupyter descartes
pip install --upgrade git+https://github.com/oarcher/sentinelrequest.git
```
## CLI usage
```
!sentinelrequest --help
```
### "One shot" from command line:
`
% sentinelrequest --user=xxxx --password=xxxxx --date='2018-09-23 00:00' --date='2018-09-23 12:00' --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --wkt='POLYGON ((-10 75, -10 86, 12 86, 12 84, -10 75))'
`
```
INFO:sentinelRequest:from 2018-09-23 00:00:00 to 2018-09-23 12:00:00 : 11 SAFES
INFO:sentinelRequest:Total : 11 SAFES
filename
S1B_EW_GRDM_1SDH_20180923T071854_20180923T071954_012839_017B47_17F2.SAFE
S1B_EW_GRDM_1SDH_20180923T071954_20180923T072054_012839_017B47_1E6F.SAFE
S1B_EW_GRDM_1SDH_20180923T072054_20180923T072154_012839_017B47_CD41.SAFE
S1B_EW_GRDM_1SDH_20180923T072154_20180923T072254_012839_017B47_3682.SAFE
S1A_EW_GRDM_1SDH_20180923T081003_20180923T081107_023823_02997B_049A.SAFE
S1A_EW_GRDM_1SDH_20180923T081107_20180923T081207_023823_02997B_6EA6.SAFE
S1B_EW_GRDM_1SDH_20180923T085656_20180923T085756_012840_017B4E_B07B.SAFE
S1B_EW_GRDM_1SDH_20180923T085756_20180923T085856_012840_017B4E_6CAD.SAFE
S1B_EW_GRDM_1SDH_20180923T085856_20180923T085956_012840_017B4E_1CCD.SAFE
S1B_EW_GRDM_1SDH_20180923T103504_20180923T103604_012841_017B54_DBBC.SAFE
S1B_EW_GRDM_1SDH_20180923T103604_20180923T103704_012841_017B54_B267.SAFE
```
### From csv file
`
% cat test.csv
`
```
index;startdate;stopdate;geometry
area1;2018-10-02 00:00;2018-10-02 21:00;POLYGON ((-12 35, -5 35, -5 45, -12 45, -12 35))
area2;2018-10-13 06:00;2018-10-13 21:00;POLYGON ((-10 32, -3 32, -3 42, -10 42, -10 32))
area3;2018-10-13 00:00;2018-10-13 18:00;POLYGON ((12 35, 5 35, 5 45, 12 45, 12 35))
```
`
% sentinelRequest --user=xxxx --password=xxxx --infile=test.csv --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --cols=index,filename
`
```
INFO:sentinelRequest:req 1/2 from 2018-10-02 00:00:00 to 2018-10-02 21:00:00 : 9/21 SAFES
INFO:sentinelRequest:req 2/2 from 2018-10-13 00:00:00 to 2018-10-13 21:00:00 : 30/35 SAFES
INFO:sentinelRequest:Total : 39 SAFES
index;filename
area1;S1A_IW_GRDH_1SDV_20181002T061827_20181002T061852_023953_029DA0_C61E.SAFE
area1;S1B_IW_GRDH_1SDV_20181002T181105_20181002T181130_012977_017F7D_FE88.SAFE
area1;S1B_IW_GRDH_1SDV_20181002T181130_20181002T181155_012977_017F7D_93FF.SAFE
area1;S1B_IW_GRDH_1SDV_20181002T181155_20181002T181222_012977_017F7D_CD9A.SAFE
area3;S1A_IW_GRDH_1SDV_20181013T053545_20181013T053610_024113_02A2DB_D121.SAFE
area3;S1A_IW_GRDH_1SDV_20181013T053815_20181013T053840_024113_02A2DB_7D53.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062502_20181013T062527_013130_018428_1E77.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062527_20181013T062552_013130_018428_82AB.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062642_20181013T062707_013130_018428_AB0E.SAFE
area2;S1B_IW_GRDH_1SDV_20181013T062707_20181013T062732_013130_018428_8210.SAFE
```
If `--date` is specified 2 times with `--infile`, it will superseeds ones founds in infile :
`
sentinelRequest --user oarcher --password nliqt6u3 --infile=test.csv --date=last-monday-7days --date=now --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --cols=index,filename
`
## API usage
```
%matplotlib inline
import geopandas as gpd
import datetime
import matplotlib.pyplot as plt
import shapely.wkt as wkt
# get your own credential from https://scihub.copernicus.eu/dhus
import pickle
user,password = pickle.load(open("credential.pkl","rb"))
import sentinelrequest as sr
# set default values, so we don't have to pass them at every requests
sr.default_user = user
sr.default_password = password
sr.default_cachedir='/tmp/scihub_cache'
sr.default_filename='S1?_?W_GRD*.SAFE'
# optional : debug messages
#import logging
#sr.logger.setLevel(logging.DEBUG)
help(sr.scihubQuery)
```
### Simplest api usage
Just a startdate and a stopdate are given, with no geometry
```
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
startdate=datetime.datetime(2018,10,2),
stopdate=datetime.datetime(2018,10,3),
fig=fig)
```
The result is a geodataframe with most information from scihub:
```
safes.iloc[0]
```
Most fields are converted from str to python type (geometry, datetime, int ...)
```
safes.iloc[1:4]['footprint'].plot()
print('safe was ingested %s after aquisition' % (safes.iloc[0]['ingestiondate']-safes.iloc[0]['endposition']))
```
### Using a geodataframe with geometries
As an example, two areas are defined. Note that the index is named with the area name
```
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2018,10,2,0) , datetime.datetime(2018,10,13,0) ],
"endposition" : [ datetime.datetime(2018,10,2,21) ,datetime.datetime(2018,10,13,18) ],
"geometry" : [ wkt.loads("POINT (-7.5 53)").buffer(4), wkt.loads("POLYGON ((-12 35, -5 35, -5 45, -12 45, -12 35))")]
},index=["Irland","Portugal"])
gdf
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
min_sea_percent=20,
fig=fig)
```
User requested area are in green, and found safes are in blue.
Index from original request are preserved, so it's easy to know the area that belong to a safe. (See end of example 2 for advanced index handling).
```
safes.loc['Portugal']
```
### Working with projection
SentinelRequest works with projections, by defining crs in gdf.
The colocalisation is done using this crs.
get safes around 1000km, at 84° (North pole included)
```
import pyproj
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2019,12,1,0) ],
"endposition" : [ datetime.datetime(2019,12,4,0)],
"geometry" : [ wkt.loads("POINT (0 84)")]
},index=["Artic"], crs=pyproj.CRS('epsg:4326'))
# to polar projection (units in meters)
gdf.to_crs(pyproj.CRS('epsg:3408'), inplace=True)
gdf.loc["Artic","geometry"]=gdf.loc["Artic"].geometry.buffer(1000 * 1000)
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
min_sea_percent=20,
fig=fig)
```
### Cyclone track colocalization
```
import pandas as pd
#ibtracs=gpd.read_file('tmp/IBTrACS.NA.list.v04r00.points.shp')
#gdf_track=ibtracs[ibtracs['SID'] == '2019235N10324']
#gdf_track=gdf_track[['ISO_TIME','USA_WIND','geometry']]
#gdf_track['ISO_TIME']=pd.to_datetime(gdf_track['ISO_TIME'],format="%Y-%m-%d %H:%M:%S")
#gdf_track.reset_index(inplace = True,drop=True)
#gdf_track.to_file("track.gpkg", driver="GPKG")
gdf_track = gpd.read_file('track.gpkg')
gdf_track['ISO_TIME']=pd.to_datetime(gdf_track['ISO_TIME'],format="%Y-%m-%d %H:%M:%S")
gdf_track
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf_track,
date='ISO_TIME', # no startdate/stopdate, but a date ans a dtime
dtime=datetime.timedelta(hours=1.5),
datatake=1, # take adjacents safes, up to one.
fig=fig)
```
#### datatake
Here, `datatake=1` is specified to retrieve adjacents safes from colocated ones (in cyan). When specified, the result contain a `datatake_index` column. 0 means the colocated one, and other values are the range of the adjacent safe (up to -n..n with `datatake=n`)
Positive `datatake_index` are for safes *after* the colocated one, and negative index are fo safes *before* the colocated one.
```
safes[['filename','datatake_index']]
```
#### Time slicing with timedelta_slice
One can see on previous figure that 3 requests are done. gdf rows are grouped to reduce the amount of scihub requests with the `timedelta_slice` parameter (default to `datetime.timedelta(weeks=1)` )
If we reduce `timedelta_slice`, we can see that more scihub request are done, with less uncolocated safes (ie yellow). (be warned with a big `timedelta_slice` : this can produce scihub timeouts).
(with `timedelta_slice=None`, this feature is *disabled* : a scihub request is done for *every* geometry).
```
# same request as above, but with reduced timedelta_slice
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf_track,
date='ISO_TIME',
dtime=datetime.timedelta(hours=1.5),
timedelta_slice=datetime.timedelta(days=1),
datatake=1,
full_fig = True, # to show internals requests and colocs
fig=fig)
```
#### Merging source and result with shared index
As seen before, the result (safes) share the same index as the source. So we can merge the two geodataframe, to associate a wind speed from the cyclone track with the safe, and compute distance from the eye to the safe.
```
# here, we merge the result with the source request, to associate wind speed to each safe.
merged=safes[['filename','datatake_index','footprint']].merge(
gdf_track[['USA_WIND','geometry']],left_index=True,right_index=True)
merged['eye_dist'] = merged.set_geometry('geometry').distance(merged.set_geometry('footprint').exterior)
# negative dist if safe contains eye
merged['eye_dist']=merged['eye_dist']*(((~merged.set_geometry('footprint').contains(merged.set_geometry('geometry'))+1)*2)-3)
merged[['filename','datatake_index','USA_WIND','eye_dist']]
```
## Annexes
### Antimeridian handling: small geometry vs large one
Given 2 points on the earth, there is two possible paths: one short, and one long that wrap around the earth.
Note: only longitude is wrapped, as if earth was a cylinder (epgs 4326 used for computation)
By default, geometry are the smallest ones. To preserve a large geometry, GeometryCollection must be used.
```
from shapely.geometry import GeometryCollection
# the polygon is more than 180 deg wide. It will be wrapped, and will cross antimeridian
large_poly = wkt.loads("POLYGON ((-140 -14, 140 -14, 140 -20, -140 -20, -140 -14))")
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2018,10,1)],
"endposition" : [ datetime.datetime(2018,10,31) ],
"geometry" : [ large_poly ]
},index=[0])
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
fig=fig)
plt.show()
# same polygon, but encapsulated in a GeometryCollection : it will not be wrapped
gdf = gpd.GeoDataFrame({
"beginposition" : [ datetime.datetime(2018,10,1)],
"endposition" : [ datetime.datetime(2018,10,31) ],
"geometry" : [ GeometryCollection([large_poly]) ]
},index=[0])
fig = plt.figure(figsize=(10,7))
safes = sr.scihubQuery(
gdf=gdf,
fig=fig)
plt.show()
gdf
import shapely.ops
len(shapely.ops.unary_union(gdf_track.geometry).buffer(2).simplify(1.9).wkt)
```
| github_jupyter |
# Reproduct Autopilot Architecture
The Autopilot has the following Architecture:
~ ResNet50-like backbone
~ FPN - DeepLabV3- UNet - like heads
~ 15 tasks
~ subtasks i.e. if task is car detection, then the sub task is what kind of car, is it stationary? Parked, broken down?
For later exploration:
Stitching up of images across space and time happens inside RNNs.
Also explore Faster R-CNNs but they have lower inference rate in real-time detection etc.
## First Step:
Use transfer learning to load ResNet-50 model because training it and loading it that way has been a hassle.
We are most likely using feature extraction techniques to get the data from resnet backbone and passing them onto the tasks
```
import torch
import torch.nn as nn
import torch.optim as optim
import torchvision
from torchvision import models, transforms
import matplotlib.pyplot as plt
import numpy as np
batch_size = 50
# get the CIFAR-10 images:
train_data_transform = transforms.Compose([
transforms.Resize(224),
transforms.RandomHorizontalFlip(),
transforms.RandomVerticalFlip(),
transforms.ToTensor(),
transforms.Normalize((0.4914, 0.4821, 0.4465), (0.2470, 0.2435, 0.2616))
])
train_set = torchvision.datasets.CIFAR10(root='./data',
train=True, download = True,
transform=train_data_transform)
train_loader = torch.utils.data.DataLoader(train_set, batch_size=batch_size,
shuffle=True, num_workers=2)
```
## CIFAR10 Data
10000 x 3072 numpy arrays. i.e 1024 values i.e. 32x32 image and since there are 3 channels, we get 3072 shaped numpy arrays.
Each row in the array stores a 32x32 colour image
The first 1024 entries contain the red channel values, the next 1024 are green and the next 1024 are blue.
```
def imshow(img):
img = img/2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1,2,0)))
plt.show()
dataiter = iter(train_loader)
images, labels = dataiter.next()
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
imshow(torchvision.utils.make_grid(images))
print(' '.join('%5s'%classes[labels[j]] for j in range(50)))
imshow(images[49])
len(train_loader)
val_data_transform = transforms.Compose([
transforms.Resize(224),
transforms.ToTensor(),
transforms.Normalize((0.4914, 0.4821, 0.4465), (0.2470, 0.2435, 0.2616))
])
val_set = torchvision.datasets.CIFAR10(root='./data',
train=False, download=True,
transform=val_data_transform)
val_order = torch.utils.data.DataLoader(val_set,
batch_size=batch_size,
shuffle=False, num_workers=2
)
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
def train_model(model, loss_function, optimizer, data_loader):
overall_start = timer()
# set the model mode
# model.train()
for epoch in range(n_epochs):
current_acc = 0
current_loss =0
model.train()
start = timer()
# iterate over the examples in the dataset:
for i, (inputs, labels) in enumerate(data_loader):
# send them to the GPU first
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
with torch.set_grad_enabled(True):
# forward
outputs = model(inputs)
_, predictions = torch.max(outputs, 1)
loss = loss_function(outputs, labels)
# backward
loss.backward()
optimizer.step()
# statistics
current_loss += loss.item() * inputs.size(0)
current_acc += torch.sum(predictions == labels.data)
total_loss = current_loss / len(data_loader.dataset)
total_acc = current_acc.double() / len(data_loader.dataset)
print('Train Loss: {:.4f}; Accuracy {:.4f}'.format(total_loss, total_acc))
def test_model(model, loss_function, data_loader):
# set model in evaluation mode
model.eval()
current_loss = 0.0
current_acc = 0
# iterate over the validation data
for i, (inputs, labels) in enumerate(data_loader):
inputs = inputs.to(device)
labels = labels.to(device)
with torch.set_grad_enabled(False):
outputs = model(inputs)
_, predictions = torch.max(outputs, 1)
loss = loss_function(outputs, labels)
# statistics
current_loss += loss.item() * inputs.size(0)
current_acc += torch.sum(predictions == labels.data)
total_loss = current_loss / len(data_loader.dataset)
total_acc = current_acc.double()/ len(data_loader.dataset)
print('Test Loss: {:.4f}; Accuracy {:.4f}'.format(total_loss, total_acc))
return total_loss, total_acc
```
Now, onto the transfer learning scenario where we are going to use the pretrained network as a feature extractor.
1. Let's use ResNet50
2. Replace last layer of the model with a new layer with 10 outputs
3. Exclude the existing network layers from the backward pass and only pass the newly added fully-connected layer to the Adam optimizer.
4. Run the training for epochs and evaluate the network accuracy after each epoch.
5. Plot the test accuracy
# Load Resnet50
```
model = torchvision.models.resnet50(pretrained=True)
model.eval()
def train(model, criterion,
optimizer,
train_loader,
valid_loader,
save_file_name,
max_epochs_stop=3,
n_epochs = 20,
print_every=2):
"""Train the Pytorch model while including the """
def tl_feature_extractor(epochs=5):
# load the pretrained model
model = torchvision.models.resnet50(pretrained=True)
# exclude the existing parameters from backward pass
# for performance
for param in model.parameters():
param.requires_grad = False
# newly constructed layers have requires_grad=True by default
num_features = model.fc.in_features
model.fc = nn.Sequential(
nn.Linear(num_features, 1000),
nn.ReLU(),
nn.Dropout(0.4),
nn.Linear(1000, 10),
nn.LogSoftmax(dim=1))
# model.fc = nn.Linear(1000, 10)
# transfer to GPU
model = model.to(device)
loss_function = nn.CrossEntropyLoss()
# only parameters of the final layer are being optimized
optimizer = optim.Adam(model.fc.parameters()) # otherwise, it would be just model.parameters()
# setting timing
overall_start = timer()
# train
test_acc = list()
for epoch in range(epochs):
print('Epoch {}/{}'.format(epoch+1, epochs))
train_model(model, loss_function, optimizer, train_loader)
_, acc = test_model(model, loss_function, val_order)
test_acc.append(acc)
plot_accuracy(test_acc)
```
```
tl_feature_extractor()
```
| github_jupyter |
<a href="https://colab.research.google.com/github/Amberineee/ecommerce_covid_analysis/blob/main/BA_775_Team_Assignment_Team_4b.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from google.colab import auth
auth.authenticate_user()
from google.colab import drive
drive.mount('/content/drive')
```
#E-commerce Dataset 2020
**For this project, we want to enhance the webpage with the provided dataset of e-Commerce dataset in February and April by taking in consideration COVID-19 pandemic.**
Goal: Our goal is analyzing customer online purchasing behavior during pre covid-19 and post covid-19 using February and April dataset, respectively. Which allows us to understand the traffic of the website and conversion made by consumers to potential solve E-commerce’s dataset problems, and expand the opportunities for marketing campaigns, target promotions and optimizing the inventory level.
Dataset: https://www.kaggle.com/mkechinov/ecommerce-behavior-data-from-multi-category-store
##First, Let's have a look for the month of February (Pre-COVID):
###To begin, we will identify the Top 10 popular brands that are purchased by the consumers for February 2020.
```
%%bigquery --project ba775-team-4b
SELECT brand, count(brand) AS total_brand_purchase
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE event_type = 'purchase'
GROUP BY brand
ORDER BY count(brand) DESC
LIMIT 10
```
Results: We noticed that Samsung is consumer’s favorite brand, followed by Apple and Xiaomi. All electronic brands!
###Next, let’s give a look to the conversion rate for the month of February from 2020.
```
%%bigquery --project ba775-team-4b
SELECT
COUNTIF(event_type = 'purchase')/COUNT(event_type) AS conversion_rate,
category_code,
ROUND(sum(price), 2) AS total_Price
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
GROUP BY category_code
ORDER BY Conversion_rate DESC
limit 10
```
Results: It shows that the highest conversion rate is the construction tools with a rate of 0.0397, followed by apparel with a rate of 0.0334.
###We also want to identify who are the Top consumers in the ecommerce webpage, and how much do they spend for this month, February.
```
%%bigquery --project ba775-team-4b
SELECT user_id, ROUND(SUM(price)) AS total_spending_FEB
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE event_type = 'purchase'
GROUP BY user_id
ORDER BY total_spending_FEB DESC
LIMIT 10
```
Result: our Top consumer identified as user_id: '563051763' spent around $302,726.0 on the webpage.
###Now, let’s check the average of all users’ spendings for the month of February.
```
%%bigquery --project ba775-team-4b
SELECT ROUND(AVG(price), 2) as avg_spending_feb
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE event_type = 'purchase'
LIMIT 10
```
Result: The average of users’ spendings for the month of February is $317.57.
###After identifying the highest consumer as user_id: '563051763', we want to compare their spending with the average of all users’ spendings for the month of February.
```
%%bigquery --project ba775-team-4b
SELECT user_id AS USER_ID, ROUND(SUM(price)) AS Total_spending, COUNT(user_session) as Sessions,
(
SELECT round(AVG(price), 2) as avg_spending
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE event_type = 'purchase'
) as Avg_spending_of_all_users
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE user_id = 563051763 and event_type = 'purchase'
GROUP BY user_id
```
This query shows the top buyer who have spend the most in this ecommerce website, number of sessions, and the average of all users' spendings.
## Now, we would check for the month of April (post-COVID):
###For the month of April, 2020. We will identify the Top 10 popular brands that were purchased by the consumers during post COVID.
```
%%bigquery --project ba775-team-4b
SELECT brand, count(brand) AS total_brand_purchase
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE event_type = 'purchase'
GROUP BY brand
ORDER BY count(brand) DESC
LIMIT 10
```
Results: We noticed that the results are the same as pre-COVID (February). Samsung is still consumer’s favorite brand, followed by Apple and Xiaomi. All electronic brands!
###Let’s give a look to the conversion rate for the month of April.
```
%%bigquery --project ba775-team-4b
SELECT countif(event_type = 'purchase')/count(event_type) AS Conversion_rate, category_code, sum(price) Total_Price
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
GROUP BY category_code
ORDER BY Conversion_rate DESC
limit 10
```
Results: It shows that the highest conversion rate is the stationery paper with a rate of 0.038, followed by kitchen appliances with a small difference of 0.001.
###Next, we want to identify who are the Top consumers in the ecommerce webpage, and how much they spend.
```
%%bigquery --project ba775-team-4b
SELECT user_id, round(SUM(price), 2) as total_spending
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE event_type = 'purchase'
GROUP BY user_id
ORDER by total_spending DESC
LIMIT 10
```
Results: our Top consumer identified as user_id: '553446649' spent around $122,525.19 on the webpage, which is way more lower than in February. This leads to a better understanding of consumers' behaviour that due the pandemic consumers had shifted their values, and cutting down unneed purchases to essentials.
###Now, let’s check the average of all users’ spendings for the month of April.
```
%%bigquery --project ba775-team-4b
SELECT ROUND(AVG(price), 2) as avg_spending_of_all_users
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE event_type = 'purchase'
```
Result: As we expected, the average of users’ spendings also decreased during pandemic. Average for April is `$252.93`, while for Feb was `$317.57`.
###After identifying the highest consumer as user_id: '553446649', we want to compare their spending with the average of all users’ spendings for the month of April.
```
%%bigquery --project ba775-team-4b
SELECT user_id AS USER_ID, ROUND(SUM(price)) AS TOTAL_SPENDING,COUNT(user_session) AS TOTA_SESSIONS,
(
SELECT round(AVG(price), 2) as avg_spending
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE event_type = 'purchase'
) as AVG_SPENDING_OF_ALL_USERS
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE user_id = 553446649 and event_type = 'purchase'
GROUP BY user_id
```
This query shows the top buyer who have spend the most in this ecommerce website, and the average of all users' spendings
##Let's check deeper the insights by combining both datasets!
###We also want to know how many times they shop on this website in February and April.
```
%%bigquery --project ba775-team-4b
SELECT
user_id,
COUNT(user_id) AS returning
FROM
(
SELECT user_id, event_type
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020` AS FEB
UNION ALL
SELECT user_id, event_type
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020` AS APR
)
WHERE event_type = 'purchase'
GROUP BY user_id
HAVING COUNT(user_id) > 1
ORDER BY returning DESC
LIMIT 10
```
Results: Our top consumer identified as user_id: ‘609817194’ returned back to the website 573 times. Surprisingly, our top consumer from the month of February and April are not in the Top 10.
###Now we want to see the impact of COVID-19 by finding the percentage change in sales for popular categories in February
```
%%bigquery --project ba775-team-4b
SELECT
category_code,
(total_sales_apr - total_sales_feb)/total_sales_feb AS percent_change
FROM
(
SELECT *
FROM
(SELECT
category_code,
SUM(price) AS total_sales_apr
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE category_code IS NOT NULL AND event_type = 'purchase'
GROUP BY category_code
ORDER BY total_sales_apr DESC
) AS apr
INNER JOIN
(SELECT
category_code AS cat,
SUM(price) AS total_sales_feb,
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE category_code IS NOT NULL and event_type = 'purchase'
GROUP BY category_code
ORDER BY total_sales_feb DESC) AS feb
ON apr.category_code = feb.cat
)
ORDER BY total_sales_feb DESC
LIMIT 10
```
Results: Most popular categories have lower sales in April than in February. But headphone, massager and refrigerators categories have higher sales in April. It might casued by working from home.
# Are views correlated with the number of speding in each month?
```
%%bigquery --project ba775-team-4b
select count(event_type) as feb_visits,
(select round(sum(price),2) as profit_from_feb from `ba775-team-4b.4b_dataset.ecommerce_feb_2020` where event_type = 'purchase')feb_profit,
(select count(event_type) as total_visits_april FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where event_type = 'view'
group by event_type) april_visits,
(select round(sum(price),2) as profit_from_april from `ba775-team-4b.4b_dataset.ecommerce_april_2020` where event_type = 'purchase') april_profit
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
where event_type = 'view'
group by event_type
```
There is a negative correlation between the number of views and consumer's spending on each month.
Results: April visits times increased by 9.8% since February, but decreased sales by 21.84%.
###Also, we wants to investigate how well samsung's top selling categories in February performed in April
```
%%bigquery --project ba775-team-4b
SELECT
A.*,FEB_total_sales,round(APR_total_sales-FEB_total_sales,2) FEB_APR_SalesDifferences,
round(((APR_total_sales-FEB_total_sales)/FEB_total_sales),2) FEB_APR_SalesChanges_percentage
FROM
(SELECT A.*,B.APR_total_sales
FROM
(SELECT A.brand,A.category_code,count(A.brand) AS count_APR_brand_sales, count_FEB_brand_sales
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`A
LEFT JOIN
(SELECT brand,category_code,count(brand) AS count_FEB_brand_sales
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE event_type = 'purchase' and brand = 'samsung' AND category_code IS NOT NULL
GROUP BY category_code,brand
) B
USING(category_code)
WHERE event_type = 'purchase' and A.brand = 'samsung' AND category_code IS NOT NULL
GROUP BY category_code,brand,count_FEB_brand_sales
ORDER BY count_APR_brand_sales DESC
limit 15)
A
LEFT JOIN
(SELECT category_code,round(sum(price),2) as APR_total_sales
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE brand = 'samsung' AND event_type = 'purchase' AND category_code IS NOT NULL
GROUP BY category_code
) B
USING(category_code)
) A
LEFT JOIN
(
SELECT category_code,round(sum(price),2) as FEB_total_sales
FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`
WHERE brand = 'samsung' AND event_type = 'purchase' AND category_code IS NOT NULL
GROUP BY category_code
) B
USING(category_code)
```
Result: most of the February's top-selling categories for samsung increased a lot in April, especially for air conditioner category.
# Number of products they purchased in different category.
```
%%bigquery --project ba775-team-4b
SELECT user_id,num_of_accessories,num_of_apparel,num_of_appliances,num_of_auto,num_of_computers,num_of_construction,num_of_country_yard,num_of_electronics,num_of_furniture,num_of_kids,num_of_medicine,num_of_sport,num_of_stationery from
(SELECT user_id, count(user_id) as num_of_accessories
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "accessories%" and event_type="purchase"
group by user_id) as accessories
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_apparel
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "apparel%" and event_type="purchase"
group by user_id) as apparel
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_appliances
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "appliances%" and event_type="purchase"
group by user_id) as appliances
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_auto
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "auto%" and event_type="purchase"
group by user_id) as auto
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_computers
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "computers%" and event_type="purchase"
group by user_id) as computers
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_construction
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "construction%" and event_type="purchase"
group by user_id) as construction
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_country_yard
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "country_yard%" and event_type="purchase"
group by user_id) as country_yard
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_electronics
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "electronics%" and event_type="purchase"
group by user_id) as electronics
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_furniture
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "furniture%" and event_type="purchase"
group by user_id) as furniture
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_kids
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "kids%" and event_type="purchase"
group by user_id) as kids
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_medicine
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "medicine%" and event_type="purchase"
group by user_id) as medicine
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_sport
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "sport%" and event_type="purchase"
group by user_id) as sport
using(user_id)
FULL OUTER JOIN
(SELECT user_id, count(user_id) as num_of_stationery
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
where category_code like "stationery%" and event_type="purchase"
group by user_id) as stationery
using(user_id)
order by num_of_computers desc
```
# Abandon Rate
```
%%bigquery --project ba775-team-4b
SELECT a.user_id,
a.cart_count,
ifnull(b.purchase_count,0) as purchase_count,
round((ifnull(b.purchase_count,0)/a.cart_count),3)as purchase_rate,
round((1-(ifnull(b.purchase_count,0)/a.cart_count)),3) as abandon_rate
FROM `ba775-team-4b.4b_dataset.cart_count_per_user_id` a
left join
`ba775-team-4b.4b_dataset.purchase_count_per_user_id` b
using (user_id)
where a.cart_count>=b.purchase_count and round((1-(ifnull(b.purchase_count,0)/a.cart_count)),3) > 0.9
order by abandon_rate desc
%%bigquery --project ba775-team-4b
SELECT a.user_id,
a.cart_count,
ifnull(b.purchase_count,0) as purchase_count,
round((ifnull(b.purchase_count,0)/a.cart_count),3)as purchase_rate,
round((1-(ifnull(b.purchase_count,0)/a.cart_count)),3) as abandon_rate
FROM `ba775-team-4b.4b_dataset.purchase_per_userid _april` a
left join
`ba775-team-4b.4b_dataset.purchase_count_per_userid_april` b
using (user_id)
where a.cart_count>=b.purchase_count and round((1-(ifnull(b.purchase_count,0)/a.cart_count)),3) > 0.9
order by abandon_rate desc
```
The abandon rate can be used as the metric to evaluate the purchase power of the users. The abandon rate of a certain user = 1-(number of 'purchase' event /number of 'cart' event). Only the user info with high abandon rate(#>0.9) is included and have the potential to work on increasing sales conversion, such as sending target promotions, poping promotional codes, running market campaigns, etc.
Note: We noticed that for some users purchased more than they carted and will make abandon rate calculated less than 0, which can be high indication of purchase power. Hereby we dropped those users as they are not target audience for this part.
# Machine Learning
```
# Using product features
CREATE MODEL
model.products
OPTIONS
( model_type = 'kmeans',
num_clusters = 2,
distance_type = 'euclidean') AS
SELECT
category_code, brand, price, count(product_id)
FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`
WHERE event_type='purchase'
GROUP BY category_code, product_id, brand, price
# Using users features
## have already create a new table `ba775-team-4b.4b_dataset.different_categories_april_2020`-only use large categories-13 distinct categories
CREATE MODEL
model.users
OPTIONS
( model_type = 'kmeans',
num_clusters = 2,
distance_type = 'euclidean') AS
SELECT
num_of_accessories,num_of_apparel,num_of_appliances,num_of_auto,num_of_computers,num_of_construction,num_of_country_yard,num_of_electronics,num_of_furniture,num_of_kids,num_of_medicine,num_of_sport,num_of_stationery
FROM `ba775-team-4b.4b_dataset.different_categories_april_2020`
```
# Tableau Dashboard
https://prod-useast-a.online.tableau.com/t/soltaniehha/views/BA775Team4bProject_16040742108920/EcommerceDashboard?:origin=card_share_link&:embed=n
```
from IPython.display import Image
# Top Purchase Brand of Feberary and April
Image(filename='Top Brand.png')
# Abandon Rate of Feberary and April
Image(filename='Abandon Rate.png')
```
| github_jupyter |
```
import sys
import importlib
import blockworld_helpers as utils
from Box2D import *
import copy
import numpy as np
world = b2World(gravity=(0,-10), doSleep=True)
groundBody = world.CreateStaticBody(
position=(0,-10),
shapes=b2PolygonShape(box=(50,10)),
)
body = world.CreateDynamicBody(position=(0,1))
box = body.CreatePolygonFixture(box=(1,1), density=1, friction=0.3)
start_positions = np.array([body.position for body in world.bodies])
timeStep = 1.0 / 60
vel_iters, pos_iters = 6, 2
# This is our little game loop.
for i in range(60):
# Instruct the world to perform a single step of simulation. It is
# generally best to keep the time step and iterations fixed.
world.Step(timeStep, vel_iters, pos_iters)
# Clear applied body forces. We didn't apply any forces, but you
# should know about this function.
world.ClearForces()
# Now print the position and angle of the body.
print(body.position, body.angle)
end_positions = np.array([body.position for body in world.bodies])
epsilon = 0.1
#start_positions = np.array([body.position for body in b2world_start.bodies])
#end_positions = np.array([body.position for body in b2world_end.bodies])
print(start_positions)
print(end_positions)
sum(sum(np.absolute(np.subtract(start_positions, end_positions))))
)
print(len(world.bodies))
# Helper functions for interacting between stimulus generation and pybox2D
def b2_x(block):
'''
Takes a block from stimulus generation and returns the x value of the center of the block
'''
return ((block.x) + (block.width / 2))
def b2_y(block):
'''
Takes a block from stimulus generation and returns the y value of the center of the block
'''
return ((block.y) + (block.height / 2))
def add_block_to_world(block, b2world):
'''
Add block from stimulus generation to b2world
'''
body = b2world.CreateDynamicBody(position=(b2_x(block),b2_y(block)))
world_block = body.CreatePolygonFixture(box=(block.width/2,block.height/2), density=1, friction=0.3)
importlib.reload(utils)
# Create world for stability check
b2world = b2World(gravity=(0,-10), doSleep=True)
groundBody = b2world.CreateStaticBody( #add ground
position=(0,-10),
shapes=b2PolygonShape(box=(50,10)),
)
# Add blocks
world = utils.World(world_width = 4,world_height = 4)
world.fill_world()
for block in world.blocks:
b2block = add_block_to_world(block, b2world)
b2world.bodies[1].fixtures[0].shape
# Run world
timeStep = 1.0 / 60
vel_iters, pos_iters = 6, 2
# This is our little game loop.
for i in range(60):
# Instruct the world to perform a single step of simulation. It is
# generally best to keep the time step and iterations fixed.
b2world.Step(timeStep, vel_iters, pos_iters)
# Clear applied body forces. We didn't apply any forces, but you
# should know about this function.
b2world.ClearForces()
# Now print the position and angle of the body.
print(world.blocks[1].x,world.blocks[1].y)
print(b2world.bodies[2].position, body.angle)
b2world.bodies
importlib.reload(display_world)
display_world.display_world()
help(b2World)
```
| github_jupyter |
```
library(data.table)
library(dplyr)
library(Matrix)
library(BuenColors)
library(stringr)
library(cowplot)
library(SummarizedExperiment)
library(chromVAR)
library(BSgenome.Hsapiens.UCSC.hg19)
library(JASPAR2016)
library(motifmatchr)
library(GenomicRanges)
library(irlba)
library(cicero)
library(umap)
library(cisTopic)
library(prabclus)
library(BrockmanR)
library(jackstraw)
library(RColorBrewer)
```
#### define functions
```
read_FM <- function(filename){
df_FM = data.frame(readRDS(filename),stringsAsFactors=FALSE,check.names=FALSE)
rownames(df_FM) <- make.names(rownames(df_FM), unique=TRUE)
df_FM[is.na(df_FM)] <- 0
return(df_FM)
}
run_pca <- function(mat,num_pcs=50,remove_first_PC=FALSE,scale=FALSE,center=FALSE){
set.seed(2019)
mat = as.matrix(mat)
SVD = irlba(mat, num_pcs, num_pcs,scale=scale,center=center)
sk_diag = matrix(0, nrow=num_pcs, ncol=num_pcs)
diag(sk_diag) = SVD$d
if(remove_first_PC){
sk_diag[1,1] = 0
SVD_vd = (sk_diag %*% t(SVD$v))[2:num_pcs,]
}else{
SVD_vd = sk_diag %*% t(SVD$v)
}
return(SVD_vd)
}
elbow_plot <- function(mat,num_pcs=50,scale=FALSE,center=FALSE,title='',width=3,height=3){
set.seed(2019)
mat = data.matrix(mat)
SVD = irlba(mat, num_pcs, num_pcs,scale=scale,center=center)
options(repr.plot.width=width, repr.plot.height=height)
df_plot = data.frame(PC=1:num_pcs, SD=SVD$d);
# print(SVD$d[1:num_pcs])
p <- ggplot(df_plot, aes(x = PC, y = SD)) +
geom_point(col="#cd5c5c",size = 1) +
ggtitle(title)
return(p)
}
run_umap <- function(fm_mat){
umap_object = umap(t(fm_mat),random_state = 2019)
df_umap = umap_object$layout
return(df_umap)
}
plot_umap <- function(df_umap,labels,title='UMAP',colormap=colormap){
set.seed(2019)
df_umap = data.frame(cbind(df_umap,labels),stringsAsFactors = FALSE)
colnames(df_umap) = c('umap1','umap2','celltype')
df_umap$umap1 = as.numeric(df_umap$umap1)
df_umap$umap2 = as.numeric(df_umap$umap2)
options(repr.plot.width=4, repr.plot.height=4)
p <- ggplot(shuf(df_umap), aes(x = umap1, y = umap2, color = celltype)) +
geom_point(size = 1) + scale_color_manual(values = colormap) +
ggtitle(title)
return(p)
}
```
### Input
```
workdir = '../output/'
path_umap = paste0(workdir,'umap_rds/')
system(paste0('mkdir -p ',path_umap))
path_fm = paste0(workdir,'feature_matrices/')
metadata <- read.table('../input/metadata.tsv',
header = TRUE,
stringsAsFactors=FALSE,quote="",row.names=1)
list.files(path_fm,pattern="^FM*")
# read in feature matrices and double check if cell names of feature matrices are consistent with metadata
flag_identical = c()
for (filename in list.files(path_fm,pattern="^FM*")){
filename_split = unlist(strsplit(sub('\\.rds$', '', filename),'_'))
method_i = filename_split[2]
if(method_i == 'chromVAR'){
method_i = paste(filename_split[2],filename_split[4],sep='_')
}
print(paste0('Read in ','fm_',method_i))
assign(paste0('fm_',method_i),read_FM(paste0(path_fm,filename)))
#check if column names are the same
flag_identical[[method_i]] = identical(colnames(eval(as.name(paste0('fm_',method_i)))),
rownames(metadata))
}
flag_identical
all(flag_identical)
labels = metadata$label
num_colors = length(unique(labels))
colormap = colorRampPalette(brewer.pal(8, "Dark2"))(num_colors)
names(colormap) = unique(metadata$label)
head(labels)
```
### SnapATAC
```
df_umap_SnapATAC <- run_umap(fm_SnapATAC)
head(df_umap_SnapATAC)
p_SnapATAC <- plot_umap(df_umap_SnapATAC,labels = labels,colormap = colormap,title='SnapATAC')
p_SnapATAC
```
### SCRAT
```
df_umap_SCRAT <- run_umap(fm_SCRAT)
p_SCRAT <- plot_umap(df_umap_SCRAT,labels = labels,colormap = colormap,title='SCRAT')
p_SCRAT
```
#### Save feature matrices and UMAP coordinates
```
dataset = 'cusanovich2018subset_no_blacklist_filtering'
saveRDS(df_umap_SnapATAC,paste0(path_umap,'df_umap_SnapATAC.rds'))
saveRDS(df_umap_SCRAT,paste0(path_umap,'df_umap_SCRAT.rds'))
save.image(file = 'run_umap_cusanovich2018subset_no_blacklist_filtering.RData')
fig_width = 8
fig_height = 4
options(repr.plot.width=fig_width, repr.plot.height=fig_height)
combined_fig = cowplot::plot_grid(p_SnapATAC+theme(legend.position = "none"),
p_SCRAT+theme(legend.position = "none"),
labels = "",nrow = 1)
combined_fig
cowplot::ggsave(combined_fig,filename = "Cusanovich_2018_ssubset_no_blacklist_filtering.pdf", width = fig_width, height = fig_height)
cowplot::ggsave(p_SCRAT ,filename = "cusanovich_legend.pdf", width = fig_width, height = fig_height)
```
| github_jupyter |
```
# default_exp learner
```
# Learner
> This contains fastai Learner extensions.
```
#export
from tsai.imports import *
from tsai.data.core import *
from tsai.data.validation import *
from tsai.models.all import *
from tsai.models.InceptionTimePlus import *
from fastai.learner import *
from fastai.vision.models.all import *
from fastai.data.transforms import *
#export
@patch
def show_batch(self:Learner, **kwargs):
self.dls.show_batch(**kwargs)
# export
@patch
def remove_all_cbs(self:Learner, max_iters=10):
i = 0
while len(self.cbs) > 0 and i < max_iters:
self.remove_cbs(self.cbs)
i += 1
if len(self.cbs) > 0: print(f'Learner still has {len(self.cbs)} callbacks: {self.cbs}')
#export
@patch
def one_batch(self:Learner, i, b): # this fixes a bug that will be managed in the next release of fastai
self.iter = i
# b_on_device = tuple( e.to(device=self.dls.device) for e in b if hasattr(e, "to")) if self.dls.device is not None else b
b_on_device = to_device(b, device=self.dls.device) if self.dls.device is not None else b
self._split(b_on_device)
self._with_events(self._do_one_batch, 'batch', CancelBatchException)
#export
@patch
def save_all(self:Learner, path='export', dls_fname='dls', model_fname='model', learner_fname='learner', verbose=False):
path = Path(path)
if not os.path.exists(path): os.makedirs(path)
self.dls_type = self.dls.__class__.__name__
if self.dls_type == "MixedDataLoaders":
self.n_loaders = (len(self.dls.loaders), len(self.dls.loaders[0].loaders))
dls_fnames = []
for i,dl in enumerate(self.dls.loaders):
for j,l in enumerate(dl.loaders):
l = l.new(num_workers=1)
torch.save(l, path/f'{dls_fname}_{i}_{j}.pth')
dls_fnames.append(f'{dls_fname}_{i}_{j}.pth')
else:
dls_fnames = []
self.n_loaders = len(self.dls.loaders)
for i,dl in enumerate(self.dls):
dl = dl.new(num_workers=1)
torch.save(dl, path/f'{dls_fname}_{i}.pth')
dls_fnames.append(f'{dls_fname}_{i}.pth')
# Saves the model along with optimizer
self.model_dir = path
self.save(f'{model_fname}', with_opt=True)
# Export learn without the items and the optimizer state for inference
self.export(path/f'{learner_fname}.pkl')
pv(f'Learner saved:', verbose)
pv(f"path = '{path}'", verbose)
pv(f"dls_fname = '{dls_fnames}'", verbose)
pv(f"model_fname = '{model_fname}.pth'", verbose)
pv(f"learner_fname = '{learner_fname}.pkl'", verbose)
def load_all(path='export', dls_fname='dls', model_fname='model', learner_fname='learner', device=None, pickle_module=pickle, verbose=False):
if isinstance(device, int): device = torch.device('cuda', device)
elif device is None: device = default_device()
if device == 'cpu': cpu = True
else: cpu = None
path = Path(path)
learn = load_learner(path/f'{learner_fname}.pkl', cpu=cpu, pickle_module=pickle_module)
learn.load(f'{model_fname}', with_opt=True, device=device)
if learn.dls_type == "MixedDataLoaders":
dls_fnames = []
_dls = []
for i in range(learn.n_loaders[0]):
_dl = []
for j in range(learn.n_loaders[1]):
l = torch.load(path/f'{dls_fname}_{i}_{j}.pth', map_location=device, pickle_module=pickle_module)
l = l.new(num_workers=0)
l.to(device)
dls_fnames.append(f'{dls_fname}_{i}_{j}.pth')
_dl.append(l)
_dls.append(MixedDataLoader(*_dl, path=learn.dls.path, device=device, shuffle=l.shuffle))
learn.dls = MixedDataLoaders(*_dls, path=learn.dls.path, device=device)
else:
loaders = []
dls_fnames = []
for i in range(learn.n_loaders):
dl = torch.load(path/f'{dls_fname}_{i}.pth', map_location=device, pickle_module=pickle_module)
dl = dl.new(num_workers=0)
dl.to(device)
first(dl)
loaders.append(dl)
dls_fnames.append(f'{dls_fname}_{i}.pth')
learn.dls = type(learn.dls)(*loaders, path=learn.dls.path, device=device)
pv(f'Learner loaded:', verbose)
pv(f"path = '{path}'", verbose)
pv(f"dls_fname = '{dls_fnames}'", verbose)
pv(f"model_fname = '{model_fname}.pth'", verbose)
pv(f"learner_fname = '{learner_fname}.pkl'", verbose)
return learn
load_learner_all = load_all
#export
@patch
@delegates(subplots)
def plot_metrics(self: Recorder, nrows=None, ncols=None, figsize=None, final_losses=True, perc=.5, **kwargs):
n_values = len(self.recorder.values)
if n_values < 2:
print('not enough values to plot a chart')
return
metrics = np.stack(self.values)
n_metrics = metrics.shape[1]
names = self.metric_names[1:n_metrics+1]
if final_losses:
sel_idxs = int(round(n_values * perc))
if sel_idxs >= 2:
metrics = np.concatenate((metrics[:,:2], metrics), -1)
names = names[:2] + names
else:
final_losses = False
n = len(names) - 1 - final_losses
if nrows is None and ncols is None:
nrows = int(math.sqrt(n))
ncols = int(np.ceil(n / nrows))
elif nrows is None: nrows = int(np.ceil(n / ncols))
elif ncols is None: ncols = int(np.ceil(n / nrows))
figsize = figsize or (ncols * 6, nrows * 4)
fig, axs = subplots(nrows, ncols, figsize=figsize, **kwargs)
axs = [ax if i < n else ax.set_axis_off() for i, ax in enumerate(axs.flatten())][:n]
axs = ([axs[0]]*2 + [axs[1]]*2 + axs[2:]) if final_losses else ([axs[0]]*2 + axs[1:])
for i, (name, ax) in enumerate(zip(names, axs)):
if i in [0, 1]:
ax.plot(metrics[:, i], color='#1f77b4' if i == 0 else '#ff7f0e', label='valid' if i == 1 else 'train')
ax.set_title('losses')
ax.set_xlim(0, len(metrics)-1)
elif i in [2, 3] and final_losses:
ax.plot(np.arange(len(metrics) - sel_idxs, len(metrics)), metrics[-sel_idxs:, i],
color='#1f77b4' if i == 2 else '#ff7f0e', label='valid' if i == 3 else 'train')
ax.set_title('final losses')
ax.set_xlim(len(metrics) - sel_idxs, len(metrics)-1)
# ax.set_xticks(np.arange(len(metrics) - sel_idxs, len(metrics)))
else:
ax.plot(metrics[:, i], color='#1f77b4' if i == 0 else '#ff7f0e', label='valid' if i > 0 else 'train')
ax.set_title(name if i >= 2 * (1 + final_losses) else 'losses')
ax.set_xlim(0, len(metrics)-1)
ax.legend(loc='best')
ax.grid(color='gainsboro', linewidth=.5)
plt.show()
@patch
@delegates(subplots)
def plot_metrics(self: Learner, **kwargs):
self.recorder.plot_metrics(**kwargs)
#export
@patch
@delegates(subplots)
def show_probas(self:Learner, figsize=(6,6), ds_idx=1, dl=None, one_batch=False, max_n=None, **kwargs):
recorder = copy(self.recorder) # This is to avoid loss of recorded values while generating preds
if one_batch: dl = self.dls.one_batch()
probas, targets = self.get_preds(ds_idx=ds_idx, dl=[dl] if dl is not None else None)
if probas.ndim == 2 and probas.min() < 0 or probas.max() > 1: probas = nn.Softmax(-1)(probas)
if not isinstance(targets[0].item(), Integral): return
targets = targets.flatten()
if max_n is not None:
idxs = np.random.choice(len(probas), max_n, False)
probas, targets = probas[idxs], targets[idxs]
fig = plt.figure(figsize=figsize, **kwargs)
classes = np.unique(targets)
nclasses = len(classes)
vals = np.linspace(.5, .5 + nclasses - 1, nclasses)[::-1]
plt.vlines(.5, min(vals) - 1, max(vals), color='black', linewidth=.5)
cm = plt.get_cmap('gist_rainbow')
color = [cm(1.* c/nclasses) for c in range(1, nclasses + 1)][::-1]
class_probas = np.array([probas[i,t] for i,t in enumerate(targets)])
for i, c in enumerate(classes):
plt.scatter(class_probas[targets == c] if nclasses > 2 or i > 0 else 1 - class_probas[targets == c],
targets[targets == c] + .5 * (np.random.rand((targets == c).sum()) - .5), color=color[i], edgecolor='black', alpha=.2, s=100)
if nclasses > 2: plt.vlines((targets == c).float().mean(), i - .5, i + .5, color='r', linewidth=.5)
plt.hlines(vals, 0, 1)
plt.ylim(min(vals) - 1, max(vals))
plt.xlim(0,1)
plt.xticks(np.linspace(0,1,11), fontsize=12)
plt.yticks(classes, [self.dls.vocab[x] for x in classes], fontsize=12)
plt.title('Predicted proba per true class' if nclasses > 2 else 'Predicted class 1 proba per true class', fontsize=14)
plt.xlabel('Probability', fontsize=12)
plt.ylabel('True class', fontsize=12)
plt.grid(axis='x', color='gainsboro', linewidth=.2)
plt.show()
self.recorder = recorder
#export
all_archs = [FCN, FCNPlus, InceptionTime, InceptionTimePlus, InCoordTime, XCoordTime,
InceptionTimePlus17x17, InceptionTimePlus32x32, InceptionTimePlus47x47, InceptionTimePlus62x62,
InceptionTimeXLPlus, MultiInceptionTimePlus, MiniRocketClassifier, MiniRocketRegressor,
MiniRocketVotingClassifier, MiniRocketVotingRegressor, MiniRocketFeaturesPlus, MiniRocketPlus, MiniRocketHead,
InceptionRocketFeaturesPlus, InceptionRocketPlus, MLP, MultiInputNet, OmniScaleCNN, RNN, LSTM, GRU, RNNPlus, LSTMPlus, GRUPlus,
RNN_FCN, LSTM_FCN, GRU_FCN, MRNN_FCN, MLSTM_FCN, MGRU_FCN, ROCKET, RocketClassifier, RocketRegressor, ResCNNBlock, ResCNN,
ResNet, ResNetPlus, TCN, TSPerceiver, TST, TSTPlus, MultiTSTPlus, TSiTPlus, TSiT, InceptionTSiTPlus, InceptionTSiT,
TabFusionTransformer, TSTabFusionTransformer, TabModel, TabTransformer, TransformerModel, XCM, XCMPlus,
xresnet1d18, xresnet1d34, xresnet1d50, xresnet1d101, xresnet1d152, xresnet1d18_deep,
xresnet1d34_deep, xresnet1d50_deep, xresnet1d18_deeper, xresnet1d34_deeper, xresnet1d50_deeper,
XResNet1dPlus, xresnet1d18plus, xresnet1d34plus, xresnet1d50plus, xresnet1d101plus,
xresnet1d152plus, xresnet1d18_deepplus, xresnet1d34_deepplus, xresnet1d50_deepplus,
xresnet1d18_deeperplus, xresnet1d34_deeperplus, xresnet1d50_deeperplus, XceptionTime, XceptionTimePlus
]
all_archs_names = [arch.__name__ for arch in all_archs]
def get_arch(arch_name):
assert arch_name in all_archs_names, "confirm the name of the architecture"
idx = all_archs_names.index(arch_name)
return all_archs[idx]
arch_name = 'InceptionTimePlus'
test_eq(get_arch('InceptionTimePlus').__name__, arch_name)
#export
@delegates(build_ts_model)
def ts_learner(dls, arch=None, c_in=None, c_out=None, seq_len=None, d=None, splitter=trainable_params,
# learner args
loss_func=None, opt_func=Adam, lr=defaults.lr, cbs=None, metrics=None, path=None,
model_dir='models', wd=None, wd_bn_bias=False, train_bn=True, moms=(0.95,0.85,0.95),
# other model args
**kwargs):
if arch is None: arch = InceptionTimePlus
elif isinstance(arch, str): arch = get_arch(arch)
model = build_ts_model(arch, dls=dls, c_in=c_in, c_out=c_out, seq_len=seq_len, d=d, **kwargs)
try:
model[0], model[1]
subscriptable = True
except:
subscriptable = False
if subscriptable: splitter = ts_splitter
if loss_func is None:
if hasattr(dls, 'loss_func'): loss_func = dls.loss_func
elif hasattr(dls, 'train_ds') and hasattr(dls.train_ds, 'loss_func'): loss_func = dls.train_ds.loss_func
elif hasattr(dls, 'cat') and not dls.cat: loss_func = MSELossFlat()
learn = Learner(dls=dls, model=model,
loss_func=loss_func, opt_func=opt_func, lr=lr, cbs=cbs, metrics=metrics, path=path, splitter=splitter,
model_dir=model_dir, wd=wd, wd_bn_bias=wd_bn_bias, train_bn=train_bn, moms=moms, )
# keep track of args for loggers
store_attr('arch', self=learn)
return learn
#export
@delegates(build_tsimage_model)
def tsimage_learner(dls, arch=None, pretrained=False,
# learner args
loss_func=None, opt_func=Adam, lr=defaults.lr, cbs=None, metrics=None, path=None,
model_dir='models', wd=None, wd_bn_bias=False, train_bn=True, moms=(0.95,0.85,0.95),
# other model args
**kwargs):
if arch is None: arch = xresnet34
elif isinstance(arch, str): arch = get_arch(arch)
model = build_tsimage_model(arch, dls=dls, pretrained=pretrained, **kwargs)
learn = Learner(dls=dls, model=model,
loss_func=loss_func, opt_func=opt_func, lr=lr, cbs=cbs, metrics=metrics, path=path,
model_dir=model_dir, wd=wd, wd_bn_bias=wd_bn_bias, train_bn=train_bn, moms=moms)
# keep track of args for loggers
store_attr('arch', self=learn)
return learn
#export
@patch
def decoder(self:Learner, o): return L([self.dls.decodes(oi) for oi in o])
# export
@patch
@delegates(GatherPredsCallback.__init__)
def get_X_preds(self: Learner, X, y=None, bs=64, with_input=False, with_decoded=True, with_loss=False, **kwargs):
if with_loss and y is None:
print('cannot find loss as y=None')
with_loss = False
dl = self.dls.valid.new_dl(X, y=y)
dl.bs = ifnone(bs, self.dls.bs)
output = list(self.get_preds(dl=dl, with_input=with_input, with_decoded=with_decoded, with_loss=with_loss, reorder=False))
if with_decoded and hasattr(self.dls, 'vocab'):
output[2 + with_input] = L([self.dls.vocab[p] for p in output[2 + with_input]])
return tuple(output)
from tsai.data.all import *
from tsai.data.core import *
from tsai.models.FCNPlus import *
dsid = 'OliveOil'
X, y, splits = get_UCR_data(dsid, verbose=True, split_data=False)
tfms = [None, [Categorize()]]
dls = get_ts_dls(X, y, splits=splits, tfms=tfms)
learn = ts_learner(dls, FCNPlus)
for p in learn.model.parameters():
p.requires_grad=False
test_eq(count_parameters(learn.model), 0)
learn.freeze()
test_eq(count_parameters(learn.model), 1540)
learn.unfreeze()
test_eq(count_parameters(learn.model), 264580)
learn = ts_learner(dls, 'FCNPlus')
for p in learn.model.parameters():
p.requires_grad=False
test_eq(count_parameters(learn.model), 0)
learn.freeze()
test_eq(count_parameters(learn.model), 1540)
learn.unfreeze()
test_eq(count_parameters(learn.model), 264580)
learn.show_batch();
learn.fit_one_cycle(2, lr_max=1e-3)
dsid = 'OliveOil'
X, y, splits = get_UCR_data(dsid, split_data=False)
tfms = [None, [Categorize()]]
dls = get_ts_dls(X, y, tfms=tfms, splits=splits)
learn = ts_learner(dls, FCNPlus, metrics=accuracy)
learn.fit_one_cycle(2)
learn.plot_metrics()
learn.show_probas()
learn.save_all()
del learn
learn = load_all()
test_probas, test_targets, test_preds = learn.get_X_preds(X[0:10], with_decoded=True)
test_probas, test_targets, test_preds
test_probas2, test_targets2, test_preds2 = learn.get_X_preds(X[0:10], y[0:10], with_decoded=True)
test_probas2, test_targets2, test_preds2
test_eq(test_probas, test_probas2)
test_eq(test_preds, test_preds2)
learn.fit_one_cycle(1, lr_max=1e-3)
#hide
from tsai.imports import create_scripts
from tsai.export import get_nb_name
nb_name = get_nb_name()
create_scripts(nb_name);
```
| github_jupyter |
# "Price Charts with Technical Indicators"
> "Calculating Stock Price Indicators using FINTA python library, and visualizing using plotly python library."
- toc: false
- branch: master
- badges: true
- comments: true
- author: Ijeoma Odoko
- categories: [stocks, python, finta, pandas, plotly, ipywidgets]

Image credits to Markus Spiske - Unsplash photos
## About
There are a couple of libraries to use to calculate technical indicators for stocks. In previous posts, we had tried out the following python libraries:
> mplfinance
> TA-lib
In this post we will be looking at the [finta library](https://https://pypi.org/project/finta/).
## Install required dependencies on Google Colab
```
!pip install finta
```
## Load required libraries
```
from pandas_datareader import data
import numpy as np
from datetime import datetime
from datetime import date, timedelta
import pandas as pd
# import ipwidgets library and functions
from __future__ import print_function
from ipywidgets import interact, interactive, fixed
import ipywidgets as widgets
from IPython.display import display
from finta import TA
```
## Create widgets and dataframe for the stock data
*Instructions for use. *
1. Insert tuple of stock list.
2. Select stock from dropdown.
3. Select number of calendar days for dates from the last trading day.
4. Rerun all code after.
```
# Insert a tuple of unique tickers into the options variables.
#tickers = ('MMM', 'AOS', 'AAN', 'ABB', 'ABT', 'ABBV', 'ABM', 'ACN', 'AYI', 'GOLF', 'ADCT', 'ADT', 'AAP', 'ADSW', 'WMS', 'ACM', 'AEG', 'AER', 'AJRD', 'AMG', 'AFL', 'AGCO', 'A', 'AEM', 'ADC', 'AL', 'APD', 'AGI', 'ALK', 'ALB', 'ACI', 'AA', 'ALC', 'ARE', 'AQN', 'BABA', 'Y')
#tickers = ('ARKF', 'ARKG', 'ARKK', 'ARKW', 'QQQ','TQQQ', 'VCR', "KARS", 'ZNGA')
tickers = ('SOXX', 'SOXL', 'TQQQ', 'QQQ', 'ARKK', 'ARKW', 'FDN', 'XLY', 'VCR', 'FPX', 'SMH')
# create dropdown for selected stocks
stock_ticker = widgets.Dropdown(
options= tickers,
description='Select Stock Ticker',
disabled=False,
style = {'description_width': 'initial'},
layout = {'width': '200px'}
)
# create selection slider for days
w = widgets.IntSlider(
value=90,
min=5,
max=365,
step=1,
description = 'Calendar days',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style = {'description_width': 'initial','handle_color' : 'blue'},
layout = {'width': '400px'}
)
# create function for time frame of selected calendar days from today
def timeframe(w):
days = timedelta(w)
start = date.today() - days
today = date.today()
print('Start Date: ',start, ' ' ,'Last Date: ',today)
dates = widgets.interactive_output(timeframe, {'w': w} )
display(stock_ticker, w, dates)
```
## Download data for the stock
```
# create text to show stock ticker
v = widgets.Text(
value=stock_ticker.value,
description='Stockticker:',
disabled=True
)
# create function to load stock data from yahoo
def load_stock_data(stock_ticker, w):
start = date.today() - timedelta(w)
today = date.today()
stock_data = data.DataReader(stock_ticker, start=start, end=today, data_source='yahoo')
return stock_data
# create dataframe for selected stock
stock = load_stock_data(stock_ticker.value, w.value)
# display ticker and dataframe
display(v, stock)
# format dataframe in the format required by finta
ohlcv = stock[['Open', 'High', 'Low', 'Close', 'Volume']] # select the columns in the order required
ohlcv.columns = ['open', 'high', 'low', 'close', 'volume'] # rename the columns
ohlcv
```
## Calculate some Stock Price Indicators
```
# create example dataframe to try out the functions
ex_df = ohlcv.copy()
ex_df['RSI'] = TA.RSI(ex_df)
ex_df['Simple_Moving_Average_50'] = TA.SMA(ex_df, 50)
ex_df[['macd', 'macd_s']] = TA.MACD(ex_df)
ex_df
```
## Create a function to create a dataframe that captures some stock technical indicators
```
# create function to create the Stock Indicator dataframe
def create_dataframe(df):
"""
This function creates a Dataframe for key indicators
"""
df['Daily_Returns'] = df['close'].pct_change() # create column for daily returns
df['Price_Up_or_Down'] = np.where(df['Daily_Returns'] < 0, -1, 1) # create column for price up or down
# add columns for the volatility and volume indicators
df['Average_True_Range'] = TA.ATR(df)
df['On_Balance_Volume'] = TA.OBV(df)
df['Volume_Flow_Indicator'] = TA.VFI(df)
## add column for moving averages
df['Simple_Moving_Average_50'] = TA.SMA(df, 50)
#df['Simple_Moving_Average_200'] = TA.SMA(df, 200)
df['Volume Weighted Average Price'] = TA.VWAP(df)
df['Exponential_Moving_Average_50'] = TA.EMA(df, 50)
# add columns for momentum indicators
df['ADX'] = TA.ADX(df) #create column for ADX assume timeperiod of 14 days
df['RSI'] = TA.RSI(df) #create column for RSI assume timeperiod of 14 days
df['William %R'] = TA.WILLIAMS(df) #create column for William %R use high, low and close, and assume timeperiod of 14 days
df['MFI'] = TA.MFI(df) #create column for MFI use high, low and close, and assume timeperiod of 14 days
df['MOM'] = TA.MOM(df)
df[['macd', 'macd_signal']] = TA.MACD(df)
return df # return the dataframe
# Create a dataframe with all the stock indicators you indicated in the create dataframe function
stocks_df = create_dataframe(df = ohlcv)
stocks_df
```
## VISUALIZATIONS USING PLOTLY
## Price Action Chart
```
# create OHLC charts with Plotly
import plotly.graph_objects as go
fig_ohlc = go.Figure(data=[go.Ohlc(x=stocks_df.index,
open=stocks_df['open'],
high=stocks_df['high'],
low=stocks_df['low'],
close=stocks_df['close'], showlegend=False)])
fig_ohlc.update_layout(title = 'Price Action Chart', yaxis_title = 'Stock Price', template = 'presentation')
fig_ohlc.update(layout_xaxis_rangeslider_visible=False)
display(v)
fig_ohlc.show()
# create Candlestick charts with Plotly
import plotly.graph_objects as go
fig_candle = go.Figure(data=[go.Candlestick(x=stocks_df.index,
open=stocks_df['open'],
high=stocks_df['high'],
low=stocks_df['low'],
close=stocks_df['close'], showlegend=False)])
fig_candle.update_layout(title = 'Price Action Chart', yaxis_title = 'Stock Price', template = 'presentation')
fig_candle.update(layout_xaxis_rangeslider_visible=False)
display(v)
fig_candle.show()
```
## Momentum Indicators
```
#hide
import plotly.graph_objects as go
import plotly.offline as pyo
trace1 = go.Scatter(x=stocks_df.index, y=stocks_df['macd'], mode='lines', marker=dict(color="green"), showlegend=True, name='macd')
trace2 = go.Scatter(x=stocks_df.index, y=stocks_df['macd_signal'], mode='lines', marker=dict(color="blue"), showlegend=True, name='macd_signal')
data= [trace1, trace2]
layout = go.Layout(title = 'MACD indicator')
fig = go.Figure(data=data, layout=layout)
pyo.plot(fig, filename='MACD_indicator.html')
import plotly.graph_objects as go
trace1 = go.Scatter(x=stocks_df.index, y=stocks_df['macd'], mode='lines', marker=dict(color="green"), showlegend=True, name='macd')
trace2 = go.Scatter(x=stocks_df.index, y=stocks_df['macd_signal'], mode='lines', marker=dict(color="blue"), showlegend=True, name='macd_signal')
data= [trace1, trace2]
layout = go.Layout(title = 'MACD indicator')
fig = go.Figure(data=data, layout=layout)
fig.show()
```
# References
[Plotly Figure Reference](https://plotly.com/python/reference/index/) accessed October 22, 2020.
[FinTA (Financial Technical Analysis) python library](https://github.com/peerchemist/finta) accessed October 22, 2020.
| github_jupyter |
# Experiment with variables of given high correlation structure
This notebook is meant to address to a shared concern from two referees. The [motivating example](motivating_example.html) in the manuscript was designed to be a simple toy for illustrating the novel type of inference SuSiE offers. Here are some slightly more complicated examples, based on the motivating example, but with variables in high (rather than perfect) correlations with each other.
## $x_1$ and $x_2$ are highly correlated
Following a reviewer's
suggestion, we simulated two variables, $x_1$ and $x_2$, with high but
not perfect correlation ($0.9$). Specifically, we simulated $n = 600$
samples stored as an $X_{600 \times 2}$ matrix, in which each row was
drawn *i.i.d.* from a normal distribution with mean zero and
$\mathrm{cor}(x_1, x_2) = 0.9$.
We then simulated $y_i = x_{i1} \beta_1 + x_{i2} \beta_2 + \varepsilon_i$,
with $\beta_1 = 1, \beta_2 = 0$,
and $\varepsilon_i$ *i.i.d.* normal with zero mean and standard
deviation of 3. We performed 1,000 replicates of this simulation
(generated with different random number seeds).
In this simulation, the correlation between $x_1$ and $x_2$ is still
sufficiently high (0.9) to make distinguishing between the two
variables somewhat possible, but not non entirely straightforward. For
example, when we run lasso (using `cv.glmnet` from the `glmnet`
R package) on these data it wrongly selected $x_2$ as having
non-zero coefficient in about 10% of the simulations (95 out of
1,000), and correctly selected $x_1$ in about 96% of simulations (956
out of 1,000). Note that the lasso does not assess uncertainty in
variable selection, so these results are not directly comparable
with SuSiE CSs below; however, the lasso results demonstrate that
distinguishing the correct variable here is possible, but not so easy
that the example is uninteresting.
Ideally, then, SuSiE should identify variable $x_1$ as an effect
variable and drop $x_2$ as often as possible. However, due to the high
correlation between the variables, it is inevitable that some
95% SuSiE credible sets (CS) will also contain $x_2$. Most
important is that we should avoid, as much as possible, reporting a CS
that contains *only* $x_2$, since the goal is that 95% of CSs
should contain at least one effect variable. The SuSiE results (SuSiE version 0.9.1 on R 3.5.2)
are summarized below. The code used for the simulation [can be found here](https://github.com/stephenslab/susie-paper/blob/master/src/ref_3_question.R).
| CSs | count |
| :---- | ----: |
| (1) | 829 |
| (1,2) | 169 |
| **(2)** | 2 |
Highlighted in **bold** are CSs that do *not* contain
the true effect variable --- there are 2 of them out of 1,000 CSs
detected. In summary, SuSiE precisely identifies the effect
variable in a single CS in the majority (83%) of the simulations, and
provides a "valid" CS (*i.e.*, one containing an effect
variable) in almost all simulations (998 out of 1,000). Further, even
when SuSiE reports a CS including both variables, it consistently
assigns higher posterior inclusion probability (PIP) to the correct
variable, $x_1$: among the 169 CSs that contain both variables, the
median PIPs for $x_1$ and $x_2$ were 0.86 and 0.14, respectively.
## When an additional non-effect variable highly correlated with both variable groups
Another referee suggested the following:
> Suppose
we have another predictor $x_5$, which is both correlated with $(x_1,
x_2)$ and $(x_3, x_4)$. Say $\mathrm{cor}(x_1, x_5) = 0.9$,
$\mathrm{cor}(x_2, x_5) = 0.7$, and $\mathrm{cor}(x_5, x_3)
= \mathrm{cor}(x_5, x_4) = 0.8$. Does the current method assign $x_5$
to the $(x_1, x_2)$ group or the $(x_3, x_4)$ group?
Following the suggestion, we simulated $x_1, \ldots, x_5$ from a
multivariate normal with zero mean and the covariance matrix
approximately as given in Table below. (Since this matrix is
not quite positive definite, in our R code we used `nearPD` from
the `Matrix` package to generate the nearest positive definite
matrix --- the entries of the resulting covariance matrix differ only
very slightly from those in Table below, with a maximum
absolute difference of 0.0025 between corresponding elements in the
two matrices).
| | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ |
| ------: | ------: | ------: | ------: | ------: | ------: |
| $x_1$ | 1.00 | 0.92 | 0.70 | 0.70 | 0.90 |
| $x_2$ | 0.92 | 1.00 | 0.70 | 0.70 | 0.70 |
| $x_3$ | 0.70 | 0.70 | 1.00 | 0.92 | 0.80 |
| $x_4$ | 0.70 | 0.70 | 0.92 | 1.00 | 0.80 |
| $x_5$ | 0.90 | 0.70 | 0.80 | 0.80 | 1.00 |
We simulated $n = 600$ samples from this
multivariate normal distribution, then we simulated $n = 600$
responses $y_i$ from the regression model $y_i = x_{i1} \beta_1 + \cdots x_{i5} \beta_5 + \varepsilon_i$,
with $\beta = (0, 1, 1, 0, 0)^T$, and $\varepsilon_i$ *i.i.d.* normal with zero mean and
standard deviation of 3. We repeated this simulation procedure 1,000
times with different random seeds, and each time we fit a SuSiE
model to the simulated data by running the IBSS algorithm. To
simplify the example, we ran the IBSS algorithm with $L = 2$, and
fixed the $\sigma_0^2 = 1$. Similar results were obtained when we used
larger values of $L$, and when $\sigma_0^2$ was estimated. For more
details on how the data were simulated and how the SuSiE models
were fitted to the data sets, [see this script](https://github.com/stephenslab/susie-paper/blob/master/src/ref_4_question.R).
Like the toy motivating example given in the paper, in this simulation
the first two predictors are strongly correlated with each other, so
it may be difficult to distinguish among them, and likewise for the
third and fourth predictors. The fifth predictor, which has no effect
on $y$, potentially complicates matters because it is also strongly
correlated with the other predictors. Despite this complication, our
basic goal remains the same: the Credible Sets inferred by SuSiE
should capture the true effects most of the time, while also
minimizing "false positive" CSs that do not contain any true
effects. (Further, each CS should, ideally, be as small as possible.)
Table below summarizes the results of these simulations:
the left-hand column gives a unique result (a combination of CSs), and
the right-hand column gives the number of times this unique result
occurred among the 1,000 simulations. The CS combinations are ordered
by the frequency of their occurrence in the simulations. We highlight
in **bold** CSs that do not contain a true effect.
| CSs | count |
| :------------- | ----: |
| (2), (3) | 551 |
| (2), (3,4) | 212 |
| (1,2), (3) | 176 |
| (1,2), (3,4) | 38 |
| (2), (3,4,5) | 9 |
| **(1)**, (3,4) | 3 |
| (2), **(4)** | 3 |
| (1,2), (3,4,5) | 2 |
| **(1)**, (3) | 1 |
| (1,2), **(4)** | 1 |
| (2), (3,5) | 1 |
| (3), (1,2,5) | 1 |
| (3), (1,2,3) | 1 |
| (3,4), (1,2,4) | 1 |
In the majority (551) of the simulations, SuSiE precisely identiied
the true effect variables, and no others. In most other cases,
SuSiE identified two CSs, each containing a correct effect variable, and
with one or more other variables included due to high correlation with
the true-effect variable. The referee asks specifically about how the
additional variable $x_5$ behaves in this example. In practice, $x_5$
was rarely included in a CS. In the few cases where $x_5$ *was*
included in a CS, the results were consistent with the simulation
setting; $x_5$ was included more frequently with $x_3$ and/or $x_4$
(12 times) rather than $x_2$ and/or $x_1$ (only once). In no
simulations did SuSiE form a large group that contains all five
predictors.
This example actually highlights the benefits of SuSiE compared to
alternative approaches (e.g., hierinf) that *first* cluster the
variables into groups based on the correlation structure, then test
the groups. As we pointed out in the manuscript, this alternative
approach (first cluster variables into groups, then test groups) would
work well in the toy example in the paper, but in general it requires
*ad hoc* decisions about how to cluster variables. In this more
complex example raised by the referee, it is far from clear how to
cluster the variables. SuSiE avoids this problem because there is
no pre-clustering of variables; instead, the SuSiE CSs are computed
directly from an (approximate) posterior distribution (which takes
into account how the variables $x$ are correlated with each other, as
well as their relationship with $y$).
| github_jupyter |
## Analyzing Hamlet
```
%load_ext autoreload
%autoreload 2
import src.data
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
from collections import OrderedDict
from IPython.display import display
pd.options.display.max_rows = 999
pd.options.display.max_columns = 999
pd.set_option("display.max_columns", None)
import itertools
file_names = {
'aurora':'2020-01-01-till-2021-02-24-aurora.csv',
'hamlet':'2020-01-01-till-2021-02-24-hamlet.csv',
'mercandia':'2020-01-01-till-2021-02-24-mercandia-iv.csv',
'tycho-brahe':'2020-01-01-till-2021-02-24-tycho-brahe.csv',
}
dfs = OrderedDict()
for ship_name, file_name in file_names.items():
file_path = os.path.join(src.data.path_ships,file_name)
reader = pd.read_csv(file_path, chunksize=1000, iterator=True) # Loading a small part of the data
dfs[ship_name] = next(reader)
for ship_name, df in dfs.items():
display(df.describe())
file_path = os.path.join(src.data.path_ships,file_names['aurora'])
reader = pd.read_csv(file_path, chunksize=1000000, iterator=True) # Loading a small part of the data
df_raw = next(reader)
df_raw.set_index('Tidpunkt [UTC]', inplace=True)
df_raw.index = pd.to_datetime(df_raw.index)
mask = df_raw['Fart över grund (kts)']>1
df = df_raw.loc[mask].copy()
df.hist(column='Kurs över grund (deg)', bins=1000)
mask = df_raw['Kurs över grund (deg)'] < 150
df_direction_1 = df.loc[mask]
df_direction_1.describe()
df_direction_1.plot(x='Longitud (deg)', y = 'Latitud (deg)', style='.', alpha=0.005)
deltas = []
for i in range(1,5):
sin_key = 'Sin EM%i ()' % i
cos_key = 'Cos EM%i ()' % i
delta_key = 'delta_%i' % i
deltas.append(delta_key)
df_direction_1[delta_key] = np.arctan2(df_direction_1[sin_key],df_direction_1[cos_key])
df_plot = df_direction_1.loc['2020-01-01 01:00':'2020-01-01 02:00']
df_plot.plot(y=['Kurs över grund (deg)','Stävad kurs (deg)'],style='.')
df_plot.plot(y='Fart över grund (kts)',style='.')
df_plot.plot(y=deltas,style='.')
df_direction_1.head()
(df_direction_1['Sin EM1 ()']**2 + df_direction_1['Cos EM1 ()']**2).hist()
df_direction_1.columns
descriptions = pd.Series(index = df_direction_1.columns.copy())
descriptions['Latitud (deg)'] = 'Latitud (deg) (WGS84?)'
descriptions['Longitud (deg)'] = 'Longitud (deg) (WGS84?)'
descriptions['Effekt DG Total (kW)'] = '?'
descriptions['Effekt EM Thruster Total (kW)'] = ''
descriptions['Sin EM1 ()'] = ''
descriptions['Sin EM2 ()'] = ''
descriptions['Sin EM3 ()'] = ''
descriptions['Sin EM4 ()'] = ''
descriptions['Cos EM1 ()'] = ''
descriptions['Cos EM2 ()'] = ''
descriptions['Cos EM3 ()'] = ''
descriptions['Cos EM4 ()'] = ''
descriptions['Fart över grund (kts)'] = 'GPS fart'
descriptions['Stävad kurs (deg)'] = 'Kompas kurs'
descriptions['Kurs över grund (deg)'] = 'GPS kurs'
descriptions['Effekt hotell Total (kW)'] = ''
descriptions['Effekt Consumption Total (kW)'] = ''
descriptions['Förbrukning GEN alla (kg/h)'] = '?'
descriptions['delta_1'] = 'Thruster angle 1'
descriptions['delta_2'] = 'Thruster angle 2'
descriptions['delta_3'] = 'Thruster angle 3'
descriptions['delta_4'] = 'Thruster angle 4'
df_numenclature = pd.DataFrame(descriptions, columns=['Description'])
df_numenclature
```
| github_jupyter |
# Train mnist with Tensorflow
**Requirements** - In order to benefit from this tutorial, you will need:
- A basic understanding of Machine Learning
- An Azure account with an active subscription - [Create an account for free](https://azure.microsoft.com/free/?WT.mc_id=A261C142F)
- An Azure ML workspace with computer cluster - [Configure workspace](../../configuration.ipynb)
- A python environment
- Installed Azure Machine Learning Python SDK v2 - [install instructions](../../../README.md) - check the getting started section
**Learning Objectives** - By the end of this tutorial, you should be able to:
- Connect to your AML workspace from the Python SDK
- Define different `CommandComponent` using YAML
- Create `Pipeline` load these components from YAML
**Motivations** - This notebook explains how to run a pipeline with distributed training component.
# 1. Connect to Azure Machine Learning Workspace
The [workspace](https://docs.microsoft.com/en-us/azure/machine-learning/concept-workspace) is the top-level resource for Azure Machine Learning, providing a centralized place to work with all the artifacts you create when you use Azure Machine Learning. In this section we will connect to the workspace in which the job will be run.
## 1.1 Import the required libraries
```
# import required libraries
from azure.identity import DefaultAzureCredential, InteractiveBrowserCredential
from azure.ai.ml import MLClient
from azure.ai.ml.dsl import pipeline
from azure.ai.ml.entities import ResourceConfiguration
```
## 1.2 Configure credential
We are using `DefaultAzureCredential` to get access to workspace.
`DefaultAzureCredential` should be capable of handling most Azure SDK authentication scenarios.
Reference for more available credentials if it does not work for you: [configure credential example](../../configuration.ipynb), [azure-identity reference doc](https://docs.microsoft.com/en-us/python/api/azure-identity/azure.identity?view=azure-python).
```
try:
credential = DefaultAzureCredential()
# Check if given credential can get token successfully.
credential.get_token("https://management.azure.com/.default")
except Exception as ex:
# Fall back to InteractiveBrowserCredential in case DefaultAzureCredential not work
credential = InteractiveBrowserCredential()
```
## 1.3 Get a handle to the workspace
We use config file to connect to a workspace. The Azure ML workspace should be configured with computer cluster. [Check this notebook for configure a workspace](../../configuration.ipynb)
```
# Get a handle to workspace
ml_client = MLClient.from_config(credential=credential)
# Retrieve an already attached Azure Machine Learning Compute.
cluster_name = "cpu-cluster"
print(ml_client.compute.get(cluster_name))
```
# 2. Define command component
We defined sample component using `command_component` decorator in [component.py](src/component.py).
```
with open("src/component.py") as fin:
print(fin.read())
%load_ext autoreload
%autoreload 2
from src.component import train_tf
help(train_tf)
```
# 3. Pipeline job
## 3.1 Build pipeline
```
@pipeline()
def train_mnist_with_tensorflow():
"""Train using TF component."""
tf_job = train_tf(epochs=1)
tf_job.compute = "cpu-cluster"
tf_job.resources = ResourceConfiguration(instance_count=2)
tf_job.distribution.worker_count = 2
tf_job.outputs.trained_model_output.mode = "upload"
# create pipeline instance
pipeline_job = train_mnist_with_tensorflow()
```
## 3.2 Submit pipeline job
```
# submit job to workspace
pipeline_job = ml_client.jobs.create_or_update(
pipeline_job, experiment_name="pipeline_samples"
)
pipeline_job
# Wait until the job completes
ml_client.jobs.stream(pipeline_job.name)
```
# Next Steps
You can see further examples of running a pipeline job [here](../)
| github_jupyter |
```
import sys
sys.path.append(r'C:\Users\moallemie\EMAworkbench-master')
sys.path.append(r'C:\Users\moallemie\EM_analysis')
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from ema_workbench import load_results, ema_logging
from ema_workbench.em_framework.salib_samplers import get_SALib_problem
from SALib.analyze import morris
# Set up number of scenarios, outcome of interest, and number of parallel processors
sc = 500 # Specify the number of scenarios where the convergence in the SA indices occures
t = 2100
outcome_var = 'GWP per Capita Indicator' # Specify the outcome of interest for SA ranking verification
nprocess = 100
```
## Loading the model, uncertainities, and outcomes and generate experiments
```
# Here we only generate experiments for loading the necessary components.
#The actual results will be loaded in the next cell.
# Open Excel input data from the notebook directory before runnign the code in multi-processing.
# Close the folder where the results will be saved in multi-processing.
# This line must be at the beginning for multi processing.
if __name__ == '__main__':
ema_logging.log_to_stderr(ema_logging.INFO)
#The model must be imoorted as .py file in parallel processing.
from Model_init import vensimModel
from ema_workbench import (TimeSeriesOutcome,
perform_experiments,
RealParameter,
CategoricalParameter,
ema_logging,
save_results,
load_results)
directory = 'C:/Users/moallemie/EM_analysis/Model/'
df_unc = pd.read_excel(directory+'ScenarioFramework.xlsx', sheet_name='Uncertainties')
# 0.5/1.5 multiplication is added to previous Min/Max cells for parameters with Reference values 0
#or min/max manually set in the spreadsheet
df_unc['Min'] = df_unc['Min'] + df_unc['Reference'] * 0.75
df_unc['Max'] = df_unc['Max'] + df_unc['Reference'] * 1.25
# From the Scenario Framework (all uncertainties), filter only those top 20 sensitive uncertainties under each outcome
sa_dir='C:/Users/moallemie/EM_analysis/Data/'
mu_df = pd.read_csv(sa_dir+"MorrisIndices_{}_sc5000_t{}.csv".format(outcome_var, t))
mu_df.rename(columns={'Unnamed: 0': 'Uncertainty'}, inplace=True)
mu_df.sort_values(by=['mu_star'], ascending=False, inplace=True)
mu_df = mu_df.head(20)
mu_unc = mu_df['Uncertainty']
mu_unc_df = mu_unc.to_frame()
# Remove the rest of insensitive uncertainties from the Scenario Framework and update df_unc
keys = list(mu_unc_df.columns.values)
i1 = df_unc.set_index(keys).index
i2 = mu_unc_df.set_index(keys).index
df_unc2 = df_unc[i1.isin(i2)]
vensimModel.uncertainties = [RealParameter(row['Uncertainty'], row['Min'], row['Max']) for index, row in df_unc2.iterrows()]
df_out = pd.read_excel(directory+'ScenarioFramework.xlsx', sheet_name='Outcomes')
vensimModel.outcomes = [TimeSeriesOutcome(out) for out in df_out['Outcome']]
from ema_workbench import MultiprocessingEvaluator
from ema_workbench.em_framework.evaluators import (MC, LHS, FAST, FF, PFF, SOBOL, MORRIS)
import time
start = time.time()
with MultiprocessingEvaluator(vensimModel, n_processes=nprocess) as evaluator:
results = evaluator.perform_experiments(scenarios=sc, uncertainty_sampling=MORRIS)
end = time.time()
print("took {} seconds".format(end-start))
experiments, outcomes = results
r_dir = 'D:/moallemie/EM_analysis/Data/'
save_results(results, r_dir+"SDG_experiments_ranking_verification_{}_sc{}.tar.gz".format(outcome_var, sc))
```
## Calculating SA (Morris) metrics
```
# Morris mu_star index calculation as a function of number of scenarios and time
def make_morris_df(scores, problem, outcome_var, sc, t):
scores_filtered = {k:scores[k] for k in ['mu_star','mu_star_conf','mu','sigma']}
Si_df = pd.DataFrame(scores_filtered, index=problem['names'])
indices = Si_df[['mu_star','mu']]
errors = Si_df[['mu_star_conf','sigma']]
return indices, errors
problem = get_SALib_problem(vensimModel.uncertainties)
X = experiments.iloc[:, :-3].values
Y = outcomes[outcome_var][:,-1]
scores = morris.analyze(problem, X, Y, print_to_console=False)
inds, errs = make_morris_df(scores, problem, outcome_var, sc, t)
```
## Where to draw the line between important and not important?
```
'''
Modifed from Waterprogramming blog by Antonia Hadgimichael: https://github.com/antonia-had/SA_verification
The idea is that we create 2 additiopnal Sets (current SA samples are Set 1).
We can create a Set 2, using only the T most important factors from our Set 1 sample,
and fixing all other factors to their default values.
We can also create a Set 3, now fixing the T most important factors to defaults
and using the sampled values of all other factors from Set 1.
If we classified our important and unimportant factors correctly,
then the correlation coefficient between the model outputs of Set 2 and Set 1 should approximate 1
(since we’re fixing all factors that don’t matter),
and the correlation coefficient between outputs from Set 3 and Set 1 should approximate 0
(since the factors we sampled are inconsequential to the output).
'''
# Sort factors by importance
inds_mu = inds['mu_star'].reindex(df_unc2['Uncertainty']).values
factors_sorted = np.argsort(inds_mu)[::-1]
# Set up DataFrame of default values to use for experiment
nsamples = len(experiments.index)
defaultvalues = df_unc2['Reference'].values
X_defaults = np.tile(defaultvalues,(nsamples, 1))
# Create Set 1 from experiments
exp_T = experiments.drop(['scenario', 'policy', 'model'], axis=1).T.reindex(df_unc2['Uncertainty'])
exp_ordered = exp_T.T
X_Set1 = exp_ordered.values
# Create initial Sets 2 and 3
X_Set2 = np.copy(X_defaults)
X_Set3 = np.copy(X_Set1)
# Define a function to convert your Set 2 and Set 3 into experiments structure in the EMA Workbench
def SA_experiments_to_scenarios(experiments, model=None):
'''
"Slighlty modifed from the EMA Workbench"
This function transform a structured experiments array into a list
of Scenarios.
If model is provided, the uncertainties of the model are used.
Otherwise, it is assumed that all non-default columns are
uncertainties.
Parameters
----------
experiments : numpy structured array
a structured array containing experiments
model : ModelInstance, optional
Returns
-------
a list of Scenarios
'''
from ema_workbench import Scenario
# get the names of the uncertainties
uncertainties = [u.name for u in model.uncertainties]
# make list of of tuples of tuples
cases = []
cache = set()
for i in range(experiments.shape[0]):
case = {}
case_tuple = []
for uncertainty in uncertainties:
entry = experiments[uncertainty][i]
case[uncertainty] = entry
case_tuple.append(entry)
case_tuple = tuple(case_tuple)
cases.append(case)
cache.add((case_tuple))
scenarios = [Scenario(**entry) for entry in cases]
return scenarios
# Run the models for the top n factors in Set 2 and Set 3 and generate correlation figures
if __name__ == '__main__':
ema_logging.log_to_stderr(ema_logging.INFO)
#The model must be imoorted as .py file in parallel processing.
from Model_init import vensimModel
from ema_workbench import (TimeSeriesOutcome,
perform_experiments,
RealParameter,
CategoricalParameter,
ema_logging,
save_results,
load_results)
vensimModel.outcomes = [TimeSeriesOutcome(outcome_var)]
from ema_workbench import MultiprocessingEvaluator
coefficient_S1_S3 = 0.99
for f in range(1, len(factors_sorted)+1):
ntopfactors = f
if coefficient_S1_S3 >= 0.1:
for i in range(ntopfactors): #Loop through all important factors
X_Set2[:,factors_sorted[i]] = X_Set1[:,factors_sorted[i]] #Fix use samples for important
X_Set3[:,factors_sorted[i]] = X_defaults[:,factors_sorted[i]] #Fix important to defaults
X_Set2_exp = pd.DataFrame(data=X_Set2, columns=df_unc2['Uncertainty'].tolist())
X_Set3_exp = pd.DataFrame(data=X_Set3, columns=df_unc2['Uncertainty'].tolist())
scenarios_Set2 = SA_experiments_to_scenarios(X_Set2_exp, model=vensimModel)
scenarios_Set3 = SA_experiments_to_scenarios(X_Set3_exp, model=vensimModel)
#experiments_Set2, outcomes_Set2 = perform_experiments(vensimModel, scenarios_Set2)
#experiments_Set3, outcomes_Set3 = perform_experiments(vensimModel, scenarios_Set3)
with MultiprocessingEvaluator(vensimModel, n_processes=nprocess) as evaluator:
experiments_Set2, outcomes_Set2 = evaluator.perform_experiments(scenarios=scenarios_Set2)
experiments_Set3, outcomes_Set3 = evaluator.perform_experiments(scenarios=scenarios_Set3)
# Calculate coefficients of correlation
data_Set1 = Y
data_Set2 = outcomes_Set2[outcome_var][:,-1]
data_Set3 = outcomes_Set3[outcome_var][:,-1]
coefficient_S1_S2 = np.corrcoef(data_Set1,data_Set2)[0][1]
coefficient_S1_S3 = np.corrcoef(data_Set1,data_Set3)[0][1]
# Plot outputs and correlation
fig = plt.figure(figsize=(14,7))
ax1 = fig.add_subplot(1,2,1)
ax1.plot(data_Set1,data_Set1, color='#39566E')
ax1.scatter(data_Set1,data_Set2, color='#8DCCFC')
ax1.set_xlabel("Set 1",fontsize=14)
ax1.set_ylabel("Set 2",fontsize=14)
ax1.tick_params(axis='both', which='major', labelsize=10)
ax1.set_title('Set 1 vs Set 2 - ' + str(f) + ' top factors',fontsize=15)
ax1.text(0.05,0.95,'R= '+"{0:.3f}".format(coefficient_S1_S2),transform = ax1.transAxes,fontsize=16)
ax2 = fig.add_subplot(1,2,2)
ax2.plot(data_Set1,data_Set1, color='#39566E')
ax2.scatter(data_Set1,data_Set3, color='#FFE0D5')
ax2.set_xlabel("Set 1",fontsize=14)
ax2.set_ylabel("Set 3",fontsize=14)
ax2.tick_params(axis='both', which='major', labelsize=10)
ax2.set_title('Set 1 vs Set 3 - ' + str(f) + ' top factors',fontsize=15)
ax2.text(0.05,0.95,'R= '+"{0:.3f}".format(coefficient_S1_S3),transform = ax2.transAxes,fontsize=16)
plt.savefig('{}/{}_{}_topfactors.png'.format(r'C:/Users/moallemie/EM_analysis/Fig/sa_verification', outcome_var, str(f)))
plt.close()
```
| github_jupyter |
##### Imports
```
import numpy as np
import pandas as pd
import os
import time
from itertools import permutations, combinations
from IPython.display import display
```
##### Prompts to choose which store you want
```
print("Welcome to Apriori 2.0!")
store_num = input("Please select your store \n 1. Amazon \n 2. Nike \n 3. Best Buy \n 4. K-Mart \n 5. Walmart\n")
print(store_num)
support_percent = input("Please enter the percentage of Support you want?\n")
print(support_percent)
confidence_percent = input("Please enter the percentage of Confidence you want?\n")
print(confidence_percent)
```
##### These are my dictionaries to choose which store to get based in Key-Value Pairs
```
def number_to_store(store_number):
switcher = {
1: "data/amazon_transactions.csv",
2: "data/nike_transaction.csv",
3: "data/best_buy_transaction.csv",
4: "data/k_mart_transaction.csv",
5: "data/walmart_transaction.csv"
}
return switcher.get(store_number)
def number_to_item_list_of_store(store_number):
switcher_dict = {
1: "data/amazon_item_names.csv",
2: "data/nike_item_names.csv",
3: "data/best_buy_item_names.csv",
4: "data/k_mart_item_names.csv",
5: "data/walmart_item_names.csv"
}
return switcher_dict.get(store_number)
def ns(store_number):
switcher_store = {
1: "Amazon",
2: "Nike",
3: "Best Buy",
4: "K-Mart",
5: "Walmart"
}
return switcher_store.get(store_number)
```
##### We first have to read in the csv files and make sure that the inputs received from the user are valid
```
def a_priori_read(item_list, transaction, support_percentage, confidence_percentage):
# Create two different functions one that is solo for read in the file data and the other that is algorithmic with the data
if support_percentage > 100 or confidence_percentage > 100 or support_percentage < 0 or confidence_percentage < 0:
print("Support Percent or Confidence Percent is Invalid. \n Enter a valid number between 0 and 100.\n")
print("Restarting Apriori 2.0.....\n")
time.sleep(2)
os.system("python Aprior_Algo")
if support_percentage >= 0 and support_percentage <= 100 and confidence_percentage >= 0 and confidence_percentage <= 100:
df_item_list = pd.read_csv(item_list)
df_transactions = pd.read_csv(transaction)
print(df_transactions.head())
print(df_item_list.head())
trans = np.array(df_transactions["transaction"])
items_names = np.array(df_item_list["item_name"])
k_value = 1
return items_names, trans, support_percentage, confidence_percentage, k_value
```
##### The first go around of the Apriori Algorithm we find the items that are most frequent when K=1
##### This is so that we can find the most frequent items given the transactions
```
def ap_1(items_names, trans, support_percentage, confidence_percentage, k_value):
counter = np.zeros(len(items_names), dtype=int)
for i in trans:
i = list((map(str.strip, i.split(','))))
s1 = set(i)
nums = 0
for x in items_names:
s2 = set()
s2.add(x)
if s2.issubset(s1):
counter[nums] += 1
nums += 1
counter = list(map(lambda x: int((x / len(trans)) * 100), counter))
df3 = pd.DataFrame({"item_name": items_names, "support": counter,"k_val" : np.full(len(items_names),k_value)})
rslt_df = df3[df3['support'] >= support_percentage]
print("When K = " + str(k_value))
print(rslt_df)
items = np.array(rslt_df["item_name"])
support_count = np.array(rslt_df["support"])
k_value += 1
return items, support_count, k_value, rslt_df
```
##### Then we use this function below to find item sets that are most frequent when K > 1
```
def ap_2(item_comb, k_value, trans, support_percentage):
boo = True
comb = combinations(item_comb, k_value)
comb = list(comb)
counter = np.zeros(len(comb), dtype=int)
if k_value > 1:
for i in trans:
i = list((map(str.strip, i.split(','))))
s1 = set(i)
nums = 0
for x in comb:
s2 = set()
x = np.asarray(x)
for q in x:
s2.add(q)
if s2.issubset(s1):
counter[nums] += 1
nums += 1
counter = list(map(lambda x: int((x / len(trans)) * 100), counter))
df3 = pd.DataFrame({"item_name": comb, "support": counter,"k_val":np.full(len(comb),k_value)})
#Making sure that user parameters are met for support
rslt_df = df3[df3['support'] >= support_percentage]
print("When K = " + str(k_value))
print(rslt_df)
items = np.array(rslt_df["item_name"])
supp = np.array(rslt_df["support"])
if len(items) == 0:
boo = False
return rslt_df, boo
return rslt_df, boo
```
##### Calls of functions and variable saving
```
frames = []
items_names, trans, support_percent, confidence_percent, k_value = a_priori_read(
str(number_to_item_list_of_store(int(store_num))), str(number_to_store(int(store_num))),
int(support_percent), int(confidence_percent))
items, supp, k_value, df = ap_1(items_names, trans, support_percent, confidence_percent, k_value)
frames.append(df)
boo = True
```
##### Increasing K by 1 until we can longer support the support value
```
while boo:
df_1, boo = ap_2(items, k_value, trans, support_percent)
frames.append(df_1)
k_value += 1
```
##### Combine the dataframes we have from when we increase K
```
print("results of item-sets that meet support are below")
display(pd.concat(frames))
df_supp = pd.concat(frames)
# df_supp.head()
```
##### Reset the index just to organize it and the results after we find the most frequent sets in the list of transactions
```
df_supp = df_supp.reset_index().drop('index',axis=1)
df_supp
```
##### This is the FUNCTION that genrerates the Associations (Permutations) and calculating the Confidence of the item sets
```
def confidence(val):
#Since we already have our support for our items what we need to worry about is the confidence levels
#item_set before the arrow
df_before = df_supp.loc[df_supp['k_val'] == val]
stuff_name_before = np.array(df_before["item_name"])
support_arr_before = np.array(df_before['support'])
#item_set of the overall set
df_overall = df_supp.loc[df_supp['k_val'] == val+1]
df_ov = np.array(df_overall["item_name"])
suppport_ov = np.array(df_overall['support'])
#variables to save
sup_ov = list()
sup_sing = list()
perm_item = list()
#When the item set is k =1 and the comparison is k = 2
if val == 1:
for i_set in df_ov:
temp_list = list(df_ov)
#I want to select the support of that overall set
ov_sup = suppport_ov[temp_list.index(i_set)]
temp = set()
#This is where we generate our permutations
for indiv_item in i_set:
temp.add(indiv_item)
perm = permutations(temp)
perm_lst = list(perm)
# for each permutation in the perm_list
for perm_item_set in perm_lst:
perm_item.append(perm_item_set)
sup_ov.append(ov_sup)
sup_sing.append(int(support_arr_before[np.where(stuff_name_before == perm_item_set[0])]))
#When the item set is k > 1 and the comparison is k += k + 1
if val > 1:
for i_set in df_ov:
temp_list = list(df_ov)
ov_sup = suppport_ov[temp_list.index(i_set)]
temp = set()
for indiv_item in i_set:
temp.add(indiv_item)
perm = permutations(temp)
perm_lst = list(perm)
for perm_item_set in perm_lst:
try:
temp_set = []
for dex in range(0,val):
temp_set.append(perm_item_set[dex])
item_set_before = tuple(temp_set)
tp_lst = list(stuff_name_before)
ss = support_arr_before[tp_lst.index(item_set_before)]
sup_ov.append(ov_sup)
sup_sing.append(ss)
perm_item.append(perm_item_set)
except:
# print("itemset below does not exist...")
# print(y)
sup_ov.append(ov_sup)
sup_sing.append(0)
perm_item.append(perm_item_set)
df_main = pd.DataFrame({"association":perm_item,"support_ov":sup_ov,"support_sing":sup_sing})
df_main = df_main.assign(confidence = lambda x:round(((x.support_ov/x.support_sing)*100),0))
return df_main
```
#### Finding the max k value in the given set
```
try:
max(df_supp["k_val"])
except:
print("No max was found...")
```
#### This is where I iteratively call the confidence() function
```
df_frames = []
try:
if len(df_supp["k_val"]) != 0 :
for lp in range(1,max(df_supp["k_val"])+1):
#print(lp)
df_0 = confidence(lp)
df_0 = df_0[df_0.support_sing != 0]
df_frames.append(df_0)
df_associations = pd.concat(df_frames)
display(df_associations.head())
except:
print("No items or transactions meet the user requirements!")
```
###### Concat the Dataframes
```
try:
df_associations = pd.concat(df_frames)
display(df_associations)
except:
print("No items or transactions meet the user requirements!")
```
##### Making sure that user parameters are met for confidence
```
try:
df_associations = df_associations[df_associations['confidence'] >= confidence_percent]
display(df_associations)
except:
print("No items or transactions meet the user requirements!")
```
##### Formatting the Dataframe Final
```
try:
df_final = df_associations.reset_index().drop(['index','support_sing'],axis=1)
df_final.columns = ["Association","Support","Confidence"]
except:
print("No items or transactions meet the user requirements!")
```
#### Final Associations
```
try:
print("Store Name: "+ str(ns(int(store_num))))
print("\nFinal Associations that meet the user standards....")
print("Support: " + str(support_percent) + "%" + "\t" + "Confidence: " + str(confidence_percent) + '%')
#this will display the max column width so we can see the associations involved....
pd.set_option('display.max_colwidth', 0)
display(df_final)
except:
print("\nNo Associations were generated based on the parameters set!")
import re
samp = np.array(df_final.Association)
with_arrow = list()
for i in samp:
left = str(i[0:(len(i)-number_of_assoications)])
left = re.sub('[\(\)\{\}<>\'''\,]', '', left)
right = i[(len(i)-number_of_assoications)]
rslt = left + " ==> "+right
with_arrow.append(rslt)
df_final.Association = with_arrow
df_final
```
| github_jupyter |
# Neural Transfer
## Input images
```
%matplotlib inline
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from PIL import Image
import matplotlib.pyplot as plt
import torchvision.transforms as transforms
import torchvision.models as models
import copy
np.random.seed(37)
torch.manual_seed(37)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
def get_device():
return torch.device('cuda' if torch.cuda.is_available() else 'cpu')
def get_image_size():
imsize = 512 if torch.cuda.is_available() else 128
return imsize
def get_loader():
image_size = get_image_size()
loader = transforms.Compose([
transforms.Resize((image_size, image_size)),
transforms.ToTensor()])
return loader
def get_unloader():
unloader = transforms.ToPILImage()
return unloader
def image_loader(image_name):
device = get_device()
image = Image.open(image_name)
# fake batch dimension required to fit network's input dimensions
loader = get_loader()
image = loader(image).unsqueeze(0)
return image.to(device, torch.float)
def imshow(tensor, title=None):
image = tensor.cpu().clone() # we clone the tensor to not do changes on it
image = image.squeeze(0) # remove the fake batch dimension
unloader = get_unloader()
image = unloader(image)
plt.imshow(image)
if title is not None:
plt.title(title)
plt.pause(0.001)
style_img = image_loader("./styles/picasso-01.jpg")
content_img = image_loader("./styles/dancing.jpg")
input_img = content_img.clone()
assert style_img.size() == content_img.size(), \
f'size mismatch, style {style_img.size()}, content {content_img.size()}'
plt.ion()
plt.figure()
imshow(input_img, title='Input Image')
plt.figure()
imshow(style_img, title='Style Image')
plt.figure()
imshow(content_img, title='Content Image')
```
## Loss functions
### Content loss
```
class ContentLoss(nn.Module):
def __init__(self, target,):
super(ContentLoss, self).__init__()
# we 'detach' the target content from the tree used
# to dynamically compute the gradient: this is a stated value,
# not a variable. Otherwise the forward method of the criterion
# will throw an error.
self.target = target.detach()
def forward(self, input):
self.loss = F.mse_loss(input, self.target)
return input
```
### Style loss
```
def gram_matrix(input):
a, b, c, d = input.size() # a=batch size(=1)
# b=number of feature maps
# (c,d)=dimensions of a f. map (N=c*d)
features = input.view(a * b, c * d) # resise F_XL into \hat F_XL
G = torch.mm(features, features.t()) # compute the gram product
# we 'normalize' the values of the gram matrix
# by dividing by the number of element in each feature maps.
return G.div(a * b * c * d)
class StyleLoss(nn.Module):
def __init__(self, target_feature):
super(StyleLoss, self).__init__()
self.target = gram_matrix(target_feature).detach()
def forward(self, input):
G = gram_matrix(input)
self.loss = F.mse_loss(G, self.target)
return input
```
## Model
```
device = get_device()
cnn = models.vgg19(pretrained=True).features.to(device).eval()
```
## Normalization
```
class Normalization(nn.Module):
def __init__(self, mean, std):
super(Normalization, self).__init__()
# .view the mean and std to make them [C x 1 x 1] so that they can
# directly work with image Tensor of shape [B x C x H x W].
# B is batch size. C is number of channels. H is height and W is width.
self.mean = torch.tensor(mean).view(-1, 1, 1)
self.std = torch.tensor(std).view(-1, 1, 1)
def forward(self, img):
# normalize img
return (img - self.mean) / self.std
cnn_normalization_mean = torch.tensor([0.485, 0.456, 0.406]).to(device)
cnn_normalization_std = torch.tensor([0.229, 0.224, 0.225]).to(device)
```
## Loss
```
content_layers_default = ['conv_4']
style_layers_default = ['conv_1', 'conv_2', 'conv_3', 'conv_4', 'conv_5']
def get_style_model_and_losses(cnn, normalization_mean, normalization_std,
style_img, content_img,
content_layers=content_layers_default,
style_layers=style_layers_default):
cnn = copy.deepcopy(cnn)
# normalization module
normalization = Normalization(normalization_mean, normalization_std).to(device)
# just in order to have an iterable access to or list of content/syle
# losses
content_losses = []
style_losses = []
# assuming that cnn is a nn.Sequential, so we make a new nn.Sequential
# to put in modules that are supposed to be activated sequentially
model = nn.Sequential(normalization)
i = 0 # increment every time we see a conv
for layer in cnn.children():
if isinstance(layer, nn.Conv2d):
i += 1
name = 'conv_{}'.format(i)
elif isinstance(layer, nn.ReLU):
name = 'relu_{}'.format(i)
# The in-place version doesn't play very nicely with the ContentLoss
# and StyleLoss we insert below. So we replace with out-of-place
# ones here.
layer = nn.ReLU(inplace=False)
elif isinstance(layer, nn.MaxPool2d):
name = 'pool_{}'.format(i)
elif isinstance(layer, nn.BatchNorm2d):
name = 'bn_{}'.format(i)
else:
raise RuntimeError('Unrecognized layer: {}'.format(layer.__class__.__name__))
model.add_module(name, layer)
if name in content_layers:
# add content loss:
target = model(content_img).detach()
content_loss = ContentLoss(target)
model.add_module("content_loss_{}".format(i), content_loss)
content_losses.append(content_loss)
if name in style_layers:
# add style loss:
target_feature = model(style_img).detach()
style_loss = StyleLoss(target_feature)
model.add_module("style_loss_{}".format(i), style_loss)
style_losses.append(style_loss)
# now we trim off the layers after the last content and style losses
for i in range(len(model) - 1, -1, -1):
if isinstance(model[i], ContentLoss) or isinstance(model[i], StyleLoss):
break
model = model[:(i + 1)]
return model, style_losses, content_losses
```
## Optimizer
```
def get_input_optimizer(input_img):
# this line to show that input is a parameter that requires a gradient
optimizer = optim.LBFGS([input_img.requires_grad_()])
return optimizer
```
## Transfer
```
import warnings
from collections import namedtuple
RESULTS = namedtuple('RESULTS', 'run style content')
results = []
def run_style_transfer(cnn, normalization_mean, normalization_std,
content_img, style_img, input_img, num_steps=600,
style_weight=1000000, content_weight=1):
model, style_losses, content_losses = get_style_model_and_losses(cnn,
normalization_mean, normalization_std, style_img, content_img)
optimizer = get_input_optimizer(input_img)
run = [0]
while run[0] <= num_steps:
def closure():
# correct the values of updated input image
input_img.data.clamp_(0, 1)
optimizer.zero_grad()
model(input_img)
style_score = 0
content_score = 0
for sl in style_losses:
style_score += sl.loss
for cl in content_losses:
content_score += cl.loss
style_score *= style_weight
content_score *= content_weight
loss = style_score + content_score
loss.backward()
run[0] += 1
results.append(RESULTS(run[0], style_score.item(), content_score.item()))
if run[0] % 10 == 0:
s_score = style_score.item()
c_score = content_score.item()
print(f'[{run[0]}/{num_steps}] Style Loss {s_score:.4f}, Content Loss {c_score}')
return style_score + content_score
optimizer.step(closure)
# a last correction...
input_img.data.clamp_(0, 1)
return input_img
with warnings.catch_warnings():
warnings.simplefilter('ignore')
output = run_style_transfer(cnn, cnn_normalization_mean, cnn_normalization_std,
content_img, style_img, input_img)
```
## Results
```
x = [r.run for r in results]
y1 = [r.style for r in results]
y2 = [r.content for r in results]
fig, ax1 = plt.subplots(figsize=(10, 5))
color = 'tab:red'
ax1.plot(x, y1, color=color)
ax1.set_ylabel('Style Loss', color=color)
ax1.tick_params(axis='y', labelcolor=color)
color = 'tab:blue'
ax2 = ax1.twinx()
ax2.plot(x, y2, color=color)
ax2.set_ylabel('Content Loss', color=color)
ax2.tick_params(axis='y', labelcolor=color)
```
## Visualize
```
plt.figure()
imshow(output, title='Output Image')
# sphinx_gallery_thumbnail_number = 4
plt.ioff()
plt.show()
```
| github_jupyter |
# Lab: Working with a real world data-set using SQL and Python
## Introduction
This notebook shows how to work with a real world dataset using SQL and Python. In this lab you will:
1. Understand the dataset for Chicago Public School level performance
1. Store the dataset in an Db2 database on IBM Cloud instance
1. Retrieve metadata about tables and columns and query data from mixed case columns
1. Solve example problems to practice your SQL skills including using built-in database functions
## Chicago Public Schools - Progress Report Cards (2011-2012)
The city of Chicago released a dataset showing all school level performance data used to create School Report Cards for the 2011-2012 school year. The dataset is available from the Chicago Data Portal: https://data.cityofchicago.org/Education/Chicago-Public-Schools-Progress-Report-Cards-2011-/9xs2-f89t
This dataset includes a large number of metrics. Start by familiarizing yourself with the types of metrics in the database: https://data.cityofchicago.org/api/assets/AAD41A13-BE8A-4E67-B1F5-86E711E09D5F?download=true
__NOTE__: Do not download the dataset directly from City of Chicago portal. Instead download a more database friendly version from the link below.
Now download a static copy of this database and review some of its contents:
https://ibm.box.com/shared/static/0g7kbanvn5l2gt2qu38ukooatnjqyuys.csv
### Store the dataset in a Table
In many cases the dataset to be analyzed is available as a .CSV (comma separated values) file, perhaps on the internet. To analyze the data using SQL, it first needs to be stored in the database.
While it is easier to read the dataset into a Pandas dataframe and then PERSIST it into the database as we saw in the previous lab, it results in mapping to default datatypes which may not be optimal for SQL querying. For example a long textual field may map to a CLOB instead of a VARCHAR.
Therefore, __it is highly recommended to manually load the table using the database console LOAD tool, as indicated in Week 2 Lab 1 Part II__. The only difference with that lab is that in Step 5 of the instructions you will need to click on create "(+) New Table" and specify the name of the table you want to create and then click "Next".
##### Now open the Db2 console, open the LOAD tool, Select / Drag the .CSV file for the CHICAGO PUBLIC SCHOOLS dataset and load the dataset into a new table called __SCHOOLS__.
<a href="https://cognitiveclass.ai"><img src = "https://ibm.box.com/shared/static/uc4xjh1uxcc78ks1i18v668simioz4es.jpg"></a>
### Connect to the database
Let us now load the ipython-sql extension and establish a connection with the database
```
%load_ext sql
# Enter the connection string for your Db2 on Cloud database instance below
# %sql ibm_db_sa://my-username:my-password@my-hostname:my-port/my-db-name
%sql ibm_db_sa://
```
### Query the database system catalog to retrieve table metadata
#### You can verify that the table creation was successful by retrieving the list of all tables in your schema and checking whether the SCHOOLS table was created
```
# type in your query to retrieve list of all tables in the database for your db2 schema (username)
#In Db2 the system catalog table called SYSCAT.TABLES contains the table metadata
%sql SELECT * from SYSCAT.TABLES where TABNAME = 'SCHOOLS'
#OR
%sql select TABSCHEMA, TABNAME, CREATE_TIME from SYSCAT.TABLES where TABSCHEMA='YOUR-DB2-USERNAME'
#OR
%sql select TABSCHEMA, TABNAME, CREATE_TIME from SYSCAT.TABLES \
where TABSCHEMA not in ('SYSIBM', 'SYSCAT', 'SYSSTAT', 'SYSIBMADM', 'SYSTOOLS', 'SYSPUBLIC')
```
### Query the database system catalog to retrieve column metadata
#### The SCHOOLS table contains a large number of columns. How many columns does this table have?
```
#In Db2 the system catalog table called SYSCAT.COLUMNS contains the column metadata
%sql SELECT COUNT(*) FROM SYSCAT.COLUMNS WHERE TABNAME = 'SCHOOLS'
#Correct answer: 78
```
Now retrieve the the list of columns in SCHOOLS table and their column type (datatype) and length.
```
%sql SELECT COLNAME, TYPENAME, LENGTH FROM SYSCAT.COLUMNS WHERE TABNAME = 'SCHOOLS'
#OR
%sql SELECT DISTINCT(NAME), COLTYPE, LENGTH FROM SYSIBM.SYSCOLUMNS WHERE TABNAME = 'SCHOOLS'
```
### Questions
1. Is the column name for the "SCHOOL ID" attribute in upper or mixed case?
1. What is the name of "Community Area Name" column in your table? Does it have spaces?
1. Are there any columns in whose names the spaces and paranthesis (round brackets) have been replaced by the underscore character "_"?
## Problems
### Problem 1
##### How many Elementary Schools are in the dataset?
```
%sql select count(*) from SCHOOLS where "Elementary, Middle, or High School" = 'ES'
#Correct answer: 462
```
### Problem 2
##### What is the highest Safety Score?
```
%sql SELECT MAX("Safety_Score") AS MAX_SAFETY FROM SCHOOLS
#Correct answer: 99
```
### Problem 3
##### Which schools have highest Safety Score?
```
%sql SELECT NAME_OF_SCHOOL FROM SCHOOLS WHERE "Safety_Score" = 99
#OR
%sql SELECT NAME_OF_SCHOOL FROM SCHOOLS WHERE "Safety_Score" = (SELECT MAX("Safety_Score") FROM SCHOOLS)
```
### Problem 4
##### What are the top 10 schools with the highest "Average Student Attendance"?
```
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS ORDER BY Average_Student_Attendance DESC LIMIT 10
```
### Problem 5
#### Retrieve the list of 5 Schools with the lowest Average Student Attendance sorted in ascending order based on attendance
```
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS ORDER BY Average_Student_Attendance LIMIT 5
```
### Problem 6
#### Now remove the '%' sign from the above result set for Average Student Attendance column
```
%sql SELECT NAME_OF_SCHOOL, REPLACE(Average_Student_Attendance, '%', '') FROM SCHOOLS ORDER BY Average_Student_Attendance LIMIT 5
```
### Problem 7
#### Which Schools have Average Student Attendance lower than 70%?
```
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS WHERE CAST(REPLACE(Average_Student_Attendance, '%', '') AS DOUBLE) < 70
#OR
%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS WHERE DECIMAL(REPLACE(Average_Student_Attendance, '%', '')) < 70 ORDER BY Average_Student_Attendance
```
### Problem 8
#### Get the total College Enrollment for each Community Area
```
%sql SELECT COMMUNITY_AREA_NAME, SUM(COLLEGE_ENROLLMENT) AS TOTAL_ENROLLMENT FROM SCHOOLS GROUP BY COMMUNITY_AREA_NAME
```
### Problem 9
##### Get the 5 Community Areas with the least total College Enrollment sorted in ascending order
```
%sql SELECT COMMUNITY_AREA_NAME, SUM(COLLEGE_ENROLLMENT) AS TOTAL_ENROLLMENT FROM SCHOOLS GROUP BY COMMUNITY_AREA_NAME ORDER BY TOTAL_ENROLLMENT LIMIT 5
```
## Summary
#### In this lab you learned how to work with a real word dataset using SQL and Python. You learned how to query columns with spaces or special characters in their names and with mixed case names. You also used built in database functions and practiced how to sort, limit, and order result sets.
Copyright © 2018 [cognitiveclass.ai](cognitiveclass.ai?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).
| github_jupyter |
# 검색
wihle loop 를 이용한 선형 검색
```
from typing import Any,List
def linear_search_while(lst:List, value:Any) -> int:
i = 0
while i != len(lst) and lst[i] != value:
i += 1
if i == len(lst):
return -1
else:
return 1
l = [1,2,3,4,5,6,7,8,9]
linear_search_while(l,9)
def linear_search_for(lst:List, value:Any) -> int:
for i in lst:
if lst[i] == value:
return 1
return -1
l = [1,2,3,4,5,6,7,8,9]
linear_search_for(l,9)
def linear_search_sentinal(lst:List, value:Any) -> int:
lst.append(value)
i=0
while lst[i] != value:
i += 1
lst.pop()
if i == len(lst):
return -1
else:
return 1
l = [1,2,3,4,5,6,7,8,9]
linear_search_sentinal(l,9)
import time
from typing import Callable, Any
def time_it(search: Callable[[list,Any],Any],L:list,v:Any):
t1 = time.perf_counter()
search(L,v)
t2 = time.perf_counter()
return (t2-t1) *1000.0
l = [1,2,3,4,5,6,7,8,9]
time_it(linear_search_while,l,5)
```
## 이진 검색
반절씩 줄여나가며 탐색하는 방법
```
def binary_search(lst:list,value:Any) -> int:
i = 0
j = len(lst)-1
while i != j+1:
m = (i+j)//2
if lst[m]<v:
i = m+1
else:
j=m-1
if 0<= i< len(lst) and lst[i]==i:
return i
else :
return -1
if __name__ == '__main__':
import doctest
doctest.testmod()
```
## Selection sort - 선택정렬
정렬되지 않은 부분 전체를 순회하며 가장 작은 값을 찾아 정렬된 부분 우측에 위치시킨다. 이것을 모든 값이 정렬될 때까지 반복한다. n길이의 선형 자료형을 n번 반복하게 되므로 n^2
```
def selection_sort(l:list):
for i in range(len(l)):
idx = l.index(min(l[i:]),i)
dummy = l[i]
l[i] = l[idx]
l[idx] = dummy
return l
l = [7,16,3,25,2,6,1,7,3]
print(selection_sort(l))
```
## Insertion sort - 삽입정렬
전체를 순회하며 현재 값이 정렬된 부분에서 올바른 위치에 삽입하는 방식.
```
# 기 정렬된 영역에 L[:b+1] 내 올바른 위치에 L[b]를 삽입
def insert(L: list, b: int) -> None:
i = b
while i != 0 and L[i - 1] >= L[b]:
i = i - 1
value = L[b]
del L[b]
L.insert(i, value)
def insertion_sort(L: list) -> None:
i = 0
while i != len(L):
insert(L, i)
i = i + 1
L = [ 3, 4, 6, -1, 2, 5 ]
print(L)
insertion_sort(L)
print(L)
```
## Merge sort - 병합정렬
```
# 2개의 리스트를 하나의 정렬된 리스트로 반환
def merge(L1: list, L2: list) -> list:
newL = []
i1 = 0
i2 = 0
# [ 1, 1, 2, 3, 4, 5, 6, 7 ]
# [ 1, 3, 4, 6 ] [ 1, 2, 5, 7 ]
# i1
# i2
while i1 != len(L1) and i2 != len(L2):
if L1[i1] <= L2[i2]:
newL.append(L1[i1])
i1 += 1
else:
newL.append(L2[i2])
i2 += 1
newL.extend(L1[i1:])
newL.extend(L2[i2:])
return newL
def merge_sort(L: list) -> None: # [ 1, 3, 4, 6, 1, 2, 5, 7 ]
workspace = []
for i in range(len(L)):
workspace.append([L[i]]) # [ [1], [3], [4], [6], [1], [2], [5], [7] ]
i = 0
while i < len(workspace) - 1:
L1 = workspace[i] # [ [1], [3], [4], [6], [1], [2], [5], [7], [1,3],[4,6],[1,2],[5,7], [1,3,4,6],[1,2,5,7],[1,1,2,3,4,5,6,7] ]
L2 = workspace[i + 1]
newL = merge(L1, L2)
workspace.append(newL)
i += 2
if len(workspace) != 0:
L[:] = workspace[-1][:]
import time, random
def built_in(L: list) -> None:
L.sort()
def print_times(L: list) -> None:
print(len(L), end='\t')
for func in (selection_sort, insertion_sort, merge_sort, built_in):
if func in (selection_sort, insertion_sort, merge_sort) and len(L) > 10000:
continue
L_copy = L[:]
t1 = time.perf_counter()
func(L_copy)
t2 = time.perf_counter()
print("{0:7.1f}".format((t2 - t1) * 1000.0), end="\t")
print()
for list_size in [ 10, 1000, 2000, 3000, 4000, 5000, 10000 ]:
L = list(range(list_size))
random.shuffle(L)
print_times(L)
```
# 객체지향 프로그래밍
```isinstance(object,class)``` 해당 객체가 클래스에 해당하는지 아닌지를 반환.
```
from typing import List,Any
class Book:
def num_authors(self) -> int:
return len(self.authors)
def __init__(self,title:str,authors:List[str],publisher:str,isbn:str,price:float) : # 생성자.
self.title = title
self.authors = authors[:] # [:] 를 적지 않고 직접 넘겨주면 참조형식이기 때문에 외부에서 값이 바뀌면 해당 값도 바뀜. 때문에 새로 만들어서 복사하는 방법을 채택.
self.publisher = publisher
self.isbn = isbn
self.price = price
def print_authors(self) -> None:
for authors in self.authors:
print(authors)
def __str__(self) -> str:
return 'Title : {}\nAuthors : {}'.format(self.title,self.authors)
def __eq__(self,other:Any) -> bool:
if isinstance(other,Book):
return True if self.isbn == other.isbn else False
return False
book = Book('My book',['aaa','bbb','ccc'],'한빛출판사','123-456-789','300000.0')
book.print_authors()
print(book.num_authors())
print(book)
newBook = Book('My book',['aaa','bbb','ccc'],'한빛출판사','123-456-789','300000.0')
print(book==newBook)
```
레퍼런스 타입을 넘겨줄때 값을 참조하는 형식이 아닌 값을 직접 받는 형식으로 취하게 해야 한다.
캡슐화 : 데이터와 그 데이터를 사용하는 코드를 한곳에 넣고 정확히 어떻게 동작하는ㄴ지 상세한 내용은 숨기는 것
다형성 : 하나 이상의 형태를 갖는 것. 어떤 변수를 포함하는 표현식이 변수가 참조하는 객체의 타입에 따라 서로 다른 일을 하는 것
상속 : 새로운 클래스는 부모 클래스(object 클래스 또는 사용자 정의 속성을 상속)
```
class Member:
def __init__(self,name:str,address:str,email:str):
self.name = name
self.address = address
self.email = email
class Faculty(Member):
def __init__(self,name:str,address:str,email:str,faculty_num:str):
super().__init__(name,address,email)
self.faculty_number = faculty_num
self.courses_teaching = []
class Atom:
'''번호, 기호, 좌표(X, Y, Z)를 갖는 원자'''
def __init__(self, num: int, sym: str, x: float, y: float, z: float) -> None:
self.num = num
self.sym = sym
self.center = (x, y, z)
def __str__(self) -> str:
'''(SYMBOL, X, Y, Z) 형식의 문자열을 반환'''
return '({}, {}, {}, {}'.format(self.sym, self.center[0], self.center[1], self.center[2])
def translate(self, x: float, y: float, z: float) -> None:
self.center = (self.center[0] + x, self.center[1] + y, self.center[2] + z)
class Molecule:
''' 이름과 원자 리스트를 갖는 분자 '''
def __init__(self, name: str) -> None:
self.name = name
self.atoms = []
def add(self, a: Atom) -> None:
self.atoms.append(a)
def __str__(self) -> str:
'''(NAME, (ATOM1, ATOM2, ...)) 형식의 문자열을 반환'''
atom_list = ''
for a in self.atoms:
atom_list = atom_list + str(a) + ', '
atom_list = atom_list[:-2] # 마지막에 추가된 ', ' 문자를 제거
return '({}, ({}))'.format(self.name, atom_list)
def translate(self, x: float, y: float, z: float) -> None:
for a in self.atoms:
a.translate(x, y, z)
ammonia = Molecule("AMMONIA")
ammonia.add(Atom(1, "N", 0.257, -0.363, 0.0))
ammonia.add(Atom(2, "H", 0.257, 0.727, 0.0))
ammonia.add(Atom(3, "H", 0.771, -0.727, 0.890))
ammonia.add(Atom(4, "H", 0.771, -0.727, -0.890))
ammonia.translate(0, 0, 0.2)
#assert ammonia.atoms[0].center[0] == 0.257
#assert ammonia.atoms[0].center[1] == -0.363
assert ammonia.atoms[0].center[2] == 0.2
print(ammonia)
```
| github_jupyter |
<center>
<img src="https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/Logos/organization_logo/organization_logo.png" width="300" alt="cognitiveclass.ai logo" />
</center>
# Access DB2 on Cloud using Python
Estimated time needed: **15** minutes
## Objectives
After completing this lab you will be able to:
- Create a table
- Insert data into the table
- Query data from the table
- Retrieve the result set into a pandas dataframe
- Close the database connection
**Notice:** Please follow the instructions given in the first Lab of this course to Create a database service instance of Db2 on Cloud.
## Task 1: Import the `ibm_db` Python library
The `ibm_db` [API ](https://pypi.python.org/pypi/ibm_db?cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ) provides a variety of useful Python functions for accessing and manipulating data in an IBM® data server database, including functions for connecting to a database, preparing and issuing SQL statements, fetching rows from result sets, calling stored procedures, committing and rolling back transactions, handling errors, and retrieving metadata.
We import the ibm_db library into our Python Application
```
import ibm_db
```
When the command above completes, the `ibm_db` library is loaded in your notebook.
## Task 2: Identify the database connection credentials
Connecting to dashDB or DB2 database requires the following information:
- Driver Name
- Database name
- Host DNS name or IP address
- Host port
- Connection protocol
- User ID
- User Password
**Notice:** To obtain credentials please refer to the instructions given in the first Lab of this course
Now enter your database credentials below
Replace the placeholder values in angular brackets <> below with your actual database credentials
e.g. replace "database" with "BLUDB"
```
#Replace the placeholder values with the actuals for your Db2 Service Credentials
dsn_driver = "{IBM DB2 ODBC DRIVER}"
dsn_database = "database" # e.g. "BLUDB"
dsn_hostname = "hostname" # e.g.: "dashdb-txn-sbox-yp-dal09-04.services.dal.bluemix.net"
dsn_port = "port" # e.g. "50000"
dsn_protocol = "protocol" # i.e. "TCPIP"
dsn_uid = "username" # e.g. "abc12345"
dsn_pwd = "password" # e.g. "7dBZ3wWt9XN6$o0J"
```
## Task 3: Create the database connection
Ibm_db API uses the IBM Data Server Driver for ODBC and CLI APIs to connect to IBM DB2 and Informix.
Create the database connection
```
#Create database connection
#DO NOT MODIFY THIS CELL. Just RUN it with Shift + Enter
dsn = (
"DRIVER={0};"
"DATABASE={1};"
"HOSTNAME={2};"
"PORT={3};"
"PROTOCOL={4};"
"UID={5};"
"PWD={6};").format(dsn_driver, dsn_database, dsn_hostname, dsn_port, dsn_protocol, dsn_uid, dsn_pwd)
try:
conn = ibm_db.connect(dsn, "", "")
print ("Connected to database: ", dsn_database, "as user: ", dsn_uid, "on host: ", dsn_hostname)
except:
print ("Unable to connect: ", ibm_db.conn_errormsg() )
```
## Task 4: Create a table in the database
In this step we will create a table in the database with following details:
<img src = "https://ibm.box.com/shared/static/ztd2cn4xkdoj5erlk4hhng39kbp63s1h.jpg" align="center">
```
#Lets first drop the table INSTRUCTOR in case it exists from a previous attempt
dropQuery = "drop table INSTRUCTOR"
#Now execute the drop statment
dropStmt = ibm_db.exec_immediate(conn, dropQuery)
```
## Dont worry if you get this error:
If you see an exception/error similar to the following, indicating that INSTRUCTOR is an undefined name, that's okay. It just implies that the INSTRUCTOR table does not exist in the table - which would be the case if you had not created it previously.
Exception: [IBM][CLI Driver][DB2/LINUXX8664] SQL0204N "ABC12345.INSTRUCTOR" is an undefined name. SQLSTATE=42704 SQLCODE=-204
```
#Construct the Create Table DDL statement
createQuery = "create table INSTRUCTOR(ID INTEGER PRIMARY KEY NOT NULL, FNAME VARCHAR(20), LNAME VARCHAR(20), CITY VARCHAR(20), CCODE CHAR(2))"
#Execute the statement
createStmt = ibm_db.exec_immediate(conn,createQuery)
```
<details><summary>Click here for the solution</summary>
```python
createQuery = "create table INSTRUCTOR(ID INTEGER PRIMARY KEY NOT NULL, FNAME VARCHAR(20), LNAME VARCHAR(20), CITY VARCHAR(20), CCODE CHAR(2))"
createStmt = ibm_db.exec_immediate(conn,createQuery)
```
</details>
## Task 5: Insert data into the table
In this step we will insert some rows of data into the table.
The INSTRUCTOR table we created in the previous step contains 3 rows of data:
<img src="https://ibm.box.com/shared/static/j5yjassxefrjknivfpekj7698dqe4d8i.jpg" align="center">
We will start by inserting just the first row of data, i.e. for instructor Rav Ahuja
```
#Construct the query
insertQuery = "insert into INSTRUCTOR values (1, 'Rav', 'Ahuja', 'TORONTO', 'CA')"
#execute the insert statement
insertStmt = ibm_db.exec_immediate(conn, insertQuery)
```
<details><summary>Click here for the solution</summary>
```python
insertQuery = "insert into INSTRUCTOR values (1, 'Rav', 'Ahuja', 'TORONTO', 'CA')"
insertStmt = ibm_db.exec_immediate(conn, insertQuery)
```
</details>
Now use a single query to insert the remaining two rows of data
```
#Inerts the remaining two rows of data
insertQuery2 = "insert into INSTRUCTOR values (2, 'Raul', 'Chong', 'Markham', 'CA'), (3, 'Hima', 'Vasudevan', 'Chicago', 'US')"
#execute the statement
insertStmt2 = ibm_db.exec_immediate(conn, insertQuery2)
```
<details><summary>Click here for the solution</summary>
```python
insertQuery2 = "insert into INSTRUCTOR values (2, 'Raul', 'Chong', 'Markham', 'CA'), (3, 'Hima', 'Vasudevan', 'Chicago', 'US')"
insertStmt2 = ibm_db.exec_immediate(conn, insertQuery2)
```
</details>
## Task 6: Query data in the table
In this step we will retrieve data we inserted into the INSTRUCTOR table.
```
#Construct the query that retrieves all rows from the INSTRUCTOR table
selectQuery = "select * from INSTRUCTOR"
#Execute the statement
selectStmt = ibm_db.exec_immediate(conn, selectQuery)
#Fetch the Dictionary (for the first row only)
ibm_db.fetch_both(selectStmt)
```
<details><summary>Click here for the solution</summary>
```python
#Construct the query that retrieves all rows from the INSTRUCTOR table
selectQuery = "select * from INSTRUCTOR"
#Execute the statement
selectStmt = ibm_db.exec_immediate(conn, selectQuery)
#Fetch the Dictionary (for the first row only)
ibm_db.fetch_both(selectStmt)
```
</details>
```
#Fetch the rest of the rows and print the ID and FNAME for those rows
while ibm_db.fetch_row(selectStmt) != False:
print (" ID:", ibm_db.result(selectStmt, 0), " FNAME:", ibm_db.result(selectStmt, "FNAME"))
```
<details><summary>Click here for the solution</summary>
```python
#Fetch the rest of the rows and print the ID and FNAME for those rows
while ibm_db.fetch_row(selectStmt) != False:
print (" ID:", ibm_db.result(selectStmt, 0), " FNAME:", ibm_db.result(selectStmt, "FNAME"))
```
</details>
Bonus: now write and execute an update statement that changes the Rav's CITY to MOOSETOWN
```
updateQuery = "update INSTRUCTOR set CITY='MOOSETOWN' where FNAME='Rav'"
updateStmt = ibm_db.exec_immediate(conn, updateQuery))
```
<details><summary>Click here for the solution</summary>
```python
updateQuery = "update INSTRUCTOR set CITY='MOOSETOWN' where FNAME='Rav'"
updateStmt = ibm_db.exec_immediate(conn, updateQuery))
```
</details>
## Task 7: Retrieve data into Pandas
In this step we will retrieve the contents of the INSTRUCTOR table into a Pandas dataframe
```
import pandas
import ibm_db_dbi
#connection for pandas
pconn = ibm_db_dbi.Connection(conn)
#query statement to retrieve all rows in INSTRUCTOR table
selectQuery = "select * from INSTRUCTOR"
#retrieve the query results into a pandas dataframe
pdf = pandas.read_sql(selectQuery, pconn)
#print just the LNAME for first row in the pandas data frame
pdf.LNAME[0]
#print the entire data frame
pdf
```
Once the data is in a Pandas dataframe, you can do the typical pandas operations on it.
For example you can use the shape method to see how many rows and columns are in the dataframe
```
pdf.shape
```
## Task 8: Close the Connection
We free all resources by closing the connection. Remember that it is always important to close connections so that we can avoid unused connections taking up resources.
```
ibm_db.close(conn)
```
## Summary
In this tutorial you established a connection to a database instance of DB2 Warehouse on Cloud from a Python notebook using ibm_db API. Then created a table and insert a few rows of data into it. Then queried the data. You also retrieved the data into a pandas dataframe.
## Author
<a href="https://www.linkedin.com/in/ravahuja/" target="_blank">Rav Ahuja</a>
## Change Log
| Date (YYYY-MM-DD) | Version | Changed By | Change Description |
| ----------------- | ------- | ---------- | ---------------------------------- |
| 2020-08-28 | 2.0 | Lavanya | Moved lab to course repo in GitLab |
<hr>
## <h3 align="center"> © IBM Corporation 2020. All rights reserved. <h3/>
| github_jupyter |
# Building a Machine Translation System with Forte
## Overview
This tutorial will walk you through the steps to build a machine translation system with Forte. Forte allows users to breaks down complex problems into composable pipelines and enables inter-operations across tasks through a unified data format. With Forte, it's easy to compose a customized machine translation management system that is able to handle practical problems like new feature requests.
In this tutorial, you will learn:
* How to read data from source
* How to create a simple NLP pipeline
* How to maintain and store the input data
* How to process data in pipeline
* How to perform sentence segmentation
* How to annotate and query the data
* How to translate the input text with a pre-trained model
* How to manage multiple data objects
* How to handle new practical requests
* How to handle structures like HTML data
* How to select a single data object for processing
* How to replace the translation model with remote translation services
* How to save and load the pipeline
Run the following command to install all the required dependencies for this tutorial:
```
!pip install forte==0.2.0 forte.nltk transformers==4.16.2 torch==1.7.0 requests sentencepiece
```
## Start with the Reader
### Overview
* **How to read data from source**
* **How to create a simple pipeline**
* How to maintain and store the input data
* How to process data in pipeline
* How to handle new practical requests
In this section, you will learn
* What is a reader and why we need it
* How to compose a simple pipeline with a pre-built reader
```
from forte import Pipeline
from forte.data.readers import TerminalReader
pipeline: Pipeline = Pipeline()
```
All pipelines need a reader to read and parse input data. To make our pipeline read queries from the user’s command-line terminal, use the `TerminalReader` class provided by Forte. `TerminalReader` transforms the user’s query into a DataPack object, which is a unified data format for NLP that makes it easy to connect different NLP tools together as Forte Processors.
```
pipeline.set_reader(TerminalReader())
```
To run the pipeline consisting of the single `TerminalReader`, call `process_dataset` which will return an iterator of DataPack objects. The second line in the following code snippet retrieves the first user query from the TerminalReader.
```
pipeline.initialize()
datapack = next(pipeline.process_dataset())
print(datapack.text)
```
### DataPack
#### Overview
* **How to read data from source**
* How to create a simple pipeline
* **How to maintain and store the input data**
* How to process data in pipeline
* How to handle new practical requests
In this section, you will learn
* What is a DataPack object and why we need it
Forte helps demystify data lineage and increase the traceability of how data flows along the pipeline and how features are generated to interface data to model. Similar to a cargo ship that loads and transports goods from one port to another, a data pack carries information when passing each module and updates the ontology states along the way.

#### DataPack and Multi-Modality
DataPack not only supports text data but also audio and image data.

## Add a pre-built Forte processor to the pipeline
### Overview
* How to read data from source
* **How to process data in pipeline**
* **How to perform sentence segmentation**
* How to annotate and query the data
* How to translate the input text with a pre-trained model
* How to manage multiple data objects
* How to handle new practical requests
In this section, you will learn
* What is a processor and why we need it
* How to add a pre-built processor to the pipeline
A Forte Processor takes DataPacks as inputs, processes them, and stores its outputs in DataPacks. The processors we are going to use in this section are all PackProcessors, which expect exactly one DataPack as input and store its outputs back into the same DataPack. The following two lines of code shows how a pre-built processor `NLTKSentenceSegmenter` is added to our pipeline.
```
from fortex.nltk.nltk_processors import NLTKSentenceSegmenter
pipeline.add(NLTKSentenceSegmenter())
```
When we run the pipeline, the `NLTKSentenceSegmenter` processor will split the user query into sentences and store them back to the DataPack created by TerminalReader. The code snippet below shows how to get all the sentences from the first query.

```
from ft.onto.base_ontology import Sentence
pipeline.initialize()
for sent in next(pipeline.process_dataset()).get(Sentence):
print(sent.text)
```
### Ontology
#### Overview
* How to read data from source
* **How to process data in pipeline**
* How to perform sentence segmentation
* **How to annotate and query the data**
* How to translate the input text with a pre-trained model
* How to manage multiple data objects
* How to handle new practical requests
In this section, you will learn
* What is the ontology system and why we need it
* How to write a customized ontology and how to use it
`Sentence` is a pre-defined ontology provided by Forte and it is used by `NLTKSentenceSegmenter` to annotate each sentence in text. Forte is built on top of an Ontology system, which defines the relations between NLP annotations, for example, the relation between words and documents, or between two words. This is the core for Forte. The ontology can be specified via a JSON format. And tools are provided to convert the ontology into production code (Python).
We can also define customized ontologies:
```
from dataclasses import dataclass
from forte.data.ontology.top import Annotation
from typing import Optional
@dataclass
class Article(Annotation):
language: Optional[str]
def __init__(self, pack, begin: int, end: int):
super().__init__(pack, begin, end)
self.language: Optional[str] = None
```
Below is a simple example showing how we can query sentences through the new ontology we just create:
```
from forte.data import DataPack
sentences = [
"Do you want to get better at making delicious BBQ?",
"You will have the opportunity, put this on your calendar now.",
"Thursday, September 22nd join World Class BBQ Champion, Tony Balay from Lonestar Smoke Rangers."
]
datapack: DataPack = DataPack()
# Add sentences to the DataPack and annotate them
for sentence in sentences:
datapack.set_text(datapack.text + sentence)
datapack.add_entry(
Sentence(datapack, len(datapack.text) - len(sentence), len(datapack.text))
)
# Annotate the whole text with Article
article: Article = Article(datapack, 0, len(datapack.text))
article.language = "en"
datapack.add_entry(article)
for article in datapack.get(Article):
print(f"Article (language - {article.language}):")
for sentence in article.get(Sentence):
print(sentence.text)
```
In our previous example, we have the following ontologies inheritance. Sentence and Article both inherit from Annotation which is used to represent text data. In Article, we have `langauge` field to represent the text language.
Actually, we not only supports text ontology but also audio, image and link which represent relationships between two entries.
* `Annotation` is inherited by all text entries which usually has a span to retrieve partial text from the full text.
* `Article`, as shown in our previous example, inherits annotation and contains `language` field to differentiate English and Germany. In the single DataPack example, English article has a span of English text in the DataPack. Likewise, Germany article has a span of Germany text in the DataPack.
* `Sentence` in our example is used to break down article, and we pass sentences into MT pipeline.
* `AudioAnnotation` is inherited by all audio entries which usually has an audio span to retrieve partial audio from the full audio.
* `Recording` is an example subclass of `AudioAnnotation`, and it has extra `recording_class` field denoting the classes the audio belongs to.
* `ImageAnnotation` is inherited by all image entries which usually has payload index pointing to a loaded image array.
* `BoundingBox` is an example subclass of `ImageAnnotation`. As the picture shows, it has more inheritance relationships than other ontology classes due to the nature of CV objects. The advantage of forte ontology is that it supports complex inheritance, and users can inherit from existing ontology and add new ontology features for their needs.
* `Link` is inherited by all link-like entries which has parent and child.
* `RelationLink` is an example subclass of `Link`, and it has a class attribute specifying the relation type.
## Create a Machine Translation Processor
### Overview
* How to read data from source
* **How to process data in pipeline**
* How to perform sentence segmentation
* How to annotate and query the data
* **How to translate the input text with a pre-trained model**
* How to manage multiple data objects
* How to handle new practical requests
In this section, you will learn
* The basics of machine translation process
* How to wrap a pre-trained machine translation model into a Forte processor
Translation converts a sequence of text from one language to another. In this tutorial we will use `Huggingface` Transformer model to translate input data, which consists of several steps including subword tokenization, input embedding, model inference, decoding, etc.
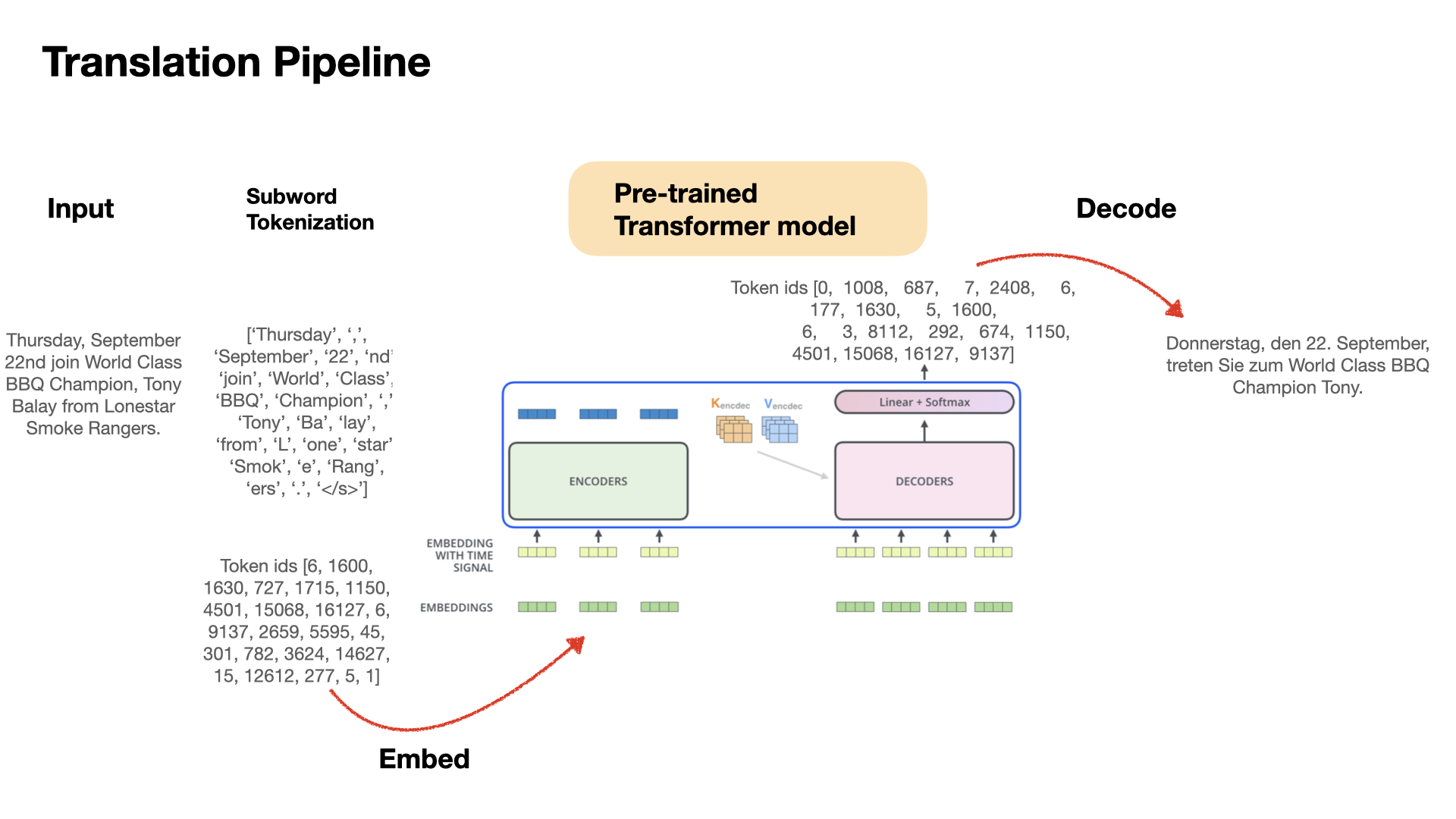
In Forte, we have a generic class `PackProcessor` that wraps model and inference-related components and behaviors to process `DataPack`. Therefore, we need to create a class that inherits the generic method from `PackProcessor`. Then we have a class definition
`class MachineTranslationProcessor(PackProcessor)`.
```
from forte.data import DataPack
from forte.data.readers import StringReader
from forte.processors.base import PackProcessor
from transformers import T5Tokenizer, T5ForConditionalGeneration
class MachineTranslationProcessor(PackProcessor):
"""
Translate the input text and output to a file.
"""
def initialize(self, resources, configs):
super().initialize(resources, configs)
# Initialize the tokenizer and model
model_name: str = self.configs.pretrained_model
self.tokenizer = T5Tokenizer.from_pretrained(model_name)
self.model = T5ForConditionalGeneration.from_pretrained(model_name)
self.task_prefix = "translate English to German: "
self.tokenizer.padding_side = "left"
self.tokenizer.pad_token = self.tokenizer.eos_token
def _process(self, input_pack: DataPack):
# en2de machine translation
inputs = self.tokenizer([
self.task_prefix + sentence.text
for sentence in input_pack.get(Sentence)
], return_tensors="pt", padding=True)
output_sequences = self.model.generate(
input_ids=inputs["input_ids"],
attention_mask=inputs["attention_mask"],
do_sample=False,
)
output = ''.join(self.tokenizer.batch_decode(
output_sequences, skip_special_tokens=True
))
src_article: Article = Article(input_pack, 0, len(input_pack.text))
src_article.language = "en"
input_pack.set_text(input_pack.text + '\n\n' + output)
tgt_article: Article = Article(input_pack, len(input_pack.text) - len(output), len(input_pack.text))
tgt_article.language = "de"
@classmethod
def default_configs(cls):
return {
"pretrained_model": "t5-small"
}
```
* Initialization of needed components:
* Users need to consider initializing all needed NLP components for the inference task such as tokenizer and model.
* Users also need to specify all configuration in `configs`, a dictionary-like object that specifies configurations of all components such as model name.
* MT operations on datapack
* After the initialization, we already have the needed NLP components. We need to consider several MT behaviors based on Forte DataPack.
* Pre-process text data
* retrieve text data from datapack (given that it already reads data from the data source).
* since T5 has a better performance given a task prompt, we also want to include the prompt in our data.
* Tokenization that transforms input text into sequences of tokens and token ids.
* Generate output sequences from model.
* Decode output token ids into sentences using the tokenizer.
The generic method to process `DataPack` is `_process(self, input_pack: DataPack)`. It should tokenize the input text, use the model class to make an inference, decode the output token ids, and finally writes the output to a target file.
Now we can add it into the pipeline and run the machine translation task.
```
input_string: str = ' '.join(sentences)
pipeline: Pipeline = Pipeline[DataPack]()
pipeline.set_reader(StringReader())
pipeline.add(NLTKSentenceSegmenter())
pipeline.add(MachineTranslationProcessor())
pipeline.initialize()
for datapack in pipeline.process_dataset([input_string]):
for article in datapack.get(Article):
print([f"\nArticle (language - {article.language}): {article.text}"])
```
#### Ontology in DataPack
Here we provide an illustration so that users can better understand the internal storage of DataPack. As we can see, text data, sentence and articles, are stored as span in `Annotations`. Their text data can be easily and efficiently retrieved by their spans.

## A better way to store source and target text: MultiPack
### Overview
* How to read data from source
* **How to process data in pipeline**
* How to perform sentence segmentation
* How to annotate and query the data
* How to translate the input text with a pre-trained model
* **How to manage multiple data objects**
* How to handle new practical requests
In this section, you will learn
* What is a MultiPack and why we need it
* How to use a Multipack
The above step outputs a DataPack which is good for holding data about one specific piece of text. A complicated pipeline like the one we are building now may need multiple DataPacks to be passed along the pipeline and this is where MultiPack can help. MultiPack manages a set of DataPacks that can be indexed by their names.
`MultiPackBoxer` is a simple Forte processor that converts a DataPack into a MultiPack by making it the only DataPack in there. A name can be specified via the config. We use it to wrap DataPack that contains source sentence.

```
from forte.data import MultiPack
from forte.processors.base import MultiPackProcessor
from forte.data.caster import MultiPackBoxer
class MachineTranslationMPProcessor(MultiPackProcessor):
"""
Translate the input text and output to a file.
"""
def initialize(self, resources, configs):
super().initialize(resources, configs)
# Initialize the tokenizer and model
model_name: str = self.configs.pretrained_model
self.tokenizer = T5Tokenizer.from_pretrained(model_name)
self.model = T5ForConditionalGeneration.from_pretrained(model_name)
self.task_prefix = "translate English to German: "
self.tokenizer.padding_side = "left"
self.tokenizer.pad_token = self.tokenizer.eos_token
def _process(self, input_pack: MultiPack):
source_pack: DataPack = input_pack.get_pack("source")
target_pack: DataPack = input_pack.add_pack("target")
# en2de machine translation
inputs = self.tokenizer([
self.task_prefix + sentence.text
for sentence in source_pack.get(Sentence)
], return_tensors="pt", padding=True)
output_sequences = self.model.generate(
input_ids=inputs["input_ids"],
attention_mask=inputs["attention_mask"],
do_sample=False,
)
# Annotate the source article
src_article: Article = Article(source_pack, 0, len(source_pack.text))
src_article.language = "en"
# Annotate each sentence
for output in self.tokenizer.batch_decode(
output_sequences, skip_special_tokens=True
):
target_pack.set_text(target_pack.text + output)
text_length: int = len(target_pack.text)
Sentence(target_pack, text_length - len(output), text_length)
# Annotate the target article
tgt_article: Article = Article(target_pack, 0, len(target_pack.text))
tgt_article.language = "de"
@classmethod
def default_configs(cls):
return {
"pretrained_model": "t5-small",
}
```
Then `MachineTranslationMPProcessor` writes the output sentence into a target DataPack.
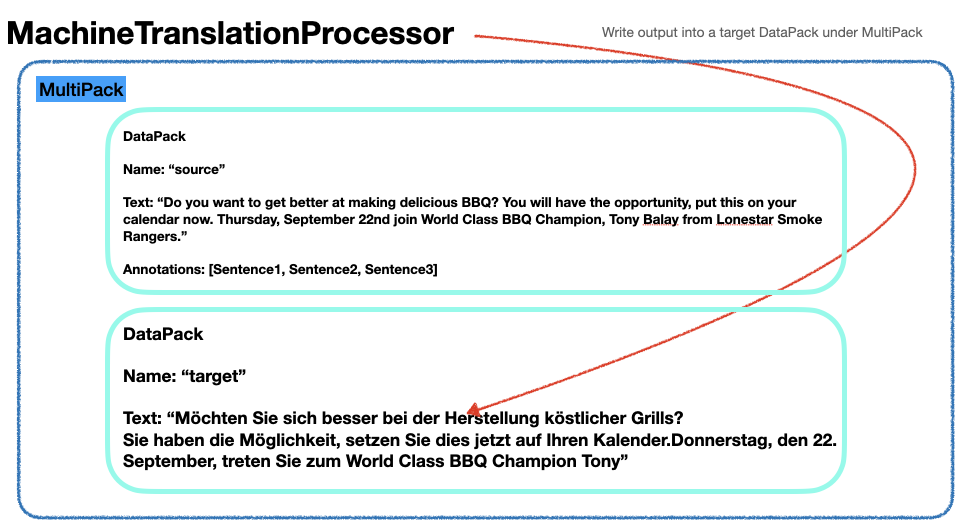
Now let's try to create a new pipeline that utilizes `MultiPack` to manage text in different languages.
```
nlp: Pipeline = Pipeline[DataPack]()
nlp.set_reader(StringReader())
nlp.add(NLTKSentenceSegmenter())
nlp.add(MultiPackBoxer(), config={"pack_name": "source"})
nlp.add(MachineTranslationMPProcessor(), config={
"pretrained_model": "t5-small"
})
nlp.initialize()
for multipack in nlp.process_dataset([input_string]):
for pack_name in ("source", "target"):
for article in multipack.get_pack(pack_name).get(Article):
print(f"\nArticle (language - {article.language}): ")
for sentence in article.get(Sentence):
print(sentence.text)
```
#### Ontology in MultiPack
For comparison, here is an illustration of the internal storage of MultiPack. We can see that MultiPack wraps one source DataPack and one target DataPack. Article spans are based on two separate DataPack text.
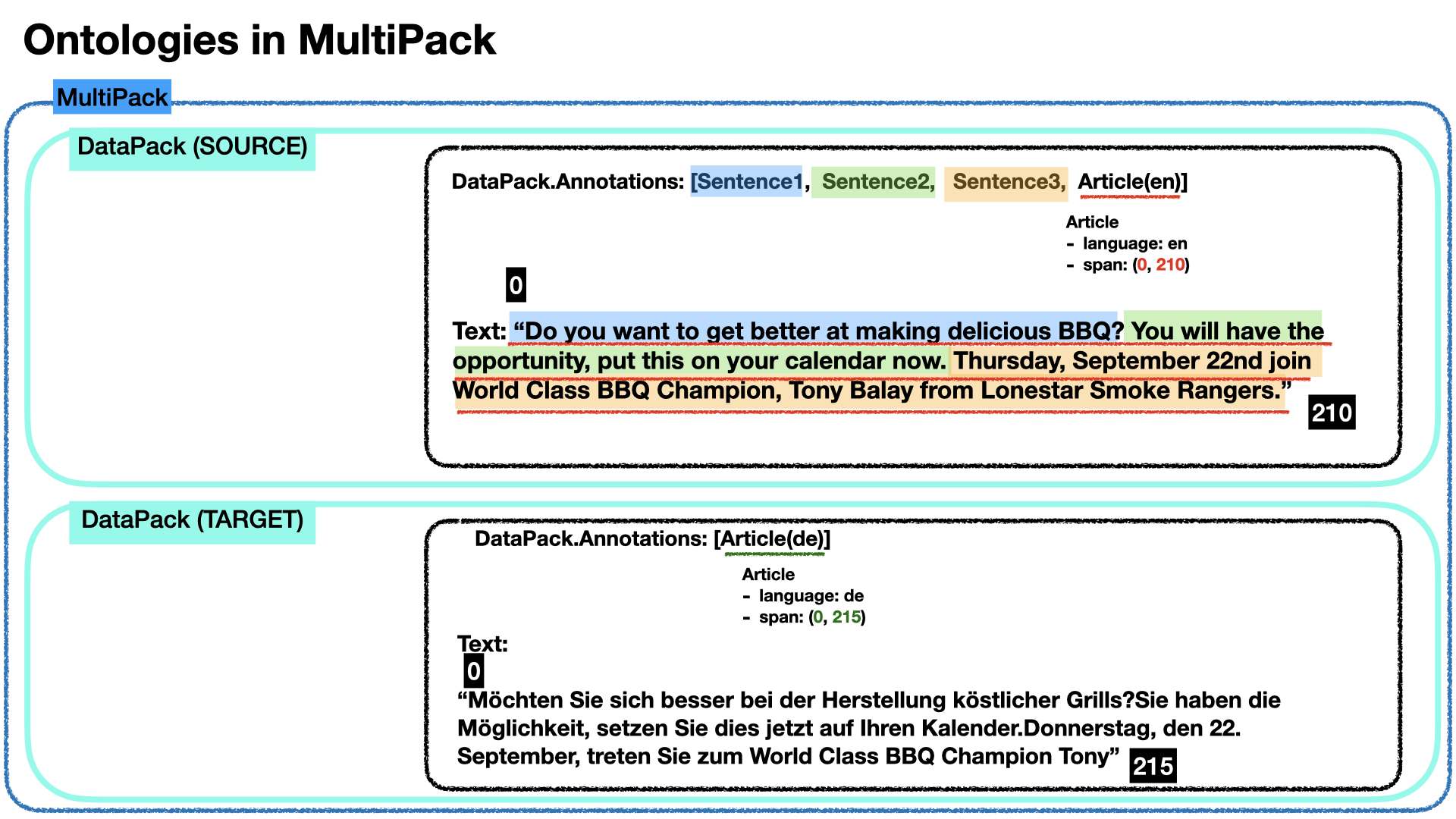
## New Requirement: Handle HTML data
### Overview
* How to read data from source
* How to process data in pipeline
* **How to handle new practical requests**
* **How to handle structures like HTML data**
* **How to select a single data object for processing**
* How to replace the translation model with remote translation services
* How to save and load the pipeline
In this section, you will learn
* How to build a translation management system
* How to preserve the structure like HTML in machine translation
* How to select a specific DataPack from MultiPack for processing
In the previous step, the input string is just a simple paragraph made up of several sentences. However, in many cases, we might need to handle data with structural information, such HTML or XML. When the input is a string of raw HTML data, the machine translation pipeline above may not work as expected:
```
html_input: str = """
<!DOCTYPE html>
<html>
<head><title>Beginners BBQ Class.</title></head>
<body>
<p>Do you want to get better at making delicious BBQ? You will have the opportunity, put this on your calendar now. Thursday, September 22nd join World Class BBQ Champion, Tony Balay from Lonestar Smoke Rangers.</p>
</body>
</html>
"""
nlp.initialize()
for multipack in nlp.process_dataset([html_input]):
print("Source Text: " + multipack.get_pack("source").text)
print("\nTarget Text: " + multipack.get_pack("target").text)
```
We can see that the original HTML structure is broken in the translated output.
## How to preserve HTML tags/structures
In order to handle structured data like HTML, we will need to update our current design of pipeline. Luckily, Forte pipelines are highly modular, we can simply insert two new processors without updating the previous pipeline.
We first need a HTML cleaner to parse all the HTML tags from input string. Picture below shows the effect of tag remover.

After the translation is finished, we will also need to recover the HTML structure from the unstructured translation output. Picture below shows replace one source sentence with one target sentence given the target sentence is ready.

```
from forte.data import NameMatchSelector
from forte.data.readers.html_reader import ForteHTMLParser
class HTMLTagCleaner(MultiPackProcessor):
def initialize(self, resources, configs):
super().initialize(resources, configs)
self._parser = ForteHTMLParser()
def _process(self, input_pack: MultiPack):
raw_pack: DataPack = input_pack.get_pack("raw")
source_pack: DataPack = input_pack.add_pack("source")
self._parser.feed(raw_pack.text)
cleaned_text: str = raw_pack.text
for span, _ in self._parser.spans:
cleaned_text = cleaned_text.replace(
raw_pack.text[span.begin:span.end], ''
)
source_pack.set_text(cleaned_text)
class HTMLTagRecovery(MultiPackProcessor):
def _process(self, input_pack: MultiPack):
raw_pack: DataPack = input_pack.get_pack("raw")
source_pack: DataPack = input_pack.get_pack("source")
target_pack: DataPack = input_pack.get_pack("target")
result_pack: DataPack = input_pack.add_pack("result")
result_text: str = raw_pack.text
for sent_src, sent_tgt in zip(source_pack.get(Sentence), target_pack.get(Sentence)):
result_text = result_text.replace(sent_src.text, sent_tgt.text)
result_pack.set_text(result_text)
```
Now we are able to create a translation management system by inserting the two processors introduced above into our previous machine translation pipeline.
```
# Pipeline with HTML handling
pipeline: Pipeline = Pipeline[DataPack]()
pipeline.set_reader(StringReader())
pipeline.add(MultiPackBoxer(), config={"pack_name": "raw"})
pipeline.add(HTMLTagCleaner())
pipeline.add(
NLTKSentenceSegmenter(),
selector=NameMatchSelector(),
selector_config={"select_name": "source"}
)
pipeline.add(MachineTranslationMPProcessor(), config={
"pretrained_model": "t5-small"
})
pipeline.add(HTMLTagRecovery())
pipeline.initialize()
for multipack in pipeline.process_dataset([html_input]):
print(multipack.get_pack("raw").text)
print(multipack.get_pack("result").text)
```
### Selector
In the code snippet above, we utilize a `NameMatchSelector` to select one specific DataPack from the MultiPack based on its reference name `select_name`. This allows `NLTKSentenceSegmenter` to process only the specified DataPack.
## Replace our MT model with online translation API
### Overview
* How to read data from source
* How to process data in pipeline
* **How to handle new practical requests**
* How to handle structures like HTML data
* **How to replace the translation model with remote translation services**
* How to save and load the pipeline
In this section, you will learn
* How to use a different translation service
Forte also allows us to update the translation model and integrate it seamlessly to the original pipeline. For example, if we want to offload the translation task to an online service, all we need to do is to update the translation processor. There is no need to change other components in the pipeline.
```
# You can get your own API key by following the instructions in https://docs.microsoft.com/en-us/azure/cognitive-services/translator/
api_key = input("Enter your API key here:")
import requests
import uuid
class OnlineMachineTranslationMPProcessor(MultiPackProcessor):
"""
Translate the input text and output to a file use online translator api.
"""
def initialize(self, resources, configs):
super().initialize(resources, configs)
self.url = configs.endpoint + configs.path
self.from_lang = configs.from_lang
self.to_lang = configs.to_lang
self.subscription_key = configs.subscription_key
self.subscription_region = configs.subscription_region
def _process(self, input_pack: MultiPack):
source_pack: DataPack = input_pack.get_pack("source")
target_pack: DataPack = input_pack.add_pack("target")
params = {
'api-version': '3.0',
'from': 'en',
'to': ['de']
}
# Build request
headers = {
'Ocp-Apim-Subscription-Key': self.subscription_key,
'Ocp-Apim-Subscription-Region': self.subscription_region,
'Content-type': 'application/json',
'X-ClientTraceId': str(uuid.uuid4())
}
# You can pass more than one object in body.
body = [{
'text': source_pack.text
}]
request = requests.post(self.url, params=params, headers=headers, json=body)
result = request.json()
target_pack.set_text("".join(
[trans['text'] for trans in result[0]["translations"]]
)
)
@classmethod
def default_configs(cls):
return {
"from_lang" : 'en',
"to_lang": 'de',
"endpoint" : 'https://api.cognitive.microsofttranslator.com/',
"path" : '/translate',
"subscription_key": None,
"subscription_region" : "westus2",
'X-ClientTraceId': str(uuid.uuid4())
}
nlp: Pipeline = Pipeline[DataPack]()
nlp.set_reader(StringReader())
nlp.add(NLTKSentenceSegmenter())
nlp.add(MultiPackBoxer(), config={"pack_name": "source"})
nlp.add(OnlineMachineTranslationMPProcessor(), config={
"from_lang" : 'en',
"to_lang": 'de',
"endpoint" : 'https://api.cognitive.microsofttranslator.com/',
"path" : '/translate',
"subscription_key": api_key,
"subscription_region" : "westus2",
'X-ClientTraceId': str(uuid.uuid4())
})
nlp.initialize()
for multipack in nlp.process_dataset([input_string]):
print("Source Text: " + multipack.get_pack("source").text)
print("\nTarget Text: " + multipack.get_pack("target").text)
```
## Save the whole pipeline with save()
### Overview
* How to read data from source
* How to process data in pipeline
* **How to handle new practical requests**
* How to handle structures like HTML data
* How to replace the translation model with remote translation services
* **How to save and load the pipeline**
In this section, you will learn
* How to export and import a Forte pipeline
Forte also allow us to save the pipeline into disk. It serializes the whole pipeline and generates an intermediate representation, which can be loaded later maybe on a different machine.
```
import os
save_path: str = os.path.join(os.path.dirname(os.path.abspath('')), "pipeline.yml")
nlp.save(save_path)
with open(save_path, 'r') as f:
print(f.read())
```
Now that the pipeline is saved, we can try to re-load the pipeline to see if it still functions as expect.
```
new_nlp: Pipeline = Pipeline()
new_nlp.init_from_config_path(save_path)
new_nlp.initialize()
for multipack in new_nlp.process_dataset([input_string]):
print("Source Text: " + multipack.get_pack("source").text)
print("\nTarget Text: " + multipack.get_pack("target").text)
```
| github_jupyter |
```
import folium
import branca
import geopandas
from folium.plugins import Search
print(folium.__version__)
```
Let's get some JSON data from the web - both a point layer and a polygon GeoJson dataset with some population data.
```
states = geopandas.read_file(
'https://rawcdn.githack.com/PublicaMundi/MappingAPI/master/data/geojson/us-states.json',
driver='GeoJSON'
)
cities = geopandas.read_file(
'https://d2ad6b4ur7yvpq.cloudfront.net/naturalearth-3.3.0/ne_50m_populated_places_simple.geojson',
driver='GeoJSON'
)
```
And take a look at what our data looks like:
```
states.describe()
```
Look how far the minimum and maximum values for the density are from the top and bottom quartile breakpoints! We have some outliers in our data that are well outside the meat of most of the distribution. Let's look into this to find the culprits within the sample.
```
states_sorted = states.sort_values(by='density', ascending=False)
states_sorted.head(5).append(states_sorted.tail(5))[['name','density']]
```
Looks like Washington D.C. and Alaska were the culprits on each end of the range. Washington was more dense than the next most dense state, New Jersey, than the least dense state, Alaska was from Wyoming, however. Washington D.C. has a has a relatively small land area for the amount of people that live there, so it makes sense that it's pretty dense. And Alaska has a lot of land area, but not much of it is habitable for humans.
<br><br>
However, we're looking at all of the states in the US to look at things on a more regional level. That high figure at the top of our range for Washington D.C. will really hinder the ability for us to differentiate between the other states, so let's account for that in the min and max values for our color scale, by getting the quantile values close to the end of the range. Anything higher or lower than those values will just fall into the 'highest' and 'lowest' bins for coloring.
```
min, max = states['density'].quantile([0.05,0.95]).apply(lambda x: round(x, 2))
mean = round(states['density'].mean(),2)
print(f"Min: {min}", f"Max: {max}", f"Mean: {mean}", sep="\n\n")
```
This looks better. Our min and max values for the colorscale are much closer to the mean value now. Let's run with these values, and make a colorscale. I'm just going to use a sequential light-to-dark color palette from the [ColorBrewer](http://colorbrewer2.org/#type=sequential&scheme=Purples&n=5).
```
colormap = branca.colormap.LinearColormap(
colors=['#f2f0f7','#cbc9e2','#9e9ac8','#756bb1','#54278f'],
index=states['density'].quantile([0.2,0.4,0.6,0.8]),
vmin=min,
vmax=max
)
colormap.caption="Population Density in the United States"
colormap
```
Let's narrow down these cities to United states cities, by using GeoPandas' spatial join functionality between two GeoDataFrame objects, using the Point 'within' Polygon functionality.
```
us_cities = geopandas.sjoin(cities, states, how='inner', op='within')
pop_ranked_cities = us_cities.sort_values(
by='pop_max',
ascending=False
)[
[
'nameascii',
'pop_max',
'geometry'
]
].iloc[:20]
```
Ok, now we have a new GeoDataFrame with our top 20 populated cities. Let's see the top 5.
```
pop_ranked_cities.head(5)
```
Alright, let's build a map!
```
m = folium.Map(location=[38,-97], zoom_start=4)
style_function = lambda x: {
'fillColor': colormap(x['properties']['density']),
'color': 'black',
'weight':2,
'fillOpacity':0.5
}
stategeo = folium.GeoJson(
states,
name='US States',
style_function=style_function,
tooltip=folium.GeoJsonTooltip(
fields=['name', 'density'],
aliases=['State', 'Density'],
localize=True
)
).add_to(m)
citygeo = folium.GeoJson(
pop_ranked_cities,
name='US Cities',
tooltip=folium.GeoJsonTooltip(
fields=['nameascii','pop_max'],
aliases=['','Population Max'],
localize=True)
).add_to(m)
statesearch = Search(
layer=stategeo,
geom_type='Polygon',
placeholder='Search for a US State',
collapsed=False,
search_label='name',
weight=3
).add_to(m)
citysearch = Search(
layer=citygeo,
geom_type='Point',
placeholder='Search for a US City',
collapsed=True,
search_label='nameascii'
).add_to(m)
folium.LayerControl().add_to(m)
colormap.add_to(m)
m
```
| github_jupyter |
# NumPy Tutorial: Data analysis with Python
[Source](https://www.dataquest.io/blog/numpy-tutorial-python/)
NumPy is a commonly used Python data analysis package. By using NumPy, you can speed up your workflow, and interface with other packages in the Python ecosystem, like scikit-learn, that use NumPy under the hood. NumPy was originally developed in the mid 2000s, and arose from an even older package called Numeric. This longevity means that almost every data analysis or machine learning package for Python leverages NumPy in some way.
In this tutorial, we'll walk through using NumPy to analyze data on wine quality. The data contains information on various attributes of wines, such as pH and fixed acidity, along with a quality score between 0 and 10 for each wine. The quality score is the average of at least 3 human taste testers. As we learn how to work with NumPy, we'll try to figure out more about the perceived quality of wine.
The wines we'll be analyzing are from the Minho region of Portugal.
The data was downloaded from the UCI Machine Learning Repository, and is available [here](https://archive.ics.uci.edu/ml/datasets/Wine+Quality). Here are the first few rows of the winequality-red.csv file, which we'll be using throughout this tutorial:
``` text
"fixed acidity";"volatile acidity";"citric acid";"residual sugar";"chlorides";"free sulfur dioxide";"total sulfur dioxide";"density";"pH";"sulphates";"alcohol";"quality"
7.4;0.7;0;1.9;0.076;11;34;0.9978;3.51;0.56;9.4;5
7.8;0.88;0;2.6;0.098;25;67;0.9968;3.2;0.68;9.8;5
```
The data is in what I'm going to call ssv (semicolon separated values) format -- each record is separated by a semicolon (;), and rows are separated by a new line. There are 1600 rows in the file, including a header row, and 12 columns.
Before we get started, a quick version note -- we'll be using Python 3.5. Our code examples will be done using Jupyter notebook.
If you want to jump right into a specific area, here are the topics:
* Creating an Array
* Reading Text Files
* Array Indexing
* N-Dimensional Arrays
* Data Types
* Array Math
* Array Methods
* Array Comparison and Filtering
* Reshaping and Combining Arrays
Lists Of Lists for CSV Data
Before using NumPy, we'll first try to work with the data using Python and the csv package. We can read in the file using the csv.reader object, which will allow us to read in and split up all the content from the ssv file.
In the below code, we:
* Import the csv library.
* Open the winequality-red.csv file.
* With the file open, create a new csv.reader object.
* Pass in the keyword argument delimiter=";" to make sure that the records are split up on the semicolon character instead of the default comma character.
* Call the list type to get all the rows from the file.
* Assign the result to wines.
```
import csv
with open("winequality-red.csv", 'r') as f:
wines = list(csv.reader(f, delimiter=";"))
# print(wines[:3])
headers = wines[0]
wines_only = wines[1:]
# print the headers
print(headers)
# print the 1st row of data
print(wines_only[0])
# print the 1st three rows of data
print(wines_only[:3])
```
The data has been read into a list of lists. Each inner list is a row from the ssv file. As you may have noticed, each item in the entire list of lists is represented as a string, which will make it harder to do computations.
As you can see from the table above, we've read in three rows, the first of which contains column headers. Each row after the header row represents a wine. The first element of each row is the fixed acidity, the second is the volatile acidity, and so on.
## Calculate Average Wine Quality
We can find the average quality of the wines. The below code will:
* Extract the last element from each row after the header row.
* Convert each extracted element to a float.
* Assign all the extracted elements to the list qualities.
* Divide the sum of all the elements in qualities by the total number of elements in qualities to the get the mean.
```
# calculate average wine quality with a loop
qualities = []
for row in wines[1:]:
qualities.append(float(row[-1]))
sum(qualities) / len(wines[1:])
# calculate average wine quality with a list comprehension
qualities = [float(row[-1]) for row in wines[1:]]
sum(qualities) / len(wines[1:])
```
Although we were able to do the calculation we wanted, the code is fairly complex, and it won't be fun to have to do something similar every time we want to compute a quantity. Luckily, we can use NumPy to make it easier to work with our data.
# Numpy 2-Dimensional Arrays
With NumPy, we work with multidimensional arrays. We'll dive into all of the possible types of multidimensional arrays later on, but for now, we'll focus on 2-dimensional arrays. A 2-dimensional array is also known as a matrix, and is something you should be familiar with. In fact, it's just a different way of thinking about a list of lists. A matrix has rows and columns. By specifying a row number and a column number, we're able to extract an element from a matrix.
If we picked the element at the first row and the second column, we'd get volatile acidity. If we picked the element in the third row and the second column, we'd get 0.88.
In a NumPy array, the number of dimensions is called the **rank**, and each dimension is called an **axis**. So
* the rows are the first axis
* the columns are the second axis
Now that you understand the basics of matrices, let's see how we can get from our list of lists to a NumPy array.
## Creating A NumPy Array
We can create a NumPy array using the numpy.array function. If we pass in a list of lists, it will automatically create a NumPy array with the same number of rows and columns. Because we want all of the elements in the array to be float elements for easy computation, we'll leave off the header row, which contains strings. One of the limitations of NumPy is that all the elements in an array have to be of the same type, so if we include the header row, all the elements in the array will be read in as strings. Because we want to be able to do computations like find the average quality of the wines, we need the elements to all be floats.
In the below code, we:
* Import the ```numpy``` package.
* Pass the ```list``` of lists wines into the array function, which converts it into a NumPy array.
* Exclude the header row with list slicing.
* Specify the keyword argument ```dtype``` to make sure each element is converted to a ```float```. We'll dive more into what the ```dtype``` is later on.
```
import numpy as np
np.set_printoptions(precision=2) # set the output print precision for readability
# create the numpy array skipping the headers
wines = np.array(wines[1:], dtype=np.float)
# If we display wines, we'll now get a NumPy array:
print(type(wines), wines)
# We can check the number of rows and columns in our data using the shape property of NumPy arrays:
wines.shape
```
## Alternative NumPy Array Creation Methods
There are a variety of methods that you can use to create NumPy arrays. It's useful to create an array with all zero elements in cases when you need an array of fixed size, but don't have any values for it yet. To start with, you can create an array where every element is zero. The below code will create an array with 3 rows and 4 columns, where every element is 0, using ```numpy.zeros```:
```
empty_array = np.zeros((3, 4))
empty_array
```
Creating arrays full of random numbers can be useful when you want to quickly test your code with sample arrays. You can also create an array where each element is a random number using ```numpy.random.rand```.
```
np.random.rand(2, 3)
```
### Using NumPy To Read In Files
It's possible to use NumPy to directly read ```csv``` or other files into arrays. We can do this using the ```numpy.genfromtxt``` function. We can use it to read in our initial data on red wines.
In the below code, we:
* Use the ``` genfromtxt ``` function to read in the ``` winequality-red.csv ``` file.
* Specify the keyword argument ``` delimiter=";" ``` so that the fields are parsed properly.
* Specify the keyword argument ``` skip_header=1 ``` so that the header row is skipped.
```
wines = np.genfromtxt("winequality-red.csv", delimiter=";", skip_header=1)
wines
```
Wines will end up looking the same as if we read it into a list then converted it to an array of ```floats```. NumPy will automatically pick a data type for the elements in an array based on their format.
## Indexing NumPy Arrays
We now know how to create arrays, but unless we can retrieve results from them, there isn't a lot we can do with NumPy. We can use array indexing to select individual elements, groups of elements, or entire rows and columns.
One important thing to keep in mind is that just like Python lists, NumPy is **zero-indexed**, meaning that:
* The index of the first row is 0
* The index of the first column is 0
* If we want to work with the fourth row, we'd use index 3
* If we want to work with the second row, we'd use index 1, and so on.
We'll again work with the wines array:
|||||||||||||
|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|
|7.4 |0.70 |0.00 |1.9 |0.076 |11 |34 |0.9978 |3.51 |0.56 |9.4 |5|
|7.8 |0.88 |0.00 |2.6 |0.098 |25 |67 |0.9968 |3.20 |0.68 |9.8 |5|
|7.8 |0.76 |0.04 |2.3 |0.092 |15 |54 |0.9970 |3.26 |0.65 |9.8 |5|
|11.2|0.28 |0.56 |1.9 |0.075 |17 |60 |0.9980 |3.16 |0.58 |9.8 |6|
|7.4 |0.70 |0.00 |1.9 |0.076 |11 |34 |0.9978 |3.51 |0.56 |9.4 |5|
Let's select the element at **row 3** and **column 4**.
We pass:
* 2 as the row index
* 3 as the column index.
This retrieves the value from the **third row** and **fourth column**
```
wines[2, 3]
wines[2][3]
```
Since we're working with a 2-dimensional array in NumPy we specify 2 indexes to retrieve an element.
* The first index is the row, or **axis 1**, index
* The second index is the column, or **axis 2**, index
Any element in wines can be retrieved using 2 indexes.
```
# rows 1, 2, 3 and column 4
wines[0:3, 3]
# all rows and column 3
wines[:, 2]
```
Just like with ```list``` slicing, it's possible to omit the 0 to just retrieve all the elements from the beginning up to element 3:
```
# rows 1, 2, 3 and column 4
wines[:3, 3]
```
We can select an entire column by specifying that we want all the elements, from the first to the last. We specify this by just using the colon ```:```, with no starting or ending indices. The below code will select the entire fourth column:
```
# all rows and column 4
wines[:, 3]
```
We selected an entire column above, but we can also extract an entire row:
```
# row 4 and all columns
wines[3, :]
```
If we take our indexing to the extreme, we can select the entire array using two colons to select all the rows and columns in wines. This is a great party trick, but doesn't have a lot of good applications:
```
wines[:, :]
```
## Assigning Values To NumPy Arrays
We can also use indexing to assign values to certain elements in arrays. We can do this by assigning directly to the indexed value:
```
# assign the value of 10 to the 2nd row and 6th column
print('Before', wines[1, 4:7])
wines[1, 5] = 10
print('After', wines[1, 4:7])
```
We can do the same for slices. To overwrite an entire column, we can do this:
```
# Overwrites all the values in the eleventh column with 50.
print('Before', wines[:, 9:12])
wines[:, 10] = 50
print('After', wines[:, 9:12])
```
## 1-Dimensional NumPy Arrays
So far, we've worked with 2-dimensional arrays, such as wines. However, NumPy is a package for working with multidimensional arrays.
One of the most common types of multidimensional arrays is the **1-dimensional array**, or **vector**. As you may have noticed above, when we sliced wines, we retrieved a 1-dimensional array.
* A 1-dimensional array only needs a single index to retrieve an element.
* Each row and column in a 2-dimensional array is a 1-dimensional array.
Just like a list of lists is analogous to a 2-dimensional array, a single list is analogous to a 1-dimensional array.
If we slice wines and only retrieve the third row, we get a 1-dimensional array:
```
third_wine = wines[3,:]
third_wine
```
We can retrieve individual elements from ```third_wine``` using a single index.
```
# display the second item in third_wine
third_wine[1]
```
Most NumPy functions that we've worked with, such as ```numpy.random.rand```, can be used with multidimensional arrays. Here's how we'd use ```numpy.random.rand``` to generate a random vector:
```
np.random.rand(3)
```
Previously, when we called ```np.random.rand```, we passed in a shape for a 2-dimensional array, so the result was a 2-dimensional array. This time, we passed in a shape for a single dimensional array. The shape specifies the number of dimensions, and the size of the array in each dimension.
A shape of ```(10,10)``` will be a 2-dimensional array with **10 rows** and **10 columns**. A shape of ```(10,)``` will be a **1-dimensional** array with **10 elements**.
Where NumPy gets more complex is when we start to deal with arrays that have more than 2 dimensions.
## N-Dimensional NumPy Arrays
This doesn't happen extremely often, but there are cases when you'll want to deal with arrays that have greater than 3 dimensions. One way to think of this is as a list of lists of lists. Let's say we want to store the monthly earnings of a store, but we want to be able to quickly lookup the results for a quarter, and for a year. The earnings for one year might look like this:
``` python
[500, 505, 490, 810, 450, 678, 234, 897, 430, 560, 1023, 640]
```
The store earned \$500 in January, \$505 in February, and so on. We can split up these earnings by quarter into a list of lists:
```
year_one = [
[500,505,490], # 1st quarter
[810,450,678], # 2nd quarter
[234,897,430], # 3rd quarter
[560,1023,640] # 4th quarter
]
```
We can retrieve the earnings from January by calling ``` year_one[0][0] ```. If we want the results for a whole quarter, we can call ``` year_one[0] ``` or ``` year_one[1] ```.
We now have a 2-dimensional array, or matrix. But what if we now want to add the results from another year? We have to add a third dimension:
```
earnings = [
[ # year 1
[500,505,490], # year 1, 1st quarter
[810,450,678], # year 1, 2nd quarter
[234,897,430], # year 1, 3rd quarter
[560,1023,640] # year 1, 4th quarter
],
[ # year =2
[600,605,490], # year 2, 1st quarter
[345,900,1000],# year 2, 2nd quarter
[780,730,710], # year 2, 3rd quarter
[670,540,324] # year 2, 4th quarter
]
]
```
We can retrieve the earnings from January of the first year by calling ``` earnings[0][0][0] ```.
We now need three indexes to retrieve a single element. A three-dimensional array in NumPy is much the same. In fact, we can convert earnings to an array and then get the earnings for January of the first year:
```
earnings = np.array(earnings)
# year 1, 1st quarter, 1st month (January)
earnings[0,0,0]
# year 2, 3rd quarter, 1st month (July)
earnings[1,2,0]
# we can also find the shape of the array
earnings.shape
```
Indexing and slicing work the exact same way with a 3-dimensional array, but now we have an extra axis to pass in. If we wanted to get the earnings for **January of all years**, we could do this:
```
# all years, 1st quarter, 1st month (January)
earnings[:,0,0]
```
If we wanted to get first quarter earnings from both years, we could do this:
```
# all years, 1st quarter, all months (January, February, March)
earnings[:,0,:]
```
Adding more dimensions can make it much easier to query your data if it's organized in a certain way. As we go from 3-dimensional arrays to 4-dimensional and larger arrays, the same properties apply, and they can be indexed and sliced in the same ways.
## NumPy Data Types
As we mentioned earlier, each NumPy array can store elements of a single data type. For example, wines contains only float values.
NumPy stores values using its own data types, **which are distinct from Python types** like ```float``` and ```str```.
This is because the core of NumPy is written in a programming language called ```C```, **which stores data differently than the Python data types**. NumPy data types map between Python and C, allowing us to use NumPy arrays without any conversion hitches.
You can find the data type of a NumPy array by accessing the dtype property:
```
wines.dtype
```
NumPy has several different data types, which mostly map to Python data types, like ```float```, and ```str```. You can find a full listing of NumPy data types [here](https://www.dataquest.io/blog/numpy-tutorial-python/), but here are a few important ones:
* ```float``` -- numeric floating point data.
* ```int``` -- integer data.
* ```string``` -- character data.
* ```object``` -- Python objects.
Data types additionally end with a suffix that indicates how many bits of memory they take up. So ```int32``` is a **32 bit integer data type**, and ```float64``` is a **64 bit float data type**.
### Converting Data Types
You can use the numpy.ndarray.astype method to convert an array to a different type. The method will actually **copy the array**, and **return a new array with the specified data type**.
For instance, we can convert wines to the ```int``` data type:
```
# convert wines to the int data type
wines.astype(int)
```
As you can see above, all of the items in the resulting array are integers. Note that we used the Python ```int``` type instead of a NumPy data type when converting wines. This is because several Python data types, including ```float```, ```int```, and ```string```, can be used with NumPy, and are automatically converted to NumPy data types.
We can check the name property of the ```dtype``` of the resulting array to see what data type NumPy mapped the resulting array to:
```
# convert to int
int_wines = wines.astype(int)
# check the data type
int_wines.dtype.name
```
The array has been converted to a **64-bit integer** data type. This allows for very long integer values, **but takes up more space in memory** than storing the values as 32-bit integers.
If you want more control over how the array is stored in memory, you can directly create NumPy dtype objects like ```numpy.int32```
```
np.int32
```
You can use these directly to convert between types:
```
# convert to a 64-bit integer
wines.astype(np.int64)
# convert to a 32-bit integer
wines.astype(np.int32)
# convert to a 16-bit integer
wines.astype(np.int16)
# convert to a 8-bit integer
wines.astype(np.int8)
```
## NumPy Array Operations
NumPy makes it simple to perform mathematical operations on arrays. This is one of the primary advantages of NumPy, and makes it quite easy to do computations.
### Single Array Math
If you do any of the basic mathematical operations ```/```, ```*```, ```-```, ```+```, ```^``` with an array and a value, it will apply the operation to each of the elements in the array.
Let's say we want to add 10 points to each quality score because we're feeling generous. Here's how we'd do that:
```
# add 10 points to the quality score
wines[:,-1] + 10
```
*Note: that the above operation won't change the wines array -- it will return a new 1-dimensional array where 10 has been added to each element in the quality column of wines.*
If we instead did ```+=```, we'd modify the array in place:
```
print('Before', wines[:,11])
# modify the data in place
wines[:,11] += 10
print('After', wines[:,11])
```
All the other operations work the same way. For example, if we want to multiply each of the quality score by 2, we could do it like this:
```
# multiply the quality score by 2
wines[:,11] * 2
```
### Multiple Array Math
It's also possible to do mathematical operations between arrays. This will apply the operation to pairs of elements. For example, if we add the quality column to itself, here's what we get:
```
# add the quality column to itself
wines[:,11] + wines[:,11]
```
Note that this is equivalent to ```wines[:,11] * 2``` -- this is because NumPy adds each pair of elements. The first element in the first array is added to the first element in the second array, the second to the second, and so on.
```
# add the quality column to itself
wines[:,11] * 2
```
We can also use this to multiply arrays. Let's say we want to pick a wine that maximizes alcohol content and quality. We'd multiply alcohol by quality, and select the wine with the highest score:
```
# multiply alcohol content by quality
alcohol_by_quality = wines[:,10] * wines[:,11]
print(alcohol_by_quality)
alcohol_by_quality.sort()
print(alcohol_by_quality, alcohol_by_quality[-1])
```
All of the common operations ```/```, ```*```, ```-```, ```+```, ```^``` will work between arrays.
## NumPy Array Methods
In addition to the common mathematical operations, NumPy also has several methods that you can use for more complex calculations on arrays. An example of this is the ```numpy.ndarray.sum``` method. This finds the sum of all the elements in an array by default:
```
# find the sum of all rows and the quality column
total = 0
for row in wines:
total += row[11]
print(total)
# find the sum of all rows and the quality column
wines[:,11].sum(axis=0)
# find the sum of the rows 1, 2, and 3 across all columns
totals = []
for i in range(3):
total = 0
for col in wines[i,:]:
total += col
totals.append(total)
print(totals)
# find the sum of the rows 1, 2, and 3 across all columns
wines[0:3,:].sum(axis=1)
```
We can pass the ```axis``` keyword argument into the sum method to find sums over an axis.
If we call sum across the wines matrix, and pass in ```axis=0```, we'll find the sums over the first axis of the array. This will give us the **sum of all the values in every column**.
This may seem backwards that the sums over the first axis would give us the sum of each column, but one way to think about this is that **the specified axis is the one "going away"**.
So if we specify ```axis=0```, we want the **rows to go away**, and we want to find **the sums for each of the remaining axes across each row**:
```
# sum each column for all rows
totals = [0] * len(wines[0])
for i, total in enumerate(totals):
for row_val in wines[:,i]:
total += row_val
totals[i] = total
print(totals)
# sum each column for all rows
wines.sum(axis=0)
```
We can verify that we did the sum correctly by checking the shape. The shape should be 12, corresponding to the number of columns:
```
wines.sum(axis=0).shape
```
If we pass in axis=1, we'll find the sums over the second axis of the array. This will give us the sum of each row:
```
# sum each row for all columns
totals = [0] * len(wines)
for i, total in enumerate(totals):
for col_val in wines[i,:]:
total += col_val
totals[i] = total
print(totals[0:3], '...', totals[-3:])
# sum each row for all columns
wines.sum(axis=1)
wines.sum(axis=1).shape
```
There are several other methods that behave like the sum method, including:
* ```numpy.ndarray.mean``` — finds the mean of an array.
* ```numpy.ndarray.std``` — finds the standard deviation of an array.
* ```numpy.ndarray.min``` — finds the minimum value in an array.
* ```numpy.ndarray.max``` — finds the maximum value in an array.
You can find a full list of array methods [here](http://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html).
## NumPy Array Comparisons
NumPy makes it possible to test to see if rows match certain values using mathematical comparison operations like ```<```, ```>```, ```>=```, ```<=```, and ```==```. For example, if we want to see which wines have a quality rating higher than 5, we can do this:
```
# return True for all rows in the Quality column that are greater than 5
wines[:,11] > 5
```
We get a Boolean array that tells us which of the wines have a quality rating greater than 5. We can do something similar with the other operators. For instance, we can see if any wines have a quality rating equal to 10:
```
# return True for all rows that have a Quality rating of 10
wines[:,11] == 10
```
### Subsetting
One of the powerful things we can do with a Boolean array and a NumPy array is select only certain rows or columns in the NumPy array. For example, the below code will only select rows in wines where the quality is over 7:
```
# create a boolean array for wines with quality greater than 15
high_quality = wines[:,11] > 15
print(len(high_quality), high_quality)
# use boolean indexing to find high quality wines
high_quality_wines = wines[high_quality,:]
print(len(high_quality_wines), high_quality_wines)
```
We select only the rows where ```high_quality``` contains a ```True``` value, and all of the columns. This subsetting makes it simple to filter arrays for certain criteria.
For example, we can look for wines with a lot of alcohol and high quality. In order to specify multiple conditions, we have to place each condition in **parentheses** ```(...)```, and separate conditions with an **ampersand** ```&```:
```
# create a boolean array for high alcohol content and high quality
high_alcohol_and_quality = (wines[:,11] > 7) & (wines[:,10] > 10)
print(high_alcohol_and_quality)
# use boolean indexing to select out the wines
wines[high_alcohol_and_quality,:]
```
We can combine subsetting and assignment to overwrite certain values in an array:
```
high_alcohol_and_quality = (wines[:,10] > 10) & (wines[:,11] > 7)
wines[high_alcohol_and_quality,10:] = 20
```
## Reshaping NumPy Arrays
We can change the shape of arrays while still preserving all of their elements. This often can make it easier to access array elements. The simplest reshaping is to flip the axes, so rows become columns, and vice versa. We can accomplish this with the ```numpy.transpose``` function:
```
np.transpose(wines).shape
```
We can use the ```numpy.ravel``` function to turn an array into a one-dimensional representation. It will essentially flatten an array into a long sequence of values:
```
wines.ravel()
```
Here's an example where we can see the ordering of ```numpy.ravel```:
```
array_one = np.array(
[
[1, 2, 3, 4],
[5, 6, 7, 8]
]
)
array_one.ravel()
```
Finally, we can use the numpy.reshape function to reshape an array to a certain shape we specify. The below code will turn the second row of wines into a 2-dimensional array with 2 rows and 6 columns:
```
# print the current shape of the 2nd row and all columns
wines[1,:].shape
# reshape the 2nd row to a 2 by 6 matrix
wines[1,:].reshape((2,6))
```
## Combining NumPy Arrays
With NumPy, it's very common to combine multiple arrays into a single unified array. We can use ```numpy.vstack``` to vertically stack multiple arrays.
Think of it like the second arrays's items being added as new rows to the first array. We can read in the ```winequality-white.csv``` dataset that contains information on the quality of white wines, then combine it with our existing dataset, wines, which contains information on red wines.
In the below code, we:
* Read in ```winequality-white.csv```.
* Display the shape of white_wines.
```
white_wines = np.genfromtxt("winequality-white.csv", delimiter=";", skip_header=1)
white_wines.shape
```
As you can see, we have attributes for 4898 wines. Now that we have the white wines data, we can combine all the wine data.
In the below code, we:
* Use the ```vstack``` function to combine wines and white_wines.
* Display the shape of the result.
```
all_wines = np.vstack((wines, white_wines))
all_wines.shape
```
As you can see, the result has 6497 rows, which is the sum of the number of rows in wines and the number of rows in red_wines.
If we want to combine arrays horizontally, where the number of rows stay constant, but the columns are joined, then we can use the ```numpy.hstack``` function. The arrays we combine need to have the same number of rows for this to work.
Finally, we can use ```numpy.concatenate``` as a general purpose version of ```hstack``` and ```vstack```. If we want to concatenate two arrays, we pass them into concatenate, then specify the axis keyword argument that we want to concatenate along.
* Concatenating along the first axis is similar to ```vstack```
* Concatenating along the second axis is similar to ```hstack```:
```
x = np.concatenate((wines, white_wines), axis=0)
print(x.shape, x)
```
## Broadcasting
Unless the arrays that you're operating on are the exact same size, it's not possible to do elementwise operations. In cases like this, NumPy performs broadcasting to try to match up elements. Essentially, broadcasting involves a few steps:
* The last dimension of each array is compared.
* If the dimension lengths are equal, or one of the dimensions is of length 1, then we keep going.
* If the dimension lengths aren't equal, and none of the dimensions have length 1, then there's an error.
* Continue checking dimensions until the shortest array is out of dimensions.
For example, the following two shapes are compatible:
``` python
A: (50,3)
B (3,)
```
This is because the length of the trailing dimension of array A is 3, and the length of the trailing dimension of array B is 3. They're equal, so that dimension is okay. Array B is then out of elements, so we're okay, and the arrays are compatible for mathematical operations.
The following two shapes are also compatible:
``` python
A: (1,2)
B (50,2)
```
The last dimension matches, and A is of length 1 in the first dimension.
These two arrays don't match:
``` python
A: (50,50)
B: (49,49)
```
The lengths of the dimensions aren't equal, and neither array has either dimension length equal to 1.
There's a detailed explanation of broadcasting [here](http://docs.scipy.org/doc/numpy/user/basics.broadcasting.html), but we'll go through a few examples to illustrate the principle:
```
wines * np.array([1,2])
```
The above example didn't work because the two arrays don't have a matching trailing dimension. Here's an example where the last dimension does match:
```
array_one = np.array(
[
[1,2],
[3,4]
]
)
array_two = np.array([4,5])
array_one + array_two
```
As you can see, array_two has been broadcasted across each row of array_one. Here's an example with our wines data:
```
rand_array = np.random.rand(12)
wines + rand_array
```
| github_jupyter |
```
%matplotlib inline
import numpy as np
import yt
```
This notebook shows how to use yt to make plots and examine FITS X-ray images and events files.
## Sloshing, Shocks, and Bubbles in Abell 2052
This example uses data provided by [Scott Randall](http://hea-www.cfa.harvard.edu/~srandall/), presented originally in [Blanton, E.L., Randall, S.W., Clarke, T.E., et al. 2011, ApJ, 737, 99](https://ui.adsabs.harvard.edu/abs/2011ApJ...737...99B). They consist of two files, a "flux map" in counts/s/pixel between 0.3 and 2 keV, and a spectroscopic temperature map in keV.
```
ds = yt.load(
"xray_fits/A2052_merged_0.3-2_match-core_tmap_bgecorr.fits",
auxiliary_files=["xray_fits/A2052_core_tmap_b1_m2000_.fits"],
)
```
Since the flux and projected temperature images are in two different files, we had to use one of them (in this case the "flux" file) as a master file, and pass in the "temperature" file with the `auxiliary_files` keyword to `load`.
Next, let's derive some new fields for the number of counts, the "pseudo-pressure", and the "pseudo-entropy":
```
def _counts(field, data):
exposure_time = data.get_field_parameter("exposure_time")
return data["fits", "flux"] * data["fits", "pixel"] * exposure_time
ds.add_field(
("gas", "counts"),
function=_counts,
sampling_type="cell",
units="counts",
take_log=False,
)
def _pp(field, data):
return np.sqrt(data["gas", "counts"]) * data["fits", "projected_temperature"]
ds.add_field(
("gas", "pseudo_pressure"),
function=_pp,
sampling_type="cell",
units="sqrt(counts)*keV",
take_log=False,
)
def _pe(field, data):
return data["fits", "projected_temperature"] * data["gas", "counts"] ** (-1.0 / 3.0)
ds.add_field(
("gas", "pseudo_entropy"),
function=_pe,
sampling_type="cell",
units="keV*(counts)**(-1/3)",
take_log=False,
)
```
Here, we're deriving a "counts" field from the "flux" field by passing it a `field_parameter` for the exposure time of the time and multiplying by the pixel scale. Second, we use the fact that the surface brightness is strongly dependent on density ($S_X \propto \rho^2$) to use the counts in each pixel as a "stand-in". Next, we'll grab the exposure time from the primary FITS header of the flux file and create a `YTQuantity` from it, to be used as a `field_parameter`:
```
exposure_time = ds.quan(ds.primary_header["exposure"], "s")
```
Now, we can make the `SlicePlot` object of the fields we want, passing in the `exposure_time` as a `field_parameter`. We'll also set the width of the image to 250 pixels.
```
slc = yt.SlicePlot(
ds,
"z",
[
("fits", "flux"),
("fits", "projected_temperature"),
("gas", "pseudo_pressure"),
("gas", "pseudo_entropy"),
],
origin="native",
field_parameters={"exposure_time": exposure_time},
)
slc.set_log(("fits", "flux"), True)
slc.set_log(("gas", "pseudo_pressure"), False)
slc.set_log(("gas", "pseudo_entropy"), False)
slc.set_width(250.0)
slc.show()
```
To add the celestial coordinates to the image, we can use `PlotWindowWCS`, if you have a recent version of AstroPy (>= 1.3) installed:
```
from yt.frontends.fits.misc import PlotWindowWCS
wcs_slc = PlotWindowWCS(slc)
wcs_slc.show()
```
We can make use of yt's facilities for profile plotting as well.
```
v, c = ds.find_max(("fits", "flux")) # Find the maximum flux and its center
my_sphere = ds.sphere(c, (100.0, "code_length")) # Radius of 150 pixels
my_sphere.set_field_parameter("exposure_time", exposure_time)
```
Such as a radial profile plot:
```
radial_profile = yt.ProfilePlot(
my_sphere,
"radius",
["counts", "pseudo_pressure", "pseudo_entropy"],
n_bins=30,
weight_field="ones",
)
radial_profile.set_log("counts", True)
radial_profile.set_log("pseudo_pressure", True)
radial_profile.set_log("pseudo_entropy", True)
radial_profile.set_xlim(3, 100.0)
radial_profile.show()
```
Or a phase plot:
```
phase_plot = yt.PhasePlot(
my_sphere, "pseudo_pressure", "pseudo_entropy", ["counts"], weight_field=None
)
phase_plot.show()
```
Finally, we can also take an existing [ds9](http://ds9.si.edu/site/Home.html) region and use it to create a "cut region", using `ds9_region` (the [pyregion](https://pyregion.readthedocs.io) package needs to be installed for this):
```
from yt.frontends.fits.misc import ds9_region
reg_file = [
"# Region file format: DS9 version 4.1\n",
"global color=green dashlist=8 3 width=3 include=1 source=1 fk5\n",
'circle(15:16:44.817,+7:01:19.62,34.6256")',
]
f = open("circle.reg", "w")
f.writelines(reg_file)
f.close()
circle_reg = ds9_region(
ds, "circle.reg", field_parameters={"exposure_time": exposure_time}
)
```
This region may now be used to compute derived quantities:
```
print(
circle_reg.quantities.weighted_average_quantity("projected_temperature", "counts")
)
```
Or used in projections:
```
prj = yt.ProjectionPlot(
ds,
"z",
[
("fits", "flux"),
("fits", "projected_temperature"),
("gas", "pseudo_pressure"),
("gas", "pseudo_entropy"),
],
origin="native",
field_parameters={"exposure_time": exposure_time},
data_source=circle_reg,
method="sum",
)
prj.set_log(("fits", "flux"), True)
prj.set_log(("gas", "pseudo_pressure"), False)
prj.set_log(("gas", "pseudo_entropy"), False)
prj.set_width(250.0)
prj.show()
```
## The Bullet Cluster
This example uses an events table file from a ~100 ks exposure of the "Bullet Cluster" from the [Chandra Data Archive](http://cxc.harvard.edu/cda/). In this case, the individual photon events are treated as particle fields in yt. However, you can make images of the object in different energy bands using the `setup_counts_fields` function.
```
from yt.frontends.fits.api import setup_counts_fields
```
`load` will handle the events file as FITS image files, and will set up a grid using the WCS information in the file. Optionally, the events may be reblocked to a new resolution. by setting the `"reblock"` parameter in the `parameters` dictionary in `load`. `"reblock"` must be a power of 2.
```
ds2 = yt.load("xray_fits/acisf05356N003_evt2.fits.gz", parameters={"reblock": 2})
```
`setup_counts_fields` will take a list of energy bounds (emin, emax) in keV and create a new field from each where the photons in that energy range will be deposited onto the image grid.
```
ebounds = [(0.1, 2.0), (2.0, 5.0)]
setup_counts_fields(ds2, ebounds)
```
The "x", "y", "energy", and "time" fields in the events table are loaded as particle fields. Each one has a name given by "event\_" plus the name of the field:
```
dd = ds2.all_data()
print(dd["io", "event_x"])
print(dd["io", "event_y"])
```
Now, we'll make a plot of the two counts fields we made, and pan and zoom to the bullet:
```
slc = yt.SlicePlot(
ds2, "z", [("gas", "counts_0.1-2.0"), ("gas", "counts_2.0-5.0")], origin="native"
)
slc.pan((100.0, 100.0))
slc.set_width(500.0)
slc.show()
```
The counts fields can take the field parameter `"sigma"` and use [AstroPy's convolution routines](https://astropy.readthedocs.io/en/latest/convolution/) to smooth the data with a Gaussian:
```
slc = yt.SlicePlot(
ds2,
"z",
[("gas", "counts_0.1-2.0"), ("gas", "counts_2.0-5.0")],
origin="native",
field_parameters={"sigma": 2.0},
) # This value is in pixel scale
slc.pan((100.0, 100.0))
slc.set_width(500.0)
slc.set_zlim(("gas", "counts_0.1-2.0"), 0.01, 100.0)
slc.set_zlim(("gas", "counts_2.0-5.0"), 0.01, 50.0)
slc.show()
```
| github_jupyter |
tobac example: Tracking deep convection based on OLR from geostationary satellite retrievals
==
This example notebook demonstrates the use of tobac to track isolated deep convective clouds based on outgoing longwave radiation (OLR) calculated based on a combination of two different channels of the GOES-13 imaging instrument.
The data used in this example is downloaded from "zenodo link" automatically as part of the notebooks (This only has to be done once for all the tobac example notebooks).
```
# Import libraries:
import iris
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import iris.plot as iplt
import iris.quickplot as qplt
import zipfile
from six.moves import urllib
from glob import glob
%matplotlib inline
# Import tobac itself:
import tobac
# Disable a few warnings:
import warnings
warnings.filterwarnings('ignore', category=UserWarning, append=True)
warnings.filterwarnings('ignore', category=RuntimeWarning, append=True)
warnings.filterwarnings('ignore', category=FutureWarning, append=True)
warnings.filterwarnings('ignore',category=pd.io.pytables.PerformanceWarning)
```
**Download example data:**
This has to be done only once for all tobac examples.
```
data_out='../'
# # Download the data: This only has to be done once for all tobac examples and can take a while
# file_path='https://zenodo.org/record/3195910/files/climate-processes/tobac_example_data-v1.0.1.zip'
# tempfile='temp.zip'
# print('start downloading data')
# request=urllib.request.urlretrieve(file_path,tempfile)
# print('start extracting data')
# zf = zipfile.ZipFile(tempfile)
# zf.extractall(data_out)
# print('example data saved in')
```
**Load data:**
```
data_file=os.path.join(data_out,'*','data','Example_input_OLR_satellite.nc')
data_file = glob(data_file)[0]
print(data_file)
# Load Data from downloaded file:
OLR=iris.load_cube(data_file,'OLR')
# Display information about the input data cube:
display(OLR)
#Set up directory to save output and plots:
savedir='Save'
if not os.path.exists(savedir):
os.makedirs(savedir)
plot_dir="Plot"
if not os.path.exists(plot_dir):
os.makedirs(plot_dir)
```
**Feature identification:**
Identify features based on OLR field and a set of threshold values
```
# Determine temporal and spatial sampling of the input data:
dxy,dt=tobac.get_spacings(OLR,grid_spacing=4000)
# Keyword arguments for the feature detection step
parameters_features={}
parameters_features['position_threshold']='weighted_diff'
parameters_features['sigma_threshold']=0.5
parameters_features['min_num']=4
parameters_features['target']='minimum'
parameters_features['threshold']=[250,225,200,175,150]
# Feature detection and save results to file:
print('starting feature detection')
Features=tobac.feature_detection_multithreshold(OLR,dxy,**parameters_features)
Features.to_hdf(os.path.join(savedir,'Features.h5'),'table')
print('feature detection performed and saved')
```
**Segmentation:**
Segmentation is performed based on the OLR field and a threshold value to determine the cloud areas.
```
# Keyword arguments for the segmentation step:
parameters_segmentation={}
parameters_segmentation['target']='minimum'
parameters_segmentation['method']='watershed'
parameters_segmentation['threshold']=250
# Perform segmentation and save results to files:
Mask_OLR,Features_OLR=tobac.segmentation_2D(Features,OLR,dxy,**parameters_segmentation)
print('segmentation OLR performed, start saving results to files')
iris.save([Mask_OLR],os.path.join(savedir,'Mask_Segmentation_OLR.nc'),zlib=True,complevel=4)
Features_OLR.to_hdf(os.path.join(savedir,'Features_OLR.h5'),'table')
print('segmentation OLR performed and saved')
```
**Trajectory linking:**
The detected features are linked into cloud trajectories using the trackpy library (http://soft-matter.github.io/trackpy). This takes the feature positions determined in the feature detection step into account but does not include information on the shape of the identified objects.
```
# keyword arguments for linking step
parameters_linking={}
parameters_linking['v_max']=20
parameters_linking['stubs']=2
parameters_linking['order']=1
parameters_linking['extrapolate']=1
parameters_linking['memory']=0
parameters_linking['adaptive_stop']=0.2
parameters_linking['adaptive_step']=0.95
parameters_linking['subnetwork_size']=100
parameters_linking['method_linking']= 'predict'
# Perform linking and save results to file:
Track=tobac.linking_trackpy(Features,OLR,dt=dt,dxy=dxy,**parameters_linking)
Track.to_hdf(os.path.join(savedir,'Track.h5'),'table')
```
**Visualisation:**
```
# Set extent of maps created in the following cells:
axis_extent=[-95,-89,28,32]
# Plot map with all individual tracks:
import cartopy.crs as ccrs
fig_map,ax_map=plt.subplots(figsize=(10,10),subplot_kw={'projection': ccrs.PlateCarree()})
ax_map=tobac.map_tracks(Track,axis_extent=axis_extent,axes=ax_map)
# Create animation of tracked clouds and outlines with OLR as a background field
animation_test_tobac=tobac.animation_mask_field(Track,Features,OLR,Mask_OLR,
axis_extent=axis_extent,#figsize=figsize,orientation_colorbar='horizontal',pad_colorbar=0.2,
vmin=80,vmax=330,cmap='Blues_r',
plot_outline=True,plot_marker=True,marker_track='x',plot_number=True,plot_features=True)
# Display animation:
from IPython.display import HTML, Image, display
HTML(animation_test_tobac.to_html5_video())
# # Save animation to file:
# savefile_animation=os.path.join(plot_dir,'Animation.mp4')
# animation_test_tobac.save(savefile_animation,dpi=200)
# print(f'animation saved to {savefile_animation}')
# Lifetimes of tracked clouds:
fig_lifetime,ax_lifetime=plt.subplots()
tobac.plot_lifetime_histogram_bar(Track,axes=ax_lifetime,bin_edges=np.arange(0,200,20),density=False,width_bar=10)
ax_lifetime.set_xlabel('lifetime (min)')
ax_lifetime.set_ylabel('counts')
```
| github_jupyter |
# Transfer Learning Template
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
import os, json, sys, time, random
import numpy as np
import torch
from torch.optim import Adam
from easydict import EasyDict
import matplotlib.pyplot as plt
from steves_models.steves_ptn import Steves_Prototypical_Network
from steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper
from steves_utils.iterable_aggregator import Iterable_Aggregator
from steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig
from steves_utils.torch_sequential_builder import build_sequential
from steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader
from steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)
from steves_utils.PTN.utils import independent_accuracy_assesment
from torch.utils.data import DataLoader
from steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory
from steves_utils.ptn_do_report import (
get_loss_curve,
get_results_table,
get_parameters_table,
get_domain_accuracies,
)
from steves_utils.transforms import get_chained_transform
```
# Allowed Parameters
These are allowed parameters, not defaults
Each of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)
Papermill uses the cell tag "parameters" to inject the real parameters below this cell.
Enable tags to see what I mean
```
required_parameters = {
"experiment_name",
"lr",
"device",
"seed",
"dataset_seed",
"n_shot",
"n_query",
"n_way",
"train_k_factor",
"val_k_factor",
"test_k_factor",
"n_epoch",
"patience",
"criteria_for_best",
"x_net",
"datasets",
"torch_default_dtype",
"NUM_LOGS_PER_EPOCH",
"BEST_MODEL_PATH",
"x_shape",
}
from steves_utils.CORES.utils import (
ALL_NODES,
ALL_NODES_MINIMUM_1000_EXAMPLES,
ALL_DAYS
)
from steves_utils.ORACLE.utils_v2 import (
ALL_DISTANCES_FEET_NARROWED,
ALL_RUNS,
ALL_SERIAL_NUMBERS,
)
standalone_parameters = {}
standalone_parameters["experiment_name"] = "STANDALONE PTN"
standalone_parameters["lr"] = 0.001
standalone_parameters["device"] = "cuda"
standalone_parameters["seed"] = 1337
standalone_parameters["dataset_seed"] = 1337
standalone_parameters["n_way"] = 8
standalone_parameters["n_shot"] = 3
standalone_parameters["n_query"] = 2
standalone_parameters["train_k_factor"] = 1
standalone_parameters["val_k_factor"] = 2
standalone_parameters["test_k_factor"] = 2
standalone_parameters["n_epoch"] = 50
standalone_parameters["patience"] = 10
standalone_parameters["criteria_for_best"] = "source_loss"
standalone_parameters["datasets"] = [
{
"labels": ALL_SERIAL_NUMBERS,
"domains": ALL_DISTANCES_FEET_NARROWED,
"num_examples_per_domain_per_label": 100,
"pickle_path": os.path.join(get_datasets_base_path(), "oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl"),
"source_or_target_dataset": "source",
"x_transforms": ["unit_mag", "minus_two"],
"episode_transforms": [],
"domain_prefix": "ORACLE_"
},
{
"labels": ALL_NODES,
"domains": ALL_DAYS,
"num_examples_per_domain_per_label": 100,
"pickle_path": os.path.join(get_datasets_base_path(), "cores.stratified_ds.2022A.pkl"),
"source_or_target_dataset": "target",
"x_transforms": ["unit_power", "times_zero"],
"episode_transforms": [],
"domain_prefix": "CORES_"
}
]
standalone_parameters["torch_default_dtype"] = "torch.float32"
standalone_parameters["x_net"] = [
{"class": "nnReshape", "kargs": {"shape":[-1, 1, 2, 256]}},
{"class": "Conv2d", "kargs": { "in_channels":1, "out_channels":256, "kernel_size":(1,7), "bias":False, "padding":(0,3), },},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features":256}},
{"class": "Conv2d", "kargs": { "in_channels":256, "out_channels":80, "kernel_size":(2,7), "bias":True, "padding":(0,3), },},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features":80}},
{"class": "Flatten", "kargs": {}},
{"class": "Linear", "kargs": {"in_features": 80*256, "out_features": 256}}, # 80 units per IQ pair
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm1d", "kargs": {"num_features":256}},
{"class": "Linear", "kargs": {"in_features": 256, "out_features": 256}},
]
# Parameters relevant to results
# These parameters will basically never need to change
standalone_parameters["NUM_LOGS_PER_EPOCH"] = 10
standalone_parameters["BEST_MODEL_PATH"] = "./best_model.pth"
# Parameters
parameters = {
"experiment_name": "tl_1v2:oracle.run1.framed-oracle.run2.framed",
"device": "cuda",
"lr": 0.0001,
"n_shot": 3,
"n_query": 2,
"train_k_factor": 3,
"val_k_factor": 2,
"test_k_factor": 2,
"torch_default_dtype": "torch.float32",
"n_epoch": 50,
"patience": 3,
"criteria_for_best": "target_accuracy",
"x_net": [
{"class": "nnReshape", "kargs": {"shape": [-1, 1, 2, 256]}},
{
"class": "Conv2d",
"kargs": {
"in_channels": 1,
"out_channels": 256,
"kernel_size": [1, 7],
"bias": False,
"padding": [0, 3],
},
},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features": 256}},
{
"class": "Conv2d",
"kargs": {
"in_channels": 256,
"out_channels": 80,
"kernel_size": [2, 7],
"bias": True,
"padding": [0, 3],
},
},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm2d", "kargs": {"num_features": 80}},
{"class": "Flatten", "kargs": {}},
{"class": "Linear", "kargs": {"in_features": 20480, "out_features": 256}},
{"class": "ReLU", "kargs": {"inplace": True}},
{"class": "BatchNorm1d", "kargs": {"num_features": 256}},
{"class": "Linear", "kargs": {"in_features": 256, "out_features": 256}},
],
"NUM_LOGS_PER_EPOCH": 10,
"BEST_MODEL_PATH": "./best_model.pth",
"n_way": 16,
"datasets": [
{
"labels": [
"3123D52",
"3123D65",
"3123D79",
"3123D80",
"3123D54",
"3123D70",
"3123D7B",
"3123D89",
"3123D58",
"3123D76",
"3123D7D",
"3123EFE",
"3123D64",
"3123D78",
"3123D7E",
"3124E4A",
],
"domains": [32, 38, 8, 44, 14, 50, 20, 26],
"num_examples_per_domain_per_label": 2000,
"pickle_path": "/root/csc500-main/datasets/oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl",
"source_or_target_dataset": "target",
"x_transforms": ["unit_power"],
"episode_transforms": [],
"domain_prefix": "ORACLE.run1_",
},
{
"labels": [
"3123D52",
"3123D65",
"3123D79",
"3123D80",
"3123D54",
"3123D70",
"3123D7B",
"3123D89",
"3123D58",
"3123D76",
"3123D7D",
"3123EFE",
"3123D64",
"3123D78",
"3123D7E",
"3124E4A",
],
"domains": [32, 38, 8, 44, 14, 50, 20, 26],
"num_examples_per_domain_per_label": 2000,
"pickle_path": "/root/csc500-main/datasets/oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl",
"source_or_target_dataset": "source",
"x_transforms": ["unit_power"],
"episode_transforms": [],
"domain_prefix": "ORACLE.run2_",
},
],
"dataset_seed": 500,
"seed": 500,
}
# Set this to True if you want to run this template directly
STANDALONE = False
if STANDALONE:
print("parameters not injected, running with standalone_parameters")
parameters = standalone_parameters
if not 'parameters' in locals() and not 'parameters' in globals():
raise Exception("Parameter injection failed")
#Use an easy dict for all the parameters
p = EasyDict(parameters)
if "x_shape" not in p:
p.x_shape = [2,256] # Default to this if we dont supply x_shape
supplied_keys = set(p.keys())
if supplied_keys != required_parameters:
print("Parameters are incorrect")
if len(supplied_keys - required_parameters)>0: print("Shouldn't have:", str(supplied_keys - required_parameters))
if len(required_parameters - supplied_keys)>0: print("Need to have:", str(required_parameters - supplied_keys))
raise RuntimeError("Parameters are incorrect")
###################################
# Set the RNGs and make it all deterministic
###################################
np.random.seed(p.seed)
random.seed(p.seed)
torch.manual_seed(p.seed)
torch.use_deterministic_algorithms(True)
###########################################
# The stratified datasets honor this
###########################################
torch.set_default_dtype(eval(p.torch_default_dtype))
###################################
# Build the network(s)
# Note: It's critical to do this AFTER setting the RNG
###################################
x_net = build_sequential(p.x_net)
start_time_secs = time.time()
p.domains_source = []
p.domains_target = []
train_original_source = []
val_original_source = []
test_original_source = []
train_original_target = []
val_original_target = []
test_original_target = []
# global_x_transform_func = lambda x: normalize(x.to(torch.get_default_dtype()), "unit_power") # unit_power, unit_mag
# global_x_transform_func = lambda x: normalize(x, "unit_power") # unit_power, unit_mag
def add_dataset(
labels,
domains,
pickle_path,
x_transforms,
episode_transforms,
domain_prefix,
num_examples_per_domain_per_label,
source_or_target_dataset:str,
iterator_seed=p.seed,
dataset_seed=p.dataset_seed,
n_shot=p.n_shot,
n_way=p.n_way,
n_query=p.n_query,
train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),
):
if x_transforms == []: x_transform = None
else: x_transform = get_chained_transform(x_transforms)
if episode_transforms == []: episode_transform = None
else: raise Exception("episode_transforms not implemented")
episode_transform = lambda tup, _prefix=domain_prefix: (_prefix + str(tup[0]), tup[1])
eaf = Episodic_Accessor_Factory(
labels=labels,
domains=domains,
num_examples_per_domain_per_label=num_examples_per_domain_per_label,
iterator_seed=iterator_seed,
dataset_seed=dataset_seed,
n_shot=n_shot,
n_way=n_way,
n_query=n_query,
train_val_test_k_factors=train_val_test_k_factors,
pickle_path=pickle_path,
x_transform_func=x_transform,
)
train, val, test = eaf.get_train(), eaf.get_val(), eaf.get_test()
train = Lazy_Iterable_Wrapper(train, episode_transform)
val = Lazy_Iterable_Wrapper(val, episode_transform)
test = Lazy_Iterable_Wrapper(test, episode_transform)
if source_or_target_dataset=="source":
train_original_source.append(train)
val_original_source.append(val)
test_original_source.append(test)
p.domains_source.extend(
[domain_prefix + str(u) for u in domains]
)
elif source_or_target_dataset=="target":
train_original_target.append(train)
val_original_target.append(val)
test_original_target.append(test)
p.domains_target.extend(
[domain_prefix + str(u) for u in domains]
)
else:
raise Exception(f"invalid source_or_target_dataset: {source_or_target_dataset}")
for ds in p.datasets:
add_dataset(**ds)
# from steves_utils.CORES.utils import (
# ALL_NODES,
# ALL_NODES_MINIMUM_1000_EXAMPLES,
# ALL_DAYS
# )
# add_dataset(
# labels=ALL_NODES,
# domains = ALL_DAYS,
# num_examples_per_domain_per_label=100,
# pickle_path=os.path.join(get_datasets_base_path(), "cores.stratified_ds.2022A.pkl"),
# source_or_target_dataset="target",
# x_transform_func=global_x_transform_func,
# domain_modifier=lambda u: f"cores_{u}"
# )
# from steves_utils.ORACLE.utils_v2 import (
# ALL_DISTANCES_FEET,
# ALL_RUNS,
# ALL_SERIAL_NUMBERS,
# )
# add_dataset(
# labels=ALL_SERIAL_NUMBERS,
# domains = list(set(ALL_DISTANCES_FEET) - {2,62}),
# num_examples_per_domain_per_label=100,
# pickle_path=os.path.join(get_datasets_base_path(), "oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl"),
# source_or_target_dataset="source",
# x_transform_func=global_x_transform_func,
# domain_modifier=lambda u: f"oracle1_{u}"
# )
# from steves_utils.ORACLE.utils_v2 import (
# ALL_DISTANCES_FEET,
# ALL_RUNS,
# ALL_SERIAL_NUMBERS,
# )
# add_dataset(
# labels=ALL_SERIAL_NUMBERS,
# domains = list(set(ALL_DISTANCES_FEET) - {2,62,56}),
# num_examples_per_domain_per_label=100,
# pickle_path=os.path.join(get_datasets_base_path(), "oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl"),
# source_or_target_dataset="source",
# x_transform_func=global_x_transform_func,
# domain_modifier=lambda u: f"oracle2_{u}"
# )
# add_dataset(
# labels=list(range(19)),
# domains = [0,1,2],
# num_examples_per_domain_per_label=100,
# pickle_path=os.path.join(get_datasets_base_path(), "metehan.stratified_ds.2022A.pkl"),
# source_or_target_dataset="target",
# x_transform_func=global_x_transform_func,
# domain_modifier=lambda u: f"met_{u}"
# )
# # from steves_utils.wisig.utils import (
# # ALL_NODES_MINIMUM_100_EXAMPLES,
# # ALL_NODES_MINIMUM_500_EXAMPLES,
# # ALL_NODES_MINIMUM_1000_EXAMPLES,
# # ALL_DAYS
# # )
# import steves_utils.wisig.utils as wisig
# add_dataset(
# labels=wisig.ALL_NODES_MINIMUM_100_EXAMPLES,
# domains = wisig.ALL_DAYS,
# num_examples_per_domain_per_label=100,
# pickle_path=os.path.join(get_datasets_base_path(), "wisig.node3-19.stratified_ds.2022A.pkl"),
# source_or_target_dataset="target",
# x_transform_func=global_x_transform_func,
# domain_modifier=lambda u: f"wisig_{u}"
# )
###################################
# Build the dataset
###################################
train_original_source = Iterable_Aggregator(train_original_source, p.seed)
val_original_source = Iterable_Aggregator(val_original_source, p.seed)
test_original_source = Iterable_Aggregator(test_original_source, p.seed)
train_original_target = Iterable_Aggregator(train_original_target, p.seed)
val_original_target = Iterable_Aggregator(val_original_target, p.seed)
test_original_target = Iterable_Aggregator(test_original_target, p.seed)
# For CNN We only use X and Y. And we only train on the source.
# Properly form the data using a transform lambda and Lazy_Iterable_Wrapper. Finally wrap them in a dataloader
transform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only
train_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)
val_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)
test_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)
train_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)
val_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)
test_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)
datasets = EasyDict({
"source": {
"original": {"train":train_original_source, "val":val_original_source, "test":test_original_source},
"processed": {"train":train_processed_source, "val":val_processed_source, "test":test_processed_source}
},
"target": {
"original": {"train":train_original_target, "val":val_original_target, "test":test_original_target},
"processed": {"train":train_processed_target, "val":val_processed_target, "test":test_processed_target}
},
})
from steves_utils.transforms import get_average_magnitude, get_average_power
print(set([u for u,_ in val_original_source]))
print(set([u for u,_ in val_original_target]))
s_x, s_y, q_x, q_y, _ = next(iter(train_processed_source))
print(s_x)
# for ds in [
# train_processed_source,
# val_processed_source,
# test_processed_source,
# train_processed_target,
# val_processed_target,
# test_processed_target
# ]:
# for s_x, s_y, q_x, q_y, _ in ds:
# for X in (s_x, q_x):
# for x in X:
# assert np.isclose(get_average_magnitude(x.numpy()), 1.0)
# assert np.isclose(get_average_power(x.numpy()), 1.0)
###################################
# Build the model
###################################
# easfsl only wants a tuple for the shape
model = Steves_Prototypical_Network(x_net, device=p.device, x_shape=tuple(p.x_shape))
optimizer = Adam(params=model.parameters(), lr=p.lr)
###################################
# train
###################################
jig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)
jig.train(
train_iterable=datasets.source.processed.train,
source_val_iterable=datasets.source.processed.val,
target_val_iterable=datasets.target.processed.val,
num_epochs=p.n_epoch,
num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,
patience=p.patience,
optimizer=optimizer,
criteria_for_best=p.criteria_for_best,
)
total_experiment_time_secs = time.time() - start_time_secs
###################################
# Evaluate the model
###################################
source_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)
target_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)
source_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)
target_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)
history = jig.get_history()
total_epochs_trained = len(history["epoch_indices"])
val_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))
confusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)
per_domain_accuracy = per_domain_accuracy_from_confusion(confusion)
# Add a key to per_domain_accuracy for if it was a source domain
for domain, accuracy in per_domain_accuracy.items():
per_domain_accuracy[domain] = {
"accuracy": accuracy,
"source?": domain in p.domains_source
}
# Do an independent accuracy assesment JUST TO BE SURE!
# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)
# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)
# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)
# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)
# assert(_source_test_label_accuracy == source_test_label_accuracy)
# assert(_target_test_label_accuracy == target_test_label_accuracy)
# assert(_source_val_label_accuracy == source_val_label_accuracy)
# assert(_target_val_label_accuracy == target_val_label_accuracy)
experiment = {
"experiment_name": p.experiment_name,
"parameters": dict(p),
"results": {
"source_test_label_accuracy": source_test_label_accuracy,
"source_test_label_loss": source_test_label_loss,
"target_test_label_accuracy": target_test_label_accuracy,
"target_test_label_loss": target_test_label_loss,
"source_val_label_accuracy": source_val_label_accuracy,
"source_val_label_loss": source_val_label_loss,
"target_val_label_accuracy": target_val_label_accuracy,
"target_val_label_loss": target_val_label_loss,
"total_epochs_trained": total_epochs_trained,
"total_experiment_time_secs": total_experiment_time_secs,
"confusion": confusion,
"per_domain_accuracy": per_domain_accuracy,
},
"history": history,
"dataset_metrics": get_dataset_metrics(datasets, "ptn"),
}
ax = get_loss_curve(experiment)
plt.show()
get_results_table(experiment)
get_domain_accuracies(experiment)
print("Source Test Label Accuracy:", experiment["results"]["source_test_label_accuracy"], "Target Test Label Accuracy:", experiment["results"]["target_test_label_accuracy"])
print("Source Val Label Accuracy:", experiment["results"]["source_val_label_accuracy"], "Target Val Label Accuracy:", experiment["results"]["target_val_label_accuracy"])
json.dumps(experiment)
```
| github_jupyter |
# Building a Fraud Prediction Model with EvalML
In this demo, we will build an optimized fraud prediction model using EvalML. To optimize the pipeline, we will set up an objective function to minimize the percentage of total transaction value lost to fraud. At the end of this demo, we also show you how introducing the right objective during the training results in a much better than using a generic machine learning metric like AUC.
```
import evalml
from evalml import AutoMLSearch
from evalml.objectives import FraudCost
```
## Configure "Cost of Fraud"
To optimize the pipelines toward the specific business needs of this model, we can set our own assumptions for the cost of fraud. These parameters are
* `retry_percentage` - what percentage of customers will retry a transaction if it is declined?
* `interchange_fee` - how much of each successful transaction do you collect?
* `fraud_payout_percentage` - the percentage of fraud will you be unable to collect
* `amount_col` - the column in the data the represents the transaction amount
Using these parameters, EvalML determines attempt to build a pipeline that will minimize the financial loss due to fraud.
```
fraud_objective = FraudCost(retry_percentage=.5,
interchange_fee=.02,
fraud_payout_percentage=.75,
amount_col='amount')
```
## Search for best pipeline
In order to validate the results of the pipeline creation and optimization process, we will save some of our data as the holdout set.
```
X, y = evalml.demos.load_fraud(n_rows=5000)
```
EvalML natively supports one-hot encoding. Here we keep 1 out of the 6 categorical columns to decrease computation time.
```
cols_to_drop = ['datetime', 'expiration_date', 'country', 'region', 'provider']
for col in cols_to_drop:
X.ww.pop(col)
X_train, X_holdout, y_train, y_holdout = evalml.preprocessing.split_data(X, y, problem_type='binary', test_size=0.2, random_seed=0)
X.ww
```
Because the fraud labels are binary, we will use `AutoMLSearch(X_train=X_train, y_train=y_train, problem_type='binary')`. When we call `.search()`, the search for the best pipeline will begin.
```
automl = AutoMLSearch(X_train=X_train, y_train=y_train,
problem_type='binary',
objective=fraud_objective,
additional_objectives=['auc', 'f1', 'precision'],
allowed_model_families=["random_forest", "linear_model"],
max_batches=1,
optimize_thresholds=True,
verbose=True)
automl.search()
```
### View rankings and select pipelines
Once the fitting process is done, we can see all of the pipelines that were searched, ranked by their score on the fraud detection objective we defined.
```
automl.rankings
```
To select the best pipeline we can call `automl.best_pipeline`.
```
best_pipeline = automl.best_pipeline
```
### Describe pipelines
We can get more details about any pipeline created during the search process, including how it performed on other objective functions, by calling the `describe_pipeline` method and passing the `id` of the pipeline of interest.
```
automl.describe_pipeline(automl.rankings.iloc[1]["id"])
```
## Evaluate on holdout data
Finally, since the best pipeline is already trained, we evaluate it on the holdout data.
Now, we can score the pipeline on the holdout data using both our fraud cost objective and the AUC (Area under the ROC Curve) objective.
```
best_pipeline.score(X_holdout, y_holdout, objectives=["auc", fraud_objective])
```
## Why optimize for a problem-specific objective?
To demonstrate the importance of optimizing for the right objective, let's search for another pipeline using AUC, a common machine learning metric. After that, we will score the holdout data using the fraud cost objective to see how the best pipelines compare.
```
automl_auc = AutoMLSearch(X_train=X_train, y_train=y_train,
problem_type='binary',
objective='auc',
additional_objectives=['f1', 'precision'],
max_batches=1,
allowed_model_families=["random_forest", "linear_model"],
optimize_thresholds=True,
verbose=True)
automl_auc.search()
```
Like before, we can look at the rankings of all of the pipelines searched and pick the best pipeline.
```
automl_auc.rankings
best_pipeline_auc = automl_auc.best_pipeline
# get the fraud score on holdout data
best_pipeline_auc.score(X_holdout, y_holdout, objectives=["auc", fraud_objective])
# fraud score on fraud optimized again
best_pipeline.score(X_holdout, y_holdout, objectives=["auc", fraud_objective])
```
When we optimize for AUC, we can see that the AUC score from this pipeline performs better compared to the AUC score from the pipeline optimized for fraud cost; however, the losses due to fraud are a much larger percentage of the total transaction amount when optimized for AUC and much smaller when optimized for fraud cost. As a result, we lose a noticable percentage of the total transaction amount by not optimizing for fraud cost specifically.
Optimizing for AUC does not take into account the user-specified `retry_percentage`, `interchange_fee`, `fraud_payout_percentage` values, which could explain the decrease in fraud performance. Thus, the best pipelines may produce the highest AUC but may not actually reduce the amount loss due to your specific type fraud.
This example highlights how performance in the real world can diverge greatly from machine learning metrics.
| github_jupyter |
```
#Always Pyspark first!
ErhvervsPath = "/home/svanhmic/workspace/Python/Erhvervs"
from pyspark.sql import functions as F, Window, WindowSpec
from pyspark.sql import Row
from pyspark.sql.types import StringType,ArrayType,IntegerType,DoubleType,StructField,StructType
sc.addPyFile(ErhvervsPath+"/src/RegnSkabData/ImportRegnskabData.py")
sc.addPyFile(ErhvervsPath+'/src/RegnSkabData/RegnskabsClass.py')
sc.addPyFile(ErhvervsPath+'/src/cvr/GetNextJsonLayer.py')
import sys
import re
import os
import ImportRegnskabData
import GetNextJsonLayer
import itertools
import functools
cvrPath = "/home/svanhmic/workspace/Python/Erhvervs/data/cdata/parquet"
cvrfiles = os.listdir(cvrPath)
#import crv data
cvrDf = (sqlContext
.read
.parquet(cvrPath+"/"+cvrfiles[0])
)
#cvrDf.show(1)
#print(cvrDf.select("cvrNummer").distinct().count())
#Extract all Aps and A/S companies
virkformCols = ("cvrNummer","virksomhedsform")
virkformDf = GetNextJsonLayer.createNextLayerTable(cvrDf.select(*virkformCols),[virkformCols[0]],virkformCols[1])
virkformDf = GetNextJsonLayer.expandSubCols(virkformDf,mainColumn="periode")
virkformDf = (virkformDf
.drop("sidstOpdateret")
.withColumn(col=F.col("periode_gyldigFra").cast("date"),colName="periode_gyldigFra")
.withColumn(col=F.col("periode_gyldigTil").cast("date"),colName="periode_gyldigTil")
)
#virkformDf.show(1)
checkCols = ["kortBeskrivelse","langBeskrivelse","virksomhedsformkode"]
#Consistencycheck is kortBeskrivelse and virksomhedsformkode always mapped the same way
#check1 = virkformDf.select(checkCols+["cvrNummer"]).distinct().groupby(*checkCols).count()
#check1.orderBy("kortBeskrivelse","count").show(check1.count(),truncate=False)
#Second test does any companies go from Aps or A/S to other or vice versa?
joinCols = ["cvrNummer","langBeskrivelse","rank"]
cvrCols = ["cvrNummer"]
gyldigCol = ["periode_gyldigFra"]
statusChangeWindow = (Window
.partitionBy(F.col(*cvrCols))
.orderBy(F.col("periode_gyldigFra").desc()))
#virkformDf.select(checkCols).distinct().show(50,truncate=False)
#Extract APS and AS here and latest status...
aggregationCols = [F.max(i) for i in gyldigCol]
groupsCol = [i for i in virkformDf.columns if i not in gyldigCol]
companyByAsApsDf = (virkformDf
.where((F.col("virksomhedsformkode") == 60) | (F.col("virksomhedsformkode") == 80))
.withColumn(col=F.rank().over(statusChangeWindow),colName="rank")
.filter(F.col("rank") == 1)
)
#Get the medarbejdstal
fastCols = ["cvrNummer","aar"]
regCols = ["intervalKodeAntalAarsvaerk","intervalKodeAntalInklusivEjere"]
reg2Cols = ["intervalKodeAntalAarsvaerk","intervalKodeAntalAnsatte"]
fCols = [F.split(F.regexp_replace(F.col(i),pattern=r'ANTAL_',replacement=""),"_").alias(i) for i in regCols]
mkCols = [F.split(F.regexp_replace(F.col(i),pattern=r'ANTAL_',replacement=""),"_").alias(i) for i in reg2Cols]
#kvartCols = [F.split(F.regexp_replace(F.col(i),pattern=r'ANTAL_',replacement=""),"_").alias(i) for i in cols]
def getLower(x):
try:
return int(x[0])
except:
return None
def getUpper(x):
try:
return int(x[0])
except:
return None
getLowerBound = F.udf(lambda x: getLower(x),IntegerType())
getUpperBound = F.udf(lambda x: getUpper(x),IntegerType())
aarsDf = (GetNextJsonLayer
.createNextLayerTable(cvrDf,["cvrNummer"],"aarsbeskaeftigelse")
.select(fastCols+fCols)
.select(fastCols+[getLowerBound(i).alias("lower_"+i) for i in regCols])
)
maanedsDf = (GetNextJsonLayer
.createNextLayerTable(cvrDf,["cvrNummer"],"maanedsbeskaeftigelse")
.select(["cvrNummer","aar","maaned"]+mkCols)
.select(["cvrNummer","aar","maaned"]+[getLowerBound(i).alias("lower_"+i) for i in reg2Cols])
)
kvartDf = (GetNextJsonLayer
.createNextLayerTable(cvrDf,["cvrNummer"],"kvartalsbeskaeftigelse")
.select(["cvrNummer","aar","kvartal"]+mkCols)
.select(["cvrNummer","aar","kvartal"]+[getLowerBound(i).alias("lower_"+i) for i in reg2Cols])
)
#maanedsDf.show()
#cvrDf.unpersist()
#maanedsDf.show()
#kvartDf.show()
#print(aarsDf.count())
#print(aarsDf.na.drop(how="all",subset=["lower_"+i for i in cols]).count())
# OK how many are represented in both or all three groups?
distinctMaanedDf = (maanedsDf
.join(companyByAsApsDf,on=(maanedsDf["cvrNummer"]==companyByAsApsDf["cvrNummer"]),how="right")
.drop(companyByAsApsDf["cvrNummer"])
.distinct()
)
#distinctMaanedDf.show()
distinctKvartalDf = (kvartDf
.join(companyByAsApsDf,on=(kvartDf["cvrNummer"]==companyByAsApsDf["cvrNummer"]),how="right")
.drop(companyByAsApsDf["cvrNummer"])
.distinct()
)
distinctAarDf = (aarsDf
.join(companyByAsApsDf,on=(aarsDf["cvrNummer"]==companyByAsApsDf["cvrNummer"]),how="right")
.drop(companyByAsApsDf["cvrNummer"])
.distinct()
)
distinctMaanedDf.write.parquet(mode="overwrite",path=cvrPath+"/MaanedsVaerker.parquet")
distinctKvartalDf.write.parquet(mode="overwrite",path=cvrPath+"/KvartalsVaerker.parquet")
distinctAarDf.write.parquet(mode="overwrite",path=cvrPath+"/AarsVaerker.parquet")
#print("månedsbeskæftigelse: "+str(distinctMaanedDf.count()))
#print("kvartalsbeskæftigelse: "+str(distinctKvartalDf.count()))
#print("årsbeskæftigelse: "+str(distinctAarDf.count()))
#print("Årsbeskæftigelse til kvartalsbeskæftigelse: "+str(distinctAarDf.select(F.col("cvrNummer")).distinct()
# .join(distinctKvartalDf.select(F.col("cvrNummer")).distinct(),(distinctKvartalDf["cvrNummer"]==distinctAarDf["cvrNummer"]),how="inner")
# .drop(distinctAarDf["cvrNummer"]).distinct().count()
#))
#print("Årsbeskæftigelse til månedsbeskæftigelse: "+str(distinctAarDf.select(F.col("cvrNummer")).distinct()
# .join(distinctMaanedDf.select(F.col("cvrNummer")).distinct(),(distinctMaanedDf["cvrNummer"]==distinctAarDf["cvrNummer"]),how="inner")
# .drop(distinctAarDf["cvrNummer"]).distinct().count()
#))
#print("Kvartalsbeskæftigelse til månedsbeskæftigelse: "+str(distinctKvartalDf.select(F.col("cvrNummer")).distinct()
# .join(distinctMaanedDf.select(F.col("cvrNummer")).distinct(),(distinctMaanedDf["cvrNummer"]==distinctKvartalDf["cvrNummer"]),how="inner")
# .drop(distinctAarDf["cvrNummer"]).distinct().count()
#))
AllThreeCount = (distinctAarDf
.select(F.col("cvrNummer")).distinct()
.join(distinctKvartalDf.select(F.col("cvrNummer")).distinct(),(distinctAarDf["cvrNummer"]==distinctKvartalDf["cvrNummer"]),how="inner")
.drop(distinctKvartalDf["cvrNummer"])
.join(distinctMaanedDf.select(F.col("cvrNummer")).distinct(),(distinctAarDf["cvrNummer"]==distinctMaanedDf["cvrNummer"]),how="inner")
.drop(distinctMaanedDf["cvrNummer"])
.distinct()
)
#print("Aarsbeskæftigelse til kvartalsbeskæftigelse til månedsbeskæftigelse: "+str(AllThreeCount.count()))
```
| github_jupyter |
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/guide/keras/custom_callback"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/keras/custom_callback.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/guide/keras/custom_callback.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/keras/custom_callback.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
# Keras custom callbacks
A custom callback is a powerful tool to customize the behavior of a Keras model during training, evaluation, or inference, including reading/changing the Keras model. Examples include `tf.keras.callbacks.TensorBoard` where the training progress and results can be exported and visualized with TensorBoard, or `tf.keras.callbacks.ModelCheckpoint` where the model is automatically saved during training, and more. In this guide, you will learn what Keras callback is, when it will be called, what it can do, and how you can build your own. Towards the end of this guide, there will be demos of creating a couple of simple callback applications to get you started on your custom callback.
## Setup
```
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# %tensorflow_version only exists in Colab.
%tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
```
## Introduction to Keras callbacks
In Keras, `Callback` is a python class meant to be subclassed to provide specific functionality, with a set of methods called at various stages of training (including batch/epoch start and ends), testing, and predicting. Callbacks are useful to get a view on internal states and statistics of the model during training. You can pass a list of callbacks (as the keyword argument `callbacks`) to any of `tf.keras.Model.fit()`, `tf.keras.Model.evaluate()`, and `tf.keras.Model.predict()` methods. The methods of the callbacks will then be called at different stages of training/evaluating/inference.
To get started, let's import tensorflow and define a simple Sequential Keras model:
```
# Define the Keras model to add callbacks to
def get_model():
model = tf.keras.Sequential()
model.add(tf.keras.layers.Dense(1, activation = 'linear', input_dim = 784))
model.compile(optimizer=tf.keras.optimizers.RMSprop(lr=0.1), loss='mean_squared_error', metrics=['mae'])
return model
```
Then, load the MNIST data for training and testing from Keras datasets API:
```
# Load example MNIST data and pre-process it
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()
x_train = x_train.reshape(60000, 784).astype('float32') / 255
x_test = x_test.reshape(10000, 784).astype('float32') / 255
```
Now, define a simple custom callback to track the start and end of every batch of data. During those calls, it prints the index of the current batch.
```
import datetime
class MyCustomCallback(tf.keras.callbacks.Callback):
def on_train_batch_begin(self, batch, logs=None):
print('Training: batch {} begins at {}'.format(batch, datetime.datetime.now().time()))
def on_train_batch_end(self, batch, logs=None):
print('Training: batch {} ends at {}'.format(batch, datetime.datetime.now().time()))
def on_test_batch_begin(self, batch, logs=None):
print('Evaluating: batch {} begins at {}'.format(batch, datetime.datetime.now().time()))
def on_test_batch_end(self, batch, logs=None):
print('Evaluating: batch {} ends at {}'.format(batch, datetime.datetime.now().time()))
```
Providing a callback to model methods such as `tf.keras.Model.fit()` ensures the methods are called at those stages:
```
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
epochs=1,
steps_per_epoch=5,
verbose=0,
callbacks=[MyCustomCallback()])
```
## Model methods that take callbacks
Users can supply a list of callbacks to the following `tf.keras.Model` methods:
#### [`fit()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit), [`fit_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit_generator)
Trains the model for a fixed number of epochs (iterations over a dataset, or data yielded batch-by-batch by a Python generator).
#### [`evaluate()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#evaluate), [`evaluate_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#evaluate_generator)
Evaluates the model for given data or data generator. Outputs the loss and metric values from the evaluation.
#### [`predict()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#predict), [`predict_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#predict_generator)
Generates output predictions for the input data or data generator.
```
_ = model.evaluate(x_test, y_test, batch_size=128, verbose=0, steps=5,
callbacks=[MyCustomCallback()])
```
## An overview of callback methods
### Common methods for training/testing/predicting
For training, testing, and predicting, following methods are provided to be overridden.
#### `on_(train|test|predict)_begin(self, logs=None)`
Called at the beginning of `fit`/`evaluate`/`predict`.
#### `on_(train|test|predict)_end(self, logs=None)`
Called at the end of `fit`/`evaluate`/`predict`.
#### `on_(train|test|predict)_batch_begin(self, batch, logs=None)`
Called right before processing a batch during training/testing/predicting. Within this method, `logs` is a dict with `batch` and `size` available keys, representing the current batch number and the size of the batch.
#### `on_(train|test|predict)_batch_end(self, batch, logs=None)`
Called at the end of training/testing/predicting a batch. Within this method, `logs` is a dict containing the stateful metrics result.
### Training specific methods
In addition, for training, following are provided.
#### on_epoch_begin(self, epoch, logs=None)
Called at the beginning of an epoch during training.
#### on_epoch_end(self, epoch, logs=None)
Called at the end of an epoch during training.
### Usage of `logs` dict
The `logs` dict contains the loss value, and all the metrics at the end of a batch or epoch. Example includes the loss and mean absolute error.
```
class LossAndErrorPrintingCallback(tf.keras.callbacks.Callback):
def on_train_batch_end(self, batch, logs=None):
print('For batch {}, loss is {:7.2f}.'.format(batch, logs['loss']))
def on_test_batch_end(self, batch, logs=None):
print('For batch {}, loss is {:7.2f}.'.format(batch, logs['loss']))
def on_epoch_end(self, epoch, logs=None):
print('The average loss for epoch {} is {:7.2f} and mean absolute error is {:7.2f}.'.format(epoch, logs['loss'], logs['mae']))
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
steps_per_epoch=5,
epochs=3,
verbose=0,
callbacks=[LossAndErrorPrintingCallback()])
```
Similarly, one can provide callbacks in `evaluate()` calls.
```
_ = model.evaluate(x_test, y_test, batch_size=128, verbose=0, steps=20,
callbacks=[LossAndErrorPrintingCallback()])
```
## Examples of Keras callback applications
The following section will guide you through creating simple Callback applications.
### Early stopping at minimum loss
First example showcases the creation of a `Callback` that stops the Keras training when the minimum of loss has been reached by mutating the attribute `model.stop_training` (boolean). Optionally, the user can provide an argument `patience` to specify how many epochs the training should wait before it eventually stops.
`tf.keras.callbacks.EarlyStopping` provides a more complete and general implementation.
```
import numpy as np
class EarlyStoppingAtMinLoss(tf.keras.callbacks.Callback):
"""Stop training when the loss is at its min, i.e. the loss stops decreasing.
Arguments:
patience: Number of epochs to wait after min has been hit. After this
number of no improvement, training stops.
"""
def __init__(self, patience=0):
super(EarlyStoppingAtMinLoss, self).__init__()
self.patience = patience
# best_weights to store the weights at which the minimum loss occurs.
self.best_weights = None
def on_train_begin(self, logs=None):
# The number of epoch it has waited when loss is no longer minimum.
self.wait = 0
# The epoch the training stops at.
self.stopped_epoch = 0
# Initialize the best as infinity.
self.best = np.Inf
def on_epoch_end(self, epoch, logs=None):
current = logs.get('loss')
if np.less(current, self.best):
self.best = current
self.wait = 0
# Record the best weights if current results is better (less).
self.best_weights = self.model.get_weights()
else:
self.wait += 1
if self.wait >= self.patience:
self.stopped_epoch = epoch
self.model.stop_training = True
print('Restoring model weights from the end of the best epoch.')
self.model.set_weights(self.best_weights)
def on_train_end(self, logs=None):
if self.stopped_epoch > 0:
print('Epoch %05d: early stopping' % (self.stopped_epoch + 1))
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
steps_per_epoch=5,
epochs=30,
verbose=0,
callbacks=[LossAndErrorPrintingCallback(), EarlyStoppingAtMinLoss()])
```
### Learning rate scheduling
One thing that is commonly done in model training is changing the learning rate as more epochs have passed. Keras backend exposes get_value API which can be used to set the variables. In this example, we're showing how a custom Callback can be used to dynamically change the learning rate.
Note: this is just an example implementation see `callbacks.LearningRateScheduler` and `keras.optimizers.schedules` for more general implementations.
```
class LearningRateScheduler(tf.keras.callbacks.Callback):
"""Learning rate scheduler which sets the learning rate according to schedule.
Arguments:
schedule: a function that takes an epoch index
(integer, indexed from 0) and current learning rate
as inputs and returns a new learning rate as output (float).
"""
def __init__(self, schedule):
super(LearningRateScheduler, self).__init__()
self.schedule = schedule
def on_epoch_begin(self, epoch, logs=None):
if not hasattr(self.model.optimizer, 'lr'):
raise ValueError('Optimizer must have a "lr" attribute.')
# Get the current learning rate from model's optimizer.
lr = float(tf.keras.backend.get_value(self.model.optimizer.lr))
# Call schedule function to get the scheduled learning rate.
scheduled_lr = self.schedule(epoch, lr)
# Set the value back to the optimizer before this epoch starts
tf.keras.backend.set_value(self.model.optimizer.lr, scheduled_lr)
print('\nEpoch %05d: Learning rate is %6.4f.' % (epoch, scheduled_lr))
LR_SCHEDULE = [
# (epoch to start, learning rate) tuples
(3, 0.05), (6, 0.01), (9, 0.005), (12, 0.001)
]
def lr_schedule(epoch, lr):
"""Helper function to retrieve the scheduled learning rate based on epoch."""
if epoch < LR_SCHEDULE[0][0] or epoch > LR_SCHEDULE[-1][0]:
return lr
for i in range(len(LR_SCHEDULE)):
if epoch == LR_SCHEDULE[i][0]:
return LR_SCHEDULE[i][1]
return lr
model = get_model()
_ = model.fit(x_train, y_train,
batch_size=64,
steps_per_epoch=5,
epochs=15,
verbose=0,
callbacks=[LossAndErrorPrintingCallback(), LearningRateScheduler(lr_schedule)])
```
### Standard Keras callbacks
Be sure to check out the existing Keras callbacks by [visiting the API doc](https://www.tensorflow.org/api_docs/python/tf/keras/callbacks). Applications include logging to CSV, saving the model, visualizing on TensorBoard and a lot more.
| github_jupyter |
# Imports
```
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Dense, Dropout, Flatten, Input, Concatenate
from tensorflow.keras.optimizers import Adam, RMSprop
import numpy as np
import matplotlib.pyplot as plt
import copy
```
# Global Variables
```
epochs = 500
batch_size = 16
number_of_particles = epochs * 2 * batch_size
dt = 0.1
```
# Classes
```
class Particle:
def __str__(self):
return "Position: %s, Velocity: %s, Accleration: %s" % (self.position, self.velocity, self.acceleration)
def __repr__(self):
return "Position: %s, Velocity: %s, Accleration: %s" % (self.position, self.velocity, self.acceleration)
def __init__(self):
self.position = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Position X, Y
self.velocity = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Velocity X, Y
self.acceleration = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Acceleration X, Y
def apply_physics(self,dt):
nextParticle = copy.deepcopy(self) # Copy to retain initial values
nextParticle.position += self.velocity * dt
nextParticle.velocity += self.acceleration * dt
return nextParticle
def get_list(self):
return [self.position[0],self.position[1],self.velocity[0], self.velocity[1], self.acceleration[0], self.acceleration[1]]
def get_list_physics(self,dt):
n = self.apply_physics(dt)
return [self.position[0],self.position[1],self.velocity[0], self.velocity[1], self.acceleration[0],
self.acceleration[1], n.position[0], n.position[1], n.velocity[0], n.velocity[1]]
class GAN:
def __init__(self,input_size,output_size,dropout=0.4):
self.input_size = input_size
self.output_size = output_size
self.dropout = dropout
self.generator = self.generator_network()
self.discriminator = self.discriminator_network()
self.adverserial = self.adverserial_network()
def discriminator_trainable(self, val):
self.discriminator.trainable = val
for l in self.discriminator.layers:
l.trainable = val
def generator_network(self): # Generator : Object(6) - Dense - Object(4)
self.g_input = Input(shape=(self.input_size,), name="Generator_Input")
g = Dense(128, activation='relu')(self.g_input)
g = Dropout(self.dropout)(g)
g = Dense(256, activation='relu')(g)
g = Dropout(self.dropout)(g)
g = Dense(512, activation='relu')(g)
g = Dropout(self.dropout)(g)
g = Dense(256, activation='relu')(g)
g = Dropout(self.dropout)(g)
g = Dense(128, activation='relu')(self.g_input)
g = Dropout(self.dropout)(g)
self.g_output = Dense(self.output_size, activation='tanh', name="Generator_Output")(g)
m = Model(self.g_input, self.g_output, name="Generator")
return m
def discriminator_network(self): # Discriminator : Object(10) - Dense - Probability
d_opt = RMSprop(lr=0.000125,decay=6e-8)
d_input = Input(shape=(self.input_size+self.output_size,), name="Discriminator_Input")
d = Dense(128, activation='relu')(d_input)
d = Dense(256, activation='relu')(d)
d = Dense(512, activation='relu')(d)
d = Dense(256, activation='relu')(d)
d = Dense(128, activation='relu')(d)
d_output = Dense(1, activation='sigmoid', name="Discriminator_Output")(d)
m = Model(d_input, d_output, name="Discriminator")
m.compile(loss='binary_crossentropy', optimizer=d_opt)
return m
def adverserial_network(self): # Adverserial : Object(6) - Generator - Discriminator - Probability
a_opt = RMSprop(lr=0.0001,decay=3e-8)
d_input = Concatenate(name="Generator_Input_Output")([self.g_input,self.g_output])
m=Model(self.g_input, self.discriminator(d_input))
m.compile(loss='binary_crossentropy', optimizer=a_opt)
return m
def train_discriminator(self,val):
self.discriminator.trainable = val
for l in self.discriminator.layers:
l.trainable = val
def train(self, adverserial_set, discriminator_set, epochs, batch_size):
losses = {"d":[], "g":[]}
for i in range(epochs):
batch = discriminator_set[int(i/2*batch_size/2):int((i/2+1)*batch_size/2)] # Gets a batch of real data
for j in adverserial_set[int(i/2*batch_size/2):int((i/2+1)*batch_size/2)]: # Gets a batch of generated data
n = copy.deepcopy(j)
p = self.predict(j)
for e in p:
n.append(e)
batch.append(n)
#self.train_discriminator(True) # Turns on discriminator weights
output = np.zeros(batch_size) # Sets output weight 0 for real and 1 for fakes
output[int(batch_size/2):] = 1
losses["d"].append(self.discriminator.train_on_batch(np.array(batch), np.array(output))) # Train discriminator
batch = adverserial_set[(i*batch_size):((i+1)*batch_size)] # Gets real data to train generator
output = np.zeros(batch_size)
#self.train_discriminator(False) # Turns off discriminator weights
losses["g"].append(self.adverserial.train_on_batch(np.array(batch), np.array(output))) # Train generator
print('Epoch %s - Adverserial Loss : %s, Discriminator Loss : %s' % (i+1, losses["g"][-1], losses["d"][-1]))
self.generator.save("Generator.h5")
self.discriminator.save("Discriminator.h5")
return losses
def predict(self, pred):
return self.generator.predict(np.array(pred).reshape(-1,6))[0]
```
# Training Data
```
training_set = []
actual_set = []
for i in range(number_of_particles):
p = Particle()
if(i%2==0):
training_set.append(p.get_list())
else:
actual_set.append(p.get_list_physics(dt))
```
# Training
```
network = GAN(input_size=6,output_size=4,dropout=0)
loss = network.train(adverserial_set=training_set,discriminator_set=actual_set,epochs=epochs,batch_size=batch_size)
fig = plt.figure(figsize=(13,7))
plt.title("Loss Function over Epochs")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.plot(loss["g"], label="Adversarial Loss")
plt.plot(loss["d"], label="Discriminative Loss")
plt.legend()
plt.show()
network.predict([0.1,0.2,0.1,0.1,0.1,0.1])
network.generator.summary()
network.discriminator.summary()
network.adverserial.summary()
```
| github_jupyter |
# Field operations
There are several convenience methods that can be used to analyse the field. Let us first define the mesh we are going to work with.
```
import discretisedfield as df
p1 = (-50, -50, -50)
p2 = (50, 50, 50)
n = (2, 2, 2)
mesh = df.Mesh(p1=p1, p2=p2, n=n)
```
We are going to initialise the vector field (`dim=3`), with
$$\mathbf{f}(x, y, z) = (xy, 2xy, xyz)$$
For that, we are going to use the following Python function.
```
def value_function(pos):
x, y, z = pos
return x*y, 2*x*y, x*y*z
```
Finally, our field is
```
field = df.Field(mesh, dim=3, value=value_function)
```
## 1. Sampling the field
As we have shown previously, a field can be sampled by calling it. The argument must be a 3-length iterable and it contains the coordinates of the point.
```
point = (0, 0, 0)
field(point)
```
However if the point is outside the mesh, an exception is raised.
```
point = (100, 100, 100)
try:
field(point)
except ValueError:
print('Exception raised.')
```
## 2. Extracting the component of a vector field
A three-dimensional vector field can be understood as three separate scalar fields, where each scalar field is a component of a vector field value. A scalar field of a component can be extracted by accessing `x`, `y`, or `z` attribute of the field.
```
x_component = field.x
x_component((0, 0, 0))
```
Default names `x`, `y`, and (for dim 3) `z` are only available for fields with dimensionality 2 or 3.
```
field.components
```
It is possible to change the component names:
```
field.components = ['mx', 'my', 'mz']
field.mx((0, 0, 0))
```
This overrides the component labels and the old `x`, `y` and `z` cannot be used anymore:
```
try:
field.x
except AttributeError as e:
print(e)
```
We change the component labels back to `x`, `y`, and `z` for the rest of this notebook.
```
field.components = ['x', 'y', 'z']
```
Custom component names can optionally also be specified during field creation. If not specified, the default values are used for fields with dimensions 2 or 3. Higher-dimensional fields have no defaults and custom labes have to be specified in order to access individual field components:
```
field_4d = df.Field(mesh, dim=4, value=[1, 1, 1, 1], components=['c1', 'c2', 'c3', 'c4'])
field_4d
field_4d.c1((0, 0, 0))
```
## 3. Computing the average
The average of the field can be obtained by calling `discretisedfield.Field.average` property.
```
field.average
```
Average always return a tuple, independent of the dimension of the field's value.
```
field.x.average
```
## 4. Iterating through the field
The field object itself is an iterable. That means that it can be iterated through. As a result it returns a tuple, where the first element is the coordinate of the mesh point, whereas the second one is its value.
```
for coordinate, value in field:
print(coordinate, value)
```
## 5. Sampling the field along the line
To sample the points of the field which are on a certain line, `discretisedfield.Field.line` method is used. It takes two points `p1` and `p2` that define the line and an integer `n` which defines how many mesh coordinates on that line are required. The default value of `n` is 100.
```
line = field.line(p1=(-10, 0, 0), p2=(10, 0, 0), n=5)
```
## 6. Intersecting the field with a plane
If we intersect the field with a plane, `discretisedfield.Field.plane` will return a new field object which contains only discretisation cells that belong to that plane. The planes allowed are the planes perpendicular to the axes of the Cartesian coordinate system. For instance, a plane parallel to the $yz$-plane (perpendicular to the $x$-axis) which intesects the $x$-axis at 1, can be written as
$$x = 1$$
```
field.plane(x=1)
```
If we want to cut through the middle of the mesh, we do not need to provide a particular value for a coordinate.
```
field.plane('x')
```
## 7. Cascading the operations
Let us say we want to compute the average of an $x$ component of the field on the plane $y=10$. In order to do that, we can cascade several operation in a single line.
```
field.plane(y=10).x.average
```
This gives the same result as for instance
```
field.x.plane(y=10).average
```
## 8. Complex fields
`discretisedfield` supports complex-valued fields.
```
cfield = df.Field(mesh, dim=3, value=(1+1.5j, 2, 3j))
```
We can extract `real` and `imaginary` part.
```
cfield.real((0, 0, 0))
cfield.imag((0, 0, 0))
```
Similarly we get `real` and `imaginary` parts of individual components.
```
cfield.x.real((0, 0, 0))
cfield.x.imag((0, 0, 0))
```
Complex conjugate.
```
cfield.conjugate((0, 0, 0))
```
Phase in the complex plane.
```
cfield.phase((0, 0, 0))
```
## 9. Applying `numpys` universal functions
All numpy universal functions can be applied to `discretisedfield.Field` objects. Below we show a different examples. For available functions please refer to the `numpy` [documentation](https://numpy.org/doc/stable/reference/ufuncs.html#available-ufuncs).
```
import numpy as np
f1 = df.Field(mesh, dim=1, value=1)
f2 = df.Field(mesh, dim=1, value=np.pi)
f3 = df.Field(mesh, dim=1, value=2)
np.sin(f1)
np.sin(f2)((0, 0, 0))
np.sum((f1, f2, f3))((0, 0, 0))
np.exp(f1)((0, 0, 0))
np.power(f3, 2)((0, 0, 0))
```
## Other
Full description of all existing functionality can be found in the [API Reference](https://discretisedfield.readthedocs.io/en/latest/_autosummary/discretisedfield.Field.html).
| github_jupyter |
## Search with Options
- Piece or Corpus
- Actual or Incremental Durations
- Chromatic or Diatonic
- Exact or Close
- Classify
***
```
from crim_intervals import *
import pandas as pd
import ast
import matplotlib
from itertools import tee, combinations
```
### The Complete Corpus
```
work_list = ['CRIM_Mass_0001_1.mei','CRIM_Mass_0001_2.mei','CRIM_Mass_0001_3.mei','CRIM_Mass_0001_4.mei','CRIM_Mass_0001_5.mei','CRIM_Mass_0002_1.mei','CRIM_Mass_0002_2.mei','CRIM_Mass_0002_3.mei','CRIM_Mass_0002_4.mei','CRIM_Mass_0002_5.mei','CRIM_Mass_0003_1.mei','CRIM_Mass_0003_2.mei','CRIM_Mass_0003_3.mei','CRIM_Mass_0003_4.mei','CRIM_Mass_0003_5.mei','CRIM_Mass_0004_1.mei','CRIM_Mass_0004_2.mei','CRIM_Mass_0004_3.mei','CRIM_Mass_0004_4.mei','CRIM_Mass_0004_5.mei','CRIM_Mass_0005_1.mei','CRIM_Mass_0005_2.mei','CRIM_Mass_0005_3.mei','CRIM_Mass_0005_4.mei','CRIM_Mass_0005_5.mei','CRIM_Mass_0006_1.mei','CRIM_Mass_0006_2.mei','CRIM_Mass_0006_3.mei','CRIM_Mass_0006_4.mei','CRIM_Mass_0006_5.mei','CRIM_Mass_0007_1.mei', 'CRIM_Mass_0007_2.mei', 'CRIM_Mass_0007_3.mei', 'CRIM_Mass_0007_4.mei', 'CRIM_Mass_0007_5.mei', 'CRIM_Mass_0008_1.mei', 'CRIM_Mass_0008_2.mei', 'CRIM_Mass_0008_3.mei', 'CRIM_Mass_0008_4.mei', 'CRIM_Mass_0008_5.mei', 'CRIM_Mass_0009_1.mei', 'CRIM_Mass_0009_2.mei', 'CRIM_Mass_0009_3.mei', 'CRIM_Mass_0009_4.mei', 'CRIM_Mass_0009_5.mei', 'CRIM_Mass_0010_1.mei', 'CRIM_Mass_0010_2.mei', 'CRIM_Mass_0010_3.mei', 'CRIM_Mass_0010_4.mei', 'CRIM_Mass_0010_5.mei', 'CRIM_Mass_0011_1.mei', 'CRIM_Mass_0011_2.mei', 'CRIM_Mass_0011_3.mei', 'CRIM_Mass_0011_4.mei', 'CRIM_Mass_0011_5.mei', 'CRIM_Mass_0012_1.mei', 'CRIM_Mass_0012_2.mei', 'CRIM_Mass_0012_3.mei', 'CRIM_Mass_0012_4.mei', 'CRIM_Mass_0012_5.mei', 'CRIM_Mass_0013_1.mei', 'CRIM_Mass_0013_2.mei', 'CRIM_Mass_0013_3.mei', 'CRIM_Mass_0013_4.mei', 'CRIM_Mass_0013_5.mei', 'CRIM_Mass_0014_1.mei', 'CRIM_Mass_0014_2.mei', 'CRIM_Mass_0014_3.mei', 'CRIM_Mass_0014_4.mei', 'CRIM_Mass_0014_5.mei', 'CRIM_Mass_0015_1.mei', 'CRIM_Mass_0015_2.mei', 'CRIM_Mass_0015_3.mei', 'CRIM_Mass_0015_4.mei', 'CRIM_Mass_0015_5.mei', 'CRIM_Mass_0016_1.mei', 'CRIM_Mass_0016_2.mei', 'CRIM_Mass_0016_3.mei', 'CRIM_Mass_0016_4.mei', 'CRIM_Mass_0016_5.mei', 'CRIM_Mass_0017_1.mei', 'CRIM_Mass_0017_2.mei', 'CRIM_Mass_0017_3.mei', 'CRIM_Mass_0017_4.mei', 'CRIM_Mass_0017_5.mei', 'CRIM_Mass_0018_1.mei', 'CRIM_Mass_0018_2.mei', 'CRIM_Mass_0018_3.mei', 'CRIM_Mass_0018_4.mei', 'CRIM_Mass_0018_5.mei', 'CRIM_Mass_0019_1.mei', 'CRIM_Mass_0019_2.mei', 'CRIM_Mass_0019_3.mei', 'CRIM_Mass_0019_4.mei', 'CRIM_Mass_0019_5.mei', 'CRIM_Mass_0020_1.mei', 'CRIM_Mass_0020_2.mei', 'CRIM_Mass_0020_3.mei', 'CRIM_Mass_0020_4.mei', 'CRIM_Mass_0020_5.mei', 'CRIM_Mass_0021_1.mei', 'CRIM_Mass_0021_2.mei', 'CRIM_Mass_0021_3.mei', 'CRIM_Mass_0021_4.mei', 'CRIM_Mass_0021_5.mei', 'CRIM_Mass_0022_2.mei', 'CRIM_Model_0001.mei', 'CRIM_Model_0008.mei', 'CRIM_Model_0009.mei', 'CRIM_Model_0010.mei', 'CRIM_Model_0011.mei', 'CRIM_Model_0012.mei', 'CRIM_Model_0013.mei', 'CRIM_Model_0014.mei', 'CRIM_Model_0015.mei', 'CRIM_Model_0016.mei', 'CRIM_Model_0017.mei', 'CRIM_Model_0019.mei', 'CRIM_Model_0020.mei', 'CRIM_Model_0021.mei', 'CRIM_Model_0023.mei', 'CRIM_Model_0025.mei', 'CRIM_Model_0026.mei',
]
```
### Short Corpus
```
work_list = ['CRIM_Mass_0002_1.mei',
'CRIM_Mass_0002_2.mei',
'CRIM_Mass_0002_3.mei',
'CRIM_Mass_0002_4.mei',
'CRIM_Mass_0002_5.mei',
'CRIM_Model_0001.mei']
# work_list = [
# 'CRIM_Model_0008.mei']
```
## Load File and Correct the MEI Metadata
```
work_list = [el.replace("CRIM_", "https://crimproject.org/mei/MEI_4.0/CRIM_") for el in work_list]
corpus = CorpusBase(work_list)
import xml.etree.ElementTree as ET
import requests
MEINSURI = 'http://www.music-encoding.org/ns/mei'
MEINS = '{%s}' % MEINSURI
for i, path in enumerate(work_list):
try:
if path[0] == '/':
mei_doc = ET.parse(path)
else:
mei_doc = ET.fromstring(requests.get(path).text)
# Find the title from the MEI file and update the Music21 Score metadata
title = mei_doc.find('mei:meiHead//mei:titleStmt/mei:title', namespaces={"mei": MEINSURI}).text
print(path, title)
corpus.scores[i].metadata.title = title
except:
continue
for s in corpus.scores:
print(s.metadata.title)
```
## Select Actual or Incremental Durations
#### About Rhythmic Durations
- For `find_close_matches` and `find_exact_matches`, rhythmic variation/duration is displayed, but **not** factored into the calculation of matching.
- **Incremental Offset** calculates the intervals using a **fixed offset between notes**, no matter their actual duration. Use this to ignore passing tones or other ornaments. The offsets are expressed in multiples of the quarter note (Offset = 1 samples at quarter note; Offset = 2 at half note, etc). Set with `vectors = IntervalBase(corpus.note_list_incremental_offset(2))`
```
vectors = IntervalBase(corpus.note_list)
#vectors = IntervalBase(corpus.note_list_incremental_offset(2))
```
***
## Select Generic or Semitone Scale:
- **Length of the Soggetto**: `into_patterns([vectors.semitone_intervals], 5)`
- The **number** in this command represents the **minimum number of vectors to find**. 5 vectors is 6 notes.
```
patterns = into_patterns([vectors.generic_intervals], 8)
#patterns = into_patterns([vectors.semitone_intervals], 4)
```
***
## Select Exact Matches Here, or Close Below
#### (Use comment feature to select screen preview or CSV output)
- **Exact** is exact in *all* ways `find_exact_matches(patterns, 2)`
- The **number** in this command represents the **minimum number of matching melodies needed before reporting**. This allows us to filter for common or uncommon soggetti.
```
exact_matches = find_exact_matches(patterns, 3)
# Use this for exact screen preview
#for item in exact_matches:
#item.print_exact_matches()
output_exact = export_pandas(exact_matches)
pd.DataFrame(output_exact).head()
output_exact["pattern_generating_match"] = output_exact["pattern_generating_match"].apply(tuple)
results = pd.DataFrame(output_exact)
results["pattern_generating_match"] = results["pattern_generating_match"].apply(tuple)
results
#export_to_csv(exact_matches)
```
### A Quick Overview of the Results
```
total_matches = len(output_exact)
unique_sogetti = output_exact.pattern_generating_match.apply(str).nunique()
summary = 'There are {} unique soggetti and {} total matches in this search'.format(unique_sogetti, total_matches)
summary
```
### Group by the Pattern Generating Match and Check Distribution of Results
- Report Top Ten and Bottom Ten Results)
```
pattern_inventory = pd.DataFrame(output_exact.groupby("pattern_generating_match").size().sort_values(ascending=False)[:10])
pattern_inventory
pattern_inventory = pd.DataFrame(output_exact.groupby("pattern_generating_match").size().sort_values(ascending=True)[:10])
pattern_inventory
```
***
### Select Close Matches Here
#### (Comment out the 'for item iteration' in order to skip screen preview)
- **Close** matches allow for melodic variation (see more below). `find_close_matches(patterns, 2, 1)`
- The **first number** in this command is the **minimum number of melodies** needed before reporting
- The **second number** is **threshold of similarity** needed in order to find a match.
- Lower number = very similar; higher number = less similar
##### More about Close Matches
- The **threshold for close matches** is determined by the **second number** called in the method.
- We select two patterns, then compare *each vector in each pattern successively*.
- The *differences between each vector are summed*.
- If that value is **below the threshold specified**, we consider the **two patterns closely matched**.
- The format of the method call is `find_close_matches(the array you get from into_patterns, minimum matches needed to be displayed, threshold for close match)`.
```
close_matches = find_close_matches(patterns, 2, 1)
#for item in close_matches:
#item.print_close_matches()
#return pd.DataFrame(close_matches)
output_close = export_pandas(close_matches)
output_close["pattern_generating_match"] = output_close["pattern_generating_match"].apply(tuple)
results = pd.DataFrame(output_close)
results["pattern_generating_match"] = results["pattern_generating_match"].apply(tuple)
results.head(50)
#export_to_csv(close_matches)
```
### How Many Unique Soggetti? How many instances?
```
total_matches = len(output_close)
unique_sogetti = output_close.pattern_generating_match.apply(str).nunique()
summary = 'There are {} unique soggetti and {} total matches in this search'.format(unique_sogetti, total_matches)
summary
```
### Top and Bottom Ten Soggetti
```
pattern_inventory = pd.DataFrame(output_close.groupby("pattern_generating_match").size().sort_values(ascending=False)[:50])
pattern_inventory
pattern_inventory = pd.DataFrame(output_close.groupby("pattern_generating_match").size().sort_values(ascending=True)[:10])
pattern_inventory
```
***
### Classify Patterns Here
#### Note: depends on choice of Close or Exact above! Must choose appropriate one below!
#### Enable "export_to_csv" line to allow this within Notebook (must answer "Y" and provide filename)
```
%%capture
classify_matches(close_matches, 2)
#classify_matches(exact_matches, 2)
cm = classify_matches(close_matches, 2)
#pd.DataFrame(classified_matches)
output_cm = export_pandas(cm)
#pd.DataFrame(output).head()
## For CSV export, use the following (and follow prompts for file name)
#export_to_csv(cm)
short_out = output_cm.drop(columns=["ema_url"])
short_out
def classified_matches_to_pandas(matches):
soggetti_matches = []
for i, cm in enumerate(matches):
for j, soggetti in enumerate(cm.matches):
soggetti_matches.append({
"piece": soggetti.first_note.metadata.title,
"type": cm.type,
"part": soggetti.first_note.part.strip("[] "),
"bar": soggetti.first_note.note.measureNumber,
"entry_number": j + 1,
"pattern": cm.pattern,
"match_number": i + 1
})
return pd.DataFrame(soggetti_matches)
df = classified_matches_to_pandas(cm)
pd.set_option('display.max_rows', 50)
df.head(10)
wide_df = df.pivot_table(index=["match_number", "piece", "type"],
columns="entry_number",
values=["part", "bar"],
aggfunc=lambda x: x)
wide_df.columns = [f"{a}_{b}" for a, b in wide_df.columns]
wide_df.head().reset_index()
wide_df.shape
wide_df
```
## Read CSV of Classified Matches
- Update file name to match the output of previous cells for Classifier
```
results = pd.read_csv('Sandrin_Classified.csv')
results.rename(columns=
{'Pattern Generating Match': 'Pattern_Generating_Match',
'Pattern matched':'Pattern_Matched',
'Classification Type': 'Classification_Type',
'Piece Title': 'Piece_Title',
'First Note Measure Number': 'Start_Measure',
'Last Note Measure Number': 'Stop_Measure',
'Note Durations': 'Note_Durations'
},
inplace=True)
results['note_durations'] = results['note_durations'].apply(ast.literal_eval)
durations = results['note_durations']
results.head()
```
# Durational Ratios
#### This Function Calculates the Ratios of the Durations in each Match
```
# makes pairs of ratio strings
def pairwise(iterable):
"s -> (s0,s1), (s1,s2), (s2, s3), ..."
a, b = tee(iterable)
next(b, None)
return zip(a, b)
def get_ratios(input_list):
ratio_pairs = []
for a, b in pairwise(input_list):
ratio_pairs.append(b / a)
return ratio_pairs
```
#### Now call the function to operate on the RESULTS file from earlier
```
# calculates 'duration ratios' for each soggetto, then adds this to the DF
results["duration_ratios"] = results.note_durations.apply(get_ratios)
short_results = results.drop(columns=["ema_url"])
short_out.head(10)
```
## Group by the Pattern Generating Match
- Each has its own string of durations, and duration ratios
- and then we compare the ratios to get the differences
- the "list(combinations)" method takes care of building the pairs, using data from our dataframe 'results'
```
def compare_ratios(ratios_1, ratios_2):
## division of lists
# using zip() + list comprehension
diffs = [i - j for i, j in zip(ratios_1, ratios_2)]
abs_diffs = [abs(ele) for ele in diffs]
sum_diffs = sum(abs_diffs)
return sum_diffs
#results["Pattern_Generating_Match"] = results["Pattern_Generating_Match"].apply(tuple)
def get_ratio_distances(results, pattern_col, output_cols):
matches = []
for name, group in results.groupby(pattern_col):
ratio_pairs = list(combinations(group.index.values, 2))
for a, b in ratio_pairs:
a_match = results.loc[a]
b_match = results.loc[b]
sum_diffs = compare_ratios(a_match.duration_ratios, b_match.duration_ratios)
match_dict = {
"pattern": name,
"sum_diffs": sum_diffs
}
for col in output_cols:
match_dict.update({
f"match_1_{col}": a_match[col],
f"match_2_{col}": b_match[col]
})
matches.append(match_dict)
return pd.DataFrame(matches)
```
### Now Run the Function to get the 'edit distances' for the durations of matching patterns
```
ratio_distances = get_ratio_distances(results, "pattern_generating_match", ["piece_title", "part", "start_measure", "end_measure"])
ratio_distances.head()
```
### And FILTER the results according to any threshold we like
```
ratios_filtered = ratio_distances[ratio_distances.sum_diffs <= 1]
ratios_filtered
```
### Now Group the Duration-Filter Results by the Pattern (which shows us very closely related soggetti in sets)
```
grouped = ratios_filtered.groupby("pattern")
grouped.head()
ratios_filtered.to_csv("filtered_sample_pair.csv")
```
| github_jupyter |
# Use the Shirt Class
You've seen what a class looks like and how to instantiate an object. Now it's your turn to write code that insantiates a shirt object.
# Explanation of the Code
This Jupyter notebook is inside of a folder called 1.OOP_syntax_shirt_practice. You can see the folder if you click on the "Jupyter" logo above the notebook. Inside the folder are three files:
- shirt_exercise.ipynb, which is the file you are currently looking at
- answer.py containing answers to the exercise
- tests.py, tests for checking your code - you can run these tests using the last code cell at the bottom of this notebook
# Your Task
The shirt_exercise.ipynb file, which you are currently looking at if you are reading this, has an exercise to help guide you through coding with an object in Python.
Fill out the TODOs in each section of the Jupyter notebook. You can find a solution in the answer.py file.
First, run this code cell below to load the Shirt class.
```
class Shirt:
def __init__(self, shirt_color, shirt_size, shirt_style, shirt_price):
self.color = shirt_color
self.size = shirt_size
self.style = shirt_style
self.price = shirt_price
def change_price(self, new_price):
self.price = new_price
def discount(self, discount):
return self.price * (1 - discount)
### TODO:
# - insantiate a shirt object with the following characteristics:
# - color red, size S, style long-sleeve, and price 25
# - store the object in a variable called shirt_one
#
#
###
shirt_one = Shirt(shirt_color='red', shirt_size= 'S', shirt_style='long-sleeve', shirt_price=25)
### TODO:
# - print the price of the shirt using the price attribute
# - use the change_price method to change the price of the shirt to 10
# - print the price of the shirt using the price attribute
# - use the discount method to print the price of the shirt with a 12% discount
#
###
print(shirt_one.price)
shirt_one.change_price(10)
print(shirt_one.price)
print(shirt_one.discount(12))
### TODO:
#
# - instantiate another object with the following characteristics:
# . - color orange, size L, style short-sleeve, and price 10
# - store the object in a variable called shirt_two
#
###
shirt_two = Shirt('orange', 'L', 'short-sleeve', 10)
### TODO:
#
# - calculate the total cost of shirt_one and shirt_two
# - store the results in a variable called total
#
###
total = shirt_two.price + shirt_one.price
### TODO:
#
# - use the shirt discount method to calculate the total cost if
# shirt_one has a discount of 14% and shirt_two has a discount
# of 6%
# - store the results in a variable called total_discount
###
total_discount = shirt_one.discount(.14) + shirt_two.discount(.06)
```
# Test your Code
The following code cell tests your code.
There is a file called tests.py containing a function called run_tests(). The run_tests() function executes a handful of assert statements to check your work. You can see this file if you go to the Jupyter Notebook menu and click on "File->Open" and then open the tests.py file.
Execute the next code cell. The code will produce an error if your answers in this exercise are not what was expected. Keep working on your code until all tests are passing.
If you run the code cell and there is no output, then you passed all the tests!
As mentioned previously, there's also a file with a solution. To find the solution, click on the Jupyter logo at the top of the workspace, and then enter the folder titled 1.OOP_syntax_shirt_practice
```
# Unit tests to check your solution
from tests import run_tests
run_tests(shirt_one, shirt_two, total, total_discount)
```
| github_jupyter |
# [NTDS'19] tutorial 5: machine learning with scikit-learn
[ntds'19]: https://github.com/mdeff/ntds_2019
[Nicolas Aspert](https://people.epfl.ch/nicolas.aspert), [EPFL LTS2](https://lts2.epfl.ch).
* Dataset: [digits](https://archive.ics.uci.edu/ml/datasets/Pen-Based+Recognition+of+Handwritten+Digits)
* Tools: [scikit-learn](https://scikit-learn.org/stable/), [numpy](http://www.numpy.org), [scipy](https://www.scipy.org), [matplotlib](https://matplotlib.org)
*scikit-learn* is a machine learning python library. Most commonly used algorithms for classification, clustering and regression are implemented as part of the library, e.g.
* [Logistic regression](https://en.wikipedia.org/wiki/Logistic_regression)
* [k-means clustering](https://en.wikipedia.org/wiki/K-means_clustering)
* [Support vector machines](https://en.wikipedia.org/wiki/Support-vector_machine)
* ...
The aim of this tutorial is to show basic usage of some simple machine learning techniques.
Check the official [documentation](https://scikit-learn.org/stable/documentation.html) for more information, especially the [tutorials](https://scikit-learn.org/stable/tutorial/index.html) section.
```
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
import sklearn
```
## Data loading
We will use a dataset named *digits*.
It is made of 1797 handwritten digits images (of size 8x8 pixels each) acquired from 44 different writers.
Each image is labelled according to the digit present in the image.
You can find more information about this dataset [here](https://archive.ics.uci.edu/ml/datasets/Pen-Based+Recognition+of+Handwritten+Digits).
Load the dataset.
```
from sklearn.datasets import load_digits
digits = load_digits()
```
The `digits` variable contains several fields.
In `images` you have all samples as 2-dimensional arrays.
```
print(digits.images.shape)
print(digits.images[0])
plt.imshow(digits.images[0], cmap=plt.cm.gray);
```
In `data`, the same samples are represented as 1-d vectors of length 64.
```
print(digits.data.shape)
print(digits.data[0])
```
In `target` you have the label corresponding to each image.
```
print(digits.target.shape)
print(digits.target)
```
Let us visualize the 20 first entries of the dataset (image display kept small on purpose)
```
fig = plt.figure(figsize=(15, 0.5))
for index, (image, label) in enumerate(zip(digits.images[0:20], digits.target[0:20])):
ax = fig.add_subplot(1, 20, index+1)
ax.imshow(image, cmap=plt.cm.gray)
ax.set_title(label)
ax.axis('off')
```
### Training/Test set
Before training our model, the [`train_test_split`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html) function will separate our dataset into a training set and a test set. The samples from the test set are never used during the training phase. This allows for a fair evaluation of the model's performance.
```
from sklearn.model_selection import train_test_split
train_img, test_img, train_lbl, test_lbl = train_test_split(
digits.data, digits.target, test_size=1/6) # keep ~300 images as test set
```
We can check that all classes are well balanced in the training and test sets.
```
np.histogram(train_lbl, bins=10)
np.histogram(test_lbl, bins=10)
```
## Supervised learning: logistic regression
### Linear regression reminder
Linear regression is used to predict an dependent value $y$ from an n-dimensional vector $x$.
The assumption made here is that the output depends linearly on the input components, i.e. $y = mx + b$.
Given a set of input and output values, the goal is to compute $m$ and $b$ minimizing the [mean squared error (MSE)](https://en.wikipedia.org/wiki/Mean_squared_error) between the predicted and actual outputs.
In scikit-learn this method is available through [`LinearRegression`](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html).
### Logistic regression
Logistic regression is used to predict categorical data (e.g. yes/no, member/non-member, ham/spam, benign/malignant, ...).
It uses the output of a linear predictor, and maps it to a probability using a [sigmoid function](https://en.wikipedia.org/wiki/Sigmoid_function), such as the logistic function $s(z) = \frac{1}{1+e^{-z}}$.
The output is a probability score between 0 and 1, and using a simple thresholding the class output will be positive if the probability is greater than 0.5, negative if not.
A [log-loss cost function](http://wiki.fast.ai/index.php/Logistic_Regression#Cost_Function) (not just the MSE as for linear regression) is used to train logistic regression (using gradient descent for instance).
[Multinomial logistic regression](https://en.wikipedia.org/wiki/Multinomial_logistic_regression) is an extension of the binary classification problem to a $n$-classes problem.
We can now create a logistic regression object and fit the parameters using the training data.
NB: as the dataset is quite simple, default parameters will give good results. Check the [documentation](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html) for fine-tuning possibilities.
```
from sklearn.linear_model import LogisticRegression
# All unspecified parameters are left to their default values.
logisticRegr = LogisticRegression(verbose=1, solver='liblinear', multi_class='auto') # set solver and multi_class to silence warnings
logisticRegr.fit(train_img, train_lbl)
```
## Model performance evaluation
For a binary classification problem, let us denote by $TP$, $TN$, $FP$, and $FN$ the number of true positives, true negatives, false positives and false negatives.
### Accuracy
The *accuracy* is defined by $a = \frac{TP}{TP + TN + FP + FN}$
NB: in scikit-learn, models may have different definitions of the `score` method. For multi-class logistic regression, the value is the mean accuracy for each class.
```
score = logisticRegr.score(test_img, test_lbl)
print(f'accuracy = {score:.4f}')
```
### F1 score
Accuracy only provides partial information about the performance of a model. Many other [metrics](https://scikit-learn.org/stable/modules/model_evaluation.html#classification-metrics) are part of scikit-learn.
A metric that provides a more complete overview of the classification performance is the [F1 score](https://en.wikipedia.org/wiki/F1_score). It takes into account not only the valid predictions but also the incorrect ones, by combining precision and recall.
*Precision* is the number of positive predictions divided by the total number of positive class values predicted, i.e. $p=\frac{TP}{TP+FP}$. A low precision indicates a high number of false positives.
*Recall* is the number of positive predictions divided by the number of positive class values in the test data, i.e. $r=\frac{TP}{TP+FN}$. A low recall indicates a high number of false negatives.
Finally the F1 score is the harmonic mean between precision and recall, i.e. $F1=2\frac{p.r}{p+r}$
Let us compute the predicted labels in the test set:
```
pred_lbl = logisticRegr.predict(test_img)
from sklearn.metrics import f1_score, classification_report
from sklearn.utils.multiclass import unique_labels
```
The [`f1_score`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html#sklearn.metrics.f1_score) function computes the F1 score. The `average` parameter controls whether the result is computed globally over all classes (`average='micro'`) or if the F1 score is computed for each class then averaged (`average='macro'`).
```
f1_score(test_lbl, pred_lbl, average='micro')
f1_score(test_lbl, pred_lbl, average='macro')
```
`classification_report` provides a synthetic overview of all results for each class, as well as globally.
```
print(classification_report(test_lbl, pred_lbl))
```
### Confusion matrix
In the case of a multi-class problem, the *confusion matrix* is often used to present the results.
```
from sklearn.metrics import confusion_matrix
def plot_confusion_matrix(y_true, y_pred, classes,
normalize=False,
title=None,
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if not title:
if normalize:
title = 'Normalized confusion matrix'
else:
title = 'Confusion matrix, without normalization'
# Compute confusion matrix
cm = confusion_matrix(y_true, y_pred)
# Only use the labels that appear in the data
classes = classes[unique_labels(y_true, y_pred)]
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
fig, ax = plt.subplots()
im = ax.imshow(cm, interpolation='nearest', cmap=cmap)
ax.figure.colorbar(im, ax=ax)
# We want to show all ticks...
ax.set(xticks=np.arange(cm.shape[1]),
yticks=np.arange(cm.shape[0]),
# ... and label them with the respective list entries
xticklabels=classes, yticklabels=classes,
title=title,
ylabel='True label',
xlabel='Predicted label')
# Rotate the tick labels and set their alignment.
plt.setp(ax.get_xticklabels(), rotation=45, ha="right",
rotation_mode="anchor")
# Loop over data dimensions and create text annotations.
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i in range(cm.shape[0]):
for j in range(cm.shape[1]):
ax.text(j, i, format(cm[i, j], fmt),
ha="center", va="center",
color="white" if cm[i, j] > thresh else "black")
fig.tight_layout()
return ax
plot_confusion_matrix(test_lbl, pred_lbl, np.array(list(map(lambda x: str(x), range(10)))), normalize=False)
```
## Supervised learning: support-vector machines
[Support-vector machines (SVM)](https://en.wikipedia.org/wiki/Support-vector_machine) are also used for classification tasks.
For a binary classification task of $n$-dimensional feature vectors, a linear SVM try to return the ($n-1$)-dimensional hyperplane that separate the two classes with the largest possible margin.
Nonlinear SVMs fit the maximum-margin hyperplane in a transformed feature space.
Although the classifier is a hyperplane in the transformed feature space, it may be nonlinear in the original input space.
The goal here is to show that a method (e.g. the previously used logistic regression) can be substituted transparently for another one.
```
from sklearn import svm
```
Default parameters perform well on this dataset.
It might be needed to adjust $C$ and $\gamma$ (e.g. via a grid search) for optimal performance (cf. [SVC documentation](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html#sklearn.svm.SVC)).
```
clf = svm.SVC(gamma='scale') # default kernel is RBF
clf.fit(train_img, train_lbl)
```
The classification accuracy improves with respect to logistic regression (here `score` also computes mean accuracy, as in logistic regression).
```
clf.score(test_img, test_lbl)
```
The F1 score is also improved.
```
pred_lbl_svm = clf.predict(test_img)
print(classification_report(test_lbl, pred_lbl_svm))
```
## Unsupervised learning: $k$-means
[$k$-means](https://en.wikipedia.org/wiki/K-means_clustering) aims at partitioning a samples into $k$ clusters, s.t. each sample belongs to the cluster having the closest mean. Its implementation is iterative, and relies on a prior knowledge of the number of clusters present.
One important step in $k$-means clustering is the initialization, i.e. the choice of initial clusters to be refined.
This choice can have a significant impact on results.
```
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=10)
kmeans.fit(digits.data)
km_labels = kmeans.predict(digits.data)
digits.target
km_labels
```
Since we have ground truth information of classes, we can check if the $k$-means results make sense.
However as you can see, the labels produced by $k$-means and the ground truth ones do not match.
An agreement score based on [mutual information](https://scikit-learn.org/stable/modules/clustering.html#clustering-evaluation), insensitive to labels permutation can be used to evaluate the results.
```
from sklearn.metrics import adjusted_mutual_info_score
adjusted_mutual_info_score(digits.target, kmeans.labels_)
```
## Unsupervized learning: dimensionality reduction
You can also try to visualize the clusters as in this [scikit-learn demo](https://scikit-learn.org/stable/auto_examples/cluster/plot_kmeans_digits.html). Mapping the input features to lower dimensional embeddings (2D or 3D), e.g. using PCA otr tSNE is required for visualization. [This demo](https://scikit-learn.org/stable/auto_examples/manifold/plot_lle_digits.html) provides an overview of the possibilities.
```
from matplotlib import offsetbox
def plot_embedding(X, y, title=None):
"""Scale and visualize the embedding vectors."""
x_min, x_max = np.min(X, 0), np.max(X, 0)
X = (X - x_min) / (x_max - x_min)
plt.figure()
ax = plt.subplot(111)
for i in range(X.shape[0]):
plt.text(X[i, 0], X[i, 1], str(y[i]),
color=plt.cm.Set1(y[i] / 10.),
fontdict={'weight': 'bold', 'size': 9})
if hasattr(offsetbox, 'AnnotationBbox'):
# only print thumbnails with matplotlib > 1.0
shown_images = np.array([[1., 1.]]) # just something big
for i in range(X.shape[0]):
dist = np.sum((X[i] - shown_images) ** 2, 1)
if np.min(dist) < 4e-3:
# don't show points that are too close
continue
shown_images = np.r_[shown_images, [X[i]]]
imagebox = offsetbox.AnnotationBbox(
offsetbox.OffsetImage(digits.images[i], cmap=plt.cm.gray_r),
X[i])
ax.add_artist(imagebox)
plt.xticks([]), plt.yticks([])
if title is not None:
plt.title(title)
from sklearn import manifold
tsne = manifold.TSNE(n_components=2, init='pca', random_state=0)
X_tsne = tsne.fit_transform(digits.data)
plot_embedding(X_tsne, digits.target,
"t-SNE embedding of the digits (ground truth labels)")
plot_embedding(X_tsne, km_labels,
"t-SNE embedding of the digits (kmeans labels)")
```
| github_jupyter |
```
%matplotlib inline
```
Introduction to artifacts and artifact detection
================================================
Since MNE supports the data of many different acquisition systems, the
particular artifacts in your data might behave very differently from the
artifacts you can observe in our tutorials and examples.
Therefore you should be aware of the different approaches and of
the variability of artifact rejection (automatic/manual) procedures described
onwards. At the end consider always to visually inspect your data
after artifact rejection or correction.
Background: what is an artifact?
--------------------------------
Artifacts are signal interference that can be
endogenous (biological) and exogenous (environmental).
Typical biological artifacts are head movements, eye blinks
or eye movements, heart beats. The most common environmental
artifact is due to the power line, the so-called *line noise*.
How to handle artifacts?
------------------------
MNE deals with artifacts by first identifying them, and subsequently removing
them. Detection of artifacts can be done visually, or using automatic routines
(or a combination of both). After you know what the artifacts are, you need
remove them. This can be done by:
- *ignoring* the piece of corrupted data
- *fixing* the corrupted data
For the artifact detection the functions MNE provides depend on whether
your data is continuous (Raw) or epoch-based (Epochs) and depending on
whether your data is stored on disk or already in memory.
Detecting the artifacts without reading the complete data into memory allows
you to work with datasets that are too large to fit in memory all at once.
Detecting the artifacts in continuous data allows you to apply filters
(e.g. a band-pass filter to zoom in on the muscle artifacts on the temporal
channels) without having to worry about edge effects due to the filter
(i.e. filter ringing). Having the data in memory after segmenting/epoching is
however a very efficient way of browsing through the data which helps
in visualizing. So to conclude, there is not a single most optimal manner
to detect the artifacts: it just depends on the data properties and your
own preferences.
In this tutorial we show how to detect artifacts visually and automatically.
For how to correct artifacts by rejection see `tut-artifact-rejection`.
To discover how to correct certain artifacts by filtering see
`tut-filter-resample` and to learn how to correct artifacts
with subspace methods like SSP and ICA see
`tut-artifact-ssp` and `tut-artifact-ica`.
Artifacts Detection
-------------------
This tutorial discusses a couple of major artifacts that most analyses
have to deal with and demonstrates how to detect them.
```
import numpy as np
import mne
from mne.datasets import sample
from mne.preprocessing import create_ecg_epochs, create_eog_epochs
# getting some data ready
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True)
```
Low frequency drifts and line noise
```
(raw.copy().pick_types(meg='mag')
.del_proj(0)
.plot(duration=60, n_channels=100, remove_dc=False))
```
we see high amplitude undulations in low frequencies, spanning across tens of
seconds
```
raw.plot_psd(tmax=np.inf, fmax=250)
```
On MEG sensors we see narrow frequency peaks at 60, 120, 180, 240 Hz,
related to line noise.
But also some high amplitude signals between 25 and 32 Hz, hinting at other
biological artifacts such as ECG. These can be most easily detected in the
time domain using MNE helper functions
See `tut-filter-resample`.
ECG
---
finds ECG events, creates epochs, averages and plots
```
average_ecg = create_ecg_epochs(raw).average()
print('We found %i ECG events' % average_ecg.nave)
joint_kwargs = dict(ts_args=dict(time_unit='s'),
topomap_args=dict(time_unit='s'))
average_ecg.plot_joint(**joint_kwargs)
```
we can see typical time courses and non dipolar topographies
not the order of magnitude of the average artifact related signal and
compare this to what you observe for brain signals
EOG
---
```
average_eog = create_eog_epochs(raw).average()
print('We found %i EOG events' % average_eog.nave)
average_eog.plot_joint(**joint_kwargs)
```
Knowing these artifact patterns is of paramount importance when
judging about the quality of artifact removal techniques such as SSP or ICA.
As a rule of thumb you need artifact amplitudes orders of magnitude higher
than your signal of interest and you need a few of such events in order
to find decompositions that allow you to estimate and remove patterns related
to artifacts.
Consider the following tutorials for correcting this class of artifacts:
- `tut-filter-resample`
- `tut-artifact-ica`
- `tut-artifact-ssp`
| github_jupyter |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from scipy.stats import kurtosis
from sklearn.decomposition import PCA
import seaborn as sns
from scipy.stats import pearsonr
%matplotlib
gov_pop_area_data = pd.read_excel('/Users/Rohil/Documents/iGEM/yemen/gov_area_pop_data.xlsx')
gov_pop_area_data = gov_pop_area_data[gov_pop_area_data.iso != 'YE-HD']
gov_pop_area_data.head()
cholera_case_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\cholera_epi_data\yemen_cholera_case_data_differenced.csv', dayfirst = True)
cholera_case_crosstab.tail()
norm_cholera_case_crosstab = cholera_case_crosstab
for index, row in gov_pop_area_data[['iso', 'population']].iterrows():
norm_cholera_case_crosstab[row.iso] = (norm_cholera_case_crosstab[row.iso] * 10000) / row.population
norm_cholera_case_crosstab.tail()
cholera_death_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\cholera_epi_data\yemen_cholera_death_data_differenced.csv', dayfirst = True)
cholera_death_crosstab.head()
norm_cholera_death_crosstab = cholera_death_crosstab
for index, row in gov_pop_area_data[['iso', 'population']].iterrows():
norm_cholera_death_crosstab[row.iso] = (norm_cholera_death_crosstab[row.iso] * 10000) / row.population
norm_cholera_death_crosstab.head()
mean_rainfall_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\rainfall\yemen_daily_mean_rainfall_crosstab.csv', dayfirst = True)
max_rainfall_crosstab = pd.read_csv(r'C:\Users\Rohil\Documents\iGEM\yemen\rainfall\yemen_daily_max_rainfall_crosstab.csv', dayfirst = True)
mean_rainfall_crosstab.head()
max_rainfall_crosstab.head()
cases_unstacked = norm_cholera_case_crosstab.set_index('date').unstack().reset_index()
cases_unstacked.columns = ['gov_iso', 'date', 'new_cases']
deaths_unstacked = norm_cholera_death_crosstab.set_index('date').unstack().reset_index()
deaths_unstacked.columns = ['gov_iso', 'date', 'new_deaths']
max_rainfall_unstacked = max_rainfall_crosstab.set_index('date').unstack().reset_index()
max_rainfall_unstacked.columns = ['gov_iso', 'date', 'max_rainfall']
mean_rainfall_unstacked = mean_rainfall_crosstab.set_index('date').unstack().reset_index()
mean_rainfall_unstacked.columns = ['gov_iso', 'date', 'mean_rainfall']
cases_unstacked.shape
cases_unstacked.head()
deaths_unstacked.shape
deaths_unstacked.head()
mean_rainfall_unstacked.shape
## date formatting has been fixed
mean_rainfall_unstacked.head()
mean_rainfall_unstacked.date.tail()
cases_unstacked.date.tail()
deaths_unstacked.date.tail()
case_death_rainfall_data = cases_unstacked.merge(deaths_unstacked, on =['date', 'gov_iso']).merge(mean_rainfall_unstacked, on =['date', 'gov_iso'], how = 'left')
case_death_rainfall_data.date = pd.to_datetime(case_death_rainfall_data.date, dayfirst = True)
case_death_rainfall_data.sort_values(by = 'date')
# YE-HD-AL refers to Al Mukulla
neighboring_gov_dict = {"YE-SA" : ["YE-SN"],
"YE-AB" : ["YE-LA", "YE-SH", "YE-BA"],
"YE-AD" : ["YE-LA"],
"YE-DA" : ["YE-LA", "YE-TA", "YE-IB", "YE-BA"],
"YE-BA" : ["YE-DH", "YE-IB", "YE-DA", "YE-AB", "YE-SH", "YE-MA", "YE-SN"],
"YE-HU" : ["YE-HJ", "YE-MW", "YE-SN", "YE-RA", "YE-DH", "YE-TA"],
"YE-JA" : ["YE-MA", "YE-SN", "YE-AM", "YE-SD"],
"YE-MR" : ["YE-HD-AL"],
"YE-MW" : ["YE-HU", "YE-HJ", "YE-AM", "YE-SN"],
"YE-AM" : ["YE-HJ", "YE-SD", "YE-JA", "YE-SN", "YE-MW"],
"YE-DH" : ["YE-IB", "YE-RA", "YE-SN", "YE-BA"],
"YE-HD-AL" : ["YE-SH", "YE-MR"],
"YE-HJ" : ["YE-MW", "YE-HU", "YE-MR"],
"YE-IB" : ["YE-TA", "YE-HU", "YE-DH", "YE-BA", "YE-DA"],
"YE-LA" : ["YE-AD", "YE-TA", "YE-DA", "YE-BA", "YE-AB"],
"YE-MA" : ["YE-BA", "YE-SN", "YE-JA", "YE-SH"],
"YE-RA" : ["YE-DH", "YE-HU", "YE-SN"],
"YE-SD" : ["YE-HJ", "YE-AM", "YE-JA"],
"YE-SN" : ["YE-BA", "YE-DH", "YE-RA", "YE-MW", "YE-AM", "YE-JA", "YE-MA"],
"YE-SH" : ["YE-AB", "YE-BA", "YE-MA", "YE-HD-AL"],
"YE-TA" : ["YE-LA", "YE-DA", "YE-IB", "YE-HU"]}
def get_past_days_features(row, var, daysback):
stock_data = full_data[full_data.stock_id == row.stock_id].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_stock_data = stock_data.loc[(stock_data.index >= x_days_date) & (stock_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_stock_data[var]), np.max(relevant_stock_data[var]), kurtosis(relevant_stock_data[var])]))
def get_past_days_features(row, var, daysback):
other_stock_data = full_data[full_data.stock_id.isin(neighboring_stocks[row.stock_id])].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_other_stock_data = other_stock_data.loc[(other_stock_data.index >= x_days_date) & (other_stock_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_other_stock_data[var]), np.max(relevant_other_stock_data[var]), kurtosis(relevant_other_stock_data[var])]))
def get_past_days_features(row, var, daysback):
if 'rainfall' in var:
rainfall_df = mean_rainfall_unstacked
rainfall_df.date = pd.to_datetime(rainfall_df.date, dayfirst = True)
gov_data = rainfall_df[rainfall_df.gov_iso == row.gov_iso].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_gov_data = gov_data.loc[(gov_data.index >= x_days_date) & (gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_gov_data[var]), np.max(relevant_gov_data[var]), kurtosis(relevant_gov_data[var])]))
else:
gov_data = case_death_rainfall_data[case_death_rainfall_data.gov_iso == row.gov_iso].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_gov_data = gov_data.loc[(gov_data.index >= x_days_date) & (gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_gov_data[var]), np.max(relevant_gov_data[var]), kurtosis(relevant_gov_data[var])]))
def get_neighbor_past_days_features(row, var, daysback):
if 'rainfall' in var:
rainfall_df = mean_rainfall_unstacked
rainfall_df.date = pd.to_datetime(rainfall_df.date, dayfirst = True)
other_gov_data = rainfall_df[rainfall_df.gov_iso.isin(neighboring_gov_dict[row.gov_iso])].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_other_gov_data = other_gov_data.loc[(other_gov_data.index >= x_days_date) & (other_gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_other_gov_data[var]), np.max(relevant_other_gov_data[var]), kurtosis(relevant_other_gov_data[var])]))
else:
other_gov_data = case_death_rainfall_data[case_death_rainfall_data.gov_iso.isin(neighboring_gov_dict[row.gov_iso])].set_index('date')
x_days_date = row.date - pd.to_timedelta(daysback, unit='d')
relevant_other_gov_data = other_gov_data.loc[(other_gov_data.index >= x_days_date) & (other_gov_data.index < row.date)].sort_index()
return (pd.Series([np.mean(relevant_other_gov_data[var]), np.max(relevant_other_gov_data[var]), kurtosis(relevant_other_gov_data[var])]))
past_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 7), axis = 1)
past_week_cases.columns = ['mean_past_week_cases', 'max_past_week_cases', 'kurtosis_past_week_cases']
neighbor_past_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 7), axis = 1)
neighbor_past_week_cases.columns = ['neighbor_mean_past_week_cases', 'neighbor_max_past_week_cases', 'neighbor_kurtosis_past_week_cases']
past_2_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 14), axis = 1)
past_2_week_cases.columns = ['mean_past_2_week_cases', 'max_past_2_week_cases', 'kurtosis_past_2_week_cases']
neighbor_past_2_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 14), axis = 1)
neighbor_past_2_week_cases.columns = ['neighbor_mean_past_2_week_cases', 'neighbor_max_past_2_week_cases', 'neighbor_kurtosis_past_2_week_cases']
past_3_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 21), axis = 1)
past_3_week_cases.columns = ['mean_past_3_week_cases', 'max_past_3_week_cases', 'kurtosis_past_3_week_cases']
neighbor_past_3_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 21), axis = 1)
neighbor_past_3_week_cases.columns = ['neighbor_mean_past_3_week_cases', 'neighbor_max_past_3_week_cases', 'neighbor_kurtosis_past_3_week_cases']
past_month_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 30), axis = 1)
past_month_cases.columns = ['mean_past_month_cases', 'max_past_month_cases', 'kurtosis_past_month_cases']
neighbor_past_month_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 30), axis = 1)
neighbor_past_month_cases.columns = ['neighbor_mean_past_month_cases', 'neighbor_max_past_month_cases', 'neighbor_kurtosis_past_month_cases']
past_6_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 42), axis = 1)
past_6_week_cases.columns = ['mean_past_6_week_cases', 'max_past_6_week_cases', 'kurtosis_past_6_week_cases']
neighbor_past_6_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 42), axis = 1)
neighbor_past_6_week_cases.columns = ['neighbor_mean_past_6_week_cases', 'neighbor_max_past_6_week_cases', 'neighbor_kurtosis_past_6_week_cases']
past_week_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 7), axis = 1)
past_week_deaths.columns = ['mean_past_week_deaths', 'max_past_week_deaths', 'kurtosis_past_week_deaths']
neighbor_past_week_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 7), axis = 1)
neighbor_past_week_deaths.columns = ['neighbor_mean_past_week_deaths', 'neighbor_max_past_week_deaths', 'neighbor_kurtosis_past_week_deaths']
past_2_week_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 14), axis = 1)
past_2_week_deaths.columns = ['mean_past_2_week_deaths', 'max_past_2_week_deaths', 'kurtosis_past_2_week_deaths']
neighbor_past_2_week_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 14), axis = 1)
neighbor_past_2_week_deaths.columns = ['neighbor_mean_past_2_week_deaths', 'neighbor_max_past_2_week_deaths', 'neighbor_kurtosis_past_2_week_deaths']
past_month_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 30), axis = 1)
past_month_deaths.columns = ['mean_past_month_deaths', 'max_past_month_deaths', 'kurtosis_past_month_deaths']
neighbor_past_month_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 30), axis = 1)
neighbor_past_month_deaths.columns = ['neighbor_mean_past_month_deaths', 'neighbor_max_past_month_deaths', 'neighbor_kurtosis_past_month_deaths']
past_week_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 7), axis = 1)
past_week_rainfall.columns = ['mean_past_week_rainfall', 'max_past_week_rainfall', 'kurtosis_past_week_rainfall']
neighbor_past_week_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 7), axis = 1)
neighbor_past_week_rainfall.columns = ['neighbor_mean_past_week_rainfall', 'neighbor_max_past_week_rainfall', 'neighbor_kurtosis_past_week_rainfall']
past_2_week_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 14), axis = 1)
past_2_week_rainfall.columns = ['mean_past_2_week_rainfall', 'max_past_2_week_rainfall', 'kurtosis_past_2_week_rainfall']
neighbor_past_2_week_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 14), axis = 1)
neighbor_past_2_week_rainfall.columns = ['neighbor_mean_past_2_week_rainfall', 'neighbor_max_past_2_week_rainfall', 'neighbor_kurtosis_past_2_week_rainfall']
past_month_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 30), axis = 1)
past_month_rainfall.columns = ['mean_past_month_rainfall', 'max_past_month_rainfall', 'kurtosis_past_month_rainfall']
neighbor_past_month_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 30), axis = 1)
neighbor_past_month_rainfall.columns = ['neighbor_mean_past_month_rainfall', 'neighbor_max_past_month_rainfall', 'neighbor_kurtosis_past_month_rainfall']
training_data = pd.concat([case_death_rainfall_data[['gov_iso', 'date', 'weekly_cases']],
past_week_cases, past_2_week_cases, past_month_cases, neighbor_past_week_cases, neighbor_past_2_week_cases, neighbor_past_month_cases,
past_week_deaths, past_2_week_deaths, past_month_deaths, neighbor_past_week_deaths, neighbor_past_2_week_deaths, neighbor_past_month_deaths,
past_week_rainfall, past_2_week_rainfall, past_month_rainfall, neighbor_past_week_rainfall, neighbor_past_2_week_rainfall, neighbor_past_month_rainfall], axis = 1)
training_data.to_csv('/Users/Rohil/Documents/iGEM/yemen/full_feature_data.csv', index = False)
col_list = []
for col in training_data.columns:
if ('max' not in col) and ('kurtosis' not in col) & ('deaths' not in col):
col_list.append(col)
# want to have at least 7 days of data for most of these examples
trunc_training_data = training_data[col_list]
trunc_training_data = trunc_training_data[(trunc_training_data['date'] > '2017-05-30')].sort_values('date')
features = trunc_training_data.iloc[:,3:].columns.tolist()
target = trunc_training_data.iloc[:,2].name
correlations = {}
for f in features:
data_temp = trunc_training_data[[f,target]]
x1 = data_temp[f].values
x2 = data_temp[target].values
key = f + ' vs ' + target
correlations[key] = pearsonr(x1,x2)[0]
data_correlations = pd.DataFrame(correlations, index=['Value']).T
data_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]
trunc_training_data = pd.concat([trunc_training_data, pd.get_dummies(trunc_training_data.gov_iso).sort_index()], axis=1)
trunc_training_data.to_csv('/Users/Rohil/Documents/iGEM/yemen/prelim_training_data.csv', index = False)
trunc_training_data[trunc_training_data.isnull().any(axis=1)]
trunc_training_data.shape
trunc_training_data.head()
whole_standard_scaler = StandardScaler()
trunc_training_features = trunc_training_data.iloc[:,3:]
trunc_training_features.shape
norm_features = whole_standard_scaler.fit_transform(trunc_training_features)
pca = PCA(n_components = 33)
pca.fit(norm_features)
pca.explained_variance_ratio_
pd.DataFrame(pca.components_, columns = trunc_training_features.columns)
pca.components_
sns.heatmap(np.log(pca.inverse_transform(np.eye(12))))
# plots of normalized cases x days back vs today
for column in trunc_training_features.columns:
fig, ax = plt.subplots(1,1)
ax.scatter(trunc_training_features[column], trunc_training_data['weekly_cases'])
ax.set_ylabel('weekly cholera cases')
ax.set_xlabel(column)
fig.savefig('/Users/Rohil/Documents/iGEM/yemen/feature_engineering/old/' + column + '_vs_cases.png')
plt.close()
norm_features = pd.DataFrame(data=norm_features, columns = trunc_training_features.columns)
norm_features
```
| github_jupyter |
# Implement image blending
We will start by importing libraries and defining a couple of functions for displaying images using matplotlib.
```
import cv2
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
plt.rcParams['figure.figsize'] = [20, 10]
def showResult(title, img):
# Colour images in OpenCV are given in BGR, but matplotlib expects RGB.
# We therefore need to convert the OpenCV images.
plt.imshow(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))
plt.title(title)
plt.show()
def showResultsSideBySide(title1, img1, title2, img2):
# Display the original and the transformed image
axes = plt.subplots(1, 2)[1]
ax1, ax2 = axes
ax1.set_title(title1)
ax2.set_title(title2)
ax1.imshow(cv2.cvtColor(img1, cv2.COLOR_BGR2RGB))
ax2.imshow(cv2.cvtColor(img2, cv2.COLOR_BGR2RGB))
plt.show()
```
## 1. Load and convert the source images
Now let's do some image processing!
First, we need to read the two images we want to blend.
<table cellspacing="0" cellpadding="0"><tr>
<td> <img src="img/thumb_lion.png" alt="Lion" style="width: 200px;"/> </td>
<td> <img src="img/thumb_tiger.png" alt="Tiger" style="width: 200px;"/> </td>
<td> <img src="img/thumb_white_tiger.png" alt="White tiger" style="width: 200px;"/> </td>
</tr></table>
- Read two images (choose two of the images given in the `data` directory)
- Hint: [cv::imread(...)](https://docs.opencv.org/4.0.1/d4/da8/group__imgcodecs.html#ga288b8b3da0892bd651fce07b3bbd3a56)
- Convert the images to `float32` and scale the pixel values so that they will lie in the interval [0, 1]
- Hint: [numpy.float32](https://numpy.org/doc/stable/reference/arrays.scalars.html?#numpy.float32)
```
# Load the images.
img_01_fname = "data/tiger.png"
img_01 = cv2.imread(img_01_fname)
img_01 = np.float32(img_01) * (1.0/255.0)
img_02_fname = "data/white_tiger.png"
img_02 = cv2.imread(img_02_fname)
img_02 = np.float32(img_02) * (1.0/255.0)
# Show the loaded images.
showResultsSideBySide(img_01_fname, img_01, img_02_fname, img_02)
```
## 2. Create an image with weights for blending
Now we need to define how the two images should be blended together.
We will do this by constructing a weight image with weights $w(u, v) \in [0, 1].$
A weight of 1 means that the blended pixel will be equal to the corresponding pixel in image 1, while a weight of 0.5 means that the resulting pixel is an equally large mix of both images.
### a) Create the weight image:
- The size is equal to the size of the input images
- It should have 3 channels given in 32-bit floating point
- The left half of the image should be black (pixel value 0.0 in all channels)
- The right half of the image should be white (pixel value 1.0 in all channels)
### b) Make a ramp (a smooth gradient) in the transition between black and white
- Hint: [cv::blur](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#ga8c45db9afe636703801b0b2e440fce37)

```
# Construct a half black, half white image.
weights = np.zeros(img_01.shape, dtype=np.float32)
half_image_width = int(0.5 * weights.shape[1])
weights[:, :half_image_width] = (1., 1., 1.)
# Create a ramp between the two halves.
ramp_width = 50
weights = cv2.blur(weights, (ramp_width+1, ramp_width+1))
# Visualise the weights.
showResult("weights", weights)
```
## 3. Simple linear blending
The next step is to implement functionality for simple linear blending, where the two images are mixed according to the weight image.
### a) Implement linear blending of two images using the weights
- $res = w \cdot img_1 + (1-w) \cdot img_2$
- Tip: You can solve this step using only image operations, without writing any loops.
### b) Run the code and check that the result looks reasonable
- Try changing the ramp size `ramp_width`. What happens?
- Try making the blend as smooth and visually pleasing as possible.
```
def linearBlending(img1, img2, mask):
return img1 * mask + img2 * (1.-mask)
# Test linear blending.
linear_blend = linearBlending(img_01, img_02, weights)
showResult('linear_blend', linear_blend)
```
## 4. Laplace blending
To demonstrate the difference between simple linear blending, and scale-aware blending, we will now implement and test Laplace blending.
We will even get to play around with scale pyramids!
Recall from the lecture that Laplace blending performs linear blending at different stages in the laplacian pyramid for an image (at different scales):

First we convert the image to a laplacian pyramid, then we perform the linear blending, and finally we reconstruct the blended laplacian into the resulting blended image.
### a) Construct a Gaussian pyramid

- Hint: Use [cv::pyrDown()](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#gaf9bba239dfca11654cb7f50f889fc2ff)
```
def constructGaussianPyramid(img):
# Construct the pyramid starting with the original image.
pyr = [img]
# Add new downscaled images to the pyramid
# until image width is <= 16 pixels
while pyr[-1].shape[1] > 16:
pyr.append(cv2.pyrDown(pyr[-1]))
return pyr
```
### b) Construct a Laplacian pyramid

- Hint: Use [cv::pyrUp()](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#gada75b59bdaaca411ed6fee10085eb784)
```
def constructLaplacianPyramid(img):
pyr = constructGaussianPyramid(img)
for i in range(len(pyr)-1):
pyr[i] -= cv2.pyrUp(pyr[i+1], dstsize=pyr[i].shape[0:2])
return pyr
```
### c) Implement function for collapsing the Laplacian pyramid

- Hint: Use [cv::pyrUp()](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#gada75b59bdaaca411ed6fee10085eb784)
```
def collapsePyramid(pyr):
for i in range(len(pyr)-2, -1, -1):
pyr[i] += cv2.pyrUp(pyr[i+1], dstsize=pyr[i].shape[0:2])
return pyr[0]
```
### d) Implement Laplacian blending
- Construct a Gaussian pyramid for the weights.
- Construct Laplacian pyramids for the images.
- Blend the images using `linearBlending()` on each pyramid level.
- Reconstruct the blended image by collapsing the blended pyramid.
```
def laplaceBlending(img1, img2, mask):
# Construct a gaussian pyramid of the mask image.
pyr_mask = constructGaussianPyramid(mask)
# Construct a laplacian pyramid of each of the images.
pyr_img1 = constructLaplacianPyramid(img1)
pyr_img2 = constructLaplacianPyramid(img2)
# Blend the laplacian pyramids according to the corresponding weight pyramid.
pyr_blend = []
for img1_lvl, img2_lvl, mask_lvl in zip(pyr_img1, pyr_img2, pyr_mask):
pyr_blend.append(
linearBlending(img1_lvl, img2_lvl, mask_lvl)
)
# Collapse the blended Laplacian pyramid.
return collapsePyramid(pyr_blend)
```
### e) Check that the results look reasonable
- Test Laplace blending
- Compare the results with linear blending.
- What happens when you reduce the ramp size down to a very steep gradient?
```
lap_blend = laplaceBlending(img_01, img_02, weights)
showResult('blend', lap_blend)
```
### f) Experiments
Try other images
- Capture images using the camera
- Download images from the internet
Try other weight masks
- Circles
- Other shapes
| github_jupyter |
<center>
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png" width="300" alt="cognitiveclass.ai logo" />
</center>
# 2D Numpy in Python
Estimated time needed: **20** minutes
## Objectives
After completing this lab you will be able to:
* Operate comfortably with `numpy`
* Perform complex operations with `numpy`
<h2>Table of Contents</h2>
<div class="alert alert-block alert-info" style="margin-thttps://op/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01: 20px">
<ul>
<li><a href="https://create/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01">Create a 2D Numpy Array</a></li>
<li><a href="https://access/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01">Accessing different elements of a Numpy Array</a></li>
<li><a href="op">Basic Operations</a></li>
</ul>
</div>
<hr>
<h2 id="create">Create a 2D Numpy Array</h2>
```
# Import the libraries
import numpy as np
import matplotlib.pyplot as plt
```
Consider the list <code>a</code>, which contains three nested lists **each of equal size**.
```
# Create a list
a = [[11, 12, 13], [21, 22, 23], [31, 32, 33]]
a
```
We can cast the list to a Numpy Array as follows:
```
# Convert list to Numpy Array
# Every element is the same type
A = np.array(a)
A
```
We can use the attribute <code>ndim</code> to obtain the number of axes or dimensions, referred to as the rank.
```
# Show the numpy array dimensions
A.ndim
```
Attribute <code>shape</code> returns a tuple corresponding to the size or number of each dimension.
```
# Show the numpy array shape
A.shape
```
The total number of elements in the array is given by the attribute <code>size</code>.
```
# Show the numpy array size
A.size
```
<hr>
<h2 id="access">Accessing different elements of a Numpy Array</h2>
We can use rectangular brackets to access the different elements of the array. The correspondence between the rectangular brackets and the list and the rectangular representation is shown in the following figure for a 3x3 array:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoEg.png" width="500" />
We can access the 2nd-row, 3rd column as shown in the following figure:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoFT.png" width="400" />
We simply use the square brackets and the indices corresponding to the element we would like:
```
# Access the element on the second row and third column
A[1, 2]
```
We can also use the following notation to obtain the elements:
```
# Access the element on the second row and third column
A[1][2]
```
Consider the elements shown in the following figure
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoFF.png" width="400" />
We can access the element as follows:
```
# Access the element on the first row and first column
A[0][0]
```
We can also use slicing in numpy arrays. Consider the following figure. We would like to obtain the first two columns in the first row
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoFSF.png" width="400" />
This can be done with the following syntax:
```
# Access the element on the first row and first and second columns
A[0][0:2]
```
Similarly, we can obtain the first two rows of the 3rd column as follows:
```
# Access the element on the first and second rows and third column
A[0:2, 2]
```
Corresponding to the following figure:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/2D_numpy.png" width="550"><br />
<h2 id="op">Basic Operations</h2>
We can also add arrays. The process is identical to matrix addition. Matrix addition of <code>X</code> and <code>Y</code> is shown in the following figure:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoAdd.png" width="500" />
The numpy array is given by <code>X</code> and <code>Y</code>
```
# Create a numpy array X
X = np.array([[1, 0], [0, 1]])
X
# Create a numpy array Y
Y = np.array([[2, 1], [1, 2]])
Y
```
We can add the numpy arrays as follows.
```
# Add X and Y
Z = X + Y
Z
```
Multiplying a numpy array by a scaler is identical to multiplying a matrix by a scaler. If we multiply the matrix <code>Y</code> by the scaler 2, we simply multiply every element in the matrix by 2, as shown in the figure.
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoDb.png" width="500" />
We can perform the same operation in numpy as follows
```
# Create a numpy array Y
Y = np.array([[2, 1], [1, 2]])
Y
# Multiply Y with 2
Z = 2 * Y
Z
```
Multiplication of two arrays corresponds to an element-wise product or <em>Hadamard product</em>. Consider matrix <code>X</code> and <code>Y</code>. The Hadamard product corresponds to multiplying each of the elements in the same position, i.e. multiplying elements contained in the same color boxes together. The result is a new matrix that is the same size as matrix <code>Y</code> or <code>X</code>, as shown in the following figure.
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoMul.png" width="500" />
We can perform element-wise product of the array <code>X</code> and <code>Y</code> as follows:
```
# Create a numpy array Y
Y = np.array([[2, 1], [1, 2]])
Y
# Create a numpy array X
X = np.array([[1, 0], [0, 1]])
X
# Multiply X with Y
Z = X * Y
Z
```
We can also perform matrix multiplication with the numpy arrays <code>A</code> and <code>B</code> as follows:
First, we define matrix <code>A</code> and <code>B</code>:
```
# Create a matrix A
A = np.array([[0, 1, 1], [1, 0, 1]])
A
# Create a matrix B
B = np.array([[1, 1], [1, 1], [-1, 1]])
B
```
We use the numpy function <code>dot</code> to multiply the arrays together.
```
# Calculate the dot product
Z = np.dot(A,B)
Z
# Calculate the sine of Z
np.sin(Z)
```
We use the numpy attribute <code>T</code> to calculate the transposed matrix
```
# Create a matrix C
C = np.array([[1,1],[2,2],[3,3]])
C
# Get the transposed of C
C.T
```
<h2>Quiz on 2D Numpy Array</h2>
Consider the following list <code>a</code>, convert it to Numpy Array.
```
# Write your code below and press Shift+Enter to execute
a = [[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]]
```
<details><summary>Click here for the solution</summary>
```python
A = np.array(a)
A
```
</details>
Calculate the numpy array size.
```
# Write your code below and press Shift+Enter to execute
```
<details><summary>Click here for the solution</summary>
```python
A.size
```
</details>
Access the element on the first row and first and second columns.
```
# Write your code below and press Shift+Enter to execute
```
<details><summary>Click here for the solution</summary>
```python
A[0][0:2]
```
</details>
Perform matrix multiplication with the numpy arrays <code>A</code> and <code>B</code>.
```
# Write your code below and press Shift+Enter to execute
B = np.array([[0, 1], [1, 0], [1, 1], [-1, 0]])
```
<details><summary>Click here for the solution</summary>
```python
X = np.dot(A,B)
X
```
</details>
<hr>
<h2>The last exercise!</h2>
<p>Congratulations, you have completed your first lesson and hands-on lab in Python.
<hr>
## Author
<a href="https://www.linkedin.com/in/joseph-s-50398b136/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01" target="_blank">Joseph Santarcangelo</a>
## Other contributors
<a href="https://www.linkedin.com/in/jiahui-mavis-zhou-a4537814a?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01">Mavis Zhou</a>
## Change Log
| Date (YYYY-MM-DD) | Version | Changed By | Change Description |
| ----------------- | ------- | ---------- | ----------------------------------------------------------- |
| 2022-01-10 | 2.1 | Malika | Removed the readme for GitShare |
| 2021-01-05 | 2.2 | Malika | Updated the solution for dot multiplication |
| 2020-09-09 | 2.1 | Malika | Updated the screenshot for first two rows of the 3rd column |
| 2020-08-26 | 2.0 | Lavanya | Moved lab to course repo in GitLab |
| | | | |
| | | | |
<hr/>
## <h3 align="center"> © IBM Corporation 2020. All rights reserved. <h3/>
| github_jupyter |
```
import torch
from torch import optim
import torch.nn as nn
import torch.nn.functional as F
import torch.autograd as autograd
from torch.autograd import Variable
from sklearn.preprocessing import OneHotEncoder
import os, math, glob, argparse
from utils.torch_utils import *
from utils.utils import *
from mpradragonn_predictor_pytorch import *
import matplotlib.pyplot as plt
import utils.language_helpers
#plt.switch_backend('agg')
import numpy as np
from models import *
from wgan_gp_mpradragonn_analyzer_quantile_cutoff import *
use_cuda = torch.cuda.is_available()
device = torch.device('cuda:0' if use_cuda else 'cpu')
from torch.distributions import Normal as torch_normal
class IdentityEncoder :
def __init__(self, seq_len, channel_map) :
self.seq_len = seq_len
self.n_channels = len(channel_map)
self.encode_map = channel_map
self.decode_map = {
nt: ix for ix, nt in self.encode_map.items()
}
def encode(self, seq) :
encoding = np.zeros((self.seq_len, self.n_channels))
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
return encoding
def encode_inplace(self, seq, encoding) :
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
def encode_inplace_sparse(self, seq, encoding_mat, row_index) :
raise NotImplementError()
def decode(self, encoding) :
seq = ''
for pos in range(0, encoding.shape[0]) :
argmax_nt = np.argmax(encoding[pos, :])
max_nt = np.max(encoding[pos, :])
seq += self.decode_map[argmax_nt]
return seq
def decode_sparse(self, encoding_mat, row_index) :
raise NotImplementError()
class ActivationMaximizer(nn.Module) :
def __init__(self, generator_dir, batch_size=1, seq_len=145, latent_size=128, sequence_template=None):
super(ActivationMaximizer, self).__init__()
self.generator = Generator_lang(4, seq_len, batch_size, 512)
self.predictor = DragoNNClassifier(batch_size=batch_size).cnn
self.load_generator(generator_dir)
self.use_cuda = torch.cuda.is_available()
self.x_mask = None
self.x_template = None
if sequence_template is not None :
onehot_mask = np.zeros((seq_len, 4))
onehot_template = np.zeros((seq_len, 4))
for j in range(len(sequence_template)) :
if sequence_template[j] == 'N' :
onehot_mask[j, :] = 1.
elif sequence_template[j] == 'A' :
onehot_template[j, 0] = 1.
elif sequence_template[j] == 'C' :
onehot_template[j, 1] = 1.
elif sequence_template[j] == 'G' :
onehot_template[j, 2] = 1.
elif sequence_template[j] == 'T' :
onehot_template[j, 3] = 1.
self.x_mask = Variable(torch.FloatTensor(onehot_mask).unsqueeze(0))
self.x_template = Variable(torch.FloatTensor(onehot_template).unsqueeze(0))
if self.use_cuda :
self.x_mask = self.x_mask.to(device)
self.x_template = self.x_template.to(device)
self.predictor.eval()
if self.use_cuda :
self.generator.cuda()
self.predictor.cuda()
self.cuda()
def load_generator(self, directory, iteration=None) :
list_generator = glob.glob(directory + "G*.pth")
generator_file = max(list_generator, key=os.path.getctime)
self.generator.load_state_dict(torch.load(generator_file))
def forward(self, z) :
x = self.generator.forward(z)
if self.x_mask is not None :
x = x * self.x_mask + self.x_template
return self.predictor.forward(x.unsqueeze(2).transpose(1, 3))
def get_pattern(self, z) :
x = self.generator.forward(z)
if self.x_mask is not None :
x = x * self.x_mask + self.x_template
return x
#Sequence length
seq_len = 145
batch_size = 64
#Sequence decoder
acgt_encoder = IdentityEncoder(seq_len, {'A':0, 'C':1, 'G':2, 'T':3})
#Sequence template
sequence_template = 'N' * 145
#Activation maximization model (pytorch)
act_maximizer = ActivationMaximizer(batch_size=batch_size, seq_len=seq_len, generator_dir='./checkpoint/' + 'mpradragonn_sample' + '/', sequence_template=sequence_template)
#Function for optimizing n sequences for a target predictor
def optimize_sequences(act_maximizer, n_seqs, batch_size=1, latent_size=128, n_iters=100, eps1=0., eps2=0.1, noise_std=1e-6, use_adam=True, run_name='default', store_intermediate_n_seqs=None, store_every_iter=100) :
z = Variable(torch.randn(batch_size, latent_size, device="cuda"), requires_grad=True)
norm_var = torch_normal(0, 1)
optimizer = None
if use_adam :
optimizer = optim.Adam([z], lr=eps2)
else :
optimizer = optim.SGD([z], lr=1)
z.register_hook(lambda grad, batch_size=batch_size, latent_size=latent_size, noise_std=noise_std: grad + noise_std * torch.randn(batch_size, latent_size, device="cuda"))
seqs = []
fitness_histo = []
n_batches = n_seqs // batch_size
for batch_i in range(n_batches) :
if batch_i % 4 == 0 :
print("Optimizing sequence batch " + str(batch_i))
#Re-initialize latent GAN seed
z.data = torch.randn(batch_size, latent_size, device="cuda")
fitness_scores_batch = [act_maximizer(z)[:, 0].data.cpu().numpy().reshape(-1, 1)]
for curr_iter in range(n_iters) :
fitness_score = act_maximizer(z)[:, 0]
fitness_loss = -torch.sum(fitness_score)
z_prior = -torch.sum(norm_var.log_prob(z))
loss = None
if use_adam :
loss = fitness_loss
else :
loss = eps1 * z_prior + eps2 * fitness_loss
optimizer.zero_grad()
loss.backward()
optimizer.step()
fitness_scores_batch.append(fitness_score.data.cpu().numpy().reshape(-1, 1))
if store_intermediate_n_seqs is not None and batch_i * batch_size < store_intermediate_n_seqs and curr_iter % store_every_iter == 0 :
onehot_batch = act_maximizer.get_pattern(z).data.cpu().numpy()
seq_batch = [
acgt_encoder.decode(onehot_batch[k]) for k in range(onehot_batch.shape[0])
]
with open(run_name + "_curr_iter_" + str(curr_iter) + ".txt", "a+") as f :
for i in range(len(seq_batch)) :
seq = seq_batch[i]
f.write(seq + "\n")
onehot_batch = act_maximizer.get_pattern(z).data.cpu().numpy()
seq_batch = [
acgt_encoder.decode(onehot_batch[k]) for k in range(onehot_batch.shape[0])
]
seqs.extend(seq_batch)
fitness_histo.append(np.concatenate(fitness_scores_batch, axis=1))
fitness_histo = np.concatenate(fitness_histo, axis=0)
return seqs, fitness_histo
n_seqs = 4096#960
n_iters = 1000
run_name = 'killoran_mpradragonn_' + str(n_seqs) + "_sequences" + "_" + str(n_iters) + "_iters_sample_wgan"
seqs, fitness_scores = optimize_sequences(
act_maximizer,
n_seqs,
batch_size=64,
latent_size=128,
n_iters=n_iters,
eps1=0.,
eps2=0.1,
noise_std=1e-6,
use_adam=True,
run_name="samples/killoran_mpradragonn/" + run_name,
store_intermediate_n_seqs=None,#960,
store_every_iter=100
)
#Plot fitness statistics of optimization runs
#Plot k trajectories
plot_n_traj = 100
f = plt.figure(figsize=(8, 6))
for i in range(min(plot_n_traj, n_seqs)) :
plt.plot(fitness_scores[i, :], linewidth=2, alpha=0.75)
plt.xlabel("Training iteration", fontsize=14)
plt.ylabel("Fitness score", fontsize=14)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlim(0, n_iters)
plt.ylim(-3, 3)
plt.tight_layout()
plt.show()
#Plot mean trajectory
f = plt.figure(figsize=(8, 6))
plt.plot(np.mean(fitness_scores, axis=0), linewidth=2, alpha=0.75)
plt.xlabel("Training iteration", fontsize=14)
plt.ylabel("Fitness score", fontsize=14)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlim(0, n_iters)
plt.ylim(-3, 3)
plt.tight_layout()
plt.show()
#Save sequences to file
with open(run_name + ".txt", "wt") as f :
for i in range(len(seqs)) :
seq = seqs[i]
f.write(seq + "\n")
```
| github_jupyter |
# Plotting and Functions
This notebook will work trough how to plot data and how to define functions. Throughout the lecture we will take a few moments to plot different functions and see how they depend on their parameters
## Plotting in Python: Matplot
```
import matplotlib.pyplot as plt
import numpy as np
import scipy as sp
```
Pyplot is a powerful plotting library that can be used to make publication quaility plots. It is also useful for quikly plotting the results of a calcualtion.
This is a quick demonstration of its use
Note: when you call al library `import matplotlib.pyplot as plt` the way that use it is to do the following `plt.function()` where `function()` is whatever you are trying to call from the library
```
# Define x and y values for some function
x = [i for i in range(20)]
y1 = [i**2 for i in x]
y2 = [i**3 for i in x]
```
The methods used above to make the lists is considered very *pythonic*. It works the same as a loop, but outputs all the results into a list. The left-hand most argument is what the list elements will be and the right hand side is the the way the loop will work.
When you use pyplot to make a plot, you can add more than one data set to the figure until you render the plot. Once you render the plot it resets
```
plt.plot(x,y1)
plt.plot(x,y2)
plt.xlabel('X', fontsize=24)
plt.ylabel('Y', fontsize=24)
plt.legend(['Quadratic', 'Cubic'], loc=0)
plt.show()
```
We can call also use numpy fucntions to make our plots. Numpy is a very powerful math library
```
# linspace will make a list of values from initial to final with however many increments you want
# this example goes from 0-2.5 with 20 increments
x=numpy.linspace(0,1.0,20)
print(x)
exp_func=np.exp(-2*np.pi*x)
print(exp_func)
plt.plot(x,exp_func, color="black")
plt.xlabel('x', fontsize=24)
plt.ylabel("y(x)", fontsize=24)
plt.show()
```
All aspects of the plot can be changed. The best way to figure out what you want to do is to go to the Matplotlib gallery and choose an image that looks like what you are trying to do.
https://matplotlib.org/gallery/index.html
### Example: Scatter plot with histograms
```
import numpy as np
#Fixing random state for reproducibility
np.random.seed(19680801)
# the random data
x = np.random.randn(1000)
y = np.random.randn(1000)
# definitions for the axes
left, width = 0.1, 0.65
bottom, height = 0.1, 0.65
spacing = 0.005
rect_scatter = [left, bottom, width, height]
rect_histx = [left, bottom + height + spacing, width, 0.2]
rect_histy = [left + width + spacing, bottom, 0.2, height]
# start with a rectangular Figure
plt.figure(figsize=(8, 8))
ax_scatter = plt.axes(rect_scatter)
ax_scatter.tick_params(direction='in', top=True, right=True)
ax_histx = plt.axes(rect_histx)
ax_histx.tick_params(direction='in', labelbottom=False)
ax_histy = plt.axes(rect_histy)
ax_histy.tick_params(direction='in', labelleft=False)
# the scatter plot:
ax_scatter.scatter(x, y)
# now determine nice limits by hand:
binwidth = 0.25
lim = np.ceil(np.abs([x, y]).max() / binwidth) * binwidth
ax_scatter.set_xlim((-lim, lim))
ax_scatter.set_ylim((-lim, lim))
bins = np.arange(-lim, lim + binwidth, binwidth)
ax_histx.hist(x, bins=bins)
ax_histy.hist(y, bins=bins, orientation='horizontal')
ax_histx.set_xlim(ax_scatter.get_xlim())
ax_histy.set_ylim(ax_scatter.get_ylim())
plt.show()
```
I don't have to be an expert in making that kind of plot. I just have to understand and guess enough to figure out. I also google things I don't know
https://www.google.com/search?client=firefox-b-1-d&q=pyplot+histogram+change+color
https://stackoverflow.com/questions/42172440/python-matplotlib-histogram-color?rq=1
https://matplotlib.org/examples/color/named_colors.html
Then I can make small changes to have the plot look how I want it to look
Notice below I changed
`ax_scatter.scatter(x, y, color="purple")`,
`ax_histx.hist(x, bins=bins, color = "skyblue")`,
`ax_histy.hist(y, bins=bins, orientation='horizontal', color="salmon")`
```
#Fixing random state for reproducibility
np.random.seed(19680801)
# the random data
x = np.random.randn(1000)
y = np.random.randn(1000)
# definitions for the axes
left, width = 0.1, 0.65
bottom, height = 0.1, 0.65
spacing = 0.005
rect_scatter = [left, bottom, width, height]
rect_histx = [left, bottom + height + spacing, width, 0.2]
rect_histy = [left + width + spacing, bottom, 0.2, height]
# start with a rectangular Figure
plt.figure(figsize=(8, 8))
ax_scatter = plt.axes(rect_scatter)
ax_scatter.tick_params(direction='in', top=True, right=True)
ax_histx = plt.axes(rect_histx)
ax_histx.tick_params(direction='in', labelbottom=False)
ax_histy = plt.axes(rect_histy)
ax_histy.tick_params(direction='in', labelleft=False)
# the scatter plot:
ax_scatter.scatter(x, y, color="purple")
# now determine nice limits by hand:
binwidth = 0.25
lim = np.ceil(np.abs([x, y]).max() / binwidth) * binwidth
ax_scatter.set_xlim((-lim, lim))
ax_scatter.set_ylim((-lim, lim))
bins = np.arange(-lim, lim + binwidth, binwidth)
ax_histx.hist(x, bins=bins, color = "skyblue")
ax_histy.hist(y, bins=bins, orientation='horizontal', color="salmon")
ax_histx.set_xlim(ax_scatter.get_xlim())
ax_histy.set_ylim(ax_scatter.get_ylim())
plt.show()
```
Notice how I changed the colors on the plot based off of what I found on the stack exchange. The way to solve issues in the course and computational work is to google them.
## Plotting Exersice 1
Find a plot from the gallery that you like. Then make some sort of noticable change to it.
```
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
points = np.ones(5) # Draw 5 points for each line
marker_style = dict(color='tab:blue', linestyle=':', marker='o',
markersize=15, markerfacecoloralt='tab:red')
fig, ax = plt.subplots()
# Plot all fill styles.
for y, fill_style in enumerate(Line2D.fillStyles):
ax.text(-0.5, y, repr(fill_style),
horizontalalignment='center', verticalalignment='center')
ax.plot(y * points, fillstyle=fill_style, **marker_style)
ax.set_axis_off()
ax.set_title('fill style')
plt.show()
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
points = np.ones(5) # Draw 5 points for each line
marker_style = dict(color='tab:green', linestyle=':', marker='o',
markersize=15, markerfacecoloralt='tab:purple')
fig, ax = plt.subplots()
# Plot all fill styles.
for y, fill_style in enumerate(Line2D.fillStyles):
ax.text(-0.5, y, repr(fill_style),
horizontalalignment='center', verticalalignment='center')
ax.plot(y * points, fillstyle=fill_style, **marker_style)
ax.set_axis_off()
ax.set_title('fill style')
plt.show()
```
## Plotting Exersice 2
Plot a the following functions on the same plot from $ -2\pi $ to $2\pi$
$$ \sin(2\pi x+\pi)$$
$$ \cos(2\pi x+\pi)$$
$$\sin(2\pi x+\pi)+\cos(2\pi x+\pi)$$
This might be useful:
https://docs.scipy.org/doc/numpy/reference/generated/numpy.sin.html
https://docs.scipy.org/doc/numpy/reference/generated/numpy.cos.html#numpy.cos
```
np.sin(np.pi/2.)
np.sin(np.array((0., 30., 45., 60., 90.)) * np.pi / 180. )
import matplotlib.pylab as plt
x = np.linspace(-np.pi, np.pi, 201)
plt.plot(x, np.sin(x))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-2*np.pi, 2*np.pi, 201)
plt.plot(x, np.sin(x))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-2*np.pi, 2*np.pi, 201)
plt.plot(x, np.sin(2*np.pi*x+np.pi))
plt.plot(x, np.cos(2*np.pi*x+np.pi))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-2*np.pi, 2*np.pi, 201)
plt.plot(x, np.cos(2*np.pi*x+np.pi))
plt.xlabel('Angle [rad]')
plt.ylabel('cos(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-2*np.pi, 2*np.pi, 201)
plt.plot(x, np.sin(2*np.pi*x+np.pi), color="blue")
plt.plot(x, np.cos(2*np.pi*x+np.pi), color="red")
plt.plot(x, np.sin(2*np.pi*x+np.pi)+np.cos(2*np.pi*x+np.pi), color="green")
plt.xlabel('x')
plt.ylabel('y(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-2*np.pi, 2*np.pi, 201)
plt.plot(x, np.sin(2*np.pi*x+np.pi), color="black")
plt.plot(x, np.cos(2*np.pi*x+np.pi), color="red")
plt.plot(x, np.sin(2*np.pi*x+np.pi)+np.cos(2*np.pi*x+np.pi), color="gray")
plt.xlabel('x')
plt.ylabel('y(x)')
plt.axis('tight')
plt.show()
```
# Lecture plots
Periodically during lecture we will take a pause to plot some of the interesting functions that we use in class.
## Classical wavefunctions
The following plot shows the the spacial component of the standard wavefunction with a wavelength of $\lambda=\text{1.45 m}$ and a relative amplitude of $A=1$ when the time, $t=0$ and the phase $\phi=1.0$.
```
x=np.linspace(0,3.0,100)
sinx=np.sin(2*np.pi*x+0+1)
plt.plot(x,sinx, color="black")
plt.xlabel('x', fontsize=24)
plt.ylabel("y(x)", fontsize=24)
plt.show()
```
Make a new figure where you plot the same wave function at three time points in the future. Assume the frequency is $\nu=.1 \text{ ms / s} $ Use a different color for each plot
## Orthogonality
Graphically show that the the following two functions are orthogonal on the interval $-3\pi$ to $3\pi$
$$ \sin(x) \text{ and } \cos(3x)$$
Plot both functions together, then plot the product of both functions and explain why it is orthogonal
```
import matplotlib.pylab as plt
x = np.linspace(-3*np.pi, 3*np.pi, 201)
plt.plot(x, np.sin(x))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-3*np.pi, 3*np.pi, 201)
plt.plot(x, np.sin(x))
plt.plot(x, np.cos(3*x))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-3*np.pi, 3*np.pi, 201)
prod=np.sin(x)*np.cos(3*x)
plt.plot(x, np.sin(x))
plt.plot(x, np.cos(3*x))
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
import matplotlib.pylab as plt
x = np.linspace(-3*np.pi, 3*np.pi, 201)
prod=np.sin(x)*np.cos(3*x)
plt.plot(x, prod, color="blue")
plt.xlabel('Angle [rad]')
plt.ylabel('sin(x)')
plt.axis('tight')
plt.show()
prod=np.sin(x)*np.cos(3*x)
prod=np.sin(x)*np.cos(3*x)
x = np.linspace(-3*np.pi, 3*np.pi, 201)
exp_func=prod
np.trapz(exp_func,x)
```
Use the numpy trapezoid rule integrator to show the the two functions are orthogonal
`np.trapz(y,x)`
https://docs.scipy.org/doc/numpy/reference/generated/numpy.trapz.html
```
# Example code
x=numpy.linspace(0,1.0,20)
exp_func=np.exp(-2*np.pi*x)
np.trapz(exp_func,x)
# Your code here
```
| github_jupyter |
```
import os
import sys
sys.path.append('../')
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from pprint import pprint
from scipy.optimize import curve_fit
import src.io as sio
import src.preprocessing as spp
import src.fitting as sft
AFM_FOLDER = sio.get_folderpath("20200818_Akiyama_AFM")
AFM_FOLDER1 = sio.get_folderpath("20200721_Akiyama_AFM")
AFM_FOLDER2 = sio.get_folderpath("20200824_Akiyama_AFM")
AFM_FOLDER3 = sio.get_folderpath("20200826_TFSC_Preamp_AFM/11613_Tip_5/Akiyama_Tip_Stage")
AFM_FOLDER4 = sio.get_folderpath("20200826_TFSC_Preamp_AFM/11613_Tip_5/Custom_Tip_Stage")
AFM_FOLDER5 = sio.get_folderpath("20200828_Tip_Approach1")
AFM_FOLDER6 = sio.get_folderpath("20200901_Tip_Approach_2/Actual_tip_approach")
```
# Approach
```
params, data = sio.read_dat(AFM_FOLDER6 + "HistoryData001.dat")
amplitude = data["Amplitude (m)"].values
fig, ax = plt.subplots()
ax.plot(amplitude*1e9)
ax.set_ylabel("Amplitude (nm)")
ax.set_xlabel("Time (a.u.)")
#plt.savefig("snap.jpg", dpi=600)
```
## 20200721_Akiyama_AFM
```
params, data = sio.read_dat(AFM_FOLDER1 + "frq-sweep002.dat")
freq_shift = data["Frequency Shift (Hz)"].values
amplitude = data["Amplitude (m)"].values
phase = data["Phase (deg)"].values
amp_freq_sweep = sft.fit_fano(freq_shift, amplitude, linear_offset=True)
phase_freq_sweep = sft.fit_fano(freq_shift, phase)
%matplotlib inline
fig, (ax1, ax2) = plt.subplots(nrows=2, sharex=True)
ax1.plot(freq_shift, amplitude*1e12)
#ax1.plot(freq_shift, amp_freq_sweep.best_fit)
ax1.set_ylabel("Amplitude (pm)")
ax2.plot(freq_shift, phase)
#ax2.plot(freq_shift, phase_freq_sweep.best_fit)
ax2.set_ylabel(data.columns[3])
ax2.set_xlabel(data.columns[0])
#plt.savefig("second.jpg", dpi=600)
```
Quality factor can be calculated as $ Q = \frac{f_R}{\Delta f} $
```
print(f'Q-factor= {params["f_res (Hz)"] / amp_freq_sweep.params["fwhm"].value}')
```
## 20200818_Akiyama_AFM
```
params, data = sio.read_dat(AFM_FOLDER + "frq-sweep001.dat")
#pprint(params, sort_dicts=False)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
fano = sft.fit_fano(freq_shift, amplitude)
lorentzian = sft.fit_fano(freq_shift, phase)
params
```
## Equations for calculating Q factor
$$ Q = \frac{f_R}{\Delta f} $$
$$ Q = \frac{A(\omega_0)}{A_{in}} $$
```
f_res = 44379.7064
sigma = 62.2841355
print(f_res/sigma)
A_drive = 50e-3
A_res = 28.3e-6 * 1 / 500e-6
print(A_res/A_drive)
# Calibration
A_drive = 50e-3
osc_amp = 50e-9
print(osc_amp/A_drive)
```
## Plot frequency sweep curves
```
fig, (ax1, ax2) = plt.subplots(nrows=2, sharex=True)
ax1.plot(freq_shift, amplitude)
ax1.plot(freq_shift, fano.best_fit)
ax1.set_ylabel(data.columns[2])
ax2.plot(freq_shift, phase)
ax2.plot(freq_shift, lorentzian.best_fit)
ax2.set_ylabel(data.columns[3])
ax2.set_xlabel(data.columns[1])
```
## Extract fit values
```
print("{} = {:.1f} +- {:.1f}".format(fano.params["sigma"].name, fano.params["sigma"].value, fano.params["sigma"].stderr))
print("{} = {:.2e} +- {:.0e}".format(fano.params["amplitude"].name, fano.params["amplitude"].value, fano.params["amplitude"].stderr))
```
# 20200824_Akiyama_AFM
## Automatically read files from disk
Reads all files stored in **AFM_FOLDER2 = "20200824_Akiyama_AFM/"** and plots the amplitude and phase data.
Optionally, the data can be fit to Fano resonances by setting the variable
```python
fit = True
```
The Q-factor is calculated as:
$$ Q = \frac{f_R}{\Delta f} = \frac{f_R}{2 \sigma} $$
Errors are calculated as (this also gives an estimate of the SNR):
$$ \frac{\Delta Q}{Q} = \sqrt{ \left( \frac{\Delta (\Delta f)}{\Delta f} \right)^2 + \left( \frac{\Delta (\sigma)}{\sigma} \right)^2 } $$
Another estimate of the SNR, is the Chi square or weighted sum of squared deviations (lower is better):
$$ \chi^2 = \sum_{i} {\frac{(O_i - C_i)^2}{\sigma_i^2}} $$
```
%matplotlib inline
fit = False # Setting to True will take slightly longer due to the fitting protocols
files = []
for file in os.listdir(AFM_FOLDER2):
if file.endswith(".dat"):
files.append(file)
fig, ax = plt.subplots(nrows=len(files), ncols=2)
for idx, file in enumerate(files):
params, data = sio.read_dat(AFM_FOLDER2 + file)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
ax[idx, 0].plot(freq_shift, amplitude)
ax[idx, 0].set_ylabel(data.columns[2])
ax[idx, 0].set_title(file)
ax[idx, 1].plot(freq_shift, phase)
ax[idx, 1].set_ylabel(data.columns[3])
ax[idx, 1].set_title(file)
if fit:
fano1 = sft.fit_fano(freq_shift, amplitude)
q_factor = (params["Center Frequency (Hz)"] + fano1.params["center"].value) / (2 * fano1.params["sigma"].value)
q_factor_err = q_factor * np.sqrt((fano1.params["center"].stderr/fano1.params["center"].value)**2 + (fano1.params["sigma"].stderr/fano1.params["sigma"].value)**2)
ax[idx, 0].plot(freq_shift, fano1.best_fit, label="Q={:.0f}$\pm{:.0f}$".format(q_factor, q_factor_err))
ax[idx, 0].legend()
fano2 = sft.fit_fano(freq_shift, phase, linear_offset=True)
ax[idx, 1].plot(freq_shift, fano2.best_fit)
print("chi-square ({}) = {:.2e}".format(file, fano1.chisqr))
fig.tight_layout()
fig.text(0.5, 0.02, data.columns[1], ha='center', va='center')
```
## 20200826_TFSC_Preamp_AFM
### 11613_Tip_5
```
%matplotlib inline
fit = False # Setting to True will take slightly longer due to the fitting protocols
files = []
for file in os.listdir(AFM_FOLDER4):
if file.endswith(".dat"):
files.append(file)
fig, ax = plt.subplots(nrows=len(files), ncols=2)
for idx, file in enumerate(files):
params, data = sio.read_dat(AFM_FOLDER4 + file)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
ax[idx, 0].plot(freq_shift, amplitude)
ax[idx, 0].set_ylabel(data.columns[2])
ax[idx, 0].set_title(file)
ax[idx, 1].plot(freq_shift, phase)
ax[idx, 1].set_ylabel(data.columns[3])
ax[idx, 1].set_title(file)
if fit:
fano1 = sft.fit_fano(freq_shift, amplitude)
q_factor = (params["Center Frequency (Hz)"] + fano1.params["center"].value) / (fano1.params["sigma"].value)
q_factor_err = q_factor * np.sqrt((fano1.params["center"].stderr/fano1.params["center"].value)**2 + (fano1.params["sigma"].stderr/fano1.params["sigma"].value)**2)
ax[idx, 0].plot(freq_shift, fano1.best_fit, label="Q={:.0f}$\pm{:.0f}$".format(q_factor, q_factor_err))
ax[idx, 0].legend()
fano2 = sft.fit_fano(freq_shift, phase, linear_offset=True)
ax[idx, 1].plot(freq_shift, fano2.best_fit)
print("chi-square ({}) = {:.2e}".format(file, fano1.chisqr))
fig.tight_layout()
fig.text(0.5, 0.02, data.columns[1], ha='center', va='center')
omega_0 = 1
omega = np.linspace(0, 2, 1000)
Q = 1
ratio = omega / omega_0
phi = np.arctan(-ratio / (Q * (1 - ratio**2)))
fid, ax = plt.subplots()
ax.plot(ratio, phi)
```
# Calibration from Thermal Noise density
From Atomic Force Microscopy, Second Edition by Bert Voigtländer
Section 11.6.5 Experimental Determination of the Sensitivity and Spring Constant in AFM Without Tip-Sample Contact
Eq. 11.28 and 11.26
```
%matplotlib widget
file = "SignalAnalyzer_Spectrum001"
params, data = sio.read_dat(AFM_FOLDER4 + file)
calibration_params = sft.find_afm_calibration_parameters(data, frequency_range=[40000, 48000], Q=1000, f_0_guess=44000)
fig, ax = plt.subplots()
ax.plot(calibration_params["Frequency (Hz)"], calibration_params["PSD squared (V**2/Hz)"])
ax.plot(calibration_params["Frequency (Hz)"], calibration_params["PSD squared fit (V**2/Hz)"])
print("Calibration (m/V) =", calibration_params["Calibration (m/V)"])
%matplotlib inline
fit = False # Setting to True will take slightly longer due to the fitting protocols
files = []
for file in os.listdir("../../Data/" + AFM_FOLDER4):
if file.endswith(".dat"):
files.append(file)
files = ["frq-sweep002.dat"]
fig, ax = plt.subplots(nrows=len(files), ncols=2)
for idx, file in enumerate(files):
params, data = sio.read_dat(AFM_FOLDER4 + file)
freq_shift = data["Frequency Shift (Hz)"]
amplitude = data["Amplitude (m)"]
phase = data["Phase (deg)"]
if len(files) == 1:
ax[0].plot(freq_shift, amplitude)
ax[0].set_ylabel(data.columns[2])
ax[1].plot(freq_shift, phase)
ax[1].set_ylabel(data.columns[3])
else:
ax[idx, 0].plot(freq_shift, amplitude)
ax[idx, 0].set_ylabel(data.columns[2])
ax[idx, 0].set_title(file)
ax[idx, 1].plot(freq_shift, phase)
ax[idx, 1].set_ylabel(data.columns[3])
ax[idx, 1].set_title(file)
if fit:
fano1 = sft.fit_fano(freq_shift, amplitude)
#q_factor = (params["Center Frequency (Hz)"] + fano1.params["center"].value) / (fano1.params["sigma"].value)
#q_factor_err = q_factor * np.sqrt((fano1.params["center"].stderr/fano1.params["center"].value)**2 + (fano1.params["sigma"].stderr/fano1.params["sigma"].value)**2)
ax[idx, 0].plot(freq_shift, fano1.best_fit)
ax[idx, 0].legend()
fano2 = sft.fit_fano(freq_shift, phase, linear_offset=True)
ax[idx, 1].plot(freq_shift, fano2.best_fit)
print("chi-square ({}) = {:.2e}".format(file, fano1.chisqr))
fig.tight_layout()
fig.text(0.5, 0.02, data.columns[1], ha='center', va='center')
```
| github_jupyter |
<a href="https://colab.research.google.com/github/yarusx/cat-vs-dogo/blob/main/cat_vs_dog_0_0_2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import matplotlib.pyplot as plt
import numpy as np
import os
import tensorflow as tf
from tensorflow.keras.preprocessing import image_dataset_from_directory
_URL = 'https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip'
path_to_zip = tf.keras.utils.get_file('cats_and_dogs.zip', origin=_URL, extract=True)
PATH = os.path.join(os.path.dirname(path_to_zip), 'cats_and_dogs_filtered')
train_dir = os.path.join(PATH, 'train')
validation_dir = os.path.join(PATH, 'validation')
BATCH_SIZE = 32
IMG_SIZE = (160, 160)
train_dataset = image_dataset_from_directory(train_dir,
shuffle=True,
batch_size=BATCH_SIZE,
image_size=IMG_SIZE)
validation_dataset = image_dataset_from_directory(validation_dir,
shuffle=True,
batch_size=BATCH_SIZE,
image_size=IMG_SIZE)
class_names = train_dataset.class_names
# plt.figure(figsize=(10, 10))
# for images, labels in train_dataset.take(1):
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# plt.imshow(images[i].numpy().astype("uint8"))
# plt.title(class_names[labels[i]])
# plt.axis("off")
val_batches = tf.data.experimental.cardinality(validation_dataset)
test_dataset = validation_dataset.take(val_batches // 5)
validation_dataset = validation_dataset.skip(val_batches // 5)
AUTOTUNE = tf.data.experimental.AUTOTUNE
train_dataset = train_dataset.prefetch(buffer_size=AUTOTUNE)
validation_dataset = validation_dataset.prefetch(buffer_size=AUTOTUNE)
test_dataset = test_dataset.prefetch(buffer_size=AUTOTUNE)
data_augmentation = tf.keras.Sequential([
tf.keras.layers.experimental.preprocessing.RandomFlip('horizontal'),
tf.keras.layers.experimental.preprocessing.RandomRotation(0.2),
])
# for image, _ in train_dataset.take(1):
# plt.figure(figsize=(10, 10))
# first_image = image[0]
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# augmented_image = data_augmentation(tf.expand_dims(first_image, 0))
# plt.imshow(augmented_image[0] / 255)
# plt.axis('off')
rescale = tf.keras.layers.experimental.preprocessing.Rescaling(1./127.5, offset= -1)
# Create the base model from the pre-trained model MobileNet V2
IMG_SHAPE = IMG_SIZE + (3,)
base_model = tf.keras.applications.MobileNetV2(input_shape=IMG_SHAPE,
include_top=False,
weights='imagenet')
image_batch, label_batch = next(iter(train_dataset))
feature_batch = base_model(image_batch)
print(feature_batch.shape)
base_model.trainable = False
global_average_layer = tf.keras.layers.GlobalAveragePooling2D()
feature_batch_average = global_average_layer(feature_batch)
print(feature_batch_average.shape)
prediction_layer = tf.keras.layers.Dense(1)
prediction_batch = prediction_layer(feature_batch_average)
print(prediction_batch.shape)
inputs = tf.keras.Input(shape=(160, 160, 3))
x = data_augmentation(inputs)
x = rescale(x)
x = base_model(x, training=False)
x = global_average_layer(x)
x = tf.keras.layers.Dropout(0.4)(x)
outputs = prediction_layer(x)
model = tf.keras.Model(inputs, outputs)
base_learning_rate = 0.0001
model.compile(optimizer=tf.keras.optimizers.Adam(lr=base_learning_rate),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
loss0, accuracy0 = model.evaluate(validation_dataset)
print("initial loss: {:.2f}".format(loss0))
print("initial accuracy: {:.2f}".format(accuracy0))
initial_epochs = 1
history = model.fit(train_dataset,
epochs=initial_epochs,
validation_data=validation_dataset)
val_acc = history.history['val_accuracy']
while np.mean(val_acc)*100 < 98.5:
initial_epochs = 3
history = model.fit(train_dataset,
epochs=initial_epochs,
validation_data=validation_dataset)
val_acc = history.history['val_accuracy']
try:
!mkdir -p saved_model
except:
pass
model.save('saved_model/dvc/')
!zip -r dvc.zip saved_model/dvc/
from google.colab import files
files.download("dvc.zip")
from google.colab import drive
drive.mount('/content/drive')
!unzip -q /content/drive/MyDrive/dvc.zip
dvc = tf.keras.models.load_model('/content/saved_model/dvc')
try:
!mkdir -p saved_model
except:
pass
model.save('saved_model/dvc/')
!zip -r dvc.zip saved_model/dvc/
from google.colab import files
files.download("dvc.zip")
from keras.preprocessing.image import load_img, img_to_array
# load and prepare the image
def load_image(filename):
# load the image
img = load_img(filename, target_size=(160, 160))
# convert to array
img = img_to_array(img)
# reshape into a single sample with 3 channels
img = img.reshape(1, 160, 160, 3)
return img
img = load_image('/content/drive/MyDrive/dayana_1.JPG')
categories = ["Cat", "Dog"]
prediction = dvc_model.predict(img)
prediction = tf.nn.sigmoid(prediction)
print(prediction)
plt.figure()
plt.imshow(img[0]/255)
plt.title(categories[int(np.round_(prediction))])
loss0, accuracy0 = model.evaluate(validation_dataset)
# #Retrieve a batch of images from the test set
# image_batch, label_batch = test_dataset.as_numpy_iterator().next()
# predictions = model.predict_on_batch(image_batch).flatten()
# # Apply a sigmoid since our model returns logits
# predictions = tf.nn.sigmoid(predictions)
# predictions = tf.where(predictions < 0.5, 0, 1)
# print('Predictions:\n', predictions.numpy())
# print('Labels:\n', label_batch)
# plt.figure(figsize=(10, 10))
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# plt.imshow(image_batch[i].astype("uint8"))
# plt.title(class_names[predictions[i]])
# plt.axis("off")
```
| github_jupyter |
```
# some_file.py
import sys
# insert at 1, 0 is the script path (or '' in REPL)
sys.path.insert(1, "/Users/dhruvbalwada/work_root/sogos/")
import os
from numpy import *
import pandas as pd
import xarray as xr
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
from xgcm import Grid
from xgcm.autogenerate import generate_grid_ds
import sogos.download_product as dlp
import sogos.load_product as ldp
import sogos.time_tools as tt
import sogos.geo_tools as gt
import sogos.download_file as df
import gsw
import cmocean as cmocean
```
# Download Latest Data
```
data_dir = "/Users/dhruvbalwada/work_root/sogos/data/raw/climatology/"
```
FTP ADDRESS: ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/
Data prior to 2017 (till Dec 2016) is in a single file
```
# download the big climatology files
wget.download(
"ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/RG_ArgoClim_Salinity_2017.nc.gz",
data_dir,
)
wget.download(
"ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/RG_ArgoClim_Temperature_2017.nc.gz",
data_dir,
)
from ftplib import FTP
ftp_address = "ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/RG_ArgoClim_2019"
url_root = "/pub/gilson/argo_climatology/"
ftp_root = "kakapo.ucsd.edu"
ftp = FTP(ftp_root)
ftp.login()
ftp.cwd(url_root)
contents = ftp.nlst("RG_ArgoClim_2017*")
contents = ftp.nlst("RG_ArgoClim_201*")
for i in contents:
print("Downloading" + i)
wget.download("ftp://kakapo.ucsd.edu/pub/gilson/argo_climatology/" + i, data_dir)
```
## Load some data
```
Tclim = xr.open_dataset(data_dir + "RG_ArgoClim_Temperature_2017.nc", decode_times=False)
Sclim = xr.open_dataset(data_dir + "RG_ArgoClim_Salinity_2017.nc", decode_times=False)
Climextra = xr.open_mfdataset(data_dir+ 'RG_ArgoClim_201*', decode_times=False)
RG_clim = xr.merge([Tclim, Sclim, Climextra])
# Calendar type was missing, and giving errors in decoding time
RG_clim.TIME.attrs['calendar'] = '360_day'
RG_clim = xr.decode_cf(RG_clim)
## Add density and other things
SA = xr.apply_ufunc(gsw.SA_from_SP, RG_clim.ARGO_SALINITY_MEAN+RG_clim.ARGO_SALINITY_ANOMALY, RG_clim.PRESSURE ,
RG_clim.LONGITUDE, RG_clim.LATITUDE,
dask='parallelized', output_dtypes=[float,]).rename('SA')
CT = xr.apply_ufunc(gsw.CT_from_t, SA, RG_clim.ARGO_TEMPERATURE_MEAN+RG_clim.ARGO_SALINITY_ANOMALY, RG_clim.PRESSURE,
dask='parallelized', output_dtypes=[float,]).rename('CT')
SIGMA0 = xr.apply_ufunc(gsw.sigma0, SA, CT, dask='parallelized', output_dtypes=[float,]).rename('SIGMA0')
RG_clim = xr.merge([RG_clim, SIGMA0])
T_region = RG_clim.ARGO_TEMPERATURE_ANOMALY.groupby('TIME.season').mean() + RG_clim.ARGO_TEMPERATURE_MEAN
S_region = RG_clim.ARGO_SALINITY_ANOMALY.groupby('TIME.season').mean() + RG_clim.ARGO_SALINITY_MEAN
rho_region = RG_clim.SIGMA0.groupby('TIME.season').mean()
plt.figure(figsize=(18,3))
plt.subplot(141)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='DJF').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.subplot(142)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='MAM').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.subplot(143)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='JJA').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.subplot(144)
T_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='SON').plot.contourf(levels=11, vmin=-9, vmax=9);
plt.figure(figsize=(18,3))
plt.subplot(141)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='DJF').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.subplot(142)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='MAM').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.subplot(143)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='JJA').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.subplot(144)
S_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='SON').plot.contourf(levels=11, vmin=33.7, vmax=34.2)
plt.tight_layoutout()
plt.figure(figsize=(18,3))
plt.subplot(141)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='DJF').plot.contourf(levels=11)
plt.subplot(142)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='MAM').plot.contourf(levels=11)
plt.subplot(143)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='JJA').plot.contourf(levels=11)
plt.subplot(144)
rho_region.sel(LATITUDE=slice(-65,-45), LONGITUDE=slice(20,60)).isel(PRESSURE=0).sel(season='SON').plot.contourf(levels=11)
```
### Some Climatological Mean Sections
```
dens_section30 = RG_clim.SIGMA0.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).load()
dens_section40 = RG_clim.SIGMA0.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).load()
glider = {"start_month": 4.99, "end_month":7.8, "start_lat": -51.5, "end_lat": -53, "max_depth": 1000}
plt.figure(figsize=(15,4))
plt.subplot(121)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)
).plot.contourf(vmin=-10, levels=24)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(linestyles='-.',levels=[1])
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C0',
linestyles='dashed', linewidths=4)
plt.plot([glider['start_lat'], glider['start_lat']], [4, glider['max_depth']], color='C2', alpha=0.5)
plt.plot([glider['end_lat'], glider['end_lat']], [4, glider['max_depth']], color='C2', alpha=0.5)
plt.gca().invert_yaxis()
plt.subplot(122)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(vmin=33.77, vmax=35.21, levels=24, cmap=cmocean.cm.haline)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[34.2], linestyles='dashdot')
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.plot([glider['start_lat'], glider['start_lat']], [4, 1900], color='C3', alpha=0.5)
plt.gca().invert_yaxis()
plt.tight_layout()
#plt.savefig('../figures/clim_TS_30E.png')
plt.figure(figsize=(15,4))
plt.subplot(121)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)
).plot.contourf(vmin=-10,levels=24)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(linestyles='-.',levels=[1])
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C0',
linestyles='dashed', linewidths=4)
plt.gca().invert_yaxis()
plt.subplot(122)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(vmin=33.77, vmax=35.21,levels=24, cmap=cmocean.cm.haline)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[34.2], linestyles='dashdot')
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section40.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.gca().invert_yaxis()
plt.tight_layout()
plt.savefig('../figures/clim_TS_40E.png')
plt.figure(figsize=(12,4))
plt.subplot(121)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(levels=24)
RG_clim.ARGO_TEMPERATURE_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[1])
plt.gca().invert_yaxis()
plt.subplot(122)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contourf(levels=24)
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=40, method='nearest').sel(LATITUDE=slice(-70,-40)).plot.contour(levels=[34.2])
plt.gca().invert_yaxis()
plt.tight_layout()
# Not much seasonality below 200m
dens_section.groupby('TIME.month').mean().mean('month').sel(PRESSURE=slice(0,1000)).plot.contourf(cmap='Blues')
dens_section.groupby('TIME.month').mean().isel(month=0).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
dens_section.groupby('TIME.month').mean().isel(month=3).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
dens_section.groupby('TIME.month').mean().isel(month=6).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
dens_section.groupby('TIME.month').mean().isel(month=9).sel(PRESSURE=slice(0,1000)).plot.contour(levels=[27.2])
plt.gca().invert_yaxis()
```
## N2
\begin{equation}
N^2 = db/dz
\end{equation}
$b = -\frac{g}{\rho_0} \rho'$
$b = g(\alpha \triangle T - \beta \triangle S)$
```
RG_clim
RG_clim = generate_grid_ds(RG_clim,
{'Z':'PRESSURE', 'X':'LONGITUDE', 'Y':'LATITUDE'})
grid = Grid(RG_clim, periodic='X')
g = 9.81
rho0 = 1000
dens_clim_monthly = RG_clim.SIGMA0.groupby('TIME.month').mean()
dens_clim_monthly
N2_clim_monthly = grid.interp(-g/rho0* grid.diff(dens_clim_monthly, 'Z', boundary='extend') / -(grid.diff(RG_clim.PRESSURE, 'Z', boundary='extend')), 'Z', boundary='extend')
N2_clim_monthly_SO = N2_clim_monthly.sel(LATITUDE=slice(-70, -30)).load()
N2_clim_monthly_SO = N2_clim_monthly_SO.rename('N2')
plt.figure(figsize=(18,12))
plt.subplot(221)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=1).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=1).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('January')
plt.gca().invert_yaxis()
plt.subplot(222)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('April')
plt.gca().invert_yaxis()
plt.subplot(223)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=7).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=7).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('July')
plt.gca().invert_yaxis()
plt.subplot(224)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=10).plot(vmin=-5e-5)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=10).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('October')
plt.gca().invert_yaxis()
plt.savefig('../figures/clim_N2_30E.png')
plt.figure(figsize=(8,3))
plt.subplot(121)
#plt.pcolormesh(N2_clim_monthly_SO.LATITUDE.sel(LATITUDE=slice(-70, -40)),
# N2_clim_monthly_SO.LATITUDE.sel(=slice(-70, -40)),
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot(vmin=-5e-5,
rasterized=True,add_colorbar=False)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=4).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('April')
plt.gca().invert_yaxis()
plt.ylim([1500, 0])
plt.xlabel('Latitude')
plt.ylabel('Depth (m)')
plt.subplot(122)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=9).plot(vmin=-5e-5,
rasterized=True,add_colorbar=False)
N2_clim_monthly_SO.sel(LATITUDE=slice(-70, -40)).sel(LONGITUDE=30, method='nearest').sel(month=9).plot.contour(levels=[2e-5])
RG_clim.ARGO_SALINITY_MEAN.sel(LONGITUDE=30, method='nearest').sel(
LATITUDE=slice(-70,-40)).plot.contour(levels=[34.35], linestyles='dashdot')
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(colors='k', levels=7)
dens_section30.groupby('TIME.month').mean().mean('month').plot.contour(levels=[27.189, 27.752], colors='C1',
linestyles='dashed', linewidths=4)
plt.title('September')
plt.gca().invert_yaxis()
plt.ylim([1500, 0])
plt.xlabel('Latitude')
plt.ylabel('Depth (m)')
plt.tight_layout()
plt.savefig('N2_climatology.pdf')
N2.sel(LATITUDE=slice(-60, -40)).sel(LONGITUDE=30, method='nearest').isel(TIME=-1).plot()
```
The apply ufunc way, not working yet.
```
CT_clim = CT.groupby('TIME.month').mean()
SA_clim = SA.groupby('TIME.month').mean()
CT_clim_region = CT_clim.sel(LATITUDE=slice(-65,-35), LONGITUDE=slice(20,50)).load()
SA_clim_region = SA_clim.sel(LATITUDE=slice(-65,-35), LONGITUDE=slice(20,50)).load()
(N2, pmid) = xr.apply_ufunc(gsw.Nsquared, SA_clim_region, CT_clim_region, RG_clim.PRESSURE,
dask='parallelized',
input_core_dims=[['PRESSURE'],['PRESSURE'],['PRESSURE']],
output_core_dims=[['PRESSURE'],['PRESSURE']], exclude_dims=set(['PRESSURE']))
```
### Gestrophic Velocities
```
psi = xr.apply_ufunc(gsw.geo_strf_dyn_height, SA, CT , RG_clim.PRESSURE,
dask='parallelized', output_dtypes=[float,]).rename('psi')
psi
vels = xr.apply_ufunc(gsw.geostrophic_velocity, psi, psi.LONGITUDE, psi.LATITUDE,
dask='parallelized', output_core_dims=[4,4], output_dtypes=[float,]).rename('vels')
vels
```
### Mixed Layer Depth
Ended up going with Holte's climatology for MLD work
```
delta_dens = RG_clim.SIGMA0 - RG_clim.SIGMA0.isel(PRESSURE=0)
import nc_time_axis
RG_clim.SIGMA0.sel(LONGITUDE=30, method='nearest').sel( LATITUDE=slice(-63,-45)).isel(TIME=-1).plot.contourf()
plt.gca().invert_yaxis()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-1).plot()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-7).plot()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-60, method='nearest').isel(TIME=-1).plot()
RG_clim.SIGMA0.sel(LONGITUDE=30, LATITUDE=-60, method='nearest').isel(TIME=-7).plot()
delta_dens.sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-1).plot()
temp = delta_dens.where(delta_dens>0.03).sel(LONGITUDE=30, LATITUDE=-50, method='nearest').isel(TIME=-1).plot()
temp = delta_dens.where(delta_dens>0.03)
MLD = temp.PRESSURE.where(temp == temp.min('PRESSURE')).min('PRESSURE')
MLD_clim = temp.PRESSURE.where(temp == temp.min('PRESSURE')).min('PRESSURE').groupby('TIME.month').mean()
MLD_clim.load()
MLD_clim.month
plt.figure(figsize=(12,4))
plt.subplot(121)
MLD_clim.sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,90)).sel(month=1).plot(vmin=0, vmax=120)
plt.subplot(122)
MLD_clim.sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,90)).sel(month=7).plot(vmin=0, vmax=120)
MLD_clim.max('month').sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,380)).plot(vmin=0, vmax=150)
deltaH = MLD_clim.max('month') - MLD_clim.min('month')
deltaH.sel(LATITUDE=slice(-75,-25), LONGITUDE=slice(20,380)).plot(vmin=0, vmax=80)
MLD_clim.sel(LATITUDE=-45, LONGITUDE=35, method='nearest').plot(label='45S')
MLD_clim.sel(LATITUDE=-50, LONGITUDE=35, method='nearest').plot(label='50S')
MLD_clim.sel(LATITUDE=-55, LONGITUDE=35, method='nearest').plot(label='55S')
MLD_clim.sel(LATITUDE=-60, LONGITUDE=35, method='nearest').plot(label='60S')
plt.legend()
MLD_clim.sel(LATITUDE=-45, LONGITUDE=45, method='nearest').plot(label='45S')
MLD_clim.sel(LATITUDE=-50, LONGITUDE=45, method='nearest').plot(label='50S')
MLD_clim.sel(LATITUDE=-55, LONGITUDE=45, method='nearest').plot(label='55S')
MLD_clim.sel(LATITUDE=-60, LONGITUDE=45, method='nearest').plot(label='60S')
plt.legend()
```
| github_jupyter |
```
#Add needed imports
import numpy as np
import pandas as pd
from imblearn.over_sampling import SMOTE
import seaborn as sns
from sklearn.preprocessing import OrdinalEncoder
from sklearn.dummy import DummyClassifier
from imblearn.over_sampling import SMOTENC
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import accuracy_score,confusion_matrix, precision_score, recall_score,f1_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.neural_network import MLPClassifier
from xgboost import XGBClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.model_selection import RepeatedStratifiedKFold, GridSearchCV
from sklearn import svm
import shap
import os
#Read data
proccessed_data_path =os.path.join(os.path.pardir,os.path.pardir,'data','processed')
train_path = os.path.join(proccessed_data_path,'dataset10.csv')
df = pd.read_csv(train_path)
labels=df['Churn']
x = df.drop(columns=['Churn','Unnamed: 0'],axis = 'columns')
y=np.ravel(labels)
x_train,x_test,y_train,y_test = train_test_split(x,y,test_size=0.3)
oversample = SMOTENC(categorical_features=[2])
oversample = SMOTE()
x_train, y_train = oversample.fit_resample(x_train, y_train)
sc = StandardScaler()
x_train = sc.fit_transform(x_train)
x_test = sc.transform(x_test)
svm_model = svm.SVC(random_state=0,gamma='auto')
rf_model=RandomForestClassifier(random_state=0)
dt_model=DecisionTreeClassifier(random_state=0,criterion='entropy',max_depth = 7,min_samples_leaf=30)
lr_model= LogisticRegression(random_state=0, max_iter=300)
mlp_model =MLPClassifier(random_state=0,activation='relu', solver='sgd',learning_rate='adaptive')
xgb_model = XGBClassifier(random_state=0 ,learning_rate=0.05, max_depth=7,eval_metric='mlogloss',use_label_encoder =False)
gmb_model= GradientBoostingClassifier(random_state=0,n_estimators=20,learning_rate=0.75,max_features=4,max_depth=5)
model_params = {
'svm': {
'model': svm_model,
'params' : {
'C': [15,10],
'kernel': ['rbf','linear']
}
},
'rf': {
'model': rf_model,
'params' : {
'n_estimators': [1,5,10]
}
},
'dt': {
'model': dt_model,
'params' : {}
},
'lr' : {
'model':lr_model,
'params': {
'C': [1,5,10]
}
},
'mlp' : {
'model':mlp_model,
'params': {}
},
'xg_boost' : {
'model':xgb_model,
'params': {}
},
'gbm' : {
'model':gmb_model,
'params': {}
}
}
scores = []
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=100)
for model_name, mp in model_params.items():
clf = GridSearchCV(mp['model'], mp['params'], cv=cv, return_train_score=False)
clf.fit(x_train,y_train)
conf_matrix =confusion_matrix(y_test,clf.predict(x_test))
scores.append({
'model': model_name,
'best_score': clf.best_score_,
'best_params': clf.best_params_,
'precision':precision_score(y_test,clf.predict(x_test)),
'recall':recall_score(y_test,clf.predict(x_test)),
'f1_score':f1_score(y_test,clf.predict(x_test)),
'true positives':conf_matrix[0][0],
'true negatives':conf_matrix[1][1],
'false postives':conf_matrix[0][1],
'false negatives':conf_matrix[1][0]
})
df = pd.DataFrame(scores,columns=['model','best_score','precision','recall','f1_score','true positives','true negatives','false postives','false negatives','best_params'])
print(df)
import shap
xgb_model = XGBClassifier(random_state=0 ,learning_rate=0.05, max_depth=7,eval_metric='mlogloss',use_label_encoder =False)
xgb_model.fit(x_train,y_train)
explainer = shap.TreeExplainer(xgb_model)
shap_values = explainer.shap_values(x)
shap.summary_plot(shap_values, x)
```
| github_jupyter |
```
# import dependencies
import pandas as pd
import requests
import json
# read csv on covid-19 covid vulnerability index data and convert to dataframe
ccvi = pd.read_csv('../resources/ccvi.csv')
# drop rows that contain any null values (there are 655 of them)
ccvi = ccvi.dropna(how='any')
# display dataframe
ccvi
# get covid data for each race by state
covid = pd.read_csv('../resources/CRDT_Data.csv')
# display dataframe
covid
# dictionary for convertying state names to corresponding numbers or abbreviations
states = {
'southcarolina': {'num': '45', 'abbr': 'SC'},
'southdakota': {'num': '46', 'abbr': 'SD'},
'tennessee': {'num': '47', 'abbr': 'TN'},
'texas': {'num': '48', 'abbr': 'TX'},
'vermont': {'num': '50', 'abbr': 'VT'},
'utah': {'num': '49', 'abbr': 'UT'},
'virginia': {'num': '51', 'abbr': 'VA'},
'washington': {'num': '53', 'abbr': 'WA'},
'westvirginia': {'num': '54', 'abbr': 'WV'},
'wisconsin': {'num': '55', 'abbr': 'WI'},
'wyoming': {'num': '56', 'abbr': 'WY'},
'puertorico': {'num': '72', 'abbr': 'PR'},
'alabama': {'num': '01', 'abbr': 'AL'},
'alaska': {'num': '02', 'abbr': 'AK'},
'arizona': {'num': '04', 'abbr': 'AZ'},
'arkansas': {'num': '05', 'abbr': 'AR'},
'california': {'num': '06', 'abbr': 'CA'},
'colorado': {'num': '08', 'abbr': 'CO'},
'delaware': {'num': '10', 'abbr': 'CT'},
'districtofcolumbia': {'num': '11', 'abbr': 'DE'},
'connecticut': {'num': '09', 'abbr': 'DC'},
'florida': {'num': '12', 'abbr': 'FL'},
'georgia': {'num': '13', 'abbr': 'GA'},
'idaho': {'num': '16', 'abbr': 'ID'},
'hawaii': {'num': '15', 'abbr': 'HI'},
'illinois': {'num': '17', 'abbr': 'IL'},
'indiana': {'num': '18', 'abbr': 'IN'},
'iowa': {'num': '19', 'abbr': 'IA'},
'kansas': {'num': '20', 'abbr': 'KS'},
'kentucky': {'num': '21', 'abbr': 'KS'},
'louisiana': {'num': '22', 'abbr': 'LA'},
'maine': {'num': '23', 'abbr': 'ME'},
'maryland': {'num': '24', 'abbr': 'MD'},
'massachusetts': {'num': '25', 'abbr': 'MA'},
'michigan': {'num': '26', 'abbr': 'MI'},
'minnesota': {'num': '27', 'abbr': 'MN'},
'mississippi': {'num': '28', 'abbr': 'MS'},
'missouri': {'num': '29', 'abbr': 'MO'},
'montana': {'num': '30', 'abbr': 'MT'},
'nebraska': {'num': '31', 'abbr': 'NE'},
'nevada': {'num': '32', 'abbr': 'NV'},
'newhampshire': {'num': '33', 'abbr': 'NH'},
'newjersey': {'num': '34', 'abbr': 'NJ'},
'newmexico': {'num': '35', 'abbr': 'NM'},
'newyork': {'num': '36', 'abbr': 'NY'},
'northcarolina': {'num': '37', 'abbr': 'NC'},
'northdakota': {'num': '38', 'abbr': 'ND'},
'oregon': {'num': '41', 'abbr': 'OR'},
'pennsylvania': {'num': '42', 'abbr': 'PA'},
'rhodeisland': {'num': '44', 'abbr': 'RI'}
}
# all statistical categories to to be queried
pops = 'B01003_001E,B02001_002E,B02001_003E,B02001_004E,B02001_005E,B02001_006E,B03001_003E'
# create list of racial groups to iterate through
races = ['total','white','black','native','asian','pacific','hispanic']
# dictionary with all data to be used from all states that made the data avaliable
stateData = {}
# all states without necessary data
error_states = []
# iterate through states
for state in states:
try:
# get census state number
state_num = states[state]['num']
# create url to request data from api
url = f'https://api.census.gov/data/2019/acs/acs5?get=NAME,{pops}&for=tract:*&in=state:{state_num}'
# set returned data to a variable
response = requests.get(url).json()
# create list to store dictionaries with data for each census tract
tracts = []
# create dictionaries with population data for each census tract
# (with properly formatted fips code)
for r in response:
if r[0] != 'NAME':
tracts.append({
'FIPS': int(f'{r[8]}{r[9]}{r[10]}'),
'total': int(r[1]),
'white': int(r[2]),
'black': int(r[3]),
'native': int(r[4]),
'asian': int(r[5]),
'pacific': int(r[6]),
'hispanic': int(r[7])
})
# create dataframe with census population data
populations = pd.DataFrame(tracts)
# merge population data and ccvi data on census tract fips code
ccvi_and_pop = pd.merge(populations, ccvi, on='FIPS')
# create dictionary to hold data for each racial demographic
demogs = {
'total': {},
'white': {},
'black': {},
'native': {},
'asian': {},
'pacific': {},
'hispanic': {}
}
# iterate through list of races
for race in races:
# calculate total population for each race
demogs[race]['population'] = int(ccvi_and_pop[race].sum())
# calculate average ccvi for each race
demogs[race]['ccvi'] = (ccvi_and_pop[race]*ccvi_and_pop['ccvi']).sum()/demogs[race]['population']
# calculate population of each race as a percentage of total population
demogs[race]['population_percent'] = (demogs[race]['population']/demogs['total']['population'])*100
# get covid data for each race by state
covid = pd.read_csv('../resources/CRDT_Data.csv')
# filter to only include data for selected state
covid = covid.loc[covid['State'] == states[state]['abbr'],:]
# filter to only include data from 2020
covid = covid.loc[covid['Date'] < 20210000,:]
# create dataframe with only relevant columns for covid cases
cases = covid[['Cases_Total','Cases_White','Cases_Black','Cases_AIAN','Cases_Asian','Cases_NHPI','Cases_Ethnicity_Hispanic']]
# create dataframe with only relevant columns for covid deaths
deaths = covid[['Deaths_Total','Deaths_White','Deaths_Black','Deaths_AIAN','Deaths_Asian','Deaths_NHPI','Deaths_Ethnicity_Hispanic']]
# iterate through covid data for selected races and place data in a dictionary
for i in range(0, len(cases.columns)):
# total cases for each race
demogs[races[i]]['cases'] = int(cases[cases.columns[i]].values[0])
# number of cases for each race as a percentage of total cases
demogs[races[i]]['percent_of_cases'] = (demogs[races[i]]['cases']/demogs['total']['cases'])*100
# percent discrepancy between percent of total cases and percent of total population for by each race
# (theoretically each race should account for the same percent of cases as their percent of the population)
demogs[races[i]]['discrepancy_percent'] = (demogs[races[i]]['percent_of_cases']/demogs[races[i]]['population_percent'])*100
# total deaths for each race
demogs[races[i]]['deaths'] = int(deaths[deaths.columns[i]].values[0])
# chance of an infection resulting in death for each race
demogs[races[i]]['chance_of_death'] = (demogs[races[i]]['deaths']/demogs[races[i]]['cases'])*100
# number of deaths for each race as a percentage of total deaths
demogs[races[i]]['percent_of_deaths'] = (demogs[races[i]]['deaths']/demogs['total']['deaths'])*100
# create dataframe without total population values
demographics = pd.DataFrame(demogs).drop(columns=['total'])
# create dictionary to hold calculated values to be used in max patch
for_max = {}
# iterate through statistical categories
for row in list(demographics.index):
# create a list that holds all values within the row of a statistical category
values = demographics.loc[row].values
# iterate through races
for i in range(1, len(races)):
# get population numbers
if row == 'population':
for_max[races[i]] = {}
for_max[races[i]][row] = int(values[i-1])
# calculate inverted ccvi values
elif row == 'ccvi':
for_max[races[i]]['inverted_ccvi'] = round(100-(values[i-1])*100, 2)
# calculate chances for where next infection will occure
elif row == 'discrepancy_percent':
for_max[races[i]]['chance_of_infection'] = round((values[i-1]/values.sum())*100, 2)
# get values for chance of infection resulting in death
elif row == 'chance_of_death':
for_max[races[i]][row] = round(values[i-1], 2)
# create keys to hold number of cases and deaths generated by Max alogrithm
for key in for_max:
for_max[key]['generated_cases'] = 0
for_max[key]['generated_deaths'] = 0
stateData[state] = for_max
except:
error_states.append(state)
# display avaliable states in alphabetical order
stateData = sorted(stateData)
for state in stateData:
print(state)
# display unavaliable states
for state in error_states:
print(state)
stateData
with open("../resources/stateData.json", "w") as outfile:
json.dump(stateData, outfile)
```
| github_jupyter |
# User testing for for Scikit-Yellowbrick
### Using data that was recorded from sensors during Data Science Certificate Program at GW
https://github.com/georgetown-analytics/classroom-occupancy
Data consist of temperature, humidity, CO2 levels, light, # of bluetooth devices, noise levels and count of people in the room.
```
import pandas as pd
%matplotlib inline
dataset = pd.read_csv('dataset.csv')
dataset.head(5)
dataset.count_total.describe()
#add a new column to create a binary class for room occupancy
countmed = dataset.count_total.median()
dataset['room_occupancy'] = dataset['count_total'].apply(lambda x: 'occupied' if x > 4 else 'empty')
# map room occupancy to a number
dataset['room_occupancy_num'] = dataset.room_occupancy.map({'empty':0, 'occupied':1})
dataset.head(5)
dataset.room_occupancy.describe()
import os
import sys
# Modify the path
sys.path.append("..")
import pandas as pd
import yellowbrick as yb
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (12, 8)
g = yb.anscombe()
```
## Feature Analysis
Feature analysis visualizers are designed to visualize instances in data space in order to detect features or targets that might impact downstream fitting. Because ML operates on high-dimensional data sets (usually at least 35), the visualizers focus on aggregation, optimization, and other techniques to give overviews of the data. It is our intent that the steering process will allow the data scientist to zoom and filter and explore the relationships between their instances and between dimensions.
At the moment we have three feature analysis visualizers implemented:
Rank2D: rank pairs of features to detect covariance
RadViz: plot data points along axes ordered around a circle to detect separability
Parallel Coordinates: plot instances as lines along vertical axes to detect clusters
Feature analysis visualizers implement the Transformer API from Scikit-Learn, meaning they can be used as intermediate transform steps in a Pipeline (particularly a VisualPipeline). They are instantiated in the same way, and then fit and transform are called on them, which draws the instances correctly. Finally show or show is called which displays the image.
```
from yellowbrick.features.rankd import Rank2D
from yellowbrick.features.radviz import RadViz
from yellowbrick.features.pcoords import ParallelCoordinates
```
### Rank2D
Rank1D and Rank2D evaluate single features or pairs of features using a variety of metrics that score the features on the scale [-1, 1] or [0, 1] allowing them to be ranked. A similar concept to SPLOMs, the scores are visualized on a lower-left triangle heatmap so that patterns between pairs of features can be easily discerned for downstream analysis.
```
# Load the classification data set
data = dataset
# Specify the features of interest
features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
# Extract the numpy arrays from the data frame
X = data[features].as_matrix()
y = data['count_total'].as_matrix()
# Instantiate the visualizer with the Covariance ranking algorithm
visualizer = Rank2D(features=features, algorithm='covariance')
visualizer.fit(X, y) # Fit the data to the visualizer
visualizer.transform(X) # Transform the data
visualizer.show() # Draw/show/show the data
# Instantiate the visualizer with the Pearson ranking algorithm
visualizer = Rank2D(features=features, algorithm='pearson')
visualizer.fit(X, y) # Fit the data to the visualizer
visualizer.transform(X) # Transform the data
visualizer.show() # Draw/show/show the data
```
### RadViz
RadViz is a multivariate data visualization algorithm that plots each feature dimension uniformely around the circumference of a circle then plots points on the interior of the circle such that the point normalizes its values on the axes from the center to each arc. This meachanism allows as many dimensions as will easily fit on a circle, greatly expanding the dimensionality of the visualization.
Data scientists use this method to dect separability between classes. E.g. is there an opportunity to learn from the feature set or is there just too much noise?
```
# Specify the features of interest and the classes of the target
features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
classes = ['empty', 'occupied']
# Extract the numpy arrays from the data frame
X = data[features].as_matrix()
y = data.room_occupancy_num.as_matrix()
# Instantiate the visualizer
visualizer = visualizer = RadViz(classes=classes, features=features)
visualizer.fit(X, y) # Fit the data to the visualizer
visualizer.transform(X) # Transform the data
visualizer.show() # Draw/show/show the data
```
For regression, the RadViz visualizer should use a color sequence to display the target information, as opposed to discrete colors.
## Parallel Coordinates
### !!! On this step notebook crashes and has to be restarted
```
# Specify the features of interest and the classes of the target
#features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
#classes = ['empty', 'occupied']
# Extract the numpy arrays from the data frame
#X = data[features].as_matrix()
#y = data.room_occupancy_num.as_matrix()
# Instantiate the visualizer
#visualizer = visualizer = ParallelCoordinates(classes=classes, features=features)
#visualizer.fit(X, y) # Fit the data to the visualizer
#visualizer.transform(X) # Transform the data
#visualizer.show() # Draw/show/show the data
```
## Regressor Evaluation
Regression models attempt to predict a target in a continuous space. Regressor score visualizers display the instances in model space to better understand how the model is making predictions. We currently have implemented two regressor evaluations:
Residuals Plot: plot the difference between the expected and actual values
Prediction Error: plot expected vs. the actual values in model space
Estimator score visualizers wrap Scikit-Learn estimators and expose the Estimator API such that they have fit(), predict(), and score() methods that call the appropriate estimator methods under the hood. Score visualizers can wrap an estimator and be passed in as the final step in a Pipeline or VisualPipeline.
```
# Regression Evaluation Imports
from sklearn.linear_model import Ridge, Lasso
from sklearn.model_selection import train_test_split
from yellowbrick.regressor import PredictionError, ResidualsPlot
```
### Residuals Plot
A residual plot shows the residuals on the vertical axis and the independent variable on the horizontal axis. If the points are randomly dispersed around the horizontal axis, a linear regression model is appropriate for the data; otherwise, a non-linear model is more appropriate.
```
# Load the data
df = data
feature_names = ['temperature','humidity','co2','light','noise','bluetooth_devices']
target_name = 'count_total'
# Get the X and y data from the DataFrame
X = df[feature_names].as_matrix()
y = df[target_name].as_matrix()
# Create the train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Instantiate the linear model and visualizer
ridge = Ridge()
visualizer = ResidualsPlot(ridge)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
### Prediction Error Plot
Plots the actual targets from the dataset against the predicted values generated by our model. This allows us to see how much variance is in the model. Data scientists diagnose this plot by comparing against the 45 degree line, where the prediction exactly matches the model.
```
# Load the data
df = data
feature_names = ['temperature','humidity','co2','light','noise','bluetooth_devices']
target_name = 'count_total'
# Get the X and y data from the DataFrame
X = df[feature_names].as_matrix()
y = df[target_name].as_matrix()
# Create the train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Instantiate the linear model and visualizer
lasso = Lasso()
visualizer = PredictionError(lasso)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
## Classifier Evaluation
Classification models attempt to predict a target in a discrete space, that is assign an instance of dependent variables one or more categories. Classification score visualizers display the differences between classes as well as a number of classifier-specific visual evaluations. We currently have implemented three classifier evaluations:
ClassificationReport: Presents the confusion matrix of the classifier as a heatmap
ROCAUC: Presents the graph of receiver operating characteristics along with area under the curve
ClassBalance: Displays the difference between the class balances and support
Estimator score visualizers wrap Scikit-Learn estimators and expose the Estimator API such that they have fit(), predict(), and score() methods that call the appropriate estimator methods under the hood. Score visualizers can wrap an estimator and be passed in as the final step in a Pipeline or VisualPipeline.
```
# Classifier Evaluation Imports
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from yellowbrick.classifier import ClassificationReport, ROCAUC, ClassBalance
```
### Classification report
The classification report visualizer displays the precision, recall, and F1 scores for the model. Integrates numerical scores as well color-coded heatmap in order for easy interpretation and detection.
```
# Load the classification data set
data = dataset
# Specify the features of interest and the classes of the target
features = ['temperature','humidity','co2','light','noise','bluetooth_devices']
classes = ['empty', 'occupied']
# Extract the numpy arrays from the data frame
X = data[features].as_matrix()
y = data.room_occupancy_num.as_matrix()
# Create the train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# Instantiate the classification model and visualizer
bayes = GaussianNB()
visualizer = ClassificationReport(bayes, classes=classes)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
### ROCAUC
Plot the ROC to visualize the tradeoff between the classifier's sensitivity and specificity.
```
# Instantiate the classification model and visualizer
logistic = LogisticRegression()
visualizer = ROCAUC(logistic)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
### ClassBalance
Class balance chart that shows the support for each class in the fitted classification model.
```
# Instantiate the classification model and visualizer
forest = RandomForestClassifier()
visualizer = ClassBalance(forest, classes=classes)
visualizer.fit(X_train, y_train) # Fit the training data to the visualizer
visualizer.score(X_test, y_test) # Evaluate the model on the test data
g = visualizer.show() # Draw/show/show the data
```
| github_jupyter |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.